Abstract
As the primary phagocytic cells of the central nervous system, microglia exquisitely regulate their lysosomal activity to facilitate brain development and homeostasis. However, mechanisms that coordinate lysosomal activity with microglia development, chemotaxis, and function remain unclear. Here, we show that embryonic macrophages require the lysosomal guanosine triphosphatase (GTPase) RagA and the GTPase-activating protein Folliculin to colonize the brain in zebrafish. We demonstrate that embryonic macrophages in rraga mutants show increased expression of lysosomal genes but display significant down-regulation of immune- and chemotaxis-related genes. Furthermore, we find that RagA and Folliculin repress the key lysosomal transcription factor Tfeb and its homologs Tfe3a and Tfe3b in the macrophage lineage. Using RNA sequencing, we establish that Tfeb and Tfe3 are required for activation of lysosomal target genes under conditions of stress but not for basal expression of lysosomal pathways. Collectively, our data define a lysosomal regulatory circuit essential for macrophage development and function in vivo.
The degradation machinery of the cell must be carefully controlled for the normal function of immune cells in the brain.
INTRODUCTION
Microglia, the primary resident macrophages of the central nervous system, govern multiple aspects of brain architecture and function. During development, microglia promote synapse formation and maturation, stimulate neurogenesis, and eliminate excess or apoptotic cells (1–4). To maintain adult brain homeostasis, microglia surveil the brain for aberrations, mediate repair and regeneration, and facilitate immune clearance of pathogens and cellular debris (5–10). As professional phagocytic cells, nearly all these activities of microglia require lysosomes for either degradation of engulfed material or mounting immune responses (11–13). Although many studies have found that aberrant lysosomal activity in microglia is associated with neurodegeneration (14–16), much less is known about the mechanisms that regulate lysosomal pathways during microglia development. A better understanding of genes regulating lysosomal function during development and homeostasis has the potential to illuminate how these critical organelles might be disrupted in neurodevelopmental and neurodegenerative diseases.
Central to lysosomal functions are transcription factor EB (Tfeb) and other members of the microphthalmia-associated transcription factor (MiTF) protein family. In cell culture and some cell types in vivo, Tfeb and the related transcription factor Tfe3 have been reported to activate diverse lysosomal processes, such as lysosomal biogenesis, autophagy, and exocytosis (11, 17–20). In recent years, there has been an increasing appreciation of the dysregulation of Tfeb in neurodegenerative diseases (21–25), which has led to attempts toward enhancing cellular clearance by overexpressing Tfeb to activate autophagy and lysosomal pathways in cell culture or animal models of neurodegeneration (24, 26–31). Although exogenous expression of Tfeb improves behavioral deficits and reduces pathology due to misfolded proteins in some models of neurodegenerative diseases (29–31), the identity of cells that must overexpress Tfeb to generate these beneficial effects is unclear. Furthermore, the role of Tfeb in microglia, cells that depend on lysosomal pathways for executing many of their functions, is unknown.
The importance of investigating the development and function of microglia in vivo is evident from experiments showing that microglia rapidly lose their transcriptional and epigenetic identity when separated from their niche (32–36). Accordingly, studies of microglia in zebrafish using live imaging and mutational studies have led to many important insights into the development and function of these critical glial cells (37–42). Here, we capitalize on the experimental advantages of zebrafish to functionally define a lysosomal regulatory circuit that is essential in development and function of microglia. We show that the lysosomal guanosine triphosphatase (GTPase) RagA (encoded by rraga) is necessary for the normal development and function of both early microglia and peripheral macrophages. We demonstrate that macrophages in rraga mutants significantly up-regulate lysosomal genes but down-regulate immune- and chemotaxis-related genes, revealing a previously unknown role for RagA in the innate immune system. We provide genetic evidence that the lysosomal GTPase-activating protein Folliculin (Flcn) is functionally coupled to RagA in macrophages and microglia. Furthermore, we show that simultaneous loss of tfeb and tfe3 (including zebrafish tfe3 paralogs, tfe3a and tfe3b) is sufficient to restore microglia numbers in rraga and flcn mutants; correspondingly, overexpression of tfe3b in the macrophage lineage recapitulates all the rraga or flcn microglia phenotypes we examined. Last, we establish that Tfeb and Tfe3 are dispensable for basal levels of lysosomal gene expression but demonstrate that these transcription factors are required to activate lysosomal pathways in response to cellular stress. Together, our observations reveal a lysosomal pathway that regulates development and function of microglia and macrophages in vivo. In addition, our observation that Tfeb and Tfe3 must be repressed for normal development and function of microglia has important implications for therapeutic strategies that modulate Tfeb activity in the context of neurodegeneration.
RESULTS
Embryonic macrophages fail to colonize the brain in rraga mutants
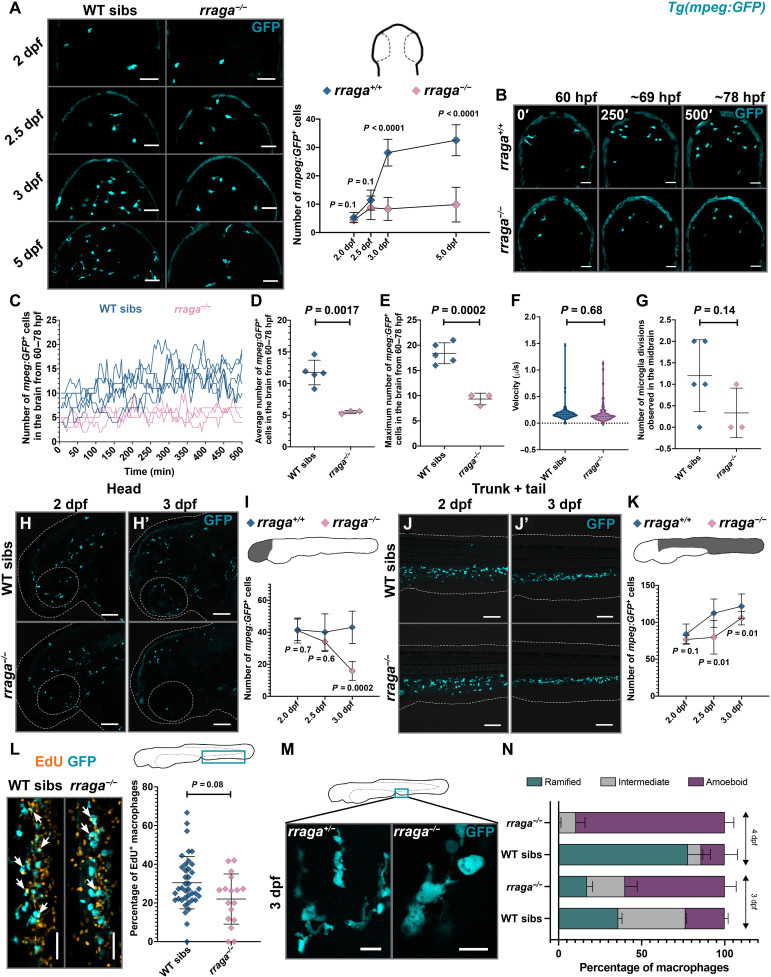
We have previously shown that zebrafish mutants for the gene rraga, which encodes the lysosomal GTPase RagA, have fewer microglia than their wild-type siblings (43). Previous studies have not, however, addressed the developmental timing or cause of this reduction in microglial cell number in rraga mutants. To define the disrupted developmental processes underlying the rraga mutant phenotype, we first examined the specification of embryonic macrophages that give rise to microglia. We quantified these cells in embryos between 24 and 26 hours post-fertilization (hpf), using the Tg(mpeg:GFP) transgene, which drives the expression of green fluorescent protein (GFP) in the macrophage lineage (44). We observed no significant difference in the number of GFP-labeled cells between rraga mutants and their wild-type siblings in either the yolk, where these macrophages are initially specified (fig. S1, A and B) (45, 46), or the rest of the body (fig. S1, A and C), indicating that the initial specification and colonization of embryonic macrophages is normal at this stage in rraga mutants.
To define the developmental window during when reduction of microglia numbers becomes apparent in rraga mutants, we quantified macrophages in the dorsal midbrain, where microglia are easily visualized, at 2, 2.5, 3, and 5 days post-fertilization (dpf) (Fig. 1A). There was a significant reduction in the number of developing microglia in the midbrain of rraga mutants at 3 dpf, but not in earlier stages (Fig. 1A). To investigate how the reduction of macrophage numbers in the brain occurs in rraga mutants, we performed time-lapse imaging starting at 60 hpf and ending at ~78 hpf (Fig. 1B). Analysis of the time-lapse movies showed that fewer macrophages migrate into the brain in rraga mutants (Fig. 1, C to E, and movies S1 and S2). Time-lapse imaging did not reveal any significant difference between rraga mutants and wild-type siblings in the velocity of macrophage migration (Fig. 1F), proliferation of macrophages in the midbrain (Fig. 1G), or apoptosis of macrophages (the absence of pycnotic macrophage nuclei in time-lapse movies). Although we cannot rule out subtle defects in proliferation, apoptosis, or specification that could affect microglia cell numbers, our observations suggest that embryonic macrophages in rraga mutants are unable to respond appropriately to chemotactic cues in the brain that direct the colonization of macrophages to form microglia (39, 47).
Fig. 1. Defective macrophages in rraga mutants fail to colonize the brain.
(A) Dorsal views of the midbrain showing mpeg:GFP expression at the indicated stages along with the corresponding quantification of developing microglia in the midbrain. WT, wild-type. (B) Representative images from time-lapse movies between 60 and ~78 hpf (dorsal views, anterior on top). Images show maximum intensity projections of z-slices at indicated times. (C to G) Quantitative analysis of time-lapse movies. In (C), each line shows macrophage cell counts from a single embryo over time. In (D), (E), and (G), each point represents counts from a single animal. (H to K) Lateral views of peripheral macrophages and quantification in rraga mutants and their siblings. (H, H′, and I) Macrophages in the head and (J, J′, and K) macrophages in the trunk + tail. Graphs in (A), (D) to (G), (I), and (K) show mean + SD; significance was determined using parametric unpaired t test. (L) EdU labeling to determine the percentage of proliferating macrophages in rraga mutants and wild-type siblings. Arrows note colocalization of EdU label with macrophages. Each point in the graph represents percentage of EdU+ macrophages from a single animal. Graph in (L) shows mean + SD; significance was determined using nonparametric Mann-Whitney U test. (M) High-magnification images showing the difference in the morphology of macrophages between rraga mutants and heterozygous animals at 3 dpf. (N) Quantification of amoeboid morphology of macrophages in rraga mutants. Graph in (N) shows mean + SD; significance was determined using nonparametric Mann-Whitney U test. The number of animals analyzed for each experiment is listed in table S1; all the panels are representative of at least two independent experiments. Scale bars, 50 μm.
Peripheral macrophages are disrupted in rraga mutants
Previous studies have not examined the extent to which macrophages outside the brain are affected in rraga mutants (43). We used the Tg(mpeg:GFP) transgene to visualize macrophages at 2, 2.5, and 3 dpf. In lateral views of the head, the numbers of macrophages were similar between rraga mutants and their wild-type siblings at 2 dpf (Fig. 1, H and I, and fig. S1H), and rraga mutants showed a slight but insignificant reduction of macrophages in this region at 2.5 dpf (Fig. 1I and fig. S1, D and H). There was, however, a notable and significant reduction of macrophage numbers in the head in rraga mutants by 3 dpf (Fig. 1, H′ and I, and fig. S1H), with very few macrophages remaining in the mutants. Although macrophage numbers were reduced in rraga mutants beginning at 2.5 dpf in the trunk and tail regions (Fig. 1, J, J′, and K, and fig. S1, D and H), many macrophages persisted in regions outside the head in rraga mutants at all stages we examined up to 8 dpf (Fig. 1, J, J′, and K, and fig. S1, E to G). Next, to determine whether the reduced macrophage numbers in rraga mutants result from reduced proliferation of these cells, we used in vivo 5-ethynyl-2’-deoxyuridine (EdU) labeling by pulsing the animals with EdU at 2.5 dpf and chasing up to 3 dpf. We observed a moderate but insignificant decrease in the number of proliferating macrophages in rraga mutants (Fig. 1L).
