Abstract
Background
Breast cancer remains the most common malignancy in females around the world. Recently, a growing number of studies have focused on gene dysregulation. In our previous study, Krüppel‐like factors (KLFs) were found to play essential roles in breast cancer development, among which KLF2 could function as a tumor suppressor. Nevertheless, the underlying molecular mechanism remains unclear.
Methods
miR‐92a‐3p was identified as the upstream regulator of KLF2 by starBase v.3.0. The regulation of KLF2 by miR‐92a‐3p was verified by a series of in vitro and in vivo assays. Further exploration revealed that Baculoviral IAP Repeat Containing 5 (BIRC5) was the target of KLF2. ChIP assay, dual‐luciferase reporter analysis, quantitative real‐time PCR, and western blot were performed for verification.
Results
miR‐92a‐3p functioned as a tumor promoter by inhibiting KLF2 by binding to its 3'‐untranslated region (3'‐UTR). In addition, KLF2 could transcriptionally suppress the expression of BIRC5.
Conclusion
Collectively, our results uncovered the miR‐92a‐3p/KLF2/BIRC5 axis in breast cancer and provided a potential mechanism for breast cancer development, which may serve as promising strategies for breast cancer therapy.
Keywords: BIRC5, breast cancer, KLF2, miR‐92a‐3p, proliferation
miR‐92a‐3p promotes breast cancer proliferation both in vitro and in vivo.
KLF2 is a target of miR‐92a‐3p in breast cancer.
KLF2 transcriptionally repressed BIRC5 expression.
A model for the miR‐92a‐3p/KLF2/BIRC5 axis in breast cancer proliferation
![]()
INTRODUCTION
Breast cancer is a major life‐threatening disease for women worldwide, with an increasing annual incidence. 1 Breast cancer is divided into four subtypes according to the specific gene expression pattern and histological features: basal‐like subtypes, HER2 enriched, luminal A, and luminal B. 2 With recent advances in treatment strategies, the prognosis of breast cancer patients has greatly improved. 3 Studies have revealed the critical roles of gene dysregulation in breast cancer, which can identify novel biomarkers for prognosis and cancer therapy. 4 , 5 , 6
Krüppel‐like factors (KLFs) are transcription factors belonging to the zinc finger family. 7 Previous studies have demonstrated their roles in cell differentiation, proliferation, invasion, and survival. 8 , 9 , 10 KLF members are involved in a variety of cancer‐related biological processes, especially in colon cancer and rectal cancer. 11 KLFs can behave as both oncogenes and tumor suppressors. 8 In our previous study, we analyzed the expression pattern of KLFs and identified KLF2/15 as tumor suppressors that inhibited cell invasion and proliferation in breast cancer. 12 However, the underlying mechanism of KLFs is still uncertain and requires further investigation.
MicroRNAs (miRNAs) are a category of RNAs that do not encode proteins but have essential roles in cell biological processes. 13 Growing evidence has revealed that coding genes in many types of cancer are regulated by miRNAs. 14 In our previous study, miR‐1271 and miR‐543 were identified as tumor suppressors and functioned through Transforming Growth Factor β (TGF‐β) signaling. 15 , 16 Furthermore, some of the miRNA‐targeted therapeutics such as miR‐122 and miR‐34 have reached the clinical experiment stage. 13 Dysregulation of miR‐92a‐3p is involved in the initiation and development of many types of cancer. 17 , 18 For instance, miR‐92a‐3p promotes cell invasion, proliferation, and migration of esophageal squamous carcinoma by regulating phosphatase and tensin homolog. 19 In addition, miR‐92a‐3p was reported to be involved in the radiosensitivity of Non‐small‐cell lung cancer by targeting Mitogen‐Activated Protein Kinase Kinase (MAP2K) via c‐Jun N‐terminal Kinase signaling. 20 Nonetheless, the functional pattern of miR‐92a‐3p is uncertain.
In the current study, the mechanism of KLF2 in breast cancer progression is further explored, identifying miR‐92a‐3p as its upstream regulator, which can bind to its 3'‐UTR region. In addition, KLF2 could transcriptionally inhibit the expression of BIRC5 by interacting with its promoter region. These findings uncovered the existence of the miR‐92a‐3p/KLF2/BIRC5 axis in breast cancer regulation, which elucidated new mechanisms for breast cancer proliferation.
MATERIALS AND METHODS
Cell culture
MCF10A (normal breast epithelial cell line), MCF‐7, T47D, SKBR3, CAL‐51, and MDA‐MB‐231 (breast cancer cell lines) were obtained from American Type Culture Collection. All cells were cultured in their corresponding medium with 1% penicillin/streptomycin (Gibco, Thermo Fisher Scientific) and 10% fetal bovine serum (Gibco) at 37°C in a humidified 5% CO2 atmosphere cell incubator.
