Abstract
Background & Aims
Rev-erbα represents a powerful transcriptional repressor involved in immunity. However, the regulation, function, and clinical relevance of Rev-erbα in Helicobacter pylori infection are presently unknown.
Methods
Rev-erbα was examined in gastric samples from H pylori-infected patients and mice. Gastric epithelial cells (GECs) were isolated and infected with H pylori for Rev-erbα regulation assays. Gastric tissues from Rev-erbα–/– and wild-type (littermate control) mice or these mice adoptively transferred with CD4+ T cells from IFN-γ–/– and wild-type mice, bone marrow chimera mice and mice with in vivo pharmacological activation or inhibition of Rev-erbα were examined for bacteria colonization. GECs, CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells and CD4+ T cells were isolated, stimulated and/or cultured for Rev-erbα function assays.
Results
Rev-erbα was increased in gastric mucosa of H pylori-infected patients and mice. H pylori induced GECs to express Rev-erbα via the phosphorylated cagA that activated ERK signaling pathway to mediate NF-κB directly binding to Rev-erbα promoter, which resulted in increased bacteria colonization within gastric mucosa. Mechanistically, Rev-erbα in GECs not only directly suppressed Reg3b and β-defensin-1 expression, which resulted in impaired bactericidal effects against H pylori of these antibacterial proteins in vitro and in vivo; but also directly inhibited chemokine CCL21 expression, which led to decreased gastric influx of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells by CCL21-CCR7-dependent migration and, as a direct consequence, reduced bacterial clearing capacity of H pylori-specific Th1 cell response.
Conclusions
Overall, this study identifies a model involving Rev-erbα, which collectively ensures gastric bacterial persistence by suppressing host gene expression required for local innate and adaptive defense against H pylori.
Keywords: Helicobacter pylori, Rev-erbα, Gastric Epithelial Cells, Host Defense
Abbreviations used in this paper: Ab, antibody; BM, bone marrow; ChIP, chromatin immunoprecipitation; CFU, colony-forming units; DMSO, dimethyl sulfoxide; ELISA, enzyme-linked immunosorbent assay; ERK, extracellular signal-regulated kinase; FACS, fluorescence-activated cell sorter; FBS, fetal bovine serum; GEC, gastric epithelial cell; HRP, horseradish peroxidase; IFN-γ, interferon gamma; IL, interleukin; M, monocyte/macrophage; MOI, multiplicity of infection; mRNA, messenger RNA; NC, nonspecific control small interfering RNA; NF-κB, nuclear factor kappa B; p.i., postinfection; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; rDNA, recombinant DNA; siRNA, small interfering RNA; Th1, T helper type 1; WT, wild-type
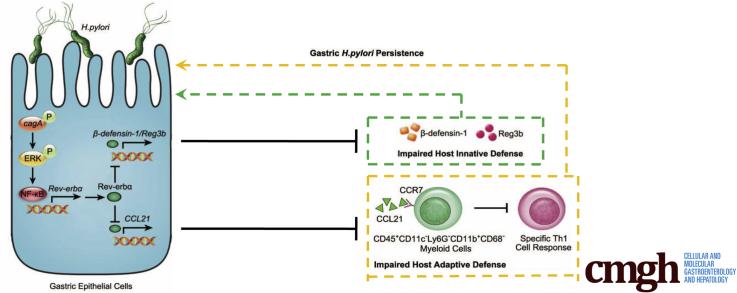
Graphical abstract
Summary.
Helicobacter pylori infection stimulates a novel regulator Rev-erbα that fosters gastric bacterial persistence by suppressing host gene expression required for local innate and adaptive defense against H pylori.
Helicobacter pylori is a human pathogen that infects nearly half of the world’s population and produces a persistent infection that can lead to gastric ulcers and gastric cancer.1 Although the persistent colonization of H pylori in gastric mucosa remains poorly understood, it is believed that the impaired host defense induced by H pylori is a key contributing factor. Gastric epithelial cells (GECs) are not only the first-contacted cell type to exert host defense in gastric mucosa during H pylori infection, but also the targets modulated by H pylori to re-create gastric microenvironment that may favor gastric H pylori persistence. Among the many altered molecules in GECs in response to H pylori infection are the nuclear receptors.2
Rev-erbα (also called nuclear receptor subfamily 1 group D member 1 [NR1D1]), an orphan nuclear receptor, encoded by Rev-erbα and belonged to the thyroid hormone receptor-like superfamily nuclear receptors,3 is part of the clock-keeping machinery4 and plays an important role in regulating metabolism5 and immunity.6 It can physiologically modulate genes involved in lipid,7 bile acid,8 and glucose metabolism9 in liver and adipose tissues. Under noninfectious conditions, Rev-erbα controls excessive inflammatory responses in liver4 or pulmonary10 inflammation. In infectious diseases, Rev-erbα plays roles in antimycobacterial function by repressing interleukin (IL)-10 expression11,12 and has protective effects in vesicular stomatitis virus–induced encephalitis model through the inhibition of CCL2 expression,13 respectively. To date, virtually nothing is known about the regulation, function, and clinical relevance of Rev-erbα in GECs during H pylori infection in either humans or mice.
In the current study, we have, for the first time, demonstrated a procolonization role of Rev-erbα in H pylori infection. Increased Rev-erbα is detected in gastric mucosa of H pylori–infected patients and mice, and Rev-erbα expression is induced in GECs by H pylori in a cagA-dependent manner via extracellular signal-regulated kinase (ERK)–nuclear factor kappa B (NF-κB) pathway activation. On the one hand, Rev-erbα impairs host innate defense to promote gastric H pylori infection by directly suppressing the expression of antibacterial proteins Reg3b and β-defensin-1. On the other hand, Rev-erbα directly inhibits CCL21 production, which in turn reduces CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell chemotaxis, as a direct consequence, impairs H pylori–specific T helper type 1 (Th1) cell response leading to increasing H pylori colonization. Collectively, these data highlight a pathological role for Rev-erbα in persistent H pylori infection.
Results
Rev-erbα Is Increased in Gastric Mucosa of H pylori–Infected Patients and Mice
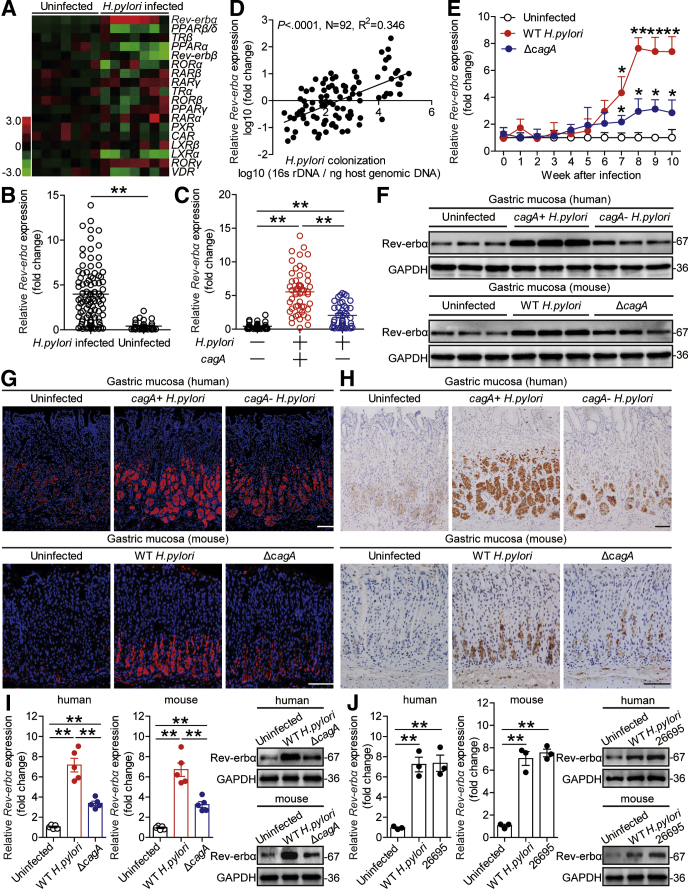
Although Rev-erbα was reported to play antimycobacterial role,11 it is currently not known whether it does also play a role during H pylori infection. To evaluate the potential role of Rev-erbα, we first compared the messenger RNA (mRNA) expression profiles of thyroid hormone receptor-like superfamily nuclear receptors in human primary gastric mucosa of H pylori–infected and uninfected donors. Among them, Rev-erbα was the most increased one in gastric mucosa infected with H pylori compared with paired uninfected counterparts (Figure 1A). We then confirmed that, compared with uninfected donors, the overall Rev-erbα mRNA level was higher in gastric mucosa of H pylori–infected patients (Figure 1B), suggesting an increased Rev-erbα in gastric mucosa of H pylori–infected patients.
Figure 1.
Rev-erbα is increased in gastric mucosa of H pylori–infected patients and mice. (A) The mRNA expression profiles of thyroid hormone receptor-like superfamily nuclear receptors in human primary gastric mucosa of H pylori–infected patients (n = 7) and uninfected donors (n = 7) was analyzed by real-time PCR. (B) Rev-erbα mRNA expression in gastric mucosa of H pylori–infected (n = 92) and uninfected donors (n = 32) was compared. (C) Rev-erbα mRNA expression in gastric mucosa of cagA+H pylori–infected (n = 51), cagA–H pylori–infected (n = 41), and uninfected donors (n = 32) was compared. (D) The correlation between Rev-erbα mRNA expression and H pylori colonization in gastric mucosa of H pylori–infected patients was analyzed with the Pearson r analyze (R2 = 0.3460, P value < .0001) (E) Dynamic changes of Rev-erbα mRNA expression in gastric mucosa of WT H pylori–infected, ΔcagA–infected, and uninfected mice. n = 5 per group per time point in E. (F–H) Rev-erbα protein in gastric corpus of cagA+H pylori–infected, cagA–H pylori–infected, and uninfected donors or in gastric corpus of WT H pylori–infected, ΔcagA-infected, and uninfected mice 8 weeks p.i. was analyzed by (F) Western blot, (G) immunohistochemical staining, and (H) immunofluorescence staining. Scale bars: 100 μm. (I) Rev-erbα mRNA expression and Rev-erbα protein in human or mouse primary gastric mucosa from uninfected donors infected with WT H pylori or ΔcagA ex vivo analyzed by real-time PCR and Western blot (n = 5). (J) Rev-erbα mRNA expression and Rev-erbα protein in human or mouse primary gastric mucosa from uninfected donors infected with WT H pylori or H pylori 26695 ex vivo analyzed by real-time PCR and Western blot (n = 3). ∗P < .05, ∗∗P < .01 for groups connected by horizontal lines, or compared with uninfected mice.
The presence of cagA is strongly associated with the pathogenicity of H pylori.14 Notably, we found that Rev-erbα expression in cagA-positive patients was significantly higher than that in cagA-negative individuals (Figure 1C). Furthermore, a positive correlation between Rev-erbα expression and H pylori colonization in gastric mucosa of H pylori–infected patients (Figure 1D) suggested Rev-erbα induction by H pylori. Consistent with our findings in humans, Rev-erbα expression was also much higher in wild-type (WT) H pylori–infected but not in ΔcagA-infected mice, reaching a peak from 8 weeks postinfection (p.i.) (Figure 1E), indicating a key role for cagA to induce Rev-erbα during H pylori infection in vivo. Furthermore, Western blot analysis (Figure 1F), immunohistochemical staining (Figure 1G) and immunofluorescence staining (Figure 1H) also showed that the level of Rev-erbα protein was higher in gastric mucosa of cagA-positive H pylori–infected patients and WT H pylori–infected mice, compared with either uninfected or cagA-negative patients and ΔcagA-infected counterparts. Furthermore, infection with WT H pylori ex vivo, the levels of Rev-erbα mRNA and protein in human primary gastric mucosa were also significantly increased compared with those in the samples either not infected or infected with ΔcagA (Figure 1I). Similar observations were made when infecting with H pylori 26695 (Figure 1J). Taken together, these findings suggest that Rev-erbα is increased in H pylori–infected gastric mucosa of patients and mice.
GECs Infected by H pylori Express Rev-erbα
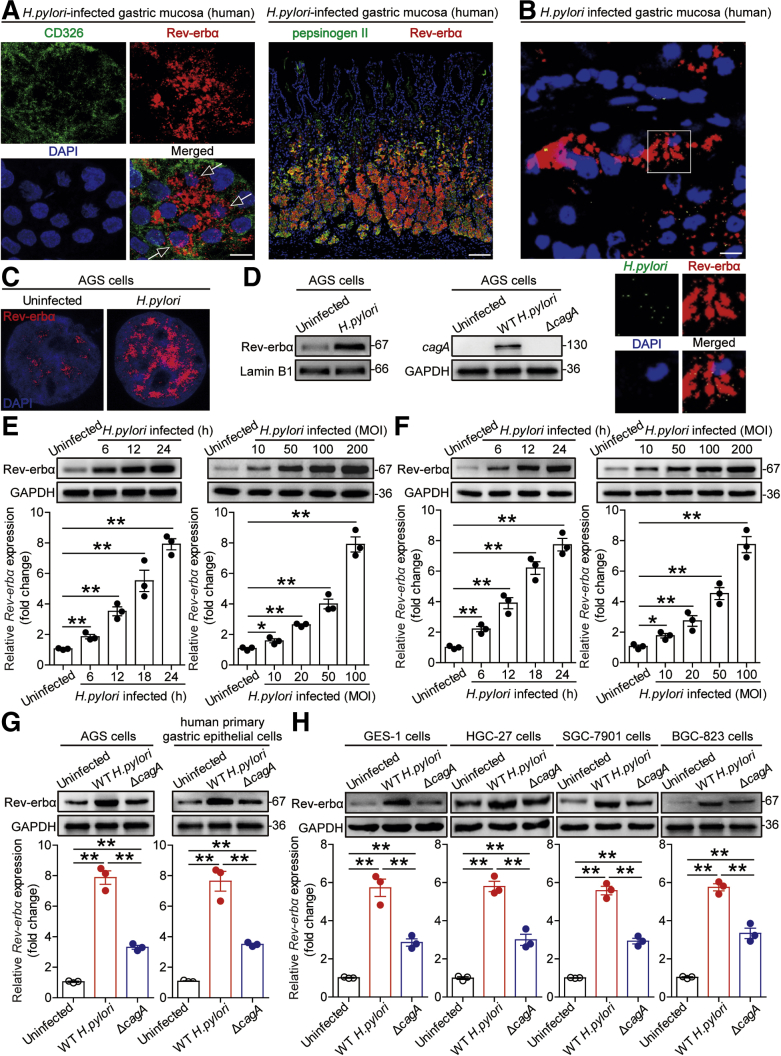
GECs, the first-contacted cells in gastric mucosa during H pylori infection,14 might be responsible for Rev-erbα expression after H pylori infection. Notably, within gastric mucosa of H pylori–infected patients, Rev-erbα was expressed in the cytoplasm and nucleus of CD326+ GECs and in the pepsinogen II+ chief cells in gastric corpus (Figure 2A), suggesting that GECs express Rev-erbα in gastric mucosa during H pylori infection. We also found that H pylori was in contact to the Rev-erbα–expressing cells in gastric mucosa of H pylori–infected patients (Figure 2B).
Figure 2.
