Abstract
Yeast Rcl1 is a potential endonuclease that mediates pre-RNA cleavage at the A2-site to separate 18S rRNA from 5.8S and 25S rRNAs. However, the biological function of Rcl1 in opisthokonta is poorly defined. Moreover, there is no information regarding the exact positions of 18S pre-rRNA processing in zebrafish. Here, we report that zebrafish pre-rRNA harbours three major cleavage sites in the 5′ETS, namely –477nt (A′-site), –97nt (A0-site) and the 5′ETS and 18S rRNA link (A1-site), as well as two major cleavage regions within the ITS1, namely 208–218nt (site 2) and 20–33nt (site E). We also demonstrate that depletion of zebrafish Rcl1 mainly impairs cleavage at the A1-site. Phenotypically, rcl1–/– mutants exhibit a small liver and exocrine pancreas and die before 15 days post-fertilization. RNA-seq analysis revealed that the most significant event in rcl1–/– mutants is the up-regulated expression of a cohort of genes related to ribosome biogenesis and tRNA production. Our data demonstrate that Rcl1 is essential for 18S rRNA maturation at the A1-site and for digestive organogenesis in zebrafish. Rcl1 deficiency, similar to deficiencies in other ribosome biogenesis factors, might trigger a common mechanism to upregulate the expression of genes responsible for ribosome biogenesis.
INTRODUCTION
Biogenesis of ribosomes, the machinery for protein biosynthesis, starts with transcription of the pre-rRNA from the rDNA loci (1–4). The full length eukaryotic pre-rRNA transcript contains the 5′-external transcribed sequence (5′ETS), 18S rRNA, internal transcribed spacer 1 (ITS1), 5.8S rRNA, internal transcribed spacer 2 (ITS2), 28S rRNA and 3′-external transcribed sequence (3′ETS). Maturation of 18S rRNA is mediated by a protein-RNA complex called the small subunit (SSU) processome which is formed stepwise along with the progression of the pre-rRNA transcription. The SSU processome can be divided into three sub-complexes according to their sequential binding to the 5′ETS and 18S rRNA during pre-RNA transcription, namely UTP-A, UTP-B and Mpp10–Imp3–Imp4 (5,6). Once the SSU processome (also called the 90S pre-ribosome) is formed, the properly positioned endonucleases, including Utp23, Utp24, MRP and NOB1, cleave the human pre-rRNA at the sites of A′ (A′ in mouse), A0 (A0 in yeast and mouse), 1 (A1 in yeast and 1 in mouse), 3 (D in yeast and 2 in mouse), E/2a (A2 in yeast and 2b in mouse) and 2 (A3 in yeast and 2c in mouse) to yield the mature 18S rRNA (1,7–12). In humans, UTP23 is proposed to cleave at the A0 site, MRP at site 2, UTP24 at sites 1 and E and NOB1 at site 3 (10,11,13).
RNA terminal phosphate cyclase like 1 (Rcl1) proteins form a protein subfamily that share sequence identity/similarity with the RNA 3′-terminal phosphate cyclase (Rtc) subfamily. However, compared with Rtc proteins, Rcl1 proteins lack the conserved histidine residue that is required for phosphate cyclization, suggesting that these protein families have distinct biochemical functions (14). The function of Rcl1 protein was first studied in yeast, where genetic studies have shown that rcl1 is an essential survival gene (14,15). Immunostaining has shown that Rcl1 is localized in the nucleolus. Molecular studies have shown that Rcl1 is associated with U3 snoRNP, and that depletion of Rcl1 leads to the inhibition of 18S rRNA processing at sites A0, A1 and A2, although processing at the A0 site is less affected than that at the other sites (15). Moreover, biochemical studies suggested that Rcl1 acts as an endonuclease to cleave the pre-RNA at the A2-site (E/2a in human) to separate the 18S rRNA from the 5.8S and 25S rRNAs in yeast (16). However, in vivo RNA-protein crosslinking (CRAC) experiments showed that yeast Rcl1 cross-linked poorly to RNA, and no pre-rRNA target sequence was significantly enriched in the recovered protein products (13). Furthermore, structural analysis and site-directed mutagenesis assay also casted doubt on the endonuclease activity of yeast Rcl1 (17). Despite the fact that the role of RCL1 as an endonuclease has not been fully established in humans and mouse, knockdown of RCL1 in cultured human Hela cells and mouse 3T3 fibroblasts both led to the elevated expression of 18S rRNA intermediates including 30S and 26S in human and 34S and 29S in mouse, suggesting that A0-site and site 1 cleavage was impaired in these cases (11–13). Interestingly, human RCL1 is highly expressed in a subclass of astrocytes in the brain. Linkage and haplotype analyses identified a rare missense in the human rcl1 gene which appears to be associated with depression across multiple generations in extended families (18).
Rcl1 forms a complex with Bms1, a GTPase, and the interaction between Rcl1 and Bms1 is conserved in yeast, humans and zebrafish (19–21). The domains I, II, and III of Bms1 are positioned between Rcl1 and helices 15 and 17 of the 5′ domain of the pre-18S rRNA whereas the domain IV of Bms1l interacts with helix 43 of the 3′ domain in the SSU processome (5,6,22). Rcl1 is imported to the nucleolus for assembly into the SSU processome in a Bms1-dependent manner, as a mutation disrupting the interaction between Rcl1 and Bms1 reduces the nuclear import of Rcl1 and the loss of stable incorporation into the SSU processome (20). In zebrafish, Bms1l is essential for the digestive system development and 18S rRNA processing (16), and a mutation in human BMS1 is (23) associated with aplasia. We ask whether Rcl1, as a partner of Bms1, has similar a biological function in organ development in zebrafish. Moreover, given that the Rcl1-Bms1 complex occupies a specific position in the SSU processome (5,6,22), it would be interesting to explore which cleavage site(s) would be affected during 18S rRNA processing in cells lacking Rcl1, especially at the whole organism level in a metazoan. In this report, we characterized two rcl1 null alleles (rcl1−/−) and found that Rcl1 is essential for digestive system development. We also found that depletion of Rcl1 resulted in the production of aberrant 18S rRNA intermediates. Through a detailed comparison of the profiles of the 18S rRNA intermediates between wild type (WT) and rcl1−/− mutant embryos, we found that depletion of Rcl1 compromised the cleavage of the 5′ETS at the A1-site. Furthermore, through an RNA-seq analysis we found that depletion of Rcl1 up-regulated the expression of a cohort of genes related to ribosome biogenesis, suggesting that Rcl1 plays a role in coordinating ribosomal gene expression.