Plasmids, miRNA, and transfection
The KLF2 expression plasmid was purchased from Miaolingbio. The miR‐92a‐3p mimic, inhibitor, and their corresponding controls were purchased from Ribobio. For transfection, Lipofectamine 3000 (Invitrogen) was utilized following the manufacturer's instructions.
Quantitative real‐time PCR
TRIZOL (Thermo) was utilized for the extraction of total RNA from cells in accordance with protocols. The RNA quality was measured by NanoDrop 2000 spectrophotometer (Thermo), and quantitative real‐time PCR (qRT‐PCR) was conducted using GoTaq qPCR Master Mix (Promega). The 2−ΔΔCt method was used for mRNA level analysis, 21 which was presented as a relative expression level compared with the control group. The primers utilized for qRT‐PCR are listed in Supporting Information Tables S1–S4.
Western blot
The transfected cells were treated with SDS buffer (supplemented with 1% protease inhibitor cocktail) for protein extraction. The protein concentration was measured by Bicinchoninicacid kit (Thermo Scientific) and SDS‐PAGE was performed to separate proteins, which were transferred onto polyvinylidene fluoride (PVDF) membranes (Thermo Scientific). The PVDF membranes were incubated with 5% bovine serum albumin and treated with primary antibodies at 4°C overnight. After washing three times with TBST and incubation with secondary antibodies, the signal intensity of the complexes was visualized with enhanced chemiluminescence reagent (Millipore). The details of the antibodies are shown in Tables S1–S4.
miRNA pull‐down assay
The biotinylated miR‐92a‐3p mimic or control RNA (Ribo bio) were transfected into MCF‐7 cells at a concentration of 100 nM. The cells were harvested after incubation for 48 h. The biotinylated RNA complex was pulled down by magnetic beads at 4°C overnight. Then, RNAs from the input and pull‐down beads were extracted by TRIZOL (Thermo), and the abundance of GAPDH and KLF2 was detected by qRT‐PCR assay. The primers utilized for qRT‐PCR are detailed in Tables S1–S4.
Dual‐luciferase reporter assay
The wild‐type and mutant oligonucleotides of the KLF 3'‐UTR region with miR‐92a‐3p binding sites or BIRC5 promoter region with KLF2 binding sites were cloned into the luciferase plasmid. The luciferase reporter was then transfected into 293FT cells with miR‐92a‐3p mimic or KLF2 plasmid and their negative controls, as previously mentioned. The luciferase activities were estimated through a Dual‐Luciferase Reporter Assay Kit (Transgene) according to the manufacturer's instructions.
Cell proliferation assay
For the MTT assay, a total of 2 × 103 cells were planted into 96‐well plates and incubated for 5 days. At the same time each day, 20 μl of MTT solution (5 mg/ml) was added to the cells and they were then incubated in the dark for 4 h. After dissolving the formazan crystals into 150 μl of DMSO, the absorbance at 570 nm wavelength was detected by a microplate reader (Bio‐Rad).
For colony formation assay, 800 cells were seeded in six‐well plates and incubated for the appropriate time period. When colonies grew to the appropriate size, the cells were fixed with 4% paraformaldehyde. Finally, 1% crystal violet was utilized for staining.
For the EdU assay, 4 × 104 cells were planted into 24‐well plates and treated with 25 μM 5‐ethynyl‐2'‐deoxyuridine (EdU) for 12 h. Subsequently, the cells were fixed, permeabilized, and stained following the protocol of the EdU labeling/detection kit (Ribobio). The percentage of EdU‐positive cells was captured and analyzed.
Flow cytometry
After transfection and incubation for 48 h, cells were harvested and fixed overnight with 95% ethanol at −20°C. After washing with phosphate buffer saline (PBS) three times, the cells were stained with 5 μl of propodeum iodide solution (Thermo) for 15 min at room temperature. The distribution of cell cycles was analyzed by a flow cytometer (BD Biosciences).
Xenograft
A total of 4 × 106 stable miR‐92a‐3p‐overexpressed MDA‐231 cells were subcutaneously injected into female server combined immune‐deficiency (SCID) mice (5–6 weeks old). The tumor volume was observed and recorded once a week. The mice were executed when the tumor reached the appropriate size. The paraffin‐embedded tumors were sectioned for IHC staining. The animal protocols were sanctioned by the Animal Ethics Committee of Tianjin Medical University Cancer Institute & Hospital.
Immunohistochemistry
The deparaffinized sections of mice tumors underwent heat‐induced antigen retrieval and endogenous peroxidase activity block. After treatment with 5% Bovine Serum Albumin the sections were treated with the primary antibodies at 4°C overnight. The sections were washed with PBS and treated with secondary antibodies, and then visualized with 3,3′‐diaminobenzidine reagent. Eventually, the sections were dehydrated and covered by neutral gum.
