Abstract
In C. elegans, PUF proteins promote germline stem cell self-renewal. Their functions hinge on partnerships with two proteins that are redundantly required for stem cell maintenance. Here we focus on understanding how the essential partner protein, LST-1, modulates mRNA regulation by the PUF protein, FBF-2. LST-1 contains two nonidentical sites of interaction with FBF-2, LST-1 A and B. Our crystal structures of complexes of FBF-2, LST-1 A, and RNA visualize how FBF-2 associates with LST-1 A versus LST-1 B. One commonality is that FBF-2 contacts the conserved lysine and leucine side chains in the KxxL motifs in LST-1 A and B. A key difference is that FBF-2 forms unique contacts with regions N- and C-terminal to the KxxL motif. Consequently, LST-1 A does not modulate the RNA-binding affinity of FBF-2, whereas LST-1 B decreases RNA-binding affinity of FBF-2. The N-terminal region of LST-1 B, which binds near the 5′ end of RNA elements, is essential to modulate FBF-2 RNA-binding affinity, while the C-terminal residues of LST-1 B contribute strong binding affinity to FBF-2. We conclude that LST-1 has the potential to impact which mRNAs are regulated depending on the precise nature of engagement through its functionally distinct FBF binding sites.
INTRODUCTION
Sequence-specific RNA-binding proteins play a crucial role in the regulation of networks of mRNAs. PUF proteins, named for Drosophila melanogaster PUmilio and Caenorhabditis elegans fem-3 Binding Factor (FBF), are an exemplary model with exquisite RNA sequence specificity (1–4). Classical PUF proteins contain a crescent-shaped RNA-binding domain composed of eight α-helical repeats (5–14). The prototypical PUF proteins, like Pumilio, recognize an 8-nt sequence with each repeat binding to one base (9). Some PUF proteins, including FBF, contain eight α-helical repeats but bind to RNA sequences longer than eight nucleotides and accommodate additional nucleotides by flipping them away from the RNA-binding surface (7,8,11,13). C. elegans FBF-1 and FBF-2 (referred to collectively as FBF), are functionally redundant in controlling self-renewal and differentiation in the germline (15,16). FBF binds to RNAs with a 9-nt canonical FBF binding element (FBE) (17–19). In the FBE, the 5′ and 3′ nucleotides are specifically recognized and the central nucleotides are flipped away from the RNA-binding surface (11). We recently found that FBF can also bind to shorter 8-nt sequences, called the compact FBE (cFBE), where no bases are flipped outward (19,20). The cFBE sequence was a highly ranked sequence from in vitro selection experiments (20), and it was identified in ∼30% of peaks from FBF iCLIP (individual-nucleotide resolution UV crosslinking and immunoprecipitation) experiments (19). Although it has not been demonstrated directly that FBF regulates a target mRNA via a cFBE in vivo, the iCLIP results suggest association of FBF with many mRNAs bearing cFBEs in the C. elegans germline.
RNA-binding proteins do not work in isolation, and PUF proteins have been demonstrated to collaborate with other proteins that can alter their RNA-binding specificity and affinity. In C. elegans, FBF collaborates with two proteins, LST-1 (Lateral Signaling Target-1) and SYGL-1 (SYnthetic GermLine proliferation defective-1). Notch signaling induces expression of LST-1 and SYGL-1 in germline stem cells (GSCs) (21). Both proteins form partnerships with FBF to repress translation of the germline defective 1 (gld-1) transcript (22), a well-established FBF target mRNA whose expression is required for differentiation (23,24). LST-1 and SYGL-1 are functionally redundant. Expression of either LST-1 or SYGL-1 is required to maintain GSC self-renewal (22), and both proteins physically interact with FBF despite their unrelated amino acid sequences (20,22,25). Here we focus on the partnership between LST-1 and FBF.
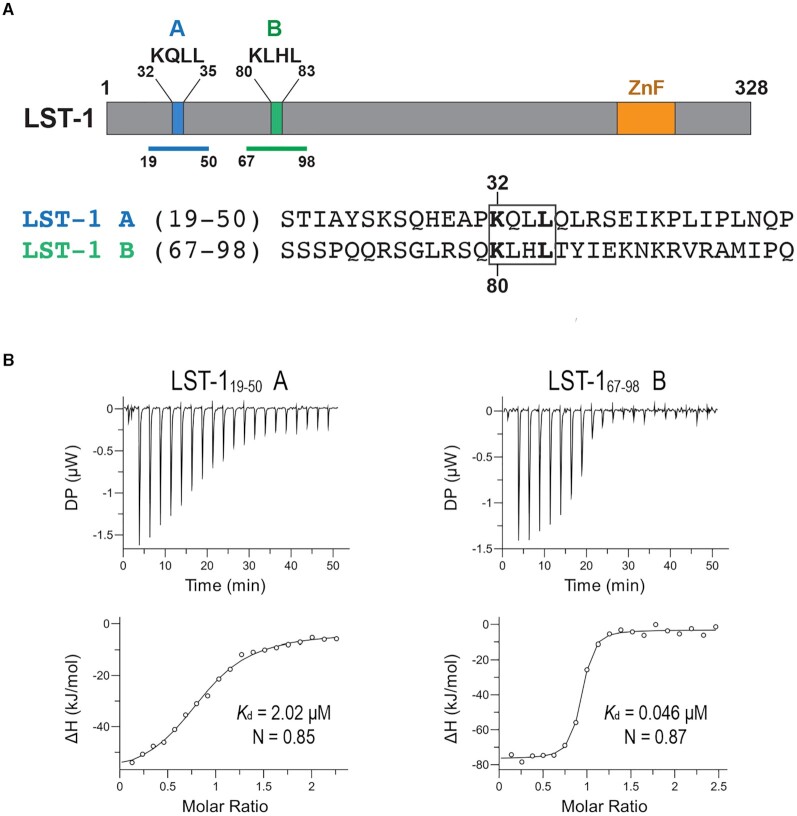
LST-1 expression is normally restricted to GSCs (22). Expression of LST-1 throughout the germline drives formation of tumors, and this tumor formation depends on the presence of FBF (22). LST-1 comprises an intrinsically-disordered N terminus and a C-terminal zinc finger (Figure 1A) (25). The N-terminal region is sufficient for stem cell self-renewal and interaction with FBF, and the C-terminal region controls the spatial restriction of LST-1 protein to the GSC pool (25). FBF-2 interacts with LST-1 via two consensus KxxL motifs (LST-1 A and B) (20,25). LST-1 with either intact KxxL motif can maintain GSCs, and worms with both KxxL motifs mutated fail to maintain GSCs and are sterile (25).
Figure 1.
FBF-2 binds to LST-1 site B with higher affinity than to site A. (A) LST-1 contains two essential FBF-interaction sites A and B, each bearing a conserved KxxL motif. Schematic drawing (top) illustrates the KxxL motifs and a C-terminal zinc finger (ZnF). The regions of the LST-1 peptides used for calorimetry are indicated with blue (A) and green (B) lines, and the sequences are shown (bottom) with the KxxL motif boxed. (B) FBF-2 binds to LST-1 B with higher affinity than to LST-1 A. Representative isothermal titration calorimetry thermograms (top, differential power [DP] vs time) and corresponding titration curve-fitting graphs (bottom) are shown. Kd and N for the experiment shown are indicated. Thermodynamic parameters are summarized in Table 1.
We previously identified an FBF-interacting region of LST-1 containing the KxxL motif B, and we determined a crystal structure of FBF-2 bound to the LST-1 B peptide and cFBE RNA (20). FBF-2 recognizes the conserved lysine and leucine residues in the KxxL motif as well as flanking regions that define a longer LST-1 B FBF binding site. The presence of an LST-1 B peptide weakens the in vitro RNA-binding affinity of FBF-2, suggesting that LST-1 could finetune mRNA target selection by promoting regulation of mRNAs that are more abundant or bind with higher affinity. Haupt, et al. identified two FBF-interacting KxxL motifs A and B in LST-1 (25). In yeast 2-hybrid analyses, the interaction between FBF-2 and LST-1 appears to be substantially weaker when mediated by the A motif than via the B motif (25). Biochemically, little is known about the effect of the LST-1 A motif on FBF function, and it has not been established whether the two FBF binding sites in LST-1 harbor functional differences.