MATERIALS AND METHODS
Experimental fish and maintenance
A wild-type (WT) zebrafish (Danio rerio) AB strain was used in this study, and all genetic mutants were generated on this background. The animals were raised and maintained according to standard procedures described in ZFIN (http://zfin.org/). The zebrafish line rcl1hi2452 was obtained from a large-scale insertional mutagenesis screen using mouse retroviral vectors as the mutagen (24) and was kindly provided by Professor Nancy Hopkins at Massachusetts Institute of Technology (Cambridge, MA, USA).
We generated another rcl1 mutant allele using the CRISPR-Cas9 method with a specific gRNA (5′-GACGACAATCCAGGACTGAGAGg -3′) targeting exon1 of the zebrafish rcl1 gene according to a protocol described previously (25). The zebrafish transgenic line Tg(bhmt:EGFP) that was used has been described previously (26).
Full-length rcl1 cDNA cloning, mRNA synthesis and microinjection
Total cDNA was synthesized from total RNA extracted from zebrafish embryos, using oligo-dT as the primer and the Super Scriptkit (Invitrogen). Full-length rcl1 cDNA was amplified (Supplementary Table S1) and cloned into the pCS2+ vector. For mRNA injection, mRNA of rcl1 was transcribed in vitro from the linearized pCS2+-rcl1 plasmid with the mMESSAGE mMACHINE SP6 Transcription Kit (Ambion, AM1340) according to the manufacturer's instructions. To overexpress Rcl1 protein, 1 nl of 100ng/μl mRNA was injected into one-cell stage zebrafish embryos.
Western blot analysis
Embryos at desired stages were first deyolked and centrifuged at 2000 × g at 4°C for 2 min. The precipitates were lysed in SDS lysis buffer (50 mM Tris–HCl, pH 6.8, 10% (v/v) glycerol, 5% (v/v) β-ME, 1% (w/v) SDS, 0.01% (w/v) bromophenol blue) supplemented with 1 × cOmplete Protease Inhibitor Cocktail (Roche, Cat#11836145001). Zebrafish adult livers were dissected under a microscope and homogenized in PBS supplemented with 1 × cOmplete Protease Inhibitor Cocktail (Roche, 16829800). After centrifugation at 2000 × g at 4°C for 5 min, the pellet was lysed in SDS lysis buffer supplemented with 1× cOmplete. The protein samples were used immediately for western blot analysis or stored at −20°C for later use.
Northern blot analysis
Total RNA was extracted from fish samples using TRIpure Reagent (Aidlab, RN0102) according to the manufacturer's instructions. Digoxigenin (DIG)-labelled 5′ETS-1, 5′ETS-2, 5′ETS-3, ITS1, ITS2 and rcl1 probes were obtained by PCR with specific primers (Supplementary Table S1) and the corresponding plasmid DNA as the template, together with the DIG DNA Labeling Mix (Roche Diagnostics, 11277065910). Biotin-labelled 5′ETS-5, ITS1-2 and ITS1-3 probes (Supplementary Figure S1A–C) were synthesized by GeneRay Biotech (China). Northern blot hybridization was performed as previously described (27). DIG- and biotin-labelled probes (28) were detected with CDP-Star Chemiluminescent Substrate (Roche, Cat#12041677001) and the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo, Cat#89880), respectively, according to the manufacturers’ instructions.
Whole-mount in situ hybridization (WISH)
WISH probes were DIG-labeled with DIG RNA Labeling Mix (Roche Diagnostics, 11277073910) according to manufacturers’ instructions. For anti-sense and sense rcl1 probes, primers were designed as shown in Supplementary Table S1, and the probes fabp10a, fabp2, trypsin, foxa3, gata6, hhex and prox1 were used as previously described (26).
Immunofluorescence staining
Cryosectioning and immunofluorescence staining of zebrafish larvae were performed as previously described (26,29,30). A rabbit polyclonal antibody against zebrafish Rcl1 (1:200) and a mouse monoclonal antibody against zebrafish Betaine-homocysteine methyltransferase (Bhmt) (1:500) (26) were generated by Hangzhou Hua An Biotechnology Company. Antibodies against Fibrillarin (Fib) (Abcam, Cat#ab4566, Clone#38F3) and phosphorylated-Histone 3(P-H3) (SantaCruz, sc-8656-R) were purchased from Abcam and Santa Cruz Biotechnology, respectively.
Terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay
TUNEL staining was performed using the in situ cell death detection kit, TMR red (Roche Diagnostics, 12156792910) as described previously (31) according to the manufacturer's instructions.
