Abstract
Inflammation and oxidative stress are key steps in the progression of non-alcoholic steatohepatitis (NASH). Intervention in these two processes will therefore benefit NASH treatment. Peroxisome proliferator-activated receptor γ (PPARγ), as a multiple functional transcription factor, has been reported to be involved in the prevention of NASH progression. However, the mechanism by which PPARγ prevents NASH remains to be elucidated. The present study demonstrated that the level of PPARγ was inversely correlated with that of microRNA (miRNA/miRs)-21-5p in both mice and humans with NASH. Activation of PPARγ inhibited lipid droplet accumulation, hepatic inflammation and oxidative stress by downregulating miR-21-5p in an in vitro model. Luciferase reporter and chromatin immunoprecipitation assays demonstrated that PPARγ suppressed transcriptional activity of miR-21-5p and bound to miR-21-5p promoter region. Furthermore, PPARγ downregulated miR-21-5p while miR-21-5p upregulated secreted frizzled-related protein 5 (SFRP5) by targeting the 3′-UTR of its mRNA. In vivo experiments revealed that PPARγ repressed inflammation and oxidative stress and miR-21-5p expression while increased SFRP5 level in a NASH mouse model. In summary, PPARγ attenuates inflammation and oxidative stress in NASH by modulating the miR-21-5p/SFRP5 pathway, thus holding promise of a new target for NASH treatment.
Keywords: peroxisome proliferator-activated receptor γ, miR-21-5p, secreted frizzled-related protein 5, nonalcoholic steatohepatitis, inflammation, oxidative stress
Introduction
Non-alcoholic steatohepatitis (NASH) is the advanced form of non-alcoholic fatty liver disease (NAFLD) and characterized by intrahepatic lipid accumulation, steatosis and inflammation (1). NASH can further progress to cirrhosis and even hepatocellular carcinoma (HSC). NASH also significantly increases the risk of cardiovascular disease and diabetes and has been identified as an increasing public health burden (2). Inflammation and oxidative stress serve a critical role in the progression of NASH. Hence, intervention in inflammation and oxidative stress will benefit NASH treatment.
Peroxisome proliferator-activated receptor γ (PPARγ), a ligand-activated transcription factor, belongs to nuclear receptor subfamily that is involved in the regulation of lipid metabolism, glucose homeostasis, inflammation and cellular growth (3). It has been reported that in an animal model of NASH, the activation of PPARγ regulates the polarization of the macrophages to M2 subtype, thus preventing development of NASH (4). Rosiglitazone, the widely investigated PPARγ agonist, has been documented to be able to prevent NASH development (5,6). However, the mechanism by which PPARγ prevents NASH remains to be elucidated.
MicroRNAs (miRNAs/miRs) are a family of small (19–25-nucleotide) endogenous noncoding RNAs that regulate gene expression by pairing interactions with mRNAs, which can lead to degradation or translation repression of target mRNAs. miR-21-5p has been reported to be involved in liver lipid metabolism through regulating a variety of targets, such as fatty acid binding protein 7, phosphate and tension homolog and SMAD family member 7 (SMAD7) (7–9). Previous studies have also demonstrated that miR-21-5p is highly expressed in NASH (9,10), suggesting that miR-21-5p may be a useful target for NASH treatment. However, the mechanism of miR-21-5p participation in regulating NASH remains to be elucidated.
Secreted frizzled-related protein 5 (SFRP5) is an anti-inflammatory adipokine that regulates metabolic homeostasis via regulating the Wnt signaling pathways (11,12). In a previous study, SFRP5 protein levels were significantly downregulated in NASH (13). However, the effect of SFRP5 on NASH remains unknown.
The present study revealed for the first time, to the best of the authors' knowledge, that PPARγ downregulated the expression of miR-21-5p, leading to the upregulation of SFRP5, which contributed to the inhibition of hepatic inflammation and oxidative stress of nonalcoholic steatohepatitis, suggesting that the PPARγ/miR-21-5p//SFRP5 signaling pathway may be a promising target for NASH treatment.
Materials and methods
Reagents and plasmids
Rosiglitazone (Rosi) was purchased from Cayman Chemical Company. miR-21-5p mimic, inhibitor and negative control (NC) were synthesized by Shanghai GenePharma Co., Ltd. Superoxide dismutase (SOD) assay kit, methane dicarboxylic aldehyde (MDA) assay kit, glutathione peroxidase (GSH) assay kit and oxidized glutathione (GSSG) assay kit were purchased from Beijing Solarbio Science & Technology Co., Ltd. miRNA Detection kit was purchased from GeneCopoeia, Inc. ChIP kit was from Merck KGaA. Dual-luciferase reporter assay system was from Promega Corporation. Human miR-21-5p promoter region or mutations in PPARγ binding sites were synthesized by Sangon Biotech Co., Ltd. The 3′-UTR of SFRP5 mRNA or mutant binding site was synthesized by Sangon Biotech Co., Ltd.
Clinical samples
A total of 20 liver tissue samples from patients with NASH and 20 normal individuals were obtained from Banan People's Hospital(Chongqing, China) between April 2020 and November 2020. All procedures were conducted in accordance with the ethical rules stated in the Declaration of Helsinki and were approved by the Ethics Committee of Banan People's Hospital (approval number BNRMYY20200016). A written informed consent was obtained from each subject before their participation. The clinical characteristics of the patients are summarized in Table I.
Table I.
Characteristics of the study individuals.
| Characteristic | Control (n=20) | NASH (n=20) | P-value |
|---|---|---|---|
| Age (years) | 43.5±3.6 | 44.2±4.3 | n.s. |
| Sex (male/female) | 11/9 | 13/7 | n.s. |
| BMI (kg/m2) | 22.3±2.7 | 25.8±3.5 | <0.05 |
| Fasting glucose (mmol/l) | 5.3±1.7 | 5.9±1.2 | <0.05 |
| Fasting insulin (mIU/l) | 10.4±2.9 | 15.3±1.8 | <0.05 |
| Triglycerides (mmol/l) | 1.55±1.32 | 3.48±1.09 | <0.05 |
| Total cholesterol (mmol/l) | 4.15±2.48 | 5.02±2.18 | n.s. |
| LDL-C (mmol/l) | 2.24±1.21 | 2.97±1.18 | n.s. |
| AST (IU/l) | 31.5±12.3 | 39.7±11.4 | <0.05 |
| ALT (IU/l) | 35.2±10.8 | 44.8±10.2 | <0.05 |
| Current smokers (%) | 2.5 | 3 | n.s |
Data are expressed as mean ± standard deviation. NASH, non-alcoholic steatohepatitis; n.s., not significant; BMI, body mass index; LDL-C, low-density lipoprotein cholesterol; AST, aspartate transaminase; ALT, alanine transaminase.
Cell culture and treatment
Liver cancer cell line HepG2 was purchased from China Center for Type Culture Collection and the cell line was authenticated by STR profiling. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc.), streptomycin (100 mg/ml) and penicillin (100 U/ml) at 37°C in a 5% CO2 humid incubator. HepG2 cells were treated with 1 mm free fatty acid for 16 h to establish a NASH cell model and then the cells were treated with DMSO, 10 M Rosi or 10 µm Rosi in the presence of miR-21-5p mimic or the combination of miR-21-5p mimic and inhibitor for 24 h, the cells were harvested for further experiment.
