SUMMARY
Muscleblind (mbl) is an essential muscle and neuronal splicing regulator. Mbl hosts multiple circular RNAs (circRNAs), including circMbl, which is conserved from flies to humans. Here, we show that mbl-derived circRNAs are key regulators of MBL by cis- and trans-acting mechanisms. By generating fly lines to specifically modulate the levels of all mbl RNA isoforms, including circMbl, we demonstrate that the two major mbl protein isoforms, MBL-O/P and MBL-C, buffer their own levels by producing different types of circRNA isoforms in the eye and fly brain, respectively. Moreover, we show that circMbl has unique functions in trans, as knockdown of circMbl results in specific morphological and physiological phenotypes. In addition, depletion of MBL-C or circMbl results in opposite behavioral phenotypes, showing that they also regulate each other in trans. Together, our results illuminate key aspects of mbl regulation and uncover cis and trans functions of circMbl in vivo.
Graphical Abstract
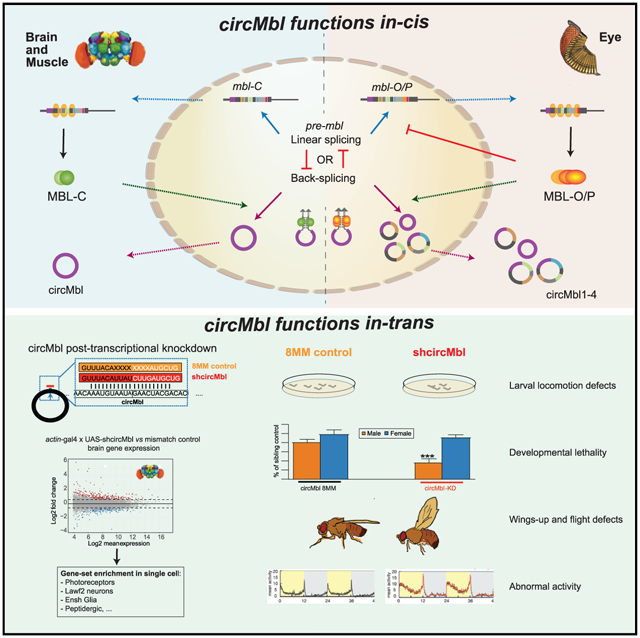
In brief
Pamudurti et al. show that splicing factor mbl is expressed and promotes circularization of its own locus in a cell-type-specific manner in vivo. In photoreceptors, circMbl2-4 regulates in trans MBL-O/P, while in the brain, circMbl regulates MBL-C. Moreover, circMbl has functions in trans related to locomotor behavior.
INTRODUCTION
Muscleblind (mbl) is an essential post-transcriptional regulator involved in muscle and photoreceptor development (Begemann et al., 1997; Vicente-Crespo et al., 2008). MBL functions by regulating alternative splicing, mRNA localization, and cleavage and polyadenylation (Batra et al., 2014; Wang et al., 2012). In Drosophila, several mRNAs generated from the mbl locus are expressed at different developmental stages (Vicente et al., 2007). There are three mammalian homologs of the Drosophila mbl gene: MBNL1, MBNL2, and MBNL3 (Fardaei et al., 2002; Huang et al., 2008). Importantly, MBNL1 and MBNL2 have a key role in the etiology of myotonic dystrophy (Jiang et al., 2004; Kanadia et al., 2003; Lee et al., 2013; Lukáš et al., 2012). MBL protein sequence and function is strongly conserved through evolution, and the human and fly orthologs can even be interchanged in rescue experiments (Vicente et al., 2007). Work in flies determined functions for mbl in vivo in eye tissue and muscle (Artero et al., 1998; Begemann et al., 1997). However, there are few insights into potential functions in the brain. Among them is a recent work which showed that MBL has functions during neuronal development (Li and Millard, 2019). Moreover, work performed in mice showed that loss of MBNL1 and/or MBNL2 leads to abnormal rapid eye movement sleep and memory (Charizanis et al., 2012) as well as abnormal development of synapses and spines in cortical neurons (Lee et al., 2019).
The mbl gene structure is strikingly conserved between very distant species such as Drosophila and humans (Ashwal-Fluss et al., 2014). This includes the presence of circular RNAs (circRNAs) (Ashwal-Fluss et al., 2014; Memczak et al., 2013; Westholm et al., 2014). circRNAs are a recently rediscovered form of RNA produced by the spliceosome through circularization of specific exons in a process known as backsplicing (Jeck and Sharpless, 2014; Wang et al., 2014). circRNA biosynthesis is promoted by the presence of complementary sequences in flanking introns and/or by specific splicing factors (Aktas et al., 2017; Ashwal-Fluss et al., 2014; Conn et al., 2015; Errichelli et al., 2017; Ivanov et al., 2015; Knupp et al., 2021; Kramer et al., 2015; Zhang et al., 2014). Most of the circRNAs contain only exons and are transported to the cytoplasm by a specific mechanism (Huang et al., 2018; Patop et al., 2019). As circRNA production competes with canonical RNA splicing, highly produced circRNAs can alter gene expression in cis by competing with the production of linear gene products from the same locus (Ashwal-Fluss et al., 2014). Interestingly, some circRNAs also produce proteins (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017).
Recent work has uncovered a handful of circRNAs that function in trans. For example, the circRNAs derived from CDR1as and sry bind to and likely regulate microRNA (miRNA) function (Hansen et al., 2013; Kleaveland et al., 2018; Memczak et al., 2013). Other circRNAs might titrate or transport proteins and regulate rRNA biogenesis (Ashwal-Fluss et al., 2014; Du et al., 2016; Guarnerio et al., 2016; Holdt et al., 2016). circRNAs can also mediate responses to viral infections by differentiating between splicing of endogenous and virus-related transcripts (Cadena and Hur, 2017; Chen et al., 2017, 2019; Li et al., 2017; Liu et al., 2019). circRNAs are particularly enriched in the brain (Rybak-Wolf et al., 2015; Veno et al., 2015; Westholm et al., 2014; You et al., 2015), and their levels increase with age in the brains of mice, worms, and flies (Cortes-Lopez et al., 2018; Gruner et al., 2016; Westholm et al., 2014). These observations suggest important roles for circRNAs in the brain. Indeed, knockout of the most abundant mouse circRNA, CDR1as, results in specific behavioral defects (Piwecka et al., 2017). Moreover, circRNAs are involved in brain-related diseases and neurodegeneration (Hanan et al., 2020; Lukiw, 2013; Zimmerman et al., 2020).
As stated above, mbl hosts a highly expressed and evolutionary conserved circRNA: circMbl. This circRNA originates from the second exon of the mbl gene in flies (circMbl) and MBNL1 or MBNL2 in mouse and humans (Ashwal-Fluss et al., 2014). Previous work showed that at least in cell culture, MBL seems to regulate its own levels by promoting the generation of circMbl (Ashwal-Fluss et al., 2014). This circRNA contains multiple binding sites for MBL protein as well as part of the open reading frame for MBL, and it has been shown to be translated (Pamudurti et al., 2017). Interestingly, only head-specific MBL isoforms can promote circRNA formation (Ashwal-Fluss et al., 2014). These isoforms also can bind to the circRNA at least in cell culture. Hence, if this regulation also exists in vivo, it might buffer the levels of MBL proteins in tissues such as the brain where both the circular and linear isoforms of the gene are highly expressed. Here, we show that different MBL isoforms are expressed in a tissue-specific manner and regulate their own levels by generating different circRNA isoforms from their own locus. In addition, we show that circMbl also displays functions in trans in vivo.
RESULTS
The mbl locus generates several RNA and protein isoforms
The Drosophila mbl locus has a complex structure with almost 20 mRNA and protein isoforms. To fully characterize the RNA transcripts and proteins generated from this locus, we analyzed RNA sequencing (RNA-seq) and ribosome footprinting datasets we previously generated from fly heads (Martin Anduaga et al., 2019; Pamudurti et al., 2017). Previous work identified four proteins produced from the mbl locus: MBL-A, MBL-B, MBL-C, and MBL-D (Vicente et al., 2007). Our genomic data indicates that in fly heads, most of the RNA isoforms include the 3′-most distal exon, which encodes a region of MBL-C that is not included in the other three isoforms (Figures 1A and S1A). The data indicate that mbl-A and mbl-B mRNAs are not produced in fly heads (Figures 1A and S1A) and that the mbl-C mRNA accounts for approximately half of the mbl mRNAs (Figure S1B). Two other mbl mRNA isoforms, mbl-O and mbl-P, generated due to alternative splicing, together account for about 45% of the mbl transcripts in fly heads (Figure S1B). In addition, a small proportion of the transcripts (Figures 1A and S1A) end in an upstream exon; these isoforms are collectively called mbl-MI hereafter.
Figure 1. The mbl locus generates several RNA and protein isoforms.
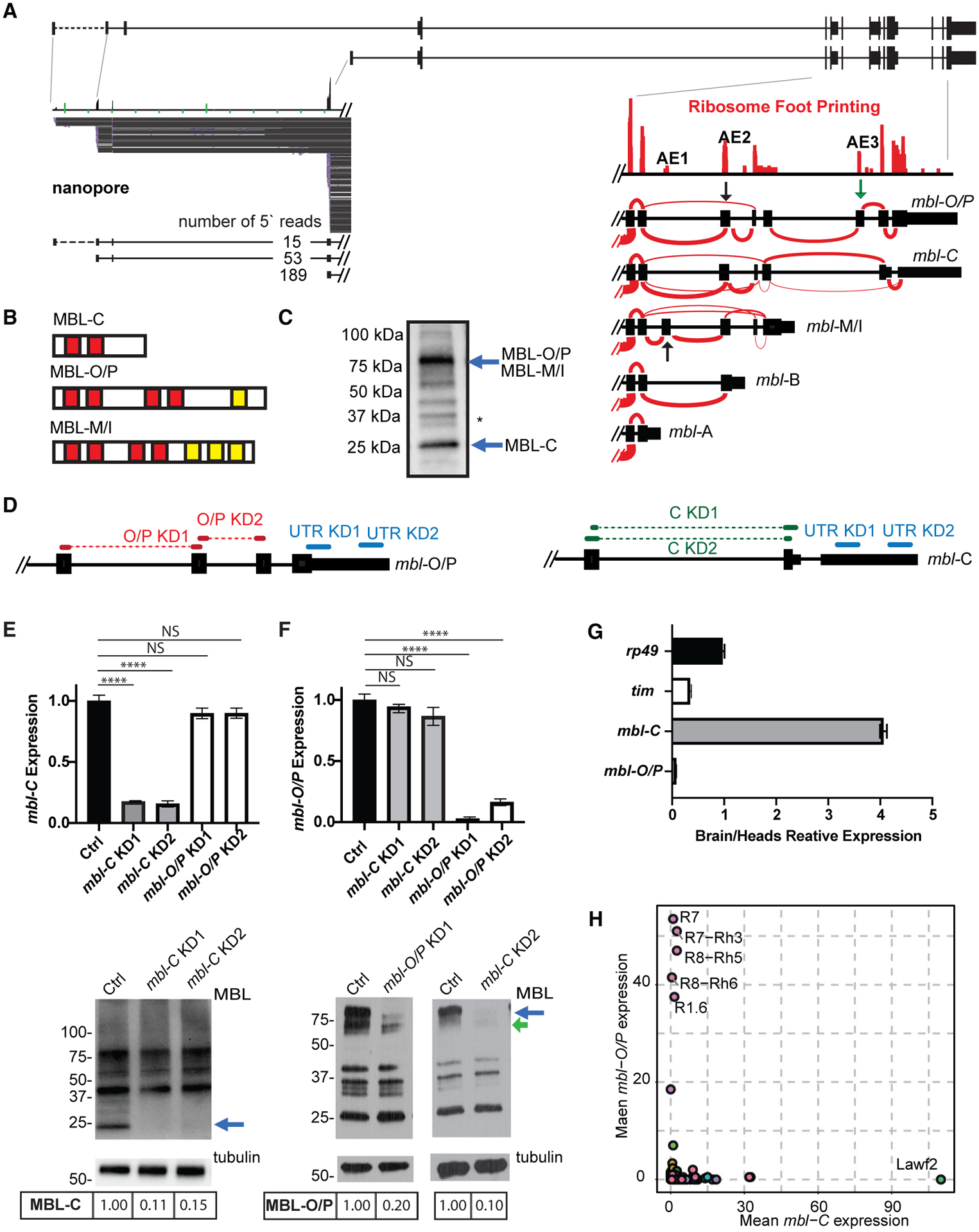
(A) Upper: schematic representation of mbl locus. Bottom left: nanopore RNA-seq reads from three distinct promoter regions. Bottom right: ribosome footprinting data. Sashimi plots show the different mbl isoforms 3′ annotation.
(B) Scheme of the protein domains in the different MBL isoforms. Red boxes indicate zinc fingers. Yellow boxes indicate intrinsically disordered regions.
(C) Western blot showing MBL-C and MBL-O/P protein isoforms (blue arrows); membrane was blotted using anti-MBL. Asterisk denotes non-specific band (based on the fact that none of the shRNAs or the available KK RNAi line affected the band consistently and that the band is not labeled when performing a western blot in an endogenously FLAG-tag MBL fly [Michela Zaffagni, personal communication]).
(D) shRNAs used to knock down mbl-C and mbl-O/P isoforms either independently or together.
(E and F). qRT-PCR (top) and western blot (bottom) from heads of KD for mbl-C (E) or mbl-O/P (F) isoform. Arrow indicates the MBL-C or MBL-O/P bands.
(G) qRT-PCR of rp49, tim, and mbl isoform expression levels in fly brain and heads.
(H) mbl-C and mbl-O/P mean expression in total RNA-seq data from sorted cells (n = 2, data from Davis et al., 2020). Each circle represents a cell type.
In all qRT-PCR analyses, tubulin was used as normalization control (n = 3, standard error of the mean [SEM], two-tailed t test performed for significant difference: ****p < 0.0001, ***p < 0.0002, **p < 0.0021, *p < 0.0332). In all western blot images, the quantification of MBL isoforms is stated below the images.
