SUMMARY
Beneficial associations with bacteria are widespread across animals, spanning a range of symbiont localizations, transmission routes, and functions. While some of these associations have evolved into obligate relationships with permanent symbiont localization within the host, the majority require colonization of every host generation from the environment or via maternal provisions. Across the broad diversity of host species and tissue types that beneficial bacteria can colonize, there are some highly specialized strategies for establishment yet also some common patterns in the molecular basis of colonization. This review focuses on the mechanisms underlying the early stage of beneficial bacterium-invertebrate associations, from initial contact to the establishment of the symbionts in a specific location of the host’s body. We first reflect on general selective pressures that can drive the transition from a free-living to a host-associated lifestyle in bacteria. We then cover bacterial molecular factors for colonization in symbioses from both model and nonmodel invertebrate systems where these have been studied, including terrestrial and aquatic host taxa. Finally, we discuss how interactions between multiple colonizing bacteria and priority effects can influence colonization. Taking the bacterial perspective, we emphasize the importance of developing new experimentally tractable systems to derive general insights into the ecological factors and molecular adaptations underlying the origin and establishment of beneficial symbioses in animals.
KEYWORDS: animal-microbe interactions, symbiosis, molecular factors, host colonization, beneficial bacteria
INTRODUCTION
Symbiosis is at least as old as eukaryotic cells (1) and has played a major role in the evolution of micro- and macro-organisms. In animals, there is compelling evidence that beneficial microorganisms can fuel major ecological innovations by conferring a diverse spectrum of functional benefits to the host (2–7). A range of bacteria are prone to interact with eukaryotes, as host associations have evolved independently in multiple different bacterial phyla (8, 9). Among these, beneficial bacteria have evolved from either environmental (8) or parasitic ancestors (10, 11). Both ecological and genomic preadaptations have likely been important for the evolution of tight interactions with eukaryotes (12).
Notably, the benefits to engage in symbiosis are often discussed from the perspective of the host, while the microbe’s ecological drivers of mutualism with an animal are poorly understood (13–15). From the symbiont’s perspective, a series of challenges must be overcome to successfully engage in a persistent association with a host (15–17). First, bacteria need to reach and establish contact. Second, the microbes must make their way to the housing tissues, persist in these, successfully compete with other microorganisms, and protect themselves against host immune reactions (Text Box 1). And third, they need to disperse or relocate to new host individuals.
TEXT BOX 1: THE ANIMAL IMMUNE SYSTEM AS A KEY CHALLENGE FOR BACTERIAL COLONIZERS
Along with diverse microbial factors influencing the establishment of beneficial interactions, host immunity plays a crucial role in determining which microbes can colonize and persist. Importantly, shifts between a free-living and a host-associated lifestyle, as well as transitions across the parasite-mutualism continuum, usually require that bacteria evolve adaptations to cope with the host immune system.
The innate immune system of invertebrates comprises two different components for controlling antagonists (18, 19). Through cellular immunity, hemocytes that are constantly circulating in the host body become activated, resulting in encapsulation or phagocytosis of microorganisms (20, 21). By contrast, humoral immunity describes the ability of pattern recognition receptors (PRRs) to recognize microorganism-associated molecular patterns (MAMPs) and subsequently trigger the expression of antimicrobial peptides (AMPs) and other effectors through complex signaling cascades (20). Notably, some invertebrates have evolved mechanisms to recognize previously encountered microorganisms (18, 22). However, the memory component is more pronounced and fine-tuned in the vertebrate adaptive immune system, which relies on specialized lymphocyte populations forming a living record of pathogen encounters that can be rapidly reactivated upon recurrent infections.
Similar to pathogens, endosymbionts have evolved multiple strategies to cope with their hosts’ immune defenses. Some can manipulate the host’s immune system, for example, by downregulating genes of defense pathways (23). Alternatively, symbionts can become more resistant to the host’s antimicrobial activity, as is the case in the secondary symbiont of tsetse flies, Sodalis glossinidius, which is more resistant to an antimicrobial compound produced by the host than are closely related nonsymbiotic bacteria (24). Another widespread mechanism for immune evasion is to hinder recognition by the host. For example, bacterial peptidoglycan is generally recognized by PRRs, but bacteria can elude detection by modifying their MAMPs or by losing the peptidoglycan machinery altogether (25).
In addition to immune evasion by the symbionts, the host can evolve mechanisms to confine the symbionts, e.g., by compartmentalization in specific tissues, like bacteriomes or specialized gut regions, and downregulation of immune effectors in these compartments (26–29). Alternatively, some hosts develop defenses that spare beneficial symbionts, e.g., by reducing specific immune factors that target a desired symbiont while keeping other components unchanged (30). Furthermore, some eusocial insects have fewer immune-related genes, resulting in a reduced repertoire of antimicrobial peptides, and instead rely on social immunity (31, 32).
The interaction between beneficial symbionts and the host’s immune system has been covered in great depth elsewhere, and interested readers are referred to previous reviews on this topic (for examples, see references 18 and 33 to 34).
Both symbiont localization and transmission strongly affect the bacterial traits required for colonization of a host. Since symbionts can reside intra- or extracellularly in a variety of host tissues (8), gaining access to the final destination requires adaptations to achieve translocation and specific molecular interactions with host cells, as well as the ability to cope with stressful conditions in the host environment. While the original establishment of symbiotic interactions necessarily relies on acquisition from the environment (8), transfer over generations—or vertical transmission—has evolved in numerous symbioses (16, 35). For highly integrated partners involving consistent intracellular localization and a strict vertical transmission route, the microbial symbiont is likely to have a passive role in colonization, especially after the massive loss of functions due to genome erosion commonly associated with an ancient host-associated lifestyle (36). We will henceforth refer to these as closed symbioses and to those relying on recurrent acquisition from the environment or unrelated hosts as open symbioses, based on categories of symbiosis proposed elsewhere (37). A combination of both vertical and horizontal transmission, called mixed-mode transmission, is likely a common phenomenon balancing some of the benefits and challenges of each mode (17, 38) (Text Box 2). Accordingly, we will here refer to such associations as mixed symbioses (37). A plethora of mechanisms has evolved across different bacteria to navigate these transmission routes, enabling colonization in taxonomically diverse hosts with various morphologies and lifestyles (12, 39–41).
TEXT BOX 2: IMPLICATIONS OF SYMBIONT TRANSMISSION MODE
The mode of symbiont transmission is usually related to the degree of dependence in the association, with vertical transmission being more likely in symbioses with increased dependence (42). Still, there are also systems in which obligate symbionts are acquired horizontally (43), and each transmission mode—vertical, horizontal, or mixed mode—entails a different set of trade-offs for both host and symbiont (Table 1). Vertical transmission can result in an obligate association involving partner coevolution and codiversification, here referred to as a closed symbiosis. The symbiont can benefit from this through an ensured passage from one host generation to the next, a sheltered and nutritious environment, reduced competition, and protection against antagonists. Faithful transmission is a mechanism to maintain specificity through partner fidelity (44). The aligned interests of host and symbiont in a system with vertical transmission also promote a shift from pathogenic to beneficial interaction. However, it is not clear whether the reduction in pathogenicity favors the evolution of vertical transmission or vice versa. Symbionts with a strict vertical transmission can suffer from genome erosion due to relaxed selection on genes unnecessary in the host environment in conjunction with repeated population bottlenecks (45–47).
TABLE 1.
Overview of potential benefits and downsides for hosts and symbionts depending on the symbiont transmission mode
| Symbiotic partner |
 Vertical |
 Mixed |
 Horizontal |
|---|---|---|---|
| Host | |||
| Benefits | Symbionts guaranteed in offspring Maximum control Selects for beneficial symbionts |
Symbionts guaranteed in offspring Flexible acquisition of novel symbionts when environment changes |
Flexible acquisition of novel symbionts when environment changes |
| Downsides | Potentially costly adaptations for transmission Inflexible to changes Accumulation of mildly deleterious mutations can reduce symbiont benefits (Muller’s ratchet) |
Symbionts can become harmful Filtering and control mechanisms necessary Competition between symbionts may reduce benefit to the host |
Symbionts may not always be available (resulting in uninfected offspring) Symbionts can become harmful Filtering and control mechanisms necessary Competition between symbionts may reduce benefit to the host |
| Symbiont | |||
| Benefits | No competition Reliable nutrients from the host Safe niche |
Priority for vertically transmitted symbiont HGTa possible Biphasic lifestyle can expand ecological potential Colonization of unrelated host individuals or new host species possible Reliable nutrients |
HGT possible Biphasic lifestyle can expand ecological potential Colonization of unrelated host individuals or new host species possible |
| Downsides | Genome erosion and Muller’s ratchet Increased dependence on host Limited or no possibility for spreading to unrelated hosts |
High competition In cases of a biphasic lifestyle, costly adaptations to host and environment (trade-offs) Transition between environments potentially challenging |
High competition Hosts may not always be available In cases of a biphasic lifestyle, costly adaptations to host and environment (trade-offs) Transition between environments potentially challenging |
HGT, horizontal gene transfer.
By contrast, in systems relying on horizontal transmission, or open symbioses, the symbiont needs to be adapted to the host, while also maintaining the ability to survive in and transition to/from free-living conditions outside of the host or other unrelated hosts throughout the symbiont’s life cycle. The evolutionary trajectory of a symbiont in such a scenario depends on many factors, including the adaptations necessary for colonization of the host and the nonhost environment(s), the frequency of encounter with these environments, trade-offs between environments, and the probability of transitioning between environments (15, 48). Also, horizontally transmitted symbionts must deal with partner choice mechanisms imposed by the host to promote specificity and ensure cooperation (44). Advantages for bacteria with horizontal transmission include the potential to spread to other host individuals, populations, or even species (49). Furthermore, secondary hosts might serve as a refuge if the primary host is not available (50). As a result of the capabilities needed to persist in various environments, horizontally transmitted symbionts generally show less dramatic genome erosion (42), and contacts with other microorganisms can enable the acquisition of novel functions through horizontal gene transfer (51).
Symbionts experiencing mixed-mode transmission, i.e., in a mixed symbiosis, potentially reap the benefits of both transmission modes. Ebert (17) speculates that this may be the most common mode of transmission and can sometimes occur as an intermediate stage in the transition toward strict vertical transmission. Strikingly, even occasional interactions with other bacteria can facilitate horizontal gene transfer and thus mitigate genome erosion caused by strict vertical transmission (52, 53).
The increasing availability of molecular data has now opened new avenues to investigate factors facilitating symbiosis and to evaluate common patterns across model and nonmodel systems. Host adaptations to accommodate symbionts and in particular host immunity have been actively investigated (Text Box 1) and are expanding subfields within symbiosis research, as highlighted in a number of studies and reviews (18, 22, 33, 34, 54–57). In the present review, bacterial factors are in the spotlight. We aim to synthesize the current knowledge on molecular mechanisms enabling beneficial bacterial symbionts to successfully colonize invertebrate animal hosts. We chose to focus on symbionts that must colonize the host tissues each generation, either after maternal provisioning or environmental acquisition, i.e., open and mixed symbioses.
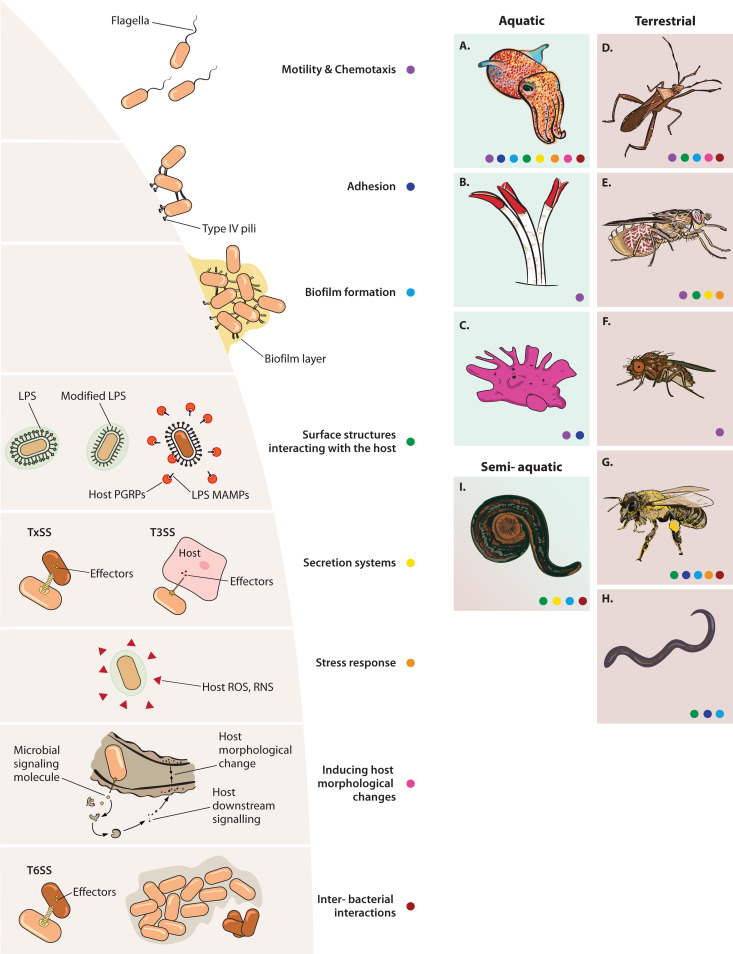
The review is structured in three sections that integrate ecological, molecular, and evolutionary perspectives on the establishment of beneficial bacterial symbionts. First, we reflect on the selective pressures that could generally drive the transition from a free-living lifestyle to a consistent association with a host. Then, we review the prevalent mechanisms that facilitate symbiont entry and the underlying genetic factors orchestrating these early phases: initial contact with the host and establishment in the housing tissues (Fig. 1). In a third and final section, we mention relevant ecological components of symbiont establishment, discussing multipartite interactions between bacteria and their prominent role in host colonization. The review does not aim to be exhaustive in covering the numerous studies on systems with a complex microbiota. We deliberately focus on a number of invertebrate animals from terrestrial, freshwater, and marine environments in which bacterial molecular factors for establishment in the host have been described (Table 2; Fig. 1) and refer to a few examples in vertebrates and plants to complement key points.
FIG 1.
Overview of known bacterial molecular mechanisms used for colonization of animal hosts in symbioses that require reentry of the symbionts into the host tissues every generation from the parents or the environment. The mechanisms addressed in the review are depicted on the left and invertebrate-bacterium symbioses described in the review are depicted on the right. The colored dots below the figures of the hosts indicate the mechanisms that their respective symbionts are known to use during colonization (color legend shown in the middle) The symbiotic systems include hosts living in marine, terrestrial, and freshwater habitats, as follows: bobtail squids (A), giant tubeworms (B), sponges (C), broad-headed bean bugs (D), tsetse flies (E), fruit flies (F), honey bees (G), entomopathogenic nematodes (H), and leeches (I). RNS, reactive nitrogen species; PGRPs, peptidoglycan recognition proteins; TxSS, any type secretion system.
TABLE 2.
