Abstract
At presynaptic active zones (AZs), conserved scaffold protein architectures control synaptic vesicle (SV) release by defining the nanoscale distribution and density of voltage-gated Ca2+ channels (VGCCs). While AZs can potentiate SV release in the minutes range, we lack an understanding of how AZ scaffold components and VGCCs engage into potentiation. We here establish dynamic, intravital single-molecule imaging of endogenously tagged proteins at Drosophila AZs undergoing presynaptic homeostatic potentiation. During potentiation, the numbers of α1 VGCC subunit Cacophony (Cac) increased per AZ, while their mobility decreased and nanoscale distribution compacted. These dynamic Cac changes depended on the interaction between Cac channel’s intracellular carboxyl terminus and the membrane-close amino-terminal region of the ELKS-family protein Bruchpilot, whose distribution compacted drastically. The Cac-ELKS/Bruchpilot interaction was also needed for sustained AZ potentiation. Our single-molecule analysis illustrates how the AZ scaffold couples to VGCC nanoscale distribution and dynamics to establish a state of sustained potentiation.
Intravital single molecule imaging pictures how scaffold proteins and Ca2+ channels execute synaptic plasticity.
INTRODUCTION
Synapses are key sites of information processing and storage in the brain. Synaptic transmission strength is not hardwired but adapts during synaptic plasticity to provide adequate input-output relationships, maintain or restore transmission when compromised, and store information (1). Mechanisms of long-term (more than seconds) postsynaptic plasticity have been extensively studied, and processes targeting postsynaptic neurotransmitter receptors have been convincingly connected to learning and memory processes (2). Presynaptic long-term plasticity, however, is also prominent, not only at mammalian hippocampal mossy fiber bouton synapses but also at many other synapse types (3). However, as we still lack a coherent understanding of how long-term presynaptic plasticity manifests itself at the molecular level, analyzing its computational and behavioral role remains challenging. Voltage-gated Ca2+ channels (VGCCs) at the presynaptic membrane are the fundamental and conserved triggers of evoked synaptic vesicle (SV) release and modulate SV release probability through the precise distance between VGCCs [within 10 to 200 nm around the active zone (AZ) center] and release-ready SVs. AZ scaffold proteins can cluster VGCCs with nanoscale precision to release sites within the membrane (4–6).
Homeostatic plasticity is observed from invertebrates to humans and actively stabilizes synaptic transmission in response to neural activity perturbation. Notably, homeostatic mechanisms control both presynaptic and postsynaptic function and likely functionally intersect with Hebbian plasticity mechanisms in stable information encoding such as learning and memory (7). Meanwhile, elucidating the conserved mechanistic basis of presynaptic homeostatic plasticity at the highly accessible neuromuscular junction (NMJ) of Drosophila larvae has provided important insights (8). Here, presynaptic homeostatic potentiation (PHP) can be acutely triggered by blocking postsynaptic glutamate receptors with Philanthotoxin (PhTx). Within minutes, PHP precisely counterbalances the reduced postsynaptic responsiveness through enhanced neurotransmitter release, involving both increased release probability for docked SVs at existing release sites and the addition of functional release sites at AZs (9, 10). During PHP, Ca2+ influx increases, and image analysis based on confocal microscopy suggests that additional VGCCs are physically recruited to preexisting AZs at NMJ synapses (9–11). Moreover, intensity levels of munc13-family release factor Unc13A and ELKS-family Bruchpilot scaffold protein (BRP) also both increase during PHP (12, 13), implying that additional release sites might get recruited at preexisting AZs (4). Notably, at the AZs of plastically remodeling rodent mossy fiber boutons, the addition of release sites and accumulation of additional VGCCs were also found to promote functional potentiation (3). Despite such progress, the exact molecular mechanisms by which AZ scaffolds intersect with VGCCs to mediate PHP still remain enigmatic (5).
In this study, we explore the in vivo dynamics of individual Drosophila Cav2–related α1 VGCCs called Cacophony (Cac) at AZs undergoing homeostatic remodeling over tens of minutes in Drosophila. At potentiating AZs, the numbers of Cac channels increased, while their mobility decreased, and the overall nanoscale distribution of the channels compacted. Mechanistically, we found that RIM-1 and RimBP (RIM binding protein) scaffold proteins and their conserved binding sites within the Cac channel’s C terminus were dispensable for Cac immobilization and compaction. In contrast, the N terminus of BRP, which also binds the Cac C terminus, was critical for Cac channel single-molecule motility and plasticity and was specifically needed to allow for sustained, but not immediate, release potentiation. BRP single-molecule imaging also revealed an extensive homeostatic plasticity-driven compaction coinciding with the Cac compaction.
Together, we present in vivo evidence at a single-molecule level for a remodeling process that compacts and immobilizes VGCCs and BRP/ELKS scaffold proteins to drive AZs into sustained potentiation.
RESULTS
Quantifying the in vivo mobility of individual VGCCs at Drosophila AZs
Cac (Dmca1A), the only Drosophila Cav2–related VGCC, is essential for evoked release, equally at not only NMJ but also central Drosophila synapses, which means that it represents the collective function of mammalian N- and P/Q-type VGCCs in this model. We investigated the in vivo dynamic distribution of individual Cac channels at remodeling NMJ AZs undergoing potentiation using single-molecule imaging techniques.
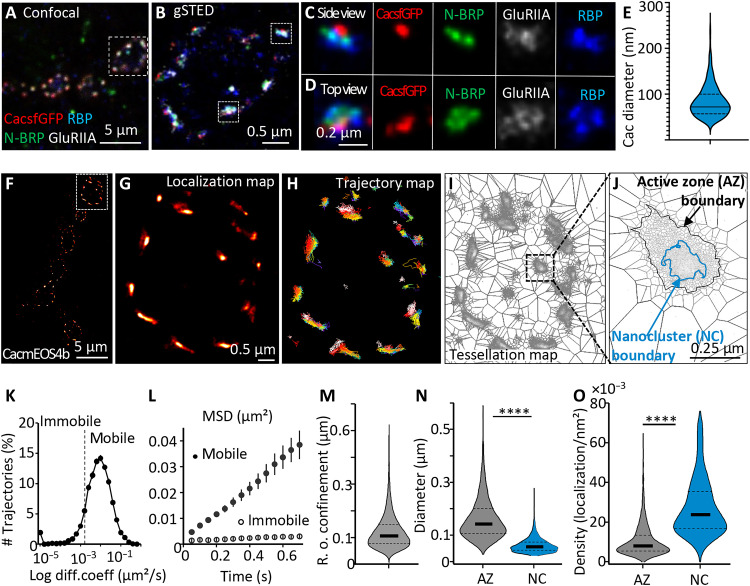
In a recent study, the “on-locus” tagging of Cac with a green fluorescent protein (CacsfGFP) close to its intracellular N terminus was shown to render Cac fully functional, conferring normal transmission and PHP at NMJs (11). We started by subjecting the CacsfGFP channel distribution at fixed larval NMJs to Stimulated emission depletion (STED) microscopy superresolution imaging. Under STED, CacsfGFP typically formed a single discrete cluster within a STED-resolved ring-like structure of the AZ scaffold protein BRP, which aligns with a postsynaptic glutamate receptor field, as expected (14). The membrane-proximal BRP N terminus, again as expected, directly overlays the Cac clusters and, in turn, was surrounded by a STED-resolved RimBP ring (Fig. 1, A to E). Quantification revealed that the AZ Cac channel cluster diameter was, on average, 85.3 ± 1.3 nm (Fig. 1E). The positioning and sizing of CacsfGFP clusters within AZs deduced by STED concur well with previous observations (6, 15, 16) and form a frame for our further investigations.
Fig. 1. Live in vivo mobility and distribution of individual VGCCs at Drosophila NMJ synapses.
(A to D) Representative four-channel confocal (A) and gSTED (B to D) images of NMJs (muscle 4) of third-instar larvae. Differential labeling of AZs with CacsfGFP (red), BRP N terminus (green), and RBP C terminus (blue) and of postsynaptic GluRIIA (gray) is evident on the side and top views (C and D). (E) Sizing of Cac signal diameter by measuring the Feret’s diameter. N = 441 planar AZs from five animals, 24 images derived from n = 2 independent sets. (F to J) Live Cac channel dynamics based on sptPALM of CacmEOS4b in 1.5 mM Ca2+ extracellular imaging buffer. Localization, trajectory, and tessellation maps (G to I) of the boxed bouton in (F) illustrate the stepwise analysis leading to the identification of Cac nanoclusters (NCs) inside AZs (I and J). (K to O) Quantification of live dynamics of Cac sptPALM imaging from 24 animals, N = 70 regions of interest (ROIs), 1166 AZs, where 14,013 trajectories were analyzed. (K to M) Tessellation analysis revealed the diameters of AZs and NCs (N) and the density of Cac channel localizations within AZ and NC boundaries (O). Data distribution was statistically tested by the Kolmogorov-Smirnov test. Statistical significance is denoted as asterisks: ****P < 0.0001. Scale bars, 5 μm (A and F), 0.5. μm (B and F), and 200 nm (C and D).
We intended to study the behavior of individual Cac channel molecules within AZs. Thus, to use live single-particle tracking via photoactivation localization microscopy (sptPALM) (17, 18), we exchanged the GFP for a mEOS4b tag, resulting in CacmEOS4b (19). CacmEOS4b flies were fully viable (note that Cac null mutants are embryonically lethal) (20). We then used sptPALM to follow functional Cac channels in AZs over time. For this, we modified a preparation described previously (21), allowing for the use of the oblique illumination profiles of a total internal reflection fluorescence (TIRF) microscope to induce stochastic blinking of individual CacmEOS4b molecules in a focal plane across several boutons of a given NMJ (Fig. 1, F to J, and fig. S1, A to C). Time windows for observation were limited by the bleaching of the fluorophore, which was irreversible after 3 to 5 min of continuous illumination. Functional properties of the synapse were monitored by imaging of the Ca2+ sensor SynapGCaMP6f (see the Supplementary Materials and fig. S3) (22).
For further analysis, we focused on the fluorescent signals of CacmEOS4b whenever robust characteristic blinking behavior indicated the excitation of individual fluorophores and the stability of the focal plane. At an acquisition frequency of 20 Hz, signal intensities were sufficient to gain an average localization accuracy of ~30 nm for individual Cac molecules (fig. S1D).
To gain access to the local distribution and dynamics of individual Cac channels, we first localized individual events and reconnected consecutive detections to create single-channel trajectories (see Materials and Methods for details). We selected trajectories longer than 8 points (400 ms) as suitable for direct extraction of both Cac channel motility and radius of confinement by calculating their mean square displacements (MSDs; Fig. 1, L, and M).
Most Cac channels were confined but not static within the synapse. We calculated a median diffusion coefficient of 0.0075 μm2/s, and from the MSD analysis, we calculated that Cac channels were distributed within an average confinement radius of 120 nm (Fig. 1M). Our data show mobile and immobile Cac channel trajectories within the AZs, where about 25% of the channels were in the immobile fraction of Cac channels, while 75% of the channels stayed mobile but confined over the observation time window (Fig. 1K).
To further characterize the biophysical milieu in which Cac channels operate at AZs, we applied statistical methods to include all trajectories as short as two time points, increasing the statistical power of the analysis. As a result, we extracted additional parameters, such as the drift field and attraction energy for Cac channels confined within the AZs (fig. S4G). We here observed that the trajectories were mostly located in high-density regions (“energy wells”), which also resemble an elliptic Cac channel cluster geometry (short axis, 95 nm, and long axis, 145 nm; fig. S4, G, and K), like the results observed by STED imaging (Fig. 1, A to E and N).
Nanodomains of high and low VGCC density within individual NMJ AZs
We find that the Cac channels at wild-type NMJs are strongly confined within the AZs (Fig. 1). This observation allowed us to exploit further the total population of Cac channel localizations obtained from the sptPALM imaging to probe whether the channels would preferentially localize to specific “nanodomains” within AZs, as previously proposed for mammalian synapses (23, 24). To this end, we used dual-Delaunay triangulation tessellation, a segmentation method, to define local density distributions based on cumulative localizations (25). This method allowed the identification of the preferred region within which most Cac channel localizations in an AZ occur, defining a boundary for each AZ (Fig. 1, J, N, and O). Furthermore, the tessellation process also robustly retrieved a subregion within the AZ clusters, wherein a significant increase in localization density was determined. Henceforth, we call these AZ-central areas “nanoclusters” (Fig. 1, J, N, and O). In addition, we used filtering and segmentation parameters similar to those previously reported in super-resolved localization microscopy of the scaffold protein BRP at Drosophila NMJ AZs (15, 16). Using these strict parameters, we robustly found AZ borders of the best in-focus AZs and found that these AZs typically form a single nanocluster of Ca2+ channels in their center (for further details, see Supplementary Materials).
The Cac channel localizations formed AZ clusters with a diameter of about 160 nm, which contained a central nanocluster with a diameter of ~60 nm (Fig. 1, N, and O), resembling the results of previous STED-based quantifications (14). On average, these Cac nanoclusters harbored about 50% of all Cac localizations found within an AZ (Fig. 1, N, and O). In addition, the diameter of Cac nanoclusters derived from tessellation analysis of our sptPALM data also falls within the 85-nm signal obtained from nanobody-based detection by two-dimensional (2D) STED microscopy, at a lateral resolution of 40 nm (Fig. 1, A to E). STED-based quantification of Cac cluster size thus apparently emphasizes the central AZ nanocluster derived from our sptPALM data. In addition, these results concur with previous STED data, which determined a Cac enrichment zone size of about ~70 nm in the AZ center, which is in the same range as the nanoscopic spacing between SV release sites marked by Unc13A clusters and Cac channels, ensuring high release probability at the NMJ and also central Drosophila synapses (26). Notably, the VGCC cluster sizes observed here are similar to those previously measured in rodent hippocampal synapses by immunohistochemistry, gated STED (gSTED) (∼70 to 100 nm), and electron microscopy (27–29).
Homeostatic remodeling triggers VGCC nanodomain compaction
As mentioned above, at Drosophila NMJs, 10 min of PhTx application suffice to induce PHP. We triggered PHP in CacmEOS4b larvae by the application of 50 μM PhTx for 10 min and performed a live fluorescent horseradish peroxidase (HRP) antibody labeling to outline the NMJ for better orientation during imaging. After 5 min, the animals were washed with plain imaging buffer to remove the unbound label before transferring the larva into the imaging chamber. Notably in this regard, PhTx treatment was shown previously to irreversibly block postsynaptic GluRIIA receptors (9, 10).
We began live sptPALM imaging of Cac channel populations at NMJ Ib boutons (muscles 4, 6, and 7) 20 min after the start of the PhTx application and imaged for 8.5 min per image at an acquisition rate of 20 Hz (Fig. 2, A to H). Thus, we analyzed the behavior of individual VGCCs within a time window of sustained PHP expression (after 20 to 30 min of PhTx treatment) (30), which follows the induction of acute (PHP) (after 10 minutes of PhTx treatment) (9–11, 13).
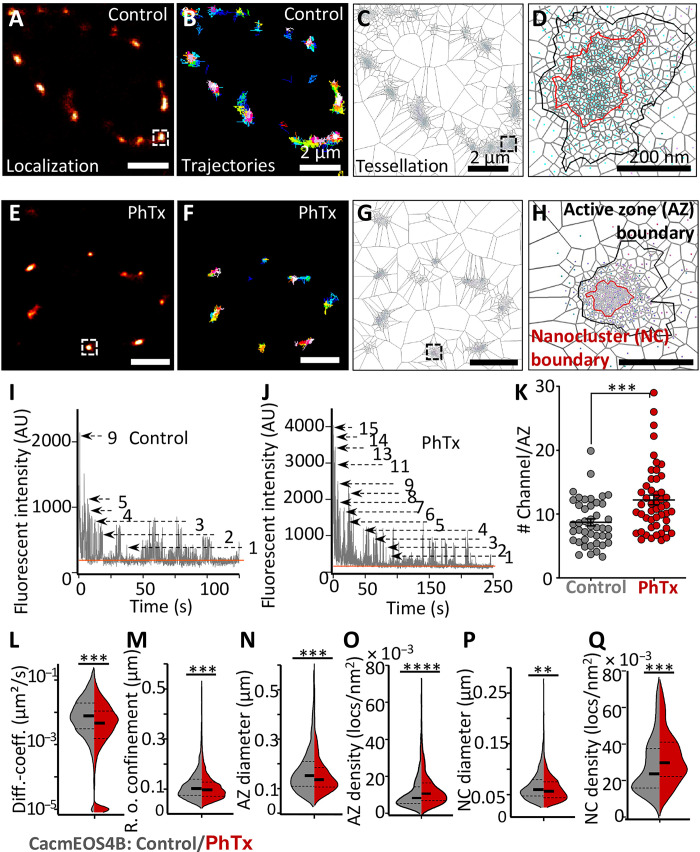
Fig. 2. Live in vivo imaging of Cac channels at remodeling AZs.
