Abstract
In many animal species, germ cell specification requires the inheritance of germ plasm, a biomolecular condensate containing maternally-derived RNAs and proteins. Most studies of germ plasm composition and function have been performed in widely evolutionarily divergent model organisms, such as C. elegans, Drosophila, Xenopus laevis, and Danio rerio (zebrafish). In zebrafish, 12 RNAs localize to germ plasm at the furrows of the early embryo. Here, we tested for the presence of these RNAs in three additional species within the Danionin clade: Danio kyathit, Danio albolineatus, and Devario aequipinnatus. By visualizing nanos RNA, we find that germ plasm segregation patterns during early embryogenesis are conserved across these species. Ten additional germ plasm RNAs exhibit localization at the furrows of early embryos in all three non-zebrafish Danionin species, consistent with germ plasm localization. One component of zebrafish germ plasm, ca15b, lacked specific localization in embryos of the more distantly related Devario aequipinnatus. Our findings show that within a subset of closely related Danionin species, the vast majority of germ plasm RNA components are conserved. At the same time, the lack of ca15b localization in Devario aequipinnatus germ plasm highlights the potential for the divergence of germ plasm composition across a restricted phylogenetic space.
Introduction
Germ line specification begins with the establishment of primordial germ cells, a small subset of embryonic cells that will give rise to the germline of the newly developing organism. This critical developmental process is achieved through two general pathways: induction and preformation (Bertocchini & Chuva de Sousa Lopes, 2016; Extavour & Akam, 2003; Magnúsdóttir & Surani, 2014; Swartz & Wessel, 2015; Wylie, 1999). Some animals, such as eutherian mammals and urodele amphibians, specify primordial germ cells (PGCs) as a result of an inductive mechanism involving cell-cell signaling driven by zygotic genes in a tissue-specific context (Lawson et al., 1999; Ying et al., 2001). Other animals, such as teleost fish, anuran amphibians, insects, and some birds, rely on a mechanism of preformation, in which PGCs are specified through the germ plasm, a membraneless cytoplasmic structure that contains maternally-inherited ribonucleoparticles (RNPs) and becomes incorporated into PGCs (Bontems et al., 2009; Eddy, 1975; Hashimoto et al., 2004; Miranda-Rodríguez et al., 2017; Strome & Updike, 2015; Tada et al., 2012). During Danio rerio (zebrafish) embryonic development, germ plasm RNPs proceed through a series of relatively well-characterized stages of localization and behavior prior to germ cell differentiation. First, as a newly fertilized zygote begins to proceed through its initial cell cycles, maternally-derived germ plasm RNPs progressively aggregate into the embryonic cleavage furrows and compact toward the blastodisc periphery (Eno et al., 2018; Miranda-Rodríguez et al., 2017; Pelegri et al., 1999). This process, largely driven through interactions with dynamic cytoskeletal elements (reviewed in (Moravec & Pelegri, 2020)), results in four stable germ plasm masses at the distal ends of each of the first two cleavage furrows. At the 16-32 cell stage, as the embryo begins the process of cellularization, these four germ plasm aggregates become internalized into newly cellularized PGCs and are subsequently asymmetrically transmitted during each successive cell division into a single daughter cell (Braat et al., 1999; Knaut et al., 2000; Pelegri et al., 1999; Yoon et al., 1997). These first four germ plasm aggregates are maintained largely intact throughout the late cleavage, blastula, and early gastrula stages, resulting in embryos with a small subset of cells containing germ plasm (approximately ~4-12 out of ~4000-8000 total embryonic cells during the late blastula period) (Braat et al., 1999; Eno et al., 2019; Knaut et al., 2000; Yoon et al., 1997). Beginning at dome stage (4.3 hours post-fertilization at 28.5°C) in zebrafish embryos, RNAs within the intact germ plasm aggregates initiate dispersal, a process during which they appear to be released from the phase-separated mass into the host cells’ cytoplasm (Braat et al., 1999; D’Orazio et al., 2021; Knaut et al., 2000; Yoon et al., 1997). The occurrence of germ plasm RNP cytoplasmic dispersal, which roughly coincides with the bulk of zygotic genome activation in the embryo, is closely followed by the appearance of perinuclear Vasa protein-containing germinal granules (Hartwig et al., 2014; Houwing et al., 2007; Knaut et al., 2000; Strasser et al., 2008). During this period PGCs are also thought to initiate the germ cell-specific gene expression program and continue to proliferate and undergo directed migratory movements towards the prospective gonadal region (Blaser et al., 2005; Bontems et al., 2009).
In zebrafish, 12 RNAs (11 transcripts and one microRNA) have been identified as components of the germ plasm. These include askopos (kop) (Blaser et al., 2005), carbonic anhydrase 15b (ca15b) (Hartwig et al., 2014), celf-1/bruno-like (celf1) (Hashimoto et al., 2004), deleted in azoospermia-like (dazl) (Hashimoto et al., 2004), dead end (dnd) (Weidinger et al., 2003), granulito (gran) (Strasser et al., 2008), hook microtubule-tethering protein 2 (hook2) (Roovers et al., 2018), microRNA 202-5p (miR-202-5p) (Zhang et al., 2017), nanos3 (nos3) (Köprunner et al., 2001), regulator of G-protein signaling 14a (rgs14a) (Hartwig et al., 2014), tudor domain protein 7 (tdrd7a) (Mishima et al., 2006), and vasa/DEADbox polypeptide 4 (vasa/ddx4) (Yoon et al., 1997) (Table 1).
Table 1.
RNA components of germ plasm in zebrafish
| RNA | Product | Functions | References |
|---|---|---|---|
| ca15b | Carbonic anhydrase 15b | Elevates pH to maintain polarity of migrating PGCs | (Hartwig et al., 2014; Wang et al., 2013) |
| dnd1 | DND microRNA-mediated repression inhibitor 1 | Binds target mRNA to prevent miRNA-mediated repression; controls actomyosin contractility, cell cortex rigidity, & cell-cell adhesion for PGC migration; inhibits miR-430 and somatic differentiation | (Goudarzi et al., 2012; Gross-Thebing et al., 2017; Weidinger et al., 2003) |
| gra | Granulito | Unknown; no knockdown phenotype; predicted negative regulator of translation initiation | (Strasser et al., 2008) |
| hook2 | Hook microtubule-tethering protein 2 | Binds to & promotes dynein-dynactin assembly during mitosis; regulates microtubule nucleation at the centrosome | (Dwivedi et al., 2019; Roovers et al., 2018) |
| miR-202-5p | microRNA 202-5p | PGC migration; KO phenotype in Medaka = significantly decreased fertility, dysregulation of key ovarian genes, & impaired egg production | (Gay et al., 2018; Jin et al., 2020; Zhang et al., 2017) |
| nanos3 | Nanos homolog 3 | RNA-binding zinc finger protein; migration & survival of PGCs; maintenance of germline stem cells | (Beer & Draper, 2013; Köprunner et al., 2001) |
| kop | Askopos | unknown; no knockdown phenotype; potential member of S100 protein-binding protein family | (Blaser et al., 2005) |
| rgs14a | Regulator of G-protein signaling 14a | Controls onset of PGC migration, polar protrusion formation, & E-cadherin expression | (Hartwig et al., 2014) |
| tdrd7a | Tudor domain containing 7a | Impacts germ granule size; germ cell differentiation; predicted RNA binding & translation regulator; predicted necessary for spermatogenesis | (D’Orazio et al., 2021; Mishima et al., 2006) |
| vasa/ddx4 | Dead box polypeptide 4 | Likely ATP-dependent DEAD box helicase; germ cell differentiation & maintenance | (Hartung et al., 2014; Olsen et al., 1997; Yoon et al., 1997) |
| celf1/brul | Cugbp, Elav-like family member 1; bruno-like | RNA-binding protein; post-transcriptionally regulates genes necessary for embryonic patterning; necessary for organogenesis of endoderm-derived tissues | (Hashimoto et al., 2004; Suzuki et al., 2000) |
| dazl | Deleted in azoospermia-like | RNA-binding protein (GUUC); controls polyA tail length; predicted association with polysomes & translation promotion; germline stem cell specification | (Bertho et al., 2021; Hashimoto et al., 2004; Maegawa et al., 2002; Takeda et al., 2009) |
The composition of germ plasm appears to vary across animal species, as indicated by studies in other model organisms, such as Caenorhabditis elegans, Drosophila melanogaster, and Xenopus laevis, with a majority of germ plasm RNAs present in some lineages but not others (Houston & King, 2000; Ikenishi, 1998). Only one RNA, nanos, is known to be a conserved germ plasm RNA component across these broadly divergent species (Köprunner et al., 2001; Subramaniam & Seydoux, 1999). Within vertebrate lineages, zebrafish germ plasm shares four RNAs, dazl, nanos, hook2, and dnd, with germ plasm in Xenopus laevis (Horvay et al., 2006; Houston et al., 1998; MacArthur et al., 1999; Owens et al., 2017) whereas the remaining nine zebrafish germ plasm RNAs are apparently absent in Xenopus laevis germ plasm (Butler et al., 2018; Karimi et al., 2018; Owens et al., 2017). Conversely, some RNA components found in the germ plasm of Xenopus laevis, such as pgat/xpat, germes/uncharacterized LOC779566, and ddx25, appear to be absent in its zebrafish counterpart (Berekelya et al., 2003; Hudson & Woodland, 1998; MacArthur et al., 2000).
Within fishes and consistent with their greater phylogenetic relatedness, RNA components of germ plasm appear to be more robustly conserved. At least four RNAs identified as zebrafish germ plasm components (dnd, nanos, vasa/ddx4, and dazl) have also been reported to localize to germ plasm in one or more additional teleost fish species, such as Scophthalmus maximus (turbot), Gadus morhua (Atlantic cod), or Salmo salmar (Atlantic salmon) (Lin et al., 2012; Nagasawa et al., 2013; Presslauer et al., 2012). In these fish species, germ plasm appears to exhibit a conserved pattern of aggregation in the forming furrows for the first several cell cycles. However, there is also evidence of germ plasm heterogeneity amongst fish, as demonstrated by the lack of furrow localization of maternally-inherited vasa/ddx4 RNA in early medaka and seabream embryos (Herpin et al., 2007; Shinomiya et al., 2000; Zhou et al., 2020).