Animal-bacterium symbioses in which symbionts must recolonize host tissues each generation and for which bacterial molecular factors involved in colonization have been identifieda
| Figure panel | Host | Symbiont group (phylum/family) | Symbiont species | Localization | Section(s) in this article | References |
|---|---|---|---|---|---|---|
| A | Euprymna scolopes (squid) | Proteobacteria, Vibrionaceae | Vibrio fischeri | Light organ | “Reaching the Host: Motility and Chemotaxis,” “Surface structures interacting with the host,” “Adhesion,” “Biofilm formation,” “Secretion systems,” “Making Space: Induction of Host Morphological Traits Relevant to Colonization,” “Coping with a New Environment: Stress Response,” “Dealing with Third Parties: Competition and Facilitation among Symbionts,” “Impact of Priority Effects on Symbiont Colonization” | 85, 100, 104, 107, 119, 131, 133, 151, 165, 184, 187, 209, 213, 250, 251 |
| B | Riftia pachyptila (giant tubeworm) | Proteobacteria, sulfur-oxidizing symbionts | “Candidatus Endoriftia persephone” | Trophosome | “Reaching the Host: Motility and Chemotaxis” | 16, 109 |
| C | Petrosia ficiformis (sponge) | Cyanobacteria, Synechococcaceae | “Candidatus Synechococcus feldmannii” | Bacteriocytes | “Reaching the Host: Motility and Chemotaxis,” “Adhesion” | 110, 155 |
| Cyanobacteria, Synechococcaceae | “Candidatus Synechococcus spongiarum” | Extracellularly | “Reaching the Host: Motility and Chemotaxis,” “Adhesion” | 110 | ||
| D | Riptortus pedestris (broad-headed bugs) | Proteobacteria, Burkholderiaceae | Caballeronia insecticola | Midgut crypts | “Reaching the Host: Motility and Chemotaxis,” “Surface structures interacting with the host,” “Biofilm formation,” “Making Space: Induction of Host Morphological Traits Relevant to Colonization,” “Coping with a New Environment,” “Dealing with Third Parties: Competition and Facilitation among Symbionts,” “Impact of Priority Effects on Symbiont Colonization” | 44, 114, 115, 119, 136, 167, 193, 194, 226 |
| E | Glossina spp. (tsetse flies) | Proteobacteria, Erwiniaceae | Wigglesworthia glossinidia | Milk glands, intrauterine larva, bacteriome | “Reaching the Host: Motility and Chemotaxis” | 122, 123 |
| Proteobacteria, Pectobacteriaceae | Sodalis glossinidius | Body lumen, milk glands, intrauterine larva, gut | “Surface structures interacting with the host,” “Secretion systems,” “Coping with a New Environment: Stress Response” | 21, 177–179, 199, 208 | ||
| F | Drosophila melanogaster(fruit fly) | Proteobacteria, Acetobacteraceae | Acetobacter thailandicus | Gut | “Reaching the Host: Motility and Chemotaxis” | 128 |
| G | Apis mellifera (honey bee) | Proteobacteria, Neisseriaceae | Snodgrassella alvi | Gut | “Surface structures interacting with the host,” “Adhesion,” “Biofilm formation,” “Coping with a New Environment: Stress Response,” “Dealing with Third Parties: Competition and Facilitation among Symbionts,” “Impact of Priority Effects on Symbiont Colonization” | 84, 145, 230 |
| H | Heterorhabditis bacteriophora (nematodes) | Proteobacteria, Morganellaceae | Photorhabdus | Gut, rectal gland cells | “Surface structures interacting with the host,” “Adhesion,” “Biofilm formation” |
138, 148, 152–154, 166 |
| I | Hirudo verbana (leech) | Bacteroidota, Rikenellaceae | Rikenella-like | Gut | “Dealing with Third Parties: Competition and Facilitation among Symbionts” | 169 |
| Proteobacteria, Aeromonadaceae | Aeromonas veronii | Gut | “Surface structures interacting with the host,” “Biofilm formation,” “Secretion systems,” “Dealing with Third Parties: Competition and Facilitation among Symbionts” | 139, 169, 174 |
Taxonomic affiliation of each partner, localization, section occurrence in this review, and associated references are included.
SWITCHING BETWEEN FREE-LIVING AND HOST-ASSOCIATED LIFESTYLES
Symbionts taking part in open or mixed systems, which includes the majority of described symbioses (58), often experience short or extended periods outside of the host. The environmental conditions during this phase can be starkly different, and relevant challenges must be overcome for successful transition in either direction, from the host to the environment or vice versa, as well as for persistence in the environment (15) (Text Box 2). What scenarios can be envisioned for the initial transition from a free-living to a host-associated lifestyle? How can recolonization in open and mixed symbioses persist at evolutionary timescales? From the symbiont’s perspective, the following scenarios are possible: (i) a selection-driven transition, where the host as a habitat or a means of dispersal provides an overall advantage in comparison to a free-living lifestyle; (ii) capture, where living in the host has a neutral or negative impact on the fitness of the symbiont but an immediate benefit for the host; and (iii) a neutral start, where none of the partners experience benefits in early stages of symbiosis.
Selection-Driven Transition
In the first case, where the microorganism benefits from being housed or vectored, symbiont adaptations favoring effective host colonization are under positive selection. These will, however, vary in strength according to the availability of host individuals and the presence of microbial competitors, since these will challenge successful colonization of the host as a valuable niche. At the same time, traits for survival in a temporary host or persistence in the transitional free-living stage, like nutritional independence, might also be favored. An open or mixed symbiosis will also give room for genetic replenishment and the maintenance of variation (for example, see reference 53). From the host’s perspective, environmental symbiont uptake entails the risk of failing to acquire beneficial symbionts or experiencing costly infections by symbionts shifting toward the detrimental end of the parasite-mutualism continuum (17, 35). However, an open symbiosis can also allow for rapid adaptation to changing conditions if the source of potential symbionts is reliable. Thus, while recurrent switching between host and environment can be a stage in the evolutionary transition leading to a closed system, it can also become established as a long-term viable strategy for symbiosis. This is possibly the case for the symbionts of squids, entomopathogenic nematodes, stinkbugs (Table 2), and potentially many other open and mixed symbioses. Notably, while adaptations on the host side to maintain environmental acquisition over generations can occur, these are not always necessary (15). For instance, plant-pathogenic microorganisms like phytoplasmas benefit from being vectored by various sap-sucking insects (59). While the insect does not necessarily benefit from hosting the microbe and acquires it passively, the microbe hitchhikes via the insect’s feeding habits and is likely to drive this insect-microbe association. In other words, the persistence of an open or mixed symbiosis can be driven solely by microbial traits. As recently discussed by Obeng and coauthors (15), the prolonged recurrence of host-environment switching enables selection to act on the different stages of the microbe’s biphasic life cycle. In this framework, evolutionary processes will promote a microbial lifestyle that is optimized for entry and persistence in the host but also for dispersal and survival under free-living conditions.
Capture
In a second scenario in which a host-associated lifestyle does not bring a net advantage to the microbial partner, symbiosis establishment is most likely asymmetrically driven by selection on the host level, favoring the capture of symbionts. Each transmission event between host individuals is thus an opportunity for the symbiont to escape and proliferate in the environment or other hosts. Therefore, traits that facilitate release into the environment are likely under selection in such a scenario. In this case, microbial adaptations enhancing establishment are unlikely, unless host-driven evolution of strict vertical transmission occurs, which would tie the symbiont’s fitness to its host’s. At this point, synergistic adaptations of host and symbiont could lead to a highly integrated association, as observed in many obligate symbioses (45). For instance, in cases where bacteria are incorporated into specialized tissues or cells, the symbionts might be alleviated from challenges imposed by the host immune system (for examples, see references 60 and 61) (Text Box 1). Under these conditions the host can also regulate bacterial metabolism by controlling access to substrates or by the coupling of metabolic pathways (25, 45, 62, 63). However, the bacteria are most often no longer capable of a free-living lifestyle, as in symbioses that are currently highly integrated. For example, Sitophilus cereal weevils exert a strong control over their intracellular symbiont “Candidatus Sodalis pierantonius.” In this symbiosis, knocking down host genes associated with the production of antimicrobial peptides in and around the symbiotic organ results in symbionts escaping the bacteriocytes, i.e., the cells usually harboring the symbionts (60, 64). Thus, the symbionts break free in the absence of host control. This suggests that in this case, the insect host and not the symbiont warrants maintenance in the symbiotic organ and might have driven symbiont confinement in the first place. More generally, it has been argued that intracellular symbioses are most often associations with unilateral benefits that are in many cases controlled by the host (14).
Neutral Start
A third case has been recently discussed in which host-microbe symbiosis can initially evolve without specific benefits for any of the partners (65). A modeling-based analysis focusing on Drosophila melanogaster and its microbiota highlighted the relevance of dispersal via the host and the impact of the microbes on host habitat, which in turn influences host development. While these aspects can in fact be considered indirect benefits, this approach is nonetheless a useful standpoint to experimentally assess whether a mutualistic association including bacterial adaptations enhancing host colonization can evolve under initially neutral conditions.
A spectrum spanning the three scenarios—selection-driven transition, capture, or neutral start—likely exists, based on the observations on dependence and genomic architecture across symbiotic systems. However, we know little about the general benefits of colonization traits for beneficial bacteria and even less about their costs. Our understanding of molecular mechanisms for bacterial colonization in hosts has been heavily influenced by pathogen research and has only begun to actively expand to beneficial associations, or bacteria shifting across the parasite-mutualist continuum (57). While the phenotypic effects of successful colonization have been intensively studied for both host and bacteria, the ecological consequences and evolutionary drivers of initiating a host-associated lifestyle in beneficial bacteria remain poorly understood. However, there is considerable work on the molecular underpinnings of symbiosis establishment in a few symbiotic model systems and growing capacities to explore similar questions in nonmodel organisms (Text Box 3). As outlined in the section Molecular Mechanisms for Symbiont Colonization, this work is paramount for better understanding the drivers and constraints of transitioning to a host-associated lifestyle.
TEXT BOX 3: GENETIC MANIPULATION TO UNRAVEL SYMBIONT COLONIZATION FACTORS
Mutations inducing loss or gain of functions are key for the identification of specific genes involved in a biological process, like host colonization. Targeted (66) and random mutagenesis techniques (67) have revealed a plethora of molecular mechanisms involved in bacterial infection and pathogenesis (68–70). For a number of beneficial bacterial symbionts, however, the inability to cultivate these in vitro hinders genetic manipulation (71). Cultivation has been nonetheless successful for several invertebrate symbionts, like Vibrio fischeri (72), Aeromonas veronii (73), Photorhabdus luminescens (74), Cabelleronia insecticola (75), Snodgrassella alvi (76), and Sodalis glossinidius (77). In these systems, a combination of targeted and/or random mutagenesis techniques are now available. For example, a versatile toolkit for genetic manipulation using broad-host-range plasmids focusing on the honey bee and bumblebee gut microbiome was recently developed (78). This includes a set of tailored constructs allowing for disruption of specific genes using CRISPR-Cas9, as well as heterologous expression of fluorescent reporters, antibiotic resistance markers, or other genes of interest in various proteobacteria. The toolkit has also helped in engineering S. alvi symbionts to activate the host’s internal RNA interference (RNAi) machinery and modulate host gene expression (79).
Another method widely used in studying symbiont molecular mechanisms is the generation of random mutant libraries using transposon insertions. Transposons allow for individually disrupting genes across the genome through a “cut-and-paste” mechanism, and many mutants can be generated in a single transformation event. Once a diverse library of mutants is available, the fitness of all mutants can be assessed simultaneously under the desired experimental conditions. Based on this approach, the composition of mutants in a library infected into a host organism will change in diversity and frequency in comparison to that grown in vitro. Additionally, screening or negative selection can be used to separate individual mutants. For example, by plating the library on low-percentage agar media and screening for mutants that are less motile or hypermotile, novel genes involved in bacterial motility can be discovered (80, 81). Similar approaches have been used in a number of invertebrate symbionts to identify other phenotypes, including a study in S. glossinidius in tsetse flies (82) and one in Caballeronia in R. pedestris bean bugs (83). The advent of next-generation sequencing technologies has furthered the development of efficient approaches to track relevant genes in these random mutant libraries. In particular, the use of transposon insertion sequencing (Tn-seq) is valuable in assessing the changes in individual mutant frequencies by insertion-directed sequencing in a high-throughput manner. Tn-seq experiments to identify symbiont genes essential for host colonization have been performed in various beneficial bacteria, including S. alvi symbionts of honey bees (84) and V. fischeri symbionts of squids (85), as well as in associates of vertebrates (86) and plants (87–89). Recent advances combining droplet-based microfluidics and Tn-seq have proven useful in studies where population effects and cell-cell competition may interfere with the identification of specific single-cell phenotypes under certain conditions (90). Another advancement is the use of CRISPR/Cas9 genome editing for targeting genes in nonmodel microbes. For example, this approach was effective in knocking out the ompA gene and also in integrating fluorescence and gentamicin resistance markers in Cedecea neteri gut symbionts of Aedes mosquitos (91). CRISPR interference (CRISPRi) is an emerging method yet to be applied in beneficial symbiosis. It enables the transcription repression of target genes without modifying the target site (92, 93). In CRISPR, a guide RNA complementary to a DNA sequence points to the target where the Cas9 nuclease protein must cleave. In contrast, in CRISPRi, the Cas9 protein is catalytically deactivated (dCas9) to downregulate the expression of the target gene. Since CRISPRi is inducible, growth essential genes (94, 95) and temporally essential genes (95, 96) may be studied, which is not possible with targeted or transposon mutagenesis techniques (93, 97, 98).
MOLECULAR MECHANISMS FOR SYMBIONT COLONIZATION
Reaching the Host: Motility and Chemotaxis
In most open symbioses, initial contact and entry into the host can depend on the ability of the microbe to sense and move toward a chemical cue (99). In fact, from land to the deep-sea waters, most described bacterial symbionts having a free-living stage retain the genes essential for motility at least until they migrate into the symbiotic structures of the host. The role of these machineries has been experimentally addressed in Vibrio fischeri bacteria associated with Euprymna scolopes, the Hawaiian bobtail squid, a key model system for the molecular cross talk in establishment of environmentally acquired symbionts. Bioluminescent V. fischeri is considered a beneficial symbiont, as it provides counterillumination, a form of camouflage, for the host to evade predators under the moonlight (100). During the first steps of the colonization process in the squids, V. fischeri cells from the seawater aggregate in mucus secreted by the squid host. Once V. fischeri cells have aggregated on the epithelial surface of the squid, they navigate along a chitobiose gradient, which is a chemoattractant established by the host, and move toward the crypts of the light organ (101–103), where the bioluminescent cells are finally housed (Fig. 2A) (104–106). Concordantly, mutations in the cheY and cheR genes impair colonization of the light organs, supporting the hypothesis that chemotaxis is required for Vibrio to navigate to its final destination in the squid (107). Motility is also key in this stage, as V. fischeri mutants that are immotile or contain a disrupted putative homolog of the V. cholerae flagellar response regulator, flrC, cannot colonize the host (107). In other systems, mucins and glycoproteins that are constituents of host mucus can act as chemoattractants, enabling pathogens to colonize the mammalian mucus-lined intestine (108). Also similar to the V. fischeri symbionts, related pathogens like Vibrio cholerae, Helicobacter pylori, and Campylobacter jejuni rely on motility to penetrate the mucus layer of the intestinal epithelium in mammalian hosts (108).
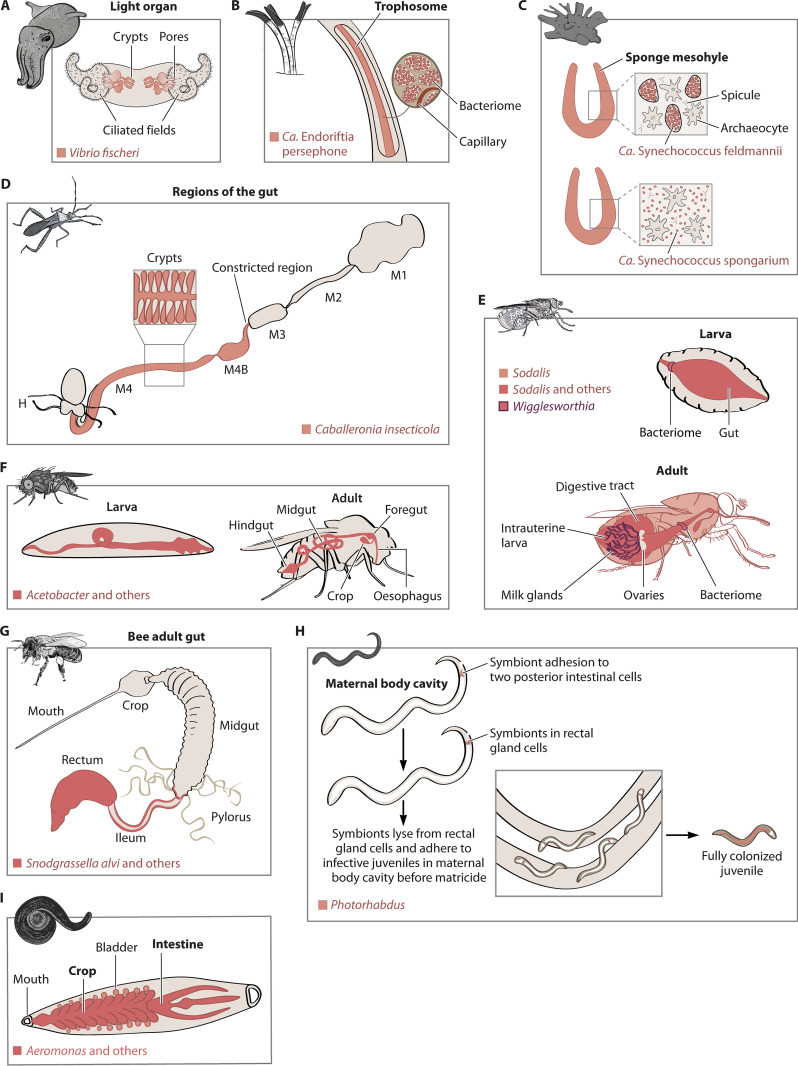
FIG 2.