Inverted reverse transcription polymerase chain reaction (RT-PCR)
Inverted RT-PCR was performed on circularized total RNA extracted from 5 days post-fertilisation (dpf) WT and rcl1hi2452 mutant embryos, as described previously (32). Briefly, 5 μg total RNA and T4 RNA Ligase Reaction Buffer (New England Biolabs, M0204S) in a total volume of 15 μl were first incubated for 4 min at 75°C, followed by rapid freezing in pre-cooled ethanol at −80°C for 1 min. Frozen samples were then placed on ice to thaw slowly before PEG8000 was added. Subsequently, ATP, RNase inhibitor (New England Biolabs, M0314) and 2μl T4 RNA Ligase 1 (New England Biolabs, M0204S) were added to yield a final reaction volume of 20 μl according to the manufacturer's instructions. The reaction mixture was incubated at 25°C for 2 h and then boiled for 2 min to terminate the reaction. The circularized total RNA was purified using RNeasy MinElute Cleanup Kit (QIAGEN, 74204) and then subjected to reverse transcription (RT) using Reverse Transcriptase M-MLV (RNase H-) (Takara, 2641A) with random primers. The cDNA was used as the template for RT-PCR with the primer pairs 18S-R1 and 18S-F1 (P1), 18S-R1 and 18S-F2 (P2), 18S-R2 and 18S-F1 (P3), and 18S-R1 and 28S-F (P0) (Supplementary Table S1). The PCR products were ligated into the pGEM-T Easy Vector (Promega, A1360) and single colonies were picked for sequencing.
RNA sequencing (RNA-seq) and data analysis
Total RNA was extracted from WT and rcl1−/− mutant embryos at 4.5 dpf, respectively. RNA library construction and high-throughput sequencing were performed by Beijing Annoroad Gene Technology Company. Briefly, multiplexed libraries were sequenced for 150bp at both ends using an Illumina HiSeq4000 platform. The sequencing data were deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra); the submission ID is SUB6195548, and the BioProject ID is PRJNA561138. Clean reads were mapped to the zebrafish genome (Danio_rerio.GRCz10.84 from ENSEMBL) using TopHat (v2.0.12) with default parameters (33,34). The raw counts generated by TopHat were concatenated together and fed to DEseq to obtain normalized expression levels and screen differentially expressed genes (DEGs) (35). The threshold parameters for DEGs were an absolute fold change ≥2 and q-value <0.05. A Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment analysis was performed using DAVID (version 6.8) (36). A Hierarchical cluster analysis was performed using the R package (v3.1.1).
Real-time quantitative PCR (qPCR)
qPCR analysis was performed to validate the RNA-seq data. Total RNA was treated with DNaseI (Thermo Scientific, EN0521) prior to reverse transcription. First-strand complementary DNA was synthesized using M-MLV Reverse Transcriptase (Invitrogen, 28025-021) and oligo-dT as the primer. qPCR was performed on a CFX96 Real-Time System (Bio-Rad, C1000 Thermal Cycler) with AceQq PCR SYBR Green Master Mix (Vazyme, Q111-02) according to the manufacturer's instructions. All reactions were run in triplicate, and gapdh (glyceraldehyde-3-phosphate dehydrogenase, gi|92097970) was used as the internal reference to normalize gene expression. The primer pairs for genes under investigation are listed in Supplementary Table S1.
Statistical analysis
Student's t-test was used for statistical comparisons (*P < 0.05; **P < 0.01; n.s.: no significant difference). We used SPSS software was used to determine the correlation between the RNA-seq data and the qPCR data through a Spearman bivariate correlation analysis.
RESULTS
Expression of rcl1 is enriched in digestive organs during organogenesis
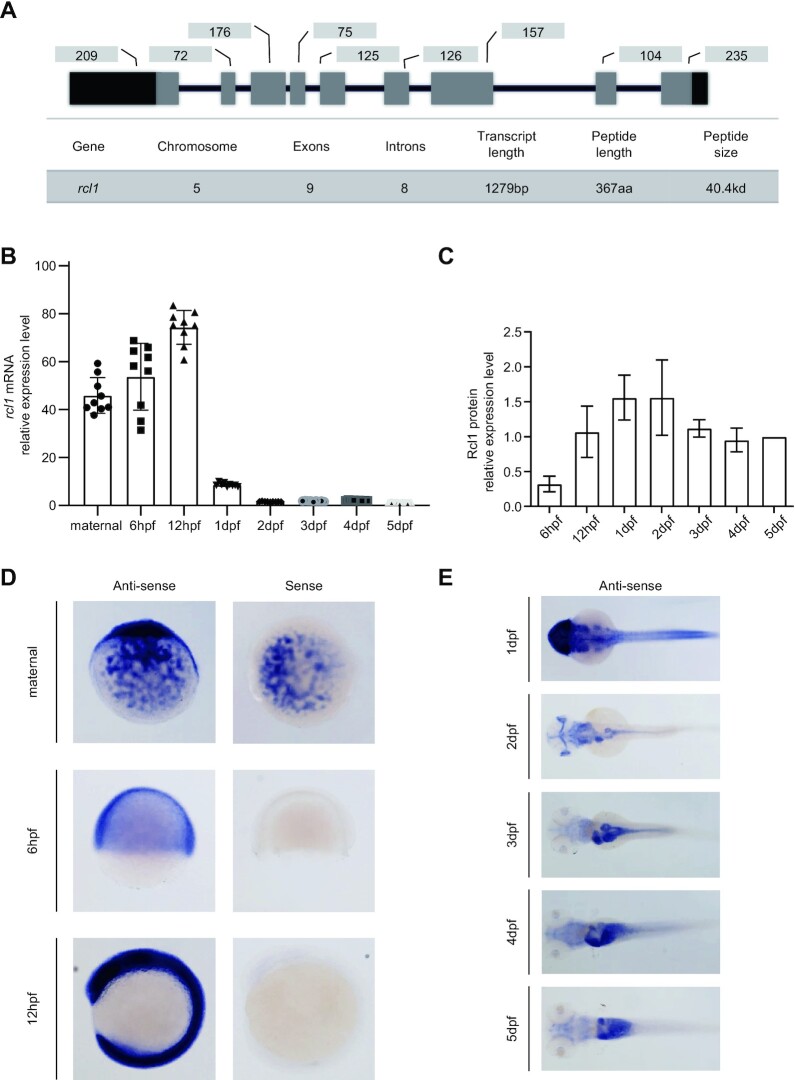