There are three exons alternatively spliced in the 3′ portion of the mbl gene (named AE1–3, right inset in Figure 1A). mbl-C mRNA is generated by exclusion of AE3 while mbl-O and -P mRNAs contain this exon (green arrow in Figure 1A, right inset). Inclusion of this exon results in a change on the reading frame and in a notable larger protein. which contains two additional zinc fingers (red boxes in Figure 1B). In addition, mbl-P mRNA contains the exon AE2 while mbl-O does not. Due to their similarity in their 3′ end region we refer to these isoforms collectively as mbl-O/P. The other alternative isoform (mbl-MI) seems to be less variable, with most (or all) mbl-M/H/J/K/S/T/R mRNAs containing the exon AE1.
Western blot of fly heads using an anti-MBL antibody revealed that the most abundant bands were of the predicted size of MBL-C (27 kDa) and MBL-O/P (72 kDa) (Figure 1C). The relative levels of these two isoforms change slightly between strains, with CantonS strain flies having more MBL-C and yw flies slightly higher levels of MBL-O/P (Figure S1C). We also observed additional less strong bands, including proteins that could be translated from the mbl-MI isoforms (70–90 kDa; Figure 1C). The MBL-C protein contains a single pair of zinc fingers, while MBL-O/P and MBL-M/I contain two. These longer proteins also have predicted intrinsically disorder domains (yellow boxes in Figure 1B).
The RNA-seq data also suggest that a fraction of mbl mRNAs originate from alternative promoters (Figure 1A). To verify the existence of these isoforms, we purified mbl mRNAs using biotinylated oligonucleotides complementary to the second exon of mbl (present in all RNA isoforms) and sequenced these RNAs using nanopore technology. This experiment confirmed that approximately one-fourth of the mbl transcripts have alternative 5′ exons (Figure 1A).
Levels of individual mbl RNA isoforms can be specifically reduced by the use of shRNAs
To further understand the role of the different mbl RNA isoforms, we generated 22 fly strains that express short hairpin RNAs (shRNAs) directed specifically against the different mbl RNA isoforms including circMbl. As a control we utilized a line expressing an shRNA similar to the one used to target circMbl but with eight mismatches in the center of the shRNA. This shRNA does not have complementarity to any sequence in the Drosophila transcriptome. Expression of most of these shRNAs with the strong actin-gal4 driver resulted in viable flies (Figure S1D). The exceptions are lines expressing shRNAs targeting the second exon (E2), the junction of the second and third exons (E2–3), or the last exon, which are shared by all mbl isoforms expressed in fly heads, which resulted in total or almost total developmental lethality (Figures 1D and S1D). This was expected, as flies with deletions in the mbl locus do not survive the pupal stage (Artero et al., 1998).
Constitutive expression of any of the two shRNAs directed against mbl-C downregulated the levels of this isoform in fly heads by more than 5-fold (Figures S1E and 1E). Upon expression of either of the two shRNAs that target the transcript that encodes MBL-C, the protein of 25 kDa was not observed by western blot (Figure 1E, bottom). In addition, expression of the shRNAs directed against mbl-O/P resulted in a strong and specific downregulation of these mRNAs and of the 75 kDa protein band (Figures 1F and S1F). Expression of the shRNAs targeting the transcripts that encode MBL-A or MBL-B did not alter the levels of any of the measured mRNA transcripts (including mbl-C, Figure S1E); this was expected, as these isoforms were not detected in fly heads.
The effect of the shRNA directed against the mbl-MI isoforms was difficult to assess due to the high levels of pre-mRNA. Indeed, most of the qRT-PCR product obtained when quantifying the levels of these isoforms corresponds to nascent RNA (as most of the RNA is in the chromatin-bound fraction, Figure S1G). This does not mean the isoform is not expressed, as it can be detected in the cytoplasmic fraction upon cell fractionation (Figure S1G) and in the ribosome footprinting data (Figure 1A). Expression of one of the shRNAs directed against mbl-MI resulted in a decrease of these transcripts by 30% (Figure S1H) and does not alter the expression of mbl-C (Figure S1E) or mbl-O/P (Figure S1F). The remaining signal likely originates from intronic or nascent RNA rather than mature transcripts.
We also generated shRNAs directed against two RNA regions which are common to several isoforms. We first tested two shRNAs targeting the last exon of mbl and hence the mbl-C, mbl-O, and mbl-P isoforms (lines UTR-KD1 and UTR-KD2, Figure 1D). The UTR-KD1 shRNA line is significantly stronger than the UTR-KD2 line and when expressed from the actin-gal4 driver, the UTR-KD1 shRNA is almost completely lethal (Figure S1D). Restricting expression to neurons resulted in a decrease of about 50% of mbl-C and mbl-O/P mRNAs and of a more dramatic decrease of the corresponding protein isoforms (Figures S1I and S1J). The UTR-KD2 shRNA line is weaker, but we still observed a decrease of MBL-C (Figures S1I and S1J, respectively).
In addition, we generated and tested two shRNAs targeting the most used first exon of mbl. Expression of any of these shRNAs (named E1-KD1 and E1-KD2) almost completely eliminated the expression of mbl-O/P and resulted in a strong decrease in the levels of mbl-C (Figures S2A and S2B). The difference in the knockdown of mbl-C and mbl-O/P could be due to the possibility that a substantial fraction of the mbl-C transcripts is generated from the distal mbl promoter (named mbl′ promoter). Therefore, we determined the levels of the transcripts generated from the upstream promoter on the different knockdown strains (see PCR amplicons in Figure S2C). Expression of the E1-KD shRNAs did not change the levels of the upstream exons 1′ and 2′ (Figure S2D). In addition, we did not observe changes on the mRNAs generated from the upstream promoter when knocking down mbl-O/P (Figure S2E). On the other hand, knockdown of mbl-C resulted in a strong (40%) decrease of the signal originating from the upstream promoter (Figure S2E). These results show that a significant fraction of mbl-C but not mbl-O/P is generated from the upstream promoter.
Mbl-C and mbl-O/P are expressed in different cells in the fly head
The results displayed above show that there is no detectable crosstalk between the levels of mbl-C and mbl-O/P, a remarkable result given the strong autoregulation operating within the mbl locus (see below). The latter could be due to tissue-specific expression of the different isoforms. We then measured the levels of mbl-C and mbl-O/P in fly heads and in dissected fly brains. A housekeeping gene such as ribosomal protein 49 (rp49) is not enriched in one part (brain/head ratio approximately 1; Figure 1G). The mRNA encoding the circadian regulator timeless (tim) is slightly enriched outside the brain (due to high expression in the eye and fat body, brain/head ratio approximately 0.3). Interestingly, mbl-C is highly enriched in the brain (brain/head ratio >4), while the levels of mbl-O/P in the brain are extremely low (Figures 1G and S2F), demonstrating that the latter isoform is mainly expressed in head tissues other than the brain (e.g., the eye) while mbl-C is highly brain enriched.
To determine where within the fly head the different mbl isoforms are expressed, we utilized data from a recent study that generated RNA-seq data from purified nuclei of 100 driver lines comprising 67 cell types sorted from fly heads (Davis et al., 2020). Strikingly, the expression of mbl-C and mbl-O/P is highly specific and not overlapping (Figure 1H). In brief, mbl-O/P mRNA is mainly expressed in the eye (photoreceptor cells) while mbl-C is expressed in different neuronal subtypes and in muscle cells (Figure 1H and Table S1).
Interestingly, cells expressing mbl-O/P tend to express mbl from the downstream/proximal promoter (e.g., photoreceptors in Figure S2G). Many of the cell types that express predominantly mbl-C display higher levels of the upstream/distal mbl promoter (e.g., Lawf2 cells in Figure S2G; Table S1). In sum, our findings demonstrate the existence of cell-specific expression of the two main mbl isoforms, which are also generated from alternative promoters.
Cells expressing circMbl can be identified using single-cell sequencing
The mbl locus expresses several circRNAs. The one originating from the second exon (circMbl) is the most abundant circRNA in flies (Ashwal-Fluss et al., 2014). However, there is no information on the cell specificity of this circRNA. Recently, the fly brain has been sequenced at the single-cell level (Davie et al., 2018). As single-cell sequencing is based on poly(A) amplification, circRNAs are not represented in this type of dataset. However, there are three short poly(A) tracks in circMbl which could potentially extend the oligo(dT) primers utilized for reverse transcription (Figure 2A, right). To determine whether circMbl could be detected in poly(A+)-based libraries, we utilized a standard circRNA identification pipeline (find_circ [Memczak et al., 2013]) in newly generated poly(A+) 3′ RNA-seq datasets from dissected fly brains. We detected high signal corresponding to the backsplice junction of circMbl in these libraries (Table S2 and Figure S3A). We then proceeded to determine whether we could identify circMbl in the fly brain single-cell sequencing dataset. Indeed, we observed a high peak in the second exon in the aggregated signal from the single-cell sequencing dataset (Figure 2A, left). Moreover, we found more than 2,000 normalized backsplicing reads for circMbl (for a few examples see Figure S3A and Table S2).
Figure 2. Levels of circMbl and mbl in the fly brain.
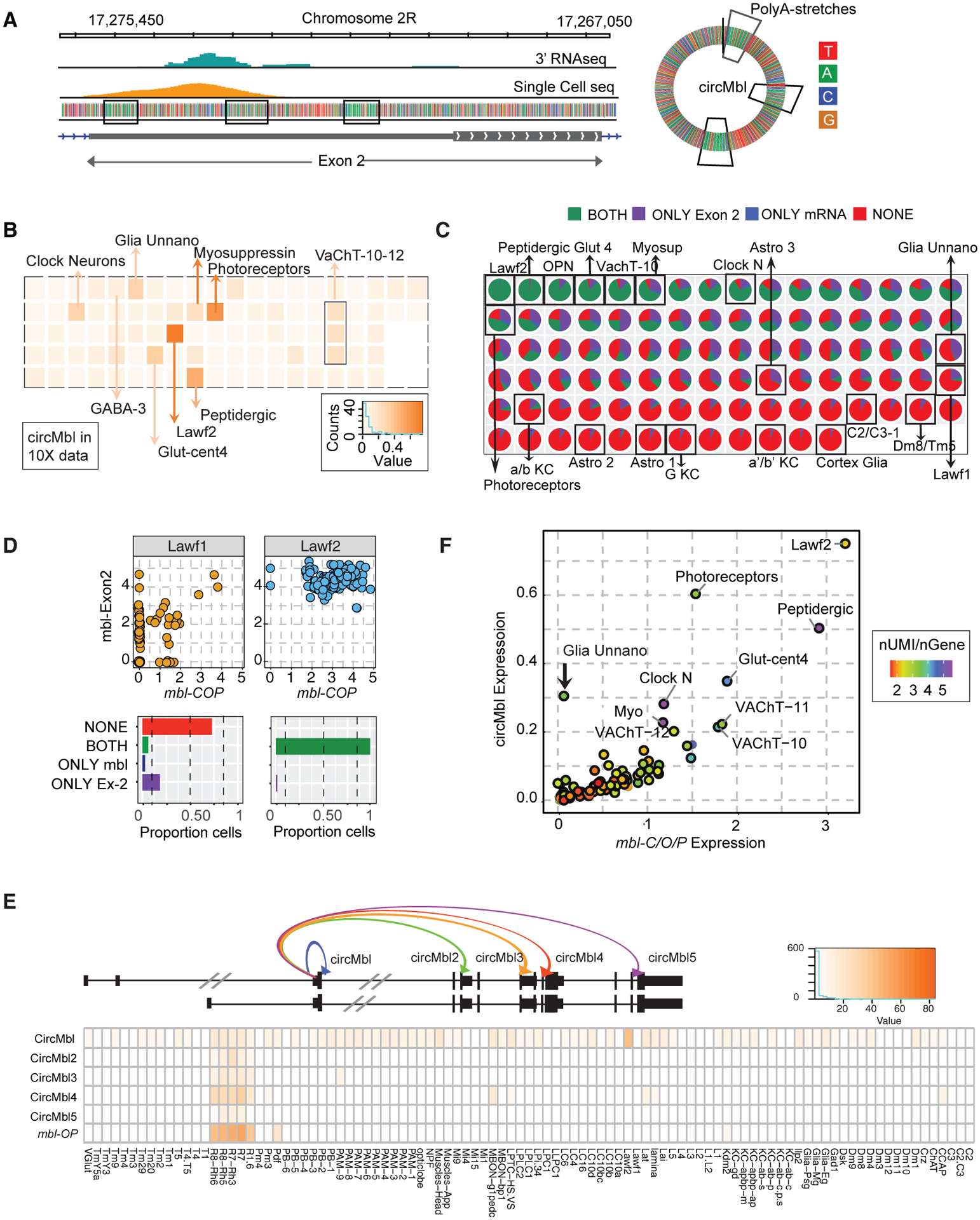
(A) Left: integrative genome viewer (IGV) snapshot of mbl exon 2 aligned reads from bulk 3′ RNA-seq and single-cell 10X RNA-seq. Right: circMbl scheme. Marked in squares are the polyA stretches.
(B) Heatmap of mean circMbl normalized expression in each single-cell cluster.
(C) Pie charts with proportion of cells with mbl exon 2 and/or mbl-C/O/P UTR signal.
(D) Top: dot plot of mbl exon 2 versus mbl-C/O/P normalized expression in single cells from Lawf2 and Lawf1 neuronal clusters. Bottom: proportion of cells expressing one, both, or no mbl exon 2 and mbl-C/OP.
(E) Heatmap of normalized expression for the different circMbl isoforms and mbl-O/P in sorted cells.
(F) Single-cell cluster mean circMbl versus mbl-C/O/P UTR normalized expression. Color represents UMIs/genes ratio.