Here we identified differences in how the LST-1 A and B peptide regions bind FBF-2 and the resulting consequences on the binding affinity of FBF-2 for RNA substrates. We demonstrated that LST-1 B binds to FBF-2 with ∼40-fold higher affinity than LST-1 A. Crystal structures of ternary complexes of FBF-2, an LST-1 A peptide, and different RNA sequences collectively illustrate how FBF-2/LST-1 partnership accommodates interaction with both 9-nt and 8-nt RNA elements. Comparing our crystal structures of complexes with LST-1 A or B peptides revealed different contacts of FBF-2 with LST-1 peptide regions N- and C-terminal to the KxxL motif. Mutational analysis identified key residues in the flanking regions that explain the differences in FBF-2 binding affinity to the A versus B peptide. We found that the N-terminal region of the LST-1 B peptide, which binds to FBF-2 near the 5′ end of target RNA elements, is essential to modulate FBF-2 RNA-binding affinity. In contrast, the LST-1 A peptide, whose N-terminal region binds differently to FBF-2, did not modulate the RNA-binding affinity of FBF-2. We conclude that the two FBF interaction sites in LST-1 offer more than redundancy and have the potential to control which mRNAs are regulated depending on which site is engaged.
MATERIAL AND METHODS
Protein expression and purification
A cDNA fragment encoding the RNA-binding domain of C. elegans FBF-2 (residues 164–575) was cloned into the pSMT3 vector (kindly provided by Dr. Christopher Lima), which encodes an N-terminal His6-SUMO tag (26). This cDNA fragment also included an N-terminal TEV protease cleavage site that was encoded in a previous pGEX6p FBF-2 construct (11). A codon-optimized synthetic DNA fragment encoding amino acid residues 19–50 of LST-1 (LST-1 A) was cloned into the pGEX4T-3 vector with a TEV site after the glutathione S-transferase (GST) tag. E. coli BL21-CodonPlus (DE3)-RIL competent cells (Agilent) were transformed with the two recombinant plasmids, and bacteria transformed with both plasmids were selected for both kanamycin and ampicillin resistance. A 5-mL culture was grown from bacterial colonies overnight at 37°C and then used to inoculate 1 L of LB medium with 50 μg/mL kanamycin and 100 μg/mL ampicillin. The culture was grown at 37°C. Protein expression was induced at OD600 of ∼0.6 with 0.1 mM IPTG, and the culture was grown at 16°C for 16–20 h.
The FBF-2/LST-1 A (residues 19–50) protein complex was purified using a similar protocol as described previously for the FBF-2/LST-1 B (residues 74–98) protein complex (20). Briefly, the soluble fraction of E. coli cell lysate in a buffer containing 20 mM Tris, pH 8.0; 0.5 M NaCl; 20 mM imidazole; 5% (v/v) glycerol; and 0.1% (v/v) β-mercaptoethanol was mixed with 5 mL Ni-NTA resin (Qiagen) for 1 h at 4°C. After washing the beads, the His6-SUMO-FBF-2 and GST-LST-1 A fusion proteins were co-eluted with elution buffer (20 mM Tris, pH 8.0; 50 mM NaCl; 200 mM imidazole, pH 8.0; 1 mM dithiothreitol [DTT]). TEV protease was added to the eluent and incubated at 4°C overnight to cleave the His6-SUMO fusion from FBF-2 and the GST fusion from LST-1 A. The FBF-2/LST-1 A protein complex was purified with a Hi-Trap Heparin column (Cytiva), eluting with a 5–100% gradient of buffer B. Heparin column buffer A contained 20 mM Tris, pH 8.0 and 1 mM DTT, and buffer B contained an additional 1 M NaCl. The expression levels of FBF-2 and LST-1 from the two plasmids were not equal, and more FBF-2 was produced stoichiometrically than LST-1. During the FBF-2/LST-1 B complex purification, two peaks eluted from the Heparin column with free FBF-2 and the FBF-2/LST-1 B complex, separately. However, during the FBF-2/ LST-1 A complex purification, one Heparin column peak eluted corresponding to a mixture of the FBF-2/ LST-1 A complex and free FBF-2. The peak fractions were concentrated, and additional purified LST-1 A protein was added and incubated at 4°C overnight to promote more FBF-2/LST-1 A complex formation. The protein complex was purified using a HiLoad 16/60 Superdex 75 column (Cytiva) in a buffer of 20 mM HEPES, pH 7.4, 0.15 M NaCl and 2 mM DTT. The FBF-2/LST1 A protein complex was concentrated to OD280 of ∼4.0 for crystallization. FBF-2 and LST-1 proteins were also prepared individually. FBF-2 protein was purified as described previously (20).
cDNAs encoding LST-1 A (residues 19–50) and LST-1 B (residues 67–98) were cloned into the pSMT3 vector. A 5-mL culture was grown overnight at 37°C and then used to inoculate 1 L of TB media with 50 μg/mL kanamycin. The culture was grown at 37°C. Protein expression was induced at OD600 of ∼1.0 with 0.4 mM IPTG, and the culture was grown at 22°C for ∼20 h. The soluble fraction of E. coli cell lysate in a buffer containing 20 mM Tris, pH 8.0; 0.5 M NaCl; 20 mM imidazole; and 5% (v/v) glycerol was mixed with 5 mL Ni-NTA resin for 1 h at 4°C. After extensive washing the LST-1 proteins were eluted with a buffer of 20 mM Tris, 50 mM NaCl and 200 mM imidazole, pH 8. The Ulp1 protease was added to the eluant and incubated at 4°C for 2 h or overnight to cleave the His6-SUMO tag from LST-1. LST-1 A protein was separated from His6-SUMO with a HiTrap Q column and the column flow-through containing LST-1 A was collected and concentrated using Amicon Ultra-15 filters (3K MWCO). LST-1 B protein was purified with a HiTrap Heparin column and eluted with a 5–100% NaCl gradient (buffer A: 20 mM Tris, pH 8.0; buffer B: 20 mM Tris, pH 8.0, 1 M NaCl). The peak fractions containing LST-1 B were concentrated. Both LST-1 A and B were further purified with a HiLoad 16/60 Superdex 75 column.
LST-1 B mutants were generated using PCR with primers containing the mutated sequences: Y85L, LRSQ76-79 to HEAP, YIEK85-88 to LRSE. The mutant proteins were purified using the same protocol as for the wild-type LST-1 B.
Crystallization, data collection and structure determination
The concentrated FBF-2/LST-1 A protein complex was mixed with cFBE RNA (5′-CUGUGAAUG-3′) or FBE RNA (5′-pUGUACUAUA-3′) at a molar ratio of 1:1.2 and incubated on ice for 1 h prior to crystallization screening. Crystals of the FBF-2/LST-1 A/cFBE complex were obtained with a crystallization solution of 30% (v/v) PEG 400, 0.1 M CHES, pH 9.5. Crystals of the ternary complex of FBF-2/LST-1 A/FBE were obtained with a crystallization solution of 25% (v/v) PEG 400, 0.1 M Tris, pH 8.5 by hanging drop vapor diffusion at 20°C with a 1:1 ratio of sample:reservoir solution. Crystals were cryoprotected by transferring them into 35% (v/v) PEG 400 with CHES or Tris pH buffer and flash freezing them in liquid nitrogen.
X-ray diffraction data were collected at a wavelength of 1.0 Å at beamline 22-ID of the Advanced Photon Source. Data sets were scaled with HKL2000 (27). Both crystals belonged to the P61 space group. An asymmetric unit contained one ternary complex. To determine the crystal structure of the FBF-2/LST-1 A/cFBE complex, the structure of an FBF-2/FBE binary complex (PDB code: 3K5Q) was used as a search model for molecular replacement with Phaser (28). The LST-1 A peptide was built manually into the electron density. The model was improved through iterative refinement and manual building with Phenix and Coot (29,30). The structure of the FBF-2/LST-1 A/FBE complex was determined similarly. Residues 27–41 and 31–41 of LST-1 were built in the ternary complex models with cFBE and FBE RNAs, respectively. Data collection and refinement statistics are shown in Table 2.
Table 2.