Mice and treatment
Male C57BL/6J mice (six weeks old) were purchased from Beijing Huafukang Bioscience Co., Ltd. and housed in specific-pathogen-free (SPF) laboratory. To induce NASH (14,15), mice were fed with methionine and choline deficient (MCD) diet (Research Diets A02082002B). Normal mice were fed with a control diet (Research Diets A02082003B). After MCD treatment for 5 weeks, mice were randomly divided into three groups (n=5): i) NASH + DMSO: These mice were fed with MCD diet and a treated with vehicle saline containing 0.1% DMSO intraperitoneally daily for 3 weeks; ii) NASH + Rosi: These mice were fed with MCD diet and treated with 10 mg/kg Rosi intraperitoneally daily for 3 weeks; iii) NASH + Rosi + miR-21-5p mimic: These mice were fed with MCD diet and were received intraperitoneal injection of 10 mg/kg Rosi daily and were injected via the tail vein daily with 8 mg/kg miR-21-5p mimic for 3 weeks (16,17). The experiment lasted for 8 weeks in total and mice were euthanized by cervical dislocation following mild (4 ml) diethyl ether (Sigma Aldrich, USA) anesthesia. The liver tissues were removed for hematoxylin and eosin (H&E) staining and the blood was collected from abdominal aorta for biochemical detections. All animal experiments were performed according to Guide for the Care and Use of Laboratory Animals 8th edition (18) and approved by the Animal Care and Use Committee of Banan People's Hospital (approval no. BNRMYY20200029).
Transfections and luciferase assays
HepG2 were seeded into 24-well plates at a density of 1×105 cells/well. The cells were grown to 70% confluence in 24-well plates before transfection at 37°C. Next, cells were transfected with the pmirGLO reporter plasmid (empty plasmid) (Promega Corporation), pmirGLO reporter plasmid containing 3′-UTR of SFRP5 mRNA, miR-21-5p mimic (50 nm; 5′-GUCUCAAUAUUAGGCUAAGCUAU-3′), miR-21-5p inhibitor (50 nm; 5′-AUAGCUUAGCCUAAUAUUGAGAC-3′), mimic negative control (NC; 5′-UUCUCCGAACGUGUCACGUUU-3′) or inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′) (Shanghai GenePharma Co., Ltd), using Lipofectamine® 2000 (Invitrogen, Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions at room temperature. Subsequently, the transfected cells were cultured for 24 h at 37°C in a 5% CO2 humid incubator. Then, the luciferase activity was detected according to the instructions of the Dual-Luciferase reporter gene assay kit (Thermo Fisher Scientific, Inc.). Renilla luciferase activity was normalized to firefly luciferase activity and the result was shown as relative luciferase activity. Transfection experiments were performed 3 times in triplicate. Data were represented as the ratios of luciferase activities/Renilla activities.
RNA interference assays
The HepG2 cells were seeded into 6-well plates, when the confluence of HepG2 cells was 70%, small interfering RNA (siRNA) against PPARγ mRNA (100 nm; cat. no. sc-29455) and NC siRNA (100 nm; scrambled siRNA, sc-37007; both Santa Cruz Biotechnology) were transfected into HepG2 cells using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C according to the instructions of the manufacture. Subsequently, 24 h after cell transfection different treatments and examinations were performed.
Reverse transcription-quantitative (RT-q) PCR
Total RNAs were extracted from cells (1×106 cells/well) or liver tissues (100 mg) with TRIzol reagent (Thermo Fisher Scientific, Inc.). Reverse transcription of total RNA was performed using Primescript RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. Subsequently, qPCR was performed with SYBR green qPCR Master Mix (Promega Corporation) according to the manufacturer's protocol using the ABI 7500 system (Thermo Fisher Scientific, Inc.), and the relative mRNA expression levels were normalized to β-actin. miRNAs were extracted with E.Z.N.A. microRNA kit (Omega Bio-Tek, Inc.) and then the expression of miR-21-5p was determined using All-in-One miRNA qRT-PCR Detection kit (GeneCopoeia, Inc.) according to the manufacturer's instructions and the relative expression level was normalized against U6 small nuclear RNA (U6 snRNA). The PCR reaction conditions were as follows: Initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The experiments were performed three times. miRNA and mRNA expression levels were quantified using the 2−ΔΔCq method (19). Primer sequences are listed in Table II.
Table II.
Primer sequences used for reverse transcription-quantitative PCR.
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| mouse PPARγ | 5′-GGAAGACCACTCGCATTCCTT-3′ | 5′-TCGCACTTTGGTATTCTTGGAG-3′ |
| human PPARγ | 5′-TCGAGGACACCGGAGAGG-3′ | 5′-GTGTCAACCATGGTCATTTCGTT-3′ |
| mouse miR-21-5p | 5′-TAGCTTATCAGACTGATGTTGA-3′ | Universal adaptor (supplied in the kit) |
| human miR-21-5p | 5′-TAGCTTATCAGACTGATGTTGA-3′ | Universal adaptor (supplied in the kit) |
| mouse SFRP5 | 5′-AAGTTCCCCCTGGACAACGA-3′ | 5′-AATGCGCATCTTGACCACAAA-3′ |
| human SFRP5 | 5′-CCACAAGTTCCCCCTGGACA-3′ | 5′-TGCGCATTTTGACCACAAAGTCA-3′ |
| mouse TNF-α | 5′-CAAACCACCAAGTGGAGGAG-3′ | 5′-GTGGGTGAGGAGCACGTAGT-3′ |
| human TNF-α | 5′-CCCTCCTTCAGACACCCT-3′ | 5′-GGTTGCCAGCACTTCACT-3′ |
| mouse IL-6 | 5′-AGTTGCCTTCTTGGGACTGA-3′ | 5′-TCCACGATTTCCCAGAGAAC-3′ |
| human IL-6 | 5′-CAATAACCACCCCTGACC-3′ | 5′-GCGCAGAATGAGATGAGTT-3′ |
| mouse MCP-1 | 5′-ACCTTTTCCACAACCACCT-3′ | 5′-GCATCACAGTCCGAGTCA-3′ |
| human MCP-1 | 5′-TTTTCCCCTAGCTTTCCC-3′ | 5′-GCAATTTCCCCAAGTCTCT-3′ |
| mouse β-actin | 5′-TGTTACCAACTGGGACGACA-3′ | 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
| human β-actin | 5′-GTGAAGGTGACAGCAGTCGGTT-3′ | 5′-GAAGTGGGGTGGCTTTTAGGA −3′ |
| mouse U6 snRNA | 5′-ACTAAGCGGCCTGACTGAAG-3′ | 5′-GCCATTGTCCTTGTGACGTG-3′ |
| human U6 snRNA | 5′-CGCTTCGGCAGCACATATACTAA-3′ | 5′-TATGGAACGCTTCACGAATTTGC-3′ |
PPARγ, peroxisome proliferator-activated receptor γ; miR, microRNA; SFRP5, secreted frizzled-related protein 5; MCP, monocyte chemotactic protein.
