Summary
The global pandemic of coronavirus disease 2019 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has become a severe global health problem because of its rapid spread. Both Ace2 and NRP1 provide initial viral binding sites for SARS-CoV-2. Here, we show that cysteine residues located in the vestigial plasminogen-apple-nematode (PAN) domain of NRP1 are necessary for SARS-CoV-2 spike protein internalization. Mutating novel cysteine residues in the PAN altered NRP1 stability and downstream activation of extracellular signal-regulated kinase (ERK) signaling pathway and impaired its interaction with the spike protein. This resulted in a significant reduction in spike protein abundance in Vero-E6 cells for the original, alpha, and delta SARS-CoV-2 variants even in the presence of the Ace2. Moreover, mutating these cysteine residues in NRP1 significantly lowered its association with Plexin-A1. As the spike protein is a critical component for targeted therapy, our biochemical study may represent a distinct mechanism to develop a path for future therapeutic discovery.
Subject areas: Virology, Biochemistry
Graphical abstract
Highlights
-
•
Vestigial PAN domain of NRP1 is essential for its function and stability
-
•
Two novel cysteines identified in NRP1 directly impact SARS-CoV-2 binding
-
•
Mutated PAN domain reduces spike protein abundance in cells for viral variants
-
•
Mutations in PAN domain modulate pERK signaling and signature gene expression
Virology; Biochemistry
Introduction
Neuropilin (NRP) is a single membrane-spanning, type-I transmembrane, non-tyrosine kinase receptor protein.1 In humans, it has two homologs, NRP1 and NRP2, with a 44% of sequence identity.2,3 Both contain five extracellular domains which are two cubilin homology (CUB) (a1/a2), two coagulation factor FV/FVIII (b1/b2), a MAM (meprin, A5, and μ-phosphatase; also known as c), and a domain that contains a transmembrane and a short cytoplasmic region.4,5 NRP1 was identified as a co-receptor for several extracellular ligands, including class III semaphorins, a specific isoform of the vascular endothelial growth factor-A (VEGF-A), heparin-binding proteins, placental growth factor 2, and fibroblast growth factor 2.6,7 Both CUB and FV/FVIII domains provide the binding sites for secreted semaphorins and VEGF.8 NRP1 plays an essential role in performing key physiological processes like angiogenesis and semaphorin-dependent axon guidance signaling.9,10
An interesting aspect of NRP1 is that its ligand, semaphorin, is synthesized as inactive precursor forms and requires proteolytic cleavage by furin or related endoproteolytic pro-protein convertases, a key feature that is also utilized by the viral coat proteins.11 For example, NRP1 has been reported to function as an entry receptor for Epstein-Barr virus (EPV) and more recently for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus.12,13,14 To promote NRP1-mediated cell internalization via tissue penetration, cleaved peptides utilize the R/K/XXR/K motif at their C-terminus (C-end rule, CendR).15 CendR peptides are known to be useful for cancer drug delivery as NRP1 is frequently overexpressed in various tumor types.16 This similar phenomenon is used by SARS-CoV-2 while binding with NRP1 for virus-host membrane fusion via the multibasic site at the S1-S2 boundary of the viral spike (S) protein.14 S protein usually gets activated by cellular proteases, either by furin and/or TMPRSS2, which ensure cleavage at the S1/S2 site.17 The S1 subunit of the S protein of coronaviruses facilitates viral entry into target cells by providing a receptor-binding domain.17 Since the alarming spread of the COVID-19 pandemic, a major focus of ongoing research has been to define the mechanism behind viral entry. For SARS-CoV-2, the CendR pocket is usually present within the extracellular b1b2 domain of NRP1.14 However, the dynamics of S protein binding to the NRP1 receptor at the specific amino acid level are yet to be established.
Results
Novel cysteine controls NRP1 stability
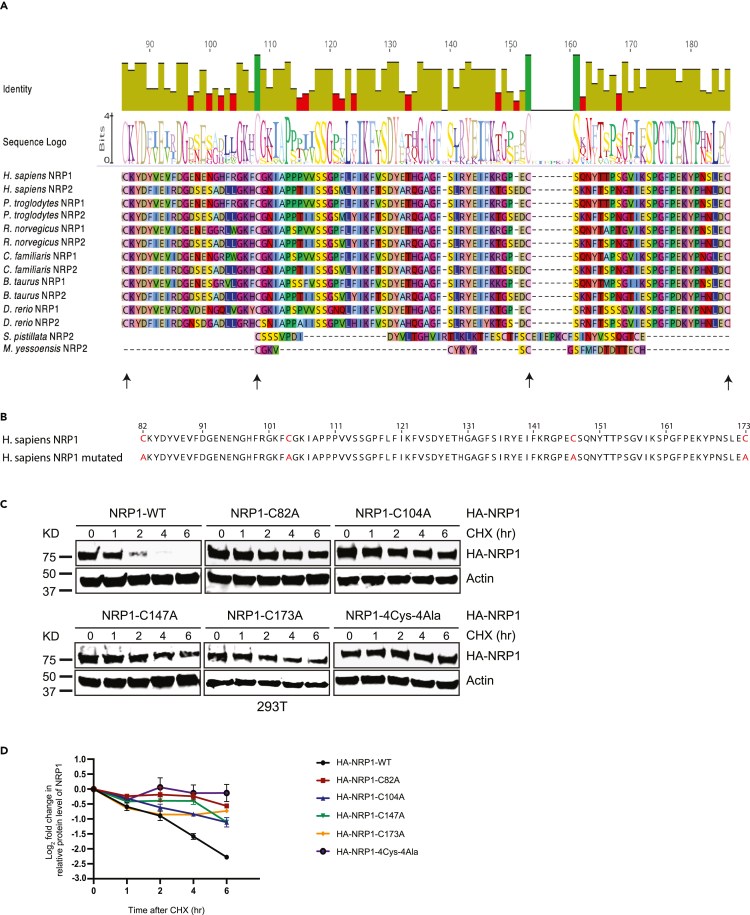
We recently identified orthologs of NRP1 and NRP2 that carry a plasminogen-apple-nematode (PAN) domain.18 Our Gene Ontology (GO) enrichment analyses using InterPro (https://www.ebi.ac.uk/interpro/) and UniProt (https://G-LecRK.uniprot.org) databases identified 28,300 PAN domains containing proteins across the 2,496 organisms.19 Surprisingly, these proteins are predominantly enriched in immune signaling processes as the majority of them were associated with the extracellular matrix (ECM) as cell surface receptors.18,19 A key feature of the PAN domain is the presence of 4-6 strictly conserved cysteine residues forming its core. We reported that this protein domain was involved in receptor binding and proteolysis of two unrelated proteins, a hepatocyte growth factor in human and a G-type lectin receptor-like kinase in plants.18,19,20 Mutation of core cysteines in the PAN domain of both proteins led to dramatic changes in immune signaling in both organisms without having a detectable impact on their structure.18,19 Because the human NRP1 did not have a reported conserved PAN domain, we sought to evaluate if any domain features could be detected by assessing residual homology with NRP1 and NRP2 from other organisms. After our careful analysis, we determined that ancestral NRP2 from Stylophora pistillata (coral reefs) have survived more than 400 million years21,22 and M. yessoensis (mollusca) around 545 million years23,24,25 carries an intact PAN domain. We aligned the human NRP1 with several NRP1 and NRP2 from other organisms including chimpanzee, rat, dog, cow, and zebrafish and the NRP2 containing functional PAN domain from S. pistillata and M. yessoensis. Based on this analysis, we identified a region of NRP1 that exhibited homology to the ancestral PAN domain. Specifically, we identified four cysteine residues at amino acid positions C82, C104, C147, and C173 in two extracellular CUB domains of NRP1. The second cysteine C104 and third cysteine C147 aligned well, while the first and fourth cysteines at both terminals did not (Figure 1A). Although the distance between the conserved cysteines in S. pistillata and M. yessonensis is less than the human and mammalian counterpart, there is sufficient sequence similarity between human NRP1 and the PAN domain in S. pistillata to suggest homology. Therefore, we classified human NRP1 as a protein with a vestigial PAN domain. Based on the previous research focusing on the PAN domain,18,26 we hypothesize that these cysteine residues may play a role in NRP1 function and, therefore, could modify its interaction with SARS-CoV-2 S protein.
Figure 1.
Novel cysteine controls NRP1 stability
(A) Multiple alignment of the sequences of PAN domain of representative proteins from seven different organisms highlights the position of conserved cysteines.
(B) Schematic diagram of amino acid sequence represents NRP1 vestigial PAN domain along with the four marked cysteines (Cys82, Cys104, Cys147, and Cys173) and the subsequent mutant version where conserved cysteines were mutated to alanine (Ala82, Ala104, Ala147, and Ala173).
(C) Immunoblot analysis of whole-cell lysates derived from 293T cells, transfected with HA-NRP1 WT and different single cysteine mutants of HA-NRP1 and HA-NRP1-4Cys-4Ala constructs as indicated. 24 h post-transfection, cells were split, and cells were treated with 100 μg/mL cycloheximide (CHX) 20 h later. At the indicated time points, whole-cell lysates were prepared for immunoblot analysis. Representative image of n = 3 biological replicates.
(D) Quantification of the band intensities in (c). The intensities of HA-NRP1 (WT and mutants) bands were normalized to actin and then normalized to HA-NRP1 WT. Data are represented as mean ± SD, n = 3, and ∗, p < 0.05; ∗∗, p < 0.005; ∗∗∗, p < 0.0005. were calculated with one-way ANOVA.
To evaluate the functional significance of these four cysteines, we introduced single cysteine-to-alanine mutations and simultaneously mutated all four cysteines to alanine in NRP1. (Figure 1B). Like most other transmembrane receptors, upon binding with its ligand, NRP1 undergoes internalization and is targeted for lysosomal degradation.27 Based on this established model, we first evaluated the overall abundance of cysteine-to-alanine NRP1 proteins in cells compared to the wild type. With the cycloheximide (CHX) protein stability assay, we observed that mutations in any of the four cysteines increased protein stability of exogenously expressed hemagglutinin (HA)-tagged NRP1 in HEK293T cells (Figures 1C and 1D). The HA-NRP1 4Cys-4Ala mutant showed a cumulative impact on the protein stability by all the four-cysteines mutation as expected (Figure 1D). We hypothesized that the higher NRP1 expression of the NRP1 mutants might result from its inability to interact with its ligand and, therefore, not following the downstream receptor degradation pathway preventing the NRP1 degradation. However, it was intriguing to ask whether these mutations lead to localization defects in NRP1. We attempted its detection in Vero-E6 cells following transient transfection. Clear staining of non-permeabilized Vero cells with an antibody against the N-terminus HA tag of the NRP1 indicates that no large-scale perturbation of cellular localization occurred in the NRP1 mutants compared to the wild-type NRP1 (Figure S1).Taken together, these data suggested that all four cysteines could be involved in the receptor-mediated endolysosomal degradation pathway, although at varying extents.