The zebrafish genome contains a single copy of the rcl1 gene located on chromosome 5. The rcl1 gene harbours nine exons and eight introns and spans ∼38 kb of genomic DNA (Figure 1A). The zebrafish rcl1 full length cDNA encodes a peptide of 367 amino acids (aa) (21). Phylogenetic analysis revealed that zebrafish Rcl1 is closely related to its mouse and human counterparts (sharing 74% similarity with both) but is distantly related to its counterparts in yeast (34–41% similarity) and other opisthokonta (Supplementary Figure S2). We first examined the temporal and spatial expression patterns of rcl1 during embryogenesis. The qPCR analysis using primer pair rcl1-F and rcl1-R and northern blot analysis showed that unfertilized eggs contain a high level of the rcl1 transcripts (Figure 1B; Supplementary Figure S3A and B), suggesting that rcl1 is a maternal gene. The zygotic expression of rcl1 peaked at 12hpf and then gradually decreased to a lower level at 5 dpf (Figure 1B; Supplementary Figure S3A and B). Western blot analysis showed that Rcl1 protein displayed an expression pattern consistent with the RNA expression profile (Figure 1C; Supplementary Figure S3C-D). WISH revealed the ubiquitous distribution of rcl1 transcripts in unfertilized eggs and in embryos at 6hpf and 12hpf (Figure 1D). At 1dpf, rcl1 transcripts were highly enriched in the head region and were also detected in the trunk region (Figure 1E). Starting from 3dpf, the expression of rcl1 was gradually enriched in the digestive system, including the oesophagus, intestinal tube, liver, pancreas and swim bladder (Figure 1E).
Figure 1.
Expression of rcl1 is enriched in the digestive organs during organogenesis. (A) Upper panel: a schematic diagram showing the genomic structure of the zebrafish rcl1 gene on chromosome 5. Lower table: summarizing the genomic and transcript information for the rcl1 gene. Dark bar, exon, 5′- and 3′-untranslated region (UTR); gray bar, exon, protein coding region; dark line, introns. Numbers above each exon indicate the length (bp) of reach corresponding exon. (B) Real-time quantitative PCR (qPCR) analysis of rcl1 transcripts at different developmental stages. Total RNA was extracted from a clutch of 100–300 eggs/embryos for each time point. Three repeats, each with triplicate qPCR analysis, were performed. Relative expression level of rcl1 was presented after normalization against gapdh. (C) Statistical analysis of Rcl1 protein levels at different developmental stages from three western blots (Supplementary Figure S3C–E). Total proteins were extracted from a clutch of 100–200 embryos for each time point. The relative signal intensity of the Rcl1 protein band was presented after normalization against the Tubulin control for each time point. (D and E) Whole-mount in situ hybridization (WISH) analysis of the rcl1 expression patterns at different developmental stages using an anti-sense rcl1 probe. The rcl1 sense probe was used as a negative control (D). hpf, hours-post-fertilization; dpf, days-post-fertilization.
Rcl1 is essential for the development of digestive organs
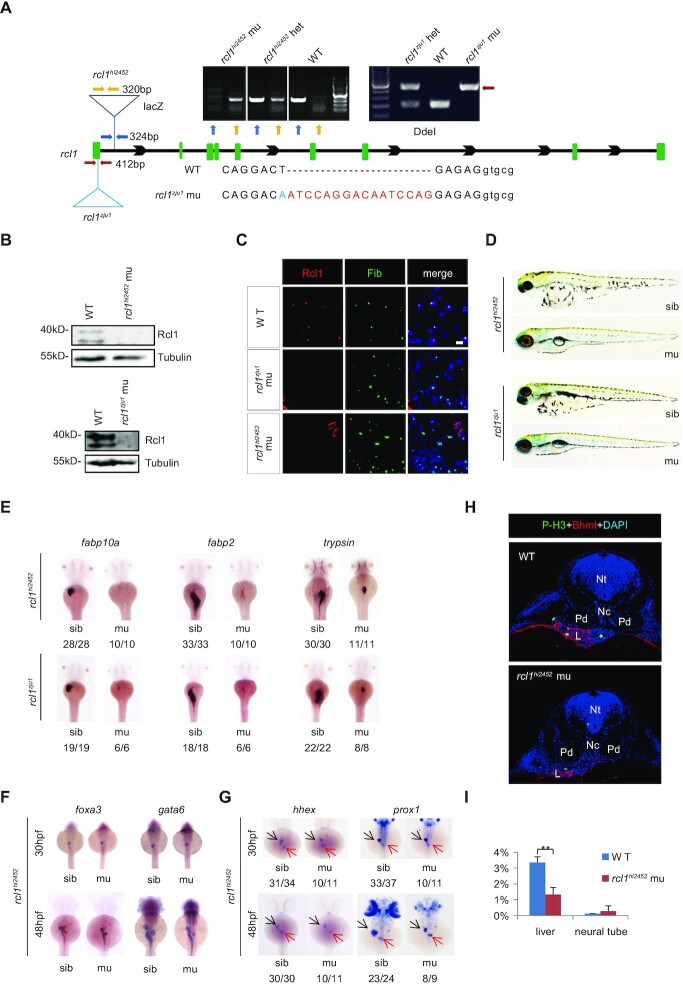
The rcl1hi2452 mutant generated by retroviral insertion mutagenesis was used to study the biological function of rcl1 (24). The retroviral vector was inserted in the first intron of the rcl1 gene (Figure 2A). We also generated a second mutant allele, rcl1zju1, which harbours a 16 base pair (bp) insertion and one base substitution in the first exon, using the CRISPR-Cas9 technique (Figure 2A) (25). This 16bp insertion created an early stop codon in the rcl1 open reading frame (ORF), with a predicted peptide containing 36 N-terminal aa and an additional 18 aa derived from the shift of the rcl1 ORF (Supplementary Figure S4A). We previously showed that both zebrafish and human Rcl1 proteins are conserved nucleolar-localized proteins (21). Western blot analysis failed to detect Rcl1 protein in the rcl1hi2452 and rcl1zju1 mutant embryos at 5dpf (Figure 2B). Immunostaining of Rcl1 revealed that hepatocytes in both the rcl1hi2452 and rcl1zju1 mutants lacked the Rcl1 signal (Figure 2C), confirming that the rcl1 gene is disrupted in these two mutant alleles. Both rcl1hi2452 and rcl1zju1 homozygous mutant embryos displayed smaller eyes, heart oedema and absence of the swim-bladder but a relatively normal body length at 5dpf (Figure 2D). Neither rcl1hi2452 nor rcl1zju1 mutants could survive beyond 15dpf, as no homozygous mutants were identified at this time point (Supplementary Figure S4B). WISH using the fabp10a (liver marker) and fabp2 (intestine marker) probes revealed that the liver and intestine tube were almost absent in rcl1hi2452 and rcl1zju1 mutant embryos at 3dpf (Figure 2E). WISH using the trypsin (exocrine pancreas marker) probe showed that the size of the exocrine pancreas was greatly reduced (Figure 2E). An allelism test showed that the rcl1hi2452/zju1 embryos exhibited defective development of the liver, exocrine pancreas, and intestine tube (Supplementary Figure S4C). These data demonstrate that rcl1 is essential for the development of digestive organs.
