New SNP Attributes
New attributes have been added to dbSNP to allow searching and filtering of human variation by the following characteristics.
Please contact [email protected] if you have any questions or comments.
| Attribute | RS Count (Build 132) |
|
423 |
|
13105 |
|
14946243 |
|
105630 |
Below are examples of the attributes shown on web pages and schema changes.
RefSNP Summary (Example)
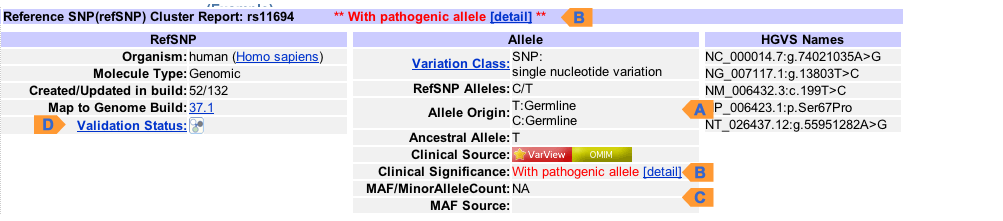
| A. Allele Origin | Indicated as Germline or Somatic for each allele |
| B. Clinical Significance | Click on "VarView" or "OMIM" to view phenotype |
| C. Global MAF | The minor allele (G), frequency (0.003), and allele count (3) is shown |
| D. Suspected | A red "?" icon is shown for suspected SNP in the "Validation" row |
SNP GeneView (Example)
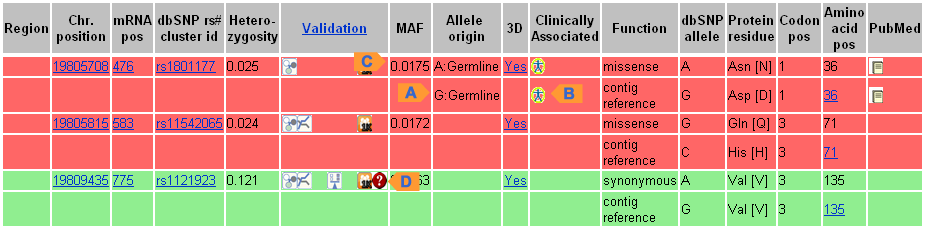
| A. Allele Origin | Indicated as Germline or Somatic for each allele |
| B. Clinical Significance | Click on icon under "Clinical Source" to view effect in Variation Viewer |
| C. Global MAF | MAF is shown for the corresponding allele |
| D. Suspected | A red "?" icon is shown for suspected SNP under the "Validation" column |
Variation Viewer (Example)
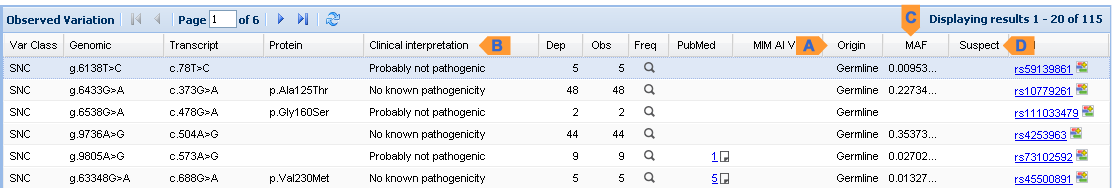
| A. Allele Origin | Indicated as Germline or Somatic for the variation under "Origin" column |
| B. Clinical Significance | shown under "Clinical Intrepretation" column |
| C. Global MAF | frequency is shown under minor allele frequency (MAF) column |
| D. Suspected | A red "?" icon is shown for suspected SNP under the "Suspect" column |
RS Docsum (XML Schema)
| SNP Attribute | XML Element |
| A. Allele Origin | Rs/AlleleOrigin |
| B. Clinical Significance | Rs/Phenotype |
| C. Global MAF | Rs/Frequency |
| D. Suspected | Rs/Validation/@name='suspect' |
Entrez Search and Eutils Retrieval

The SNP attributes can be search from the web using the field and terms below or selected from the limit page. The search can also be performed programatically using eUtils eSearch. Note: eUtils eFetch is currently being updated to support retrieval of variations with the new attributes. We'll let you know when the update is done.
| SNP Attribute | Search Field (type) | Search Terms |
| A. Allele Origin | ALLELE_ORIGIN (text) | somatic |
| germline | ||
| B. Clinical Significance | CLINICAL_SIGNIFICANCE (text) | non pathogenic |
| other | ||
| pathogenic | ||
| probable non pathogenic | ||
| probable pathogenic | ||
| unknown | ||
| C. Global MAF | GLOBAL_MAF (floating-point) | exact 0.01[GLOBAL_MAF] or range 0.01:0.05[GLOBAL_MAF]) |
| D. Suspected | SUSPECTED (text) | paralog |
| SNP Attribute | VCF Tag |
| A. Allele Origin | ##INFO=<ID=SAO,Number=1,Type=Integer,Description="SNP Allele Origin: 0 - unspecified, 1 - Germline, 2 - Somatic, 3 - Both"> |
| B. Clinical Significance | ##INFO=<ID=SCS,Number=1,Type=Integer,Description="SNP Clinical Significance, 0 - unknown, 1 - untested, 2 - non-pathogenic, 3 - probable-non-pathogenic, 4 - probable-pathogenic, 5 - pathogenic, 6 - drug-response, 7 - histocompatibility, 255 - other"> |
| C. Global MAF | ##INFO=<ID=GMAF,Number=1,Type=Float,Description="Global Minor Allele Frequency [0, 0.5]; global population is 1000GenomesProject phase 1 genotype data from 629 individuals, released in the 08-04-2010 dataset"> |
| D. Suspected | ##INFO=<ID=SSR,Number=1,Type=Integer,Description="SNP Suspect Reason Code, 0 - unspecified, 1 - Paralog, 2 - byEST, 3 - Para_EST, 4 - oldAlign, 5 - other"> |