GECs express Rev-erbα during H pylori infection. (A) Representative immunofluorescence staining images showing Rev-erbα expression (red) in cytoplasm and nucleus (arrow) of CD326+ cells (green) and Rev-erbα–expressing (red) pepsinogen II+ chief cells (green) in gastric corpus of H pylori–infected patients. Scale bars: 10 μm or 100 μm. (B) Representative immunofluorescence staining images showing Rev-erbα–expressing (red) cells and H pylori (green) colonization in gastric mucosa of H pylori–infected patients. Scale bars: 10 μm. (C) Representative immunofluorescence staining images showing Rev-erbα expression (red) in WT H pylori–infected, and uninfected AGS cells (MOI = 100, 6 hours). Scale bars: 1 μm. (D) Rev-erbα expression in the nucleus of WT H pylori–infected, and uninfected AGS cells (MOI = 100, 24 hours) was analyzed by Western blot. The expression of cagA protein in AGS cells infected with WT H pylori or ΔcagA (MOI = 100, 24 hours) was analyzed by Western blot. (E, F) Rev-erbα mRNA expression and Rev-erbα protein in WT H pylori–infected and (E) uninfected AGS cells or (F) human primary GECs at different time point (MOI = 100) or with different MOI (24 hours) were analyzed by real-time PCR or Western blot (n = 3). (G) Rev-erbα mRNA expression and Rev-erbα protein in WT H pylori–infected, ΔcagA-infected, and uninfected AGS cells or human primary GECs (MOI = 100, 24 hours) were analyzed by real-time PCR and Western blot (n = 3). (H) Rev-erbα expression in WT H pylori–infected, ΔcagA-infected, and uninfected GES-1 cells, HGC-27 cells, SGC-7901 cells, and BGC-823 cells (MOI = 100, 24 hours) was analyzed by real-time PCR and Western blot (n = 3). ∗P < .05, ∗∗P < .01 for groups connected by horizontal lines.
Next, to explore the Rev-erbα induction in GECs, we confirmed increased Rev-erbα expression in the nucleus of AGS cells, the cells used to investigate the effects of H pylori infection on GECs,15 infected with H pylori (Figure 2C and D). We further demonstrated that H pylori–infected AGS cells (Figure 2E) and human primary GECs (Figure 2F) increased Rev-erbα expression in a time-dependent and infection dose-dependent manner. Notably, compared with uninfected or ΔcagA-infected, WT H pylori–infected AGS cells and human primary GECs potently increased Rev-erbα expression (Figure 2G). Similar observations were made when other human GEC lines were infected with H pylori (Figure 2H). Collectively, these results demonstrate that H pylori infection induces Rev-erbα expression in GECs.
H pylori Induces GECs to Express Rev-erbα via ERK-NF-κB Pathway
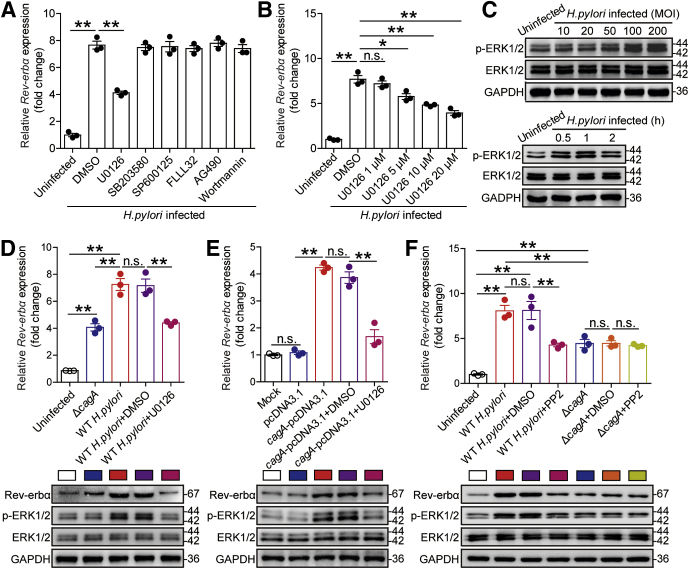
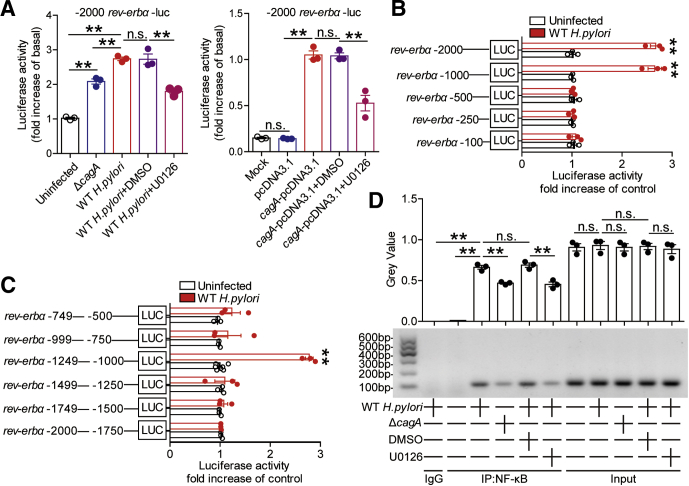
To further explore which signaling pathways might operate in the induction of Rev-erbα in GECs by H pylori, we first pretreated AGS cells with corresponding pathway inhibitors and then infected them with H pylori. The results showed that only blocking the signal transduction of ERK pathway with inhibitor U0126 effectively decreased Rev-erbα expression (Figure 3A and B). Furthermore, ERK1/2, a direct ERK pathway downstream substrate, was phosphorylated in the infected AGS cells in an infection dose-dependent and time-dependent manner (Figure 3C), which was abolished when pretreated with inhibitor U0126 (Figure 3D). Next, to confirm whether cagA could induce Rev-erbα via ERK pathway, we infected AGS cells with WT H pylori or ΔcagA, or transfected AGS cells with cagA-pcDNA3.1. We found increased Rev-erbα mRNA and protein and increased ERK1/2 phosphorylation in AGS cells either infected with WT H pylori or transfected with cagA-pcDNA3.1 compared with those infected with ΔcagA or transfected with the vector (pcDNA3.1) (Figure 3D–F). Importantly, these were abrogated by pretreatment with the ERK pathway inhibitor U0126 or with cagA EPIYA motif phosphorylation inhibitor PP2 (Figure 3D–F).16 To further investigate the effect of H pylori on Rev-erbα gene transcription, we synthesized a series of Rev-erbα-luc promoter constructs and cloned them into pGL3-basic vectors. Compared with ΔcagA or pcDNA3.1, WT H pylori infection or cagA-pcDNA3.1 transfection enhanced luciferase activity in AGS cells transfected with full length (–2000/0) Rev-erbα-luc promoter construct plasmid, which was abolished when pretreatment with the ERK pathway inhibitor U0126 (Figure 4A). Interestingly, WT H pylori infection only enhanced luciferase activity in cells transfected with the Rev-erbα-luc promoter construct flanking regions –2000 to –500 (Figure 4B). Further detailed luciferase reporter assay revealed that only the Rev-erbα promoter construct flanking regions –1249 to –1000 produced an increase in cellular luciferase activity upon WT H pylori infection (Figure 4C). The PROMO tool V.8.3 of TRANSFAC (Beverly, MA) showed that Rev-erbα promoter (–1249/–1000) contains a NF-κB binding site (comprising the sequence: GGGAAATGAC). Subsequently, chromatin immunoprecipitation (ChIP) assay showed that, compared with no infection or ΔcagA infection, WT H pylori infection significantly increased binding to the Rev-erbα promoter in AGS cells, which was abolished when pretreated with the ERK pathway inhibitor U0126 (Figure 4D). Taken together, these findings clearly demonstrate that cagA-mediated ERK signaling pathway activation modulates NF-κB’s transcriptional regulation on Rev-erbα expression in GECs during H pylori infection.
Figure 3.
H pylori induces Rev-erbα expression via the phosphorylated cagA-activated ERK signaling pathway. (A, B) AGS cells were pretreated with signal pathway inhibitors and then stimulated with WT H pylori (MOI = 100) for 24 hours. Rev-erbα mRNA expression in AGS cells was compared (n = 3). (C) AGS cells were stimulated with WT H pylori with different MOI (6 hours) or at different time point (MOI = 100). ERK1/2 and p-ERK1/2 proteins were analyzed by Western blot. (D) AGS cells were pretreated with or without U0126 and then infected with WT H pylori or ΔcagA (MOI = 100) for 24 hours. Rev-erbα mRNA expression in these cells was compared (n = 3). Rev-erbα, ERK1/2, and p-ERK1/2 proteins were analyzed by Western blot. (E) AGS cells were transfected with plasmids pcDNA3.1 or cagA-pcDNA3.1 for 24 hours, then treated with or without U0126 for 2 hours and cultured for an additional 24 hours. Rev-erbα mRNA expression in these cells was compared (n = 3). Rev-erbα, ERK1/2, and p-ERK1/2 proteins were analyzed by Western blot. (F) AGS cells were pretreated with or without PP2 and then infected with WT H pylori or ΔcagA (MOI = 100) for 24 hours. Rev-erbα mRNA expression in these cells was compared (n = 3). Rev-erbα, ERK1/2 and p-ERK1/2 proteins were analyzed by Western blot. ∗P < .05, ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
Figure 4.
H pylori induces Rev-erbα expression via ERK-NF-κB pathway. (A) AGS cells were transfected with luciferase reporter constructs containing the Rev-erbα-luc promoter for 4 hours. Luciferase activity was measured to assess promoter activity after WT H pylori (pretreated with or without U0126) or ΔcagA infection (MOI = 100) for 24 hours (n = 3). AGS cells were cotransfected with Rev-erbα-luc construct and cagA-pcDNA3.1 (pretreated with or without U0126) or pcDNA3.1 (control vector) for 48 hours. Luciferase activity was measured to assess Rev-erbα promoter activity (n = 3). (B, C) AGS cells were transfected with Rev-erbα-luc constructs for 4 hours. Luciferase activity was measured to assess promoter activity after WT H pylori infection (MOI = 100) for 24 hours (n = 3). (D) Representative data and statistical analysis of ChIP assay in AGS cells infected with WT H pylori (pretreated with or without U0126) or ΔcagA, followed by regular PCR with primers designed for NF-κB binding site of Rev-erbα promoter region (n = 3). ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines, or compared with uninfected cells.
Rev-erbα Suppresses Antibacterial Proteins Reg3b and β-Defensin-1, Leading to Increased Bacterial Burden in Gastric Mucosa During H pylori Infection
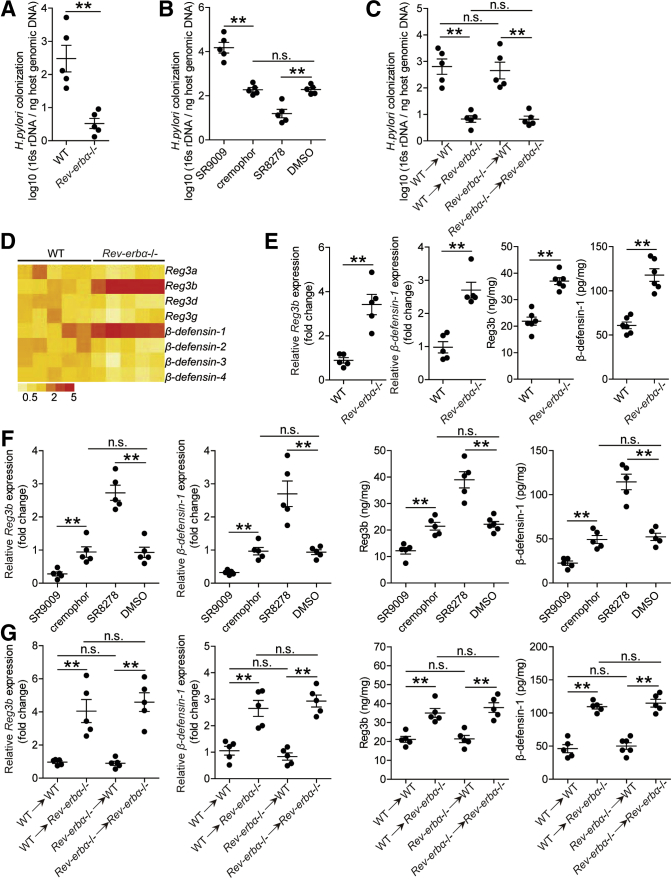
To evaluate the possible biological effects of Rev-erbα in H pylori–associated pathogenesis in vivo, we compared the levels of bacterial colonization in gastric mucosa 8 weeks p.i. in WT and Rev-erbα–/– mice, and found that lacking endogenous Rev-erbα in Rev-erbα–/– mice effectively reduced H pylori colonization when compared with that in WT mice (Figure 5A). Next, we again compared bacterial colonization in mice with in vivo pharmacological activation or inhibition of Rev-erbα by injecting exogenous Rev-erbα agonist or Rev-erbα antagonist and found that activation of Rev-erbα significantly increased H pylori colonization; conversely, inhibition of Rev-erbα significantly reduced gastric colonization when compared with that in control mice (Figure 5B). Finally, consistent with our previous findings that GECs are the source cells that express Rev-erbα in gastric mucosa during H pylori infection (Figure 2), by generating bone marrow (BM) chimera mice, we found that non–BM-derived Rev-erbα–expressing cells (including GECs) largely contribute to increasing bacterial colonization in this model (Figure 5C). Taken together, our data demonstrate that Rev-erbα plays an essential role in promoting bacterial colonization in vivo.
Figure 5.
Rev-erbα suppresses antibacterial proteins Reg3b and β-defensin-1 and increases bacterial burden in gastric mucosa during H pylori infection. (A–C) The bacteria colonization in (A) gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice, (B) gastric mucosa of WT H pylori–infected mice with in vivo by injecting Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control, or (C) gastric mucosa of WT H pylori–infected BM chimera mice was compared 8 weeks p.i. (n = 5). (D) The mRNA expression profiles of β-defensins and Reg3 proteins in gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice 8 weeks p.i. was analyzed by real-time PCR (n = 5). (E–G) The mRNA expression or the concentrations of Reg3b and β-defensin-1 in (E) gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice, (F) gastric mucosa of WT H pylori–infected mice with in vivo by injecting Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control, or (G) gastric mucosa of WT H pylori–infected BM chimera mice was compared 8 weeks p.i. (n = 5). ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
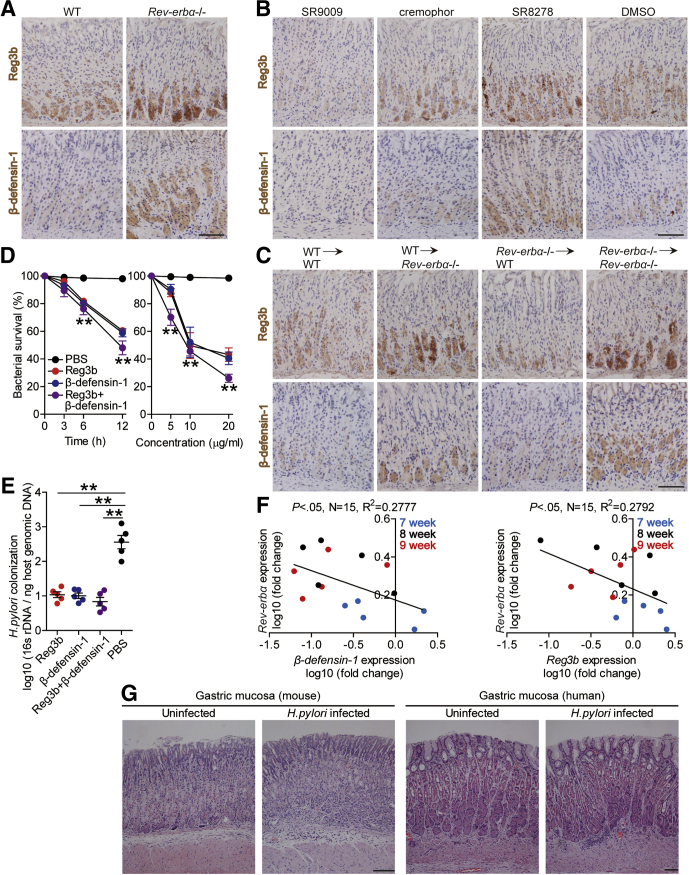
Given powerful transcriptional repressing effects of Rev-erbα was observed in other infectious diseases,11,13 we hypothesized that Rev-erbα might exert inhibiting effects on genes encoding proteins contributing to host defense. The β-defensins17 and Reg3 proteins18 play roles in mucosal defense during H pylori infection. We therefore screened these proteins in gastric mucosa 8 weeks p.i. in WT and Rev-erbα–/– mice, and found that lacking Rev-erbα in Rev-erbα–/– mice led to increased Reg3b and β-defensin-1 expression when compared with that in WT mice (Figure 5D and E). Similar observations were made when using in vivo pharmacological activation or inhibition of Rev-erbα experiments (Figure 5F). Again, BM chimera experiments confirmed that non–BM-derived Rev-erbα–expressing cells were largely responsible for inhibiting Reg3b and β-defensin-1 in gastric mucosa during H pylori infection (Figure 5G). Furthermore, similar observations were made when analyzing Reg3b and β-defensin-1 protein by immunohistochemical staining (Figure 6A–C).
