Abstract
Aims
Elevated brain natriuretic peptide (BNP) and the N‐terminal fragment of its pro‐hormone (NT‐proBNP) have become established biomarkers for heart failure and are associated with cardiovascular morbidity and mortality. Investigating sources of inter‐individual heterogeneity, particularly genetic factors, could help better identify patients at risk of future cardiovascular disease. The aim of this study was to estimate the heritability of circulating NT‐proBNP levels, to perform a genome‐wide association study (GWAS) and gene‐candidate analysis focused on NPPB–NPPA genes on these levels, and to examine their association with cardiovascular or metabolic outcomes.
Methods and results
A total of 1555 individuals from the STANISLAS study were included. The heritability of circulating NT‐proBNP levels was estimated at 15%, with seven single nucleotide polymorphisms (SNPs) reaching the significant threshold in the GWAS. All above SNPs were located on the same gene cluster constituted of MTHFR, CLCN6, NPPA, NPPB, and C1orf167. NPPA gene expression was also associated with NT‐proBNP levels. Moreover, six other SNPs from NPPA–NPPB genes were associated with diastolic function (lateral e′ on echocardiography) and metabolic features (glycated haemoglobin).
Conclusions
The heritability of natriuretic peptides appears relatively low (15%) and mainly based on the same gene cluster constituted of MTHFR, CLCN6, NPPA, NPPB, and C1orf167. Natriuretic peptide polymorphisms are associated with natriuretic peptide levels and diastolic function. These results suggest that natriuretic peptide polymorphisms may have an impact in the early stages of cardiovascular and metabolic disease.
Keywords: NT‐proBNP, NPPB, NPPA, Genome‐wide association study, Polymorphism, Cardiovascular diseases
Introduction
Natriuretic peptides are cardiac hormones produced primarily by the atria and ventricles and are deeply involved in the pathophysiology of heart failure (HF). These peptides play a critical role in the regulation of circulatory volume status, plasma renin‐aldosterone concentrations, natriuresis, and the maintenance of blood pressure levels. 1 Plasma levels of brain natriuretic peptides (BNPs) and the N‐terminal fragment of its pro‐hormone (NT‐proBNP) have become established biomarkers to facilitate the diagnosis and risk stratification of HF and are associated with cardiovascular (CV) morbidity and mortality in the general population. 1 , 2 , 3 Therefore, most guidelines for the management of HF recommend their use to assist in the diagnosis of HF. 4 , 5 , 6 In addition, natriuretic peptides are important prognostic markers in the setting of primary prevention as they are associated with clinical outcome in patients with hypertension and/or chronic kidney disease.
Natriuretic peptides variability has been perceived until now as the result of a pathophysiological process. Yet, inter‐individual heterogeneity is a known limitation of natriuretic peptides variability, the source of which has yet to be extensively studied. While some studies have focused on genetic factors involved in natriuretic peptides variability in subjects without HF, 7 , 8 , 9 , 10 their role in CV function has never been assessed. A better understanding of genetic risk factors involved in natriuretic peptides variability could help better identify patients at risk of future CV disease.
The present study focused on the genetic bases of NT‐proBNP plasma levels in a population‐based family cohort. The aims of the study were to (i) estimate the heritability of plasma NT‐proBNP levels; (ii) perform a genome‐wide association study (GWAS) analysis of these levels; (iii) examine potential associations between CV and metabolic phenotypes and top single nucleotide polymorphisms (SNPs) from the GWAS on NT‐proBNP levels; (iv) run a more specific gene‐candidate analyses for SNPs located within the NPPB and NPPA genes (the coding gene of NT‐proBNP and BNP) and cardiometabolic phenotypes; and (v) assess the association between the involved gene expressions and NT‐proBNP levels and polymorphisms highlighted in both the GWAS analysis and NPPB gene‐candidate analysis.
Methods
Study population
The data that support the findings of this study are available from the corresponding author upon reasonable request. The design of the STANISLAS (Suivi Temporaire Annuel Non‐Invasif de la Santé des Lorrains Assurés Sociaux) cohort has been previously described. 11 In brief, the STANISLAS cohort is a family‐based longitudinal cohort, initially including 4598 healthy individuals of French origin from 1006 families living in the Lorraine region (North‐East of France). Participants were examined every 5 to 10 years, during 20 years. A total of 1705 participants returned for the fourth visit (V4), held between 2011 and 2016, consisting of an interview by trained nurses using a structured questionnaire, including items pertaining to socio‐demographic characteristics, medical and family history, smoking status, lifestyle, diet, and anthropometric data. Beyond the detailed clinical and laboratory assessment, CV echography and routine laboratory measurements include a spot urine and blood samples. Measurement reproducibility was assessed from a duplicate reading by experienced echocardiographers blinded to each other and to demographic parameters. These results have been previously published 12 , 13 and suggest a good reproducibility of all considered echo variables.
The study protocols for all examinations were reviewed and approved by the local ethics committee of CPP Est 3, France. All participants provided written informed consent to participate in the study.
Biomarker measurements
All blood draws were taken in the morning on fasting subjects. The samples were then centrifuged at 1500 g for 15 min at room temperature and frozen in the 4 h following the blood draw at −196°C. Circulating levels of multiple proteins (including NT‐proBNP) were assessed both at baseline (Visit 1) and at V4, using the Olink method with PEA as described in Ferreira et al. 14 NT‐proBNP levels are expressed in arbitrary unit because they were measured with the Olink® technology standardized log2 NPX values. A subset of 428 subjects has also been measured for NT‐proBNP levels using Elisa methods in order to test for comparability between methods, the two measured are highly correlated (coef cor = 0.89, P < 0.001, Supporting Information, Figure S1 ).
Genotyping
Genotyping of the STANISLAS V4 participants was conducted at the Centre National de Recherche en Génomique Humaine (Evry, France) using two chips: the Illumina Global Screening Array, which is composed of 687 572 intronic and exonic markers, and the Illumina Exome Array, which is composed of 244 330 SNPs, mostly exonic. After QC steps on raw genotyped data [exclusion of extremely rare markers with a minor allele frequency (maf) < 0.005, call rate for markers and individuals set up at 95%, and Hardy–Weinberg equilibrium with P value < 10−8], 1576 individuals and 520 773 polymorphic autosomal markers were included in the GWAS.
Selection of single nucleotide polymorphisms from the NPPB–NPPA genes for gene‐candidate analysis
In addition to GWAS analysis, a gene‐candidate approach focused on NPPB and NPPA genes has been run. Firstly, intervals encompassing the boundaries (±20 kb) of the two genes (NPPA and NPPB) were defined, based on the reference genome built 37 from the Ensembl database. NPPB and NPPA genes were both located on chromosome 1; boundaries were respectively chr1: 11917521–11918988 and chr1: 11905766–11908402 (http://grch37.ensembl.org). All SNPs comprised between these boundaries in the two chips were subsequently selected, including 21 SNPs from the GSA chip and 8 from the Exome chip. However, five markers were duplicated between the two chips, resulting in one of each being excluded. Hence, 24 SNPs were ultimately selected (Supporting Information, Table S1 ).