(A to H) Live sptPALM imaging of CacmEOS4b at muscle 4 or 6/7 NMJs, performed in 1.5 mM Ca2+ 5 min after a 10-min incubation in HL3 with PhTx or in plain HL3 (Control). Images show representative PALM recordings (A and E), trajectory maps (B and F), and tessellation analysis representations of CacmEOS4B (C, D, G, and H). Scale bars, 2 μm (A to C and E to G) and 200 nm (D and H). (I and J) Recording of stepwise photobleaching as a reference for counting the relative number of CacmEOS4b molecules within AZs of control and PhTx-treated NMJs during a period of continuous illumination (405 nm/561 nm). (K) Quantification of channel numbers from bleaching curves (I and J), derived from the same sptPALM recordings used in (L to Q). A total of 43 AZs (control) and 49 AZs (PhTx) were analyzed based on 12 to 16 images/NMJ from three to four animals for each condition, P < 0.0003 (unpaired t-test). (L and M) Quantification of diffusion coefficients and radii of confinement from live Cac channel sptPALM imaging. Control NMJs: NMJs from 18 animals, N = 54 ROIs; 8013 trajectories; PhTx-treated NMJs: 20 animals, N = 58 ROIs; 9139 trajectories. (N to Q) Tessellation analysis from the same sptPALM dataset was analyzed in (A) to (K) for diameters and densities of Cac channel localizations within AZ and NC cluster boundaries. Here, 54/58 ROIs from 18/19 animals generated 644 AZs (control) and 892 AZs (PhTx), respectively, that were analyzed. Statistical significance is denoted as asterisks: **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data distribution was statistically tested with a Kolmogorov-Smirnov test. AU, arbitrary units.
Concerning the analysis of the sptPALM data, we first counted the relative Cac channel molecule numbers per AZ at both PhTx-treated and untreated control animals. To do so, we analyzed the bleaching behavior of the CacmEOS4b signals at individual AZs upon steady illumination. Discrete bleach steps were identified, which indicate the bleaching of individual mEOS4b-tagged Cac channel fluorophores, allowing us to calculate the fluorescent intensity of single-channel molecules (for details, see figs. S1 and S2). Under control conditions, an average of nine channel molecules per AZ was measured (9.2 ± 0.5). In contrast, after PhTx treatment, an average of 12 channels per AZ was found (12.2 ± 0.8) (Fig. 2, I to K). This analysis thus provides direct single molecule–derived evidence that the numbers of VGCCs per AZ increase upon PhTx treatment, consistent with previous interpretations based on confocal and Ca2+ imaging (9–11). We would like to emphasize that this approach does not depend on the use of antibody labeling, which has the inherent issues of inhomogeneous labeling and limitation in the accessibility of the relevant epitopes. Still, the Cac channel numbers that we retrieved might well represent an underestimation of the total channel number, as we calculated them from the live imaging sptPALM data with limited control over fluorophore conversion and initial photobleaching.
The sptPALM data were further used to characterize the dynamics and distribution of Cac channels of animals undergoing PhTx-triggered plasticity (see details in Materials and Methods). We first analyzed the Cac channel dynamics. Notably, PhTx treatment reduced Cac channel mobility, evident in a profound decrease of the median diffusion coefficient (median diffusion coefficient PhTx treated: 0.0027 μm2/s; untreated controls: 0.0074 μm2/s) (Fig. 2L). In addition, the mean radius of confinement of Cac trajectories was slightly but significantly decreased for PhTx-treated compared to the untreated animals (control, 113 ± 0.6 nm SEM; PhTx treated, 107 ± 0.5 nm SEM) (Fig. 2M). Thus, PhTx treatment seemingly promotes immobilization and confines the spread of Cac channels within the AZ membrane during PHP.
We also used tessellation analysis to calculate nanoscale spread and densities of Cac channels within the broader AZ cluster and the central nanocluster from PALM localization data (Fig. 2, C, D, G, H, and N to Q). Concerning AZ clusters, sizes decreased (by about 10%; Fig. 2N), while channel localization densities significantly increased (by about 20%) in PhTx-treated animals compared to controls (Fig. 2O). Similarly, nanoclusters also displayed a significant increase (about 16%) in Cac channel density, and the nanocluster diameter decreased significantly (by about 8%; Fig. 2, P, and Q). Thus, the high-density, central nanocluster of Cac channels gets further compacted during PHP.
Together, by observing individual VGCCs within 20 to 30 min after the acute induction of PHP (via 10 min of PhTx treatment), i.e., during the phase of sustained PHP, we observed an immobilization of individual Cac channels and a compaction of the overall channel distribution at AZs undergoing stable potentiation of SV release.
RIM or RimBP binding is dispensable for VGCC nanodomain compaction
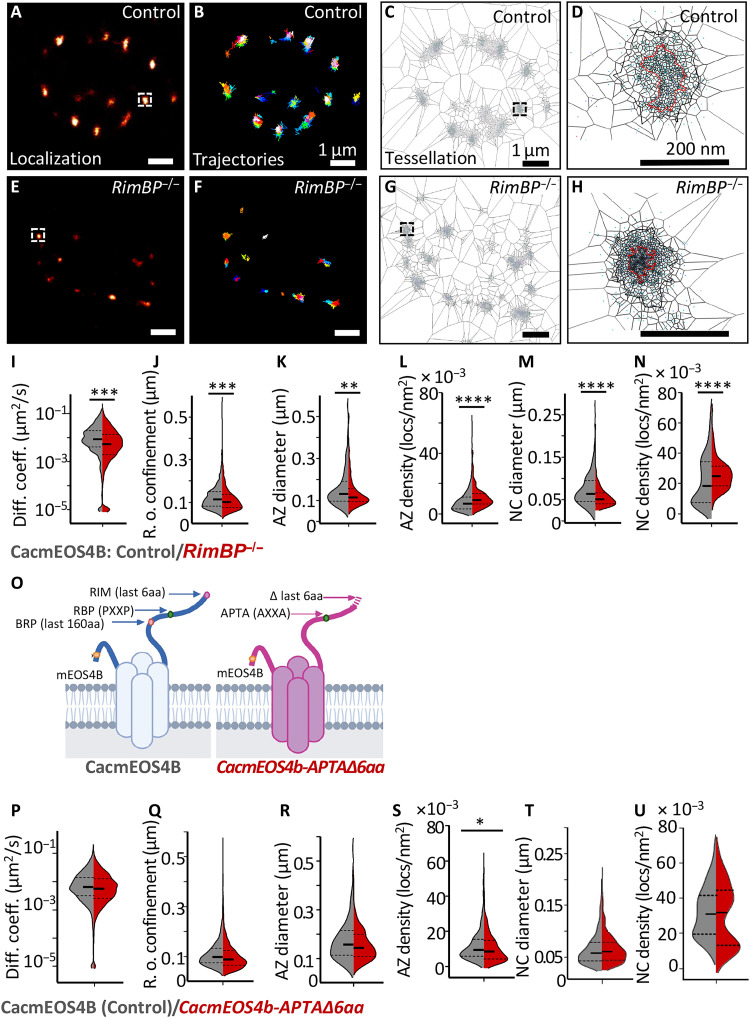
We determined how this VGCC nanoscale plasticity could be molecularly orchestrated and, if so, which AZ scaffold proteins might contribute to this process. Cav2 class voltage–gated VGCCs (including Cac channels) bind to the AZ scaffold via conserved binding motifs within their intracellular C termini and thus form contacts to RIM and RimBP (31–36). These two highly conserved multidomain proteins are responsible for orchestrating SV release and organizing the functional SV release site architectures across evolution (37). To our surprise, however, in null mutants of both proteins, Cac channels showed significantly lower diffusion coefficients compared to controls, with effects being more pronounced for RimBP mutants (Fig. 3I, Control: 0.0071 μm2/s versus RimBP−/−: 0.0037 μm2/s; fig. S5I, Control: 0.0068 μm2/s versus RIM−/−: 0.0044 μm2/s). Thus, neither RimBP nor RIM seems per se essential for the stable AZ anchoring of Cac channels in this system. At the single AZ level, radii of Cac confinement were also significantly reduced (Fig. 3J and fig. S5J). Cac nanocluster sizes were significantly reduced as well, with RimBP again displaying the stronger phenotype (Fig. 3M and fig. S5M). These changes resulted in increased Cac localization densities within their AZs and nanocluster boundaries, again particularly pronounced for RimBP mutants (Fig. 3, L, and N, and fig. S5, L, and N). Similar trends in the distribution of Cac channel localizations within the AZ boundary were observed in the background of RIM−/− (fig. S5, K, and L) and RimBP−/− mutant backgrounds (Fig. 3, K, and L). Thus, instead of increasing Cac motility, as might have been expected, the lack of RimBP and RIM surprisingly provoked a “pre-compaction” of Cac distributions even in the absence of the PhTx application.
Fig. 3. Cac channels at CacmEOS4b;;RimBP−/− null mutant AZs and CacmEOS4b-APTA∆6aa mutants visualized for their live mobility and distribution at AZs.
(A to H) Live sptPALM imaging of CacmEOS4b and CacmEOS4b;;RimBP−/− mutants imaged in 1.5 mM Ca2+ extracellular imaging buffer. Representative boutons displaying PALM images (A and E), trajectory maps (B and F), and representation of tessellation analysis (C, D, G, and H) of individual Cac channels in a bouton of CacmEOS4B (Control or Con) and CacmEOS4B;;RimBP−/− mutant animals. Scale bars, 1 μm (A to C and E to G) and 200 nm (D and H). (I and J) Diffusion coefficient and confinement quantification of live sptPALM data of CacmEOS4B (Con) and CacmEOS4B;;RimBP−/− NMJs, 10 animals (Con) and 9 animals (RBP−/−), N = 28 ROIs (Con) and 26 ROIs (RBP−/−), where 2212 trajectories (Con) and 1753 trajectories (RBP−/−) of individual channels were analyzed. (K to N) Tessellation analysis of the diameter and density of Cac channel localizations within AZs (K and L) and NCs (M and N) from the same dataset, from which 229 (Con) and 264 (RBP−/−) AZs were analyzed. (O) Schematic diagram of on-locus deletion mutant CacmEOS4BAPTA∆6aa animals generated from endogenously tagged CacmEOS4B control flies. (P to U) Live sptPALM imaging analysis of CacmEOS4b (Con) and CacmEOS4B-APTA∆6aa mutants (APTA∆6aa) imaged in 1.5 mM Ca2+ extracellular imaging buffer. Diffusion coefficients (P) and radii of confinement quantifications (Q) are based on 10 (Con) and 6 (APTA∆6aa) animals, N = 28 ROIs (Con) and 17 ROIs (APTA∆6aa), from which 2320 trajectories (Con) and 1890 trajectories (APTA∆6aa) were analyzed. (R to U) Tessellation analysis of the diameter and density of Cac channel localizations within AZ (Q and R) and NC (S and T) cluster boundaries from the same dataset (P to Q), from which 441 (Con) and 276 (APTA∆6aa) AZs were analyzed. PALM data distribution was statistically tested with the Kolmogorov-Smirnov test. Statistical significance is denoted as asterisks: *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Rather than the physical binding of RimBP or RIM to the Cac channels, other mechanistic means might explain these phenotypes, particularly when considering the severe release deficits observed in both RimBP and, to a lesser degree, RIM mutants (31, 32, 36). Thus, to discretely test for the role of the binding sites, we specifically disrupted the conserved RIM and RimBP binding sites of Cac in the background of CacmEOS4b. Concretely, we eliminated, on the locus, the last six amino acids of the Cac channel C terminus to preclude binding to the RIM-PDZ domain and introduced a double point mutation (PPTP to APTA at the second PxxP motif within the Cac C terminus at amino acid positions 1690 to 1693) to disturb the Cac/RimBP interaction (Fig. 3, O to U) (33–35). The on-locus engineered Cac channel mutant (“CacAPTAΔlast6aa”) still displayed nearly normal baseline transmission and normal levels of action potential (AP)–evoked SV release (quantal content) and allowed for both acute (10-min PhTx treatment) and sustained PHP (30-min PhTx treatment) (figs. S6, B to M, and S7, A to L). Following suit, the dynamic localization and distribution parameters of individual Cac channels derived from the live sptPALM data obtained at these mutant AZs were essentially identical to controls (Fig. 3, O to U). In detail, neither the mobility within AZs nor the nanodomain organization was different for the CacAPTAΔlast6aa mutant channels.
These results are consistent with the analysis of RimBP/RIM mutant AZs (Fig. 3 and fig. S5) regarding the notion that the discrete “classical” binding motifs connecting the Cav2 class type VGCCs with these two scaffold proteins are dispensable for AZ tethering and immobilization in this context.
The BRP/ELKS scaffold is essential for stable tethering of VGCCs at AZs
We proceeded by genetically approaching the compaction process from the perspective of the Cac channel. Recent studies demonstrate that the distal C-terminal interaction surface of VGCCs harbors alargenumber of channel-scaffold interactions, some of which were shown to mediate VGCC localization in cultured rodent neurons (38–40). Thus, we generated an on-locus Cac channel C-terminal deletion mutant that removes the C-terminal 160 amino acids of CacmEOS4B (CacΔlast160aa) (Fig. 4 and fig. S6A). To evaluate the functional consequences of this deletion, we performed two-electrode voltage clamp (TEVC) recordings in mutant and control animals. The CacΔlast160aa mutants suffered from a severe reduction of AP-evoked response and, consequently, had significantly lower quantal contents compared to controls (fig. S6). Moreover, the CacΔlast160aa mutants could support neither acute nor sustained PHP (fig. S7).
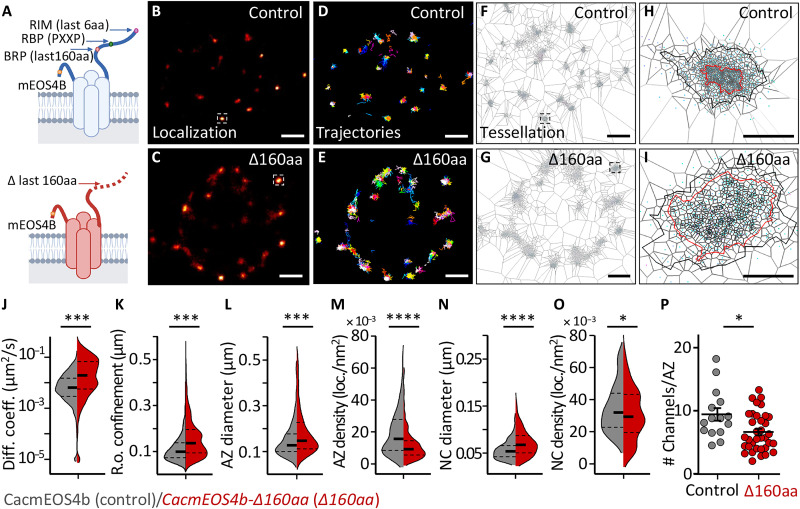
Fig. 4. Live in vivo imaging of Cac channels in CacmEOS4B∆160aa mutants.
(A) Schematic diagram of on-locus deletion mutant CacmEOS4B-∆160aa animals generated from endogenously tagged CacmEOS4B control flies. (B to I) Representative live sptPALM imaging of CacmEOS4B (Control or Con) and CacmEOS4B-∆160aa (∆160aa) mutants imaged in 1.5 mM Ca2+ extracellular imaging buffer displaying the PALM localization (B and C), trajectory map (D and E), and a visualization of the tessellation analysis (F to I) in CacmEOS4B/Control and ∆160aa boutons (F and G) and individual AZs (H and I). Scale bars, 1 μm (A to C and E to G) and 200 nm (D and H). (J and K) Live sptPALM diffusion coefficient (J), the radius of confinement quantification of CacmEOS4B and CacmEOS4B∆160aa NMJs (K), 8 (Con) and 6 (∆160aa) animals, N = 23 ROIs (Con) and 15 ROIs (∆160aa), 2996 trajectories (Con) and 2436 trajectories (∆160aa). (L to O) Tessellation analysis of the diameter and density of Cac channel localizations within AZ (L and M) and NC (N and O) cluster boundaries based on the same data, containing 303 (Con) and 371 (∆160aa) AZs analyzed in n = 3 independent sptPALM ∆160aa experiments that were conducted with concurrent controls. PALM data distribution was statistically tested by the Kolmogorov-Smirnov test. Statistical significance is denoted as asterisks: *P < 0.05; ***P < 0.001; ****P < 0.0001. (P) For comparison, quantification of Cac channel numbers extracted from the same live sptPALM data was further characterized within control conditions [for CacmEOS4b (Con) and CacmEOS4b∆160aa (∆160aa) mutants]. The data were obtained from three to four ROIs out of three to four animals per condition from which 43 AZs (Con) and 49 AZs (PhTx) were analyzed. P < 0.0003 (unpaired t-test). N = 12 to 16 ROIs from four animals per condition from which 15 AZs (Con) and 35 AZs (∆160aa) were analyzed. P < 0.0003 (unpaired t test).