Figure 6.
Antibacterial proteins Reg3b and β-defensin-1 exerted killing activity against H pylori in vitro and in vivo. (A–C) Representative immunohistochemical staining images showing Reg3b or β-defensin-1 expression (brown) in (A) gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice, (B) gastric mucosa of WT H pylori–infected mice injected with Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control, or (C) or in gastric mucosa of WT H pylori–infected BM chimera mice 8 weeks p.i.. Scale bars: 100 μm. (D, E) In vitro and in vivo bactericidal assay was performed as described in the Materials and Methods and statistically analyzed (n = 3). (F) The correlation between Rev-erbα expression and Reg3b or β-defensin-1 expression in gastric mucosa of WT H pylori–infected WT mice 7, 8, and 9 weeks p.i. was analyzed with the Pearson r analyzer (R2 = 0.2777 and 0.2792, P value < .05). (G) Representative hematoxylin and eosin staining images showed the histology of gastric mucosa of uninfected and H pylori–infected human and mice. Scale bars: 100 μm. ∗∗P < .01, for groups connected by horizontal lines, or compared with samples treated with PBS.
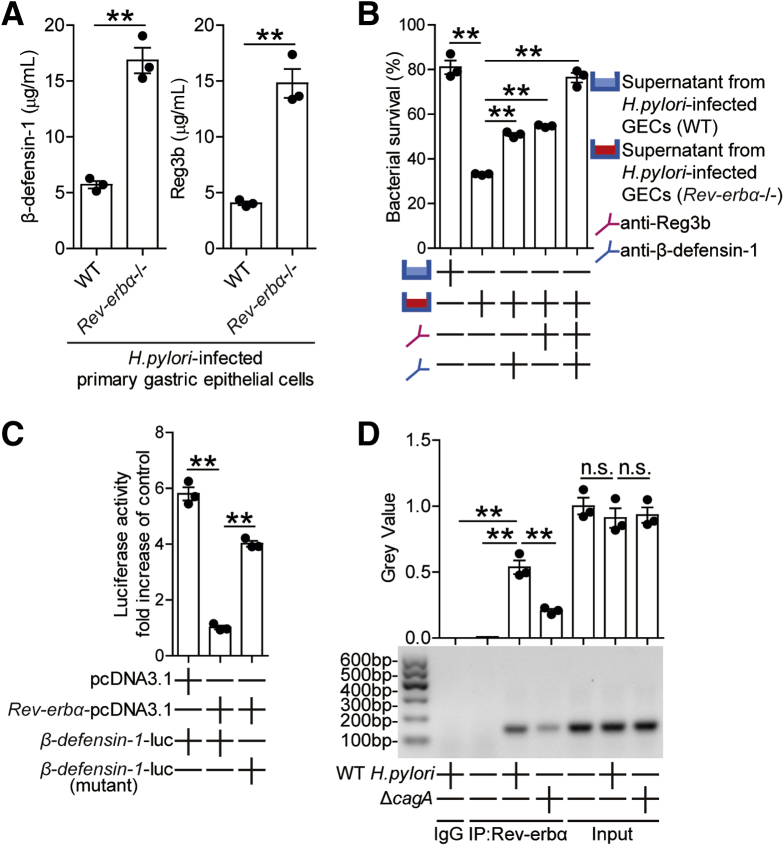
To further investigate the roles of Reg3b and β-defensin-1 in H pylori infection, we performed in vitro and in vivo bactericidal assay. First, in vitro bactericidal assay showed that Reg3b and β-defensin-1 exerted killing activity against H pylori in a time-dependent and infection dose-dependent manner (Figure 6D). Next, in vivo gain-of-function experiments showed that Reg3b or β-defensin-1 administration significantly reduced H pylori colonization in gastric mucosa of WT mice (Figure 6E). As for the negative correlations between Rev-erbα and Reg3b/β-defensin-1 (Figure 6F) in gastric mucosa of H pylori–infected mice and host defense against H pylori from GEC-derived antibacterial proteins,17 we next found that, upon H pylori infection, GECs derived from Rev-erbα–/– mice produced more Reg3b and β-defensin-1 compared with those from WT mice (Figure 7A), and that supernatants from H pylori–infected GECs of Rev-erbα–/– mice exerted more pronounced killing activity against H pylori than those of WT mice (Figure 7B), which could be abrogated by blocking Reg3b or β-defensin-1 (Figure 7B). Finally, to explore the molecule mechanism of β-defensin-1 inhibition in GECs by Rev-erbα (human have no Reg3b gene), we performed luciferase reporter assays, and the results showed decreased activity of β-defensin-1-luc when transfected together with the Rev-erbα-pcDNA3.1, and retrieved activity when the Rev-erbα binding site on the β-defensin-1 promoter was mutated (Figure 7C); further ChIP assays showed that WT H pylori infection significantly increased binding activity to the β-defensin-1 promoter with the Rev-erbα antibodies (Abs) (Figure 7D), together suggesting that in WT H pylori–infected AGS cells, Rev-erbα directly inhibited β-defensin-1 expression. Taken together, our data demonstrate that, in H pylori–infected gastric mucosa, Rev-erbα in GECs inhibits Reg3b and β-defensin-1 directly, which likely contributes to bacterial persistence.
Figure 7.
Rev-erbα in GECs inhibits Reg3b and β-defensin-1 directly, which contributes to H pylori persistence. (A) Primary gastric epithelial cells from uninfected Rev-erbα–/– and WT mice were stimulated with WT H pylori (MOI = 100) for 24 hours. Reg3b and β-defensin-1 production was measured in cell culture supernatants by ELISA (n = 3). (B) In vitro bactericidal assay was performed as described in the Materials and Methods and statistically analyzed (n = 3). (C) AGS cells were cotransfected with β-defensin-1-luc construct or a mutant construct and Rev-erbα-pcDNA3.1 or pcDNA3.1 (control vector) for 48 hours. Luciferase activity was measured to assess β-defensin-1 promoter activity (n = 3). (D) Representative data and statistical analysis of chromatin immunoprecipitation assay in AGS cells infected with WT H pylori or ΔcagA, followed by regular PCR with primers designed for Rev-erbα binding site of β-defensin-1 promoter region (n = 3). ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
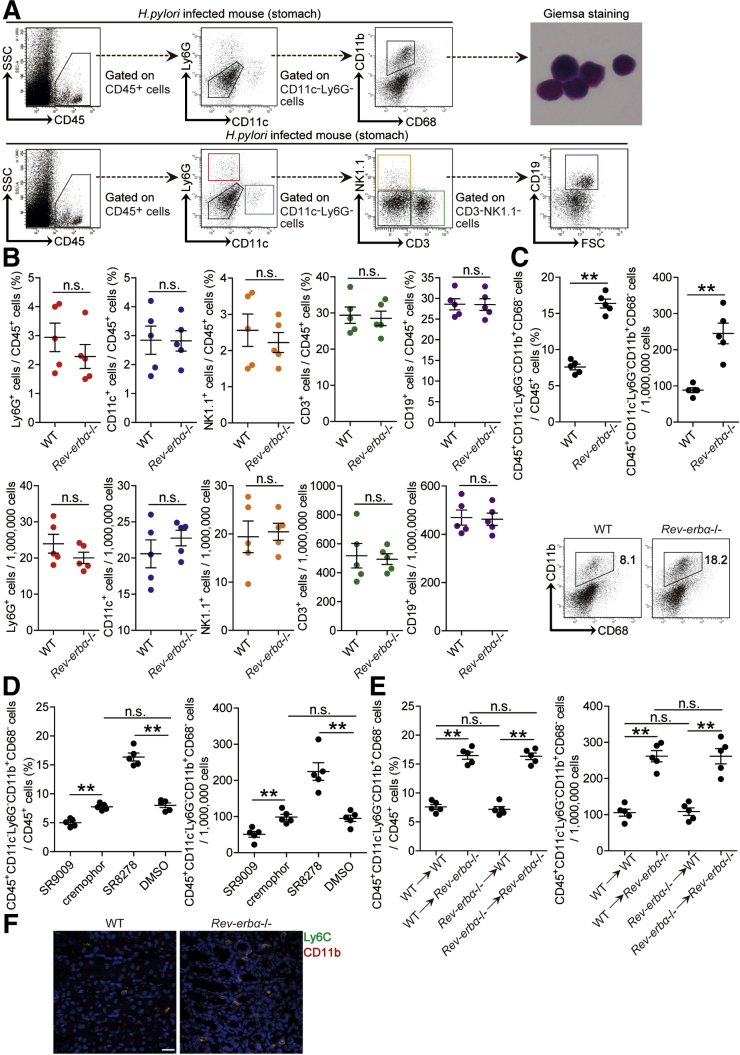
Rev-erbα Inhibits CD45+CD11c–Ly6G–CD11b+CD68– Myeloid Cell Accumulation and CCL21 Production in Gastric Mucosa During H pylori Infection
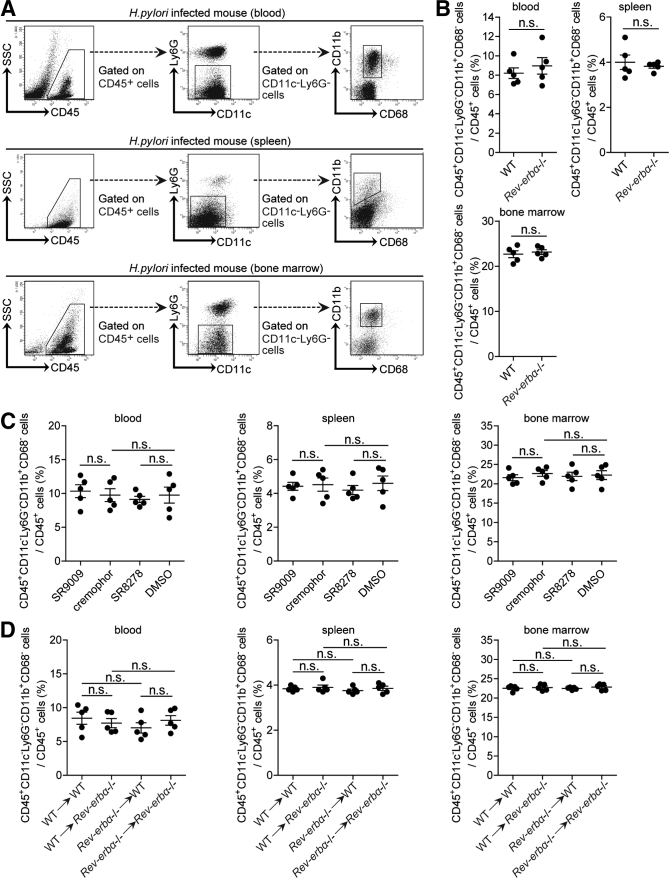
To investigate whether increased Rev-erbα regulated immune cell infiltration into the gastric mucosa during H pylori infection, we compared the levels of CD45+CD11c+Ly6G– dendritic cells, CD45+CD11c–Ly6G+ neutrophils, CD45+CD11c–Ly6G–CD3+NK1.1– T cells, CD45+CD11c–Ly6G–CD3–NK1.1+ natural killer cells, CD45+CD11c–Ly6G–CD3–NK1.1–CD19+ B cells, and CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells in gastric mucosa 8 weeks p.i. in WT and Rev-erbα–/– mice, and found that lacking Rev-erbα in Rev-erbα–/– mice only led to increased CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells (Figure 8A–C). These results were also confirmed by our in vivo pharmacological activation or inhibition of Rev-erbα experiments and BM chimera experiments in which non–BM-derived Rev-erbα–expressing cells were largely responsible for this myeloid cell reduction in gastric mucosa during H pylori infection (Figure 8D and E). Similar observations were made when analyzing the number of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells per million total cells in gastric mucosa (Figure 8C–E). Notably, compared with WT mice, Ly6C+CD11b+ myeloid cell accumulation was higher in gastric mucosa of H pylori–infected Rev-erbα–/– mice 8 weeks p.i. (Figure 8F). Furthermore, blood, spleen, or BM CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell levels were not altered in these mouse models above (Figure 9). Taken together, our data demonstrate that Rev-erbα plays an essential role in the inhibition of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell accumulation in gastric mucosa during H pylori infection.
Figure 8.
Rev-erbα inhibits CD45+CD11c–Ly6G–CD11b+CD68–myeloid cell accumulation in gastric mucosa during H pylori infection. (A) Representative dot plots of CD45+CD11c+Ly6G– dendritic cells, CD45+CD11c–Ly6G+ neutrophils, CD45+CD11c–Ly6G–CD3+NK1.1– T cells, CD45+CD11c–Ly6G–CD3–NK1.1+ natural killer cells, CD45+CD11c–Ly6G–CD3–NK1.1–CD19+ B cells, and CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells by gating on CD45+ cells in gastric mucosa of WT H pylori–infected mice 8 weeks p.i.. Representative Giemsa staining images showing FACS-sorted CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells in gastric mucosa of WT H pylori–infected WT mice 8 weeks p.i.. (B) The levels or numbers of CD45+CD11c+Ly6G– dendritic cells, CD45+CD11c–Ly6G+ neutrophils, CD45+CD11c–Ly6G–CD3+NK1.1– T cells, CD45+CD11c–Ly6G–CD3–NK1.1+ natural killer cells, CD45+CD11c–Ly6G–CD3–NK1.1–CD19+ B cells, and CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells in gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice were compared 8 weeks p.i. (n = 5). (C–E) CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell levels or CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell numbers in (C) gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice, (D) gastric mucosa of WT H pylori–infected mice injected with Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control, or (E) gastric mucosa of WT H pylori–infected BM chimera mice were compared 8 weeks p.i. (n = 5). (F) Representative immunofluorescence staining images showed Ly6C+CD11b+ myeloid cell infiltration in gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice 8 week p.i.. Scale bars: 20 μm. ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
Figure 9.
Rev-erbα has no effects on blood, spleen, or BM CD45+CD11c–Ly6G–CD11b+CD68–myeloid cells during H pylori infection. (A) Representative dot plots of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells by gating on CD45+ cells in peripheral blood, spleen or BM of WT H pylori–infected WT mice 8 weeks p.i.. (B–D) The levels of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells in (B) peripheral blood, spleen, or BM of WT H pylori–infected WT and Rev-erbα–/– mice; (C) gastric mucosa of WT H pylori–infected mice injected with Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control; or (D) gastric mucosa of WT H pylori–infected BM chimera mice were compared 8 weeks p.i. (n = 5). n.s. P > .05 for groups connected by horizontal lines.
