Abstract
Cln Three Requiring 9 (CTR9), a scaffold protein of the polymerase-associated factor-1 (PAF1) complex (PAF1c), is primarily localized in the nucleus of cells. Recent studies show that CTR9 plays essential roles in the development of various human cancers and their occurrence; however, its regulatory roles and precise mechanisms in hepatocellular carcinoma (HCC) remain unclear. In this study, we investigated the roles of CTR9 using in vitro assays and a xenograft mouse model. We found that CTR9 protein is upregulated in tumor tissues from HCC patients. Knockdown of CTR9 substantially reduced HCC cell proliferation, invasion, and migration, whereas its overexpression promoted these activities. In addition, in vitro results revealed that CTR9 silencing dramatically increased cell cycle regulators, p21 and p27, but markedly decreased matrix metalloproteinases, MMP2 and MMP9, with these outcomes reversed upon CTR9 overexpression. Furthermore, the underlying molecular mechanism suggests that CTR9 promoted the oncogene paternally expressed gene 10 (PEG10) transcription via its promoter region. Finally, the oncogenic roles of CTR9 were confirmed in a xenograft mouse model. This study confirms that CTR9, an oncoprotein that promotes HCC cell proliferation, invasion, and migration, increases tumor growth in a xenograft mouse model. CTR9 could be a novel therapeutic target. Further investigation is warranted to verify CTR9 potential in novel therapies for HCC.
Keywords: hepatocellular carcinoma, Cln Three Requiring 9, paternally expressed gene 10, cell cycle, matrix metalloproteinase
Introduction
Hepatocellular carcinoma (HCC), a malignant tumor of the digestive system that is common worldwide, is the third leading cause of death among malignant tumors of the digestive system [1, 2]. Its mortality rate is the third-highest among malignant tumors [3]. Despite considerable advances in diagnosis and treatment, the prognosis of HCC patients remains poor [4]; therefore, it is crucially important to study the mechanisms underlying HCC to prolong patient survival.
Cln Three Requiring 9 (CTR9) is a scaffold protein for the polymerase-associated factor-1 (PAF1) complex (PAF1c), which comprises five core proteins: PAF1, RTF1, CTR9, CDC73, and LEO1 [5–7]. Studies show that CTR9 plays significant roles in cellular activities, including transcriptional regulation, methylation modification, protein transport, and cellular differentiation [8–10]. A recent study reported that CTR9 is involved in maintaining normal telomerase RNA levels and regulating telomere length, with a subsequent study suggesting that CTR9 plays an important role in retaining low levels of telomeric repeat-containing RNA [11, 12]. In addition, CTR9 participates in regulating the immune response by inhibiting T helper 17 cell differentiation and regulating the expression of interleukin-17 [13–15]. Although CTR9 reportedly affects the progression of Wilms’ tumor and breast cancer [16, 17], its roles in tumorigenesis and development, as well as the underlying mechanisms, remain poorly understood, particularly in HCC.
Paternally expressed gene 10 (PEG10) is a genetic imprinting gene that participates in the development of several tumors by promoting cell proliferation, inhibiting apoptosis, and inducing tumor cell invasion and migration [18–21]. Indeed, elevated PEG10 expression has been reported in multiple malignancies, including B-cell chronic lymphocytic leukemia, lung cancer, pancreatic cancer, and HCC [20, 22–24], and PEG10 upregulation in such malignancies are associated with cell proliferation and metastasis.
In this study, we investigated the expression of CTR9 in HCC tissues and the effect of CTR9 protein level on the proliferation, invasion, and migration of HCC cells. Results suggest that CTR9 accelerates HCC cell proliferation, invasion, and migration by facilitating PEG10 transcription.
Materials and methods
Tissues
Human non-tumorous liver and HCC tissue samples were provided by the Affiliated Hospital of Xuzhou Medical College (Xuzhou, China). HCC tissue specimens were obtained from patients undergoing surgical resection due to HCC, and normal liver specimens were obtained from patients undergoing partial hepatectomy to treat liver rupture due to trauma. All tissue samples were immediately frozen in liquid nitrogen and stored at −80 °C. This study was approved by the ethics research committee of the Affiliated Hospital of Xuzhou Medical University (approval no. XYFY2019-KL129-01), and all samples were obtained with patient consent.
Antibodies and plasmids
Antibodies against CTR9 were purchased from Cell Signaling Technology (Cell Signaling Technology, Danvers, MA, USA). Antibodies against PEG10, p21, p27, MMP2, MMP9, and β-actin were purchased from Proteintech (Wuhan, Hubei, China). Scramble short-hairpin (sh)RNA, shRNA targeting CTR9 (shCTR9), and the 3 × FLAG-Vector, 3 × FLAG-CTR9, Myc-Vector, and Myc-PEG10 plasmids were purchased from Youbio (Changsha, Hunan, China).
Cell culture
The HEK293T cell line and the HepG2 and Huh7 HCC cell lines were purchased from the Cell Bank of the Shanghai Institutes of Chinese Academy of Sciences (Shanghai, China), and were cultured in an incubator at 37 °C and 5% CO2 with (Minimum Essential Medium or Dulbecco’s Modified Eagle’s Medium) (Yuanpei, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Gibco, Shanghai, China).
Lentivirus construction
Three plasmids were constructed using primers to silence CTR9: shCTR9#1-F, 5ʹ gat ccg CCT CCA GAG ATT CTC AAT AAT TTC AAG AGA ATT ATT GAG AAT CTC TGG AGG TTT TTT g 3ʹ, and shCTR9#1-R, 5ʹ aat tcA AAA AAC CTC CAG AGA TTC TCA ATA ATT CTC TTG AAA TTA TTG AGA ATC TCT GGA GGc g 3ʹ; shCTR9#2-F, 5ʹ gat ccg CGT GCA AAT GAG ACT ATT CTT TTC AAG AGA AAG AAT AGT CTC ATT TGC ACG TTT TTT g 3ʹ, and shCTR9#2-R, 5ʹ aat tcA AAA AAC GTG CAA ATG AGA CTA TTC TTT CTC TTG AAA AGA ATA GTC TCA TTT GCA CGcg 3ʹ; shCTR9#3-F, 5ʹ gat ccG CCA TAA TTT CAT CAA GTG ATT TCA AGA GAA TCA CTT GAT GAA ATT ATG GCT TTT TTg 3ʹ, and shCTR9#3-R, 5ʹ aat tcA AAA AAG CCA TAA TTT CAT CAA GTG ATT CTC TTG AAA TCA CTT GAT GAA ATT ATG GCg 3ʹ. To produce the viruses, HEK293T cells were cotransfected with the corresponding plasmids and helper plasmids using Polyjet (SignaGen, Gaithersburg, MD, USA).
