This publication is provided for historical reference only and the information may be out of date.
Overview of PMC
PubMed Central (PMC) is NLM’s digital archive of medical and life sciences journal articles and an extension of NLM’s permanent print collection. It was launched in early 2000 with a single issue each of two journals, and has grown steadily since. As of June 2013, it contained over 2.7 million articles; more than 1200 journals were depositing all their published content in PMC, and a few thousand other journals were depositing selected articles. (See http://www.ncbi.nlm.nih.gov/pmc/ for current counts and titles.) Almost all the articles in PMC have a corresponding citation in PubMed. The exceptions are a few types of material, such as book reviews, that PubMed does not cover.
In its early years, PMC received all its content from journals that deposited complete issues. In 2006, NLM began offering publishers the additional option to deposit just selected articles from an issue. In both cases, the publisher provides PMC with the final published versions of articles; deposits are covered by formal participation agreements that address copyright and other rights and responsibilities. Participating publishers must deposit full-text XML and PDFs, along with high resolution image files and any supplementary data that is published with an article. Details about these participation agreements are available on the Publisher Information webpage (1).
Although publishers began providing material to PMC just a few months before its launch in 2000, the archive contains a substantial number of articles that were published long before that. Publishers often include several years of back files when they begin participating in PMC. In addition, in 2002, NLM undertook a project to scan and digitize the complete print archives of journals that were depositing current content in PMC. Two years later, the Wellcome Trust and the Joint Information Systems Committee in the UK joined in supporting the effort. The project ran for about 6 years and resulted in the addition of more than 1.2 million articles to PMC, dating back to the early 1800s.
Article Data Formats
Newly published articles, and much of the material in PMC that has been published since the late 1990s, are archived as full-text XML. For the articles from the print issue digitization project, most of which were published before 2000, PMC has PDFs of scanned page images, along with automatically extracted OCR text that is used to support full-text searching, and abstracts in XML form (e.g., http://www.ncbi.nlm.nih.gov/pmc/articles/PMC361653/). For some journals, there may also be a relatively short period when they were making the transition from print to electronic publication. Before they moved to creating full-text XML (or SGML) they had electronically formatted PDFs. For this period (generally the mid-1990s, but extending to the mid-2000s for some journals) PMC has a PDF and an XML abstract for each article (e.g., http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148429/).
Full-text XML is at the heart of PMC’s design philosophy and therefore is a requirement for all current content. XML files are machine- and human-readable and are not technology dependent. This makes XML easy to migrate as technology changes and, therefore, an excellent archival format. Regardless of the source DTD to which incoming content is structured, all full-text XML in PMC is converted to a common archival format, the “NLM DTD,” which is now a NISO standard (see “NLM DTD to NISO JATS Z39.96-2012”). Full-text HTML pages (e.g., http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3382486/?report=classic) are created dynamically from the XML at retrieval time. This allows article presentation styles to be changed relatively quickly and easily. Even a completely new format like the PubReader display (e.g., http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3382486/?report=reader) can be introduced without any changes to, or preprocessing of, the source article record in the PMC database.
Access and Copyright
Everything in PMC is free to read, much of it from the time of publication, the rest after a delay that generally is 12 months or less. Participating journals are expected to deposit their articles at time of publication even if they are not made available publicly in PMC immediately. A growing number of articles in PMC are available under a Creative Commons (2) or similar license that generally allows more liberal redistribution and reuse than a traditional copyrighted work. However, the majority of the material in PMC is still protected by standard copyright, held by the respective publishers. NLM does not hold copyright on any of the material.
Author manuscripts
Since 2005, PMC also has been the designated repository for NIH’s Public Access Policy (3) and similar policies of other research funders in the US and abroad. Researchers supported by these agencies are required to deposit in PMC the accepted manuscript of any peer-reviewed journal article that arises from the funding they have received. Some journals deposit the final published versions of such articles directly in PMC on behalf of their authors under one of the PMC participation agreements mentioned above. In the remaining cases, the author or publisher deposits manuscript files (e.g., a Word document or a minimally formatted PDF that has not yet gone through final copy editing by the journal) in a Manuscript Submission system—the NIHMS in the US or similar, derivative systems in the UK and Canada. The manuscript is converted to JATS XML format and reviewed and approved by the author before being transferred to PMC.
PMC International
NLM supports the operation of journal archives similar to PMC in the UK and Canada. These two archives have a copy of the majority of the articles in PMC, based on agreements with the respective publishers. Information about the PMC international collaboration is available on the PMC International webpage (4).
Architecture Overview
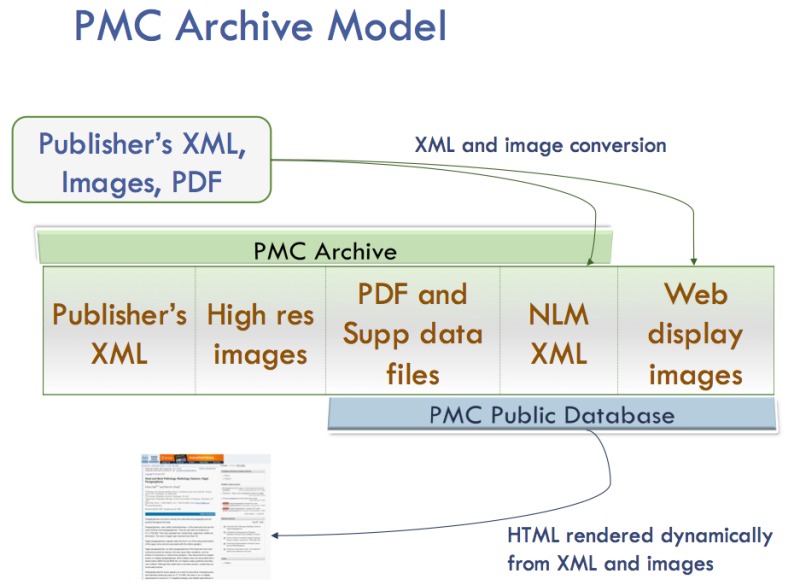
The PMC processing model is diagrammed in Figure 1. For each article, we receive a set of files that includes the text in SGML or XML, the highest resolution figures available, a PDF file if one has been created for the article, and any supplementary material or supporting data. The text is converted to the current version of the NISO Z39.96-2012 Journal Article Tag Suite (JATS) Archiving and Interchange article model, and the images are converted to a web-friendly format. The source SGML or XML, original images, supplementary data files, PDFs, and NLM XML files are stored in the archive. Articles are rendered online using the NLM XML, PDFs, supplementary data files, and the web-friendly images.
Figure 1.
PMC Processing Model.
Ingest
Participating PMC publishers submit the full text of each article in some "reasonable" SGML or XML format along with the highest-resolution images available, PDF files (if available), and all supplementary material. Complete details on the PMC’s file requirements are available (5).
A reasonable SGML or XML format is one where there is sufficient granularity in the source model to map those elements critical to the understanding of the article (and/or its functioning in the PMC system) from the original article to the appropriate place in the PMC XML model. Currently we convert all incoming articles into the NISO Z39.96-2012 Journal Article Tag Suite (JATS) version 1.0 Archiving and Interchange article model, but we have articles in nearly every version of the NLM DTD/NISO JATS article model in the PMC database.
The Journal Evaluation Process
Journals joining PMC must pass two tests. First, the content must be approved for the NLM collection (6).
Next the journal must go through a technical evaluation to "be sure that the journal can routinely supply files of sufficient quality to generate complete and accurate articles online without the need for human action to correct errors or omissions in the data." (1)
For the technical evaluation, a journal supplies a sample set of articles. These articles are put through a series of automated and human checks to ensure that the XML is valid and that it accurately represents the article content. There is a set of "Minimum Data Requirements" that must be met before the evaluation proceeds to the more human-intense content accuracy checking (7). These minimum criteria are listed below briefly:
- Each sample package must be complete: all required data files (XML/SGML, PDF if available, image files, supplementary data files) for every article in the package must be present and named correctly.
- All XML files must conform to an acceptable journal article schema.
- All XML/SGML files must be valid according to their schema.
- Regardless of the XML/SGML schema used, the following metadata information must be present and tagged with correct values in every sample file:
- 1.
Journal ISSN or other unique Journal ID
- 2.
Journal Publisher
- 3.
Copyright statement (if applicable)
- 4.
License statement (if applicable)
- 5.
Volume number
- 6.
Issue number (if applicable)
- 7.