Crystallographic data collection and refinement statistics
| FBF-2/LST-1 A/cFBE | FBF-2/LST-1 A/FBE | |
|---|---|---|
| Resolution range1 | 35.7 - 2.39 (2.48 - 2.39) | 36.1 - 2.34 (2.42 - 2.34) |
| Space group | P61 | P61 |
| Unit cell dimensions | ||
| a, b,c (Å) | 93.2 93.2 111.3 | 93.8 93.8 113.2 |
| α, β, γ (°) | 90 90 120 | 90 90 120 |
| Unique reflections2 | 21602 (2118) | 23,811 (2393) |
| Multiplicity | 5.1 | 5.0 |
| Completeness (%) | 99.5 (99.4) | 99.7 (100) |
| Mean I/sigma(I) | 14.5 | 11.5 |
| Wilson B-factor | 42.7 | 46.6 |
| R-meas | 0.071 (0.420) | 0.090 (0.853) |
| R-pim | 0.031 (0.186) | 0.039 (0.370) |
| CC1/2 | 1.0 (0.882) | 1.0 (0.711) |
| CC* | 1.0 (0.968) | 1.0 (0.911) |
| Refinement | ||
| Reflections used in refinement | 21,623 (2118) | 23,809 (2393) |
| Reflections used for R-free | 1,991 (191) | 2,020 (197) |
| R-work | 0.171 (0.230) | 0.178 (0.272) |
| R-free | 0.215 (0.264) | 0.225 (0.304) |
| Number of atoms | ||
| protein | 3277 | 3267 |
| RNA | 171 | 171 |
| solvent | 95 | 136 |
| RMSD bonds (Å) | 0.002 | 0.002 |
| RMSD angles (°) | 0.446 | 0.386 |
| Ramachandran favoured (%) | 98.0 | 99.0 |
| Ramachandran outliers (%) | 0.0 | 0.0 |
| Average B-factors (Å2) | ||
| protein | 50.7 | 57.1 |
| RNA | 56.8 | 63.2 |
| solvent | 49.9 | 55.7 |
1The highest-resolution shell is shown in parentheses.
2Statistics for the highest-resolution shell are shown in parentheses.
Isothermal Titration Calorimetry (ITC)
Experiments were performed at 20°C using a MicroCal PEAQ-ITC Automated calorimeter (Malvern Instruments). FBF-2 and LST-1 variants were prepared in the same buffer of 20 mM HEPES pH 7.4, 150 mM NaCl and 1 mM TCEP by gel filtration. LST-1 variants (100–400 μM) were titrated from the syringe into the cell containing FBF-2 (10–30 μM) in 2 μl aliquots with 20 injections. Experiments were performed in duplicate due to the limitations on the amount of protein needed. Data were analyzed with the one-site model using the MicroCal PEAQ-ITC Analysis Software provided by the manufacturer.
Microscale Thermophoresis (MST) RNA-binding assay
3′-Cy5 labeled gld-1 FBEa (5′-AUCAUGUGCCAUAC-Cy5-3′) and cFBE (5′-AUCUGUGAAUGA-Cy5-3′) RNAs were synthesized by Horizon Dharmacon. FBF-2 protein was serially diluted two-fold to prepare 12 concentrations with 40 μM as the highest concentration. Purified LST-1 A or LST-1 B protein (200 μM) was added to the FBF-2 protein dilution series at 1:1 (v/v) ratio and preincubated at 4°C for 2 h. After the preincubation, 2 μl of protein complex solution was mixed with 18 μl binding buffer (10 mM HEPES, pH 7.4, 50 mM NaCl, 0.01% Tween-20, 0.1 mg/ml BSA, 0.1 mg/ml yeast tRNA and 2 mM DTT) containing 5 nM fluorescently labeled RNA. The final FBF-2 concentration ranged from 2000 nM to 1.95 nM, and the LST-1 A or B concentration was constant at 10 μM. Reactions were incubated at 4°C overnight in the dark.
The samples were loaded into MonolithNT.Automated Capillary Chips and the measurements were performed at 25°C using a MonolithNT.Automated picoRed/nanoBlue instrument (NanoTemper Technologies). Instrument parameters were adjusted to 6% (for gld-1 FBEa RNA) or 10% (for cFBE RNA) LED excitation power and 40% MST power. An MST-on time of 20 s was used. Data from three independently pipetted measurements were analyzed with MO.Affinity Analysis software (NanoTemper Technologies), and Kd values were derived using the signal from an MST-on time of 9–10 s.
RESULTS
LST-1 interacts with FBF-2 via two peptide regions with different binding affinities
To begin to explore potential similarities or differences in activity of the two FBF-interacting regions of LST-1, we used isothermal titration calorimetry (ITC) to determine the affinity of interaction between the FBF-2 RNA-binding domain and LST-1 peptides containing motif A (residues 19–50) or B (residues 67–98) (Figure 1A). For simplicity, we will refer to the regions of LST-1 containing the LST-1 A or B KxxL motifs as LST-1 A or B, respectively, and note the residues that are included for specific experiments, because the LST-1 FBF interaction sites extend beyond the KxxL motifs (below and (20)). We titrated LST-1 peptides to an FBF-2 protein solution to measure binding affinities of the A and B peptides (Figure 1, Table 1). The LST-1 A peptide, LST-119–50, bound to FBF-2 with a dissociation constant, Kd, of 2.0 μM, whereas the LST-1 B peptide, LST-167–98, bound to FBF-2 > 40-fold more tightly than the A peptide with a Kd of 0.046 μM (Figure 1B). This is consistent with previous yeast two-hybrid analyses showing that mutation of the higher-affinity motif B in full-length LST-1 has a greater effect on overall interaction than mutation of the lower-affinity motif A (25). The stoichiometry of binding to FBF-2 was 1:1 for both the LST-1 A and B peptides, as the number of sites (N) was ∼0.9. Binding of both peptides to FBF-2 was driven by negative enthalpy change, which offset the unfavorable decrease in entropy (Table 1). The negative enthalpy change for the LST-1 B peptide was larger than for the LST-1 A peptide, resulting in a more negative binding free energy, and thus increased binding affinity of LST-1 B vs LST-1 A.
Table 1.
Binding affinities, stoichiometries and thermodynamic parameters for the interaction between LST-1 peptides and FBF-2 measured by ITC
| LST-1 variant | Sequence1 | K d (μM)2 | N (# of sites)3 | ΔH (kJ/mol) | ΔG (kJ/mol) | -TΔS (kJ/mol) |
|---|---|---|---|---|---|---|
| LST-119–50 A | QHEAPKQLLQLRSEIK | 2.13 | 0.88 | -57.3 | -31.9 | 25.4 |
| 2.02 | 0.85 | -57.1 | -32.0 | 25.1 | ||
| LST-167–98 B | GLRSQKLHLTYIEKNK | 0.046 | 0.87 | -73.4 | -41.2 | 32.2 |
| 0.045 | 0.88 | -71.3 | -41.3 | 30.0 | ||
| LST-119–50 A | QHEAPKQLLQLRSEIK | 2.41 | 1.0 | -50.6 | -31.6 | 19.0 |
| 2.29 | 1.0 | -50.4 | -31.7 | 18.7 | ||
| LST-167–98 B | GLRSQKLHLTYIEKNK | 0.052 | 1.0 | -64.2 | -40.9 | 23.3 |
| 0.051 | 1.0 | -62.5 | -41.0 | 21.5 | ||
| LST-1 B mutN | GHEAPKLHLTYIEKNK | 0.21 | 1.0 | -56.3 | -37.6 | 18.7 |
| 0.24 | 1.0 | -53.7 | -37.2 | 16.5 | ||
| LST-1 B mutC | GLRSQKLHLTLRSENK | 1.51 | 1.0 | -42.7 | -32.7 | 10.0 |
| 1.45 | 1.0 | -43.5 | -32.8 | 10.7 | ||
| LST-1 B Y85L | GLRSQKLHLTLIEKNK | 0.74 | 1.0 | -50.3 | -34.4 | 15.8 |
| 0.85 | 1.0 | -50.3 | -34.1 | 16.2 |
1The relevant portions of the amino acid sequences of LST-1 A (residues Q27-K42) and LST-1 B (residues G75-K90) are shown. Complete peptide sequences are in Figure 1. The lysine and leucine residues in the KxxL motifs are in boldface. The substitutions in the mutant peptides are blue.
2Two technical replicates were performed for each analysis, and the values for each replicate are shown.
3For the analysis of LST-1 A and LST-1 B, we determined the LST-1 titrant concentrations by A280. For the analysis of LST-1 B mutants, the number of sites, N, was set at 1 during curve fitting while the LST-1 B concentration was fit, because we could not accurately determine the titrant concentration for LST-1 B mutC and Y85L due to a lack of aromatic residues. For consistency, we also show the binding affinities and thermodynamic parameters for LST-119–50 A and LST-167–98 B when N was set at 1. The concentrations of titrants were fit to values similar to those that could be determined by A280. The LST-1 A19-50 concentration was fit to 0.24 mM vs 0.28 mM by A280, the LST-1 B67-98 concentration was fit to 0.14 mM vs 0.12 mM by A280, and the LST-1B mutN concentration was fit to 0.16 mM v 0.12 mM by A280.