Chromatin Immuno‐Precipitation (ChIP) analysis
ChIP analysis was conducted using Upstate Biotechnology following the manufacturer's instruction as explained previously. 22 The fold change of percentage input was used to calculate the enrichment of the analyzed DNA region. The sequences of primers used for ChIP are listed in Tables S1–S4.
Statistical analysis
The data were analyzed by SPSS software 22.0 and compared by Student's t‐test or one‐way analysis of variance. The value of p < 0.05 was considered statistically significant.
RESULTS
KLF2 is a direct target of miR‐92a‐3p
StarBase v.3.0 was utilized to investigate the potential upstream regulators of KLF2 and provided several candidate miRNAs through multiple target‐predicting programs (Figure 1a). The expression of KLF2 and candidate miRNAs was analyzed by starBase v.3.0 to filter the targets, which showed that miR‐92a‐3p had the most significant negative correlation with KLF2 (Figure 1b). KLF2 was identified as a tumor suppressor in our previous study; however, the function of miR‐92a‐3p in breast cancer remains unclear. KM plotter was used to investigate the correlation between miR‐92a‐3p and prognosis, and showed that the overexpression of miR‐92a led to poor clinical outcomes (Figure S1). KLF2 overexpression was related to better prognosis in our previous study, 12 which was contrary to the result of miR‐92a. The expression level of miR‐92a‐3p was upregulated in all breast cancer cell lines compared with normal breast cells (Figure 1c). However, KLF2 was found to be remarkably downregulated in breast cancer cell lines compared with MCF‐10A cells in our previous study. 12 Moreover, qRT‐PCR (Figure 1d) and western blot (Figure 1e) revealed that the expression level of KLF2 was significantly decreased in miR‐92a‐3p‐overexpressed MCF‐7 and T47D cell lines but increased in miR‐92a‐3p‐depleted CAL‐51 and MDA‐231 cells. The putative binding site of miR‐92a‐3p with KLF2 was predicted by TargetScan (Figure 1f). Furthermore, the pull‐down assay revealed a notably higher KLF2 enrichment ratio of biotin‐miR‐92a‐3p (Figure 1g). To confirm the regulation of miR‐92a‐3p on KLF2, the predicted binding site wild‐type or mutant 3' UTR was cloned into the luciferase reporter plasmid. Overexpressed miR‐92a‐3p resulted in an approximately 40% decrease in luciferase activity, while no change was found in the mutant group (Figure 1h). These findings revealed that KLF2 is a direct target of miR‐92a‐3p.
FIGURE 1.
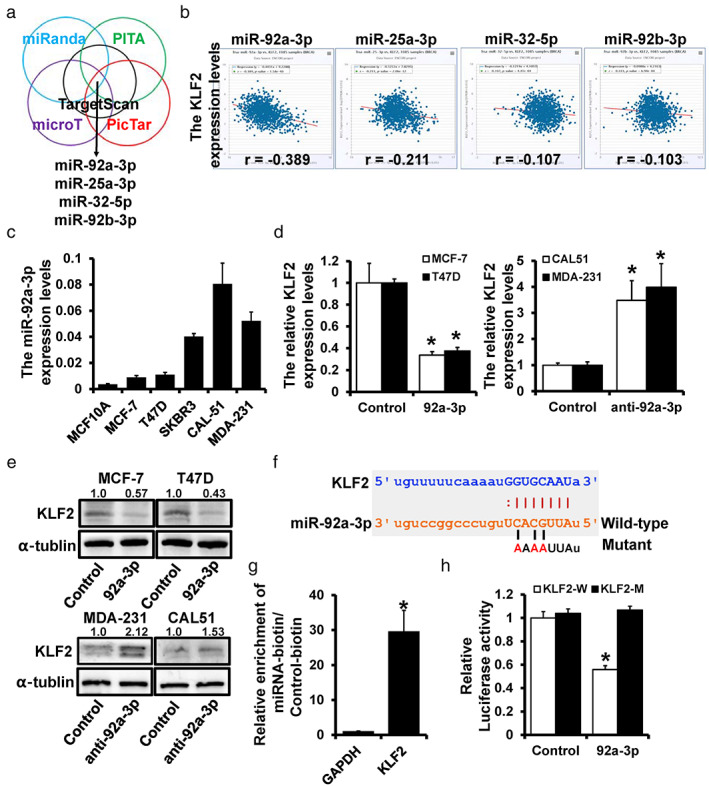
KLF2 is a direct target of miR‐92a‐3p. (a) The potential regulators of KLF2 predicted by starBase v.3.0. (b) The correlation between candidate miRNAs and KLF2 expression. (c) The relative miR‐92a‐3p expression levels in breast cell lines were determined by qRT‐PCR. (d) The mRNA expression levels of KLF2 in miR‐92a‐3p overexpressed MCF‐7 cells and T47D cells (left), and in miR‐92a‐3p‐depleted CAL‐51 cells and MDA‐231 cells (right) detected by qRT‐PCR. (e) The protein levels of KLF2 in miR‐92a‐3p overexpressed MCF‐7 cells and T47D cells, and in miR‐92a‐3p‐depleted CAL‐51 cells and MDA‐231 cells were detected by western blot. (f) The predicted binding of miR‐92a‐3p with KLF2 3′ UTR. (g) Enrichment of KLF2 in MCF‐7 cells after pull‐down assay with biotinylated miR‐92a‐3p. (h) The regulation of miR‐92a‐3p on KLF2 was detected by a dual‐luciferase reporter assay. *p < 0.05