Pagination/article sequence number
- 8.
Issue-based or Article-based publication dates. Articles submitted to PMC must contain publication dates that accurately reflect the journal’s publication model.
- All image files for figures must be legible, and submitted in high-resolution TIFF or EPS format, according to the PMC Image File Requirements.
These seem like simple and obvious things—XML files must be valid—but the minimum data requirements have greatly reduced the amount of rework that the PMC Data Evaluation group has to do. It helps to be explicit about even the most obvious of things.
PMC’s XML Philosophy
PMC’s XML philosophy is a balance between strictness and flexibility that enables control of the quality of data being loaded into the system without being too restrictive on submitters.
PMC does a complete review of any new schema in which content is being submitted, as described above. We do not take articles in HTML. We also do a complete review of sample articles for each new journal to be sure that the content provider is able to provide content that is structurally and semantically correct.
Another thing we are strict about is that all content must be valid according to the schema in which it was submitted—not just during data evaluation but in the ongoing production process as well. This seems obvious, but there was a surprising amount of controversy about this in the early days of PMC, and we still receive invalid files. Problems usually arise now because the submitter has made a schema change (as simple as adding a new character entity to the DTD or a new required element) without telling us or sending an updated schema.
Also, we do not fix text; all content changes must be made by the submitter, and the content must be resubmitted.
Some things we are more flexible about, which reduces some of the burden on our submitters. First, we don’t require all content to be in our format or to follow our tagging rules. We don’t force updates of content to the latest DTD version, and we can generally follow journal style where it does not interfere with processing.
PMC Internal DTD
We use the JATS Archiving and Interchange DTD ("out of the box") as the format for all articles loaded to the PMC database. This model was created specifically for archiving article content. It was designed to be an "easy target to hit" when transforming content from the over 40 different input models that we receive content in. Currently we are writing content into version NISO JATS version 1.0.
We do not migrate all content to each new version of the JATS DTD when one is released. The system is robust enough to handle content from versions 1.0 through 3.0 of the NLM DTD and version 1.0 of the NISO JATS DTD, so we are not constantly churning the data.
All of the versions of the DTD are managed with an XML Catalog (8), which we also use to manage all of the input DTDs (SGML and XML). We maintain all mappings of PUBLIC and SYSTEM IDs for any DTD that we use in the XML catalog on our Linux machines and then create other catalogs from it each time it is updated. We create an SGML Catalog for the SGML tools that we use; a single "Oxygen" catalog that everyone on the team can use over the network with the XML editor; and a copy of the catalog that refers to http-based copies of the DTDs for PMC International sites. The XML Catalog is an essential piece of the PMC system.
PMC Tagging Style
Next, we’ve defined a set of rules for objects within articles that is more restrictive than the DTD. This allows us to have normalized structures (figures, tables, contributors) in articles for ease of processing and rendering. We call these rules the PMC Tagging Style, and all articles must "pass style" before being loaded to the database. They are documented in the PMC Tagging Guidelines (9).
(Re)Usability of XML
Finally, our XML must be useable by others. The NLM XML that we create from whatever was submitted to us is always available to the submitting publisher (the content owner), and a subset of the articles that are Open Access are available to anyone for download through the PMC Open Archives Service (10). This keeps us honest. We can’t allow ourselves to take shortcuts with the data. All articles must be valid according to the JATS schema version that they reference, and we only use Processing Instructions for instructions about processing.
Text Processing
There are four main principles to PMC text processing:
First, we expect to receive marked-up versions of an article that are well-formed, valid, and accurately represent the article as it was published, (i.e., that it represents the version of record). The question of what is the "Version of Record" is left up to the publisher. It may be a printed copy, a PDF version, or the journal’s website.
We do not correct articles or files. That is, we will not fix something that is wrong in the version of record nor will we make a correction to an XML file. All problems found in either processing or QA of files are reported to the publisher to be corrected and resubmitted.
The goal of PMC is to represent the content of the article and not the formatting of the printed page, the PDF, or the journal’s website.
Finally, we run a Quality Assessment on content coming into PMC to ensure that the content rendered in PMC accurately reflects the article as it was published. Our QA is a combination of automated checks and manual checking of articles. To help ensure that the content we are spending time ingesting to PMC is likely to be worthwhile, journals must pass through an evaluation process before they can send content to PMC in a regular production workflow.
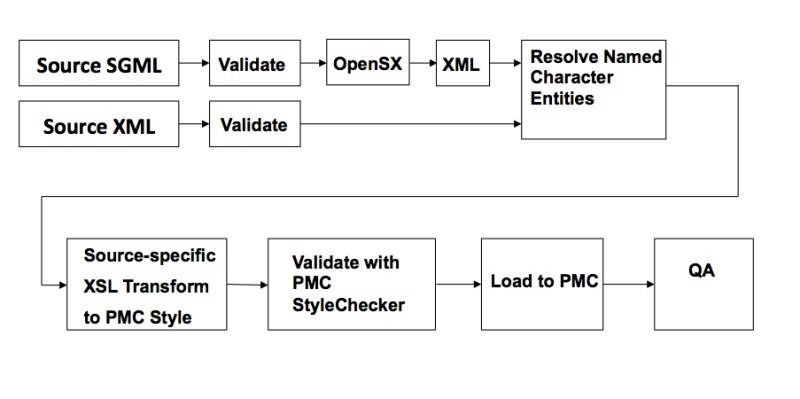
Figure 2 shows the workflow for text coming into PMC.
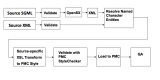
Figure 2.
PMC Text Processing.
Images
Generally, authors submit raw image data files to a publishing house in various formats (PPT, PDF, TIF, JPG, XML, etc.). The files are then normalized to produce print or electronic output. PMC requires the normalized output, which is high-resolution, and of sufficient width and quality to be considered archival. Images generated at low resolution for display purposes are not acceptable.
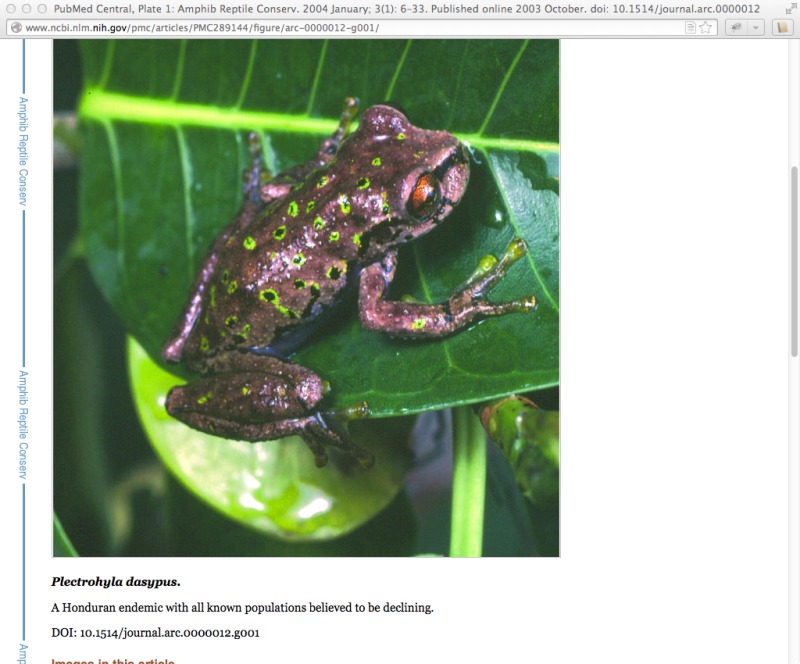
During ingest, the submitted images are converted to web-friendly thumbnail (Figure 3) and full-sized (Figure 4) versions for display within the article.
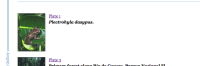
Figure 3.
An image thumbnail.
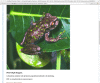
Figure 4.
Full sized image displayed in a new window.
The thumbnail from "Plate 1" links to a full view of the figure including the caption.