Interactions between FBF-2 and the KxxL motifs of LST-1 A and B peptides are indistinguishable
To compare the molecular details of FBF-2 interaction with the LST-1 A and B peptides, we determined a crystal structure of a ternary complex of the FBF-2 RNA-binding domain with an LST-1 A peptide (residues 27–41) and cFBE RNA (Table 2, Supplementary Figure S1A). We previously determined a crystal structure of FBF-2, LST-1 B peptide (residues 76–90), and the cFBE RNA (5′-CUGUGAAUG-3′) (20), which allowed direct comparisons of the two structures. Both crystal structures indicated that LST-1 interacts via the KxxL motif with FBF-2 on the surface opposite its concave RNA-binding surface in a region between repeats 7 and 8 of FBF-2 (Figure 2A). Notably, mutation of this region of FBF-2 impairs its binding with additional protein partners (CPB-1 and GLD-3) (31–33), indicating that it is a common platform for protein-protein interaction.
Figure 2.
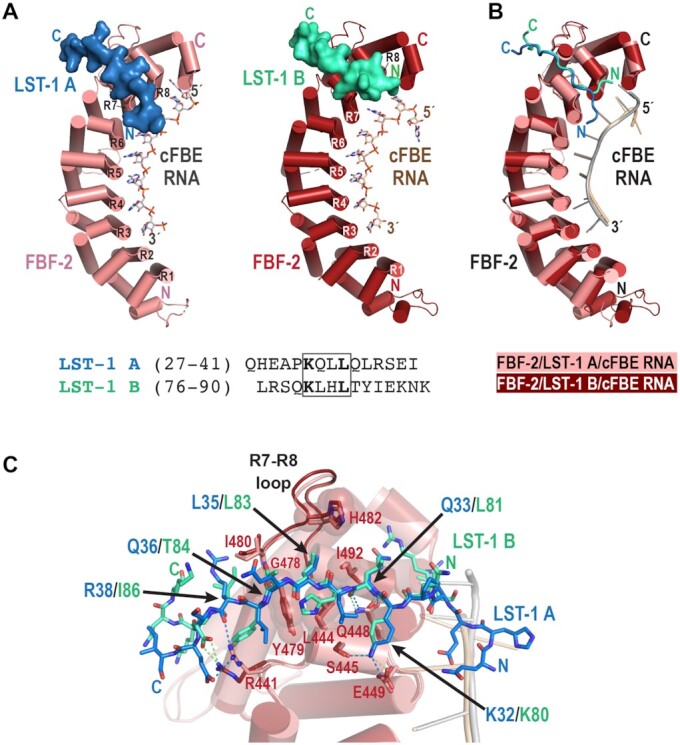
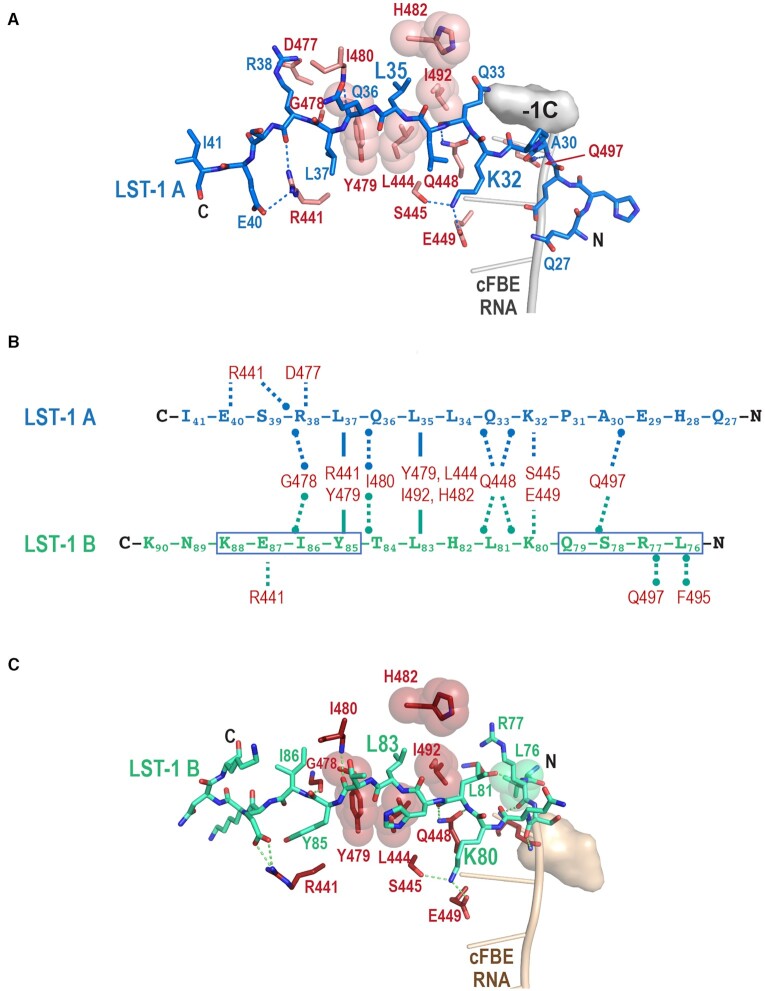
LST-1 A and B peptides interact with the same binding site on FBF-2 using the conserved KxxL motif. (A) Crystal structures of FBF-2 in complex with LST-1 A or B peptides and cFBE RNA. FBF-2 is shown as a cartoon with cylindrical α helices, cFBE RNA is shown as a stick model, and LST-1 peptides are shown as surface representations (LST-1 A, blue; LST-1 B, green). FBF-2 in complex with LST-1 A is pink and FBF-2 with LST-1 B is red. The RNA-binding helices of FBF-2 repeats 1–8 (R1-R8) are labelled. Although present in the crystallized RNA, the last nucleotide G8 was disordered, indicating lack of specific interaction with FBF-2 repeat 1. The sequences of LST-1 A and B peptides visible in the crystal structures are shown with the KxxL motifs boxed. (B) Superposition of crystal structures of FBF-2 in complex with LST-1 A or B peptides and cFBE RNA. FBF-2/cFBE RNA in complex with LST-1 A is pink and FBF-2/cFBE RNA with LST-1 B is red. FBF-2 and cFBE RNA are shown as cartoon models, and LST-1 peptides are shown as ribbons (LST-1 A, blue; LST-1 B, green). Residues 313–567 of FBF-2 were aligned (RMSD 0.57 Å over 1588 atoms). Although it is difficult to quantitate the FBF-2 conformational change, the qualitative curvature difference of FBF-2 in the complexes with LST-1 A or B peptide is apparent in the misalignment of the N-terminal repeats. (C) FBF-2 interacts with six-residue sequences containing the KxxL motifs in LST-1 A and B peptides. The LST-1 peptides are shown as stick models, and cFBE RNAs are shown as cartoons. FBF-2 residues that interact with LST-1 are displayed as stick models. Side chains that form a hydrophobic pocket for L35/L83 are shown with transparent spheres. Hydrogen bond and salt bridge interactions are indicated with dashed lines (LST-1 A, blue; LST-1 B, green).
FBF-2 interacts with six-residue sequences containing the KxxL motifs of LST-1 A (KQLLQL32-37) and B (KLHLTY80-85) using a similar set of protein-protein interactions (Figures 2 and 3, Supplementary Figure S1B). FBF-2 makes direct contacts with the two conserved residues of the LST-1 KxxL motif: lysine in the first position (K32 in the A motif and K80 in the B motif) and leucine in the fourth position (L35 in the A motif and L83 in the B motif). FBF-2 S445 and E449 in the first α helix of repeat 7 interact with the amine group of the K32/K80 side chain, and FBF-2 interacts with L35/L83 using a hydrophobic binding pocket at the base of the R7-R8 loop between repeats 7 and 8, formed by Y479, L444, I492, and H482. The R7-R8 loop of FBF-2 adopts the same conformation in the two structures (Figure 2B, C). The six-residue LST-1 A and B interaction sequences follow a similar path in both structures despite different residues at several positions (RMSD = 0.5 Å over 16 backbone atoms). As a result, FBF-2 uses similar interactions to recognize backbone atoms of the second and fifth residues. FBF-2 Q448 forms hydrogen bonds with the main chain amino and carboxyl groups of LST-1 Q33/L81, and the main chain amino group of FBF-2 I480 contacts the main chain carboxyl group of LST-1 Q36/T84 (Figure 2C). In the previous FBF-2/LST-1 B76-90/cFBE RNA structure, two ternary complexes are present in an asymmetric unit, and interactions are similar in both complexes (20) (Supplementary Figure S2). We previously showed that FBF-2 Q448G weakened interaction with LST-1 B (20), and the structure reported here predicts a similar effect on interaction with LST-1 A.
Figure 3.
