Abstract
Specialized cells of the somatic gonad primordium of nematodes play important roles in the final form and function of the mature gonad. Caenorhabditis elegans hermaphrodites are somatic females that have a two-armed, U-shaped gonad that connects to the vulva at the midbody. The outgrowth of each gonad arm from the somatic gonad primordium is led by two female distal tip cells (fDTCs), while the anchor cell (AC) remains stationary and central to coordinate uterine and vulval development. The bHLH protein HLH-2 and its dimerization partners LIN-32 and HLH-12 had previously been shown to be required for fDTC specification. Here, we show that ectopic expression of both HLH-12 and LIN-32 in cells with AC potential transiently transforms them into fDTC-like cells. Furthermore, hlh-12 was known to be required for the fDTCs to sustain gonad arm outgrowth. Here, we show that ectopic expression of HLH-12 in the normally stationary AC causes displacement from its normal position and that displacement likely results from activation of the leader program of fDTCs because it requires genes necessary for gonad arm outgrowth. Thus, HLH-12 is both necessary and sufficient to promote gonadal regulatory cell migration. As differences in female gonadal morphology of different nematode species reflect differences in the fate or migratory properties of the fDTCs or of the AC, we hypothesized that evolutionary changes in the expression of hlh-12 may underlie the evolution of such morphological diversity. However, we were unable to identify an hlh-12 ortholog outside of Caenorhabditis. Instead, by performing a comprehensive phylogenetic analysis of all Class II bHLH proteins in multiple nematode species, we found that hlh-12 evolved within the Caenorhabditis clade, possibly by duplicative transposition of hlh-10. Our analysis suggests that control of gene regulatory hierarchies for gonadogenesis can be remarkably plastic during evolution without adverse phenotypic consequence.
Keywords: cell migration, cell fate specification, phylogenetics, evolution, bHLH transcription factors
Introduction
Female gonadal morphology varies greatly among nematodes, and understanding how these interspecific variations are generated is a fundamental question at the interface of developmental and evolutionary biology. Caenorhabditis elegans hermaphrodites are somatic females that have a two-armed, U-shaped gonad that connects to the vulva at the midbody. The specification and positioning of two specialized cells in the somatic gonad primordium, here called female distal tip cells (fDTCs; in previous publications, hDTCs), and their subsequent function in leading the outgrowth of the gonad arm, is foundational for determining the final gonad shape (Kimble and Hirsh 1979; Kimble and White 1981). In other nematode species, variations in female gonad morphology, such as the length, trajectory, and number of the gonad arms, can be attributed to differences in the fate or migratory properties of the fDTCs (e.g., Sternberg and Horvitz 1981; Félix and Sternberg 1996; Rudel et al. 2005).
The gonad primordium in both sexes, and as far as we know in all species, forms during embryogenesis and contains four cells: the somatic progenitors Z1 and Z4, and the germline progenitors Z2 and Z3. In C. elegans, the first phase of hermaphrodite gonadogenesis begins during the first larval (L1) stage with the division of Z1 and Z4 and ends in the L2 stage, when their 12 descendants rearrange to form the somatic gonad primordium (Kimble and Hirsh 1979; Figure 1A). At that point, there are three differentiated “regulatory cells”: two fDTCs, which not only lead gonad arm extension but also serve as the niche for the germline stem cells in each arm, and the anchor cell (AC), which organizes patterning of the vulva and uterus in the L3 stage and the connection between them in the L4 stage. The other nine cells are precursors to the structural cells of the gonad (ovary, spermatheca, and uterus). When the somatic gonad primordium forms in the L2 stage, the regulatory cells occupy strategic positions for their various roles: the fDTCs are located at the distal ends for the leading outgrowth of the two gonad arms, and the AC remains located centrally to induce and connect to the vulva. In males, the somatic gonad primordium forms with the two male DTCs positioned posteriorly, where they remain stationary and serve as the germline stem cell niche, and a regulatory cell called the linker cell (LC) that takes up an anterior position and leads outgrowth of a single, J-shaped gonad arm that connects to the cloaca in the tail.
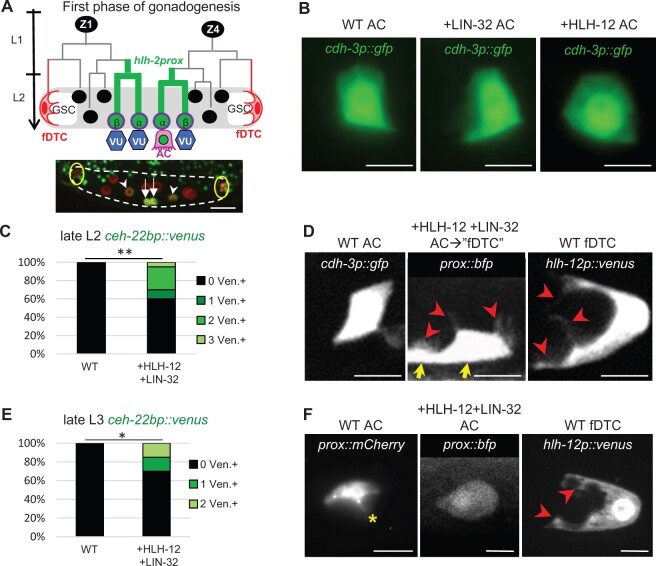
Figure 1.
Transient reprogramming of cells with AC potential into fDTCs by concomitant expression of HLH-12 and LIN-32. The designation “WT” represents animals lacking the +HLH-12 and/or +LIN-32 transgenes, but other markers are present as noted. (A) First phase of gonadogenesis. Lineage diagram (top) shows the first phase of hermaphrodite somatic gonad development. This phase encompasses the early lineages of the somatic gonad progenitors, Z1 and Z4, and culminates with the formation of the somatic gonad primordium, cartooned here. Z1 and Z4 each give rise to an α and a β cell; all four cells initially have the potential to be an AC (Seydoux et al. 1990). The β cells invariably adopt the VU fate, while the α cells interact with each other via LIN-12/Notch to specify one AC and one VU (the “AC/VU decision,” reviewed in (Greenwald, 2012) ) . HLH-2 appears to act as a homodimer for conferring AC potential and during the AC/VU decision (Karp and Greenwald 2003, 2004; Sallee and Greenwald 2015; Attner et al. 2019). hlh-2prox, a regulatory element derived from hlh-2, begins to drive expression (green) in the parents of these cells and continues to drive expression in the α and β cells (Sallee and Greenwald 2015). After the AC/VU decision has occurred, hlh-2prox expression is upregulated in the AC (represented by the green nucleus) and downregulated in the VUs. The fDTCs are highlighted in red. GSC, germline stem cells. Photomicrograph (bottom): All somatic gonad cells are labeled in red by arTi145[ckb-3p::mcherry::h2b], and the transcriptional reporter arTi22[hlh-2prox::gfp::h2b] labels the four proximal cells green, at a time when transcription has begun to diminish in the β cells but remains strong in the α cells undergoing the AC/VU decision. White dashed line outlines the gonad, yellow circles mark the fDTCs, arrows indicate the α cells, and arrowheads indicate the β cells. The scale bar is 10 µm. (B) Ectopic expression of individual genes of the fDTC code. In wild-type hermaphrodites, there is a single AC, which has characteristic morphology and is marked by arIs51[cdh-3p::gfp]. The AC appears similar to wild type in late L2 animals expressing LIN-32 (“+LIN-32”) (22/22) or HLH-12 (“+HLH-12”) (26/26). Scale bar is 5 µm. (C) Ectopic expression of both genes of the fDTC code (+HLH-12+LIN-32), scored in the late L2 stage. ceh-22bp::venus is a marker for fDTCs and in a WT background is never expressed in cells with AC potential (left bar). By contrast, when HLH-12 and LIN-32 are concomitantly expressed in cells with AC potential, ectopic expression of ceh-22bp::venus suggests that they are transformed into cells with fDTC character. When two proximal gonadal cells express the marker, we infer that the two α cells were transformed prior to the AC/VU decision; when one cell expresses the marker, we infer that the presumptive AC was transformed. In one individual, three Venus-expressing cells were observed, suggesting that a β cell was also transformed. n = 20 for each genotype, **P < 0.01, Fisher’s exact test. (D) Photomicrographs showing the transformation of cells with AC potential into fDTC-like cells in the late L2 stage. Cellular processes that project into the germ line are characteristic of the fDTC (right, visualized with hlh-12p::venus) and not the AC (left, visualized with cdh-3p::gfp). When both LIN-32 and HLH-12 are expressed, fDTC-like projections are observed proximally, where one AC normally is seen; in this photomicrograph, two cells, presumably derived from the two α cells, have fDTC-like projections, as visualized by BFP expression from the +HLH-12 bicistronic transgene (the two cell bodies are indicated with yellow arrows). 9/20 +HLH-12+LIN-32 animals displayed DTC-like projections (indicated by red arrowheads) into the germ line from at least one proximal somatic gonadal cell. The scale bar is 5 µm. As described in the text, fDTC-like marker expression and morphology were highly correlated, supporting the inference that there was a cell fate transformation. (E) Ectopic expression of both genes of the fDTC code (+HLH-12+LIN-32), scored in the late L3 stage. At this time, some +HLH-12+LIN-32 ACs still display ceh-22bp::venus fDTC marker expression. Twenty individuals were scored for each genotype, *P < 0.05, Fisher’s exact test. (F) +HLH-12+LIN-32 ACs lack AC-associated invadopodia as well as fDTC-associated projections in the late L3 stage. The asterisk indicates an apically directed invadopodium as is normally observed in an AC (left), red arrowheads indicate projections toward the germline that are normally observed in an fDTC (right). 14/14 +HLH-12+LIN-32 ACs lack both of these features. The scale bar is 5 µm.