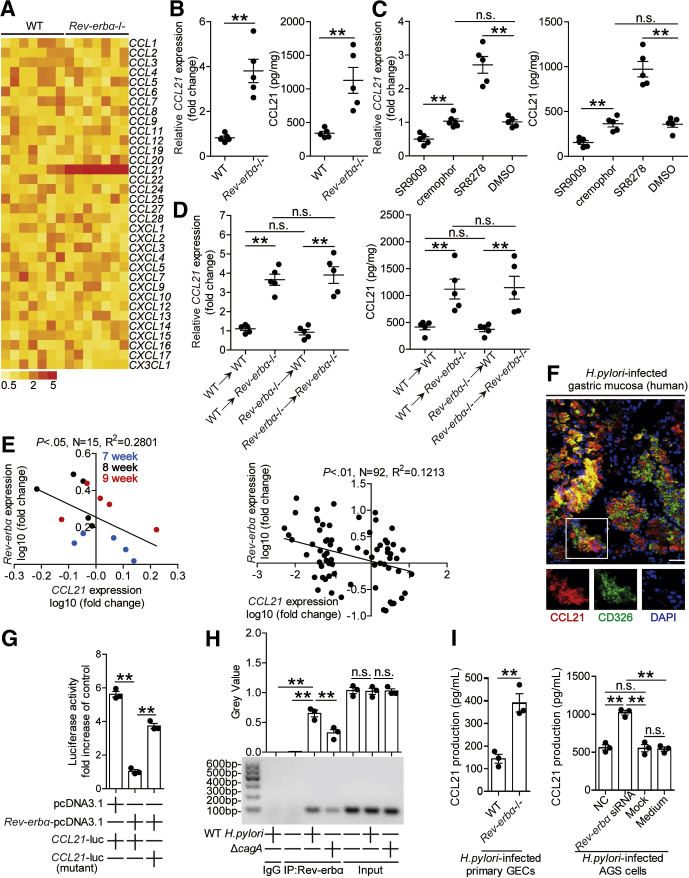
Chemotaxis plays important roles in immune cell migration. We were therefore interested to know if Rev-erbα regulated chemokine production in gastric mucosa. We screened chemokines in gastric mucosa 8 weeks p.i. in WT and Rev-erbα–/– mice, and found that only CCL21 expression was increased in Rev-erbα–/– mice (Figure 10A and B). Again, these results were confirmed by our in vivo pharmacological activation or inhibition of Rev-erbα experiments and BM chimera experiments in which non–BM-derived Rev-erbα–expressing cells were largely responsible for CCL21 inhibition in gastric mucosa during H pylori infection (Figure 10C and D). Taken together, our data demonstrate that Rev-erbα plays an essential role in inhibiting CCL21 production in gastric mucosa during H pylori infection.
Figure 10.
Rev-erbα inhibits CCL21 in gastric mucosa during H pylori infection. (A) The mRNA expression profiles of chemokines in gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice 8 weeks p.i. was analyzed by real-time PCR (n = 5). (B–D) The mRNA expression or the concentrations of CCL21 in (B) gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice, (C) gastric mucosa of WT H pylori–infected mice injected with Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control, or (D) gastric mucosa of WT H pylori–infected BM chimera mice were compared 8 weeks p.i. (n = 5). (E) The correlation between Rev-erbα expression and CCL21 expression in gastric mucosa of WT H pylori–infected WT mice 7, 8, and 9 weeks p.i. or in gastric mucosa of H pylori–infected patients was analyzed. (F) Representative immunofluorescence staining images showing CCL21-expressing (red) CD326+ cells (green) in gastric mucosa of H pylori–infected patients. Scale bars: 20 μm. (G) AGS cells were cotransfected with CCL21-luc construct or a mutant construct and Rev-erbα-pcDNA3.1 or pcDNA3.1 (control vector) for 48 hours. Luciferase activity was measured to assess CCL21 promoter activity (n = 3). (H) Representative data and statistical analysis of chromatin immunoprecipitation assay in AGS cells infected with WT H pylori or ΔcagA, followed by regular PCR with primers designed for Rev-erbα binding site of CCL21 promoter region (n = 3). (I) Primary GECs from uninfected Rev-erbα–/– and WT mice, and Rev-erbα siRNA, NC, or lipo2000-only (Mock) pretreated AGS cells or AGS cells without treatment (medium) were infected with WT H pylori (MOI = 100) for 24 hours. CCL21 production was measured in cell culture supernatants by ELISA (n = 3). ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
To understand the negative correlation between Rev-erbα and CCL21 (Figure 10E) and CCL21 expression in CD326+ cells (Figure 10F) in gastric mucosa of H pylori–infected patients and mice, we then explored the molecular mechanism by which Rev-erbα down-modulate CCL21 expression in GECs. Luciferase reporter assay showed decreased activity of CCL21-luc when transfected together with the Rev-erbα-pcDNA3.1, and retrieved activity when the Rev-erbα binding site on the CCL21 promoter was mutated (Figure 10G); further ChIP assays showed that WT H pylori infection significantly increased binding activity to the CCL21 promoter by the Rev-erbα Abs (Figure 10H), together suggesting that in WT H pylori–infected AGS cells, Rev-erbα directly inhibited CCL21 expression. Finally, CCL21 production from AGS cells as well as from mouse primary GECs was negatively regulated in a Rev-erbα–dependent manner (Figure 10I). Taken together, our data demonstrate that Rev-erbα inhibits CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell accumulation and CCL21 production in gastric mucosa during H pylori infection.
Rev-erbα Inhibits CD45+CD11c–Ly6G–CD11b+CD68– Myeloid Cell Accumulation In Vivo and Migration In Vitro During H pylori Infection via CCL21-CCR7 Axis
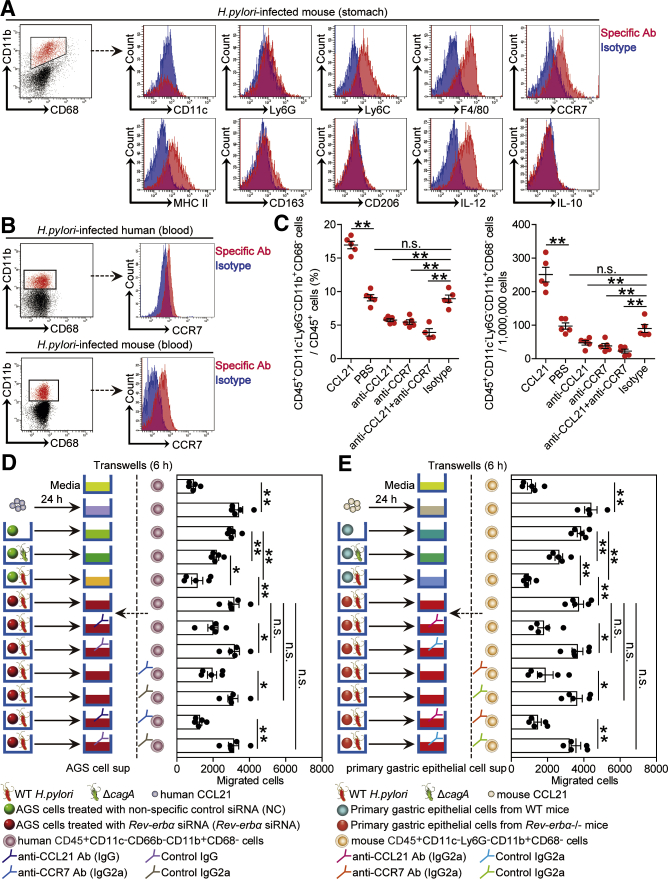
Next, we tried to determine whether CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell migration and accumulation during H pylori infection was regulated by the Rev-erbα-CCL21 axis. We first showed that CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells expressed high levels of the monocyte/macrophage (M)-associated markers Ly6C and F4/80, M1-polarized marker major histocompatibility complex II, and interferon gamma (IFN-γ)–producing T cell function–associated cytokine IL-12, but expressed little M2-polarized markers CD163 and CD206, or monocyte-derived myeloid-derived suppressor cells–associated cytokine IL-10 (Figure 11A), and exhibited a monolobar nucleus characteristic of monocyte/macrophage by Giemsa staining (Figure 8A). Next, gastric (Figure 11A) or blood (Figure 11B) CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells from H pylori–infected humans and mice showed higher expression of CCR7, the chemokine receptor of CCL21. Finally, we conducted a series of loss- and gain-of-function experiments in vivo involving CCL21 or CCR7, and evaluated the CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells in gastric mucosa 8 weeks p.i. CCL21 administration significantly increased CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell accumulation; conversely, neutralization of CCL21 or CCR7 significantly reduced CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell accumulation in gastric mucosa (Figure 11C).
Figure 11.
Rev-erbα inhibits CD45+CD11c–Ly6G–CD11b+CD68–myeloid cell accumulation in vivo and migration in vitro during H pylori infection via CCL21-CCR7 axis. (A) Expression of CD11c, Ly6G, Ly6C, F4/80, CCR7, MHC II, CD163, CD206, IL-12, and IL-10 on/in gastric CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells from WT H pylori–infected WT mice 8 weeks p.i.. Red histograms represent staining of the molecules of interest; blue histograms represent isotype control. (B) Representative dot plots of CCR7 expression on blood CD45+CD11c–CD66b–CD11b+CD68– myeloid cells in blood of H pylori–infected patients or CCR7 expression on CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells of WT H pylori–infected mice 8 weeks p.i.. (C) CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell levels or numbers in gastric mucosa of WT H pylori–infected mice injected with CCL21 or PBS control, or Abs against CCL21 or CCR7 or corresponding isotype control Ab were compared 8 weeks p.i. (n = 5). (D, E) Human CD45+CD11c–CD66b–CD11b+CD68– myeloid cell migration and mouse CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell migration were assessed via a transwell assays as described in the Materials and Methods and statistically analyzed (n = 5). ∗P < .05, ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
To further evaluate the contribution of a Rev-erbα-CCL21-CCR7 axis to the migration of these monocyte-like myeloid cells in vitro, human CD45+CD11c–CD66b–CD11b+CD68– myeloid cell chemotaxis assays were performed. It was demonstrated that culture supernatants from ΔcagA-infected AGS cells pretreated with nonspecific control small interfering RNA (NC) or from WT H pylori–infected AGS cells pretreated with Rev-erbα siRNA induced significantly more CD45+CD11c–CD66b–CD11b+CD68– myeloid cell migration than those from WT H pylori–infected AGS cells pretreated with NC; and this effect was lost upon pretreatment with neutralizing Abs against CCL21 or CCR7 (Figure 11D).
Similarly, culture supernatant from ΔcagA-infected primary GECs of WT mice or from WT H pylori–infected primary GECs of Rev-erbα–/– mice also induced significantly more mouse CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell migration than those from WT H pylori–infected primary GECs of WT mice; and this effect was also lost upon pretreatment with neutralizing Abs against CCL21 or CCR7 (Figure 11E). Collectively, these results therefore suggest that a Rev-erbα-CCL21-CCR7 axis contributes to CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell accumulation within gastric mucosa during H pylori infection.
Rev-erbα Impairs Specific Th1 Cell Response to Promote Bacterial Colonization in Gastric Mucosa During H pylori Infection
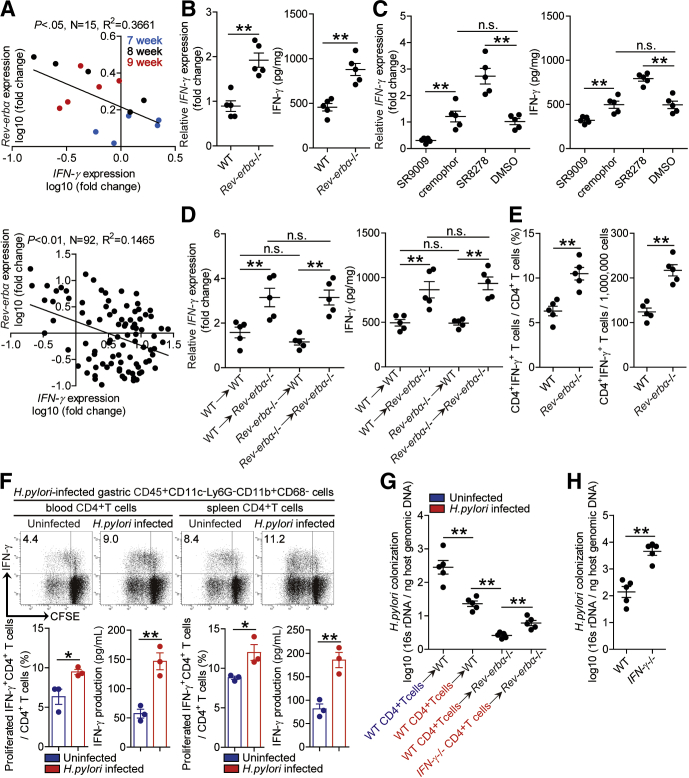
Th1 cell response and its effector IFN-γ play key roles in control H pylori infection.19,20 First, there were negative correlations between Rev-erbα and IFN-γ in gastric mucosa of H pylori–infected patients and mice (Figure 12A), and IFN-γ was increased in gastric mucosa 8 weeks p.i. in Rev-erbα–/– mice when compared with that in WT mice (Figure 12B). Again, these results were confirmed by our in vivo pharmacological activation or inhibition of Rev-erbα experiments and BM chimera experiments in which non–BM-derived Rev-erbα–expressing cells were largely responsible for IFN-γ inhibition in gastric mucosa during H pylori infection (Figure 12C and D). Next, Th1 cell response level was also increased in Rev-erbα–/– mice (Figure 12E). Notably, myeloid cell/T cell co-cultures showed that gastric CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells from H pylori–infected mice induced more Th1 cells from blood or spleen CD4+T cells of H pylori–infected mice to proliferate and produce IFN-γ than those from uninfected counterparts (Figure 12F), suggesting a promoting effect of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells on antigen-specific Th1 cell response during H pylori infection.
Figure 12.
Rev-erbα impairs specific Th1 cell response to promote bacterial colonization in gastric mucosa during H pylori infection. (A) The correlation between Rev-erbα expression and IFN-γ expression in gastric mucosa of WT H pylori–infected WT mice 7, 8, and 9 weeks p.i. or in gastric mucosa of H pylori–infected patients was analyzed with the Pearson r analyze (R2 = 0.3661 and 0.1485, P value < .05 and < .01). (B–D) The mRNA expression and the concentrations of IFN-γ in (B) gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice, (C) gastric mucosa of WT H pylori–infected mice injected with Rev-erbα agonist SR9009 or cremophor control, or Rev-erbα antagonist SR8278 or DMSO control, or (D) gastric mucosa of WT H pylori–infected BM chimera mice were compared 8 weeks p.i. (n = 5). (E) CD4+IFN-γ+ T cell levels or numbers in gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice were compared 8 weeks p.i. (n = 5). (F) Carboxyfluorescein succinimidyl ester (CFSE)–labeled peripheral or spleen CD4+ T cells from uninfected or WT H pylori–infected mice (8 weeks p.i.) were co-cultured for 5 days with FACS-sorted gastric CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells from WT H pylori–infected mice (8 weeks p.i.). Representative data and statistical analysis of CD4+IFN-γ+ T cell level and IFN-γ production were shown (n = 3). (G) The bacteria colonization in gastric mucosa of WT H pylori–infected WT and Rev-erbα–/– mice adoptively transferred with spleen CD4+ T cells from uninfected or WT H pylori–infected WT or IFN-γ–/– mice (8 weeks p.i.) was compared 8 weeks p.i. (n = 5). (H) The bacteria colonization in gastric mucosa of WT H pylori–infected WT and IFN-γ–/– mice was compared 8 weeks p.i. (n = 5). ∗P < .05, ∗∗P < .01, n.s. P > .05 for groups connected by horizontal lines.
To determine the potential contributions of Rev-erbα–mediated inhibition of specific Th1 cells to H pylori colonization, we conducted a series of in vivo adoptive transfer experiments and evaluated bacterial colonization in gastric mucosa 8 weeks p.i. First, CD4+ T cells from H pylori–infected WT donors into WT recipients effectively reduced H pylori colonization when compared with WT mice received CD4+ T cells from uninfected WT donors, suggesting specific CD4+ T cells contribute to reduced bacterial colonization (Figure 12G). Next, transferring CD4+ T cells from H pylori–infected WT donors into uninfected (or naïve) Rev-erbα–/– recipients effectively reduced H pylori colonization when compared with those in the uninfected WT mice received the same CD4+ T cells, suggesting Rev-erbα–mediated inhibition on H pylori–specific CD4+ T cells leading to increased bacterial colonization (Figure 12G). Finally, transferring CD4+ T cells from H pylori–infected WT donors into uninfected Rev-erbα–/– recipients effectively reduced H pylori colonization when compared with those in the uninfected Rev-erbα–/– recipients received CD4+ T cells from H pylori–infected IFN-γ–/– donors, suggesting Rev-erbα–mediated inhibition on IFN-γ production of H pylori–specific CD4+ T cells (Th1 cells) leading to increased bacterial colonization (Figure 12G), which was also reflected by increased colonization in IFN-γ–/– mice (Figure 12H). Overall, these results indicate that Rev-erbα promotes gastric H pylori colonization, most likely through inhibiting H pylori–specific Th1 cells’ IFN-γ response.