FBF-2 makes distinct interactions with the N- and C-terminal regions flanking the KxxL motifs of LST-1 A and B peptides. (A) The path of N-terminal residues of LST-1 A allowed interaction of FBF-2 with the upstream −1C in cFBE RNA. A surface representation of the −1C nucleotide is shown. FBF-2 residues that interact with LST-1 are displayed as stick models. Side chains that form a hydrophobic pocket for L35 are shown with transparent spheres. Hydrogen bond and salt bridge interactions are indicated with dashed lines. (B) Schematic drawing of interactions of FBF-2 with LST-1 A or B peptides. The direction of the LST-1 peptide sequences matches the structures in panels A and C with the N-termini to the right. FBF-2 interactions with both A and B peptides (middle), specific to LST-1 A (top), or specific to LST-1 B (bottom) are indicated by dashed lines. Interactions via main chain atoms end with circles. The LST-1 B LRSQ sequence that was mutated to HEAP and YIEK that was mutated to LRSE are boxed. (C) The path of N-terminal residues of LST-1 B placed L76 (shown with green spheres) in the FBF-2 upstream C binding pocket, displacing −1C. FBF-2 side chains that interact with LST-1 are displayed as stick models. Side chains that form a hydrophobic pocket for L83 are shown with transparent spheres. Hydrogen bond and salt bridge interactions are indicated with dashed lines. This figure shows interactions observed in one of two complexes in the asymmetric unit (PDB ID 6PUN, FBF-2 chain A). The interactions in the second complex are shown in Supplementary Figure S2.
Distinct flanking regions of LST-1 A and B peptides produce differences in affinity for FBF-2
We examined our crystal structures of complexes of FBF-2 with LST-1 A or B to identify molecular explanations for the 40-fold tighter binding of LST-1 B than LST-1 A. The crystal structures revealed that the residues of LST-1 A and B N-terminal to the KxxL motif take divergent paths on the surface of FBF-2 (Figure 2). As a result, FBF-2 forms additional interactions with the N-terminal residues of the LST-1 B peptide that are not formed with LST-1 A (Figure 3B). LST-1 B interaction buries a larger surface area than LST-1 A (839 Å2 vs. 726 Å2), consistent with tighter binding of LST-1 B than LST-1 A. Notably, the path of the LST-1 B peptide also places L76 in the pocket on FBF-2 that binds to a cytosine upstream of the core RNA recognition sequence, and the −1C base is either disordered or flipped away from the upstream cytosine binding pocket (Figure 3C, Supplementary Figure S2) (8,20). In contrast, the path of the LST-1 A peptide leaves the upstream cytosine binding pocket of FBF-2 fully available to bind the −1C base of the cFBE. Consequently, we observed interaction with the −1C base in the crystal structure of the FBF-2/LST-1 A27-41/cFBE complex (Figure 3A).
C-terminal to the KxxL motif, the side chains of L37 in LST-1 A and Y85 in LST-1 B form CH-π interactions with FBF-2 R441 and Y479 (Figure 3). However, slightly different interactions of FBF-2 with the aliphatic L37 of LST-1 A versus the aromatic Y85 of LST-1 B may contribute to the weaker peptide binding affinity of the site A peptide. We previously showed that mutation of FBF-2 Y479 disrupted interaction with LST-1 B (20). Based on conservation of the hydrophobic interactions, we hypothesize that mutation of FBF-2 Y479 would affect LST-1 A binding as well.
Given that interactions of FBF-2 with the KxxL motifs in LST-1 A and B are similar, we hypothesized that the differences in interaction of FBF-2 with the N- and C-terminal sequences flanking the KxxL motifs account for the differences in binding affinity. To determine which residues in the LST-1 A and B peptides contribute to the 40-fold weaker binding affinity of LST-1 A versus LST-1 B, we generated mutant peptides where we substituted residues of LST-1 B with the equivalent residues from LST-1 A. Residues N-terminal to the KxxL motif of LST-1 A and B take divergent paths (Figure 2B, C), so we mutated LRSQ76-79 of LST-1 B67-98 to HEAP, corresponding to LST-1 A residues 28–31 (Figure 3B). Residues C-terminal to the KxxL motif, including L37 versus Y85, form slightly different interactions with FBF-2 (Figure 3), so we mutated YIEK85-88 of LST-1 B67-98 to LRSE, corresponding to LST-1 A residues 37–40 (Figure 3B). We measured binding affinities of these LST-1 B67-98 mutants to FBF-2 by ITC (Supplementary Figure S3, Table 1). The N-terminal LST-1 B mutant (mutN) bound to FBF-2 with a Kd of 0.2 μM, ∼4-fold lower than the LST-1 B wild type affinity of 0.05 μM. The C-terminal LST-1 B mutant (mutC) bound to FBF-2 with a Kd of 1.5 μM, ∼30-fold weaker than wild type, indicating the importance of these C-terminal residues. As noted above, our crystal structures indicated differences in interaction of FBF-2 with Y85 in LST-1 B versus L37 in LST-1 A. We therefore tested an LST-1 B67-98 peptide with a single Y85L mutation and found that this substitution alone weakened binding to FBF-2 ∼15 fold. Taken together, we conclude that Y85 of LST-1 B is a major contributor to the tighter binding of the LST-1 B peptide versus L37 of LST-1 A, and both the N-terminal and C-terminal sequences flanking the KxxL motif make additional contributions.
Adaptability of the FBF-2 structural scaffold permits distinct RNA recognition modes
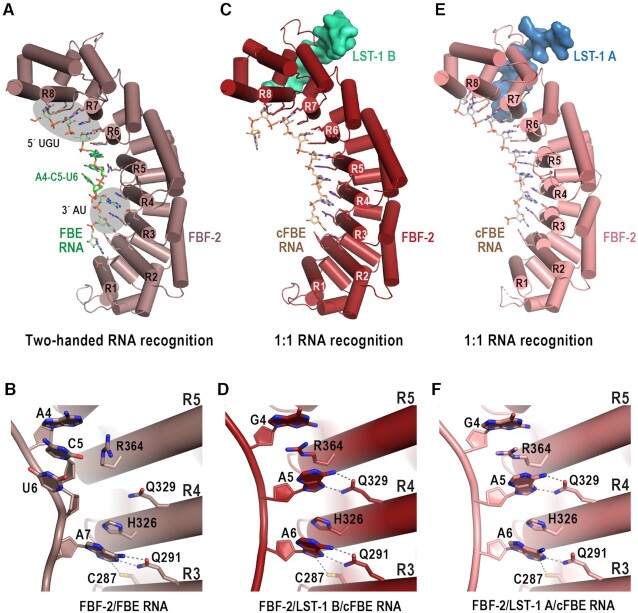
In addition to identifying differences in FBF-2 interaction with LST-1 A and B peptides, we compared the modes of RNA recognition in complexes of FBF-2 with LST-1 A or B peptides and found that FBF-2 used different strategies to recognize the central nucleotides of cFBE RNA. We previously showed that FBF-2 uses two distinct RNA recognition modes to recognize 9-nt versus 8-nt RNA elements. A two-handed recognition mode is used to bind to a consensus 9-nt RNA sequence, 5′-UGURnnAUn-3′ (R = purine, n = any nucleotide), by the eight repeats of FBF-2 (34). We observed this mode of binding to a variety of RNA target sequences that match this consensus (11). Using this mode, the 5′ UGU (nts 1–3) is bound by repeats 6–8 and the 3′ AU (nt 7–8) is bound by repeats 2 and 3 (Figure 4A). The bases of the central nucleotides 4–6 are directly stacked and flipped away from the RNA-binding surface, which allows binding to RNAs with variable sequences at these positions (Figure 4A, B). We previously noted that the RNA-binding surface of FBF-2 is somewhat flatter than that of other PUF proteins, like human Pumilio1 or C. elegans PUF-8, that recognize 8-nt sequences (11,35). In addition to the two-handed recognition mode, FBF-2 also uses a 1-repeat-to-1-nucleotide RNA recognition mode that we first identified in a crystal structure of the ternary complex of FBF-2 with an LST-1 B peptide and cFBE RNA (5′-CUGUGAAU-3′) (20). In this 1:1 recognition mode FBF-2 repeats 2–8 each recognize one RNA base, and no nucleotides are flipped away from the RNA-binding surface (Figure 4C, D). The crystal structure of the FBF-2/LST-1 B/cFBE RNA complex also shows that the overall curvature of the RNA-binding surface of FBF-2 is increased relative to previous FBF-2 complexes with 9-nt RNAs and appears to mold the RNA-binding surface to directly recognize the cFBE sequence (20). Our crystal structure of FBF-2 with LST-1 A peptide and cFBE RNA revealed a modified 1:1 RNA recognition mode (Figure 4E, F), and the FBF-2 curvature was reduced compared to the complex with LST-1 B (Figure 2B).