miR‐92a‐3p‐deleption inhibits breast cancer proliferation
Although previous studies indicated that miR‐92a‐3p performed as a tumor promoter in multiple cancers, the role of miR‐92a‐3p in breast cancer remains uncertain. We constructed stable miR‐92a‐3p‐depleted MDA‐231 and CAL‐51 cells, which were verified by qRT‐PCR (Figure 2a). The MTT assay indicated that downregulated miR‐92a‐3p remarkably inhibited cell viability both in MDA‐231 and CAL‐51 cells (Figure 2b). The consistent results were observed by colony formation assay (Figure 2c) and EdU assay (Figure 2d). To further explore the performance of miR‐92a‐3p in cell cycle progression, a flow cytometry assay was conducted and showed a decreased proportion of S phase cells in miR‐92a‐3p‐depleted MDA‐231 cells and CAL‐51 cells (Figure 2e). In vivo, the stable miR‐92a‐3p‐depleted MDA‐231 cells were subcutaneously injected into SCID mice, demonstrating a significant decrease in tumors in the miR‐92a‐3p‐depleted group compared with the control group. In addition, the expression of Ki‐67 was also decreased in miR‐92a‐3p‐depleted MDA‐231 cells (Figure 2f). Thus, miR‐92a‐3p‐depletion can inhibit cell proliferation both in vitro and in vivo.
FIGURE 2.
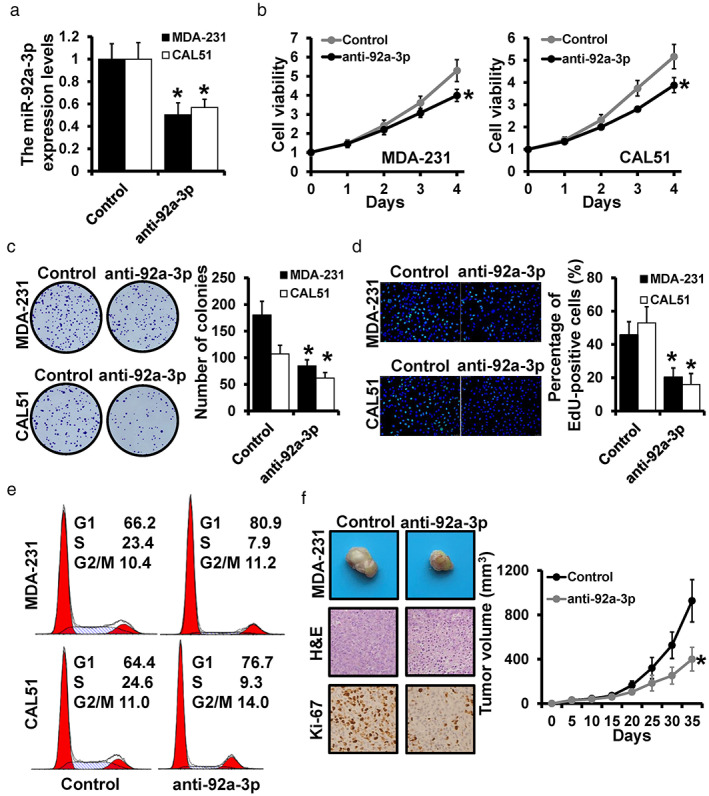
miR‐92a‐3p‐depletion inhibits breast cancer progression. (a) The expression levels of miR‐92a‐3p in miR‐92a‐3p‐depleted CAL‐51 cells and MDA‐231 cells were detected by qRT‐PCR. For cell proliferation assay, MTT (b), colony formation assay (c), and EdU assay (d) were performed in miR‐92a‐3p‐depleted CAL‐51 cells and MDA‐231 cells. (e) The distribution of the cell cycle was detected by flow cytometry. (f) Representative photos and volumes of tumors, as well as H&E staining or Ki‐67 expression of xenograft tumors. *p < 0.05
miR‐92a‐3p promotes breast cancer proliferation by regulation of KLF2
To investigate whether miR‐29a‐3p functioned through regulating KLF2, we transfected KLF2 expression plasmid into miR‐92a‐3p‐overexpressed MCF‐7 and T47D cells, which was verified by qRT‐PCR (Figure 3a). The upregulation of KLF2 remarkably decreased the cell viability (Figure 3b), the number of colonies (Figure 3c), and the percentage of EdU‐positive cells (Figure 3d) compared with miR‐92a‐3p‐overexpressed MCF‐7 and T47D cells. As for cell cycle progression, we observed that overexpression of KLF2 significantly decreased the proportion of cells in the S phase in miR‐92a‐3p‐overexpressed cells (Figure 3e). Briefly, the upregulation of KLF2 reversed the promotion effect of miR‐92a‐3p on breast cancer cells, which illustrated that miR‐92a‐3p functioned by regulating KLF2.