An interesting feature of macrophages and microglia is that the morphology of these cells is frequently used as a readout of their functional state; highly ramified microglia are typically thought to be surveilling, whereas rounded microglia are considered to be activated in response to infection or injury or are dysfunctional in some way (48). Starting at 3 dpf, macrophages in the tail region of rraga mutants displayed an abnormal, rounded morphology (Fig. 1, M and N), similar to microglia in these mutants (43). To test the hypothesis that macrophages in rraga mutants may be hyperactivated by the presence of excess cellular debris in their environment, we sought to reduce the number of apoptotic cells in rraga larvae using a combination of caspase-3 morpholino injection and ZVAD treatment (fig. S2, A to C) (37). In addition, to inhibit the uptake of debris by macrophages in rraga mutants, we used BAI1 + TIM4 morpholinos (fig. S2, A to C) (49), or we separately treated larvae with phagocytosis inhibitors including cytochalasin B (50) and latrunculin A (fig. S2, D and E) (51). We found that these treatments do not ameliorate the amoeboid, activated appearance of macrophages in rraga mutants, indicating that the macrophage defects in rraga mutants are not likely to be caused by the presence or uptake of apoptotic debris but instead result from macrophage-intrinsic genes and pathways regulated by RagA.
Macrophages in rraga mutants show a significant up-regulation of lysosomal pathways
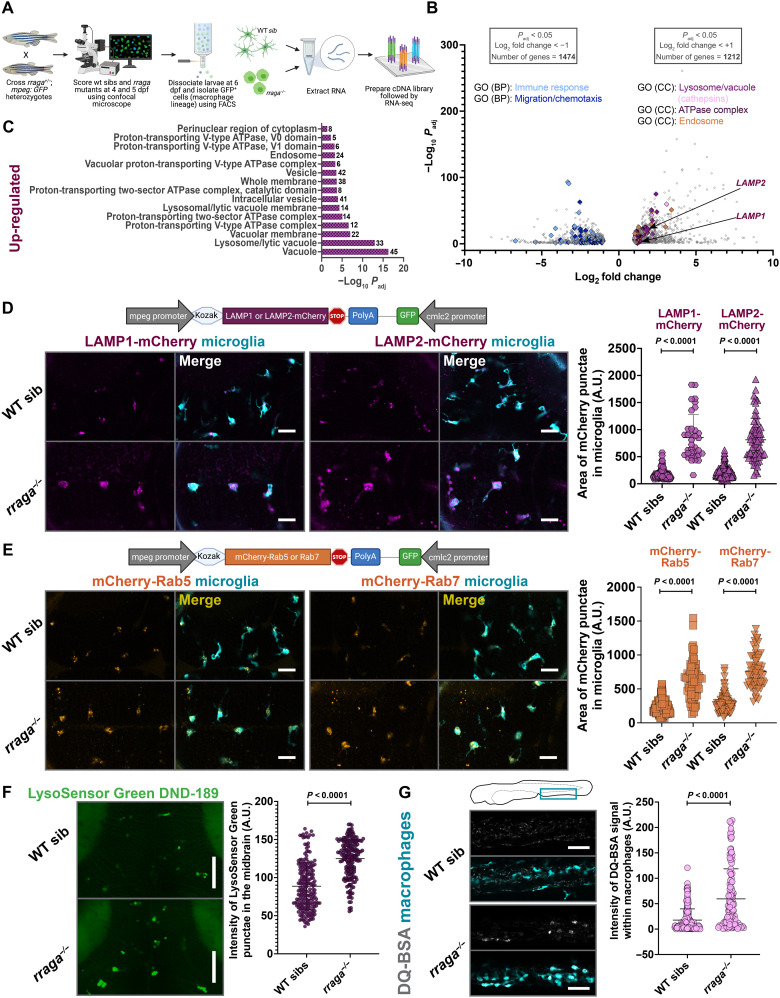
To further investigate the molecular mechanisms underlying the defects in the macrophage lineage in rraga mutants, we performed RNA sequencing (RNA-seq) on macrophages and microglia isolated from these mutants. We crossed rraga heterozygotes carrying the mpeg:GFP transgene, scored the mutant and wild-type siblings using confocal microscopy [rraga mutants are not adult viable (43)], dissociated the larvae, sorted GFP+ cells at 6 dpf, extracted RNA, and performed RNA-seq (Fig. 2A). Gene Ontology (GO) term enrichment analysis of the genes up-regulated in macrophages from rraga mutants revealed a strong enrichment of the terms Vacuole, Lysosome, Lytic Vacuole, and Endosome (Fig. 2, B and C; fig. S3A; and table S3). To validate these transcriptomic observations, we generated genetic tools and used chemical probes to assess the endolysosomes in macrophages and microglia in vivo. LAMP1 and LAMP2 are estimated to contribute 50% of all proteins in lysosomal membrane (52), and these genes were significantly up-regulated in our dataset (Fig. 2B). To examine the lysosomal compartments in macrophages, we generated zebrafish LAMP1-mCherry and LAMP2-mCherry constructs and used the mpeg promoter to express these transgenes in the macrophage lineage (Fig. 2D). In parallel, we also generated mCherry-Rab5 or mCherry-Rab7 constructs, driven using the mpeg promoter, to label early and late endosomes, respectively (53). Consistent with the up-regulation of transcripts associated with the endolysosomal pathway in our RNA-seq data, we found a significant expansion of LAMP1-mCherry, LAMP2-mCherry, mCherry-Rab5, and mCherry-Rab7 punctae in the microglia of rraga mutants relative to their wild-type siblings (Fig. 2, D and E).
Fig. 2. Lysosomal genes are significantly up-regulated and endolysosomes are expanded in macrophages isolated from rraga mutants.
(A) Experimental schematic for RNA-seq. Macrophages were isolated using FACS from N = 90 larvae for each biological replicate RNA sample from rraga mutants or wild-type siblings. Four biological replicate samples of each genotype were processed for RNA-seq. (B) Volcano plot showing significantly differentially up-regulated and down-regulated genes in the macrophages of rraga mutants relative to wild-type siblings. (C) GO (Cellular Component) enrichment analysis showing an up-regulation of lysosome-associated terms in macrophages from rraga mutants. (D) Imaging and quantification of LAMP1 or LAMP2-mCherry transgene expression in rraga mutants and wild-type siblings at 4 dpf. Scale bars, 20 μm. (E) Imaging and quantification of mCherry-Rab5 or Rab7 transgene expression in rraga mutants and wild-type siblings at 4 dpf. Scale bars, 20 μm. (F) LysoSensor Green labeling and quantification of LysoSensor Green intensity at 4 dpf. Scale bars, 50 μm. Graphs in (D) to (F) show mean + SD; significance was determined using parametric unpaired t test. (G) DQ-BSA labeling at 4 dpf and quantification of DQ-BSA intensity inside macrophages. Scale bars, 50 μm. Graph shows mean + SD; significance was determined using nonparametric Mann-Whitney U test. The number of animals analyzed for each experiment is listed in table S1; all the panels (except RNA-seq) are representative of at least two independent experiments. A.U., arbitrary units.
Our analysis also revealed a significant expansion of proton-transporting vATPase complex genes (Fig. 2, B and C, and fig. S3A), which are central players in intracellular acidification and organelle pH control (54). To observe the intracellular acidification of microglia, we treated rraga mutant larvae with the dye LysoSensor Green DND-189, which becomes more fluorescent in acidic compartments of the cell (55). As suggested by our RNA-seq data, we found a significant increase of LysoSensor Green intensity in the midbrain (where microglia are enriched) of rraga mutants relative to their wild-type siblings (Fig. 2F). Last, several cathepsin genes, which encode lysosomal proteases activated by acidic pH, were up-regulated in rraga mutants (Fig. 2B and table S3) (56). To assay this apparent increase in proteolytic activity in the macrophages of rraga mutants, we used DQ-BSA (57) and Magic Red (MR)–Cathepsin (58), dyes that are nonfluorescent in neutral pH but fluoresce brightly when proteases cleave to release the quenching residues in these dyes. Once again, in accord with our RNA-seq data, we found an increased fluorescence of both DQ-BSA and MR-Cathepsin in macrophages of rraga mutants (Fig. 2G and fig. S3B). Thus, our results demonstrate that endolysosomal gene expression is expanded and acid-activated protease activity is increased in macrophages of rraga mutants in vivo.
Immune- and chemotaxis-related genes are significantly down-regulated in macrophages of rraga mutants
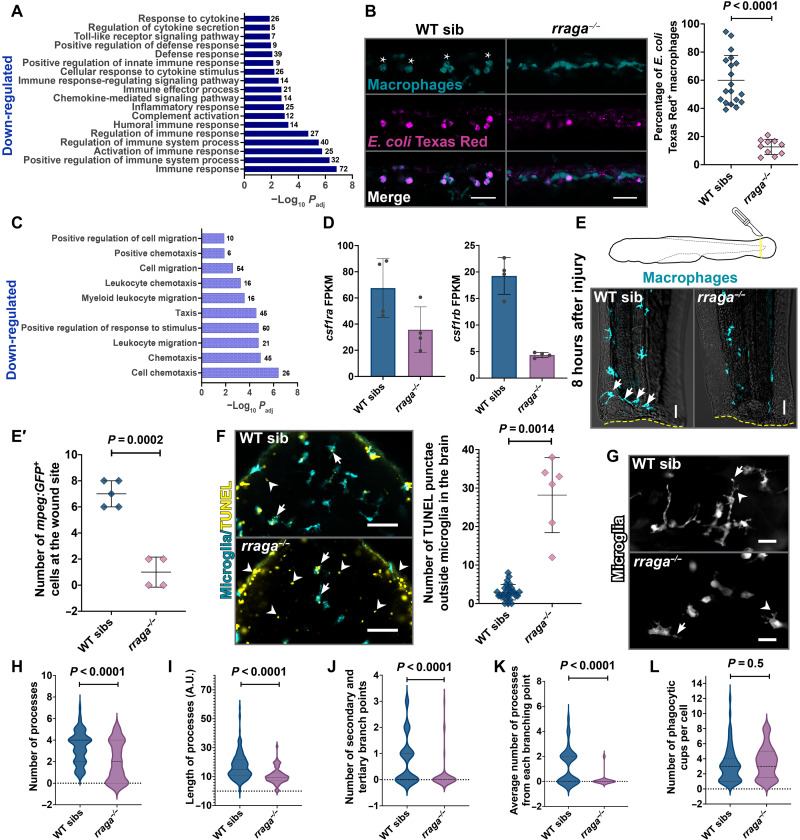
The gene enrichment terms most significantly down-regulated in macrophages from rraga mutants were immune response and migration/taxis, followed by lipid metabolism and other metabolic pathways (Figs. 2B and 3, A and C; fig. S4, A and B; and table S4). To examine the immune response of macrophages to microbial debris, we challenged the tail macrophages of rraga larvae with Escherichia coli particles labeled with Texas Red. There was a marked reduction in the uptake of these E. coli particles by rraga mutants relative to their wild-type siblings (Fig. 3B and fig. S4C). In contrast, macrophages in rraga mutants can sense and ingest weakly immunogenic molecules such as Dextran Texas Red and Ovalbumin 555 (fig. S4, D and E), suggesting that RagA is differentially required for engulfment or processing of different substrates (59).
Fig. 3. Macrophages and microglia in rraga mutants show defective clearance of debris.