Discussion
Our results from both in vivo and in vitro gain- and loss-of-function experiments identify Rev-erbα as a pathological regulator that benefits bacterial colonization and contributes to gastric H pylori persistence. This is supported by several observations. First, Rev-erbα is preferentially expressed in GECs during H pylori infection, such as gastric pepsinogen II+ cells, and Rev-erbα–/– mice have notably decreased H pylori colonization associated with a marked increase in antibacterial protein content, chemokine production and Th1 cell response within gastric mucosa. We observed similar alterations upon knocking down Rev-erbα in AGS cells (higher Reg3b and β-defensin-1 content and impaired CCL21-derived chemotaxis function). By contrast, pharmacological activation of Rev-erbα in vivo in mice infected with H pylori and in vitro in H pylori–infected AGS cells resulted in the opposite phenotype, highlighting direct Rev-erbα effect on GECs. Second, our data indicate that H pylori induces Rev-erbα expression in GECs through the ERK-NF-κB signaling pathway, and that H pylori virulence factor cagA plays an essential role in this process. Finally, we show that GEC Rev-erbα regulates several genes involved in different steps of the host defense process, including genes more specifically dedicated to innate (Reg3b and β-defensin-1) and adaptive defense (CCL21 and IFN-γ). Together, these data support the concept that Rev-erbα acts through a two-pronged mechanism, involving both suppression of host innate defense and inhibition of host adaptive defense. Thus, during H pylori infection, when Rev-erbα is induced, there is a progressive impaired host defense within gastric mucosa, allowing for gastric H pylori persistence. And the role of Rev-erbα during H pylori–associated carcinogenesis process needs further investigation.
In this study, we not only identified a previously unrecognized role for Rev-erbα during H pylori infection, but also found that Rev-erbα expression was readily induced upon H pylori infection ex vivo in primary gastric tissues. This response is consistent with previous observations on Rev-erbα expression in inflamed tissues, such as liver21 and skeletal muscle.22 The regulation of Rev-erbα by H pylori provides a molecular mechanism whereby H pylori controls Rev-erbα expression through its virulence factor cagA. The cagA is crucial in the pathogenicity of H pylori that is injected into GECs via the type IV secretion system (T4SS) to modulate the phenotype and function of GECs.14 Lack of cagA activity in H pylori is associated with impaired expression of Rev-erbα in GECs, attenuated inhibition of Rev-erbα–mediated CCL21 expression and decreased CD45+CD11c–Ly6G–CD11b+CD68– myeloid cell chemotaxis. Thus, our findings that H pylori–associated virulence factor cagA was necessary to induce maximal Rev-erbα expression may suggest that intrinsic factors encoded by the infection itself are likely to be important in influencing the role of Rev-erbα. In addition, we revealed that ERK pathway activation–mediated Rev-erbα promoter transcription initiation is essential for the induction of Rev-erbα in GECs in response to H pylori. Of note, it is reported that several inflammatory factors such as TGF-β23 and transcription factors such as NF-κB24 are involved in Rev-erbα induction in hepatic stellate cells or under oxidative stress, which resembles our data on Rev-erbα regulation by NF-κB in H pylori–infected GECs.
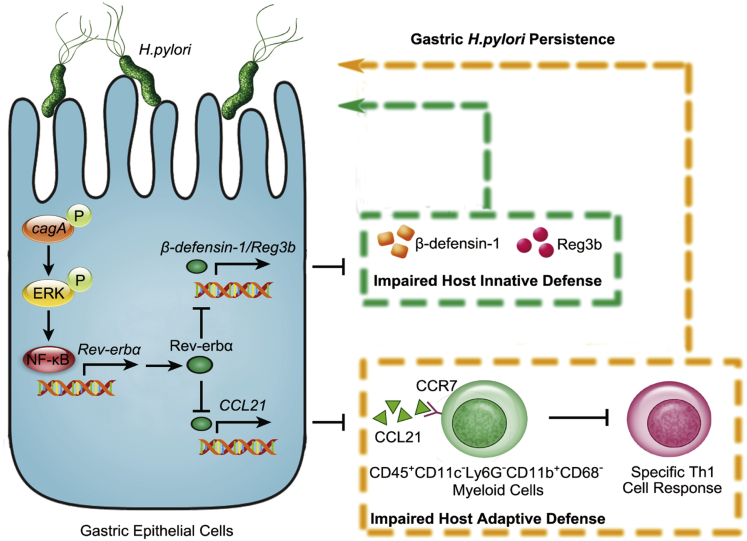
Host defense is a dynamic host-pathogen interaction process that ensures the selective clearance of invading pathogens, which is particularly important in infected cells such as GECs. It has been reported that Rev-erbα ameliorates Mycobacterium tuberculosis clearance through regulating autophagy-related genes,11 whereas our data demonstrate that Rev-erbα impedes H pylori clearance by regulating several genes involved in innate and adaptive defense. Several antibacterial proteins including LL-3725 and β-defensin-217 exerts antimicrobial activity against H pylori, here we add Reg3b and β-defensin-1 onto that list which are selectively suppressed by H pylori–induced Rev-erbα26 that represses β-defensin-1 gene directly triggering decreased bactericidal effect against H pylori to maintain gastric bacterial persistence. Several data have shown that Rev-erbα represses many cytokine and chemokine gene expression such as IL-6,27 IL-17,28 and CCL229 in inflammatory diseases. In our model, Rev-erbα downregulates CCL21 expression, leading to impaired chemotaxis of CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells and therefore decreased stimulation on H pylori–specific Th1 cells. IFN-γ, one of the key effector molecules of the Th1 cells, is reported to play key roles in controlling H pylori infection in mice19 and humans.30 Moreover, IFN-γ deficiency indeed results in uncontrolled colonization of H pylori in gastric mucosa.20 Notably, our findings mechanistically demonstrate for the first time that H pylori–induced Rev-erbα in GECs impairs host H pylori–specific Th1 response resulting in increased bacterial colonization. Together, these data support a prominent role for Rev-erbα in H pylori infection on connecting innate and adaptive host defense. Given the apparent relationship between Rev-erbα levels and the extent of bacterial colonization in H pylori–infected patients observed in this study, Rev-erbα should be considered as a novel diagnostic biomarker for H pylori infection–associated diseases. Specifically, our in vitro and in vivo data together provide a multistep model of H pylori persistent infection involving direct and indirect interactions among H pylori, GECs, Rev-erbα, Reg3b, β-defensin-1, CCL21, CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells, and Th1 cells within the gastric mucosa (Figure 13).
Figure 13.
A proposed model of crosstalk among H pylori, GECs, Rev-erbα, Reg3b, β-defensin-1, CCL21, CD45+CD11c–Ly6G–CD11b+CD68–myeloid cells, and Th1 cell response leading to Rev-erbα-mediated procolonization in gastric mucosa during H pylori infection.
Although eradication therapy for H pylori by oral antibiotics progressed in recent years,31,32 it is noteworthy that H pylori colonization commonly persists because of increased antimicrobial resistance and impaired host defense. In this regard, our findings suggest a possible therapeutic target, Rev-erbα, for decreasing gastric H pylori persistence. In addition to the direct action of Rev-erbα on genes associated with host defense, Rev-erbα deletion, or overexpression causes disturbed circadian rhythmicity33 may contribute to the observed phenotype. In conclusion, Rev-erbα is a novel pathological regulator of the function of host defense during H pylori infection. Thus, pharmacological inhibition of Rev-erbα may be a promising approach for the treatment of gastric diseases caused by H pylori infection.
Materials and Methods
Patients and Specimens
The gastric biopsy specimens and blood were collected from 92 H pylori–infected and 32 uninfected patients who underwent upper esophagogastroduodenoscopy for dyspeptic symptoms at XinQiao Hospital (Table 1). H pylori infection was determined by [14C] urea breath test and rapid urease test of biopsy specimens taken from the antrum, and subsequently conformed by real-time polymerase chain reaction (PCR) for 16s recombinant DNA (rDNA) and serology test for specific anti-H pylori Abs.34 For isolation of human primary GECs, fresh nontumor gastric tissues (at least 5 cm distant from the tumor site) were obtained from gastric cancer patients who underwent surgical resection and were determined as H pylori–negative individuals (conformed by real-time PCR for 16s rDNA and serology test for specific anti-H pylori Abs) as previous at the Southwest Hospital. None of these patients had received chemotherapy or radiotherapy before sampling. Individuals with atrophic gastritis, hypochlorhydria, antibiotics treatment, autoimmune disease, infectious diseases and multiprimary cancer were excluded. The study was approved by the Ethics Committee of XinQiao Hospital and Southwest Hospital of Third Military Medical University. A written informed consent was obtained from each subject.
Table 1.
Clinical Characteristics of Patients
| Variables | H pylori Infected | Uninfected |
|---|---|---|
| Age, y | 46 (21–73) | 45 (24–68) |
| Male/female | 53/39 | 18/14 |
NOTE. Values are median (interquartile range) or n. Exclusion criteria were previous treatment for H pylori infection, use of inhibitors of acid secretion or antibiotics during the 2 months before the study, use of anticoagulant drugs in the last week, gastrointestinal malignancy, severe concomitant cardiovascular, respiratory or endocrine diseases, clinically significant renal or hepatic disease, hematological disorders, previous gastroesophageal surgery, history of allergy to any of the drug used in the study, pregnancy or lactation, alcohol abuse, drug addiction, severe neurological or psychiatric disorders, and long-term use of corticosteroids or anti-inflammatory drugs.
Antibodies and Other Reagents
For information on antibodies and other reagents, see Table 2.
Table 2.
Antibodies and Other Reagents
| Antibodies and Reagents | Manufacturer |
|---|---|
| Antibodies for flow cytometry | |
| anti-mouse CD45-PE-Cy7(103114) | BioLegend |
| anti-mouse Ly6G-FITC (127605) | BioLegend |
| anti-mouse CD11c-PerCP-Cy5.5(117327) | BioLegend |
| anti-mouse CD3-APC (300311) | BioLegend |
| anti-mouse NK1.1-PE (108707) | BioLegend |
| anti-mouse CD19-APC-Cy7(115529) | BioLegend |
| anti-mouse CD11b-PE (101207) | BioLegend |
| anti-mouse CD68-APC-Cy7(137023) | BioLegend |
| anti-mouse CCR7-APC (120108) | BioLegend |
| anti-mouse Ly6C-APC (128016) | BioLegend |
| anti-mouse F4/80-APC (123116) | BioLegend |
| anti-human CD45-PE-Cy7(368532) | BioLegend |
| anti-human CD66b-FITC (305104) | BioLegend |
| anti-human CD11c-PerCP-Cy5.5(301624) | BioLegend |
| anti-human CD11b-PE (301306) | BioLegend |
| anti-human CD68-APC-Cy7(333822) | BioLegend |
| anti-human CCR7-APC (353214) | BioLegend |
| anti-mouse CD4-PerCP-Cy5.5(100433) | BioLegend |
| anti-mouse IFN-γ-APC (505810) | BioLegend |
| anti-mouse CCR7-Brilliant Violet 605 (120125) | BioLegend |
| anti-mouse CD163-Brilliant Violet 421 (155309) | BioLegend |
| anti-mouse CD206-Alexa Fluor 647 (141711) | BioLegend |
| anti-mouse MHC II Brilliant Violet 510 (107635) | BioLegend |
| anti-mouse IL-12-APC (505205) | BioLegend |
| anti-mouse IL-10-Brilliant Violet 421 (505021) | BioLegend |
| Antibodies for immunohistochemical staining | |
| rabbit anti-human/mouse Rev-erbα(ab174309, 1:100) | Abcam |
| horseradish peroxidase anti-rabbit IgG (ZB-2301) | Zhongshan Biotechnology |
| sheep anti-mouse Reg3b (AF5110) | R&D Systems |
| horseradish peroxidase anti-sheep IgG (ZB-2306) | Zhongshan Biotechnology |
| rabbit anti-mouse β-defensin-1(PA5-75666) | Thermo Fisher Scientific |
| Antibodies for immunofluorescence | |
| rabbit anti-human/mouse Rev-erbα(ab174309;ab251057, 1:100) | Abcam |
| mouse anti-human/mouse CD326 (EpCAM) (ab212580, 1:100) | Abcam |
| rabbit anti-human CCL21(ab231116;ab248260, 1:100) | Abcam |
| rabbit anti-human/mouse pepsinogen II (ab259997, 1:100) | Abcam |
| rabbit anti-mouse CD11b (ab232427, 1:100) | Abcam |
| rat anti-mouse Ly6C (ab15627, 1:100) | Abcam |
| mouse anti-H pylori (188-10881-1, 1:100) | RayBiotech |
| goat anti-rabbit-TRITC (ZF-0316, 1:50) | Zhongshan Biotechnology |
| goat anti-rabbit-FITC (ZF-0311, 1:50) | Zhongshan Biotechnology |
| goat anti-mouse-TRITC (ZF-0313, 1:50) | Zhongshan Biotechnology |
| goat anti-mouse-FITC (ZF-0312, 1:50) | Zhongshan Biotechnology |
| goat anti-rat-FITC (ZF-0315, 1:50) | Zhongshan Biotechnology |
| Antibodies for neutralizing and blocking | |
| anti-human CCL21 (Goat IgG) (AF366) | R&D Systems |
| Goat IgG Control (AB-108-C) | R&D Systems |
| anti-human CCR7 (Mouse IgG2a) (MAB197) | R&D Systems |
| Mouse IgG2a Isotype Control (MAB003) | R&D Systems |
| anti-mouse CCL21 (Rat IgG2a) (MAB457) | R&D Systems |
| anti-mouse CCR7 (Rat IgG2a) (MAB3477) | R&D Systems |
| Rat IgG2a Isotype Control (MAB006) | R&D Systems |
| anti-mouse Reg3b (MAB5110) | R&D Systems |
| anti-mouse β-defensin-1(PA5-75666) | Thermo Fisher Scientific |
| Antibodies for Western blot | |
| rabbit anti-human/mouse Rev-erbα(ab174309, 1:1000) | Abcam |
| rabbit anti-human ERK1/2(4695, 1:1000) rabbit anti-human ERK1/2(ab218017, 1:1000) | Cell Signaling Technology Abcam |
| rabbit anti-human p-ERK1/2(4370S, 1:2000) | Cell Signaling Technology |
| rabbit anti-human p-ERK1/2(ab242418, 1:2000) | Abcam |
| rabbit anti-human/mouse GAPDH (BL042F, 1:1000) | Beijing Ray Antibody Biotech |
| rabbit anti-human/mouse GAPDH (ab186930;ab199554, 1:1000) | Abcam |
| rabbit anti-human/mouse Lamin B1(ab133741;ab220797, 1:1000) | Abcam |
| rabbit anti-cagA(sc-28368, 1:100) | Santa Cruz Biotechnology |
| ELISA kits | |
| mouse Reg3b (ml058487-C) | ML Bio |
| mouse β-defensin-1(ml002270-C) | ML Bio |
| human CCL21(ELH-6Ckine-1) | RayBiotech |
| mouse CCL21(ELM-6Ckine) | RayBiotech |
| mouse IFN-γ(1210002) | Dakewe Biotech Co |
| Reagents for signaling pathways inhibition | |
| MEK-1 and MEK-2 inhibitor U0126(U120) | Merk Millipore |
| MAPK inhibitor SB203580(S8307) | Calbiochem |
| JNK inhibitor SP600125(S5567) | Calbiochem |
| STAT3 phosphorylation inhibitor FLLL32(406111) | MedKoo Biosciences |
| JAK inhibitor AG490(200121) | Merk Millipore |
| PI3K-AKT inhibitor Wortmannin (406447) | Merk Millipore |
| Reagents for Luciferase Reporter Assay | |
| Dual-luciferase reporter assay system (E1910) | Promega |
| Endo-free Plasmid Mini Kit (D6950) | Omega |
| Pierce Magnetic CHIP Kit (26157) | Thermo Fisher Scientific |
| truChIP Chromatin Shearing Kit (520127) | Covaris |
| 16% Formaldehyde, Methanol-Free (12606) | Cell Signaling Technology |
| Antibodies for CHIP | |
| Rev-erbα rabbit mAb (13418s, 1:100) | Cell Signaling Technology |
| NF-κB p65 rabbit mAb (8242s, 1:100) | Cell Signaling Technology |
| SimpleChip human NR1D1 promoter primers (13413s) | Cell Signaling Technology |
| Purified anti-CD3 antibodies (100201) | BioLegend |
| Purified anti-CD28 antibodies (102101) | BioLegend |
| Human CD326 microbeads (130-061-101) | Miltenyi Biotec |
| Mouse CD326 microbeads (130-105-958) | Miltenyi Biotec |
| EasySep Mouse CD4+ T Cell Isolation Kit (19852) | StemCell Technologies |
| Dynabeads FlowComp Mouse CD4 Kit | Invitrogen |
| 5-μm pore size Transwells (3421) | Corning |
| 0.4-μm pore size Transwells (3413) | Corning |
| Collagenase IV (17104019) | Gibco |
| DNase I (AMPD1) | Sigma-Aldrich |
| Phorbol myristate acetate (P8139) | Sigma-Aldrich |
| Ionomycin (407951) | Sigma-Aldrich |
| Golgistop (554724) | BD Pharmingen |
| Perm/Wash solution (554714) | BD Pharmingen |
| Carboxylfluorescein succinimidyl ester (CFSE) (65-0850-84) | eBioscience |
| Rev-erbα agonist SR9009(554726) | Merck |
| Rev-erbα antagonist SR8278(554718) | Merck |
| Cremophor (61791-12-6) | BASF |
| DMSO (D2650) | Sigma-Aldrich |
| Protein Extraction Reagent (89901) | Pierce |
| SuperSignal West Dura Extended Duration Substrate kit (34075) | Thermo |
| Fetal bovine serum (FBS) (10091148) | Gibco |
| Fetal bovine serum (FBS) (10099141C) | Invitrogen |
| Penicillin/Streptomycin (15140122) | Gibco |
| RPMI-1640(SH30809.01) | Hyclone |
| DMEM/F12 (1:1) (SH30023.01) | Hyclone |
| Ficoll-Paque Plus (17-1440-02) | GE Healthcare |
| lyses solution (RT122) | TIANGEN |
| TRIzol reagent (15596026) | Invitrogen |
| Lipofectamine 2000 Transfection Reagent (11668027) | Invitrogen |
| Lipofectamine RNAiMAX Transfection Reagent (13778150) | Invitrogen |
| QIAamp DNA Mini Kit (51304) | QIAGEN |
| PrimeScript RT reagent Kit (R037A) | TaKaRa |
| Real-time PCR Master Mix (QPK-201) | Toyobo |
| pGL3-basic vector (E1751) | Promega |
| Recombinant mouse Reg3b (5110-RG) | R&D Systems |
| Recombinant mouse β-defensin-1(228-11913-1) | RayBiotech |
| Recombinant mouse CCL21(457-6C) | R&D Systems |
| Recombinant human CCL21(366-6C) | R&D Systems |
| All other recombinant human/mouse cytokines and chemokines | PeproTech |
APC, allophycocyanin; APC-Cy7, allophycocyanin-cyanin 7; ChIP, chromatin immunoprecipitation; FITC, fluorescein isothiocyanate; IFN, interferon; PE, phycoerythrin; PE-Cy7, phycoerythrin-cyanin 7; PerCP-Cy5.5, peridin chlorophyl protein-cyanin 5.5.