FIGURE 3.
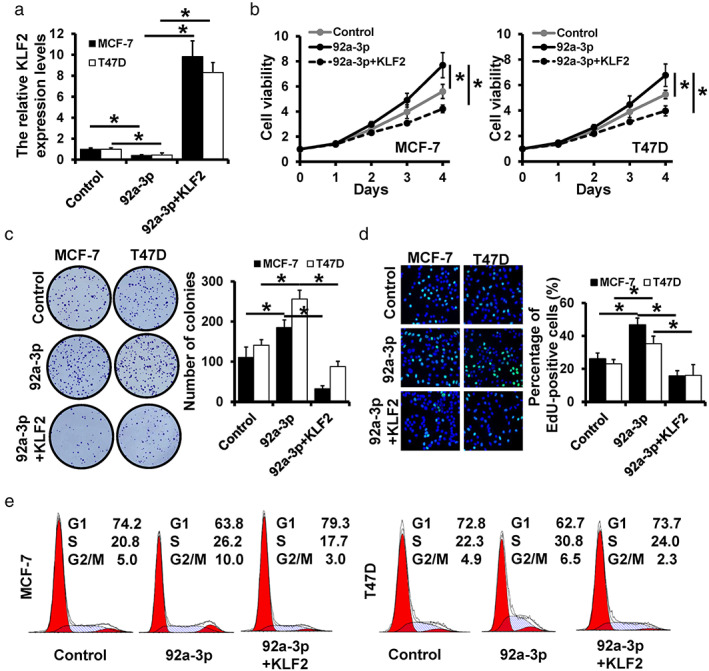
miR‐92a‐3p promotes breast cancer progression by regulation of KLF2. (a) The expression level of KLF2 in miR‐92a‐3p‐ or KLF2/miR‐92a‐3p‐overexpressed MCF‐7 and T47D cells detected by qRT‐PCR. Cell proliferation assays, including MTT (b), colony formation (c), and EdU assay (d), were performed. (e) The distribution of cell cycle in miR‐92a‐3p‐overexpressed and KLF2/miR‐92a‐3p‐overexpressed cells, as well as the control cells detected by flow cytometry. *p < 0.05
KLF2 transcriptionally repressed BIRC5 expression
In our previous study, overexpression of KLF2 led to downregulation of Survivin (BIRC5) and Cyclin D1 (CCND1). 12 To further clarify the mechanism of KLF2, the expression levels of KLF2 with BIRC5 and CCND1 were explored with starBase v.3.0. BIRC5 showed a stronger negative correlation and perhaps interacted with KLF2 (Figure 4a). The transcription factor profiles were obtained from the JASPAR website and the binding site of KLF2 on the BIRC5 promoter region (−5000 to +1) was predicted (Figure 4b). The occupancy of KLF2 in the BIRC5 promoter region was detected in MCF‐7 and MDA‐231 cells through ChIP analysis (Figure 4c). To investigate the regulation of KLF2 on BIRC5, the wild‐type and mutant predicted binding sites were cloned into luciferase reporter plasmids, revealing notably decreased luciferase activity in the wild‐type group (Figure 4d). The expression correlation of KLF2 and BIRC5 was further verified, and the results indicated that KLF2 overexpression notably downregulated the expression of BIRC5. In contrast, KLF2 depletion increased the expression level of BIRC5, as evidenced by the qRT‐PCR (Figure 4e) and western blot (Figure 4f) results. These findings indicate that KLF2 transcriptionally inhibits BIRC5 expression.
FIGURE 4.