We then performed live sptPALM imaging on CacΔlast160aa mutant NMJs and analyzed Cac dynamics, distribution, and relative channel numbers (Fig. 4, J, K, L to O, and P, respectively). The sptPALM imaging revealed a significantly higher Cac mobility (median diffusion coefficient of 0.0163 μm2/s for CacΔlast160aa compared to 0.0062 μm2/s for controls; Fig. 4J) and a significantly larger radius of confinement (control: 111.5 ± 0.9 nm; CacΔlast160aa: 155.8 ± 1.7 nm; Fig. 4K). Tessellation analysis of Cac localization distribution within the AZ cluster and nanocluster boundaries showed that the CacΔlast160aa mutant channels appear to be dispersed over a larger area, as nanocluster diameters were significantly larger (Fig. 4, L, and N) and the localization densities within AZs and nanoclusters were concomitantly reduced (Fig. 4, M; and O). In addition, bleach curve analysis of CacΔlast160aa PALM data uncovered a significant reduction of the AZ channel number in CacΔlast160aa mutants (Fig. 4P). Collectively, the distal 160 amino acids of the Cac C terminus are critical to effectively tether Cac channels at the AZ presynaptic membrane and to densely pack them into the AZ-central nanocluster. Besides being fewer, the remaining Cac channels at CacΔlast160aa mutant AZs were also atypically mobile (Fig. 4, J, K, and P).
We went on to address the molecular nature of this tethering activity within AZs. BRP is the only representative of the evolutionarily generic Cast/ELKS protein family in Drosophila and the major building block of the NMJ AZ ultrastructural scaffold (also known as the “T-bar”). Combinations of electron and superresolution light microscopy show high Cac densities beneath the AZ center, i.e., the BRP-based T-bar, and the Cac channel levels within AZs are reduced in BRP−/− animals (14, 41–44). Similarly, the reduction of ELKS proteins decreased the presynaptic Ca2+ influx at hippocampal synapses (42) and reduced the clustering of Cav2.1 channels at the Calyx of Held (38).
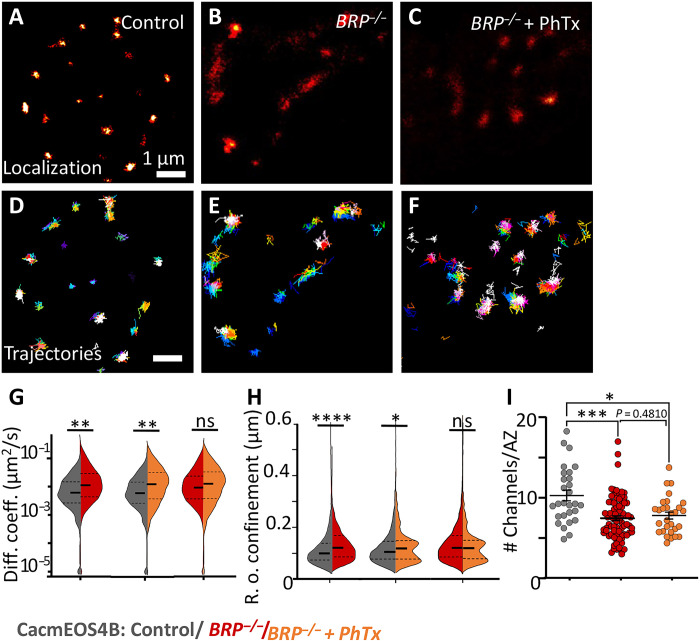
To first test for a principal role of BRP in Cac dynamics, CacmEOS4b channels were imaged at BRP null mutant AZs using sptPALM (Fig. 5). Notably, individual Cac molecules were atypically mobile at BRP null mutant AZs and less confined than at control AZs (Fig. 5, A to H). Furthermore, the mobility of Cac channels and their confinement radius were increased compared to controls, and this increase even persisted after PhTx treatment (median diffusion coefficient: control, 0.0054 μm2/s; BRP−/−, 0.0104 μm2/s; and BRP−/− after PhTx, 0.0104 μm2/s; the radius of confinement: control, 113.9 ± 1 nm SEM; BRP−/−, 136.6 ± 1 nm SEM; and BRP−/− after PhTx, 121.2 ± 2 nm SEM; Fig. 5, G, and H). This demonstrates a pivotal role of BRP in clustering Cac channels into their nanodomain localization within AZs, as previously observed in BRP−/− mutant AZs (41). We would like to mention that we used the same control data for both experimental conditions, BRP−/− mutant and BRP−/− mutant after PhTx, as these conditions were concurrently imaged in one experiment (Fig. 5, A to H).
Fig. 5. Live in vivo imaging of Cac channels in BRP mutant AZs.
(A to F) Live sptPALM imaging of CacmEOS4B (Control or Con) (A and B) and CacmEOS4B;BRP−/− (BRP−/−) mutants in plain HL3 (B and E) or preincubated in 10-min PhTx in HL3 (C and F), imaged in 1.5 mM Ca2+ extracellular imaging buffer and visualized in PALM images (A to C) and trajectory maps (D to F). Scale bars, 1 μm (A to C and E to G) and 200 nm (D and H). (G and H) Live sptPALM diffusion coefficient and confinement quantification of CacmEOS4B, CacmEOS4B;BRP−/−, and PhTx-treated CacmEOS4B;BRP−/− NMJs. N = 17 (Con) and 13 (BRP−/−) or 12 (BRP−/− + PhTx) ROIs; 2192 (Con), 1644 (BRP−/−), and 1001 (BRP−/− + PhTx) trajectories from six animals per condition. PALM data distribution was statistically tested by a Kolmogorov-Smirnov test. Statistical significance is denoted as asterisks: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (I) Quantification of Cac channel numbers extracted from live sptPALM data further characterized within control conditions [for CacmEOS4b (Con) and CacmEOS4b; BRP−/−] and after induction of homeostatic plasticity with a 10-min incubation of the larvae in PhTx in extracellular imaging solution. The data are from 12 to 16 NMJs from three to four animals for each condition from which 72 AZs/ROIs (Con), 80 AZs (BRP−/−), and 26 AZs (BRP−/− + PhTx) were analyzed. P < 0.0003 (unpaired t-test).
We also attempted to use the localization data from the sptPALM recording for the tessellation analysis of Cac in AZs of BRP−/− mutants. Here, however, far fewer AZs and nanoclusters could be retrieved per NMJ terminal area using the standard tessellation parameters (fig. S9). Consequently, we refrained from this type of analysis. Instead, we extracted the average numbers of Cac molecules from BRP−/− mutant AZs from the local mEOS bleaching profiles. Consistent with previous work using a cDNA-derived, GFP-tagged Cac construct (14, 43), the endogenously tagged CacmEOS4B line also displayed reduced Cac numbers in BRP null mutant AZs (Fig. 5I). Moreover, unlike the situation in wild-type NMJs (Fig. 2, I to K), PhTx treatment failed to increase Cac channel numbers in BRP−/− mutant AZs (Fig. 5I). In short, the BRP/ELKS scaffold is a precondition to stably tether Cac channels to NMJ AZs.
In vivo sptPALM identifies a pronounced BRP scaffold compaction upon PhTx treatment
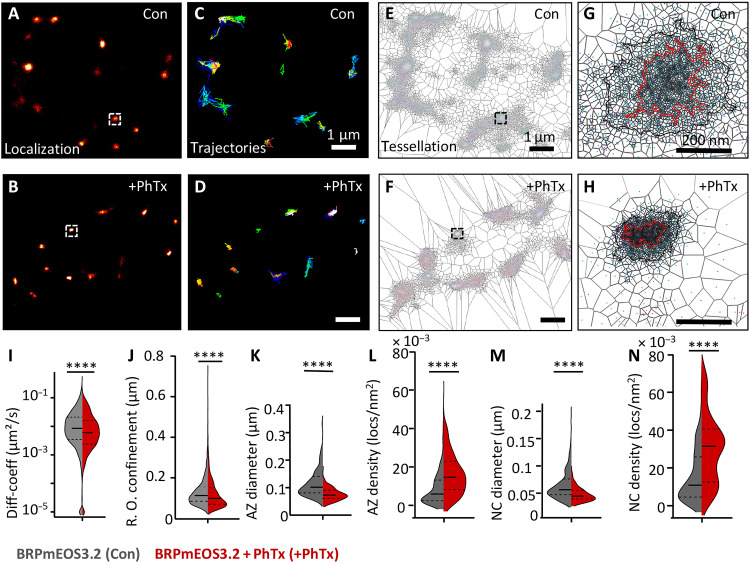
As Cac channel compaction is eliminated in BRP null mutants (Fig. 5), we reasoned that the BRP scaffold might drive the PhTx-triggered Cac compaction (41). To directly investigate this in vivo, we generated on-locus tagged BRP labeled with mEOS3.2 integrated into a site found to preserve functionality and predicted to label both the 190- and 170-kDa BRP isoforms (45). We first confirmed that the mEOS3.2 tag did not interfere with AZ functionality (fig. S10). We then subjected the BRPmEOS3.2 flies to sptPALM imaging under similar conditions as used for CacmEOS4b imaging and analyzed BRP single-molecule dynamics, again in a time window of 20 to 30 min after PhTx treatment (Fig. 6, A to H). After PhTx treatment, individual BRP molecules became significantly less mobile and more confined (control, 0.0087 μm2/s versus PhTx treatment, 0.0049 μm2/s; the radius of confinement: control, 127.5 ± 1 nm SEM versus PhTx treatment, 116.7 ± 1 nm SEM; Fig. 6, I, and J). Tessellation analysis revealed a pronounced compaction of the BRP scaffold upon PhTx application, observed as significantly smaller AZ and nanocluster diameters with significantly higher localization densities (Fig. 6, K to N). Thus, we can show that, in live animals, the BRP scaffold within the NMJ undergoes a strong immobilization and compaction in response to PhTx treatment, which might well drive the nanoscale compaction and immobilization of Cac channels and, consequently, be responsible for the functional potentiation of SV release. We went on testing this idea.
Fig. 6. Live in vivo imaging of BRP at remodeling AZs.
(A to N) Live sptPALM imaging of endogenously tagged BRPmEOS3.2 line containing a mEOS3.2 tag at the stable MiMIC insertion site (brp[MI02987]), visualized at muscle 4 or 6/7 NMJs, performed in 1.5 mM Ca2+ 5 min after a 10-min incubation in HL3 with PhTx or in plain HL3 (Con) and imaging at 30 min after PhTx treatment. Images show representative PALM localizations (A and B), trajectory maps (C and D), and the application of tessellation analysis (E to H). Scale bars, 1 μm (B to G) and 200 nm (H and I). (J and K) Quantification of diffusion coefficients and radius of confinement from live BRPmEOS3.2 sptPALM imaging. Control NMJs: N = 7 images; 1443 trajectories from three animals; PhTx-treated NMJs: N = 9 images; 1761 trajectories from three animals. (K to N) Tessellation analysis from the same sptPALM dataset analyzed the diameters (K and M) and densities (L and N) of BRPmEOS3.2 localizations within AZ (K and L) and NC cluster (M and N) boundaries. Control: N = 26 ROIs, 406 AZs (Con) from 11 animals; PhTx: N = 26 ROIs, 226 AZs from 7 animals included for either condition. n = 2 sets with concurrent controls. Statistical significance is denoted as asterisks: ****P < 0.0001. Data distribution was statistically tested with a Kolmogorov-Smirnov test.
The BRP N-terminal couples VGCC nanoscale plasticity to sustaining potentiation
To further dissect these findings, we functionally analyzed the role of BRP/Cac channel interaction. In mammals, loss of CAST/ELKS reduces CaV2.1-dependent currents and channel abundance (38, 42). Previously, yeast two-hybrid and pull-down assays revealed that the BRP N terminus, which is conserved between ELKS family members, interacts with the Cac C terminus (14). We thus tested whether the distal 160 amino acids of Cac, critical for its AZ immobilization (Fig. 4), would also be critical for binding the BRP N-terminal region using detailed yeast two-hybrid mapping (fig. S8). Binding here was attenuated by deleting the last 60 amino acids and further weakened upon removal of the last 130 amino acids from the Cac channel C terminus (fig. S8). Thus, binding of the AZ membrane proximal BRP N-terminal region to the Cac C-terminal region might mediate the immobilization.
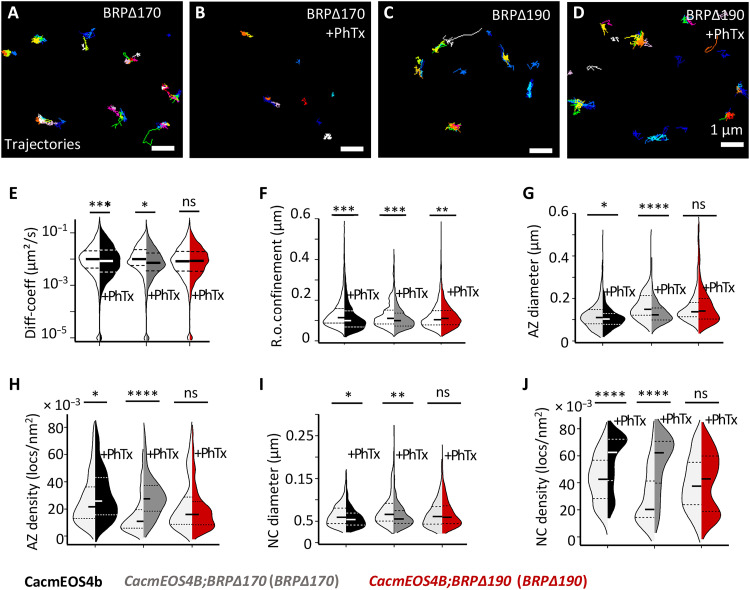
Notably, it is exactly this N-terminal region that distinguishes the longer (190 kDa) from the shorter (170 kDa) BRP isoform, while per se both isoforms are present in about equal abundance to collectively form the “T-bar structure” at NMJ AZs (46). STED imaging previously showed that this BRP N-terminal domain specific to the 190-kDa isoform is in close proximity to the AZ plasma membrane (41). We thus determined whether the 190-kDa isoform might play a specific role in Cac channel nanoscale plasticity and, consequently, potentiation. Notably, in contrast to observations in BRP null mutants, the principal VGCC clustering in the AZs of the BRP∆170 or BRP∆190 isoform mutants was not impaired (46), enabling tessellation analysis (Fig. 7, G to J) and a specific analysis of the plasticity behavior here.
Fig. 7. Live in vivo imaging of Cac channels in BRP∆170 and BRP∆190 mutant in CacmEOS4b background at normal and remodeling AZs.