(A) Bar graph showing GO terms related to immune signaling significantly down-regulated in macrophages from rraga mutants. (B) Injection of E. coli Texas Red in rraga mutants and siblings at 4 dpf and corresponding quantification. Wild-type macrophages responding to E. coli become activated and display amoeboid morphology (asterisks). Scale bars, 50 μm. (C) GO terms related to migration/taxis significantly down-regulated in macrophages from rraga mutants. (D) Graphs showing fragments per kilobase of exon per million mapped reads (FPKM) values of csf1ra and csf1rb transcripts in macrophages from rraga mutants; each point represents FPKM value from a single RNA-seq biological replicate. FPKM, fragments per kilobase of exon per million mapped reads. (E and E′) Macrophage response to tail injury at 4 dpf. Arrows in the wild-type image show macrophages at the wound site (yellow dotted line). Scale bars, 20 μm. (F) TUNEL assay and quantification on rraga mutants and their wild-type siblings. Arrows indicate TUNEL+ microglia, and arrowheads indicate TUNEL signal outside microglia. Scale bars, 50 μm. (G to L) Time-lapse imaging and analysis of cellular characteristics of microglia at 5 dpf. (G) Images of rraga mutants and their siblings, with arrows showing phagocytic cup formation and arrowheads showing branching points. Scale bars, 20 μm. All graphs show mean + SD. Significance in (B) was determined using nonparametric Mann-Whitney U test; significance in (E′), (F), and (H) to (L) was determined using parametric unpaired t test. The number of animals analyzed for each experiment is listed in table S1; all the panels are representative of at least two independent experiments.
The down-regulation of genes corresponding to chemotaxis in macrophages from rraga mutants (Fig. 3C) corroborates our earlier observation that early macrophages do not migrate into the brain of rraga mutants to become microglia (Fig. 1, B to G). Furthermore, we found that transcripts of csf1ra and csf1rb, which encode key macrophage lineage receptors essential for the colonization of microglia and other tissue macrophages (39, 60), were down-regulated in our RNA-seq data (Fig. 3D and fig. S4F). To assess the ability of macrophages to appropriately respond to environmental stimuli, we used time-lapse imaging to track the directional movement of macrophages following injury of the tail fin (Fig. 3E). Significantly fewer macrophages in rraga mutants moved to the wound site at 8 hours after injury (Fig. 3E′). When combined with our RNA-seq analysis, the abnormalities in brain colonization by embryonic macrophages (Fig. 1, B to G) and aberrant wound response of peripheral macrophages (Fig. 3, E and E′) indicate that cells of the macrophage lineage in rraga mutants are unable to respond appropriately to developmental signals (37, 47) or injury cues. Together, our RNA-seq data illuminate heretofore unappreciated functions of RagA in the macrophage lineage, particularly the contribution of RagA in activation of innate immune response and cell chemotaxis.
To examine whether microglia can detect and clear cellular debris in the brain, we performed TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) assay to detect apoptotic neurons (Fig. 3F). We found that whereas microglia in both rraga mutants and their wild-type siblings can engulf neuronal debris (arrows in Fig. 3F), rraga mutants display a significantly higher TUNEL signal outside the microglia in the brain (arrowheads in Fig. 3F). To account for this reduced debris clearance, we inspected various characteristics of microglia in rraga mutants using time-lapse imaging (movies S3 and S4). Microglia processes in rraga mutants are fewer in number (Fig. 3, G and H), shorter (Fig. 3, G and I), and less complex (Fig. 3, G, J, and K) than in their wild-type siblings. Notably, among the microglia that have processes in rraga mutants, phagocytic cup formation is not significantly different in rraga mutants relative to their wild-type siblings (Fig. 3L). Collectively, these experiments indicate that microglia in rraga mutants are able to sense and engulf apoptotic debris but that scavenging of neuronal debris in these mutants is severely compromised.
Simultaneous loss of lysosomal transcription factors tfeb, tfe3a, and tfe3b rescues microglia numbers in rraga mutants
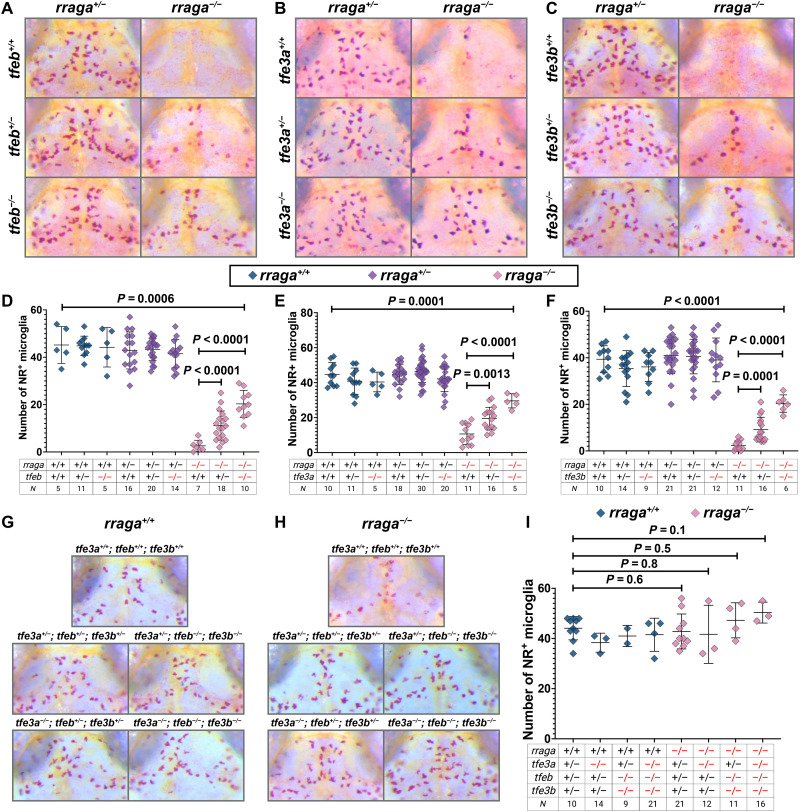
Tfeb activates most lysosomal functions including lysosomal biogenesis, autophagy, and exocytosis (17–19). Several lines of evidence indicate that RagA represses Tfeb and other related transcription factors (11, 20), although the extent to which the RagA-Tfeb lysosomal pathway functions in the macrophage lineage in vivo is unclear. To address whether RagA represses Tfeb in microglia, we crossed rraga+/−; tfeb+/− double heterozygotes (61) and labeled microglia using neutral red, a dye that preferentially accumulates in microglia (46). Imaging and quantification of microglia in the dorsal midbrain showed that rraga mutants have significantly fewer neutral red–labeled microglia, as shown previously (Fig. 4A) (43). Loss of tfeb in rraga mutants partially restored microglia numbers (Fig. 4, A and D). These data provided evidence that hyperactivated Tfeb contributes to, but does not solely cause, microglial defects in rraga mutants.
Fig. 4. Microglia numbers are restored in rraga mutants with simultaneous mutation of tfeb, tfe3a, or tfe3b.
Images show dorsal views of the midbrain (anterior on top) of neutral red (NR)–stained larvae of indicated genotypes at 5 dpf, and graphs show corresponding quantification. (A and D) Progeny from rraga+/−; tfeb+/− intercross, (B and E) Progeny from rraga+/−; tfe3a+/− intercross. (C and F) Progeny from rraga+/−; tfe3b+/− intercross. (G to I) Progeny from rraga+/−; tfeb+/−; tfe3a+/−; tfe3b+/− quadruple heterozygous intercross. Images of a rraga mutant and tfeb and tfe3 genotypes that result in complete rescue of NR+ microglia in rraga mutants are shown in (H). In (G), the same genotypes, but in a background wild-type for rraga, are included for comparison. All graphs show mean + SD; significance was determined using parametric unpaired t test. The number of animals analyzed for each experiment is listed in the tables below each graph; (A) to (F) are representative of at least two independent experiments.
One hypothesis for the incomplete rescue we observed in rraga; tfeb double mutants is that RagA might also repress other members of the MiTF family, which consists of Mitf, Tfeb, Tfe3, and Tfec (11). Notably, Tfeb and Tfe3 have been shown to act cooperatively to regulate cytokine production in the RAW 264.7 macrophage cell line (62), although the extent of functional overlap between these transcription factors in macrophages in vivo remains uncharacterized. To determine whether Tfe3 functions in macrophages and microglia, we generated mutants for the two zebrafish tfe3 paralogs tfe3a and tfe3b (fig. S5 and table S2) (63). Neutral red assay revealed that mutations in tfe3a (Fig. 4, B and E) or tfe3b (Fig. 4, C and F) can partially rescue microglia numbers in rraga mutants, much as observed in rraga; tfeb double mutants (Fig. 4, A and D). The loss of even a single copy of tfeb, tfe3a, or tfe3b can partially rescue microglia in rraga mutants (rraga−/−; tfeb+/− in Fig. 4D; rraga−/−; tfe3a+/− in Fig. 4E; and rraga−/−; tfe3b+/− in Fig. 4F), indicating that microglia are highly sensitive to increased levels of each of these transcription factors.
To further explore the dosage-dependent rescue of Tfeb and Tfe3, as well as to better understand the extent of functional overlap between Tfeb, Tfe3a, and Tfe3b, we generated rraga+/−; tfeb+/−; tfe3a+/−; tfe3b+/− quadruple heterozygous animals. Neutral red assay and quantification (Fig. 4, G to I) revealed that combined loss of a single copy of tfeb, tfe3a, and tfe3b was sufficient to completely restore microglia numbers in rraga mutants (Fig. 4H). We also observed a full rescue of microglia in rraga mutants when four, five, or six copies of tfeb, tfe3a, and tfe3b were mutated (Fig. 4, H and I). We did not observe any obvious defects in microglia in tfeb, tfe3a, or tfe3b mutants alone (Fig. 4, A to F) or in combination, including in tfeb; tfe3a; tfe3b triple mutants (Fig. 4, G to I). Collectively, these observations indicate that RagA represses Tfeb, Tfe3a, and Tfe3b in microglia, that hyperactivity of these transcription factors causes the microglia defects in rraga mutants, and that tfeb and its homologs are not required for normal microglial development.
flcn and rraga mutants have similar phenotypes
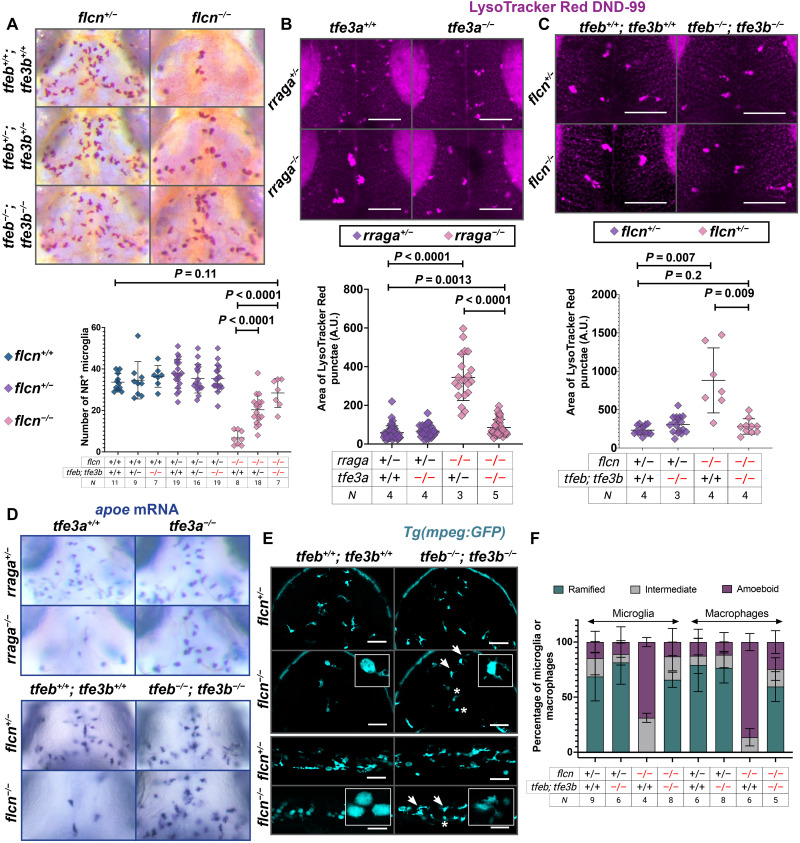
Several studies link the protein Flcn to the repression of Tfe3 (64–66) and, in some cases, Tfeb (67); however, the mechanism through which Flcn regulates the activation of Tfe3 (or Tfeb) is not fully understood. To assess the functional interaction between Flcn and Tfeb/Tfe3, we generated flcn mutants using CRISPR-Cas9 (fig. S6A). We found that mutations in flcn or rraga caused a markedly similar phenotype in macrophages and microglia. flcn mutants have significantly fewer neutral red–labeled microglia in their dorsal midbrain (Fig. 5A and fig. S6B), as in the case of rraga mutants (Fig. 4) (43). Furthermore, this reduction in microglia numbers in flcn mutants was rescued by simultaneous loss of tfeb and tfe3b (Fig. 5A) and partially rescued in flcn; tfe3a double mutants (fig. S6B). Once again, as in the case of rraga mutants (Fig. 4), we observed a dosage-dependent effect, and flcn mutants that are heterozygous for the tfeb; tfe3b double mutant chromosome or tfe3a display a partial, intermediate level of rescue of microglia number in the neutral red assay (Fig. 5A and fig. S6B).