The bHLH transcription factor HLH-2 is the sole C. elegans ortholog of mammalian E proteins and Drosophila Daughterless (Krause et al. 1997), and hlh-2 activity is required for the development of all regulatory cell types in both sexes (Karp and Greenwald 2003, 2004; Tamai and Nishiwaki 2007; Chesney et al. 2009). HLH-2 is a Class I bHLH protein, and as such can form homodimers or heterodimers with Class II bHLH proteins (Murre et al. 1994; Massari and Murre 2000). For specification and function of the AC, HLH-2 appears to act as a homodimer (Sallee and Greenwald 2015). Each of the other regulatory cell types expresses HLH-2 together with a distinct complement of Class II bHLH proteins, which we proposed constitute a “bHLH code” for the specification and functions of the distinct regulatory cells (Sallee et al. 2017). We supported this hypothesis by finding predicted phenotypic changes when we genetically altered the bHLH complement of the AC by expressing the Class II proteins associated with the mDTCs, LIN-32, and HLH-8. In this paper, we will be primarily concerned with testing the code for the fDTC, for which the Class II genes are lin-32 and hlh-12.
hlh-2 and hlh-12 (also called mig-24) are required for the process of gonad arm outgrowth in both C. elegans sexes, led via fDTCs in hermaphrodites and the LC in males. Null alleles of hlh-12 do not affect leader cell specification or most other aspects of gonadogenesis, but cause defects in leader cell migration (Tamai and Nishiwaki 2007). Two direct transcriptional targets of the HLH-2:HLH-12 heterodimer are necessary for the execution of migration: gon-1, which encodes an ADAMTS protease (Blelloch and Kimble 1999; Tamai and Nishiwaki 2007), and ina-1, which encodes an α-integrin (Meighan et al. 2015). Many other genes have also been shown to be necessary for stereotyped leader cell migration, including components and modifiers of basement membranes, pathfinding information provided by the Netrin signaling system and stage-specific cues (reviewed in Sherwood and Plastino 2018). Most of these genes have only been studied for roles in fDTC leader function, but there may be a single “leader program” coregulated in both sexes.
Here, we show that ectopic expression of both LIN-32 and HLH-12 in cells with AC potential transiently transforms them into fDTC-like cells, providing further support for the bHLH code hypothesis. Furthermore, we show that ectopic expression of only HLH-12 in the normally stationary AC causes its displacement along the anterior–posterior axis from its stereotyped position. This displacement likely results from activation of the leader program of fDTCs because it requires genes necessary for fDTC migration: ina-1/α-integrin (Meighan et al. 2015), cdc-42/CDC42, a Rho family GTPase (Cram et al. 2006), and pfd-1/Prefoldin, a microtubule chaperone (Lundin et al. 2008). Thus, HLH-12 is both necessary and sufficient to promote gonadal regulatory cell migration.
Given the essential roles of hlh-12 in fDTC specification and leader function in C. elegans, we hoped to extend our work to other genera where female gonads have different morphologies that might be attributed to differences in fDTC leader function, such as longer or shorter gonad arms. Furthermore, in some species, the AC itself displays migratory behavior, generally in association with a more posterior vulva (e.g., Félix and Sternberg 1996; Félix 2006; Sommer 2006; Kiontke et al. 2007; Haag et al. 2018). We hypothesized that evolutionary changes in the expression of hlh-12 in the fDTCs and AC of these species could account for the evolution of these morphological differences. However, surprisingly, we were unable to identify an hlh-12 ortholog outside of Caenorhabditis. This absence led us to perform a comprehensive phylogenetic analysis of all Class II bHLH proteins, including the other bHLH code proteins (HLH-3, HLH-8, and LIN-32), in selected nematode species. We confirmed that, while all other bHLH code proteins are generally conserved, HLH-12 is restricted to the Caenorhabditis clade. Our phylogenetic analysis further suggests that hlh-12 evolved within Caenorhabditis by duplicative transposition of hlh-10 after the divergence of Caenorhabditis parvicauda. Thus, HLH-12 was recently co-opted to control the anciently conserved fDTC migratory behavior and took over this function from a different pre-existing factor. Such co-option suggests that control of gene regulatory hierarchies can be remarkably plastic during evolution without adverse phenotypic consequence.
Materials and methods
Caenorhabditis elegans genetics
Strain names and full genotypes are listed in Table 1. Strains were analyzed at 25°C. The allele fos-1(ar105) contains an early termination codon and is a likely null allele (Seydoux et al. 1990; Sherwood et al. 2005). The fos-1 allele bmd138[gfp::fos-1] (Medwig-Kinney et al. 2020) is an endogenously tagged allele generated through CRISPR/Cas9. The allele ina-1(ar639[ina-1::gfp]) was generated using CRISPR/Cas9 as described below.
Table 1.
Strains used in this study
| Strains | Source | Figures |
|---|---|---|
| GS9309: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; qIs90[ceh-22bp::venus]; arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry]; arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 1C and E |
| GS8958: arIs51[cdh-3p::gfp]; arIs222[lag2p::2xnls-tagRFP] him-5(e1490) | This paper | 1B and D and 2C |
| OS8909: nsIs497[hlh-12p::venus + nhr-67p::mCherry + unc-119(+)]; unc-119(ed3); him-5(e1490) | Shai Shaham, pers. comm | 1D and F |
| GS7894: qIs90[ceh-22bp::venus]; arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | Maria Sallee and I.G., unpublished | 1C and E |
| DQM497: fos-1(bmd138[fos-1>LoxP::GFP::FOS-1]) | Medwig-Kinney et al. (2020) | 2B |
| GS9491: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; fos-1(bmd138[fos-1>LoxP::GFP::FOS-1]) | This paper | 2B |
| GS9150: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; arIs51[cdh-3p::gfp]; arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 1B and 2C |
| GS9165: ina-1(ar639[ina-1::gfp]); arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 2D |
| GS9497: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; ina-1(ar639[ina-1::gfp]); arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 2D |
| GS9306: qyIs176[zmp-1p::mCherry-moesinABD]; arIs131[lag-2p::2xnls-yfp] | This paper | 2A and S1A |
| GS9308: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; qyIs176[zmp-1p::mCherry-moesinABD]; arIs131[lag-2p::2xnls-yfp] | This paper | 2A and S1A |
| GS9500: fos-1(ar105)/tmC16[unc-60(tmIs1210)] | This paper | 2F |
| GS9501: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; fos-1(ar105)/tmC16[unc-60(tmIs1210)] | This paper | 2F |
| GS8195: arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 2C and E, S1B and C, and S2B–D |
| GS9084: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2] | This paper | 2F |
| GS9218: qIs90[ceh-22bp::venus]; arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry]; arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | |
| GS8518: arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry]; ccIs4443[arg-1p::gfp] | Sallee et al. (2017) | 1F |
| GS9148: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry]; arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 1D–F and S1C |
| GS9116: arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]; arIs222[lag-2p::2xnls-tagRFP] him-5(e1490) | This paper | 2C and E, S1B and C, and S2B–D |
| GS9109: arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry]; arIs51[cdh-3p::gfp] | This paper | 1B |
The following transgenes were used in this study.
arIs51[cdh-3p::gfp] (Karp and Greenwald 2003) is expressed in the AC after its specification in late L2 and through L3, and later in the AC and developing uterine seam cell (utse) in L4.
arIs222[lag-2p::2xlns-tagRFP] and arIs131[lag-2p::2xnls-YFP] (Li and Greenwald 2010; Sallee and Greenwald 2015) are expressed in all somatic gonad regulatory cells except for the mDTCs, as well as P6.p and its descendants.
qIs90[ceh-22bp::venus] (Lam et al. 2006) is expressed in the fDTCs and their sisters, and the mDTCs.
qyIs176[zmp-1p::mcherry-moesinABD] (Schindler and Sherwood 2011) contains the moesin actin-binding domain and localizes to the AC invasive membrane. It is driven by zmp-1p, which is expressed in the AC beginning in L3, as well as in the LC.
arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry] and arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2] are single-copy insertion transgenes which cause ectopic expression of LIN-32 (arTi148) or HLH-12 (arSi45), respectively, in the hermaphrodite proximal gonad. For arTi148, see Sallee et al. (2017). arSi45 was generated for this study using CRISPR/Cas9-mediated insertion in a defined LGI (Linkage Group I) site (Pani and Goldstein 2018).