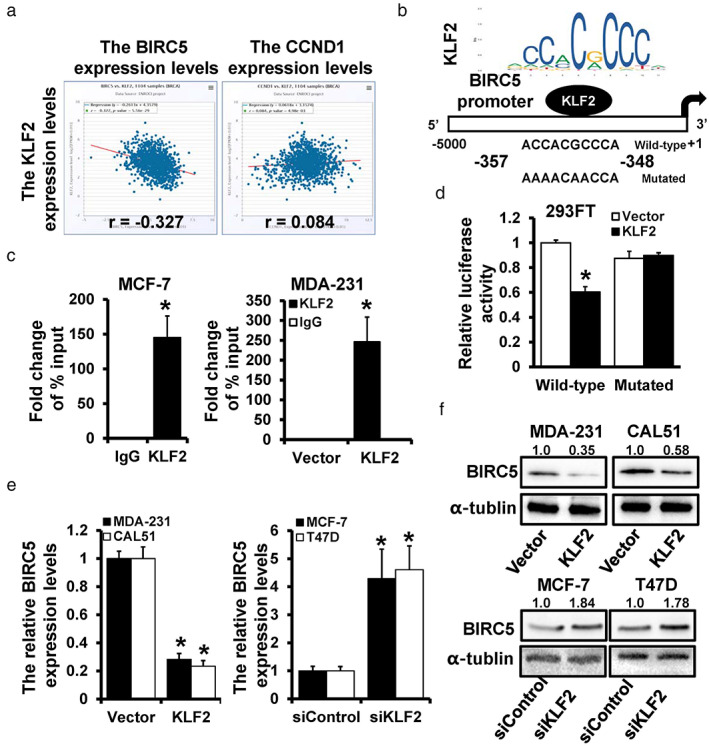
KLF2 repressed BIRC5 expression transcriptionally. (a) The correlation expression levels of KLF2 with BIRC5 and CCND1. (b) The potential binding site of KLF2 on the BIRC5 promoter region was predicted by JASPAR. (c) ChIP analysis for investing the interaction between KLF2 and BIRC5 promoter, including endogenous interaction (left) and exogenous interaction (right). (d) Dual‐luciferase reporter analysis of BIRC5 promoter activity in KLF2‐transfected 293FT cells. The expression level of BIRC5 in KLF2‐overexpressed MDA‐231 and CAL‐51 cells, as well as in KLF2‐depleted MCF‐7 and T47D cells, was detected by qRT‐PCR (e) and western blot (f). *p < 0.05
DISCUSSION
KLFs participate in diversified cellular biological processes and human diseases, including multiple types of human cancers. In our previous study, we demonstrated the abnormal expression of KLFs in breast cancer and identified KLF2 and KLF15 as prognostic predictors. The critical roles of KLF2 in cancers have been extensively studied. For instance, KLF2 can suppress cell invasion and migration by targeting GPX4 in clear cell renal cell carcinoma. 23 In addition, KLF2 acted as a tumor suppressor in hepatocellular carcinoma via a negative feedback loop over TGF‐β signaling. 24 Consistent with our previous study, KLF2 inhibited tumor proliferation and induced cell cycle arrest. However, the regulation mechanism remains unclear.
miRNAs serve as noncoding RNAs, which participate in gene expression regulation through binding with the 3'‐UTR region of target genes. In the current study, we identified miR‐92a‐3p as the upstream regulator of KLF2. The abnormal expression level of miR‐92a‐3p has been observed in a variety of cancer types. However, miR‐92a‐3p can act as both a tumor suppressor and a promoter. Li et al. reported decreased miR‐92a‐3p levels in Wilms' tumor, which inhibited tumor progression by targeting FRS2. 25 Previous studies have demonstrated the regulation of miR‐92a‐3p on KLFs. Yang et al. reported that miR‐92a‐3p overexpression was related to cisplatin resistance and regulated KLF4‐mediated EMT and cell apoptosis in cervical cancer. 26 Moreover, Mao et al. revealed the negative association between miR‐92a‐3p and KLF2 in gastric cancer. 27 Wang et al. reported that downregulated miR‐92a‐3p could inhibit apoptosis and promote cell proliferation by enhancing KLF2 expression. 28 In this study, miR‐92a‐3p promoted breast cancer progression in vivo and in vitro. Pull‐down assay with biotined‐miR‐92a‐3p and dual‐luciferase reporter assay indicated the regulation of miR‐92a‐3p on KLF2. In addition, KLF2 overexpression reversed the promotion of miR‐92a‐3p on breast cancer cell lines. Thus, miR‐92a‐3p promotes breast cancer progression by directly targeting KLF2.