Construction of stable cell lines
To stably silence CTR9, HepG2 and Huh7 cells were infected with Scramble and shCTR9 viruses, respectively. After 72 h, 2.5 μg/mL puromycin (Beyotime, Shanghai, China) was added to the medium for continuous culture to obtain cell lines stably expressing Scramble and shCTR9.
5-Ethynyl-2ʹ-deoxyuridine (EdU) assay
To examine the effect of CTR9 on the proliferation of HCC cells, we performed EdU-incorporation assays using an EdU assay kit (RiboBio, Guangzhou, China). Briefly, cells were seeded in 96-well plates at a density of 1 × 104 cells/well and cultured for 24 h. The cells were then incubated with 50 μM EdU at 37 °C for 2 h, washed with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde for 30 min. After permeabilization with 0.5% Triton X-100 for another 10 min, the cells were washed with PBS three times and treated with a 1 × Apollo® reaction cocktail at 25 °C for 30 min. Finally, nuclei were stained with 5 μg/mL Hoechst 33342 for 20 min and imaged using a fluorescence microscope (IX71; Olympus, Tokyo, Japan).
Colony formation assay
Cells were seeded into a 60 mm dish at a density of 1000 cells/dish, supplemented with medium to 8 mL, and cultured continuously for 2 weeks, followed by fixation with 4% paraformaldehyde, staining with 0.3% Crystal Violet solution (Beyotime, Nantong, China), washing with tap water, air drying, and imaging using a digital camera. Colony formation was confirmed by manual counting, with colonies defined as >50 cells.
Transwell invasion and migration assays
Transwell assays were carried out using Transwell inserts with an 8 μm aperture size to investigate cell invasion and migration. Briefly, 3 × 104 cells per well suspended in serum-free medium were added to the upper compartment of a 24-well Transwell culture plate, and medium containing 10% FBS was added to the lower compartment. After incubation at 37 °C for 48 h, cells that had not traversed the upper compartment were removed using cotton‐tipped swabs, and cells that migrated to the lower side of the insert were fixed with 4% paraformaldehyde for 30 min, dyed with 0.3% Crystal Violet for 30 min, and counted using a microscope. Cell invasion was assessed using a similar method, except that the inserts were pre-covered with 10 μg of Matrigel (BD, Shanghai, China).
Western blotting
For Western blotting, cells were lysed using RIPA buffer and centrifuged at 12,000 × g and 4 °C for 10 min, and supernatants were collected. Equal amounts of proteins were subjected to 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes with a pore size of 0.45 μm (Millipore, Billerica, MA, USA). After the membranes had been blocked with 5% non-fat milk, they were incubated with primary antibodies (CTR9, FLAG, PEG10, p21, p27, MMP2/9, and β-actin) at 4 °C overnight, followed by incubation with secondary antibodies at 25 °C for 1 h. Bound antibodies were detected using ECL Plus Western blotting substrate (Thermo Fisher Scientific, Waltham, MA, USA) and visualized using a chemiluminescence detection system (Tanon, Shanghai, China). Band density was quantified using ImageJ software (NIH, Bethesda, MD, USA). Relative protein levels were determined by standardizing the optical density values of the target proteins and the loading control.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from HepG2 and Huh7 cells using TRIzol reagent (Thermo Fisher Scientific) according to manufacturer instructions. The RNA was reverse transcribed using a FastQuant RT kit (with gDNase; Tiangen, Beijing, China) to obtain a single-stranded cDNA template. PEG10 expression in HCC cells was examined using SYBR Green qPCR master mix (MCE, Shanghai, China) using the following program: with 40 cycles at 95 °C for 10 s and 60 °C for 30 s. The following primers were used for amplification: PEG10-F, 5ʹ AGA GGA AGA GTG CCC AGA AGA CC 3ʹ, and PEG10-R, 5ʹ AAG CAG ACA CGG ACA CGA TCA AC 3ʹ; CTR9-F, 5ʹ CTG CTT CTG CCT ACT TGA GGG 3ʹ, and CTR9-R, 5ʹ CCT GGG TTA GTA CGC AAT GCT 3ʹ; GAPDH-F, 5ʹ GAT GAC CTT GCC CAC AGC CT 3ʹ, and GAPDH-R, 5ʹ ATC TCT GCC CCC TCT GCT GA 3ʹ.
Luciferase reporter assay
HCC cells were cultured in a 6-well plate at 37 °C and then cotransfected with an empty plasmid (3 × FLAG-Vector) or expression plasmid (3 × FLAG-CTR9) along with the PEG10 reporter plasmid. After 48 h, Firefly and Renilla luciferase activities were detected using a Dual-Lumi II luciferase reporter gene assay kit (Beyotime). The ratio of the relative luciferase activities was calculated for each sample. The reporter gene was synthesized by Youbio [chr7: 94654560-94656567; ~−1810 to −1 (promoter), +1 to ~+198 (5ʹ untranslated region)].
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out using a ChIP assay kit (Cell Signaling Technology) according to manufacturer instructions. Briefly, cells treated with 1% formaldehyde for cross-linking were resuspended in 1% SDS lysis buffer, and fragmented DNA was incubated with anti-CTR9 or negative control anti-IgG in ChIP dilution buffer at 4 °C with rotation overnight. Immunocomplexes were washed, eluted, and purified, and the ChIP products were evaluated by qPCR using PEG10 primers designed to cover the predicted CTR9-binding site in the PEG10 promoter region: Q-F, 5ʹ-AAT AAT CCG TAG CAA TGT TAC-3ʹ and Q-R, 5ʹ-TGG GAA ACT GAC AAT CTG-3ʹ.