Localization of beneficial symbionts in the following invertebrate systems discussed in the review: bobtail squids (based on reference 105) (A), giant tubeworms (based on reference 264) (B), sponges (based on references 265 and 266) (C), broad-headed bean bugs (based on reference 267) (D), tsetse flies (based on reference 268 and 269) (E), fruit flies (based on reference 270) (F), honey bees (based on reference 271) (G), entomopathogenic nematodes (based on reference 152) (H), and leeches (based on reference 272) (I). Red highlights indicate the specific location of a symbiont in an organ/body of the host. Multiple symbiotic strains or species may sometimes colocalize together, which is indicated in darker red.
In symbionts of other marine invertebrates, functional genomics and comparative studies suggest that motility and chemotaxis are also important for colonization initiation. A metagenome analysis of “Candidatus Endoriftia persephone” endosymbionts that colonize bacteriocytes in the giant tubeworm Riftia pachyptila (Fig. 2B) revealed that the symbionts carry a wide array of chemoreception and motility genes (16, 109). In Petrosia ficiformis sponges, the facultative intracellular symbiont “Candidatus Synechococcus feldmannii” (Fig. 2C) carries a motility-related pilus retraction ATPase gene, pilT (110). Strikingly, this gene is absent in the obligate extracellular symbiont “Candidatus Synechococcus spongiarum” (Fig. 2C), pointing to a different way of colonization (110). However, direct evidence for the role of motility is lacking and challenging to obtain in this and other systems that are not amenable to manipulation.
Terrestrial environments present considerable limitations to bacterial movement. Hence, on land, the first encounter of free-living symbiotic bacteria with a host might be more spatially restricted. To initiate contact under these conditions, bacteria can benefit from alternative mechanisms, such as their host’s behavior and biology. This is particularly observed in microbe-insect interactions, where the symbionts can take advantage of trophallaxis, coprophagy, and egg surface smearing (35, 111). Presumably, symbionts hitchhike via these behaviors that serve nutrient exchange, although the evolutionary drivers could be both nutrition and bacterial transmission. In fact, the presence of bacteria might promote these behaviors in some cases, as observed in cockroaches. Feces from artificially created axenic individuals are less attractive to conspecifics due to differences in the volatile blend, usually containing aggregation pheromones. Thus, the bacteria promote this gregarious behavior and thereby increase their chances for transmission among cockroach hosts (112).
After gaining entry, bacteria must still migrate to and/or enter the specific symbiotic organs or target niches to establish an association in terrestrial hosts. In Riptortus pedestris bean bugs, Caballeronia (previously Burkholderia) symbionts are orally acquired by nymphs (113) and they require functional flagellar motility to enter the M4 midgut crypts where they are housed (114, 115) (Fig. 2D). Related opportunistic pathogens, like Burkholderia cepacia and Burkholderia glumae, also utilize flagellar motility to invade the host (116, 117), which is common across many pathogenic bacteria (118). Interestingly, in vitro assays demonstrate that the bug’s Caballeronia symbionts have developed a specialized corkscrew-like motion (119) in which the flagellum, wrapped around the cell body, thrusts the cell forward. The same study identified a similar mechanism in a Vibrio fischeri strain, implying that this type of flagellar motility might also be relevant for other bacteria (119). The wrapped flagellum is not essential for penetrating the constriction region, but it is speculated that it improves the efficiency of movement through the mucus in this region (119).
While being motile can be, unsurprisingly, important for beneficial bacteria that recurrently colonize the host, it is widely observed that vertically transmitted endosymbionts lose motility genes or, as in Buchnera aphidicola, flagellar genes may be involved in protein transport (120, 121). There are exceptions, however, like the Wigglesworthia symbionts of tsetse flies. These bacteria reside intracellularly in bacteriocytes near the anterior midgut and are also found extracellularly in the milk gland lumen, from which they are vertically transmitted to the intrauterine progeny via a milk gland (122) (Fig. 2E). Here, expression of motility and flagellar genes is upregulated during maternal transmission and larval intrauterine development, suggesting motility as an important factor for colonization of the offspring (122, 123).
In the case of obligate, vertically transmitted endosymbionts, symbiont translocation is often achieved through transport mechanisms of the host (124, 125). Therefore, the endosymbionts might lose genes essential for motility and chemotaxis as a more consistent association with the host evolves (98). For example, free-living Serratia symbiotica bacteria that are transitioning from pathogenic to beneficial lifestyles in aphids have lost swimming motility and chemotaxis while retaining some genes important for interaction with the host (126). Similarly, a defensive symbiont of Lagria villosa beetles facing ongoing genome reduction lacks the genes required for chemotaxis and flagellar motility (127). In a relatively open symbiotic association, a comparative analysis shows that flagellar motility is lost in Acetobacter bacteria that reside in the gut of laboratory-reared Drosophila fruit flies (Fig. 2F), while it is retained in those strains isolated from wild-caught flies. This suggests that motility is not advantageous in the laboratory, where strains remain in close contact with the host through several generations of lab rearing, but it may facilitate colonization and establishment in a more heterogeneous natural environment (128). Loss of flagellar motility or the structural components may also be an evolutionary adaptation to avoid activation of host innate and adaptive immune defenses, since flagellins are recognized as immune activators in plants and mammalian cells (129, 130).
In summary, motility appears to be a valuable tool for beneficial bacterial colonizers both outside and inside an animal yet can quickly become dispensable for symbionts evolving toward tighter associations with the host.
Engaging with the Host
Surface structures interacting with the host.
Bacterial surface structures are key during colonization, as they mediate the cell’s contact with the external environment. These structures are involved in partner recognition and signaling, symbiont immune evasion, and mediation of adhesion to host tissues (see also Text Box 1). Surface components like lipopolysaccharide (LPS), tracheal cytotoxin (TCT) from the peptidoglycan layer, and exopolysaccharides are termed microbe-associated molecular patterns (MAMPs), as they can interact with MAMP recognition proteins of the host (131, 132).
In the squid symbiont, V. fischeri, MAMPs are important for signaling the bacterial presence to the squid host. Although unspecifically, bacterial surface peptidoglycans in the seawater trigger mucus secretion by the host even before colonization (104). Also, despite being part of the cell wall peptidoglycan, TCT can be released and act at a distance. TCT triggers hemocyte infiltration into the ciliated epithelial fields and induces apoptosis of the epithelial cells that make up the fields of the squid light organ (133), leading to the restructuring of host ciliated appendages (131, 133), as also mentioned in “Making Space: Induction of Host Morphological Traits Relevant to Colonization.”
In other symbionts, surface structures and, in particular, LPS are crucial for colonizers to avoid detection by the host immune components during entry. Anchored to the outer membrane of the cell, the LPS consists of three regions—lipid A, core oligosaccharide, and O antigen—and acts as a protective barrier in pathogens and mutualists against a harsh external environment (134). The host immune system recognizes LPS when it is released from the outer membrane due to cell death or removed from the outer membrane by the host lipid A binding protein (LBP). To avoid recognition by the host immune system, some bacteria exhibit changes in the chemical structure of LPS, such as alterations in acylation patterns and phosphorylation of the lipid A structure, or variations in the O-antigen polysaccharide structure (135). Phosphorylation changes the net charge exposed on the cell surface and thus affects the interaction of bacterial cells with the environment, AMPs, and antibiotics (134). In the Caballeronia symbionts of the Riptortus bean bug, an intact O antigen helps protect Caballeronia symbionts from cationic antimicrobial peptides of the host until the symbionts reach the symbiotic midgut region (136). It is speculated that after colonization, the host induces loss of the O antigen to maintain control over symbiont titers (137), implying that modification of the surface structure may be essential for symbiosis establishment. The LPS core, O antigens, and genes involved in their assembly are also speculated to help in immune evasion or to aid in colonization in other beneficial symbionts including Snodgrasella alvi in bees (84), Photorhabdus in entomopathogenic nematodes (138), Sodalis glossinidius in tsetse flies (20), and Aeromonas in leeches (139).
Adhesion.
Surface structures that help in attachment to the host are widely important for horizontally transmitted symbionts. This is especially true in the marine environment, where free-swimming cells face the threat of being washed away by currents, but also in the gut, where the same occurs with the passage of food by peristaltic movement (140). Therefore, adhesions mediated by pili or fimbriae, curli proteins, or trimeric autotransporter adhesin (TAA) or through biofilm formation (see “Coping with a New Environment: Stress Response”) are important for mutualists and pathogens alike (141).
Pili or fimbriae are hairlike appendages that are found on the bacterial cell surface. While these terms are often used interchangeably, there are some general distinctions: fimbriae are shorter than pili and may not be involved in transfer of DNA (142) (see the GLOSSARY in the APPENDIX). Among these structures, type IV pili are especially relevant for adhesion to host cells, biofilm formation, twitching motility, and protein transport. These structures have been widely studied in pathogens like Pseudomonas aeruginosa, Neisseria gonorrhoeae, Vibrio cholerae, and Clostridium sp. (143, 144). In a similar fashion, type IV pili may aid host colonization in a number of host-beneficial bacteria of invertebrates. They are predicted to be essential for colonization by S. alvi symbionts in the gut of the honey bee, Apis mellifera (Fig. 2G) (84, 145), akin to the related pathogen Neisseria gonorrhoeae, which also employs the type IV pilus to adhere to the host epithelium (146). Among beneficial symbionts, the role of pili in adhesion to host tissues extends to those associated with earthworms (147), entomopathogenic nematodes (148), humans (149, 150), and sponges (110). Interestingly, in “Ca. Synechococcus” symbionts of sponges, the relevance of pili likely relates to the transmission route of the bacteria. Although intracellular, the facultative and presumably horizontally acquired “Ca. Synechococcus feldmannii” (Fig. 2C) retains type IV pilus genes (110). By contrast, these are absent in the congeneric “Ca. Synechococcus spongiarum,” which is an obligate and vertically transmitted symbiont despite its extracellular location (110). It is likely that for the latter, the stable transfer over generations obviates the need for attachment mechanisms. A putative pilus-encoding gene in V. fischeri offers a competitive advantage during cocolonization with other strains in the squid light organ, while the absence of the gene does not affect individual mutants during colonization (151). This diversity of systems underlines the relevance of pilus-mediated adhesion for beneficial bacteria that must repeatedly colonize the host and thus retain this feature from free-living or pathogenic lifestyles.
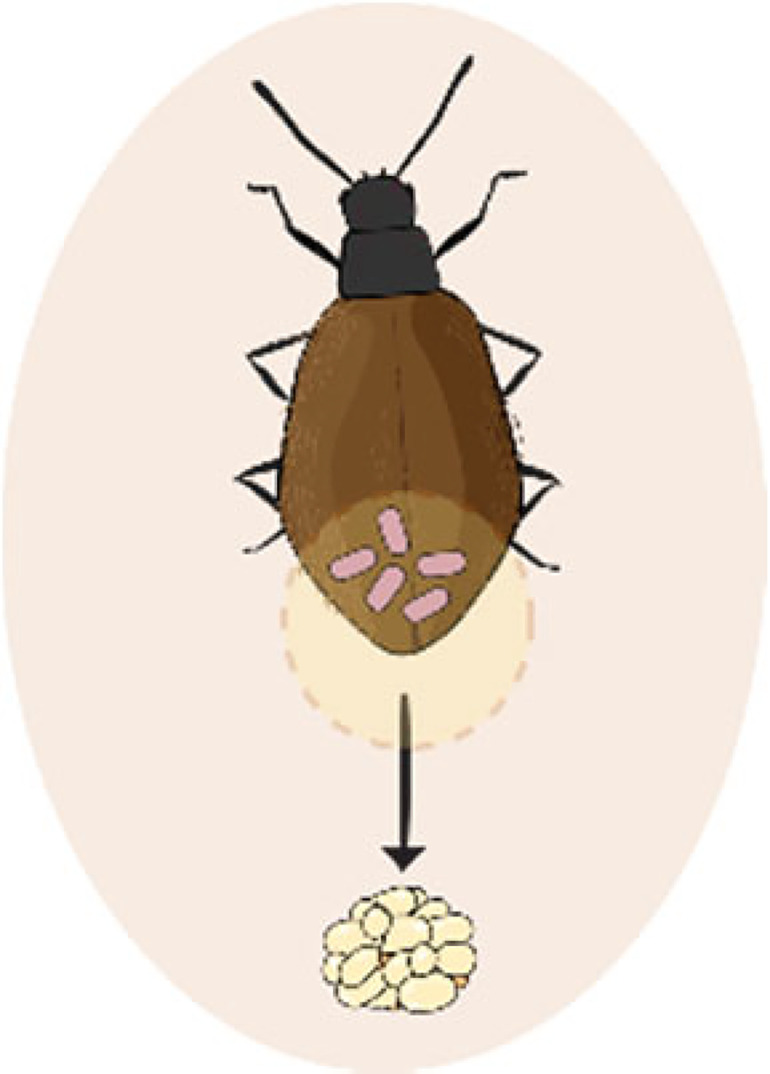
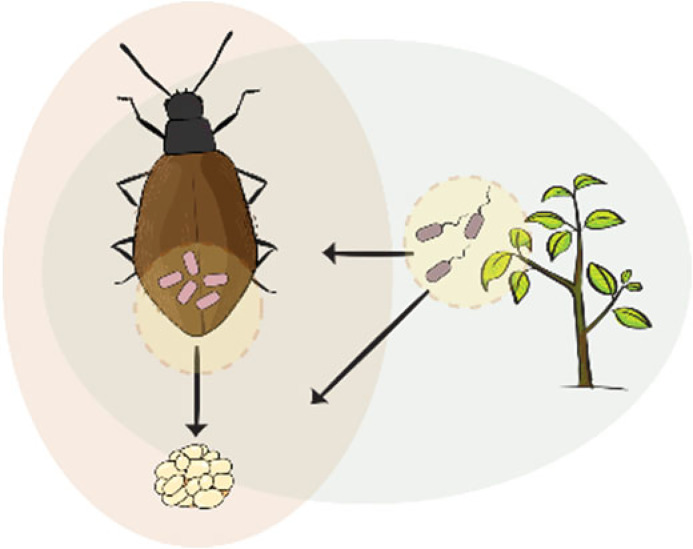
In addition to type IV pili, the role of fimbriae, chaperone-usher pili, curli fibers, and TAA for adhesion in host-associated bacteria is well characterized (141). A relevant case study occurs in Heterorhabditis bacteriophora entomopathogenic nematodes, where the symbiont Photorhabdus luminescens is maternally transmitted to the developing juveniles in the mother’s body cavity (Fig. 2H). The symbionts use the mad (maternal adhesion defective) fimbrial locus for binding to the intestine of the maternal nematode (152). Cells that can adhere to and invade the adult rectal gland cells are preferentially transmitted to the offspring developing inside the body cavity (153). Notably, an invertible promoter controls an ON-and-OFF switch that regulates this fimbrial locus. This switch mediates the bacterial transition between a mutualistic form in the host nematode and an insect-pathogenic form in prey insects, which is key for the entomopathogenic phase of the nematode’s life cycle (154).
Other relevant adhesion factors in the context of mutualistic bacteria include type Vc secretion system adhesins and eukaryote-like proteins such as fibronectin type III (FN3), cadherin, and leucine-rich repeat domain-containing proteins, which have been reported in bacteria from the sponge microbiome, although evidence for their role in symbiont colonization remains indirect (110, 155). For example, FN3 domains are enriched in sponge-associated cyanobacteria in comparison to free-living strains of the same group (110). FN3-containing proteins are possibly involved in binding to glycoproteins and structural proteins on host cells (156). Fibronectin-like proteins are also known to be essential for attaching to the host epithelial cells in pathogenic bacteria like C. jejuni (157), as well as in probiotic Lactobacillus attaching to the mammalian gut (158).