Mice
All breeding and experiments were undertaken with review and approval from the Animal Ethical and Experimental Committee of Third Military Medical University. C57BL/6 Rev-erbα+/– mice were obtained from the Jackson Laboratory (Bar Harbor, ME). C57BL/6 Rev-erbα–/– mice and their littermate control (WT) mice were generated by breeding between C57BL/6 Rev-erbα+/– mice. C57BL/6 interferon-γ–/– (IFN-γ–/–) mice were kindly provided by Dr. Richard A. Flavell (Yale University). All mice used in experiments were viral Ab negative for pathogenic murine viruses, negative for pathogenic bacteria including Helicobacter spp. and parasites, and were maintained under specific pathogen–free conditions in a barrier sustained facility and provided with sterile food and water.
Bacteria Culture and Infection of Mice With Bacteria
H pylori NCTC 11637 (cagA positive) (WT H pylori) and cagA-knockout mutant H pylori NCTC 11637 (ΔcagA) or H pylori 26695 were grown in brain-heart infusion plates containing 10% rabbit blood at 37°C under microaerophilic conditions. For infecting mouse, bacteria were propagated in Brucella broth with 5% fetal bovine serum (FBS) with gentle shaking at 37°C under microaerobic conditions. After culture for 1 day, live bacteria were collected and adjusted to 109 colony-forming units (CFU)/mL. The mice were fasted overnight and orogastrically inoculated twice at a 1-day interval with 3 × 108 CFU bacteria. H pylori infection status and H pylori–induced gastritis in murine experiments were confirmed using real-time PCR of H pylori 16s rDNA, urease biopsy assays, Warthin-Starry staining, and immunohistochemical staining for H pylori.34,35
Generation of BM Chimera Mice
The following BM chimeric mice were created: male WT BM→female WT mice, male WT BM→female Rev-erbα–/– mice, male Rev-erbα–/– BM→female WT mice, and male Rev-erbα–/– BM→female Rev-erbα–/– mice. BM cells were collected from the femurs and tibia of donor mice by aspiration and flushing, and were suspended in phosphate-buffered saline (PBS) at the concentration of 5 × 107/mL. The BM in recipient mice was ablated with lethal irradiation (8 Gy). Then, the animals received intravenously 1.5 × 107 BM cells from donor mice in a volume of 300 μL sterile PBS under the anesthesia. Thereafter, the transplanted BM was allowed to reconstitute for 8 weeks before subsequent experimental procedures. To verify successful engraftment and reconstitution of the BM in the host mice, genomic DNA was isolated from tail tissues of each chimera mouse 8 weeks after BM transplantation. Quantitative PCR was performed to detect the Sry gene present in the Y chromosome (primers seen in Table 3) and mouse β2-microglobulin gene as an internal control. The chimeric rates were calculated on the assumption that the ratio of the Sry to β2-microglobulin gene was 100% in male mice. We confirmed that the chimeric rates were consistently higher than 90%. After BM reconstitution was confirmed, mice were infected with bacteria as described previously.
Table 3.
Primer and Probe Sequences for Real-Time PCR Analysis
| Gene | Primer or Probe | Sequence 5′→3′ |
|---|---|---|
| H pylori 16s rDNA | forward | TTTGTTAGAGAAGATAATGACGGTATCTAAC |
| reverse | CATAGGATTTCACACCTGACTGACTATC | |
| probe | CGTGCCAGCAGCCGCGGT | |
| Mouse β2-microglobulin | forward | CCTGCAGAGTTAAGCATGCCAG |
| reverse | TGCTTGATCACATGTCTCGATCC | |
| probe | TGGCCGAGCCCAAGACCGTCTAC | |
| H pylori cagA | forward | GAGTCATAATGGCATAGAACCTGAA |
| reverse | TTGTGCAAGAAATTCCATGAAA | |
| Mouse Sry | forward | TGGGACTGGTGACAATTGTC |
| reverse | GAGTACAGGTGTGCAGCTCT | |
| Human Rev-erbα | forward | TCAGCTGGTGAAGACATGACGAC |
| reverse | GGAGCCACTGGAGCCAATGTA | |
| Human GAPDH | forward | ACCCAGAAGACTGTGGATGG |
| reverse | CAGTGAGCTTCCCGTTCAG | |
| Human CCL21 | forward | TATCCTGGTTCTGGCCTTTG |
| reverse | CAGCCTAAGCTTGGTTCCTG | |
| Human β-defensin-1 | forward | CCAGTCGCCATGAGAACTTCC |
| reverse | GTGAGAAAGTTACCACCTGAGGC | |
| Mouse Rev-erbα | forward | GCTTCTCTCAGTTCCCACAAC |
| reverse | GGTGAAGATTTCTCGATGGGC | |
| Mouse β-actin | forward | AGTGTGACGTTGACATCCGT |
| reverse | GCAGCTCAGTAACAGTCCGC | |
| Mouse CCL1 | forward | ATGGCACTGATGTGCCTGCT |
| reverse | GGTGGAGGACTGAGGGAAA | |
| Mouse CCL2 | forward | TCACCTGCTGCTACTCATTCA |
| reverse | CACTGTCACACTGGTCACTCC | |
| Mouse CCL3 | forward | TTCTCTGTACCATGACACTCTGC |
| reverse | CGTGGAATCTTCCGGCTGTAG | |
| Mouse CCL4 | forward | TGTCTGCCCTCTCTCTCCTCT |
| reverse | AGCAAGGACGCTTCTCAGTGA | |
| Mouse CCL5 | forward | GCTGCTTTGCCTACCTCTCC |
| reverse | TCGAGTGACAAACACGACTGC | |
| Mouse CCL6 | forward | CCAAGACTGCCATTTCATTC |
| reverse | AAGCAATGACCTTGTTCCCA | |
| Mouse CCL7 | forward | ATGGAAGTCTGCGCTGAAG |
| reverse | ACATGAGGTCTCCAGAGCTTT | |
| Mouse CCL8 | forward | ACGCTAGCCTTCACTCCAAAA |
| reverse | TTCCAGCTTTGGCTGTCTCTT | |
| Mouse CCL9 | forward | TGGCATATCTGGCTTTGTCA |
| reverse | ATGGCTGTAGCTCAAGATGGT | |
| Mouse CCL11 | forward | TCCACAGCGCTTCTATTCCT |
| reverse | GCAGTTCTTAGGCTCTGGGTT | |
| Mouse CCL12 | forward | TCGAAGTCTTTGACCTCAACA |
| reverse | GGGAACTTCAGGGGGAAATA | |
| Mouse CCL19 | forward | ACTTGCACTTGGCTCCTGAA |
| reverse | AGTCTTCCGCATCATTAGCA | |
| Mouse CCL20 | forward | GCAAGCGTCTGCTCTTCCTT |
| reverse | TTAGGCTGAGGAGGTTCACA | |
| Mouse CCL21 | forward | GATGATGACTCTGAGCCTCCT |
| reverse | TTCTGCACCCAGCCTTCCT | |
| Mouse CCL22 | forward | TGGCAATTCAGACCTCTGATG |
| reverse | TTGCTGGAATGGCAGAAGAA | |
| Mouse CCL24 | forward | TCATCTTGCTGCACGTCCTTT |
| reverse | TAAACCTCGGTGCTATTGCCA | |
| Mouse CCL25 | forward | TCTCAGGACCAGAAAGGCATT |
| reverse | TGGCGGAAGTAGAATCTCACA | |
| Mouse CCL27 | forward | AGGCTGAGTGAGCATGATGGA |
| reverse | TTGGCGTTCTAACCACCGA | |
| Mouse CCL28 | forward | GCTGTGTGTGTGGCTTTTCAA |
| reverse | TACCTCTGAGGCTCTCATCCA | |
| Mouse CX3CL1 | forward | TGGCTTTGCTCATCCGCTATCAG |
| reverse | CGTCTGTGCTGTGTCGTCTCC | |
| Mouse CXCL1 | forward | ACCCAAACCGAAGTCATAG |
| reverse | TTGTATAGTGTTGTCAGAAGC | |
| Mouse CXCL2 | forward | ACTTCAAGAACATCCAGAG |
| reverse | CTTTCCAGGTCAGTTAGC | |
| Mouse CXCL3 | forward | CAGCCACACTCCAGCCTA |
| reverse | CACAACAGCCCCTGTAGC | |
| Mouse CXCL4 | forward | AGCGATGGAGATCTTAGCTGTGT |
| reverse | CCAGGCTGGTGATGTGCTTAA | |
| Mouse CXCL5 | forward | AGTCAAGAATCATTGGTTGTTAACCTT |
| reverse | TCCGGAGACAATGCAATAGTCA | |
| Mouse CXCL7 | forward | GGAGTTCACTGTGCTGATGTGGA |
| reverse | CACAGATGAAGCAGCTGGTCAGTAA | |
| Mouse CXCL9 | forward | ACAAATCCCTCAAAGACCTCAAACAG |
| reverse | ATCTCCGTTCTTCAGTGTAGCAATG | |
| Mouse CXCL10 | forward | TGAAAGCGTTTAGCCAAAAAAGG |
| reverse | AGGGGAGTGATGGAGAGAGG | |
| Mouse CXCL12 | forward | CCTCCAAACGCATGCTTCA |
| reverse | ACTCTCCTCCCTTCCATTGCA | |
| Mouse CXCL13 | forward | CAGGCCACGGTATTCTGGA |
| reverse | CAGGGGGCGTAACTTGAATC | |
| Mouse CXCL14 | forward | GCTTCATCAAGTGGTACAAT |
| reverse | CTGGCCTGGAGTTTTTCTTTCCAT | |
| Mouse CXCL15 | forward | CTAGGCATCTTCGTCCGTCC |
| reverse | TTGGGCCAACAGTAGCCTTC | |
| Mouse CXCL16 | forward | AAACATTTGCCTCAAGCCAGT |
| reverse | GTTTCTCATTTGCCTCAGCCT | |
| Mouse CXCL17 | forward | ATGAAGCTTCTAGCCTCTCCC |
| reverse | CTATAAGGGCAGCGCAAAGCTTGC | |
| Mouse β-defensin-1 | forward | GAACACGGTACACAGGCTTCC |
| reverse | CCTGAATCACAGATGTCCAAG | |
| Mouse β-defensin-2 | forward | CTCTCTGGAGTCTGAGTGCCC |
| reverse | AGGACGCCTGGCAGAAGGAGG | |
| Mouse β-defensin-3 | forward | TGCTGCTGTCTCCACCTGC |
| reverse | AGTGTTGCCAATGCACCGAT | |
| Mouse β-defensin-4 | forward | ACATGCATGACCAATGGAGCC |
| reverse | CATCTTGCTGGTTCTTC | |
| Mouse Reg3a | forward | CTGCTCTCCTGCCTGTTGTT |
| reverse | GGAGCGATAAGCCTTGTAACC | |
| Mouse Reg3b | forward | AGGCTTATGGCTCCTACTGCT |
| reverse | GAAGCCTCAGCGCTATTGAG | |
| Mouse Reg3g | forward | TGCCTATGGCTCCTATTGCT |
| reverse | CATGGAGGACAGGAAGGAAG | |
| Mouse Reg3d | forward | CTGTCTTCTCCACGCATCAG |
| reverse | CTGCTCCACTTCCATCCATT |
NOTE. For the probes, a FAM fluorescent reporter is coupled to the 5' end, and a TAMRA quencher is coupled to the 3' end.
Antibodies/CCL21/Reg3b/β-Defensin-1/Rev-erbα Agonist/Rev-erbα Antagonist Administration
One day after infection with WT H pylori as described previously, WT mice were injected intraperitoneally with recombinant mouse CCL21 (25 μg) or Reg3b or β-defensin-1 (50 μg), or anti-mouse CCL21 or anti-mouse CCR7 or isotype control Abs (100 μg), or Rev-erbα agonist SR9009 (100 mg/kg, dissolved in cremophor) or cremophor control, or Rev-erbα antagonist SR8278 (25 mg/kg, dissolved in dimethyl sulfoxide [DMSO]) or DMSO control and repeated every week until the mice were sacrificed.
T Cell Adoptive Transfer
One day before infection with WT H pylori, WT or Rev-erbα–/– mice were injected intravenously (1 × 106 cells/mouse) with purified spleen CD4+ T cells (StemCell Technologies, Vancouver, Canada) from uninfected WT mice, or WT H pylori–infected WT mice or WT H pylori–infected IFN-γ–/– mice (8 week p.i.). Then, the recipient mice were infected with bacteria as described previously and sacrificed for bacteria colonization evaluation at week 8 p.i..
Evaluation of Bacteria Colonization
The mice were sacrificed at the indicated time points. The stomach was cut open from the greater curvature and half of the tissue was cut into 4 parts for RNA extraction, DNA extraction, protein extraction, and tissue fixation for immunohistochemistry or immunofluorescence staining, respectively. DNA of the biopsy specimens were extracted with QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). As previously described,36 H pylori colonization was quantified by measuring H pylori–specific 16s rDNA using specific primer and probe (Table 3) by the TaqMan method. The amount of mouse β2-microglobulin DNA in the same specimen was used to normalize the data. According to a previous study,37 the density of H pylori was shown as the number of bacterial genomes per nanogram of host genomic DNA. Another half of stomach was used for isolation of single cells as described subsequently. The isolated single cells were collected and analyzed by flow cytometry staining.