Novel cysteines in NRP1 facilitate its interaction with SARS-CoV-2 S protein
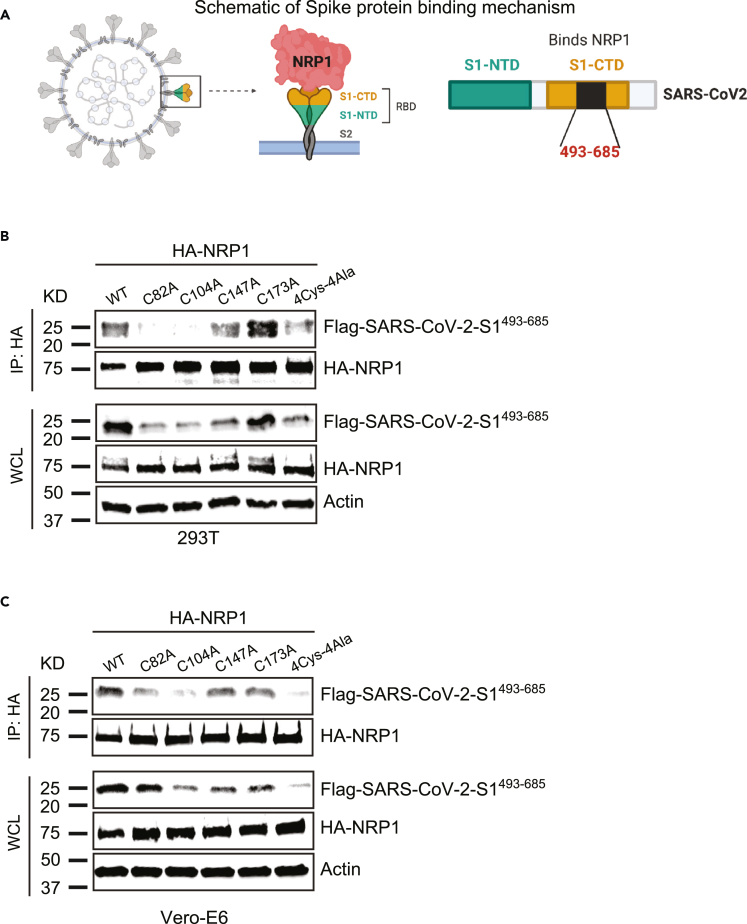
Secondly, we sought to evaluate impact of these mutations on NRP1 interaction with SARS-CoV-2 S1 protein. SARS-CoV-2 S1 protein residues 493-685 have been reported to bind with NRP1.14 We made a FLAG-SARS-CoV-2-S1493−685 construct based on the sequence available in NCBI (Figure 2A). To assess the impact of these four novel cysteines in NRP1 interaction with COVID-19, we carried out a direct co-immunoprecipitation assay between HA-NRP1-WT, HA-NRP1-C82A, HA-NRP1-C104A, HA-NRP1-C147, HA-NRP1-C173A, HA-NRP1-4Cys-4Ala, and FLAG-SARS-CoV-2-S1493−685 following expression in HEK293T cells. Mutations in any of the first three cysteines (C82A, C104A, and C147A) in NRP1 had a significantly negative impact on FLAG-SARS-CoV-2-S1493−685 protein binding as evidenced by quantification of the relative amount of FLAG-SARS-CoV-2-S1493−685 pulled down with different variants of NRP1 in 293T cells (Figures 2B, S2A, and S2B). We also took advantage of repeating the experiment in Vero-E6 cell lines as they are relevant for SARS-CoV-2 research and observed notably reduced interaction for the first two cysteines. The C147A mutant shows diminished interactions in Vero cells, but the binding was not statistically significant (Figures 2C, S2C, and S2D).
Figure 2.
Novel cysteines in NRP1 facilitate its interaction with SARS-CoV-2 S protein
(A) Schematic representation of SARS-CoV-2 S protein binding with NRP1 receptor. Contacting residues on S protein is from 493-685.
(B and C) NRP1 binding with S protein is novel cysteines dependent. 293T and Vero-E6 cells were transfected with the indicated HA-NRP1 (wild type and mutants) and FLAG-SARS-CoV-2-S1493−685 constructs. Cells were lysed 30 h post-transfection, and the interaction between HA-NRP1 and FLAG-SARS-CoV-2-S1493−685 was analyzed.
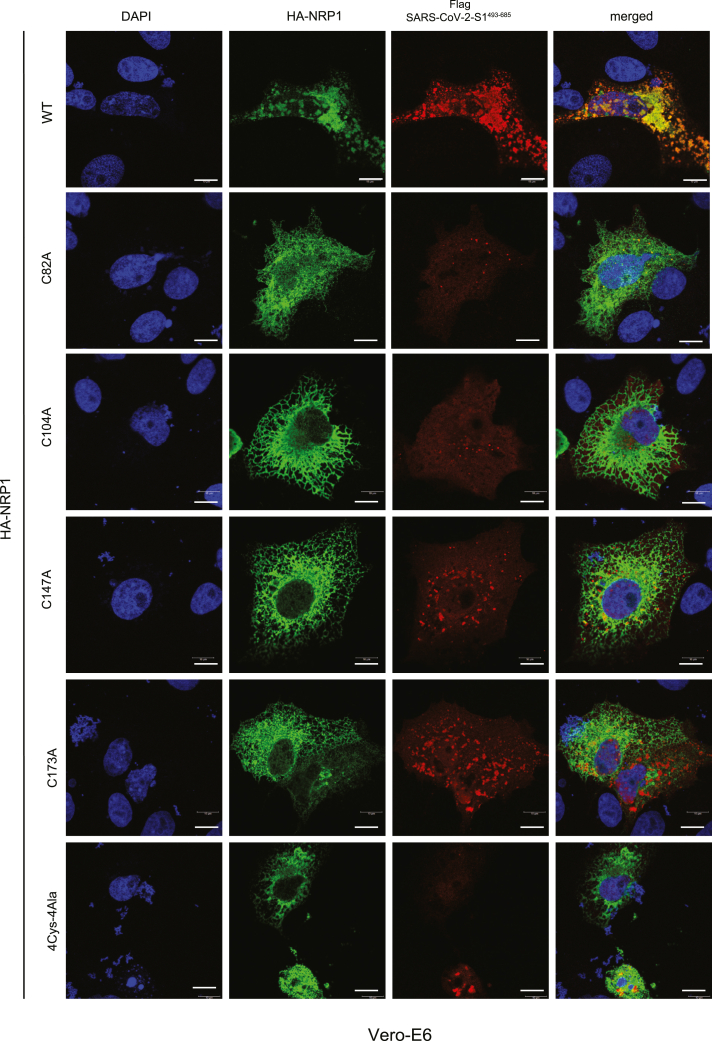
To bolster our conclusion that these novel single-cysteine residues in NRP1 are critical for SARS-CoV-2 interaction, we used both Vero-E6 and HeLa cells to perform co-immunolocalization assays between NRP1(s) and the FLAG-SARS-CoV-2-S1493-685. Our results corroborated these findings as they showed markedly reduced colocalization between HA-NRP1-C82A, HA-NRP1-C104A, HA-NRP1-C147A, HA-NRP1-4Cys-4Ala, and FLAG-SARS-CoV-2-S1493-685 compared to HA-NRP1-WT in both Vero-E6 and HeLa cells (Figures 3 and S3A). Remarkably, in cells with transfected HA-NRP1-C82A, HA-NRP1-C104A, and HA-NRP1-C147A the SARS-CoV-2-S1493−685 protein shows significant reduction in overall abundance (Figures S3B and S4). These results could be due to the weaker association between the S protein and those NRP1 mutants altering the S protein expressions in cells. Previous studies show that NRP1 binds with SARS-Cov-2 S protein with greater affinity and stabilizes the S1 (c-terminal region), which remains in folded conformation14,28,29 and, thereby, restores the S protein expression. In the absence of NRP1, published data show that SARS-CoV-2 S protein gets stretched and unfolded.30 Our data show a reduced abundance of S protein in cells transfected with mutant NRP1. Taken together, we propose that in the presence of a mutated version of NRP1, the S protein shows reduced binding and becomes more stretched, unfolded, and non-functional. Unfolded or non-functional proteins are less stable and are more susceptible to degradation pathways.31,32 Co-transfection of HA-NRP1-4Cys-4Ala with S1 might express some unfolded S proteins targeted by the secretory pathway components for its degradation and thereby altering the overall S protein abundance in cells.
Figure 3.
Detection of interaction of NRP1 and SARS-CoV-2 S protein in cells
Representative images of colocalization studies between FLAG-SARS-CoV-2-S1493−685 and different HA-NRP1 constructs by confocal immunofluorescence microscopy in Vero-E6 cells. The cells were transiently transfected with FLAG-SARS-CoV-2-S1493−685 and different mutants of HA-NRP1 as indicated. 30 h post-transfection, cells were fixed and mounted, and protein expression patterns were visualized using a Leica SP8 White Light Laser Confocal System. Scale bars represent 10 μm. The images shown are representative from three independent biological experiments (average 100 cells were observed per experimental condition per replicate).