In our previous study, we illustrated that the expression of tumor promotors cyclin D1 and survivin was downregulated in KLF2‐overexpressed breast cancer cells. Here, we further investigate the regulation of KLF2 on BIRC5. BIRC5 (survivin) is a well‐known dual cellular function protein that can directly regulate mitosis and apoptosis in cancer cells during tumorigenesis and tumor metastasis. 29 Thus, BIRC5 is commonly considered a biomarker in human cancer, and many studies have uncovered the critical role of BIRC5 in breast cancer. Nina et al. demonstrated that BIRC5 is a sensitive survival marker that functions independently of estrogen receptor (ER) and nodal status, and expression levels should be taken into consideration while formulating clinical treatment strategies. 30 In addition, extensive studies have identified BIRC5 as an interacted target in cancer progression. Cao et al. reported that OCT4 could increase the expression of BIRC5 and CCND1, and promote cancer progression in hepatocellular carcinoma. 31 Wang et al. clarified that miR‐218 promoted apoptosis of U2OS osteosarcoma cells by targeting BIRC5. 32 In addition, it has been reported that BIRC5 was the direct target of KLF2 and miR‐126‐5p could promote KLF2 expression to inhibit BIRC5 activation in lung adenocarcinoma cells. 33 In the current study, the potential binding site of KLF2 on the BIRC5 promoter region was identified and verified through ChIP analysis and dual‐luciferase reporter analysis. The results indicate that KLF2 could suppress BIRC5 expression transcriptionally.
CONCLUSION
In summary, the miR‐92a‐3p/KLF2/BIRC5 regulation axis was identified in breast cancer proliferation, which may serve as emerging cancer biomarkers and provide promising strategies for breast cancer therapy.
AUTHOR CONTRIBUTIONS
C.X.C. and W.X. designed the study. Y.Z.H., C.Z.H., Z.G.L., Z.X.J., and M.H.Y. performed the experiments and statistical analysis. Y.Z.H., W.X., and C.X.C. drafted the article. W.X. and C.X.C. reviewed and revised the paper. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
CONSENT FOR PUBLICATION
Not applicable.
DECLARATIONS
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
DATA AVAILABILITY
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Supporting information
Supporting Information Table S1Oligonucleotides of miRNAs and siRNAs
Supporting Information Table S2 Oligonucleotides used for RT‐qPCR
Supporting Information Table S3 Antibodies used for study
Supporting Information Table S4 Oligonucleotides used for ChIP
ACKNOWLEDGMENT
This study was supported by the grants from National Natural Science Foundation of China (No. 82172827 and No.82172835).
Yu Z‐H, Chen Z‐H, Zhou G‐L, Zhou X‐J, Ma H‐Y, Yu Y, et al. miR‐92a‐3p promotes breast cancer proliferation by regulating the KLF2/BIRC5 axis. Thorac Cancer. 2022;13(21):2992–3000. 10.1111/1759-7714.14648
Funding information National Natural Science Foundation of China, Grant/Award Numbers: 82172827, 82172835
Contributor Information
Xin Wang, Email: wangxin@tjmuch.com.
Xu‐Chen Cao, Email: caoxuchen@tmu.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–50. [DOI] [PubMed] [Google Scholar]
- 4. Hammerl D, Smid M, Timmermans AM, Sleijfer S, Martens J, Debets R. Breast cancer genomics and immuno‐oncological markers to guide immune therapies. Semin Cancer Biol. 2018;52(Pt 2):178–88. [DOI] [PubMed] [Google Scholar]
- 5. Zeng X, Liu C, Yao J, Wan H, Wan G, Li Y, et al. Breast cancer stem cells, heterogeneity, targeting therapies and therapeutic implications. Pharmacol Res. 2021;163:105320. [DOI] [PubMed] [Google Scholar]
- 6. Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, et al. CXCL1 derived from tumor‐associated macrophages promotes breast cancer metastasis via activating NF‐κB/SOX4 signaling. Cell Death Dis. 2018;9(9):880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rane MJ, Zhao Y, Cai L. Krϋppel‐like factors (KLFs) in renal physiology and disease. EBioMedicine. 2019;40:743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tetreault MP, Yang Y, Katz JP. Krüppel‐like factors in cancer. Nat Rev Cancer. 2013;13(10):701–13. [DOI] [PubMed] [Google Scholar]
- 9. Yang Y, Nakagawa H, Tetreault MP, Billig J, Victor N, Goyal A, et al. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res. 2011;71(20):6475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tetreault MP, Wang ML, Yang Y, et al. Klf4 overexpression activates epithelial cytokines and inflammation‐mediated esophageal squamous cell cancer in mice. Gastroenterology. 2010;139(6):2124–2134.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang Z, He H, Qiu F, Qian H. Expression and prognosis value of the KLF family members in colorectal cancer. J Oncol. 2022;2022:6571272–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu KY, Tian Y, Li YX, Meng QX, Ge J, Cao XC, et al. The functions and prognostic value of Krüppel‐like factors in breast cancer. Cancer Cell Int. 2022;22(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. [DOI] [PubMed] [Google Scholar]