Gene expression analysis
Whole blood RNAs were automatically extracted from PAXgene Blood RNA Tubes (Qiagen, Hilden, Germany) using MagMAX for Stabilized Blood Tubes RNA Isolation Kit (Life Technologies, Villebon‐sur‐Yvette, France) on a KingFisher Duo Prime automated purification system (ThermoFisher Scientific, Dardilly, France). Extracted RNAs were quantified using a Nanodrop spectrophotometer, and their quality was assessed using the RNA ScreenTape system (Agilent, les Ulis, France). Transcriptome analysis was conducted at the Ingénierie Moléculaire et Physiopathologie Articulaire (IMoPA) (Vandoeuvre‐les‐Nancy, France) using Clariom D® assays (Affymetrix, ThermoFisher Scientific, Dardilly, France) following the manufacturer's recommendations. Gene expression intensities were normalized with ‘Limma’ (Version 3.44.3) 15 implemented in the Transcription Analysis Console Software 4.0.2 by SST‐RMA and then extracted as normalized intensity values (in log2) obtained from Clariom D® microarrays for each individual. Gene expressions were assessed on a subgroup of 553 individuals having available transcriptomic data.
Statistical analyses
For heritability estimation, a genetic relationship matrix was used in a linear mixed model to estimate the variance captured by additive genetic effects via average information restricted maximum likelihood analysis. 16 An additional random effect was defined in order to take into account the common household effect resulting from nuclear families (i.e. parental couple and children aged less than 20 years old). In addition to usual covariates (sex and age), estimated glomerular filtration rate (eGFR), heart rate, systolic blood pressure, urinary sodium, and body mass index (BMI) were also tested for their association with plasma NT‐proBNP levels using a multivariate linear model. GWAS were run using a linear mixed under an additive model in order to take into account pedigree data, with age and sex used as covariates. Ethnicity was not included in covariates, but all subjects were found to be genetically homogenous in QC steps.
Both models were performed using R (Version 3.4.1.) and were implemented in the ‘gaston’ R package (Version 1.5.6). 17 For the GWAS, the statistical threshold was set at 10−7. For the top SNP association tests, multivariate linear models, with age and sex as covariates, were used and the statistical significance level was fixed at an FDRq < 0.05, after applying a Benjamini–Hochberg correction for multiple testing [i.e. false discovery rate (FDR) set at 5%].
Results
Characteristics of study participants
A total of 1555 individuals were included in the present study; their characteristics are displayed in Table 1 . A total of 150 individuals were not included because 26 had no NT‐proBNP measurements and 124 were unsuccessfully genotyped (the characteristics of these 150 subjects are not sizeably different from the ones of the subjects considered in our subsequent analyses; Supporting Information, Table S2 ). The participants belonged to 664 families, comprised from 1 to 6 individuals [173 individuals (11.1%) were sole member of their family in this fourth visit]. A subgroup of 553 participants had available gene expression data. They were slightly younger (mean age 44.43 years, range 18–74; t‐test P value < 0.001), but the sex ratio was statistically not different (281 women, 50.8%). NT‐proBNP levels are lower in healthy subjects than in hypertensive or in subjects with any CV conditions (Table 1 —all P value < 0.001). In subsequent analysis, we classified subjects according to healthy vs. unhealthy status (combination of hypertensive and any CV conditions) to perform stratified analyses.
Table 1.
Characteristics of the study subjects (n = 1555)
| Characteristics | Mean ± SD/n (%) |
|---|---|
| Age (years) | 48.81 ± 14.11 |
| Sex female | 800 (51.45%) |
| Current smoker | 328 (21.15%) |
| BMI (kg/m2) | 25.89 ± 4.78 |
| Diabetes | 72 (4.63%) |
| Fasting glycaemia (g/L) | 0.91 ± 0.16 |
| Glycated haemoglobin (%) | 5.64 ± 0.56 |
| eGFR (mL/min/1.73 m2) | 96.42 ± 15.47 |
| Urinary sodium (mmol/L) | 115.47 ± 47.20 |
| Heart rate (b.p.m.) | 62.49 ± 9.77 |
| SBP (mmHg) | 120.12 ± 10.22 |
| DBP (mmHg) | 74.25 ± 7.21 |
| Hypertension | 487 (31.68%) |
| Any CV conditions a | 70 (4.50%) |
| Heart failure | 13 (0.84%) |
| Myocardial infarction | 12 (0.77%) |
| Stroke | 21 (1.35%) |
| Valvular disease | 41 (2.64%) |
| Septal e′ (cm/s) | 9.92 ± 2.95 |
| Lateral e′ (cm/s) | 12.88 ± 4.11 |
| e/a | 1.19 ± 0.42 |
| e/e′ | 6.47 ± 1.87 |
| LAVI (mL) | 22.73 ± 7.31 |
| Deceleration time | 211.00 ± 53.22 |
| NT‐proBNP levels according to category of subjects (arbitrary PEA units) | |
| All study subjects | 3.58 ± 1.01 |
| Healthy (n = 1042) | 3.45 ± 0.94 |
| Hypertension (n = 443) | 3.74 ± 1.04 |
| Any CV conditions a (n = 70) | 4.51 ± 1.24 |
| NPPA expression (arbitrary unit) (n = 501) | 3.88 ± 0.32 |
| NPPB expression (arbitrary unit) (n = 501) | 3.73 ± 0.22 |
BMI, body mass index; CV, cardiovascular; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; LAVI, left atrial volume index; NT‐proBNP, N‐terminal pro‐brain natriuretic peptide; SBP, systolic blood pressure; SD, standard deviation.
Any CV condition category includes heart failure, myocardial infarction, stroke, and valvular disease.
Heritability estimation
Among potentially associated variables, spot urinary sodium, heart rate, and eGFR were significantly associated with plasma NT‐proBNP levels (Supporting Information, Table S3 ) and were included in the heritability estimations as covariates, along with sex and age. When all covariates were included in the model, the heritability of circulating plasma NT‐proBNP was estimated at 15.1% (Figure 1 ); common environmental effects due to nuclear family accounted for less than 1%, effects due to covariates accounted for 23%, while 61.3% of the variance remained unexplained. When only age and sex were considered as covariates, the heritability estimate was similar, whereas the effect of common environment accounted for 1.6% while covariates accounted for 19.8% (Figure 1 and Supporting Information, Table S4 ). Using plasma levels of NT‐proBNP at baseline (V1), heritability estimations are similar with those at V4 (data not shown). Sensibility analysis that includes only healthy subjects (i.e. participants without hypertension, HF, stroke, myocardial infarction, or valvular disease) gave similar results for NT‐proBNP heritability (Supporting Information, Table S5 ).
Figure 1.