Western blotting
Whole proteins from cell/tissue samples were lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology). Protein concentrations were determined using the BCA assay(Beyotime Institute of Biotechnology). The proteins (50 µg) were separated by 12% SDS- polyacrylamide gel electrophoresis and transferred onto PVDF membranes (EMD Millipore). Subsequently, the membranes were blocked with 5% bull serum albumin (Beyotime Institute of Biotechnology) at room temperature and then incubated at 4°C overnight with primary antibody for SFRP5 (1:1,000; Abcam; cat. no. ab230425), PPARγ (1:500; Santa Cruz Biotechnology, Inc.; cat. no. sc-271392) or β-actin (1:2,000; Abcam; cat. no. ab8227). Following washing with TBST (10 M Tris, 150 mm NaCl, 0.05% Tween-20), the membranes were incubated with HRP-conjugated secondary antibody (goat anti Rabbit IgG; Abcam; cat. no. ab205718, 1:10,000) and incubated at 37°C for 1 h. Finally, the blots were visualized using the enhanced chemiluminescence detection reagents (Thermo Fisher Scientific, Inc.) The grey value of protein bands was analyzed using ImageJ software 1.46r (National Institutes of Health).
Bioinformatics analysis
The putative PPARγ response element (PPARE) in miR-21-5p promoter region was predicted by NUBIScan (nubiscan.unibas.ch). The potential binding sites of miR-21-5p in 3′-UTR of human SFRP5 mRNA were predicted using the online databases miRBase (mirbase.org), TargetScan (targetscan.org) and miRanda (microrna.org).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed using EZ-ChIP kit (EMD Millipore) according to the manufacturer's protocol. Briefly, HepG2 cells (1×107) were treated with 0.1% DMSO or 5 µm Rosi at 37°C for 24 h and then harvested after fixation with 1% formaldehyde for 10 min at room temperature to cross-link the nuclear proteins to DNA. Then, the cells were lysed in SDS lysis buffer at room temperature for 10 min and the chromatin was sonicated with Ultrasonic Sonicator at 30% of maximum power 10 times for 10-sec pulses on ice, with 2 min interval. to shear the DNA to an average length between 200 and 1,000 bp, followed by immunoprecipitation with the antibody directed against PPARγ (Santa Cruz Biotechnology, Inc.; cat. no. sc-271392×), taking IgG as a negative control. The precipitated DNAs were purified and subjected to PCR amplification that cover the PPARγ response element (PPARE) in miR-21-5p promoter region (−276 to −127), taking ‘input’ (the total DNA extract) as a positive control while ‘no DNA’ as a negative control (20).
Oil Red O staining
The treated cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with PBS. Next, cells were soaked in 60% Oil Red O stock solution (0.25 g Oil Red O/100 ml isopropanol) diluted by distilled water for 30 min at 25°C. The stained cells were washed using PBS until the background became clear (21). Finally, images were captured under a light microscope (Olympus Corporation) 10 randomly selected fields of view were evaluated at ×200 magnification.
Hematoxylin and eosin (H&E) staining
The mouse liver tissues were harvested and gently cut into 0.3×0.5×0.5 cm cubes and fixed with 4% polyoxymethylene at 4°C for 24 h. The fixed liver tissues were dehydrated with gradient ethanol solutions at 25°C. After washing with xylene two times at 25°C, 1.5 h each, the samples were embedded in paraffin. The paraffin blocks were sectioned at 5 µm. Sections from each paraffin block were stained with hematoxylin for 3 min and counterstained with 1% Eosin Y for 10 min at 25°C to examine the pathologic structures of the tissues. Histological steatosis, inflammation and fibrosis were graded according to the NASH activity score (NAS) (22,23) by a certified pathologist. Images were captured by light microscopy (200× magnification; Olympus Corporation); ≥10 randomly selected fields of view were assessed.
Statistical analysis
Statistical analyses and graphical representations were performed using GraphPad Prism 5.0 software (GraphPad Software Inc). Data are presented as the mean ± standard deviation. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. Comparisons between two groups were analyzed using a paired t-test. The correlation analysis was performed using Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
The level of PPARγ is inversely correlated with that of miR-21-5p in NASH in both mice and humans
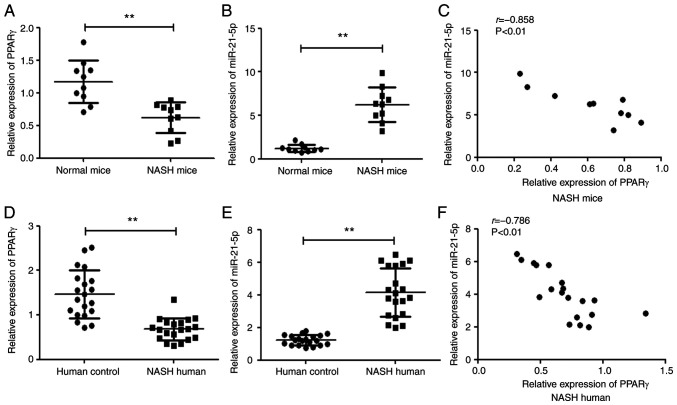
It has been reported that PPARγ and miR-21-5p are associated with progression of NASH and have been proposed as potential targets for NASH treatment. Hence, the relationship between PPARγ and miR-21-5p was explored in NASH in mice and humans. As shown Fig. 1A and B, PPARγ was lowly expressed while miR-21-5p was highly expressed in the liver tissue of mouse model of NASH and the human tissue sample from patients with NASH (Fig. 1D and E). Furthermore, there was a significantly inverse correlation between the level of PPARγ and miR-21-5p in NASH in both mice and humans (Fig. 1C and F).
Figure 1.
PPARγ expression is inversely correlated with miR-21-5p in NASH in both mice and humans. The level of (A) PPARγ and (B) miR-21-5p in liver tissues of NASH mice and normal mice was detected by qPCR (n=10). (C) The correlation between the PPARγ and miR-21-5p mRNA levels in NASH mice was analyzed using Pearson's correlation coefficient (n=10; r=−0.858, P<0.01). The level of (D) PPARγ and (E) miR-21-5p in 20 liver tissue samples from NASH patients and 20 normal individuals was examined by qPCR. (F) Correlation between the PPARγ and miR-21-5p mRNA levels in NASH patients using Pearson's correlation coefficient (n=20; r=−0.786, P<0.01). **P<0.01 vs. normal group. PPARγ, peroxisome proliferator-activated receptor γ; miR, microRNA; NASH, non-alcoholic steatohepatitis; qPCR, reverse transcription-quantitative PCR.