Isolation of Single Cells From Tissues
Fresh tissues were washed three times with Hank’s solution containing 1% FBS, cut into small pieces, collected in RPMI 1640 containing 1 mg/mL collagenase IV and 10 mg/mL DNase I, and then mechanically dissociated by using the gentle MACS Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). Dissociated cells were further incubated for 0.5–1 hour at 37°C under continuous rotation. The cell suspensions were then filtered through a 70-μm cell strainer (BD Labware; Becton Dickinson, Franklin Lakes, NJ).
Cell/Tissue Culture, Transfection, and Stimulation
Primary GECs were purified from gastric tissue single-cell suspensions from uninfected donors or mice with a MACS column purification system using anti-human or mouse CD326 magnetic beads. The sorted primary GECs were used only when their viability was determined >90% and their purity was determined >95%. For human GEC lines (AGS cells, GES-1 cells, HGC-27 cells, SGC-7901 cells, BGC-823 cells), 3 × 105 cells per well in 12-well cell culture plate (for real-time PCR) or 1 × 106 cells per well in 6-well cell culture plate (for Western blot and enzyme-linked immunosorbent assay [ELISA]) were starved in Dulbecco’s modified Eagle medium/F-12 medium supplemented with penicillin (100 U/mL) and streptomycin (100 μg/mL) for 6 hours in a humidified environment containing 5% CO2 at 37 °C. Then the cells were incubated in antibiotic-free Dulbecco’s modified Eagle medium/F-12 medium supplemented with 10% FBS instead. The cell lines were used when their viability was determined >90%. Human GEC lines, primary GECs, or primary gastric mucosa tissues from uninfected donors or mice were infected with WT H pylori, ΔcagA or H pylori 26695 at a multiplicity of infection (MOI) of 100 for 24 hours. AGS cells were also infected with WT H pylori at different MOI (24 hours) or at the indicated time points (MOI = 100). For signal pathway inhibition experiments, AGS cells were pretreated with 5 μL (20 μM) U0126 (an MEK-1 and MEK-2 inhibitor), SB203580 (a p38 MAPK inhibitor), SP600125 (a JNK inhibitor), FLLL32 (an STAT3 inhibitor), AG490 (a JAK inhibitor), Wortmannin (a PI3K-AKT inhibitor), PP2 (a cagA EPIYA motif phosphorylation inhibitor), or DMSO control for 2 hours. For Rev-erbα activation or inhibition experiments, AGS cells were pretreated with Rev-erbα siRNA, NC (40 nM), or lipofectamine 2000 only (Mock) for 24 hours. In some cases, AGS cells were transfected with plasmids pcDNA3.1 or cagA-pcDNA3.1 by using lipofectamine 2000 according to the manufacturer’s protocols. At 24 hours after transfection, cells were treated with or without U0126 (5 μL, 20 μM) or DMSO control for 2 hours and cultured for an additional 24 hours. After co-culture, cells were collected for immunofluorescence, real-time PCR, and Western blot, and the culture supernatants were harvested for ELISA.
In Vitro Bactericidal Assay
Two hundred microliters of 5 × 106 CFU/mL WT H pylori suspension was incubated with 20 μg/mL mouse Reg3b or β-defensin-1 for 3, 6, or 12 hours, or incubated with 5, 10, or 20 μg/mL mouse Reg3b or β-defensin-1 for 24 hours. PBS was used as control. In another set of experiments, primary GECs from WT or Rev-erbα–/– mice were infected with WT H pylori (MOI = 100) for 24 hours. The culture supernatants were filtered through 0.4-μm filters and collected. Then 200 μL of 5 × 106 CFU/mL WT H pylori suspension were incubated with the above collected culture supernatants with or without anti-Reg3b or anti-β-defensin-1 Abs (20 μg/mL) for 24 hours. Bacteria were serially diluted and plated on brain-heart infusion plates containing 10% rabbit blood and incubated for 3–5 days at 37°C under microaerophilic conditions and CFU was enumerated. The results was determined by counting CFU of alive bacteria with agar plating and expressed as the survival rate of WT H pylori after incubation with Reg3b/β-defensin-1 or PBS, or was determined by counting CFU of alive bacteria with agar plating and expressed as the survival rate of WT H pylori after incubation with supernatants from H pylori–infected GECs of Rev-erbα–/– mice or WT mice with or without anti-Reg3b or anti-β-defensin-1 Abs.
In Vitro T Cell Culture System
Purified mouse peripheral or spleen CD4+ T cells from uninfected or WT H pylori–infected WT mice (8 week p.i.) were labeled with carboxyfluorescein succinimidyl ester and co-cultured (1 × 105 cells/well) with fluorescence-activated cell sorter (FACS)–sorted gastric CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells from WT H pylori–infected WT mice (8 week p.i.) at 2:1 ratio in 200 μL RPMI 1640 medium containing mouse recombinant IL-2 (20 IU/mL), anti-CD3 (2 μg/mL), and anti-CD28 (1 μg/mL) Abs. After a 5-day incubation, cells were collected and analyzed by intracellular cytokine staining, and the culture supernatants were harvested for ELISA.
Chemotaxis Assay
Human CD45+CD11c–CD66b–CD11b+CD68– myeloid cells or mouse CD45+CD11c–Ly6G–CD11b+CD68– myeloid cells from blood of H pylori–infected donors or WT H pylori–infected mice (8 week p.i.) were sorted by FACS (FACSAria II; BD Biosciences, Franklin Lakes, NJ). AGS cells were pretreated with Rev-erbα siRNA or NC (both at 40 nM) for 24 hours, and then infected with WT H pylori or ΔcagA (MOI = 100) for 24 hours. The culture supernatants were filtered through 0.4-μm filters, collected and used as source of chemoattractants in a human myeloid cell chemotaxis assay. In another set of experiments, mouse primary GECs were purified from gastric tissue single-cell suspensions of uninfected WT or Rev-erbα–/– mice with anti-mouse CD326-conjugated MACS magnetic beads, and then infected with WT H pylori or ΔcagA (MOI = 100) for 24 hours. The culture supernatants were then collected mentioned above. These culture supernatants were then used as source of chemoattractants in a mouse myeloid cell chemotaxis assay.
In a chemotaxis assay, FACS-sorted myeloid cells (1 × 105) were transferred into the upper chambers of the transwells (5-μm pore). CCL21 (100 ng/mL) and culture supernatants from various cultures were placed in the lower chambers. After 6 hours culture, migration was quantified by counting cells in the lower chamber and cells adhering to the bottom of the membrane. In some cases, blocking Ab for human/mouse CCL21 (20 μg/mL) or corresponding control IgG/IgG2a (20 μg/mL) were added into the culture supernatants, and blocking Ab for human/mouse CCR7 (20 μg/mL) or corresponding control IgG2a (20 μg/mL) were added into myeloid cell suspensions and incubated for 2 hours before chemotaxis assays.
Immunohistochemistry
Paraformaldehyde-fixed and paraffin-embedded samples were cut into 5-μm sections. For immunohistochemical staining, the sections were incubated with rabbit anti-human/mouse Rev-erbα, sheep anti-mouse Reg3b, or rabbit anti-mouse β-defensin-1 followed by horseradish peroxidase (HRP)–conjugated anti-rabbit IgG or HRP-conjugated anti-sheep IgG and later its substrate diaminobenzidine. All the sections were finally counterstained with hematoxylin and examined using a microscope (Nikon Eclipse 80i; Nikon, Tokyo, Japan).
Immunofluorescence
Paraformaldehyde-fixed cryostat tissue sections or AGS cells were washed in PBS, blocked for 30 minutes with 20% goat serum in PBS, stained for Rev-erbα, Rev-erbα and CD326, CCL21 and CD326, Rev-erbα and pepsinogen II, Ly6C and CD11b, or Rev-erbα and H pylori. Slides were examined with a confocal fluorescence microscope (LSM 510 META, Zeiss, Oberkochen, Germany).
Real-Time PCR
DNA of the biopsy specimens were extracted with QIAamp DNA Mini Kit and RNA of biopsy specimens and cultured cells were extracted with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). The RNA samples were reversed transcribed into complementary DNA with PrimeScript RT reagent Kit (Takara Bio, Mountain View, CA). Real-time PCR was performed on an IQ5 (Bio-Rad, Hercules, CA) with Real-time PCR Master Mix according to the manufacturer's specifications. The mRNA expression of 16s rDNA, cagA, Rev-erbα, chemokine, IFN-γ, β-defensin and Reg3 genes was measured using the TaqMan or SYBR green method with the relevant primers (Table 3). For mouse samples, mouse β2-microglobulin mRNA level served as a normalizer, and its level in the stomach of uninfected or WT mice served as a calibrator. For human samples, human β-actin mRNA level served as a normalizer, and its level in the uninfected cells/tissues or stomach of uninfected donors served as a calibrator. The relative gene expression was expressed as fold change of relevant mRNA calculated by the ΔΔCt method.
Luciferase Reporter Assay
Promoter constructs containing the region from –2000 to 0, –1000 to 0, –500 to 0, –250 to 0, –100 to 0, and other 250-bp fragments of Rev-erbα gene were amplified from human genomic DNA using specifically designed primers (Table 4) by PCR. The amplified full-length or fragmented sequences were cloned into the NheI and HindIII sites of the pGL3-basic vector respectively. Promoter constructs containing the region from -2000 to 0 of CCL21 and β-defensin-1 gene, and mutants of predicted Rev-erbα binding site of CCL21 and β-defensin-1 gene sequences were also synthesized with primers (Table 5) and cloned into the NheI and HindIII sites of the pGL3-basic vector respectively. The above constructs were sequence verified. Plasmids overexpressing Rev-erbα (Rev-erbα-pcDNA3.1) were constructed by inserting target gene into plasmid pcDNA3.1, these plasmids were constructed and produced by Sangon Biotech (Shanghai, China). Cells were transfected with various combinations of the reporter plasmid, which contains the internal control pRL-TK (Promega, Madison, WI) or the expression plasmid. Cells were harvested at 24 hours (H pylori infection assay) or 48 hours (cagA-pcDNA3.1 or Rev-erbα-pcDNA3.1 plasmid transfection assay) after transfection. Luciferase activities of the lysates were measured using the Dual-Luciferase Reporter Assay Kit (Promega) following the manufacturer’s protocol. Luciferase activity was normalized to Renilla luciferase activity.
Table 4.
Primers for Amplify Full-Length and the Different Fragment Sequences of Rev-erbα Gene for PCR
| Name | Primer | Sequence 5′→3′ |
|---|---|---|
| Human Rev-erbα (–2000/0) | forward | GCTCTTACGCGTGCTAGCTGCCTGTGGAGAAGGGCTTC |
| reverse | TACCGGAATGCCAAGCTTGTCTTCACCAGCTGAGAGCG | |
| Human Rev-erbα (–1000/0) | forward | GCTCTTACGCGTGCTAGCCCCGGTCACCAGTAACCTC |
| reverse | CAGTACCGGAATGCCAAGC | |
| Human Rev-erbα (–500/0) | forward | GCTCTTACGCGTGCTAGCTTGGCAGAGTGAAATATTACTGC |
| reverse | CAGTACCGGAATGCCAAGC | |
| Human Rev-erbα (–250/0) | forward | GCTCTTACGCGTGCTAGCTGATTCCCCCTACACACTCTC |
| reverse | CAGTACCGGAATGCCAAGC | |
| Human Rev-erbα (–100/0) | forward | GCTCTTACGCGTGCTAGCTCCCCGCCCTTAGCCAGT |
| reverse | CAGTACCGGAATGCCAAGC | |
| Human Rev-erbα (–2000/–1750) | forward | GCTCTTACGCGTGCTAGCATCAATACCATCCCAGGAGCTGCCTGTGGAGAAGGG |
| reverse | TACCGGAATGCCAAGCTTAAAACAGCCAGTATTTCGCTTC | |
| Human Rev-erbα (–1749/–1500) | forward | GCTCTTACGCGTGCTAGCTTGTTTGTTTGTTTTGGAGACAG |
| reverse | TACCGGAATGCCAAGCTTCTGAGGTGGGTTGATCACCTG | |
| Human Rev-erbα (–1499/–1250) | forward | GCTCTTACGCGTGCTAGCCCTCACAAAGTGCTGGGATT |
| reverse | TACCGGAATGCCAAGCTTAATTCCACAGAAGATTACTACTCAG | |
| Human Rev-erbα (–1249/–1000) | forward | GCTCTTACGCGTGCTAGCCATATTTTATCCTCCAGCACCG |
| reverse | TACCGGAATGCCAAGCTTTCCAGGGGAAGGAGTTCC | |
| Human Rev-erbα (–999/–750) | forward | GCTCTTACGCGTGCTAGCGGTGTTCTCCCTAAGGCGAG |
| reverse | TACCGGAATGCCAAGCTTGCAAGCTTGTTTGCTGTCTG | |
| Human Rev-erbα (–749/–500) | forward | GCTCTTACGCGTGCTAGCCAGTCCCTCCCCAGAAATTC |
| reverse | TACCGGAATGCCAAGCTTCAGTGACACACTTTTCCAACAGC |
Table 5.
Primers for Amplify Full-Length Sequences of CCL21 and β-Defensin-1 Gene for PCR and Sequences of Heterologous Reporter Constructs
| Name | Sequence 5′→3′ |
|---|---|
| CCL21 promoter | GCTCTTACGCGTGCTAGCAAAGACTGAAGAAAAAAAAAAGTCAAAC |
| TACCGGAATGCCAAGCTTTTGGGGGTCTGTGCGTGG | |
| Original sequence | 682 ttg agg tca ggc 694 |
| CCL21 promoter mutant | GCTCTTACGCGTGCTAGCAAAGACTGAAGAAAAAAAAAAGTCAAAC |
| CCGTCTAGAGGAGCAGTGAGATTGTGGAAAG | |
| ATCTCACTGCTCCTCTAGACGGCATGGCTCAGCCTCTTC | |
| CCGTCTAGAGGAGCAGTGAGATTGTGGAAAG | |
| Mutant sequence | 682 ttg cta gac ggc 694 |
| β-defensin-1 promoter | GCTCTTACGCGTGCTAGCGGTGGGATTACAGGTGTGTGC |
| TACCGGAATGCCAAGCTTGGTGAACTTCTAATCGCTAACCCC | |
| Original sequence | 754 cct agg tca ggc 766 |
| β-defensin-1 promoter mutant | GCTCTTACGCGTGCTAGCAAAGACTGAAGAAAAAAAAAAGTCAAAC |
| CCGTCTAGAGGAGCAGTGAGATTGTGGAAAG | |
| ATCTCACTGCTCCTCTAGACGGCATGGCTCAGCCTCTTC | |
| CCGTCTAGAGGAGCAGTGAGATTGTGGAAAG | |
| Mutant sequence | 754 cct cta gac ggc 766 |
PCR, polymerase chain reaction.
Chromatin Immunoprecipitation
AGS cells infected with H pylori (MOI = 100, 24 hours) were treated at room temperature for 10 minutes with 1% formaldehyde in cell culture media. Glycine (11% in media) solution was then gently mixed in at room temperature for 5 minutes to terminate cross-linking. Cells were washed twice with ice-cold PBS and palleted at 3000 g for 5 minutes. Membrane Extraction Buffer containing protease/phosphatase inhibitors was then added to each palleted sample. Cell lysates were pulse-sonicated on ice; supernatants containing the digested chromatin were collected into two tubes for input and immunoprecipitation. Anti-NF-κB p65 or anti-Rev-erbα Ab was added and IP reactions conducted overnight at 4°C with agitation. ChIP grade protein A/G magnetic beads were then added to each IP reaction. Two hours later beads were collected, washed, bounded IP materials eluted with 5 M NaCl containing 20 μg/mL Proteinase K. The cross-linking was reversed by heating up to 65°C for 1.5 hours and DNA was purified. Purified DNA samples were analyzed by PCR with designed primers (Table 6).