Figure 2.
Depletion of Rcl1 causes hypoplastic digestive organs. (A) Schematic drawing shows the genomic DNA structure of the rcl1 gene and relative positions of the rcl1hi2452 (the viral vector insertion in intron 1) and rcl1zju1 (16bp insertion in the exon1) mutation sites. Primer pairs used for genotyping the two mutant alleles are indicated by different color arrowheads, and a gel photo of their corresponding PCR products is shown above the drawing. Yellow arrowheads, primer pair for checking the viral insertion in heterozygous (+/−) or homozygous (−/−) mutants; blue arrowheads, for checking the wild-type gene in wild-type sibling (+/+) or heterozygous mutant (+/−); red arrowheads, for genotyping the rcl1zju1 mutant by checking the PCR product digested by Dde1 (WT but not the mutant PCR product can be cleaved by Dde1). Lower panel: showing the DNA sequence changes in rcl1zju1 mutant (red letters, insertion; blue letters, substitution; capital letters, exon; small letters: intron). (B) Western blot of Rcl1 protein in embryos at 5dpf. Upper panels: WT and rcl1hi245; lower panels: WT and rcl1zju1. (C) Immunostaining of Rcl1 and Fibrillarin (Fib) in the liver of WT, rcl1hi2452 and rcl1zju1 mutant at 5dpf. Nuclei are stained with DAPI; Scale bars: 50 μm. (D) Comparison of overall morphology between rcl1hi2452and rcl1zju1embryos with their corresponding sibling at 5dpf. (E–G) WISH using the fabp10a, fabp2 and trypsin probes on 3dpf-old rcl1hi2452 and rcl1zju1 intercross embryos (E). WISH using the foxa3 and gata6 probes (F) and hhex and prox1 (G) on rcl1hi2452 intercross embryos at 30hpf and 48hpf, respectively. Denominator vs numerator: number of embryos exhibiting the phenotype vs number of embryos of corresponding genotype examined. Black arrow, liver bud; red arrow, exocrine pancreas. (H) Immunostaining of P-H3 (green) and Bhmt (red) on cryosections obtained from 2.5dpf-old WT and rcl1hi2452 mutant embryos (H). Nuclei are stained with DAPI; Nt: neural tube; Nc: notochord; Pd: pronephric duct; L: liver. (I) Quantitative analysis of the P-H3 positive cells in the liver and neural tube in WT and rcl1zju1 mutants from three embryos, respectively. **P < 0.01.
We next performed WISH using the early pan-endodermal markers foxa3 and gata6 and the early hepatic markers prox1 and hhex to determine the point at which the mutant liver phenotype is discernible. The result showed that all four markers did not reveal obvious phenotypic differences between WT and rcl1hi2452 embryos at 30hpf (Figure 2F and G). However, at 48hpf, the four markers revealed that the mutant liver bud and intestinal bulb were much smaller than those observed in the WT (Figure 2F and G). Therefore, similar to the observations of other nucleolar factors, including pes (31), def (31,37,38), bms1l (21,39), sas10 and mpp10 (30), mutation in rcl1 did not affect the initiation of digestive organogenesis but did retard their growths.
We then performed TUNEL staining to compare the level of apoptosis between WT and rcl1hi2452 but failed to find any significant difference (Supplementary Figure S4D). In contrast, staining to detect phosphorylated-Histone 3 (P-H3) (indicative of the G2 to M transition) revealed that the ratio of P-H3-positive cells in WT (3.36%) was much higher than that in rcl1hi2452 (1.33%) (Figure 2H and I; Supplementary Table S2). Therefore, the small liver phenotype of rcl1hi2452 is due to impaired hepatoblast proliferation rather than elevated apoptosis.