Activation of PPARγ inhibits lipid droplet accumulation, hepatic inflammation and oxidative stress by downregulating miR-21-5p in vitro
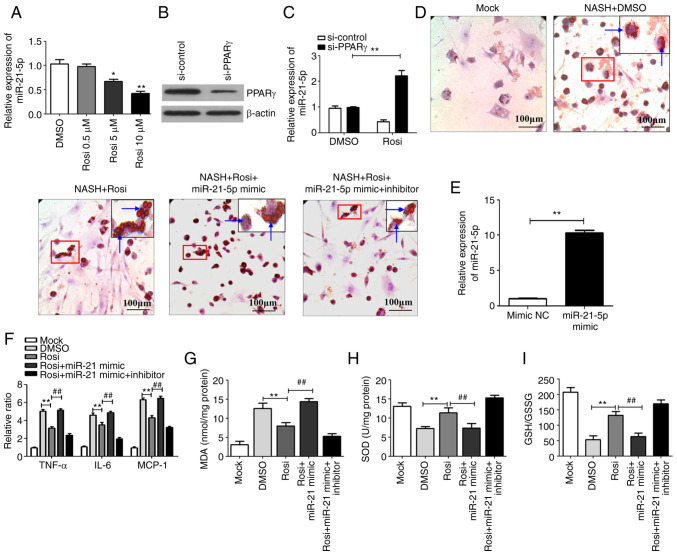
To investigate whether PPARγ is involved in the regulation of miR-21-5p, HepG2 cells were treated with the PPARγ agonist Rosi and the expression of miR-21-5p was detected. As shown in Fig. 2A, activation of PPARγ with Rosi decreased miR-21-5p expression in a dose-dependent manner. Knockdown of PPARγ by siRNA (cat. no. sc-29455; Santa Cruz Biotechnology, Inc.), in which the silencing effect was high (Fig. 2B), significantly attenuated the Rosi-mediated downregulation of miR-21-5p (Fig. 2C). It is well established that lipid droplet accumulation, hepatic inflammation and oxidative stress serve important roles in the progression of NASH (24–26). Therefore, the levels of lipogenesis, inflammatory cytokines and oxidative stress were evaluated in an in vitro model of NASH, in which the cells were treated with PPARγ agonist Rosi and miR-21-5p mimic or miR-21-5p inhibitor. As shown in Fig. 2D, the number of lipid particles was markedly decreased in cells treated with Rosi, in which PPARγ was activated, relative to the cells treated with DMSO. However, the number of lipid particles was increased in the cells treated with miR-21-5p mimic, which the miR-21-5p was overexpressed after transfected with miR-21-5p mimic (Fig. 2E). In addition, the expression levels of inflammatory markers TNF-α, IL-6 and monocyte chemotactic protein (MCP)-1 (Fig. 2F) and oxidative stress (expression levels of oxidative stress markers including MDA, SOD and GSH/GSSG ratio; Fig. 2G-I) in NASH cells was inhibited by activation of PPARγ, but the inhibitory effect was attenuated by miR-21-5p mimic (Fig. 2F-I). Inhibition of miR-21-5p by miR-21-5p inhibitor significantly enhanced the PPARγ-mediated inhibition of lipogenesis (Fig. 2D), inflammation (Fig. 2F) and oxidative stress in NASH cells (Fig. 2G-I). Taken together, these results indicated that PPARγ repressed progression of NASH via downregulation of miR-21-5p.
Figure 2.
Activation of PPARγ inhibits lipid droplet accumulation, hepatic inflammation and oxidative stress by downregulating miR-21-5p. (A) HepG2 cells were treated with Rosi (0.5, 5 and 10 µm) or control (0.1% DMSO) for 48 h and then the expression of miR-21-5p was detected by qPCR. (B) siRNA against PPARγ (si-PPARγ), or scrambled siRNA negative control (si-control) was transfected into HepG2 cells and the expression of PPARγ was detected by western blotting. (C) Following transfection with si-control or si-PPARγ, HepG2 cells were treated with DMSO or 10 µm Rosi for 24 h. Then the level of miR-21-5p was assayed by qPCR. Data are expressed as means ± SD. *P<0.05, **P<0.01 vs. DMSO. (D) HepG2 cells were treated with 1 mm free fatty acid for 16 h to establish a NASH cell model and then the cells were treated with DMSO, Rosi (10 µm), Rosi (10 µM) + miR-21-5p mimic (50 nm) or 10 µm Rosi + miR-21-5p (50 nm) mimic + miR-21-5p inhibitor (50 nm) for 24 h. The effect of Rosi on lipid accumulation in NASH cells was assayed by using oil Red O staining (magnification, ×200), the arrows indicate lipid particles. (E) miR-21-5p mimic (50 nm) or mimic NC (50 nm) were transfected into HepG2 cells. After 24 h, the cells were harvested and the miR-21-5p was detected by qPCR. (F) The inflammatory cytokines TNF-α, IL-6 and MCP-1 were detected by qPCR. (G) The lipid peroxidation product MDA was tested using an MDA assay kit. (H) The activity of SOD was measured using a SOD assay kit. (I) The ratio of GSH/GSSG was evaluated. Data are expressed as means ± SD. **P<0.01 vs. Rosi, ##P<0.01 vs. Rosi + miR-21-5p mimic. PPARγ, peroxisome proliferator-activated receptor γ; miR, microRNA; Rosi, rosiglitazone; qPCR, reverse transcription-quantitative PCR; siRNA, small interfering RNA; NC, negative control; NASH, non-alcoholic steatohepatitis; MCP, monocyte chemotactic protein; MDA, methane dicarboxylic aldehyde; SOD, superoxide dismutase; GSH, glutathione peroxidase; GSSG, oxidized glutathione.
PPARγ suppresses the transcriptional activity of miR-21-5p gene promoter
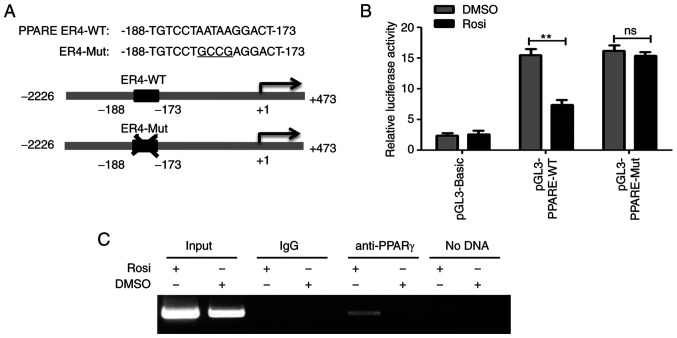
As a transcription factor, PPARγ usually regulates gene transcription by binding to the PPARE in the promoter region of target gene (27). The present study found a putative PPARE ER4 (TGTCCTAATAAGGACT, −188 to −173) in miR-21-5p promoter region by online bioinformatics analysis (NUBIScan; Fig. 3A). To investigate the mechanism by which PPARγ downregulates miR-21-5p, the luciferase reporter plasmids containing wild-type (pGL3-PPARE-WT) or mutant PPARγ (pGL3-PPARE-Mut) binding sites were constructed (Fig. 3A) and the luciferase reporter assays were performed. As shown in Fig. 3B, activation of PPARγ significantly decreased the luciferase activity of construct pGL3-PPARE-WT, whereas the luciferase activity of construct pGL3-PPARE-Mut was not affected, indicating that PPARγ suppressed the transcriptional activity of miR-21-5p promoter. In addition, the ChIP assay demonstrated that PPARγ could directly bind to PPARE ER4 of the miR-21-5p promoter region (Fig. 3C). These results reveal that PPARγ downregulated miR-21-5p by binding to its promoter region.