Table 6.
Primers for ChIP PCR analysis
| Gene | Primer | Sequence 5′→3′ |
|---|---|---|
| β-defensin-1 | forward | AGCAAGGAAAGCTGTGTTTCGG |
| reverse | GATGCAGTGGGATCCTGTAGCT | |
| CCL21 | forward | GGAGCGTAGTGAGGAGACAGT |
| reverse | GGGCCTGAGCCTTCCTTCTAT |
NOTE. Primers for ChIP analysis of β-defensin-1 and CCL21 were designed and produced by Sangon Biotech (Shanghai, China). Primers for Rev-erbα purchased in Cell Signaling Technology pathway (Cat#:13413).
ChIP, chromatin immunoprecipitation; PCR, polymerase chain reaction.
Flow Cytometry
Cell surface markers were stained with specific or isotype control Abs. For intracellular molecules measurements, the cells were stimulated for 5 hours with phorbol myristate acetate (50 ng/mL) plus ionomycin (1 μg/mL) in the presence of Golgistop (BD Biosciences). Intracellular cytokine staining was performed after fixation and permeabilization, using Perm/Wash solution. Then, the cells were analyzed by multicolor flow cytometry on FACSCanto II and FACSCanto (BD Biosciences). Data were analyzed with FlowJo software (TreeStar, Ashland, OR) or FACSDiva software (BD Biosciences).
Enzyme-Linked Immunosorbent Assay
Isolated human and mouse gastric tissues were homogenized in 1 mL sterile Protein Extraction Reagent, and centrifuged. Tissue supernatants were collected for ELISA. Concentrations of CCL21, Reg3b, β-defensin-1, or IFN-γ in the tissue supernatants; concentrations of CCL21, Reg3b, or β-defensin-1 in the gastric epithelial cell culture supernatants; and concentrations of IFN-γ in the T cell culture supernatants were determined using ELISA kits according to the manufacturer’s instructions.
Western Blot Analysis
Western blots were performed on 10%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel transferred polyvinylidene difluoride membranes with equivalent amounts of cell or tissue lysate protein for each sample. Five percent skim milk was used for blocking the polyvinylidene difluoride membranes. Mouse Rev-erbα, ERK1/2, and p-ERK1/2 were detected with rabbit anti-Rev-erbα Ab, rabbit anti-ERK1/2 Ab, and rabbit anti-p-ERK1/2 Ab; human Rev-erbα, ERK1/2, and p-ERK1/2 were detected with rabbit anti-Rev-erbα Ab, rabbit anti-ERK1/2 Ab, and rabbit anti-p-ERK1/2 Ab; cagA was detected with rabbit anti-cagA Ab respectively. This was followed by incubation with HRP-conjugated secondary Abs. Bound proteins were visualized by using SuperSignal West Dura Extended Duration Substrate kit (Thermo Fisher Scientific).
Statistical Analysis
Results are expressed as mean ± SEM. Student’s t test was generally used to analyze the differences between two groups, but when the variances differed, the Mann-Whitney U test was used. For multiple comparisons, the 1-way analysis of variance was used. Correlations between parameters were assessed using Pearson correlation analysis and linear regression analysis, as appropriate. SPSS statistical software (version 13.0; SPSS Inc, Chicago, IL) was used for all statistical analysis. All data were analyzed using 2-tailed tests, and P < .05 was considered statistically significant. Microarray data analysis was performed with the assistance of Genminix Informatics (Shanghai, China). Raw data from each array were analyzed using TwoClassDif.
CRediT Authorship Contributions
Fang-Yuan Mao (Investigation: Equal)
Yi-Pin Lv (Investigation: Supporting)
Chuan-Jie Hao (Investigation: Supporting)
Yong-Sheng Teng (Investigation: Supporting)
Yu-Gang Liu (Investigation: Supporting)
Ping Cheng (Investigation: Supporting)
Shi-Ming Yang (Resources: Supporting)
Weisan Chen (Writing – review & editing: Supporting)
Tao Liu (Investigation: Supporting)
Quan-Ming Zou (Investigation: Supporting)
Rui Xie (Investigation: Supporting)
Jing-Yu Xu (Investigation: Supporting)
Yuan Zhuang (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Writing Assistance None.
Conflicts of Interest The authors disclose no conflicts.
Funding This work was supported by the National Natural Science Foundation of China (Grant Nos. 81870394 and 82070578), the Chongqing National Science Fund for Distinguished Young Scholars (Grant No. cstc2019jcyjjqX0003), the National Key Research and Development Program of China (Grant No. 2016YFC1302200), and the Science Innovation Capacity Promotion Project of Army Medical University (Grant No. 2019XQY03).
References
- 1.Hooi J.K.Y., Lai W.Y., Ng W.K., Suen M.M.Y., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W.S., Wu J.C.Y., Chan F.K.L., Sung J.J.Y., Kaplan G.G., Ng S.C. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Jin H.S., Kim T.S., Jo E.K. Emerging roles of orphan nuclear receptors in regulation of innate immunity. Arch Pharm Res. 2016;39:1491–1502. doi: 10.1007/s12272-016-0841-6. [DOI] [PubMed] [Google Scholar]
- 3.Mazaira G.I., Zgajnar N.R., Lotufo C.M., Daneri-Becerra C., Sivils J.C., Soto O.B., Cox M.B., Galigniana M.D. The nuclear receptor field: a historical overview and future challenges. Nucl Receptor Res. 2018;5:101320. doi: 10.11131/2018/101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pourcet B., Zecchin M., Ferri L., Beauchamp J., Sitaula S., Billon C., Delhaye S., Vanhoutte J., Mayeuf-Louchart A., Thorel Q., Haas J.T., Eeckhoute J., Dombrowicz D., Duhem C., Boulinguiez A., Lancel S., Sebti Y., Burris T.P., Staels B., Duez H.M. Nuclear receptor subfamily 1 group D member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology. 2018;154:1449–1464.e20. doi: 10.1053/j.gastro.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kojetin D.J., Burris T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis A.M., Bellet M.M., Sassone-Corsi P., O'Neill L.A. Circadian clock proteins and immunity. Immunity. 2014;40:178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Feng D., Liu T., Sun Z., Bugge A., Mullican S.E., Alenghat T., Liu X.S., Lazar M.A. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duez H., van der Veen J.N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Baugé E., Havinga R., Bloks V.W., Wolters H., van der Sluijs F.H., Vennström B., Kuipers F., Staels B. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 9.Cho H., Zhao X., Hatori M., Yu R.T., Barish G.D., Lam M.T., Chong L.W., DiTacchio L., Atkins A.R., Glass C.K., Liddle C., Auwerx J., Downes M., Panda S., Evans R.M. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pariollaud M., Gibbs J.E., Hopwood T.W., Brown S., Begley N., Vonslow R., Poolman T., Guo B., Saer B., Jones D.H., Tellam J.P., Bresciani S., Tomkinson N.C., Wojno-Picon J., Cooper A.W., Daniels D.A., Trump R.P., Grant D., Zuercher W., Willson T.M., MacDonald A.S., Bolognese B., Podolin P.L., Sanchez Y., Loudon A.S., Ray D.W. Circadian clock component REV-ERBalpha controls homeostatic regulation of pulmonary inflammation. J Clin Invest. 2018;128:2281–2296. doi: 10.1172/JCI93910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandra V., Bhagyaraj E., Nanduri R., Ahuja N., Gupta P. NR1D1 ameliorates Mycobacterium tuberculosis clearance through regulation of autophagy. Autophagy. 2015;11:1987–1997. doi: 10.1080/15548627.2015.1091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra V., Bhagyaraj E., Nanduri R., Ahuja N., Gupta P. Human IL10 gene repression by Rev-erbalpha ameliorates Mycobacterium tuberculosis clearance. J Biol Chem. 2013;288:10692–10702. doi: 10.1074/jbc.M113.455915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagnidze K., Hajdarovic K.H., Moskalenko M., Karatsoreos I.N., McEwen B.S., Bulloch K. Nuclear receptor REV-ERBalpha mediates circadian sensitivity to mortality in murine vesicular stomatitis virus–induced encephalitis. Proc Natl Acad Sci U S A. 2016;113:5730–5735. doi: 10.1073/pnas.1520489113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amieva M., Peek R.M., Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150:64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki M., Mimuro H., Kiga K., Fukumatsu M., Ishijima N., Morikawa H., Nagai S., Koyasu S., Gilman R.H., Kersulyte D., Berg D.E., Sasakawa C. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Mueller D., Tegtmeyer N., Brandt S., Yamaoka Y., De Poire E., Sgouras D., Wessler S., Torres J., Smolka A., Backert S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest. 2012;122:1553–1566. doi: 10.1172/JCI61143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hase K., Murakami M., Iimura M., Cole S.P., Horibe Y., Ohtake T., Obonyo M., Gallo R.L., Eckmann L., Kagnoff M.F. Expression of LL-37 by human gastric epithelial cells as a potential host defense mechanism against Helicobacter pylori. Gastroenterology. 2003;125:1613–1625. doi: 10.1053/j.gastro.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Yoshino N., Ishihara S., Rumi M.A., Ortega-Cava C.F., Yuki T., Kazumori H., Takazawa S., Okamoto H., Kadowaki Y., Kinoshita Y. Interleukin-8 regulates expression of Reg protein in Helicobacter pylori-infected gastric mucosa. Am J Gastroenterol. 2005;100:2157–2166. doi: 10.1111/j.1572-0241.2005.41915.x. [DOI] [PubMed] [Google Scholar]
- 19.Akhiani A.A., Pappo J., Kabok Z., Schon K., Gao W., Franzen L.E., Lycke N. Protection against Helicobacter pylori infection following immunization is IL-12-dependent and mediated by Th1 cells. J Immunol. 2002;169:6977–6984. doi: 10.4049/jimmunol.169.12.6977. [DOI] [PubMed] [Google Scholar]
- 20.Sayi A., Kohler E., Hitzler I., Arnold I., Schwendener R., Rehrauer H., Muller A. The CD4+ T cell-mediated IFN-gamma response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J Immunol. 2009;182:7085–7101. doi: 10.4049/jimmunol.0803293. [DOI] [PubMed] [Google Scholar]
- 21.Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Lo Sasso G., Moschetta A., Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woldt E., Sebti Y., Solt L.A., Duhem C., Lancel S., Eeckhoute J., Hesselink M.K., Paquet C., Delhaye S., Shin Y., Kamenecka T.M., Schaart G., Lefebvre P., Nevière R., Burris T.P., Schrauwen P., Staels B., Duez H. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T., Eheim A.L., Klein S., Uschner F.E., Smith A.C., Brandon-Warner E., Ghosh S., Bonkovsky H.L., Trebicka J., Schrum L.W. Novel role of nuclear receptor Rev-erbalpha in hepatic stellate cell activation: potential therapeutic target for liver injury. Hepatology. 2014;59:2383–2396. doi: 10.1002/hep.27049. [DOI] [PubMed] [Google Scholar]
- 24.Yang G., Wright C.J., Hinson M.D., Fernando A.P., Sengupta S., Biswas C., La P., Dennery P.A. Oxidative stress and inflammation modulate Rev-erbalpha signaling in the neonatal lung and affect circadian rhythmicity. Antioxid Redox Signal. 2014;21:17–32. doi: 10.1089/ars.2013.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Wu W.K., Gallo R.L., Fang E.F., Hu W., Ling T.K., Shen J., Chan R.L., Lu L., Luo X.M., Li M.X., Chan K.M., Yu J., Wong V.W., Ng S.C., Wong S.H., Chan F.K., Sung J.J., Chan M.T., Cho C.H. Critical role of antimicrobial peptide cathelicidin for controlling Helicobacter pylori survival and infection. J Immunol. 2016;196:1799–1809. doi: 10.4049/jimmunol.1500021. [DOI] [PubMed] [Google Scholar]
- 26.Sulli G., Rommel A., Wang X., Kolar M.J., Puca F., Saghatelian A., Plikus M.V., Verma I.M., Panda S. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature. 2018;553:351–355. doi: 10.1038/nature25170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbs J.E., Blaikley J., Beesley S., Matthews L., Simpson K.D., Boyce S.H., Farrow S.N., Else K.J., Singh D., Ray D.W., Loudon A.S. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X., Rollins D., Ruhn K.A., Stubblefield J.J., Green C.B., Kashiwada M., Rothman P.B., Takahashi J.S., Hooper L.V. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato S., Sakurai T., Ogasawara J., Takahashi M., Izawa T., Imaizumi K., Taniguchi N., Ohno H., Kizaki T. A circadian clock gene, Rev-erbalpha, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J Immunol. 2014;192:407–417. doi: 10.4049/jimmunol.1301982. [DOI] [PubMed] [Google Scholar]
- 30.Chen L., Li B., Yang W.C., He J.L., Li N.Y., Hu J., He Y.F., Yu S., Zhao Z., Luo P., Zhang J.Y., Li H.B., Zeng M., Lu D.S., Li B.S., Guo H., Yang S.M., Guo G., Mao X.H., Chen W., Wu C., Zou Q.M. A dominant CD4(+) T-cell response to Helicobacter pylori reduces risk for gastric disease in humans. Gastroenterology. 2013;144:591–600. doi: 10.1053/j.gastro.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Molina-Infante J., Romano M., Fernandez-Bermejo M., Federico A., Gravina A.G., Pozzati L., Garcia-Abadia E., Vinagre-Rodriguez G., Martinez-Alcala C., Hernandez-Alonso M., Miranda A., Iovene M.R., Pazos-Pacheco C., Gisbert J.P. Optimized nonbismuth quadruple therapies cure most patients with Helicobacter pylori infection in populations with high rates of antibiotic resistance. Gastroenterology. 2013;145:121–128.e1. doi: 10.1053/j.gastro.2013.03.050. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W., Chen Q., Liang X., Liu W., Xiao S., Graham D.Y., Lu H. Bismuth, lansoprazole, amoxicillin and metronidazole or clarithromycin as first-line Helicobacter pylori therapy. Gut. 2015;64:1715–1720. doi: 10.1136/gutjnl-2015-309900. [DOI] [PubMed] [Google Scholar]
- 33.Gerhart-Hines Z., Feng D., Emmett M.J., Everett L.J., Loro E., Briggs E.R., Bugge A., Hou C., Ferrara C., Seale P., Pryma D.A., Khurana T.S., Lazar M.A. The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature. 2013;503:410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang Y., Cheng P., Liu X.F., Peng L.S., Li B.S., Wang T.T., Chen N., Li W.H., Shi Y., Chen W., Pang K.C., Zeng M., Mao X.H., Yang S.M., Guo H., Guo G., Liu T., Zuo Q.F., Yang H.J., Yang L.Y., Mao F.Y., Lv Y.P., Zou Q.M. A pro-inflammatory role for Th22 cells in Helicobacter pylori-associated gastritis. Gut. 2015;64:1368–1378. doi: 10.1136/gutjnl-2014-307020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv Y.P., Cheng P., Zhang J.Y., Mao F.Y., Teng Y.S., Liu Y.G., Kong H., Wu X.L., Hao C.J., Han B., Ma Q., Yang S.M., Chen W., Peng L.S., Wang T.T., Zou Q.M., Zhuang Y. Helicobacter pylori-induced matrix metallopeptidase-10 promotes gastric bacterial colonization and gastritis. Sci Adv. 2019;5:eaau6547. doi: 10.1126/sciadv.aau6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roussel Y., Wilks M., Harris A., Mein C., Tabaqchali S. Evaluation of DNA extraction methods from mouse stomachs for the quantification of H. pylori by real-time PCR. J Microbiol Methods. 2005;62:71–81. doi: 10.1016/j.mimet.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Mikula M., Dzwonek A., Jagusztyn-Krynicka K., Ostrowski J. Quantitative detection for low levels of Helicobacter pylori infection in experimentally infected mice by real-time PCR. J Microbiol Methods. 2003;55:351–359. doi: 10.1016/s0167-7012(03)00166-0. [DOI] [PubMed] [Google Scholar]