This publication is provided for historical reference only and the information may be out of date.
Summary
PubMed is a free resource developed and maintained by the National Center for Biotechnology Information (NCBI), a division of the U.S. National Library of Medicine (NLM), at the National Institutes of Health (NIH).
PubMed comprises over 22 million citations and abstracts for biomedical literature indexed in NLM’s MEDLINE database, as well as from other life science journals and online books. PubMed citations and abstracts include the fields of biomedicine and health, and cover portions of the life sciences, behavioral sciences, chemical sciences, and bioengineering. PubMed also provides access to additional relevant websites and links to other NCBI resources, including its various molecular biology databases.
PubMed uses NCBI's Entrez search and retrieval system. PubMed does not include the full text of the journal article; however, the abstract display of PubMed citations may provide links to the full text from other sources, such as directly from a publisher’s website or PubMed Central (PMC).
Data Sources
MEDLINE
The primary component of PubMed is MEDLINE, NLM’s premier bibliographic database, which contains over 19 million references to journal articles in life sciences, with a concentration on biomedicine.
The majority of journals selected for MEDLINE are based on the recommendation of the Literature Selection Technical Review Committee (LSTRC), an NIH-chartered advisory committee of external experts analogous to the committees that review NIH grant applications. Some additional journals and newsletters are selected based on NLM-initiated reviews in areas that are special priorities for NLM or other NIH components (e.g., history of medicine, health services research, AIDS, toxicology and environmental health, molecular biology, and complementary medicine). These reviews generally also involve consultation with an array of NIH and outside experts or, in some cases, external organizations with which NLM has special collaborative arrangements.
Non-MEDLINE
In addition to MEDLINE citations, PubMed also contains:
- In-process citations, which provide a record for an article before it is indexed with NLM Medical Subject Headings (MeSH) and added to MEDLINE or converted to out-of-scope status.
- Citations that precede the date that a journal was selected for MEDLINE indexing.
- Some OLDMEDLINE citations that have not yet been updated with current vocabulary and converted to MEDLINE status.
- Citations to articles that are out-of-scope (e.g., covering plate tectonics or astrophysics) from certain MEDLINE journals, primarily general science and general chemistry journals, for which the life sciences articles are indexed with MeSH for MEDLINE.
- Citations to some additional life science journals that submit full-text articles to PubMed Central and receive a qualitative review by NLM.
Journal Selection Criteria
Journals that are included in MEDLINE are subject to a selection process. The Fact Sheet on Journal Selection for Index Medicus®/MEDLINE® describes the journal selection policy, criteria, and procedures for data submission.
History
PubMed was first released in January 1996 as an experimental database under the Entrez retrieval system with full access to MEDLINE. The word "experimental" was dropped from the website in April 1997, and on June 26, 1997, free MEDLINE access via PubMed was announced at a Capitol Hill press conference. Use of PubMed has grown exponentially since its introduction: PubMed searches numbered approximately 2 million for the month of June 1997, while current usage typically exceeds 3.5 million searches per day.
PubMed was significantly redesigned in 2000 to integrate new features such as LinkOut, Limits, History, and Clipboard. PubMed began linking to PubMed Central full-text articles and the Bookshelf's initial book, Molecular Biology of the Cell. The Entrez Programming Utilities, E-Utilities, and the Cubby (My NCBI subsequently replaced the Cubby) also were released.
In 2002, the PubMed database programming was completely redesigned to work directly from XML files, and two new NCBI databases, Journals (now the NLM Catalog) and MeSH, were created to provide additional search capabilities for PubMed.