Figure 3.
PPARγ suppresses transcriptional activity of miR-21-5p and upregulates SFRP5. (A) Diagram of the putative wild-type PPARE ER4-WT and mutant ER4 (ER4-Mut, the mutated bases are underlined) in miR-21-5p gene promoter region. (B) HepG2 cells were transfected with luciferase reporter plasmids pGL3-PPARE-WT, or pGL3-PPARE-Mut or pGL3-Basic and pRL-TK and cultured for 12 h and then treated with control (0.1% DMSO) or 5 µm Rosi for 24 h. Dual-luciferase reporter assays were performed. The firefly luciferase activity was normalized to Renilla luciferase activity and the result was shown as relative luciferase activity. Data are expressed as mean ± SD of three assays performed in triplicate. **P<0.01. (C) HepG2 cells were treated with 0.1% DMSO or 5 µm Rosi for 24 h and the cells harvested for chromatin immunoprecipitation assay. PPARγ, peroxisome proliferator-activated receptor γ; miR, microRNA; SFRP5, secreted frizzled-related protein 5; PPARE, PPARγ response element; PPARγ, peroxisome proliferator-activated receptor γ; WT, wild-type; Mut, mutant; Rosi, rosiglitazone.
PPARγ downregulates miR-21-5p while upregulates SFRP5
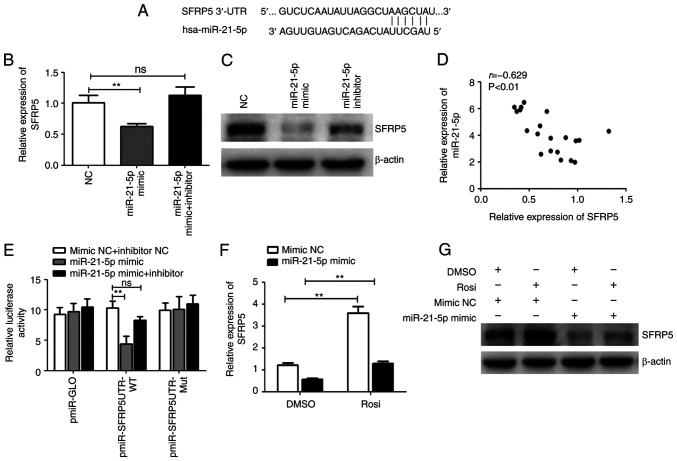
miRNAs regulate gene expression by pairing interactions with mRNAs, which can lead to degradation or translation repression of target mRNAs (28). The online databases (TargetScan, miRBase, miRanda) predicted SFRP5, an anti-inflammatory adipokine that regulates progression of NASH, as a potential target gene of miR-21-5p (Fig. 4A). To investigate whether miR-21-5p is involved in the regulation of SFRP5, HepG2 cells were treated with miR-21-5p mimic or inhibitor. Treatment with miR-21-5p mimic significantly reduced the mRNA and protein levels of SFRP5, which was clearly attenuated by miR-21-5p inhibitor (Fig. 4B and C). In addition, SFRP5 was lowly expressed while miR-21-5p highly expressed in human liver tissue samples from NASH patients and there was a significant inverse correlation between SFRP5 and miR-21-5p levels (Fig 4D). miR-21-5p mimic notably decreased the luciferase activity of pmir-SFRP5-UTR-WT (the luciferase reporter plasmid containing the predicted miR-21-5p wild-type binding sites but not the binding site mutant 3′-UTR), which was alleviated by miR-21-5p inhibitor (Fig. 4E). These results suggested that SFRP5 is one of the target genes of miR-21-5p. It was found that activation of PPARγ enhanced the expression of SFRP5 and overexpression of miR-21-5p markedly inhibited the PPARγ-induced SFRP5 expression (Fig. 4F and G). These results indicated that activation of PPARγ suppresses expression of miR-21-5p and upregulates its target gene SFRP5.
Figure 4.
PPARγ downregulates miR-21-5p but upregulates SFRP5. (A) Diagrams of the predicted potential binding sites of miR-21-5p in 3′-UTR of SFRP5 mRNA. HepG2 cells were transfected with miR-21-5p mimic, inhibitor, or NC and cultured for 24 h and the (B) mRNA and (C) protein levels of SFRP5 were examined by qPCR or western blotting. (D) The correlation between the miR-21-5p and SFRP5 mRNA levels in 20 liver tissue samples from NASH patients was analyzed using Pearson's correlation coefficient (r=−0.629, P<0.01). (E) HepG2 cells were transfected with the luciferase reporter plasmid in the presence of miR-21-5p mimic, inhibitor, or mimic NC and inhibitor NC and cultured for 24 h. The luciferase activities were measured by dual-luciferase reporter assays. The firefly luciferase activity was normalized to Renilla luciferase activity. HepG2 cells were transfected with miR-21-5p mimic or mimic NC and cultured for 12 h and then cells were treated with control (0.1% DMSO) or 5 µm Rosi. After treatment for 24 h, the (F) mRNA and (G) protein levels of SFRP5 were examined by qPCR or western blotting. Data are expressed as means ± SD. **P<0.01 vs. Rosi. PPARγ, peroxisome proliferator-activated receptor γ; miR, microRNA; SFRP5, secreted frizzled-related protein 5; NC, negative control; qPCR, reverse transcription-quantitative PCR; NASH, non-alcoholic steatohepatitis; Rosi, rosiglitazone.
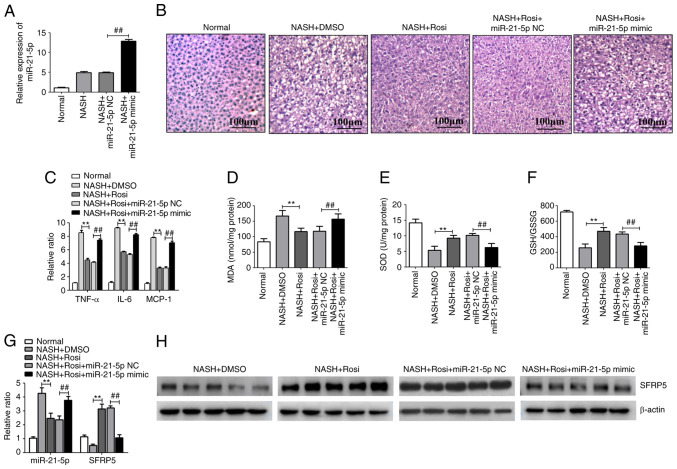
Activation of PPARγ represses the progression of NASH by regulating the miR-21-5p/SFRP5 axis in mice
NASH was induced in C57Bl/6 mice by the MCD diet and this NASH model was used for evaluating the effects of the PPARγ/miR-21-5p/SFRP5 pathway on NASH progression. Hepatic histopathology analysis with H&E staining demonstrated that various degrees of liver steatosis and numerous lipid droplets of different sizes were observed in MCD-fed mice. However, these pathological conditions were significantly alleviated by Rosi treatment. In addition, treatment of miR-21-5p mimic, which the miR-21-5p was overexpressed in the liver by caudal vein injection (Fig. 5A), clearly reduced the PPARγ-mediated inhibition of liver steatosis and lipid accumulation in NASH model (Fig. 5B). In addition, the levels of inflammatory cytokines and oxidative related genes were tested in the liver tissues of mice with MCD-induced NASH. As shown in Fig. 5C, activation of PPARγ dramatically suppressed the expression of proinflammatory cytokines such as TNF-α, IL-6 and MCP-1. Similar trends were observed in the levels of oxidative stress-related genes including MDA, SOD and GSH/GSSG ratio in the liver tissue of mice with MCD-induced NASH (Fig. 5D-F). However, overexpression of miR-21-5p markedly suppressed PPARγ-mediated inhibition of hepatic inflammation and oxidative stress (Fig. 5D-F). In addition, activation of PPARγ markedly decreased miR-21-5p while increased SFRP5 in MCD-fed mice (Fig. 5G and H). These results imply that activation of PPARγ inhibited the progression of NASH by manipulating the miR-21-5p/SFRP5 axis in vivo.