To date, no study has determined if within a more restricted phylogenetic space, germ plasm RNA components are broadly conserved as a core set, or whether some RNAs but not others are conserved within related yet distinct sets of varying composition. To begin to gain some insight into this question, we use the Danio genus as a “model genus” (Irion & Nüsslein-Volhard, 2019; McCluskey & Postlethwait, 2015a), additionally taking advantage of the limited set of RNAs evidently present in the germline of this species (Eno et al., 2019). We systematically test for the presence of known zebrafish RNAs in three other species that span the phylogenetic depth of the genus Danio, with the orange-finned danio, D. kyathit, being most closely related to zebrafish (most common recent ancestor (MRCA) ca. 6.5 MYA), the pearl danio, D. albolineatus, as an intermediate relative (MRCA ca. 9.5 MYA), and the giant danio, Devario aequipinnatus, as a representative of the sister genus Devario (MRCA ca. 13 MYA) (McCluskey & Postlethwait, 2015a; Rüber et al., 2007) (Sup. Fig. 1). We find that 11 of 12 germ plasm RNAs in zebrafish are also germ plasm components in all three of the tested non-zebrafish Danionin species, Danio kyathit, Danio albolineatus, and Devario aequipinnatus, indicating a high level of conservation of germ plasm components at this phylogenetic scale. The sole exception, ca15b, is not observed at the furrows of Devario aequipinnatus and therefore is unlikely to be a germ plasm component in this species. Our data also indicate that the patterns of germ plasm localization, and likely the underlying cellular mechanisms for germ plasm aggregation and segregation, are conserved across these species.
Results
Identification of presumptive germ plasm in three non-zebrafish Danionin species
We used transcripts of the zebrafish nanos3 (previously known as nanos1, the ortholog of germline-associated nanos in other species) (Köprunner et al., 2001) gene as a marker of germ plasm at selected developmental stages in embryos from the Danionin species, Danio kyathit, Danio albolineatus, and Devario aequipinnatus, which as a set spans the evolutionary depth of the Danio genus and its closest sister genus Devario (Fig. 1A, Fig. 2, Sup. Fig. 1) (McCluskey & Braasch, 2020; McCluskey & Postlethwait, 2015b). The developmental stages were chosen to be representative of major steps in the germ plasm life cycle established in zebrafish, including aggregation at furrow distal tips during cleavage, asymmetric segregation into a subset of cells during blastula stages, and cytoplasmic dispersal into host PGCs. Due to the lack of reference genomic sequence information for many non-zebrafish species, and as an initial test of the extent of sequence conservation of germline-associated genes within the Danionin subfamily, we used a hapten-labeled RNA probe designed to complement zebrafish nanos3 (Fig. 2A) for the in situ hybridization analysis. We predicted that a probe targeting zebrafish nanos3 could potentially allow detection of the homologous gene in other Danio species and closely related genera such as Devario through cross-hybridization due to the broadly conserved nature of the nanos3 gene across animal lineages, including relatively high conservation at the sequence level across cyprinid fishes (Sup. Fig. 2) (Vallée et al., 2006). Indeed, we found that the zebrafish nanos3 probe detected transcripts in embryonic regions consistent with germ plasm aggregation, suggesting that the zebrafish probe successfully bound homologous transcripts in all three non-zebrafish species. Specifically, we found that at the four-cell stage of each species tested, nanos3-containing RNA particles form large aggregates at the distal ends of both cleavage furrows (Fig. 2A). In all species and similar to zebrafish (Eno et al., 2018; Miranda-Rodríguez et al., 2017; Pelegri et al., 1999; Yoon et al., 1997) the compacted RNAs that form the aggregates appear to form distinctive cylindrical masses within cleavage furrows, which begin as extended structures but gradually compact toward the distal ends. This suggests that the spatiotemporal pattern of germ plasm localization during the initial aggregation period as originally determined in the zebrafish model system is conserved in multiple species across the Danio and Devario genera.
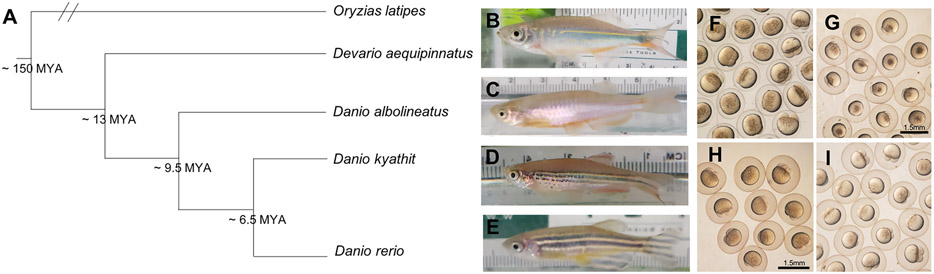
Figure 1.
Phylogeny and morphology of four Danionin species. A) Relationship between Danio and Devario species used in this study, with the Japanese rice fish, medaka (Oryzias latipes), as an outgroup. Simplified and redrawn from: (McCluskey & Postlethwait, 2015a). Branch lengths are arbitrary. See Supplementary Figure 1 for relationship to other lineages and expanded Danionin phylogeny. B-E) Representative adult male fish from each species in our laboratory colonies used in this study, including B) Devario aequipinnatus (giant danio), C) Danio albolineatus (pearl danio), D) Danio kyathit (orange-finned danio), and E) Danio rerio (zebrafish). F-I: Live cleavage stage embryos from each of the four species of interest, including F) Devario aequipinnatus, G) Danio albolineatus, H) Danio kyathit, and I) Danio rerio.
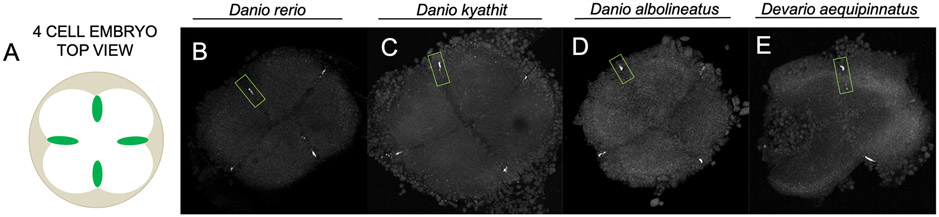
Figure 2.
Identification of furrow-localized germ plasm marker nanos3 in multiple Danionin species. Diagram (A) and micrographs (B-E) visualizing top view (blastodisc) of 4-cell Danionin fish embryos with nanos3 mRNA labeled via fluorescence in situ hybridization. The general appearance and localization pattern of the germ plasm marker nanos3 at the distal end of the first two cleavage furrows is consistent across all four species, including Danio rerio (B), Danio kyathit (C), Danio albolineatus (D), and Devario aequipinnatus (E). A nanos3 germ plasm aggregate in each embryo is highlighted by a green box outline. Micrographs are 2D maximum projections of confocal Z-stacks.
Conservation of dynamic germ plasm localization patterns throughout embryogenesis
We also assessed germ plasm localization using nanos3 as a marker at later stages of embryogenesis, including stages that in zebrafish comprise the asymmetric segregation (128-cell to sphere) and RNA dispersal (dome to mid-epiboly) periods of germ plasm dynamics (Eno et al., 2019; Knaut et al., 2000). In order to distinguish individual cells at these later stages of embryogenesis, visualization of DAPI-stained DNA and cell boundaries labeled with anti-ß-catenin antibodies was carried out simultaneously with fluorescent in situ hybridization (Figure 3). As with the results at the four-cell stage, signal detection at the sphere stage of embryogenesis from the species Danio kyathit, Danio albolineatus, and Devario aequipinnatus revealed similar patterns to what has been previously established for segregating germ plasm in zebrafish. In each species, the putative germ plasm aggregates appear as intact, phase-separated cytoplasmic masses that occupy a substantial portion of host cells, often near the nucleus. At the dome stage in each species, which occurs post-zygotic genome activation in zebrafish (Aanes et al., 2011; Harvey et al., 2013; Heyn et al., 2014; Kane & Kimmel, 1993; Lee et al., 2014; Mathavan et al., 2005) and can be easily distinguished by the bulging appearance of the yolk cell under a concave blastoderm, the germ plasm undergoes a dramatic morphological transition from intact mass to “leaky” appearance, with the RNA particles dispersing to fill the cytoplasm (Figure 3) (D’Orazio et al., 2021). Another hallmark of this transition in zebrafish is the subsequent appearance of perinuclear Vasa protein-containing germ granules, which are retained throughout PGC migration to the genital ridge during somitogenesis (Blaser et al., 2005; Knaut et al., 2000; Strasser et al., 2008). Further confirming our identification of germ plasm in these non-zebrafish Danionin species, we observed that nanos3 RNA-containing cells after the dome stage also exhibited the tell-tale perinuclear Vasa protein granules in Danio kyathit, Danio albolineatus, and Devario aequipinnatus (Figure 4). Together, the observed localization patterns of nanos3 and Vasa suggest that both maternally-derived germ plasm (labeled by nanos3) and germ granules at later stages of development (labeled by Vasa) are expressed and localized consistently across multiple species in the Danio and Devario genera.