(A to J) Live sptPALM imaging of CacmEOS4B (Con), CacmEOS4B;BRP∆170/brp69 (BRP∆170) mutants (A and B), and CacmEOS4B;BRP∆190/brp69 (BRP∆190) mutants (C and D) that were preincubated in 10-min PhTx in HL3 (B and D) or plain HL3 (A and C). These were imaged in 1.5 mM Ca2+ extracellular imaging buffer images via sptPALM from which trajectory maps (A to D) and tessellation analysis were calculated. Scale bars, 1 μm (A to D). (E and F) Analysis of live sptPALM diffusion coefficient (E) and radius of confinement (F) quantification of CacmEOS4B, BRP∆190, and BRP∆170 mutants with and without PhTx treatment. (G) N = 25 ROIs, 10 animals, and 4861 trajectories (Con); N = 21 ROIs, 9 animals, and 3428 trajectories (Con + PhTx); N = 9 ROIs, 4 animals, and 1096 trajectories (BRP∆170); N = 22 ROIs, 6 animals, and 3942 trajectories (BRP∆170 + PhTx); N = 22 ROIs, 8 animals, and 3951 trajectories (BRP∆190); and N = 18 ROIs, 7 animals, and 5657 trajectories (BRP∆190 + PhTx). ROIs from n = 2 experiment conducted with concurrent controls. (G to J) Tessellation analysis of the diameter and density of Cac channel localizations within AZ (G and H) and NC (I and J) cluster boundaries from N = 37 ROIs, 10 animals, and 471 AZs (Con); N = 33 ROIs, 9 animals, and 470 AZs (Con + PhTx); N = 25 ROIs, 7 animals, and 120 AZs (BRP∆170); N = 23 ROIs, 6 animals, and 236 AZs (BRP∆170 + PhTx); N = 26 ROIs, 8 animals, and 273 AZs (BRP∆190); and N = 24, 7 animals, and 445 AZs (BRP∆190+ PhTx). n = 2 individual experiments conducted with concurrent controls. PALM data distribution was statistically tested by the Mann-Whitney U test followed by the Kolmogorov-Smirnov test. All PALM data distribution was statistically tested by a Kolmogorov-Smirnov test. Statistical significance is denoted as asterisks: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Upon PhTx treatment of BRP∆170 (BRP∆170/brp69) mutants, Cac channel mobility parameters showed a normal reduction in diffusion coefficients and confinement area, similar to the expected reductions in treated controls compared to untreated controls (Fig. 7, E, and F). In contrast, the BRP∆190 (BRP∆190/brp69) mutants (Fig. 7, E, and F) no longer reduced Cac mobility and even presented a larger confinement area after PhTx treatment (median diffusion coefficients: control, 0.009882 μm2/s; control after PhTx, 0.008384 μm2/s; BRP∆170, 0.01007 μm2/s; BRP∆170 after PhTx, 0.007244 μm2/s; BRP∆190, 0.008174 μm2/s; and BRP∆190 after PhTx, 0.008373 μm2/s; the radius of confinement: control, 116.4 ± 1 nm SEM; control after PhTx, 101.2 ± 1 nm SEM; BRP∆170, 109.5 ± 1 nm SEM; BRP∆170 after PhTx, 99.1 ± 1 nm SEM; BRP∆190, 103.1 ± 1 nm SEM; and BRP∆190 after PhTx, 108.8 ± 1 nm SEM).
In addition, analysis of this same sptPALM data showed that PhTx treatment resulted in the expected provoked reduction of AZ and nanocluster diameter size, in both control and BRP∆170 mutant animals (Fig. 7, G, and I). This was concomitantly followed by increased channel localization density within AZs and nanoclusters after PhTx treatment in control and BRP∆170 mutant conditions (Fig. 7, H, and J). These data show that the normal Cac channel compaction observed during PHP in control animals is maintained in BRP∆170 mutant conditions. In BRP∆190 mutants, however, PhTx treatment neither increased the Cac channel density nor reduced the distribution width of channel localizations, in stark contrast to controls and BRP∆170 mutants (Fig. 7, G to J). This finding shows that the BRP∆190 mutants failed to perform the normal compaction of Cac channels within the presynaptic AZ membrane associated with PHP and allowed us to finally test whether Cac nanoscale plasticity was causally connected to functional potentiation. Here, BRP null mutants still allow for functional PHP induction (measured 10 min after PhTx application) but failed in the sustained expression of PHP at 30 min (13, 30). Thus, we analyzed both BRP isoforms for PHP induction and sustained PHP via current-clamp electrophysiological recordings, comparing flies expressing either a single gene copy of the 190- or 170-kDa isoform to controls.
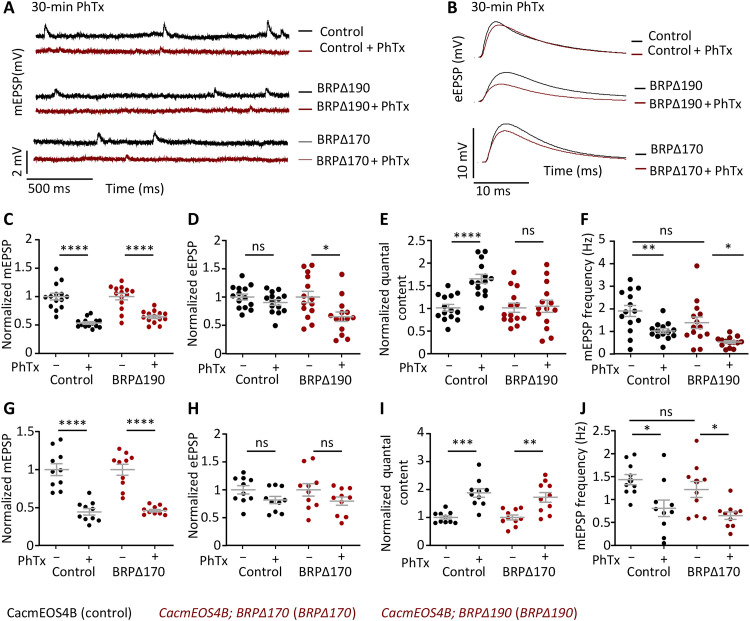
As expected for controls, and similarly observed in the BRP∆170 mutant, PhTx treatment stably reduced postsynaptic sensitivity in response to PhTx treatment, evidenced by a significant miniature excitatory postsynaptic potential (mEPSP) amplitude reduction and an increase in the quantal content, observable even after 30 min of treatment (Fig. 8, G to I). When testing PHP induction (i.e., 10-min PhTx application), all groups showed significant PHP, obvious in their ability to acutely compensate and stabilize evoked excitatory postsynaptic potential (eEPSP) size by increasing the numbers of SVs released per AP (quantal content) (Con, BRP∆170, and BRP∆190 with and without PhTx application; fig. S11). In stark contrast, only the BRP∆190 mutants specifically failed to sustain PHP measured after 30 min of PhTx treatment (Fig. 8, C to E). Thus, the BRP N-terminal region, binding the intracellular C-terminal region of Cac, seems to causally couple Cac nanoscale compaction and immobilization to sustain PHP, suggesting that the BRP scaffold compaction could directly drive nanoscale plasticity to potentiate release.
Fig. 8. Electrophysiological characterization of BRP∆170 and BRP∆190 mutants in CacmEOS4B background at normal or 30-min PhTx-treated remodeling AZs.
(A to I) Electrophysiological recordings of CacmEOS4B (Con), CacmEOS4B;BRP∆170/brp69 (BRP∆170), and CacmEOS4B;BRP∆190/brp69 (BRP∆190) mutants in CacmEOS4B (Con) background. Electrophysiological recordings of Con, BRP∆170, and BRP∆190 animals after 30-min incubation in HL3 or 50 μM PhTx in HL3 solution using current-clamp recordings. Representative traces of mEPSP and eEPSP in Control, BRP∆190, and BRP∆170 (A and B) animal before (− PhTx; black line) and after 30 min of PhTx (+ PhTx; red line) treatment. Scale bars, 10 mV and 10 ms (eEPSP); 2 mV and 500 ms (mEPSP). Quantifications of mEPSP amplitude, eEPSP amplitude, quantal content, and mEPSP frequency in PhTx-treated, untreated Control, and BRP∆190 (C to F) or BRP∆170 (G to J) mutant cells are normalized on the measurement obtained without PhTx for each genotype. Measurements were performed at third-instar larval muscle 6 NMJs of abdominal segments 2 and 3. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Together, this suggests that specifically, the long BRP190 isoform is responsible for the coordinated compaction of BRP with Cac channels into AZ and nanocluster boundaries, likely through interaction at the BRP–N terminus::Cac C terminus interface. Given that the BRP∆190 mutants could not perform the typical PhTx-mediated compaction of Cac channels, the conserved N-terminal domain unique to the BRP190 isoform seems to drive the immobilization and compaction of Cac channels to allow for sustained PHP. This would also directly link the compaction of the BRP scaffold to the Cac channel compaction. In conclusion, the conserved N-terminal BRP/ELKS scaffold regulates the sustained phase of PHP (Fig. 8), is likely responsible for the immobilization of Cac channels close to release sites (Fig. 7), and may thereby promote synaptic release at individual AZs.
DISCUSSION
Given that the tuning of VGCCs is crucially important in defining SV release, a deeper understanding of the exact physiological control of these channels, in terms of their numbers, density, and nanoscale positioning (relative to release sensors), is most critical. VGCCs are confined to specific positions at the presynaptic membrane as a function of the AZ protein scaffold (24, 28, 29, 47). In parallel, recent evidence suggests that AZ scaffolds, associated SV release sites, and VGCC populations can remodel on the minute time scale at not only hippocampal AZs (28, 48, 49) but also Drosophila NMJ AZs (11–13, 16). How these different processes are dynamically integrated has remained largely unknown. Thus, to explore potential connections between AZ scaffold remodeling and VGCC dynamics, we here established in vivo live imaging of individual VGCCs at remodeling AZs over several minutes, exploiting the unique advantages of Drosophila NMJs for in vivo imaging.
A principal finding of our analysis is that the lateral dynamics of individual VGCCs (Cac channels) of the Drosophila NMJ AZs decline in the context of homeostatic potentiation, accompanied by a compaction of the channel distribution within the AZs, which coincides with and most likely directly contributes to the sustained increase in presynaptic SV release. Here, Cac channels became significantly less mobile at AZs during homeostatic remodeling (Fig. 2L). At the same time, their overall confinement area became compacted, evident in a decreased size of both the AZ cluster and AZ-central nanocluster boundaries (Fig. 2, M, N, and P) of their distribution. In addition, the (relative) numbers of VGCCs per AZ, estimated via bleach curves from in vivo sptPALM movies, increased significantly (Fig. 2, I to K). Consequently, the density of individual Cac channels per unit area increased strongly, particularly in the center of the AZ (Fig. 2, N to Q). The Cac channel number increase, compaction, and immobilization during potentiation coincide with and most likely directly contribute to the sustained increase in presynaptic SV release (Fig. 2, I to K).
Notably, constitutive GluRIIA mutant larvae have been extensively used to study long-term plasticity at NMJ AZs (13, 50, 51). Similar to the PhTx treatment, sptPALM analysis of Cac in the GluRIIA mutant background (fig. S12, A and B) showed a Cac channel compaction and density increase for the AZ-central nanocluster (fig. S12, G, and H). At the same time, Cac mobility was unchanged in GluRIIA mutants compared to controls (fig. S12C). Thus, while Cac density increases the VGCC compaction, which seems to contribute to both sustained PHP and chronic GluRIIA mutant plasticity scenarios, the chronic GluRIIA mutants might have already consolidated the dynamic phase of Cac channel redistribution. Our study identifies BRP/ELKS as the critical scaffold protein mediating these changes in Cac channel number, mobility, and compaction, thereby promoting sustained and stable functional potentiation (for a model, see Fig. 9). We conclude this as we can show that (i) BRP is essential for Cac channel single-molecule immobilization and compaction under PhTx (Fig. 5); (ii) the distal Cac channel C terminus (which includes the BRP/ELKS binding region) is essential for the immobilization and compaction of Cac channels at NMJ AZs (Fig. 4) (13, 30); (iii) the BRP AZ scaffold undergoes PhTx-triggered compaction and becomes immobile at remodeling NMJ AZs, as evidenced by live sptPALM (Fig. 6), a result consistent with previous dSTORM analysis of fixed NMJs (16); and (iv) the BRP190 isoform, which contains the unique N-terminal region that binds to the Cac C terminus, is specifically needed for tuning Cac channel mobility and compaction as well as functional potentiation (sustained PHP) under PhTx-triggered remodeling (Figs. 7 and 8). Notably, the 170-kDa BRP isoform lacking this N-terminal region, but present at AZs in similar amounts as the 190-kDa isoform, is dispensable for Cac compaction and sustained PHP.
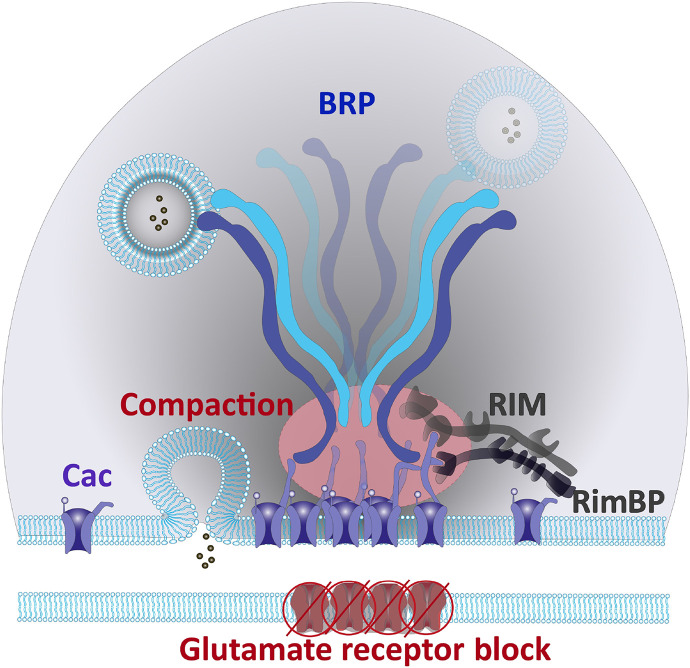
Fig. 9. A schematic representation of NMJ AZ scaffold after 20 to 30 min of PHP.
Here, the N-terminal BRP and C-terminal Cac Ca2+ channel interaction surface sequesters Cac channels into the AZ center and promotes a coordinated compaction of the BRP scaffold and Cac Ca2+ channel distribution, during sustained PHP.
As mentioned above, during PHP, Ca2+ influx is increased, and previous confocal microscopic analyses have shown Cac channel levels to increase during PHP already after 10 min of PhTx treatment, while here we subject the Cac channels to even longer PHP sustained at time scales of 20 to 30 min (Figs. 2 to 5 and 7 and figs. S5 and S12) (10). Biophysical analysis has suggested that SV release dominantly operates at a distance less than 100 nm from the AZ center, i.e., the nanocluster center at Drosophila NMJ AZs (4, 6). It thus appears most plausible that the increased Ca2+ channel density accounts for the PhTx-triggered increase of Ca2+ influx and, consequently, directly contributes to PHP expression (8, 11). An increase in the total number of channels has been proposed as a first-order mechanism (8, 11), assuming that VGCCs will behave independently with respect to their voltage-dependent activation. However, it has also been shown that VGCCs can act cooperatively, depending on their proximity to each other and on their activation history (52, 53). It is thus conceivable that the AZ and nanocluster compaction of VGCCs within the AZ serves to promote channel-scaffold interactions in favor of more reliable Ca2+ signaling, particularly within the identified nanocluster of each AZ. The condensation of Cac channels into the central nanocluster of an AZ supports the idea that beyond the increased channel number, an increase in channel density entails a cooperative modulation of VGCC activity, wherein immobile and likely active Cac channels clustered in close proximity to release sites deliver an impact on the increase of SV release required for PHP. The compaction of Cac channel distribution may also add to the isolation of Ca2+ influx dedicated to SV release from peri-AZ Ca2+ influx related to SV endocytosis (54). Because of the supra-linear dependence of the SV release on intracellular Ca2+ concentration as shown for many synapses, even the nanoscale change in Cac channel organization could already be very effective and work as fast feedback for acute activity changes. Per se, several mechanisms could be envisioned to support and maintain the compaction of the central Cac channel cluster over time, for example, differences in local molecular crowding within the membrane prolonging the dwell time of channels in particular membrane domains. In addition, specific interactions with particular intracellular scaffold proteins (14, 31–34, 36, 41, 42) or through interactions with scaffold protein complexes by liquid-liquid phase separation (LLPS) are per se possible (see below) (55–58). Given our results, we do favor the idea that the compaction of the BRP/ELKS scaffold increases the local density of Cac channels via the BRP–N terminus::Cac C terminus interaction surface. We show that the BRP scaffold undergoes a compaction under sustained PhTx treatment (Fig. 6), concurring with recent observations using dSTORM analysis of a C-terminal BRP antibody epitope in fixed samples subjected to PhTx for 10 min (16). It thus appears that BRP scaffold compaction extends over the plasticity sequence. This scaffold compaction might, during the early induction phase, not involve increases in AZ scaffold proteins, as reported by Mrestani et al. (16), while at a later phase of the plasticity sequence, Cac channel numbers increase (Figs. 2 and 6). It cannot, however, in our eyes, be excluded that crowding effects from using antibody complexes might have confounded the detection of AZ molecule increases in this previous study (16).
A rather surprising result of our analysis is that the absence of RimBP and RIM scaffold proteins rendered the AZs into a “pre-compacted” state of high Cac channel density and immobility irrespective of PhTx treatment (Fig. 3 and fig. S5). Obviously, these mutant AZs operate under conditions of chronically inefficient evoked SV release (31–34, 36), a scenario converging with PhTx treatment in terms of reduced evoked postsynaptic glutamate receptor currents. Thus, loss of RimBP and RIM might trigger a compensation that might well bear mechanistic similarities to PhTx treatment. Notably, RIM and RimBP mutants are also defective for immediate release potentiation upon PhTx treatment (31–34, 36), which likely operates by tuning existing release sites. Thus, release deficits resulting from improper setup of the release machinery might directly trigger VGCC compaction, and particularly, RimBP mutant AZs might be at their limit concerning their ability to use this mechanistic module. This result is now to be kept in mind when interpreting mutants interfering with PHP expression while also suffering from baseline-evoked release deficits.
