| enter query directly into CDART home page as a protein sequence, set of conserved domains, or multiple queries |
| retrieve sequence record from Entrez Protein → follow "Domain Relatives" link |
 

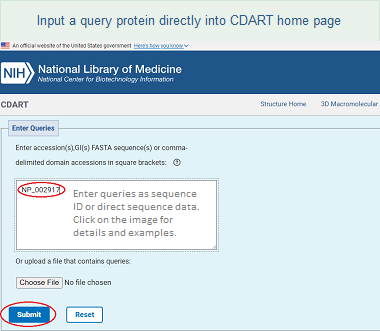

One way to retrieve proteins with similar domain architectures is to enter your query as a protein sequence, or as a set of conserved domains, directly into the CDART home page in any of the following formats:
Protein sequence  
You can submit a protein sequence as:
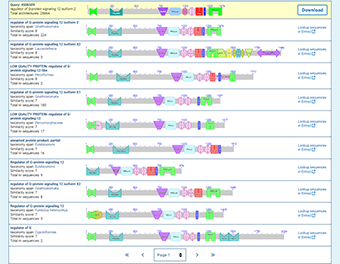
The CDART results will show the functional domains found in the query protein and will list proteins with a similar domain architecture. The similar proteins must include at least one of the conserved domain superfamilies in the query sequence. The similarity score of each domain architecture indicates the number of domain superfamilies in the architecture that match domain superfamilies in the query protein, and is used to rank the search results.
| |
Note: The proteins that are returned by CDART will include at least one of the domains you have specified. The similarity score of each domain architecture indicates the number of domain superfamilies in the architecture that match domain superfamilies in the query protein, and is used to rank the search results.
Regardless of how you specify the conserved domains in your query (as superfamily cluster IDs or as the accessions or PSSM IDs of individual domain models), the CDART search results will display the superfamilies to which those models belong, and not the individual domain models themselves. However, you can see superfamilies and individual domain models by following the "domain details" link that appears in the expanded view of any domain architecture.
If you enter a single conserved domain as a query, you will retrieve all the domain architectures that contain the domain, ranked by the number of non-redundant proteins that have a given architecture.
|
|
|
Multiple queries  
You can enter multiple queries using any of the formats above (i.e., as protein sequences or sets of conserved domains) or a mix of those formats. Note that:
- Each protein Accession or GI number should be on a separate line
- FASTA formatted sequence data and bare sequences can occupy multiple lines. (The FASTA format definition line, however, should occupy a single line).
- If one of your queries is a set of conserved domains, they should be entered on a single line, separated by commas, and surrounded by square brackets [], as in the third line of the example below.
- If you include a bare sequence as one of the queries, use a blank line to separate it from the query that precedes it, as in the last part of the example below.
