Abstract
Introduction
Acute myocardial infarction (AMI) is a ubiquitous cardiovascular disease ensuing adverse prognosis caused by myocardial necrosis. Effective and rapid diagnosis of AMI is essential to following treatment in clinical practice while the existed biomarkers have inherent limitations. Consequently, exploration of novel biomarkers is needed. Long noncoding RNA (lncRNA) emerges as the upcoming biomarkers adopted in clinical use, and we aim at investigating the diagnostic power of lncRNA TTTY15 and HULC in AMI patients.
Method
We measured lncRNA level in 80 AMI patients and 36 healthy volunteers in discovering cohort and 50 AMI patients and 20 healthy volunteers in verification cohort with quantitative RT-PCR method. Receiver operating characteristic (ROC) analysis was administered to detect the diagnostic power of selected lncRNAs. Regression and correlation analyses were performed to explore the related factors.
Results
ROC analysis reveals the superiority of TTTY15 and HULC as biomarkers against conventional AMI biomarkers CKMB (AUC of TTTY15: 0.915 versus CKMB: 0.768 versus TnT: 0.869); AUC of HULC: 0.905 versus CKMB: 0.768 versus TnT: 0.869). Regression and correlation analysis indicates that TTTY15 and HULC may be one of the contributing factors to AMI and related to accepted risk factors.
Conclusion
Our results revealed the diagnostic potency of lncRNA TTTY15 and HULC, and they could also be treated as novel therapeutic targets in AMI therapy, hinting inspiration to the cardiologist in clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12872-022-02529-5.
Keywords: Acute myocardial infarction, Diagnosis, lncRNA, TTTY15, HULC
Introduction
Acute myocardial infarction (AMI), as a ubiquitous cardiovascular disease ensuing adverse prognosis, is caused by myocardial necrosis induced by unstable ischemic syndrome [1]. Contemporary treatment for AMI is percutaneous coronary intervention (PCI) or coronary artery bypass graft (CABG), with attendant thrombolytic therapy, respectively [2, 3]. Effective and rapid diagnosis of AMI is essential for the selection of a corresponding treatment strategy as it could prevent the progressively deleterious ravage to the myocardium and accrue considerable prognosis [4, 5]. Conventional biomarkers like cardiac troponin (cTns) and creatinine kinase MB (CKMB) are adopted as the golden standard in AMI diagnosis, yet the inherent limitations that other cardiovascular diseases may present exaggeration of cTns and CKMB level still exist [5, 6]. Consequently, exploration of novel biomarkers with adequate accuracy is imminent.
With the advanced development of genomic techniques, enormous biomarkers are prone to be applicable in the clinical diagnosis of AMI [7]. Specifically, with plausible RNA-sequencing techniques, long non-coding RNAs (lncRNA) of more than 200 nucleotides are proved to play a vital role in regulating gene expression by various mechanisms including gene transcription, translation, epigenetic inheritance, etc. [8, 9]. Moreover, plasma lncRNA level has been reported to be altered in diseases including the cardiovascular system, so leveraging lncRNA as diagnostic markers is feasible [10–12].
lncRNA highly up-regulated in liver cancer (HULC) is known as abnormal secretion in cancer cells and is reported to be protective against myocardial I/R injury and H/R cardiomyocyte apoptosis by inhibiting NLRP3/Caspase-1/1L-1β signaling pathway [13, 14]. lncRNA testis-specific transcript Y-linked 15 (TTTY15) is found to be upregulated in AMI patients and H2O2-stimulated myocardial cells, and the theory has been verified utilizing knockdown experiment [15]. Thus, HULC and TTTY15 are conceivable to be potential diagnostic biomarkers in AMI patients.
In this study, we aim at exploring the feasibility of adopting HULC and TTTY15 as novel biomarkers compared with CKMB in AMI diagnosis.