To quantify the levels of circMbl in single cells, we realigned the single-cell RNA-seq to a modified version of the genome in which the original annotation of the mbl gene was replaced by a circMbl gene (which consists of the regions encompassing the circRNA backsplicing junction) and the 3′ end of the different mbl linear RNA isoforms (Figure S3B). We observed that circMbl is expressed in many neuronal clusters within the brain and a few glial subclusters (Figure 2B and Table S2). We also performed a separate analysis in which we aligned reads to the mbl second exon. The circRNA is the main molecule produced from the second exon (>70% when comparing circRNA and linear reads [Ashwal-Fluss et al., 2014; Westholm et al., 2014] or when evaluating the remaining signal upon circMbl-KD; see below). Hence, the signal originating from the second exon in this library constitutes a good proxy for circMbl expression (compare Figure S3C with Figure 2B). The last exon provides accurate information regarding mbl-C mRNA levels (as mbl-O/P is not expressed in the brain, Figures 1G and S2F). We used this analysis to identify the cells that express mbl mRNA, circMbl, or both. By integrating these two comparisons (circMbl versus mbl mRNA and exon 2 versus mbl mRNA), we were able to identify three types of clusters of cells: those that express very low levels (or no) mbl and circMbl; those that express high levels of both mbl and circMbl; and those that express only circMbl as determined by exon2 signal (Figure 2C and Table S2). Interestingly, related cell types have a quite different expression pattern of mbl expression. For example, almost every Lawf2 cell expresses circMbl and mbl mRNA, but most of the closely related Lawf1 cells express neither linear nor circular mbl RNA (Figures 2C and 2D). Many neuronal cell types express both mbl and circMbl (Figure 2C and Table S2), and several neuronal groups do not express any of the mbl gene products. To the best of our knowledge, this is the first time that the cell specificity of a circRNA has been determined from a whole tissue at the single-cell level.
We also utilized the nuclei data from 100 cell types in the fly head and determined independently the expression of circMbl. As in the single-cell RNA-seq data, we observed high expression of circMbl across the fly brain with very high levels in Lawf2 (but not Lawf1) cells as well as in the groups described above (i.e., Ilp2 and photoreceptors; Figure S3D and Table S2). Interestingly, we generally detected high levels of circMbl on cells expressing either mbl-C or mbl-O/P (Figures 2E, S3D, and S3E; Table S2).
While the circRNA generated from the second exon of mbl (circMbl) is the most abundant isoform, there are other additional abundant circular isoforms (Ashwal-Fluss et al., 2014; Westholm et al., 2014). While circMbl was the most abundant circRNA isoform in all the sequenced brain cell populations (in which mbl-C is the main/unique linear isoform, Table S1), eye cell populations (e.g., R1, R7, R8) express very high levels of the other circMbl isoforms (Figures 2E and S3F). Indeed, in photoreceptors the levels of circMbl4 are double those of circMbl, and circMbl3 is expressed at levels similar to those of circMbl (Figure S3F). Interestingly, these cells express predominantly mbl-O/P (Figure 2E and Table S1). Expression of the most abundant of those (circMbl4) strongly correlates with the levels of mbl-O/P but not mbl-C (Figure S3G). These results strongly suggest that circMbl2-5 contribute to the regulation of mbl-O/P and/or are regulated by mbl-O/P but not mbl-C in cis.
The levels of circMbl and mbl-C correlate in individual clusters in the fly brain
If MBL protein regulates circMbl production, we expect a strong correlation between the levels of the mbl mRNA and circMbl within the clusters in the fly brain. We thus quantified levels of mbl (mbl-C predominantly) and circMbl in the individual clusters of the single-cell brain data. Indeed, we observed a very strong correlation between the levels of mbl and circMbl in the brain cell clusters (Figure 2F, R2 = 0.82, p < 2.2e–16). This correlation is not due to the total number of unique molecular identifiers per gene (UMIs, see color code in Figures 2F and S4A). Moreover, there was no correlation between either mbl mRNA or circMbl and the neuronal marker elav or the glial marker repo, showing that the correlation is not a data artifact (Figure S4B). A notable exception to the correlation between mbl-C and circMbl was observed in one group of glial cells that express circMbl and not mbl-C (arrow in Figure 2F).
mbl-C and circMbl regulate each other in cis
To determine the relationship between MBL-C and circMbl in the brain, we up- or downregulated the levels of this protein. First, we utilized an available UAS-MBL-C fly line in combination with a broad (actin-gal4) or neuronal-cell-specific (elav-gal4) driver. Overexpression of MBL-C using the broad driver resulted in complete developmental lethality (Figure S5A). By limiting the expression of MBL-C to the nervous system, we still observed some lethality (56%) but were able to obtain enough flies for follow-up molecular experiments (Figure S5A).
Overexpression of MBL-C in the nervous system (Figure S5B) resulted in a 2-fold increase in the levels of circMbl in dissected brains (Figures 3A and 3B). Importantly, the effect of MBL-C overexpression is specific to circMbl, as it did not increase the levels of other two abundant circRNAs (Figure 3B). We observed a small but significant decrease in the levels of circHaspin, likely due to indirect effects. As we did not observe an increase in mbl pre-mRNA upon MBL-C overexpression (Ex2-In2 amplicon, Figures 3A and 3C), it is possible that the newly produced circMbl is made at the expense of the linear RNA. Therefore, we determined the level of the two major endogenously expressed isoforms of mbl using primers that do not overlap with the overexpressed MBL-C open reading frame (Figure 3C). Overexpression of MBL-C decreased the levels of the endogenous mbl-C/O/P qPCR target by almost 40% (Figure 3D). As mbl-O/P levels in the brain are very low (Cq value approximately 33) and are not affected by MBL-C overexpression (Figure 3D), the decrease in 40% of the endogenous mbl expression is likely due to the 2-fold increase in circMbl production. Moreover, the lower levels of mbl are unlikely to be due to other regulatory processes such as alternative splicing or nonsense-mediated decay, as all the identified isoforms expressed in the head at considerable levels contain the last exon and we could not identify a combination of splicing events that would generate an isoform with a premature stop codon. In any case, these results demonstrate the existence of competition between circMbl and mbl-C expression in vivo and a role in cis for circMbl production in the fly brain.
Figure 3. mbl-C and circMbl regulate each other in cis.
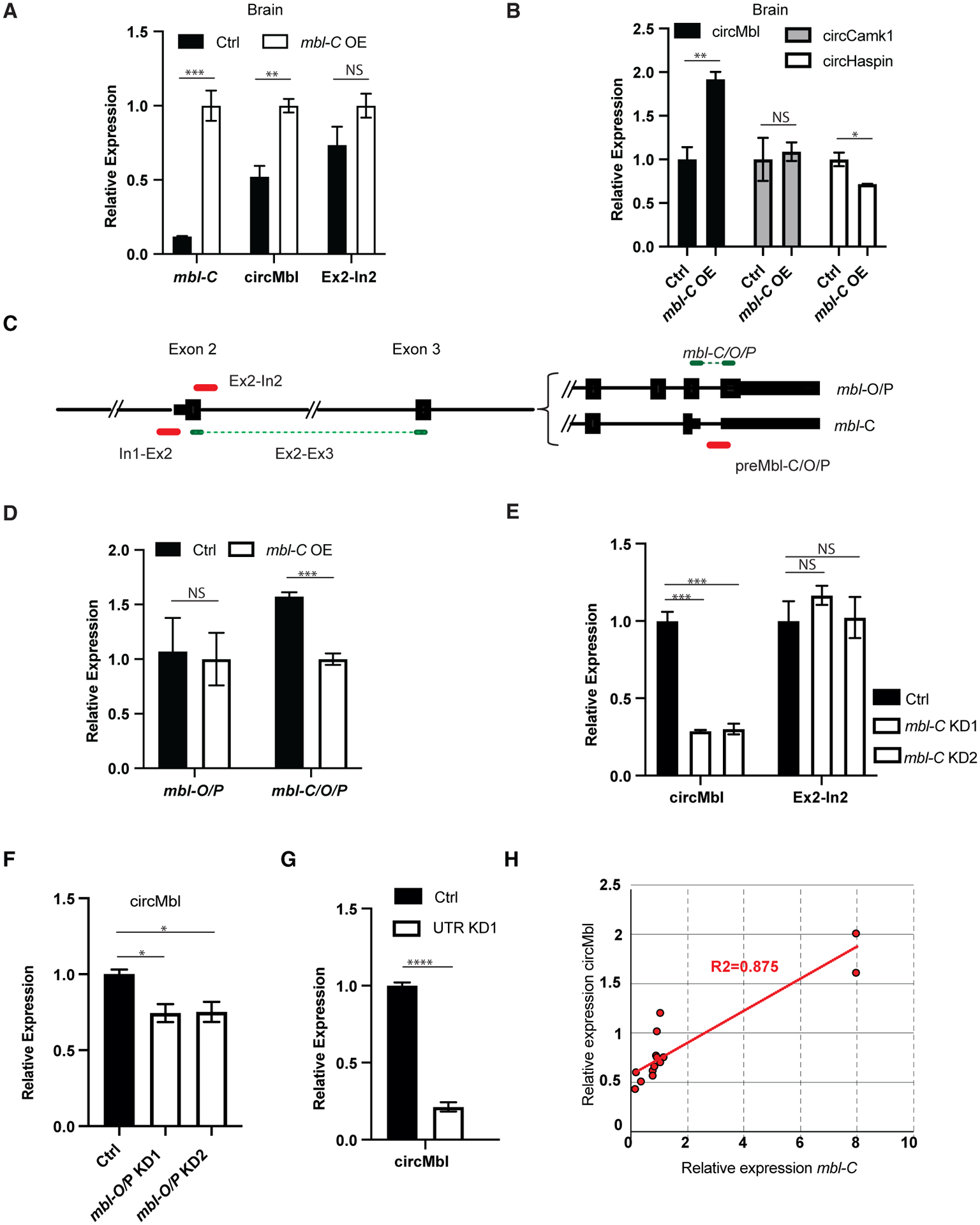
(A and B) qRT-PCR of the indicated targets in mbl-C-OE fly brains.
(C) Scheme of the primer sets used to quantify the mbl locus expression.
(D) qRT-PCR of mbl-O/P and mbl-C/O/P in mbl-C-OE fly brain.
(E) qRT-PCR of the indicated targets in mbl-C-KD fly heads.
(F and G) qRT-PCR of circMbl in heads of mbl-O/P (F) and UTR-KD flies (G).
(H) mbl-C and circMbl expression levels correlation plot in various mbl isoform KD flies.
In all cases tubulin was used as a normalization control (n = 3, error bars represent SEM, two-tailed t test performed for significant difference: ****p < 0.0001, ***p < 0.0002, **p < 0.0021, *p < 0.0332).
Moreover, specific downregulation of mbl-C provoked a 4-fold reduction in circMbl levels in fly brains (Figure 3E). The decrease in circMbl was not due to lower levels of mbl transcription (see Ex-In2 signal, Figure 3E). The levels of MBL-C seem to be a key determinant of circMbl levels in the fly head, as expression of the shRNA targeting mbl-O/mbl-P resulted in a decrease of only 25% of the levels of circMbl (likely in the eye; Figure 3F). Moreover, expression of the shRNA targeting the 3′ UTR shared by mbl-C, -O, and -P led to a 4-fold downregulation in the levels of circMbl in the fly brain (Figure 3G). Indeed, when we combined all the qPCR measurements, we observed a strong correlation between the levels of MBL-C and circMbl in both the head and the brain tissue (Figure 3H). These results together with the strong correlation observed in the brain between mbl-C and circMbl (Figure 2F) demonstrate that circMbl production by itself has a role in regulating the expression on mbl-C. This cis-regulatory mechanism limits the levels of MBL-C protein and mRNA.
mbl-O/P regulates its own production by two different mechanisms
As shown above, cells in the fly eye express only mbl-O/P as well as high levels of other circRNAs generated from the mbl locus: circMbl2, circMbl3, and circMbl4. This raises the possibility that in the fly eye the production of those circRNAs (and not the canonical circMbl) is the main driver of the mbl self-regulatory loop. Indeed, knockdown of mbl-O/P resulted in a significant decrease in circMbl2, circMbl3, and circMbl4 (Figure 4A), demonstrating that MBL-O/P is required for the expression of these circRNAs.
Figure 4. MBL-O/P regulates its own production by two different mechanisms.
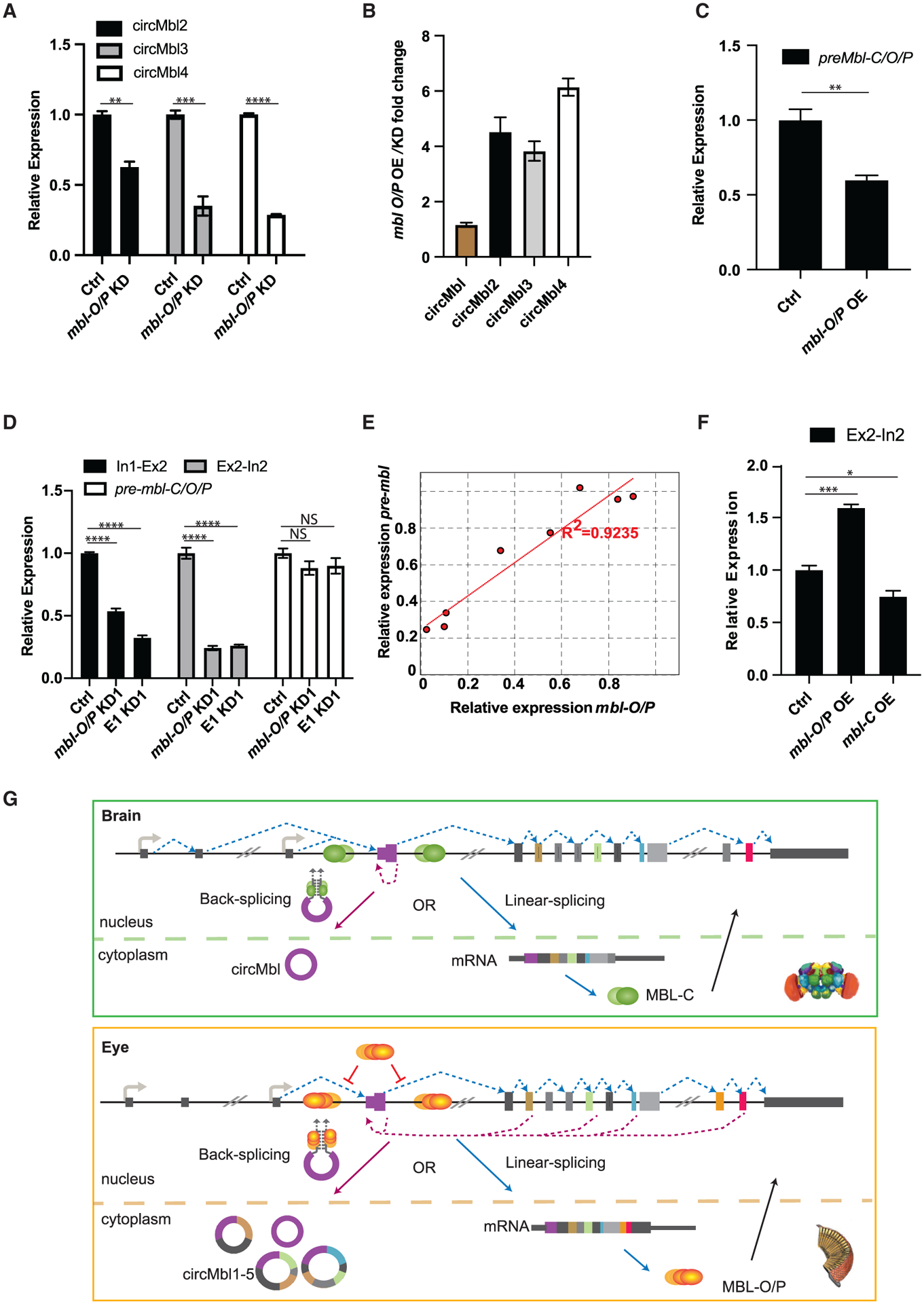
(A) qRT-PCR of circMbl isoforms in mbl-O/P-KD fly heads.