Nonfimbrial and nonpilus adhesins like TAA are secreted through the outer membrane in Gram-negative bacteria, specifically by the type Vc secretion system (159). TAA is a common family of adhesion factors often associated with virulence in bacteria (160). In one of the gut symbionts of honey bees, S. alvi, TAA possibly aids in binding to the bee gut epithelium, as predicted by a genome-wide screening analysis using a mutant library (84). TAA could be more commonly implicated in symbiont establishment, as homologous proteins have also been found in the genome of Burkholderia Lv-StB symbionts of Lagria villosa beetles (127), although the importance of TAA as colonization factors in these systems remains unexplored.
Biofilm formation.
Adhesion often precedes the formation of a biofilm, an important strategy for microbial colonization on surfaces, including those within a host (161). Subsistence in a biofilm can entail important benefits such as increased protection from antimicrobial substances, including host immune factors (Text Box 1), occupation of nutrient-rich areas, and facilitated cooperation (162, 163).
Both pathogenic and beneficial bacteria in a broad range of animal hosts—within and beyond invertebrates—rely on the formation of biofilms upon attachment to settle in the host tissues. In Euprymna squids, for instance, formation of a biofilm is crucial for the V. fischeri symbionts to pass through the squid ciliated epithelial area before entering the light organ pore (Fig. 2A). The production of the extracellular “symbiosis polysaccharide” (Syp) by the bacteria (164), as well as several regulator molecules (85, 165), enables biofilm formation. In entomopathogenic nematodes, the formation of a biofilm allows Photorhabdus bacteria to become established in the posterior end of the gut before invading cells in the rectal gland epithelium (166) (Fig. 2H). Similarly, there are indications that S. alvi gut symbionts of honey bees also form biofilms that facilitate host colonization and rely on adhesion factors for their formation (84). Caballeronia symbionts of the bean bug R. pedestris also rely on biofilm formation for proper establishment in the insect gut (167), similar to related pathogens of the genus Burkholderia that produce a biofilm for successful infection (168).
Polymicrobial settlement in biofilms can be another key phenomenon in the gut or on other host surfaces. Several studies indicate that the presence of a polysaccharide matrix formed by already existing bacteria can facilitate the recruitment of new associates, even when they are themselves impaired in biofilm formation. The possibility of attaching to other bacteria instead of directly to the host can thus spare the need of host-directed adhesins (161). As observed in leeches, and further discussed in “Dealing with Third Parties: Competition and Facilitation among Symbionts,” the bacterial polymeric matrix might also promote important cross talk between coexisting symbionts (169).
Secretion systems.
Secretion systems have been widely studied in beneficial and pathogenic bacteria and are known to enhance communication with the host or to mediate interbacterial warfare.
The type II secretion system (T2SS) has several evolutionary similarities to the type IV pilus, flagella in archaea, and competence pili in Gram-positive bacteria (170). Unlike the type III and type VI secretion systems (T3SS and T6SS), the T2SS secretes proteins into the extracellular environment for adhesion and biofilm formation, lysis of host tissue, or to remodel the environmental niche (171). An example of the use of T2SS by mutualistic bacteria includes Aeromonas veronii symbionts found in the leech crop (Fig. 2I). A. veronii secretes hemolysin via a T2SS to lyse a part of the ingested erythrocytes in the blood meal of the leech and utilize it as a heme source. The absence of hemolysis activity in A. veronii renders them incapable of colonizing the leech crop. Similarly, T2SS in a related pathogen, Aeromonas hydrophila, is important for pathogenesis by secretion of putative virulence proteins, including hemolysin (172).
The T3SS is known in symbiosis as an important colonization determinant. Several T3SS components are similar to those of the flagellar proteins, and the T3SS apparatus spans the bacterial cell membrane. When in contact with the host cell membrane, the T3SS injects effector molecules into the eukaryotic cell (173). It helps bacteria gain entry into the host cells, and modulate signaling processes in the host or avoid host innate immune factors. An example of a microbial symbiont that uses the T3SS for immune evasion is A. veronii. There, the T3SS helps avoid phagocytosis by leech hemocytes circulating in the intraluminal fluid of the crop and is additionally involved in pathogenesis in mammalian hosts, showing that the T3SS has a dual role as a colonization factor and a virulence factor in different hosts (174). Among the Aeromonadaceae, apart from the beneficial symbiont of leeches, pathogens such as Aeromonas salmonicida and A. hydrophila carry a T3SS (175). The use of the T3SS for modulation of the host immune response is also observed in systems involving taxonomically distant hosts, like leguminous plants. There, nitrogen-fixing rhizobia use the T3SS to inject Nops (nodulation outer protein) effectors to suppress host immune responses and become established in the root nodules of host plants. However, in incompatible rhizobium-legume combinations, Nops effectors can also affect nodulation negatively and prevent infection, showing their role in specific colonization and their similarity to plant-pathogenic effectors (176). Other bacteria are capable of inducing changes to the host cytoskeleton and interrupting host signaling processes for immune suppression using the T3SS, including enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli (EHEC), and Yersinia pestis (173, 177).
S. glossinidius symbionts of tsete flies use the T3SS machinery to gain entry into host cells. There, the T3SS regulated by the PhoP-PhoQ two-component system affects the ability of the bacteria to colonize the host fly (178, 179). Pathogens like Salmonella, Shigella, and Chlamydia (173) employ a factor similar to that of the S. glossinidius symbiont of tsetse flies in establishing an intracellular lifestyle using the T3SS. Interestingly, S. glossinidius has only recently transitioned to an endosymbiotic lifestyle in tsetse flies, and it retains a T3SS machinery that shares a common ancestry with Salmonella enterica and Shigella flexneri invasins. This machinery now facilitates the symbiont’s vertical transmission from the hemolymph to the intrauterine progeny (82). The T3SS also seems to be a conserved feature in a close relative of S. glossinidius, S. pierantonius. This intracellular symbiont of cereal weevils uses the T3SS for translocation during host metamorphosis. The endosymbiont undergoes a transient extracellular phase, as a few cells migrate from larval bacteriocytes to stem cells that are precursors of adult bacteriocytes (61).
Other than the T3SS, in the squid symbiont, V. fischeri, the two-component secretion system TamAB is likely to be involved in host colonization (85). Furthermore, as addressed in detail in “Dealing with Third Parties: Competition and Facilitation among Symbionts,” the T6SS is also now recognized as important machinery for animal-associated microbes. It mediates interactions with both the host and cocolonizers in mutualistic and pathogenic bacteria (180).
Making Space: Induction of Host Morphological Traits Relevant to Colonization
In many symbioses, spatial confinement or compartmentalization of the microbial partners in host tissues or organs is important for isolating and regulating the symbionts, maintaining partner fidelity, and/or avoiding direct conflict between different symbionts in a single host (28, 181). In some hosts, the formation of symbiont-housing organs is hardwired, potentially due to coadaptations during the evolution of the symbiosis, and does not require cross talk with the symbionts to trigger its development (for examples, see references 182 and 183). However, in other cases, such morphological alterations are induced in the presence of a specific symbiont enabling successful establishment. Over two decades ago, it was shown that V. fischeri symbionts trigger the postembryonic development of the light organ, where the bacteria will later reside (184) (Fig. 2C). The colonization of the crypts induces major changes in the light organ tissues, including swelling of the epithelial cells, increased microvillar density, and later apoptosis and regression of the ciliated field, where symbionts are recruited. This process begins within hours after first contact between the host and the bacteria and is orchestrated through an impressive molecular dialogue between symbiont and host, which has been characterized in detail (105, 185–187). During this process, actin rearrangements trigger the light organ formation in squids by V. fischeri (185, 187). Similarly, some pathogens of mammals (188, 189) or plants (190, 191) also target actin in the host cell cytoskeleton by secreting proteases and other effectors that manipulate the cellular framework, promoting invasion and dissemination (192). While defined and in some cases highly specialized symbiont-housing organs are present in many other animal and plant hosts (181), the role of symbiont molecular factors that might induce their formation is known for only a few model systems.
Symbiont-induced morphogenetic changes can also enhance partner specificity during colonization. In the bean bug R. pedestris, the passage of Caballeronia symbionts or related strains through a constricted region leading to the crypt-bearing section of the midgut provokes its closure (Fig. 2D). This blocks the subsequent entry of nonspecific bacteria and thus reinforces partner choice during colonization (193, 194). As noted above, a similar finding was reported in Euprymna squids, where V. fischeri triggers the constriction of a passage through which symbionts must navigate to reach the crypts. As in the bugs, this bacterium-induced mechanism is relevant for temporal and spatial regulation of colonization, in addition to increased specificity and compartmentalization of the symbionts (185).
Coping with a New Environment: Stress Response
Even though the host might offer a stable environment for some symbionts, colonizing bacterial partners can experience extreme and/or fluctuating conditions in terms of temperature, pH, oxygen concentration, nutrient supply, and oxidative stress. Bacteria can mitigate stress with the help of chaperones (195, 196). In intracellular symbionts, genes encoding chaperones are commonly the most highly expressed genes (197), which indicates that symbionts experience a stressful environment. However, extracellular symbionts that experience some degree of genome erosion and related suboptimal codon usage also seem to require increased assistance of protein refolding via chaperones (198). For example, the tsetse fly symbiont S. glossinidius must react to rapid fluctuations in temperature due the intake of warm blood by the fly, which causes thermal stress. As a response to elevated temperatures, the symbiont upregulates the expression of the chaperones dnaK, dnaJ, and grpE (199), which are known to aid bacterial cells in coping with raised temperatures (200) by the stabilization and refolding of denatured proteins (201).
A common threat to bacteria residing within host tissues are reactive oxygen species (ROS) that can originate from the environment, the host, or the bacteria themselves (202). Highly reactive molecules such as hydrogen peroxide (H2O2), superoxide anions (O2•−), and hydroxyl radicals (OH•) have a dramatic effect on the structure and activity of proteins, DNA, and membrane lipids (203). Consequently, most organisms utilize enzymes to transform ROS into a nonharmful state and repair cellular damage (204–206). For the tsetse fly symbiont, the high temperatures related to blood intake could additionally result in such oxidative stress (207), to which S. glossinidius reacts with the upregulation of genes involved in the breakdown of ROS, repair of oxidative damage, transport of iron and manganese, and protein refolding (208), under the control of the regulatory N-(3-oxohexanoyl)homoserine lactone (OHHL).
In a second example, the Hawaiian bobtail squid E. scolopes regulates bacterial colonization by releasing nitric oxide synthase (NOS) and its product, nitric oxide (NO), at the epithelia of the superficial ciliated fields, ducts, and crypt antechambers (209). Furthermore, it provides aggregation mucus with vesicles containing NO and NOS to limit the number of bacteria (209). V. fischeri uses the flavohemoglobin Hmp for protection against the inhibition of aerobic respiration caused by NO (210). In the absence of NO, Hmp is expressed at a low level but is upregulated up to 120-fold in its presence (211). Flavorubredoxin produced by V. fischeri can also combat NO, albeit to a lesser extent and only under anaerobic conditions (210). Additionally, V. fischeri detoxifies ROS-damaged peroxidized membrane lipids by upregulating multiple genes during colonization stages inside the host (212). Once inside the light organ crypts, high expression of multidrug efflux pump genes suggests that the symbiont might actively expel antimicrobial compounds present in the crypts, a process that is probably coordinated by quorum sensing (212, 213). The aforementioned NO also plays a role during colonization in other symbiotic systems, e.g., in the legume-rhizobium symbiosis (214) and possibly in the beewolf-Streptomyces symbiosis (215). This suggests that withstanding oxidative stress is likely important for establishment in several host-associated microbes.
S. alvi bacteria in the honey bee gut must also cope with various environmental challenges. Powell et al. (84) deduced that during the first 5 days of colonization, multiple stress response mechanisms are likely important and speculate on candidate genes that might enable these mechanisms based on infections with a random mutant library. This includes genes involved in modifying rRNA and tRNA, which could improve translation efficiency and fidelity and thereby mitigate nutritional and temperature stress (84). Furthermore, the expression of different enzymes responsible for amino acid synthesis and amino acid transporters can potentially be adjusted to help safeguard cell survival during nutrient limitation, a strategy previously shown in E. coli (216, 217). Additionally, factors involved in protein recycling and stabilization might be essential to ensure protein quality. S. alvi might also counteract ROS-inflicted DNA damage and repair DNA breaks by expressing a pathway that includes genes for the SOS response, recombinational repair, D-loop extension, resolution of Holliday junctions, and postrepair chromosomal separation. Furthermore, the antioxidant glutathione might play an important role in gut colonization, as suggested by the expression of genes for its synthesis and activity (84, 218).
Cell wall integrity can also play an important role in stress tolerance during establishment in the host. In S. alvi, outer membrane stress appears to be mitigated by the regulation of LPS synthesis and export (137). In the case of Caballeronia symbionts of Riptortus bean bugs, the deletion of a crucial gene for cell wall biosynthesis (uppP) results in an altered peptidoglycan structure, a higher sensitivity to lysozyme activity, and environmental stressors in vitro and failed initiation of symbiosis in vivo (219). This implies that the absence of the gene may expose the cells to bactericidal agents in the gut, although direct evidence is lacking (219). However, other examples of cell wall integrity changes related to stress in beneficial bacteria while colonizing extracellularly are to our knowledge lacking. In highly integrated intracellular symbionts of some insects, the host has horizontally acquired bacterial genes for cell wall metabolism and thereby gains increased control over the host-microbe interaction (for examples, see references 220 and 221).
MULTIPARTITE MICROBIAL INTERACTIONS AND THEIR ROLE IN COLONIZATION
Dealing with Third Parties: Competition and Facilitation among Symbionts
Besides the interaction with the host, colonization success and its specificity also depend on the interplay between bacterial competitors. Inter- or intraspecific interactions between bacterial colonizers can be facilitating, when one individual positively influences another directly, for example, by supplying nutrients or collective protection in a shared biofilm (222) or indirectly by modifying hosts resources, behavior, or immune responses (223). Alternatively, antagonistic interactions can involve exploitative competition through higher growth rate, enhanced motility or resource utilization, or interference competition in the form of antibiotic production, signaling disruption, or predation (224, 225). Importantly, as with host-microbe interactions, the outcome of an interaction between bacterial players can be context dependent and vary along a continuum from mutualism to antagonism (10).
Work in the Riptortus-Caballeronia symbiosis directly demonstrates how competition can play an important role in preventing other bacteria from colonizing and increasing specificity in the association (44, 226). While some closely related Pandoraea bacteria are able to bypass the selective machinery imposed by the host, these bacteria are quickly outcompeted by the Caballeronia symbiont in the M4 gut upon coinfection (226). While the specific mechanisms are unclear, Itoh et al. (226) speculate that the host environment is tailored toward housing its beneficial Caballeronia symbiont, providing it with a competitive advantage over other microbes.
In other cases, molecular mechanisms mediating interference competition by cocolonizing bacteria have been identified. For example, effector proteins associated with the T6SS are known to facilitate antagonistic interactions between bacterial strains in some symbiotic systems (227). The T6SS is a contractile apparatus built by a sheath tube of protein subunits (TssB and TssC), within which are stacks of hemolysin coregulated protein (Hcp) that help to transport effector molecules. At the tip, the Valine-glycine repeat protein G (VgrG) syringe serves to puncture bacterial competitors and deliver effectors (227). Direct competition between coinfecting bacteria influenced by the T6SS has been described at the molecular level in the squid-Vibrio symbiosis. Here, multiple strains compete over establishing a symbiosis within the crypts of the nascent light organ. At least three strains utilize a T6SS to eliminate competitors (228). This is promoted by upregulating the expression of Hcp and controlled by the alternative sigma factor σ54 and the bacterial enhancer binding protein VasH (229). In bees, S. alvi and coinfecting Gilliamella apicola symbionts conceivably engage in interbacterial competition during colonization, as suggested by upregulation of genes coding for a T6SS and various recombination hot spot (Rhs) toxins with antimicrobial activity (84, 230). However, other S. alvi and G. apicola strains lack the T6SS or Rhs genes, indicating that interbacterial interactions of core symbionts in the bee gut may also involve alternate mechanisms. The T6SS machinery also interacts with the host and cocolonizing bacteria in pathogenic bacteria. Pathogenic relatives of beneficial symbionts, such as Burkholderia thailandensis, Pseudomonas aeruginosa, and Vibrio cholerae, target host macrophages during infection and/or mediate bacterial warfare using different classes of T6SS (180).