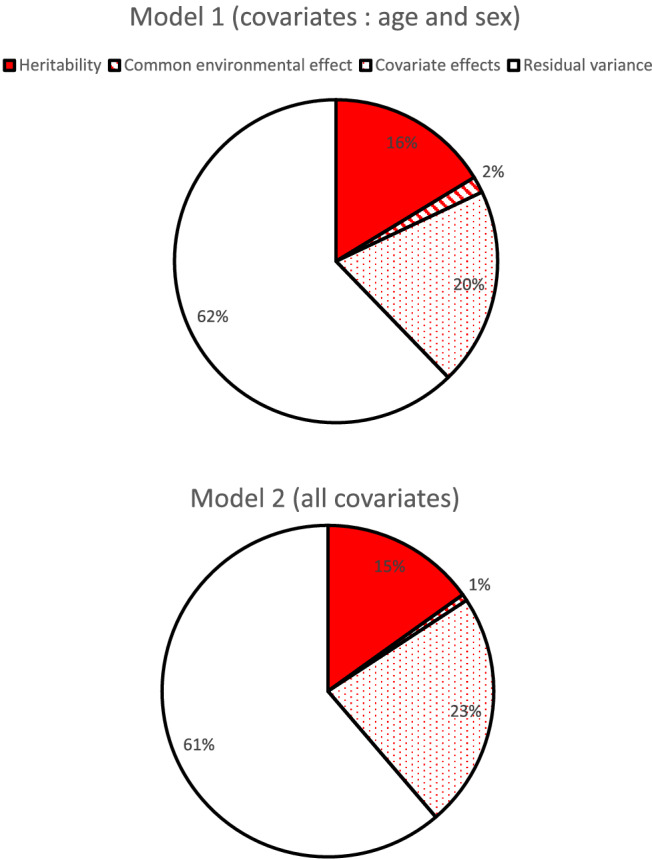
Variance decomposition of circulating N‐terminal pro‐brain natriuretic peptide with age, sex, estimated glomerular filtration rate, and heart rate as covariates (Model 1: age and sex as covariates; Model 2: age, sex, estimated glomerular filtration rate, heart rate, and urinary sodium as covariates).
Genome‐wide association study for circulating N‐terminal pro‐brain natriuretic peptide levels
Seven SNPs reached the GWAS significant threshold with a P value < 10−7 (Table 2 , Figure 2 ). All were located on chromosome 1: the top SNP, rs198389, was located on the NPPB gene, rs4845881 and rs4845877 were located on C1orf167, rs1801131 and rs1476413 were located on the MTHFR gene, rs1023252 was located on CLCN6, and rs6676300 was located on the intergenic region upstream of NPPB (Figure 3 ). Minor alleles were associated with a higher level of circulating NT‐proBNP for all seven SNPs.
Table 2.
Characteristics of the seven single nucleotide polymorphisms significantly associated with N‐terminal pro‐brain natriuretic peptide (P value < 10−7) in the genome‐wide association study analysis
| Chr | pos | Rs name | A1 | A2 | Freq A2 | Beta | SD | P value | Gene location |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 11919271 | rs198389 | G | A | 0.563 | −0.257 | 0.034 | 1.76e‐14 | NPPB |
| 1 | 11828319 | rs4845881 | G | A | 0.674 | −0.230 | 0.036 | 1.02e‐10 | C1orf167 |
| 1 | 11854476 | rs1801131 | G | T | 0.692 | −0.230 | 0.036 | 1.18e‐10 | MTHFR |
| 1 | 11925300 | rs6676300 | G | A | 0.621 | −0.212 | 0.035 | 9.59e‐10 | None |
| 1 | 11852300 | rs1476413 | T | C | 0.732 | −0.213 | 0.037 | 1.09e‐08 | MTHFR |
| 1 | 11899033 | rs1023252 | T | G | 0.728 | −0.209 | 0.037 | 1.65e‐08 | CLCN6 |
| 1 | 11824303 | rs4845877 | T | C | 0.664 | −0.194 | 0.035 | 2.17e‐08 | C1orf167 |
A1, allele 1; A2, allele 2; beta, effect per A2 under the additive model; Chr, chromosome; Freq, allele 2 frequency; pos, position; SD, standard deviation.
Figure 2.
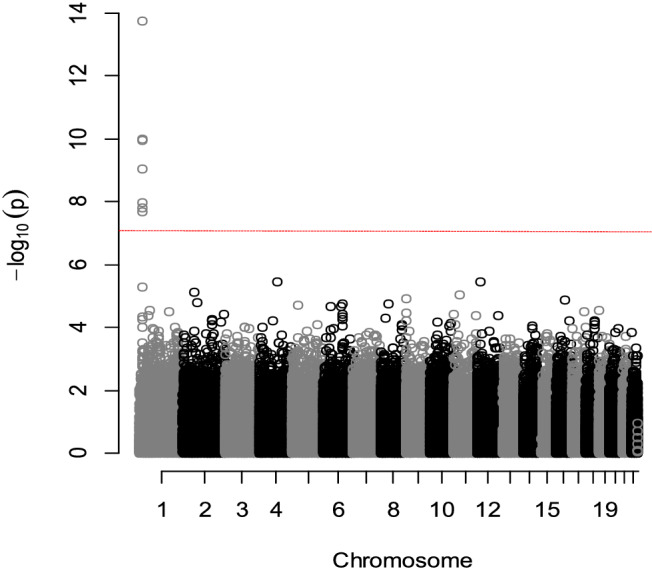
Manhattan plot of the genome‐wide association study analysis of circulating plasma N‐terminal pro‐brain natriuretic peptide levels (the horizontal red line indicates the statistically significant threshold at P value = 10−7). Sex and age were used as covariates.
Figure 3.
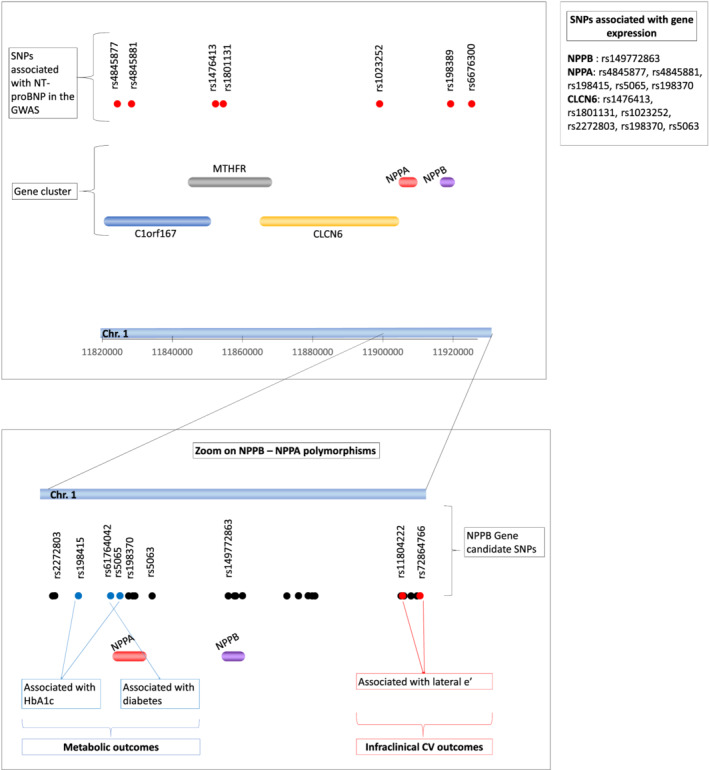
Illustration of the study's main findings. The upper part of the figure shows the localization of the seven GWAS SNPs (red points) and the genes along chromosome 1. The lower part of the figure shows a zoom on SNPs from the NPPB–NPPA genes. SNPs indicated with blue points are associated with metabolic outcomes, SNPs indicated with red points are associated with infraclinical CV outcomes, and the other SNPs are represented with black points. The SNPs, from either the GWAS study or the gene‐specific analysis, which associate with gene expression, are listed close to the name of the gene they are associated with. CV, cardiovascular; GWAS, genome‐wide association study; HbA1c, glycated haemoglobin; NT‐proBNP, N‐terminal pro‐brain natriuretic peptide; SNPs, single nucleotide polymorphisms.