Electronic Data Submission
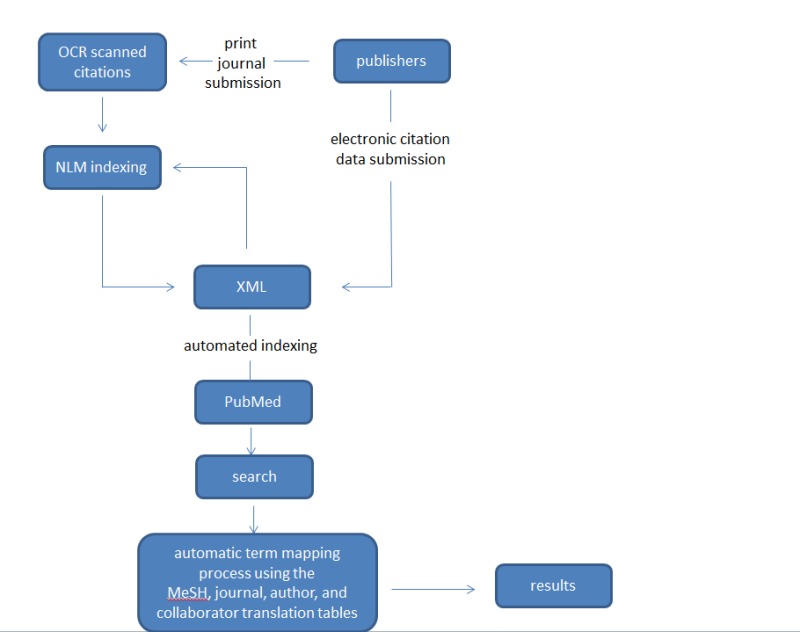
Publishers of journals indexed for MEDLINE are encouraged to submit citation and abstract data electronically for inclusion in PubMed. Electronic submissions ensure that citations and abstracts are available to the public within 48 hours of uploading a properly formatted XML file. See Figure 1 for information about the PubMed data flow.
Figure 1.
A schematic representation of PubMed data flow
NCBI works with many publishers and commercial data providers to prepare and submit electronic citation and abstract data. In 2012, over 90% of the citations added to PubMed were submitted electronically.
Electronic citation data submission also fulfills one of the requirements to add an icon to PubMed citations that links to full text of the articles available on the journal website. Linking can be achieved using LinkOut, which is the feature that generates a direct link from a PubMed citation to the journal website.
NLM manually creates citations for journals uninterested or unable to submit electronic citation data by scanning the print copy of the journal and using optical character recognition (OCR). This process can take significantly longer than electronic submissions due to the manual nature of the process.
Electronic Data Submission Process
Electronic citation and abstract data are submitted via File Transfer Protocol (FTP) in XML according to the PubMed Document Type Definition (DTD). Details about the PubMed XML tagged format, XML tag descriptions, sample XML files, and how to handle special characters are available in the online documentation.
NCBI staff guide new data providers through the approval process for file submission. New data providers are asked to submit a sample XML file, which is reviewed for XML formatting and syntax and for bibliographic accuracy and completeness. The file is revised and resubmitted until all criteria are met. Once approved, a private account is set up on the NCBI FTP site to receive XML citation data for future journal issues.
NCBI loads publisher-supplied data daily, Monday through Friday, at approximately 8:30 a.m. Eastern Time. New citations are assigned a PubMed ID (PMID), a confirmation report is sent to the data provider, and the citations become available in PubMed after 6:00 a.m. the next day, Tuesday through Saturday. Citation data submitted Friday after 8:30 a.m. through Sunday will be available in PubMed after 6:00 a.m. the following Tuesday.
After posting in PubMed, the citations are sent to NLM’s Index Section for bibliographic data verification and the addition of MeSH terms from NLM’s controlled vocabulary. The indexing process can take up to a few months, after which time the completed citations flow back into PubMed, replacing the original data.
Publishers or others interested in submitting electronic data may view the XML Data Provider Help or write to vog.hin.mln.ibcn@rehsilbup for more information.
Database Management and Hardware
PubMed uses its own proprietary Web-based search engine. The Web server software is the open-source Apache HTTP server.