Immunohistochemistry (IHC)
IHC analysis was performed using a kit (Zhongshan Goldenbridge Biotech, Beijing, China) according to manufacturer’s instructions. Tissue sections were deparaffinized, blocked using 10% goat serum, and incubated at 25 °C with primary antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibodies. After incubation in 3,3′-diaminobenzidine solution, the sections were counterstained with hematoxylin, dehydrated, and mounted. Images were obtained using a microscope and analyzed using Image-Pro-Plus software (v.6.0; Media Cybernetics, Bethesda, MD, USA). Protein expression was represented as the mean optical density.
Establishment of the mouse xenograft model
The animal studies have been conducted in accordance with the Institutional Animal Care and Use Committee of Xuzhou Medical University. Ten male nude mice (Vital River Laboratory Animal Technology, Beijing, China) with no significant difference in body weight (at 4 weeks of age) were reared in a sterile environment for 1 week. The mice were then randomly divided into two groups (n = 5/group). After disinfecting their skin, 100 µL of cell suspension (1 × 106/mL) was injected into the dorsal side of the right hind limb. The mice were then returned to their cages and housed under the same conditions. Tumor length and width were measured every other day using Vernier calipers, and tumor volume was calculated as follows: length × (width2/2). After 5 weeks, all mice were euthanized, and their subcutaneous tumors were removed and weighed.
Statistical analysis
Data represent the results of experiments repeated at least three times, and all quantitative data represent the mean ± standard deviation. Statistical analysis was performed using SPSS (v.22.0; IBM Corp., Armonk, NY, USA). Student’s t tests were used to compare two samples with normality, homogeneity of variance, and independence. Non-parametric tests were used to analyze measurement or count data that did not meet these requirements. P < 0.05 was considered significant.
Results
CTR9 is overexpressed in human HCC tissues
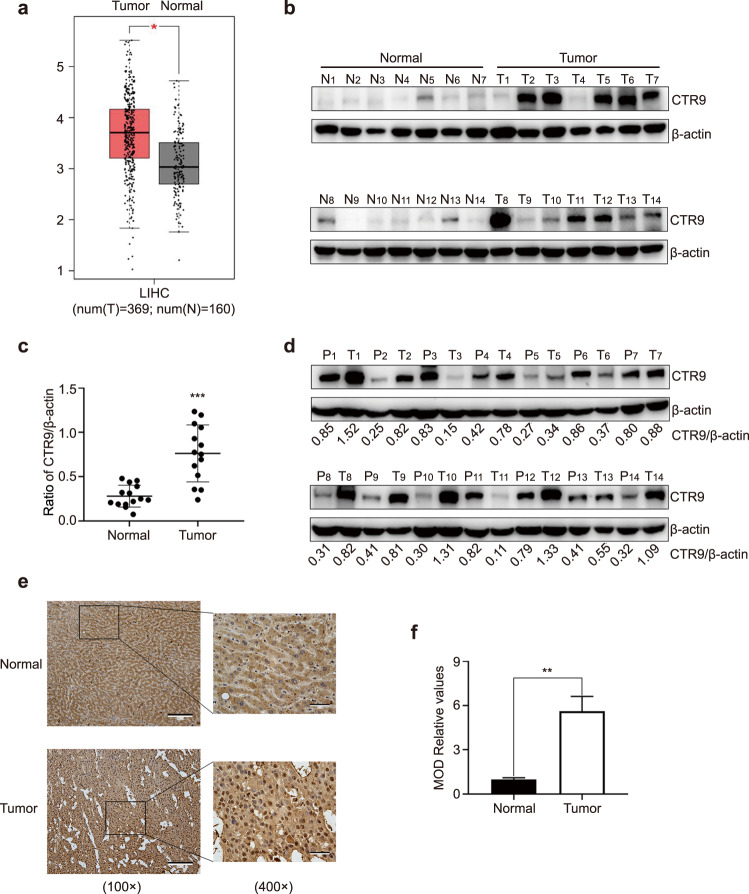
To determine the clinical significance of CTR9, we analyzed CTR9 mRNA levels in HCC using the Gene Expression Profiling Interactive Analysis database. Results indicated that CTR9 was upregulated in HCC patients (Fig. 1a). We then measured CTR9 protein levels in total lysates extracted from 14 normal liver tissue specimens and 14 human HCC tissue specimens using a Western blotting blot. The results showed that CTR9 levels were upregulated in HCC samples compared to non-tumor samples (Fig. 1b and c). In addition, measurement of CTR9 levels in total lysates extracted from 14 HCC tissue specimens and corresponding para-cancerous tissue specimens revealed that levels in cancer tissue specimens were higher than those in para-cancerous tissues (Fig. 1d). Consistently, IHC analysis confirmed that CTR9 expression was stronger in HCC tissues than in normal tissues (Fig. 1e and f). These data indicated that CTR9 is overexpressed in human HCC tissues and might affect HCC progression.
Fig. 1. CTR9 expression in hepatocellular carcinoma (HCC) patients.