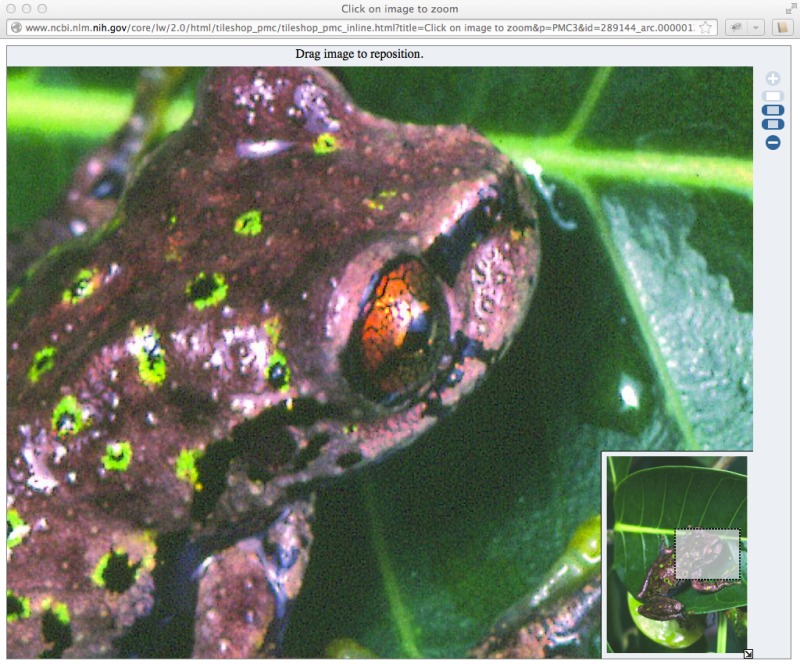
A very large version of the image is also created so that users can zoom in and inspect the image up close. Linking from the full image view takes you to the Tileshop view (Figure 5). The image index in the lower right corner shows which part of the whole image is available on the screen.
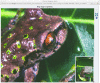
Figure 5.
Very large representation of the image that allows zooming.
With the original high-resolution images stored in the archive, when these display technologies become out of date, the images can be generated in whatever the latest image display technology is available.
PDFs and Supplemental Data
PDF versions of the article may be supplied to accompany the XML version in PMC, but they are not required.
PMC requires all available supplementary material to be submitted in a portable format, such as PDF, DOC, CSV, etc. Supplementary material should not be externally linked to a www location from the article text as a substitute for submission. Supplementary material includes the following:
- Voluminous material that was used to support the conclusions of the narrative, such as a genomic database or the multiple data sets for an article that presents the highlights, which can never accompany a paper based on sheer mass.
- "Extra" tables that do not display with the work, but that record the measurements on which the article is based, for example, that need to be available so the peer reviewers can check the article.
- Material added to the work for enhancement purposes, such as a quiz, an instructional video, the 3-minute version of the reaction that was described in the work with narrative and a few still images, a form that can be filled out, etc.
Quality Assessment and Workflows
Quality Assessment (QA) is done for all content coming into PMC to ensure that the content in PMC accurately reflects the article as it was published. Our QA is a combination of automated and manual checks and is managed by a team of Journal Managers (JM’s) who are each assigned responsibility for a large number of journals. JM’s are also responsible for ensuring that content is deposited on schedule, passes successfully through the automated workflow, and is released to the live site in a timely manner. JM’s interact regularly with the publishers and content providers to resolve problems and answer questions.
For journals in our regular production workflow, an automated workflow is set up so that new content uploaded to our FTP site for the journal is picked up and processed automatically, usually within several hours of the upload. A notification email is sent to the responsible JM indicating whether the session succeeded or failed. If the session succeeds, the content has been successfully ingested and processed and an entry will be added into our QA system. If the session resulted in an "error," the logs will be reviewed by the responsible JM and resolved often after updated files are sent from the publisher or content provider. For this automated system to work, it is important the publishers and content providers adhere to the File Submission Specifications (5) and follow a consistent naming scheme. Submissions that don’t adhere to a consistent naming scheme will remain on the FTP site and must be reviewed by the JM before they can proceed through the automated workflow.
Automated QA checks that occur as part of this workflow include checking that the XML/SGML is valid according to its schema, that all images and supplementary files referred to in the files are present and properly named, and that volume and issue information in the filename of the submission package (typically a ZIP file) correspond correctly to the volume and issue tagging in the XML/SGML files. All content that is not well-formed (if XML) or valid is returned to the provider to be corrected and resubmitted. Furthermore, the PMC Style Checker (9) is used during the automated workflow processing to ensure that all content flowing into PMC is in the PMC common XML format for loading to the database. The errors reported by the Style Checker provide us with a level of automated checking on the content itself that can highlight problems, but it only goes so far. For example, the Style Checker can tell if an electronic publication date is tagged completely to PMC Style (contains values in year, month, and day elements) in a file, but it can’t tell if the values themselves are correct and actually represent the electronic publication date of the article.
Manual QA is done by the JM’s after the automated workflow has successfully completed. PMC’s QA system shows each JM the journals that are assigned to her, and which articles need to be checked. The QA System marks a percentage of articles from each "batch" of new content deposited by the journal for manual QA. By default, new journals that come out of data evaluation and move into production are set with a higher percentage of articles selected for manual QA. Once the JM is confident in the journal’s ability to provide good, clean data, the percentages are lowered. If the JM begins to see problems on an ongoing basis, the percentage of articles checked may be increased. QA errors are grouped into eight major categories: Article Information, Article Body, Back Matter, Figures and Tables, Special Characters and Math, Generic Errors, Image Quality, and PDF Quality. Within each of these major categories, there may be one or more sub-categories. For example, in the "Article Body" section there is a subcategory for "Sections and Subsections," containing errors for missing sections, or sections that have been nested incorrectly in the flow of the body text. The JM looks at each article selected for QA and goes through all the categories and subcategories in this checklist that apply, and records any problems found. Error reports are then sent to the publisher or content provider and revisions are requested.
PMC also has a series of automated data integrity checks that run nightly for articles that have successfully passed through the automated workflow and have been loaded to the database. The integrity checks can identify, among other things, problems like duplicate articles submitted to the system, and potential discrepancies in issue publication dates for a group of articles in the same issue.
Article Identifiers and Version Numbers
There are several different types of identifiers that are used within the PMC system. This section describes these various IDs, and how they are related. Conversion among some of these IDs is available through the PMCID - PMID - Manuscript ID - DOI Converter tool (12), which is described in more detail below and summarized in Table 1.
PMCIDs, Article IDs, and UIDs
The most basic type of identifier used in the PMC system is the PMCID, which uniquely identifies an article. The PMCID is composed of the letters "PMC" followed by a string of decimal digits, for example, "PMC1868567." This is also sometimes referred to as the "PMC accession number." Once assigned, the PMCID is permanent, and can be used to unambiguously refer to a particular article within PMC, from that point on.
The numeric portion of the PMCID (without the "PMC" prefix) is referred to as the Article ID, or AID. For a given article, this numeric identifier is the same across all PMCI sites (see PMCI, below).
The NCBI Entrez system refers to items in any of its databases by a numeric identifier that is known, in that system, as the UID. Every Entrez database defines a numeric UID that identifies a record in that system. In the case of PMC, the UID is the same as the AID.
Versions
A recently added enhancement to the PMC system is the ability to handle multiple versions of the same article. Each version is a distinct instance of an article, which is archived separately and made permanently available for retrieval. Versions of an article can be accessed through a URI that uses the same form as that for a canonical article URI, but with a PMCID + version number. For example, here are the three versions of a PLoS Currents article that were available at the time of this writing:
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3283037.1/
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3283037.2/
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3283037.3/
Note that there are actually two URIs that can be used to access the latest version of an article. Each of these URIs refers to the same resource, but with different semantics. For example,
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3283037/ - This URI will always point to the latest version of this article
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3283037.3/ - This URI will always point to version number 3 of this article
Every article in PMC has a version number, whether or not it actually has multiple versions. In other words, an article that only has a single version has the version number "1".
PubMed IDs
PMC articles are often identified by their PubMed IDs, or PMIDs. This is the numeric identifier in the PubMed database (see “PubMed: The Bibliographic Database”) corresponding to this article, and is independent of the PMCID. Note that not every PMC article has a PMID (although most do). Articles can be accessed by URIs using their PMID, and these cause a redirect to the canonical URI for that article. For example,
• http://www.ncbi.nlm.nih.gov/pmc/articles/pmid/17401604/ redirects to → http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1868567/
Manuscript IDs
PMC also maintains a Manuscript ID, or MID, for those articles that arrive as manuscripts, most often through the NIHMS system. In general, these manuscripts continue to be available even after the final published version of the article arrives. As with article versions (described above) these are unique article instances that are archived separately.