PubMed runs on approximately 62 standard Linux servers, each with two quad core 2.6-3.6 GHz Intel Nehalem CPUs, 48-64 GB of memory, 1TB of local storage, and a Gigabit Ethernet connection. Fourteen NCBI Portal servers render the PubMed Web interface and eight servers search the PubMed index. PubMed records are retrieved from four proprietary XML article servers, while the "Related Articles" service relies on 20 servers running a specialized non-relational database system. The remaining servers support services such as links to related information in other NCBI databases (e.g., PubMed Central, Nucleotide, and the Sequence Read Archive), LinkOut, History, and My NCBI.
To accommodate the volume of data output by PubMed and other Web-based services, the NLM has a 3-Gbps connection to the commercial Internet as well as a 20-Gbps connection to Internet2, the non-commercial network used by many leading research universities at the NIH campus, and similar connectivity at a second redundant data center.
Indexing
The Automatic Indexing Process
The indexing process automatically generates access points for each field of a PubMed citation.
- Terms within a citation are initially extracted (stopwords are ignored) and matched against a list of useful phrases.
- Individual terms are added to the corresponding field, e.g., title words are added to the title field index and the title/abstract field index.
- All terms are also added to the all fields index (except for the terms found in the place of publication (Country) and transliterated title fields).
- Some fields use a special set of rules for extracting data:
- Several index points are created for authors, e.g., indexes for the author Harold Varmus will include Varmus H; Varmus, Harold; and Varmus.
- Indexes for MeSH terms include the MeSH term and MeSH major term fields, subheading fields, etc.
Field indexes may be browsed using the PubMed Advanced Search Builder “Show index list” feature.
How PubMed Queries Are Processed
Automatic Term Mapping
Untagged terms that are entered in the search box are matched against the following translation tables and indexes in this order:
- 1.
a MeSH (Medical Subject Headings) translation table,
- 2.
a journals translation table,
- 3.
the full author translation table,
- 4.
author index,
- 5.
the full investigator (collaborator) translation table, and
- 6.
an investigator (collaborator) index.
On the right side of a search results page there are a number of tools designed to enhance user discovery, including one called “Search details” that shows the search term translations.
When a match is found for a term or phrase in a translation table the mapping process is complete and does not continue on to the next translation table.
1. MeSH Translation Table
The MeSH Translation Table contains:
- MeSH Terms
- See-reference mappings (also known as entry terms) for MeSH terms
- MeSH Subheadings
- Publication Types
- Pharmacologic Actions
- Terms derived from the Unified Medical Language System (UMLS) that have equivalent synonyms or lexical variants in English
- Supplementary Concepts (chemical, protocol or disease terms) and their synonyms
If a match is found in the MeSH translation table, the term will be searched as MeSH (that includes the MeSH term and any specific terms indented under that term in the MeSH hierarchy), and in all fields.
For example, if you enter “multiple sclerosis” in the search box, PubMed will translate this search to:
"multiple sclerosis"[MeSH Terms] OR ("multiple"[All Fields] AND "sclerosis"[All Fields]) OR "multiple sclerosis"[All Fields]
If you enter a MeSH Term that is also a Pharmacologic Action, PubMed will search the term as [MeSH Terms], [Pharmacologic Action], and [All Fields].
If you enter a synonym for a MeSH term, the translation will also include an all fields search for the MeSH term associated with the synonym.
Search term: ear infection
“ear infection” is an synonym for the MeSH term “otitis” in the MeSH translation table.
Search translated to: "otitis"[MeSH Terms] OR "otitis"[All Fields] OR ("ear"[All Fields] AND "infection"[All Fields]) OR "ear infection"[All Fields]
When a term is searched as a MeSH term, PubMed automatically searches that term plus the more specific terms underneath in the MeSH hierarchy:
Search term: breast cancer
“Breast cancer” is an entry term for the MeSH term “breast neoplasms” in the MeSH translation table.
“Breast neoplasms” includes the specific headings below, all of which are also searched:
Breast Neoplasms, Male
Carcinoma, Ductal, Breast
Hereditary Breast and Ovarian Cancer Syndrome
Inflammatory Breast Neoplasms
2. Journals Translation Table
If the search term(s) is not found in the MeSH translation table, the process continues on to look for a match in the journals translation table, which contains full journal title, journal title abbreviation, and International Standard Serial Numbers (ISSNs). Search term(s) automatically map to the journal abbreviation:
Search term: New England Journal of Medicine
“New England Journal of Medicine” maps to N Engl J Med.