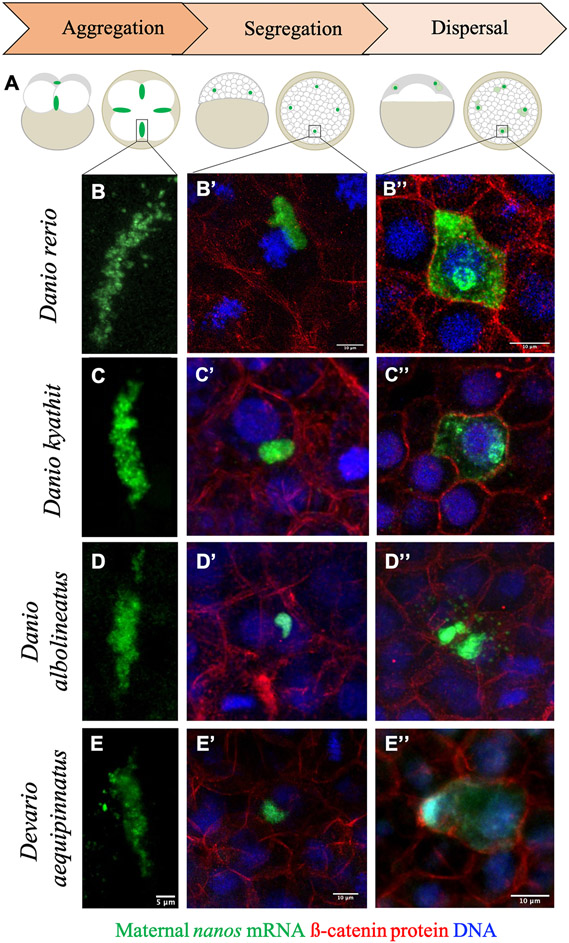
Figure 3.
Key stages of germ plasm localization dynamics across Danionin species. A) Diagram depicting an overview of germ plasm localization during early embryogenesis in zebrafish. Germ plasm ribonucleoparticles (green) in zebrafish form extended aggregates at the distal end of the first two embryonic cleavage furrows (1 hpf; aggregation; left), which become cellularized and segregate asymmetrically during cell divisions throughout the late cleavage through mid-blastula period of development (~1.5 hpf – 4 hpf; asymmetric segregation; middle), before dispersing to fill the cytoplasm of PGCs beginning at the dome stage through early gastrulation (~4.3 hpf – 6 hpf; dispersal; right). B-B”) Micrographs of germ plasm localization and morphology in zebrafish (Danio rerio) embryos during each of the highlighted stages, represented by the germ plasm marker nanos RNA (green) visualized via fluorescence in situ hybridization and confocal microscopy. B’ and B’’ also depict cell boundaries with beta catenin immunolabeling (red) and nuclei with DAPI stain (blue). The general appearance and localization patterns of germ plasm at these stages are consistent across species, including Danio kyathit (C-C”), Danio albolineatus (D-D”), and Devario aequipinnatus (E-E”).
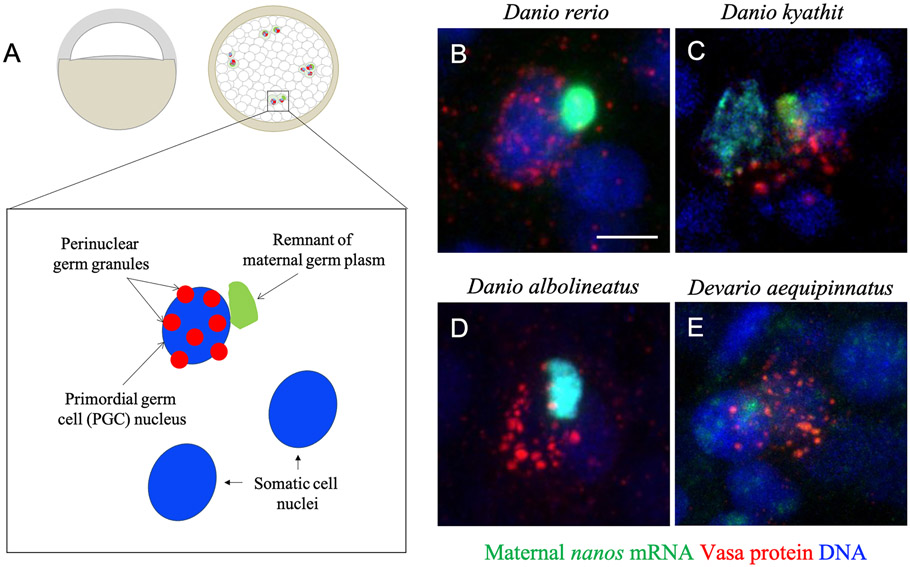
Figure 4.
Co-occurrence of maternally inherited germ plasm RNA and germline protein granules. Diagram (A) and micrographs (B-E) of germ plasm RNA and perinuclear Vasa granules during epiboly in four Danionin species. Remnants of maternally inherited germ plasm RNA (nanos RNA: green), and newly formed perinuclear germ granules (Vasa protein: red; nuclei: blue) co-exist in primordial germ cells during early epiboly/gastrulation in (B) zebrafish, (C) Danio kyathit, (D) Danio albolineatus, and (E) Devario aequipinnatus embryos. Scale bar: 10 μm.
Most zebrafish germ plasm RNAs are localized to the cleavage furrows in Danio kyathit, Danio albolineatus, and Devario aequipinnatus
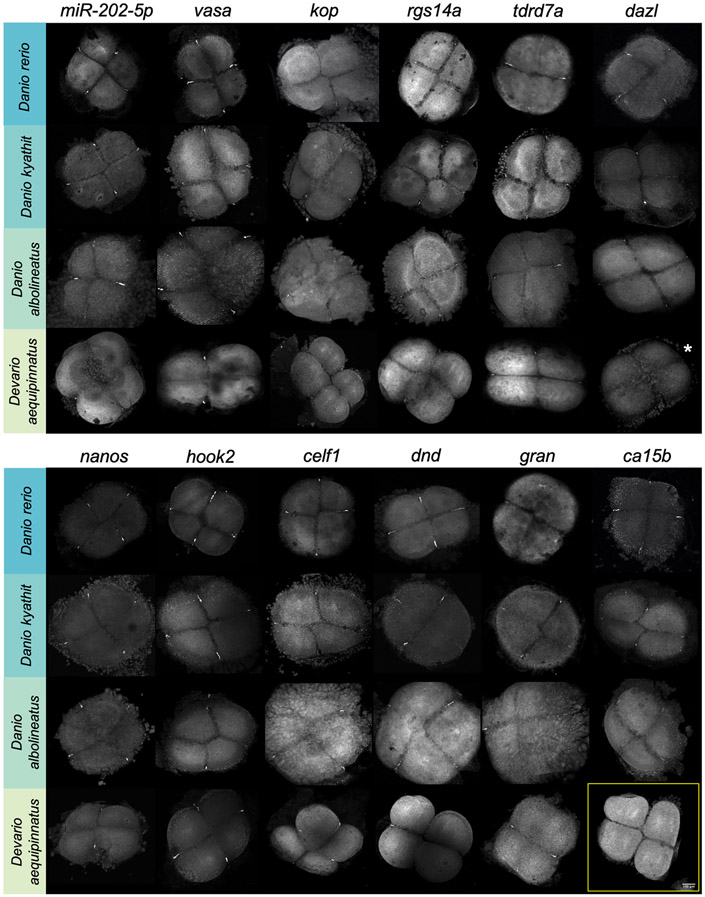
Since there was no published reference genome for other Danio and Devario species at the time of this work, we expanded a similar cross-hybridization approach in select species to also assess the presence and localization of all other germ plasm RNAs as originally identified in zebrafish. Using RNA probes against the zebrafish gene sequence, we performed fluorescent in situ hybridization at the four-cell stage of embryonic development (approximately one-hour post-fertilization at 28.5°C for all species) in Danio kyathit, Danio albolineatus, and Devario aequipinnatus. At this developmental stage in zebrafish, germ plasm RNAs and associated proteins have compacted into four distinct aggregates at the distal ends of cleavage furrows and are readily identifiable at low magnification (Fig. 2), making it a convenient timepoint to assess germ plasm localization across the selected species. In addition to nanos3, the set of tested germ plasm RNAs included askopos (kop) (Blaser et al., 2005), carbonic anhydrase 15b (ca15b) (Hartwig et al., 2014), celf-1/bruno-like (celf1) (Hashimoto et al., 2004), deleted in azoospermia-like (dazl) (Hashimoto et al., 2004), dead end (dnd) (Weidinger et al., 2003), granulito (gran) (Strasser et al., 2008), hook microtubule-tethering protein 2 (hook2) (Roovers et al., 2018), microRNA 202-5p (Zhang et al., 2017), regulator of G-protein signaling 14a (rgs14a) (Hartwig et al., 2014), tudor domain protein 7 (tdrd7a) (Mishima et al., 2006), and vasa/DEADbox polypeptide 4 (vasa/ddx4) (Yoon et al., 1997). We first verified that, as with nanos3 (Fig. 2-3), our other 11 RNA probes (see Methods and Materials) recapitulated germ plasm localization for all established germ plasm RNAs in zebrafish embryos (Figure 5). After verifying the probes in zebrafish, we proceeded to test them in the other three Danionin species. Probes for each of the 12 zebrafish germ plasm RNAs hybridized to the target RNA in germ plasm at the furrow region of four-cell stage Danio kyathit and Danio albolineatus embryos, while we observed furrow localization of all but one RNA (ca15b) in Devario aequipinnatus embryos (Figure 5, Figure 6). Another RNA, dazl, was difficult to observe in low magnification images of Devario aequipinnatus embryos, but germ plasm localization in the distal furrow region was confirmed with increased magnification. Since dazl appeared faint even at this increased magnification (perhaps due to interspecies probe mismatch), we also checked blastula-stage embryos and found dazl-positive germ plasm in putative PGCs (Sup. Fig. 3). This result underscored that, like nanos3, the majority of germ plasm genes originally identified in zebrafish are also maternally-expressed and, furthermore, germ plasm-localized, in closely related fish species.
Figure 5.
RNA localization at the distal ends of cleavage furrows in four cell stage Danio rerio, D. kyathit, D. albolineatus, and Devario aequipinnatus embryos as visualized by fluorescence in situ hybridization and confocal microscopy. All 12 known RNA components of zebrafish germ plasm are also germ plasm localized in Danio kyathit and Danio albolineatus, while all except ca15b are germ plasm localized in Devario aequipinnatus (yellow box; see also Figure 6). Furrow localization and/or labeling of dazl RNA in Devario aequipinnatus during the early cleavage stages was weak and difficult to visualize at 10x magnification (indicated by white asterisk) but could be observed at higher magnification (Sup. Fig. 3). All micrographs are 2D maximum projections of confocal stacks.