Constructs and generation of single-copy insertion transgenes
arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2]
hlh-12 cDNA (synthesized by genewiz.com) and hlh-2prox regulatory sequences amplified from pHL4 (Sallee et al. 2017) were combined using fusion PCR (Hobert 2002), and inserted into SmaI/XbaI-digested pBS SK+ (Mayer 1995) using Gibson assembly (Gibson et al. 2009) to create pHL48. The unc-54 3ʹUTR was amplified from pHL4; 2xtagBFP2 (hereafter “BFP”) was amplified from pZW109 (Dickinson et al. 2015) with the addition 3ʹ of the first 15 base pairs of the 2xGGGGS linker sequence plus an artificial ochre stop codon to allow for better amplification of the repetitive sequence. The two pieces were inserted into SmaI/XbaI-digested pBS SK+ using Gibson assembly to create pHL50. The sl2 trans-splicing sequence was amplified from pHL3 (Sallee et al. 2017) and fused together with BFP::unc-54 3ʹUTR amplified from pHL50 to create sl2::BFP::unc-54 3ʹUTR. hlh-2prox::hlh-12cDNA was amplified from pHL48. hlh-2prox::hlh-12cDNA and sl2::BFP::unc-54 3ʹUTR were inserted into SpeI/AvrII-digested pZW111 (Pani and Goldstein 2018) using Gibson assembly to create pHL53.
The insert from pHL53 was inserted using CRISPR/Cas9 into a defined site on LGI using the method of Pani and Goldstein (2018). Hermaphrodites were injected with 55 ng/µL pAP82, 10 ng/µL pGH8, 2.5 ng/µL pCFJ90, and 10 ng/µL pHL53. Injected P0 were placed at 25°C and treated with 400 µL 5 mg/mL hygromycin 3 days after injection. Approximately 100 L1 from plates with homozygous “dark rollers” were heat-shocked at 34°C for 4 h to excise the roller cassette, and the line containing arSi45 was established from a single non-Roller L4 progeny of heat-shocked animals.
ina-1(ar639[ina-1::gfp])
The homology repair template pHL57 was cloned into pDD282 digested with AvrII and SpeI using the methods described in Dickinson et al. (2015). pHL58 contained the sgRNA and was constructed by digesting pJW1285 (Ward 2015) with NdeI and SpeI. PCR fragments containing the missing portion of pJW1285, but with the desired ina-1 sgRNA replacing the original pha-1 sgRNA contained in pJW1285, were inserted into this digested backbone using Gibson assembly. This resulted in a new vector identical in sequence to pJW1285, with the exception of the desired ina-1 sgRNA sequence in place of the original pha-1 sgRNA. Microinjections were performed using the method described in Dickinson et al. (2015); template and sgRNA constructs were each injected at 50 ng/µL, and lines were isolated as described above.
Scoring cell fate transformation and function
Worms were synchronized for scoring by L1 arrest (Stiernagle 2006) and recovering on Escherichia coli OP50-seeded plates at 25°C. Experiments were scored using widefield microscopy, with either a Zeiss Axio Imager D1 microscope (40x or 63x PlanNeofluar objectives) and a Zeiss AxioCam MRm camera or a Zeiss Axio Imager Z1 microscope (40x Plan-Neofluar and 63x Plan-Apochromat objectives) and a Hamamatsu Orca-ER camera. In addition, we used a Cell Observer SD spinning disk confocal microscope (Carl Zeiss) with a 63x Plan-Apochromat objective to better visualize cellular morphology and a Zeiss Axio Imager M2 (40x Plan-Neofluar objective) equipped with an ApoTome and an Axiocam 506 camera for capturing images in Figure 1F.
Reprogramming pro-ACs into fDTCs
Worms carrying arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry] and/or arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2] were evaluated at two stages. “Late L2” is equivalent to 24–28 h post egg prep or 8–10 h post-L1 arrest at 25°C, and “late L3” is equivalent to 32–34 h post egg prep or 22–25 h post-L1 arrest at 25°C. Markers were evaluated using widefield microscopy and considered “ON” at the following exposures: qIs90[ceh-22bp::venus]: 800 ms (Axio Imager D1 microscope, 63x objective) or 50% laser power and 120 ms, spinning disk confocal (63x). arIs51[cdh-3::gfp]: 250 ms, both widefield microscopes (63x objective). Morphological changes were assessed using the confocal microscope, and cells were considered to have fDTC-like morphology if they displayed flat cell bodies and long projections toward the germline. Visualization of cell morphology was facilitated by fluorescence from the hlh-2prox::lin-32::sl2::2xtagBFP2 transgene, imaged at 40% laser power and 800 ms exposure time.
To examine the correlation between fDTC-like morphology and ceh-22bp::venus expression, we examined hermaphrodites carrying both arTi148[hlh-2prox::lin-32cDNA::sl2::mCherry] and arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2] using spinning disk confocal microscopy; to test statistical significance, we generated a 2 × 2 contingency table and used a Fisher’s exact test, with morphology scored as fDTC-like or WT and ceh-22bp::venus expression scored as ON or OFF under the conditions described above.
+HLH-12 AC
The ACs of hermaphrodites carrying arSi45[hlh-2prox::hlh-12cDNA::sl2::2xtagBFP2] were scored for features characteristic of the wild-type AC using markers and morphology indicated here.
Invadopodia
Invadopodia were defined by colocalization of membrane protrusions and mCherry-moeABD puncta. Worms were imaged using the spinning disk confocal microscope at 63x in late L3, 24–28 h after plating as arrested L1. For WT, qyIs176[zmp-1p::mcherry-moeABD] was imaged at 40% power and 800 ms; for +HLH-12, it was imaged at 50% power and 1200 ms. arIs131[lag-2p::2xnls-YFP] was imaged at 20% power and 500 ms in both backgrounds.
Vertical displacement scoring
Vertical displacement was measured using ImageJ (fiji.sc).
Quantification of displacement distance (see Figure 2C and Supplementary Figure S1C)
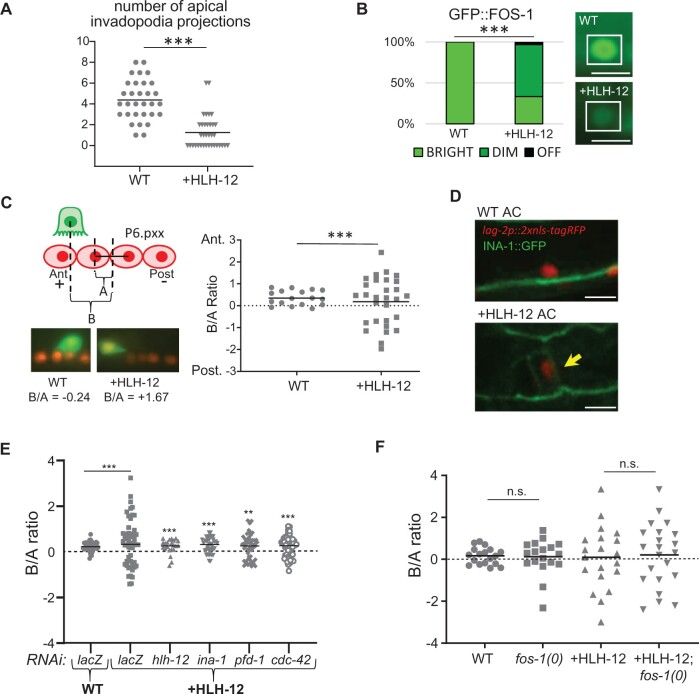
Figure 2.
Ectopic expression of HLH-12 in the AC does not change its fate but compromises its function. WT represents animals lacking the +HLH-12 transgene but containing other markers as noted. (A) The number of apical invadopodia is reduced in ACs that ectopically express HLH-12. Invadopodia were examined in WT and +HLH-12 animals using lag-2p::2xNLS-yfp to mark the nucleus of the AC and zmp-1p::mcherry::MoesinABD to visualize invadopodia. ***P < 0.001, Kolmogorov–Smirnov; n = 31 (WT) or 35 (+HLH-12). (B) Endogenous GFP::FOS-1 expression is downregulated in ACs that ectopically express HLH-12. WT and +HLH-12 animals carry gfp::fos-1(bmd138) (Medwig-Kinney et al. 2020). In the bar graph, ***P < 0.001, Fisher’s exact test; n = 17 (WT) or 29 (+HLH-12). In the photomicrographs, the AC is surrounded by white boxes. Scale bar is 5 µm. (C) ACs are displaced horizontally by ectopic expression of HLH-12. The cartoon depicts the method used to quantify displacement of the AC with respect to P6.pxx nuclei as a ratio of two measurements in the anterior–posterior axis, B/A, as depicted in the cartoon (see Materials and Methods and Supplementary Figure S2A for further details); no dorsal-ventral displacement was seen at this stage of development (Supplementary Figure S1B). Photomicrographs show representative ACs for WT and +HLH-12; lag-2p::2xnls-tagRFP marks AC and P6.pxx nuclei; cdh-3p::gfp marks AC cytoplasm. (D) Expression of endogenous INA-1::GFP is activated in the AC by ectopic expression of HLH-12. The photomicrographs show early L2 hermaphrodites, with arIs222[lag-2p::2xnls-rfp] marking the nucleus of the AC; in the +HLH-12 hermaphrodite, the yellow arrow points to cell membrane of the AC showing ectopic INA-1::GFP (bottom). 9/16 +HLH-12 and 0/15 WT hermaphrodites showed ectopic INA-1::GFP expression. Scale bar is 5 µm. (E) RNAi against factors known to be required for fDTC migration suppresses displacement of ACs caused by ectopic expression of HLH-12. In +HLH-12 animals treated with negative control RNAi (lacZ), the AC is displaced compared to animals lacking the +HLH-12 transgene [genotype arIs222[lag-2p::2xnls-tagrfp] him-5(e1490), labeled “WT”]; treatment of animals carrying the +HLH-12 transgene with the positive control RNAi (hlh-12), or RNAi against ina-1, pdf-1, or cdc-42 abrogates this displacement. **P < 0.01, ***P < 0.001, Fisher’s exact test. The statistics shown for the positive control and experimental RNAis are compared to +HLH-12 treated with lacZ(RNAi). n = 20–52 for each genotype. This panel shows one trial; an additional trial for each RNAi is shown in Supplementary Figure S2. (F) Loss of FOS-1 does not affect AC displacement in +HLH-12 animals at the L3 stage (Pn.pxx). n = 18–22 for each genotype. Note that fos-1(0) hermaphrodites were segregated from balanced heterozygotes (see Materials and Methods). Bars show mean.