Figure 4.
FBF-2 recognizes different edges of the A6 base of the cFBE RNA. (A) FBF-2 recognizes 9-nt FBE RNA using the two-handed recognition mode where FBF-2 recognizes 5′-UGU and AU-3′ elements (grey ovals). A crystal structure of FBF-2 in complex with consensus FBE RNA (5′-UGUACUAUA-3′) is shown (PDB ID 3K5Q). The three central nucleotides, A4, C5, and U6 (green) are directly stacked and flipped away from the RNA-binding surface of FBF-2. The RNA-binding helices of FBF-2 repeats 1–8 (R1-R8) are labelled. The N-terminal FBF-2 repeat binds to the 3′ nucleotide of the RNA. (B) FBF-2 interacts with the Watson-Crick edge of A7 in a complex with consensus FBE RNA. Hydrogen bond and van der Waals interactions are indicated with dashed lines. (C) FBF-2/LST-1 B recognizes a cFBE RNA (5′-CUGUGAAU-3′) using a 1:1 recognition mode where each nucleotide is recognized by an FBF-2 repeat (PDB ID: 6PUN). (D) FBF-2 interacts with the Hoogsteen edges of both A5 and A6 in the complex with LST-1 B peptide. (E) FBF-2/LST-1 A recognizes a cFBE RNA using a 1:1 recognition mode but with a flatter curvature than in the complex with LST-1 B (see Figure 2B). (F) FBF-2 interacts with the Hoogsteen edge of A5 and the Watson-Crick edge of A6 in the complex with LST-1 A peptide.
By comparing the crystal structures of FBF-2 with either LST-1 A or B, we found that FBF-2 in the complex with LST-1 A recognized the Hoogsteen edge of A5 and Watson-Crick edge of A6 in the cFBE, whereas in the complex with LST-1 B, FBF-2 recognized the Hoogsteen edges of both A5 and A6 (Figure 4D, F). Therefore, a 1:1 RNA recognition mode does not require FBF-2 curvature change, because the RNA can also change conformation. Although we observed distinct FBF-2 curvatures in the crystal structures with LST-1 A or B, the crystal structures do not indicate whether LST-1 A or B peptide binding induced the differences in curvature. It is noteworthy that the two ternary complexes were crystallized in different space groups. The crystals of the FBF-2/LST-1 A/cFBE RNA complex belonged to the P61 space group (Table 2), which matches that of previous FBF-2/RNA structures (11), and in all cases, the FBF-2 curvature is in the flatter conformation. The crystals of the FBF-2/LST-1 B/cFBE RNA complex belonged to the P1 space group with two complexes per asymmetric unit (20). Although the FBF-2 curvature is increased in the two independent LST-1 B complexes, crystal contacts may favor the greater curvature versus the P61 space group. Our crystal structures have captured unique FBF-2 conformations in LST-1 A and B complexes, but in solution it is quite possible that FBF-2 may use either 1:1 RNA recognition mode when binding to cFBE RNA.
The degree of PUF protein curvature appears to correspond to the pattern of base edge recognition for A5 and A6 in other PUF protein crystal structures. We observed that human PUM1 (36) and S. cerevisiae Puf3 (14) also recognize the Hoogsteen edges of consecutive A5 and A6 nucleotides, while human PUM2 recognizes the Hoogsteen edge of A5 and Watson-Crick edge of A6 (36). The protein scaffolds of human PUM1 and yeast Puf3 are subtly more curved than that of PUM2. Therefore, for several PUF proteins, increased curvature is associated with recognizing the Hoogsteen edges of both A5 and A6, and decreased curvature is associated with recognizing the Hoogsteen edge of A5 and the Watson-Crick edge of A6.
We also determined a crystal structure of a ternary complex of FBF-2 and LST-1 A31-41 with a consensus 9-nt FBE RNA (5′-UGUACUAUA-3′), which illustrated that LST-1 partnership is also used with the two-handed mode of RNA recognition (Table 2, Supplementary Figure S4A). The conformation of FBF-2 was similar when compared to its structure in the ternary complex of FBF-2/LST-1 A/cFBE (RMSD of 0.49 Å over 396 FBF-2 Cα atoms) or a previous structure of the binary FBF-2/FBE complex (PDB ID: 3K5Q) (RMSD of 0.77 Å over 389 FBF-2 Cα atoms) (11). In crystal structures of the ternary and binary complexes, FBF-2 bound to FBE RNA using the two-handed RNA recognition mode (Supplementary Figure S4A) with a flatter overall curvature (Supplementary Figure S4B). We have not obtained crystals of a ternary complex of FBF-2 with LST-1 B and an RNA that binds via the two-handed mode, but RNA-binding assays indicate that FBF-2/LST-1 B binds with high affinity to such RNAs (20). Thus, the conformations of both the FBF-2 scaffold and the RNAs appear to be dynamic.
LST-1 B, not LST-1 A, modulates the RNA-binding affinity of FBF-2
We previously demonstrated that LST-1 B weakens the interaction of FBF-2 with RNAs containing either 8-nt or 9-nt sequence motifs in vitro and thereby may finetune target mRNA selection (20). We therefore sought to determine whether LST-1 A also modulates the RNA-binding affinity of FBF-2. To do so, we used Microscale Thermophoresis (MST) technology (37) to measure the binding affinities of FBF-2 for fluorescently-labeled RNAs in the presence or absence of LST-1 A (Figure 5 and Supplementary Figure S5). We first established that FBF-2 bound to RNAs containing a 9-nt element (gld-1 FBEa, 5′-AUCAUGUGCCAUAC-Cy5-3′) or an 8-nt element (cFBE, 5′-AUCUGUGAAUGA-Cy5-3′) with similar affinity, consistent with our previous measurements by electrophoretic mobility shift assay (EMSA). Using different techniques, we note that the absolute Kd values obtained using the equilibrium MST method (110 nM for FBEa and 67 nM for cFBE) were higher than the values we previously determined by EMSA (12 nM for FBEa and 10 nM for cFBE), but relative changes in affinity in the presence of LST-1 B were in agreement (20). We next examined the effect of LST-1 A or LST-1 B on the RNA-binding affinity of FBF-2 by preincubating LST-1 peptide with titrations of FBF-2 before mixing with RNA. The protein-RNA mixtures were allowed to come to equilibrium by incubating at 4°C overnight prior to MST analysis. LST-1 A or B peptides alone did not bind to the RNAs, even at the highest peptide concentration of 20 μM. Consistent with what we demonstrated previously, FBF-2 in the presence of LST-1 B bound to both RNAs with ∼3-fold lower affinity than FBF-2 alone (Figure 5, Table 3, Supplementary Figure S5). Surprisingly, LST-1 A had no effect on FBF-2 affinity: FBF-2 in the presence of LST-1 A bound to both RNAs with slightly higher affinity than FBF-2 alone, but the 1.3 to 1.4-fold difference was not statistically significant (Table 3, Supplementary Figure S5). We reasoned that this difference in ability to modulate FBF-2 RNA-binding affinity could be due to the weaker overall interaction between FBF-2 and LST-1 A versus LST-1 B or the different interactions made by the N- and C-terminal sequences flanking the KxxL motifs of the two peptides.
Figure 5.
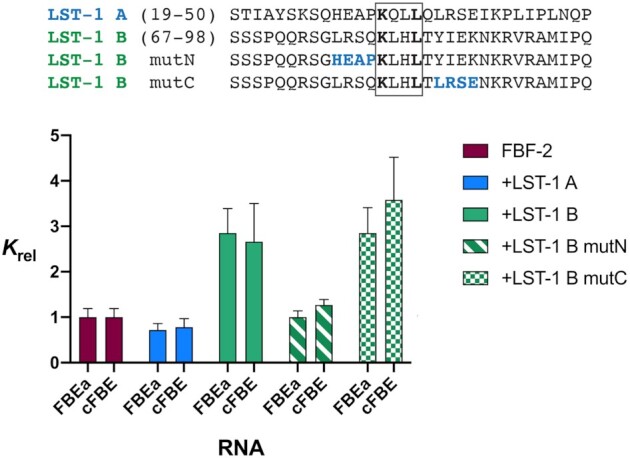
LST-1 B, but not LST-1 A, weakens the RNA-binding affinity of FBF-2. Relative binding affinities (Krel) and standard deviations of FBF-2 for FBEa and cFBE RNAs measured by MST are shown. Krel values are normalized to the binding affinity of FBF-2 alone to the respective RNA. Kd values are in Table 3. The addition of LST-1 B (+LST-1 B) increased the Krel, but addition of LST-1 A (+LST-1 A) had no effect. Substitution of N-terminal residues in LST-1 B (+LST-1 B mutN) abrogated the ability of LST-1 B to modulate FBF-2 RNA-binding affinity, while substitution of C-terminal residues in LST-1 B (+LST-1 B mutC) had no effect.