Figure 6.
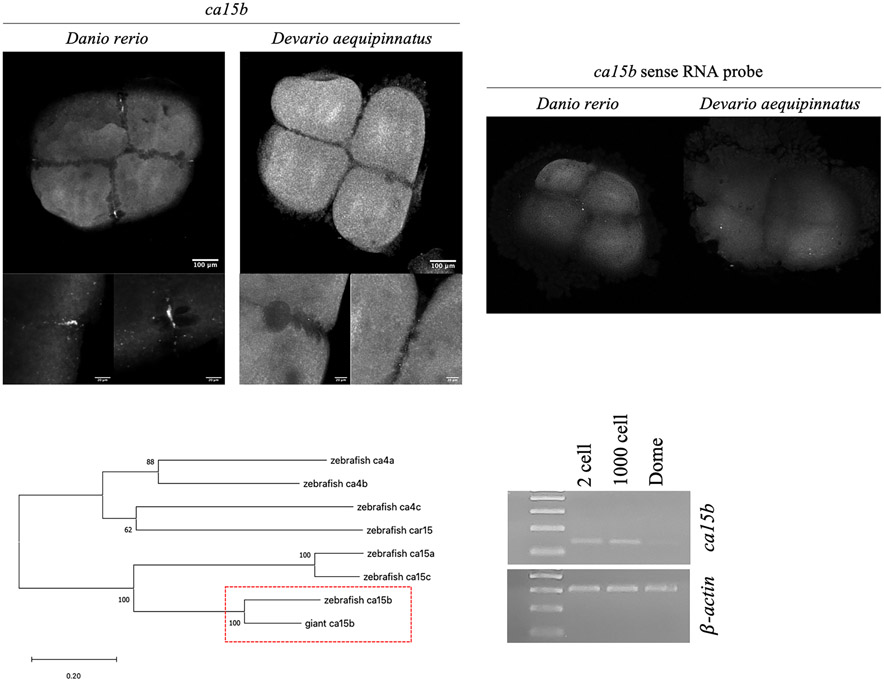
ca15b RNA is maternally expressed but not germ plasm-localized in Devario aequipinnatus embryos. A) ca15b RNA localization in zebrafish (Danio rerio) and Devario aequipinnatus four-cell embryos as visualized by fluorescence in situ hybridization and confocal microscopy. In zebrafish, ca15b is specifically localized to the germ plasm aggregates at the distal ends of cleavage furrows. In contrast, ca15b does not localize to germ plasm in Devario aequipinnatus cleavage furrows, and instead appears to be distributed throughout the blastodisc. This is in contrast to the generalized background fluorescence exhibited when using a sense RNA probe for ca15b (B). All micrographs are maximum projections of three-dimensional stacks, with lower insets showing increased magnification of furrow regions. C) A gene tree depicting the evolutionary relationship between the putative Devario aequipinnatus ca15b sequence and all known members of the carbonic anhydrase 4/15 family in the zebrafish genome. The tree was built using the Maximum Likelihood method and Tamura-Nei model (Tamura & Nei, 1993) with evolutionary analysis conducted in MEGA X (Kumar et al., 2018; Stecher et al., 2020) (also see Methods). D) Expression of ca15b during the 2-cell (45 mpf), 1000 cell (3 hpf), and dome (4.3 hpf) stages of Devario aequipinnatus as assayed by reverse-transcriptase PCR. Transcripts of ca15b are maternally expressed, with positive signal at the 2-cell and 1000 cell stages and decreased signal at the dome stage. Beta-actin was used as a control.
ca15b mRNA is not a germ plasm component in the giant danio Devario aequipinnatus
Animal genomes, including fish, often encode multiple carbonic anhydrases (Gilmour & Perry, 2009; Howe et al., 2021; Tashian, 1989) (Sup. Fig. 4); however, ca15b is the sole member of the carbonic anhydrase family known to localize to germ plasm in any species, as reported in zebrafish (Wang et al., 2013), Danio kyathit, and D. albolineatus (this report). The apparent lack of germ plasm-specific ca15b localization in Devario aequipinnatus embryos, in contrast to its presence in at least three Danio species, suggested that either the zebrafish and D. aequipinnatus ca15b mRNA sequences have diverged enough to prevent interspecies probe hybridization or that ca15b is not a component of D. aequipinnatus germ plasm. We identified the putative ca15b sequence in Devario aequipinnatus from data contributed to the NCBI Sequence Read Archive by the Vertebrate Genomes Project (Leinonen et al., 2011; Rhie et al., 2021) and, in addition to confirming that it shares a closer evolutionary history with zebrafish ca15b than other members of the carbonic anhydrase IV/XV family (Figure 6C, Sup. File 1), used the sequence information to repeat the fluorescent in situ labeling after amplifying exonic template DNA for RNA probe synthesis directly from Devario aequipinnatus, rather than zebrafish cDNA (Sup. Fig. 5). Even with a probe that is precisely homologous to D. aequipinnatus ca15b and as in our initial experiments, ca15b was not observed in germ plasm and instead appeared to distribute uniformly throughout the blastodisc. We extracted total RNA at various times during early embryonic development in Devario aequipinnatus embryos and performed RT-PCR to assess for ca15b expression. We found that Devario aequipinnatus ca15b transcripts are present at the 2-cell (45 mpf) and 1000-cell (3 hpf) but greatly reduced at dome stage (4.3 hpf), suggestive of maternal-specific expression for this gene and supporting our conclusion that ca15b transcripts are present in the early embryo but not specifically-localized during this time. Together with the previous expression data, this demonstrates that the RNA composition of germ plasm, as it is currently known, is largely but not completely shared across multiple species spanning the Danionin phylogenetic clade.
Discussion
Stages of germ plasm localization are conserved in multiple Danionin species
Our findings suggest that prior to PGC proliferation and migration to the presumptive gonads, patterns of germ plasm localization and morphology are robustly conserved across the Danionin lineage. Using a probe designed to target zebrafish nanos3, we first tracked germ plasm localization at characteristic stages of development in Danio rerio, D. kyathit, D. albolineatus, and Devario aequipinnatus embryos with the finding that germ plasm segregation behavior is consistent throughout these Danionin species. In each species, the germ plasm morphology progressed from four, rodlike aggregates to irregular, compact masses, albeit with discrete borders, before dispersing into submicron-units that flood the cytoplasm of the host cell. Moreover, the developmental timing of germ plasm dispersal is tightly conserved across species, with each species initiating dispersal approximately at dome stage. Also, as visualized using combined fluorescent in situ hybridization and immunolabeling, germ plasm-containing cells in zebrafish and each of the three tested non-zebrafish species acquire perinuclear Vasa protein granules soon after germ plasm dispersal.
All RNA germ plasm components previously identified in zebrafish are also present in the germ plasm of Danio kyathit and Danio albolineatus
The full set of 12 known germ plasm components previously characterized in zebrafish are present and conserved in two other Danio species. Probes designed for zebrafish were sufficient in both Danio kyathit and Danio albolineatus, suggesting a high degree of sequence conservation at the nucleotide level. Mismatch tolerance/specificity of ribonucleotide probes depends on many variables, such as hybridization conditions, probe length, and transcript accessibility, but some estimates suggest that probes generally require at least 85% complementarity between oligonucleotide probe and target sequence (He et al., 2005). The conserved nature of this set of germ plasm RNAs across species and genera indicates the importance of these factors in early development and germline differentiation, and could potentially suggest a conserved core set of RNA components required for PGC specification among Danionin fish. Due to the similarities in localization patterns between species, we also predict that the mechanisms underlying germ plasm aggregation, such as interactions with the cytoskeleton and cell division apparatus, are also shared between these species.
Although reference genomes are not yet available for most non-zebrafish Danionin species, including the three tested in this work, several are expected to be included in the Vertebrate Genomes Project (there is now an assembled version of the Danio kyathit genome and raw reads of Danio albolineatus and Devario aequipinnatus available in the NCBI Sequence Read Archive). We anticipate that reference genome availability will open up additional resources and investigative paths, such as determining the precise extent of sequence conservation between germ plasm genes and if any are present as duplicate copies in the genome.
Divergence of germ plasm RNA composition between Danio and Devario genera
We found that mRNA encoded by the carbonic anhydrase gene ca15b is located in the germ plasm of three Danio species but not in Devario aequipinnatus, a member of the sister genus. Carbonic anhydrase genes encode enzymes that reversibly catalyze the hydration of carbon dioxide into bicarbonate and a proton in most living organisms, including animals, plants, bacteria, and archaea, thus mediating CO2 transport, ion regulation, and acid-base homeostasis (Henry, 1996; Hewett-Emmett & Tashian, 1996). Carbonic anhydrases have nearly maximal catalytic efficiencies that approach the diffusion limit, with different isozymes exhibiting different kinetic properties (Boone et al., 2013; Silverman & Lindskog, 1988). Functional specialization in regard to subcellular localization, tissue-type localization (Hewett-Emmett & Tashian, 1996), and even during life mode transitions such as in lamprey metamorphosis (Ferreira-Martins et al., 2016) may have driven the appearance of a large number (greater than 3,200 in vertebrates according to the Ensembl genome database) of carbonic anhydrase genes.