Method
Participants
We enrolled all AMI patients presented to Guangdong Second Provincial General Hospital (Guangzhou, China) from September 2020 to November 2021 and a total of 80 AMI patients were enrolled with 36 recruited healthy controls. A verification cohort with a total of 50 AMI patients and 20 controls were recruited from Guangdong Provincial People’s Hospital to verify the results. All enrolled patients were negative for COVID-19. Inclusion criteria were AMI diagnosis conforming to 2017 ESC guideline [16], elevated conventional cardiac biomarkers above the upper limit, and abnormal echocardiogram (ECG) findings. Exclusion criteria were patients complicated with other advanced or serious diseases such as malignancy and organ failure. Healthy controls were volunteers without a history of cardiovascular diseases or other organ issues. Conventional biomarkers, CKMB and troponin-T (TnT), were measured routinely in the first-meet. For the hypothetic biomarkers, the lncRNAs were measured just after admission (pre-PCI) and after the PCI procedure in the AMI group, while they were measured just after recruiting in healthy controls.
To verify the diagnostic value of selected lncRNA, another cohort with a total of 50 AMI patients receiving PCI procedure and 20 controls were recruited in another center, Guangdong Provincial People’s Hospital. Conventional biomarkers and the selected lncRNA were measured as above.
Blood sample collection
Regarding the AMI group, peripheral blood samples were obtained just after admission (pre-PCI) and after the PCI procedure, while for healthy controls, blood samples were obtained after recruiting. All blood samples were collected in the plain tube (BD Vacutainer®, 369714) containing EDTA anticoagulant without coagulation and hemolysis. The collected blood samples were centrifuged at 3500 rpm for 10 min, the supernatant was carefully transferred into an RNase-free tube and then immediately frozen at − 80 °C. Since the two IncRNAs are also expressed in blood cells, sample processing is carried out immediately after qualified samples are collected to minimize the contamination caused by the death and destruction of blood cells.
RNA isolation and Quantitative RT-PCR
Total RNA was extracted from plasma samples using Plasma/Serum RNA Purification Maxi Kit (Norgen, Product #56200) as described by the manufacturer. iScript® cDNA Synthesis Kit (Bio-Rad) was adopted to perform reverse-transcription of cDNA (component: total 20 μl reaction system containing 200 ng RNA template, 4 μl 5 × iScript Reaction mix, 1 μl iScript Reverse Transcriptase, and Nuclease-free water; reaction protocol: 5 min at 25 °C, 30 min at 42 °C, 5 min at 85 °C, and then hold at 4 °C). RNase-Free DNase I Kit (Norgen, Product #25710) was adopted to on-colomn DNA removal process to avoid genomic DNA contamination as described by the manufacturer. Green PCR Kit (Takara, Dalian, China) was administered in qRT-PCR procedure with specifically designed primers for lncRNA HULC and TTTY15. When performing qRT-PCR of HULC and TTTY15, GAPDH was treated as the internal control. Specific primers used in this study are as follows: HULC: forward 5ʹ-ATGGGGGTGGAACTCATGATGG-3ʹ, reverse 5ʹ-AAGAATGGACATCATTT ATTTCA-3ʹ; TTTY15: TTTY15, forward 5′-TGAGGGAGGGATGTAGCTTT-3′, reverse 5′-GAAGTCAAGCAGGCAACTGA-3′; GAPDH: forward 5ʹ-TGCACCACCAACTGCTTAGC-3ʹ, reverse 5ʹ-GGCAT GGACTGTGGTCATGAG-3ʹ. The relative expression level of detected lncRNA was measured following 2−ΔΔcq methods. Boxplot showing the cq value of the selected lncRNA was displayed in Additional file 1: Figure S1.
Statistical analysis
All data were presented with mean ± SD or number (percentage), and involved data analyses were performed in SPSS 23.0 and R (Version 3.6.2) software. Independent sample t-test, spearman correlation test, and chi-square test were performed in SPSS. The receiver operating characteristic (ROC) curve was employed to evaluate the specificity and sensitivity of selected lncRNA and the area under the ROC curve (AUC) was used to evaluate the predictive power. Both ROC and AUC were obtained via R with the pROC package.