Mutation in the novel cysteines in NRP1 universally impairs its interaction with different SARS-CoV-2 S protein variants
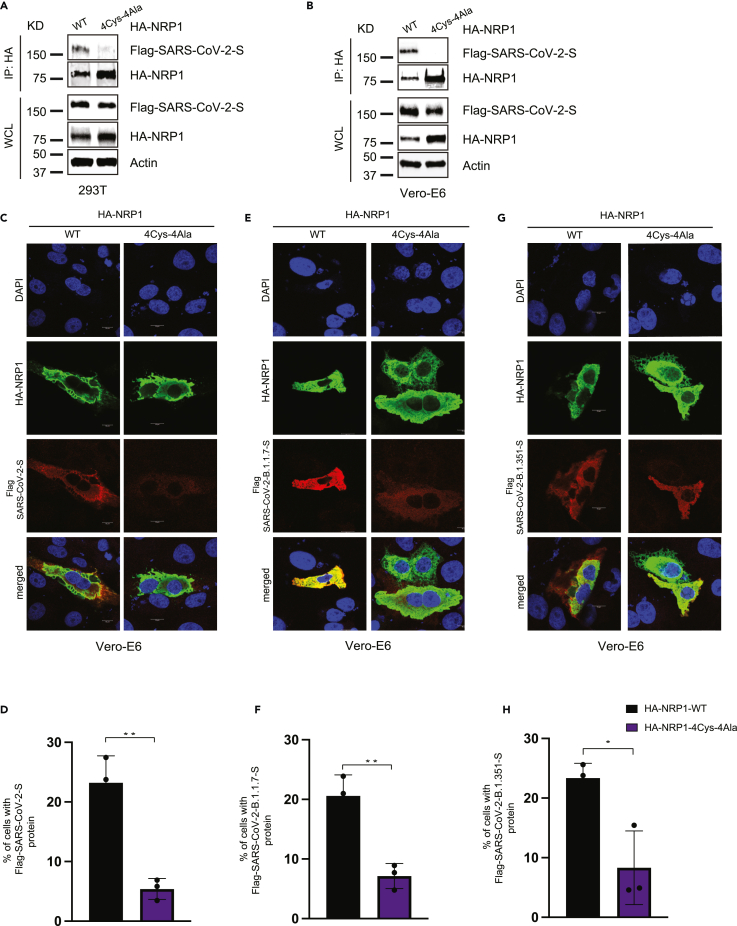
We then examined whether the full-length SARS-CoV-2-S protein could influence the interaction with NRP1. The interaction between HA-NRP1-4Cys-4Ala and SARS-CoV-2-S was impaired when co-expressed in 293T and Vero-E6 cells and subjected to immunoprecipitation (Figures 4A and 4B). We concluded that full-length SARS-CoV-2-S protein is equally susceptible to NRP1 cysteine mutations. The immediate development of a vaccine against COVID-19 has provided a vital tool to fight against the rapid spread of the SARS-CoV-2 virus. However, constantly evolving mutations in the S protein imposes a challenge for effective deployment of vaccines.
Figure 4.
Mutation in the novel cysteines in NRP1 universally impairs its interaction with different SARS-CoV-2 S protein variants
(A and B) NRP1 PAN domain cysteines play crucial role in SARS-CoV-2 S protein binding. Both 293T and Vero-E6 cells were transfected with the indicated HA-NRP1 (wild type and 4Cys-4Ala mutant) and FLAG-SARS-CoV-2-S constructs. Cells were lysed 30 h post-transfection, and the interaction between HA-NRP1 and FLAG-SARS-CoV-2-S was analyzed.
(C, E, and G) Representative images of colocalization studies between FLAG-tagged different SARS-CoV-2 S protein variants and indicated HA-NRP1 constructs by confocal immunofluorescence microscopy in Vero-E6 cells. The cells were transiently transfected with different FLAG-SARS-CoV-2-S and different constructs of HA-NRP1 as indicated. 30 h post-transfection, cells were fixed and mounted, and protein expression patterns were visualized using a Leica SP8 White Light Laser Confocal System. Scale bars represent 10 μm. The images shown are representative from three independent biological experiments (average 100 cells were observed per experimental condition per replicate).
(D, F, and H) Quantification of the Vero-E6 cells expressing FLAG-SARS-CoV-2-S, FLAG-SARS-CoV-2-B.1.1.7-S, and FLAG-SARS-CoV-2-B.1351-S in the presence of indicated HA-NRP1 constructs. Percentage of cells with FLAG-SARS-CoV-2-S protein was calculated for each of the variant separately and normalized compared to their respective control set (cells with HA-NRP1-WT). Data are represented as mean ± SD, n = 3 (average 100 cells were observed for each condition per experiment), and ∗, p < 0.05; ∗∗, p < 0.005; ∗∗∗, p < 0.0005 were calculated by Student’s t test.
To establish NRP1 as a potential universal candidate for targeted therapy, we used full-length FLAG-tagged SARS-CoV-2-S, SARS-CoV-2-B.1.1.7-S, and FLAG-tagged SARS-CoV-2-B1.351-S, representing the original, alpha, and beta variants, respectively, for colocalization assays. HA-NRP1-4Cys-4Ala was equally incapable of interacting with the variants as confirmed from the immunofluorescence data (Figures 4C, 4E, and 4G). In addition, viral S protein load in cells co-transfected with HA-NRP1-4Cys-4Ala was reduced significantly as compared to the NRP1-WT (Figures 4D, 4F, and 4H). This study provides the first experimental evidence supporting our hypothesis that targeting these specific amino acids in NRP1 PAN domain could be effective in reducing viral spread irrespective of the S protein variants, although emerging variants will need to be evaluated.
NRP1 interaction with co-receptor Plexin-A1 is cysteine dependent
As mentioned earlier, NRP1 acts as a ligand for the secreted semaphorins. However, NRP1 cannot transduce the semaphorin-induced downstream signal in the cytoplasm due to their short intracellular domain.33 Regular interaction with type-A plexins is required to form the functionally active receptor for secreted semaphoring 3A.34,35 Both NRP1 and plexins are well-known prognostic markers for several cancers, including glioblastoma and pediatric brain tumors causing poor survival in the patients.36,37,38 Therefore, we decided to test whether these cysteine residues in the vestigial PAN domain of NRP1 can be targeted against the plexin-A1 binding. HA-NRP1-WT and HA-NRP1-4Cys-4Ala variants were co-expressed with FLAG-Plexin-A1 in HEK293T cells, and complexes were immunoprecipitated using anti-FLAG. Our results indicated that the 4Cys-4Ala mutant showed significantly reduced binding with Plexin-A1 (Figures S5A and S5B). In agreement with our biochemical data, we reconfirmed that mutating these cysteine residues reduced the colocalization signal for NRP1 and Plexin-A1 when transfected in HeLa cells (Figure S5C). Plexin-NRP1-4Cys-4Ala reduced interaction reveals that the PAN domain in NRP1 might be helpful to targeted therapy for cancers caused by aberrant activation of the NRP1 signaling pathway. Several NRP1-directed therapies such as anti-NRP1 mAb, NRP1 antagonist, and anti-NRP1 nanobodies showed reduced size tumors without toxicity.39,40,41,42 Above studies reveal that targeting NRP1 can be highly effective without side effects. It could also be considered as a path in treating the severe SARS-CoV-2 infection in cancer patients.
Attenuated association of NRP1 mutant with endolysosomal marker lysosome-associated membrane protein 1 (LAMP1)
Upon binding with its substrate, efficient transmembrane receptor internalization and trafficking are essential for the proper progression of several cellular processes.27 Various pathogenic viruses utilize the host endocytic pathway to get access to cells including SARS-CoV-2 as it follows the conventional endosomal secretory route for its subsequent maturation and release from the infected cells.43,44 Therefore, numerous antiviral drugs targeting this pathway have been demonstrated against pathogenic viruses, namely Ebola (EBOV), African swine fever virus (ASFV), and SARS-CoV-2.44 Cellular proteins promoting this endocytic trafficking could provide a potential target against the spreading of the pathogenic virus. Thus, we determined the vesicular distribution of NRP1-WT and NRP1-4Cys-4Ala by staining for endolysosomal membrane protein marker, LAMP1. We transfected Vero-E6 cells with HA-tagged NRP1-WT and NRP1-4Cys-4Ala. Immunofluorescence analysis revealed a substantially low level of colocalization between the NRP1-4Cys-4Ala mutant and LAMP1 compared with NRP1-WT in cells (Figure S6). Thus, our study supports the notion that an overall reduction in viral S protein loads in the cells co-transfected with NRP1-4Cys-4Ala mutant could result from a widespread alteration in the endolysosomal pathway.
Reduced expression of SARS-CoV-2 S protein is related to NRP1 functional mutation
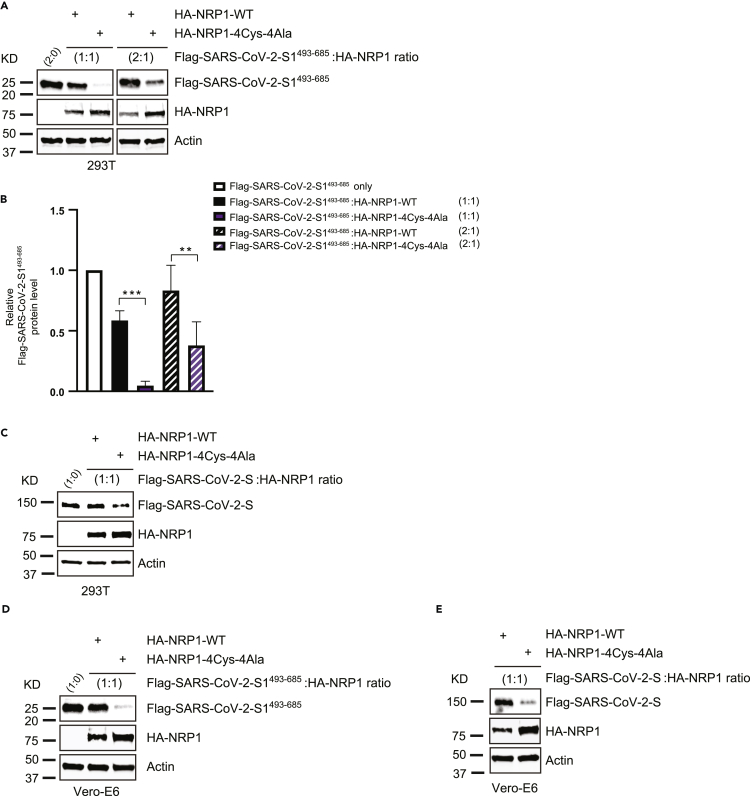
To ascertain our observations for lower S protein density in cells related to the NRP1 PAN domain mutation, we co-transfected 293T cells with HA-NRP1-WT and NRP1-4Cys-4Ala mutant with FLAG-SARS-CoV-2-S1493−685 at different ratios and measured the expressed S protein levels (Figure 5A), and the outcome further supported our previous observation that reduced S protein density was solely due to the lack of functional NRP1 receptor in cells (Figure 5B). In fact, increasing the transfection ratio for FLAG-SARS-CoV-2-S1493−685 could not fully rescue the S protein yield in cells transfected with HA-NRP1-4Cys-4Ala mutant (Figures 5A and 5B). Full-length SARS-CoV-2-S protein also exhibited reduced S protein density when co-transfected in a 1:1 ratio together with the HA-NRP1-4Cys-4Ala mutant compared with wild type (Figure 5C). Similar impact on experimental outcome was also noted while using Vero-E6 cells (Figures 5D and 5E). These results demonstrate that NRP1 4Cys-4Ala mutation at the PAN domain plays a critical role in diminishing both initial S protein recognition and overall S protein abundance in transfected cells.