a GEPIA database analysis showing that CTR9 mRNA levels are higher in HCC tissues (Tumor) than in non-tumorous liver tissues (Normal) (P < 0.05). b Analysis of total proteins isolated from non-tumorous liver tissues (Normal) and HCC tissues (Tumor) were analyzed using Western blot to assess CTR9 levels. c Statistical analysis of CTR9 expression. Ratios represent CTR9 normalized against β-actin levels in each sample. d Analysis of total proteins isolated from tumor tissue and paired adjacent normal tissues of 14 patients using Western blot to assess CTR9 levels. e Representative images of CTR9 in normal and tumor tissues according to IHC analysis (magnification, ×100 and scale bar, 200 μm; magnification, ×400 and scale bar, 50 μm). f Histogram showing quantitative analysis of relative CTR9 levels. *P < 0.05, **P < 0.01, ***P < 0.001
CTR9 knockdown inhibits HCC cell proliferation, invasion, and migration
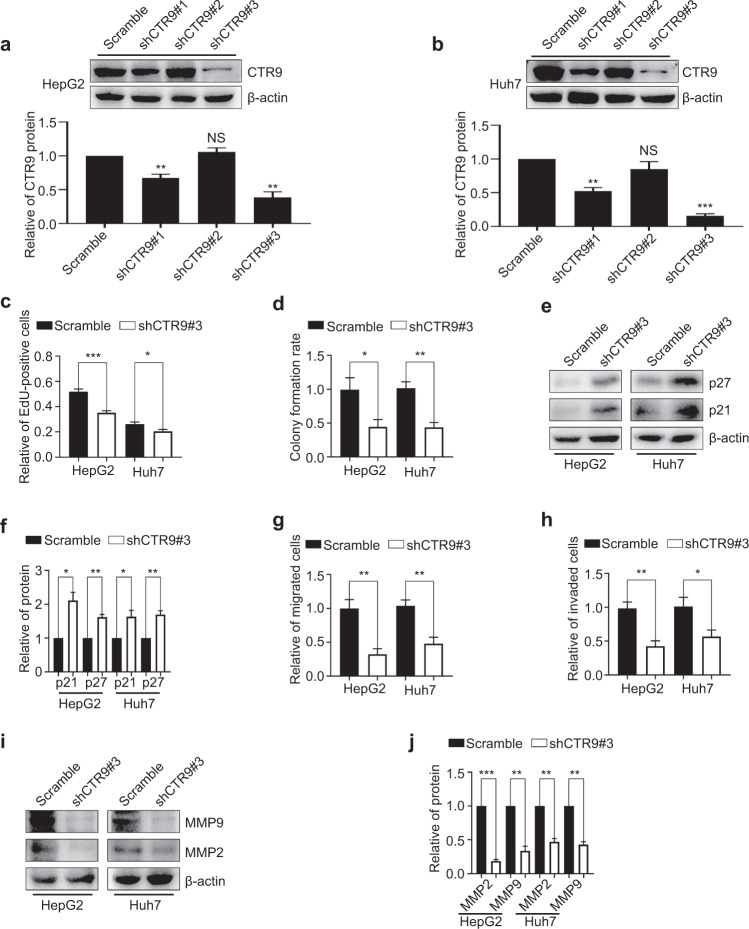
To examine whether CTR9 plays an important role in the proliferation, invasion, and migration of HCC cells, we performed loss- and gain-of-function experiments. First, we screened three shRNA sequences (shCTR9#1, shCTR9#2, and shCTR9#3) for their ability to inhibit CTR9 expression, with non-targeting shRNA (Scramble) as a negative control. The shCTR9s and Scramble were packaged into lentiviruses, which were used to infect HCC cells to develop stable cell lines deficient in CTR9. Notably, shCTR9#3 displayed a silencing efficiency of ~60% and ~80% in HepG2 and Huh7 cells, respectively (Fig. 2a and b). We then performed cell growth and motility assays to determine whether CTR9 silencing influences the proliferation, invasion, and/or migration of HCC cells. Compared with results in the Scramble, CTR9 silencing considerably reduced the number of EdU-positive cells by 32.3% and 22.1% in HepG2 and Huh7 cells, respectively (Fig. 2c and S1a). In addition, CTR9 knockdown clearly reduced the colony formation ability of the cells (Fig. 2d and S1b) and increased the expression of cell cycle regulators, such as p21 and p27 (Fig. 2e and f). Moreover, Transwell migration assays revealed that the percentage of migratory cells was 67.6% and 55.9% lower in the CTR9-silenced groups, respectively, relative to that in cells infected with Scramble (Fig. 2g and S1c). Transwell invasion assays with Matrigel showed that CTR9 silencing reduced cell invasion (Fig. 2h and S1d). Furthermore, we found significantly decreased MMP2 and MMP9 expression following CTR9 silencing (Fig. 2i and j). These results indicated that CTR9 silencing exerted critical inhibitory effects on HCC cell proliferation, invasion, and migration.
Fig. 2. CTR9 downregulation suppresses HCC cell proliferation, invasion, and migration.
a, b Representative blots and quantification of shCTR9 efficiency in HepG2 and Huh7 cells. c Quantification of the EdU-incorporation results in HepG2 and Huh7 cells after CTR9 knockdown. d Quantification of colony formation assay results in CTR9-silenced HepG2 and Huh7 cells. e Representative blots and f quantification of p21 and p27 levels in CTR9-silenced HepG2 and Huh7 cells. g Quantification of Transwell migration h invasion assay results in CTR9-silenced HepG2 and Huh7 cells. i Representative blots and j quantification of MMP2 and MMP9 levels in CTR9-silenced HepG2 and Huh7 cells. *P < 0.05, **P < 0.01, ***P < 0.001
CTR9 overexpression enhances HCC cell proliferation, invasion, and migration
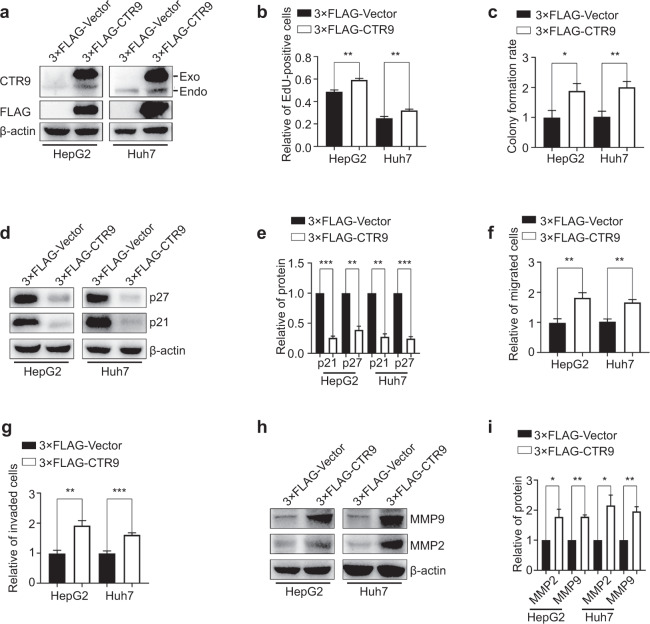
To further elucidate the specific roles of CTR9, we transiently transfected HCC cells with 3 × FLAG-tagged CTR9 cDNA and analyzed the consequences of CTR9 overexpression by Western blot (Fig. 3a). In addition, to detect the effect of CTR9 upregulation on cell proliferation, invasion, and migration, we evaluated cell growth and motility at 24 h post-transfection. EdU-incorporation assays revealed that CTR9 overexpression increased the number of EdU-positive HepG2 and Huh7 cells by ~21.3% and ~27.2%, respectively (Fig. 3b and S2a). Moreover, CTR9 upregulation enhanced colony formation (Fig. 3c and S2b) and reduced p21 and p27 expression (Fig. 3d and e). Furthermore, cell migration and invasion assays revealed that CTR9 upregulation increased the percentage of migratory cells by ~83.2% and ~64.4%, respectively (Fig. 3f and S2c) and increased cell invasion (Fig. 3g and S2d). Similarly, we observed clear increases in MMP2 and MMP9 expression following CTR9 upregulation (Fig. 3h and i). These results suggested that elevated CTR9 expression strongly promoted HCC cell proliferation, invasion, and migration.