Our analysis consistently showed that both the Cac channel C-terminal interaction motifs of RIM and RimBP were dispensable for PhTx-triggered Cac channel immobilization, measured via live sptPALM imaging (Fig. 3 and fig. S5). In contrast, strong synaptic transmission defects were observed in C-terminal Cac channel mutant Cac-Δlast160aa mutant, which potentially derives its impact from the BRP-Cac interaction being disrupted along with disruption of RIM and RimBP binding sites (Figs. 3 and 4 and figs. S5 to S7). Here, the importance of the tight control of Cac localization via the Cac C-terminal interaction surface is also made evident, as the AZs of Cac-Δlast160aa mutant could neither compact Cac channels correctly nor achieve a normal distribution of Cac channels (Fig. 4). Improper Cac localization may thus cause increases in average distances between Cac channels and release sites, which would lead to lowered release probabilities for SVs, which, in turn, would explain the facilitation observed in the paired-pulse ratio and the increased rise time of the evoked excitatory postsynaptic currents (eEPSCs) (fig. S6). Another possibility is that the Cac-Δlast160aa mutation, while located in the cytosolic part of the channel and not within the pore region, could still affect the gating characteristics of the channel, which might also explain the observed changes in the eEPSC kinetics.
BRP mutant AZs also suffer from reduced evoked release levels (14, 41, 44), although not as severely as the release deficits reported in RimBP mutants (33), but did not form pre-compacted Cac channel clusters as is the case in RIM and RimBP mutant AZs (Fig. 3 and fig. S5). In contrast, in the BRP mutant AZs, Cac channels were completely unable to either form normal high-density nanoclusters and maintain the normal distribution of Cac channels or trigger immobilization of Cac channels within AZs after PhTx treatment (Fig. 5). Here, the ELKS homolog BRP is essential, serving as an AZ anchor and immobilizing the Cac channels. Given that BRP is essential for stable PHP expression and maintenance of PHP (but not PHP induction) (13), our results further suggest that Cac channels need to be stably anchored to maintain PHP or at least to deliver their full impact in triggering SV release. The BRP/ELKS scaffold protein might thus be an important conserved AZ scaffold interaction surface that sequesters Cac channels into their proper nanoscale distribution within the AZ and thus acts as the master scaffold responsible for Cac compaction.
How then might BRP mediate VGCC compaction? We show that in live NMJs, the BRP AZ scaffold also undergoes a compaction-like process in response to PhTx (Fig. 6), concurring with previous observations from dSTORM analysis of the C-terminal BRP antibody epitope (16). In addition, we find that in live AZs, the mobility of BRP molecules is also significantly reduced, with its spatial compaction occurring during the sustained phase of PHP (Fig. 6). The biophysical nature of the BRP scaffold and its ability to undergo compaction, like Cac channels, might contribute to a consequent connection of BRP to Cac channels, which, in turn, likely drives the distribution and compaction of Cac channels during PHP. The BRP scaffold has recently been shown to regulate the later sustained phase of PHP, rather than its early induction phase (30). A detailed look into the BRP regulation of PHP reveals that the BRP190 isoform, which contains the unique N-terminal region that forms a conserved interaction surface with Cac channel C terminus, likely acts as the regulator of the sustained phase of PHP through alterations in the Cac channel mobility and compaction during PHP (Figs. 7 and 8). Thus, the BRP190 isoform N-terminal and the Cac channel C-terminal regions seemingly form critical interaction surfaces, through which a coordination of the compaction of both scaffolds during homeostatic plasticity might be regulated.
LLPS-like processes are, in our eyes, prime candidates for the regulation of the interaction between Cac channels and the BRP/ELKS scaffold. Notable in this regard, ELKS homologs and the VGCC intracellular C-terminus were shown in vitro to form LLPS-like condensates (55–58). This implicates an additional role for the interaction surface between the N-terminal ELKS/BRP190 isoform and the Cac channel C terminus to promote a common LLPS phase condensate, which might be part of the remodeling process of AZs under PHP. Notable in terms of behavioral relevance, the two BRP isoforms, 170 and 190, are differentially regulated in different populations of synapses, and the 190 isoform was specifically found to contribute to synaptic circadian rhythm plasticity (59). Given the apparently collective nature of this previously unidentified form of AZ plasticity, relating its mechanisms to other state switches, particularly LLPS-like processes, is a subject warranting future research.
MATERIALS AND METHODS
Experimental model and subject details
Fly husbandry, stocks, and handling
Fly strains and crosses were reared under standard laboratory conditions and raised at 25°C on semi-defined medium (Bloomington recipe). Male third-instar larvae were used for experiments. RimBP, RIM, and BRP null or isoform-specific mutants are specified below and were used upon recombination with CacsfGFP or CacmEOS4b on an otherwise wild-type background: RimBPNull, RimBPStop1/RimBPS2.01; BRPNull, brpΔ6.1/brp69; RIMNull, rimex1.103/Df(3R)ED5785; BRP∆170 mutant, CacsfGFP or CacmEOS4b; BRPΔ170/brp69; BRP∆190 mutant, CacsfGFP or mEOS4b; and BRPΔ190/brp69, GluRIIAnull; GluRIIAsp16.
Stocks were provided by the Bloomington Drosophila Stock Center (BDSC) and previously described as follows: RimBPStop1, RimBPS2.01 6; rimex1.103 (32), brpΔ6.1 (14), brp69 (41), BRPΔ190 and BRPΔ790 (46), GluRIIAsp16 (BDSC stock: 64202), and SynapGCaMP6f (MHC-CD8-GCaMP6f-Sh) (22) lines were provided.
Drosophila genetics and genome engineering
On-locus tagging and mutagenesis of Cac
The endogenously tagged fly lines CacsfGFP and CacmEOS4b were generated in the O’Connor Giles laboratory as previously described (11). Briefly, N-terminally tagged Cac was generated using a scarless CRISPR/piggyBac-based approach (https://flycrispr.org/).
The following transgenic animals were generated by Well Genetics Inc. (Taipei City, Taiwan), using modified methods of Kondo and Ueda (60). The following lines were generated using the CRISPR-Cas9–mediated genome editing approach by homology-dependent repair using one guide RNA (gRNA) and a double-stranded DNA plasmid donor.
CacmEOS4bΔlast160AA
We generated a deletion allele of the Cac gene by truncating its distal C-terminal sequence of the last 160 amino acids. Briefly, the gRNA sequences GACGGTTTGCTGGGAGTCGG[AGG] (same gRNA used for CacmEOAS4b-APTA) and AGGAGGATTGGTGCTAGCAA[AGG] were cloned into U6 promoter plasmids. Cassette PBacDsRed containing two PBac terminals, 3xP3 DsRed, and two homology arms, cloned into pUC57 Kan was used as a donor template for repair. Cac/CG43368 targeting gRNAs and hs-Cas9 were supplied in DNA plasmids, together with a donor plasmid for microinjection into embryos of control strain CacmEOS4b/FM7a. F1 flies carrying the selection marker of 3xP3 DsRed were further validated by genomic polymerase chain reaction (PCR) and sequencing, and CRISPR-driven insertion of the PBacDsRed cassette generated a 734–base pair (bp) deletion, resulting in a 160–amino acid C-terminal truncation of CacmEOS4B. Last, the PBacDsRed used for screening was eventually removed by Flippase Recognition Target (FRT)-mediated excision to generate the final CacmEOS4BΔlast160aa fly line.
CacmEOS4bAPTAΔlast6AA
Briefly, the gRNA sequence AGGAGGATTGGTGCTAGCAA[AGG] was cloned into U6 promoter plasmids. The cassette PBacDsRed containing two PBac terminals, 3xP3 DsRed, and two homology arms, cloned into pUC5 7 Kan was used as a donor template for repair. Cac/CG43368 targeting gRNAs and hs-Cas9 were supplied in DNA plasmids, together with donor plasmid for microinjection into embryos of control strain CacmEOS4B-APTA CRISPR/FM7a. F1 flies carrying the selection marker 3xP3 DsRed were further validated by genomic PCR and sequencing. The resulting 21-bp deletion truncates CacmEOS4B-APTA for the last six amino acids (DEEDWC). Last, the PBacDsRed used for screening was eventually removed by FRT-mediated excision to generate the final CacmEOS4B-APTAΔlast6aa fly line.
On-locus tagging of brp
The endogenously tagged BRPmEOS3.2 fly was generated at WellGenetics Inc. using PhiC31-mediated recombination-mediated cassette exchange to insert a mEOS3.2 exon cassette (pBS-KS-attB1-2-PT-SA-mEos3.2-SD-0) into the stable MiMIC insertion site of BRP[MI02987] that is positioned between exons 6 and 7 of the BRP-RI transcript isoform and thereby tags all nine annotated BRP isoforms including the long and short BRP190 and BRP170 isoforms (45, 46). The mEos3.2-C1 sequence was obtained from Addgene plasmid #54550. Because of the high density of the BRP molecules in the AZ and the sptPALM imaging requirement to get to individual blinking events of BRPmEOS3.2, we expressed only one copy of BRPmEOS3.2 in the wandering third-instar male larvae that were used in experiments in Fig. 6 and fig. S10. In addition, we tested and confirmed that the mEOS3.2 tag did not interfere with the functionality of these AZs in fig. S10.
Yeast two-hybrid assay
In principle, all experiments were conducted according to the yeast two-hybrid protocols of Clontech using the strain AH109 with minor adjustments meeting the requirements of the interaction-domain mapping experiments. Cotransformation of the yeast strain AH109 with both prey and bait constructs (Clontech) was conducted using the following protocol: The yeast was plated on a YPDA (yeast, peptone, dextrose, adenine) medium agar plate and cultured for 2 days at 30°C. An appropriate amount of YPDA medium was inoculated with a single clone and cultured at 30°C overnight while shaking at approximately 200 rpm. After dilution to an optical density at 600 nm (OD600) of 0.2 (10-ml culture volume per transformation), the culture was grown until OD600 reached 0.6. The cells were pelleted, washed in 1× TE (tris-EDTA) medium at pH 8.0, re-pelleted, and resuspended in TE/LiAc (lithium acetate; 100 μl per 10-ml culture volume). Resuspended yeast (100 μl) was mixed with the transformation mix, which was composed as follows, 500 ng of each of the two plasmids and 10 μl (= 100 μg) of Herring Testes carrier DNA (Clontech). After adding 600 μl of polyethylene glycol/LiAc, the mix was vortexed and left to incubate at 30°C for 30 min in a shaker. Then, 70 μl of dimethyl sulfoxide (10% final concentration) was added, and the cells were heat-shocked for 15 min at 42°C. After chilling on ice, the yeast was carefully sedimented, the pellet was resuspended in 100 μl of 1× TE medium (pH 8.0), and the transformation was plated on minimal SD (synthetic defined)/-Leu/-Trp medium plates to ensure the presence of both plasmids. After growing for 2 to 3 days, at least 10 clones each were analyzed on SD/-Ade/-His/-Leu/-Trp/X-α-gal medium plates to select for positive interaction. If >70% of the clones plated on SD/-Ade/-His/-Leu/-Trp/X-α-gal medium plates grew, this was regarded as weak positive interaction (+), >80% as intermediate interaction (++), and >90% and blue clones as strong interaction (+++). Negative controls consisted of both laminin as bait with the prey to be tested and the corresponding bait with the empty prey vector. In the positive control, pGBKT7 p53 was transformed with pGADT7 containing the SV40 large T antigen.
The BRP D1 and Cac fragments depicted in fig. S8 were cloned into pGADT7 using the following primer pairs: Cac C-term_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac C-term_Rev, 5′-GTCTCTCGAGTTATCAGCACCAATCCTCCTCATC-3′; Cac∆304 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆304 aa_Rev, 5′-CAAGCTCGAGTTATCACAAGCTAGGCGATCGCCGATG-3′; Cac∆130 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆130 aa_Rev, 5′-GTCTCTCGAGTTATCAGTGTGTATGTGTGGGACTGG-3′; Cac∆81 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆81 aa_Rev, 5′-GTCTCTCGAGTTATCACAGGCGATCGCGATCCCTAT-3′; Cac∆60 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆60 aa_Rev, 5′-CAAGCTCGAGTTATCATTCGCGTTCGTACAACTCTCGT-3′; Cac∆31 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆31 aa_Rev, 5′-GTCTCTCGAGTTATCAACCTGTTCGGCCCATAGCCA-3′; BRP D1_Fwd, 5′-GTCTATCGGAATTCATGGGCAGTCCATACTAC-3′; and BRP D1_Rev, 5′-GGAATTCATCGATGAGCTCGTCCTCTAGGTAC-3′. The BRP D1 and Cac fragments depicted in fig. S8 were cloned into pGBKT7 using the following primer pairs: Cac C-term_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac C-term_Rev, 5′-GTCTGTCGACTTATCAGCACCAATCCTCCTCATCCGAA-3′; Cac∆304 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆304 aa_Rev, 5′-GTCTGTCGACCAAGCTAGGCGATCGCCGATG-3′; Cac∆130 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆130 aa_Rev, 5′-GTCTGGATCCTTATCAGTGTGTATGTGTGGGACTGG-3′; Cac∆81 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆60 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆60 aa_Rev, 5′-GTCTGTCGACTTATCATTCGCGTTCGTACAACTCTCGT-3′; Cac∆31 aa_Fwd, 5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGAG-3′; Cac∆31 aa_Rev, 5′-GTCTGGATCCTTATCAACCTGTTCGGCCCATAGCCA-3′; BRP D1_Fwd, 5′-GTCTATCGGAATTCATGGGCAGTCCATACTAC-3′; and BRP D1_Rev, 5′-GGAATTCGTCGACGAGCTCGTCCTCTAGGTAC-3′.
Immunofluorescence histochemistry of larval NMJs
Dissections were performed following standard protocols (61) and are described here: Third-instar larvae were opened in HL3 by opening dorsally along the midline and removing the innards. Filets were fixated with ice-cold methanol (MeOH) for 10 min for all experiments to quench the autofluorescence of sfGFP/mEOS4b to subsequently boost them with an antibody or nanobody booster, after testing all antibodies function optimally in MeOH. After fixation, the filets were washed with phosphate-buffered saline (PBS) plus 0.05% Triton X-100 and blocked for 60 min in 5% normal goat serum. The larvae were incubated with primary antibodies at 4°C overnight and subsequently washed in a PBS plus 0.05% Triton X-100 solution for 2 hours at room temperature for immunostaining. Larvae were then incubated for 4 hours with secondary antibodies at room temperature followed by the same washing procedures. Immunocytochemistry was done the same way for both conventional confocal and STED microscopy except for the mounting medium. Larvae were finally mounted in either Mowiol (Sigma-Aldrich) for confocal microscopy or ProLong Gold (Thermo Fisher Scientific) for STED. The following antibodies were used at the indicated concentrations: mouse anti-Bruchpilot (Brp) Nc82/BRPC-term (Developmental Studies Hybridoma Bank, catalog no. nc82; RRID: AB_2314866) and BRPN-term (1:500) (62), rabbit RIMBPC-term (1:500) (63), mEOS4B nanobody coupled to Atto 580 or GFP nanobody coupled to STAR635P at 1:300 (NanoTag, catalog no. N0304 or N3102), rabbit or guinea pig anti-GluRIIA (generated in house), anti-mEOS4b nanobody coupled to Atto 647 (1:300; NanoTag, catalog no. 31200501), and anti-HRP conjugated to Alexa Fluor 405 at 1:200 to 1:500 (Jackson ImmunoResearch). Species-specific ATTO490LS (Hypermol), Alexa Fluor 488, and Alexa Fluor 594 secondary antibodies (Invitrogen, Jackson ImmunoResearch) were used at 1:300.
Short acute PHP induction and longer PHP sustained phase via PhTx assay
Third-instar control and mutant larvae in CacsfGFP/CacmEOS4b backgrounds were treated with a 10-min PHP assay using Philantotoxin-433 drug (PhTx; AOBIOUS). Larvae were carefully pinned down at mouth and tail while avoiding any stretching of body wall muscles and minimally dissected in 50 μl of the solution containing 50 μM PhTx in Ca2+- and Mg2+-free HL3 or No PhTx-HL3. Animal’s hemolymph was mixed with the treatment solution by regularly mixing the solutions during the 10 minute treatment window undertaken with care to not stretch or pull the larvae muscles. For live-sptPALM assays, treated animals were moved into the imaging solution containing HL3 with 4 mM Mg2+ (to reduce muscle contractions) and basal 1.5 mM Ca2+. For Figs. 1 to 7 and figs. S5 to S12, the induction phase of PHP was generated with a 10-min PhTx drug application and the later sustained phase of PHP was targeted with a 30-min PhTx drug application. We would like to emphasize that the 30-min PhTx drug treatment also undergoes an induction phase during the first 10 min of this treatment and that the AZs undergoing 30 min of sustained PHP have undergone first an acute induction of the PHP.