(B) qRT-PCR of circMbl isoforms fold change in mbl-O/P transgenic fly heads.
(C) qRT-PCR of pre-mbl in MBL-O/P-OE fly heads.
(D) qRT-PCR evaluation of the levels of preRNA in mbl-O/P and Exon1-KD fly heads.
(E) mbl-O/P and preMbl (Ex2-In2) expression levels correlation plot in various mbl isoforms KD flies.
(F) qRT-PCR in mbl-O/P and -C-OE fly heads.
(G) Representation of MBL-C and MBL-O/P regulation in cis by circMbl isoforms in different tissues. In the brain (green), MBL-C binds to pre-mRNA in order to facilitate backsplicing (as described in Ashwal-Fluss et al., 2014). In the eye, MBL-O/P regulates its own levels by two different mechanisms: inhibiting the splicing of the first and second introns (red inhibition symbols) and promoting backsplicing (dashed violet lines).
Tubulin was used as a normalization control (n = 3, error bars represent SEM, two-tailed t test performed for significant difference: ****p < 0.0001, ***p < 0.0002, **p < 0.0021, *p < 0.0332).
We generated flies expressing MBL-O/P. As for MBL-C, overexpression of MBL-O/P utilizing the constitutive driver actin-gal4 resulted in developmental lethality (Figure S5A). Overexpression of MBL-O/P in the eye using the eye-specific gmr-gal4 driver (see Figures S5C and S5D) provoked a significant increase in the levels of circMbl2 and circMbl4 but not circMbl and circMbl3 (Figure S5E). By combining these data, we were able to determine that MBL-O/P can dramatically modulate the levels of circMbl2, circMbl3, and circMbl4 but not circMbl (Figure 4B). We attribute the small/lack of effect on circMbl levels mainly to the fact that circMbl expression is lower in the eye and that it is also produced in many cells that do not express MBL-O/P.
Moreover, we observed a decrease in the levels of the pre-mRNA of mbl-C/O/P (Figure 4C), demonstrating that the newly formed circRNA likely comes at the expense of the levels of the endogenous mbl. The lower levels of pre-mbl-C/O/P following MBL-O/P overexpression are due to production of the circRNAs and not transcription, as the signal in the other tested introns is not decreased (see below). These results demonstrate that the levels of mbl-O/P are regulated mainly by the production of the longer circRNAs instead of circMbl.
As MBL regulates splicing of introns carrying MBL binding sites (Goers et al., 2010; Li and Millard, 2019), and the introns in the mbl gene contain several of those sites (Ashwal-Fluss et al., 2014), we determined the levels of mbl unspliced introns in fly head samples depleted of different MBL isoforms. As shown above, a decrease in MBL-C levels did not change amounts of pre-mRNA as measured in the intron-exon boundaries flanking the circularizable exon 2 (Figure 3E, and see scheme in Figure 3C). This result suggests that MBL-C promotes circMbl backsplicing directly and not by modulating the splicing efficiency of the flanking introns. Interestingly, downregulation of mbl-O/P resulted in a strong decrease in the intronic signal flanking the main circularizable exon of mbl (exon 2; see In1-Ex2 and Ex2-In2 signals in Figures 4D and S5F) with no effect on the levels of the last intron (preMbl-C/O/P primer pair; Figures 4D and S5F). We obtained similar results when performing a similar experiment from nascent RNA using heads of control, mbl-C, and mbl-P/mbl-O knockdown strains (Figures S5G–S5I). This suggests that MBL-O/P inhibits the splicing of the first and second mbl introns. Indeed, flies expressing the E1KD1 shRNA (which resulted in a 10-fold reduction of mbl-O/P mRNA, Figure S2A) display a 3-fold reduction on the levels of the first and second mbl introns (Figure 4D). Moreover, we observed a strong correlation between the levels of unspliced mbl second intron and mbl-O/P mRNA when we combined all the silencing experiments of mbl isoforms in fly heads (Figure 4E). Additionally, overexpression of MBL-O/P but not MBL-C in the fly eye provoked a 1.5-fold increase in the pre-mRNA of mbl (Figure 4F), demonstrating that MBL-O/P regulates its own levels by inhibiting its own splicing. Interestingly, similar to MBL-O/P, overexpression of MBL-C in the eye increased the levels of the eye-specific circRNAs without changing the levels of the introns flanking the second mbl exon (Figures S5J and 4F). This result strongly suggests that the effect of MBL-O/P on the levels of the circular RNAs is independent of the activity of this protein on the splicing efficiency of the first and second mbl introns. These results suggest the existence of an additional (circRNA-independent) mechanism by which MBL-O/P regulate their own production by inhibiting the splicing of the first and second mbl introns.
The results presented in these sections show that MBL directly limits/regulates itself. In the fly brain, MBL-C is the main protein isoform and limits its own mRNA and protein amounts by promoting the formation of circMbl (see scheme in Figure 4G). As MBL-C is required to bind to both introns flanking the circularizable exon (Ashwal-Fluss et al., 2014), it likely promotes circRNA biosynthesis directly by bringing those introns together through protein-protein interaction and recruitment of the spliceosome. In the fly eye, where MBL-O/P is the predominantly expressed protein isoform, MBL limits/buffers its own expression by two independent mechanisms: inhibition of its own splicing and promoting expression of several circMbl isoforms (circMbl 1–4; see Figure 4G). These results demonstrate a gene expression cis-regulatory role for the biosynthesis of the circRNAs derived from the mbl locus in vivo.
circMbl can be specifically downregulated in vivo by microRNA-derived shRNAs
To determine potential roles of circMbl in trans, we depleted circMbl by using shRNAs directed against the circMbl-specific backspliced junction (Ni et al., 2011; Pamudurti et al., 2020). We generated flies expressing the shRNA against circMbl under the control of the constitutive actin-gal4 driver. We then analyzed gene expression in controls and circMbl-KD flies using total and poly(A+) RNA-seq. We observed a specific and strong reduction in circMbl levels in fly heads (Figures 5A and 5B). The effect was highly specific for the circular molecule, as we did not observe a reduction of any of the linear mbl mRNA isoforms (Figures 5B, 5C, and S6A). We followed by confirming that the second exon signal mainly originates from the circRNA. Indeed, we observed a 70% decrease in the signal originating from the second exon of mbl in the poly(A+) RNA-seq library from flies expressing the shRNA targeting circMbl compared with the control library (Figures 5A–5C and S6A).
Figure 5. circMbl can be specifically downregulated in vivo.
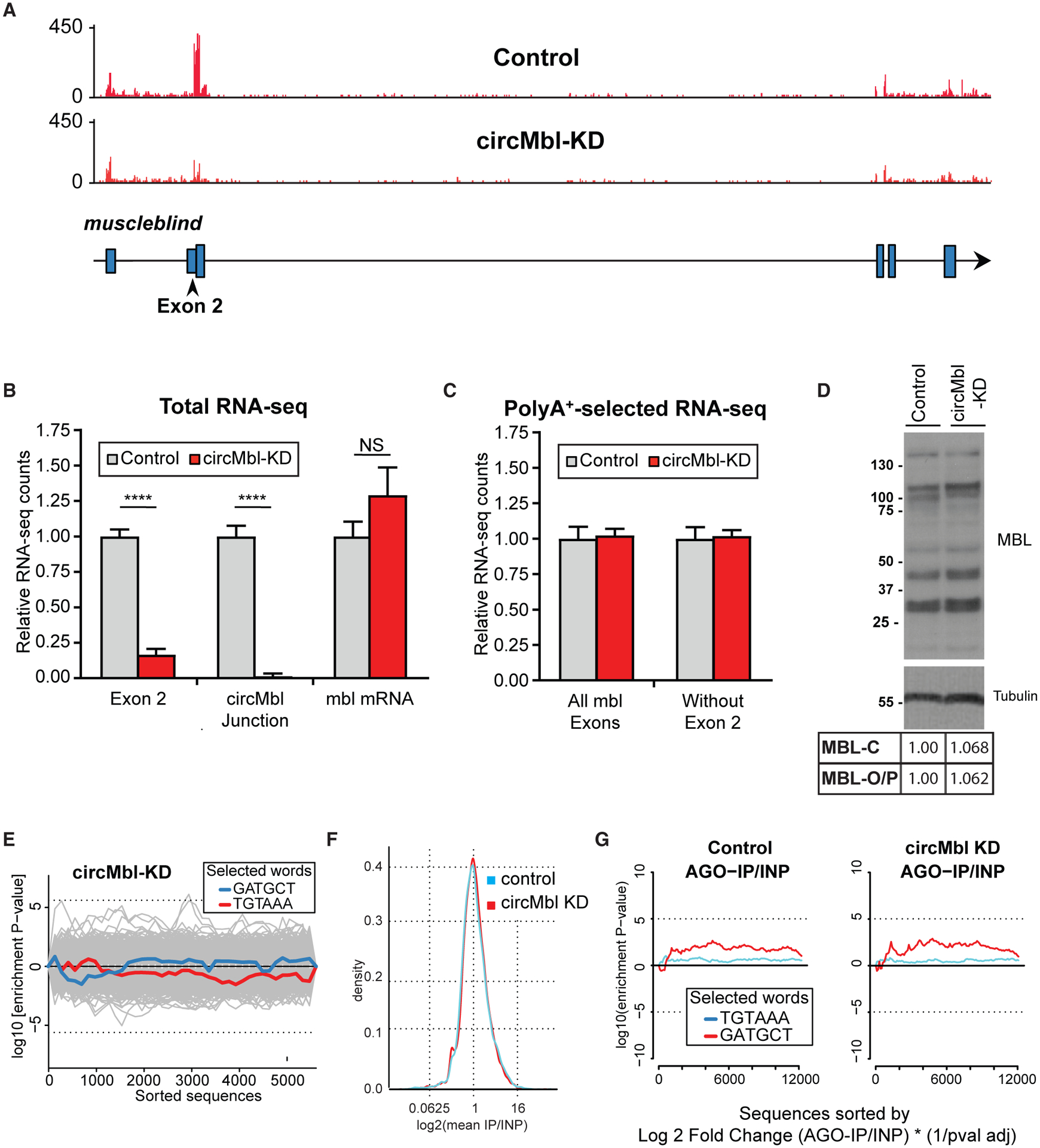
(A) IGV snapshot, showing a specific reduction of exon 2 in respect to a control strain.
(B and C) Quantification of the indicated mbl regions from total (B) and poly(A+) (C) RNA-seq data (n = 3, error bars represent SEM, two-tailed t test performed for significant difference: ****p < 0.0001, ***p < 0.0002, **p < 0.0021, *p < 0.0332).
(D) Western blot of control and circMbl-KD flies using anti-MBL immunosera.
(E) Assessment of off-targets by Sylamer. Traces show the seed enrichment for the genes differentially expressed upon downregulation of circMbl. shRNA and shRNA* seed sequences shown in blue and red, respectively.
(F) General binding of mRNAs to AGO1 in shRNA and control line.
(G) Sylamer enrichment landscape plot for sh-circMbl and sh-circMbl* 6-mers. The x axis represents the genes sorted from the most to the least enriched in the AGO1 immunoprecipitation (IP) sequencing. INP, input.
The shRNA that targets circMbl does not have off-target effects
To rule out the possibility that expression of the circMbl shRNA also affects the protein product expressed from the linear RNAs, we compared the MBL protein expression in control flies with that in the circMbl-KD line. No difference was detected, demonstrating that expression of the shRNA against circMbl is specific and does not alter the levels of any of the MBL isoforms (Figure 5D). As the knockdown by the shRNA is post-transcriptional and only is suitable for determining functions of the circRNA once in the cytoplasm, this does not contradict the results demonstrating a role for circRNA production on the expression of mbl mRNA.
The shRNA could also have off-target effects on other mRNAs by acting as an miRNA. Therefore, we determined whether the downregulated mRNAs in the shRNA-expressing strain were enriched for the seed of the shRNA or shRNA* utilizing Sylamer (van Dongen et al., 2008). None of the downregulated mRNAs were enriched for these seed sequences (Figure 5E).