Vertebrate carbonic anhydrase genes can be categorized according to subcellular localization, such as cytosolic, mitochondrial, membrane associated and secreted (Frost, 2014). Bony fish encode seven subgroups of carbonic anhydrase isoenzymes with predicted transmembrane or extracellular function: ca4, ca6, ca9, ca12, ca14, ca15, and ca16 (Gilmour & Perry, 2009), with gene members from each of these subgroups broadly present across vertebrate lineages. Interestingly, the ca15 subgroup, including the ca15b gene, though also widely distributed across vertebrates, is present in egg-laying mammals (monotremes) but absent in other mammals (marsupials and placentals) (Sup. Fig. 4) (Howe et al., 2021). The association of ca15 genes with egg-laying may be indicative of a role for Ca15b in oogenesis and/or embryonic development specifically in egg-laying animals. Indeed, in the zebrafish Ca15b protein localizes to the plasma membrane of oocytes (Wang et al., 2013), suggesting a likely function during oogenesis. Despite the broad distribution of genes from the ca15 subgroup across egg-laying vertebrates, not all egg-laying vertebrates have a ca15 gene (Sup. Fig. 4) and the presence of a ca15 gene, including ca15b, additionally does not precisely correspond to the use of germ plasm for germ cell induction found in vertebrate phylogeny (Hansen & Pelegri, 2021b; A. D. Johnson et al., 2011). Phylogenetic analysis of the ca15b gene thus suggests a maternal function, such as during oogenesis and/or early embryonic development in some egg-laying species, that is broadly distributed but not universal within vertebrates. In the Danio lineage, this gene’s maternal function may have been co-opted to incorporate its transcripts as a germ plasm component to facilitate primordial gem cell developmental processes, such as migration to the prospective gonad (see below). Further analysis of the distribution of ca15b and other germ plasm components across phylogenetic space will provide further insights into the function and evolutionary history of this gene.
In addition to a potential function in oogenesis, zebrafish Ca15b has been proposed to play a role in primordial germ cell migration by establishing a pH gradient within PGCs as a response to a chemokine-sensitive signaling cascade and is thought to promote actin polymerization at the leading edge of migrating cells (Tarbashevich et al., 2015). In zebrafish, Ca15b knockdown impaired the ability of PGCs to migrate to the prospective gonad. It is unclear if Ca15b protein is able to be restricted to the PGC lineage in Devario aequipinnatus and similarly function in PGC migration despite the absence of ca15b transcripts in germ plasm. Alternatively, Devario aequipinnatus maternal ca15b may not function in in germ cell migration, and other cellular components or pathways could perform a redundant role. We also note that prior to this study zebrafish was the only known species with germ plasm-localized ca15b; therefore, other than species within the Danio genus as shown in this work, other animal species may be able to achieve PGC migration and establish their germline without it. Generally speaking, migrating cells, particularly bleb-forming migratory cells, rely on polarization, often generated by concentration gradients of various molecules, including proteins or ions (Martin et al., 2010; Richardson & Lehmann, 2010). It is possible that PGC migration in Devario aequipinnatus employs a different cell polarization mechanism, and/or that the Ca15b-mediated pH gradient in embryos of zebrafish and presumably other Danios represents an evolutionarily novel (potentially more efficient) means to achieve PGC migration.
In conclusion, we identify shared dynamics of germ plasm segregation and a set of a largely, but not absolutely, conserved germ plasm RNAs within the relatively narrow phylogenetic confines of the related Danio and Devario genera. By leveraging multiple species within the Danionin clade, this study laid the groundwork for further investigation into the function of individual molecular components of germ plasm in specific species and the processes through which germ plasm composition is adaptable across phylogeny.
Methods
Fish Maintenance
Stocks of wild-type AB Danio rerio fish were raised and maintained in standard conditions at 28.5°C (Brand et al., 2002). Stocks of the other Danionin fish species (Danio kyathit, Danio albolineatus, and Devario aequipinnatus) were also raised and maintained in standard conditions at 28.5°C. All procedures involving live fish were conducted according to the University of Wisconsin-Madison and Institutional Animal Care and Use Committee (IACUC) guidelines.
Embryo Collection and Fixation
Embryos from all species were collected and developed in E3 embryonic medium and were staged according to age and morphological characteristics corresponding to those in zebrafish (Kimmel et al., 1995). Embryos were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) while still in their chorions for at least 4 hours at room temperature to overnight at 4°C. All experiments involved mixed clutches of embryos from between 2 and 5 females per experiment for both zebrafish and non-zebrafish species.
RNA extraction and cDNA synthesis
A pool of embryos was collected from three Devario aequipinnatus clutches. Total RNA was extracted from 50 dechorionated embryos at the two-cell stage (45 mpf), the 1,000-cell stage (3 hpf), and the dome stage (4.3 hpf) using the standard TRizol (Invitrogen) protocol. Isolated total RNA was quantified using a Nandrop 2000 (Thermo Scientific). Samples (~10 ng total RNA each) were reverse transcribed into first-strand cDNA using SuperScript III RNase H- reverse transcriptase and oligo dT primers (SuperScript III First-Strand Synthesis SuperMix, Invitrogen).
Reverse transcription-PCR
Reverse transcription PCR (RT-PCR) was used to assay maternal gene expression at multiple stages of early development in Devario aequipinnatus embryos. Samples of first-strand cDNA synthesized from total RNA were used as templates for RT-PCR. Reaction mixes containing cDNA template, 150nM each primer, and EconoTaq DNA Polymerase ran with the following thermocycler parameters: 55°C for 5min; 95°C for 2min; 40 cycles of 95°C for 15s, 55°C for 15s, and 72°C for 30s. Results were visualized by running the RT-PCR products on 1.7% agarose gels via gel electrophoresis.
Primers used for Devario aequipinnatus RT-PCR:
ca15b_F: CATGGCCCATCATTGCAGAA
ca15b_R: GAAATTAATACGACTCACTATAGGGCCAACGTATGTGAAAGAGGT
beta-actin_F: GCCCATCTATGAGGGTTACG
beta-actin_R: AGGAAGGAAGGCTGGAAGAG
Fluorescent in situ hybridization (FISH)
Antisense probes targeting germ plasm mRNAs were synthesized using linearized DNA purified from plasmids established for use in zebrafish or amplified from cDNA made from 4 cell stage Danio rerio embryos (except where noted). Template DNA for the probes targeting askopos (Blaser et al., 2005), dazl (Maegawa et al., 1999), dnd (Weidinger et al., 2003), nanos3 (Köprunner et al., 2001), and vasa (Yoon et al., 1997) was linearized from plasmids as previously described. DNA templates for dnd, ca15b, celf1, gran, hook2, rgs14a, and tdrd7a were produced from 4-cell stage zebrafish cDNA and PCR amplified using a primer that included the T7 RNA polymerase promoter sequence, then transcribed into labeled RNA probes using T7 and hapten-conjugated uridine triphosphates (UTPs) in the reaction mixture. As noted in the text and in the table below, two additional ca15b probes were synthesized in this way, although using Devario aequipinnatus template DNA instead of zebrafish cDNA. The template for miR-202-5p, based off of the zebrafish sequence, was ordered as a DNA oligonucleotide with a T7 RNA polymerase promoter sequence from IDT (GAAATTAATACGACTCACTATAGGTTCCATGCATATACCTCTTTG) and transcribed into antisense labeled RNA probes using T7 and hapten-conjugated UTPs. Primers used for in situ probe synthesis listed in Table 2.
Table 2.
Primers used for in situ probe synthesis with T7 RNAP promoter underlined
| Probe target | Forward primer | Reverse primer | Source of template |
|---|---|---|---|
| ca15b_exon8-9 | TGCAATGAAGGTGTGATCTGG | GAAATTAATACGACTCACTATAGGATTGACTGGCTGAACGGATC | D. rerio cDNA (4 cell) |
| ca15b_exon8-9 *sense probe | GAAATTAATACGACTCACTATAGG TGCAATGAAGGTGTGATCTGG | TTGACTGGCTGAACGGATC | D. rerio cDNA (4 cell) |
| ca15b_exon4 (Devario) | CATGGCCCATCATTGCAGAA | GAAATTAATACGACTCACTATAGGGCCAACGTATGTGAAAGAGGT | Devario DNA (adult fin clip) |
| ca15b_exon8-9 (Devario) | TGCAATGAAGGTGTGATCTGG | GAAATTAATACGACTCACTATAGGATTGACTGGCTGAACGGATC | Devario cDNA (2 cell) |
| celf1 | GACCAGCCCGACATTGATTC | GAAATTAATACGACTCACTATAGGTCTGTGACTGGTGCATGGAT | D. rerio cDNA (4 cell) |
| granulito | GACCAATAGAAAAACCCGACAAC | GAAATTAATACGACTCACTATAGGTGCCATCTTAACTAGGAAATCCC | D. rerio cDNA (4 cell) |
| hook2 | CGCAGACAAAACACACTCGA | GAAATTAATACGACTCACTATAGGTCTTTCTCCGACAGCTGGTT | D. rerio cDNA (4 cell) |
| rgs14a | ATCAGACAGCAAGACCAGC | GAAATTAATACGACTCACTATAGGGATCCAGACACCAACTCCAG | D. rerio cDNA (4 cell) |
| tdrd7a | AATCACCAGTGCCTGAGAAG | GAAATTAATACGACTCACTATAGGATTCGACCCTGAACCAACTC | D. rerio cDNA (4 cell) |
Fixed embryos were permeabilized overnight with 100% methanol at −20°C prior to treatment with prehybridization buffer for at least 4 hours at 69°C. Embryos were allowed to hybridize overnight at 69°C with RNA probes labeled with either fluorescein- (FLU-), digoxigenin- (DIG-), or dinitrophenol- (DNP-) conjugated UTPs. FLU, DIG, and DNP haptens were detected and fluorescent signal was developed using the TSA Plus Fluorescein/Cy3/Cy5 system (PerkinElmer) following a modified “Triple fluorescent in situ” protocol (Hansen & Pelegri, 2021a). Following completion of the FISH procedure, embryos were stained to detect DNA with 1:200 dilution of 100 ug/ml DAPI in PBS for 10 minutes followed by PBS washes. All embryos were deyolked after labeling and mounted flat (blastodiscs facing up) in 50% glycerol:PBS on slides for imaging.
Combined FISH and immunofluorescence for simultaneous RNA and protein visualization
For combined fluorescent in situ hybridization (FISH) and protein immunofluorescence, embryos were collected, staged at the selected developmental time points, and fixed as described above. FISH was performed first with a fluorescein-labeled antisense nanos3 probe. At the end of the FISH protocol and prior to embryo deyolking and DAPI labeling, the embryos were washed in PBS-Tween (Tween 0.1%) 3 × 10 minutes and then incubated at room temperature for at least 2 hours in casein blocking reagent. Embryos were then incubated with primary antibody solution (rabbit anti-Vasa, 1:200, GeneTex, and mouse anti-β-catenin, 1:1,000, in blocking reagent) overnight at 4°C, washed with PBS-Tween the following day and incubated with secondary antibodies overnight at 4°C. After PBS-Tween washes, the embryos were deyolked, DAPI-labeled to detect DNA (1:200 dilution of 100ug/ml DAPI in PBS for 10 minutes followed by PBS washes), and mounted flat as described above.