Bacterial competition also operates as a screening process during symbiont colonization. Such a scenario has been modeled (231) and experimentally supported (232) for the defensive symbionts of attine ants. In this system, antibiotic-producing actinomycete bacteria are hosted on the insect cuticle and protect the ants’ fungus garden from parasites (233). While the beneficial actinobacterial symbionts can be vertically transmitted, this mixed symbiosis allows for the entry of multiple other bacterial strains. Nonetheless, rich host-provided resources may fuel interference competition, and the priority effects of vertical transmission also play an important role, as discussed in the following section on Impact of Priority Effects on Symbiont Colonization. The antibiotic producers thereby likely prevent nonproducers from successful colonization (232). However, other antibiotic producers can be picked up horizontally and subsequently benefit the host by adding their metabolites to the antibiotic cocktail. This shows that at least for antibiotic-based defensive symbioses, competition not only drives community assembly but also may screen for beneficial symbionts.
In contrast to the antagonistic scenarios described above, facilitation aids cocolonization of A. veronii and a Rikenella-like bacterium in the digestive tract of the medical leech Hirudo verbana (169) (Fig. 2I). Mixed microcolonies of the two bacteria are covered by a polysaccharide matrix that provides an oxygen-depleted environment for the obligate anaerobe Rikenella-like bacterium. In addition, the matrix promotes nutrient transfer between the two cocolonizing bacteria (234, 235). Thus, both facilitative and competitive interactions can shape the assembly, composition, and specificity of host-associated microbial communities.
Impact of Priority Effects on Symbiont Colonization
While competition can heavily influence colonization success, other factors like the order and timing of strain arrival, or priority effects, are also relevant for open and mixed symbioses. The recurring entry of microbes might make hosts more susceptible to the entry of various nonsymbiotic or harmful microorganisms, demanding regulation mechanisms that favor a stable symbiosis. In this context, the temporal aspect of symbiont colonization can be decisive for the structure and function of the microbiota, as is true for biological communities in general (236), and can have important ecological and evolutionary consequences (231).
Microbes colonizing specialized glands or crypts are likely to experience such impacts intensively, since priority effects are usually stronger in environments that promote rapid changes in the size or structure of a local population, i.e., fast local population dynamics (236). How does that pertain to symbiotic organs? High growth rates in relation to immigration rate are usually tied to fast local population dynamics, as likely occurs in partially confined symbiotic organs. Several factors relevant in comparable scenarios might affect dynamics in symbiotic organs. First, stable conditions can promote fast growth (237). Second, the carrying capacity is reached more quickly because of the small habitat size in comparison to alternative environments (238, 239). Third, in systems in which symbionts are acquired during a specific host life stage or time point, immigration is usually bounded. Also, priority effects might be especially influential during symbiont colonization due to the capability for fast adaptation to local conditions after arrival (240). Experiments testing sequential infection of Hydra vulgaris by two types of the same bacterial symbiont, distinguished by fluorescent labeling, reveal a clear impact of priority effects (241). This study demonstrates that provided a sufficient time lag in colonization, a secondary colonizer will be at a disadvantage, given that the primary colonizer is already closer to reaching the total carrying capacity. Other studies have put forward specific mechanisms underlying such effects in the case of unrelated colonizers. In the human gut, for example, priority effects during community assembly in early life are thought to have long-lasting consequences (242). An early entry of E. coli can deplete oxygen levels, hindering colonization by facultative aerobes and facilitating colonization of obligate anaerobes such as Bacteroides spp. (243). In newborns, Bifidobacterium spp. deplete breast milk oligosaccharides and thereby limit the colonization of other species requiring these carbon sources (244). Similar effects might hamper the establishment of pathogenic bacteria, as suggested by manipulative experiments in model animals. Work by Litvak et al. (245) in mice and chicks suggests that the combined effects of carbohydrate breakdown by Clostridia—which stimulates oxygen consumption in the gut epithelium—and oxygen consumption by gut-associated Enterobacteriaceae result in colonization resistance against the pathogen Salmonella enteritidis. Also in mice, gut-associated Clostridia and Erysipelotrichia deplete sugar alcohols, which are required for the colonization of some opportunistic enteric pathogens (246).
Priority effects can impact symbiotic communities at the species or strain level and should be especially common among symbiont strains with a high phenotypic overlap. This is particularly relevant if recurrent exposure to a free-living phase prevents sympatric evolution and thereby niche differentiation (247). Notably, priority effects at the strain level might be often overlooked. In honey bees, assessment of the gut bacterial community composition using the customary 97% similarity threshold in the 16S rRNA gene shows an apparently high consistency across bees. However, a more rigorous assessment of the strain-level composition revealed that each bee carries a single strain of the core bacterial taxa but that there is a high variation in strain identity across individuals. Strikingly, most of the strains were found to be both dominant and rare in the guts of different bees from the same apiary (248). This suggests that priority effects play an important role in this system, although the mechanisms underlying the dominance of single strains in the honey bee microbiota are not fully understood. Along similar lines, controlled experiments in mice show that higher taxonomic levels remained unchanged independent of arrival order, but specific bacterial types were over- or underrepresented depending on the timing of host exposure (249). In Euprymna squids, the earlier arrival of so-called dominant beneficial strains can inhibit the entry of other strains. However, secondary colonization by a dominant strain occasionally persists (250). Therefore, other factors in addition to priority effects are likely to play a role in determining strain dominance in this system. These might be genetically encoded in V. fischeri strains, yet none of the candidate genes have been conclusively linked to colonization dominance so far (186, 251). In summary, priority effects may have a strong impact on establishment success and specificity in open and mixed symbioses and should be considered when investigating the dynamics and molecular mechanisms underlying host colonization by multipartite microbial communities.
CONCLUDING REMARKS AND OUTLOOK
There is now a wealth of knowledge on the identity of beneficial symbiotic bacteria and their fitness impact on the host. However, the field has yet to catch up on understanding key factors that drive the establishment of these associations, especially from the perspective of the microbe. By spanning a number of symbionts from invertebrate hosts, we aimed to provide an overview of the known strategies used by beneficial bacteria to colonize invertebrates (Fig. 1). While doing so, parallels between the bacterial factors regulating the establishment of beneficial and pathogenic infections in animal hosts become readily apparent. The examples of related bacteria that engage in these two different lifestyles using similar strategies for colonization are useful for comparative studies investigating shifts across the mutualism-parasitism continuum and their mechanistic basis.
There are yet a few challenges and outstanding questions in the field that merit attention. First, the difficulty in culturing many host-associated bacteria acts as a barrier in advancing molecular symbiosis studies. In some symbioses, multiple symbiotic species or strains coexist in the host and probably mediate interbacterial cross talk, and studies with singular symbiont taxa and host may not represent the true nature of these associations or the factors involved in their establishment (248, 252). Second, being able to separate and genetically modify both the symbiont(s) and the host to experimentally determine the mediators of colonization and establishment has so far been possible in only a handful of systems (57, 71). Symbioses that enable us to cross this barrier are those in which host-symbiont integration is mild or the microbial partner retains its free-living abilities. Due to the limited systems that can be studied, there is a potential bias. Already known molecular factors like flagellar genes, modifications to the LPS, type IV pilus adhesion factors, and genes related to biofilm formation arise in many host-associated bacteria as important colonization features. Therefore, we risk overlooking other relevant factors in beneficial bacteria that may sometimes be particular to a group of microbes or hosts. The syp locus in V. fischeri and the mad fimbrial locus in Photorhabdus are examples of such factors unique to a system, and there is likely more to be explored in other beneficial symbionts. Another promising aspect for future research is the role of phages aiding in colonization through genetic innovation and dynamic transfer between strains, as well as their impact on the establishment of microbial communities by providing competitive advantages to some bacteria while hindering others (for examples, see reference 253). So far, a few recent reports discuss the effects of phages in animal microbiomes, including those of bees (254), aphids (255), corals (256), sponges (257, 258), and avian and mammalian pathogens (253). While prophages are increasingly recognized as relevant players in host-associated microbiomes, they remain an underexplored aspect of microbial evolution in the context of animal colonization.
The expanding availability of sequencing and imaging tools to study complex microbial communities, including nonculturable symbionts, will certainly be fundamental to the further exploration of the diversity of microbial molecular factors involved in animal colonization. For most microbes that still experience both free-living and host-associated conditions, a direct comparison of the implications of each lifestyle and the knowledge of the drivers to transition between these are lacking. Experimental evolution and modeling are valuable tools to address this knowledge gap (15, 259) and have been used to evaluate associated questions, such as host fitness consequences of horizontal versus vertical transmission (260–262). Monitoring systems that allow us to track symbionts in their free-living state and quantify fitness have also been proposed as a promising approach (263). Importantly, understudied symbioses need to be transformed into experimentally tractable systems, with targeted or genome-wide genetic manipulation representing particularly promising approaches (Text Box 3). Thereby, we are likely to derive detailed and generalizable insights into the microbial adaptations for establishing as beneficial or pathogenic symbionts in animal hosts.
ACKNOWLEDGMENTS
We are grateful to Aurélien Vigneron and three anonymous reviewers for valuable comments on the manuscript.
This research was supported by funding from German Science Foundation (DFG) research grant no. FL1051/1-1 (to L.V.F.) and KA2846/6-1 (to M.K.) and the Max Planck Society.
We declare that there are no conflicts of interests.
Biographies

Ramya Ganesan pursued a bachelor's degree in biotechnology at SASTRA University, Tanjore, India. In 2016, she did her thesis work at the Max Planck Institute for Chemical Ecology in Germany and spent a year after graduation studying the genetic basis of nanotube formation in bacteria. She began her doctoral studies in 2017 at the Johannes Gutenberg University Mainz and recently moved to the Max Planck Institute for Chemical Ecology. She works on understanding molecular factors that mediate colonization in a defensive symbiosis between Burkholderia bacteria and Lagria beetles.

Jürgen C. Wierz graduated from the Johannes Gutenberg University in Mainz, Germany, in 2016 with a B.Sc. in Biology studying personality traits in ants. He moved on to do an M.Sc. at the same university, investigating transmission and pathogenicity of bacterial symbionts of insect pests to crop plants. Since 2019, he has been a doctoral student working on the evolutionary history of tyrosine-supplementing symbionts in selected beetle families and recently moved to continue this research project in the Department of Insect Symbiosis at the Max Planck Institute for Chemical Ecology in Jena, Germany.

Martin Kaltenpoth obtained his M.Sc. and Ph.D. degrees from the University of Würzburg in Germany, followed by postdoctoral research at the University of Regensburg and the University of Utah in Salt Lake City. In 2009, he established the Insect Symbiosis Research Group at the Max Planck Institute for Chemical Ecology in Jena. In 2015, he became a full professor for Evolutionary Ecology at the University of Mainz. Since 2021, he has been back at the Max Planck Institute for Chemical Ecology as the director of the Department of Insect Symbiosis. His research focuses on the evolution, chemical, and molecular ecology of insect-microbe symbioses.

Laura V. Flórez finished her B.Sc. studies in Biology and Chemistry in 2011 at Universidad de Los Andes in Bogotá, Colombia. She then carried out her Ph.D. at the Max Planck Institute for Chemical Ecology on a symbiotic association between beetles and antibiotic-producing bacteria. In 2016, she became a project leader at the Johannes Gutenberg University in Mainz, Germany, further investigating the ecological and molecular factors underlying the interaction between insects and defensive symbionts. Since 2021, she has been a postdoctoral fellow at the Department for Plant and Environmental Sciences at the University of Copenhagen. Her research focuses on the ecology, evolution, and molecular interactions between entomopathogenic fungi, bacterial symbionts, and insect hosts.
APPENDIX
GLOSSARY
- Bacteriocyte/bacteriome
Bacteriocytes are animal cells that are specialized to house endosymbiotic microbes. Bacteriomes are organs formed by bacteriocytes.
- Biofilm
Assemblage of microbial cells attached to a surface and embedded in a self-produced polymer matrix composed predominantly of extracellular polysaccharides.
- Closed symbioses
Symbioses involving strict vertical transmission of microbial symbionts, often via a transovarial route.
- Facultative symbiosis
Symbioses in which one or both of the partners involved can live independently without a drastic effect on fitness or only a context-dependent impact on survival.
- Fimbriae
Term that is often interchangeable with “pili” for historical reasons (142). Fimbriae are hairlike structures mediating adhesion and biofilm formation and are usually distinguishable from pili because they are shorter and not directly involved in DNA transfer.
- Flagella
Filamentous structures found in some microorganisms that help in locomotion.
- In vivo
An experiment (or process) carried out in a living organism, in this case in an animal host.
- In vitro
An experiment (or process) performed under laboratory culture conditions, not in the animal host.
- Lipopolysaccharide (LPS)
LPS comprises outer membrane components of Gram-negative bacteria that induce an immune response in eukaryotic hosts. The components include (i) O antigen, (ii) core oligosaccharide, and (iii) lipid A components.
- Local population
A set of individuals within a delimited area that is smaller than the geographic range of the species, which is often within a population or is a disconnected population.
- Mixed symbioses
Symbioses involving both vertical and horizontal symbiont transmission.
- Obligate symbiosis
Symbiosis in which one or both of the organisms involved cannot survive without the other.
- Open symbioses
Symbioses in which microbial symbionts are acquired horizontally, i.e., from the environment or unrelated hosts.
- Peptidoglycan
Polymer forming a layer around the cell membrane of bacteria that supports cellular integrity. Gram-positive bacteria have a thicker peptidoglycan layer than do Gram-negative bacteria.
- Pili
Tubular appendages found on the surface of many Gram-negative and Gram-positive bacteria that are important for adhesion, twitching motility, transfer of DNA, and protein secretion. They are formed by pilin protein subunits.
- Population dynamics
How and why populations change in size and structure over time. Key factors affecting these changes are reproduction, death, and migration rates.
- Secretion system
Complex protein structure found embedded in the external membrane of Gram-negative and Gram-positive bacteria. Secretion systems help in the transport of substances from the cytosol to the extracellular environment or the delivery of effector molecules to other organisms.
- Sympatry
The condition in which species or populations share the same habitat or geographical range. In the context of this review, it refers to microbial strains or species persistently sharing the same host tissue or symbiotic organ.
- Tracheal cytotoxin (TCT)
Soluble peptidoglycan components in the cell wall of Gram-negative bacteria comprised of a disaccharide and a peptide chain. TCTs released by some pathogens cause damage to ciliated epithelial tissue and hinder the removal of foreign microbes and mucus from the tissue surface.
Contributor Information
Martin Kaltenpoth, Email: kaltenpoth@ice.mpg.de.
Laura V. Flórez, Email: lvf@plen.ku.dk.