For example, the following article does not have a "final published version", and is available through two URIs that refer to the same document:
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3159421/
• http://www.ncbi.nlm.nih.gov/pmc/articles/mid/NIHMS311352/
Whereas the following article has both a manuscript and a final published version, so these two URIs refer to different documents (different article instances):
• http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1434700/
• http://www.ncbi.nlm.nih.gov/pmc/articles/mid/NIHMS5786/
DOIs
A well-known external identification system that is used to specify articles is the Digital Object Identifier (DOI). PMC does not assign DOIs, but records them when they are supplied to us, and makes articles available using these identifiers. Articles in PMC can be accessed with URIs using the DOI, which then causes a redirect to the canonical URI for that article. For example,
http://www.ncbi.nlm.nih.gov/pmc/articles/doi/10.1172/JCI62818 redirects to → http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3484440/
At the time of this writing, PMC does not support DOIs that refer to specific article versions, but support is planned in the near future.
ISSN, Volume, Issue, and Page
Finally, articles can also be identified by their citation information: ISSN of the journal, volume, issue, and page. For example, the Journal of Clinical Investigation has ISSN 0021-9738. To access an article from that journal’s volume 117, issue #9, page 2380, you could construct a URI using the "ivip" path segment. For example:
For electronic journals that don’t have pagination, the e-ID replaces that page number. For example,
Summary
Table 1.
Summary of PMC Identifiers and URIs.
Retrieval / Data Processing
Indexing
Every day, the contents of the PMC database are indexed so that they are available via the NCBI Entrez interface. The Entrez interface is used by the PMC home page search facility, as well as by the Entrez Programming Utilities (EUtils; (13)). EUtils allows for third-party tools to provide discovery and search capabilities that mirror those provided by the NCBI website. In this respect, PMC is just one of the approximately 50 NCBI databases (at the time of this writing) that provide data to the public via this interface.
Fields, Filters, and Links
Every NCBI database has its own indexing criteria, including a unique set of fields, filters, and links to other databases. The PMC Help book (14) describes these for the PMC database, and gives information about how to use them to perform effective searches using Entrez.
Entrez searches allow you to enter search criteria with complex boolean expressions, using text phrases that are (optionally) qualified with either fields or filters. For example:
wilson eo[author] OR (eusociality AND author manuscript[filter])
Search fields are entered into a query with a text string followed by the field name in square brackets. For example,
wilson eo[author]
Consult the PMC Help book to get the list of available search fields. Search fields are added or changed occasionally, and the most up-to-date list of fields can be retrieved from one of two places:
- 1.
The PMC Advanced Search Builder (15). Click the "All Fields" drop-down option box, for a list of all the available search field names.
- 2.
The EInfo utilty, at http://eutils
.ncbi.nlm .nih.gov/entrez/eutils/einfo .fcgi?db=pmc (the result will be in XML). The <FieldList> element includes content describing each of the fields.
Filters are actually a special kind of field (the field named "filter") and are similar to the "tags" or "categories" that are used in many social media sites. (For example, just as a blog post might have several tags, a given record in the PMC database might correspond with several filters). There are two types of filters: standard (built-in) and custom.
Examples of built-in filters are "author manuscript," "reply," "retraction," "open access," and "CC BY NC license." To find records corresponding to a built-in filter value, enter the value in quotes, followed by the word "filter" in square brackets. For example, to find all the author manuscripts in PMC that are in the open access subset, enter the search phrase
"author manuscript"[filter] AND "open access"[filter]
You can get the complete list of filter values available by going to the PMC Advanced Search Builder, selecting "filter" in the first drop-down option box, and then clicking "Show index list".
Filters can be used by setting up your MyNCBI account to specify specific filter values that will appear as links on every Entrez search results page. This is done through the MyNCBI Filters webpage (16), selecting the "PMC" database. In the panel on the right, you can select any of the built-in filters. Clicking the checkbox enables that filter, so that it appears on every Entrez search results page, for easy access. Managing filters with MyNCBI is described in more detail in the MyNCBI Help Book under Working with Filters (17).
From the MyNCBI page, you can also manage custom filters, which are simply named Entrez queries. That is, any arbitrary Entrez query can be set up as a custom filter. A given record matches a custom filter value if it would be found by the corresponding Entrez query.
Links allow you to discover records in other NCBI databases that correspond to PMC documents. You can find the list of PMC-related links at the master Entrez Link Description webpage (18).
The data that correlates fields, filters, and links with particular objects in the PMC database are produced by our internal indexing tasks.
Indexing Tasks
There are two types of indexing tasks: full and merge. Merge indexing occurs daily. In the PMC database, an IndexStatus field is maintained, which keeps track of which articles have been indexed, and when. Merge indexing only operates on those articles that are new or that have been updated since the last time they were indexed.
Full indexing is scheduled to occur once per week, but might also occur if there is a specific need to re-index all of the PMC content; for example, if there is a change to the database structure, or to a search field or filter.
Note that, currently, indexing is done on the basis of UID (as described above) not individual versions of an article. Therefore, Entrez search results will always display links to the most up-to-date version of an article.
The PMC indexing tasks are integrated with the new NCBI CIDX indexing system, which is an automated workflow driven by Ergatis (19), a Web-based tool that allows workflows to be defined as pipelines, composed of reusable steps.
In addition to full text indexing, which generates the data required for searching by fields from within Entrez, the indexing tasks also generate data for filters, links, a spell-checker, and for auto-complete dictionaries. The indexing script accesses the article full text and the metadata from the PMC database, and produces XML files that are fed into the Entrez system. The XML gives full-text search strings and keywords, for each article, that are broken down by the Entrez fields that are defined for PMC.
For built-in filters, there are three possible sources of data, depending on how the filter is defined in the system:
- Explicit Filters—The indexing script produces these as explicit lists of article identifiers that match the given filter.
- Derived Filters—Like user-defined custom filters, these are based on Entrez queries.
- Links Filters—Any article that is the subject of at least one link of a given type automatically matches the filter with the same name. For example, any PMC article that cites another article in PMC matches the filter "pmc pmc cites."
For a base set of Entrez links, the indexing task generates the link from PMC records to other NCBI Entrez databases, and writes these into the Entrez links database. This database then produces some derived links automatically. For example, the link data for pmc_pmc_cites is generated by querying the PMC database to find, for a given article, all of the other articles in PMC that this article cites. The links database utilities then automatically generate the reciprocal link pmc_pmc_citedby, and stores that data.
The results of the linking processes are available from the discovery column of a PMC article display, as described in the Entrez Help book, or through the Entrez utility ELink. For example, to find all of the articles in PMC that are cited by a given PMC article, you could retrieve the URI
http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pmc&id=14909&linkname=pmc_pmc_cites.
Text Mining
PMC mines the full text of articles in its collection for references to specific types of entities, as agreed to with PMC participating publishers. These references are to entities that are stored in other NCBI databases. The results of this text mining are stored in the TagServer database, described below. When an article is requested from a Web browser, the TagServer software then retrieves this data, and uses it to enrich the presentation.
The text mining script is technically part of the TagServer software. It is implemented as a Perl script written in modern Perl style, and based on PMC-specific Perl modules that are, in turn, based on Perl Moose (20).
The text mining process is scheduled to run every day, and it continuously re-indexes all of the articles in PMC. During each daily iteration, it processes articles in this order:
- new articles never-before mined,
- articles updated since the last time they were mined,
- all others
Thus, all of the PMC articles are re-mined on a periodic basis. (Currently the time between successive mining of a given article is about two months.) It is necessary to continuously re-index all of the PMC articles, even if they have not changed, because the databases that the mining software uses to determine referents are not static, they are constantly being updated. Therefore, re-mining the same article some period of time later can find results that were not found before.
The text-mining software currently mines the articles for references to other journal articles (in PMC and PubMed), as well as these types of data that are stored in other Entrez databases:
- taxonomy
- nucleotide
- unists
- protein
- snp
- geo
- structure
In addition to merely recognizing these terms in the articles, the software validates the results, by verifying that each of the terms actually exists in the target database.
The text mining software is used in NCBI Books as well as in PMC. The software is very configurable. Among the things that can be configured are:
- The types of terms to search for, and where (which logical sections) in the articles to search.
- What parts of articles to ignore.
The text mining software is organized as a set of plugins, with each plugin implemented as a Perl module, and mining the article for a particular kind of data.
Web Services and APIs
PMC provides several Web services and APIs to facilitate programmatic access to our resources. Among these are the OAI-PMH service, an FTP server, and the Open Access (OA) Web service.