Figure 5.
Reduced expression of SARS-CoV-2 S protein is related to NRP1 functional mutation
(A) 293T cells were transfected with the indicated HA-NRP1 (wild type and 4Cys-4Ala mutant) and FLAG-SARS-CoV-2-S1493−685 constructs at different ratio. Cells were lysed 30 h post transfection, and the protein expression was analyzed.
(B) Quantification of the band intensities (n = 3). FLAG-SARS-CoV-2-S1493−685 band intensities were normalized to the actin band and further normalized to FLAG-SARS-CoV-2-S1493−685 level in presence of NRP1-WT for each experimental set up. Mean and SD are indicated. Data were analyzed by one-way ANOVA: ∗,p < 0.05; ∗∗,p < 0.005; ∗∗∗, p < 0.0005.
(C) 293T cells were transfected with the indicated HA-NRP1 (wild type and 4Cys-4Ala mutant) and FLAG-SARS-CoV-2-S constructs at 1:1 ratio. Cells were lysed 30 h post transfection, and the protein expression was analyzed.
(D and E) Immunoblot analysis showing altered expression of exogenously expressed SARS-CoV-2-S1493−685 or (e) SARS-CoV-2-S in Vero-E6 cells in presence of NRP1-4Cys-4Ala mutant.
Transcriptome analysis of 293T cells shows differential gene expression following PAN mutation
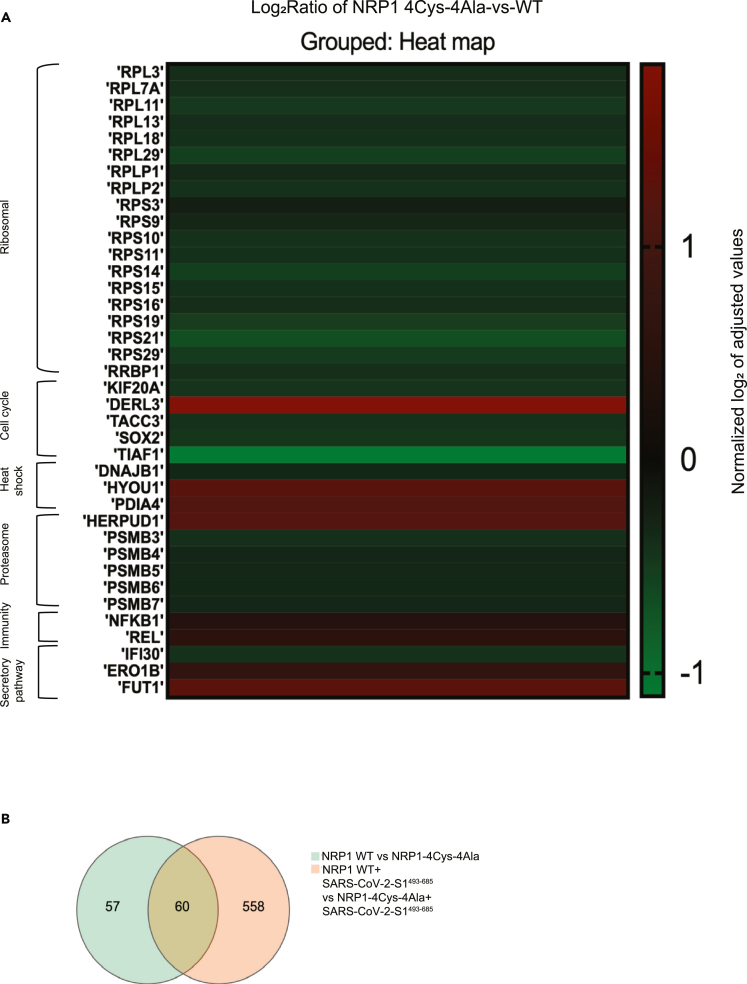
As we have pinpointed specific NRP1 cysteines necessary for S1 binding, we aimed to identify the downstream pathways that could be activated resulting from this interaction. The goal was to find whether mutating NRP1 results in altered expression of target genes. To that end, we characterized transcriptional responses in cells transiently co-transfected either with HA-NRP1-WT and FLAG-SARS-CoV-2-S1493−685 or HA-NRP1-4Cys-4Ala and FLAG-SARS-CoV-2-S1493−685. Total RNA was extracted from HEK293T cells 30 h post-transfection. Transcriptional profiling demonstrated differential expression of the genes takes part in ribosomal biogenesis, immune signaling, proteasomal degradation, and cell cycle pathway (Figure 6A). Specifically, we observed reduced expression for multiple ribosomal proteins (RPs) in cells transfected with HA-NRP1-4Cys-4Ala. RPs have been shown in several studies to be an important candidate in immune signaling.
Figure 6.
Transcriptome analysis in 293T cells
(A) Heatmap of selected genes. Heatmap displaying pattern of expression for the candidates showed an altered expression pattern following transfection with different NRP1 constructs (WT and 4Cys-4Ala) and FLAG-SARS-CoV-2-S1493−685. Genes are depicted based on their expression ratios across three RNA seq comparison. Colors range from bright red (upregulation; log2 ratio over control) to bright green (downregulation; log2 ratio over control).
(B) Venn diagram is to visualize the overlap in the genes found to be differentially expressed in the NRP1-WT vs NRP1- 4Cys-4Ala mutant and NRP1-WT+ SARS-CoV-2-S1493−685 vs NRP1- 4Cys-4Ala+ SARS-CoV-2-S1493−685. Each circle represents a group of gene sets, and the areas superimposed by different circles represent the intersection of these gene sets. The non-overlapping part indicates the uniquely expressed genes, and the numbers on the figure indicate the number of genes in the corresponding area.
We also observed reduced expression of RPL13 in cells transfected with mutated HA-NRP1-4Cys-4Ala. RPL13 overexpression promotes the induction and activation of promoters for the nuclear factor-κB (NF-κB) and interferon-β (IFN-β) genes.45 The NF-κB pathway is already considered a potential therapeutic target to treat COVID-19 disease.46 Another candidate of RP involvement in immune signaling is ribosomal Protein S3 (RPS3), which selectively modulates NF-κB gene expression. RPS3 was identified as the non-Rel component of the NF-κB complex by directly binding to the RelA subunit.47 In addition, IκB kinase B (IkkB) phosphorylates RPS3 at serine 309, which promotes RPS3 translocation to the nucleus48 and induces ubiquitination of NF-κB inhibitor. In addition, we see the high expression of NF-κB1, an inhibitor of the NF-κB pathway.49 We believe as the expression of RPS3 was downregulated, the IkkB-RPS3 could not be activated and induced the expression of NF-κB1 in HA-NRP1-4Cys-4Ala and FLAG-SARS-CoV-2-S1493−685 co-transfected cells. Some RPs also facilitate translation initiation, where the virus infects the host.
RPS9, RPLP1/2, and RPS19 are the signature genes required for viral protein synthesis.50,51,52 We could see the expression of these genes is downregulated. So, selective inhibition of the S1-HA-NRP1-4Cys-4Ala interaction would reduce SARS-Cov-2 infection when exposed to the virus. However, further investigation is needed to understand how ribosomal protein expression relates to stable S1-NRP1 interaction.
In addition, we evaluated expression patterns of genes that participate in cell cycle pathways in our transcriptome analysis since we were interested in finding out which pathway gets activated. We found considerably higher expression of some cell cycle genes in cells transfected with HA-NRP1-WT compared to the cells transfected with HA-NRP1-4Cys-4Ala in the presence of SARS-CoV-2-S1493−685 (Figure 6A). These included kinesin family member 20A (KIF20A), SRY-box 2 (SOX2), and (TACC3). KIF20A, which was initially discovered as a key protein in mitosis, was later found to be responsible for poor prognosis in cancers when it is upregulated.8 Downregulation of KIF20A induces cell-cycle arrest and apoptosis by suppressing PI3K/AKT pathway.53 SOX2 is a universal cancer stem cell marker, accountable for the maintenance of cancer stem cells and tissue homeostasis,54 and it is known to initiate cell division and is known to be a regulator of G1/S transition in cell cycle. TACC3 regulates spindle organization during mitosis and is an important candidate for cell division.55 Growth of lung cancer has been reported to be inhibited by targeting Proteasome subunit beta type6 (PSMB6).56 PSMB7 is a prognostic biomarker for breast cancer.57 On the other hand, transcriptome profiling revealed upregulated expression of several genes implicated in cellular pathways like ubiquitination, degradation, and suppression of inflammation. We observed significantly higher expression of both degradation in ER protein 3 (DERL3) and homocysteine-inducible ER protein with ubiquitin-like domain 1 (HERPUD1), both of which are known to promote degradation of misfolded proteins in the ER.58,59 Interestingly, DERL3 has been identified as a potential candidate gene that might be regulated via SARS-CoV-2-induced DNA methylation changes in COVID-19 infection.60 HERPUD1, which usually activates innate immune response, is an ER stress marker and was found to be upregulated in L cells infected with mouse hepatitis virus (MHV) or SARS-CoV.61 To our surprise, we saw higher ER oxidoreductase 1 beta (ERO1B) expression in cells transfected with HA-NRP1-4Cys-4Ala. ERO1B re-oxidizes PDIA4 leading to reactive oxygen species production.62
Abrogating the interaction between NRP1 and S1 reduces the expression of several 20S proteasomes, which includes PSMB 3,4,5,6, and 7. PSMBs are in the class of modified proteasomes, involved in processing class I major histocompatibility complex peptides, and have an enhanced ability to generate antigenic peptides.63 Moreover, Proteasomes are present throughout the eukaryotic cells and cleave peptides in an ATP/ubiquitin-dependent process.64
Inflammatory responses define the outcome of viral infection, and hyperinflammation in COVID-19 contributes to disease severity.65 One of the important factors that play a central role in the initiation and resolution of this response is the NF-κB/REL family of transcription factors. NF-κB subunit c-Rel acts as a transcriptional repressor of inflammatory genes.66 Our results indicated higher expression for both c-REL and NF-κB1 with HA-NRP1-4Cys-4Ala in the presence of SARS-CoV-S1493−685.