Microscopy and Image Analysis
Embryos were imaged using a Zeiss LSM710 or LSM780 confocal microscope and analyzed using ImageJ (Schneider et al., 2012). Unless otherwise indicated in figure legends, standard image processing in ImageJ included maximum 2D Z-projection of multiple slices from 3D stacks, signal smoothing, and adjustments to brightness and contrast (applied across entire image) as appropriate.
Phylogenetic and sequence analysis
Devario aequipinnatus raw sequence data was downloaded from NCBI Sequence Read Archive (acc. ERR3332370 and ERR3332374; Vertebrate Genomes Project). Quality was assessed with FastQC (Andrews, 2010) and reads were trimmed with fastp (Chen et al., 2018). Trimmed reads files were concatenated and gene assemblies were performed with HybPiper (M. G. Johnson et al., 2016; Tange, 2011) and SPAdes v3.14.1 (Prjibelski et al., 2020) using the zebrafish Ca15b protein sequence (NCBI RefSeq acc. XP_005169499.1) as bait. Coding regions were annotated and spliced using Exonerate (Slater & Birney, 2005).
The ca15b gene tree was built using the Maximum Likelihood method and Tamura-Nei model (Tamura & Nei, 1993). The tree with the highest log likelihood (−16318.87) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura-Nei model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 8 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. There were a total of 2960 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018; Stecher et al., 2020).
Supplementary Material
Supplementary Figure 1. Overview of phylogenetic relationships between zebrafish and other traditional model organisms and species within the Danionin fish clade. A) Evolutionary relationship with approximate times of divergence in millions of years to the most recent common ancestor for common laboratory model species. The divergence time between zebrafish and Xenopus, the two most commonly used vertebrate systems for germ plasm research, is estimated at approximately ninety million years. Figure redrawn based on tree in (Wheeler & Brändli, 2009); branch lengths not to scale. Animal icons from the Noun Project under CC license (from top to bottom: Human by Adrien Coquet, Mouse by Pham Thanh Loc, Chicken by Firza Alamsyah, Frog by Bismillah, Fish by Baboon designs, Fly by Studio GLD, and Worm by Deemak Daksina). B) Evolutionary relationship to the most recent common ancestor for selected members of the Danionin clade. The most recent common ancestor (MRCA) of the Rasbora/Danionin lineage was estimated to exist approximately 31 million years ago; the Devario and Danio genera have been diverging for approximately 13 million years according to (Rüber et al., 2007). The phylogenetic tree is adapted with permission from (McCluskey & Braasch, 2020).
Supplementary Figure 2. High degree of nucleotide sequence conservation across nanos3 mRNA sequences in zebrafish and related species. NCBI multiple sequence alignment of nanos3 mRNA sequences from cyprinid fishes (most closely related to zebrafish) with available reference sequences. Nucleotide bases are represented by different colors (T/U = red, G = gray, C = blue, A = green).
Supplementary Figure 3. dazl RNA (red) localization Devario aequipinnatus four-cell and late blastula stage embryos as visualized by fluorescence in situ hybridization and confocal microscopy. In four cell embryos, dazl particles appear to aggregate at the distal tips of cleavage furrows. In late blastula stage embryos, a small subset of cells (presumptive PGCs) contain a dazl-positive mass consistent with the typical appearance of germ plasm (DAPI-labeled nuclei in blue). Intensity of both images was increased higher than usual settings due to faint in situ signal.
Supplementary Figure 4. Radial phylogenetic trees demonstrate the distribution of selected carbonic anhydrase gene family subgroups across animal species. The gene gain/loss tree tool in the Ensembl genome database (Howe et al., 2021) was used to produce gene tree graphics for carbonic anhydrase subgroups from the Danio rerio genome with predicted extracellular or transmembrane function, including the ca15, ca4, ca16, ca6, ca9, ca12, ca14 and subgroups. No significant gene gain (expansion) or gene loss (contraction) events were detected for any of the gene subgroups selected (De Bie et al., 2006). The gene trees show that the various carbonic anhydrase gene family subgroups are widely present across vertebrates, although gene members of the ca15 subgroup, unlike others, are only present in monotreme mammals (represented by platypus) and absent in non-egg-laying (marsupial and placental) mammals. Species that do not have any members of the selected carbonic anhydrase subgroup are shaded light gray, while species with at least one gene from the selected subgroup are shaded black. The total number of genes from the selected carbonic anhydrase subgroup per each species is indicated at the node closest to the species image/name, ranging from 0 to 7.
Supplementary Figure 5. Nucleotide alignment of the ca15b coding sequence in zebrafish and Devario aequipinnatus as visualized in MView (Madeira et al., 2019). The target sequence for ca15b RNA probes used in this research are highlighted (ca15b_exon4 in orange, ca15b_exon8-9 in purple). Three different antisense ca15b RNA in situ probes were used to test ca15b localization in Devario aequipinnatus embryos in case mismatches between zebrafish and Devario aequipinnatus ca15b nucleotide sequence prevented cross-hybridization. The first was synthesized using ca15b_exon8-9 primers with zebrafish cDNA, another using ca15b_exon 4 primers with genomic Devario aequipinnatus DNA, and another using ca15b_exon8-9 primers with Devario aequipinnatus cDNA. Aligned nucleotide bases are highlighted as follows: G and A = green, T = blue, and C = yellow.
Acknowledgements
We thank past and current members of the Pelegri lab for their contributions to our research, particularly our fish husbandry staff for care of the aquatic facilities. Confocal microscopy was performed at the Newcomb Imaging Center, Department of Botany at the University of Wisconsin – Madison. This work was supported by NIH grant GM007133 to C.L.H., NIH grant GM065303 to F.P., USDA/Hatch grant to F.P., University of Wisconsin-Madison 2020 grant to F.P., and additional support from the College of Agricultural and Life Sciences, the School of Medicine and Public Health, and the Laboratory of Genetics at the University of Wisconsin – Madison.
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SGP, Lim AYM, Hajan HS, Collas P, Bourque G, Gong Z, Korzh V, Aleström P, & Mathavan S (2011). Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Research, 21(8), 1328–1338. 10.1101/gr.116012.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. http//:www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- Berekelya LA, Ponomarev MB, Luchinskaya NN, & Belyavsky AV (2003). Xenopus Germes encodes a novel germ plasm-associated transcript. Gene Expression Patterns: GEP, 3(4), 521–524. 10.1016/s1567-133x(03)00055-3 [DOI] [PubMed] [Google Scholar]
- Bertocchini F, & Chuva de Sousa Lopes SM (2016). Germline development in amniotes: A paradigm shift in primordial germ cell specification. Bioessays, 38(8), 791–800. 10.1002/bies.201600025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, & Raz E (2005). Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. Journal of Cell Science, 118(Pt 17), 4027–4038. 10.1242/jcs.02522 [DOI] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, & Dosch R (2009). Bucky ball organizes germ plasm assembly in zebrafish. Current Biology: CB, 19(5), 414–422. 10.1016/j.cub.2009.01.038 [DOI] [PubMed] [Google Scholar]
- Boone CD, Gill S, Habibzadegan A, & McKenna R (2013). Carbonic Anhydrase: An Efficient Enzyme with Possible Global Implications. International Journal of Chemical Engineering, 2013, e813931. 10.1155/2013/813931 [DOI] [Google Scholar]
- Braat AK, Zandbergen T, Water SVD, Goos HJT, & Zivkovic D (1999). Characterization of zebrafish primordial germ cells: Morphology and early distribution of vasa RNA. Developmental Dynamics, 216(2), 153–167. [DOI] [PubMed] [Google Scholar]
- Brand M, Granato M, & Nüsslein-Volhard C (2002). Keeping and raising zebrafish. Zebrafish (2002). [Google Scholar]
- Butler AM, Owens DA, Wang L, & King ML (2018). A novel role for sox7 in Xenopus early primordial germ cell development: Mining the PGC transcriptome. Development, 145(dev155978). 10.1242/dev.155978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, & Gu J (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England), 34(17), i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, & Hahn MW (2006). CAFE: A computational tool for the study of gene family evolution. Bioinformatics, 22(10), 1269–1271. 10.1093/bioinformatics/btl097 [DOI] [PubMed] [Google Scholar]
- D’Orazio FM, Balwierz PJ, González AJ, Guo Y, Hernández-Rodríguez B, Wheatley L, Jasiulewicz A, Hadzhiev Y, Vaquerizas JM, Cairns B, Lenhard B, & Müller F (2021). Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization. Developmental Cell. 10.1016/j.devcel.2021.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM (1975). Germ plasm and the differentiation of the germ cell line. International Review of Cytology, 43, 229–280. 10.1016/s0074-7696(08)60070-4 [DOI] [PubMed] [Google Scholar]
- Eno C, Gomez T, Slusarski DC, & Pelegri F (2018). Slow calcium waves mediate furrow microtubule reorganization and germ plasm compaction in the early zebrafish embryo. Development (Cambridge, England), 145(10). 10.1242/dev.156604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eno C, Hansen CL, & Pelegri F (2019). Aggregation, segregation, and dispersal of homotypic germ plasm RNPs in the early zebrafish embryo. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 248(4), 306–318. 10.1002/dvdy.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, & Akam M (2003). Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development (Cambridge, England), 130(24), 5869–5884. 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- Ferreira-Martins D, McCormick SD, Campos A, Lopes-Marques M, Osório H, Coimbra J, Castro LFC, & Wilson JM (2016). A cytosolic carbonic anhydrase molecular switch occurs in the gills of metamorphic sea lamprey. Scientific Reports, 6(1), 33954. 10.1038/srep33954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SC (2014). Physiological Functions of the Alpha Class of Carbonic Anhydrases. In Frost SC & McKenna R (Eds.), Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications (pp. 9–30). Springer Netherlands. 10.1007/978-94-007-7359-2_2 [DOI] [Google Scholar]