When analysis was performed accordingly to health status of subjects, only rs198389 reached the 10−7 threshold in both categories. Moreover, similar results are obtained where GWAS was run using NT‐proBNP levels measured at baseline for all subjects, with the same top SNP rs198389 (P = 3.4·10−16).
Associations with infraclinical cardiovascular outcomes
Cardiac function outcomes
The following infraclinical CV outcomes have been tested: diastolic function, septal e′, lateral e′, e/a, e/e′, and left atrial volume index (LAVI). Only one of the seven SNPs reaching the significant GWAS threshold showed a borderline association with one of these outcomes (minor allele of rs4845877 with lower deceleration time, P = 0.01, FDR = 0.07). Among the SNPs located within the NPPB–NPPA gene cluster from the gene‐candidate analysis, rare alleles of rs72864766 and rs11804222 alleles were significantly associated with higher lateral e′ (P = 0.0002, FDR = 0.004 and P = 0.002, FDR = 0.02, respectively) (Table 3 , Figure 3 ). In stratified analyses, these associations remained significant only in the healthy group of subjects.
Table 3.
Statistically significant association of single nucleotide polymorphisms from the NPPB–NPPA cluster gene with cardiac or metabolic functions
| Tested outcome | Rs name | All subjects (n = 1555) | Healthy subjects (n = 1042) | HTN or any CV condition (n = 513) | P interaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SD | P val | FDR | Beta | SD | P val | FDR | Beta | SD | P val | FDR | |||
| Lateral e′ | rs72864766 | 0.96 | 0.26 | <0.001 | 0.004 | 1.46 | 0.33 | <0.001 | <0.001 | 0.35 | 0.41 | 0.39 | 0.82 | <0.001 |
| rs11804222 | 0.58 | 0.18 | 0.002 | 0.020 | 0.88 | 0.23 | <0.001 | <0.001 | 0.16 | 0.28 | 0.57 | 0.85 | <0.001 | |
| Decelaration Time | rs4845877 | −4.96 | 1.98 | 0.012 | 0.070 | −5.76 | 2.19 | 0.009 | 0.05 | −5.69 | 3.74 | 0.12 | 0.19 | 0.88 |
| Glycated haemoglobin | rs198415 | −0.10 | 0.03 | <0.001 | 0.012 | −0.01 | 0.03 | 0.64 | 0.74 | −0.26 | 0.06 | <0.001 | <0.001 | 0.001 |
| rs5065 | −0.09 | 0.03 | 0.001 | 0.016 | −0.01 | 0.03 | 0.74 | 0.74 | −0.25 | 0.06 | <0.001 | <0.001 | 0.001 | |
| Diabetes | rs61764042 | <0.001 | 0.002 | 0.002 | 0.01 | 0.009 | 0.06 | <0.001 | ||||||
CV, cardiovascular; FDR, false discovery rate; HTN, hypertension; SD, standard deviation.
Sex and age were used as covariates. Diabetes occurrence was defined as a fasting plasma glucose ≥ 126 mg/dL or a glycated haemoglobin ≥ 6.5% or a random plasma glucose ≥ 200 mg/dL.
Metabolic outcomes
None of the seven SNPs from the GWAS were significantly associated with any of the metabolic outcomes (i.e. diabetes occurrence, glycated haemoglobin, and plasma glucose) tested after correction for multiple tests. Among the SNPs from the NPPB–NPPA candidate analysis, rs198415 and rs5065 were significantly associated with lower glycated haemoglobin levels (P = 0.0005, FDR = 0.01 and P = 0.001, FDR = 0.02, respectively). Lastly, rs61764042 was associated with diabetes occurrence (P = 9·10−5, FDR = 0.002) (Table 3 , Figure 3 ). In stratified analyses according to health status, SNPs association with glycated haemoglobin remained statistically significant in the unhealthy group whereas SNPs association with diabetes occurrence was present in both groups.
Gene expression analysis
The expression of the NPPB, MTHFR, CLCN6, and C1orf167 genes containing the seven SNPs highlighted from the GWAS and the NPPA gene, also members of the same gene cluster, was analysed. The gene expressions were not correlated with each other (except for C1orf167 and NPPA) (Supporting Information, Table S6 ).
The association between the five gene expressions and the seven GWAS SNPs is described in Table 4 , with rs4845877 and rs4845881 associated with NPPA expression, and rs1023252, rs1476413, and rs1801131 associated with CLCN6 expression.
Table 4.
Association between gene‐cluster expressions, the seven genome‐wide association study single nucleotide polymorphisms, and single nucleotide polymorphisms from the NPPB–NPPA analysis
| Rs name | NPPB | NPPA | CLCN6 | MTHFR | C1orf167 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SD | FDR | Beta | SD | FDR | Beta | SD | FDR | Beta | SD | FDR | Beta | SD | FDR | ||
| SNPs from the GWAS | rs1023252 | −0.029 | 0.016 | 0.224 | 0.019 | 0.023 | 0.647 | −0.102 | 0.034 | 0.009 | −0.026 | 0.023 | 0.504 | −0.003 | 0.013 | 0.968 |
| rs4845877 | −0.013 | 0.015 | 0.439 | −0.056 | 0.022 | 0.033 | 0.016 | 0.033 | 0.722 | −0.016 | 0.022 | 0.578 | −0.012 | 0.013 | 0.917 | |
| rs4845881 | −0.021 | 0.016 | 0.439 | −0.074 | 0.023 | 0.008 | 0.051 | 0.034 | 0.239 | −0.016 | 0.023 | 0.578 | −0.009 | 0.013 | 0.917 | |
| rs1476413 | −0.033 | 0.016 | 0.224 | 0.017 | 0.023 | 0.647 | −0.089 | 0.034 | 0.020 | −0.036 | 0.023 | 0.415 | 0.006 | 0.013 | 0.942 | |
| rs1801131 | −0.015 | 0.016 | 0.439 | 0.003 | 0.022 | 0.911 | −0.107 | 0.033 | 0.009 | −0.050 | 0.022 | 0.168 | 0.001 | 0.013 | 0.968 | |
| rs198389 | −0.011 | 0.015 | 0.459 | −0.016 | 0.021 | 0.647 | −0.007 | 0.032 | 0.818 | −0.010 | 0.021 | 0.621 | −0.010 | 0.012 | 0.917 | |
| rs6676300 | −0.016 | 0.015 | 0.439 | −0.005 | 0.021 | 0.911 | −0.042 | 0.032 | 0.254 | −0.022 | 0.021 | 0.504 | −0.022 | 0.012 | 0.578 | |
| SNPs from the NPPB–NPPA genes a | rs2272803 | −0.041 | 0.040 | 0.567 | −0.001 | 0.058 | 0.985 | −0.277 | 0.085 | 0.024 | −0.076 | 0.058 | 0.543 | 0.002 | 0.034 | 0.985 |
| rs198415 | 0.042 | 0.022 | 0.263 | −0.119 | 0.032 | 0.010 | 0.097 | 0.047 | 0.224 | −0.005 | 0.032 | 0.957 | −0.013 | 0.019 | 0.750 | |
| rs5065 | 0.042 | 0.022 | 0.254 | −0.098 | 0.031 | 0.030 | 0.096 | 0.046 | 0.224 | −0.020 | 0.031 | 0.802 | −0.022 | 0.018 | 0.567 | |
| rs198370 | 0.036 | 0.026 | 0.518 | −0.133 | 0.038 | 0.010 | 0.181 | 0.056 | 0.024 | 0.006 | 0.038 | 0.957 | −0.013 | 0.022 | 0.804 | |
| rs5063 | −0.04 | 0.039 | 0.567 | −0.002 | 0.056 | 0.985 | −0.261 | 0.081 | 0.024 | −0.095 | 0.056 | 0.341 | −0.002 | 0.032 | 0.985 | |
| rs149772863 | 0.406 | 0.130 | 0.030 | 0.106 | 0.190 | 0.804 | 0.020 | 0.301 | 0.985 | −0.396 | 0.187 | 0.221 | 0.123 | 0.111 | 0.567 | |
Beta, effect per allele 2 under the additive model; FDR, false discovery rate; GWAS, genome‐wide association study; SD, standard deviation; SNPs, single nucleotide polymorphisms.