Finally, Venn diagram shows overlap of genes found to be differentially expressed when cells were transfected with NRP1-WT vs. NRP1-4Cys-4Ala without the S protein as well as when cells were transfected with NRP1-WT plus SARS-CoV-2-S1493−685 vs. NRP1-4Cys-4Ala plus SARS-CoV-2-S1493−685 (Figure 6B). Interestingly, the inclusion of SARS-CoV-2-S1493−685 dramatically increased differential expression of unique genes to 10 times higher between the two groups (Figure 6B). Collectively, these data potentially revealed previously unknown downstream targets involved in NRP1 mediation of SARS-CoV-2 S1 interaction.
Reduced cellular expression of pERK was observed in cells co-transfected with SARS-CoV-2 S protein and NRP1 mutant
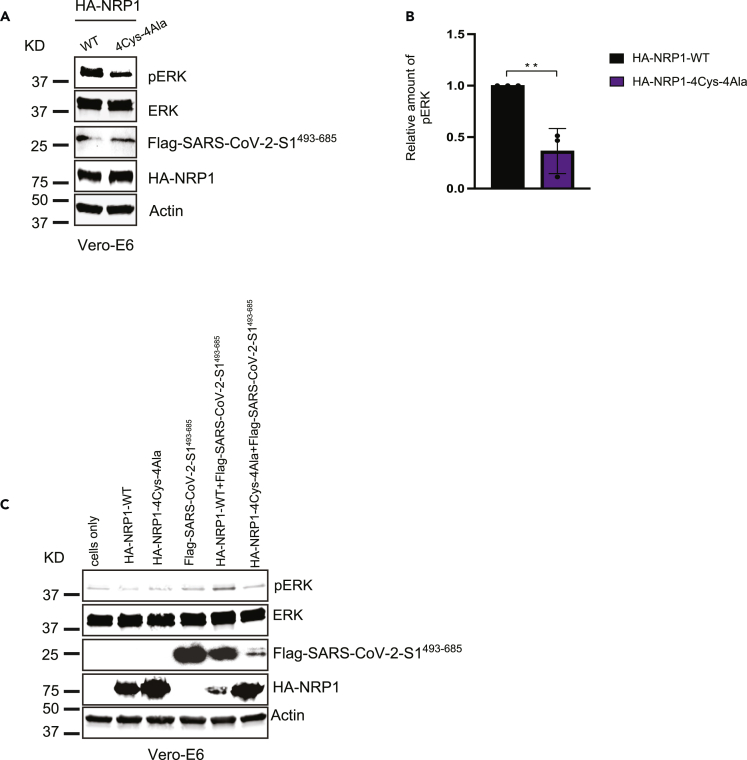
As obligate intracellular pathogen, viruses take over the complex metabolic machinery of the host to produce more viral particles and maintain viral pathogenesis. One way to do that is to manipulate the existing host cell signal transduction pathway to ensure optimal viral replication. One of the best-known pathways required by several DNA and RNA viruses to activate their replication machinery is that the mitogen-activated protein kinase (MAPK) family comprises extracellular signal-regulated kinases 1 and 2 (ERK1/2) cascade, activated in response to environmental factors and inflammatory stress.67,68,69 Upon activation by phosphorylation, ERK1/2 MAPKs translocate to the nucleus and trigger downstream gene expression required to facilitate viral infection, dysregulation of the host cell cycle, and alterations to host immune responses to upregulate viral pathogenesis.70 Previous studies have reported that PD98059, a specific inhibitor of the ERK/MAPK pathway, abolishes visna virus replication.71 Prior work has suggested that coronavirus S protein activates the MAPK pathway and downstream inflammatory responses and can be targeted by MEKi.72 In this study, we have taken a step forward in understanding the impact of novel cysteine mutations in NRP1 on the alteration of the downstream signaling cascade by evaluating the phosphorylation levels of MAPK pathway-associated proteins. First, we analyzed the downstream activation of ERK1/2 in Vero-E6 cells transfected with NRP1-WT or NRP1-4Cys-4Ala mutant together with SARS-CoV-2-S1493-685. The level of ERK phosphorylation was significantly downregulated in cells transfected with NRP1-4Cys-4Ala as determined by western blot in the presence of recombinant S protein (Figures 7A and 7B). To further investigate the correlation between reduced ERK phosphorylation and NRP1 mutation, we compared the level of endogenous phosphorylated extracellular signal-regulated kinase (pERK) among cells transfected with NRP1 without S protein. We found no changes in the pERK level in cells transfected with NRP1-WT or NRP1-4Cys-4Ala mutant in the absence of the S protein (Figure 7C). Taken together; we concluded that successful NRP1-S protein interaction is imperative for the higher expression of pERK.
Figure 7.
Reduced cellular expression of pERK was observed in cells co-transfected with SARS-CoV-2 S protein and NRP1 mutant
(A) Vero-E6 cells were transfected with the indicated HA-NRP1 constructs together with FLAG-SARS-CoV-2-S1493−685. Cells were lysed, and endogenous ERK phosphorylation was analyzed.
(B) Quantification of the band intensities (n = 3). pERK band intensities were normalized to the Actin control. Data are represented as mean ± SD, and ∗∗, p < 0.005 (Two-tailed Student’s t test).
(C) Immunoblot analysis of whole-cell lysate derived from Vero-E6 cells transfected with the indicated constructs. Upregulation of ERK phosphorylation was abrogated in cells co-transfected with SARS-CoV-2 S protein and NRP1 mutant.
However, in our study, we have not exclusively examined the functional consequences of the NRP1 mutations on the actual SARS-CoV-2 viral particle infection. Based on the biochemical data we presented so far, it might be reasonable to consider further preclinical experiments to establish a more direct approach to use these novel cysteines as a target to suppress SARS-CoV-2 infection. Although our study dealt mainly with increasing mechanistic knowledge, the observations here signify to the potential of fighting against the SARS-CoV-2 virus by selectively targeting these novel cysteines.
Discussion
PAN domain was originally discovered with a diverse biological function including a site for protein-protein or protein-carbohydrate interaction.26 Our recent discoveries have confirmed that PAN mutation is exclusively associated with the protein function rather than an overall structural change.18 In this study, we have identified the presence of an unconventional PAN domain in NRP1 containing novel cysteine residues, which when mutated, interfere with its stability as well as its ability to bind with the SARS-CoV-2 S protein. However, unlike the conventional PAN domain that we have reported earlier, with NRP1, each cysteine residue shows varying impact on NRP1 function and stability at the protein level. We also demonstrated a marked reduction in overall S protein levels even in the presence of an intact endogenous angiotensin converting enzyme 2 (ACE2) receptor in Vero-E6 cells following mutation of these residues. There was no significant association between the NRP1-4Cys-4Ala and SARS-CoV-2 variants, suggesting that these novel cysteines are critical determinants for NRP1 activity. Moreover, NRP1 has been connected to the occurrence of various cancers. Interaction between NRP1 and its ligand, VEGF165, is known to cause pathological angiogenesis.73 Considered to be a therapeutic target against cancer, NRP1 signaling could be modulated by targeting these novel cysteines since our results showed cysteine-mutated NRP1 was unable to interact with its co-receptor Plexin-A1. This might broaden a new avenue in the cancer field as the NRP-plexin receptor complex regulates tumor cell migration, angiogenesis, and proliferation and represents an interesting therapeutic target.36 Aberrant expression of NRP1 is associated with poor prognosis in cancer and is already identified as a potential diagnostic or therapeutic target to improve survival and life quality of patients.37,74 Because NRPs are considered as a prooncogenic co-receptor, these novel sites could be used as a promising target for the development of highly specific and formidable NRP1 inhibitors.
Transcriptomic analysis reveals that the attenuated interaction of S1 and NRP1 cysteines showed some downregulation of specific genes, which take part in immune signaling, proteasomal degradation, secretory pathway, and cell cycle. Targeting the cysteine residues in NRP1 could act as the multi-pathway target for SARS-CoV-2 S protein internalization. In addition, we provided experimental evidence that mutating core cysteine residues in the NRP1 PAN domain results in negative regulation of downstream ERK1/2 MAPK signaling cascade following S protein expression and altered endocytosis for the NRP1 receptor itself. Cantuti-Castelvetri et al. have identified the extracellular soluble b1/b2 domain of NRP1 as the primary binding site for SARS-CoV-2 viral S protein,13,75 or the computational model predicts a2-b1-b2 recognizes RRAR amino acid sequence of the S1 domain of SARS-CoV-2 following furin protease cleavage.76 Here, for the first time to date, we showed direct evidence that novel cysteine residues in vestigial PAN domain directly regulate the S protein interaction. Among the four cysteines identified, we showed two specific cysteine residues, C82 and C104, located in the two N terminal CUB domain (a1/a2) have an significant impact on the S protein recognition across various cell lines used even in the presence of intact b1/b2 domain and have the potential to act as a target site as mutation in any of these two reduced the S protein recognition by NRP1 (Figure 8). Considering the lack of viral particles in this study, we postulate that future examination of these newly identified sites will be necessary. However, our research provides new insights about S protein-NRP1 interaction, increasing our understanding and enhancing our ability to design more targeted strategies to counteract the differential response of COVID-19 virus variants against the vaccines.