- Gilmour KM, & Perry SF (2009). Carbonic anhydrase and acid–base regulation in fish. Journal of Experimental Biology, 212(11), 1647–1661. 10.1242/jeb.029181 [DOI] [PubMed] [Google Scholar]
- Hansen CL, & Pelegri F (2021a). Methods for Visualization of RNA and Cytoskeletal Elements in the Early Zebrafish Embryo. Methods in Molecular Biology (Clifton, N.J.), 2218, 219–244. 10.1007/978-1-0716-0970-5_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CL, & Pelegri F (2021b). Primordial germ cell specification in vertebrate embryos: Phylogenetic distribution and conserved molecular features of preformation and induction. Frontiers in Cell and Developmental Biology. 10.3389/fcell.2021.730332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig J, Tarbashevich K, Seggewiß J, Stehling M, Bandemer J, Grimaldi C, Paksa A, Groß-Thebing T, Meyen D, & Raz E (2014). Temporal control over the initiation of cell motility by a regulator of G-protein signaling. Proceedings of the National Academy of Sciences, 111(31), 11389–11394. 10.1073/pnas.1400043111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D, & Smith JC (2013). Identification of the zebrafish maternal and paternal transcriptomes. Development (Cambridge, England), 140(13), 2703–2710. 10.1242/dev.095091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Maegawa S, Nagai T, Yamaha E, Suzuki H, Yasuda K, & Inoue K (2004). Localized maternal factors are required for zebrafish germ cell formation. Developmental Biology, 268(1), 152–161. 10.1016/j.ydbio.2003.12.013 [DOI] [PubMed] [Google Scholar]
- He Z, Wu L, Li X, Fields MW, & Zhou J (2005). Empirical Establishment of Oligonucleotide Probe Design Criteria. Applied and Environmental Microbiology, 71(7), 3753–3760. 10.1128/AEM.71.7.3753-3760.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RP (1996). Multiple Roles of Carbonic Anhydrase in Cellular Transport and Metabolism. Annual Review of Physiology, 58(1), 523–538. 10.1146/annurev.ph.58.030196.002515 [DOI] [PubMed] [Google Scholar]
- Herpin A, Rohr S, Riedel D, Kluever N, Raz E, & Schartl M (2007). Specification of primordial germ cells in medaka (Oryzias latipes). BMC Developmental Biology, 7(1), 3. 10.1186/1471-213X-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett-Emmett D, & Tashian RE (1996). Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Molecular Phylogenetics and Evolution, 5(1), 50–77. 10.1006/mpev.1996.0006 [DOI] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, & Neugebauer KM (2014). The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Reports, 6(2), 285–292. 10.1016/j.celrep.2013.12.030 [DOI] [PubMed] [Google Scholar]
- Horvay K, Claußen M, Katzer M, Landgrebe J, & Pieler T (2006). Xenopus Dead End mRNA Is a Localized Maternal Determinant that Serves a Conserved Function in Germ Cell Development. Developmental Biology, 291, 1–11. 10.1016/j.ydbio.2005.06.013 [DOI] [PubMed] [Google Scholar]
- Houston DW, & King ML (2000). A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development (Cambridge, England), 127(3), 447–456. [DOI] [PubMed] [Google Scholar]
- Houston DW, Zhang J, Maines JZ, Wasserman SA, & King ML (1998). A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development (Cambridge, England), 125(2), 171–180. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RHA, Hannon GJ, Draper BW, & Ketting RF (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell, 129(1), 69–82. 10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]
- Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Charkhchi M, Cummins C, Da Rin Fioretto L, Davidson C, Dodiya K, El Houdaigui B, Fatima R, … Flicek P (2021). Ensembl 2021. Nucleic Acids Research, 49(D1), D884–D891. 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, & Woodland HR (1998). Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mechanisms of Development, 73(2), 159–168. 10.1016/s0925-4773(98)00047-1 [DOI] [PubMed] [Google Scholar]
- Ikenishi K (1998). Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Development, Growth & Differentiation, 40(1), 1–10. 10.1046/j.1440-169X.1998.t01-4-00001.x [DOI] [PubMed] [Google Scholar]
- Irion U, & Nüsslein-Volhard C (2019). The identification of genes involved in the evolution of color patterns in fish. Current Opinion in Genetics & Development, 57, 31–38. 10.1016/j.gde.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Richardson E, Bachvarova RF, & Crother BI (2011). Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction (Cambridge, England), 141(3), 291–300. 10.1530/REP-10-0474 [DOI] [PubMed] [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, & Wickett NJ (2016). HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment1. Applications in Plant Sciences, 4(7). 10.3732/apps.1600016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, & Kimmel CB (1993). The zebrafish midblastula transition. Development (Cambridge, England), 119(2), 447–456. [DOI] [PubMed] [Google Scholar]
- Karimi K, Fortriede JD, Lotay VS, Burns KA, Wang DZ, Fisher ME, Pells TJ, James-Zorn C, Wang Y, Ponferrada VG, Chu S, Chaturvedi P, Zorn AM, & Vize PD (2018). Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Research, 46(D1), D861–D868. 10.1093/nar/gkx936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, & Schilling TF (1995). Stages of embryonic development of the zebrafish. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 203(3), 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Knaut H, Pelegri F, Bohmann K, Schwarz H, & Nüsslein-Volhard C (2000). Zebrafish vasa RNA but Not Its Protein Is a Component of the Germ Plasm and Segregates Asymmetrically before Germline Specification. The Journal of Cell Biology, 149(4), 875–888. 10.1083/jcb.149.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köprunner M, Thisse C, Thisse B, & Raz E (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes & Development, 15(21), 2877–2885. 10.1101/gad.212401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, & Tamura K (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6), 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, & Hogan BL (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes & Development, 13(4), 424–436. 10.1101/gad.13.4.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, & Giraldez AJ (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annual Review of Cell and Developmental Biology, 30, 581–613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, & Shumway M (2011). The Sequence Read Archive. Nucleic Acids Research, 39(Database issue), D19–D21. 10.1093/nar/gkq1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Xu S, Ma D, Xiao Z, Zhao C, Xiao Y, Chi L, Liu Q, & Li J (2012). Germ line specific expression of a vasa homologue gene in turbot (Scophthalmus maximus): Evidence for vasa localization at cleavage furrows in euteleostei. Molecular Reproduction and Development, 79(11), 803–813. 10.1002/mrd.22120 [DOI] [PubMed] [Google Scholar]
- MacArthur H, Bubunenko M, Houston DW, & King ML (1999). Xcat2 RNA is a translationally sequestered germ plasm component in Xenopus. Mechanisms of Development, 84(1–2), 75–88. 10.1016/s0925-4773(99)00075-1 [DOI] [PubMed] [Google Scholar]
- MacArthur H, Houston DW, Bubunenko M, Mosquera L, & King ML (2000). DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mechanisms of Development, 95(1–2), 291–295. 10.1016/s0925-4773(00)00357-9 [DOI] [PubMed] [Google Scholar]
- Madeira F, Park Y. mi, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, & Lopez R (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research, 47(W1), W636–W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Yasuda K, & Inoue K (1999). Maternal mRNA localization of zebrafish DAZ-like gene. Mechanisms of Development, 81(1–2), 223–226. 10.1016/s0925-4773(98)00242-1 [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir E, & Surani MA (2014). How to make a primordial germ cell. Development (Cambridge, England), 141(2), 245–252. 10.1242/dev.098269 [DOI] [PubMed] [Google Scholar]
- Martin C, Pedersen SF, Schwab A, & Stock C (2010). Intracellular pH gradients in migrating cells. American Journal of Physiology-Cell Physiology, 300(3), C490–C495. 10.1152/ajpcell.00280.2010 [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SGP, Mak A, Miller LD, Murthy KRK, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, & Lufkin T (2005). Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genetics, 1(2), 260–276. 10.1371/journal.pgen.0010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey BM, & Braasch I (2020). Chapter 2—Zebrafish Phylogeny and Taxonomy. In Cartner SC, Eisen JS, Farmer SC, Guillemin KJ, Kent ML, & Sanders GE (Eds.), The Zebrafish in Biomedical Research (pp. 15–24). Academic Press. 10.1016/B978-0-12-812431-4.00002-6 [DOI] [Google Scholar]
- McCluskey BM, & Postlethwait JH (2015a). Phylogeny of Zebrafish, a “Model Species,” within Danio, a “Model Genus.” Molecular Biology and Evolution, 32(3), 635–652. 10.1093/molbev/msu325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey BM, & Postlethwait JH (2015b). Phylogeny of zebrafish, a “model species,” within Danio, a “model genus.” Molecular Biology and Evolution, 32(3), 635–652. 10.1093/molbev/msu325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Rodríguez JR, Salas-Vidal E, Lomelí H, Zurita M, & Schnabel D (2017). RhoA/ROCK pathway activity is essential for the correct localization of the germ plasm mRNAs in zebrafish embryos. Developmental Biology, 421(1), 27–42. 10.1016/j.ydbio.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, & Inoue K (2006). Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Current Biology: CB, 16(21), 2135–2142. 10.1016/j.cub.2006.08.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravec CE, & Pelegri F (2020). The role of the cytoskeleton in germ plasm aggregation and compaction in the zebrafish embryo. Current Topics in Developmental Biology, 140, 145–179. 10.1016/bs.ctdb.2020.02.001 [DOI] [PubMed] [Google Scholar]
- Nagasawa K, Fernandes JMO, Yoshizaki G, Miwa M, & Babiak I (2013). Identification and migration of primordial germ cells in Atlantic salmon, Salmo salar: Characterization of Vasa, Dead End, and Lymphocyte antigen 75 genes. Molecular Reproduction and Development, 80(2), 118–131. 10.1002/mrd.22142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DA, Butler AM, Aguero TH, Newman KM, Van Booven D, & King ML (2017). High-throughput analysis reveals novel maternal germline RNAs crucial for primordial germ cell preservation and proper migration. Development (Cambridge, England), 144(2), 292–304. 10.1242/dev.139220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri F, Knaut H, Maischein HM, Schulte-Merker S, & Nüsslein-Volhard C (1999). A mutation in the zebrafish maternal-effect gene nebel affects furrow formation and vasa RNA localization. Current Biology: CB, 9(24), 1431–1440. 10.1016/s0960-9822(00)80112-8 [DOI] [PubMed] [Google Scholar]