Hermaphrodites at the P6.pxx stage of the vulval lineage were scored for the degree of anterior–posterior displacement of the AC from its normal position, as shown in Figure 2C and Supplementary Figure S2. A positive ratio indicates the anterior position and a negative ratio indicating posterior position. All distances were measured using FIJI, with a macro to create perpendicular lines. For statistical analysis, datapoints were sorted into bins based on the relative position of the AC nucleus above the P6.pxx nuclei row, as shown in Supplementary Figure S2A. Note that anterior and posterior locations were combined for bins 2 and 3. The Kolmogorov–Smirnov test was used to determine if the spread of data between bins was significantly different.
Marker expression
All markers except ina-1(ar639[ina-1::gfp]) were scored using widefield microscopy at 63x and scored as “ON” if fluorescence was visible at the following exposures: arIs51[cdh-3::gfp] (250 ms), qIs90[ceh-22bp::venus] (800 ms), and fos-1(bmd138) (300 ms). Expression of ina-1(ar639[ina-1::gfp]) was scored using confocal microcopy (30% power and 500 ms, 63x).
Effect of loss of fos-1 activity
fos-1(ar105) was maintained as a heterozygote balanced by tmC16[unc-60(tmIs1210)]. tmC16 shows green fluorescence as a heterozygote and a homozygote and is Unc as a homozygote (Dejima et al. 2018). fos-1(ar105) homozygous worms were thus identified as non-Unc individuals lacking fluorescence. Worms were scored in late L3. AC displacement was scored using Nomarski optics.
Effect of RNAi directed against genes required for fDTC migration
Worms were plated after L1 arrest, grown at 25°C, and scored 24–28 h after feeding on plates seeded with feeding bacteria. Escherichia coli strains containing RNAi clones came from a commercial library and feeding RNAi was performed as described in Kamath and Ahringer (2003); RNAi targeting lacZ was used as a negative control. Worms were staged based on time after plating and scored for fDTC outgrowth defects at 63x using widefield microscopy, providing an internal positive control that RNAi had been effective. lag-2p::tagrfp expression was used to identify the AC and P6.pxx nuclei. Photomicrographs were taken at 500 ms exposure (lag-2p::tagrfp) and 750–900 ms (hlh-2prox::hlh-12::sl2::BFP).
Search for HLH-12 in the genomes of various nematode species
BLAST searches using protein and transcript sequences of HLH-12 were performed against the scaffold databases at the Caenorhabditis Genomes Project v1 (caenorhabditis.org) for Oscheius tipulae, Mesorhabditis belari, Poikilolaimus oxycercus, Diploscapter coronatus; WormBase ParaSite v9.0 (parasite.wormbase.org) for Ascaris suum, Heterorhabditis bacteriophora, and Panagrellus redivivus; and pristionchus.org (El Paco V1 and V2) for Pristionchus pacificus. For Caenorhabditis species, sequences were acquired from WormBase version WS266 (wormbase.org) and caenorhabditis.org. For some species, entire preliminary genome assemblies were downloaded from the FTP server of Caenorhabditis.org and BLAST searches performed with PrfectBLAST (Santiago-Sotelo and Ramirez-Prado 2012). Initial BLAST searches were done with an E-value threshold of 1.0 × 10−10, but in some cases, the threshold had to be raised to identify a homolog. Reciprocal BLAST was used to test whether the protein is a likely ortholog of HLH-12. Potential bHLH orthologs were also screened using NCBI’s Conserved Domain search (ncbi.nlm.nih.gov) against database CDD v3.17–55426 PSSMs with an E-value threshold of 0.01, and transcripts without predicted bHLH domains were eliminated. In the course of making alignments, we found that some annotations were incorrect. These were corrected manually for the region of the protein containing the bHLH domain. The sequences are listed in Supplementary Table S1 and are available as FASTA files in Supplementary Files S1 and S2.
Caenorhabditis bHLH protein phylogeny
The protein sequences of all C. elegans bHLH domain proteins were downloaded from WormBase and trimmed to the bHLH domain using the domain annotation at Uniprot.org. The bHLH domains were aligned using ClustalW implemented in MegaX (https://www.megasoftware.net/; Stecher et al. 2020) with the gap penalties set to 7. A phylogenetic analysis of this alignment was performed using BEAST (Suchard et al. 2018), run for 5,000,000 generations with sampling every 1000 generations and a burnin of 15% (Supplementary Figure S3). This analysis resulted in support for a clade containing HLH-12 and 20 other bHLH proteins (HLH-1, HLH-2, HLH-3, HLH-4, HLH-6, HLH-8, HLH-10, HLH-11, HLH-13, HLH-14, HLH-15, HLH-16, HLH-17, HLH-19, HLH-31, HLH-32, CND-1, HND-1, NGN-1, and LIN-32). Those were selected for further analyses along with HIF-1, which was previously proposed to be related to HLH-12 (Grove et al. 2009). The genomes of Caenorhabditis species are available at WormBase and caenorhabditis.org.
Assembling a dataset for orthologs of 22 C. elegans bHLH domain proteins in 10 species and phylogenetic analysis
We searched for potential orthologs of the above 22 proteins in proteomes of 10 species (O. tipulae, H. bacteriophora, D. coronatus, Caenorhabditis monodelphis, C. parvicauda, Caenorhabditis bovis, Caenorhabditis castelli, Caenorhabditis japonica, C. elegans, and Caenorhabditis briggsae) representing a broad range of phylogenetic lineages bracketing the lineage in which LIN-12 first appeared. Orthologs were found using BLAST searches against the protein database in caenorhabditis.org. The final dataset contained 191 operational taxonomic units, including “in-paralogs” (paralogs arising within a species lineage, e.g., HLH-17, HLH-31, and HLH-32 within C. elegans). A list of these proteins is found in Supplementary Table S1 and the sequences in Supplementary File S2.
Preliminary alignments were made to determine which amino acids were least well-aligned, and these were trimmed off of the sequences as well as well-aligned positions that were far away from the bHLH domain (e.g., in HIF-1). Using these trimmed protein sequences, three different alignments were obtained: (A) a 228-position alignment from Clustal Omega at https://www.ebi.ac.uk/Tools/msa/clustalo/ (Sievers et al. 2011) using default alignment parameters but with the maximum number of five iterations, (B) a 199-position alignment from ClustalW (Larkin et al. 2007) as implemented in MegaX using default alignment parameters, and (C) a 223-position alignment from ClustalW using a gap opening penalty of 6.0 for pairwise and multiple alignment and a gap extension penalty of 0.05 for pairwise alignment and of 0.1 for multiple alignment. These so-called “large” character sets were used to form three different data matrices. Three additional data matrices were constructed by truncating each of these alignments to leave only the most unambiguously aligned positions, corresponding to the “small” character sets of alignments A (83 characters), B (94 characters), and C (86 characters).
Each of these six data matrices was subjected to phylogenetic analysis with two different methods via the high-performance, parallelized, CIPRES (Cyberinfrastructure for Phylogenetic Research) web tools at phylo.org: Bayesian Inference (BI, using MrBayes) and bootstrapped Maximum Likelihood (ML, using RAxML). For BI, default parameters were used, except that datatype was designated as protein and differential rates across positions were modeled with a gamma distribution in four discrete categories. Also, Markov Chain Monte Carlo was run for 5,000,000 generations (2 runs of 4 chains), with trees and model parameters being sampled every 1000 generations. Trees for calculating clade credibility values were collected after a 25% “burnin” (i.e., 7502 trees). Prior to running ML, we ran all data matrices on ModelTest-NG (Darriba et al. 2020), which suggested LG+G4 (Le and Gascuel 2008) as the best model (with gamma shape parameter 0.72). For ML, we used the RAxML-BlackBox tool on CIPRES, with default parameters except specifying the LG+G4 model and using 1000 bootstrap replications for each run. We used these 12 different phylogenetic analyses to assess the robustness of the phylogenetic results to alignment, character number, and method. Three of the trees resulting from BI are shown as phylograms in Supplementary Figure S4; one of these is also depicted as a cladogram in Figure 4B. All results, as well as the data matrix input files, are available by e-mailing the authors.