Table 3.
RNA-binding affinities (Kd /nM1) of FBF-2 and FBF-2/LST-1 measured by MST
| FBF-2 | FBF-2/LST-1A | FBF-2/LST-1B wt | FBF-2/LST-1B mutN | FBF-2/LST-1B mutC | |
|---|---|---|---|---|---|
| gld-1 FBEa | 110 ± 21 | 79 ± 15 | 314 ± 59 | 110 ± 15 | 314 ± 62 |
| cFBE | 67 ± 13 | 52 ± 13 | 178 ± 56 | 85 ± 8 | 240 ± 63 |
1 K d values and standard deviations were determined from curves fit to three independently pipetted technical replicates.
The N-terminal residues of LST-1 B are required to reduce FBF-2 RNA-binding affinity
We used the N- and C-terminal LST-1 B mutants from above to probe which residues flanking the KxxL motif in LST-1 B are critical to modulate FBF-2 RNA-binding affinity. Substitution of the LST-1 B N-terminal residues with the N-terminal residues of LST-1 A (mutN) eliminated the ability to decrease FBF-2 RNA-binding affinity (Figure 5, Table 3). In contrast, substitution of the LST-1 B C-terminal residues with the C-terminal residues of LST-1 A (mutC) had no effect, and the LST-1 B mutC peptide reduced RNA-binding affinity to the same extent as wild type LST-1 B. This indicates that the LST-1 B N-terminal interactions with FBF-2 are critical to modulate RNA-binding affinity. The specific interactions appear to be more important than the overall affinity between LST-1 and FBF-2. The LST-1 B mutC peptide retained the ability to modulate FBF-2 RNA-binding affinity, even though the LST-1 B C-terminal substitutions strongly reduced affinity to FBF-2 (30-fold). The N-terminus of the LST-1 B peptide is located near the 5′ end of the RNA in our crystal structure, and the N-terminal residue L76 protrudes into the upstream cytosine pocket. Although residues N-terminal to L76 are not observed in our crystal structure, we propose that they have the potential to interfere with binding to the 5′ end of RNA elements, including the nearby U1 nucleotide. This is consistent with our previous results showing that LST-1 B reduced the affinity of FBF-2 for RNAs with or without an upstream cytosine ∼3 fold.
DISCUSSION
Protein-protein interactions via short linear interaction motifs are critical for the formation and regulation of dynamic macromolecular complexes across a wide range of biological pathways (38,39). These interactions are used to recruit additional components, and complex formation can also stabilize interactions that are typically transient and weak. PUF proteins use their conserved C-terminal RNA-binding domains to interact with target mRNAs (3,4), and Drosophila melanogaster and human Pumilio proteins have been shown to utilize their N-terminal intrinsically disordered regions to interact with deadenylation machinery (40–43), although the molecular details of these interactions remain to be fully elucidated. PUF proteins also form specific protein partnerships for mRNA target selection, such as D. melanogaster Pumilio with Nanos or FBF with LST-1, SYGL-1, CPB-1 (Cytoplasmic Polyadenylation element-Binding protein 1), and GLD-3 (GermLine development Defective-3). In these complexes, the non-RNA binding surface of the C-terminal repeats of the PUF protein interacts with the partner protein (12,20,31–33). This work expands our understanding of the functional consequences of partnerships on association with RNA and reveals a diversity of outcomes.
FBF interacts with CPB-1 and GLD-3 during spermatogenesis (44,45). Similar to LST-1, both CPB-1 and GLD-3 possess a KxxL motif. However, other than the consensus lysine and leucine residues, there is little sequence similarity among these FBF partner proteins. Our crystal structures revealed sequence-independent interactions of FBF-2 with peptide backbone atoms of LST-1 A and B as well as interactions with side chain atoms of the consensus lysine and leucine residues. We demonstrated that N-terminal and C-terminal sequences flanking the KxxL motif of the LST-1 A and B peptides contribute to FBF-2 interaction to varying degrees. Thus, a single stretch of ∼12 amino acid residues in FBF partner proteins modulates their binding affinities. CPB-1 has been shown to bind to FBF-2 with a Kd of low nanomolar range, and GLD-3 binding is weaker with a Kd about 20 times higher than CPB-1 (32,33). By adopting a similar mode of interaction via their KxxL motifs, these FBF partners might compete with one another, and the associated partner protein could induce different regulatory outcomes. Alternatively, the spatial patterning of expression within tissues may dictate the dominant regulatory modality of the protein complex. Rigorous biochemical studies are critical to assess the intricacies of the individual interactions to elucidate the properties of each complex individually.
Our study expands our molecular and quantitative understanding of FBF and LST-1 partnership. Interaction between FBF-2 and LST-1 via at least one of the two KxxL motifs is required for LST-1 activity in stem cell self-renewal (25). In C. elegans, expression of either LST-1 or SYGL-1 is sufficient to maintain GSCs, so LST-1 mutant analysis is performed in strains that do not express SYGL-1. SYGL-1 null animals expressing LST-1 with mutations disrupting either the A or B KxxL motif maintain GSCs and are fertile (25). In contrast, SYGL-1 null animals expressing LST-1 with mutations at both A and B motifs do not maintain GSCs and are infertile. This result and our finding that LST-1 A and B bind at the same location on FBF-2 suggests functional redundancy is built into LST-1. The LST-1 A and B site redundancy is intramolecular and adds another layer to the intermolecular redundancy of FBF partnership with both LST-1 and SYGL-1 in GSC self-renewal. This depth of functional redundancy both between and within partner proteins appears to reflect the crucial importance of GSC self-renewal.
We also demonstrated here that LST-1 A and B peptides bind to FBF-2 with different affinities and only LST-1 B modulates the RNA-binding affinity of FBF-2. This suggests that the two FBF interaction sites in LST-1 are more than just redundant and offer additional mechanisms for target mRNA regulation. Depending on whether the A or B site is utilized, the FBF-2/LST-1 complex may control different subsets of mRNAs. As we noted previously, the weakened RNA-binding affinity of the FBF-2/LST-1 B partnership could focus regulation on mRNAs that are more abundant or contain higher affinity binding elements (20). The tighter binding affinity of LST-1 B suggests it would be more likely to interact with FBF. However, post-translational modifications of LST-1, such as phosphorylation, could inhibit or alter the strength or availability of one of the FBF interaction sites in LST-1 and influence whether site A or B is engaged with FBF. This work provides a conceptual basis for searching for these potentially crucial modifications.
It is also possible that both LST-1 sites could be engaged simultaneously with two separate PUF proteins (25), and these higher order complexes could recognize mRNAs with two PUF binding elements. In addition to FBF, LST-1 interacts with C. elegans PUF proteins PUF-3 and PUF-11 (46). The recently discovered ‘PUF hub’ of four PUF proteins (FBF-1, FBF-2, PUF-3, and PUF-11) and two partner proteins (LST-1 and SYGL-1) that together control GSC self-renewal expands the range of different higher order complexes that may be formed. The PUF hub for C. elegans GSC self-renewal is an ideal opportunity to take advantage of the tandem LST-1 FBF interaction sites, expanding mechanisms for regulation of gld-1 as well as other target mRNAs. The PUF protein interaction sites in SYGL-1 have not yet been identified, but even a single PUF protein interaction site expands the number of different regulatory complexes. The four PUF proteins and two partner proteins in the PUF hub can form at least 18 different combinations (assuming SYGL-1 bears a single PUF interaction site). The particular PUF proteins and partners that interact could dictate which effector factors, such as deadenylases or poly(A) polymerases, are recruited and therefore lead to radically different outcomes for the bound mRNAs. It remains an intriguing possibility that similar higher order complexes might be broadly utilized to mediate collaborations between PUF and partner proteins in other tissues and organisms.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 7RZZ (the FBF-2/LST-1 A/cFBE RNA complex) and 7S02 (the FBF-2/LST-1 A/FBE RNA complex).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline staff for assistance with data collection at SER-CAT beamlines 22-ID and 22-BM at the Advanced Photon Source, Argonne National Laboratory, and Lars Pedersen and Juno Krahn for crystallographic and data collection support at NIEHS. We are grateful to Judith Kimble, Sarah Crittenden, Brian Carrick, and Ahlan Ferdous for helpful discussion and comments on the manuscript. We appreciate critical reading of this manuscript by our NIEHS colleagues, Huanchen Wang and Geoffrey Mueller.