Fig. 5. Simultaneous mutation in tfeb, tfe3a, or tfe3b rescues flcn and rraga mutant phenotypes.
(A) NR assay and quantification of microglia in larvae obtained from flcn+/−; tfeb+/−; tfe3b+/− intercross. (B and C) LysoTracker Red assay and quantification of the area of LysoTracker Red punctae in microglia of larvae from (B) rraga+/−; tfe3a+/− intercross and (C) flcn+/−; tfeb+/−; tfe3b+/− intercross. Images show LysoTracker Red signal in dorsal view of the midbrain. Scale bars, 50 μm. Graphs show mean + SD; significance was determined using parametric unpaired t test. (D) apoe mRNA expression at 4 dpf and rescue of microglia in the progeny of rraga+/−; tfe3a+/− intercross and flcn+/−; tfeb+/−; tfe3b+/− intercross. (E) mpeg:GFP expression to visualize microglia and macrophages in flcn; tfeb; tfe3b triple mutants. Arrows denote microglia and macrophages in which ramified morphology has been restored, asterisks denote cells with amoeboid morphology, and insets show magnified views of cell morphology. Scale bars, 50 μm. (F) Quantification of amoeboid morphology and rescue. Graph shows mean + SD; significance was determined using nonparametric Mann-Whitney U test. The number of animals analyzed for each experiment is listed in the tables below each graph; all the panels are representative of at least two independent experiments.
We also used the dye LysoTracker Red (38) to examine the morphology of acidic compartments within microglia in rraga or flcn mutants. As shown previously (43), rraga mutants have a significant expansion of LysoTracker Red punctae in microglia (Fig. 5B); we found that this expansion is partially rescued in rraga; tfe3a double mutants (Fig. 5B). Similar to rraga mutants, we also found that flcn mutants had enlarged LysoTracker Red punctae in their microglia and that this expansion is fully rescued in flcn; tfeb; tfe3b mutants (Fig. 5C). rraga mutants have very few apoe-expressing microglia in their dorsal midbrain (Fig. 5D) (43), and we found that this defect is partially rescued in rraga; tfe3a double mutants (Fig. 5D). Similarly, there was a strong reduction in apoe-expressing microglia in flcn mutants, and this defect was rescued in flcn; tfeb; tfe3b mutants (Fig. 5D). Last, we examined these genotypes using the Tg(mpeg:GFP) line to assess the morphology of microglia and macrophages. Both rraga and flcn mutants displayed abnormal, rounded morphology of microglia and macrophages (Fig. 5, E and F and fig. S6, C and D), a defect that was at least partially restored in rraga; tfe3a mutants (fig. S6, C and D) and flcn; tfeb; tfe3b mutants (Fig. 5, E and F). In summary, our experiments show that the key lysosomal proteins Flcn and RagA have very similar functions in microglia and macrophages in vivo and are both essential to repress Tfeb and Tfe3. Moreover, the macrophage number and morphology defects observed in rraga or flcn mutants can be rescued to varying degrees by simultaneous loss of tfeb, tfe3a, or tfe3b.
Overexpression of tfe3b in macrophages recapitulates rraga mutant phenotypes in microglia and macrophages
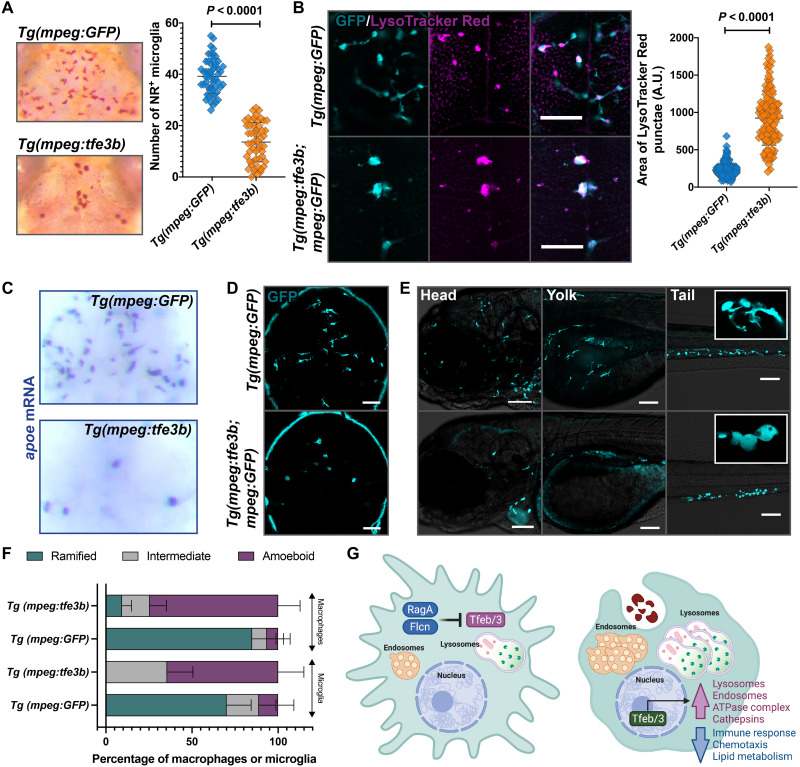
We reasoned that if Tfeb and Tfe3 are the primary downstream targets repressed by RagA and Flcn, then overexpression of Tfeb or Tfe3 in macrophages should recapitulate the rraga or flcn mutant phenotypes. We constructed transgenic fish that overexpressed Tfe3b in cells of the macrophage lineage, Tg(mpeg:tfe3b; cmlc2:mCherry), and found that these animals had defects characteristic of rraga or flcn mutants (Fig. 6, A to F). First, Tg(mpeg:tfe3b) animals have significantly fewer neutral red–labeled microglia than Tg(mpeg:GFP) animals (Fig. 6A), similar to mutants for rraga or flcn (Figs. 4 and 5A and fig. S6B) (43). Second, animals that overexpress tfe3b in macrophages have significantly enlarged LysoTracker Red punctae in microglia (Fig. 6B), as observed in rraga or flcn mutants (Fig. 5, B and C). Third, Tg(mpeg:tfe3b) animals had reduced apoe labeling in the dorsal midbrain (Fig. 6C), once again similar to both rraga and flcn mutants (Fig. 5D) (43), and in contrast to Tg(mpeg:GFP) controls (Fig. 6C). Last, by crossing Tg(mpeg:tfe3b) animals to the Tg(mpeg:GFP) fish, we found that animals overexpressing tfe3b in the macrophage lineage had fewer microglia and macrophages and that the remaining cells exhibited abnormal, rounded morphology (Fig. 6, D to F), similar to rraga (Fig. 1, M and N, and fig. S6, C and D) (43) or flcn (Fig. 5, E and F) mutants. These experiments demonstrate that the overexpression of tfe3b is sufficient to recapitulate the rraga mutant phenotype in microglia and macrophages (Fig. 6, A to F). Collectively, our data indicate that the primary essential function of RagA and Flcn in macrophages is to repress Tfeb and Tfe3 (Fig. 6G).
Fig. 6. Overexpression of tfe3b in the macrophage lineage disrupts microglia number and morphology as in rraga mutants.
Comparison of macrophages and microglia between animals overexpressing Tfe3b in the macrophage lineage, Tg(mpeg:tfe3b), and controls, Tg(mpeg:GFP), using (A) neutral red assay and quantification and (B) LysoTracker Red assay and quantification. Graphs show mean + SD; significance was determined using parametric unpaired t test. (C) apoe in situ hybridization and (D and E) live imaging with the mpeg:GFP transgene in (D) the brain and (E) the head, yolk, and tail regions of Tg(mpeg:tfe3b) and Tg(mpeg:GFP) larvae. Insets show magnified views of cell morphology. (F) Quantification of amoeboid morphology of microglia and macrophages. Graph shows mean + SD; significance was determined using nonparametric Mann-Whitney U test. Scale bars, 50 μm. The number of animals analyzed for each experiment is listed in table S1; all the panels are representative of at least two independent experiments. (G) Schematic summarizing the lysosomal regulatory circuit in microglia and macrophages.
Tfeb and Tfe3 activate lysosomal pathways under conditions of cellular stress but are dispensable for basal expression of these genes
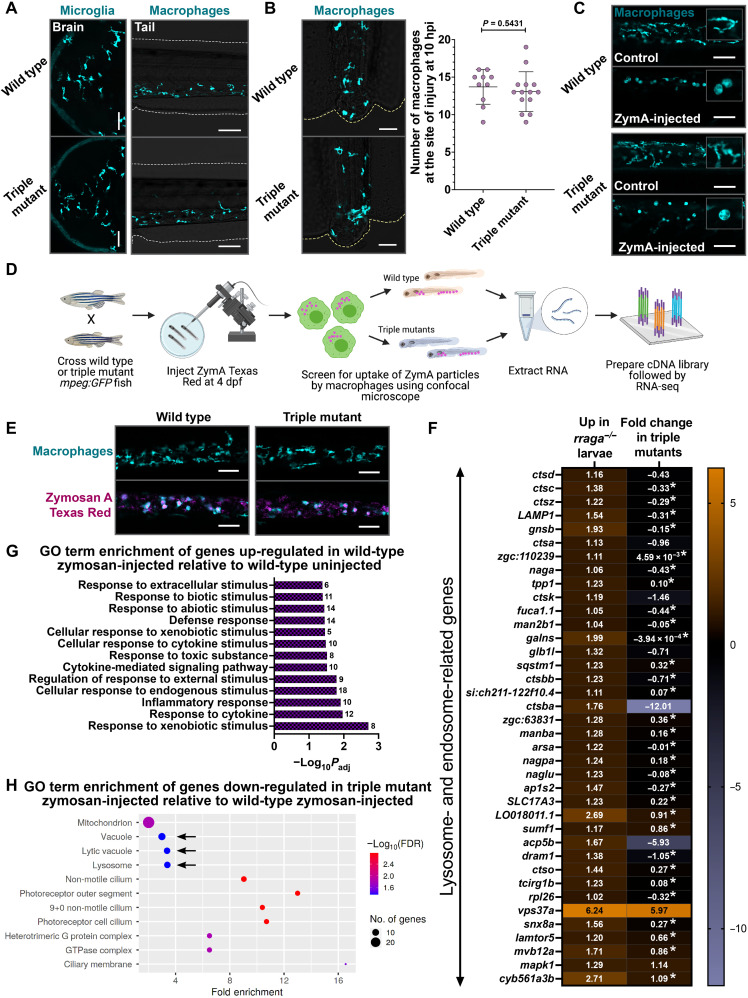
To test the requirement of Tfeb or Tfe3 for the specification, maintenance, or function of microglia and macrophages, we generated tfeb; tfe3a; tfe3b triple mutants (hereafter triple mutants). Corroborating our earlier analysis, microglia and macrophages in the triple mutants are indistinguishable from their wild-type counterparts in both number and morphology (Figs. 4 and 7A and fig. S7A). To examine the extent to which macrophages in triple mutants respond differently from their wild-type siblings to environmental challenges, we injured the tail fins of these animals and found that macrophages in triple mutants respond to tail injury similarly to their siblings (Fig. 7B). In addition, we injected zymosan A (ZymA) into the circulation of these larvae and found once again that activation of macrophages in triple mutants is comparable to that of their wild-type siblings (amoeboid morphology in Fig. 7C and fig. S7B). These observations indicate that Tfeb, Tfe3a, and Tfe3b are not necessary for the survival of microglia and macrophages or for their response to injury or inflammatory stimulus.