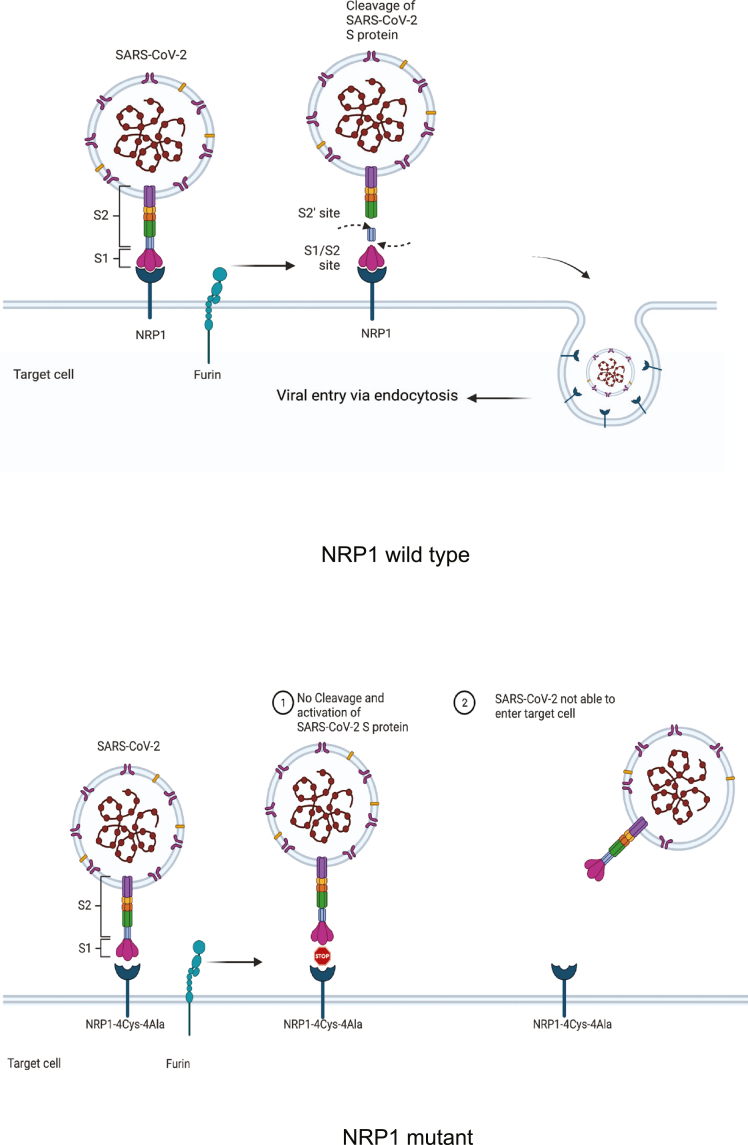
Figure 8.
A proposed model showing how novel cysteines in NRP1 extracellular domains determine the fate of SARS-CoV-2 virus entry
(Top panel) Intact novel cysteines in NRP1 determine its functional capability by providing the attachment site to SARS-CoV-2 S protein leading toward the internalization of virus in the host cells. (Bottom panel) Alteration to those cysteine residues significantly reduces the binding between viral S protein and NRP1 and blocks the virus entry into the host cells. Figure created with BioRender.com.
Limitations of the study
Though our study suggests the presence of previously unknown novel sites in NRP1 that could be targeted against the SARS-CoV-2 infection, this research has some limitations. Our study was solely based on the data that we gathered by using various in vitro cell lines. This study did not include the actual viral particle. Experimental determination of the interaction between viral particles and the cysteine residues in the vestigial PAN domain of NRP1 is necessary. We have also shown a significantly reduced abundance of SARS-CoV-2 S protein in the presence of NRP1 with the mutated PAN domain. But the direct evidence showing the exact mechanism remains unclear. Moreover, we did not conduct any animal experiments to verify some of our conclusion. However, despite these limitations, our current study provided insights into the biochemical mechanism behind the NRP1-SARS-CoV-2 S protein binding that might be useful for developing targeted therapy to mitigate the viral infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HA.C5 | Abcam | Cat #ab18181; RRID: AB_444303 |
| Lamp1 | Abcam | Cat #24170; RRID: AB_775978 |
| Flag-M2 | Sigma | Cat# SLBT7654; RRID: AB_259529 |
| β-Actin | Sigma | Cat# A5441; RRID: AB_476744 |
| HA.11 | Biolegend | Cat# 902302; RRID: AB_2565019 |
| p44/42 MAPK (Erk1/2) | Cell Signaling | Cat# 9102; RRID: AB_330744 |
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) | Cell Signaling | Cat# 4370; RRID: AB_2315112 |
| Flag (DYKDDDDK Tag D6W5B) | Cell Signaling | Cat# 2368S; RRID: AB_2572291 |
| ZO-1A12 | ThermoFisher Scientific | Cat# 33-9100; RRID: AB_2533147 |
| Chemicals, peptides, and recombinant proteins | ||
| TransIT-LT1 | Mirus Bio | Cat# MIR2305 |
| Cycloheximide | EMD Millipore | Cat# AC357420010 |
| Fluorimount G | Southern Biotech | Cat# 0100-01 |
| Hoechst 33342 | ThermoFisher Scientific | Cat# 62249 |
| TRIzol™ Reagent | ThermoFisher Scientific | Cat# 15596026 |
| Dulbecco’s Modified Eagle’s (DMEM) complete medium | Corning | Cat# 10-017-CV |
| Fetal bovine serum (FBS) | Seradigm | Cat# 1500-500GH |
| Penicillin-streptomycin | Millipore Sigma | Cat# P4333-100ML |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 15710 |
| Amersham ECL Western Blotting Detection Reagent | FisherScientific | Cat# 45-002-401 |
| Deposited data | ||
| HEK 293T RNA-seq | This paper | NCBI under BioProject ID PRJNA767890 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA767890 |
| Experimental models: Cell lines | ||
| HEK-293T (293T is an epithelial-like cell that was originally isolated from the kidney of a patient, age:fetus; available through ATCC) | ATCC | Cat# CRL-3216™; RRID: CVCL_0063 |
| HeLa (Originally isolated in 1951 from a cervical carcinoma derived from a 31-year-old black female patient; available through ATCC) | ATCC | Cat# CCL-2™; RRID: CVCL_0030 |
| Vero-E6 also known as Vero C1008 (African Green Monkey Kidney Cells with epithelial morphology; available through ATCC) | A generous gift from Carmen Foster at the Oak Ridge National Laboratory. | Cat# CRL-1586™; RRID: CVCL_0574 |
| Recombinant DNA | ||
| N-terminal tagged HA-NRP1 and different mutants of HA-NRP1 | GenScript (customized) | |
| C-terminal tagged Flag-Plexin-A1 | GenScript (customized) | |
| N-terminal tagged Flag-SARS-CoV-2-S-original (NC_045512.2), Flag-SARS-CoV-2-B.1.1.7-S, Flag-SARS-CoV-B.1.351-S | GenScript (customized) | |
| N-terminal tagged Flag-SARS-CoV-2-S1493−685 | GenScript (customized) | |
| Software and algorithms | ||
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com/ |
| ImageJ | ImageJ software | https://imagej.net/Fiji |
| DESeq2 | Bioconductor software |
https://bioconductor.org/ https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Wellington Muchero (mucherow@ornl.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mammalian cell culture, transfection, and drug treatment
Vero-E6 cells were obtained from Carmen Foster at the ORNL as a generous gift. HeLa and HEK 293T cells were obtained from ATCC and maintained in a humidified atmosphere at 5% CO2 in Dulbecco’s Modified Eagle’s (DMEM) complete medium (Corning) supplemented with 10% fetal bovine serum (FBS; Seradigm) in 37°C. Plasmid transfections were done with TransIT-LT1 (Mirus Bio) per the manufacturer’s instructions. Detailed information regarding all the cell lines used was provided in the “key resources table.”
Declaration
No human or animal subjects were used for this study.
Plasmids and recombinant proteins
N-terminal tagged HA-NRP1 and different mutants of HA-NRP1, C-terminal tagged Flag-Plexin-A1, N-terminal tagged Flag-SARS-CoV-2-S-original (NC_045512.2), Flag-SARS-CoV-2-B.1.1.7-S, Flag-SARS-CoV-B.1.351-S were all obtained from GenScript (Cloned into pcDNA 3.1 vector). N-terminal tagged Flag-SARS-CoV-2-S1493-685 was made in GenScript.
Method details
PAN domain sequence alignment
Proteins with PAN domains were identified from Uniprot and were selected from 2 model organisms.77 The PAN domain coordinates from Uniprot were used to extract the PAN domain sequences from the full-length proteins which were then aligned with MAFFT linsi.78 The alignment was visualized with Geneious.
Immunofluorescence and confocal microscopy
Vero-E6 cells were seeded on coverslips in 24 well plates. Where indicated, cells were transfected with HA-NRP1 WT and different PAN domain mutants together with different SARS-CoV-2 spike protein constructs as indicated for 30 hr, followed by fixation in 4% paraformaldehyde. Next, the cells were permeabilized with 0.5% Triton X-100 in PBS, washed and then blocked for 30 minutes at room temperature with 5% BSA in PBS. Cells were incubated with primary antibodies in 5% BSA in 1X-PBS with 0.5% Triton X-100 for 1 hr at room temperature. After washing the cells were incubated with appropriate secondary antibodies in 5% BSA in PBST for 30 minutes at room temperature. DNA was counterstained with 1 μg/mL Hoechst 33342 and mounted with Fluorimount G (Southern Biotech). Cells were imaged using a Leica SP8 White Light Laser Confocal System. HeLa cells were visualized using a Zeiss LSM 710 confocal microscope.
Membrane localization of different NRP1(s) in Vero-E6 cells were done as described previously.79,80 Briefly, cells were grown on glass coverslips and transfected with N-terminal tagged HA-NRP1 WT and different NRP1 mutants as indicated for 30 hr. All the washing, antibody incubation and 4% (w/v) paraformaldehyde fixation were done at 4 °C without incorporating any permeabilization steps. ZO-1 was used as a control membrane marker. Images were acquired on Zeiss LSM 710 confocal microscope.
Western blotting and immunoprecipitation
For immunoprecipitation, either HA-tagged NRP1 (or mutants) and Flag-tagged SARS-CoV-2-S (or S1493-685) mutant or HA-tagged NRP1 (or mutants) and Flag- Plexin-A1 were expressed where indicated in cells for 30 hr. Cell extracts were generated in EBC buffer, 50mM Tris (pH 8.0), 120mM NaCl, 0.5% NP40, 1mM DTT, and protease and phosphatase inhibitors tablets (Thermo Fisher Scientific). Equal amounts of cell lysates were incubated with the indicated antibodies conjugated to protein G beads (Invitrogen) respectively from 4h to overnight at 4°C. The beads were then washed with EBC buffer including inhibitors. Immunoprecipitation samples or equal amount of whole cell lysates were resolved by SDS-PAGE, transferred to PVDF membranes (Milipore) probed with the indicated antibodies, and visualized with either LiCor Odyssey infra-red imaging system or Chemiluminescence detection. For cycloheximide stability assay, 293T cells were transfected with the constructs encoding HA-NRP1 or HA-NRP1 mutants. 24 hr post transfection, cells were splitted and replated. 20 hr post splitting, cells were treated with 100 ug cycloheximide. Cells washed with PBS and lysed. Briefly, Cell extracts were generated on ice in EBC buffer, 50mM Tris (pH 8.0), 120mM NaCl, 0.5% NP40, 1mM DTT, and protease and phosphatase inhibitors tablets (Thermo Fisher Scientific). Extracted proteins were quantified using the PierceTM BCA Protein assay kit (Thermo Fisher). Proteins were separated by SDS acrylamide gel electrophoresis and transferred to IMMOBILON-FL 26 PVDF membrane (Milipore) probed with the indicated antibodies and visualized either by chemiluminescence (according to the manufacturer’s instruction) or using LiCor Odyssey infra-red imaging system. To detect the phosphorylation of ERK1/2, Vero cells were transfected with indicated constructs expressing NRP1 and SARS-CoV-2 spike proteins. 30 hr post transfection, cells were lysed and the expression of p-ERK and total ERK were determined by Western blot.