Figure 5.
Activation of PPARγ represses the progression of NASH by regulating the miR-21-5p/SFRP5 axis in mice. Mice were fed for 8 weeks either a methionine- and choline-sufficient diet (Normal) or a methionine- and choline-deficient (MCD) diet (NASH model). Model group mice were treated with vehicle (saline containing 0.1% DMSO), Rosi (10 mg/kg), Rosi (10 mg/kg) + miR-21-5p NC (8 mg/kg), or Rosi (10 mg/kg) + miR-21-5p mimic (8 mg/kg) daily for 3 weeks (n=5). (A) The level of miR-21-5p in liver tissues of mice which treated with miR-21-5p NC or miR-21-5p mimic were detected by qPCR. (B) The photomicrographs of liver sections stained with routine hematoxylin and eosin (magnification, ×200). (C) The inflammatory cytokines TNF-α, IL-6 and MCP-1 in the liver tissue were detected by qPCR. (D) The lipid peroxidation product MDA was tested using an MDA assay kit. (E) The activity of SOD was measured using a SOD assay kit. (F) The ratio of GSH/GSSG was evaluated. Data are expressed as means ± SD. **P<0.01 vs. DMSO, ##P<0.01 vs. miR-21-5p NC. (G) The expression of miR-21-5p and SFRP5 in the liver tissues was detected by qPCR. **P<0.01 vs. DMSO, ##P<0.01 vs. miR-21-5p NC. (H) The protein level of SFRP5 in the liver tissue was detected by western blotting. PPARγ, peroxisome proliferator-activated receptor γ; NASH, non-alcoholic steatohepatitis; miR, microRNA; SFRP5, secreted frizzled-related protein 5; Rosi, rosiglitazone; NC, negative control; qPCR, reverse transcription-quantitative PCR; MCP, monocyte chemotactic protein; MDA, methane dicarboxylic aldehyde; SOD, superoxide dismutase; GSH, glutathione peroxidase; GSSG, oxidized glutathione; SFRP5, secreted frizzled-related protein 5; NASH, non-alcoholic steatohepatitis; Rosi, rosiglitazone.
Discussion
NASH has become a global burden with an increasing prevalence and leads to cirrhosis and HSC, which results in a high rate of mortality per year worldwide. At present, no approved pharmacological treatment is available for NASH, despite development of a large variety of drugs. Currently, the detailed mechanism of drug therapies remains to be elucidated. Thus, more studies are required to explore the mechanisms of NASH and potential pharmacological treatment options (29).
PPARγ is a ligand-activated transcriptional factor that serves an important role in regulating glucose and lipid metabolism (30,31). Rosi is a synthetic ligand of PPARγ that is clinically used as a insulin sensitizer in the treatment of T2DM (32). Rosi has shown promising results in preclinical studies. Administration of Rosi improved high-fat diet induced hepatic steatosis and lipid metabolism through reducing hepatic Toll like receptor 4/NF-κB expression and M1-polarized Kupffer cells (33). In MCD-diet-induced fibrosing NASH models, Rosi may ameliorate hepatic fibrosis by activating PPARγ, which can inhibit HSC activation and suppress TGF-β1 and CTGF expression (34). In a biopsy study, Rosi treatment of NASH patients for 48 weeks results in improved hepatic steatosis, necroinflammation and ballooning (35). In patients with NASH, treatment with Rosi for 1 year improves steatosis and reduced insulin resistance in most patients (36). Although PPARγ agonists have had beneficial effects in in preclinical models of NAFLD/NASH, their effectiveness in human pathology is limited. In addition, the adverse effects or limited potency of PPARγ agonists also limit their application (37–39). Therefore, the mechanism by which PPARγ prevents NASH needs further study, which would aid development of novel anti-NASH strategies.
As previously reported, miR-21-5p is induced in NASH progression and recognized as a biomarker in patients with NASH (40,41). Lack of miR-21-5p leads to significantly less fibrogenesis, TGF-β production and downstream signaling of the TGF-β pathway (9). miR-21-5p ablation results in a progressive decrease in steatosis, inflammation and lipoapoptosis in NASH (42). Therefore, miR-21-5p may be a useful target for NASH treatment. The present study found that miR-21-5p was markedly upregulated while PPARγ was downregulated in the liver tissue of mouse model of NASH and the human tissue sample from patients with NASH. There was also a significant inverse correlation between the level of PPARγ and miR-21-5p in NASH in mice and humans. These results suggested that there may be a regulatory relationship between PPARγ and miR-21-5p in NASH progress. Previous researches have shown that PPARγ, as a transcription factor, can recognize and bind to PPARE in the promoter region of target genes to regulate the expression of target genes (43–46). Therefore, the present study found some PPAREs in the miR-21-5p promoter region by bioinformatics analysis and the reporter assay and ChIP assay demonstrated that PPARγ could directly bind to PPARE ER4 of the miR-21-5p promoter region, suggesting that PPARγ downregulated miR-21-5p through binding to its promoter region.
miRNA can regulate multiple genes by targeting mRNAs in partial sequence homology, leading to mRNA degradation or translation repression (28). Online database searches (miRBase, TargetScan and miRanda) predicted some possible target genes including secreted frizzled-related protein (SFRP)5, which belongs to the SFRP family. Studies have found that SFRP serves a regulatory role in the Wnt signaling pathways (47–49), specifically inhibiting the combination of Wnt protein with its cell membrane receptors and blocking the downstream Wnt signaling pathways by binding to extracellular Wnt-5a or Wnt-3a (12). Kupffer cell activation and intrahepatic inflammation can also be suppressed with recombinant SFRP5, thus improving NASH (14). Clinical investigations have revealed that SFRP5 protein levels are significantly lower in NASH patients than in control subjects (13), suggesting that SFRP5 could have important research significance in the progress of NASH. A previous study has shown that PPARγ directly binds to the SFRP5 promoter domain and regulates the SFRP5 in 3T3-L1 cells (50). The present study discovered that PPARγ upregulated SFRP5 via downregulating miR-21-5p in vitro. It also found that miR-21-5p expression was inversely correlated with SFRP5 expression in liver tissue samples from NASH patients and miR-21-5p directly bound to SFRP5 3′-UTR and inhibited SFRP5 expression, indicating that SFRP5 is a target gene of miR-21-5p. The present study also demonstrated that the PPARγ/miR-21-5p/SFRP5 pathway is a novel mechanism underlying the anti-NASH effects of PPARγ, indicating that PPARγ can regulate SFRP5 in direct and indirect manners. However, more in-depth research is needed to explore the role of the PPARγ/miR-21-5p/SFRP5 pathway so as to explore novel targets for treating NASH.