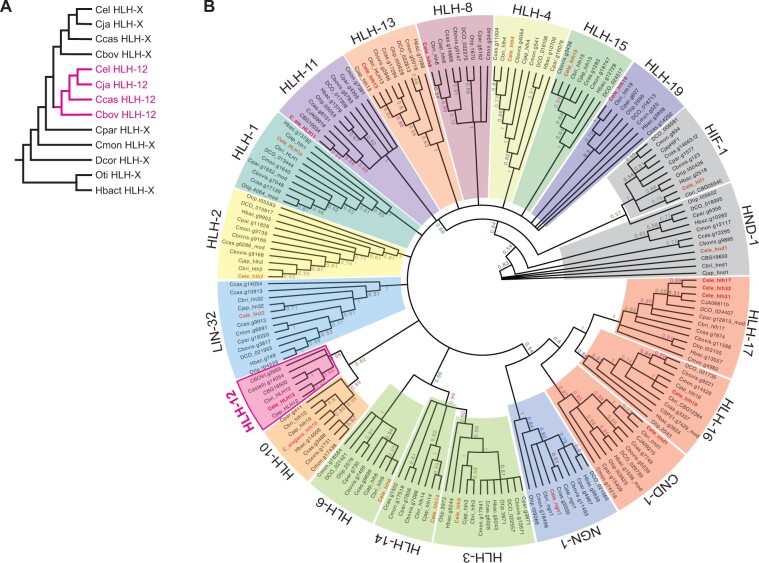
Figure 4.
Phylogenetic relationship of class II bHLH proteins in selected nematode species. (A) Hypothetical phylogeny of HLH-12 and its ancestor. (B) Unrooted circular cladogram resulting from BI on a Clustal Omega alignment of inferred protein sequences trimmed of essentially all ambiguously aligned amino acid positions (86 characters). Clades of inferred orthologs are designated in colored blocks and labeled using the C. elegans gene name. Clade support values are depicted on the branches (see Materials and Methods for more details about phylogenetic analyses). Note that, although relationships within ortholog clades are insufficiently supported due to too-few phylogenetically informative sites and changes, the ortholog groups themselves (and thus membership of proteins within those groups) are generally very highly supported (clade credibility values near 1.0). Membership within several ortholog clades is further supported by other conserved areas peculiar to those groups (data not shown). Abbreviations for the species: Cbovis = Caenorhabditis bovis, Cbri, CBG = C. briggsae, Ccas = C. castelli, Cel = C. elegans, Cjap = C. japonica, Cmon = C. monodelphis, Cpar = C. parvicauda, DCO = Diploscapter coronatus, Otip = Oscheius tipulae, Hbac = Heterorhabditis bacteriophora.
Results
Concomitant expression of HLH-12 and LIN-32 leads to transient reprogramming of cells with AC potential into fDTCs
Previously, we proposed that the distinct complement of bHLH-encoding genes constitutes a “bHLH code” for regulatory cell fate and function (Sallee et al. 2017). To test the hypothesis, we used the AC, which normally only expresses HLH-2, as tabula rasa. When we ectopically expressed the mDTC Class II bHLH complement (HLH-8 and LIN-32) in cells with AC potential, we transiently reprogrammed them into mDTC-like cells (Sallee et al. 2017). Similarly, if the bHLH code hypothesis is also correct for the fDTCs, we would expect that ectopic expression of the fDTC Class II bHLH complement (HLH-12 and LIN-32) would transform cells with AC potential into fDTC-like cells.
To achieve ectopic expression, we used the regulatory element hlh-2prox, which drives expression in the four cells that initially have the potential to become the AC (“pro-AC”) and their parents in the L2 stage, and later becomes restricted to the differentiated AC in the L3 stage (Sallee and Greenwald 2015; see Figure 1A). We generated single-copy insertion transgenes in the form of artificial bicistronic operons (see Materials and Methods), constructed as hlh-2prox::bhlh coding region::sl2acc::fluorescent protein-coding region, where “sl2acc” represents a trans-splicing acceptor sequence (Blumenthal 2012), and the “bhlh coding region” is a cDNA for hlh-12 or lin-32. Two separate mRNAs are produced from these operons via SL2-mediated trans-splicing, one encoding the bHLH gene of interest and the other a fluorescent protein. The continued production of the fluorescent protein suggested continued expression of the transgene after cell fate transformation and marked the cells to facilitate analysis. We refer to strains carrying arSi45 [hlh-2prox-driven HLH-12] as “+HLH-12,” strains carrying arTi148 [hlh-2prox-driven LIN-32] as “+LIN-32,” and strains carrying both as “+HLH-12+LIN-32.”
To test if expression of HLH-12 and LIN-32 in cells with AC potential transforms them into fDTC-like cells, we examined the expression of qIs90[ceh-22bp::venus], which is normally expressed in the fDTC but not the AC (Lam et al. 2006), and the presence of projections toward the germ line characteristic of fDTC but not AC morphology. Expression of single bHLH genes in +HLH-12 or +LIN-32 hermaphrodites did not cause fDTC-like transformation based on these criteria (n = 22–26 for each genotype). The AC marker arIs51[cdh-3p::gfp] was expressed normally (Figure 1B), and vulval induction, a core function of the AC, was also normal. We note that +HLH-12 animals displayed other interesting abnormalities in AC function that became the focus of our analysis, as described in detail below.
By contrast, concomitant expression of both HLH-12 and LIN-32 resulted in one or more proximal cells with fDTC-like properties. We observed ceh-22bp::venus fDTC marker expression in two adjacent cells or a single cell in the center of the somatic gonad in the L2 stage, soon after the somatic primordium had formed, albeit at incomplete penetrance (Figure 1C). At this time, we also observed morphological changes consistent with fDTC reprogramming, i.e., flat cell bodies with long projections reaching toward the germline (Figure 1D). There was a significant correlation (P < 0.001) between ceh-22bp::venus expression and fDTC-like morphology: for the most part, +HLH-12 +LIN-32 cells displayed either both ceh-22bp::venus expression and fDTC-like morphology (n = 11/56) or neither expression nor morphological changes (n = 41/56), supporting the inference that there was a cell fate transformation.
When we observed two fDTC-like cells in an individual, their position centrally and adjacent to each other suggested that they resulted from the transformation of the two α cells, which retain pro-AC potential and hlh-2prox expression for a longer time than the two β cells (Seydoux et al. 1990; Sallee and Greenwald 2015; Sallee et al. 2017). When we observed a single fDTC-like cell in an individual, we infer that the presumptive AC was transformed after the AC/VU decision had been completed, consistent with the continued expression driven by hlh-2prox in the AC. Regardless of whether one or two fDTC-like cells were observed, aspects of their transformation appeared to be transient: although a similar proportion of proximal cells expressed ceh-22b in both the late L2 and late L3 stages, by late L3 they no longer contained fDTC-like projections (Figure 1, E and F). Such transience was also observed for the highly penetrant transformation of cells with AC potential into mDTCs upon ectopic expression of the mDTC code genes hlh-8 and lin-32 (Sallee et al. 2017), suggesting there is a general mechanism that resists the maintenance of reprogramming.
Ectopic expression of HLH-12 in the AC does not change its fate but compromises its function
As described above, expression of LIN-32 or HLH-12 alone did not lead to fDTC-like cells in the proximal gonad or affect the number of cells expressing the AC marker arIs51[cdh-3p::gfp], suggesting that the AC/VU decision was not disrupted and that the AC had been specified normally. However, while adult +LIN-32 hermaphrodites had a normal vulva, adult +HLH-12 hermaphrodites had a fully penetrant egg-laying defect (Egl) and a high rate of vulval abnormalities, with 90% displaying abnormal vulval eversion (Evl) and about 10% lacking a vulva altogether (Vul), suggesting abnormalities in AC function. Indeed, we found that the +HLH-12 AC is defective in a critical function of the AC in the L3 stage, i.e., invasion of the basement membrane surrounding the gonad in order to contact the vulval cells (Sherwood and Plastino 2018). In addition, the AC in +HLH-12 animals was displaced from its normal position through apparent activation of the fDTC migration program (see below). These abnormalities in AC function can account for the Egl, Evl, and Vul phenotypes observed in +HLH-12 hermaphrodites and are seen at equivalent frequencies in +HLH-12+LIN-32 hermaphrodites as well (Supplementary Figure S1C).
Two morphological abnormalities suggested that the +HLH-12 AC is defective in invading the underlying vulval epithelium: the number of invadopodia was significantly reduced on the apical membrane of +HLH-12 ACs (Figure 2A), and we observed misplaced invadopodia projecting from the basolateral membrane of +HLH-12 ACs (Supplementary Figure S1A). AC invasion requires the transcription factor FOS-1, which promotes the development of multiple invadopodia on the apical membrane of the AC (Sherwood et al. 2005); invadopodia projecting instead from the basolateral membrane are a hallmark of fos-1(0) (D. Matus, personal communication). We therefore examined the expression of fos-1(bmd138), which encodes endogenous GFP::FOS-1 and is normally robustly expressed in ACs (Medwig-Kinney et al. 2020). We found that expression is reduced or absent in +HLH-12 ACs (Figure 2B). Taken together, these results indicate that impaired invasion in +HLH-12 ACs results from downregulation of fos-1. While it is possible that fos-1 is downregulated because HLH-12:HLH-2 heterodimers reduce the formation of the HLH-2:HLH-2 homodimers necessary to activate fos-1, we have not seen such an effect for LIN-32 or HLH-8, two other individual bHLH proteins we have ectopically expressed in a similar manner (Sallee et al. 2017; Figure 1B).