The shRNA could perturb miRNA-mediated gene regulation or could provoke changes in translation of mRNAs without significant effects at the RNA level. To rule out this possibility, we determined whether the expression of the shRNA against circMbl altered the population of mRNAs bound to AGO1, the key component of the miRNA effector machinery in Drosophila (Forstemann et al., 2007). To do so, we sequenced mRNAs that co-purified with AGO1 in heads of control and circMbl-KD flies. Expression of the shRNA against circMbl did not alter the general profile of RNAs bound to AGO1 (Figure 5F). Moreover, the k-mer enrichment was similar for control flies and circMbl-KD flies, and there was no significant enrichment for the 6-mers that could be generated from the processing of shRNA designed to target circMbl (Figure 5G). Furthermore, the few mRNAs that were differentially bound to AGO1 in the circMbl-KD flies were not enriched for seed sequences potentially targeted by the shRNA and shRNA* expressed in this strain (Figure S6B). All these results indicate that the shRNA designed to target circMbl is highly specific and suitable for determining the functionality of this circRNA in vivo.
Reduction of circMbl levels leads to abnormal developmental and adult phenotypes
As shown above, knockdown of individual mbl isoforms produced viable adult flies. Interestingly, circMbl-KD flies displayed male developmental lethality with high penetrance (Figure 6A). To confirm these phenotypes, we generated two additional shRNAs against the circMbl junction (Figure 6B). When the shRNAs targeting circMbl were expressed under the actin-gal4 driver, we observed significant but incomplete developmental male lethality with one of the shRNAs (circMbl-KD3) as observed with the original shRNA. Expression of the other shRNA (circMbl-KD2) resulted in complete male lethality and a very strong lethality in females, while expression of the shRNA with mismatches to the circMbl junction (circMbl-8MM) did not affect viability or the number of males (Figure 6A). Expression of the circMbl-targeted shRNAs provoked an almost complete silencing of circMbl (Figure S6C). However, expression of the two shifted shRNAs had some effects on the levels of mbl-C and mbl-O/P (Figure S6C). These effects observed on the linear RNAs are likely due to the longer stretches of complementarity resulting from the shift. In any case, the phenotypes attributed to the circMbl-KD1 and circMbl-KD3 cannot be attributed to alterations on the linear RNAs because: (1) circMbl-KD1 flies do not show changes in any of the linear mRNA isoforms; and (2) we did not see any of the phenotypes observed with the circMbl-KD lines when targeting linear mbl isoforms.
Figure 6. Knockdown of circMbl provokes specific phenotypes.
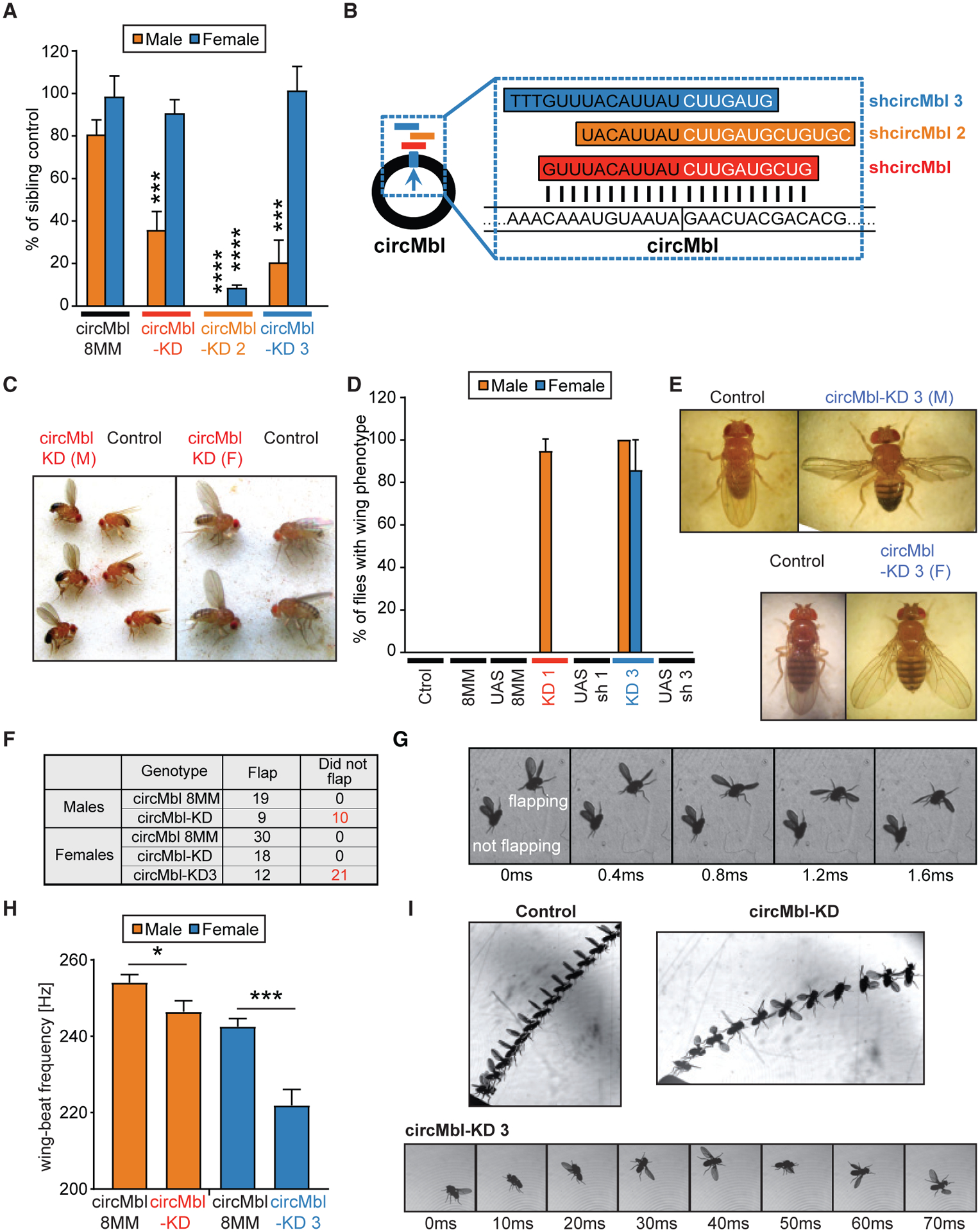
(A) Viability of males and females from control and circMbl-KD lines. We plotted the percentage of males and females of the indicated genotype against the sibling controls. The plotted results are the average of eight independent experiments for circMbl-KD flies and seven for the rest of the strains (comparison of KD lines with controls of same sex, Student’s t test: ***p < 0.0005, ****p < 0.0001).
(B) Scheme of the shRNAs against circMbl.
(C) Representative picture of circMbl-KD males with “wings up” reared at 25°C (left) or females (right) reared at 29°C next to control flies (actin-Gal4).
(D) Percentage of wing phenotype in circMbl-KD lines and its control flies. We plotted the percentage of males and females presenting wings up (circMbl-KD) or open (circMbl-KD3) phenotypes for the indicated genotypes. The plotted results are the average of eight independent experiments for circMbl-KD flies and seven for the other strains (error bars represent SEM, two-tailed t test performed for significant difference: ****p < 0.0001, ***p < 0.0002, **p < 0.0021, *p < 0.0332).
(E) Representative pictures of circMbl-KD3 males (top right) and females (bottom right) next to controls (actin-Gal4 flies).
(F) Results of the tapping assay.
(G) A sequence of side-view images from the tapping assay taken 0.4 ms apart. Images show two male circMbl-KD flies falling side by side.
(H) Mean wing-beat frequency in the free-flight assay. We measured ~30 flies from first three lines and 12 flies from the circMbl-KD3 (n = 25/32/29/12; Student’s t test: *p < 0.05, ***p < 0.0005).
(I) Representative flight events from the free-flight assay. Top left: a control male taking off normally. Superposed images are shown every 4 ms. Top right: a male circMbl-KD fly taking off. Superposed images are shown every 6 ms. Bottom: a female circMbl-KD3 fly shown shortly after take-off. Images are shown every 10 ms.
A large proportion of the circMbl-KD males that escaped the developmental lethality displayed a strong wing-posture phenotype (Figure 6C, left). Females displayed a similar phenotype when raised at 29°C (Figure 6C, right). We observed normal wing postures in all control strains. All circMbl-KD3 males and females also displayed wing-posture phenotypes (Figures 6D and 6E). These results demonstrate that downregulation of circMbl using different shRNAs provokes related phenotypes. Importantly, knockdown of the mbl linear isoforms did not provoke any of those phenotypes, demonstrating that circMbl and mbl have different functions.
The observed wing phenotypes suggest that circMbl is necessary for correct muscle function and flight. We next evaluated the flight of the different circMbl-KD strains. We first determined whether control, circMbl-KD, and circMbl-KD3 flies could flap their wings when released. In these conditions all the males and females from a control strain flapped their wings (Figure 6F). Similarly, all the female flies from the circMbl-KD strain flapped their wings (Figure 6F). In contrast, only half (9/19) of the males from the circMbl-KD strain and one-third (12/33) of circMbl-KD3 females managed to flap their wings (see Figure 6F for the summarized results and Figure 6G for an example of males from circMbl-KD).
We then performed a second type of assay in which we carefully placed the flies on a surface and allowed them to take off and fly freely. In this assay, we determined the mean wing-beat frequency per flight. Males of the circMbl-KD strain and females of the circMbl-KD3 strain displayed significantly lower wing-beat frequencies than controls (Figure 6H). In addition, many of circMbl-KD3 females could not sustain normal flight and tended to lose flight stability and “crash” even after a seemingly normal take-off and while beating their wings (Figure 6I). In sum, these physiological studies demonstrate a role for circMbl in locomotion and flight.
Modulation of circMbl alters expression of brain- and muscle-related genes
We recently generated flies that allow overexpression (OE) of circMbl (Pamudurti et al., 2017). To identify genes affected by modulation of circMbl, we generated and sequenced 3′ RNA-seq libraries from heads of control, circMbl-KD, and circMbl-OE flies. There were 39 mRNAs differentially expressed in both circMbl-KD and circMbl-OE flies (Table S3). Thirty-five of these genes showed opposite trends in the OE and KD strains (Figure S6D). This group of genes is enriched for genes involved in muscle development and function.
The low number of differentially expressed genes in this experiment might be due to masking of changes in the brain by other tissues present in the fly head. Therefore, we utilized dissected fly brains of control and circMbl-KD flies to generate and sequence 3′ RNA-seq libraries. Indeed, we found 504 differentially expressed genes (adjusted p value <0.05 and fold change >1.5; Figure S7A and Table S4). Interestingly, genes involved in signal transduction and neuropeptide receptor activity were among the most enriched gene ontology terms within the genes downregulated upon circMbl-KD (Table S4). We found genes involved in neurotransmitter-gated ion-channel clustering, RNA localization, and protein metabolism among the upregulated genes (Table S4).
To gain insights into the cell types within the brain affected upon circMbl knockdown, we utilized the single-cell brain data to determine cell clusters enriched for these differentially expressed genes (see STAR Methods). By conducting this analysis we found that knockdown of circMbl altered the gene expression of genes particularly enriched in some glial clusters, peptidergic cells, clock neurons, Lawf2 cells, and photoreceptors (Figure 7A and Table S5). Most of the clusters in which we previously detected high mean or total levels of circMbl (Table S2) are among the ones more enriched for differentially expressed genes upon circMbl knockdown (see squares in Figure 7A).
Figure 7. Knockdown of circMbl and MBL-C results in locomotor defects.
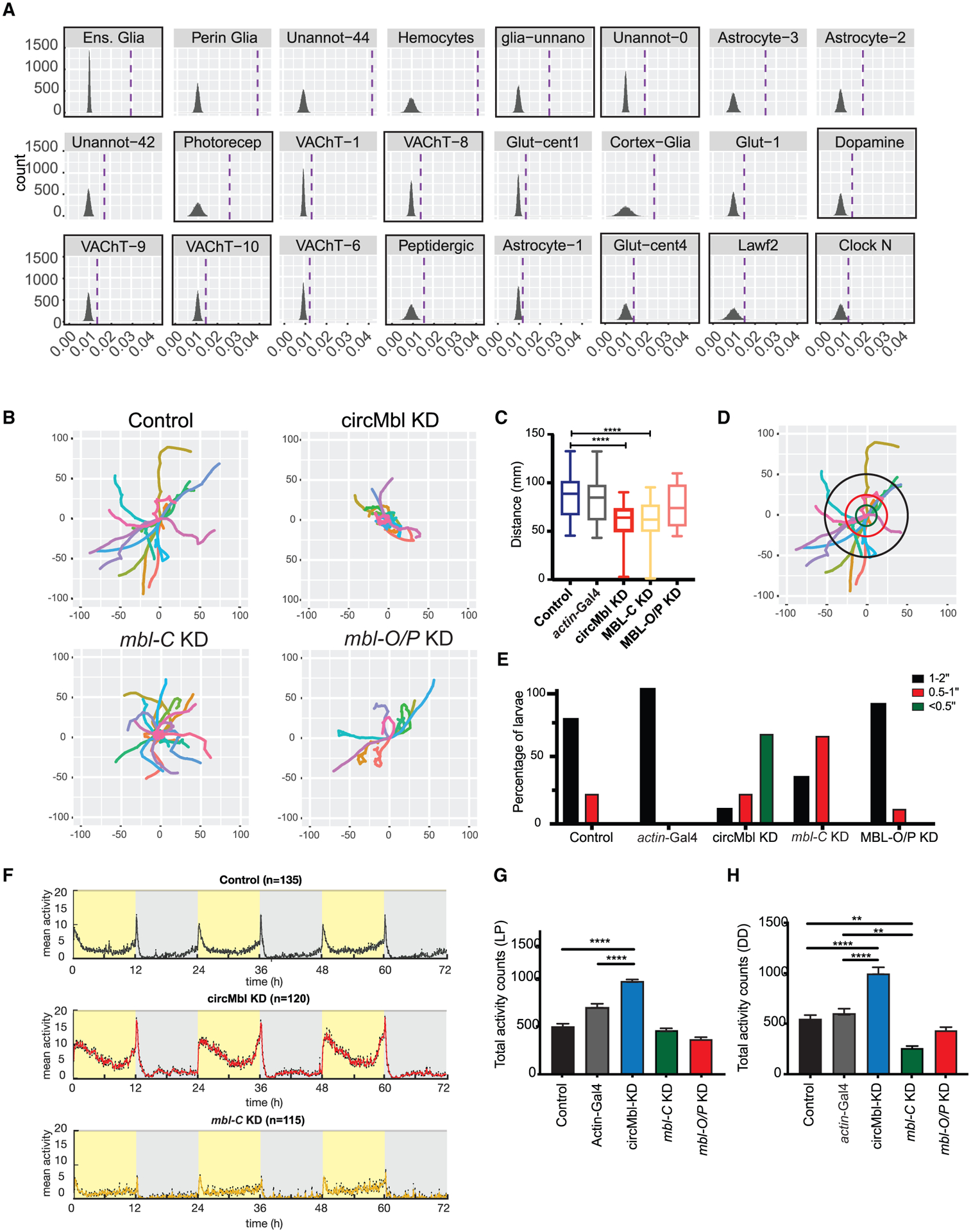
(A) Single-cell clusters with significant enrichment for genes differentially expressed in circMbl-KD brains. Dashed lines denote mean gene set enrichment in each cluster. Clusters with high levels of circMbl are highlighted with black squares.