Loss-of-function of Rcl1 impairs the maturation of 18S rRNA
In yeast, the Rcl1 null mutant shows defective processing at sites A0, A1 and A2 of 35S pre-rRNA (15). Biochemical studies have shown that yeast Rcl1, as an endonuclease, mainly targets pre-rRNA at the A2-site to separate the 18S rRNA precursor from the 5.8S and 25S precursor (16,40). However, this function of Rcl1 is controversial based on a structural and site-directed mutagenesis analysis in yeast (17), and has not been fully established in humans (13,16). To determine whether Rcl1 is also involved in pre-rRNA processing in zebrafish, we first compared the ratios of mature 28S to 18S rRNA in rcl1hi2452 and rcl1zju1homozygotes with their corresponding siblings (mixture of heterozygotes and WT genotypes) and with the WT control at 5dpf using an Agilent Bioanalyzer 2100. We found that the ratios of 28S to 18S rRNA in both mutants (4.2:1 in rcl1hi2452, 4.15:1 in rcl1zju1) were significantly higher than those in the siblings (2.8:1 in siblings containing rcl1hi2452/+ and WT, 2.5:1 in siblings containing rcl1zju1/+ and WT) or the WT control (2.65:1) (Figure 3A and B). Next, to compare the intermediates of the processed pre-rRNA in WT, rcl1zju1 and rcl1hi2452 mutants, we performed northern blot analysis using probes derived from the 5′ETS, ITS1 and ITS2 regions, respectively (Figure 3C). The result showed that the 5′ETS-1 probe mainly detected bands a and b while ITS1-1 probe detected bands a, b and d in the WT control (Figure 3D) as reported previously (40–42). In contrast, in both mutants, the 5′ETS-1 probe detected a new band c in addition to bands a and b (Figure 3D). Interestingly, the ITS1-1 probe also detected band c but failed to detect band d in either of the mutants when compared with the WT (Figure 3D). Products b, c and d migrated between mature 28S and 18S rRNA, indicating that they were 18S rRNA precursors. Importantly, the size of product c was smaller than that of product b (detected by both 5′ETS-1 and ITS1-1 probes) but larger than d (detected by the ITS1-1 probe only). No obvious aberrant intermediates were detected by the ITS2-1 probe (Figure 3D). As band c contains both 5′ETS and ITS1 sequences, we considered it likely that a major impairment of 18S rRNA processing has probably occurred at the 5′ETS region in the rcl1−/− mutants.
Figure 3.
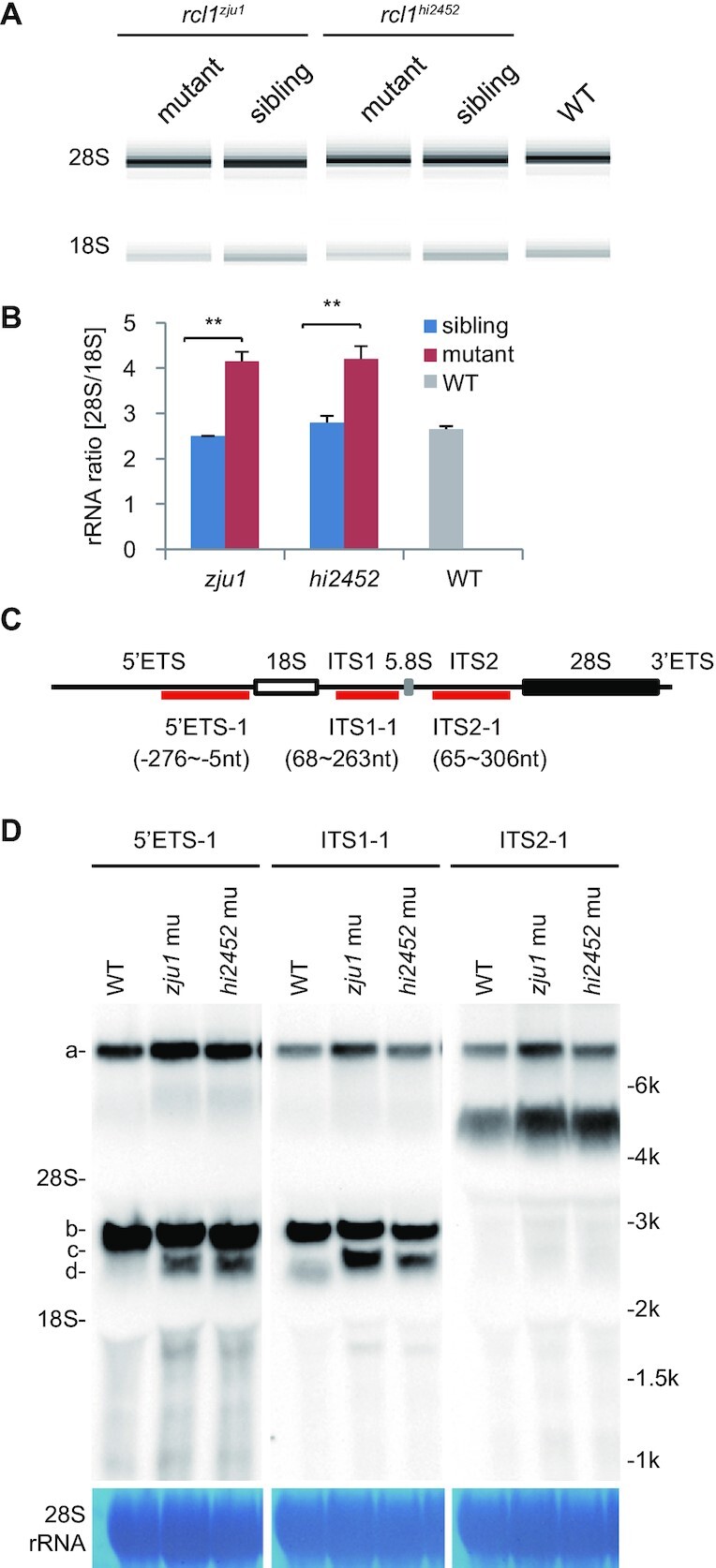