Ectopic expression of HLH-12 appears to activate leader function in the AC
The AC induces the vulval precursor cell closest to it, P6.p, to adopt the 1° vulval fate, characterized by a lineage that gives rise to a row of four granddaughters collectively called P6.pxx. The nuclei of P6.p and its descendants as well as the AC may be marked using arIs222[lag-2p::tag-rfp] to measure the position of the AC with respect to the P6.pxx row (Figure 2C and Supplementary Figure S2A). Normally, the AC is located slightly anterior to the center of the P6.pxx row. However, in +HLH-12 hermaphrodites, we observed significant displacement of the AC from its normal position, anteriorly or posteriorly at equal frequency (Figure 2C).
We considered the possibility that the AC is displaced because ectopic expression of HLH-12 alone activates the expression of genes required for fDTC migration without any cell fate transformation per se. If so, displacement would depend on the activity of these genes. To test this possibility, we analyzed ina-1, a direct target of HLH-2:HLH-12 heterodimers in fDTCs (Meighan and Schwarzbauer 2007; Meighan et al. 2015). First, to examine endogenous ina-1 expression, we used CRISPR/Cas9 to generate ina-1(ar639[ina-1::gfp]). In a wild-type background, INA-1::GFP was not present in the AC or any other proximal gonadal cells in the L2 stage (Figure 2D). However, in +HLH-12 hermaphrodites, INA-1::GFP was detected in the AC in the L2 stage, suggesting that the presence of HLH-12 in the AC causes ectopic ina-1 transcription (Figure 2E). Second, we performed ina-1(RNAi) on +HLH-12 hermaphrodites and found that loss of ina-1 suppressed the displacement of the +HLH-12 AC, indicating that +HLH-12 AC movement depends on ina-1 activity (Figure 2D and Supplementary Figure S2, B and C).
Two other lines of evidence suggest that displacement of the AC in +HLH-12 is a result of the fDTC migration program. First, we tested cdc-42 and pfd-1, two other genes required for fDTC migration (Lucanic and Cheng 2008; Lundin et al. 2008) and found that RNAi directed against them also suppressed the displacement of the AC in +HLH-12 hermaphrodites (Figure 2D and Supplementary Figure S2C). We also tested gon-1, the other known direct target of HLH-12:HLH-2 heterodimers during fDTC migration (Tamai and Nishiwaki 2007) but the results were uninterpretable because the integrity of the gonad was compromised by gon-1(RNAi) (Supplementary Figure S2D). Second, displacement by expression of HLH-2 was similar in the fos-1(+) and fos-1(0) backgrounds, suggesting that the +HLH-12 AC is not displaced as a secondary consequence of the invasion defect described above (Figure 2F).
The dependence of +HLH-12 AC displacement on ina-1, cdc-42, and pfd-1, three genes required for fDTC migration, suggests that expression of HLH-12 is sufficient to activate AC movement by the mechanism used for leader cell outgrowth. The modest nature of +HLH-12 AC displacement compared to the more extensive migration achieved by the leader cells at that point in development may stem from the AC being embedded among other uterine cells, impeding its movement or access to external guidance or stimulating cues, or be due to incomplete activation of the migration program by the +HLH-12 transgene.
hlh-12 evolved within the genus Caenorhabditis
As far as we know, all nematode gonads develop from a primordium containing homologous regulatory cells, but adult gonads can have strikingly different morphologies caused in part by different behaviors of the regulatory cells (Sternberg and Horvitz 1981; Félix and Sternberg 1996; Rudel et al. 2005). Differences in the length and trajectory of the gonad arm migration led by fDTC outgrowth results in differences in gonadal morphology in many species (Figure 3). Furthermore, the posterior vulvae of M. belari and P. redivivus results from a posterior migration of the ACs in these species (Figure 3; Sternberg and Horvitz 1981; Félix and Sternberg 1996). Our finding that HLH-12 is able to cause migratory-like behavior in the C. elegans AC led us to consider the possibility that evolutionary changes in bHLH regulation—and of HLH-12 in particular—could underlie the repeated evolution of migrating ACs in other species.
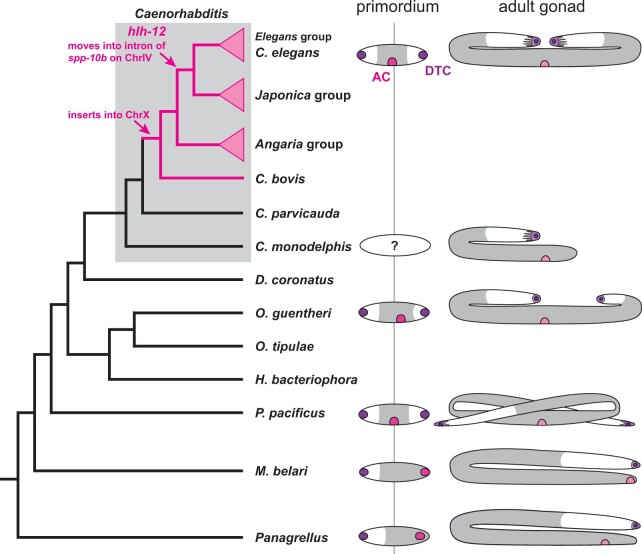
Figure 3.
Phylogenetic relationship of species discussed in the text (C. =Caenorhabditis, D. = Diploscapter, H. = Heterorhabditis, M. = Mesorhabditis, O. = Oscheius, P. = Pristionchus, Panagrellus = Panagrellus redivivus) and variation in female gonad morphologies. Phylogeny after (Kiontke et al. 2007; Stevens et al. 2020); Lineages that possess HLH-12 are marked in pink. Right: cartoons of the female/hermaphrodite gonad in a selection of species where it differs from the gross morphology of the C. elegans gonad. DTC in purple, AC in pink, white area indicates the location of the germ line. In the adult gonad, the light pink crescent indicates the position of the vulva (based on Félix and Sternberg 1996; Rudel et al. 2005; Slos et al. 2017).
As a first step to map the roles of bHLH genes in gonad development of other species, we performed reciprocal BLAST searches for hlh-12 in the genomes of 11 species (C. japonica, C. monodelphis, Diploscapter pachys, D. coronatus, H. bacteriophora, O. tipulae, P. pacificus, M. belari, P. oxycercus, P. redivivus, and A. suum) representing diverse nematode groups. Whereas BLAST readily returned hits for HLH-12 in most Caenorhabditis species, no hlh-12 ortholog could be identified in the genome of any species outside of this clade. To pinpoint the phylogenetic lineage in which hlh-12 originated, we searched for hlh-12 orthologs in genome sequences from 33 species within the Caenorhabditis clade, as well as representatives from its sister clade, Protorhabditis-Diploscapter (Kiontke et al. 2007; Fradin et al. 2017). Mapping hlh-12 presence/absence on the most recent phylogeny (Stevens et al. 2020) confirmed that hlh-12 arose within and near the base of the Caenorhabditis clade, likely just before C. bovis diverged from other Caenorhabditis species (Figure 3 and Supplementary Figure S5). We thus conclude that this protein evolved only recently within the Caenorhabditis clade, and therefore that HLH-12 cannot itself be involved in the migratory behaviors of ACs, or of the fDTCs and LC, in other rhabditid species.
The ancestor of hlh-12 might be hlh-10
Since HLH-12 clearly plays an important role in the specification and migration of gonadal regulatory cells in C. elegans, it is puzzling that it is apparently absent in species in which these cells migrate as in C. elegans. Understanding the origin and evolution of HLH-12 would help us understand how it inherited this function from its ancestor, or possibly co-opted it. We therefore tried to identify the gene that gave rise to hlh-12. We started with the assumption that at least the bHLH domain of HLH-12 must have been inherited from an ancestral bHLH gene and that a comprehensive phylogenetic analysis may reveal its ancestral relationship.
We note that different hypotheses have been put forward for the relationships of HLH-12 with other bHLH proteins, placing HLH-12 in the Achaete-Scute family (Tamai and Nishiwaki 2007), Atonal family (Stevens et al. 2008; Skinner et al. 2010), or HIF-1 family (Grove et al. 2009). However, these previous placements were based either on simple alignments of the bHLH domain without phylogenetic analysis or based on phylogenetic analysis with evolutionarily distant phyla such as Drosophila, mammals, and plants. We therefore performed a comprehensive phylogenetic analysis of all Class II bHLH proteins in 10 nematode species, including the other bHLH code proteins (HLH-2, HLH-3, HLH-8, and LIN-32; Figure 4 and Supplementary Figure S5). As described below, our analysis confirmed that, while all other bHLH code proteins are generally conserved in all nematodes examined, HLH-12 is restricted to the Caenorhabditis clade.