RNA-seq analysis
For RNA-seq analysis, Total RNA was extracted from HEK293T cell line 30 hr post transfection (HA-NRP1 and SARS-CoV-2-S1493-685 and or HA-NRP1-4Cys-4Ala and SARS-CoV-2-S1493-685) using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Quantification and quality control of isolated RNA was performed by measuring absorbance at 260 nm and 280 nm on a NANODROP ONEC spectrophotometer (Thermo Scientific, USA). The RNA-seq run was performed with four biological replicates. Library prep and sequencing was performed by BGI using the DNBSEQ-G400 platform which generated 100bp paired-end reads. The raw RNA-seq reads have been deposited at NCBI under BioProject ID PRJNA767890. Clean reads were aligned to the human reference genome GRCh38. Reads were mapped with bowtie2 v2.2.5.81 Differential expression analysis was performed with DESeq2, genes with an adjusted p-value less than 0.05 were considered differentially expressed.82
Quantification and statistical analysis
Statistical analyses were performed on individual experiments, as indicated, with GraphPad Prism 8 Software using an unpaired t-Test, equal variance for comparison between two groups and one-way ANOVA for comparisons between more than two groups. A P value of ∗P<0.05 was considered as statistically significant.
Acknowledgments
Bioinformatics analyses of PAN domain distribution and functional inference was supported by the United States Department of Energy’s Office of Science Early Career Research Program under the Biological and Environmental Research office. Biochemical, immunofluorescence, and transcriptome analyses in human cell lines were supported by the Oak Ridge National Laboratory Lab Directed Research Development program. Part of this research used resources at the Oak Ridge Leadership Computing Facility (OLCF) and the Compute and Data Environment for Science (CADES) at the Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract Number DE-AC05-00OR22725. Debjani Pal was supported by the United States Department of Energy’s Office of Science Lab Directed Research and Development fund under the Radioisotope Science and Technology Division Research office (grant number: 3130F004) during the revised version of manuscript preparation. The Vero-E6 cells were a generous gift from Carmen Foster at ORNL.
Author contributions
Conceptualization: DP, KD, and WM. Methodology: DP, KD, TBY, and JK. Funding acquisition: WM. Writing – original draft: DP, KD, and WM. Writing – review & editing: DP, KD, TBY, and WM.
Declaration of interests
The authors declare that they have no competing interests.
Published: February 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106274.
Supplemental information
Data and code availability
-
•
The published article and supplemental information include all data generated and analyzed during this study. This paper does not report original code.
-
•
RNA-seq data has been deposited at NCBI and are publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
Any additional information supporting the current study in this paper is available from the lead contact upon request.
References
- 1.Appleton B.A., Wu P., Maloney J., Yin J., Liang W.C., Stawicki S., Mortara K., Bowman K.K., Elliott J.M., Desmarais W., et al. Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. Embo j. 2007;26:4902–4912. doi: 10.1038/sj.emboj.7601906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H., Chédotal A., He Z., Goodman C.S., Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron. 1997;19:547–559. doi: 10.1016/s0896-6273(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 3.Kolodkin A.L., Levengood D.V., Rowe E.G., Tai Y.T., Giger R.J., Ginty D.D. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 4.Takagi S., Hirata T., Agata K., Mochii M., Eguchi G., Fujisawa H. The A5 antigen, a candidate for the neuronal recognition molecule, has homologies to complement components and coagulation factors. Neuron. 1991;7:295–307. doi: 10.1016/0896-6273(91)90268-5. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann G., Bork P. An adhesive domain detected in functionally diverse receptors. Trends Biochem. Sci. 1993;18:40–41. doi: 10.1016/0968-0004(93)90049-s. [DOI] [PubMed] [Google Scholar]
- 6.Mamluk R., Gechtman Z., Kutcher M.E., Gasiunas N., Gallagher J., Klagsbrun M. Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J. Biol. Chem. 2002;277:24818–24825. doi: 10.1074/jbc.M200730200. [DOI] [PubMed] [Google Scholar]
- 7.Holmes D.I.R., Zachary I.C. Vascular endothelial growth factor regulates stanniocalcin-1 expression via neuropilin-1-dependent regulation of KDR and synergism with fibroblast growth factor-2. Cell. Signal. 2008;20:569–579. doi: 10.1016/j.cellsig.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura F., Goshima Y. Structural and functional relation of neuropilins. Adv. Exp. Med. Biol. 2002;515:55–69. doi: 10.1007/978-1-4615-0119-0_5. [DOI] [PubMed] [Google Scholar]
- 9.Lampropoulou A., Ruhrberg C. Neuropilin regulation of angiogenesis. Biochem. Soc. Trans. 2014;42:1623–1628. doi: 10.1042/bst20140244. [DOI] [PubMed] [Google Scholar]
- 10.Telley L., Cadilhac C., Cioni J.M., Saywell V., Jahannault-Talignani C., Huettl R.E., Sarrailh-Faivre C., Dayer A., Huber A.B., Ango F. Dual function of NRP1 in axon guidance and subcellular target recognition in cerebellum. Neuron. 2016;91:1276–1291. doi: 10.1016/j.neuron.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Parker M.W., Linkugel A.D., Vander Kooi C.W. Effect of C-terminal sequence on competitive semaphorin binding to neuropilin-1. J. Mol. Biol. 2013;425:4405–4414. doi: 10.1016/j.jmb.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H.-B., Zhang H., Zhang J.-P., Li Y., Zhao B., Feng G.-K., Du Y., Xiong D., Zhong Q., Liu W.-L., et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015;6:6240–6313. doi: 10.1038/ncomms7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao H.Q., Soker S., Feiner L., Alonso J.L., Raper J.A., Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility. J. Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pal D., De K., Shanks C.M., Feng K., Yates T.B., Morrell-Falvey J., Davidson R.B., Parks J.M., Muchero W. Core cysteine residues in the Plasminogen-Apple-Nematode (PAN) domain are critical for HGF/c-MET signaling. Commun. Biol. 2022;5:646. doi: 10.1038/s42003-022-03582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pal D., De K., Shanks C.M., Feng K., Yates T.B., Morrell-Falvey J., Davidson R.B., Parks J.M., Muchero W. Core cysteine residues in the PAN domain are critical for HGF/c-MET signaling. Commun. Biol. 2022;5:646. doi: 10.1101/2021.09.20.460979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuntal De D.P., Shanks C.M., Feng K., Jawdy S.S., Hassan M., Prabhakar P.K., Yang J.-Y., Chapla D., Moremen K.W., Urbanowicz B., Muchero W. The Plasminogen-Apple-Nematode (PAN) domain suppresses JA/ET defense pathways in plants. Plant Cell. 2021 (under revision) [Google Scholar]
- 21.Okubo N., Taniguchi H., Motokawa T. Successful methods for transplanting fragments of Acropora formosa and Acropora hyacinthus. Coral Reefs. 2005;24:333–342. doi: 10.1007/s00338-005-0496-0. [DOI] [Google Scholar]
- 22.Douek J., Amar K.-O., Rinkevich B. Maternal-larval population genetic traits in Stylophora pistillata, a hermaphroditic brooding coral species. Genetica. 2011;139:1531–1542. doi: 10.1007/s10709-012-9653-x. [DOI] [PubMed] [Google Scholar]
- 23.Parkhaev P.Y. In: Phylogeny and Evolution of the Mollusca. Ponder W., editor. University of California Press; 2008. 33The early cambrian radiation of Mollusca. [DOI] [Google Scholar]
- 24.Parkhaev P.Y. Origin and the early evolution of the phylum Mollusca. Paleontol. J. 2017;51:663–686. doi: 10.1134/S003103011706003X. [DOI] [Google Scholar]
- 25.Haszprunar G., Wanninger A. Molluscs. Curr. Biol. 2012;22:R510–R514. doi: 10.1016/j.cub.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 26.Tordai H., Bányai L., Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 27.Okon I.S., Coughlan K.A., Zhang C., Moriasi C., Ding Y., Song P., Zhang W., Li G., Zou M.-H. Protein kinase LKB1 promotes RAB7-mediated neuropilin-1 degradation to inhibit angiogenesis. J. Clin. Invest. 2014;124:4590–4602. doi: 10.1172/JCI75371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jobe A., Vijayan R. Characterization of peptide binding to the SARS-CoV-2 host factor neuropilin. Heliyon. 2021;7:e08251. doi: 10.1016/j.heliyon.2021.e08251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaewkla M., Charoenwongpaiboon T., Mahalapbutr P. Molecular basis of the new COVID-19 target neuropilin-1 in complex with SARS-CoV-2 S1 C-end rule peptide and small-molecule antagonists. J. Mol. Liq. 2021;335:116537. doi: 10.1016/j.molliq.2021.116537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z.L., Buck M. Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of S protein S1 and S2. Biophys. J. 2021;120:2828–2837. doi: 10.1016/j.bpj.2021.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 32.Preissler S., Ron D. Early events in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth L., Koncina E., Satkauskas S., Crémel G., Aunis D., Bagnard D. The many faces of semaphorins: from development to pathology. Cell. Mol. Life Sci. 2009;66:649–666. doi: 10.1007/s00018-008-8518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohm B., Ottemeyer A., Lohrum M., Püschel A.W. Plexin/neuropilin complexes mediate repulsion by the axonal guidance signal semaphorin 3A. Mech. Dev. 2000;93:95–104. doi: 10.1016/S0925-4773(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 35.Sharma A., Verhaagen J., Harvey A.R. Receptor complexes for each of the class 3 semaphorins. Front. Cell. Neurosci. 2012;6:28. doi: 10.3389/fncel.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob L., Sawma P., Garnier N., Meyer L.A.T., Fritz J., Hussenet T., Spenlé C., Goetz J., Vermot J., et al. Inhibition of PlexA1-mediated brain tumor growth and tumor-associated angiogenesis using a transmembrane domain targeting peptide. Oncotarget. 2016;7:57851–57865. doi: 10.18632/oncotarget.11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douyère M., Chastagner P., Boura C. Neuropilin-1: a key protein to consider in the progression of pediatric brain tumors. Front. Oncol. 2021;11:665634. doi: 10.3389/fonc.2021.665634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi T., Fournier A., Nakamura F., Wang L.H., Murakami Y., Kalb R.G., Fujisawa H., Strittmatter S.M. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell. 1999;99:59–69. doi: 10.1016/s0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 39.Wild J.R.L., Staton C.A., Chapple K., Corfe B.M. Neuropilins: expression and roles in the epithelium. Int. J. Exp. Pathol. 2012;93:81–103. doi: 10.1111/j.1365-2613.2012.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weekes C.D., Beeram M., Tolcher A.W., Papadopoulos K.P., Gore L., Hegde P., Xin Y., Yu R., Shih L.M., Xiang H., et al. A phase I study of the human monoclonal anti-NRP1 antibody MNRP1685A in patients with advanced solid tumors. Invest. New Drugs. 2014;32:653–660. doi: 10.1007/s10637-014-0071-z. [DOI] [PubMed] [Google Scholar]