Bold indicates the significant associations.
Among the 24 SNPs, only those with at least one significant association are reported sex and age were used as covariates.
Among the SNPs from the NPPB–NPPA candidate analysis, NPPA gene expression was associated with rs5065 and rs198415, which were both associated with glycated haemoglobin as well as with rs198370. rs198370 was also associated with CLCN6 gene expression as well as rs2272803 and rs5063. Only rs149772863 was found to be significantly associated with NPPB gene expression (Table 4 ). A statistically significant association was found between expression of the NPPA gene and NT‐proBNP levels (beta = 0.24 ± 0.12, P = 0.04). Associations between NPPA or NPPB expressions and clinical traits were almost all not significant except for NPPB expression and diabetes occurrence (P = 0.04).
Discussion
The main findings of our study are summarized in the integrative illustration in Figure 3 . In brief, our results show that (i) plasma NT‐proBNP levels have a genetic basis with approximately 15% heritability; (ii) SNPs associated with NT‐proBNP levels are mainly located on chromosome 1, specifically on the gene cluster composed of the MTHFR, CLCN6, NPPA, NPPB, and C1orf167 genes; (iii) NPPA gene expression is associated with NT‐proBNP levels, along with two SNPs from the seven GWAS SNPs; in addition, two other SNPs from the NPPB–NPPA gene‐candidate analysis are associated with CLCN6 gene expression; and (iv) certain SNPs from NPPB–NPPA genes are associated with diastolic or metabolic functions.
In view of the above, we thus report for the first time an association between natriuretic peptide polymorphisms and diastolic and metabolic function as assessed by echocardiography.
Insights from the heritability estimation
Estimation of the heritability of plasma NT‐proBNP levels using covariate‐adjusted analysis suggests that over 15% of the NT‐proBNP variance can be attributed to additive genetic factors and seem to be consistent through lifetime and through lifetime and health status of subjects. Our results are slightly lower than in the previous published GWAS from Salo et al., 10 where they estimated that 23% of the variance of NT‐proBNP was explained by all SNPs in a population‐based (not family‐based) study. However, to our knowledge, no other family‐based study has focused on the heritability of NT‐proBNP using this type of dedicated model.
Common environmental factors do not appear to confer any substantial contribution to this variance in the present study (less than 1%) when taking into account the majority of known and measurable confounding factors (i.e. heart rate, urinary sodium, BMI, eGFR, and blood pressure) in the analysis. In addition to age and sex, these confounding factors account for approximately 20% of the variance. Despite the latter, nearly 60% of the variance in plasma NT‐proBNP remains unexplained. This can be attributed to unmeasured genetic effects such as dominance or epigenetic factors, other unmeasured environmental effects, or underlying interactions between genetic and environmental effects. 18
Genome‐wide association study of plasma N‐terminal pro‐brain natriuretic peptide levels
Results from our GWAS analysis of plasma NT‐proBNP levels are similar to previously published GWAS data. The most associated SNP, rs198389, is a functional variant located in the NPPB promoter region, with the rare allele often found to be associated with a higher NT‐proBNP level or even with higher BNP level in healthy subjects from other previous GWAS. 1 , 4 , 7 , 8 , 9 The six other SNPs that reached the significance threshold in our study were all located within the same cluster of five genes (MTHFR–CLCN6–NPPA–NPPB–C1orf167), which has also been previously identified for its association with NT‐proBNP levels, 7 , 8 in particular rs667300 and rs1023252 (CLCN6) highlighted by Del Greco et al. 7 Of note, most of the SNPs associated with higher NT‐proBNP levels were not associated with NPPB expression levels. Multiple hypothesis can account for such discrepancy such as (i) the fact that white blood cells may not account for cardiac NPPB expression and (ii) plasma NT‐proBNP is not a perfect surrogate for proBNP production. 19 , 20
Polymorphisms associated with cardiac function
The SNP rs198389 has been associated in previous population studies with reduced systolic and diastolic blood pressure and hypertension 1 as well as with diabetes occurrence. 21 However, we did not identify any significant association between this SNP and neither blood pressure nor other infraclinical CV outcomes (diastolic function, septal e′, lateral e′, e/a, e/e′, and LAVI) in the present study. However, taking advantage of the detailed echocardiography exam performed within the STANISLAS cohort, we found two SNPs (rs72864766 and rs11804222) associated with reduced lateral e′ and one SNP (rs4845877) associated with reduced deceleration time. These results suggest, for the first time, an association between natriuretic peptide polymorphisms and diastolic function.
Polymorphisms associated with metabolic function
In the present study, three SNPs (rs5065, rs198415, and rs61764042) were found associated with metabolic variables linked to diabetes (diabetes occurrence and glycated haemoglobin, but not with blood glucose). Only rs5065, located on the NPPA gene, associated with glycated haemoglobin in our results, has already been previously reported for its association with several CV outcomes, in particular hypertension. 4 While we did not find any association between rs5065 and infraclinical CV outcomes, these results nonetheless emphasize the importance of the crosstalk between the natriuretic peptide system and metabolic pathways. Metabolic abnormalities such as diabetes, which have been shown to be associated with certain natriuretic peptide polymorphisms, could be further related to the development of subsequent CV diseases (see, e.g. Kishimoto et al. 22 in which glycated haemoglobin levels were found to predict HF hospitalization).