(B) Path covered in larval assay for the indicated flies.
(C) Total distance (mm) traveled by each larva (three independent replicas, control n = 32, circMbl-KD n = 26, MBL-C-KD n = 32, MBL-O/P-KD n = 32). Boxplot shows mean, interquartile, and extreme values (two-tailed t test performed for significant difference: ****p < 0.0001, ***p < 0.0002, **p < 0.0021, *p < 0.0332).
(D) Representation of the analysis of the movement of control (8MM) larvae. The concentric circles are at distance of <0.5 inch (green), 0.5–1 inch (red), and 1–2 inches (black).
(E) Percentage of larvae that cross each of the concentric circles described in (D) (three independent experiments, number of larvae per experiment >10 when possible).
(F) Average activity over 3 days in 12:12 LD at 25°C. Light phase is represented in yellow and dark phase in gray (five independent replicas, control n = 135, circMbl-KD n = 120, MBL-C-KD n = 115, MBL-O/P-KD n = 96).
(G and H) Total activity during the light period in LD (G) and total activity over 5 days in complete darkness (DD) (H). Asterisks represent statistical significance relative to 8MM and Actin-Gal4 controls calculated by one-way ANOVA and Tukey’s multiple comparisons test (****p < 0.0001, **p < 0.005).
circMbl and MBL-C regulate locomotor activity in different ways
The potential involvement of peptidergic and circadian neurons suggests roles of circMbl in locomotor behavior. Therefore, we determined locomotor activity upon circMbl, mbl-C, or mbl-O/P knockdown. First, we used a simple assay to examine the larval locomotion behavior by placing a single larva at the center of an agar plate and following its movement. As expected, wildtype larvae move toward the edge of the plate (Figures 7B and S7B). Interestingly, knockdown of circMbl and mbl-C (but not mbl-O/P) significantly reduced the covered distance when compared with the controls (Figure 7C). Moreover, we observed that the locomotion defect observed in the circMbl- and mbl-C-KD larvae are different, with most mbl-C-KD displaying straight locomotion patterns while circMbl-KD larvae seem to “move in circles” (compare patterns in Figure 7B, and see Figures 7D and 7E). These results show that depletion of mbl-C and circMbl lead to different locomotor deficits in larvae.
We then evaluated daily locomotor activity patterns in adult flies from circMbl-KD, mbl-C, and mbl-O/P and control strains under 12 h light/12 h dark (LD) conditions. Surprisingly, depletion of circMbl resulted in a very strong (almost 2-fold) increase in locomotor activity during the light (Figures 7F, 7G, and S7C) but not during the dark period (Figure S7D). On the other hand, knockdown of mbl-C significantly diminished the activity during the dark but not the light period (Figures S7E and S7F). We did not detect significant changes in sleep or circadian rhythmicity (Table S6). In addition, we found that in constant darkness, modulation of mbl-C and circMbl alters locomotor activity in a significant and opposite way (Figures 7H, S7I, and S7J). This effect was very strong, with downregulation of circMbl increasing the levels of activity 2-fold and mbl-C-KD diminishing them more than 2-fold, while we did not see any effect when downregulating mbl-O/P (Figures 7H and S7K). These results demonstrate that mbl-C and circMbl work antagonistically to regulate the levels of locomotor activity.
DISCUSSION
Several autoregulatory mechanisms have been described in mammals for MBNL proteins: MBNL1 binds to its own first exon and prevents the generation of a fully functional protein by favoring exclusion of exon 1 (Konieczny et al., 2017) or by promoting the exclusion of exon 5, which has the nuclear localization signal (Kino et al., 2015). Here we show that the mbl locus utilizes the production of circRNAs as a way to balance/buffer its own levels in a cell-type-specific manner (which involves circMbl in the brain and circMbl2-5 in the eye).
Surprisingly, we found that MBL-C and MBL-O/P regulate circRNA production when overexpressed in the eye, despite the additional domains present in MBL-O/P. This can be explained by the fact that deletion studies in MBNL1 showed that the first two zinc fingers not only are enough to bind to the RNA but also to promote splicing of most targets (Edge et al., 2013; Purcell et al., 2012). It is possible that MBL-O/P has additional functions in the cytoplasm or in splicing regulation. In this context, it will be very useful to use CRISPR to generate flies which could only generate one of the isoforms.
In this work, we pioneer the use of single-cell sequencing data to determine the spatial expression of a circRNA. To this end, we modified available quantification tools and took advantage of the presence of a poly(A) track near the circMbl backsplicing junction. Unfortunately, this approach can be used only for few circRNAs: those that contain A tracks near the backsplicing junction. New technologies using either random or specific priming of regions close to backsplicing junctions would allow determination of single-cell expression of more circRNAs.
In addition, we found a good correlation between the levels of circMbl and mbl in the brain. We believe that this is due to the existence of a strong co-regulatory mechanism. Moreover, the fact that overexpression of MBL-C or O/P but not knockdown has such a deleterious effect strongly argues that the levels and/or activity of MBL should be maintained within a limited range. Additional perturbation experiments involving fast and transient overexpression of MBL would be very helpful to understand how quickly these mechanisms act.
While production of circMbl has a clear effect on the rate of mbl mRNA synthesis, this effect is mainly co-transcriptional, as we did not see changes in the levels of mbl mRNA and protein upon post-transcriptional knockdown of circMbl. Moreover, knocking down circMbl leads to specific phenotypes, some of them opposite to those generated by knockdown of the linear mbl counterpart. As circMbl knockdown is restricted to the cytoplasm, if circMbl has a strong role in antagonizing or promoting any function of MBL in the nucleus, it could not be assessed in this way. Further determination of this type of function needs the generation of new mbl mutants that cannot produce circMbl or which produce isoforms of circMbl that cannot bind MBL. Indeed, we attempted to mutate MBL binding sites flanking the circularizable exon in mbl but failed in obtaining viable and stable flies, likely because of deleterious changes in expression of mbl mRNA.
While the experiments presented herein demonstrate the functionality of circMbl both in cis and in trans, they provide little insight into the molecular mechanism by which this circRNA operates. However, the circMbl-KD flies constitute an excellent system to determine how circMbl potentially alters MBL function in vivo. Interestingly, the phenotypes observed upon circMbl knockdown do not directly correlate with those previously described while modulating MBL protein. All the above data suggest that while circMbl and MBL functions are tightly interconnected, the mechanism of interaction between these molecules is complex.
Limitations of the study
We acknowledge that this study has some limitations. For example, the fact we use shRNAs for knocking down circMbl precludes determining cis-related functions of circRNA production, which could be accomplished by generating flies that cannot produce circMbl. Moreover, as part of our study we could not perform rescue experiments, which could be key to identifying sequence and spatial requirements of circMbl.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Sebastian Kadener (skadener@brandeis.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed material transfer agreement.
Data and code availability
All next-generation sequencing data have been deposited at the GEO repository and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly strains and reagents
Fly strains
Wild type flies that we used in this study are yw and w1118 strains (Bloomington Drosophila Stock Center Indiana, USA). Elav-Gal4; UAS Dcr2 were generated by using elav-Gal4 (stock number 458, Bloomington Drosophila Stock Center, Indiana, USA) and UAS-Dcr2 flies. The circMbl OE strain is described in (Pamudurti et al., 2017). Mbl-C OE strain was kindly provided by Dr. Ruben D Artero (Department of Genetics, University of Valencia) Unless indicated otherwise, all crosses were performed and raised at 25°C in 12 h light-dark cycle (LD).
Generation of transgenic lines
To generate specific mbl linear and circRNA KD flies we designed oligonucleotides with perfect 21-nucleotide complementary sequence to the circRNA junction or the specific isoform targeted sequence (linear junction or exon sequence), annealed them, and ligated in to the linearized Valium20 vector with EcoR1 and Nhe1 restriction enzymes. Colonies were screened by PCR and the plasmid was purified and sequenced from positive colonies. These plasmids were sent for injection to BestGene Inc (CA, USA). A list of the RNAs targeted and a list of the oligonucleotides used for cloning is presented in Table S7. The presence of potential off-targets was verified by performing Blast against the fly genome and transcriptome. We did not observe any perfect complementary of 16 bases or more for any of the shRNAs. To generate mbl-C-FLAG OE and mbl-O/P-FLAG OE flies we used the cDNA from wildtype flies to amply the ORF of each isoform and then cloned in to the pUAS attB plasmid using the primers that mention in the Table S7. These plasmids were sent for the injection to BestGene Inc as mentioned above.
METHODS DETAILS
Molecular biology methods
Nascent RNA extraction
Nascent RNA was extracted from heads as described in (Khodor et al., 2011). Briefly, 3 days old fly heads were homogenized with a Dounce homogenizer, in the 300 uL homogenization buffer (10 mM Hepes-KOH pH 7.5, 10 mM KCL, 1.5 mm MgCl2, 0.8 M Sucrose, 0.5 mM EDTA, 1 mM DTT, 0.5 units/ul SuperaseIN, and protease inhibitor cocktail (mini complete, Roche)). Then the homogenized mixture was loaded on 350ul sucrose cushion buffer (10 mm Hepes-KOH pH 7.5, 10 mm KCl, 1.5 mM MgCl2, 1 M Sucrose, 10% Glycerol, 0.5 mM EDTA, 1 mM DTT, 0.5 units/ul SuperaseIN, and protease inhibitor cocktail (mini complete, Roche)). This mix was centrifuged at maximum speed for 10 min at 4°C. The supernatant, which will be the cytosolic fraction, was taken and prepared for RNA extraction using TRIZOL LS (ambion) as per instructions mentioned by the manufactured. The nuclei pellet was gently resuspend by pipetting 150ul of Nuclear lysis buffer (10 mM Hepes-KOH pH 7.6, 100 mM KCL, 0.1 M EDTA, 10% Glycerol, 0.15 mM Spermine, 0.5 mM Spermidine, 0.1 mM HaF, 0.1 mM Na3VO4, 0.1 mM ZnCl2, 1 mM DTT, 1 units/ul SuperaseIN, and protease inhibitor cocktail (mini complete, Roche)). Finally, 150 ul of NUN buffer were added and mixed gently. Then the sample was incubated on ice for 20 min and then centrifuged (maximum speed, 30 min 4°C). The supernatant containing the Nucleoplasm RNA was separated for RNA extraction with TRIZOL LS. The pellet containing the chromatin bound RNA was resuspended in Trizol (TRI reagent from Sigma) and incubated for 15 min at 65°C and then proceed with RNA extraction as mentioned in the manufacturer protocol. After RNA extraction from 3 different fractionations, cDNA was synthesized and qPCR was performed as mentioned in the methods above.
Nanopore sequencing
The probes mixture (1ug of each probe) was denatured at 85°C for 3 min, and placed on ice immediately. Probes mixture was added to total RNA, mixed well and incubated for 3 h at 37°C rotating. Streptavidin C1 beads from NEB (CAT# S1420S) were prepared. 100μL of streptavidin beads per samples were used, 4X washes with 10 mM Tris-HCl, pH 7.5, and then 2X washes with 1X Hybridization Buffer. Finally, the beads were resuspended in 450μL of Tris-HCl, pH 7.5. 50μL of resuspended beads were added to each sample, and incubated for 1 h at 37°C rotating. The beads were separated from the samples using a magnet, and then washed 5X in 8X bead volume (800μL) of 1X Hybridization buffer. For elution we added 100μL of elution buffer and then incubated for 10 min at 95°C. We transferred to fresh tube and extract RNA using TRI reagent (Sigma). Samples were treated with DNaseI (NEB) and performed the nanopore sequencing as instructed in the manual from oxford nanopore technology MinION Mk1B kit.
Real-time PCR
Total RNA was extracted from adult fly either heads or brains using TRI Reagent (Sigma) and treated with DNase I (NEB) following the manufacturer’s protocol. cDNA was synthesized from this RNA (using iScript and random primers, Bio-Rad) and was utilized as a template for quantitative real-time PCR performed with the C1000 Thermal Cycler Bio-Rad. The PCR mixture contained Taq polymerase (SYBR green Bio-Rad). Cycling parameters were 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 55°C for 10 s and 72°C for 30 s fluorescence intensities were plotted versus the number of cycles by using an algorithm provided by the manufacturer. Primer efficiency was determined for all primers described in this study and incorporated into the relative expression calculation. The sequences of all the primers used in this assay are detailed in Table S7.
Assessment of developmental lethality
Ten homozygous sh-circRNA male flies were crossed with 10 virgin female actin-Gal4 flies and transferred to new bottles every 3 days. The F1 progeny was separated based on their genotype (indicated by the presence of the marker/balancer CyO) and the males and females fly were counted. We performed this assessment for each bottle for 9 days or until the totality of the F1 eclose.