In summary, the findings of the present study demonstrated that the activation of PPARγ attenuated hepatic inflammation and oxidative stress in nonalcoholic steatohepatitis via downregulating miR-21-5p. Mechanism studies found that PPARγ could bind to PPARE ER4 of the miR-21-5p promoter region and inhibited miR-21-5p, which directly bound to SFRP5 3′-UTR and inhibited SFRP5 expression. These findings suggested that the PPARγ/miR-21-5p/SFRP5 axis could improve the progression of NASH and may represent a potential strategy for the treatment of NASH.
Acknowledgements
Not applicable.
Funding Statement
The study was funded by the grant from the Natural Science Foundation of Chongqing (grant no. cstc2018jcyjAX0280).
Funding
The study was funded by the grant from the Natural Science Foundation of Chongqing (grant no. cstc2018jcyjAX0280).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YJ, JC and XiyZ designed the present study, analyzed the data and wrote the manuscript. XiyZ, FD, YZ and XiaZ conducted the experiments. FD helped to analyze the data. YJ and JC supervised the present study and had full access to all the data. YJ performed the manuscript submission and revised the manuscript. YJ and JC confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experimental procedures were approved by the Ethics Committee of Banan People's Hospital. (approval no. BNRMYY20200016). A written informed consent was obtained from each subject before their participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Sanyal AJ. Past, present and future perspectives in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2019;16:377–386. doi: 10.1038/s41575-019-0144-8. [DOI] [PubMed] [Google Scholar]
- 3.Jain MR, Giri SR, Bhoi B, Trivedi C, Rath A, Rathod R, Ranvir R, Kadam S, Patel H, Swain P, et al. Dual PPAR α/γ agonist saroglitazar improves liver histopathology and biochemistry in experimental NASH models. Liver Int. 2018;38:1084–1094. doi: 10.1111/liv.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong X, Liu H. Honokiol attenuates diet-induced non-alcoholic steatohepatitis by regulating macrophage polarization through activating peroxisome proliferator-activated receptor gamma. J Gastroenterol Hepatol. 2018;33:524–532. doi: 10.1111/jgh.13853. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary NS, Kumar N, Duseja A. Peroxisome proliferator-activated receptors and their agonists in nonalcoholic fatty liver disease. J Clin Exp Hepatol. 2019;9:731–739. doi: 10.1016/j.jceh.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skat-Rordam J, Ipsen DH, Lykkesfeldt J, Tveden-Nyborg P. A role of peroxisome proliferator-activated receptor γ in non-alcoholic fatty liver disease. Basic Clin Pharmacol Toxicol. 2019;124:528–537. doi: 10.1111/bcpt.13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res. 2012;56:1665–1674. doi: 10.1002/mnfr.201200182. [DOI] [PubMed] [Google Scholar]
- 8.Wei J, Feng L, Li Z, Xu G, Fan X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother. 2013;67:387–392. doi: 10.1016/j.biopha.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Dattaroy D, Pourhoseini S, Das S, Alhasson F, Seth RK, Nagarkatti M, Michelotti GA, Diehl AM, Chatterjee S. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2015;308:G298–G312. doi: 10.1152/ajpgi.00346.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loyer X, Paradis V, Henique C, Vion AC, Colnot N, Guerin CL, Devue C, On S, Scetbun J, Romain M, et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut. 2016;65:1882–1894. doi: 10.1136/gutjnl-2014-308883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouchi N, Higuchi A, Ohashi K, Oshima Y, Gokce N, Shibata R, Akasaki Y, Shimono A, Walsh K. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science. 2010;329:454–457. doi: 10.1126/science.1188280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, Zorn AM. Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes. 2008;22:3050–3063. doi: 10.1101/gad.1687308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez-Vidal R, Vega-Badillo J, Reyes-Fermin LM, Hernandez-Perez HA, Sanchez-Munoz F, Lopez-Alvarez GS, Larrieta-Carrasco E, Fernandez-Silva I, Mendez-Sanchez N, Tovar AR, et al. SFRP5 hepatic expression is associated with non-alcoholic liver disease in morbidly obese women. Ann Hepatol. 2015;14:666–674. doi: 10.1016/S1665-2681(19)30761-6. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Zhao X, Liang G, Sun J, Lin Z, Hu R, Chen P, Zhang Z, Zhou L, Li Y. Recombinant SFRP5 protein significantly alleviated intrahepatic inflammation of nonalcoholic steatohepatitis. Nutr Metab (Lond) 2017;14:56. doi: 10.1186/s12986-017-0208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinkiko S, Pang XC, Yuan ZW, Chen SY, Zhu YZ, Xie Y. Combinational applicaton of silybin and tangeretin attenuates the progression of non-alcoholic steatohepatitis (NASH) in mice via modulating lipid metabolism. Pharmacol Res. 2020;151:104519. doi: 10.1016/j.phrs.2019.104519. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu Sh, Fan LH, Zhang XJ, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 18.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals, corp-author. National Academies Press (US); Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals, 8th edition. [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Hu C, Liu D, Zhang Y, Lou G, Huang G, Chen B, Shen X, Gao M, Gong W, Zhou P, et al. LXRα-mediated downregulation of FOXM1 suppresses the proliferation of hepatocellular carcinoma cells. Oncogene. 2014;33:2888–2897. doi: 10.1038/onc.2013.250. [DOI] [PubMed] [Google Scholar]