Depletion of Rcl1 impairs the maturation of 18S rRNA. (A) Analysis of total RNA from 5dpf-old rcl1hi2452 and rcl1zju1 mutants and their WT-phenotype siblings and also WT embryos by Agilent 2100 Bioanalyzer. (B) Statistic analysis of the ratios of mature 28S/18S rRNA in 5dpf-old rcl1hi2452 (hi2452) and rcl1zju1 (zju1) mutants and their WT-phenotype siblings (sibling) and also WT based on the data from Agilent 2100 Bioanalyzer. **P < 0.01. (C) Schematic drawing showing the structure of the zebrafish pre-rRNA and the relative positions of probes (5′ETS-1, ITS1-1, ITS2-1) designed for northern blot analysis of different pre-rRNA intermediates. The 18S, 5.8S and 28S mature rRNAs are indicated as white, gray and black boxes, respectively. (D) Northern blot analysis of pre-rRNA intermediates in 5dpf-old WT, rcl1hi2452 (hi2452) and rcl1zju1 (zju1) mutant embryos using the 5′ETS-1, ITS1-1 and ITS2-1 probes. Methylene blue staining of 28S rRNA: loading control. The pre-rRNA intermediates are highlighted to the left as a, b, c and d, respectively. To facilitate the comparison, same samples were reloaded three times on the same gel for electrophoresis. The filter was cut and subjected to the northern blot analysis using the 5′ETS-1, ITS1-1 and ITS2-1 probes, respectively.
-447nt in the 5′ETS of zebrafish pre-rRNA might serve as the A′ cleavage site
The exact steps of 18S rRNA maturation in zebrafish remains elusive, and previous reports tended to use information from yeast and humans as a reference (40–42). Before comparing the 18S rRNA intermediates between the WT and rcl1hi2452 mutant embryos, we first characterized the identities of 5′ETS and 3′ETS in the pre-RNA of WT zebrafish. To this end, we extracted the total RNA from 5dpf-old WT embryos and circularized the RNA using T4 RNA ligase1 (32) and used this as the template for inverted RT-PCR with the primer pair 18S-R1 and 28S-F1 (P0) (Figure 4A). The inverted RT-PCR products were cloned into the pGEM-T Easy vector, and colonies were randomly picked. Sequencing analysis identified –631nt and –447nt within the 5′ETS (Supplementary Figure S5A; Supplementary Table S3), suggesting that the pre-rRNA transcription start site (TSS) probably resides around the -631nt region upstream of the first base of 18S rRNA, and that –447nt likely represents the cleavage site corresponding to the A′-site in humans (11,13). We also noticed a 41nt site downstream of the last base of 28S rRNA, suggesting that this sequence probably represents the 3′ETS (Supplementary Figure S5A; Supplementary Table S3). Interestingly, the –447nt 5′ETS and 41nt 3′ETS sites were also detected in the inverted RT-PCR products (Supplementary Table S3), suggesting that the cleavage at the A′-site is the earliest event in the pre-rRNA processing, as in humans (11,13).
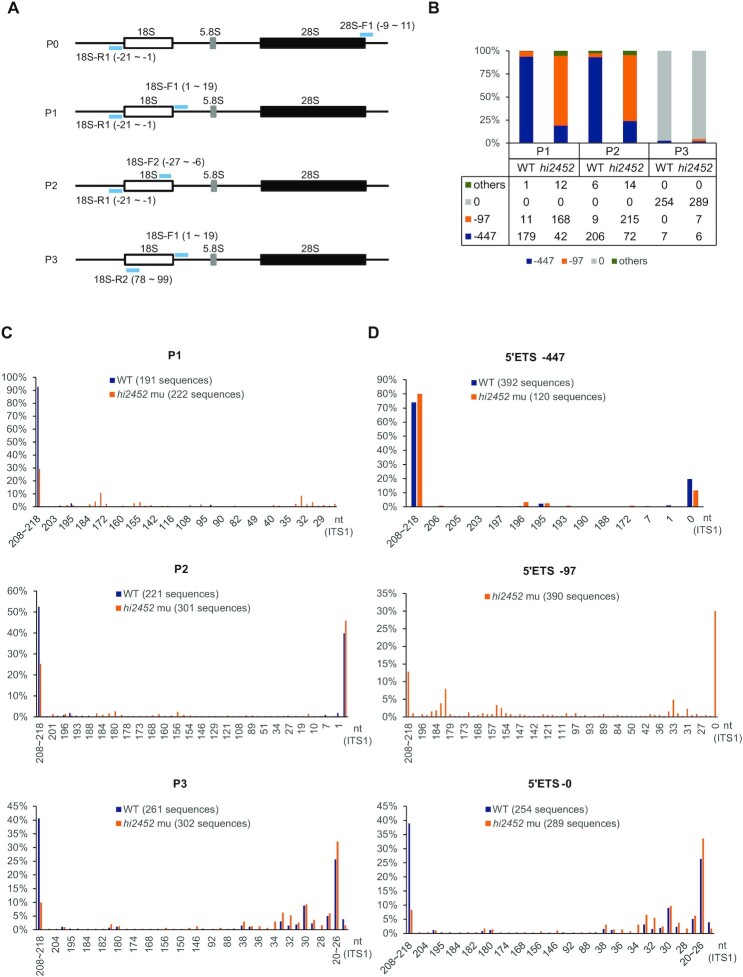
Figure 4.