Fig. 7. Tfeb and Tfe3 activate lysosomal pathways only under conditions of stress.
(A) Microglia and macrophages visualized using the mpeg:GFP transgene in wild-type animals and tfeb; tfe3a; tfe3b triple mutants at 4 dpf. Macrophage response to (B) tail injury and (C) systemic ZymA injection in wild-type and triple mutant animals at 4 dpf. hpi, hours post-injury. Insets in (C) show magnified views of macrophage morphology. Graph in (B) shows mean + SD; significance was determined using parametric unpaired t test. The number of animals analyzed for each experiment is listed in table S1; all the panels are representative of at least two independent experiments. (D) Experimental schematic for RNA-seq. (E) Microscopy-based validation of ZymA Texas Red uptake by the trunk and tail macrophages in triple mutants and wild-type animals at 4 dpf before RNA-seq. Approximately 20 larvae of each genotype were used for RNA extraction per biological replicate; three biological replicates were used for RNA-seq. (F) Heatmap depicting all the lysosomal genes significantly up-regulated (log2 fold change > +1, Padj < 0.05) in rraga mutant whole larvae and the corresponding fold change of the gene in triple mutant larval RNA preparations. The log2 fold change is shown in each cell; fold change values with asterisks are not significant (Padj > 0.05). (G) GO term enrichment analysis of genes differentially up-regulated in wild-type ZymA-injected animals relative to uninjected wild-type controls. (H) GO term enrichment of genes significantly down-regulated in ZymA-injected triple mutants. FDR, false discovery rate.
Tfeb and Tfe3 are widely regarded as master lysosomal transcription factors that are essential for the activation of lysosomal genes (19); thus, our observation that cells of the macrophage lineage are apparently functionally normal in triple mutants was unexpected. To determine the extent to which these transcription factors are required for the activation of lysosomal pathways in vivo, we decided to perform RNA-seq on triple mutant whole larvae. In addition, to investigate the hypothesis that Tfeb and Tfe3 may activate lysosomal genes only in response to cellular stress (68, 69), we developed a protocol to challenge macrophages with ZymA in vivo. Briefly, we crossed wild-type or triple mutant fish carrying the mpeg:GFP transgene and injected ZymA Texas Red into the yolk of some of these larvae at 4 dpf (Fig. 7D). Within 1 to 1.5 hours after injection, the ZymA particles are present in the peripheral blood vessels, where they are engulfed by macrophages (Fig. 7E). Because macrophages are the primary phagocytic cells in the larvae, we anticipated that the differential transcriptional changes we observed will chiefly reflect the macrophage response to ZymA. We screened for the uptake of ZymA particles by macrophages using confocal microscopy following injection (Fig. 7, D and E), pooled ZymA-injected and uninjected animals of wild-type and triple mutant genotypes separately, extracted RNA, prepared complementary DNA (cDNA) libraries, and performed RNA-seq (Fig. 7D).
Comparison of gene expression in triple mutants to wild-type animals yielded 366 up-regulated (log2 fold change > +1; Padj < 0.05) and 511 down-regulated genes (log2 fold change < −1; Padj < 0.05), but there was no GO term enrichment in genes differentially up- or down-regulated in triple mutants (table S5). We also performed whole-larvae RNA-seq on rraga mutants to compare the differential lysosomal gene expression between rraga and tfeb; tfe3a; tfe3b mutants. As expected, rraga mutants show a significant up-regulation of lysosomal genes; however, expression of most of these genes did not change significantly in the triple mutants, with some exceptions (Fig. 7F). These results revealed that Tfeb and Tfe3 are not necessary for expression of lysosomal pathways in whole animals under normal conditions.
Comparison of genes differentially up-regulated in the ZymA-injected wild-type versus uninjected wild-type animals revealed expected enrichment of GO categories corresponding to defense response, response to various kinds of stimuli, as well as an inflammatory signature (Fig. 7G). Notably, GO categories Vacuole, Lytic Vacuole, and Lysosome were significantly down-regulated in ZymA-injected triple mutants versus ZymA-injected wild-type animals (Fig. 7H). These terms are not significantly enriched in the corresponding dataset of genes down-regulated in uninjected triple mutants versus uninjected wild-type animals. In another enrichment approach, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed enrichment of the category Lysosome in the genes down-regulated in ZymA-injected triple mutants, while this category is not significantly changed in the uninjected triple mutants (fig. S7, C and D). Last, Ingenuity Pathway Analysis yielded phagosome maturation and CLEAR signaling pathway [lysosomal genes activated by Tfeb and Tfe3 (17)] as top canonical pathways when we analyzed the genes down-regulated in ZymA-injected triple mutants (fig. S7E). Together, our results indicate that Tfeb and Tfe3 are dispensable for basal levels of lysosomal gene expression under homeostatic conditions in vivo, but these transcription factors are required for activation of lysosomal pathways in response to cellular stress.
DISCUSSION
Our experiments define an antagonistic regulatory circuit—involving RagA and Flcn on one side, and Tfeb, Tfe3a, and Tfeb on the other—that is essential for the development and function of macrophages in vivo (Fig. 6G). Using transcriptomic analyses, we demonstrate that macrophages from rraga mutants display a strong up-regulation of known lysosomal target genes of Tfeb and Tfe3; correspondingly, we observe a significant expansion of the endolysosomal compartments in these mutants in vivo. Despite the observed expansion of endolysosomal compartments, our data show that rraga mutants display excess apoptotic corpses in the brain and are unable to respond appropriately to microbial debris, indicating that macrophages in rraga mutants cannot effectively respond to environmental cues. In light of recent studies showing that expansion of lysosomal pathways does not necessarily correlate with enhanced debris clearance (70), an important unanswered question that remains following our study is how the up-regulation of Tfeb and Tfe3 targets in rraga mutants affects digestion and processing of debris.
Notably, we uncover a previously unappreciated role for RagA in the regulation of genes involved in chemotaxis and innate immunity, and our cellular studies reveal disrupted colonization of embryonic macrophages in the brain. In the developing zebrafish embryo, apoptotic neurons (37, 47) and Csf1r-Il34 signaling pathway (39) serve as the primary cues directing embryonic macrophages to colonize the brain. As described previously (47), we found that macrophages in both rraga mutants and their wild-type siblings predominantly enter through the bilateral periphery of the developing zebrafish brain to colonize the midbrain. However, fewer macrophages enter the brain in rraga mutants, likely due to the reduced expression of csf1r homologs and other chemotactic signaling pathways in these mutants. Future studies will define the extent to which Csf1r-Il34 signaling and other pathways contribute to the mobility defects of macrophages in rraga mutants.
Our experiments reveal that cells of the macrophage lineage are acutely sensitive to the levels of Tfeb, Tfe3, and Tfe3b, because loss of even a single copy of one of these three genes is sufficient to partially restore the microglia numbers in rraga or flcn mutants. Overexpression of Tfe3b alone is sufficient to phenocopy the microglia number and morphology defects seen in rraga and flcn mutants, indicating that Tfeb, Tfe3a, and Tfe3b are at least partly functionally redundant in the macrophage lineage. Interesting future experiments will investigate the extent to which the targets of the MiTF family of transcription factors overlap in different cell types, and uncover the mechanisms by which cells sense the activity levels of these transcription factors.
Microglia and macrophages depend on lysosomal pathways to execute their phagocytic functions, and Tfeb and related transcription factors are widely regarded as master regulators of lysosomal activity. Furthermore, a major wave of neuronal cell death occurs in zebrafish larvae before 3 dpf (47, 71), indicating that phagocytic and lysosomal activity in microglia is required at very early stages of development. It is therefore seemingly paradoxical that microglia and macrophages appear normal in tfeb; tfe3a; tfe3b triple mutants. tfeb; tfe3a; tfe3b triple mutant animals are viable and fertile as adults, although they seem to grow at a slower rate than their wild-type siblings. Our whole-larvae transcriptomic studies revealed that Tfeb, Tfe3a, and Tfe3b are not required for basal expression of lysosomal and autophagy genes, indicating that other pathways must control lysosome biogenesis and activity during normal development. We show that Tfeb and Tfe3 are required to activate lysosomal pathways in whole larvae under conditions of stress, providing compelling in vivo evidence for the role of these transcription factors as mediators of the cellular stress response (68, 69, 72). A more detailed analysis of Tfeb and Tfe3 targets under different conditions of stress, such as apoptotic debris, misfolded proteins, or microbial infection, may reveal the entire range of lysosomal and other pathways activated by these transcription factors in response to specific environmental challenges. Future studies will also address how different cell types use RagA, Flcn, Tfeb, and Tfeb3 to detect and respond to diverse range of stressors.
Last, attempts to enhance Tfeb-mediated lysosomal pathways in animal models of neurodegenerative diseases (21, 22, 24, 25, 73), particularly Alzheimer’s disease (AD) (29–31), have led to beneficial effects, but the identity of cells that must overexpress Tfeb in vivo to generate these favorable cognitive outcomes remains unclear. Because prolonged activation of inflammatory pathways in AD results in neurotoxicity and indiscriminate loss of viable cells leading to cognitive defects (74–78), our observation that hyperactivation of Tfeb and Tfe3 may down-regulate immune pathways in the macrophage lineage may offer a key insight into the beneficial outcomes observed in animal models of AD. Moreover, while we find that repression of Tfeb and Tfe3 is necessary for the normal development of embryonic macrophages, studying the role of these transcription factors in the maintenance and function of adult microglia presents an exciting future direction (40, 79, 80). Future studies aiming to understand the regulation and functions of Tfeb and Tfe3 in microglia during development, disease, and aging will not only broaden our understanding of lysosomal pathways in microglia but also inform therapeutic strategies aimed at modulating Tfeb activity in neurodegeneration and other diseases.
MATERIALS AND METHODS
Zebrafish lines
Embryos from wild-type [Tupfel long-fin (TL) or AB] strains and Tg(mpeg:GFP) transgenic line (44) were raised at 28.5°C in embryo water with methylene blue. For all imaging experiments, embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU) to inhibit pigmentation between 10 and 24 hpf, and anesthetization was done using 0.016% MS-222 (tricaine) before experimental procedures. For neutral red assay, methylene blue was excluded from embryo water. All animal protocols have been approved by the Stanford Institutional Animal Care and Use Committee.
Time-course and time-lapse experiments
Embryos for all time-course and time-lapse experiments were staged between 0 and 12 hpf and typically dechorionated between 30 and 36 hpf. For the 24 to 26 hpf time point, embryos were dechorionated immediately before mounting. Genotyping was typically performed after image acquisition using a Zeiss LSM 700 confocal microscope. For all time-course experiments, cells were first counted, and a single, high-resolution, representative plane was subsequently imaged using the 20× objective.
For the time-lapse experiments starting at 60 hpf, a region of interest in the brain containing the midbrain, forebrain, and a portion of the hindbrain was selected. All images were acquired using the 20× objective. The depth of acquisition was determined in each case individually; the most dorsal portion of the head containing macrophages was selected as the first plane, and 10 z-slices, 2 μm apart, were acquired, with an interval of 300 s per acquisition and a total of 100 imaging cycles. Following 17 hours and 41 min of acquisition, maximum intensity projection of the z-slices was performed using the Zen Black software. For the purpose of analyses, macrophages within the brain were counted at each time point from the acquired images.
For the rraga injury time-lapse experiments, animals at 4 dpf were anesthetized and the tip of the tail fin was cut using a new scalpel, ensuring approximately the same size of incision in all animals. Immediately after injury, animals were mounted in 1.5% agarose with anesthetic, and time-lapse imaging was performed for 8 hours after injury. All imaging was done using a 10× objective, and 10 z-slices were selected, each slice 1 μm apart, at an interval of 300 s per acquisition. The number of macrophages at the site of injury (regenerated fin tissue + blastema) was counted from maximum intensity projection of the z-slices. For the triple mutant injury experiment, tail tip injury was performed in wild-type animals and triple mutants and the animals were allowed to recover in embryo water. Ten hours after injury, all larvae were mounted in 1.5% agarose with anesthetic, and the number of macrophages at the injury site was counted.
To determine the characteristics of microglia in rraga mutants, we used LSM 980 with Airyscan2 for rapid image acquisition to observe phagocytic cup formation. At 5 dpf, larvae were anesthetized and mounted in 1.5% agarose with the anesthetic. We acquired 30 z-slices (232 ms per slice) every 10 s for a total of 90 cycles. Analysis was done after Airyscan processing and orthogonal projection to collapse all the z-slices to a single frame.
Quantification of cellular characteristics of microglia and macrophages
Velocity
The velocity of each macrophage migrating into the brain was calculated using MtrackJ plugin in Fiji. Cells that appeared in a single frame but disappeared in the next frame (t = 0) were not included in the analyses.
Amoeboid morphology
Microglia and macrophages were classified as “amoeboid” if the cell bodies were rounded with no observed processes in the image, “intermediate” if the cell bodies were rounded but the cells extended processes, and “ramified” if the cells displayed elaborate processes emanating from cell bodies that appeared normal.
Length of processes
The length of each process was calculated using Fiji. Length was calculated in a single frame from the center of the cell to the extreme end of each process, including any secondary and tertiary branches. Cells with no processes were not included in the analyses.
Complexity of processes and phagocytic cup formation
All the secondary and tertiary branches as well as formation of phagocytic cup in each process of microglia were noted across the entire length of the time lapse for each cell. Cells with no processes were not included in the analyses.
Dissociation of larvae, macrophage sorting, and RNA extraction
rraga+/−; mpeg:GFP animals were intercrossed, and embryos were collected, treated with PTU before 24 hpf, and washed daily with embryo water containing PTU until 6 dpf. At 4 and 5 dpf, animals were anesthetized, mounted in a drop of embryo water on a slide containing coverslip bridges, and examined under the confocal microscope for the presence of the GFP transgene. Animals were scored as mutants or wild-type siblings based on the morphology of macrophages (amoeboid corresponding to rraga mutants) and separated into dishes. At 6 dpf, larvae were anesthetized using tricaine and left at 4°C for 30 min to euthanize the animals. Larvae were washed once with cold Hanks’ balanced salt solution (HBSS) (Thermo Fisher Scientific, 14025092), and each animal was cut into at least three pieces with a fresh scalpel on ice. Slices of 90 animals of each genotype were divided into three 15-ml conical tubes (3 × 30 for each genotype) and spun down at 800g for 5 min at 4°C. HBSS was removed, and 2 ml of freshly made collagenase (1 mg/ml) (Worthington Biochemical, LS004194) in HBSS was added to each tube. Collagenase digestion was carried out with gentle agitation in an incubator set at 32°C for 30 min; larval tissue was pipetted up and down 10× using P1000 after 15 min of incubation. Following a spin at 800g for 5 min at 4°C, 5 ml of 0.25% (5×) cold trypsin-EDTA (Santa Cruz Biotechnology, sc-363354; 1:1 of 0.5% or 10× in HBSS) was added to each tube. Trypsin digestion was carried out for 20 min with gentle agitation at room temperature; larval tissue was pipetted up and down 10× using P1000 after 10 min of trypsin digestion. Trypsin digestion was stopped by adding fetal bovine serum (FBS) at 5% (250 μl of FBS in 5 ml of trypsin-EDTA). Tubes were spun at 800g for 5 min at 4°C, trypsin was removed, and the pellet was resuspended in 2 ml of cold FACSmax dissociation solution (Genlantis, T200100). Following 10 min of incubation in FACSmax with gentle agitation at room temperature, tubes were spun at 800g for 5 min at 4°C, FACSmax was removed, and the pellet was resuspended in 2 ml of cold HBSS containing 5% FBS. Following another spin, the pellet was resuspended in 250 μl of phosphate-buffered saline (PBS) containing 2% FBS and 1 mM EDTA. The pellet in PBS was strained through a 40-μm cell strainer, and the cell suspension was transferred to a 5-ml fluorescence-activated cell sorting (FACS) tube to proceed with sorting. All cell suspensions were kept on ice during the entire duration of the sort. TO-PRO-3 (AAT Bioquest, 17572) was used to distinguish the live GFP+ cells and added at 1:1000 concentration (in PBS) 20 min before each tube was sorted. GFP+ cells (at least 10,000 cells) from rraga mutants or wild-type siblings were sorted using BD FACSAria II directly into lysis buffer (RLT + β-mercaptoethanol) from the Qiagen RNeasy Micro Kit (74004). RNA was extracted immediately after sorting using the Qiagen RNeasy Micro Kit (74004), and the eluate containing RNA was snap-frozen and stored at −80°C.
Whole-larvae sample preparation for RNA-seq
Wild-type mpeg:GFP or tfeb; tfe3a; tfe3b triple mutants or rraga+/−; mpeg:GFP heterozygotes were intercrossed to obtain wild-type, triple mutant, and rraga mutant larvae, respectively. At 4 dpf, approximately 100 each of wild-type or triple mutant larvae were injected with ZymA Texas Red. Approximately 1 hour after injection, the uptake of ZymA Texas Red particles by peripheral macrophages was visualized by temporarily mounting anesthetized larvae on slides with coverslip bridges and examining the animals using a confocal microscope. Animals with substantial Texas Red labeling in peripheral blood vessels, as well as amoeboid morphology of macrophages, were selected and allowed to recover in embryo water. Uninjected wild-type or triple mutants were not screened on the confocal. Approximately 2 hours after injection, 20 larvae of each genotype (wild-type or triple mutant) or treatment (uninjected or ZymA Texas Red–injected) were pooled into snap cap tubes in triplicates (total of 60 larvae), the excess water was removed, and larvae were frozen on dry ice. rraga mutants were screened on the basis of amoeboid morphology of macrophages at 4 dpf as described above and snap-frozen on dry ice. All the larvae were homogenized using a FastPrep-24 Classic bead beating grinder and lysis system (MP Biomedicals), RNA was extracted using a Takara NucleoSpin RNA Plus (740984.50) kit, and the eluate containing RNA was snap-frozen and stored at −80°C.
Library preparation and quality control
The subsequent steps of RNA-seq were performed by Novogene Corporation Inc. Quality control for all RNA samples was performed using an Agilent 2100 Bioanalyzer system; all samples had a sample integrity value of RNA integrity number > 9.0. For the macrophage RNA-seq experiment, five biological replicates were initially processed; following RNA quality control, four biological replicates were advanced for library preparation and sequencing. For whole-larvae sequencing, three biological replicates were used for each treatment and/or genotype. cDNA library preparation was done using the Takara SMART-Seq v4 Ultra Low Input RNA Kit for the macrophage RNA samples and the NEBNext Ultra II RNA Library Prep Kit for the whole-larvae RNA samples. Sequencing was performed using the Illumina NovaSeq platform (PE150). Data quality control was performed to confirm that the error rate was 0.02 to 0.03% for all samples and the guanine-cytosine content of paired reads was approximately 50%. At least 82 million reads were obtained for each of the macrophage samples, and more than 39 million reads were obtained from whole-larvae samples.
RNA-seq analysis
After reads containing adapters and low-quality reads were removed, the percentage of clean reads was more than 94% from macrophage samples and at least 97% for samples from whole larvae. These reads were mapped using HISAT2 v2.0.5 to Ensembl GRCz11 (Genome Reference Consortium Zebrafish Build 11), and quantification of reads was done using featureCounts v1.5.0 (81). More than 90% of the clean reads were mapped to the genome, and close to 80% of these reads mapped to exonic regions in the genome. Correlation analysis was performed to confirm that biological replicates had a Pearson correlation coefficient of >0.92, and principal components analysis was performed to ensure that biological replicates clustered together. Differential expression analysis was done using DESeq2 R package (82), and the resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling false discovery rate. A Padj cutoff of <0.05 and a log2 fold change of >+1 (for differentially up-regulated genes) or <−1 (for differentially down-regulated genes) were used for all samples with the exception of the heatmap in Fig. 7. ClusterProfiler (83) and ShinyGO (84) were used for GO and KEGG enrichment analyses. Fold enrichment in GO enrichment graphs is defined as the percentage of genes in our list belonging to a pathway, divided by the corresponding percentage in the background. Qiagen Ingenuity Pathway Analysis software was used to determine top canonical pathways enriched in differentially expressed genes.
EdU labeling and TUNEL assay
rraga+/−; mpeg:GFP animals were intercrossed and dechorionated at approximately 50 hpf. For EdU labeling, larvae were anesthetized, 1 nl of 5 mM EdU from Thermo Fisher Scientific [C10086; 10 mM EdU stock in dimethyl sulfoxide (DMSO) was diluted 1:1 in 1× PBS before injections] was injected in the duct of Cuvier at 60 hpf, and the EdU was chased overnight for 12 hours. All larvae for both EdU and TUNEL experiments were fixed at 72 hpf with 4% paraformaldehyde for 2 hours at room temperature. Fixed larvae were permeabilized using proteinase K (Thermo Fisher Scientific, 25530049) at a dilution of 1:1000 for 30 min and postfixed using 4% paraformaldehyde for 20 min at room temperature. Larvae were incubated in blocking solution [1% DMSO, 1% donkey serum, 1% bovine serum albumin (BSA), and 0.7% Triton X-100 in 1× PBS] for 2 hours at room temperature and incubated in blocking solution with anti-GFP antibody (1:500; Abcam, ab6658) overnight at 4°C. Following six 10-min washes in 1× PBS with 0.8% Triton X-100, larvae were incubated in block solution with 1:500 donkey anti-goat Alexa Fluor 488 or 594 for 2 hours at room temperature, followed by six 10-min washes in 1× PBS with 0.8% Triton X-100. For EdU labeling, Click-iT reaction was performed according to the manufacturer’s instructions (Thermo Fisher Scientific, C10086) and animals were incubated in development solution for 1 hour in the dark at room temperature. For TUNEL assay, larvae were incubated in 1:10 dilution of in situ cell death detection kit solution (Roche, SKU 12156792910) (10 μl of enzyme solution with 90 μl of label solution) and incubated at 37°C in the dark. Following washes with 1× PBS with 0.8% Triton X-100 overnight, animals were mounted in 2% agarose and imaged. Quantification and genotyping were performed after imaging.
Morpholino and drug treatments
Injection with BAI1 + TIM4 morpholinos or caspase-3 morpholino followed by ZVAD treatment was performed as described previously (37, 49). All animals were imaged at 4 dpf. For cytochalasin B or latrunculin A treatment, 1 nl of 0.5 mM stock was injected into the yolk of zebrafish larvae at 2.5 dpf (before the amoeboid morphology of macrophages in rraga mutants was apparent). Animals were imaged at 3.5 dpf.
Yolk injection of dyes or debris to assay macrophage uptake
Injections of DQ Red–BSA (Thermo Fisher Scientific, D12051; 1 mg/ml in 1× PBS), MR-Cathepsin (Immunochemistry Technologies, 937; vial 6133 suspended in 50 μl of DMSO, 1:1 dilution in 1× PBS before use), E. coli Texas Red (Thermo Fisher Scientific, E2863; 20 mg/ml in 1× PBS), zymosan (Sigma-Aldrich, Z4250; 10 mg/ml in 1× PBS, boiled to solubilize), and ZymA Texas Red (Thermo Fisher Scientific, Z2843; 20 mg/ml in 1× PBS) were all performed at 4 dpf. One nanoliter of each of the above suspensions was injected in the yolk of anesthetized larvae (85) of the required genotype. The injected dyes or debris particles migrate to the peripheral blood vessels of the larvae within approximately 1 to 1.5 hours after injection, where macrophages are easily visualized. For microscopy experiments, larvae were anesthetized, mounted in 1.5% agarose, and imaged using a Zeiss LSM 700 confocal microscope. For RNA-seq experiments, larvae injected with ZymA Texas Red were mounted in a drop of embryo water on a slide containing coverslip bridges and returned to embryo water to recover after the uptake of Texas Red particles by macrophages was confirmed.
Transgene constructs and injection
Full-length zebrafish tfe3b (accession number: NM_001045066.1) and LAMP1b (accession number: NM_001326532.1) were cloned from wild-type cDNA, and LAMP2, rab5, and rab7 were amplified and cloned from respective plasmids (53, 86). All the cloning primers are listed in table S2. Amplified polymerase chain reaction (PCR) fragments of respective genes were cloned into pCR8 vector (for tfe3b) or pCR8 vector with mCherry (for LAMP1b, LAMP2, rab5, and rab7) and verified by Sanger sequencing. Macrophage-specific expression vector was assembled using multisite Gateway Cloning (Invitrogen, 12538120) method using the mpeg promoter and a destination vector containing Tol2-transposon sites for genomic integration and Tg(cmlc2:mCherry) or Tg(cmlc2:GFP) for selection (87). The plasmid expressing the transgene was coinjected at 12 to 25 pg along with 50 to 100 pg of Tol2 transposase mRNA at one-cell stage (87). For tfe3b, injected embryos were selected for further analysis based on strong expression of mCherry in the heart by the cardiac reporter Tg(cmlc2:mCherry). The injected animals were raised and outcrossed to TL or Tg(mpeg:GFP) lines, and the presence of stable expression of mCherry in the heart was confirmed before experiments and/or imaging. Tg(cmlc2:GFP) was used as the selection marker for lamp1b-mCherry, lamp2-mCherry, mCherry-rab5, and mCherry-rab7 constructs. The Rab5 and Rab7 transgene constructs were crossed to rraga+/−; mpeg:GFP lines, and microscopy experiments were performed on F1 generation. LAMP1B and LAMP2 experiments were performed on rraga+/−; mpeg:GFP intercross animals after injection and normalization for cmlc2:GFP expression (F0 generation).
Generation of tfe3a, tfe3b, and flcn mutants with CRISPR-Cas9
Single-guide RNAs (sgRNAs) targeting tfe3a, tfe3b, and flcn were designed using CHOPCHOP (https://chopchop.cbu.uib.no/) (88, 89). Oligonucleotides containing T7 binding site and the CRISPR sequence were annealed to trans-activating CRISPR RNA template. Assembled oligonucleotides were transcribed using a HiScribe T7 Quick (New England Biolabs, E2050S) kit. Following deoxyribonuclease treatment, the RNA was purified using a mirVana microRNA isolation kit (Invitrogen, AM1561). An aliquot of the sgRNA eluate was run on agarose gel and quantified using NanoDrop 8000. CRISPR injections were performed at one-cell stage. Injection mix consisted of Cas9 protein (300 ng/μl) (Macrolab, Berkeley; http://qb3.berkeley.edu/macrolab/cas9-nls-purified-protein/) and sgRNA (300 ng/μl) in tris-HCl (pH 7.5). A small amount of phenol red was added to the mix to help with visualization during injection. Injected fish were raised to adulthood.
The F1 progeny of F0-injected fish were screened for out-of-frame insertions or deletions. Details of guide RNAs used, lesions, and restriction enzyme assays for genotyping mutants are described in table S2. The tfe3a allele, st124, is a 2–base pair (bp) deletion in exon 2. We used two mutant alleles of tfe3b: st125, a 25-bp deletion in exon 2, and st126, which is linked to tfeb (st120) and is a 2-bp deletion in exon 2. The flcn mutation has two different lesions generated by coinjecting two guide RNAs: st127 has a 7 (−8 + 1)–bp change in exon 2 (which disrupts the open reading frame), followed by a 4-bp deletion in exon 7, approximately 10,000 kb downstream.
Neutral red assays
Neutral red was dissolved in distilled water to give a stock solution (2.5 mg/ml), which is stable at room temperature for several months. Staining using neutral red was done by treating larvae at 4 dpf with solution (5 μg/ml) of neutral red in embryo water containing PTU for 3 hours (46). Animals were rinsed at least twice after neutral red staining and washed overnight in embryo water with PTU at 28.5°C. Approximately 24 hours after neutral red treatment, larvae were anesthetized and mounted in 1.5% low–melting point agarose. The number of neutral red+ microglia was counted, and images were acquired immediately after counting using a Zeiss AxioCam HRc camera with the AxioVision software. Between 100 and 150 embryos were mounted and imaged for each double, triple, and quadruple mutant experiment. After imaging, all animals were genotyped by PCR and statistical analysis was performed.
LysoTracker red and LysoSensor green assays
LysoTracker Red DND-99 (Thermo Fisher Scientific, L7528) was diluted 1:100 in embryo water + PTU, and LysoSensor Green DND-189 (Thermo Fisher Scientific, L7535) was diluted 1:1000 in embryo water + PTU. Larvae at 4 dpf were incubated in the LysoTracker Red or LysoSensor Green solution for 30 to 45 min. After staining, larvae were washed in embryo water containing PTU for 30 min, with at least three washes approximately 8 to 10 min apart. Following anesthesia and mounting in agarose, images were acquired using a Zeiss LSM 700 confocal microscope. All genotyping was done by PCR after imaging, and the intensity of LysoSensor Green or area of LysoTracker Red punctae in the midbrain, where most microglia are present, was calculated using ImageJ for at least three animals per genotype.
In situ hybridization
apoe antisense probe was generated from a pCRII clone carrying 505 bp of apoeb gene (42). In situ hybridization was performed using standard methods (90). Briefly, embryos at 4 dpf were fixed overnight in 4% paraformaldehyde, dehydrated at least overnight in 100% methanol, rehydrated in PBS–Triton X-100 and PBS, permeabilized using proteinase K (20 mg/ml) at a dilution of 1:1000 for 1 hour, and incubated overnight with antisense riboprobes at 65°C. Following washes in 2× and 0.2× SSC, and incubation in Maleic Acid Buffer block containing 10% normal sheep serum, animals were incubated overnight at 4°C in MAB block containing 1:1000 dilution of anti-digoxigenin antibody conjugated to alkaline phosphatase. The following day, after at least six 20-min washes in MAB–Triton X-100 buffer, animals were incubated in development solution containing nitro blue tetrazolium (Roche, 11383213001) and bromochloroindolyl phosphate (Roche, 11383221001). Development was stopped at the same time for all samples in a single experiment by evaluating the strength of the apoe signal in control animals. Animals were washed twice in PBS–Triton X-100, left in 100% ethanol overnight for destaining, and rehydrated in PBS–Triton X-100. Following genotyping of finclips, representative animals were mounted in 100% glycerol and images were captured using a Zeiss AxioCam HRc camera with the AxioVision software.
Acknowledgments
We thank T. Reyes and C. Hill for maintaining the fish facility. We are grateful to D. Lysko, M. Sidoli, E. Bouchard, and J. Vaughen for their comments on the manuscript and to G. Wang for his help with time-lapse microscopy. Templates for the constructs generated in this manuscript were donated by colleagues: Rab5 and Rab7 plasmids from B. Link at the Medical College of Wisconsin, LAMP2 plasmid from M. Bagnat at Duke University, and mCherry plasmid from G. Kingman at Stanford University. We acknowledge the use of Zeiss LSM 980 from the Stanford Wu Tsai Neuroscience Microscopy Service. Experimental schematics were made using Biorender.
Funding: This work was supported by Postdoctoral Fellowship (18POST33990334), American Heart Association (to H.I.); Developmental Project Award, Stanford Alzheimer’s Disease Research Center and National Institute on Aging (to H.I.); Postdoctoral Fellowship (A2021011F), BrightFocus Foundation (to H.I.); A∗STAR Singapore (to K.S.); NIH grant R35NS111584 (to W.S.T.); and National Multiple Sclerosis Society grant RG-1707-28694 (to W.S.T.).
Author contributions: Conceptualization: H.I. Methodology: H.I. and W.S.T. Investigation: H.I., K.S., and A.M.M. Visualization: H.I. Supervision: W.S.T. Writing—original draft: H.I. Writing—review and editing: H.I., W.S.T., and A.M.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data have been deposited in Gene Expression Omnibus and are available using these accession numbers: macrophage RNA-seq GSE197349 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197349) and whole-larvae RNA-seq GSE197348 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197348).
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
Movies S1 to S4
REFERENCES AND NOTES
- 1.Cunningham C. L., Martinez-Cerdeno V., Noctor S. C., Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 33, 4216–4233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafer D. P., Lehrman E. K., Kautzman A. G., Koyama R., Mardinly A. R., Yamasaki R., Ransohoff R. M., Greenberg M. E., Barres B. A., Stevens B., Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shigemoto-Mogami Y., Hoshikawa K., Goldman J. E., Sekino Y., Sato K., Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34, 2231–2243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakselman S., Bechade C., Roumier A., Bernard D., Triller A., Bessis A., Developmental neuronal death in hippocampus requires the microglial CD11b integrin and DAP12 immunoreceptor. J. Neurosci. 28, 8138–8143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davalos D., Grutzendler J., Yang G., Kim J. V., Zuo Y., Jung S., Littman D. R., Dustin M. L., Gan W. B., ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758 (2005). [DOI] [PubMed] [Google Scholar]
- 6.De Biase L. M., Schuebel K. E., Fusfeld Z. H., Jair K., Hawes I. A., Cimbro R., Zhang H. Y., Liu Q. R., Shen H., Xi Z. X., Goldman D., Bonci A., Local cues establish and maintain region-specific phenotypes of basal ganglia microglia. Neuron 95, 341–356.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagemeyer N., Hanft K. M., Akriditou M. A., Unger N., Park E. S., Stanley E. R., Staszewski O., Dimou L., Prinz M., Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134, 441–458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matcovitch-Natan O., Winter D. R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada Gonzalez F., Perrin P., Keren-Shaul H., Gury M., Lara-Astaiso D., Thaiss C. A., Cohen M., Bahar Halpern K., Baruch K., Deczkowska A., Lorenzo-Vivas E., Itzkovitz S., Elinav E., Sieweke M. H., Schwartz M., Amit I., Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Nimmerjahn A., Kirchhoff F., Helmchen F., Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Sierra A., Encinas J. M., Deudero J. J., Chancey J. H., Enikolopov G., Overstreet-Wadiche L. S., Tsirka S. E., Maletic-Savatic M., Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Napolitano G., Ballabio A., TFEB at a glance. J. Cell Sci. 129, 2475–2481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y., Xu Y., Zhang C., Gao X., Dykema K. J., Martin K. R., Ke J., Hudson E. A., Khoo S. K., Resau J. H., Alberts A. S., MacKeigan J. P., Furge K. A., Xu H. E., Identification of a lysosomal pathway that modulates glucocorticoid signaling and the inflammatory response. Sci. Signal. 4, ra44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Netea-Maier R. T., Plantinga T. S., van de Veerdonk F. L., Smit J. W., Netea M. G., Modulation of inflammation by autophagy: Consequences for human disease. Autophagy 12, 245–260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtman I. R., Raj D. D., Miller J. A., Schaafsma W., Yin Z., Brouwer N., Wes P. D., Moller T., Orre M., Kamphuis W., Hol E. M., Boddeke E. W., Eggen B. J., Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co-expression meta-analysis. Acta Neuropathol. Commun. 3, 31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plaza-Zabala A., Sierra-Torre V., Sierra A., Autophagy and microglia: Novel partners in neurodegeneration and aging. Int. J. Mol. Sci. 18, 598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sole-Domenech S., Cruz D. L., Capetillo-Zarate E., Maxfield F. R., The endocytic pathway in microglia during health, aging and Alzheimer’s disease. Ageing Res. Rev. 32, 89–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A., Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852–3866 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A., A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A., TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Settembre C., Fraldi A., Medina D. L., Ballabio A., Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortes C. J., Miranda H. C., Frankowski H., Batlevi Y., Young J. E., Le A., Ivanov N., Sopher B. L., Carromeu C., Muotri A. R., Garden G. A., La Spada A. R., Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat. Neurosci. 17, 1180–1189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J., Bjorklund A., TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. U.S.A. 110, E1817–E1826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy K., Cusack C. L., Nnah I. C., Khayati K., Saqcena C., Huynh T. B., Noggle S. A., Ballabio A., Dobrowolski R., Dysregulation of nutrient sensing and CLEARance in presenilin deficiency. Cell Rep. 14, 2166–2179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsunemi T., Ashe T. D., Morrison B. E., Soriano K. R., Au J., Roque R. A., Lazarowski E. R., Damian V. A., Masliah E., La Spada A. R., PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4, 142ra197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martini-Stoica H., Xu Y., Ballabio A., Zheng H., The autophagy-lysosomal pathway in neurodegeneration: A TFEB perspective. Trends Neurosci. 39, 221–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickrell A. M., Youle R. J., The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dehay B., Bove J., Rodriguez-Muela N., Perier C., Recasens A., Boya P., Vila M., Pathogenic lysosomal depletion in Parkinson’s disease. J. Neurosci. 30, 12535–12544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parr C., Carzaniga R., Gentleman S. M., Van Leuven F., Walter J., Sastre M., Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-β precursor protein. Mol. Cell. Biol. 32, 4410–4418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polito V. A., Li H., Martini-Stoica H., Wang B., Yang L., Xu Y., Swartzlander D. B., Palmieri M., di Ronza A., Lee V. M., Sardiello M., Ballabio A., Zheng H., Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 6, 1142–1160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Burchett J. M., Schuler D. R., Cirrito J. R., Diwan A., Lee J. M., Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 34, 9607–9620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Tripoli D. L., Czerniewski L., Ballabio A., Cirrito J. R., Diwan A., Lee J. M., Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Aβ generation and amyloid plaque pathogenesis. J. Neurosci. 35, 12137–12151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amit I., Winter D. R., Jung S., The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat. Immunol. 17, 18–25 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Bohlen C. J., Bennett F. C., Tucker A. F., Collins H. Y., Mulinyawe S. B., Barres B. A., Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94, 759–773.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butovsky O., Jedrychowski M. P., Moore C. S., Cialic R., Lanser A. J., Gabriely G., Koeglsperger T., Dake B., Wu P. M., Doykan C. E., Fanek Z., Liu L., Chen Z., Rothstein J. D., Ransohoff R. M., Gygi S. P., Antel J. P., Weiner H. L., Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosselin D., Link V. M., Romanoski C. E., Fonseca G. J., Eichenfield D. Z., Spann N. J., Stender J. D., Chun H. B., Garner H., Geissmann F., Glass C. K., Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gosselin D., Skola D., Coufal N. G., Holtman I. R., Schlachetzki J. C. M., Sajti E., Jaeger B. N., O’Connor C., Fitzpatrick C., Pasillas M. P., Pena M., Adair A., Gonda D. D., Levy M. L., Ransohoff R. M., Gage F. H., Glass C. K., An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casano A. M., Albert M., Peri F., Developmental apoptosis mediates entry and positioning of microglia in the zebrafish brain. Cell Rep. 16, 897–906 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Peri F., Nusslein-Volhard C., Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133, 916–927 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Wu S., Xue R., Hassan S., Nguyen T. M. L., Wang T., Pan H., Xu J., Liu Q., Zhang W., Wen Z., Il34-Csf1r pathway regulates the migration and colonization of microglial precursors. Dev. Cell 46, 552–563.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Xu J., Zhu L., He S., Wu Y., Jin W., Yu T., Qu J. Y., Wen Z., Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev. Cell 34, 632–641 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Meireles A. M., Shiau C. E., Guenther C. A., Sidik H., Kingsley D. M., Talbot W. S., The phosphate exporter xpr1b is required for differentiation of tissue-resident macrophages. Cell Rep. 8, 1659–1667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiau C. E., Monk K. R., Joo W., Talbot W. S., An anti-inflammatory NOD-like receptor is required for microglia development. Cell Rep. 5, 1342–1352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen K., Sidik H., Talbot W. S., The rag-ragulator complex regulates lysosome function and phagocytic flux in microglia. Cell Rep. 14, 547–559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellett F., Pase L., Hayman J. W., Andrianopoulos A., Lieschke G. J., mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herbomel P., Thisse B., Thisse C., Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Herbomel P., Thisse B., Thisse C., Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274–288 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Xu J., Wang T., Wu Y., Jin W., Wen Z., Microglia colonization of developing zebrafish midbrain is promoted by apoptotic neuron and lysophosphatidylcholine. Dev. Cell 38, 214–222 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Block M. L., Zecca L., Hong J. S., Microglia-mediated neurotoxicity: Uncovering the molecular mechanisms. Nat. Rev. Neurosci. 8, 57–69 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Mazaheri F., Breus O., Durdu S., Haas P., Wittbrodt J., Gilmour D., Peri F., Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 5, 4046 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Malawista S. E., Gee J. B., Bensch K. G., Cytochalasin B reversibly inhibits phagocytosis: Functional, metabolic, and ultrastructural effects in human blood leukocytes and rabbit alveolar macrophages. Yale J. Biol. Med. 44, 286–300 (1971). [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira C. A., Kashman Y., Mantovani B., Effects of latrunculin A on immunological phagocytosis and macrophage spreading-associated changes in the F-actin/G-actin content of the cells. Chem. Biol. Interact. 100, 141–153 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Eskelinen E. L., Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspects Med. 27, 495–502 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Clark B. S., Winter M., Cohen A. R., Link B. A., Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240, 2452–2465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colacurcio D. J., Nixon R. A., Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin H. J., Herman P., Kang J. S., Lakowicz J. R., Fluorescence lifetime characterization of novel low-pH probes. Anal. Biochem. 294, 118–125 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yadati T., Houben T., Bitorina A., Shiri-Sverdlov R., The ins and outs of cathepsins: Physiological function and role in disease management. Cell 9, 1679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song X. B., Liu G., Liu F., Yan Z. G., Wang Z. Y., Liu Z. P., Wang L., Autophagy blockade and lysosomal membrane permeabilization contribute to lead-induced nephrotoxicity in primary rat proximal tubular cells. Cell Death Dis. 8, e2863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kundu S. T., Grzeskowiak C. L., Fradette J. J., Gibson L. A., Rodriguez L. B., Creighton C. J., Scott K. L., Gibbons D. L., TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat. Commun. 9, 2731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haney M. S., Bohlen C. J., Morgens D. W., Ousey J. A., Barkal A. A., Tsui C. K., Ego B. K., Levin R., Kamber R. A., Collins H., Tucker A., Li A., Vorselen D., Labitigan L., Crane E., Boyle E., Jiang L., Chan J., Rincon E., Greenleaf W. J., Li B., Snyder M. P., Weissman I. L., Theriot J. A., Collins S. R., Barres B. A., Bassik M. C., Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat. Genet. 50, 1716–1727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuil L. E., Oosterhof N., Ferrero G., Mikulasova T., Hason M., Dekker J., Rovira M., van der Linde H. C., van Strien P. M., de Pater E., Schaaf G., Bindels E. M., Wittamer V., van Ham T. J., Zebrafish macrophage developmental arrest underlies depletion of microglia and reveals Csf1r-independent metaphocytes. eLife 9, e53403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meireles A. M., Shen K., Zoupi L., Iyer H., Bouchard E. L., Williams A., Talbot W. S., The lysosomal transcription factor TFEB represses myelination downstream of the Rag-ragulator complex. Dev. Cell 47, 319–330.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pastore N., Brady O. A., Diab H. I., Martina J. A., Sun L., Huynh T., Lim J. A., Zare H., Raben N., Ballabio A., Puertollano R., TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12, 1240–1258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lister J. A., Lane B. M., Nguyen A., Lunney K., Embryonic expression of zebrafish MiT family genes tfe3b, tfeb, and tfec. Dev. Dyn. 240, 2529–2538 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong S. B., Oh H., Valera V. A., Baba M., Schmidt L. S., Linehan W. M., Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLOS ONE 5, e15793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wada S., Neinast M., Jang C., Ibrahim Y. H., Lee G., Babu A., Li J., Hoshino A., Rowe G. C., Rhee J., Martina J. A., Puertollano R., Blenis J., Morley M., Baur J. A., Seale P., Arany Z., The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 30, 2551–2564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J., Wada S., Weaver L. K., Biswas C., Behrens E. M., Arany Z., Myeloid Folliculin balances mTOR activation to maintain innate immunity homeostasis. JCI Insight 5, e126939 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El-Houjeiri L., Possik E., Vijayaraghavan T., Paquette M., Martina J. A., Kazan J. M., Ma E. H., Jones R., Blanchette P., Puertollano R., Pause A., The transcription factors TFEB and TFE3 link the FLCN-AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 26, 3613–3628.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martina J. A., Diab H. I., Brady O. A., Puertollano R., TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35, 479–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martina J. A., Puertollano R., TFEB and TFE3: The art of multi-tasking under stress conditions. Transcription 8, 48–54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian R., Abarientos A., Hong J., Hashemi S. H., Yan R., Drager N., Leng K., Nalls M. A., Singleton A. B., Xu K., Faghri F., Kampmann M., Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat. Neurosci. 24, 1020–1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Demy D. L., Carrere M., Noche R., Tauzin M., Le Bris M., Baek C., Leshchiner I., Goessling W., Herbomel P., The cationic amino acid exporter Slc7a7 is induced and vital in zebrafish tissue macrophages with sustained efferocytic activity. J. Cell Sci. 133, jcs249037 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Raben N., Puertollano R., TFEB and TFE3: Linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol. 32, 255–278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H., Wang R., Xu S., Lakshmana M. K., Transcription factor EB is selectively reduced in the nuclear fractions of Alzheimer’s and amyotrophic lateral sclerosis brains. Neurosci. J. 2016, 4732837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong S., Beja-Glasser V. F., Nfonoyim B. M., Frouin A., Li S., Ramakrishnan S., Merry K. M., Shi Q., Rosenthal A., Barres B. A., Lemere C. A., Selkoe D. J., Stevens B., Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rivest S., Regulation of innate immune responses in the brain. Nat. Rev. Immunol. 9, 429–439 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Simon E., Obst J., Gomez-Nicola D., The evolving dialogue of microglia and neurons in Alzheimer’s disease: Microglia as necessary transducers of pathology. Neuroscience 405, 24–34 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Combs C. K., Johnson D. E., Karlo J. C., Cannady S. B., Landreth G. E., Inflammatory mechanisms in Alzheimer’s disease: Inhibition of β-amyloid-stimulated proinflammatory responses and neurotoxicity by PPARγ agonists. J. Neurosci. 20, 558–567 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin L., Liu Y., Cooper C., Liu B., Wilson B., Hong J. S., Microglia enhance β-amyloid peptide-induced toxicity in cortical and mesencephalic neurons by producing reactive oxygen species. J. Neurochem. 83, 973–983 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Mazzolini J., Le Clerc S., Morisse G., Coulonges C., Kuil L. E., van Ham T. J., Zagury J. F., Sieger D., Gene expression profiling reveals a conserved microglia signature in larval zebrafish. Glia 68, 298–315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferrero G., Mahony C. B., Dupuis E., Yvernogeau L., Di Ruggiero E., Miserocchi M., Caron M., Robin C., Traver D., Bertrand J. Y., Wittamer V., Embryonic microglia derive from primitive macrophages and are replaced by cmyb-dependent definitive microglia in zebrafish. Cell Rep. 24, 130–141 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge S. X., Jung D., Yao R., ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benard E. L., van der Sar A. M., Ellett F., Lieschke G. J., Spaink H. P., Meijer A. H., Infection of zebrafish embryos with intracellular bacterial pathogens. J. Vis. Exp. 3781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park J., Levic D. S., Sumigray K. D., Bagwell J., Eroglu O., Block C. L., Eroglu C., Barry R., Lickwar C. R., Rawls J. F., Watts S. A., Lechler T., Bagnat M., Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev. Cell 51, 7–20.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B., The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
- 88.Labun K., Montague T. G., Gagnon J. A., Thyme S. B., Valen E., CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., Valen E., CHOPCHOP: A CRISPR-Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S5
Movies S1 to S4