- 14. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu BW, Yu ZH, Chen AX, Chi JR, Ge J, Yu Y, et al. Estrogen receptor‐α‐miR‐1271‐SNAI2 feedback loop regulates transforming growth factor‐β‐induced breast cancer progression. J Exp Clin Cancer Res. 2019;38(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ji W, Mu Q, Liu XY, Cao XC, Yu Y. ZNF281‐miR‐543 feedback loop regulates transforming growth factor‐β‐induced breast cancer metastasis. Mol Ther Nucleic Acids. 2020;21:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR‐92a‐3p in gastric cancer. Mol Cancer. 2018;17(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cun J, Yang Q. Bioinformatics‐based interaction analysis of miR‐92a‐3p and key genes in tamoxifen‐resistant breast cancer cells. Biomed Pharmacother. 2018;107:117–28. [DOI] [PubMed] [Google Scholar]
- 19. Li X, Guo S, Min L, Guo Q, Zhang S. miR‐92a‐3p promotes the proliferation, migration and invasion of esophageal squamous cell cancer by regulating PTEN. Int J Mol Med. 2019;44(3):973–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Wang Z, Liu L, Du Y, Mi Y, Wang L. The HNF1A‐AS1/miR‐92a‐3p axis affects the radiosensitivity of non‐small cell lung cancer by competitively regulating the JNK pathway. Cell Biol Toxicol. 2021;37(5):715–29. [DOI] [PubMed] [Google Scholar]
- 21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 22. Yu Y, Luo W, Yang ZJ, Chi JR, Li YR, Ding Y, et al. miR‐190 suppresses breast cancer metastasis by regulation of TGF‐β‐induced epithelial‐mesenchymal transition. Mol Cancer. 2018;17(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu Y, Qin H, Jiang B, Lu W, Hao J, Cao W, et al. KLF2 inhibits cancer cell migration and invasion by regulating ferroptosis through GPX4 in clear cell renal cell carcinoma. Cancer Lett. 2021;522:1–13. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Tu S, Zeng Y, Zhang C, Deng T, Luo W, et al. KLF2 inhibits TGF‐β‐mediated cancer cell motility in hepatocellular carcinoma. Acta Biochim Biophys Sin (Shanghai). 2020;52(5):485–94. [DOI] [PubMed] [Google Scholar]
- 25. Li JL, Luo P. MiR‐140‐5p and miR‐92a‐3p suppress the cell proliferation, migration and invasion and promoted apoptosis in Wilms' tumor by targeting FRS2. Eur Rev Med Pharmacol Sci. 2020;24(1):97–108. [DOI] [PubMed] [Google Scholar]
- 26. Yang J, Hai J, Dong X, Zhang M, Duan S. MicroRNA‐92a‐3p enhances cisplatin resistance by regulating Krüppel‐like factor 4‐mediated cell apoptosis and epithelial‐to‐mesenchymal transition in cervical cancer. Front Pharmacol. 2021;12:783213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mao QQ, Chen JJ, Xu WJ, Zhao XZ, Sun X, Zhong L. miR‐92a‐3p promotes the proliferation and invasion of gastric cancer cells by targeting KLF2. J Biol Regul Homeost Agents. 2020;34(4):1333–41. [DOI] [PubMed] [Google Scholar]
- 28. Ling L, Wang HF, Li J, Li Y, Gu CD. Downregulated microRNA‐92a‐3p inhibits apoptosis and promotes proliferation of pancreatic acinar cells in acute pancreatitis by enhancing KLF2 expression. J Cell Biochem. 2020;121:3739‐3751. [DOI] [PubMed] [Google Scholar]
- 29. Lin TY, Chan HH, Chen SH, Sarvagalla S, Chen PS, Coumar MS, et al. BIRC5/Survivin is a novel ATG12‐ATG5 conjugate interactor and an autophagy‐induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy. 2020;16(7):1296–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oparina N, Erlandsson MC, Fäldt Beding A, Parris T, Helou K, Karlsson P, et al. Prognostic significance of BIRC5/Survivin in breast cancer: results from three independent cohorts. Cancers (Basel). 2021;13(9):2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao L, Li C, Shen S, Yan Y, Ji W, Wang J, et al. OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer. 2013;13:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang DZ, Jing SF, Hao SB, Huang XY, Miao QT, Gao JF. MiR‐218 promotes apoptosis of U2OS osteosarcoma cells through targeting BIRC5. Eur Rev Med Pharmacol Sci. 2018;22(20):6650–7. [DOI] [PubMed] [Google Scholar]
- 33. Han F, Huang D, Meng J, Chu J, Wang M, Chen S. miR‐126‐5p enhances radiosensitivity of lung adenocarcinoma cells by inhibiting EZH2 via the KLF2/BIRC axis. J Cell Mol Med. 2022;26(9):2529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table S1Oligonucleotides of miRNAs and siRNAs
Supporting Information Table S2 Oligonucleotides used for RT‐qPCR
Supporting Information Table S3 Antibodies used for study
Supporting Information Table S4 Oligonucleotides used for ChIP
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