External users wishing to reuse the content in the PMC Open Access subset should retrieve the articles from our FTP servers, rather than attempting to download them by other means.
If you have questions or comments about these, or any of the other services provided by PMC, please write to the ksed pleh CMP. To stay informed about new or updated tools or services provided by PMC, subscribe to the PMC-Utils-Announce mailing list.
You can read about each of these services on their respective description pages: the OAI-PMH Service, the FTP Server, and the OA Web Service.
The OAI-PMH service is implemented as a CGI program, written in C++, which uses the NCBI C++ Toolkit (21).
The FTP site is populated by a "dumper" script, which encapsulates knowledge about which articles within the Open Access subset have been updated, and how to propagate those updates to the various resources on the FTP site.
The OA web service is implemented as a Perl script deployed as a fast CGI under the Apache Web server. The OA Web service uses database tables that are maintained by the same dumper script as generates the artifacts that are available on the PMC FTP site. Those tables store the information needed by this service, including, for each article in the OA subset, the most recent dates and times that a file (of either format TAR.GZ or PDF) were updated.
Usage Statistics
Each PMC participant has password-controlled access to a website that presents usage reports for that participant’s journals at both the journal and article level. The reports, updated daily, include counts of available articles, total retrieval by format (such as full-text HTML and PDFs), total number of unique IP addresses that have accessed the content, and the most frequently retrieved articles. At the individual article level, usage statistics are available for every article beginning with a journal’s first submission to PMC.
The reports may be downloaded as a CSV file, for analysis with a spreadsheet package such as Microsoft Excel. PMC also provides a CSV file each month, with usage for the month at the article level. Article-level usage data can also be retrieved directly via a Web service call.
PMC’s usage reports generally contain all information called for in the COUNTER specification, except that PMC does not report use by specific institutions. NLM’s privacy policy prevents the reporting of use at an individual or organizational level.
Rendering
Rendering Architecture Overview
PMC implements dynamic rendering of the articles at request time, passing source XML through an XSLT transformation, and integrating external data. Journal archive pages, table-of-contents (issue) pages, and articles, as well as the various discovery portlets that are displayed alongside articles, are all generated dynamically from data in the database and external sources.
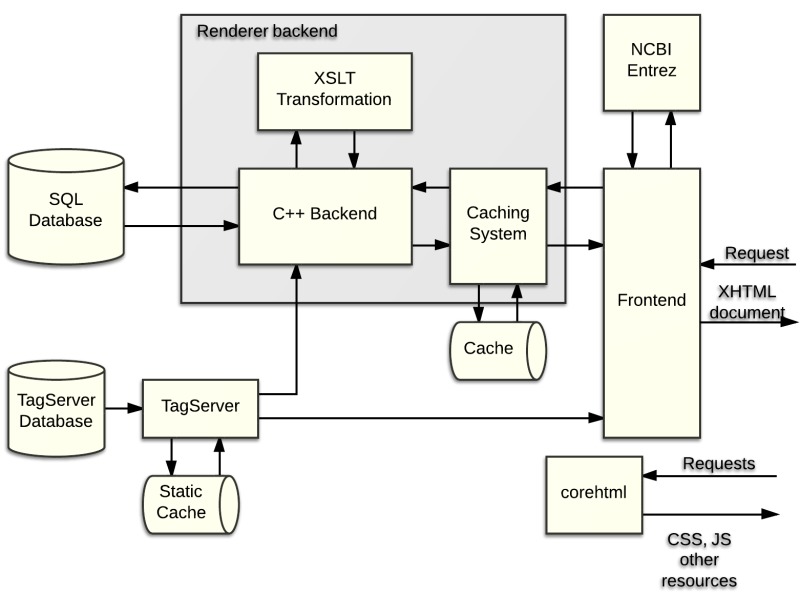
Figure 6 depicts the main components of the renderer, which handle most of the Web pages and other resources available through PMC.
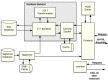
Figure 6.
Components of the PMC Renderer.
The SQL database is the core of the PMC archive, storing all the necessary information related to journals, issues, and individual articles that we receive from a variety of sources.
When a request arrives from a client browser, the frontend system (NCBI Portal) analyzes it, and determines, at a high level, how to handle it. Most requests are for dynamic PMC resources such journal archive pages, issue table-of-contents, or articles themselves, and these requests are routed to the renderer backend.
The renderer backend has a custom short-lived, filesystem-based caching system that is designed to improve performance under certain conditions. Currently, this caching system is disabled for the PMC system, but is enabled in the NCBI Bookshelf. When enabled, the caching system first checks the request to determine if it matches a previous request that has been stored in the filesystem cache. If so, then the cached value is returned. If not, then the request is processed further.
Requests that are not cache hits are parsed and transformed into a set of SQL database queries. The results from those queries are then patched with TagServer data, and then passed through a set of XSLT transformations. Those results are then stored in the filesystem cache (in case there are later requests that match this one) and delivered back to the frontend. The frontend also accesses other NCBI resources, such as Entrez, for data that is added to the display to enhance the usefulness, and the finished results are delivered back to the client.
Many resources, such as images, javascript and css files, are served directly from a static library called corehtml.
Other pages, such as the Home page, about pages, and the Entrez search results page are served through the frontend, but without accessing the renderer backend or the TagServer.
Pages, Views, and URI Structure
The PMC site provides access to many different types of Web pages and other resources. Table 2 below lists those, with hyperlinks to examples.
PMC URIs are designed to be clear, concise, and resource-oriented, as well as to be consistent with the URIs of other resources across the NCBI site. The PMC home page, at http://www.ncbi.nlm.nih.gov/pmc, is the base URI of all of the PMC resources.
The URI space is designed hierarchically, using path segments to identify resource collections, and identifiers to specify items within those collections. For example, the URI /pmc/journals/ specifies the collection of academic journals that have deposited material into the PMC archives. The URI /pmc/journals/2/ specifies one particular journal, the Proceedings of the National Academy of Sciences.
With the PMC URI scheme, there is some flexibility in accessing some types of material because, in a number of cases, more than one URI can be used for the same resource. In these instances, one URI is considered to be canonical (primary) and using any of the ancillary (secondary) URIs for that resource will cause a redirect to the canonical form.
For example, a given article has the canonical URI /pmc/articles/PMC2150930/, in which the identifier PMC2150930 is the PMCID for this article. However, the article could also be accessed through any of several other URIs, which each use a different scheme to identify the unique article, such as the PubMed ID (pmid), the DOI, or the issn-volume-issue-page (ivip). When accessed via those other URIs, the client receives an HTTP redirect to the canonical URI. See Table 2 for a list of URIs supported by the PMC site.
Table 2.
A list of URIs supported by the PMC site. Canonical URIs are given in bold.
Furthermore, we use a couple of NCBI-standard query string parameters to specify various views (report) and formats of these resources. So, for example, "?report=reader" accesses the PubReader view of a full text article.
SQL Databases
PMC uses Microsoft SQL Server to store all of the articles, supplementary material, and metadata of the archive.
Within the database, we define the term "domain," which corresponds roughly to an individual journal. In a simplified view, each publisher can have many domains, each domain can have many issues, each issue has many articles, and each article has one-to-many versions.
There is also a separate table to store information about the citations within an article, including the identifier of the item that is referenced, and another table to store a set of relationships between various articles, including, for example, links to commentary, corrections, and updates.
The actual source content for the articles are stored as "blobs" in the database, including the source XML, images, thumbnails, PDF files, media files, supplementary material, etc. These blobs are stored within dedicated database instances. Once a blob database is full, then it is closed, and never written to or modified again.
The ArticleBlobInfo table cross-references the articles to their associated blobs in the corresponding blob database. This table allows for the dereferencing of request URIs to their requested resources, at render-time. If a particular blob must be changed or deleted for whatever reason, and the blob database in which it is stored is already closed, then we simply write the new version of the blob to a new blob database, and update the pointer in the ArticleBlobInfo table.
Rendering Backend
C++ Backend
The C++ backend is based on the NCBI C++ toolkit, and is written in C++ so that its performance is as fast as possible, and so that it can take advantage of built-in features of that library for logging, database access, and XML processing.
The software runs as a Fast CGI, and provides an HTTP API to the frontend. The frontend passes several parameters to the backend in an XML document via HTTP POST. Among those parameters are the path portion of the original URI (which identifies the requested resource), a session ID, and a user ID (which identifies the MyNCBI account, if the user is logged in). The frontend also passes the Apache environment, which contains important information like the client’s user-agent string.