REFERENCES
- 1.Sagan L. 1967. On the origin of mitosing cells. J Theor Biol 14:225–274. 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraune S, Bosch TCG. 2010. Why bacteria matter in animal development and evolution. BioEssays 32:571–580. 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 4.Walter J, Britton RA, Roos S. 2011. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A 108:4645–4652. 10.1073/pnas.1000099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert SF, Sapp J, Tauber AI. 2012. A symbiotic view of life: we have never been individuals. Q Rev Biol 87:325–341. 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 6.Mcfall-Ngai M, Hadfield MG, Bosch TCG, Carey H V, Domazet-Lo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ, King N, Kremer N. 2013. Animals in a bacterial world, a new imperative for the life sciences. PNAS 110:3229–3236. 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudakaran S, Kost C, Kaltenpoth M. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol 25:375–390. 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Sachs JL, Skophammer RG, Regus JU. 2011. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci U S A 108:10800–10807. 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 10.Drew GC, Stevens EJ, King KC. 2021. Microbial evolution and transitions along the parasite–mutualist continuum. Nat Rev Microbiol 19:623–638. 10.1038/s41579-021-00550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachs JL, Skophammer RG, Bansal N, Stajich JE. 2013. Evolutionary origins and diversification of proteobacterial mutualists. Proc Biol Sci 281:20132146. 10.1098/rspb.2013.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toft C, Andersson SGE. 2010. Evolutionary microbial genomics: Insights into bacterial host adaptation. Nat Rev Genet 11:465–475. 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 13.Garcia JR, Gerardo NM. 2014. The symbiont side of symbiosis: Do microbes really benefit? Front Microbiol 5:510. 10.3389/fmicb.2014.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeling PJ, McCutcheon JP. 2017. Endosymbiosis: The feeling is not mutual. J Theor Biol 434:75–79. 10.1016/j.jtbi.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obeng N, Bansept F, Sieber M, Traulsen A, Schulenburg H. 2021. Evolution of microbiota-host associations: the microbe’s perspective. Trends Microbiol 29:779–787. 10.1016/j.tim.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Bright M, Bulgheresi S. 2010. A complex journey: Transmission of microbial symbionts. Nat Rev Microbiol 8:218–230. 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert D. 2013. The epidemiology and evolution of symbionts with mixed-mode transmission. Annu Rev Ecol Evol Syst 44:623–643. 10.1146/annurev-ecolsys-032513-100555. [DOI] [Google Scholar]
- 18.Gerardo NM, Hoang KL, Stoy KS. 2020. Evolution of animal immunity in the light of beneficial symbioses. Philos Trans R Soc Lond B Biol Sci 375:20190601. 10.1098/rstb.2019.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haney CH, Urbach J, Ausubel FM. 2014. Innate immunity in plants and animals. Biochemist 36:40–44. 10.1042/BIO03605040. [DOI] [Google Scholar]
- 20.Strand MR. 2008. The insect cellular immune response. Insect Sci. 10.1111/j.1744-7917.2008.00183.x. [DOI] [Google Scholar]
- 21.Trappeniers K, Matetovici I, Van Den Abbeele J, De Vooght L. 2019. The tsetse fly displays an attenuated immune response to its secondary symbiont, Sodalis glossinidius. Front Microbiol 10:1650. 10.3389/fmicb.2019.01650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyholm S V., Graf J. 2012. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol 10:815–827. 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horak RD, Leonard SP, Moran NA. 2020. Symbionts shape host innate immunity in honeybees: Symbionts shape honey bee immunity. Proc Biol Sci 287:20201184. 10.1098/rspb.2020.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. 2001. Tsetse immune responses and trypanosome transmission: Implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc Natl Acad Sci U S A 98:12648–12653. 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zientz E, Dandekar T, Gross R. 2004. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev 68:745–770. 10.1128/MMBR.68.4.745-770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper L V. 2009. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol 7:367–374. 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- 27.Maire J, Vincent-Monégat C, Balmand S, Vallier A, Hervé M, Masson F, Parisot N, Vigneron A, Anselme C, Perrin J, Orlans J, Rahioui I, Da Silva P, Fauvarque MO, Mengin-Lecreulx D, Zaidman-Rémy A, Heddi A. 2019. Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc Natl Acad Sci U S A 116:5623–5632. 10.1073/pnas.1821806116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chomicki G, Werner GDA, West SA, Kiers ET. 2020. Compartmentalization drives the evolution of symbiotic cooperation. Philos Trans R Soc Lond B Biol Sci 375:20190602. 10.1098/rstb.2019.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFall-Ngai M. 2007. Adaptive immunity: Care for the community. Nature 445:153–153. 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 30.Douglas AE, Bouvaine S, Russell RR. 2011. How the insect immune system interacts with an obligate symbiotic bacterium. Proc Biol Sci 278:333–338. 10.1098/rspb.2010.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cremer S, Armitage SAO, Schmid-Hempel P. 2007. Social immunity. Curr Biol 17:R693–R702. 10.1016/j.cub.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, Zhang P, Huang Z, Berger SL, Reinberg D, Wang J, Liebig J. 2010. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329:1068–1071. 10.1126/science.1192428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldhaar H, Gross R. 2008. Immune reactions of insects on bacterial pathogens and mutualists. Microbes Infect 10:1082–1088. 10.1016/j.micinf.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Gross R, Vavre F, Heddi A, Hurst GDD, Zchori-Fein E, Bourtzis K. 2009. Immunity and symbiosis. Mol Microbiol 73:751–759. 10.1111/j.1365-2958.2009.06820.x. [DOI] [PubMed] [Google Scholar]
- 35.Salem H, Florez L, Gerardo N, Kaltenpoth M. 2015. An out-of-body experience: The extracellular dimension for the transmission of mutualistic bacteria in insects. Proc Biol Sci 282:20142957. 10.1098/rspb.2014.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 37.Perreau J, Moran NA. 2021. Genetic innovations in animal–microbe symbioses. Nat Rev Genet 23:23–39. 10.1038/s41576-021-00395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wierz JC, Gaube P, Klebsch D, Kaltenpoth M, Flórez L V. 2021. Transmission of bacterial symbionts with and without genome erosion between a beetle host and the plant environment. Front Microbiol 12:715601. 10.3389/fmicb.2021.715601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruby EG. 2008. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6:752–762. 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct 4:13. 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch EJ, McFall-Ngai M. 2019. Model systems for the study of how symbiotic associations between animals and extracellular bacterial partners are established and maintained. Drug Discov Today Dis Models 28:3–12. 10.1016/j.ddmod.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher RM, Henry LM, Cornwallis CK, Kiers ET, West SA. 2017. The evolution of host-symbiont dependence. Nat Commun 8:15973. 10.1038/ncomms15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartmann AC, Baird AH, Knowlton N, Huang D. 2017. The paradox of environmental symbiont acquisition in obligate mutualisms. Curr Biol 27:3711–3716.e3. 10.1016/j.cub.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 44.Ohbayashi T, Mergaert P, Kikuchi Y. 2020. Host-symbiont specificity in insects: Underpinning mechanisms and evolution, p. 27–62. In Oliver KM, Russell JA (ed.), Advances in Insect Physiology. Academic Press, Inc, New York, NY. [Google Scholar]
- 45.McCutcheon JP, Boyd BM, Dale C. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol 29:R485–R495. 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Muller HJ. 1964. The relation of recombination to mutational advance. Mutat Res 106:2–9. 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 47.Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A 93:2873–2878. 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao M, Goodrich-Blair H. 2017. Ready or not: microbial adaptive responses in dynamic symbiosis environments. J Bacteriol 199:16. 10.1128/JB.00883-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chrostek E, Pelz-Stelinski K, Hurst GDD, Hughes GL. 2017. Horizontal transmission of intracellular insect symbionts via plants. Front Microbiol 8:2237. 10.3389/fmicb.2017.02237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frago E, Dicke M, Godfray HCJ. 2012. Insect symbionts as hidden players in insect-plant interactions. Trends Ecol Evol 27:705–711. 10.1016/j.tree.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 51.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 52.Pinto-Carbó M, Sieber S, Dessein S, Wicker T, Verstraete B, Gademann K, Eberl L, Carlier A. 2016. Evidence of horizontal gene transfer between obligate leaf nodule symbionts. ISME Journal 10:2092–2105. 10.1038/ismej.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell SL, Pepper-Tunick E, Svedberg J, Byrne A, Castillo JR, Vollmers C, Beinart RA, Corbett-Detig R. 2020. Horizontal transmission and recombination maintain forever young bacterial symbiont genomes. PLoS Genet 16:e1008935. 10.1371/journal.pgen.1008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller BM, Baumler AJ. 2021. The Habitat Filters of Microbiota-Nourishing Immunity. Annu Rev Immunol 39:1–18. 10.1146/annurev-immunol-101819-024945. [DOI] [PubMed] [Google Scholar]
- 55.Dierking K, Pita L. 2020. Receptors Mediating Host-Microbiota Communication in the Metaorganism: The Invertebrate Perspective. Front Immunol 11:1251. 10.3389/fimmu.2020.01251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McFall-Ngai M, Nyholm S V, Castillo MG. 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 22:48–53. 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt K, Engel P. 2021. Mechanisms underlying gut microbiota-host interactions in insects. Journal of Experimental Biology 224:jeb207696. 10.1242/jeb.207696. [DOI] [PubMed] [Google Scholar]
- 58.Russell SL. 2019. Transmission mode is associated with environment type and taxa across bacteria-eukaryote symbioses: A systematic review and meta-analysis. FEMS Microbiol Lett 366:fnz013. [DOI] [PubMed] [Google Scholar]
- 59.Weintraub PG, Beanland L. 2006. Insect vectors of phytoplasmas. Annu Rev Entomol 51:91–111. 10.1146/annurev.ento.51.110104.151039. [DOI] [PubMed] [Google Scholar]
- 60.Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334:362–365. 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 61.Maire J, Parisot N, Ferrarini MG, Vallier A, Gillet B, Hughes S, Balmand S, Vincent-Monégat C, Zaidman-Rémy A, Heddi A. 2020. Spatial and morphological reorganization of endosymbiosis during metamorphosis accommodates adult metabolic requirements in a weevil. PNAS 117:19347–19358. 10.1073/pnas.2007151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luan JB, Chen W, Hasegawa DK, Simmons A, Wintermantel WM, Ling KS, Fei Z, Liu SS, Douglas AE. 2015. Metabolic Coevolution in the Bacterial Symbiosis of Whiteflies and Related Plant Sap-Feeding Insects. Genome Biol Evol 7:2635–2647. 10.1093/gbe/evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ankrah NYD, Luan J, Douglasa AE. 2017. Cooperative metabolism in a threepartner insect-bacterial symbiosis revealed by metabolic modeling. J Bacteriol 199:872–888. 10.1128/JB.00872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maire J, Vincent-Monégat C, Masson F, Zaidman-Rémy A, Heddi A. 2018. An IMD-like pathway mediates both endosymbiont control and host immunity in the cereal weevil Sitophilus spp. Microbiome 6:1–10. 10.1186/s40168-017-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sieber M, Traulsen A, Schulenburg H, Douglas AE. 2021. On the evolutionary origins of host-microbe associations. Proc Natl Acad Sci U S A 118:e2016487118. 10.1073/pnas.2016487118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu J-Z, Zhang W-G. 2016. Strategies used for genetically modifying bacterial genome: site-directed mutagenesis, gene inactivation, and gene over-expression. Journal of Zhejiang Univ-Sci B (Biomed & Biotechnol) 17:83–99. 10.1631/jzus.B1500187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol. 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roden JA, Wells DH, Chomel BB, Kasten RW, Koehler JE. 2012. Hemin binding protein C is found in outer membrane vesicles and protects bartonella henselae against toxic concentrations of hemin. Infect Immun 80:929–942. 10.1128/IAI.05769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol 55:368–380. 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 70.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–663. 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masson F, Lemaitre B. 2020. Growing Ungrowable Bacteria: Overview and Perspectives on Insect Symbiont Culturability. Microbiology and Molecular Biology Reviews 84:e00089-20. 10.1128/MMBR.00089-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christensen DG, Visick KL. 2020. Vibrio fischeri: Laboratory Cultivation, Storage, and Common Phenotypic Assays. Curr Protoc Microbiol 57:e103. 10.1002/cpmc.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graf J. 1999. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect Immun 67:1–7. 10.1128/IAI.67.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forst S, Dowds B, Boemare N, Stackebrandt E. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol 51:47–72. 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 75.Takeshita K, Tamaki H, Ohbayashi T, Meng X-Y, Sone T, Mitani Y, Peeters C, Kikuchi Y, Vandamme P. 2018. Burkholderia insecticola sp. nov., a gut symbiotic bacterium of the bean bug Riptortus pedestris. Int J Syst Evol Microbiol 68:2370–2374. 10.1099/ijsem.0.002848. [DOI] [PubMed] [Google Scholar]
- 76.Kwong WK, Moran NA. 2013. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a memb. Int J Syst Evol Microbiol 63:2008–2018. 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 77.Dale C, Maudlin I. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly GIossina morsitans morsitans. Int J Syst Bacteriol 49:267–275. 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 78.Leonard SP, Perutka J, Powell JE, Geng P, Richhart DD, Byrom M, Kar S, Davies BW, Ellington AD, Moran NA, Barrick JE. 2018. Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth Biol 7:1279–1290. 10.1021/acssynbio.7b00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leonard SP, Powell JE, Perutka J, Geng P, Heckmann LC, Horak RD, Davies BW, Ellington AD, Barrick JE, Moran NA. 2020. Engineered symbionts activate honey bee immunity and limit pathogens. Science 367:573–576. 10.1126/science.aax9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kakkanat A, Phan M-D, Lo AW, Beatson SA, Schembri MA. 2017. Novel genes associated with enhanced motility of Escherichia coli ST131. PLoS One 12:e0176290. 10.1371/journal.pone.0176290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nolan LM, Whitchurch CB, Barquist L, Katrib M, Boinett CJ, Mayho M, Goulding D, Charles IG, Filloux A, Parkhill J, Cain AK. 2018. A global genomic approach uncovers novel components for twitching motility-mediated biofilm expansion in Pseudomonas aeruginosa. Microb Genom 4:e000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dale C, Young SA, Haydon DT, Welburn SC. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. PNAS 98:1883–1888. 10.1073/pnas.98.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim JK, Jang HA, Won YJ, Kikuchi Y, Heum Han S, Kim CH, Nikoh N, Fukatsu T, Lee BL. 2014. Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME Journal 8:552–563. 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. 2016. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci U S A 113:13887–13892. 10.1073/pnas.1610856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brooks JF, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. PNAS 111:17284–17289. 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wheatley RM, Ford BL, Li L, Aroney STN, Knights HE, Ledermann R, East AK, Ramachandran VK, Poole PS. 2020. Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. PNAS 117:23823–23834. 10.1073/pnas.2009094117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Do Amaral FP, Tuleski TR, Pankievicz VCS, Melnyk RA, Arkin AP, Griffitts J, Tadra-Sfeir MZ, Maltempi de Souza E, Deutschbauer A, Monteiro RA, Staceya G. 2020. Diverse bacterial genes modulate plant root association by beneficial bacteria. mBio 11:1–15. 10.1128/mBio.03078-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, Malmstrom RR, Deutschbauer AM, Dangl JL, Visel A. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 15:e2002860. 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thibault D, Jensen PA, Wood S, Qabar C, Clark S, Shainheit MG, Isberg RR, van Opijnen T. 2019. Droplet Tn-Seq combines microfluidics with Tn-Seq for identifying complex single-cell phenotypes. Nat Commun 10:5729. 10.1038/s41467-019-13719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hegde S, Nilyanimit P, Kozlova E, Anderson ER, Narra HP, Sahni SK, Heinz E, Hughes GL. 2019. CRISPR/Cas9-mediated gene deletion of the ompA gene in symbiotic Cedecea neteri impairs biofilm formation and reduces gut colonization of Aedes aegypti mosquitoes. PLoS Negl Trop Dis 13:e0007883. 10.1371/journal.pntd.0007883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang T, Guan C, Guo J, Liu B, Wu Y, Xie Z, Zhang C, Xing XH. 2018. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance-net. Nat Commun 9:2475. 10.1038/s41467-018-04899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X, Gallay C, Kjos M, Domenech A, Slager J, Kessel SP van, Knoops K, Sorg RA, Zhang J-R, Veening J-W. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13:931. 10.15252/msb.20167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wiles TJ, Schlomann BH, Wall ES, Betancourt R, Parthasarathy R, Guillemin K. 2020. Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation. PLoS Biol 18:e3000661. 10.1371/journal.pbio.3000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Todor H, Silvis MR, Osadnik H, Gross CA. 2021. Bacterial CRISPR screens for gene function. Curr Opin Microbiol 59:102–109. 10.1016/j.mib.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang R, Xu W, Shao S, Wang Q. 2021. Gene silencing through CRISPR interference in bacteria: current advances and future prospects. Front Microbiol 12:635227. 10.3389/fmicb.2021.635227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raina J-B, Fernandez V, Lambert B, Stocker R, Seymour JR. 2019. The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol 17:284–294. 10.1038/s41579-019-0182-9. [DOI] [PubMed] [Google Scholar]