Fig. 3. CTR9 upregulation enhances HCC cell proliferation, invasion, and migration.
a Western blot analysis of 3×FLAG-CTR9 overexpression in HepG2 and Huh7 cells. b Quantification of the EdU-incorporation assay results in HepG2 and Huh7 cells after CTR9 overexpression. c Quantification of colony formation results in CTR9-overexpressing HepG2 and Huh7 cells. d Representative blots and e quantification of p21 and p27 levels in CTR9-overexpressing HepG2 and Huh7 cells. f Quantification of Transwell migration and g invasion results in CTR9-overexpressing HepG2 and Huh7 cells. h Representative blots and i quantification of MMP2 and MMP9 levels in CTR9-overexpressing HepG2 and Huh7 cells. *P < 0.05, **P < 0.01, ***P < 0.001
CTR9 enhances HCC cell proliferation, invasion, and migration by actively regulating PEG10
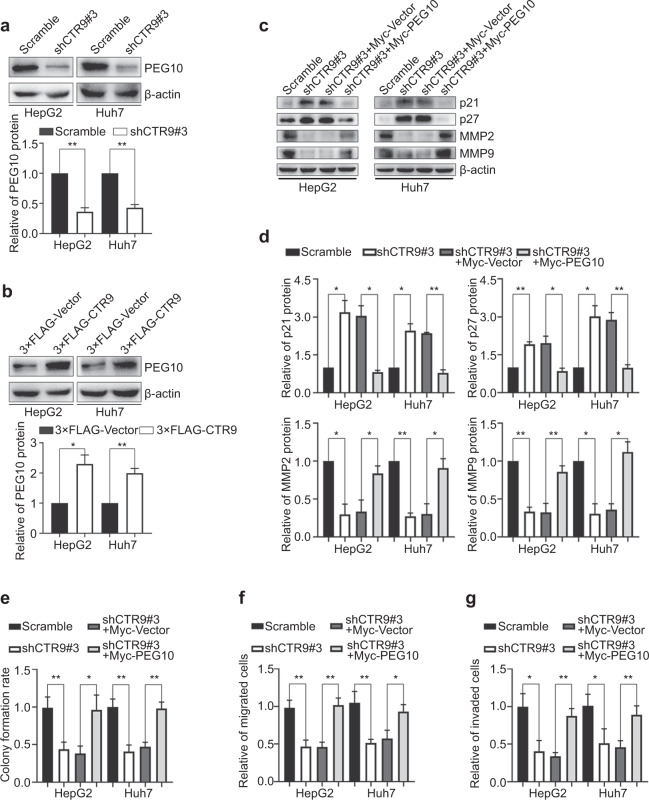
We then investigated the specific molecular mechanisms underlying the observed effects on HCC cell activities. Previous studies showed that PEG10 could promote the proliferation, invasion, and migration of various types of tumor cells [20, 22, 23, 25–27], as well as participate in the regulation of p21, p27, MMP2, and MMP9 [20, 25]. In addition, we previously demonstrated that PEG10 is upregulated in HCC patients [28], and another report indicated that E2F transcription factor 1 could directly enhance PEG10 expression via binding to its promoter. CTR9 is a key component of PAF1c and participates in regulating transcriptional processes [5–7]. Therefore, we hypothesized that CTR9 might transcriptionally regulate PEG10 to affect HCC cell proliferation, invasion, and migration. To test this hypothesis, we investigated their regulatory relationship. Western blot analysis confirmed that CTR9 knockdown decreased PEG10 protein levels, whereas CTR9 upregulation increased PEG10 expression (Fig. 4a and b). We then performed rescue experiments by overexpressing Myc-PEG10 in CTR9-downregulated cells to examine whether CTR9 accelerates HCC cell proliferation, invasion, and migration via PEG10. Notably, PEG10 overexpression effectively reversed the expression of p21, p27, and MMPs (Fig. 4c and d), as well as the effects of CTR9 silencing on cell proliferation, invasion, and migration (Fig. 4e–g and S3). These results suggest that CTR9 facilitates PEG10 expression, thereby accelerating the proliferation, invasion, and migration of HCC cells.
Fig. 4. CTR9 promotes PEG10 transcription in HCC cells.