The C++ backend always returns an XML document that encapsulates the response, that is divided into a response header (including status code, response type, error messages if appropriate), and a response body, which includes the document payload.
When the C++ backend gets a request, it first parses the URI, to determine whether or not it is of canonical form. If the URI is canonical, then the requested resource will be returned directly; if not, then a redirect will be performed. The results of parsing the URI also identify the resource that is being requested. Based on that information, the backend queries the PMC database.
If the query results in an error, or if there is no resource matching the identifiers provided, then the backend will return a response document to the frontend that indicates the error, with appropriate status code and error message.
When the request is for a full-text article, and there is no error, then the renderer backend will also make a request to the TagServer application to retrieve data related to the tags for this article, and that data is patched into the document (see the TagServer section for more information). The backend will pass an MD5 signature of the article document, and the TagServer will compare that to the signature that it has stored. If they do not match, then the request is rejected, and no tag data is returned. This ensures that the patching mechanism, which relies on precise byte offsets into the document, is robust.
The TagServer response actually contains two parts: ids and markers that are used to mark where in the document the tags occur, and the tag attributes. The tag attributes are bundled together and stored in memory, where they are made available to the XSLT converters (described in the next section) through an extension to the XSLT document() function.
The C++ backend also takes some of the metadata related to the resource, and writes that into the document as processing instructions (PIs).
The document is then passed into the XSLT transformer, which uses one of several possible "entry points" within the XSLT stylesheet library to determine how to process the document. The XSLTs are described in more detail in the next section.
XSLTs
The XSLT files that are used by the PMC Renderer are written in XSLT 1.0, since they are processed by libxslt within the NCBI C++ toolkit. They use a few extensions:
- EXSLT (22)
- A few custom XSLT extension functions written in C, to support internationalization
- A custom document() function, to allow the fast retrieval of the TagServer tag attributes.
As mentioned above, the C++ backend invokes the XSLTs at an "entry point," which is a main module that imports all of the others, defines top-level XSLT variables and parameters, and the top-level matching template (the template that matches the document root node). There are different entry points for, for example, full text articles in classic view versus PubReader view.
The XSLTs are designed in a modular fashion, such that there are a core set of templates that are included by all (or most) of the applications, and then each application imports those, but then overrides them selectively, as needed, in order to customize the result.
The XSLT processor gets its input from the following:
- The main XML document—this is passed to the processor as its input document, and comprises:
- NXML—the main article as stored in the PMC database.
- Patches inserted by the TagServer patching mechanism. These are XML elements and attributes that mark the location of tags in the NXML.
- Processing instructions (PIs) inserted by the C++ backend, with metadata about the article from the PMC database.
- XSLT parameters, which come from:
- Parameters passed in from the frontend,
- Renderer backend configuration files (ini files),
- Metadata from the PMC database.
- In-memory document—this is the tag attribute data from the TagServer. This is accessed from within the XSLTs using the document() function, which invokes a custom extension integrated with the C++ backend.
NCBI Portal
The frontend system that is used to render PMC content is an internally-developed XML-based web application server known as NCBI-Portal (referred to simply as "Portal" for the rest of this section). Portal is written in C++, based on the NCBI C++ toolkit, and uses the libxml and libxslt libraries for XSLT and the Zorba XQuery processor (23) for XQuery.
The Portal system is used throughout NCBI, and individual applications are implemented with their own applications, which are bundles of components called snapshots. Each snapshot is versioned independently. The PMC site is implemented with three snapshots: PMCViewer, PMCStatic, and the PMC Entrez snapshots.
Request Router
Since the Portal system handles requests for all of NCBI, the first order of business for the Portal system, when it receives a request, is to dispatch that request to the correct snapshot. This is done in two steps:
- 1.
The combination of the top-level domain (i.e., "www") and the topmost path segment of the URI (in our case, "pmc") is used to select a request router. (Within NCBI we use other top-level domains, for example, "test", for development and testing, and these might resolve to different request routers.)
- 2.
The request router contains a list of regular expression rules that are matched against the URI and the request environment. The first rule that matches selects the snapshot that will handle this request.
This design allows each application within NCBI to be independent, which is important in such a large and diverse organization. It also provides great flexibility in handling requests, because it permits fine-grained control over how specific URIs are handled within a site.
As mentioned above, the PMC site is handled by three separate snapshots, which are described in more detail below.
PMCViewer Snapshot
The PMCViewer snapshot is the main snapshot used by the PMC system. It handles the rendering tasks for the journal list, journal, issue, and article pages, as well as many others. This is the snapshot that interacts with the renderer backend, as shown in Figure 6. (The other snapshots produce different types of pages that do not originate with the renderer backend).
The snapshot contains a set of rules which further examine the requested URI and other HTTP headers in order to correctly dispatch the request. As the result of this, the snapshot might:
- Immediately respond with a redirect to another URI (for example, if a page has moved, or if the user has used a non-canonical URI).
- Reverse-proxy a binary resource, such as a PDF, figure or table thumbnail, etc.
- Invoke the rendering backend to retrieve page data from the database (as described above).
When invoking the rendering backend, the snapshot passes the URI and other request parameters to the rendering backend, which responds with information about how to render this particular page. The snapshot then does some further processing of the XML response from the backend, and produces the final integrated HTML page that is sent to the browser.
PMCStatic Snapshot
The PMCStatic snapshot handles the rendering of information and news pages, for example, the PMC Overview. These are generated from a set of XML, XHTML and other types of files that are stored in a specific directory within the renderer backend. The directory includes these types of resources:
- nav.xml—specifies the navigation bar menu items
- redirects.xml—this specifies any redirects that should occur (in those cases where a page has moved, the old URI will redirect to the new)
- XHTML pages—these are the content of the individual pages, and are divided into the groups about, pub (for publishers), and tools (related resources)
- Images, PDFs, and other downloadable resources
When a page is requested, the URI is translated into the pathname of an XHTML file within this directory, that file is retrieved, and then it is processed with XSLT to combine it with the navigation bar, and the other NCBI-standard components such as the header and the footer.
PMC Entrez Snapshot
The PMC Entrez snapshot is derived from the NCBI standard Entrez package of components, which allows PMC to share the same look-and-feel and many functional aspects with other applications at NCBI. In particular, the home page is delivered by this snapshot, as well as many of the search-related pages such as limits, the Advanced Search Builder, the clipboard, and search details.
Customizations to the default Entrez packages exist to provide PMC-specific behavior. For example, on the home page, a panel in the center displays up-to-date highlights on the number of articles currently archived in PMC. This display is produced from an XML file that is generated daily and pushed to the renderer backend location.
Customizations for the Entrez search results provide for, for example, displaying imagedocsum results.
PubReader
PubReader is a set of JavaScript and CSS files that provide for rendering journal articles in a way conducive to reading, especially on a tablet or a small-screen device.
The article document for the PubReader view is generated by the same mechanism as the classic view, utilizing the renderer Backend, the TagServer, and the Frontend, but the format of the XHTML document is somewhat different, in terms of the specific elements and attributes used, CSS class names, and the document structure. This difference is achieved within the renderer by invoking the XSLTs with a PubReader-specific entry point.
PubReader uses some of the latest features from the HTML5 and CSS3 standards, in order to achieve the dynamic display. Chief among these is the CSS3 multi-column layout module, in conjunction with a fixed page height.
Each JavaScript component is written as a jQuery extension, based on the "Nested Namespacing Plugin Pattern," by Doug Neiner, and described in Learning JavaScript Design Patterns (24).
Among the components of PubReader are:
- PageManager—controls and performs page turning in the PubReader.
- HistoryKeeper—monitors and controls the fragment identifier (the part after the "#" symbol) of the URI, and instructs the PageManager to turn pages to the destination defined in that fragment identifier.
- ObjectBox—this component handles the modal box that opens to display a figure, a table, or a message box.
- PageProgressBar—gives a visual indication of the current location within the document, and provides a control to allow the user to move to a new location. This component uses a modified version of the rangeinput widget from jquerytools (25).
- Links—provides the ability to hijack clicks on links.
The PubReader code is managed in the master NCBI Subversion repository, and is mirrored to the GitHub repository NCBITools/PubReader.