Example: - The example below includes six queries, in the following order: (1) protein GI number; (2) protein accession number; (3) set of conserved domain superfamily cluster IDs; (4) protein sequence in FASTA format; (5) another protein sequence in FASTA format; (6) protein as bare sequence data:
269849668
EDV04934
[75,2783]
>gi|239592572|gb|EEQ75153.1| asparaginase [Ajellomyces dermatitidis SLH14081]
MSPPIPQPRQRTRSQPLFKPAVILHGGAGNIQHSRLPPELYKQYRTSLLTYLRSTTALLNADIEEEEPSI
NAKNDAVDDNMRISPASALNAAVHAVSLMEDNELFNCGRGSVFTSAGTIEMEASVMVASLLNDEDSVDDF
NNSEVNCLASEKTPGSIKRGAGVMLVRNVRHPIQLAKEVLLRTGYASDGDGDGGNMHSQLSGEYVEGLAR
DWGMEFCPDDWFWTKKRWDEHRRGLKKGKTRGRMTDGRNMGADVEVRGEGEADDGDGLYLSQGTVGCVCL
DRWGNIAVATSTGGLTNKCPGRIGDTPTLGAGFWAEAWDVEGVEGLSNMSDSSNSVCASGRDRSKGCIQL
KRDTMNYQTQDGRDNLLAYQASSSTTTTTSSYRMGSQWRSDFDSNSAFTLIRDCFSSSPPPPGYAALEPS
KYPVEKFPLGKSTSSPHTDFNPHRYSQPQRRRILALSGTGNGDSFLRTAATRTAAAMVRFGSAQNSISLA
QAVTAVAGPGGELQRSAGRRWGKTGEGEGGIIGIEAEVETDEQTLGEGKLRRGKVVFDFNSTGMFRAWME
EKDGKDVERMMVFRDDYE
>gi|336020358|ref|NP_001229488.1| mitogen-activated protein kinase kinase kinase kinase 4 isoform 4 [Homo sapiens]
MANDSPAKSLVDIDLSSLRDPAGIFELVEVVGNGTYGQVYKGRHVKTGQLAAIKVMDVTEDEEEEIKLEI
NMLKKYSHHRNIATYYGAFIKKSPPGHDDQLWLVMEFCGAGSITDLVKNTKGNTLKEDWIAYISREILRG
LAHLHIHHVIHRDIKGQNVLLTENAEVKLVDFGVSAQLDRTVGRRNTFIGTPYWMAPEVIACDENPDATY
DYRSDLWSCGITAIEMAEGAPPLCDMHPMRALFLIPRNPPPRLKSKKWSKKFFSFIEGCLVKNYMQRPST
EQLLKHPFIRDQPNERQVRIQLKDHIDRTRKKRGEKDETEYEYSGSEEEEEEVPEQEGEPSSIVNVPGES
TLRRDFLRLQQENKERSEALRRQQLLQEQQLREQEEYKRQLLAERQKRIEQQKEQRRRLEEQQRREREAR
RQQEREQRRREQEEKRRLEELERRRKEEEERRRAEEEKRRVEREQEYIRRQLEEEQRHLEVLQQQLLQEQ
AMLLECRWREMEEHRQAERLQRQLQQEQAYLLSLQHDHRRPHPQHSQQPPPPQQERSKPSFHAPEPKAHY
EPADRAREVEDRFRKTNHSSPEAQSKQTGRVLEPPVPSRSESFSNGNSESVHPALQRPAEPQVPVRTTSR
SPVLSRRDSPLQGSGQQNSQAGQRNSTSIEPRLLWERVEKLVPRPGSGSSSGSSNSGSQPGSHPGSQSGS
GERFRVRSSSKSEGSPSQRLENAVKKPEDKKEVFRPLKPADLTALAKELRAVEDVRPPHKVTDYSSSSEE
SGTTDEEDDDVEQEGADESTSGPEDTRAASSLNLSNGETESVKTMIVHDDVESEPAMTPSKEGTLIVRQT
QSASSTLQKHKSSSSFTPFIDPRLLQISPSSGTTVTSVVGFSCDGMRPEAIRQDPTRKGSVVNVNPTNTR
PQSDTPEIRKYKKRFNSEILCAALWGVNLLVGTESGLMLLDRSGQGKVYPLINRRRFQQMDVLEGLNVLV
TISGKKDKLRVYYLSWLRNKILHNDPEVEKKQGWTTVGDLEGCVHYKVVKYERIKFLVIALKSSVEVYAW
APKPYHKFMAFKSFGELVHKPLLVDLTVEEGQRLKVIYGSCAGFHAVDVDSGSVYDIYLPTHIQCSIKPH
AIIILPNTDGMELLVCYEDEGVYVNTYGRITKDVVLQWGEMPTSVAYIRSNQTMGWGEKAIEIRSVETGH
LDGVFMHKRAQRLKFLCERNDKVFFASVRSGGSSQVYFMTLGRTSLLSW
MEQDPKPPRLRLWALIPWLPRKQRPRISQTSLPVPGPGSGPQRDSDEGVLKEISITHHVKAGSEKADPSH
FELLKVLGQGSFGKVFLVRKVTRPDSGHLYAMKVLKKATLKVRDRVRTKMERDILADVNHPFVVKLHYAF
QTEGKLYLILDFLRGGDLFTRLSKEVMFTEEDVKFYLAELALGLDHLHSLGIIYRDLKPENILLDEEGHI
KLTDFGLSKEAIDHEKKAYSFCGTVEYMAPEVVNRQGHSHSADWWSYGVLMFEMLTGSLPFQGKDRKETM
TLILKAKLGMPQFLSTEAQSLLRALFKRNPANRLGSGPDGAEEIKRHVFYSTIDWNKLYRREIKPPFKPA
VAQPDDTFYFDTEFTSRTPKDSPGIPPSAGAHQLFRGFSFVATGLMEDDGKPRAPQAPLHSVVQQLHGKN
LVFSDGYVVKETIGVGSYSECKRCVHKATNMEYAVKVIDKSKRDPSEEIEILLRYGQHPNIITLKDVYDD
GKHVYLVTELMRGGELLDKILRQKFFSEREASFVLHTIGKTVEYLHSQGVVHRDLKPSNILYVDESGNPE