Contributor Information
Chen Qiu, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Robert N Wine, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Zachary T Campbell, Department of Biological Sciences, University of Texas at Dallas, Richardson, TX 75025, USA.
Traci M Tanaka Hall, Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported in part by the National Institutes of Health [R01NS100788 and R01NS114018 to ZTC] and by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences [1ZIA50165 to TMTH]. The Advanced Photon Source used for this study was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31–109-Eng-38. Funding for open access charge: National Institutes of Health, National Institute of Environmental Health Sciences [1ZIA50165].
Conflict of interest statement. None declared.
REFERENCES
- 1. Barker D.D., Wang C., Moore J., Dickinson L.K., Lehmann R.. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992; 6:2312–2326. [DOI] [PubMed] [Google Scholar]
- 2. Macdonald P.M. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development. 1992; 114:221–232. [DOI] [PubMed] [Google Scholar]
- 3. Zamore P.D., Williamson J.R., Lehmann R.. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997; 3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M.P.. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997; 390:477–484. [DOI] [PubMed] [Google Scholar]
- 5. Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K.. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001; 105:281–289. [DOI] [PubMed] [Google Scholar]
- 6. Jenkins H.T., Baker-Wilding R., Edwards T.A.. Structure and RNA binding of the mouse Pumilio-2 PUF domain. J. Struct. Biol. 2009; 167:271–276. [DOI] [PubMed] [Google Scholar]
- 7. Miller M.T., Higgin J.J., Hall T.M.T.. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008; 15:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qiu C., Kershner A., Wang Y., Holley C.P., Wilinski D., Keles S., Kimble J., Wickens M., Hall T.M.T.. Divergence of Pumilio/fem-3 mRNA Binding Factor (PUF) protein specificity through variations in an RNA-binding pocket. J. Biol. Chem. 2012; 287:6949–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X., McLachlan J., Zamore P.D., Hall T.M.T.. Modular recognition of RNA by a human Pumilio-homology domain. Cell. 2002; 110:501–512. [DOI] [PubMed] [Google Scholar]
- 10. Wang X., Zamore P.D., Hall T.M.T.. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001; 7:855–865. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y., Opperman L., Wickens M., Hall T.M.T.. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U. S. A. 2009; 106:20186–20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weidmann C.A., Qiu C., Arvola R.M., Lou T.F., Killingsworth J., Campbell Z.T., Hall T.M.T., Goldstrohm A.C.. Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. eLife. 2016; 5:e17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilinski D., Qiu C., Lapointe C.P., Nevil M., Campbell Z.T., Hall T.M.T., Wickens M.. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat. Commun. 2015; 6:8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu D., Stumpf C.R., Krahn J.M., Wickens M., Hall T.M.T.. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U. S. A. 2009; 106:20192–20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kershner A., Crittenden S.L., Friend K., Sorensen E.B., Porter D.F., Kimble J.. Germline stem cells and their regulation in the nematode Caenorhabditis elegans. Adv. Exp. Med. Biol. 2013; 786:29–46. [DOI] [PubMed] [Google Scholar]
- 16. Wang X., Voronina E.. Diverse Roles of PUF Proteins in Germline Stem and Progenitor Cell Development in C. elegans. Front. Cell Dev. Biol. 2020; 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bernstein D., Hook B., Hajarnavis A., Opperman L., Wickens M.. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005; 11:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Opperman L., Hook B., DeFino M., Bernstein D.S., Wickens M.. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005; 12:945–951. [DOI] [PubMed] [Google Scholar]
- 19. Prasad A., Porter D.F., Kroll-Conner P.L., Mohanty I., Ryan A.R., Crittenden S.L., Wickens M., Kimble J.. The PUF binding landscape in metazoan germ cells. RNA. 2016; 22:1026–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu C., Bhat V.D., Rajeev S., Zhang C., Lasley A.E., Wine R.N., Campbell Z.T., Hall T.M.T.. A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity. eLife. 2019; 8:e48968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kershner A.M., Shin H., Hansen T.J., Kimble J.. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. U. S. A. 2014; 111:3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shin H., Haupt K.A., Kershner A.M., Kroll-Conner P., Wickens M., Kimble J.. SYGL-1 and LST-1 link niche signaling to PUF RNA repression for stem cell maintenance in Caenorhabditis elegans. PLos Genet. 2017; 13:e1007121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crittenden S.L., Bernstein D.S., Bachorik J.L., Thompson B.E., Gallegos M., Petcherski A.G., Moulder G., Barstead R., Wickens M., Kimble J.. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002; 417:660–663. [DOI] [PubMed] [Google Scholar]
- 24. Francis R., Barton M.K., Kimble J., Schedl T.. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995; 139:579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haupt K.A., Enright A.L., Ferdous A.S., Kershner A.M., Shin H., Wickens M., Kimble J.. The molecular basis of LST-1 self-renewal activity and its control of stem cell pool size. Development. 2019; 146:dev181644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mossessova E., Lima C.D.. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000; 5:865–876. [DOI] [PubMed] [Google Scholar]
- 27. Otwinowski Z., Minor W.. [20]Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 28. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 31. Campbell Z.T., Menichelli E., Friend K., Wu J., Kimble J., Williamson J.R., Wickens M.. Identification of a conserved interface between PUF and CPEB proteins. J. Biol. Chem. 2012; 287:18854–18862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Menichelli E., Wu J., Campbell Z.T., Wickens M., Williamson J.R.. Biochemical characterization of the Caenorhabditis elegans FBF.CPB-1 translational regulation complex identifies conserved protein interaction hotspots. J. Mol. Biol. 2013; 425:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu J., Campbell Z.T., Menichelli E., Wickens M., Williamson J.R.. A protein.protein interaction platform involved in recruitment of GLD-3 to the FBF.fem-3 mRNA complex. J. Mol. Biol. 2013; 425:738–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valley C.T., Porter D.F., Qiu C., Campbell Z.T., Hall T.M.T., Wickens M.. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc. Natl. Acad. Sci. U. S. A. 2012; 109:6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhat V.D., McCann K.L., Wang Y., Fonseca D.R., Shukla T., Alexander J.C., Qiu C., Wickens M., Lo T.W., Hall T.M.T.et al.. Engineering a conserved RNA regulatory protein repurposes its biological function in vivo. eLife. 2019; 8:e43788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu G., Hall T.M.T.. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011; 19:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jerabek-Willemsen M., Wienken C.J., Braun D., Baaske P., Duhr S.. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011; 9:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar M., Gouw M., Michael S., Samano-Sanchez H., Pancsa R., Glavina J., Diakogianni A., Valverde J.A., Bukirova D., Calyseva J.et al.. ELM-the eukaryotic linear motif resource in 2020. Nucleic. Acids. Res. 2020; 48:D296–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Roey K., Uyar B., Weatheritt R.J., Dinkel H., Seiler M., Budd A., Gibson T.J., Davey N.E.. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 2014; 114:6733–6778. [DOI] [PubMed] [Google Scholar]
- 40. Arvola R.M., Chang C.T., Buytendorp J.P., Levdansky Y., Valkov E., Freddolino P.L., Goldstrohm A.C.. Unique repression domains of Pumilio utilize deadenylation and decapping factors to accelerate destruction of target mRNAs. Nucleic. Acids. Res. 2020; 48:1843–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Enwerem III, Elrod N.D., Chang C.T., Lin A., Ji P., Bohn J.A., Levdansky Y., Wagner E.J., Valkov E., Goldstrohm A.C.. Human Pumilio proteins directly bind the CCR4-NOT deadenylase complex to regulate the transcriptome. RNA. 2021; 27:445–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Etten J., Schagat T.L., Hrit J., Weidmann C.A., Brumbaugh J., Coon J.J., Goldstrohm A.C.. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 2012; 287:36370–36383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weidmann C.A., Goldstrohm A.C.. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and Brain Tumor. Mol. Cell. Biol. 2012; 32:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eckmann C.R., Crittenden S.L., Suh N., Kimble J.. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004; 168:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luitjens C., Gallegos M., Kraemer B., Kimble J., Wickens M.. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000; 14:2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haupt K.A., Law K.T., Enright A.L., Kanzler C.R., Shin H., Wickens M., Kimble J.. A PUF hub drives self-renewal in Caenorhabditis elegans germline stem cells. Genetics. 2020; 214:147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 7RZZ (the FBF-2/LST-1 A/cFBE RNA complex) and 7S02 (the FBF-2/LST-1 A/FBE RNA complex).