a, b Representative blots and quantification of PEG10 levels in CTR9-silenced or CTR9-overexpressing HepG2 and Huh7 cells. c Representative blots and d quantification showing that PEG10 overexpression reversed p21, p27, and MMP expression in CTR9-silenced HepG2 and Huh7 cells. e–g Colony formation and Transwell migration and invasion assays showing that PEG10 overexpression reverses the effects of CTR9 knockdown on cell proliferation, migration, and invasion. *P < 0.05, **P < 0.01
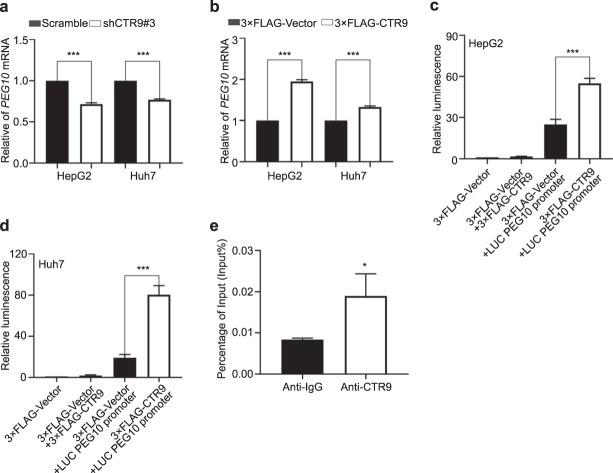
CTR9 promotes PEG10 expression by increasing its transcription
To further explore the molecular mechanism by which CTR9 regulates PEG10 level, we detected PEG10 mRNA levels. We found that CTR9 silencing significantly reduced PEG10 mRNA levels, whereas CTR9 overexpression increased these levels (Fig. 5a and b). To determine the interaction between CTR9 and the PEG10 promoter region, we performed luciferase reporter assays, finding that CTR9 overexpression enhanced the luciferase activity of HCC cells transfected with pGL3-Basic containing the PEG10 promoter (Fig. 5c and d), suggesting PEG10 as a target of CTR9 and that CTR9 positively regulates PEG10 expression by targeting its promoter. To evaluate whether CTR9 can be recruited to the putative binding site on the PEG10 promoter, we performed ChIP-qPCR assays involving the predicted CTR9-binding sequence, with the results demonstrating that CTR9 could bind to that predicted region of the PEG10 promoter (Fig. 5e).
Fig. 5. CTR9 binds and activates PEG10 promoter.
Quantification of PEG10 mRNA levels in a CTR9-silenced HepG2 and Huh7 cells and b Quantification of PEG10 mRNA levels in CTR9-overexpressing HepG2 and Huh7 cells. c, d Quantification of luminescence after CTR9 overexpression. e Binding efficiency measured using ChIP-qPCR in HepG2 cells. *P < 0.05, ***P < 0.001
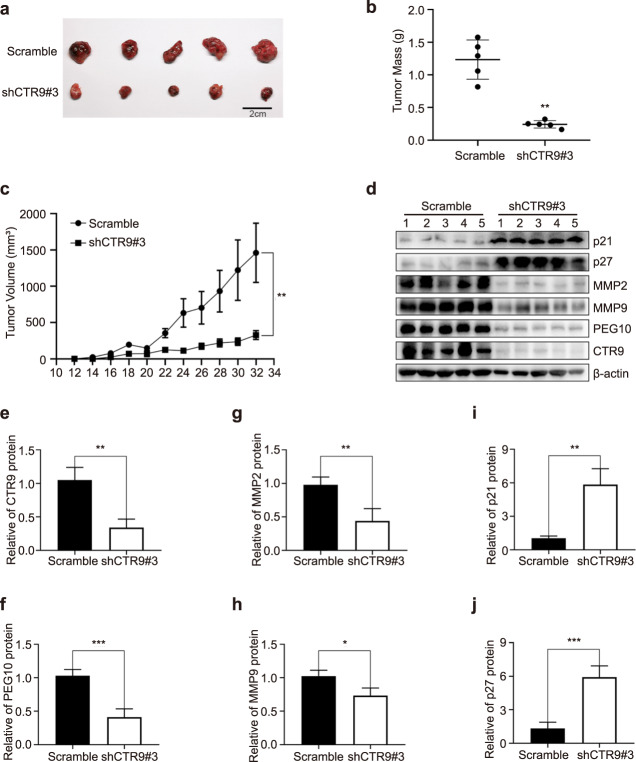
CTR9 promotes HCC cell growth by regulating PEG10 in vivo
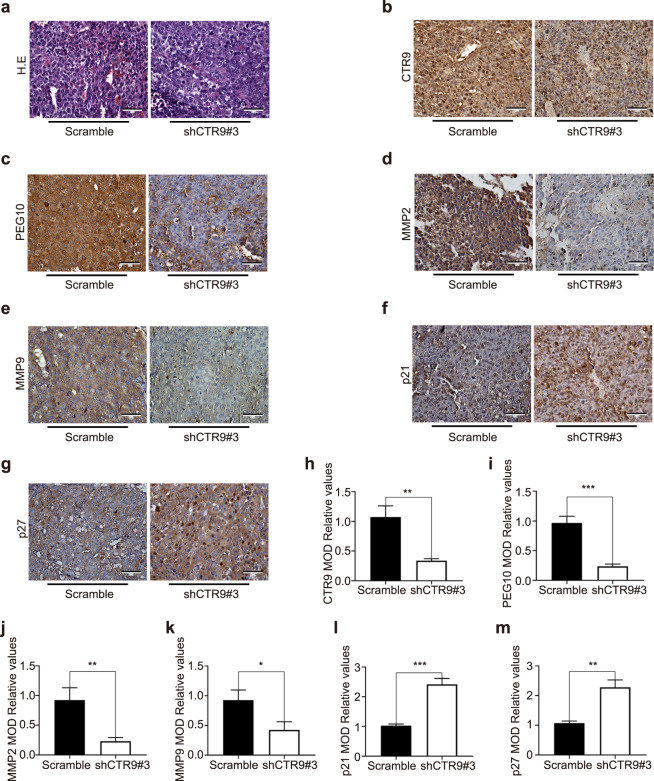
Finally, we examined the relationship between CTR9 expression and HCC development in nude mice bearing tumors formed by CTR9-silenced Huh7 cells. Consistent with the in vitro observations, CTR9 knockdown markedly inhibited tumor growth in nude mice compared with that observed in tumors seeded using cells infected with the Scramble control (Fig. 6a–c). Moreover, Western blot revealed decreased PEG10 and MMP levels and increased p21 and p27 levels in tumors formed by CTR9-silenced Huh7 cells (Fig. 6d–j), consistent with IHC results (Fig. 7). These findings suggested that CTR9 acts as a tumor promoter in HCC by regulating PEG10 expression.