Results
Baseline characteristics
A total of 80 AMI patients and 36 healthy controls were included to explore the feasibility of HULC and TTTY15 as novel biomarkers to diagnose AMI, with an average age of 58.50 and 58.25, respectively. No significant difference could be observed in age, the occurrence of diabetes mellitus, alcohol drinking, and cholesterol level between the two groups (P > 0.05). There was a significant difference in BMI (24.15 ± 2.29 versus 26.08 ± 2.06), the occurrence of hypertension (38.9% versus 67.5%), smoking population (46.1% versus 70.0%), total cholesterol level (4.22 ± 0.75 versus 4.54 ± 0.77), LDL level (2.76 ± 0.37 versus 3.34 ± 0.33), CK-MB level (53.97 ± 47.17 versus 108.39 ± 64.48) and troponin T level (0.04 ± 0.02 vs. 1.15 ± 0.56) between two groups (P < 0.05). Detail was documented in Table 1. For the verification cohort, the average age was 57.49 in the control group and 58.02 in the AMI group. Other baseline characteristics for the overall and verification cohort were shown in Additional file 2: Table S1.
Table 1.
Baseline characteristics of AMI patient group and control group
| Control group (n = 36) | AMI group (n = 80) | P value | |
|---|---|---|---|
| Age (years) | 58.50 ± 4.29 | 58.25 ± 8.99 | 0.874 |
| BMI (kg/m2) | 24.15 ± 2.29 | 26.08 ± 2.06 | < 0.001 |
| Hypertension (n) | 14 (38.9%) | 55 (67.5%) | 0.002 |
| Diabetes mellitus (n) | 16 (44.4%) | 50 (58.8%) | 0.069 |
| Alcohol drinking (n) | 16 (44.4%) | 53 (66.3%) | 0.027 |
| Smoking (n) | 13 (46.1%) | 56 (70.0%) | < 0.001 |
| Tc (mmol/L) | 4.22 ± 0.75 | 4.54 ± 0.77 | 0.039 |
| LDL (mmol/L) | 2.76 ± 0.37 | 3.34 ± 0.33 | < 0.001 |
| HDL (mmol/L) | 1.24 ± 0.12 | 1.15 ± 0.16 | 0.003 |
| CKMB (U/L) | 53.97 ± 47.17 | 108.39 ± 64.48 | < 0.001 |
| TnT (μg/L) | 0.04 ± 0.02 | 1.15 ± 0.56 | < 0.001 |
Values are presented in mean ± standard deviation (sd) or n(%)
BMI body mass index, Tc total cholesterol, LDL low density lipoprotein, HDL high density lipoprotein; CK-MB creatine kinase-MB, TnT troponin T
Plasma level of lncRNA TTTY15 and HULC
To verify the possibility of lncRNA TTTY15 and HULC being novel biomarkers in AMI diagnosis, their plasma levels in AMI patients were tested in comparison with healthy control. The difference between the group was tested employing Mann Whitney test, including lncRNA level at admission (pre-PCI) versus post-PCI and lncRNA level at pre-PCI versus post-PCI.
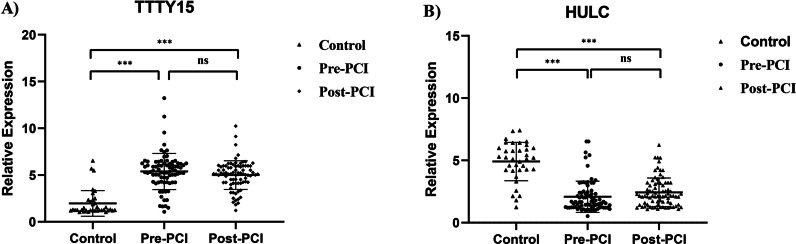
Regarding plasma TTTY15 level, a total of 80 AMI patients’ plasma and 36 healthy controls’ plasma were collected. After detecting the TTTY15 level via qRT-PCR calculated by 2−ΔΔcq methods, TTTY15 level of AMI patients were significantly up-regulated in comparison with the control group, and detailed information was displayed in the scatter plot of Fig. 1A. For plasma HULC level, the same number of blood samples was tested. Inversely, HULC levels in AMI patients were significantly down-regulated in comparison with the control group, with scatter plot shown in Fig. 1B. The expression levels of two lncRNA in the verification cohort were shown in Additional file 3: Figure S2.
Fig. 1.