- 21.Zhong D, Yan Z, Zeng YJ, Gao M, Wu GZ, Hu CJ, Huang G, He FT. MicroRNA-613 represses lipogenesis in HepG2 cells by downregulating LXRα. Lipids Health Dis. 2013;12:32. doi: 10.1186/1476-511X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang W, Menke AL, Driessen A, Koek GH, Lindeman JH, Stoop R, Havekes LM, Kleemann R, van den Hoek AM. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One. 2014;9:e115922. doi: 10.1371/journal.pone.0115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi Y, Fukusato T. Histopathology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2014;20:15539–15548. doi: 10.3748/wjg.v20.i42.15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh M, Ogawa Y, Suganami T. Chronic inflammation as a molecular basis of nonalcoholic steatohepatitis: Role of macrophages and fibroblasts in the liver. Nagoya J Med Sci. 2020;82:391–397. doi: 10.18999/nagjms.82.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smeuninx B, Boslem E, Febbraio MA. Current and future treatments in the fight against non-alcoholic fatty liver disease. Cancers (Basel) 2020;28:1714. doi: 10.3390/cancers12071714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ore A, Akinloye OA. Oxidative stress and antioxidant biomarkers in clinical and experimental models of non-alcoholic fatty liver disease. Medicina (Kaunas) 2019;55:26. doi: 10.3390/medicina55020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penvose A, Keenan JL, Bray D, Ramlall V, Siggers T. Comprehensive study of nuclear receptor DNA binding provides a revised framework for understanding receptor specificity. Nat Commun. 2019;10:2514. doi: 10.1038/s41467-019-10264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 29.Paik JM, Golabi P, Younossi Y, Mishra A, Younossi ZM. Changes in the global burden of chronic liver diseases from 2012 to 2017: The growing impact of NAFLD. Hepatology. 2020;72:1605–1616. doi: 10.1002/hep.31173. [DOI] [PubMed] [Google Scholar]
- 30.Hong F, Xu P, Zhai Y. The opportunities and challenges of peroxisome proliferator-activated receptors ligands in clinical drug discovery and development. Int J Mol Sci. 2018;19:2189. doi: 10.3390/ijms19082189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Alvares K, Huang Q, Rao MS, Reddy JK. Cloning of a new member of the peroxisome proliferator-activated receptor gene family from mouse liver. J Biol Chem. 1993;268:26817–26820. doi: 10.1016/S0021-9258(19)74184-2. [DOI] [PubMed] [Google Scholar]
- 32.Soccio RE, Chen ER, Lazar MA. Thiazolidinediones and the promise of insulin sensitization in type diabetes. Cell Metab. 2014;20:573–591. doi: 10.1016/j.cmet.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu HM, Ni XX, Xu QY, Wang Q, Li XY, Hua J. Regulation of lipid-induced macrophage polarization through modulating peroxisome proliferator-activated receptor-gamma activity a ects hepatic lipid metabolism via a Toll-like receptor 4/NF-B signaling pathway. J Gastroenterol Hepatol. 2020;35:1998–2008. doi: 10.1111/jgh.15025. [DOI] [PubMed] [Google Scholar]
- 34.Nan YM, Fu N, Wu WJ, Liang BL, Wang RQ, Zhao SX, Zhao JM, Yu J. Rosiglitazone prevents nutritional fibrosis and steatohepatitis in mice. Scand J Gastroenterol. 2009;44:358–365. doi: 10.1080/00365520802530861. [DOI] [PubMed] [Google Scholar]
- 35.Neuschwander-Tetri BA, Brunt EM, Wehmeier KR, Oliver D, Bacon BR. Improved nonalcoholic steatohepatitis after 48 weeks of treatment with the PPAR-gamma ligand rosiglitazone. Hepatology. 2003;38:1008–1017. doi: 10.1002/hep.1840380427. [DOI] [PubMed] [Google Scholar]
- 36.Ratziu V, Giral P, Jacqueminet S, Charlotte F, Hartemann-Heurtier A, Serfaty L, Podevin P, Lacorte JM, Bernhardt C, Bruckert E, et al. Rosiglitazone for nonalcoholic steatohepatitis: one-year results of the randomized placebo-controlled fatty liver improvement with rosiglitazone therapy (FLIRT) trial. Gastroenterology. 2008;135:100–110. doi: 10.1053/j.gastro.2008.03.078. [DOI] [PubMed] [Google Scholar]
- 37.Berlie HD, Kalus JS, Jaber LA. Thiazolidinediones and the risk of edema: A meta-analysis. Diabetes Res Clin Pract. 2007;76:279–289. doi: 10.1016/j.diabres.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Singh S, Loke YK, Furberg CD. Thiazolidinediones and heart failure: A teleo-analysis. Diabetes Care. 2007;30:2148–2153. doi: 10.2337/dc07-0141. [DOI] [PubMed] [Google Scholar]
- 39.Loke YK, Kwok CS, Singh S. Comparative cardiovascular effects of thiazolidinediones: Systematic review and meta-analysis of observational studies. BMJ. 2011;342:d1309. doi: 10.1136/bmj.d1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Xiao Y, Wu X, Jiang L, Yang S, Ding Z, Fang Z, Hua H, Kirby MS, Shou J. A circulating microRNA signature as noninvasive diagnostic and prognostic biomarkers for nonalcoholic steatohepatitis. BMC Genomics. 2018;19:188. doi: 10.1186/s12864-018-4575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker PP, Rau M, Schmitt J, Malsch C., Hammer C, Bantel H, Mullhaupt B, Geier A. Performance of serum microRNAs −122, −192 and −21 as biomarkers in patients with non-alcoholic steatohepatitis. PLoS One. 2015;10:e0142661. doi: 10.1371/journal.pone.0142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues PM, Afonso MB, Simao AL, Carvalho CC, Trindade A, Duarte A, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM, Castro RE. miR-21-5p ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017;8:e2748. doi: 10.1038/cddis.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicole W, Kay-Dietrich W. The role of PPARs in disease. Cells. 2020;9:2367. doi: 10.3390/cells9112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji H, Wang H, Zhang F, Li X, Xiang L, Aiguo S. PPARγ agonist pioglitazone inhibits microglia inflammation by blocking p38 mitogen-activated protein kinase signaling pathways. Inflamm Res. 2010;59:921–929. doi: 10.1007/s00011-010-0203-7. [DOI] [PubMed] [Google Scholar]
- 45.Wang D, Shi L, Xin W, Xu J, Xu J, Li Q, Xu Z, Wang J, Wang G, Yao W, et al. Activation of PPARg inhibits pro-inflammatory cytokines production by upregulation of miR-124 in vitro and in vivo. Biochem Biophys Res Commun. 2017;486:726–731. doi: 10.1016/j.bbrc.2017.03.106. [DOI] [PubMed] [Google Scholar]
- 46.Qiu Y, Yang J, Bian S, Guozhu C, Yu J. PPARγ suppresses the proliferation of cardiac myxoma cells through downregulation of MEF2D in a miR-122-dependent manner. Biochem Biophys Res Commun. 2016;474:560–565. doi: 10.1016/j.bbrc.2016.04.112. [DOI] [PubMed] [Google Scholar]
- 47.Liu W, Ji Y, Chu H, Wang M, Yang B, Yin C. SFRP5 mediates downregulation of the wnt5a/caveolin-1/JNK signaling pathway. J Endocrinol. 2020;247:263–272. doi: 10.1530/JOE-20-0328. [DOI] [PubMed] [Google Scholar]
- 48.Zou DP, Chen YM, Zhang LZ, Yuan XH, Zhang YJ, Inggawati A, Nguyet PT, Gao TW, Chen J. SFRP5 inhibits melanin synthesis of melanocytes in vitiligo by suppressing the Wnt/β-catenin signaling. Genes Dis. 2020;8:677–688. doi: 10.1016/j.gendis.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong S, Ji Q, Du Y, Zhu X, Zhu C, Zhou Y. Sfrp5/Wnt pathway: A protective regulatory system in atherosclerotic cardiovascular disease. J Interferon Cytokine Res. 2019;39:472–482. doi: 10.1089/jir.2018.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng J, Hu J, Lian Y, Jiang Y, Chen B. SFRP5 is a target gene transcriptionally regulated by PPARγ in 3T3-L1 adipocytes. Gene. 2018;641:190–195. doi: 10.1016/j.gene.2017.10.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.