Search translated to: “N Engl J Med” [Journal Name]
Journal titles are included in the all fields index; therefore, a search for a MeSH term that is also a journal title will retrieve citations for the journal as well:
Search term: nature
Search translated as: "nature"[MeSH Terms] OR "nature"[All Fields]
The search will include the journal Nature
3. Full Author Index
The full author translation table includes full author names for articles published from 2002 forward, if available.
4. Full Investigator (Collaborator) index
If the term is not found in the above tables, except for full author, and is not a single term, the full investigator index is consulted for a match. The full investigator (collaborator) translation table includes full names, if available.
5. Author Index
If the term is not found in the above tables, except for full author or full investigator, and is not a single term, PubMed checks the author index for a match.
An author name search should be entered in the form: last name (space) initials, e.g., o'malley f, smith jp, or gomez-sanchez m.
If only one initial is used, PubMed retrieves all names with that first initial, and if only an author's last name is entered, PubMed will search that name in All Fields. It will not default to the author index because an initial does not follow the last name:
Search term: o'malley f
Search retrieves authors: o'malley fa, o'malley fb, o'malley fc, o'malley fd, o'malley f jr, etc.
Search term: o'malley
Search translated as: “o'malley” [All Fields]
A history of the NLM's author indexing policy regarding the number of authors to include in a citation is outlined in Table 1.
Table 1.
History of NLM author-indexing policy
6. Investigator (Collaborator) index
If the term is not found in the above tables, except for full author, author, or full investigator, and is not a single term, PubMed checks the investigator index for a match.
7. If no match is found?
PubMed breaks apart the phrase and repeats the above automatic term mapping process until a match is found. PubMed ignores stopwords in searches.
If there is no match, the individual terms will be combined (AND-ed) together and searched in all fields (see Simple Searching section below).
One exception: PubMed interprets a sequence of numbers, e.g., 23436005 23193264, as PubMed IDs, and the IDs will be OR-ed individually rather than combined (AND-ed).
Search Rules and Field Abbreviations
It is possible to override PubMed's automatic term mapping by using search rules, syntax, and specific search field tags.
The Boolean operators AND, OR, and NOT should be entered in uppercase letters and are processed left to right. Nesting of search terms is possible by enclosing concepts in parentheses. The terms inside the set of parentheses will be processed as a unit and then incorporated into the overall strategy, e.g., therapy AND (hay fever OR asthma).
Terms may be qualified using PubMed's Search Field Descriptions and Tags. Each search term should be followed by the appropriate search field tag, which indicates which field will be searched. For example, the search term cell [jour] will only search the journal field. Specifying the field precludes the automatic term mapping process that would result in using the translation tables, e.g., MeSH.
Using PubMed
Searching
Simple Searching
A simple search can be conducted from the PubMed homepage by entering terms in the search box and clicking the Search button or pressing the Enter key.
Term suggestions will display for search terms entered in the search box.
If more than one term is entered in the search box, PubMed will go through the automatic term mapping process described in the previous section, looking for exact matches for each term. If the exact phrase is not found, PubMed clips a term off the end and repeats the automatic term mapping, again looking for an exact match, but this time to the abbreviated query. This continues until none of the words are found in any one of the translation tables. In this case, PubMed combines terms (with the AND Boolean operator) and applies the automatic term mapping process to each individual word. PubMed ignores stopwords, such as “about,” “of,” or “what.” Users may also apply their own Boolean operators (AND, OR, NOT) to multiple search terms; the Boolean operators must be in uppercase.
If a phrase of more than two terms is not found in any translation table, then the last word of the phrase is dropped, and the remainder of the phrase is sent through the entire process again. This continues, removing one word at a time, until a match is found.