LncRNA relative expression level of TTTY15 and HULC between AMI patients and healthy controls in the discovery cohort. A Relative expression level of TTTY15 with the comparison of control, pre-PCI and post-PCI procedure; B Relative expression level of HULC with the comparison of control, pre-PCI and post-PCI. ***indicated P < 0.001 and ns indicated no significance
The predictive power of lncRNA TTTY15 and HULC as novel biomarkers
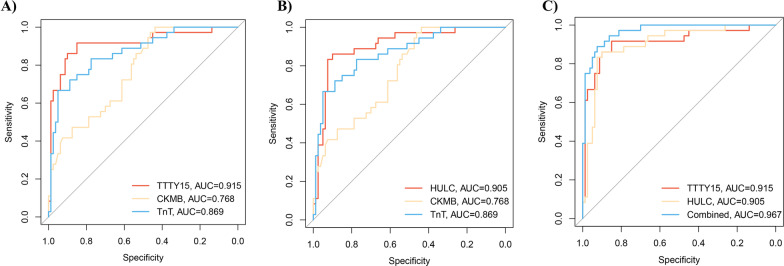
To evaluate the predictive power of TTTY15 and HULC as the novel biomarkers in AMI diagnosis, the ROC curve was preferentially adopted in verification. From Fig. 2A, ROC analysis unveiled the superiority of TTTY15 as biomarkers against conventional AMI biomarkers CKMB and TnT (AUC of TTTY 15: 0.915 versus CKMB: 0.768 versus TnT: 0.869). Similarly, as the results obtained from Fig. 2B, ROC analysis revealed the significant predictive power of HULC compared with CKMB and TnT (AUC of HULC: 0.905 versus CKMB: 0.768 versus TnT: 0.869). We also established a model with the combination of TTTY15 with HULC to test whether the predictive power would improve. As shown in Fig. 2C, the combination model revealed higher predictive power than TTTY15 or HULC alone (AUC of combination = 0.967).
Fig. 2.
AUC of selected lncRNA. A ROC curve of TTTY15 (AUC = 0.915) in comparison with CKMB (AUC = 0.768) and TnT (AUC = 0.869); B ROC curve of HULC (AUC = 0.905) in comparison with CKMB (AUC = 0.768) and TnT (AUC = 0.869); C ROC curve showing the TTTY15 (AUC = 0.915), HULC (AUC = 0.905) and combination of TTTY15 with HULC (AUC = 0.967). AUC Area under the curve; ROC curve receiver operator characteristic curve
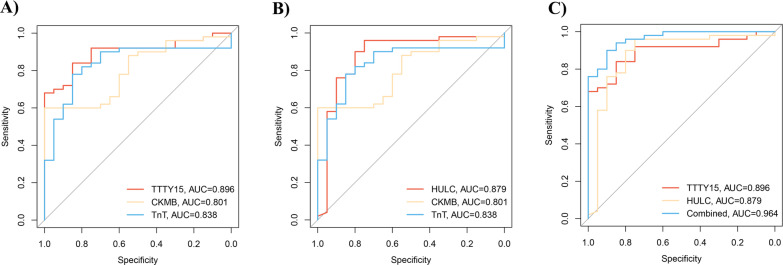
Also, to test the potential diagnostic value of the selected lncRNA, AUC was tested in a verification cohort. As shown in Fig. 3, TTTY15 (AUC = 0.896, Fig. 3A) and HULC (AUC = 0.879, Fig. 3B) were revealed to be better biomarkers in comparison with CKMB (AUC = 0.801) and TnT (AUC = 0.838). Similarly, the combination of TTTY15 and HULC shows higher predictive power (AUC = 0.964, Fig. 3C). Further, to verify the predictive power of the selected lncRNA, we combined the exsiting biomarkers in the ROC analysis, indicating that predictive power of the selected lncRNA were higher than the exsiting biomarkers (Fig. 3 and Additional file 4: Figure S3).
Fig. 3.
AUC of selected lncRNA in verification cohort. A ROC curve of TTTY15 (AUC = 0.896) in comparison with CKMB (AUC = 0.801) and TnT (AUC = 0.838); B ROC curve of HULC (AUC = 0.879) in comparison with CKMB (AUC = 0.801)and TnT (AUC = 0.838); C ROC curve showing the TTTY15 (AUC = 0.896), HULC (AUC = 0.879) and combination of TTTY15 with HULC (AUC = 0.964). AUC Area under the curve, ROC curve Receiver operator characteristic curve
Collectively, lncRNA TTTY15 and HULC are potentially valuable biomarkers in AMI diagnosis.