We reasoned that we may be able to identify the real ancestor of HLH-12 by performing a focused and thorough phylogenetic analysis involving members of the bHLH protein family from nematode species closest to the lineage where HLH-12 evolved. If this analysis could recover the true evolutionary history of HLH-12, we expect it to be nested as a new branch within the clade of its ancestor X after the divergence of the X ortholog in C. parvicauda (Figure 4A).
To focus our analysis on the bHLH proteins that are most likely candidates for the HLH-12 ancestor, we first performed a phylogenetic analysis of the bHLH domains of all C. elegans proteins using Bayesian Inference (BI). This analysis confirmed a clade of class II bHLH proteins that also included HLH-2. There was no evidence for HIF-1 to be part of this clade but for further analyses, we included HIF-1 as a potential outgroup (Supplementary Figure S3).
We next identified potential orthologs of these 22 proteins in the genomes of 7 Caenorhabditis species (C. elegans, C. briggsae, C. japonica, C. castelli, C. bovis, C. parvicauda, and C. monodelphis) and 3 representatives of other nematode taxa (D. coronatus, H. bacteriophora, and O. tipulae). A phylogenetic analysis of this dataset revealed that the HLH-12 clade is most closely related to the HLH-10 clade (clade credibility 85%; Figure 4B). However, different from what was expected if HLH-12 arose from HLH-10 within Caenorhabditis, HLH-12 was not nested within the HLH-10 clade (cf. Figure 4, A and B).
Because BI is somewhat sensitive to evolutionary model and because the data matrix was fairly small (214 characters), we tested the robustness of our phylogenetic results to differences in alignment, character number (number of alignment positions), and phylogenetic inference method (Supplementary Figure S4). We found that the relationship of HLH-12 and HLH-10 was quite robust (as were other previously found relationships, such as that between Achaete-scute family members HLH-3 and HLH-14), but that HIF-1 jumped onto the HLH-12 branch in about half of the most credible trees resulting from BI (Supplementary Figure S4). Interestingly, HIF-1 is the taxon with the longest branch in all trees and is most different from the other bHLHs. In the other half of the most credible trees, HIF-1 is attached to HND-1, another long-branch taxon. We think this unstable behavior of the HIF-1 branch is due to an artifact known as long-branch attraction, to which Bayesian analyses may be susceptible if datasets contain many ambiguously aligned characters (Kolaczkowski and Thornton 2009; Susko 2015). This possibility is consistent with HIF-1 not being closely related to HLH-12 when data matrices were reduced to only the most unambiguously aligned characters as in Qu et al. (2017).
In summary, although we find too little phylogenetic information to establish the relationships between most of the bHLH ortholog groups, the most likely ancestor of HLH-12 is HLH-10. If true, we are left to explain why our analyses place HLH-12 outside of the HLH-10 clade instead of being nested inside the Caenorhabditis part of the HLH-10 clade. One possible explanation is that, during the divergence between HLH-10 and HLH-12 after duplication (e.g., via positive selection for subfunctionalization, or specialization of their roles), changes were superimposed over the likely smaller number of phylogenetically informative changes that would have revealed HLH-12's origin within the HLH-10 clade.
If HLH-12 is derived from HLH-10, it could have originated by tandem duplication or duplicative transposition of HLH-10. If HLH-12 evolved by tandem duplication, we might expect its gene to be located near hlh-10, similar to the two copies of hlh-12 in C. briggsae, which are both close to one another on Chromosome IV. Because chromosome linkage groups are highly conserved in Caenorhabditis (Stevens et al. 2020) and indeed even between Caenorhabditis and Pristionchus (Rodelsperger et al. 2017), we might expect hlh-10 and hlh-12 to be on the same chromosome in all species. However, we find a different and more diverse picture (Supplementary Figure S5): hlh-10 is on chromosome V in all Caenorhabditis species where the genome sequences have sufficient contiguity. In C. bovis, Caenorhabditis plicata, and Caenorhabditis virilis—and thus the lineage in which it first appeared—hlh-12 is located on the X chromosome between the orthologs of C. elegans genes C64H3.2 and C25G6.3. These genes are also in close proximity in C. monodelphis and C. parvicauda, the two species that branch off before the evolution of hlh-12, and in C. elegans. Different from almost all other bHLH genes in our analysis, which are on the same chromosome in species as divergent as C. bovis and C. elegans (Supplementary Table S1), hlh-12 changed position repeatedly: in the Angaria group of Caenorhabditis, it is still likely located on the X chromosome but situated between orthologs of cest-7 and gcy-36. In Caenorhabditis uteleia, a species that branches off between the Angaria and Elegans super-groups, hlh-12 is located in an intron of the ortholog of C. elegans W09G3.6 on chromosome I. Finally, in all species of the Elegans super-group, hlh-12 is located in an intron of spp-10 on chromosome IV. In summary, this synteny pattern is most consistent with the origin of hlh-12 by duplicative transposition (and possible domain shuffling), where the bHLH-coding domain of hlh-10 was first inserted into the X chromosome and subsequently and repeatedly transposed.
Discussion
Previous work showed that all regulatory cells of the developing C. elegans gonad require the Class I bHLH protein HLH-2, that HLH-2 functions as a homodimer in specification and function of the AC, and that the other gonadal regulatory cells express a distinct complement of Class II dimerization partners for HLH-2 (Krause et al. 1997; Karp and Greenwald 2003, 2004; Tamai and Nishiwaki 2007; Chesney et al. 2009; Sallee and Greenwald 2015). A further study suggested that there is a “bHLH code” that governs the specification and function of regulatory cells and proposed that such a code provides a potential mechanism for the evolutionary plasticity of gonad form seen in nematodes (Sallee et al. 2017). Here, we extended the bHLH code hypothesis by showing that expression of genes for the fDTC in the AC leads to transient transformation into cells with fDTC character. One of these genes is hlh-12, which was known to be required for fDTC (as well as male LC) migration that leads to gonad arm outgrowth (Tamai and Nishiwaki 2007; Sallee et al. 2017). We have now shown that HLH-12 is also sufficient for coordinating a module of the cellular machinery responsible for migratory cell behavior: by expressing HLH-12 in the nonmigratory AC, where its dimerization partner HLH-2 is present, we showed that this migratory module can be activated to displace the AC from its normal position. During this displacement, the fate of the AC itself is not affected, but the FOS-1-controlled program that produces invadopodia is halted, thus preventing the formation of a connection between the vulva and the gonad.
We propose that such modularity in the genetic architecture could permit the evolution of novel positioning of the AC, as in species with posterior vulvae. Of course, additional steps must have also evolved in such species, such as signaling to guide the migratory AC to posterior positions. Our analysis also suggests that activation of the migration module itself may prevent invadopodia from forming, such that migration of the AC would concomitantly delay the formation of invadopodia until the AC is in position and the migration module deactivated.
We have also shown that the fDTC migratory module cannot be controlled by HLH-12 in all nematode species, because HLH-12 arose only within Caenorhabditis, probably from a duplication of HLH-10. We think the most conservative (most parsimonious) explanation is that HLH-12's ancestor, HLH-10, already governed the fDTC migratory module, in addition to potential roles in neurons inferred from the expression of hlh-10 in sensory neurons (Portman and Emmons 2004; Etchberger et al. 2007). Duplication would have allowed subfunctionalization of these roles, such that HLH-12 took on the fDTC role and HLH-10 maintained the other roles. Consistent with this view, hlh-10 does not play a role in fDTC specification or leader function in C. elegans (Sallee et al. 2017). In future experiments, it will be interesting to probe the expression and function of HLH-10 orthologs in basal Caenorhabditis species to ascertain if hlh-10 functions in fDTCs as we propose.
The mystery does not end there, however. Some species more distantly diverged from the Caenorhabditis clade, such as D. coronatus and O. tipulae are also missing HLH-10, but nevertheless have migrating fDTCs. Thus, although the cellular machinery responsible for migratory behavior is likely to be highly conserved, there must be substantial plasticity regarding the specific transcription factor(s) controlling it. Regulation of fDTC migration in rhabditid nematodes seems to be another case of “developmental system drift” (True and Haag 2001) in which the underlying mechanism is surprisingly plastic while the phenotypic outcome remains quite conserved. In addition, the modularity of this system appears to allow novel deployment of migratory activity and consequently the evolution of novel morphologies.
Data availability
The data underlying this article are available in the article and in its Supplementary material online. Supplementary files are available at figshare (https://doi.org/10.25386/genetics.15062469). Supplementary Figures S1–S5 contain additional information about ectopic expression and RNAi experiments, phylogenetic trees for C. elegans bHLH proteins, phylograms for the phylogenetic analysis in this manuscript, and the phylogenetic distribution of hlh-12 in Caenorhabditis. Supplementary Table S1 summarizes information about bHLH proteins and genes in Caenorhabditis species and other nematodes. Supplementary File S1 contains HLH-12 sequences in Caenorhabditis in FASTA format. Supplementary File S2 contains bHLH proteins sequences in 10 nematode species in FASTA format. Strains, plasmids, sequence alignments, and input and output files for phylogenetic analysis can be obtained from the corresponding authors upon request.