- Presslauer C, Nagasawa K, Fernandes JMO, & Babiak I (2012). Expression of vasa and nanos3 during primordial germ cell formation and migration in Atlantic cod (Gadus morhua L.). Theriogenology, 78(6), 1262–1277. 10.1016/j.theriogenology.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, & Korobeynikov A (2020). Using SPAdes De Novo Assembler. Current Protocols in Bioinformatics, 70(1), e102. 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, Lee C, Ko BJ, Chaisson M, Gedman GL, Cantin LJ, Thibaud-Nissen F, Haggerty L, Bista I, Smith M, … Jarvis ED (2021). Towards complete and error-free genome assemblies of all vertebrate species. Nature, 592(7856), 737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, & Lehmann R (2010). Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nature Reviews. Molecular Cell Biology, 11(1), 37–49. 10.1038/nrm2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers EF, Kaaij LJT, Redl S, Bronkhorst AW, Wiebrands K, de Jesus Domingues AM, Huang H-Y, Han C-T, Riemer S, Dosch R, Salvenmoser W, Grün D, Butter F, van Oudenaarden A, & Ketting RF (2018). Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification. Developmental Cell, 46(3), 285–301.e9. 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüber L, Kottelat M, Tan HH, Ng PKL, & Britz R (2007). Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world’s smallest vertebrate. BMC Evolutionary Biology, 7, 38. 10.1186/1471-2148-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya A, Tanaka M, Kobayashi T, Nagahama Y, & Hamaguchi S (2000). The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Development, Growth & Differentiation, 42(4), 317–326. 10.1046/j.1440-169x.2000.00521.x [DOI] [PubMed] [Google Scholar]
- Silverman DN, & Lindskog S (1988). The catalytic mechanism of carbonic anhydrase: Implications of a rate-limiting protolysis of water. Accounts of Chemical Research, 21(1), 30–36. 10.1021/ar00145a005 [DOI] [Google Scholar]
- Slater GSC, & Birney E (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics, 6, 31. 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher G, Tamura K, & Kumar S (2020). Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Molecular Biology and Evolution, 37(4), 1237–1239. 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser MJ, Mackenzie NC, Dumstrei K, Nakkrasae L-I, Stebler J, & Raz E (2008). Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Developmental Biology, 8, 58. 10.1186/1471-213X-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, & Updike D (2015). Specifying and protecting germ cell fate. Nature Reviews. Molecular Cell Biology, 16(7), 406–416. 10.1038/nrm4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, & Seydoux G (1999). Nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development (Cambridge, England), 126(21), 4861–4871. [DOI] [PubMed] [Google Scholar]
- Swartz SZ, & Wessel GM (2015). Germ Line Versus Soma in the Transition from Egg to Embryo. Current Topics in Developmental Biology, 113, 149–190. 10.1016/bs.ctdb.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada H, Mochii M, Orii H, & Watanabe K (2012). Ectopic formation of primordial germ cells by transplantation of the germ plasm: Direct evidence for germ cell determinant in Xenopus. Developmental Biology, 371(1), 86–93. 10.1016/j.ydbio.2012.08.014 [DOI] [PubMed] [Google Scholar]
- Tamura K, & Nei M (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10(3), 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- Tange O (2011). Gnu parallel-the command-line power tool. The USENIX Magazine, 36(1), 42–47. [Google Scholar]
- Tarbashevich K, Reichman-Fried M, Grimaldi C, & Raz E (2015). Chemokine-Dependent pH Elevation at the Cell Front Sustains Polarity in Directionally Migrating Zebrafish Germ Cells. Current Biology, 25(8), 1096–1103. 10.1016/j.cub.2015.02.071 [DOI] [PubMed] [Google Scholar]
- Tashian RE (1989). The carbonic anhydrases: Widening perspectives on their evolution, expression and function. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 10(6), 186–192. 10.1002/bies.950100603 [DOI] [PubMed] [Google Scholar]
- Vallée M, Robert C, Méthot S, Palin M-F, & Sirard M-A (2006). Cross-species hybridizations on a multi-species cDNA microarray to identify evolutionarily conserved genes expressed in oocytes. BMC Genomics, 7, 113. 10.1186/1471-2164-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Teng Y, Xie Y, Wang B, Leng Y, Shu H, & Deng F (2013). Characterization of the carbonic anhydrases 15b expressed in PGCs during early zebrafish development. Theriogenology, 79(3), 443–452. 10.1016/j.theriogenology.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, & Raz E (2003). Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Current Biology: CB, 13(16), 1429–1434. 10.1016/s0960-9822(03)00537-2 [DOI] [PubMed] [Google Scholar]
- Wheeler GN, & Brändli AW (2009). Simple vertebrate models for chemical genetics and drug discovery screens: Lessons from zebrafish and Xenopus. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 238(6), 1287–1308. 10.1002/dvdy.21967 [DOI] [PubMed] [Google Scholar]
- Wylie C (1999). Germ cells. Cell, 96(2), 165–174. 10.1016/s0092-8674(00)80557-7 [DOI] [PubMed] [Google Scholar]
- Ying Y, Qi X, & Zhao GQ (2001). Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proceedings of the National Academy of Sciences of the United States of America, 98(14), 7858–7862. 10.1073/pnas.151242798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, & Hopkins N (1997). Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development (Cambridge, England), 124(16), 3157–3165. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu W, Jin Y, Jia P, Jia K, & Yi M (2017). MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish. Scientific Reports, 7(1), 7055. 10.1038/s41598-017-07675-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang X, Du S, Wang Y, Zhao H, Du T, Yu J, Wu L, Song Z, Liu Q, & Li J (2020). Germline Specific Expression of a vasa Homologue Gene in the Viviparous Fish Black Rockfish (Sebastes schlegelii) and Functional Analysis of the vasa 3′ Untranslated Region. Frontiers in Cell and Developmental Biology, 8. 10.3389/fcell.2020.575788 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Overview of phylogenetic relationships between zebrafish and other traditional model organisms and species within the Danionin fish clade. A) Evolutionary relationship with approximate times of divergence in millions of years to the most recent common ancestor for common laboratory model species. The divergence time between zebrafish and Xenopus, the two most commonly used vertebrate systems for germ plasm research, is estimated at approximately ninety million years. Figure redrawn based on tree in (Wheeler & Brändli, 2009); branch lengths not to scale. Animal icons from the Noun Project under CC license (from top to bottom: Human by Adrien Coquet, Mouse by Pham Thanh Loc, Chicken by Firza Alamsyah, Frog by Bismillah, Fish by Baboon designs, Fly by Studio GLD, and Worm by Deemak Daksina). B) Evolutionary relationship to the most recent common ancestor for selected members of the Danionin clade. The most recent common ancestor (MRCA) of the Rasbora/Danionin lineage was estimated to exist approximately 31 million years ago; the Devario and Danio genera have been diverging for approximately 13 million years according to (Rüber et al., 2007). The phylogenetic tree is adapted with permission from (McCluskey & Braasch, 2020).
Supplementary Figure 2. High degree of nucleotide sequence conservation across nanos3 mRNA sequences in zebrafish and related species. NCBI multiple sequence alignment of nanos3 mRNA sequences from cyprinid fishes (most closely related to zebrafish) with available reference sequences. Nucleotide bases are represented by different colors (T/U = red, G = gray, C = blue, A = green).
Supplementary Figure 3. dazl RNA (red) localization Devario aequipinnatus four-cell and late blastula stage embryos as visualized by fluorescence in situ hybridization and confocal microscopy. In four cell embryos, dazl particles appear to aggregate at the distal tips of cleavage furrows. In late blastula stage embryos, a small subset of cells (presumptive PGCs) contain a dazl-positive mass consistent with the typical appearance of germ plasm (DAPI-labeled nuclei in blue). Intensity of both images was increased higher than usual settings due to faint in situ signal.
Supplementary Figure 4. Radial phylogenetic trees demonstrate the distribution of selected carbonic anhydrase gene family subgroups across animal species. The gene gain/loss tree tool in the Ensembl genome database (Howe et al., 2021) was used to produce gene tree graphics for carbonic anhydrase subgroups from the Danio rerio genome with predicted extracellular or transmembrane function, including the ca15, ca4, ca16, ca6, ca9, ca12, ca14 and subgroups. No significant gene gain (expansion) or gene loss (contraction) events were detected for any of the gene subgroups selected (De Bie et al., 2006). The gene trees show that the various carbonic anhydrase gene family subgroups are widely present across vertebrates, although gene members of the ca15 subgroup, unlike others, are only present in monotreme mammals (represented by platypus) and absent in non-egg-laying (marsupial and placental) mammals. Species that do not have any members of the selected carbonic anhydrase subgroup are shaded light gray, while species with at least one gene from the selected subgroup are shaded black. The total number of genes from the selected carbonic anhydrase subgroup per each species is indicated at the node closest to the species image/name, ranging from 0 to 7.
Supplementary Figure 5. Nucleotide alignment of the ca15b coding sequence in zebrafish and Devario aequipinnatus as visualized in MView (Madeira et al., 2019). The target sequence for ca15b RNA probes used in this research are highlighted (ca15b_exon4 in orange, ca15b_exon8-9 in purple). Three different antisense ca15b RNA in situ probes were used to test ca15b localization in Devario aequipinnatus embryos in case mismatches between zebrafish and Devario aequipinnatus ca15b nucleotide sequence prevented cross-hybridization. The first was synthesized using ca15b_exon8-9 primers with zebrafish cDNA, another using ca15b_exon 4 primers with genomic Devario aequipinnatus DNA, and another using ca15b_exon8-9 primers with Devario aequipinnatus cDNA. Aligned nucleotide bases are highlighted as follows: G and A = green, T = blue, and C = yellow.