Caching System
The caching system in PMC is used primarily to boost performance in those cases when there is a surge of requests, over a short period of time, for one or a few articles or resources. It uses the filesystem to store copies of each resource as it is requested, using a pathname that is generated by a unique key. The key value is an MD5 signature of a string that is composed of query parameters and environment variables that uniquely specify the requested resource. When another request arrives that results in the same key and before the cache entity has expired (a cache hit), then the resource is retrieved from the filesystem instead of being regenerated dynamically.
Currently, the caching system is disabled for the PMC renderer backend but is being used with NCBI Bookshelf. Each of these is each set up with an independent cache.
The types of requests that are cached are configurable. Also configurable is a setting for the minimum amount of free space on the filesystem. This prevents the caching system from filling up the disc and is typically set to four gigabytes. Once that limit is reached, no new requests will be cached until old ones are purged.
There is a purging mechanism that runs continuously and "garbage collects" cache entities on the filesystem that have expired.
The full path for each entity in the cache is derived from the MD5 signature by using the first four hex digits and directory names, and the rest of the MD5 as a filename. The disc files that make up the cache include, in a header, metadata about the resource and about this particular cache entity, including the request parameters that resulted the cache hit, the date and time the entity was stored, and metadata from the PMC database such as article id, blobname, etc.
The caching system is configured so that an entity will expire after one hour. The reason for this is that the final rendered output for a request typically changes every day.
The system is implemented in libraries in both C++ and in Perl. They are maintained independently, but both use the same format for the filesystem and the headers.
TagServer
Overview
The TagServer is a database system that contains metadata which has been mined from PMC articles. Tags are (typically) strings of text within a source document that are associated with a set of metadata attributes. For example, the gene name "D17Jcs48" might occur within an article in PMC. The text mining process would recognize this as a gene name, determine its numeric ID number in the Entrez gene database, and would store information about this instance of this gene name into the TagServer database, as a single tag, associated with the article instance.
The TagServer system was designed to be generic, and the TagServer database highly configurable, such that it can store metadata about a wide variety of tags that can occur in a variety of source documents.
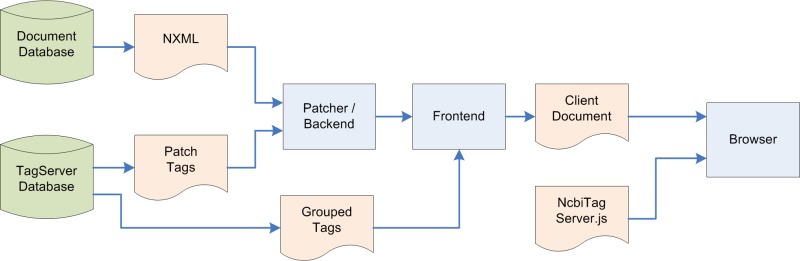
Figure 7 illustrates the data flow involved in mashing up TagServer data with an article rendered in PMC.
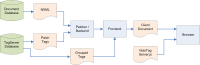
Figure 7.
TagServer data flow.
As can be seen in this illustration, during rendering, the TagServer delivers two types of tags associated with an article. The first, "patch tags," are integrated with the XML document immediately as it comes out of the document database, before it is processed as XML. These are tags that are associated with very specific parts of the document, for example, specific strings of text like gene names. The second type of tags is grouped tags, and these are associated with areas within the article that are identified by XML id attributes, for example, paragraphs. The data for these tags is integrated into the XML document by the frontend XML processing engine.
Finally, the generated XHTML document is delivered to the client browser, along with a JavaScript module that performs the final steps required for rendering, such as positioning discovery blocks, implementing tooltips, etc.
Tags
Tags in the TagServer database are classified in a number of ways.
The most basic classification is the nature of the entity within the PMC article (i.e., the subject of the tag). This is associated with whether or not, and how the tag marks are patched into the document. For example, some tags refer to short snippets of text, and these are patched in as new XML elements. Others are patched in "raw," meaning that the content of the tag is simply inserted into the document. Others are not patched in at all; for example, tags that refer to the document as a whole, rather than to a specific part.
Tags are also classified according to the semantics of the reference (i.e., the object of the tag). This classification is "tag type," and is fully configurable. Examples are entrez-gene, entrez-nucleotide, glossary (for glossary terms), and reference. New tag types can be defined as needed.
There are also special types of tags that are used to store XML id values that get patched into the instance documents, to enhance their addressability.
Every tag has a set of attributes associated with it, which are key-value pairs. These attributes are stored in the database by the text mining process, and are not constrained by the TagServer software itself. However, some tag attributes have a set meaning defined by the application layer. For example, the tag attribute term_id is used to specify the numeric ID of the object of a tag. In the case of an item in an Entrez database, term_id stores the UID of the item. Other attribute names with fixed meanings are reference_id, pubmed_id, etc.
Database Design
The TagServer data is organized around the concept of a presentation, which is like a stored query. When the database is accessed with a given presentation and an object identifier, then it will respond with a specific set of tags, in a defined format. A given presentation is specified by three parameters: object type, site, and request type.
Each tag refers to a specific object in the PMC database, which is an XML instance document. Because tags are patched into this document before they are parsed as XML, it is essential that the instance document is byte-for-byte identical with the copy that was used for text mining. To ensure this, an MD5 signature of each object is stored in the TagServer database. When the article is rendered, the MD5 is again computed, and if it doesn’t match, then the tag data is discarded, and not patched into the document.
API
The TagServer is accessed through a RESTful Web service API, with resources identified in the path portion of the URIs, and query parameters used to define the desired set of tags and their format. An example of a TagServer request is
/tags/prod/srv/pmcai/2464733/tags?site=prod&rt=backend
This specifies the PMC article instance with ID 2464733, and the site and rt (request type) parameters specify the desired presentation. The presentation, as described above, specifies exactly what tag types are desired, and the format, sorting, and grouping.
Implementation
The TagServer is implemented as a Perl Fast CGI script, and has a separate database from the main PMC database. It has a Perl Catalyst based Web interface that allows it to be configured for each application in which the TagServer is used. That configuration consists of definitions of tag types, sites, request types, and the various presentations, or collections and groupings of tags that are returned for each type of request.
TagServer Static Cache
The TagServer static cache system is used to boost the performance of the TagServer. The static cache itself is a large binary file that holds all of the possible "realistic" TagServer responses for all of the PMC articles in our archives. "Realistic" responses mean those that are actually generated by requests from production servers, given the way they are currently configured.
Currently, for every article, there are three different realistic responses, corresponding to the three request types: backend, frontend, and indexer. Multiplying those by about three million articles means that there are on the order of ten million responses that are stored in the static cache file.
The static cache file stores the responses as a large hash table, using a form of open addressing (26) collision resolution algorithm known as Cuckoo hashing (27). This file is regenerated every day from all the PMC articles, including any that have been newly mined.
This design means that the static cache system is very efficient: since it contains (nearly) all of the possible responses, the hit rate is very high (about 99.9%). The only requests that do not hit the cache are those for articles that were updated since the last time the static cache was regenerated. Also, the response time when there is a hit is on the order of 10 milliseconds, versus 150 milliseconds when there is no hit.
PMCI
PMC International (PMCI) is a collaborative effort between the NLM, publishers, and organizations in other countries, to create a network of digital archives that share content. Currently there are two active PMCI sites: Europe PubMed Central ((28); originally UKPMC), which went online in January, 2007, and PMC Canada (29), which went online in October, 2009. See the PMC International page on the PMC website (4) for more general information about this collaboration effort.
The PMCI sites use the same database architecture, the same backend software, and the same business rules as NCBI PMC. They deploy the following software developed by NCBI:
- Portable NIHMS (pNIHMS)—a portable version of NIH manuscript submission system (NIHMS) that allows authors and publisher to submit manuscripts directly to one of the PMCI sites.
- Portable PMC (pPMC)—an archiving and rendering system used to store and deliver the content to the end users.