If there is no match found during the automatic term mapping process, the individual terms will be combined with AND and searched in all fields:
Search term: heart attack bad diet
Automatic term mapping process:
heart attack bad diet => no matches
heart attack bad => no matches
heart attack => MeSH translation table match, remove "heart attack" from the query
bad diet => no matches
bad => no matches, search in all fields, remove "bad" from the query
diet => MeSH translation table match, remove "diet" from the query
Processing stops because the query string is empty
Translated as: ("myocardial infarction"[MeSH Terms] OR ("myocardial"[All Fields] AND "infarction"[All Fields]) OR "myocardial infarction"[All Fields] OR ("heart"[All Fields] AND "attack"[All Fields]) OR "heart attack"[All Fields]) AND bad[All Fields] AND ("diet"[MeSH Terms] OR "diet"[All Fields])
Consult the sidebar discovery tool “Search details” to see how PubMed translated a search.
Complex Searching
There are a variety of ways that PubMed can be searched in a more sophisticated manner than simply typing search terms into the search box and clicking Search. It is possible to construct complex search strategies using Boolean operators and the features listed below:
- Use the Advanced search page to:
- Search by a specific field
- Browse the index of terms
- Combine searches using history
- Preview the number of search results
- Filters narrow search results by article types, text availability, publication dates, species, languages, sex, subjects, journal categories, ages, and search fields.
Additional PubMed Features
The following resources are available to facilitate effective searches:
- Use the MeSH Database to find MeSH terms, including Subheadings, Publication Types, Supplementary Concepts, and Pharmacological Actions, and then build a PubMed search
- The Clinical Queries page provides searching by clinical study categories that use built-in search filters to limit retrieval to citations to articles reporting research conducted with specific methodologies, including those that report applied clinical research.
- The NLM Catalog includes information about the journals in PubMed and the other NCBI databases.
- Use the Batch Citation Matcher to retrieve PMIDs (PubMed IDs) for multiple citations in batch mode.
- My NCBI saves searches, results, bibliographies, and features an option to automatically update and email search results. Preferences include storing and changing an email address, highlighting search terms, opening the abstract display supplemental data by default, and turning off the auto suggest feature. Additional features include filtering search results, managing recent activity, and setting a LinkOut icon, document delivery services, and outside tool preferences.
- PubMed Mobile provides a simplified mobile-friendly web interface to access PubMed.
Results
PubMed search results are displayed in a summary format; citations are initially displayed 20 items per page with the most recently entered citations displayed first. (Note that this date can differ significantly from the publication date.)
A spell-checking feature suggests alternative spellings for search terms that may include misspellings.
To provide users with targeted results, query sensors may display for searches that match specific characteristics. For example, a citation sensor will display for searches that include author names, journal titles, publication dates, or article titles, e.g., zong science 2012. A gene sensor checks for queries that include gene symbols, e.g., brca1
Depending on the search, additional sensors and discovery tools, e.g., Results by year, Recent activity, Related searches, PMC Images, may also display.
Citations can be viewed in other formats and can be sorted, saved or e-mailed, and printed. The full text may also be available online or ordered from a library.
How to Create Hyperlinks to PubMed
To create Web URL links that search and retrieve PubMed data the following tools are useful:
- The Entrez Programming Utilities (E-utilities), which are a set of eight server-side programs that provide a stable interface into the Entrez query and database system at NCBI. E-Utilities provide a fast, efficient way to search and download citation data without using the front-end query engine.
- Generate the URL manually using the Creating a Web Link to PubMed online documentation.
Customer Support
If you need more assistance:
- Contact the Help Desk by selecting the Support Center link displayed on all PubMed pages
- Call the NLM Customer service desk: 1-888-FIND-NLM (1-888-346-3656)
Publication Details
Author Information and Affiliations
Authors
Kathi Canese and Sarah Weis.Publication History
Created: October 9, 2002; Last Update: March 20, 2013.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Canese K, Weis S. PubMed: The Bibliographic Database. 2002 Oct 9 [Updated 2013 Mar 20]. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.