Logistic regression and correlation analysis
To further validate the fidelity of our postulation, logistic regression was performed to investigate the contribution of selected lncRNA to AMI in cooperation with established risk factors. From logistic regression, widely accepted risk factors such as CKMB (OR 1.02, P < 0.001), TnT (OR 9.481, P < 0.001), and BMI (OR 1.487, P < 0.001) were significantly associated with AMI. In concert with established risk factors, lncRNA TTTY15 (OR 2.752, P < 0.001) and HULC (OR 0.358, P < 0.001) were revealed to be associated with AMI. Detailed information on logistic regression was documented in Table 2.
Table 2.
Logistics regression analysis for the association of lncRNA TTTY15, HULC and potential risk factors with occurrence of AMI
| Variable | B | S.E | Wald | P | OR | 95% CI | |
|---|---|---|---|---|---|---|---|
| TTTY15 | 1.012 | 0.187 | 29.174 | < 0.001 | 2.752 | 1.906 | 3.974 |
| HULC | − 1.027 | 0.193 | 28.411 | < 0.001 | 0.358 | 0.245 | 0.522 |
| Age | − 0.043 | 0.029 | 2.191 | 0.139 | 0.958 | 0.904 | 1.014 |
| DM | − 0.489 | 0.435 | 1.262 | 0.261 | 0.613 | 0.261 | 1.439 |
| Tc | 0.576 | 0.304 | 3.594 | 0.058 | 1.779 | 0.981 | 3.225 |
| CKMB | 0.015 | 0.004 | 14.166 | < 0.001 | 1.015 | 1.007 | 1.023 |
| TnT | 2.249 | 0.468 | 23.082 | < 0.001 | 9.481 | 3.787 | 23.733 |
| BMI | 0.397 | 0.111 | 12.754 | < 0.001 | 1.487 | 1.196 | 1.849 |
| Smoking | − 0.358 | 0.428 | 0.873 | 0.357 | 0.524 | 0.258 | 1.643 |
| Alcohol | 0.254 | 0.63 | 0.289 | 0.652 | 1.285 | 0.531 | 3.275 |
DM diabetes mellitus, Tc total cholesterol, LDL low density lipoprotein, CKMB creatine kinase isoenzymes, TnT troponin T, BMI body mass index
In addition, correlation analysis was performed to investigate the relationship between selected lncRNA and cardiovascular risk factors in the AMI group. For TTTY15, it was unveiled to be positively related to LDL (coefficient of 4.260, P < 0.008), CKMB (coefficient of 0.015, P < 0.001), and TnT (coefficient of 2.161, P < 0.001). To be in concordance with qRT-PCR results, HULC was revealed negatively related to CKMB (r = − 0.823, P < 0.001) and TnT (coefficient of − 1.115, P < 0.005). Detailed information about correlation analysis was shown in Table 3.
Table 3.
Correlation analysis investigating the association between lncRNA TTTY15 and HULC with potential cardiac risk factors
| TTTY15 | HULC | |||
|---|---|---|---|---|
| Coefficient | P value | Coefficient | P value | |
| Age | − 0.039 | 0.259 | 0.040 | 0.135 |
| BMI | 0.352 | 0.002 | − 0.247 | 0.005 |
| Tc | 0.576 | 0.058 | − 0.465 | 0.085 |
| LDL | 4.260 | < 0.001 | − 12.611 | < 0.001 |
| CKMB | 0.015 | < 0.001 | − 0.823 | < 0.001 |
| TnT | 2.161 | < 0.001 | − 1.115 | < 0.001 |
BMI body mass index, Tc total cholesterol, LDL low density lipoprotein, CKMB creatine kinase isoenzymes, TnT troponin T
Discussion
Incremental studies have reported the potency of lncRNA being novel biomarkers to diagnose AMI, which may increase not only the accuracy but also the efficiency of the diagnostic process [17–19]. Meanwhile, lncRNA predominates in pivotal biological processes involved in cardiovascular diseases through ceRNA regulated gene expression, so exploration of these biomarkers is also beneficial to unveil potential therapy targets and get familiar with the underlying pathological changes of the myocardium [20, 21]. More importantly, lncRNA owns acceptable stability in peripheral circulation and detection sensitivity [22, 23]. Collectively, employing lncRNA plus conventional biomarkers is pressing needed in AMI diagnosis.