RNA libraries preparation for RNA-seq analysis
Total RNA was extracted using Trizol reagent (Sigma) and treated with DNase I (NEB) following the manufacturer’s protocol. Stranded ligation-based, total-RNA libraries preparation was modified from (Engreitz et al., 2013) as follows: For PolyA + libraries, 0.5 μg of total RNA was polyA + selected (using Oligo(dT) beads, Invitrogen), fragmented in FastAP buffer (Thermo Scientific) for 3 min at 94°C, then dephosphorylated with FastAP, cleaned (using SPRI beads, Agencourt) and ligated to a linker1 (5Phos/AXXXXXXXXAGATCGGAAGAGCGTCGTGTAG/3ddC/, where XXXXXXXX is an internal barcode specific for each sample), using T4 RNA ligase I (NEB). Ligated RNA was cleaned-up with Silane beads (Dynabeads MyOne, Life Technologies) and pooled into a single tube. RT was then performed for the pooled sample, with a specific primer (5′-CCTACACGACGCTCTTCC-3′) using AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent Technologies). Then, RNA-DNA hybrids were degraded by incubating the RT mixture with 10% 1 M NaOH (e.g. 2ul to 20ul of RT mixture) at 70°C for 12 min pH was then normalized by addition of corresponding amount of 0.5 M AcOH (e.g. 4ul for 22 ul of NaOH + RT mixture). The reaction mixture was cleaned up using Silane beads and second ligation was performed, where 3′end of cDNA was ligated to linker2 (5Phos/AGATCGGAAGAGCACACGTCTG/3ddC/) using T4 RNA ligase I. The sequences of linker1 and linker2 are partially complementary to the standard Illumina read1 and read2/barcode adapters, respectively. Reaction Mixture was cleaned up (Silane beads) and PCR enrichment was set up using enrichment primers 1 and 2:
5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′, 5′-CAAGCAGAAGACGGCATACGAGATXXXXXXXXGTGACTGGAGTTCAGAC
GTGTGCTCTTCCGATCT-3′, where XXXXXXX is barcode sequence) and Phusion HF MasterMix (NEB). 12 cycles of enrichment were performed. Libraries were cleaned with 0.7X volume of SPRI beads. Libraries were characterization by Tapestation. RNA was sequenced as paired-end samples, in a NextSeq 500 sequencer (Illumina).
rRNA− libraries were similarly prepared, without the polyA + selection step: 0.25 μg of total RNA from each sample were fragmented in FastAP buffer (Thermo Scientific) for 3 min at 94°C, then dephosphorylated with FastAP, cleaned and ligated to linker1 using T4 RNA ligase I (NEB). Ligated RNA was cleaned-up with Silane beads (Dynabeads MyOne, Life Technologies) and pooled into a single tube. 1/4 of the pooled sample was rRNA depleted using Ribo-Zero rRNA removal kit (epicentre). Unbound RNA (rRNA−RNA) was cleaned (using SPRI beads) and reverse transcribed, ligated to linker2 and enriched by PCR as described above (for total PolyA + libraries).
For digital 3′ gene expression, library preparation was similar to the total RNA (PolyA+) libraries described above, with one exception: PolyA + selection was not done before fragmentation, but after linker1 ligation and samples pooling (before the RT reaction step).
Western blot analysis
Fly heads (20 heads per sample) were collected on dry ice. Heads were homogenized in RIPA lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 0.5% Sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS), 1 mM DTT, supplemented by protease inhibitor cocktail and phosphatase inhibitors) using motorized pestle. Head lysates were then centrifuged at maximum speed for 10 min and the supernatant was saved. lysates were boiled with protein sample buffer (Bio-Rad) and resolved by Criterion XT Bis-Tris gels (Bio-Rad). Antibodies used for western blotting: sheep Anti-mbl antibody was kindly provided by Prof. Darren Monckton (School of Life Sciences, University of Glascow), mouse anti-tubulin (DM1A; SIGMA, 1:30,000). Western blots quantification performed using ImageJ and provided the values at bottom of each blot.
AGO1-seq procedure
AGO1 immunoprecipitation was performed as previously described (Kadener et al., 2009; Lerner et al., 2015). The RNA-seq libraries were performed utilizing the Ovation RNA-seq System for model organisms.
Physiological and behavioral assessments
Larval locomotion
The larval assay was performed with actin Gal4/CyO GFP flies generated by crossing the actin Gal4 to UAS-CyO GFP flies. The ubiquitous knockdown of circMbl and MBL-C was achieved by crossing the UAS-shcircMbl and UAS-shMBL-C lines to actin-Gal4/CyO-GFP. The control for these experiments was 8MM and actin-Gal4.
The cross was set up in standard agar-yeast media and raised at 25C. Third-instar larvae expressing no GFP were picked up gently using a brush under a stereo microscope fitted with an external florescence filter. An individual third-instar larvae was washed with distilled water and placed in the center of a 2% agar plate (1 mm thickness). The illumination was provided from the bottom of the agar plate using a light pad pro and the activity of the larva was recorded for 2 min using an iPod. The video was then trimmed to 1 min and analyzed using MATLAB (Mathworks Inc, Natick, MA, USA) based software Ctrax (Branson et al., 2009) a multi-fly tracker (http://ctrax.sourceforge.net/). We used custom scripts on R-studio to generate the trajectories of larval crawling to the periphery of the agar plate.
Locomotor activity and sleep measurements
Male flies were monitored using Trikinetics Drosophila Activity Monitors (DAM) using 1-min bins. Each fly was placed into a glass tube containing 2% agarose and 5% sucrose food. Flies were entrained for 4 days in 12:12 Light: Dark cycles (LD) and 7 days in constant darkness (DD). All the experiments were performed at 25°C. Analyses were performed with a signal processing toolbox (Levine et al., 2002). All the activity assessments were done in LD. For the DD assessments, flies were considered rhythmic if the rhythm index was greater than 0.20 for the first 5 days in DD, weakly rhythmic if 0.1 < RI < 0.2 and arrhythmic if RI < 0.1. The sleep parameters of Activity and sleep were analyzed using the MATLAB script S.C.A.M.P.
Flight analysis
In both the tapping and free-flight assays we used a Phantom v2012 fast camera (Vision Research, NJ) oriented horizontally and back-illuminated by a near-infrared LED. The camera operated at 10,000 frames per second and resolution of 1280 × 800 pixels. Triggering was performed manually. In the tapping assay, groups of ~5 files we placed in a bottle with flat sides to allow undistorted imaging. Once the flies climbed to the top of the bottle, we tapped it down against the bench, which caused the flies to fall. Overall, we recorded 119 flies during their fall and measured whether they flapped their wings or not. In the free-flight assay we placed an individual fly on a pipette tip or a thin wire and allowed it to take off freely. The fly was released inside a transparent Plexiglas cubic container, with side length of ~20 cm, to allow free flight far from any solid boundary. The flapping frequency of 131 flies was manually extracted from the videos.
QUANTIFICATION AND STATISTICAL ANALYSIS
Computational analysis
RNAseq and ribosome footprinting data for mbl isoform annotation and quantification
Data was downloaded from GEO (Series GSE79626: Pamudurti et al., 2017 and GSE124134 all time point at 25C: Martin Anduaga et al., 2019) and aligned to Drosophila melanogaster dm6 genome and transcriptome version using STAR (Dobin et al., 2013). Gene expression and splice junctions were quantified using featurecounts implementation in Rsubread (Liao et al., 2019). Specific splice junctions were used as a proxy of each alternative exon1 usage, mblC and mblO/P. To evaluate the proportion of each 3′UTR usage (mblA, mblB, mblMi, mblC/O/P) we quantified the reads aligning to 74 bases after each stop codon. Along this paper we called mbl-C the transcript that gives origin to the reported protein MBLC to maintain clarity and consistency with the literature. However, this transcript is called mbl-RD in flybase and UCSC genome browser.
Nanopore data processing and visualization for mbl 5′UTR annotation
Nanopore reads were demultiplexed using EPI2ME software from Nanopore and aligned using minimap2 (Li, 2018) aligner and dm6 genome. Integrative genome viewer (IGV) was used for visualization.
Differential gene expression from brain data
RNA was aligned to Drosophila melanogaster dm6 genome and transcriptome version using STAR (Dobin et al., 2013). Gene expression was countified with ESAT (Derr et al., 2016). Only samples with more than 3 million reads were kept for further analysis. Deseq2 was used for normalization and differential gene expression. Genes with p value adjusted less than 0.05 and fold-change more than 1.5 were selected as significant. Actin-Gal4 UAS-8-missmatch flies were used as a control.
circMbl and mbl-C/O/P annotation for polyA selected libraries (bulk and single cell)
The sequences of relevant mbl exons were extracted from UCSC genome browser and added to the dm6 genome version of Drosophila melanogaster with the original mbl gene region deleted. This allowed us to quantify reads coming from exon2 and the 3′UTR region. To specifically annotate circRNA junction, the junction sequence was generated manually and added to the genome. The number of bases surrounding the junction was selected to guarantee the mapping of only circRNA reads, for example if the read length were 50 bases, then we selected 45 bases to each side of the circRNA junction. The proper read mapping was checked manually by inspecting the alignment in IGV genome browser.
Single cell analysis
Raw data was downloaded from geo (Series GSE107451) and aligned using the modified dm6 genome with 3′UTR region and either exon2 or circMbl sequence. To avoid counting multimapping reads we used a modified version of 10X genomic’s cellranger in which the multimapping option was removed (https://github.com/inespatop/cellranger).
Single cell clustering was done using Seurat R package version 3.0.2 (Butler et al., 2018). Data was log normalized. Clustering was done using 30 dimensions and a resolution of 6 UMAP was used for visualization. Cluster assignment was done using marker genes from previous publications and subsequent label transfer between different alignments using CCA as in (Butler et al., 2018).
Gene set enrichment in single cell data
To analyze enrichment of genes in a particular single cluster the function “AddModuleScore” from Seurat was used and then the scores were averaged per cell. To see the significance of the score a null distribution was created by randomizing the cluster labels over the cells 10.000 times. Each time the mean per cluster was taken and a distribution was created. Considering the Central Limit Theorem, the p value was calculated for a normal distribution of the mean. FDR was used to account for multiple comparison.
Head cell-type mbl and circMbl isoforms expression
Raw data was downloaded from GEO (Series GSE116969 (Davis et al., 2020), and aligned to Drosophila melanogaster dm6 genome and transcriptome version using STAR (Dobin et al., 2013). Gene expression and splice junctions were quantified using featurecounts implementation in Rsubread (Liao et al., 2019). Specific splice junctions were used as a proxy of each alternative exon1 usage, mblC and mblO/P. circRNA annotation and quantification was done using the latest version of find_circ (Memczak et al., 2013) combined with ciRcus R package for quantification and annotation (https://github.com/BIMSBbioinfo/ciRcus). All reads were normalized using DeSeq2 implementation of library depth normalization.
Fly head RNA seq data
RNA-seq reads were aligned to the genome and transcriptome (dm3) using tophat (Trapnell et al., 2009). circRNA detection in RNA-seq data was performed as previously described (Memczak et al., 2013).
CircRNA expression levels were determined by counting back-spliced reads and normalizing to total number of reads. Similarly, for linear RNA expression we used the number of reads from the linear exon-exon junctions from both sides of the circRNA boundaries. samtools-depth tool was used for counting reads within exons.
Differential exon usage analysis was performed using DEXseq (Anders et al., 2012). The analysis was done using polyA selected library data. UAS-shcircMbl flies were used as a control.
For differential gene expression analysis in circMbl KD with CNS-specific driver (elav-Gal4;UAS-Dcr2) we used total RNA-seq data. Gene expression levels were determined using HT-seq tool and differential expression analysis was performed with DEseq. Flies expressing shRNA against Luciferase (UAS-shLuc) under the same promoter were used a control. We considered genes with p value<0.05 as significantly changing.
Gene expression levels from 3′ DGE experiments were determined using ESAT tool (Derr et al., 2016) and differential expression analysis was performed with DEseq. We considered genes with fold change>1.5 and p value<0.05 as significantly changing. Actin-Gal4 flies were used as a control for the lines expressing shRNA under actin promoter. In order to clean non-specific effects, we excluded from downstream analyses genes that were changing in similar direction when comparing the actin-Gal4 control flies and circMbl 8MM KD line. elav-Gal4 flies were used as a control for the lines expressing shRNA under elav promoter.
SYLAMER algorithm (van Dongen et al., 2008) was used to check for general off-target effect of the shRNA. In order to obtain a list of potential shRNA off target genes we blast all shRNA sequences against the drosophila transcriptome. 3′ RNA-seq data was used to determine the expression level of each putative off-targets gene relative to control line.
Gene ontology analysis
For all analysis topGO package (Alexa and Rahnenfuhrer, 2021) was used for enrichment analysis of gene ontology. GO terms with p value <0.1 (after FDR correction) were considered significant.