Confocal microscopy and analysis
Figure S7 (M and N) was imaged using confocal microscopy via a Leica TCS SP8 inverted confocal microscope (Leica DMI6000, Leica Microsystems, Germany). All L3-stage Drosophila NMJ larval images were taken at room temperature using LCS AF software (Leica Microsystems, Germany). Imaging was performed using a 63× 1.4 NA (numerical aperture) oil immersion objective. Images Muscle 4 Type 1b NMJs were imaged from ice-cold MeOH fixed larval preparations. Images were acquired in line scanning mode with a pixel size of 75.16 nm × 75.16 nm and a Z-step of 0.25 μm. Representative images of NMJ AZs were processed using the ImageJ/Fiji software (1.52P; https://imagej.net/software/fiji/) for adjusting brightness with the brightness/contrast function. All images are shown in fig. S7 (M and N) were processed with the same parameters. Analysis of Cac and BRP intensities was calculated via the FIJI/Image J Analyze Particles function, and the intensity values were normalized to the untreated conditions. In fig. S7 (M and N),10 animals per condition yielding 19 to 20 images from three repeated sets with concurrent controls were used and statistically tested with a one-way analysis of variance (ANOVA) with Dunn’s multiple comparisons tests.
STED imaging and analysis
gSTED microscopy was performed using an Abberior Instruments Expert Line STED setup equipped with an inverted IX83 microscope (Olympus), two pulsed STED lasers for depletion at 775 nm (0.98-ns pulse duration, up to 80-MHz repetition rate), and at 595 nm (0.52-ns pulse duration, 40-MHz repetition rate), and pulsed excitation lasers (at 488, 561, and 640 nm).
Multichannel 2D confocal and gSTED images were acquired with a 100× oil-immersion objective lens (NA = 1.4; UPLSAPO100XO, Olympus), with a pixel dwell time of 2 μs, with 10× and 30× line accumulation, respectively, at 16-bit sampling and a field of view of 10 μm × 10 μm. The lateral pixel size was set to 20 nm. The dyes STAR 635P, Alexa Fluor 594, and ATTO490LS were depleted at 775 nm. Alexa Fluor 488 was depleted at 595 nm. Time gating was set at 750 ps with a width of 8 ns. Fluorescence signals were detected sequentially by line by avalanche photodiode detectors at appropriate spectral regions (STAR 635P and ATTO490LS, 680 to 765 nm; Alexa Fluor 594, 584 to 630 nm; and Alexa Fluor 488, 500 to 551 nm). Alexa Fluor 488 confocal and gSTED images were acquired following the acquisition of the other channels. These procedures were operated by the software Imspector (version 16.3.13367, Abberior Instruments, Germany). Raw gSTED images were processed for Richardson-Lucy deconvolution with default settings using the Imspector software (version 16.3.13367, Abberior Instruments, Germany). The point spread function was automatically computed with a 2D Lorentzian function having a full width at half maximum of 40 nm based on measurements with 40-nm crimson beads. To analyze Cac signal sizes, we first filtered out the top and side-view AZs, defined manually by the planar view of RimBP signals that forms a ring around a central Cac signal. We imaged 651 AZs obtained from two independent experiments, five animals, and 24 images from which we analyzed 441 planar AZs.
gSTED analysis to determine CacsfGFP signal diameter size (Feret’s diameter) from these images was performed by particle segmentation above the threshold signal with the function “Find maxima” in ImageJ [ImageJ-1.52g, National Institutes of Health (NIH)].
Electrophysiology
TEVC and single-electrode (“current-clamp”) recordings were performed at room temperature on muscle 6 of third-instar larval NMJs in the abdominal segments A2 and A3. Male third-instar larvae were dissected in modified Ca2+-free hemolymph-like saline [HL3; 70 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 20 mM (TEVC) or 10 mM (current clamp) MgCl2, 115 mM sucrose, 5 mM trehalose, and 5 mM Hepes]. For PhTx experiments, larvae were cut open along the midline and incubated for the indicated time (10 or 30 min) in HL3 containing either 50 μM PhTx or the equivalent volume of water for controls. The incubation solution was gently perfused into the preparation using a pipette at the start of the incubation and once more at the half-time point (after 5 or 15 min). The preparation was then finished during the last 2 min of incubation time and afterward washed three times with HL3 before being transferred to a bath solution for electrophysiological recordings. Recordings were obtained with a bath solution of HL3 with 1.5 mM (TEVC) or 0.4 mM (current clamp) CaCl2. Recordings were made from cells with an initial Vm between −50 mV (TEVC) or −40 mV (current clamp) and −80 mV, and input resistances of ≥4 megohms, using intracellular electrodes with resistances of 30 to 50 megohms, filled with 3 M KCl. Two cells were recorded per animal. Glass electrodes were pulled using a Flaming Brown Model P-97 micropipette puller (Sutter Instrument, CA, USA). Recordings were made using an Axoclamp 2 B amplifier with HS-2A x0.1 head stage (Molecular Devices, CA, USA) on a BX51WI Olympus microscope with a 40× LUMPlanFL/IR water immersion objective (Olympus Corporation, Shinjuku, Tokyo, Japan). Miniature excitatory junctional currents (mEJCs)/mEPSPs were recorded for 90 s with the voltage clamped at −80 mV (for TEVC recordings); all other recordings were performed while clamping the voltage at −60 mV (for TEVC recordings). eEJCs/eEPSPs were recorded after stimulating the appropriate motor neuron bundle with 5 V (eEJCs) or 8 V (eEPSPs) and 300 μs at 0.2 Hz using the S48 Stimulator (Grass Instruments, Astro-Med Inc., RI, USA). Signals were digitized at 10 kHz using an Axon Digidata 1322 A digitizer (Molecular Devices, CA, USA) and low pass–filtered at 1 kHz using an LPBF-48DG output filter (NPI Electronic, Tamm, Germany). The recordings were analyzed with pCLAMP 10 (Molecular Devices, Sunnyvale, CA, USA), GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA), and two Python scripts using the pyABF package for Python 3.10 [S. W. Harden. pyABF 2.3.5 (2022); available at https://pypi.org/project/pyabf]. Stimulation artifacts of eEJCs/eEPSPs were removed for clarity. mEJCs/mEPSPs were further filtered with a 500 Hz Gaussian low-pass filter. Using a single template for all cells, mEJCs/mEPSPs were identified and averaged, generating a mean mEJC/mEPSP trace for each cell. An average trace was generated from 20 eEJC/eEPSP traces per cell for 0.2 Hz stimulation and 10-ms interstimulus interval (ISI) paired-pulse recordings and from 10 traces for 30-ms ISI paired-pulse recordings. Rise time was calculated from the average trace of the 0.2-Hz stimulation recording as the time from 10 to 90% of the total amplitude before the peak. Decay constant τ was calculated by fitting a first-order decay function to the region of the average trace of the 0.2 Hz stimulation recording from 60 to 5% of the total amplitude after the peak. The amplitude of the average eEJC/eEPSP trace from the 0.2 Hz stimulation recording was divided by the amplitude of the averaged mEJC/mEPSP, for each respective cell, to determine the quantal content. 10 and 30 ms ISI paired-pulse ratios were calculated by dividing the amplitude after the second pulse by the amplitude after the first pulse. The baseline for the second amplitude was set at the last point before the second stimulation artifact onset.
Single-particle tracking PALM
All live sptPALM experiments were conducted on male third-instar larvae. Larval body walls designated for single-particle tracking were prepared according to the previous studies (21, 64), and imaging experiments were performed using an inverted TIRF setup. The recording temperature was set to 25°C. The microscope (Nikon Eclipse Ti) was equipped with a 100× NA 1.49 Apo TIRF oil objective (Nikon). Up to 10,000 images were captured using an electron-multiplying charge-coupled device camera (iXon+ 897, Andor Technology) controlled by NIS-Elements (Nikon) at a frame rate of 20 Hz. CacmEOS4b and BRPmEOS3.2 containing larvae were imaged using the live HRP-488 staining as an indicator for the Z-plane. We used a 1.6 magnification lens to reduce the pixel size to 107 nm × 107 nm. The N-terminally expressed mEOS4b on the Cac channel was excited by continuous illumination with 405/561-nm light using 2 to 5% of the initial laser power of the 405-nm laser line (100 mW) and 25 to 40% of the 561-nm laser line (100 mW) for up to 10,000 frames. For the BRPmEOS3.2 fly line, an additional pre-bleach step was required to gain individual blinking events because of the high density of the BRP molecules in the AZ. Here, 5000 frames at a 50 ms frame rate of BRPmEOS3.2 were prebleached with a 100% 561-nm laser line (100 mW) to reach an individual blinking phase.Subsequently, a 10,000-frame recording with 10% 405 and 35% 561 laser powers was used to image individually blinking BRPmEOS3.2 molecules within individual AZs of a few boutons which are available for stochastic conversion in the best oblique-TIRF Z focal plane.
Dissection chambers were assembled by gluing a magnetic foil (7.4 cm length, 5.4 cm width, 2 mm thickness) to an equally sized glass plate, both (rather than just the foil) with a central hole of 2 cm diameter. The hole was closed by a coverslip of 3.2 cm diameter and 0.17 mm thickness, glued underneath the glass plate, and the inner edge was sealed with silicon. Larvae were placed dorsal side up into the chamber, and clips consisting of stainless pins with tin-made holders on the magnetic foil were used to clamp the animals at either end. Dissections were then carried out as described (21, 64) in Ca2+- and Mg2+-free HL3.1 saline (70 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 115 mM sucrose, 5 mM trehalose, and 5 mM Hepes). Subsequently, the larvae were washed briefly in HL3.1 imaging buffer containing 4 mM Mg2+ and 1.5 mM Ca2+ for all control situations and/or 0.2 mM for the experiment in fig. S3 to remove debris of innards. Next, the larvae were live-stained in a nonstretched position in the imaging buffer solution containing Alexa Fluor 488–conjugated anti-HRP antibody (1:1000), serving as a surface marker to visualize the outline of NMJs during subsequent imaging, for 5 min. The larvae were briefly washed in the HRP-free imaging buffer to remove excess HRP label. Motor nerves were cut close to the ventral nerve cord, and the brains were removed to ensure reduced muscle contractions during live imaging. Clamped at the tip, body walls were turned around such that the muscles were facing the coverslip of the imaging chamber. Samples were straightened carefully using six clips and subjected to TIRF-illuminated PALM imaging. An optimal imaging focal plane was chosen, guided by the HRP signal, within which the highest number of boutons of one type 1b NMJ (from segments A2 to A4 on muscle 4 or 6/7) could be captured within the focal plane.
Male mutant and control larvae were subjected to 10-min PhTx or HL3 incubation, followed by a live HRP-488 stain imaged at the HRP Z-plane of type 1b NMJs (from segments A2 to A4 on muscle 4 or 6/7). Localization maps were generated from the acquired data using PALMTracer (MetaMorph, provided by J.-B. Siberita, Bordeaux). Before localization detection, the movies were drift-corrected and cropped to exclude movement artifacts by using the ImageJ plugin NanoJ (65) or Thunderstorm (66). The subsequently generated trajectories (PALMTracer) were further analyzed by calculating the MSD. Analysis of the local channel density within confined regions was performed by cluster analysis based on Voronoi tessellation constructed from localized channels using the software package SR-Tesseler (25).
Localization and trajectory reconnection of mEOS signals was carried out using a wavelet-based algorithm and a simulated annealing algorithm as previously described, which considers molecule localization and total intensity. It has been reported that mEOS4b molecules can show blinking-like behavior (67). To avoid false reconnections between trajectories, all sub-trajectories were analyzed as individual trajectories. Diffusion coefficients (D) were calculated by the exponential fitting of the first eight points of the MSD plots. MSD plots of immobilized molecules (on fixed samples) revealed that, under our imaging conditions, molecules with D ≥ 0.002 μm2/s can be considered as mobile. The offset of MSD curves from immobile molecules was taken as an additional empirical measure for the localization accuracy under the live imaging conditions [31.3 nm; interquartile range (IQR), 25.9 to 36.6 nm; 247 trajectories; fig. S1C]. The localization accuracy and the number of collected photons per individual fluorophore (fig. S1, A, and B) were calculated using the Thunderstorm plugin in ImageJ, matching the value obtained from the MSD curves of immobile molecules.
Calcium imaging
The spontaneous synaptic activity was monitored by imaging of postsynaptic calcium transients reflecting the stochastic release of SVs. We used the genetically encoded membrane-tethered SynapGCaMP6f reporter previously established in Drosophila NMJ AZs (22). Spontaneous calcium transients were recorded in an imaging buffer containing 4 mM Mg2+ and 1.5 mM Ca2+ for all control situations or 0.2 mM Ca2+ to decrease the frequency of spontaneous release events. Images were acquired with a frame rate of 20 Hz. SynapGCaMP6f was excited with a 488 nm laser, and transient fluorescent emission changes were detected at emission wavelengths between 520 and 550 nm (fig. S3). The analysis of calcium transient maxima (ΔF/F0) was performed on background-corrected maximum projections using a routine in ImageJ software (NIH) and involved the detection of synaptic puncta with SynapGCaMP6f fluorescence signal exceeding a threshold of 2× the SD (calculated 30 frames before stimulus) in response to the stimulus.
Cac channel number analysis by Bleach curves
The blinking of mEOS4b-labeled Cac channels was used as an approximate measure for the number of channels within individual AZs, due to the one-to-one stoichiometry between the pore-forming subunit and the genetically encoded fluorophore. Only image stacks were analyzed where individual blinking events could be identified. The localization of the channel population within individual AZs was defined within average projections of image stacks. Before further analysis, images were processed by background subtraction and drift correction using the Fiji plugin NanoJ (Table 1) (65). The confined localization of the channels within the AZ allowed for the counting of fluorescent units within regions of 5 × 5 pixels (pixel size, 107 nm × 107 nm), which was sufficient to cover the localization of the fluorescent signal. Individual symmetric AZs within single boutons were selected manually for analysis of the bleach curve. The fluorescent signal was completely bleached within less than 5000 frames (20-Hz acquisition rate), which allowed the identification of single fluorophores as one-step photobleaching events (see figs. S1 and S2). After background subtraction and normalization of the bleach curves to the average noise at the end of each recording, the amplitude of single events was accessed. The limitation in the acquisition frequency partially biased the precision of amplitude detection; hence, we report what is most likely an underestimated number of calcium channels within the AZ. Nevertheless, after normalizing the bleach curves to the averaged value of single steps for individual AZs, there are quantal changes in the fluorescent intensity visible that we interpret to be equivalent to the actual number of channels that are linked to individual mEOS4b fluorophores. Averaging the normalized single fluorophore responses reveals a clearly separated peak with respect to the noise (fig. S2). The quotient of the maximal fluorescence value and the averaged single fluorophore response was taken as a rough estimate for labeled channel molecules within the AZ. Because of the acquisition rate, as well as the shortcomings of live imaging parameters, and the varying positions of AZs within individual boutons, there are likely many missed bleaching steps recorded. Thus, the calculated channel numbers are most likely an underestimate of the actual number of molecules. Comparative Cac channel numbers under control conditions and after the PHP assay as well as for the BRP and the Cac C-terminal mutant (CacmEOS4B∆160aa mutants) were analyzed (Figs. 2, 4, and 5).