Importantly, our analyses stratified on health status suggest that NPPB–NPPA genes cluster are preferentially associated with cardiac function (lateral e′ and DT) in healthy subjects whereas they are preferentially associated with glycated haemoglobin in participants with hypertension and/or any CV condition. The variations in cardiac phenotypes for subjects with hypertension or CV disease may be primarily related to other factors than genetics, conversely to subjects from the healthy group. In addition, the level of variability of glycated haemoglobin might be too low in healthy subjects for genetics to be associated with it. These factors might explain the differential association identified in healthy subjects versus subjects with CV conditions in our study.
Limitations
Given that the STANISLAS cohort is composed of initially healthy subjects and in relatively good health at V4, it may be difficult to highlight certain associations of cardiometabolic conditions with genetic factors. Even if only 1705 subjects have return at V4, their characteristics have been reported to be overall similar to subjects lost to follow‐up or deceased 23 ; hence, these missing subjects are unlikely to have modify substantially our results.
Furthermore, NT‐proBNP measurement is known to be impacted by glycosylation, 20 , 24 and the estimation of heritability of natriuretic peptides in our study could consequently partly be related to glycosylation genetic variance. However, measurement of NT‐proBNP after deglycosylation of the samples (as described in references 20 , 24 ) will hamper the specificity of NT‐proBNP quantification because proBNP will also be detected. 25 Furthermore, none of the SNP tested affect any of the glycosylated sites described in proBNP. 26 Therefore, the association we observed between the SNPs tested and NT‐proBNP in heritability is unlikely to be related to NT‐proBNP glycosylation.
Moreover, our findings regarding SNPs associated with cardiac or metabolic functions should be taken with caution as they have not been replicated herein in another cohort. Yet, the seven SNPs associated with circulating NT‐proBNP levels, especially rs198389, have already been reported to be associated with natriuretic peptides, 1 , 4 , 7 , 8 , 9 which strengthen the external validity of our findings.
Lastly, NT‐proBNP levels were measured with PEA technology, providing unitless standardized normalized expression; hence, no direct conversion to standard ‘classical/mass’ values is possible at this stage.
Clinical considerations
Our study identified a significant association between natriuretic peptide polymorphisms and diastolic function (as measured by lateral e′ using echocardiography). This finding is of significant interest given the major involvement of the natriuretic peptides system in HF, including heart failure with preserved ejection fraction (HFpEF), due to diastolic abnormalities. Natriuretic peptides would appear to be related to diastolic function upstream of the aging process and environmental impacts. In addition, because diastolic function is associated with the incidence of HFpEF, this could imply that natriuretic peptide polymorphism may potentially represent a risk factor for HFpEF at later stages, even more so because they are associated with metabolic disturbances that in turn are linked to the risk for HFpEF. Our results suggest that the plasma variation in natriuretic peptides is not only the consequence of pathophysiological processes occurring because of environmental factors but could also conversely be causally (directly or indirectly) related to certain metabolic function and/or cardiac abnormalities, upstream of CV disease occurrence. These findings should modify our perspective regarding natriuretic peptides, which appear to be more than simple passive bystanders in the early stages of CV disease.
Conclusions
The heritability of natriuretic peptides appears relatively low (15%) and mainly based on the same gene cluster constituted of MTHFR, CLCN6, NPPA, NPPB, and C1orf167. Given the association of natriuretic peptide polymorphisms with natriuretic peptide levels and diastolic and metabolic function, the present results suggest that natriuretic peptide polymorphisms could potentially have an impact in the early stages of CV disease.
Conflict of interest
The authors declare not having conflicts of interest with regard to the content of this manuscript.
Funding
The fourth examination of the STANISLAS study was sponsored by the Centre Hospitalier Régional Universitaire of Nancy (CHRU) and the French Ministry of Health (Programme Hospitalier de Recherche Clinique Inter‐régional 2013), by the Contrat de Plan Etat‐Lorraine and “Fonds Européen de Développement Régional” (FEDER Lorraine), and by a public grant overseen by the French National Research Agency (ANR) as part of the second “Investissements d'Avenir” program FIGHT‐HF (reference: ANR‐15‐RHU‐0004) and by the French “Projet investissement d'avenir” (PIA) project “Lorraine Université d'Excellence” (reference: ANR‐15‐IDEX‐04‐LUE). It is also supported by the Sixth European Union‐Framework program (EU‐FP) Network of Excellence Ingenious HyperCare (#LSHM‐CT‐2006‐037093), the Seventh EU‐FP MEDIA (Européen “Cooperation”—Theme “Health”/FP7‐HEALTH‐2010‐single‐stage #261409), HOMAGE (grant agreement no. Heart “Omics” in Ageing, 7th Framework Program grant #305507), FOCUS‐MR (reference: ANR‐15‐CE14‐0032‐01), and FIBRO‐TARGETS (FP7 #602904) projects, and by ERA‐CVD EXPERT (reference: ANR‐16‐ECVD‐0002‐02). The study was supported by the F‐Clinical Research Infrastructure Network (F‐CRIN) Cardiovascular and Renal Clinical Trialists (INI‐CRCT).
Supporting information
Table S1. list of the 24 SNPs included in the NPPB – NPPA gene candidate analysis.
Table S2. Characteristics of the subjects not included in the study (n = 150).
Table S3. Covariate effects in the linear model for plasma NT‐proBNP levels.
Table S4. Variance decomposition for plasma NT‐proBNP levels for all included subjects.
Table S5. Variance decomposition for plasma NT‐proBNP levels among healthy subjects.
Table S6. Correlations between the gene expressions of the gene cluster.
Figure S1. Comparison between measured by clinical routine assays and the PEA technology.