Fig. 6. CTR9 promotes HCC cell growth in vivo by regulating PEG10.
a Tumor tissues isolated from tumors initiated with cells infected with Scramble or shCTR9#3 vectors. b Tumor mass. c Growth curve obtained by measuring tumor size on the indicated days. d Representative blots. e–j Quantification of CTR9, PEG10, MMPs, p21, and p27 levels in tumor tissues. *P < 0.05, **P < 0.01, ***P < 0.001
Fig. 7. Protein levels in tumors in vivo.
a Representative images of tumor tissues stained with hematoxylin and eosin. Scale bar, 50 μm. b–g Representative IHC images of CTR9, PEG10, p21, p27, and MMPs in tumor tissues. Scale bar, 50 μm. h–m Histogram showing the quantitative analysis of relative CTR9, PEG10, p21, p27, and MMP levels. *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
In this study, we demonstrated that CTR9 expression is upregulated in HCC. Specifically, we showed that CTR9 downregulation inhibited HCC cell proliferation, invasion, and migration, increased levels of cell cycle regulators (p21 and p27), and decreased MMP levels, whereas CTR9 overexpression exerted the opposite effects. Further study of the underlying molecular mechanism revealed that CTR9 promoted the transcription of the oncogene PEG10 by binding its promoter. These findings suggest that elevated CTR9 expression might promote HCC cell proliferation, invasion, and migration by increasing PEG10 transcription.
CTR9 is a key component of PAF1c, which plays multifunctional roles in RNA polymerase-II-related transcriptional processes and is also related to the regulation of embryonic stem cell development and pluripotency maintenance [5–7]. In addition, previous studies report that CTR9 affects multiple cellular activities, including transcriptional regulation, methylation modification, protein transport, and differentiation [8–10]. Moreover, recent studies confirmed CTR9 as a tumor promoter associated with familial Wilms tumors, whereas another study suggested CTR9 as a tumor suppressor [16, 29]. However, other studies implied that CTR9 acts independently of other PAF1c subunits to play an active regulatory role in mediating estrogen-dependent transcriptional regulation and drive the progression of E estrogen receptor-α-positive breast cancer [17, 30]. Furthermore, a previous study indicated that CTR9 promotes the proliferation, migration, and invasion of human glioma cells [31] and is involved in HCC; however, there are few studies on the direct effect of CTR9 on HCC [32]. In the present study, we found that CTR9 was upregulated in HCC, and its oncogenic effects on the growth and motility of HCC cells were verified in our study. Notably, we found that CTR9 knockdown suppressed cell proliferation, invasion, and migration, whereas CTR9 overexpression promoted these activities. These findings were further confirmed in a tumor-bearing nude mouse model, suggesting that CTR9 might promote HCC progression.
PEG10 was first identified in HCC and is closely associated with its occurrence and development [33]. Previous studies confirmed that PEG10 is upregulated in samples from patients with normal serum α-fetoprotein levels and small tumors [34], and that PEG10 is upregulated in most HCC tissues but not in matched non-cancerous liver tissues [21]. These findings suggested that PEG10 might act as a potential biomarker for HCC [23]. In the present study, we demonstrated that CTR9 participates in the transcriptional regulation of PEG10 to accelerate HCC cell proliferation, invasion, and migration by controlling levels of p21, p27, and MMPs. Consequently, we hypothesize that elevated CTR9 expression in HCC patients upregulates PEG10, thus, accelerating proliferation, invasion, and migration of HCC cells. Therefore, CTR9 upregulation could play a significant role in HCC occurrence and development.
In conclusion, these findings suggest that elevated CTR9 expression occurs in HCC tissues and exists close association with HCC occurrence and malignant development via the upregulated transcription of PEG10. Consequently, CTR9 could be a potential target for HCC diagnosis and treatment. Notably, a recent study showed that 21 tetratricopeptide repeats in CTR9, including 42 continuous α-helices, surround the PAF1 N-terminal domain in a right-handed supercoiled manner. This represents a multifunctional protein–protein-interaction domain that can serve as an interaction scaffold in multi-protein complexes involved in different cellular processes [35, 36]. This may indicate that, as a subunit of PAF1c, PAF1 may act along with PEG10 expression. Nevertheless, further studies are required to elucidate the precise mechanisms underlying CTR9-mediated PEG10 upregulation in HCC.
Supplementary information
Acknowledgements
This research was supported by the Jiangsu Provincial Commission of Health and Family Planning [H2018037 and BJ18010], the Basic Research Program of Jiangsu Province [BK20191153], the Xuzhou Institute of Technology [KC20091 and KC19035], the National Natural Science Foundation of China [81874081], the Jiangsu Provincial Qing Lan Project and Jiangsu Provincial Medical Youth Talent [QNRC2016784], the Young Science and Technology Innovation Team of Xuzhou Medical University Key Research and Development Plan of Jiangsu Province [TD202006], and the Jiangsu Postgraduate Research Innovation Program [KYCX19_2237].
Author contributions
BZ, ZYL, RW, CMZ, KC, WGS, ZL, MJ, and ZLT conducted the experiments; BZ, ZYL, GS, HLS, and RHW designed the experiments; ZYL and RW wrote the manuscript; BZ, GS, HLS, and RHW revised the manuscript. All authors participated in the study’s design, analysis of the data, interpretation of the findings, and review of the manuscript and agreed to the publication of the final version.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Bin Zhang, Zhi-yi Liu, Rui Wu.
Contributor Information
Gautam Sethi, Email: phcgs@nus.edu.sg.
Heng-liang Shi, Email: shl@xzhmu.edu.cn.