Table 1. Software and online sources used in this manuscript.
| Resource | Source | Identifier |
|---|---|---|
| Fiji | NIH (65) | https://fiji.sc/ |
| FITMASTER | HEKA Elektronik | www.heka.com/downloads/downloads_main.html#down_fitmaster |
| GraphPad Prism 6 and 7 | GraphPad Software | www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Imspector | Abberior Instruments | https://imspector.abberior-instruments.com/ |
| Microsoft Office Excel, Word, and PowerPoint 2010 | Microsoft | www.office.com/?omkt=de-de |
| MetaMorph Microscopy Automation and Image Analysis Software incl. PALM tracer built-in application | Molecular Devices | www.moleculardevices.com/systems/metamorph-research-imaging/metamorph-microscopy-automation-and-image-analysis-software |
| NanoJ-SRRF (ImageJ/Fiji plugin) | Gustafsson et al. (68) | http://sites.imagej.net/NanoJ/; http://sites.imagej.net/NanoJ-SRRF/ |
| SR-Tesseler | Levet et al. (25) | https://github.com/flevet/SR-Tesseler/releases/tag/v1.0 |
| pCLAMP 10 | Molecular Devices | www.moleculardevices.com/ |
| Python pyABF package for Python 3.10 (Harden, 2022); pyABF 2.3.5 | Python 3.10 | https://pypi.org/project/pyabf |
Well analysis
We scanned through all possible local grid bin sizes between 50 and 100 nm with a step size of 5 nm, for each bin size dx. For each value of dx, we performed the following iterative procedure starting at iteration k = 1:
1) First, we restricted the analysis to the trajectory points falling in a square region of size (2k + 1)dx × (2k + 1)dx centered at the high-density bin center (if k > 1, then the square is centered at the center of the ellipse found in the previous iteration instead). We obtained the well boundary by fitting the 95% confidence ellipse to the spatial distribution of these points.
2) Inside this ellipse, we computed the depth of the well A based on a circular estimator computed on the drift map based on a local grid centered at the well center with bin sizes dx, while the diffusion coefficient D was obtained by considering the well as a single bin.
3) The iteration was scored using the well parabolic score.
4) We go back to step 1, increasing k by 1, and stopped iterating when (2k + 1)dx > 300 nm and kept the iteration with the minimum score where the well had more than 20 trajectory points and five drift bins with at least seven trajectory displacements per bin.
The selected well for a given high-density region was chosen as the well with the minimum parabolic score overall local bin grid sizes. Last, to obtain the statistics, only the wells with a parabolic score of <0.5 and an energy of <10 kT were kept.
The ring ellipses in fig. S4 (D to F) were obtained by manually selecting wells forming an elliptic region and fitting an ellipse around their centers using a minimum volume ellipsoid algorithm with a tolerance of 0.01. The residence τ time of a molecule inside a well reported in fig. S4 (I and J) was computed for each well as follows: .
Tessellation analysis of Cac and BRP localizations within AZ cluster and nanocluster distributions
SR-Tesseler software and the ImageJ NanoJ-SRRF were used to process and segment each Cac/BRP localization recorded by live sptPALM of all mutant and control images (Table 1). Tessellation was performed on NanoJ-SRRF drift-corrected sptPALM movies from which the first 300 frames were rejected to avoid the detection of multiple molecules fluorescent at the same time. This movie was then subjected to ThunderSTORM to extract all PALM localizations of Cac channels in an image containing an NMJ with several boutons, with each bouton containing high-density localizations of Cac channels at AZs. This extracted map of all Cac/BRP localizations was processed in SR-Tesseler to generate a tessellated map of all Cac localizations.
CacmEOS4B
A list of the following parameters was used to select and analyze the highest-density Cac localizations at AZs that reflected the best TIRF Z focus and exemplified planar Cac localization clusters from the tessellation map of all Cac channel localizations in an image automatically and robustly. By this automated filtration process, we excluded the out-of-focus and side-view orientations of synapses and their Cac clusters.
Tessellation settings: AZ boundary settings: Voronoi object, 30 density factor; minimum area, 2000; and min localization, 5/10; nanocluster boundary settings: Voronoi nanocluster object, 3 density factor; minimum area, 50; and minimum localization, 5/10. List of AZ clusters and their nanoclusters: size, area, number of localizations, density, and number of clusters were exported and filtered to remove all AZ clusters lacking a nanocluster and were also within 30 to 600 nm AZ and nanocluster limits. In addition, we applied further limitations to Cac localization number = AZ boundary: 3000 and nanocluster boundary: 500, diameter size = AZ boundary: 600 to 30 nm and nanocluster boundary: 300 to 30 nm, area: AZ boundary: 50,000 nm2 and nanocluster boundary: 7000 nm2, and density = AZ and nanocluster boundary: 0.072 localizations/nm2 to define well the nanodomain of Cac channels at the presynaptic membrane of the Drosophila NMJ. We took into consideration the parameters set for BRP localizations from recent dSTORM and qPAINT imaging studies and our average diameter size and imaging of average Cac spot size (Fig. 1, A to H) (15, 16).
BRPmEOS3.2
A list of the following parameters modeled upon the parameters for BRP localizations from recent fixed NMJ studies analyzing BRP C-terminal antibody epitope with dSTORM and qPAINT imaging was used to define boundaries of cumulative BRP localization within average AZ sizes (Fig. 6) (15, 16). These parameters enabled automatic and robust selection and subsequent analysis of the highest-density BRP localizations at AZs that reflected the best TIRF Z focus.
AZ boundary settings: Voronoi object, 80 to 120 density factor; minimum area, 2000; and minimum localization, 200. Nanocluster boundary settings: Voronoi nanocluster object, 1.5 density factor; minimum area, 50; and minimum localization, 20. List of AZ clusters and their nanoclusters: size, area, number of localizations, density, and number of clusters were exported and filtered to remove all AZ clusters lacking a nanocluster and were also within 30- to 600-nm AZ and nanocluster limits. In addition, we applied further limitations to BRP localization number = AZ boundary: 8000 and nanocluster boundary: 1500, diameter size = AZ boundary: 600 to 30 nm and nanocluster boundary: 300 to 30 nm, and area: AZ boundary: 50,000 nm2 and nanocluster boundary: 10,000 nm2 to define the spread of BRP localization within an average Drosophila NMJ AZ.
Sample definition and in-laboratory replication
Confocal
Figure S7 (M and N): Quantification of Cac (O) intensity levels in CacmEOS4B∆160aa mutants, N = 40 images, four independent experimental sets.
STED
Figure 1 (A to E): N = 441 planar AZs from five animals, 24 images derived from n = 2 independent sets.
sptPALM
Diffusion coefficient, confinement, and tessellation analysis were performed on the same sptPALM raw data. Here at:
Figure 1 (K to M): 24 animals, N = 70 images were analyzed from n = 7 independent sets.
Figure 2 (L to Q): Control NMJs: 18 animals, N = 54 images and PhTx-treated NMJs: 20 animals, N = 58 images were derived from n = 5 independent sets with concurrent controls.
Figure 3 (I to N) and fig. S5 (I to L): Data of RimBP−/− experiment: CacmEOS4B (Con) and CacmEOS4B;;RBP−/− NMJs, 10 to 12 animals (Con), 9 animals (RBP−/−), and 18 animals (RIM−/−), N = 31 to 28 (Con) and 26 (RBP−/−) and 24 (RIM−/−). Images from n = 3 RimBP−/− and RIM−/− experiments were conducted together with concurrent controls.
Figure 3 (P to U): Data of CacmEOS4BAPTA∆6aa experiment: 10 (Con) and 6 (APTA∆6aa) animals. N = 28 images (con) and 17 images (APTA∆6aa) were analyzed from n = 2 independent sptPALM CacmEOS4-APTA∆6aa experiments that were conducted with concurrent controls.
Figure 4 (J to P): Data of CacmEOS4B∆160aa experiment: 8 (Con) and 6 (∆160aa) animals with N = 23 images (Con) and 15 (∆160aa). Images were analyzed from n = 2 independent sptPALM CacmEOS4B∆160aa experiments that were conducted with concurrent controls.
Figure 5 (A to H): Data of CacmEOS4B, CacmEOS4B;BRP−/−, and PhTx-treated CacmEOS4B;BRP−/− NMJs, N = 17 (Con) and 13 (BRP−/−) or 12 (BRP−/− + PhTx). Images, 2192 (Con), 1644 (BRP−/−), and 1001 (BRP−/− + PhTx) trajectories from six animals per condition. n = 3 individual BRP−/− experiments conducted with concurrent controls.
Figure 6 (J and K): Data from live BRPmEOS3.2 sptPALM imaging: control NMJs, N = 7 images from three animals; PhTx-treated NMJs, N = 9 images from three animals; tessellation analysis (Fig. 6, M and N): control, N = 26 images, from 11 animals and PhTx: N = 26 images, from seven animals included for either condition, was analyzed; n = 2 sets with concurrent controls.
Figure 7 (E to I): Control, BRP∆170, and BRP∆190 mutants: N = 37 to 25 images from 10 animals (Con), N = 33 to 21 images from 9 animals (Con + PhTx), N = 25 to 9 images from 4 to 7 animals (BRP∆170), N = 23 images from 6 animals (BRP∆170 + PhTx), N = 26 to 22 images from 8 animals (BRP∆190), and N = 24 to 18 images from 7 animals (BRP∆190 + PhTx); n = 2 sets with control, BRP∆170, and BRP∆190 mutant animals in both conditions, i.e., with and without PhTx concurrently imaged.
Figure S12 (C to H): Data of CacmEOS4B and CacmEOS4B;;GluRIIA−/− NMJs: N = 19 (Con) and 20 (GluRIIA−/−) regions of interest (ROIs) from 10 animals, respectively. n = 3 individual GluRIIA−/− experiments were conducted with concurrent controls.
Notably, here, RIM−/− and RimBP−/− mutant experiments (fig. S5 and Fig. 3, A to N) were performed in one concomitant experiment with three repetitions. The data shown in Fig. 5 for BRP−/−, BRP−/− + PhTx, and controls were done in one concomitant experiment, with three repetitions, and all the data shown in Fig. 7 were done in one concomitant experiment with two repetitions.
Electrophysiology
Figure S6 (B to I): Electrophysiological recordings of CacmEOS4B, CacmEOS4B-APTAΔ6aa, and CacmEOS4BΔ160aa animals using TEVC. N = 10 cells. Measurements were performed at third-instar larval muscle 6 NMJs of abdominal segments 2 and 3.
Figure S7: Electrophysiological recordings of CacmEOS4B, CacmEOS4B-APTAΔ6aa, and CacmEOS4BΔ160aa animals using current-clamp recording after 10 or 30 min of PhTx treatment [N = 12 cells (30 min of PhTx) and N = 10 cells (10 min of PhTx)]. Measurements were performed at third-instar larval muscle 6 NMJs of abdominal segments 2 and 3.
Figure 8 (A to I) and fig. S11: Electrophysiological recordings of CacmEOS4B(Con), CacmEOS4B;BRP∆170/brp69 (BRP∆170), and CacmEOS4B;BRP∆190/brp69 (BRP∆190) mutants in CacmEOS4B(Con) background after 30-min incubation in HL3 or 50 μM PhTx in HL3 solution using current-clamp recordings. Quantifications of mEPSP amplitude, eEPSP amplitude, quantal content, and mEPSP frequency in PhTx-treated untreated control and BRP∆190 (Fig. 8, D to F and I) or BRP∆170 (Fig. 8, G to J) mutant cells are normalized on the measurement obtained without PhTx for each genotype [N = 12 cells (BRP∆190) and N = 10 cells (BRP∆170)]. Measurements were performed at third-instar larval muscle 6 NMJs of abdominal segments 2 and 3.
Software and online resources
Quantification and statistical analysis
The statistical analysis was carried out using Prism software (GraphPad). The normality of distribution was verified by D’Agostino and Pearson omnibus normality test. To test the statistical significance of differences between two conditions, Kolmogorov-Smirnov test was used to assess the distribution of the diffusion coefficient data, and a Mann-Whitney U test was used to assess the difference in medians of confinement data and tessellation data (Figs. 2 to 7 and fig. S5). sptPALM data analysis for mobility, tessellation, well analysis, and channel numbers are shown either as median and an IQR or as mean ± SEM, as described in their respective figure legends. Channel numbers in Figs. 2K, 4O, and 5I were tested with a Mann-Whitney U test. For the confocal experiment in fig. S7 (M and N), one-way ANOVA with Dunn’s multiple comparisons test was performed. For electrophysiological experiments in Fig. 8 and figs. S6, S7, and S11, all datasets were tested for normality with the D’Agostino and Pearson tests. For datasets with three or more groups, normally distributed data were first tested to see whether SDs were comparable between groups with the Brown-Forsythe test. If they were, the data were further analyzed using one-way ANOVA followed by Tukey’s multiple comparisons tests; otherwise, they were analyzed using Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test. Non-normally distributed datasets with three or more groups were analyzed using a Kruskal-Wallis test followed by Dunn’s multiple comparisons tests. For normalized PhTx data, each genotype was normalized separately to the results of that genotype without PhTx treatment to compare the effects of PhTx more easily on that genotype. Normally distributed datasets were then analyzed using a two-tailed t-test if their SDs were similar, or with Welch’s t-test if they were not; non-normally distributed datasets were analyzed using a Mann-Whitney U test. Data are presented throughout the text as mean ± SEM or the median with the IQR as indicated. Significance levels are given as *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001; and ns, nonsignificant.
Acknowledgments
We thank the Biosupramol and supraFAB facilities at the FU, Berlin. We are grateful to J.-B. Siberita and F. Levet for providing image analysis tools (PALMTracer, SR-Tesseler) and the ERC-Adv 882673 for making D.H.’s contribution possible. We thank G. Paez-Segala and L. Looger for the mEOS4b construct.
Funding: This study was supported by the Deutsche Forschungsgemeinschaft [the Transregio (TRR)–SFB 186 (project ID 278001972)] to S.J.S. and A.M.W.; Excellence Cluster Neurocure (EXC-2049-390688087) to S.J.S.; and HE3604/11-1, SFB 1080, and Project B12 (INst 161/985-1) to M.H. We thank the funding for K.O.G. and S.G. for providing the CacmOES4B tool via the grant from the National Institute of Neurological Disorders and Stroke, NIH to K.O.G. (R01NS078179).