Acknowledgements
We acknowledge Robert Olaso and his lab ‘Production Platforms in Human Genomics’ at CNRGH for genotyping data production and Anne Boland at CNRGH for management of the genotyping study.We are highly grateful to the Vandoeuvre‐Lès Nancy Centre de Médecine Préventive staff and to Dr Sophie Visvikis‐Siest (Inserm U1122) who managed the STANISLAS cohort for the first three visits. The authors deeply thank the staff of the Clinical Investigation Center and other personnel involved in the STANISLAS cohort management: Biostatisticians: Fay R, Lamiral Z, Machu JL. Computer scientists: Boucenna N, Gallina‐Muller C, Maclot PL, Sas T. Co‐investigators: Chau K, Di Patrizio P, Dobre D, Gonthier D, Huttin O, Malingrey L, Mauffrey V, Olivier A, Poyeton T, Steyer E, Watfa G. Data managers: Cimon P, Eby E, Merckle L. Data entryoperators: Batsh M, Blanger O, Bottelin C, Haskour N, Jacquet V, Przybylski MC, Saribekyan Y, Thomas H, Vallee M. Echocardiographists, echographists: Ben Sassi M, Cario S, Camara Y, Coiro S, Frikha Z, Kearney‐Schwartz A, Selton‐Suty C, Watfa G. Imaging engineer: Bozec E. Laboratory Engineer Nuee‐Capiaumont J and Technicians: Fruminet J, Kuntz M, Ravey J, Rousseau E, Tachet C. Project manager: Bouali S, Hertz C. Quality engineer: Lepage X. Registered nurses: Giansily M, Poinsignon L, Robin N, Schmartz M, Senn M, Micor‐Patrignani E, Toutlemonde M. Hospital technician: Fleurot MT. Resident doctors: Alvarez‐Vasquez R, Amiot M, Angotti M, Babel E, Balland M, Bannay A, Basselin P, Benoit P, Bercand J, Bouazzi M, Boubel E, Boucherab‐Brik N, Boyer F, Champagne C, Chenna SA, Clochey J, Czolnowski D, Dal‐Pozzolo J, Desse L, Donetti B, Dugelay G, Friang C, Galante M, Garel M, Gellenoncourt A, Guillin A, Hariton ML, Hinsiger M, Haudiquet E, Hubert JM, Hurtaud A, Jabbour J, Jeckel S, Kecha A, Kelche G, Kieffert C, Laurie're E, Legay M, Mansuy A, Millet‐Muresan O, Meyer N, Mourton E, Naudé AL, Pikus AC, Poucher M, Prot M, Quartino A, Saintot M, Schiavi A, Schumman R, Serot M, Sert C, Siboescu R, Terrier‐de‐la‐Chaise S, Thiesse A, Thietry L, Vanesson M, Viellard M. Secretaries: De Amorin E, Villemain C, Ziegler N. Study coordinators: Dauchy E, Laurent S, and all persons not listed above who helped to the funding, initiation, accrual, management, and analysis of the fourth visit of the STANISLAS cohort. They also thank the CRB Lorrain of the Nancy CHRU for management of the biobank. Steering committee: Pierre Mutzenhardt, Mehdy Siaghy, Patrick Lacolley, Marie‐Ange Luc, Pierre Yves Marie, Jean Michel Vignaud. Advisory members: Sophie Visvikis Siest, F Zannad. Technical committee: Christiane Branlant, Isabelle Behm‐Ansmant, Jean‐Michel Vignaud, Christophe Philippe, Jacques Magdalou, Faiez Zannad, Patrick Rossignol. Scientific committee: Laurence Tiret, Denis Wahl, Athanase Benetos, Javier Diez, Maurizio Ferrari, Jean Louis Gueant, Georges Dedoussis, François Alla, François Gueyffier, Pierre‐Yves Scarabin, Claire Bonithon Kopp, Xavier Jouven, Jean‐Claude Voegel, Jan Staessen. [Correction added on 09 November 2021, after first online publication: Funding and Acknowledgement section have been updated in this version.]
Xhaard, C. , Rouget, R. , Vodovar, N. , Le Floch, E. , Dandine‐Roulland, C. , Wagner, S. , Bacq‐Daian, D. , Thuillier, Q. , Boivin, J.‐M. , Branlant, C. , Deleuze, J.‐F. , Behm‐Ansmant, I. , Zannad, F. , Rossignol, P. , and Girerd, N. (2022) Impact of natriuretic peptide polymorphisms on diastolic and metabolic function in a populational cohort: insights from the STANISLAS cohort. ESC Heart Failure, 9: 729–739. 10.1002/ehf2.13674.
Contributor Information
Constance Xhaard, Email: c.xhaard@chru-nancy.fr.
Nicolas Girerd, Email: n.girerd@chru-nancy.fr.
References
- 1. Seidelmann SB, Vardeny O, Claggett B, Yu B, Shah AM, Ballantyne CM, Selvin E, MacRae CA, Boerwinkle E, Solomon SD. An NPPB promoter polymorphism associated with elevated N‐terminal pro‐B‐type natriuretic peptide and lower blood pressure, hypertension, and mortality. J Am Heart Assoc 2017; 6: e005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Choquet H, Cavalcanti‐Proença C, Lecoeur C, Dina C, Cauchi S, Vaxillaire M, Hadjadj S, Horber F, Potoczna N, Charpentier G, Ruiz J, Hercberg S, Maimaitiming S, Roussel R, Boenhnke M, Jackson AU, Patsch W, Krempler F, Voight BF, Altshuler D, Groop L, Thorleifsson G, Steinthorsdottir V, Stefansson K, Balkau B, Froguel P, Meyre D. The T‐381C SNP in BNP gene may be modestly associated with type 2 diabetes: an updated meta‐analysis in 49 279 subjects. Hum Mol Genet 2009; 18: 2495–2501. [DOI] [PubMed] [Google Scholar]
- 3. Ledwidge M, Gallagher J, Conlon C, Tallon E, O'Connell E, Dawkins I, Watson C, O'Hanlon R, Bermingham M, Patle A, Badabhagni MR, Murtagh G, Voon V, Tilson L, Barry M, McDonald L, Maurer B, McDonald K. Natriuretic peptide‐based screening and collaborative care for heart failure: the STOP‐HF randomized trial. JAMA 2013; 310: 66–74. [DOI] [PubMed] [Google Scholar]
- 4. Ellis KL, Newton‐Cheh C, Wang TJ, Frampton CM, Doughty RN, Whalley GA, Ellis CJ, Skelton L, Davis N, Yandle TG, Troughton RW, Richards AM, Cameron VA. Association of genetic variation in the natriuretic peptide system with cardiovascular outcomes. J Mol Cell Cardiol 2011; 50: 695–701. [DOI] [PubMed] [Google Scholar]
- 5. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, Falk V, González‐Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GMC, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P, ESC Scientific Document Group . 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology. Eur Heart J 2016; 37: 2129–2200. [DOI] [PubMed] [Google Scholar]
- 6. Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE Jr, Colvin MM, Drazner MH, Filippatos GS, Fonarow GC, Givertz MM, Hollenberg SM, Lindenfeld J, Masoudi FA, McBride P, Peterson PN, Stevenson LW, Westlake C. 2017 ACCF/ACC/HFSA focused update on new pharmacological therapy for heart failure: an update of the 2013 ACCF/AHA guidelines for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Heart Failure Society of America. J Am Coll Cardiol 2017; 70: 776–803. [DOI] [PubMed] [Google Scholar]