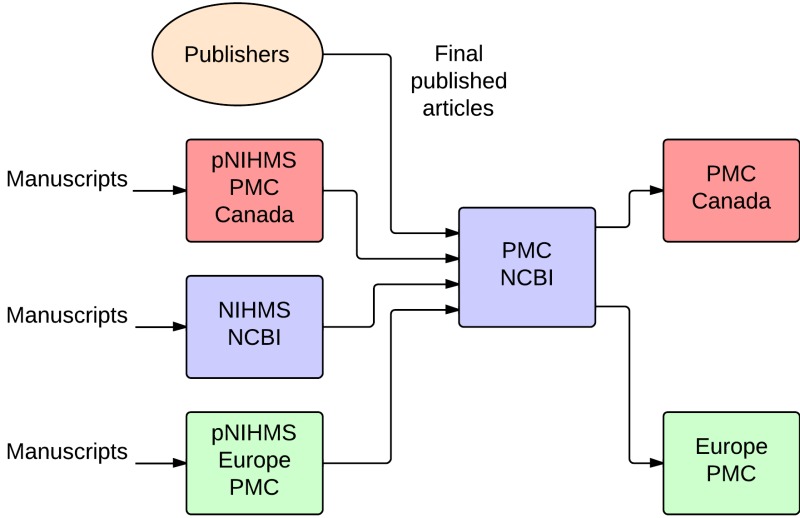
The pPMC software has a collector component that communicates with NCBI PMC to update content in the pPMC database. The collector enables the exchange of content and metadata between the PMCI site and NCBI PMC, as illustrated in Figure 8. Whereas publishers submit final published versions of all articles directly to NCBI PMC, PMCI sites receive author manuscripts through the pNIHMS system. For example, Europe PMC accepts and processes author manuscripts of journal articles funded by the Europe PMC sponsoring agencies (30).
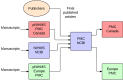
Figure 8.
Data exchange between PMCi sites and PMC NCBI.
All content is initially sent to NCBI PMC, and then redistributed to the PMCI sites. NCBI controls which new and updated content is distributed to which sites, and makes that content available through the PMCI collector system. The PMCI sites retrieve that content from the PMCI collector, as frequently as necessary.
The pPMC sites use the same database structure as NCBI PMC, and the software includes most of the same rendering components, including the rendering backend . The frontend is different for each, which allows each PMC site to customize and enrich the article display in its own way.
Note that in order to support PMCI, the PMC renderer software is internationalized, to allow for multilingual translations of various components.
Other Tools and Utilities
PMC provides a number of tools and utilities, mostly to aid publishers who deposit content in our archive, but a few others that are of general usefulness. The publisher tools and utilities are described on the File Validation Tools page [31].
Here is a list of the tools and utilities provided by PMC:
- Citation Search
- PMCID - PMID - Manuscript ID - DOI Converter
- XML validator and SGML validator
- PMC Style Checker
- Article Previewer
Most of these tools are served through the PMCStatic snapshot, described above.
Citation Search
This is a very simple form interface to the Entrez search system.
PMCID - PMID - Manuscript ID - DOI Converter
This is served by the PMCStatic snapshot of the frontend system, at the URL http://www.ncbi.nlm.nih.gov/pmc/pmctopmid/. This tool is just a wrapper for the ID converter API service. See the ID Converter API documentation page for more information.
XML and SGML Validators
These are served by the PMCStatic snapshot, which accesses a CGI backend written in Perl to handle the validation. The uploaded file is sent the CGI backend via an HTTP POST request, and a separate Perl module is engaged to validate the document, and report errors.
PMC Style Checker
The Style Checker verifies that an uploaded document conforms to the PMC XML Tagging Guidelines (9). This utility is served by the PMCStatic snapshot, via a CGI backend written in Perl. That CGI script sends the uploaded document through a set of XSLT transformations that check the document conforms to the set of rules defined by the tagging guidelines. A downloadable version of these XSLT transformations is available.
Article Previewer
The article previewer allows users to view uploaded articles the way they would appear in PMC. The tool requires users to have a MyNCBI account, and it associates any uploaded articles with that account.
The tool runs the same processes that are used during PMC production. This allows users to view an article as it would appear in PMC that is tagged in NLM XML according to PMC Style, or any XML or SGML DTD that PMC currently accepts for data submissions. See the Article Previewer Instructions and FAQ for more information.
The article previewer is currently implemented as a stand-alone CGI program (not served through any NCBI Portal frontend snapshot). It accesses a separate and independent database that is created with the same schema and tables as the main PMC production database. To render the converted articles, it uses a stand-alone version of the renderer backend.
References
- 1.
- Add a Journal to PMC [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/pub/pubinfo/. - 2.
- License your work [Internet]. Mountain View, CA: Creative Commons; 2013 [cited 2013 Nov 1]. Available from http:
//creativecommons .org/about/license/. - 3.
- Public Access [Internet]. Bethesda, MD: National Institutes of Health; 2013 [cited 2013 Nov 1]. Available from http://publicaccess
.nih.gov/. - 4.
- PMC International [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/about/pmci/. - 5.
- PMC File Submission Specifications [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/pub/filespec/ - 6.
- Fact Sheet: Technical Services Division [Internet]. Bethesda, MD: National Library of Medicine: 2013 [cited 2013 Nov 1]. Available from http://www
.nlm.nih.gov /pubs/factsheets/tsd.html. - 7.
- Minimum Criteria for PMC Data Evaluations [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2009 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/pmcdoc/mindatareq.pdf - 8.
- XML Catalogs. OASIS Standard, Version 1.1. 7 October 2005. (http://www
.oasis-open .org/committees/download .php/14810/xml-catalogs.pdf) - 9.
- PubMed Central Tagging Guidelines [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/pmcdoc/tagging-guidelines /article/style.html - 10.
- OAI-PMH Service [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/about/oai.html. - 11.
- W3C. “Processing Instructions.” Extensible Markup Language (XML) 1.0 (Fifth Edition). 2008 [cited 2013 Nov 1]. Available from (http://www
.w3.org/TR/REC-xml/#sec-pi). - 12.
- PMCID/PMID/NIHMSID Converter [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/pmctopmid/. - 13.
- Entrez Programming Utilities Help [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2010 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/books/NBK25501/. - 14.
- PMC Help [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2005 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/books/NBK3825/. - 15.
- PMC Advanced Search Builder [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/advanced/. - 16.
- My NCBI – Filters [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/sites/myncbi/filters/. - 17.
- “Working with Filters” My NCBI Help [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2010 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/books/NBK53591/. - 18.
- Entrez Link Descriptions [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://eutils
.ncbi.nlm .nih.gov/entrez/query /static/entrezlinks.html#pmc. - 19.
- Ergatis: Workflow creation and monitoring interface. [Internet]. [cited 2013 Nov 1]. Available from http://ergatis
.sourceforge.net/. - 20.
- Moose: A postmodern object system for Perl [Internet]. Infinity Interactive; 2006 [cited 2013 Nov 1]. Available from http://moose
.iinteractive.com/en/. - 21.
- The NCBI C++ Toolkit Book [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2004 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/books/NBK7160/. - 22.
- EXSLT [Internet]. [cited 2013 Nov 1]. Available from http://www
.exslt.org/. - 23.
- Zorba NoSQL Query Processor [Internet]. 2008 [cited 2013 Nov 1]. Available from http://www
.zorba.io/. - 24.
- Osmani, Addy. Learning JavaScript Design Patters [Internet]. 2012 [cited 2013 Nov 1]. Available from http://addyosmani
.com /resources/essentialjsdesignpatterns /book/. - 25.
- RANGEINPUT [Internet]. jQuery Tools [cited 2013 Nov 1]. Available from http://jquerytools
.org /documentation/rangeinput/index.html. - 26.
- Open Addressing [Internet]. Wikpedia [cited 2013 Nov 1]. Available from http://en
.wikipedia.org /wiki/Open_addressing. - 27.
- Cuckoo hashing [Internet]. Wikpedia [cited 2013 Nov 1]. Available from http://en
.wikipedia.org /wiki/Cuckoo_hashing. - 28.
- Europe PubMed Central. [cited 2013 Nov 1]. Available from http://europepmc
.org/. - 29.
- PMC Canada. [cited 2013 Nov 1]. Available from http:
//pubmedcentralcanada.ca/pmcc/. - 30.
- Europe PubMed Central Funders. [cited 2013 Nov 1]. Available from http://europepmc
.org/funders/. - 31.
- File Validation Tools [Internet]. Bethesda, MD: National Center for Biotechnology Information; 2013 [cited 2013 Nov 1]. Available from http://www
.ncbi.nlm.nih .gov/pmc/pub/validation/.
Publication Details
Author Information and Affiliations
Authors
Chris Maloney,1 Ed Sequeira,1 Christopher Kelly,1 Rebecca Orris,1 and Jeffrey Beck1.Affiliations
Publication History
Created: November 14, 2013; Last Update: December 5, 2013.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Maloney C, Sequeira E, Kelly C, et al. PubMed Central. 2013 Nov 14 [Updated 2013 Dec 5]. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.