- 100.Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144:1151–1155. 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 101.Kremer N, Philipp EER, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol 12:e1001783. 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EAC, DeLoney-Marino CR, McFall-Ngai MJ, Ruby EG. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 78:4620–4626. 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nyholm S V., Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol 68:5113–5122. 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nyholm SV, McFall-Ngai MJ. 2021. A lasting symbiosis: how the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat Rev Microbiol 19:666–679. 10.1038/s41579-021-00567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nyholm S V., Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. PNAS 97:10231–10235. 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hussa EA, O’Shea TM, Darnell CL, Ruby EG, Visick KL. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J Bacteriol 189:5825–5838. 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chaban B, Hughes HV, Beeby M. 2015. The flagellum in bacterial pathogens: For motility and a whole lot more. Semin Cell Dev Biol 46:91–103. 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 109.Robidart JC, Bench SR, Feldman RA, Novoradovsky A, Podell SB, Gaasterland T, Allen EE, Felbeck H. 2008. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ Microbiol 10:727–737. 10.1111/j.1462-2920.2007.01496.x. [DOI] [PubMed] [Google Scholar]
- 110.Burgsdorf I, Handley KM, Bar-Shalom R, Erwin PM, Steindler L. 2019. Life at home and on the roam: genomic adaptions reflect the dual lifestyle of an intracellular facultative symbiont. mSystems 4:e00057-19. 10.1128/mSystems.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Onchuru TO, Martinez AJ, Ingham CS, Kaltenpoth M. 2018. Transmission of mutualistic bacteria in social and gregarious insects. Curr Opin Insect Sci 28:50–58. 10.1016/j.cois.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 112.Wada-Katsumata A, Zurek L, Nalyanya G, Roelofs WL, Zhang A, Schal C. 2015. Gut bacteria mediate aggregation in the German cockroach. Proc Natl Acad Sci U S A 112:15678–15683. 10.1073/pnas.1504031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hosokawa T, Kikuchi Y, Fukatsu T. 2007. How many symbionts are provided by mothers, acquired by offspring, and needed for successful vertical transmission in an obligate insect-bacterium mutualism? Mol Ecol 16:5316–5325. 10.1111/j.1365-294X.2007.03592.x. [DOI] [PubMed] [Google Scholar]
- 114.Ohbayashi T, Takeshita K, Kitagawa W, Nikohc N, Koga R, Meng XY, Tago K, Hori T, Hayatsu M, Asano K, Kamagata Y, Lee BL, Fukatsu T, Kikuchi Y. 2015. Insect’s intestinal organ for symbiont sorting. Proc Natl Acad Sci U S A 112:E5179–E5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee JB, Byeon JH, Jang HA, Kim JK, Yoo JW, Kikuchi Y, Lee BL. 2015. Bacterial cell motility of Burkholderia gut symbiont is required to colonize the insect gut. FEBS Lett 589:2784–2790. 10.1016/j.febslet.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 116.Tomich M, Herfst CA, Golden JW, Mohr CD. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect Immun 70:1799–1806. 10.1128/IAI.70.4.1799-1806.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nickzad A, Lépine F, Déziel E. 2015. Quorum sensing controls swarming motility of Burkholderia glumae through regulation of rhamnolipids. PLoS One 10:e0128509. 10.1371/journal.pone.0128509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duan Q, Zhou M, Zhu L, Zhu G. 2013. Flagella and bacterial pathogenicity. J Basic Microbiol 53:1–8. 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 119.Kinosita Y, Kikuchi Y, Mikami N, Nakane D, Takayuki N. 2018. Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body. ISME Journal 12:838–848. 10.1038/s41396-017-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Militello G. 2019. Motility control of symbionts and organelles by the eukaryotic cell: the handling of the motile capacity of individual parts forges a collective biological identity. Front Psychol 10:2080. 10.3389/fpsyg.2019.02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maezawa K, Shigenobu S, Taniguchi H, Kubo T, Aizawa S, Morioka M. 2006. Hundreds of flagellar basal bodies cover the cell surface of the endosymbiotic bacterium Buchnera aphidicola sp. strain APS. J Bacteriol 188:6539–6543. 10.1128/JB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rio RVM, Symula RE, Wang J, Lohs C, neng Wu Y, Snyder AK, Bjornson RD, Oshima K, Biehl BS, Perna NT, Hattori M, Aksoy S. 2012. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. mBio 3:e00240-11. 10.1128/mBio.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pais R, Lohs C, Wu Y, Wang J, Aksoy S. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol 74:5965–5974. 10.1128/AEM.00741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koga R, Meng XY, Tsuchida T, Fukatsu T. 2012. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci U S A 109:E1230–E1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luan J, Sun X, Fei Z, Douglas AE. 2018. Maternal inheritance of a single somatic animal cell displayed by the bacteriocyte in the whitefly Bemisia tabaci. Curr Biol 28:459–465. 10.1016/j.cub.2017.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Renoz F, Champagne A, Degand H, Faber A-MM, Morsomme P, Foray V, Hance T, Renoz F, Champagne A, Degand H, Faber A-MM, Morsomme P, Foray V, Hance T. 2017. Toward a better understanding of the mechanisms of symbiosis: A comprehensive proteome map of a nascent insect symbiont. PeerJ 5:e3291. 10.7717/peerj.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Waterworth SC, Flórez L V, Rees ER, Hertweck C, Kaltenpoth M, Kwan JC. 2020. Horizontal gene transfer to a defensive symbiont with a reduced genome in a multipartite beetle microbiome. mBio 11:e02430-19. 10.1128/mBio.02430-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Winans NJ, Walter A, Chouaia B, Chaston JM, Douglas AE, Newell PD. 2017. A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Mol Ecol 26:4536–4550. 10.1111/mec.14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hajam IA, Dar PA, Shahnawaz I, Jaume JC, Lee JH. 2017. Bacterial flagellin—a potent immunomodulatory agent. Exp Mol Med 49:e373. 10.1038/emm.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Akahoshi DT, Bevins CL. 2022. Flagella at the host-microbe interface: key functions intersect with redundant responses. Front Immunol 13:828758. 10.3389/fimmu.2022.828758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koropatnick TA, Engle JT, Apicella MA, Stabb E V, Goldman WE, Mcfall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1188. 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 132.Chagnot C, Zorgani MA, Astruc T, Desvaux M. 2013. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol 4:303. 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McAnulty SJ, Nyholm SV. 2017. The role of hemocytes in the hawaiian bobtail squid, euprymna scolopes: A model organism for studying beneficial host-microbe interactions. Front Microbiol 7:2013. 10.3389/fmicb.2016.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Steimle A, Autenrieth IB, Frick JS. 2016. Structure and function: Lipid A modifications in commensals and pathogens. Int J Med Microbiol 306:290–301. 10.1016/j.ijmm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 135.Lerouge I, Vanderleyden J. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol Rev 26:17–47. 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 136.Kim JK, Park HY, Lee BL. 2016. The symbiotic role of O-antigen of Burkholderia symbiont in association with host Riptortus pedestris. Dev Comp Immunol 60:202–208. 10.1016/j.dci.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 137.Kim JK, Son DW, Kim C-HH, Cho JH, Marchetti R, Silipo A, Sturiale L, Park HY, Huh YR, Nakayama H, Fukatsu T, Molinaro A, Lee BL. 2015. Insect gut symbiont susceptibility to host antimicrobial peptides caused by alteration of the bacterial cell envelope. J Biol Chem 290:21042–21053. 10.1074/jbc.M115.651158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bennett HPJ, Clarke DJ. 2005. The pbgPE operon in Photorhabdus luminescens is required for pathogenicity and symbiosis. J Bacteriol 187:77–84. 10.1128/JB.187.1.77-84.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Silver AC, Rabinowitz NM, Küffer S, Graf J. 2007. Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J Bacteriol 189:6763–6772. 10.1128/JB.00685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the Natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 141.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592. 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 142.Ottow JC. 1975. Ecology, physiology, and genetics of fimbriae and pili. Annu Rev Microbiol 29:79–108. 10.1146/annurev.mi.29.100175.000455. [DOI] [PubMed] [Google Scholar]
- 143.Ligthart K, Belzer C, de Vos WM, Tytgat HLP. 2020. Bridging bacteria and the gut: functional aspects of type IV pili. Trends Microbiol 28:340–348. 10.1016/j.tim.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 144.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. PNAS 111:11509–11514. 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, Van Putten JPM, Koomey M. 2001. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc Natl Acad Sci U S A 98:15276–15281. 10.1073/pnas.261574998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dulla GFJ, Go RA, Stahl DA, Davidson SK. 2012. Verminephrobacter eiseniae type IV pili and flagella are required to colonize earthworm nephridia. ISME J 6:1166–1175. 10.1038/ismej.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brivio MF, Toscano A, De Pasquale SM, De Lerma Barbaro A, Giovannardi S, Finzi G, Mastore M. 2018. Surface protein components from entomopathogenic nematodes and their symbiotic bacteria: effects on immune responses of the greater wax moth, Galleria mellonella (Lepidoptera: Pyralidae). Pest Manag Sci 74:2089–2099. 10.1002/ps.4905. [DOI] [PubMed] [Google Scholar]
- 149.Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, Ventura M. 2014. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett. 10.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 150.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stabb E V., Ruby EG. 2003. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol 69:820–826. 10.1128/AEM.69.2.820-826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Somvanshi VS, Kaufmann-Daszczuk B, Kim K, Mallon S, Ciche TA. 2010. Photorhabdus phase variants express a novel fimbrial locus, mad, essential for symbiosis. Mol Microbiol 77:1021–1038. 10.1111/j.1365-2958.2010.07270.x. [DOI] [PubMed] [Google Scholar]
- 153.Clarke DJ. 2014. The genetic basis of the symbiosis between Photorhabdus and its invertebrate hosts. Adv Appl Microbiol 88:1–29. [DOI] [PubMed] [Google Scholar]
- 154.Somvanshi VS, Sloup RE, Crawford JM, Martin AR, Heidt AJ, Kim K, Clardy J, Ciche TA. 2012. A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science 337:88–93. 10.1126/science.1216641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Burgsdorf I, Slaby BM, Handley KM, Haber M, Blom J, Marshall CW, Gilbert JA, Hentschel U, Steindler L. 2015. Lifestyle evolution in cyanobacterial symbionts of sponges. mBio 6:e00391-15. 10.1128/mBio.00391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, Thomas T. 2012. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. PNAS 109:E1878–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME. 2009. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect Immun 77:2399–2407. 10.1128/IAI.01266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hymes JP, Johnson BR, Barrangou R, Klaenhammer TR. 2016. Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 82:2676–2685. 10.1128/AEM.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Leo JC, Grin I, Linke D. 2012. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philosophical Transactions of the Royal Society B 367:1088–1101. 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mil-Homens D, Fialho AM. 2011. Trimeric autotransporter adhesins in members of the Burkholderia cepacia complex: a multifunctional family of proteins implicated in virulence. Front Cell Infect Microbiol 1:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stones DH, Krachler AM. 2016. Against the tide: the role of bacterial adhesion in host colonization. Biochem Soc Trans 44:1571–1580. 10.1042/BST20160186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173. 10.1111/j.1574-6968.2004.tb09643.x. [DOI] [PubMed] [Google Scholar]
- 163.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 164.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ 54 -dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 165.Chavez-Dozal A, Gorman C, Nishiguchi MK. 2015. Proteomic and metabolomic profiles demonstrate variation among free-living and symbiotic vibrio fischeri biofilms. BMC Microbiol 15:1–11. 10.1186/s12866-015-0560-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ciche TA, Kim K-SS, Kaufmann-Daszczuk B, Nguyen KCQQ, Hall DH. 2008. Cell Invasion and Matricide during Photorhabdus luminescens Transmission by Heterorhabditis bacteriophora Nematodes. Appl Environ Microbiol 74:2275–2287. 10.1128/AEM.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim JK, Kwon JY, Kim SK, Han SH, Won YJ, Lee JH, Kim CH, Fukatsu T, Lee BL. 2014. Purine biosynthesis, biofilm formation, and persistence of an insect-microbe gut symbiosis. Appl Environ Microbiol 80:4374–4382. 10.1128/AEM.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Murphy MP, Caraher E. 2015. Residence in biofilms allows Burkholderia cepacia complex (Bcc) bacteria to evade the antimicrobial activities of neutrophil-like dHL60 cells. FEMS Pathogens and disease 73:ftv069. 10.1093/femspd/ftv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nelson MC, Graf J. 2012. Bacterial symbioses of the medicinal leech Hirudo verbana. Gut Microbes 3:322–331. 10.4161/gmic.20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Korotkov KV, Sandkvist M, Hol WGJ. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10:336–351. 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cianciotto NP, White RC. 2017. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun 85:e00014-17. 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Barger PC, Liles MR, Newton JC. 2020. Type II secretion is essential for virulence of the emerging fish pathogen, hypervirulent Aeromonas hydrophila. Front Vet Sci 7:574113. 10.3389/fvets.2020.574113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 174.Silver AC, Kikuchi Y, Fadl AA, Sha J, Chopra AK, Graf J. 2007. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. PNAS 104:9481–9486. 10.1073/pnas.0700286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tomás JM. 2012. The Main Aeromonas Pathogenic Factors. ISRN Microbiol 2012:1–22. 10.5402/2012/256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Staehelin C, Krishnan HB. 2015. Nodulation outer proteins: double-edged swords of symbiotic rhizobia. Biochemical Journal 470:263–274. 10.1042/BJ20150518. [DOI] [PubMed] [Google Scholar]
- 177.Gaytán MO, Martínez-Santos VI, Soto E, González-Pedrajo B. 2016. Type three secretion system in attaching and effacing pathogens. Front Cell Infect Microbiol. 10.3389/fcimb.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dale C, Jones T, Pontes M. 2005. Degenerative Evolution and Functional Diversification of Type-III Secretion Systems in the Insect Endosymbiont Sodalis glossinidius. Mol Biol Evol 22:758–766. 10.1093/molbev/msi061. [DOI] [PubMed] [Google Scholar]
- 179.Pontes MH, Smith KL, De Vooght L, Van Den Abbeele J, Dale C. 2011. Attenuation of the Sensing Capabilities of PhoQ in Transition to Obligate Insect–Bacterial Association. PLoS Genet 7:e1002349. 10.1371/journal.pgen.1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hachani A, Wood TE, Filloux A. 2016. Type VI secretion and anti-host effectors 29:81–93. 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 181.Stubbendieck RM, Li H, Currie CR. 2019. Convergent evolution of signal-structure interfaces for maintaining symbioses. Curr Opin Microbiol 50:71–78. 10.1016/j.mib.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. 2003. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol 1:070–076. 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.KALTENPOTH M, SCHMITT T, POLIDORI C, KOEDAM D, STROHM E. 2010. Symbiotic streptomycetes in antennal glands of the South American digger wasp genus Trachypus (Hymenoptera, Crabronidae). Physiol Entomol 35:196–200. 10.1111/j.1365-3032.2010.00729.x. [DOI] [Google Scholar]