- 7. Del Greco MF, Pattaro C, Luchner A, Pichler I, Winkler T, Hicks AA, Fuchsberger C, Franke A, Melville SA, Peters A, Wichmann HE, Schreiber S, Heid IM, Krawczak M, Minelli C, Wiedermann CJ, Pramstaller PP. Genome‐wide association analysis and fine mapping of NT‐proBNP level provide novel insight into the role of the MTHFR‐CLCN6‐NPPA‐NPPB gene cluster. Hum Mol Genet 2011; 20: 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johansson Å, Eriksson N, Lindholm D, Varenhorst C, James S, Syvänen AC, Axelsson T, Siegbahn A, Barratt BJ, Becker RC, Himmelmann A, Katus HA, Steg PG, Storey RF, Wallentin L. Genome‐wide association and Mendelian randomization study of NT‐proBNP in patients with acute coronary syndrome. Hum Mol Genet 2016; 25: 1447–1456. [DOI] [PubMed] [Google Scholar]
- 9. Musani SK, Fox ER, Kraja A, Bidulescu A, Lieb W, Lin H, Beecham A, Chen MH, Felix JF, Fox CS, Kao WH, Kardia SL, Liu CT, Nalls MA, Rundek T, Sacco RL, Smith J, Sun YV, Wilson G, Zhang Z, Mosley TH, Taylor HA, Vasan RS. Genome‐wide association analysis of plasma B‐type natriuretic peptide in blacks: the Jackson Heart Study. Circ Cardiovasc Genet 2015; 8: 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salo PP, Havulinna AS, Tukiainen T, Raitakari O, Lehtimaki T, Kahonen M, Kettunen J, Mannikko M, Eriksson JG, Jula A, Blankenberg S, Zeller T, Salomaa V, Kristiansson K, Perola M. Genome‐wide association study implicates atrial natriuretic peptide rather than B‐type natriuretic peptide in the regulation of blood pressure in the general population. Circ Cardiovasc Genet 2017; 10: e001713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferreira JP, Girerd N, Bozec E, Merckle L, Pizard A, Bouali S, Eby E, Leroy C, Machu JL, Boivin JM, Lamiral Z, Rossignol P, Zannad F. Cohort profile: Rationale and design of the fourth visit of the STANISLAS cohort: a familial longitudinal population‐based cohort from the Nancy region of France. Int J Epidemiol 2017; 183: 285–295. [DOI] [PubMed] [Google Scholar]
- 12. Coiro S, Huttin O, Bozec E, Selton‐Suty C, Lamiral Z, Carluccio E, Trinh A, Fraser AG, Ambrosio G, Rossignol P, Zannad F, Girerd N. Reproducibility of echocardiographic assessment of 2D‐derived longitudinal strain parameters in a population‐based study (the STANISLAS cohort study). Int J Cardiovasc Imaging 2017; 33: 1361–1369. [DOI] [PubMed] [Google Scholar]
- 13. Frikha Z, Girerd N, Huttin O, Courand PY, Bozec E, Olivier A, Lamiral Z, Zannad F, Rossignol P. Reproducibility in echocardiographic assessment of diastolic function in a population based study (the STANISLAS cohort study). PLoS One 2015; 10: e0122336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferreira JP, Lamiral Z, Xhaard C, Duarte K, Bresso E, Devignes MD, Lefloch E, Roulland CD, Deleuze JF, Wagner S, Guerci B, Girerd N, Zannad F, Boivin JM, Rossignol P. Circulating plasma proteins and new‐onset diabetes in a population‐based study: proteomic and genomic insights from the STANISLAS cohort. Eur J Endocrinol 2020; 47: 395–395j. [DOI] [PubMed] [Google Scholar]
- 15. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucl Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xhaard C, Dandine‐Roulland C, Villemereuil P, Floch EL, Bacq‐Daian D, Machu JL, Ferreira JP, Deleuze JF, Zannad F, Rossignol P, Girerd N. Heritability of a resting heart rate in a 20‐year follow‐up family cohort with GWAS data: insights from the STANISLAS cohort. Eur J Prev Cardiol 2019: 2047487319890763. [DOI] [PubMed] [Google Scholar]
- 17. Dandine‐Roulland C. Manipulation of genetic data (SNPs). 2017. Computation of GRM and dominance matrix, LD, heritability with efficient algorithms for linear mixed model (AIREML). Hum Hered; 18: 1–29. [Google Scholar]
- 18. Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet 2008; 9: 255–266. [DOI] [PubMed] [Google Scholar]
- 19. Semenov AG, Postnikov AB, Tamm NN, Seferian KR, Karpova NS, Bloshchitsyna MN, Koshkina EV, Krasnoselsky MI, Serebryanaya DV, Katrukha AG. Processing of pro‐brain natriuretic peptide is suppressed by O‐glycosylation in the region close to the cleavage site. Clin Chem 2009; 55: 489–498. [DOI] [PubMed] [Google Scholar]
- 20. Røsjø H, Dahl MB, Jørgensen M, Røysland R, Brynildsen J, Cataliotti A, Christensen G, Høiseth AD, Hagve TA, Omland T. Influence of glycosylation on diagnostic and prognostic accuracy of N‐terminal pro‐B‐type natriuretic peptide in acute dyspnea: data from the Akershus Cardiac Examination 2 Study. Clin Chem 2015; 61: 1087–1097. [DOI] [PubMed] [Google Scholar]
- 21. Pfister R, Sharp S, Luben R, Welsh P, Barroso I, Salomaa V, Meirhaeghe A, Khaw KT, Sattar N, Langenberg C, Wareham NJ. Mendelian randomization study of B‐type natriuretic peptide and type 2 diabetes: evidence of causal association from population studies. PLoS Med 2011; 8: e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kishimoto I, Makino H, Ohata Y, Tamanaha T, Tochiya M, Kada A, Ishihara M, Anzai T, Shimizu W, Yasuda S, Ogawa H. Hemoglobin A1c predicts heart failure hospitalization independent of baseline cardiac function or B‐type natriuretic peptide level. Diabetes Res Clin Pract 2014; 104: 257–265. [DOI] [PubMed] [Google Scholar]
- 23. Chau K, Girerd N, Magnusson M, Lamiral Z, Bozec E, Merckle L, Leosdottir M, Bachus E, Frikha Z, Ferreira JP, Despres JP, Rossignol P, Boivin JM, Zannad F. Obesity and metabolic features associated with long‐term developing diastolic dysfunction in an initially healthy population‐based cohort. Clin Res Cardiol 2018; 107: 887–896. [DOI] [PubMed] [Google Scholar]
- 24. Seferian KR, Tamm NN, Semenov AG, Tolstaya AA, Koshkina EV, Krasnoselsky MI, Postnikov AB, Serebryanaya DV, Apple FS, Murakami MAM, Katrukha AG. Immunodetection of glycosylated NT‐proBNP circulating in human blood. Clin Chem 2008; 54: 866–873. [DOI] [PubMed] [Google Scholar]
- 25. Jaffe AS, Apple FS, Mebazaa A, Vodovar N. Unraveling N‐terminal pro‐B‐type natriuretic peptide: another piece to a very complex puzzle in heart failure patients. Clin Chem 2015; 61: 1016–1018. [DOI] [PubMed] [Google Scholar]
- 26. Schellenberger U, O'Rear J, Guzzetta A, Jue RA, Protter AA, Pollitt NS. The precursor to B‐type natriuretic peptide is an O‐linked glycoprotein. Arch Biochem Biophys 2006; 451: 160–166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. list of the 24 SNPs included in the NPPB – NPPA gene candidate analysis.
Table S2. Characteristics of the subjects not included in the study (n = 150).
Table S3. Covariate effects in the linear model for plasma NT‐proBNP levels.
Table S4. Variance decomposition for plasma NT‐proBNP levels for all included subjects.
Table S5. Variance decomposition for plasma NT‐proBNP levels among healthy subjects.
Table S6. Correlations between the gene expressions of the gene cluster.
Figure S1. Comparison between measured by clinical routine assays and the PEA technology.