Author contributions: Conceptualization: T.G., M.H., and S.J.S. Investigation: T.G., M.E., K.E.,H.D. and J.L. Well and PALM analyses: P.P., D.H., and M.H. Methodology: U.T., M.M., T.M.-R., and S.R. Resources: U.T., S.G., and K.O.G. Writing (original draft): T.G., M.H., and S.J.S. Funding acquisition: A.M.W. and S.J.S. Supervision: M.H., A.M.W., and S.J.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Fly lines are available through the corresponding author.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Legends for movies S1 to S3
Other Supplementary Material for this : manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.A. Citri, R. C. Malenka, Synaptic plasticity: Multiple forms, functions, and mechanisms. Neuropsychopharmacology 33, 18–41 (2008). [DOI] [PubMed] [Google Scholar]
- 2.B. E. Herring, R. A. Nicoll, Long-term potentiation: From CaMKII to AMPA receptor trafficking. Annu. Rev. Physiol. 78, 351–365 (2016). [DOI] [PubMed] [Google Scholar]
- 3.H. R. Monday, T. J. Younts, P. E. Castillo, Long-term plasticity of neurotransmitter release: Emerging mechanisms and contributions to brain function and disease. Annu. Rev. Neurosci. 41, 299–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. Reddy-Alla, M. A. Böhme, E. Reynolds, C. Beis, A. T. Grasskamp, M. M. Mampell, M. Maglione, M. Jusyte, U. Rey, H. Babikir, A. W. McCarthy, C. Quentin, T. Matkovic, D. D. Bergeron, Z. Mushtaq, F. Göttfert, D. Owald, T. Mielke, S. W. Hell, S. J. Sigrist, A. M. Walter, Stable positioning of Unc13 restricts synaptic vesicle fusion to defined release sites to promote synchronous neurotransmission. Neuron 95, 1350–1364.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 5.C. Liu, P. S. Kaeser, Nanoscale location matters: Emerging principles of Ca2+ channel organization at the presynaptic active zone. Neuron 104, 627–629 (2019). [DOI] [PubMed] [Google Scholar]
- 6.M. A. Böhme, C. Beis, S. Reddy-Alla, E. Reynolds, M. M. Mampell, A. T. Grasskamp, J. Lützkendorf, D. D. Bergeron, J. H. Driller, H. Babikir, F. Göttfert, I. M. Robinson, C. J. O’Kane, S. W. Hell, M. C. Wahl, U. Stelzl, B. Loll, A. M. Walter, S. J. Sigrist, Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca2+ channel-vesicle coupling. Nat. Neurosci. 19, 1311–1320 (2016). [DOI] [PubMed] [Google Scholar]
- 7.J. Li, E. Park, L. R. Zhong, L. Chen, Homeostatic synaptic plasticity as a metaplasticity mechanism–A molecular and cellular perspective. Curr. Opin. Neurobiol. 54, 44–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.G. W. Davis, M. Muller, Homeostatic control of presynaptic neurotransmitter release. Annu. Rev. Physiol. 77, 251–270 (2015). [DOI] [PubMed] [Google Scholar]
- 9.C. A. Frank, M. J. Kennedy, C. P. Goold, K. W. Marek, G. W. Davis, Mechanisms underlying the rapid induction and sustained expression of synaptic homeostasis. Neuron 52, 663–677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. Müller, G. W. Davis, Transsynaptic control of presynaptic Ca2+ influx achieves homeostatic potentiation of neurotransmitter release. Curr. Biol. 22, 1102–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. J. Gratz, P. Goel, J. J. Bruckner, R. X. Hernandez, K. Khateeb, G. T. Macleod, D. Dickman, K. M. O’Connor-Giles, Endogenous tagging reveals differential regulation of Ca2+ channels at single active zones during presynaptic homeostatic potentiation and depression. J. Neurosci. 39, 2416–2429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Weyhersmüller, S. Hallermann, N. Wagner, J. Eilers, Rapid active zone remodeling during synaptic plasticity. J. Neurosci. 31, 6041–6052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. A. Böhme, A. W. McCarthy, A. T. Grasskamp, C. B. Beuschel, P. Goel, M. Jusyte, D. Laber, S. Huang, U. Rey, A. G. Petzoldt, M. Lehmann, F. Gottfert, P. Haghighi, S. W. Hell, D. Owald, D. Dickman, S. J. Sigrist, A. M. Walter, Rapid active zone remodeling consolidates presynaptic potentiation. Nat. Commun. 10, 1085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.W. Fouquet, D. Owald, C. Wichmann, S. Mertel, H. Depner, M. Dyba, S. Hallermann, R. J. Kittel, S. Eimer, S. J. Sigrist, Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 186, 129–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N. Ehmann, S. van de Linde, A. Alon, D. Ljaschenko, X. Z. Keung, T. Holm, A. Rings, A. DiAntonio, S. Hallermann, U. Ashery, M. Heckmann, M. Sauer, R. J. Kittel, Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5, 4650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Mrestani, M. Pauli, P. Kollmannsberger, F. Repp, R. J. Kittel, J. Eilers, S. Doose, M. Sauer, A.-L. Sirén, M. Heckmann, M. M. Paul, Active zone compaction correlates with presynaptic homeostatic potentiation. Cell Rep. 37, 109770 (2021). [DOI] [PubMed] [Google Scholar]
- 17.A. T. Bademosi, E. Lauwers, P. Padmanabhan, L. Odierna, Y. J. Chai, A. Papadopulos, G. J. Goodhill, P. Verstreken, B. van Swinderen, F. A. Meunier, In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat. Commun. 8, 13660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Schneider, E. Hosy, J. Kohl, J. Klueva, D. Choquet, U. Thomas, A. Voigt, M. Heine, Mobility of calcium channels in the presynaptic membrane. Neuron 86, 672–679 (2015). [DOI] [PubMed] [Google Scholar]
- 19.M. G. Paez-Segala, M. G. Sun, G. Shtengel, S. Viswanathan, M. A. Baird, J. J. Macklin, R. Patel, J. R. Allen, E. S. Howe, G. Piszczek, H. F. Hess, M. W. Davidson, Y. Wang, L. L. Looger, Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat. Methods 12, 215–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.F. Kawasaki, S. C. Collins, R. W. Ordway, Synaptic calcium-channel function in Drosophila: Analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J. Neurosci. 22, 5856–5864 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. Marter, O. Kobler, I. Erdmann, E. Soleimanpour, P. Landgraf, A. Müller, J. Abele, U. Thomas, D. C. Dieterich, Click chemistry (CuAAC) and detection of tagged de novo synthesized proteins in Drosophila. Bio. Protoc. 9, e3142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Z. L. Newman, A. Hoagland, K. Aghi, K. Worden, S. L. Levy, J. H. Son, L. P. Lee, E. Y. Isacoff, Input-specific plasticity and homeostasis at the drosophila larval neuromuscular junction. Neuron 93, 1388–1404.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Y. Nakamura, H. Harada, N. Kamasawa, K. Matsui, J. S. Rothman, R. Shigemoto, R. A. Silver, D. A. DiGregorio, T. Takahashi, Nanoscale distribution of presynaptic Ca2+ channels and its impact on vesicular release during development. Neuron 85, 145–158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N. Rebola, M. Reva, T. Kirizs, M. Szoboszlay, A. Lőrincz, G. Moneron, Z. Nusser, D. A. DiGregorio, Distinct nanoscale calcium channel and synaptic vesicle topographies contribute to the diversity of synaptic function. Neuron 104, 693–710.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 25.F. Levet, E. Hosy, A. Kechkar, C. Butler, A. Beghin, D. Choquet, J.-B. Sibarita, SR-Tesseler: A method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 12, 1065–1071 (2015). [DOI] [PubMed] [Google Scholar]
- 26.A. Fulterer, T. F. M. Andlauer, A. Ender, M. Maglione, K. Eyring, J. Woitkuhn, M. Lehmann, T. Matkovic-Rachid, J. R. P. Geiger, A. M. Walter, K. I. Nagel, S. J. Sigrist, Active zone scaffold protein ratios tune functional diversity across brain synapses. Cell Rep. 23, 1259–1274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. M. Brockmann, M. Maglione, C. G. Willmes, A. Stumpf, B. A. Bouazza, L. M. Velasquez, M. K. Grauel, P. Beed, M. Lehmann, N. Gimber, J. Schmoranzer, S. J. Sigrist, C. Rosenmund, D. Schmitz, RIM-BP2 primes synaptic vesicles via recruitment of Munc13–1 at hippocampal mossy fiber synapses. eLife 8, e43243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R. Fukaya, M. Maglione, S. J. Sigrist, T. Sakaba, Rapid Ca2+ channel accumulation contributes to cAMP-mediated increase in transmission at hippocampal mossy fiber synapses. Proc. Natl. Acad. Sci. U.S.A. 118, e2016754118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N. Holderith, A. Lorincz, G. Katona, B. Rozsa, A. Kulik, M. Watanabe, Z. Nusser, Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat. Neurosci. 15, 988–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O. Turrel, N. Ramesh, M. J. F. Escher, A. Pooryasin, S. J. Sigrist, Transient active zone remodeling in the Drosophila mushroom body supports memory. Curr. Biol. 32, 4900–4913.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 31.E. R. Graf, V. Valakh, C. M. Wright, C. Wu, Z. Liu, Y. Q. Zhang, A. DiAntonio, RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J. Neurosci. 32, 16586–16596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Müller, K. S. Liu, S. J. Sigrist, G. W. Davis, RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J. Neurosci. 32, 16574–16585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. S. Y. Liu, M. Siebert, S. Mertel, E. Knoche, S. Wegener, C. Wichmann, T. Matkovic, K. Muhammad, H. Depner, C. Mettke, J. Buckers, S. W. Hell, M. Muller, G. W. Davis, D. Schmitz, S. J. Sigrist, Rim-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science 334, 1565–1569 (2011). [DOI] [PubMed] [Google Scholar]
- 34.H. Hibino, R. Pironkova, O. Onwumere, M. Vologodskaia, A. J. Hudspeth, F. Lesage, RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca2+ channels. Neuron 34, 411–423 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. Siebert, M. A. Böhme, J. H. Driller, H. Babikir, M. M. Mampell, U. Rey, N. Ramesh, T. Matkovic, N. Holton, S. Reddy-Alla, F. Göttfert, D. Kamin, C. Quentin, S. Klinedinst, T. F. Andlauer, S. W. Hell, C. A. Collins, M. C. Wahl, B. Loll, S. J. Sigrist, A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones. eLife 4, e06935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.P. S. Kaeser, L. Deng, Y. Wang, I. Dulubova, X. Liu, J. Rizo, T. C. Sudhof, RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144, 282–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. M. Brockmann, F. Zarebidaki, M. Camacho, M. K. Grauel, T. Trimbuch, T. C. Sudhof, C. Rosenmund, A trio of active zone proteins comprised of RIM-BPs, RIMs, and Munc13s governs neurotransmitter release. Cell Rep. 32, 107960 (2020). [DOI] [PubMed] [Google Scholar]
- 38.W. Dong, T. Radulovic, R. O. Goral, C. Thomas, M. Suarez Montesinos, D. Guerrero-Given, A. Hagiwara, T. Putzke, Y. Hida, M. Abe, K. Sakimura, N. Kamasawa, T. Ohtsuka, S. M. Young Jr., CAST/ELKS proteins control voltage-gated Ca2+ channel density and synaptic release probability at a mammalian central synapse. Cell Rep. 24, 284–293.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R. G. Held, C. Liu, K. Ma, A. M. Ramsey, T. B. Tarr, G. De Nola, S. S. H. Wang, J. Wang, A. van den Maagdenberg, T. Schneider, J. Sun, T. A. Blanpied, P. S. Kaeser, Synapse and active zone assembly in the absence of presynaptic Ca2+ channels and Ca2+ entry. Neuron 107, 667–683.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Lübbert, R. O. Goral, R. Satterfield, T. Putzke, A. M. van den Maagdenberg, N. Kamasawa, S. M. Young Jr., A novel region in the CaV2.1 α1 subunit C-terminus regulates fast synaptic vesicle fusion and vesicle docking at the mammalian presynaptic active zone. eLife 6, e28412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R. J. Kittel, C. Wichmann, T. M. Rasse, W. Fouquet, M. Schmidt, A. Schmid, D. A. Wagh, C. Pawlu, R. R. Kellner, K. I. Willig, S. W. Hell, E. Buchner, M. Heckmann, S. J. Sigrist, Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 42.C. Liu, L. S. Bickford, R. G. Held, H. Nyitrai, T. C. Südhof, P. S. Kaeser, The active zone protein family ELKS supports Ca2+ influx at nerve terminals of inhibitory hippocampal neurons. J. Neurosci. 34, 12289–12303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.S. Hallermann, R. J. Kittel, C. Wichmann, A. Weyhersmüller, W. Fouquet, S. Mertel, D. Owald, S. Eimer, H. Depner, M. Schwarzel, S. J. Sigrist, M. Heckmann, Naked dense bodies provoke depression. J. Neurosci. 30, 14340–14345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. A. Wagh, T. M. Rasse, E. Asan, A. Hofbauer, I. Schwenkert, H. Durrbeck, S. Buchner, M. C. Dabauvalle, M. Schmidt, G. Qin, C. Wichmann, R. Kittel, S. J. Sigrist, E. Buchner, Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844 (2006). [DOI] [PubMed] [Google Scholar]
- 45.S. Nagarkar-Jaiswal, P. T. Lee, M. E. Campbell, K. Chen, S. Anguiano-Zarate, M. C. Gutierrez, T. Busby, W. W. Lin, Y. He, K. L. Schulze, B. W. Booth, M. Evans-Holm, K. J. Venken, R. W. Levis, A. C. Spradling, R. A. Hoskins, H. J. Bellen, A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife 4, e05338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.T. Matkovic, M. Siebert, E. Knoche, H. Depner, S. Mertel, D. Owald, M. Schmidt, U. Thomas, A. Sickmann, D. Kamin, S. W. Hell, J. Bürger, C. Hollmann, T. Mielke, C. Wichmann, S. J. Sigrist, The Bruchpilot cytomatrix determines the size of the readily releasable pool of synaptic vesicles. J. Cell Biol. 202, 667–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. A. Bohme, A. T. Grasskamp, A. M. Walter, Regulation of synaptic release-site Ca2+ channel coupling as a mechanism to control release probability and short-term plasticity. FEBS Lett. 592, 3516–3531 (2018). [DOI] [PubMed] [Google Scholar]
- 48.J. Heck, P. Parutto, A. Ciuraszkiewicz, A. Bikbaev, R. Freund, J. Mitlöhner, M. Andres-Alonso, A. Fejtova, D. Holcman, M. Heine, Transient confinement of CaV2.1 Ca2+-channel splice variants shapes synaptic short-term plasticity. Neuron 103, 66–79.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 49.M. Orlando, A. Dvorzhak, F. Bruentgens, M. Maglione, B. R. Rost, S. J. Sigrist, J. Breustedt, D. Schmitz, Recruitment of release sites underlies chemical presynaptic potentiation at hippocampal mossy fiber boutons. PLOS Biol. 19, e3001149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. P. Haghighi, B. D. McCabe, R. D. Fetter, J. E. Palmer, S. Hom, C. S. Goodman, Retrograde control of synaptic transmission by postsynaptic CaMKII at the Drosophila neuromuscular junction. Neuron 39, 255–267 (2003). [DOI] [PubMed] [Google Scholar]
- 51.P. Goel, D. Dickman, Synaptic homeostats: Latent plasticity revealed at the Drosophila neuromuscular junction. Cell. Mol. Life Sci. 78, 3159–3179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R. E. Dixon, C. M. Moreno, C. Yuan, X. Opitz-Araya, M. D. Binder, M. F. Navedo, L. F. Santana, Graded Ca2+/calmodulin-dependent coupling of voltage-gated CaV1.2 channels. eLife 4, e05608 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. M. Moreno, R. E. Dixon, S. Tajada, C. Yuan, X. Opitz-Araya, M. D. Binder, L. F. Santana, Ca2+ entry into neurons is facilitated by cooperative gating of clustered CaV1.3 channels. eLife 5, e15744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.N. Krick, S. Ryglewski, A. Pichler, A. Bikbaev, T. Gotz, O. Kobler, M. Heine, U. Thomas, C. Duch, Separation of presynaptic CaV2 and CaV1 channel function in synaptic vesicle exo- and endocytosis by the membrane anchored Ca2+ pump PMCA. Proc. Natl. Acad. Sci. U.S.A. 118, e2106621118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Emperador-Melero, P. S. Kaeser, Liquid active zones for controlling the phases of synaptic transmission. Mol. Cell 73, 859–860 (2019). [DOI] [PubMed] [Google Scholar]
- 56.M. Liang, G. Jin, X. Xie, W. Zhang, K. Li, F. Niu, C. Yu, Z. Wei, Oligomerized liprin-α promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins. Cell Rep. 34, 108901 (2021). [DOI] [PubMed] [Google Scholar]
- 57.N. A. McDonald, R. D. Fetter, K. Shen, Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature 588, 454–458 (2020). [DOI] [PubMed] [Google Scholar]
- 58.X. Wu, Q. Cai, Z. Shen, X. Chen, M. Zeng, S. Du, M. Zhang, RIM and RIM-BP form presynaptic active-zone-like condensates via phase separation. Mol. Cell 73, 971–984.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 59.O. Woznicka, A. Gorlich, S. Sigrist, E. Pyza, BRP-170 and BRP190 isoforms of Bruchpilot protein differentially contribute to the frequency of synapses and synaptic circadian plasticity in the visual system of Drosophila. Front. Cell. Neurosci. 9, 238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. Kondo, R. Ueda, Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.T. F. M. Andlauer, S. J. Sigrist, Quantitative analysis of Drosophila larval neuromuscular junction morphology. Cold Spring Harb. Protoc. 2012, 490–493 (2012). [DOI] [PubMed] [Google Scholar]
- 62.G. Qin, T. Schwarz, R. J. Kittel, A. Schmid, T. M. Rasse, D. Kappei, E. Ponimaskin, M. Heckmann, S. J. Sigrist, Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J. Neurosci. 25, 3209–3218 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.H. Depner, J. Lützkendorf, H. A. Babkir, S. J. Sigrist, M. G. Holt, Differential centrifugation-based biochemical fractionation of the Drosophila adult CNS. Nat. Protoc. 9, 2796–2808 (2014). [DOI] [PubMed] [Google Scholar]
- 64.P. Ramachandran, V. Budnik, Dissection of Drosophila larval body-wall muscles. Cold Spring Harb. Protoc. 2010, pdb.prot5469 (2010). [DOI] [PubMed] [Google Scholar]
- 65.R. F. Laine, K. L. Tosheva, N. Gustafsson, R. D. M. Gray, P. Almada, D. Albrecht, G. T. Risa, F. Hurtig, A. C. Lindas, B. Baum, J. Mercer, C. Leterrier, P. M. Pereira, S. Culley, R. Henriques, NanoJ: A high-performance open-source super-resolution microscopy toolbox. J. Phys. D Appl. Phys. 52, 163001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. Ovesny, P. Krizek, J. Borkovec, Z. Svindrych, G. M. Hagen, ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.E. De Zitter, J. Ridard, D. Thedie, V. Adam, B. Levy, M. Byrdin, G. Gotthard, L. Van Meervelt, P. Dedecker, I. Demachy, D. Bourgeois, Mechanistic investigations of green mEos4b reveal a dynamic long-lived dark state. J. Am. Chem. Soc. 142, 10978–10988 (2020). [DOI] [PubMed] [Google Scholar]
- 68.N. Gustafsson, S. Culley, G. Ashdown, D. M. Owen, P. M. Pereira, R. Henriques, Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 7, 12471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Legends for movies S1 to S3
Movies S1 to S3