In the current study, we analyzed the lncRNA TTTY15 and HULC levels in peripheral blood of AMI patients and healthy volunteers to identify their potential capacity to be adopted as novel biomarkers in AMI diagnosis. Our results recapitulated that TTTY15 and HULC were plausible biomarkers to diagnose AMI. Interestingly, TTTY15 was upregulated while HULC was downregulated in AMI patients. ROC analysis delineated acceptable predictive power of TTTY15 and HULC as biomarkers compared with CKMB and TnT in AMI diagnosis. Moreover, consistent with established cardiovascular risk factors, TTTY15 and HULC levels were identified to be associated with AMI and, they have also correlated to LDL as well as CKMB levels. This is the first study revealing the capacity of lncRNA TTTY15 and HULC to be administered in AMI diagnosis, which may inspire cardiologists in clinical practice. A single biomarker has its inherent limitation in diagnosis, yet multi-biomarkers are available to provide more information about specific pathological mechanisms [24, 25]. Consequently, the combination of lncRNA as biomarkers in clinical practice is necessary and it could hint at an effective therapeutic strategy.
In previous studies, TTTY15 has been found highly expressed in peripheral blood of AMI patients and H2O2-stimulated AC16 cells model, which are in concert with our results [15, 26]. Moreover, TTTY15 downregulation or silence suppresses H2O2-stimulated AC16 cell apoptosis, inflammatory response, and oxidative stress, and improves cell viability. Based on these theories, not surprisingly, silenced TTTY15 could reduce the size of infarction in the AMI model [15]. Apart from being a potential biomarker in AMI diagnosis, the abovementioned experimental evidence extrapolates that suppressing TTTY15 could be a plausible pathway to alleviate the progression of AMI, providing novel therapeutic targets of treatment.
Before being acknowledged the roles in the cardiovascular field, lncRNA HULC has been studied thoroughly in the cancer domain. It has been reported to be the novel biomarkers in hepatocellular carcinoma and abnormally expressed in pancreatic and gastric cancer [27–29]. Recently, HULC has been explored its function in the cardiovascular field. It has been revealed that HULC would be downregulated in the I/R-injured myocardial model and overexpressed by miR-377-5p mediation leading increase of conventional myocardial injury biomarkers (Troponin-T and CKMB) level in the H9c2 cell model [14]. In addition, HULC plays an essential role in cellular inflammation, a pivotal pathway involved in myocardial injury. Knockdown of HULC has been found to profoundly reduce inflammatory factors level (IL-6, ICAM1, VCAM1) in lipopolysaccharide (LPS) treatment induced cellular experiments, and its overexpression could significantly relieve TNF-α induced cell injury [30, 31]. Collectively, in combination with our results, HULC could be not only the diagnostic biomarkers but also the potential therapeutic target.
Several limitations should be considered in this research. Firstly, although the positive results delineate the potentially novel biomarkers in AMI diagnosis, future studies investigating these lncRNAs with larger sample sizes are imminent. Secondly, lncRNA TTTY15 and HULC are associated with other diseases so the administration of these novel biomarkers should rule out the confounding diseases. Thirdly, qRT-PCR is the preferential method to detect RNA while it is expensive and time-consuming, so quantification of lncRNA in clinical practice is challenged. Last but not the least, it should be noted that the pathophysiology of the lncRNA should be further studied due to the currently limited knowledge.
Conclusion
Our results recapitulate that lncRNA TTTY15 and HULC are significantly different between AMI patients and healthy controls, revealing the potential capacity of these lncRNAs to be novel biomarkers to assist AMI diagnosis with the combination of conventional biomarkers, while the relatedly confounding diseases should be ruled out when administering these biomarkers.
Supplementary Information
Additional file 1. Cq value or each group.
Additional file 2. Baseline characteristics of the verification cohort.
Additional file 3. LncRNA relative expression level in the verification cohort.
Additional file 4. AUC of the selected lncRNA and existed biomarkers.
Acknowledgements
Not applicable.