AGO-1 enrichment analysis
For the analysis of the AGO1-seq we aligned the RNA-seq reads to the genome and transcriptome of Drosophila melanogaster (dm6 version) using STAR (Dobin et al., 2013). Counts per gene were obtained using HTSeq. To assess the distribution of AGO1 immunoprecipitation results we did a Kernel density plot in log2 scale for the mean of normalized gen counts for IP over the normalized gen counts for the input (INP). The AGO1-IP enrichment analysis was done by a negative binomial GLM approach using DeSeq2 (Love et al., 2014). For this analysis we compared INP an IP counts for control and sh-circMbl flies independently. To further compare these results from both control and sh-Mbl we did a Likelihood Ratio Test (LTR) comparing a simpler model that only considers IP and input as factors with another more complex that includes also an interaction between the IP results and the genetic background. To see possible sh-circMbl off-target effects, we analyzed 6 nt kmer enrichment in the 3′UTR of the genes using Sylamer algorithm (van Dongen et al., 2008). For this, we ranked the gene list by Log2 fold change multiplied by the inverse of the p adjusted value (log2FC*1/(pval)) the results of the IP enrichment analysis.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-MBL | Lab of Prof. Darren Monckton PMID:17309604 | N/A |
| mouse monoclonal Anti-FLAG | Sigma-Aldrich | CAT# F3165; RRID:AB_259529 |
| mouse monoclonal DM1A anti α-tubulin | Sigma-Aldrich | CAT# T6199; RRID: AB_477583 |
| Deposited data | ||
| Ago-IP of circMbl KD and control fly heads | This paper | GEO: GSE118360 |
| 3′RNA seq of KD and OE circMbl fly heads | This paper | GEO: GSE122694 |
| RNAseq from KD of circMbl using UAS-circMbl in combination with different Gal4 | This paper | GEO: GSE122693 |
| Nanopore sequencing from Mbl-exon2 pulldown | This paper | GEO: GSE163780 |
| 3′RNA seq from sh-circMbl Actin-Gal4 KD fly brains | This paper | GEO: GSE163797 |
| Fly brain single cell RNA sequencing | Davie et al. (2018) | GEO: GSE107451 |
| nuclear total RNA-seq libraries prepared from specific cell types using INTACT/TAPIN | Davis et al., (2020) | GEO: GSE116969 |
| Ribosome footprinting data | Pamudurti et al. (2017) | GEO: GSE79626 |
| RNA sequencing | Martin Anduaga et al., 2019 | GEO: GSM3523858 to GSM3523869 (all timepoints at 25C from series GSE124136) |
| Drosophila melanogaster genome and transcriptome build 6, Dm6 | Ensembl | http://ftp.ensembl.org/pub/release-104/fasta/drosophila_melanogaster/dna/ and http://ftp.ensembl.org/pub/release-104/gtf/drosophila_melanogaster |
| Experimental models: Cell lines | ||
| D. melanogaster: Cell line S2: S2-DRSC | N/A | N/A |
| Experimental models: Organisms/strains | ||
| D. melanogaster: w1118 | Bloomington Drosophila stock center | BDSC: 5905 FlyBase:FBal0018186 |
| D. melanogaster: yv1 | Bloomington Drosophila stock center | N/A |
| D. melanogaster: Actin-Gal4: w[1118]; P{w [+mC] = AyGAL4}25/CyO | Bloomington Drosophila stock center | BDSC: 3953 FlyBase:FBti0012290 |
| D. melanogaster: Elav-Gal4 | Bloomington Drosophila stock center | BDSC: 458 FBgn0260400 |
| D. melanogaster: UAS-Dcr2 | Bloomington Drosophila stock center | N/A |
| D. melanogaster: UAS-circMbl-OE | Pamudurti et al. (2017) PMID: 28344080 | N/A |
| D. melanogaster: UAS-MBL-C-FLAG OE | This paper | N/A |
| D. melanogaster: UAS-MBL-O/P-FLAG OE | This paper | N/A |
| D. melanogaster: Scrambled Control (8MM) | This paper | N/A |
| D. melanogaster: MBL-A-KD1 | This paper | N/A |
| D. melanogaster: MBL-A-KD2 | This paper | N/A |
| D. melanogaster: MBL-B-KD1 | This paper | N/A |
| D. melanogaster: MBL-B-KD2 | This paper | N/A |
| D. melanogaster: MBL-C-KD1 | This paper | N/A |
| D. melanogaster: MBL-C-KD2 | This paper | N/A |
| D. melanogaster: MBL-O/P-KD1 | This paper | N/A |
| D. melanogaster: MBL-O/P-KD2 | This paper | N/A |
| D. melanogaster: MBL-UTR-KD1 | This paper | N/A |
| D. melanogaster: MBL-UTR-KD2 | This paper | N/A |
| D. melanogaster: MBL-M/I-KD1 | This paper | N/A |
| D. melanogaster: MBL-M/I-KD2 | This paper | N/A |
| D. melanogaster: MbL-E1-KD1 | This paper | N/A |
| D. melanogaster: MbL-E1-KD2 | This paper | N/A |
| D. melanogaster: MbL-E2-KD1 | This paper | N/A |
| D. melanogaster: MbL-E2-KD2 | This paper | N/A |
| D. melanogaster: MbL-E2–3-KD1 | This paper | N/A |
| D. melanogaster: MbL-E2–3-KD2 | This paper | N/A |
| D. melanogaster: circMbl KD1 | This paper | N/A |
| D. melanogaster: circMbl KD2 | This paper | N/A |
| D. melanogaster: circMbl KD3 | This paper | N/A |
| Oligonucleotides | ||
| Oligos for cloning, and qPCR | This paper | Table S7 |
| Recombinant DNA | ||
| UAS-MBL-C-FLAG OE | This paper | N/A |
| UAS-MBL-O/P-FLAG OE | This paper | N/A |
| Software and algorithms | ||
| tophat | Trapnell et al. (2009) | https://ccb.jhu.edu/software/tophat/index.shtml |
| Samtools | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| SYLAMER algorithm | van Dongen et al. (2008) | https://www.ebi.ac.uk/research/enright/software/sylamer |
| Find_circ | Memczak et al. (2013) | https://github.com/marvin-jens/find_circ |
| DEXseq | Anders et al. (2012) | https://bioconductor.org/packages/release/bioc/html/DEXSeq.html |
| DEseq | https://www.bioconductor.org/packages//2.10/bioc/html/DESeq.html | |
| DEseq2 | Love et al. (2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ESAT tool | Derr et al. (2016) | https://github.com/garber-lab/ESAT |
| STAR | Dobin et al. (2013) | https://github.com/alexdobin/STAR |
| Seurat R package version 3.0.2 | Butler et al. (2018) | https://satijalab.org/seurat/articles/install.html |
| Minimap2 | (Li, 2018) | https://github.com/lh3/minimap2 |
| Rsubread | (Liao et al., 2019) | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| ciRcus | https://github.com/BIMSBbioinfo/ciRcus | |
Highlights.
mbl-C and mbl-O/P are cell-type specific and mutually exclusive
mbl-C and -O/P form self-regulatory loops with different circMbl isoforms
Each circMbl functions in cis by competing with endogenous linear mbl in vivo
circMbl has functions in trans related to muscle and brain function
ACKNOWLEDGMENTS
This work was funded by the NIH R01 grant (R01GM124406) to S.K.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110740.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
REFERENCES
- Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, and Akhtar A (2017). DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119. 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- Alexa A, and Rahnenfuhrer J (2021). topGO: Enrichment analysis for Gene Ontology. R package version 2.10.0. [Google Scholar]
- Anders S, Reyes A, and Huber W (2012). Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017. 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, and Baylies MK (1998). The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev. Biol 195, 131–143. 10.1006/dbio.1997.8833. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, and Kadener S (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66. 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, Goodwin M, Zhang C, Sobczak K, Thornton CA, and Swanson MS (2014). Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell 56, 311–322. 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Paricio N, Artero R, Kiss I, Pérez-Alonso M, and Mlodzik M (1997). muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development 124, 4321–4331. [DOI] [PubMed] [Google Scholar]
- Branson K, Robie AA, Bender J, Perona P, and Dickinson MH (2009). High-throughput ethomics in large groups of Drosophila. Nat. Methods 6, 451–457. 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena C, and Hur S (2017). Antiviral immunity and circular RNA: no end in sight. Mol. Cell 67, 163–164. 10.1016/j.molcel.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia GG, et al. (2012). Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450. 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, and Chang HY (2017). Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5. 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al. (2019). N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9. 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, and Goodall GJ (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Cortes-Lopez M, Gruner MR, Cooper DA, Gruner HN, Voda AI, van der Linden AM, and Miura P (2018). Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genomics 19, 8. 10.1186/s12864-017-4386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, Aibar S, Makhzami S, Christiaens V, Bravo Gonzalez-Blas C, et al. (2018). A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174, 982–998.e20. 10.1016/j.cell.2018.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FP, Nern A, Picard S, Reiser MB, Rubin GM, Eddy SR, and Henry GL (2020). A genetic, genomic, and computational resource for exploring neural circuit function. Elife 9. 10.7554/eLife.50901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr A, Yang C, Zilionis R, Sergushichev A, Blodgett DM, Redick S, Bortell R, Luban J, Harlan DM, Kadener S, et al. (2016). End Sequence Analysis Toolkit (ESAT) expands the extractable information from single-cell RNA-seq data. Genome Res. 26, 1397–1410. 10.1101/gr.207902.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, and Yang BB (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge C, Gooding C, and Smith CW (2013). Dissecting domains necessary for activation and repression of splicing by Muscleblind-like protein 1. BMC Mol. Biol 14, 29. 10.1186/1471-2199-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, and Lander E (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X-chromosome. Science. 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, et al. (2017). FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun 8, 14741. 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, and Brook JD (2002). Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet 11, 805–814. 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, and Zamore PD (2007). Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130, 287–297. 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goers ES, Purcell J, Voelker RB, Gates DP, and Berglund JA (2010). MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 38, 2467–2484. 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, and Miura P (2016). CircRNA accumulation in the aging mouse brain. Sci. Rep 6, 38907. 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, and Pandolfi PP (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165, 289–302. 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Hanan M, Simchovitz A, Yayon N, Vaknine S, Cohen-Fultheim R, Karmon M, Madrer N, Rohrlich TM, Maman M, Bennett ER, et al. (2020). A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med 12, e11942. 10.15252/emmm.201911942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, and Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun 7, 12429. 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Wahlin KJ, McNally M, Irving ND, and Adler R (2008). Developmental regulation of muscleblind-like (MBNL) gene expression in the chicken embryo retina. Dev. Dyn 237, 286–296. 10.1002/dvdy.21408. [DOI] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, and Wilusz JE (2018). A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 32, 639–644. 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, and Rajewsky N (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177. 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Jeck WR, and Sharpless NE (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol 32, 453–461. 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, and Thornton CA (2004). Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet 13, 3079–3088. 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, and Rosbash M (2009). A role for microRNAs in the Drosophila circadian clock. Genes Dev. 23, 2179–2191. 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, and Swanson MS (2003). A muscleblind knockout model for myotonic dystrophy. Science 302, 1978–1980. 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, and Rosbash M (2011). Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 25, 2502–2512. 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Kurosawa M, Oma Y, Hattori N, Ishiura S, and Nukina N (2015). Nuclear localization of MBNL1: splicing-mediated autoregulation and repression of repeat-derived aberrant proteins. Hum. Mol. Genet 24, 740–756. 10.1093/hmg/ddu492. [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, and Bartel DP (2018). A network of non-coding regulatory RNAs acts in the mammalian brain. Cell 174, 350–362.e17. 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knupp D, Cooper DA, Saito Y, Darnell RB, and Miura P (2021). NOVA2 regulates neural circRNA biogenesis. Nucleic Acids Res. 49, 6849–6862. 10.1093/nar/gkab523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny P, Stepniak-Konieczna E, Taylor K, Sznajder LJ, and Sobczak K (2017). Autoregulation of MBNL1 function by exon 1 exclusion from MBNL1 transcript. Nucleic Acids Res. 45, 1760–1775. 10.1093/nar/gkw1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, and Wilusz JE (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29, 2168–2182. 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Li M, Manchanda M, Batra R, Charizanis K, Mohan A, Warren SA, Chamberlain CM, Finn D, Hong H, et al. (2013). Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med 5, 1887–1900. 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Chang HC, Seah C, and Lee LJ (2019). Deprivation of muscle-blind-like proteins causes deficits in cortical neuron distribution and morphological changes in dendritic spines and postsynaptic densities. Front. Neuroanat 13, 75. 10.3389/fnana.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. (2017). Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell 66, 22–37.e29. 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner I, Bartok O, Wolfson V, Menet JS, Weissbein U, Afik S, Haimovich D, Gafni C, Friedman N, Rosbash M, and Kadener S (2015). Clk post-transcriptional control denoises circadian transcription both temporally and spatially. Nat. Commun 6, 7056. 10.1038/ncomms8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, and Hall JC (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JSS, and Millard SS (2019). Deterministic splicing of Dscam2 is regulated by Muscleblind. Sci. Adv 5, eaav1678. 10.1126/sciadv.aav1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 (16), 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, and Chen LL (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. cell 67, 214–227.e7. 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, et al. (2019). Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e1. 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukáš Z, Falk M, Feit J, Souček O, Falková I, Štefančíková L, Janoušová E, Fajkusová L, Zaorálková J, and Hrabálková R (2012). Sequestration of MBNL1 in tissues of patients with myotonic dystrophy type 2. Neuromuscul. Disord 22, 604–616. 10.1016/j.nmd.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ (2013). Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet 4, 307. 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Anduaga A, Evantal N, Patop IL, Bartok O, Weiss R, and Kadener S (2019). Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. Elife 8. 10.7554/eLife.44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405–407. 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. (2017). Translation of CircRNAs. Mol. Cell 66, 9–21.e27. 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Patop IL, Krishnamoorthy A, Ashwal-Fluss R, Bartok O, and Kadener S (2020). An in vivo strategy for knockdown of circular RNAs. Cell Discov. 6, 52. 10.1038/s41421-020-0182-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patop IL, Wust S, and Kadener S (2019). Past, present, and future of circRNAs. EMBO J, e100836. 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- Purcell J, Oddo JC, Wang ET, and Berglund JA (2012). Combinatorial mutagenesis of MBNL1 zinc fingers elucidates distinct classes of regulatory events. Mol. Cell Biol 32, 4155–4167. 10.1128/MCB.00274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885. 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S, Abreu-Goodger C, and Enright AJ (2008). Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods 5, 1023–1025. 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, and Kjems J (2015). Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16, 245. 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Monferrer L, Poulos MG, Houseley J, Monckton DG, O’dell KM, Swanson MS, and Artero RD (2007). Muscleblind isoforms are functionally distinct and regulate alpha-actinin splicing. Differ. Res. Biol. Divers 75, 427–440. 10.1111/j.1432-0436.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- Vicente-Crespo M, Pascual M, Fernandez-Costa JM, Garcia-Lopez A, Monferrer L, Miranda ME, Zhou L, and Artero RD (2008). Drosophila muscleblind is involved in troponin T alternative splicing and apoptosis. PLoS One 3, e1613. 10.1371/journal.pone.0001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. (2012). Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724. 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, and Salzman J (2014). Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9, e90859. 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, and Lai EC (2014). Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980. 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641. 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci 18, 603–610. 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, and Yang L (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zimmerman AJ, Hafez AK, Amoah SK, Rodriguez BA, Dell’Orco M, Lozano E, Hartley BJ, Alural B, Lalonde J, Chander P, et al. (2020). A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol. Psychiatry 25, 2712–2727. 10.1038/s41380-020-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All next-generation sequencing data have been deposited at the GEO repository and are publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