Acknowledgments
The authors are grateful to David Matus for strain DQM497 and additional information about the fos-1 null phenotype, Shai Shaham for strain OS8909, Maria Sallee for strain GS7894, Jee Hun (Henry) Kim for help with generating some C. elegans strains, Justin Benavidez for providing the photomicrograph of the somatic gonad primordium in Figure 1A, and Ji-Sup Yang for help with phylogenetic analyses. They are also grateful to Claire de la Cova for help with statistical analyses, Oliver Hobert for discussion, and Michelle Attner for comments on the manuscript.
Funding
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Research reported in this publication was supported by the Institute of General Medicine of the National Institutes of Health under award numbers R35GM131746 (to I.G.) and R01GM141395 (to D.H.A.F.), the National Science Foundation under award number 1656736 (to D.H.A.F.), and research funds from NYU Shanghai (to D.H.A.F.).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Attner MA, Keil W, Benavidez JM, Greenwald I.. 2019. HLH-2/E2A expression links stochastic and deterministic elements of a cell fate decision during C. elegans gonadogenesis. Curr Biol. 29:3094–3100.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Kimble J.. 1999. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 399:586–590. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. 2012. Trans-splicing and operons. In: WormBook. The C. elegans Research Community, editor. WormBook. doi:10.1895/wormbook.1.5.1. [Google Scholar]
- Chesney MA, Lam N, Morgan DE, Phillips BT, Kimble J.. 2009. C. elegans HLH-2/E/Daughterless controls key regulatory cells during gonadogenesis. Dev Biol. 331:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram EJ, Shang H, Schwarzbauer JE.. 2006. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci. 119:4811–4818. [DOI] [PubMed] [Google Scholar]
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, et al. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37:291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K, Hori S, Iwata S, Suehiro Y, Yoshina S, et al. 2018. An aneuploidy-free and structurally defined balancer chromosome toolkit for Caenorhabditis elegans. Cell Rep. 22:232–241. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B.. 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200:1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, et al. 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21:1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A. 2006. Oscheius tipulae. WormBook. The C. elegans Research Community, ed. WormBook. doi:10.1895/wormbook.1.119.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Sternberg PW.. 1996. Symmetry breakage in the development of one-armed gonads in nematodes. Development. 122:2129–2142. [DOI] [PubMed] [Google Scholar]
- Fradin H, Kiontke K, Zegar C, Gutwein M, Lucas J, et al. 2017. Genome architecture and evolution of a unichromosomal asexual nematode. Curr Biol. 27:2928–2939.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, et al. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6:343–345. [DOI] [PubMed] [Google Scholar]
- Greenwald I. 2012. Notch and the awesome power of genetics. Genetics. 191:655–669. doi: 10.1534/genetics.112.141812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, et al. 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 138:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag ES, Fitch DHA, Delattre M.. 2018. From “the worm” to “the worms” and back again: the evolutionary developmental biology of nematodes. Genetics. 210:397–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 32:728–730. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. [DOI] [PubMed] [Google Scholar]
- Karp X, Greenwald I.. 2003. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17:3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Greenwald I.. 2004. Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev Biol. 272:460–469. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D.. 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 70:396–417. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG.. 1981. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 81:208–219. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Barriere A, Kolotuev I, Podbilewicz B, Sommer R, et al. 2007. Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol. 17:1925–1937. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton JW.. 2009. Long-branch attraction bias and inconsistency in Bayesian phylogenetics. PLoS One. 4:e7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Park M, Zhang JM, Yuan J, Harfe B, et al. 1997. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 124:2179–2189. [DOI] [PubMed] [Google Scholar]
- Lam N, Chesney MA, Kimble J.. 2006. Wnt signaling and CEH-22/tinman/Nkx2.5 specify a stem cell niche in C. elegans. Curr Biol. 16:287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Li J, Greenwald I.. 2010. LIN-14 inhibition of LIN-12 contributes to precision and timing of C. elegans vulval fate patterning. Curr Biol. 20:1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Cheng HJ.. 2008. A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 4:e1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin VF, Srayko M, Hyman AA, Leroux MR.. 2008. Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev Biol. 313:320–334. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C.. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 20:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 163:41–46. [DOI] [PubMed] [Google Scholar]
- Medwig-Kinney TN, Smith JJ, Palmisano NJ, Tank S, Zhang W, et al. 2020. A developmental gene regulatory network for C. elegans anchor cell invasion. Development. 147:dev185850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan CM, Kann AP, Egress ER.. 2015. Transcription factor hlh-2/E/Daughterless drives expression of alpha integrin ina-1 during DTC migration in C. elegans. Gene. 568:220–226. [DOI] [PubMed] [Google Scholar]
- Meighan CM, Schwarzbauer JE.. 2007. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors. Genes Dev. 21:1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, et al. 1994. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1218:129–135. [DOI] [PubMed] [Google Scholar]
- Pani AM, Goldstein B.. 2018. Direct visualization of a native Wnt in vivo reveals that a long-range Wnt gradient forms by extracellular dispersal. Elife. 7:e38325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman DS, Emmons SW.. 2004. Identification of C. elegans sensory ray genes using whole-genome expression profiling. Dev Biol. 270:499–512. [DOI] [PubMed] [Google Scholar]
- Qu XJ, Jin JJ, Chaw SM, Li DZ, Yi TS.. 2017. Multiple measures could alleviate long-branch attraction in phylogenomic reconstruction of Cupressoideae (Cupressaceae). Sci Rep. 7:41005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodelsperger C, Meyer JM, Prabh N, Lanz C, Bemm F, et al. 2017. Single-molecule sequencing reveals the chromosome-scale genomic architecture of the nematode model organism Pristionchus pacificus. Cell Rep. 21:834–844. [DOI] [PubMed] [Google Scholar]
- Rudel D, Riebesell M, Sommer RJ.. 2005. Gonadogenesis in Pristionchus pacificus and organ evolution: development, adult morphology and cell-cell interactions in the hermaphrodite gonad. Dev Biol. 277:200–221. [DOI] [PubMed] [Google Scholar]
- Sallee MD, Greenwald I.. 2015. Dimerization-driven degradation of C. elegans and human E proteins. Genes Dev. 29:1356–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee MD, Littleford HE, Greenwald I.. 2017. A bHLH code for sexually dimorphic form and function of the C. elegans somatic gonad. Curr Biol. 27:1853–1860.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Sotelo P, Ramirez-Prado JH.. 2012. prfectBLAST: a platform-independent portable front end for the command terminal BLAST+ stand-alone suite. Biotechniques. 53:299–300. [DOI] [PubMed] [Google Scholar]
- Schindler AJ, Sherwood DR.. 2011. The transcription factor HLH-2/E/Daughterless regulates anchor cell invasion across basement membrane in C. elegans. Dev Biol. 357:380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Schedl T, Greenwald I.. 1990. Cell-cell interactions prevent a potential inductive interaction between soma and germline in C. elegans. Cell. 61:939–951. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, Butler JA, Kramer JM, Sternberg PW.. 2005. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 121:951–962. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, Plastino J.. 2018. Invading, leading and navigating cells in Caenorhabditis elegans: insights into cell movement in vivo. Genetics. 208:53–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH.. 2010. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation. 80:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slos D, Sudhaus W, Stevens L, Bert W, Blaxter M.. 2017. Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis. BMC Zool. 2:4. [Google Scholar]
- Sommer RJ. 2006. Pristionchus pacificus. WormBook. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher G, Tamura K, Kumar S.. 2020. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 37:1237–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR.. 1981. Gonadal cell lineages of the nematode Panagrellus redivivus and implications for evolution by the modification of cell lineage. Dev Biol. 88:147–166. [DOI] [PubMed] [Google Scholar]
- Stevens JD, Roalson EH, Skinner MK.. 2008. Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation. 76:1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L, Rooke S, Falzon LC, Machuka EM, Momanyi K, et al. 2020. The genome of Caenorhabditis bovis. Curr Biol. 30:1023–1031.e4. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. In: WormBook. The C. elegans Research Community, editor. WormBook. doi:10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, et al. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4:vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susko E. 2015. Bayesian long branch attraction bias and corrections. Syst Biol. 64:243–255. [DOI] [PubMed] [Google Scholar]
- Tamai KK, Nishiwaki K.. 2007. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev Biol. 308:562–571. [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES.. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 3:109–119. [DOI] [PubMed] [Google Scholar]
- Ward JD. 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics. 199:363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary material online. Supplementary files are available at figshare (https://doi.org/10.25386/genetics.15062469). Supplementary Figures S1–S5 contain additional information about ectopic expression and RNAi experiments, phylogenetic trees for C. elegans bHLH proteins, phylograms for the phylogenetic analysis in this manuscript, and the phylogenetic distribution of hlh-12 in Caenorhabditis. Supplementary Table S1 summarizes information about bHLH proteins and genes in Caenorhabditis species and other nematodes. Supplementary File S1 contains HLH-12 sequences in Caenorhabditis in FASTA format. Supplementary File S2 contains bHLH proteins sequences in 10 nematode species in FASTA format. Strains, plasmids, sequence alignments, and input and output files for phylogenetic analysis can be obtained from the corresponding authors upon request.