Ren-hao Wang, Email: wangrenhao@xzhmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00812-3.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Chok K. Management of recurrent hepatocellular carcinoma after liver transplant. World J Hepatol. 2015;7:1142–8. doi: 10.4254/wjh.v7.i8.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, et al. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–9. doi: 10.1128/MCB.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, et al. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–74. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–80. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K, Haversat JM, Mager J. CTR9/PAF1c regulates molecular lineage identity, histone H3K36 trimethylation and genomic imprinting during preimplantation development. Dev Biol. 2013;383:15–27. doi: 10.1016/j.ydbio.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Gois S, Slama P, Pietrancosta N, Erdozain AM, Louis F, Bouvrais-Veret C, et al. Ctr9, a protein in the transcription complex Paf1, regulates dopamine transporter activity at the plasma membrane. J Biol Chem. 2015;290:17848–62. doi: 10.1074/jbc.M115.646315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo HS, Seo JH, Yoo JY. CTR9, a component of PAF complex, controls elongation block at the c-Fos locus via signal-dependent regulation of chromatin-bound NELF dissociation. PLoS One. 2013;8:e61055. doi: 10.1371/journal.pone.0061055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol Cell Biol. 2008;28:4152–61. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues J, Lydall D. Paf1 and Ctr9, core components of the PAF1 complex, maintain low levels of telomeric repeat containing RNA. Nucleic Acids Res. 2018;46:621–34. doi: 10.1093/nar/gkx1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo HS, Choi Y, Ahn N, Lee S, Kim WU, Jang MS, et al. Transcriptional regulator CTR9 inhibits Th17 differentiation via repression of IL-17 expression. J Immunol. 2014;192:1440–8. doi: 10.4049/jimmunol.1201952. [DOI] [PubMed] [Google Scholar]
- 14.Youn MY, Yoo HS, Kim MJ, Hwang SY, Choi Y, Desiderio SV, et al. hCTR9, a component of Paf1 complex, participates in the transcription of interleukin 6-responsive genes through regulation of STAT3-DNA interactions. J Biol Chem. 2007;282:34727–34. doi: 10.1074/jbc.M705411200. [DOI] [PubMed] [Google Scholar]
- 15.Lee JW, Bae E, Kwon SH, Yu MY, Cha RH, Lee H, et al. Transcriptional modulation of the T helper 17/interleukin 17 axis ameliorates renal ischemia-reperfusion injury. Nephrol Dial Transpl. 2019;34:1481–98. doi: 10.1093/ndt/gfy370. [DOI] [PubMed] [Google Scholar]
- 16.Hanks S, Perdeaux ER, Seal S, Ruark E, Mahamdallie SS, Murray A, et al. Germline mutations in the PAF1 complex gene CTR9 predispose to Wilms tumour. Nat Commun. 2014;5:4398. doi: 10.1038/ncomms5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng H, Xu W. Ctr9, a key subunit of PAFc, affects global estrogen signaling and drives ERα-positive breast tumorigenesis. Genes Dev. 2015;29:2153–67. doi: 10.1101/gad.268722.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li CM, Margolin AA, Salas M, Memeo L, Mansukhani M, Hibshoosh H, et al. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006;66:665–72. doi: 10.1158/0008-5472.CAN-05-1553. [DOI] [PubMed] [Google Scholar]
- 19.Hishida T, Naito K, Osada S, Nishizuka M, Imagawa M. peg10, an imprinted gene, plays a crucial role in adipocyte differentiation. FEBS Lett. 2007;581:4272–8. doi: 10.1016/j.febslet.2007.07.074. [DOI] [PubMed] [Google Scholar]
- 20.Peng YP, Zhu Y, Yin LD, Zhang JJ, Wei JS, Liu X, et al. PEG10 overexpression induced by E2F-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J Exp Clin Cancer Res. 2017;36:30. doi: 10.1186/s13046-017-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–8. [PubMed] [Google Scholar]
- 22.Deng X, Hu Y, Ding Q, Han R, Guo Q, Qin J, et al. PEG10 plays a crucial role in human lung cancer proliferation, progression, prognosis and metastasis. Oncol Rep. 2014;32:2159–67. doi: 10.3892/or.2014.3469. [DOI] [PubMed] [Google Scholar]
- 23.Ip WK, Lai PB, Wong NL, Sy SM, Beheshti B, Squire JA, et al. Identification of PEG10 as a progression related biomarker for hepatocellular carcinoma. Cancer Lett. 2007;250:284–91. doi: 10.1016/j.canlet.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Kainz B, Shehata M, Bilban M, Kienle D, Heintel D, Krömer-Holzinger E, et al. Overexpression of the paternally expressed gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in high-risk B-cell chronic lymphocytic leukemia. Int J Cancer. 2007;121:1984–93. doi: 10.1002/ijc.22929. [DOI] [PubMed] [Google Scholar]
- 25.Xiong J, Qin J, Zheng Y, Peng X, Luo Y, Meng X. PEG10 promotes the migration of human Burkitt’s lymphoma cells by up-regulating the expression of matrix metalloproteinase-2 and −9. Clin Invest Med. 2012;35:E117–25. doi: 10.25011/cim.v35i3.16587. [DOI] [PubMed] [Google Scholar]
- 26.Zang W, Wang T, Huang J, Li M, Wang Y, Du Y, et al. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther. 2015;22:138–44. doi: 10.1038/cgt.2014.77. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Xiao R, Tembo K, Hao L, Xiong M, Pan S, et al. PEG10 promotes human breast cancer cell proliferation, migration and invasion. Int J Oncol. 2016;48:1933–42. doi: 10.3892/ijo.2016.3406. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Tian Z, Cao K, Zhang B, Wen Q, Zhou X, et al. TSG101 promotes the proliferation, migration and invasion of hepatocellular carcinoma cells by regulating the PEG10. J Cell Mol Med. 2019;23:70–82. doi: 10.1111/jcmm.13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martins AG, Pinto AT, Domingues R, Cavaco BM. Identification of a novel CTR9 germline mutation in a family with Wilms tumor. Eur J Med Genet. 2018;61:294–99. doi: 10.1016/j.ejmg.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Zeng H, Lu L, Chan NT, Horswill M, Ahlquist P, Zhong X, et al. Systematic identification of Ctr9 regulome in ERα-positive breast cancer. BMC Genom. 2016;17:902. doi: 10.1186/s12864-016-3248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Chen J, He G, Zhang Y. CTR9-mediated JAK2/STAT3 pathway promotes the proliferation, migration, and invasion of human glioma cells. J Clin Lab Anal. 2021;35:e23943. doi: 10.1002/jcla.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Reebye V, Hitchen P, Fan J, Jiang H, Sætrom P, et al. Mechanisms involved in the activation of C/EBPα by small activating RNA in hepatocellular carcinoma. Oncogene. 2019;38:3446–57. doi: 10.1038/s41388-018-0665-6. [DOI] [PubMed] [Google Scholar]
- 33.Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–7. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- 34.Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen Y, et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:1133–9. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- 35.Deng P, Zhou Y, Jiang J, Li H, Tian W, Cao Y, et al. Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology. Proc Natl Acad Sci USA. 2018;115:9998–10003. doi: 10.1073/pnas.1812256115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–67. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.