Authors' contributions
The detailed contributions of each author are listed as followed: JX: conceptualization, methodology, data analysis, manuscript writing. WL: methodology, data analysis. WC: investigation. DL: verification cohort and investigation. QT: investigation. YL: investigation. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article. There was no any sequencing, genomic, phylogenetic data generated during this study. Request for the related data of this study should refer to Dr. Xie, the corresponding author.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Guangdong Second Provincial General Hospital and informed consents were obtained from all included patients. Also, the protocol of this study was performed in concordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
All authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD, Thygesen K, Alpert JS, White HD, Jaffe AS, et al. Third universal definition of myocardial infarction. J Am Coll Cardiol. 2012;60(16):1581–1598. doi: 10.1016/j.jacc.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Khalid MF, Khan AA, Khattak F, Ayub MT, Bagai J, Mukherjee D, Helton T, Cohen MG, Banerjee S, Paul TK. Culprit vessel only versus multivessel percutaneous coronary intervention in acute myocardial infarction with cardiogenic shock: a systematic review and meta-analysis. Cardiovasc Revascularization Med Incl Mol Interv. 2019;20(11):956–964. doi: 10.1016/j.carrev.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Anderson JL, Morrow DA. Acute myocardial infarction. N Engl J Med. 2017;376(21):2053–2064. doi: 10.1056/NEJMra1606915. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Cárceles MD, Noguera J, Jiménez JL, Martínez P, Luna A, Osuna E. Diagnostic efficacy of biochemical markers in diagnosis post-mortem of ischaemic heart disease. Forensic Sci Int. 2004;142(1):1–7. doi: 10.1016/j.forsciint.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Park KC, Gaze DC, Collinson PO, Marber MS. Cardiac troponins: from myocardial infarction to chronic disease. Cardiovasc Res. 2017;113(14):1708–1718. doi: 10.1093/cvr/cvx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Masoudi FA, Li J, Wang Q, Li X, Spertus JA, Ross JS, Desai NR, Krumholz HM, Jiang L. National assessment of early β-blocker therapy in patients with acute myocardial infarction in China, 2001–2011: The China Patient-centered Evaluative Assessment of Cardiac Events (PEACE)-Retrospective AMI Study. Am Heart J. 2015;170(3):506–515.e501. doi: 10.1016/j.ahj.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng C, Li P, Wang YG, Bi MH, Wu PS. Study on the expression of VEGF and HIF-1α in infarct area of rats with AMI. Eur Rev Med Pharmacol Sci. 2016;20(1):115–119. [PubMed] [Google Scholar]
- 8.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 9.Navickas R, Gal D, Laucevičius A, Taparauskaitė A, Zdanytė M, Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riffo-Campos AL, Perez-Hernandez J, Ortega A, Martinez-Arroyo O, Flores-Chova A, Redon J, Cortes R. Exosomal and plasma non-coding RNA signature associated with urinary albumin excretion in hypertension. Int J Mol Sci. 2022;23(2):823. doi: 10.3390/ijms23020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Nan S, Duan X, Wang L, Sun X, Zheng H. Relationship between expression of plasma lncRNA-HEIH and prognosis in patients with coronary artery disease. Dis Markers. 2021;2021:5662080. doi: 10.1155/2021/5662080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Z, Shen H, Cha W, Xia H, Liu L. Predictive value of using plasma long non-coding RNAs ANRIL and HOXA11-AS for in-stent restenosis. Exp Ther Med. 2022;23(2):115. doi: 10.3892/etm.2021.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, Liu Y, Wei C, Yang Z, Chang W, Zhang X. LncRNA HULC promotes the progression of gastric cancer by regulating miR-9–5p/MYH9 axis. Biomed Pharmacother. 2020;121:109607. doi: 10.1016/j.biopha.2019.109607. [DOI] [PubMed] [Google Scholar]
- 14.Liang H, Li F, Li H, Wang R, Du M. Overexpression of lncRNA HULC attenuates myocardial ischemia/reperfusion injury in rat models and apoptosis of hypoxia/reoxygenation cardiomyocytes via targeting miR-377-5p through NLRP3/caspase-1/IL-1β signaling pathway inhibition. Immunol Investig. 2020;50:925–938. doi: 10.1080/08820139.2020.1791178. [DOI] [PubMed] [Google Scholar]