CLRICDFGFAKQLRAENGLLMTPCYTANFVAPEVLKRQGYDEGCDIWSLGILLYTMLAGYTPFANGPSDT
PEEILTRIGSGKFTLSGGNWNTVSETAKDLVSKMLHVDPHQRLTAKQVLQHPWVTQKDKLPQSQLSHQDL
QLVKGAMAATYSALNSSKPTPQLKPIESSILAQRRVRKLPSTTL
|
 

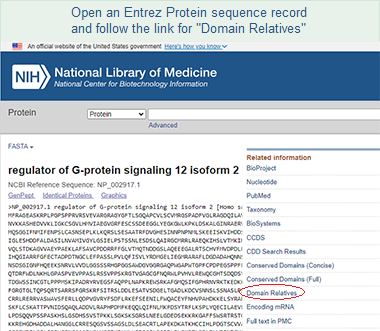

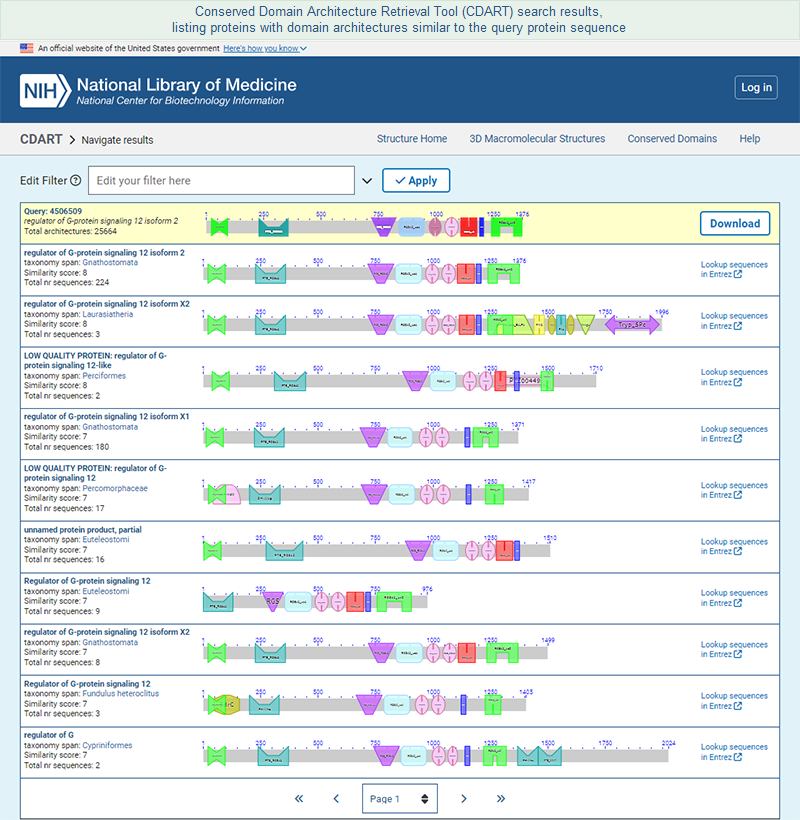
A second way to access CDART is to start by retrieving a record of interest from the Entrez Protein database, then follow the "Domain Relatives" link in the right margin of the sequence record. That will open the precalculated CDART results for the protein.
Note that the "Domain Relatives" is one of four links available from a protein sequence record to conserved domain annotations, allowing you to choose:
(a) the format in which you want to view the conserved domains (e.g., in graphical format as domain footprints aligned to the protein sequence; as a list of records from the Conserved Domain Database, each of which includes a multiple sequence alignment of the proteins used to create the domain model; or as a list of proteins with similar domain architectures), and
(b) the level of redundancy in the list of conserved domain models (e.g., a concise list of the top scoring models or a full list of all models that have a statistically significant RPS-BLAST hit to the protein).
The number of conserved domain models retrieved, and the order in which they are sorted/presented, depends upon the view you select:
- Domain Relatives -- opens a graphical display of similar domain architectures, as determined by the CDART tool. A domain architecture is defined as the sequential order of conserved domains in a protein query sequence. The score for each CDART hit represents the number of domains that match those found in the query protein. (The CDART paper provides additional details.
)
- CDD Search Results -- opens a graphical display (illustrated example) of conserved domain model footprints on the query protein, ranked by their RPS-BLAST score and hit type. A model may appear more than once if it aligns to multiple regions of the query sequence. A concise display showing only the top-scoring hits is presented by default, and it can be changed to a full display of all hits if desired. (The CDD help document provides additional details.)
- Conserved Domains (Concise) -- opens a concise list of the conserved domain models that are the top-scoring RPS-BLAST hits to the query protein. Each domain model is listed only once, even if a model had a hit to more than one region on the query sequence. (The CDD help document provides additional details.)
- Conserved Domains (Full) -- opens a full list of all the conserved domain models that have a statistically significant RPS-BLAST hit to the query protein. Each domain model is listed only once, even if a model had a hit to more than one region on the query sequence. (The CDD help document provides additional details.)
|
|
|