Depletion of Rcl1 compromised the cleavage of 18S pre-rRNA at the A1-site. (A) Schematic drawing showing the structure of the zebrafish pre-rRNA and the positions of four primer pairs (light blue bar) used in IPCR. P0: 18S-R1 (R1) and 28S-F1; P1: 18S-R1 and 18S-F1; P2: 18S-R1 and 18S-F2; P3: 18S-R2 and 18S-F1. (B) Histogram shows the ratios of -97nt, -447nt and other 5′ETS sequences based on sequencing the IPCR products obtained using the P1, P2 and P3 primer pairs, respectively. Table below the histogram shows the numbers of the sequenced single colonies (combined from three independent repeats) from WT and rcl1hi2452 for each primer pair. (C) Histogram comparing the patterns and ratios of ITS1 sequences between WT and rcl1hi2452 (hi2452) in the clones obtained by using P1, P2 and P3 primer pairs, respectively. (D) Histogram comparing the patterns and ratios of ITS1 sequences in the clones containing the –447nt 5′ETS (top panel) or without the 5′ETS sequence (5′ETS-0) (bottom panel) between WT and rcl1hi2452 (hi2452). Since there were very few (only 20 out of 673) WT clones containing the –97 5′ETS, in view of statistics, the analysis was only done for the rcl1hi2452 (hi2452) mutant (total 390 sequences available) (middle panel). In C and D, the number after WT and hi2452 represented the total number of sequences analyzed for each primer pair.
–97nt in the 5′ETS of zebrafish pre-rRNA might serve as the A0 cleavage site, and Rcl1 depletion leads to accumulation of the 18S rRNA intermediates containing –97nt 5′ETS
To characterize 18S rRNA intermediates in the WT and rcl1hi2452 mutant embryos in detail, we used the circularized RNA as the template for inverted RT-PCR with the primers pairs 18S-R1 and 18S-F1 (P1), 18S-R1 and 18S-F2 (P2) and 18S-R2 and 18S-F1 (P3) (Figure 4A; Supplementary Table S1). The inverted RT-PCR products from three independent repeats were cloned and sequenced. To summarize, sequences from 191 and 222 individual clones from P1 (Supplementary Table S4), 221 and 301 from P2 (Supplementary Table S5), and 261 and 302 from P3 (Supplementary Table S6) were obtained for the WT control and rcl1hi2452 mutant, respectively.
We first compared the patterns of 5′ETS sequences between WT and rcl1hi2452. For the P1 and P2 primer pairs (sharing the common primer 18S-R1, which targets a sequence at the 3′-end of the 5′ETS region adjacent to the 5′-end of mature 18S; Figure 4A), we identified two major types of 5′ETS sequences in both WT and rcl1hi2452, comprising 447nt (-447nt) and another 97nt (-97nt) upstream of the first base of mature 18S rRNA (Figure 4B; Supplementary Tables S4 and S5). In the WT, the ratios of the –97nt to –447nt were 11:179 (–97nt: ∼6%) and 9:206 (–97nt: ∼4%) for P1 and P2, respectively, suggesting that the –447nt 5′ETS intermediate is the primary form of the 18S rRNA precursors (Figure 4B; Supplementary Tables S4 and S5). To our surprise, the ratios of the –97nt to –447nt in rcl1hi2452 were 168:42 (–97nt: 80%) and 215:72 (–97nt: ∼75%) for P1 and P2, which were considerably higher than those observed in the WT (Figure 4B; Supplementary Tables S4 and S5). As expected, for the P3 primer pair (containing the primer 18S-R2 derived from the 5′-end of the mature 18S; Figure 4A), more than 95% of the clones lacked a 5′ETS sequence in both the WT and rcl1hi2452 (Figure 4B; Supplementary Table S6). Interestingly, we noticed that, in several of the clones containing the 5′ETS sequences, only the –447nt sequence was identified in the WT, whereas both the –447nt and –97nt sequences were identified in rcl1hi2452 (Figure 4B; Supplementary Table S6). These results suggest that: (i) following the cleavage at the A′-site (-447nt), there is a quick cut at the –97nt site prior to the cleavage at the A1-site (which separates the 5′ETS from 18S rRNA); and (ii) depletion of Rcl1 impairs A1-site cleavage. The –97nt site likely corresponds to the A0-site in humans (11,13).
The region of 208–218nt region in ITS1 serves as the cleavage site for the separation of 18S preRNA and 5.8S–28S pre-rRNAs in both WT and rcl1hi2452
Next, we analyzed the ITS1 sequences. In both WT and rcl1hi2452, for all 1,498 clones obtained from P1, P2 and P3, the ITS1 sequences ranging from 208nt to 218nt (208-to-218nt) downstream of the last base of the mature 18S rRNA were identified to be the longest product (a stretch of 11 T nucleotides between 208nt and 218nt may explain the length variation; Supplementary Figure S5A) (Figure 4C; Supplementary Tables S4–S6). The 208-to-218nt 5′ETS clones accounted for ∼93% in WT and ∼29% in rcl1hi2452 in P1 clones, and ∼41% in WT and ∼10% in rcl1hi2452 in P3 clones, respectively (Figure 4C; Supplementary Table S4 and S6). With regard to the P2 primer pair, the clones mainly contained either the 208-to-218nt ITS1 sequences (∼53% in WT, ∼25% in rcl1hi2452) or no ITS1 sequences in either WT or rcl1hi2452 (∼40% in WT, ