Summary
The intestinal epithelium undergoes continuous renewal and has an exceptional capacity to regenerate after injury. Maintenance and proliferation of intestinal stem cells (ISCs) are regulated by their surrounding niche, largely through Wnt signaling. However, it remains unclear which niche cells produce signals during different injury states, and the role of endothelial cells (ECs) as a component of the ISC niche during homeostasis and after injury has been underappreciated. Here, we show that lymphatic endothelial cells (LECs) reside in proximity to crypt epithelial cells and secrete molecules that support epithelial renewal and repair. LECs are an essential source of Wnt signaling in the small intestine, as loss of LEC-derived Rspo3 leads to a lower number of stem and progenitor cells and hinders recovery after cytotoxic injury. Together, our findings identify LECs as an essential niche component for optimal intestinal recovery after cytotoxic injury.
Graphical Abstract
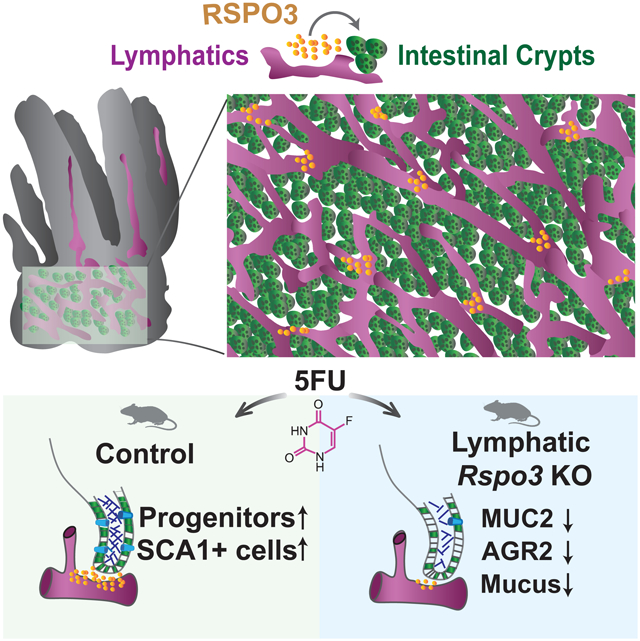
In brief
Palikuqi et al. employ imaging of cleared intestinal tissue, transcriptional analyses and genetic mouse models to investigate the role of lymphatic endothelial cells in the intestinal stem cell niche. The authors demonstrate that lymphatic endothelial cells are an essential source of Rspo3 for proper intestinal recovery after cytotoxic injury.
Introduction
Enteric blood and lymphatic networks are intimately associated with the small intestinal epithelium, including the ISCs that reside in epithelial invaginations called crypts (Bernier-Latmani et al., 2015; Bernier-Latmani and Petrova, 2017; Beumer and Clevers, 2020; Cifarelli and Eichmann, 2019). Over the last decade, several groups have shown that paracrine factors secreted by endothelial cells lining blood and lymphatic vessels, known as angiocrine factors, are important for stem cell maintenance and regeneration in many tissues (Augustin and Koh, 2017; Rafii et al., 2016). Blood endothelial cells (BECs) play a central role in the regeneration of several organs, such as liver, lung, bone marrow, and thymus (Butler et al., 2010; Ding et al., 2010; Ding et al., 2011; Ding et al., 2012; Hu et al., 2014; Wertheimer et al., 2018). Notably, LECs, through the secretion of the lymphangiocrine factor Reelin (RELN), have been linked to heart regeneration (Liu et al., 2020). In addition, the interaction of stem cells with LECs regulates stem cell cycling in the skin (Gur-Cohen et al., 2019). However, the role of intestinal ECs in ISC self-renewal and regeneration remains undetermined.
ISCs, which are marked by Lgr5, and their transit amplifying descendants self-renew and differentiate along the various intestinal epithelial lineages (Barker et al., 2007; Gehart and Clevers, 2019). Maintenance of ISCs is dependent on the niche, which is comprised of several cell types and is an essential source of Wnt ligands and modulators (McCarthy et al., 2020a; Palikuqi et al., 2021). Among these molecules, R-spondins (RSPOs), which bind to LGR receptors (LGR4 and LGR5 in the intestine), potentiate Wnt signaling (de Lau et al., 2011; Glinka et al., 2011) by stabilizing Frizzled receptors (Hao et al., 2012). RSPOs are essential for the expansion and renewal of ISCs both in vivo (Yan et al., 2017) and in vitro (Sato et al., 2009). RSPO3 is the predominant RSPO in the small intestine (Ogasawara et al., 2018), and inhibition of RSPO3 along with RSPO2 with a neutralizing antibody negatively affects epithelial regeneration after irradiation (Storm et al., 2015).
Several mesenchymal populations have been reported to be essential sources of Wnt ligands and modulators in the intestine (Degirmenci et al., 2018; Kabiri, 2014; McCarthy et al., 2020b; Stzepourginski et al., 2017; Valenta et al., 2016). For instance, mesenchymal cells marked by Foxl1+, known as telocytes, express high levels of Wnt2 and Wnt5a (Aoki et al., 2016; Kondo et al., 2019; Shoshkes-Carmel et al., 2018). The inhibition of Wnt ligand secretion by deletion of Porcupine (Porcn) in Foxl1+ telocytes results in rapid stem and progenitor cell loss (Shoshkes-Carmel et al., 2018). Another subtype of mesenchymal cells, known as trophocytes and marked by expression of Grem1 and Cd81 and low expression of Pdgfra, resides in proximity to ISCs and expresses the Wnt-family member Wnt2b as well as high levels of all R-spondins. Ablation of the trophocyte mesenchymal population leads to loss of Lgr5+ cells, while the population of transient amplifying (TA) cells remains stable (McCarthy et al., 2020b). Moreover, deletion of Rspo3 in Pdgfra+ cells does not have a major effect on epithelial cells during homeostasis (Greicius et al., 2018), suggesting that this essential signal is produced by several different niche cell types.
LECs express high levels of Rspo3 (Kalucka et al., 2020; Ogasawara et al., 2018), but it is unknown if LECs are an essential source of Wnt signaling in the small intestine during homeostasis or injury. Additionally, while several lines of evidence suggest that endothelial cells become activated and start proliferating in response to intestinal injury to drive epithelial recovery (Abel et al., 2005; Kinchen et al., 2018; Paris et al., 2001; Rehal et al., 2018), the contribution of endothelial cells and specifically LECs to intestinal maintenance and repair remains an open question. Here, we establish the spatial organization of lymphatic vessels in relation to crypt cells and utilize single cell RNA-sequencing (scRNAseq) to uncover the transcriptional composition of LECs, both during homeostasis and after cytotoxic injury. Using mouse genetics, we show that LEC-secreted Rspo3 is essential for optimal intestinal repair after cytotoxic injury, positioning LECs as a key component of the intestinal niche and a critical source of Wnt signaling.
Results
Lymphatic endothelial cells reside in proximity to crypt cells
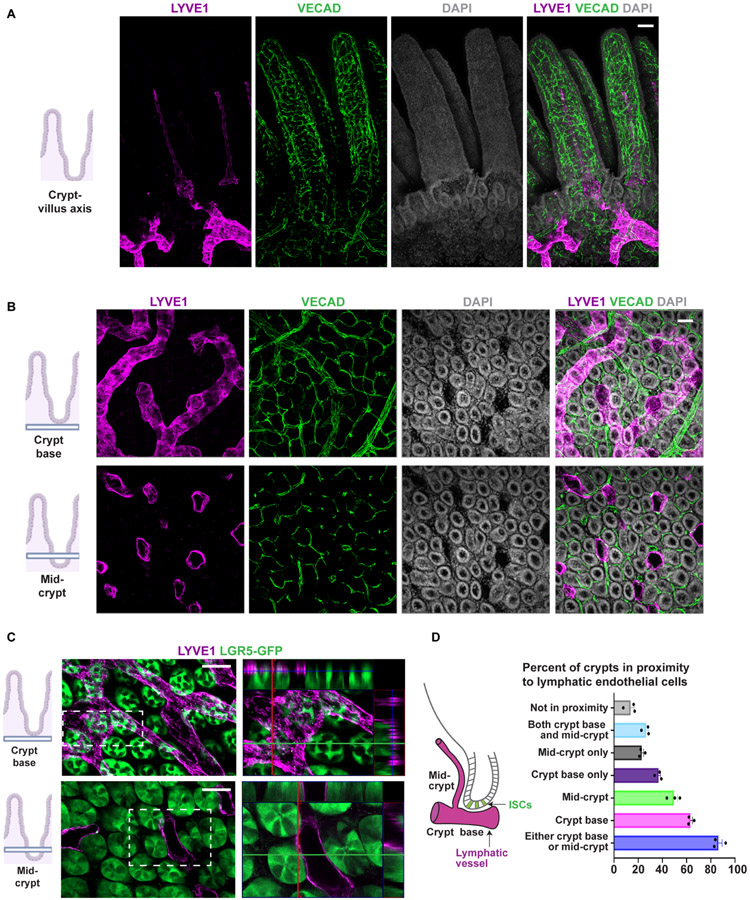
We first set out to delineate the location and molecular composition of LECs in the small intestinal niche. We utilized the CUBIC clearing method (Matsumoto et al., 2019), paired with whole mount imaging, to determine the spatial distribution of LECs in the mouse small intestine during homeostasis and to establish the relationship of LECs to BECs and mesenchymal cells in the niche (Figures 1A-B and S1A-B). VECAD+ BECs and PDGFRA+ mesenchymal cells surrounded every crypt (Figures 1B and S1A), but lymphatic vessels displayed a unique pattern of organization. We found two distinct sets of lymphatic vessels: large lymphatic vessels that encircled the base of the crypts and gave rise to narrow lymphatic vessels in the villi, known as lacteals (Figures 1A-B and S1A-B). To determine the proximity of LECs to Lgr5+ stem cells in the crypts, we performed whole mount imaging on cleared small intestinal tissue from Lgr5GFP-CreER mice. LECs were in proximity to Lgr5+ crypt cells at the base of the crypts and at the mid-crypt level (Figures 1C and S1C). More than 80 percent of crypts were in proximity to lymphatics at the base and/or mid-crypt levels, with 36 percent of crypts in proximity to LECs only at the base, 23 percent only at the mid-crypt region, and 26 percent at both (Figure 1D).
Figure 1. LECs reside in proximity to crypt cells.
A) Sagittal view of whole mount imaging of CUBIC cleared tissue from adult wild type mouse small intestine stained for LYVE1 and VECAD showing the localization of lymphatic and blood vessels within the crypt-villus axis in the small intestine. B-C) Transverse view of LYVE1 and VECAD showing (B) the organization of LECs and BECs at both the crypt base region and mid-crypt region in adult wild type mice, as well as, (C) the LECs proximity to Lgr5+ ISCs in adult Lgr5GFP-Cre mice. Insets represent orthogonal projections of the whole mounts depicting adjacency and at times contact of LECs to Lgr5+ ISCs. D) Left: schematic representation of position of lymphatic vessels in relation to crypts. Right: quantification of percentage of crypts in vicinity to LECs at the base of the crypts, at mid-crypt level or not in proximity to any LECs (n=3 mice). Scale bars = 50 μm. See also Figure S1.
Lymphatic endothelial cells express high levels of Rspo3
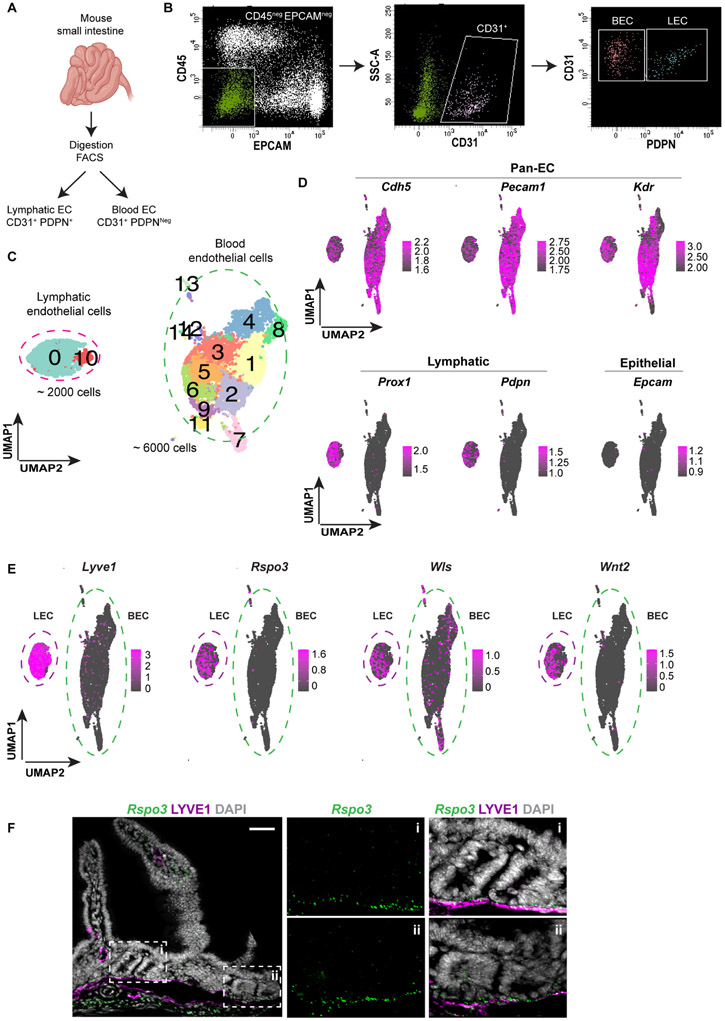
Next, we set out to establish the molecular composition of LECs in the small intestine by performing scRNAseq. BECs and LECs were enriched by fluorescence-activated cell sorting (FACS) from the small intestine of wild type mice and identified as CDH5+, PODOPLANIN (PDPN)Neg and CDH5+, PDPN+ cells, respectively (Figures 2A-B). In all, 6000 BECs and 2000 LECs were captured by scRNAseq with BECs clustering in 13 groups and LECs clustering in 2 groups (Figure 2C). LECs could be distinguished from BECs by the high expression Lyve1 and Pdpn (Figure 2D). Clustering was similar amongst 4 different mice in 2 separate experiments (Figure S2D). Notably, LECs uniquely expressed high levels of Aqp1, Cyp4b1, Il33 and Mmrn1 genes compared to BECs (Figure S1E). The transcription factor Maf was also selectively expressed in LECs (Figure S1E). Reelin (RELN), which is expressed in the heart by LECs and has been found to be essential for heart regeneration, was also highly expressed by LECs but not BECs in the small intestine (Figure S1E).
Figure 2. LEC scRNAseq data reveal expression of Rspo3 and other Wnt signaling molecules.
A) Small intestines from 4 wild-type mice in 2 independent experiments were digested, and LECs and BECs were isolated by sorting CD31+, PDPN+ and CD31+, PDPNNeg cells, respectively. B) Gating strategy to isolate LECs and BECs. First, we selected CD45Neg and EPCAMNeg cells to exclude immune and epithelial cells, then CD31+ endothelial cells were isolated, and finally LECs and BECs were distinguished as PDPN+ and PDPNNeg, respectively. C) UMAP (Uniform Manifold Approximation and Projection) plot of all ECs showing that LECs group in 2 clusters (cluster 0 and 10) and BECs group in 13 different clusters. D) UMAP plots of the expression of markers for pan-endothelial, lymphatic, and epithelial cells validating the identity of LECs and BECs. E) UMAP plots of Lyve1, Rspo3, Wls and Wnt2 expressions inside EC clusters showing that LECs in the small intestine express high levels of Rspo3 and Wls and Wnt2. F) Rspo3 In situ hybridization (RNAscope) coupled with LYVE1 immunofluorescence of adult mouse small intestine demonstrating expression of Rspo3 mRNA in LECs that are in proximity to the crypt region. Scale bar = 50 μm. See also Figure S1.
As Wnt signaling is essential for ISC maintenance and proliferation, we mined the scRNAseq data for the expression of Wnt-family members. As previously reported (Kalucka et al., 2020; Ogasawara et al., 2018), LECs in the small intestine expressed high levels of Rspo3 (Figures 2E and S1F). Additionally, LECs expressed high levels of Wntless (Wls, essential for the secretion of Wnt ligands) and the canonical Wnt-family member Wnt2 (Figures 2E and S1F). Interestingly, Rspo3 and Wnt2 were uniquely expressed by LECs, whereas Wls was also expressed by BECs (Figures 2E and S1F). Rspo3 expression by LECs in proximity to epithelial crypt cells was confirmed by in situ hybridization (RNAscope) (Figure 2F); no other Wnt-family members were expressed by LECs during homeostasis.
Loss of lymphatic Rspo3 leads to impaired intestinal recovery after cytotoxic injury
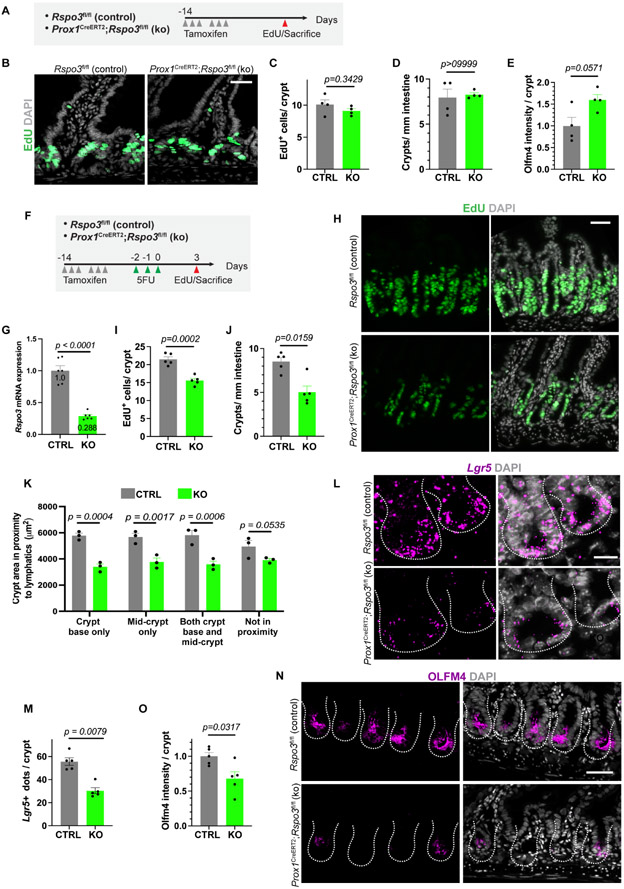
As Rspo3 is important for the proliferation of stem and progenitor cells in the crypt, we next utilized a genetic model to specifically delete Rspo3 in LECs. We bred mice carrying the lymphatic driver Prox1CreER with Rspo3fl/fl mice to generate Prox1CreERT2;Rspo3fl/fl mice (Figure 3A). Deletion of Rspo3 in adult LECs did not significantly alter crypts in the small intestine during homeostasis. Control and knock-out mice had similar levels of EdU incorporation, crypt number and OLFM4 expression after tamoxifen induction (Figures 3B-E). Thus, during homeostasis, Wnt signaling from other, redundant niche components, such as PDGFRA+ cells, is sufficient for ISC maintenance.
Figure 3. Lymphatic Rspo3 is essential for intestinal recovery after 5FU injury.
A) Tamoxifen induced Prox1CreERT2;Rspo3fl/fl knock-out mice and control Rspo3fl/fl were sacrificed 14 days after beginning of tamoxifen induction and after a 2-hour EdU pulse. B) Representative images of EdU staining from control and knock-out mice at homeostasis (n=4 mice). Scale bar = 50 μm. C) Quantification of EdU+ cells per crypt at homeostasis for control (CTRL) and knock-out (KO) mice (n=4 mice). D) Quantification of total crypts per millimeter (mm) at homeostasis for control (CTRL) and knock-out (KO) mice (n=4 mice). E) Quantification of OLFM4 staining intensity for control (CTRL) and knock-out (KO) mice at homeostasis (n=4 mice). F) After tamoxifen treatment, control (CTRL) and Rspo3 knock-out (KO) mice were treated for 3 days with 5FU at 75 mg/kg. Recovery was assessed at day 3 after injury. Mice were injected with EdU 2 hours before sacrifice. G) Deletion efficiency as quantified by Rspo3 mRNA levels analyzed by RT-qPCR in sorted LECs at day 3 after the end of 5FU treatment (n=6 mice). H) Representative images of EdU staining in the crypts of control and Rspo3 knock-out mice at day 3 and (I) quantification of EdU+ cells per crypt at day 3 (n=5 mice). Scale bar = 50 μm. J) Quantification of total crypts per millimeter (mm) at day 3 after 5FU treatment for control (CTRL) and knock-out (KO) mice (n=5 mice). K) Quantification of crypt size (area in μm2 of crypt section) in relation to their position to lymphatic vessels in control and Rspo3 knock-out mice at day 3 after 5FU (n=3 mice). L) Representative images of Lgr5+ in situ hybridization (RNAscope) for control and knock-out mice at day 3 after 5FU and (M) quantification of Lgr5 transcripts per crypt (n=5 mice). Scale bar = 20 μm. N) OLFM4 staining in control and knock mice at day 3 after 5FU (n=5 mice). Scale bar = 50 μm. O) Quantification of OLFM4 intensity staining per crypt at day 3 after 5FU (n=5 mice). Bar graph data are mean +/− s.e.m., Mann Whitney U-test, two-sided. See also Figure S2.
We then assessed whether lymphatic Rspo3 is essential for intestinal recovery after injury, when crypt epithelial cells must undergo high proliferation. To test this hypothesis, we utilized 5-fluorouracil (5FU), a chemotherapeutic that selectively kills proliferating cells. Control and Rspo3 knock-out mice underwent sublethal 5FU injury and were sacrificed at day 3 after the last 5FU injection (Figures 3F and S2A-B). Sustained deletion of Rspo3 after injury was confirmed by qPCR (Figure 3G). After 5FU injury, control crypt epithelium exhibited high levels of proliferation as measured by EdU incorporation, but knock-out mice had significantly decreased proliferation (Figures 3H-I). The total number of regenerating crypts was significantly reduced in knock-out mice (Figure 3J). Additionally, we quantified the size of crypts after 5FU and determined that, in knock-out mice, crypts in proximity to LECs were decreased in size more than those not close to LECs (Figure 3K). The total levels of Lgr5 mRNA and OLFM4 protein were also decreased in knock-out crypt cells (Figures 3L-O). Lastly, we asked whether loss of Rspo3 resulted in structural changes in LECs after 5FU injury. LEC density and percent of crypts found in proximity to LECs were similar in control and knock-out mice, suggesting that changes in crypt cells after 5FU were due to lymphangiogrine signaling rather than to loss of LEC structures (Figures S2B-D).
5FU treatment induces substantial damage to proliferating cells, potentially affecting both epithelial and niche cells. We therefore investigated whether a similar requirement for LEC-derived Rspo3 exists when Lgr5+ cells alone are ablated. We bred Prox1CreERT2;Rspo3fl/fl;Lgr5DTR mice, in which tamoxifen injection leads to deletion of Rspo3 in LECs, and Diphtheria Toxin (DT) (Tian et al., 2011) injection results in ablation of Lgr5+ cells. Tamoxifen induction was followed by Lgr5+ cell ablation for 3 consecutive days (Figure S2E). Although there was a slight decrease in levels of proliferation as measured by EdU incorporation, the effect was less pronounced compared with that observed after cytotoxic injury (Figures S2F-J). Thus, LEC-derived Rspo3 is essential for adequate intestinal recovery when there is a requirement for a marked increase in proliferation of epithelial cells and when the niche becomes activated during injury.
Loss of lymphatic Rspo3 results in lower TA and Sca1high cells after cytotoxic injury
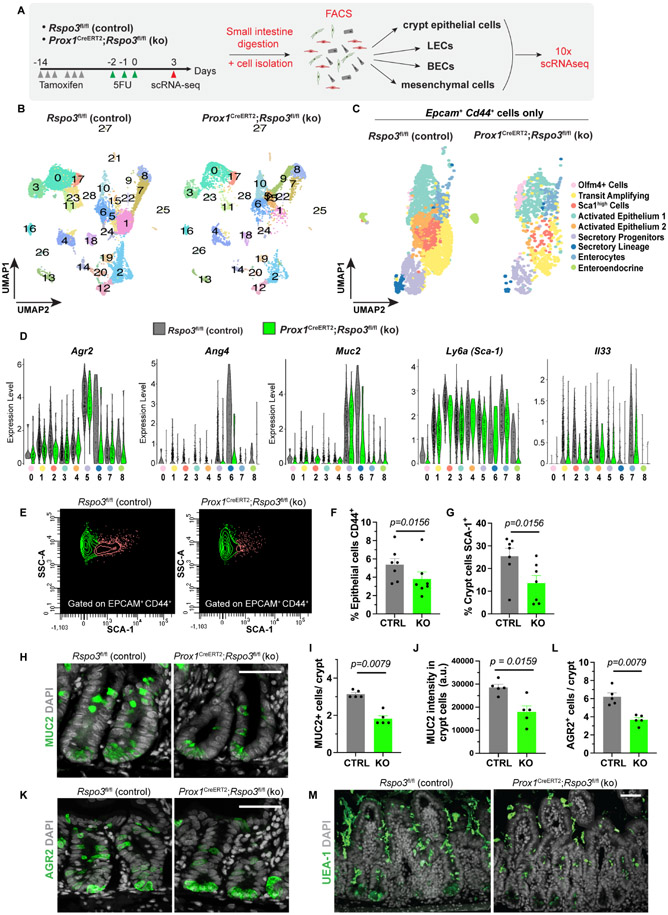
Next, we set out to further uncover molecular changes in Prox1CreERT2;Rspo3fl/fl knock-out mice after 5FU injury. At day 3 after the final 5FU injection, we isolated the following from both control and knock-out mice: crypt epithelial cells as EPCAM+, CD44+; LECs as CD31+, PDPN+; BECs as CD31+, PDPNneg; and other stromal cells as EPCAMneg, CD31neg. We then performed scRNAseq on these cells (Figures 4A and S3A). Analysis included cells from two biological replicates with a total of 20,803 cells for control mice and 9,153 for knock-out mice, with cells grouping in 29 clusters (Figures 4B and S3B). Clustering patterns were similar among the two experiments (Figure S3B). Crypt epithelial cells were identified as Epcam+, Cd44+, and LECs were identified by the pan-endothelial markers Cd31, Cdh5 or Kdr and expression of Lyve1 or Pdpn (Figures S3C-D). We also identified clusters of BECs as positive for Cd31, Cdh5 or Kdr and negative for Lyve1 and Pdpn, mesenchymal cells as Pdgfra+, and myofibroblasts as Myh11+ (Figures S3C-D). Within the Pdgfra+ mesenchymal populations, we identified telocytes as Foxl1+, Pdgfrahigh and trophocytes as Grem1+, Cd81+/Cd81neg, Pdgfralow, amongst others (Figures S4F-G).
Figure 4. Loss of LEC Rspo3 results in decreased numbers of TA, SCA1high and mucus secreting cells.
A) Schema of scRNAseq experiment. Tamoxifen treated control and knock-out mice were administered 5FU on 3 consecutive days. Crypt epithelial cells, LECs, BECs and other stromal cells were enriched by FACS, and the samples submitted for 10X scRNAseq analysis. Data are from 2 independent biological experiments. Each biological experiment included 2 control and 2 knock-out mice. B) The sequenced cells group in 29 clusters as depicted in UMAP plots. C) Crypt epithelial cells were identified as Epcam+, Cd44+ cells and re-clustered. Crypt epithelial cells group in 9 distinct clusters as shown in UMAP plots. D) Violin plot of differentially expressed genes (DEGs) in crypt epithelial cells from control (grey) versus knock-out (green) mice. E) Flow cytometry analysis of SCA-1 negative (green) and positive (red) cells, gated on crypt epithelial cells (EPCAM+ and CD44+), in control and knock-out mice at day 3 after 5FU (n=7 mice). F-G) Proportion of epithelial cells that are CD44+ (F) and crypt epithelial cells that are SCA-1+ (G) quantified by flow cytometry in control (CTRL) and Rspo3 knock-out (KO) mice at day 3 after 5FU (n=7 mice). H-J) Representative images (H) and quantification (I) of MUC2+ cells and (J) quantification of MUC2 staining intensity in the crypts of control (CTRL) and Rspo3 knock-out (KO) mice at day 3 after 5FU (n=5 mice). K-L) Representative images (K) and quantification (L) of AGR2+ cells in crypts of control (CTRL) and Rspo3 knock-out (KO) mice at day 3 after 5FU (n=5 mice). M) Representative images of mucus (UEA-1) staining in control and Rspo3 knock-out mice at day 3 after 5FU (n=4 mice). Bar graph data are mean +/− s.e.m., Wilcoxon paired t-test two-sided F-G, Mann Whitney U-test, two-sided I, J, L. See also Figures S3-4.
To compare the effect of 5FU treatment on crypt epithelial cells in control and knock-out mice, we isolated Epcam+, Cd44+ cells from the merged cell population and re-performed cluster analysis (Figure 4C). We identified 9 clusters in Epcam+, Cd44+ epithelial cells: stem cells (Olfm4+, Lgr5+), cycling transit amplifying cells (TA) (Mki67+), secretory progenitors (Atoh1+), secretory cells (including Paneth cells as Lyz1+ and goblet cells as Muc2+), enteroendocrine cells (ChgA+, ChgB+) and mature enterocytes (Alpi+) (Figures 4C and S3E). We also identified 3 other clusters of crypt epithelial cells. One was a Sca1high (Ly6a) cluster; Sca1 has been identified as a marker of dedifferentiation and of the transition to a fetal-like state in the small intestine after injury (Nusse et al., 2018). We also identified 2 clusters with elevated expression of markers such as Cd74 and Gpx2 (activated epithelium 1 and 2), which have been associated with activated epithelium in bacterial and parasitic infections (Haber et al., 2017) (Figures 4C and S3E). We determined differential cell composition and gene expression between control and knock-out samples for each cluster. Knock-out crypt epithelial cells had a lower expression of Agr2, Ang4, Muc2, Ly6a (Sca1) and Il33, amongst others (Figure 4D). Control crypt epithelial cells had a higher proportion of TA and Sca1high cells (Figure S3F). Lower numbers of TA and Sca1high cells in mutant mice than in control were accompanied by a higher proportion of Olfm4+, Lgr5+ cells compared to other crypt epithelial cells (Figure S3F). We confirmed decreased SCA-1 expression in knock-out crypt epithelial cells compared to control cells via flow-cytometry (Figure 4E-G).
Loss of lymphatic Rspo3 leads to a decrease in mucus-secreting cells
Secretory progenitors and secretory lineage cells were also decreased in knock-out mice. Crypt epithelial cells displayed a decrease in genes involved in mucus production such as Agr2 and Muc2 (Figure 4D). Agr2 is important for mucus secretion and processing, and its loss has been associated with mucus barrier dysfunction and colitis (Zheng et al., 2006). We confirmed by immunofluorescence a decrease in both MUC2 and AGR2 in crypt epithelial cells in mutant mice (Figures 4H-K). Notably, these changes in secretory cells resulted in lower mucus levels in knock-out mice compared to control mice after 5FU injury (Figure 4M).
Knock-out LECs upregulate an endothelial to mesenchymal transition program after 5FU injury
We next asked what molecular changes occurred in LECs after 5FU injury in knock-out mice compared to control mice. We isolated all endothelial cells from the scRNAseq data and performed clustering analysis. Endothelial cells grouped into 11 distinct clusters (Figures S4A-B). Amongst those, LECs were identified by the expression of Lyve1 and included 2 clusters: cluster 0 and 3 (Figures S4A-B). In order to better understand these transcriptional changes in LECs, we performed differential expression analysis of all LECs in control versus knock-out mice. Interestingly, genes related to endothelial to mesenchymal transition such as Prss23 (Bayoumi et al., 2017), Tgfb1 and Tgfb2 (Maleszewska et al., 2013; Yoshimatsu et al., 2020), and genes shown to have anti-lymphangiogenic effects such as Cavin1 (Yang et al., 2020), were expressed at significantly higher levels in Rspo3 knock-out LECs (Figures S4C-D). Cluster 3, which consists largely of knock-out LECs, was especially enriched in these genes. We confirmed these patterns of changes in LECs by performing qPCR analysis on FACS-isolated LECs (Figure S4E). Lastly, we examined the mesenchymal cell populations and observed that a new Rspo3neg myofibroblast population was present in knock-out mice, but not in control mice (Figure S4F-G).
In sum, loss of Rspo3 from LECs leads to reduced crypt proliferation after cytotoxic injury, which is reflected in a lower number of TA and Sca1high crypt epithelial cells. Rspo3 loss in LECs also causes a decrease in mucus-secreting cells and a reduction in mucus production after 5FU injury.
Discussion
The cellular components that constitute the ISC niche are currently the subject of intensive investigation, but the involvement of endothelial cells has been understudied. We report that LECs are physically part of the niche, with large vessels located just under the crypts and in proximity to ISCs, and with smaller lacteal vessels ascending alongside the crypt to the top of villi. Together, over 80% of crypts are in proximity to lymphatic vessels. Analysis of our scRNAseq data revealed that LECs secrete several components of the Wnt pathway, such as Rspo3, Wls and Wnt2. A parallel study utilizing spatial transcriptomics demonstrated that Rspo3 and Wnt2 are highly expressed in LECs in the crypt region, compared to lacteals in the villus region (Niec et al, 2022). Deleting Rspo3 specifically in LECs demonstrated that LEC-secreted Rspo3 is essential for the proper regeneration of the intestinal epithelium after injury caused by 5FU treatment. Rspo3 knock-out mice displayed a decrease in proliferative cells in the crypt and lower levels of SCA1+ cells after 5FU injury. Additionally, loss of Rspo3 in LECs resulted in a marked decrease in mucus-producing cells and in levels of mucus after 5FU injury; this could potentially result in increased vulnerability of crypt epithelial cells to pathogens.
Rspo3 produced by LECs supports regeneration of the small intestine after 5FU-induced injury, but it is dispensable during homeostasis. This indicates that Rspo3 is produced by several sources in the ISC niche during homeostasis. Consistent with this notion, other studies have shown that deletion of Rspo3 in Pdgfra+ fibroblasts (Goto et al., 2022; Greicius et al., 2018) or in myofibroblasts (Harnack et al., 2019) alone, or generalized inhibition of RSPO3 by a neutralizing antibody (Storm et al., 2015) has no obvious effect on epithelial homeostasis. Additionally, loss of LEC-derived Rspo3 seems to have a negligible effect on intestinal repair after ablation of Lgr5+ cells. We report that after a perturbation that is addressed through high levels of proliferation, such as 5FU treatment, Rspo3 secreted by LECs is essential for the timely repair of epithelial cells. In addition, LECs themselves appear relatively stable to injury and are relatively unaffected by deleterious conditions, such as 5FU (this paper) or irradiation (Goto et al., 2022).
The 5FU injury triggers a massive proliferative and damage response, as seen by the high number of EdU+ cells, and this is not seen when Lgr5+ cells alone are ablated. The number of proliferative cells per crypt stays about the same as during homeostasis after ablation of Lgr5+ cells, but it more than doubles after 5FU injury. The massive proliferation in the crypts after 5FU requires all sources of Rspo3 available including that coming from LECs. We propose that there is a hierarchy of lymphatic Rspo3 need that increases from homeostasis through ablation of Lgr5+ cells to 5FU injury. It therefore appears that intestinal maintenance and regeneration rely on distinct niche sources of Rspo3, presumably depending on the level of damage and the proliferation required to achieve repair. This also suggests that the intestinal niche should not be seen as static under all conditions, but rather that different niche sources come together depending on need and injury to facilitate proper regeneration.
Angiocrine factors support stem cells and regenerative processes of several tissues. This has led to an appreciation that ECs are more than just the plumbing that carries blood and oxygen, but rather that they provide essential molecular signaling for maintaining tissue function. Our findings demonstrate that lymphangiocrine-derived factors in the intestine are critical for intestinal regeneration after cytotoxic injury. This suggests that LECs could be an important source of secreted factors in other tissues as well. The finding of a supportive role for LECs to ISCs and regenerative cells is also relevant to diseases in the gut. In colorectal cancer (CRC), lymphangiogenesis is one cause of the development of metastasis (Li et al., 2011; Tacconi et al., 2015). Our finding that after 5FU treatment LEC-secreted Rspo3 contributes to ISC maintenance suggests that LECs could also be involved in the promotion of cancer. Furthermore, maintenance of the lymphatic vessel density of LECs after 5FU treatment raises questions about the potential involvement of LECs in cancer cell survival and escape from chemotherapy.
Limitations of the study
In our work, we determined that lymphatic derived Rspo3 contributes to intestinal regeneration after 5FU injury. However, we did not observe a phenotype after conditional deletion of Rspo3 from the lymphatics during homeostasis. In a complementary study to ours, Goto et. al explored all sources of Rspo3 in the intestine. They found that when Rspo3 is deleted from both mesenchymal and lymphatic sources, ISC maintenance at homeostasis was affected (Goto et al., 2022). We also determined that LEC derived Rspo3 is dispensable for intestinal recovery after ablation of Lgr5+ cells, but it is possible that we could have identified a phenotype if we looked at shorter or longer time points after ablation of Lgr5+ cells.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ophir Klein (Ophir.klein@ucsf.edu).
Materials availability
All the materials will be available upon request to the lead contact under material transfer agreement with UCSF.
Data availability
All scRNAseq data have been deposited on the Gene Expression Omnibus (GEO) and can be viewed under accession number GSE198469. This manuscript did not generate any new code. Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
All mouse experiments were conducted under the IACUC protocol AN176864 at the University of California San Francisco. All mice (male and female) were used at 8-12 weeks of age at the beginning of each experiment. Rspo3fl/fl mice were a gift of Christof Niehrs (Scholz et al., 2016), Prox1CreERT2 mice were purchased from Jackson labs (022075), Lgr5DTR/GFP were originally derived at Genentech and are a kind gift of Fred de Sauvage (Tian et al., 2011), Lgr5GFP-IRES-CreERT2 were purchased from Jackson labs (008875; (Barker et al., 2007)). For knock-out induction, mice were gavaged with tamoxifen in corn oil (200 mg/kg) 6 times, 2 x 3 days interspersed by 3 days of rest. For EdU proliferation experiments, mice were injected with EdU (5mg/mL) 2 hours before sacrifice. For injury experiments, mice were injected for 3 consecutive days with either 5FU at 75 mg/kg in 0.9% saline or Diphtheria toxin (DT) (50 μg/kg). For vessel visualization, mice were injected with an antibody against VECAD (25 μg/mouse in 100 μl of PBS) conjugated to Alexa-647 8 min prior to sacrifice.
METHOD DETAILS
Tissue collection and preparation for microscopy
For all microscopy experiments, the jejunum was collected and fixed in 4% PFA for 24h. For sections, tissues were incubated in 30% sucrose for 24h at 4°C and then embedded in OCT before being sectioned to a thickness of 8 μm. For whole mount, fixed tissues were cleared for 3 to 6 days in CUBIC-L solution (10% N-butyldiethanolamine, 10% Triton X-100 (Matsumoto et al., 2019).
Immunofluorescence, RNAscope, EdU labeling on sections and Whole mount staining
Frozen sections were baked at 60°C for 30min, unmasked for 30min, blocked (5% NDS, 1X Animal Free blocking, 0.3% Triton X-100) for 1h and incubated overnight with primary antibodies at 4°C (table). Then, sections were incubated with secondary antibodies for 1h at RT (table).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| Rat monoclonal anti-LYVE1 (clone ALY7) | eBioscience | Cat# 14-0443-82, RRID:AB_1633414 | |
| Rabbit polyclonal anti-LYVE1 | AngioBio | Cat# 11-034, RRID:AB_2813732 | |
| Goat polyclonal anti-PDGFRA | R&D systems | Cat# AF1062, RRID:AB_2236897 | |
| Rabbit monoclonal anti-OLFM4 (clone D6Y5A) | Cell signaling technology | Cat# 39141, RRID:AB_2650511 | |
| Rabbit polyclonal anti-MUC2 | Novus biologicals | Cat# NBP1-31231, RRID:AB_10003763 | |
| Rabbit polyclonal anti-AGR2 | Origene | Cat# TA336715 | |
| Donkey anti-Goat IgG Secondary Antibody (Alexa Fluor Plus 488) | Thermo Fischer Scientific | Cat# A32814, RRID:AB_2762838 | |
| Donkey Anti-Rat IgG Secondary Antibody (Alexa Fluor 647) | Jackson ImmunoResearch Labs | Cat# 712-605-153, RRID:AB_2340694 | |
| Donkey Anti-Rat IgG Secondary Antibody (Alexa Fluor 488) | Thermo Fischer Scientific | Cat# A-21208, RRID:AB_2535794 | |
| Donkey anti-Goat IgG Secondary Antibody (Alexa Fluor Plus 647) | Thermo Fischer Scientific | Cat# A32849, RRID:AB_2762840 | |
| Donkey Anti-Rabbit IgG Secondary Antibody (Alexa Fluor 555) | Thermo Fischer Scientific | Cat# A32794, RRID:AB_2762834 | |
| Goat Anti-Rabbit IgG Secondary Antibody (Alexa Fluor 488) | Thermo Fischer Scientific | Cat# A-11008, RRID:AB_143165 | |
| Goat Anti-Rabbit IgG Secondary Antibody (Alexa Fluor 568) | Invitrogen | Cat# A-11011, RRID:AB_143157 | |
| Alexa Fluor 647 anti-mouse CD326 (Ep-CAM) antibody | BioLegend | Cat# 118212, RRID:AB_1134101 | |
| Alexa Fluor 488 anti-mouse CD31 antibody | BioLegend | Cat# 102414, RRID:AB_493408 | |
| APC anti-mouse CD326 (Ep-CAM) antibody | BioLegend | Cat# 118214, RRID:AB_1134102 | |
| PE/Cyanine7 anti-mouse Podoplanin antibody | BioLegend | Cat# 127412, RRID:AB_10613648 | |
| PE-Cyanine7 anti-mouse LYVE1 Monoclonal Antibody (ALY7) | Thermo Fisher Scientific | Cat# 25-0443-82, RRID:AB_2802237 | |
| PE/Cyanine7 anti-mouse/human CD44 antibody | BioLegend | Cat# 103030, RRID:AB_830787 | |
| Brilliant Violet 421 anti-mouse CD45 antibody | BioLegend | Cat# 103134, RRID:AB_2562559 | |
| PE anti-mouse CD45 antibody | BioLegend | Cat# 103106, RRID:AB_312971 | |
| Alexa Fluor 647 anti-mouse CD144 (VE-cadherin) antibody | BioLegend | Cat# 138006, RRID:AB_10569114 | |
| APC anti-mouse Ly-6A/E (Sca-1) Monoclonal Antibody (D7) | eBioscience | Cat# 17-5981-82, RRID:AB_469487 | |
| Chemicals, peptides, and recombinant proteins | |||
| ProLong Gold Antifade Mounting Solution | Thermo Fischer Scientific | Cat# P36930 | |
| Tamoxifen | Sigma-Aldrich | Cat# T5648-5G | |
| Corn Oil | Sigma-Aldrich | Cat# C8267 | |
| Macs SmartStrainers | Miltenyi Biotec | Cat# 130-110-916 | |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 | |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat# 017-000-121 | |
| Animal-free Blocker | Vector Laboratories | Cat# sp-5030 | |
| PFA | Fischer Scientific | Cat# 04042 | |
| Sucrose | VWR | Cat# JT4072-7 | |
| Tissue-Tek O.C.T. Compound | Sakuraus | Cat# 4583 | |
| N-butyldiethanolamine | TCI Chemicals | Cat# TCI-B0725 | |
| Antigen Unmasking Solution, Citrate-Based | VWR | Cat# 101098-054 | |
| DAPI | Sigma-Aldrich | Cat# D9564 | |
| RPMI-1640 Medium | Sigma-Aldrich | Cat# R7509 | |
| Glutamax | Thermo Fisher Scientific | Cat# 35050061 | |
| MEM Non Essential Amino Acids Solution (100X) | Thermo Scientific | Cat# 11140050 | |
| HEPES | Thermo Fisher Scientific | Cat# 15630080 | |
| Penicillin Streptomycin | Thermo Fisher Scientific | Cat# 15140122 | |
| EDTA | Sigma-Aldrich | Cat# E5134 | |
| DTT | Sigma-Aldrich | Cat# 10197777001 | |
| HBSS | Thermo Scientific | Cat# 14175095 | |
| Collagenase A | Sigma-Aldrich | Cat# 10103586001 | |
| Dispase II | Sigma-Aldrich | Cat# 4942078001 | |
| DNAse | Worthington Biochemical | Cat# LS002007 | |
| iTaq™ Universal SYBR® Green Supermix | Biorad | Cat# 1725124 | |
| Lectin from Ulex europaeus (FITC conjugated) | Sigma-Aldrich | Cat# L9006 | |
| Critical commercial assays | |||
| RNAscope Multiplex Fluorescent V2 Assay | ACD | Cat# 323100 | |
| Click-iT™ Plus EdU Alexa Fluor™ 647 Imaging Kit | Thermo Fisher Scientific | Cat# C10640 | |
| PicoPure™ RNA Isolation Kit | Thermo Scientific | Cat# KIT0204 | |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Scientific | Cat# 4368814 | |
| Deposited data | |||
| All scRNAseq data has been deposited on GEO | This paper | GSE198469 | |
| Experimental models: Organisms/strains | |||
| Mouse strain: Rspo3fl/fl | Christof Niehrs (Institute of Molecular Biology) | ||
| Mouse strain: Prox1CreERT2 | The Jackson Laboratory | JAX: 022075 | |
| Mouse strain: Lgr5DTR/GFP | Genentech (Dr. Frederic de Sauvage) | ||
| Mouse strain: Lgr5GFP-IRES-CreERT2 | The Jackson Laboratory | JAX: 008875 | |
| Oligonucleotides | |||
| RNAscope Probe- Mm-Lgr5 | ACD | Cat# 312171 | |
| RNAscope Probe- Mm-Rspo3 | ACD | Cat# 402011 | |
| Rspo3 primers Fw: GACTCGCCTAACACCTACATAC RV GACATTAGGATGCATTCTTCGC | This paper | ||
| Apoa1 primers Fw: CGCTAATGTGTATGTGGATGCG Rv: CCCAATCTGTTTCTTTCTCCAGG | This paper | ||
| Cavin1 primers Fw: CCAGATCCAGCTGACCCAAG Rv: TTCTTTATCTGGCCGGCCTG | This paper | ||
| Prss23 primers Fw: CCGCACAGAGACTCGGGT Rv: CCAGTGCAGCCAGTAGACAA | This paper | ||
| Tgfb1 primers Fw: TACAACAGCACCCGCGAC Rv: ACAGCAATGGGGGTTCGG | This paper | ||
| Tgfb2 primers Fw: AGGGATCTTGGATGGAAATGGA Rv: CCAATGTAATAGAGAATGGTCAGTGG | This paper | ||
| Vim primers Fw: CGTTTCCAAGCCTGACCTCA Rv: GGCATCCACTTCACAGGTGA | This paper | ||
| Software and algorithms | |||
| GraphPad Prism 9 | https://www.graphpad.com | ||
| ImageJ (Fiji) | https://imagej.net/software/fiji/ | v2.1.0/1.53c; RRID:SCR_002285 | |
| Adobe Illustrator | https://www.adobe.com | v25.4 | |
| R | https://www.r-project.org/ | v.4.0.3 | |
| BD FACS Diva | https://www.bdbiosciences.com/ | v.8.0.1 | |
| ZenBlue | https://www.zeiss.com/ | v.3.1 | |
| WEB-GESTALT (WEB-based GEne SeT AnaLysis Toolkit) | https://www.webgestalt.org/ | ||
| Seurat | v.4.1.0 | ||
| Cell Ranger | https://10xgenomics.com | v.5.0.1 | |
| Biorender | https://biorender.com | ||
RNAscope was done according to the manufacturer’s protocol. Briefly, the sections were post-fixed in 4% PFA, incubated with hydrogen peroxide to inhibit endogenous peroxidase before performing target retrieval. Subsequently, the tissues were incubated with specific RNA probes (table), followed by revelation and amplification steps.
For EdU labeling, after the baking, slides were permeabilized for 5min in 1% Triton X-100 and EdU staining performed according to manufacturer protocol (table).
For whole mount staining, 1 cm tissue pieces were blocked for 1 day, incubated with primary and then secondary antibodies for 2 days and finally post-fixed for 1 day in 4% PFA (Bernier-Latmani, 2016). Each step was done at 4°C.
For all staining and labeling, tissues were counterstained with DAPI and mounted in Prolong Gold. The images were taken using a Zeiss Apotome 910 system.
Quantification of staining
All image quantifications were done using ImageJ software. The proliferative cell number was quantified by counting the number of EdU+ cells per crypt and calculating the mean for each mouse (~30 crypts per mouse). MUC2+ and AGR2+ cell numbers were quantified by counting positive cells in the crypts except Paneth cells present at the bottom (~30 crypts per mouse). For quantification of MUC2 intensity, crypts were traced and manually segmented. Yen thresholding followed by particle analysis was then performed on the segmented image and the mean grey value was measured for each region of interest (~40 crypts/mouse). The crypt number was calculated by counting OLFM4+ crypts in 20 to 30 mm of tissue length. The OLFM4 intensity was analyzed by calculating the mean of the staining in a defined area corresponding to the crypt (~50 crypts per mouse). Lgr5 RNAscope was quantified by calculating the mean of positive dots in each crypt (~20 crypts per mouse). For the quantification of crypt number in close proximity to lymphatic vessels, crypts were assigned to different categories: 1) crypt base: at least one cell of the crypt base is above a lymphatic vessel, 2) mid-crypt: at least one side of the mid-crypt is beside of a lymphatic vessel, 3) mid-crypt only, 4) crypt base only, 5) both crypt base and mid-crypt, 6) either crypt base or mid-crypt and 7) not in proximity: neither crypt-base nor mid-crypt (~200 crypts per mouse). For the quantification of lymphatic vessel density, after Z-projection of all optical sections, the area fraction of the LYVE-1 signal (LYVE-1 positive area on total area) was quantified for each mouse. For crypt size calculation, in whole mount staining, crypts were first classified as in proximity or not with lymphatics as previously, then transverse optical sections at the mid crypt position were chosen to quantify the area of the width using ImageJ (~200 crypts per mouse).
Tissue preparation and digestion
Small intestines from adult mice were collected and cut into small pieces (0.5 cm) and put in 30 mL of dissociation solution (RPMI + 1% Glutamax, 1% non-essential amino acids, 2.5% HEPES, 1% Pen-strep, 5mM EDTA, 3% FBS and 1 μM DTT) in a rotator at 37°C for 30 min. Tissues were washed twice in HBSS, spun down and further minced using scissors. The minced tissue was then digested in a rotator for 30 min at 37°C in 4 ml of digestion solution (Collagenase A (2.5 mg/ml), Dispase II (1 U/ml) and DNAse (30 μg/ml) in HBSS with 1% Glutamax, 1% non-essential amino acids and 2.5% HEPES). The cells were strained (100 μM strainer) and the left-over tissue digested for another 30 min in more 4 ml of digestion solution. Both fractions were stained with antibodies and Aria II with a 100-micron nozzle utilized to perform the sort.
For sequencing experiments during homeostasis:
Small intestines were digested and prepared as described above. Data are from two biological replicates. Each biological replicates contains cells from one adult male and one adult female mouse. Lymphatic endothelial cells were sorted as CD31+, PDPN+, EPCAMneg, CD45neg and blood endothelial cells as CD31+, PDPNneg, EPCAMneg and CD45neg. 6000 BECs and 2000 LECs were sequenced in total.
For 5FU experiments:
At day 3 after the last 5FU injection, small intestines were digested and prepared as described above. Data are from two biological replicates. Each biological replicate contains cells from one adult male and one adult female mouse. Crypt epithelial cells were sorted as EPCAM+, CD44+, CD45neg, CD31neg, lymphatic endothelial cells were sorted as CD31+, PDPN+, EPCAMneg, CD45neg, blood endothelial cells as CD31+, PDPNneg, EPCAMneg, CD45neg, epithelial cells as EPCAM+, CD45neg, CD31neg and stroma cells EPCAMneg, CD45neg, CD31neg. After sorting, the different populations were mixed and loaded into 10x Chromium.
For SCA-1 validation experiments:
At day 3 after the last 5FU injection, small intestines were digested and prepared as described above. Data are from 7 biological replicates. SCA-1+ cells were gated on the EPCAM+, CD44+, CD45neg, CD31neg population.
scRNAseq analysis
The single-cell suspension was loaded onto the 10X Chromium Single Cell instrument (10X Genomics). Single-cell barcoding, and cDNA libraries were constructed using the 10X Chromium single-cell 3’ Library Kit according to the manufacturer’s protocol. Libraries were sequenced on an Illumina NovaSeq 6000 and the sequence output processed with Cell Ranger 5.0.1 to produce filtered feature-barcode count matrices (http://10xgenomics.com).
Single-cell analyses were performed using the Seurat package in R (v.4.1.0) (Butler et al., 2018). Count matrices filtered to retain only genes expressed in three or more cells in a sample, were used for further analysis. Cells with high mitochondrial gene percentage or in the 1st or 99th percentile of transcript counts were also removed. Initially, all sequenced cell types were analyzed together. Data were processed following standard practices, including log-normalization of UMI counts, and regression of mitochondrial gene expression. Principal component analysis was subsequently performed on the most variable genes. Cluster identification (at a resolution of 0.5) and UMAP visualization was performed on the top 20 principal components. Differential gene expression for gene-marker discovery across the clusters was performed using the Wilcoxon rank-sum test.
Subsequently, for 5FU experiments the following analyses were performed. 1) For crypt epithelial cells: Epcam+, Cd44+ cells were identified as having expression of Epcam >1, Cd44 >1 and expression of Cdh5, Cd31, Kdr and Pdfgra <1. Myh11+ cells were also excluded. 2) ECs were isolated as Cdh5, Cd31, Kdr1 >1 and Epcam, Cdh1, Pdgfra1 <1. EC clusters that expressed Lyve1 were identified as LECs. 3) Mesenchymal cells were isolated as Pdgfra>1, Cdh5, Kdr, Cd31, Epcam, Cdh1 <1. For each cell type, filtering, clustering, and differential gene expression testing was done as described above. For pathway analysis, WEB-GESTALT (WEB-based Gene SeT AnaLysis Toolkit) (Liao et al., 2019) was utilized, where genes that had at least a 0.5 Log2 upregulation in control LEC vs knock-out LEC or genes that had at least a 0.5 Log2 upregulation in knock-out LECs vs. control LECs were utilized for over-representation analysis. False discovery rate (FDR)-adjusted P values of less than 0.05 were considered statistically significant.
Mucin staining
For mucin staining, the jejunum (middle third of the small intestine) was isolated 3 days after the last 5FU injection. Fragments of 1.5 cm in length containing fecal pellets were then selected and placed directly into methacarn fixative (60% methanol, 30% chloroform, 10% acetic acid). Fragments were fixed for 3 days rocking at room temperature. Following fixation, fragments were washed twice in methanol for 20 minutes each wash, twice in absolute ethanol for 20 minutes each, and twice in xylene for 10 minutes each. The tissue was then embedded in paraffin blocks following standard procedures and cut into 4, 7, and 14 μm sections. Sections were stained with 20 μg/ml UEA1-FITC.
mRNA extraction and RT-qPCR analysis
Tissues were collected and LECs were sorted as described above. 20,000 LECs were then resuspended in lysis buffer from the PicoPure™ RNA Isolation Kit. mRNA was extracted following the manufacturer’s protocol and resuspended in 13.2uL of elution water. mRNA was then used for reverse transcription using the High-Capacity cDNA Reverse Transcription Kit. The resulting cDNA was diluted (4X) before proceeding to qPCR using TaKaRa Taq DNA Polymerase mix. Relative gene expression was calculated by using ΔΔCt method and normalized to β2m expression. Results are displayed as mean +/− s.e.m., Mann Whitney U-test, two-sided.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were assessed and analyzed using appropriate statistical methods. Mann Whitney U-test was utilized unless otherwise noted. GraphPad Prism v.9 was used for all statistical analysis, unless otherwise indicated. No statistical methods were used to determine sample size. Unless otherwise stated, the experiments were not randomized. The investigators were blinded during 5FU and DT experiment quantifications.
Supplementary Material
Highlights.
Intestinal crypts are in proximity to lymphatics, which express Rspo3.
Intestinal recovery is impaired in lymphatic Rspo3 knock-out mice after 5FU injury.
Lymphatic Rspo3 knock-out mice have decreased progenitor cells after 5FU injury.
Lymphatic Rspo3 knock-out mice have reduced mucus production after 5FU injury.
Acknowledgments
This work was funded by NIH R35-DE026602 and by U01-DK103147 from the Intestinal Stem Cell Consortium, a collaborative research project funded by the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Allergy and Infectious Diseases, to O.D.K. B.P. was supported by the UCSF CIRM Scholars Training Program EDUC4-12812. We thank F. de Sauvage for critical discussion, helpful comments, and reagents. We thank B. Hoehn, A. Rathnayake, E. Sandoval, K. Pan, P. Marangoni and A. Verma for technical assistance and animal maintenance. We thank V. Nguyen at the UCSF Parnassus Flow Cytometry Core for assistance with FACS experiments and A. Poon and C. Chu for assistance with 10x Chromium and library preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Abel E, Ekman T, Warnhammar E, Hultborn R, Jennische E, and Lange S (2005). Early disturbance of microvascular function precedes chemotherapy-induced intestinal injury. Dig Dis Sci 50, 1729–1733. 10.1007/s10620-005-2926-9. [DOI] [PubMed] [Google Scholar]
- Aoki R, Shoshkes-Carmel M, Gao N, Shin S, May CL, Golson ML, Zahm AM, Ray M, Wiser CL, Wright CV, and Kaestner KH (2016). Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol 2, 175–188. 10.1016/j.jcmgh.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin HG, and Koh GY (2017). Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science 357. 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- Barker N, Es J.H.v., Kuipers J, Kujala P, Born M.v.d., Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bayoumi AS, Teoh JP, Aonuma T, Yuan Z, Ruan X, Tang Y, Su H, Weintraub NL, and Kim IM (2017). MicroRNA-532 protects the heart in acute myocardial infarction, and represses prss23, a positive regulator of endothelial-to-mesenchymal transition. Cardiovasc Res 113, 1603–1614. 10.1093/cvr/cvx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Latmani J, Cisarovsky C, Demir CS, Bruand M, Jaquet M, Davanture S, Ragusa S, Siegert S, Dormond O, Benedito R, et al. (2015). DLL4 promotes continuous adult intestinal lacteal regeneration and dietary fat transport. 10.1172/JCI82045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Latmani J, and Petrova TV (2017). Intestinal lymphatic vasculature: structure, mechanisms and functions. Nat Rev Gastroenterol Hepatol 14, 510–526. 10.1038/nrgastro.2017.79. [DOI] [PubMed] [Google Scholar]
- Beumer J, and Clevers H (2020). Cell fate specification and differentiation in the adult mammalian intestine. Nature Reviews Molecular Cell Biology 22, 39–53. doi: 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, et al. (2010). Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 6, 251–264. 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifarelli V, and Eichmann A (2019). The Intestinal Lymphatic System: Functions and Metabolic Implications. Cell Mol Gastroenterol Hepatol 7, 503–513. 10.1016/j.jcmgh.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, and Basler K (2018). GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453. 10.1038/S41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, et al. (2010). Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 468, 310–315. 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding BS, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, Crystal RG, Simons M, Sato TN, Worgall S, et al. (2011). Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell 147, 539–553. 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, and Morrison SJ (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462. 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman RA, deSchoolmeester ML, Hurst RJ, Wright SH, Pemberton AD, and Else KJ (2012). The goblet cell is the cellular source of the anti-microbial angiogenin 4 in the large intestine post Trichuris muris infection. PLoS One 7, e42248. 10.1371/journal.pone.0042248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehart H, and Clevers H (2019). Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 16, 19–34. 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, and Niehrs C (2011). LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep 12, 1055–1061. 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Imada S, Deshpande V, and Yilmaz ÖH (2022). Lymphatics constitute a novel component of the intestinal stem cell niche. Cell Stem Cell, this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, and Virshup DM (2018). pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A 115, E3173–E3181. 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur-Cohen S, Yang H, Baksh SC, Miao Y, Levorse J, Kataru RP, Liu X, Cruz-Racelis J.d.l., Mehrara BJ, and Fuchs E (2019). Stem cell–driven lymphatic remodeling coordinates tissue regeneration. 10.1126/science.aay4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al. (2012). ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- Harnack C, Berger H, Antanaviciute A, Vidal R, Sauer S, Simmons A, Meyer TF, and Sigal M (2019). R-spondin 3 promotes stem cell recovery and epithelial regeneration in the colon. Nat Commun 10, 4368. 10.1038/s41467-019-12349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Stappenbeck TS, Hong CV, and Gordon JI (2003). Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol 4, 269–273. 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- Hu J, Srivastava K, Wieland M, Runge A, Mogler C, Besemfelder E, Terhardt D, Vogel MJ, Cao L, Korn C, et al. (2014). Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science 343, 416–419. 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- Kabiri Z (2014). Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. (2020). Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779.e720. 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. (2018). Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 175, 372–386.e317. 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo A, Department of Genetics, P.S.o.M., University of Pennsylvania, Philadelphia, PA 19104, USA, Kaestner KH, and Department of Genetics, P.S.o.M., University of Pennsylvania, Philadelphia, PA 19104, USA (2019). Emerging diverse roles of telocytes. Development 146. 10.1242/dev.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu B, Xiao J, Yuan Y, Ma J, and Zhang Y (2011). Roles of VEGF-C and Smad4 in the lymphangiogenesis, lymphatic metastasis, and prognosis in colon cancer. J Gastrointest Surg 15, 2001–2010. 10.1007/s11605-011-1627-2. [DOI] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, and Zhang B (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47, W199–W205. 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, De la Cruz E, Gu X, Balint L, Oxendine-Burns M, Terrones T, Ma W, Kuo H-H, Lantz C, Bansal T, et al. (2020). Lymphoangiocrine signals promote cardiac growth and repair. Nature 588, 705–711. doi: 10.1038/s41586-020-2998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszewska M, Moonen JR, Huijkman N, van de Sluis B, Krenning G, and Harmsen MC (2013). IL-1β and TGFβ2 synergistically induce endothelial to mesenchymal transition in an NFκB-dependent manner. Immunobiology 218, 443–454. 10.1016/j.imbio.2012.05.026. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Mitani TT, Horiguchi SA, Kaneshiro J, Murakami TC, Mano T, Fujishima H, Konno A, Watanabe TM, Hirai H, and Ueda HR (2019). Advanced CUBIC tissue clearing for whole-organ cell profiling. Nature Protocols 14, 3506–3537. doi: 10.1038/s41596-019-0240-9. [DOI] [PubMed] [Google Scholar]
- McCarthy N, Kraiczy J, and Shivdasani RA (2020a). Cellular and molecular architecture of the intestinal stem cell niche. Nature Cell Biology 22, 1033–1041. doi: 10.1038/s41556-020-0567-z. [DOI] [PubMed] [Google Scholar]
- McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S, et al. (2020b). Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 26, 391–402.e395. 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niec RE, Chu T, Schernthanner M, Gur-Cohen S, Hidalgo L, Pasolli HA, Luckett KA, Wang Z, Bhalla SR, Cambuli F, Kataru RP, Ganesh K, Mehrara BJ, Pe'er D, & Fuchs E (2022). Lymphatics act as a signaling hub to regulate intestinal stem cell activity. Cell Stem Cell, 29(7), 1067–1082.e18. 10.1016/j.stem.2022.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AKM, Landman TA, de Sauvage FJ, Locksley RM, and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113. 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara R, Hashimoto D, Kimura S, Hayase E, Ara T, Takahashi S, Ohigashi H, Yoshioka K, Tateno T, Yokoyama E, et al. (2018). Intestinal Lymphatic Endothelial Cells Produce R-Spondin3. Scientific Reports 8, 1–9. doi: 10.1038/s41598-018-29100-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikuqi B, Rispal J, and Klein O (2021). Good Neighbors: The Niche that Fine Tunes Mammalian Intestinal Regeneration. 10.1101/cshperspect.a040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris F, Fuks Z, Kang A, Capodieci P, Juan G, Ehleiter D, Haimovitz-Friedman A, Cordon-Cardo C, and Kolesnick R (2001). Endothelial apoptosis as the primary lesion initiating intestinal radiation damage in mice. Science 293, 293–297. 10.1126/science.1060191. [DOI] [PubMed] [Google Scholar]
- Rafii S, Butler JM, and Ding BS (2016). Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325. 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehal S, Stephens M, Roizes S, Liao S, and von der Weid PY (2018). Acute small intestinal inflammation results in persistent lymphatic alterations. Am J Physiol Gastrointest Liver Physiol 314, G408–G417. 10.1152/ajpgi.00340.2017. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Scholz B, Korn C, Wojtarowicz J, Mogler C, Augustin I, Boutros M, Niehrs C, and Augustin HG (2016). Endothelial RSPO3 Controls Vascular Stability and Pruning through Non-canonical WNT/Ca(2+)/NFAT Signaling. Dev Cell 36, 79–93. 10.1016/j.devcel.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Durinck S, de Sousa e Melo F, Tremayne J, Kljavin N, Tan C, Ye X, Chiu C, Pham T, Hongo J-A, et al. (2015). Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 529, 97–100. doi: 10.1038/nature16466. [DOI] [PubMed] [Google Scholar]
- Stzepourginski I, Nigro G, Jacob JM, Dulauroy S, Sansonetti PJ, Eberl G, and Peduto L (2017). CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A 114, E506–E513. 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconi C, Correale C, Gandelli A, Spinelli A, Dejana E, D'Alessio S, and Danese S (2015). Vascular endothelial growth factor C disrupts the endothelial lymphatic barrier to promote colorectal cancer invasion. Gastroenterology 148, 1438–1451.e1438. 10.1053/j.gastro.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, and de Sauvage FJ (2011). A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259. 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, Hausmann G, Cantù C, Aguet M, and Basler K (2016). Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep 15, 911–918. 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
- Wertheimer T, Velardi E, Tsai J, Cooper K, Xiao S, Kloss CC, Ottmuller KJ, Mokhtari Z, Brede C, deRoos P, et al. (2018). Production of BMP4 by endothelial cells is crucial for endogenous thymic regeneration. Sci Immunol 3. 10.1126/sciimmunol.aal2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, et al. (2017). Non-equivalence of Wnt and R-spondin ligands during Lgr5. Nature 545, 238–242. 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kim J.-d., Gu Q, Yan Q, Astin J, Crosier PS, Yu P, Rockson SG, and Fang L (2020). AIBP-CAV1-VEGFR3 axis dictates lymphatic cell fate and controls lymphangiogenesis. 10.1101/2020.10.16.342998. [DOI] [Google Scholar]
- Yoshimatsu Y, Kimuro S, Pauty J, Takagaki K, Nomiyama S, Inagawa A, Maeda K, Podyma-Inoue KA, Kajiya K, Matsunaga YT, and Watabe T (2020). TGF-beta and TNF-alpha cooperatively induce mesenchymal transition of lymphatic endothelial cells via activation of Activin signals. PLoS One 15, e0232356. 10.1371/journal.pone.0232356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Rosenstiel P, Huse K, Sina C, Valentonyte R, Mah N, Zeitlmann L, Grosse J, Ruf N, Nürnberg P, et al. (2006). Evaluation of AGR2 and AGR3 as candidate genes for inflammatory bowel disease. Genes Immun 7, 11–18. 10.1038/sj.gene.6364263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scRNAseq data have been deposited on the Gene Expression Omnibus (GEO) and can be viewed under accession number GSE198469. This manuscript did not generate any new code. Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.