- 184.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–1729. 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- 185.Essock-Burns T, Bongrand C, Goldman WE, Ruby EG, McFall-Ngai MJ. 2020. Interactions of symbiotic partners drive the development of a complex biogeography in the squid-vibrio symbiosis. mBio 11:e00853-20. 10.1128/mBio.00853-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Visick KL, Stabb EV, Ruby EG. 2021. A lasting symbiosis: how Vibrio fischeri finds a squid partner and persists within its natural host. Nat Rev Microbiol 19:654–665. 10.1038/s41579-021-00557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kimbell JR, McFall-Ngai MJ. 2004. Symbiont-induced changes in host actin during the onset of a beneficial animal-bacterial association. Appl Environ Microbiol 70:1434. 10.1128/AEM.70.3.1434-1441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stradal TEB, Schelhaas M. 2018. Actin dynamics in host–pathogen interaction. FEBS Lett 592:3658. 10.1002/1873-3468.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Colonne PM, Winchell CG, Voth DE. 2016. Hijacking host cell highways: Manipulation of the host actin cytoskeleton by obligate intracellular bacterial pathogens. Front Cell Infect Microbiol 6:107. 10.3389/fcimb.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Henty-Ridilla JL, Shimono M, Li J, Chang JH, Day B, Staiger CJ. 2013. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog 9:e1003290. 10.1371/journal.ppat.1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang X, Han L, Wang Q, Zhang C, Yu Y, Tian J, Kong Z. 2019. The host actin cytoskeleton channels rhizobia release and facilitates symbiosome accommodation during nodulation in Medicago truncatula. New Phytol 221:1049–1059. 10.1111/nph.15423. [DOI] [PubMed] [Google Scholar]
- 192.Jimenez A, Chen D, Alto NM. 2016. How bacteria subvert animal cell structure and function. Annu Rev Cell Dev Biol 32:373–397. 10.1146/annurev-cellbio-100814-125227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kikuchi Y, Ohbayashi T, Jang S, Mergaert P. 2020. Burkholderia insecticola triggers midgut closure in the bean bug Riptortus pedestris to prevent secondary bacterial infections of midgut crypts. ISME J 14:1627–1638. 10.1038/s41396-020-0633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jang S, Kikuchi Y. 2020. Re-opening of the symbiont sorting organ with aging in Riptortus pedestris. J Asia Pac Entomol 23:1089–1095. 10.1016/j.aspen.2020.09.005. [DOI] [Google Scholar]
- 195.Hightower LE. 1991. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell 66:191–197. 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 196.Kupper M, Gupta SK, Feldhaar H, Gross R. 2014. Versatile roles of the chaperonin GroEL in microorganism-insect interactions. FEMS Microbiol Lett 353:1–10. 10.1111/1574-6968.12390. [DOI] [PubMed] [Google Scholar]
- 197.Fares MA, Moya A, Barrio E. 2004. GroEL and the maintenance of bacterial endosymbiosis. Trends Genet 20:413–416. 10.1016/j.tig.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 198.Nechitaylo TY, Sandoval-Calderón M, Engl T, Wielsch N, Dunn DM, Goesmann A, Strohm E, Svatoš A, Dale C, Weiss ERB, Kaltenpoth M. 2021. Incipient genome erosion and metabolic streamlining for antibiotic production in a defensive symbiont. Proc Natl Acad Sci U S A 118:e2023047118. 10.1073/pnas.2023047118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Roma JS, D’Souza S, Somers PJ, Cabo LF, Farsin R, Aksoy S, Runyen-Janecky LJ, Weiss BL. 2019. Thermal stress responses of Sodalis glossinidius, an indigenous bacterial symbiont of hematophagous tsetse flies. PLoS Negl Trop Dis 13:e0007464. 10.1371/journal.pntd.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Guisbert E, Yura T, Rhodius VA, Gross CA. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 72:545–554. 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Maleki F, Khosravi A, Nasser A, Taghinejad H, Azizian M. 2016. Bacterial heat shock protein activity. J Clin Diagn Res 10:BE01-3. 10.7860/JCDR/2016/14568.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Imlay JA. 2019. Where in the world do bacteria experience oxidative stress? Environ Microbiol 21:521–530. 10.1111/1462-2920.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ezraty B, Gennaris A, Barras F, Collet J-F. 2017. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15:385–396. 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 204.Storz G, Tartaglia LA, Farr SB, Ames BN. 1990. Bacterial defenses against oxidative stress. Trends in Genetics 6:363–368. 10.1016/0168-9525(90)90278-E. [DOI] [PubMed] [Google Scholar]
- 205.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 206.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Benov L, Fridovich I. 1995. Superoxide dismutase protects against aerobic heat shock in Escherichia coli. J Bacteriol 177:3344–3346. 10.1128/jb.177.11.3344-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pontes MH, Babst M, Lochhead R, Oakeson K, Smith K, Dale C. 2008. Quorum sensing primes the oxidative stress response in the insect endosymbiont, Sodalis glossinidius. PLoS One 3:e3541. 10.1371/journal.pone.0003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. 2004. NO means “yes” in the squid-vibrio symbiosis: Nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol 6:1139–1151. 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 210.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. 2010. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 78:903–915. 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. 2010. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci U S A 107:8375–8380. 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Thompson LR, Nikolakakis K, Pan S, Reed J, Knight R, Ruby EG. 2017. Transcriptional characterization of Vibrio fischeri during colonization of juvenile Euprymna scolopes. Environ Microbiol 19:1845–1856. 10.1111/1462-2920.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Heath-Heckman EAC, Gillette AA, Augustin R, Gillette MX, Goldman WE, Mcfall-Ngai MJ. 2014. Shaping the microenvironment: Evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-vibrio symbiosis. Environ Microbiol 16:3669–3682. 10.1111/1462-2920.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Meilhoc E, Boscari A, Bruand C, Puppo A, Brouquisse R. 2011. Nitric oxide in legume-rhizobium symbiosis. Plant Science 181:573–581. 10.1016/j.plantsci.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 215.Strohm E, Herzner G, Ruther J, Kaltenpoth M, Engl T. 2019. Nitric oxide radicals are emitted by wasp eggs to kill mold fungi. Elife 8:e43718. 10.7554/eLife.43718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, Conway T. 2011. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the “feast to famine” gradient in Escherichia coli. Mol Microbiol 79:830–845. 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cho BK, Federowicz S, Park YS, Zengler K, Palsson B. 2012. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat Chem Biol 8:65–71. 10.1038/nchembio.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Smirnova G V., Oktyabrsky ON. 2005. Glutathione in bacteria. Biochemistry 70:1199–1211. 10.1007/s10541-005-0248-3. [DOI] [PubMed] [Google Scholar]
- 219.Kim JK, Lee HJ, Kikuchi Y, Kitagawa W, Nikoh N, Fukatsu T, Lee BL. 2013. Bacterial cell wall synthesis gene uppP is required for burkholderia colonization of the stinkbug gut. Appl Environ Microbiol 79:4879–4886. 10.1128/AEM.01269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Smith TE, Lee M, Person MD, Hesek D, Mobashery S, Moran NA. 2021. Horizontal-acquisition of a promiscuous peptidoglycan-recycling enzyme enables aphids to influence symbiont cell wall metabolism. mBio 12. 10.1128/mBio.02636-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bublitz DAC, Chadwick GL, Magyar JS, Sandoz KM, Brooks DM, Mesnage S, Ladinsky MS, Garber AI, Bjorkman PJ, Orphan VJ, McCutcheon JP. 2019. Peptidoglycan production by an insect-bacterial mosaic. Cell 179:703–712.e7. 10.1016/j.cell.2019.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. 2007. Evolution of species interactions in a biofilm community. Nature 445:533–536. 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 223.Zélé F, Magalhães S, Kéfi S, Duncan AB. 2018. Ecology and evolution of facilitation among symbionts. Nat Commun 9:4869. 10.1038/s41467-018-06779-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. 2008. Rules of engagement: Interspecies interactions that regulate microbial communities. Annu Rev Microbiol 62:375–401. 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 225.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Itoh H, Jang S, Takeshita K, Ohbayashi T, Ohnishi N, Meng XY, Mitani Y, Kikuchi Y. 2019. Host–symbiont specificity determined by microbe–microbe competition in an insect gut. Proc Natl Acad Sci U S A 116:22673–22682. 10.1073/pnas.1912397116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, Miyashiro T, Septer AN. 2018. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A 115:E8528–E8537. 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Guckes KR, Cecere AG, Williams AL, McNeil AE, Miyashiro T. 2020. The bacterial enhancer binding protein vasH promotes expression of a type vi secretion system in vibrio fischeri during symbiosis. J Bacteriol 202:e00777-19. 10.1128/JB.00777-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Steele MI, Kwong WK, Whiteley M, Moranb NA. 2017. Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 8:e01630-17. 10.1128/mBio.01630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Scheuring I, Yu DW. 2012. How to assemble a beneficial microbiome in three easy steps. Ecol Lett 15:1300–1307. 10.1111/j.1461-0248.2012.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Worsley SF, Innocent TM, Holmes NA, Al-Bassam MM, Schiøtt M, Wilkinson B, Murrell JC, Boomsma JJ, Yu DW, Hutchings MI. 2021. Competition-based screening helps to secure the evolutionary stability of a defensive microbiome. BMC Biol 19:205. 10.1186/s12915-021-01142-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Currie CR, Scott JA, Summerbell RC, Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704. 10.1038/19519. [DOI] [Google Scholar]
- 234.Worthen PL, Gode CJ, Graf J. 2006. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl Environ Microbiol 72:4775–4781. 10.1128/AEM.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kikuchi Y, Graf J. 2007. Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl Environ Microbiol 73:1984–1991. 10.1128/AEM.01833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Fukami T. 2015. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst 46:1–23. 10.1146/annurev-ecolsys-110411-160340. [DOI] [Google Scholar]
- 237.Tucker CM, Fukami T. 2014. Environmental variability counteracts priority effects to facilitate species coexistence: evidence from nectar microbes. Proc Biol Sci 281:20132637. 10.1098/rspb.2013.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Fukami T. 2004. Assembly history interacts with ecosystem size to influence species diversity. Ecology 85:3234–3242. 10.1890/04-0340. [DOI] [Google Scholar]
- 239.Orrock JL, Fletcher RJ. 2005. Changes in community size affect the outcome of competition. Am Nat 166:107–111. 10.1086/430641. [DOI] [PubMed] [Google Scholar]
- 240.Urban MC, De Meester L. 2009. Community monopolization: local adaptation enhances priority effects in an evolving metacommunity. Proc Biol Sci 276:4129–4138. 10.1098/rspb.2009.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wein T, Dagan T, Fraune S, Bosch TCG, Reusch TBH, Hülter NF. 2018. Carrying capacity and colonization dynamics of Curvibacter in the hydra host habitat. Front Microbiol 9. 10.3389/fmicb.2018.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sprockett D, Fukami T, Relman DA. 2018. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol 15:197–205. 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Solís G, de los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. 2010. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16:307–310. 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 245.Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, Kutter L, Alcantara MA, Byndloss MX, Tiffany CR, Walker GT, Faber F, Zhu Y, Bronner DN, Byndloss AJ, Tsolis RM, Zhou H, Bäumler AJ. 2019. Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25:128–139.e5. 10.1016/j.chom.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 246.Tiffany CR, Lee J-Y, Rogers AWL, Olsan EE, Morales P, Faber F, Bäumler AJ. 2021. The metabolic footprint of Clostridia and Erysipelotrichia reveals their role in depleting sugar alcohols in the cecum. Microbiome 9:1–13. 10.1186/s40168-021-01123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Brown WL, Wilson EO. 1956. Character displacement. Syst Zool 5:49–64. 10.2307/2411924. [DOI] [Google Scholar]
- 248.Ellegaard KM, Engel P. 2019. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun 10:1–13. 10.1038/s41467-019-08303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Martínez I, Maldonado-Gomez MX, Gomes-Neto JC, Kittana H, Ding H, Schmaltz R, Joglekar P, Cardona RJ, Marsteller NL, Kembel SW, Benson AK, Peterson DA, Ramer-Tait AE, Walter J. 2018. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. Elife 18:e36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bongrand C, Ruby EG. 2019. Achieving a multi-strain symbiosis: strain behavior and infection dynamics. ISME Journal 13:698–706. 10.1038/s41396-018-0305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bongrand C, Moriano-Gutierrez S, Arevalo P, McFall-Ngai M, Ruby EG, Visick KL, Polz M. 2020. Using colonization assays and comparative genomics to discover symbiosis behaviors and factors in vibrio fischeri. mBio 11. 10.1128/mBio.03407-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Flórez L V., Scherlach K, Miller IJ, Rodrigues A, Kwan JC, Hertweck C, Kaltenpoth M. 2018. An antifungal polyketide associated with horizontally acquired genes supports symbiont-mediated defense in Lagria villosa beetles. Nat Commun 9:2478. 10.1038/s41467-018-04955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Chevallereau A, Pons BJ, van Houte S, Westra ER. 2021. Interactions between bacterial and phage communities in natural environments. Nat Rev Microbiol 20:49–62. 10.1038/s41579-021-00602-y. [DOI] [PubMed] [Google Scholar]
- 254.Bonilla-Rosso G, Steiner T, Wichmann F, Bexkens E, Engel P. 2020. Honey bees harbor a diverse gut virome engaging in nested strain-level interactions with the microbiota. Proc Natl Acad Sci U S A 117:7355–7362. 10.1073/pnas.2000228117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Boyd BM, Chevignon G, Patel V, Oliver KM, Strand MR. 2021. Evolutionary genomics of APSE: a tailed phage that lysogenically converts the bacterium Hamiltonella defensa into a heritable protective symbiont of aphids. Virol J 18:1–18. 10.1186/s12985-021-01685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Boilard A, Dubé CE, Gruet C, Mercière A, Hernandez-Agreda A, Derome N. 2020. Defining coral bleaching as a microbial dysbiosis within the coral holobiont. Microorganisms 8:1–26. 10.3390/microorganisms8111682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Jahn MT, Lachnit T, Markert SM, Stigloher C, Pita L, Ribes M, Dutilh BE, Hentschel U. 2021. Lifestyle of sponge symbiont phages by host prediction and correlative microscopy. The ISME Journal 15:2001–2011. 10.1038/s41396-021-00900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Jahn MT, Arkhipova K, Markert SM, Stigloher C, Lachnit T, Pita L, Kupczok A, Ribes M, Stengel ST, Rosenstiel P, Dutilh BE, Hentschel U. 2019. A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host Microbe 26:542–550.e5. 10.1016/j.chom.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 259.Bansept F, Obeng N, Schulenburg H, Traulsen A. 2021. Modeling host-associating microbes under selection. The ISME Journal 2021 15:3648–3656. 10.1038/s41396-021-01039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sachs JL, Wilcox TP. 2006. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proc Biol Sci 273:425–429. 10.1098/rspb.2005.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Herrera P, Schuster L, Wentrup C, König L, Kempinger T, Na H, Schwarz J, Köstlbacher S, Wascher F, Zojer M, Rattei T, Horn M. 2020. Molecular causes of an evolutionary shift along the parasitism–mutualism continuum in a bacterial symbiont. Proc Natl Acad Sci U S A 117:21658–21666. 10.1073/pnas.2005536117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Shapiro JW, Turner PE. 2014. The impact of transmission mode on the evolution of benefits provided by microbial symbionts. Ecol Evol 4:3350–3361. 10.1002/ece3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Douglas AE. 2018. What will it take to understand the ecology of symbiotic microorganisms? Environ Microbiol 20:1920–1924. 10.1111/1462-2920.14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hinzke T, Kleiner M, Meister M, Schlüter R, Hentschker C, Pané-Farré J, Hildebrandt P, Felbeck H, Sievert SM, Bonn F, Völker U, Becher D, Schweder T, Markert S. 2021. Bacterial symbiont subpopulations have different roles in a deep-sea symbiosis. Elife 10:e58371. 10.7554/eLife.58371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cerrano C, Giovine M, Steindler L. 2022. Petrosia ficiformis (Poiret, 1789): an excellent model for holobiont and biotechnological studies. Curr Opin Biotechnol 74:61–65. 10.1016/j.copbio.2021.10.022. [DOI] [PubMed] [Google Scholar]
- 266.Lee YK, Lee J-H, Lee HK. 2001. Microbial symbiosis in marine sponges. J Microbiol 39:254–264. [Google Scholar]
- 267.Kikuchi Y, Fukatsu T. 2014. Live imaging of symbiosis: Spatiotemporal infection dynamics of a GFP-labelled Burkholderia symbiont in the bean bug Riptortus pedestris. Mol Ecol 23:1445–1456. 10.1111/mec.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kariithi HM, Meki IK, Schneider DI, de Vooght L, Khamis FM, Geiger A, Demirbaş-Uzel G, Vlak JM, iNCE ikbal A, Kelm S, Njiokou F, Wamwiri FN, Malele II, Weiss BL, Abd-Alla AMM. 2018. Enhancing vector refractoriness to trypanosome infection: achievements, challenges and perspectives. BMC Microbiol 18 (Suppl 1):179. 10.1186/s12866-018-1280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zaidman-Rémy A, Vigneron A, Weiss BL, Heddi A. 2018. What can a weevil teach a fly, and reciprocally? Interaction of host immune systemswith endosymbionts in Glossina and Sitophilus. BMC Microbiol 18(Suppl1):150. 10.1186/s12866-018-1278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ma D, Leulier F. 2018. The importance of being persistent: The first true resident gut symbiont in Drosophila. PLoS Biol 16:e2006945. 10.1371/journal.pbio.2006945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Graf J, Kikuchi Y, Rio RVM. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol 14:365–371. [DOI] [PubMed] [Google Scholar]