- 15.Ma R, Gao L, Liu Y, Du P, Chen X, Li G. LncRNA TTTY15 knockdown alleviates H(2)O(2)-stimulated myocardial cell injury by regulating the miR-98-5p/CRP pathway. Mol Cell Biochem. 2020;476:81–92. doi: 10.1007/s11010-020-03887-4. [DOI] [PubMed] [Google Scholar]
- 16.Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, Caforio ALP, Crea F, Goudevenos JA, Halvorsen S, et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: The Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC) Eur Heart J. 2017;39(2):119–177. doi: 10.1093/eurheartj/ehx393. [DOI] [PubMed] [Google Scholar]
- 17.Qiu L, Zhao Q, Dai L, Zhu A, Xu X, Zhao S, Chen J. Long non-coding RNA DANCR alleviates hypoxia-caused H9c2 cells damage through up regulation of HIF-1α. Artif Cells Nanomed Biotechnol. 2020;48(1):533–541. doi: 10.1080/21691401.2020.1725026. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Dai Y, Yan S, Shi Y, Han B, Li J, Cha L, Mu J. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun. 2017;491(4):1026–1033. doi: 10.1016/j.bbrc.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, Zhai M, Huang Y, Yan KW, Dong YH, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9(1):29. doi: 10.1038/s41467-017-02280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Zhang Y, Ding N, Zhao Y, Ye Z, Shen L, Yi H, Zhu Y. Inhibition of lncRNA XIST improves myocardial I/R injury by targeting miR-133a through inhibition of autophagy and regulation of SOCS2. Mol Ther Nucl Acids. 2019;18:764–773. doi: 10.1016/j.omtn.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Yao T, Deng F, Yu W, Song Y, Chen J, Ruan Z. LncRNA MALAT1 Promotes oxygen-glucose deprivation and reoxygenation induced cardiomyocytes injury through sponging miR-20b to enhance beclin1-mediated autophagy. Cardiovasc Drugs Ther. 2019;33(6):675–686. doi: 10.1007/s10557-019-06902-z. [DOI] [PubMed] [Google Scholar]
- 22.Ankö ML, Neugebauer KM. Long noncoding RNAs add another layer to pre-mRNA splicing regulation. Mol Cell. 2010;39(6):833–834. doi: 10.1016/j.molcel.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55(2):941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 24.Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, Castelvecchio S, Gaetano C, Finato N, Beltrami AP, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14(1):183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, Huang Y, Zhao X, Huang L, Wang Z, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Tao W, Guo Z, Cao J, Huang X. Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-455-5p. Gene. 2019;701:1–8. doi: 10.1016/j.gene.2019.02.098. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Liao Z, Liu F, Su C, Zhu H, Li Y, Tao R, Liang H, Zhang B, Zhang X. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging. 2019;11(20):9111–9127. doi: 10.18632/aging.102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Ota Y, Kogure T, Suzuki Y, Iwamoto H, Yamakita K, Kitano Y, Fujii S, Haneda M, Patel T, et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020;111(1):98–111. doi: 10.1111/cas.14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin L, Zhou Q, Yuan YW, Zhou LQ, Liu L, Li SH, Liu C. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol. 2019;145(10):2507–2517. doi: 10.1007/s00432-019-03015-w. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Fu Y, Song YF, Li N. Increased expression of lncRNA UCA1 and HULC is required for pro-inflammatory response during LPS induced sepsis in endothelial cells. Front Physiol. 2019;10:608. doi: 10.3389/fphys.2019.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu P, Wang Q, Wang Z, Gao C. Long non-coding RNA highly up-regulated in liver cancer protects tumor necrosis factor-alpha-induced inflammatory injury by down-regulation of microRNA-101 in ATDC5 cells. Int Immunopharmacol. 2019;72:148–158. doi: 10.1016/j.intimp.2019.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Cq value or each group.
Additional file 2. Baseline characteristics of the verification cohort.
Additional file 3. LncRNA relative expression level in the verification cohort.
Additional file 4. AUC of the selected lncRNA and existed biomarkers.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article. There was no any sequencing, genomic, phylogenetic data generated during this study. Request for the related data of this study should refer to Dr. Xie, the corresponding author.