- 41.De Vlaeminck Y.D., Bonelli S., Awad R.M., Dewilde M., Rizzolio S., Lecocq Q., Bolli E., Santos A.R., Laoui D., Schoonooghe S., et al. Targeting neuropilin-1 with nanobodies reduces colorectal carcinoma development. Cancers. 2020;12 doi: 10.3390/cancers12123582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung K., Kim J.A., Kim Y.J., Lee H.W., Kim C.H., Haam S., Kim Y.S. A neuropilin-1 antagonist exerts antitumor immunity by inhibiting the suppressive function of intratumoral regulatory T cells. Cancer Immunol. Res. 2020;8:46–56. doi: 10.1158/2326-6066.Cir-19-0143. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., Takvorian P.M., Bleck C., Hsu V.W., Fehr A.R., et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galindo I., Garaigorta U., Lasala F., Cuesta-Geijo M.A., Bueno P., Gil C., Delgado R., Gastaminza P., Alonso C. Antiviral drugs targeting endosomal membrane proteins inhibit distant animal and human pathogenic viruses. Antiviral Res. 2021;186 doi: 10.1016/j.antiviral.2020.104990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H.-X., Liu Z.-X., Sun Y.-P., Zhu J., Lu S.-Y., Liu X.-S., Huang Q.-H., Xie Y.-Y., Zhu H.-B., Dang S.-Y., et al. Rig-I regulates NF-κB activity through binding to Nf-κb1 3'-UTR mRNA. Proc. Natl. Acad. Sci. USA. 2013;110:6459–6464. doi: 10.1073/pnas.1304432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kircheis R., Haasbach E., Lueftenegger D., Heyken W.T., Ocker M., Planz O. NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Front. Immunol. 2020;11:598444. doi: 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan F., Anderson D.E., Barnitz R.A., Snow A., Bidere N., Zheng L., Hegde V., Lam L.T., Staudt L.M., Levens D., et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Wan F., Weaver A., Gao X., Bern M., Hardwidge P.R., Lenardo M.J. IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 2011;12:335–343. doi: 10.1038/ni.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moorthy A.K., Savinova O.V., Ho J.Q., Wang V.Y.F., Vu D., Ghosh G. The 20S proteasome processes NF-kappaB1 p105 into p50 in a translation-independent manner. Embo J. 2006;25:1945–1956. doi: 10.1038/sj.emboj.7601081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campos R.K., Wong B., Xie X., Lu Y.F., Shi P.Y., Pompon J., Garcia-Blanco M.A., Bradrick S.S. RPLP1 and RPLP2 are essential Flavivirus host factors that promote early viral protein accumulation. J. Virol. 2017;91 doi: 10.1128/jvi.01706-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukushi S., Okada M., Stahl J., Kageyama T., Hoshino F.B., Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 52.Cheng E., Haque A., Rimmer M.A., Hussein I.T.M., Sheema S., Little A., Mir M.A. Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19) J. Biol. Chem. 2011;286:11814–11824. doi: 10.1074/jbc.M110.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin Z., Tao S., Zhang C., Xu D., Zhu Z. KIF20A promotes the development of fibrosarcoma via PI3K-Akt signaling pathway. Exp. Cell Res. 2022;420 doi: 10.1016/j.yexcr.2022.113322. [DOI] [PubMed] [Google Scholar]
- 54.Takeda K., Mizushima T., Yokoyama Y., Hirose H., Wu X., Qian Y., Ikehata K., Miyoshi N., Takahashi H., Haraguchi N., et al. Sox2 is associated with cancer stem-like properties in colorectal cancer. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-36251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Z.M., Huang C.J., Jiao X.F., Wu D., Huo L.J. The role of TACC3 in mitotic spindle organization. Cytoskeleton (Hoboken) 2017;74:369–378. doi: 10.1002/cm.21388. [DOI] [PubMed] [Google Scholar]
- 56.Lu Z., Song Q., Yang J., Zhao X., Zhang X., Yang P., Kang J. Comparative proteomic analysis of anti-cancer mechanism by periplocin treatment in lung cancer cells. Cell. Physiol. Biochem. 2014;33:859–868. doi: 10.1159/000358658. [DOI] [PubMed] [Google Scholar]
- 57.Munkácsy G., Abdul-Ghani R., Mihály Z., Tegze B., Tchernitsa O., Surowiak P., Schäfer R., Györffy B. PSMB7 is associated with anthracycline resistance and is a prognostic biomarker in breast cancer. Br. J. Cancer. 2010;102:361–368. doi: 10.1038/sj.bjc.6605478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Serra P., Marcilla M., Villanueva A., Ramos-Fernandez A., Palau A., Leal L., Wahi J.E., Setien-Baranda F., Szczesna K., Moutinho C., et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 2014;5:3608. doi: 10.1038/ncomms4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho D.V., Chan J.Y. Induction of Herpud1 expression by ER stress is regulated by Nrf1. FEBS Lett. 2015;589:615–620. doi: 10.1016/j.febslet.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muhammad J.S., Saheb Sharif-Askari N., Cui Z.-G., Hamad M., Halwani R. SARS-CoV-2 infection-induced promoter hypomethylation as an epigenetic modulator of heat shock protein A1L (HSPA1L) gene. Front. Genet. 2021;12:622271. doi: 10.3389/fgene.2021.622271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zito E., Chin K.-T., Blais J., Harding H.P., Ron D. ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 2010;188:821–832. doi: 10.1083/jcb.200911086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarthy M.K., Weinberg J.B. The immunoproteasome and viral infection: a complex regulator of inflammation. Front. Microbiol. 2015;6:21. doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka K. The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gustine J.N., Jones D. Immunopathology of hyperinflammation in COVID-19. Am. J. Pathol. 2021;191:4–17. doi: 10.1016/j.ajpath.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Jesús T.J., Ramakrishnan P. NF-κB c-Rel dictates the inflammatory threshold by acting as a transcriptional repressor. iScience. 2020;23 doi: 10.1016/j.isci.2020.100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furler R.L., Uittenbogaart C.H. Signaling through the P38 and ERK pathways: a common link between HIV replication and the immune response. Immunol. Res. 2010;48:99–109. doi: 10.1007/s12026-010-8170-1. [DOI] [PubMed] [Google Scholar]
- 68.Pleschka S. RNA viruses and the mitogenic Raf/MEK/ERK signal transduction cascade. Biol. Chem. 2008;389:1273–1282. doi: 10.1515/bc.2008.145. [DOI] [PubMed] [Google Scholar]
- 69.Brunetti J.E., Foscaldi S., Quintana V.M., Scolaro L.A., López N., Castilla V. Role of the ERK1/2 signaling pathway in the replication of junín and tacaribe viruses. Viruses. 2018;10:199. doi: 10.3390/v10040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DuShane J.K., Maginnis M.S. Human DNA virus exploitation of the MAPK-ERK cascade. Int. J. Mol. Sci. 2019;20:3427. doi: 10.3390/ijms20143427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barber S.A., Bruett L., Douglass B.R., Herbst D.S., Zink M.C., Clements J.E. Visna virus-induced activation of MAPK is required for virus replication and correlates with virus-induced neuropathology. J. Virol. 2002;76:817–828. doi: 10.1128/jvi.76.2.817-828.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou L., Huntington K., Zhang S., Carlsen L., So E.Y., Parker C., Sahin I., Safran H., Kamle S., Lee C.M., et al. MEK inhibitors reduce cellular expression of ACE2, pERK, pRb while stimulating NK-mediated cytotoxicity and attenuating inflammatory cytokines relevant to SARS-CoV-2 infection. Oncotarget. 2020;11:4201–4223. doi: 10.18632/oncotarget.27799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soker S., Miao H.Q., Nomi M., Takashima S., Klagsbrun M. VEGF165 mediates formation of complexes containing VEGFR-2 and neuropilin-1 that enhance VEGF165-receptor binding. J. Cell. Biochem. 2002;85:357–368. doi: 10.1002/jcb.10140. [DOI] [PubMed] [Google Scholar]
- 74.Niland S., Eble J.A. Neuropilins in the context of tumor vasculature. Int. J. Mol. Sci. 2019;20:639. doi: 10.3390/ijms20030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alnomasy S.F. Virus-receptor interactions of SARS-CoV-2 S receptor-binding domain and human neuropilin-1 b1 domain. Saudi J. Biol. Sci. 2021;28:3926–3928. doi: 10.1016/j.sjbs.2021.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z.-L., Buck M. Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of S protein domains S1 and S2. bioRxiv. 2021 doi: 10.1101/2021.01.06.425627. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lalioti V., González-Sanz S., Lois-Bermejo I., González-Jiménez P., Viedma-Poyatos Á., Merino A., Pajares M.A., Pérez-Sala D. Cell surface detection of vimentin, ACE2 and SARS-CoV-2 S proteins reveals selective colocalization at primary cilia. Sci. Rep. 2022;12:7063. doi: 10.1038/s41598-022-11248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carrodus N.L., Teng K.S.L., Munro K.M., Kennedy M.J., Gunnersen J.M. Differential labeling of cell-surface and internalized proteins after antibody feeding of live cultured neurons. J. Vis. Exp. 2014:e51139. doi: 10.3791/51139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The published article and supplemental information include all data generated and analyzed during this study. This paper does not report original code.
-
•
RNA-seq data has been deposited at NCBI and are publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
Any additional information supporting the current study in this paper is available from the lead contact upon request.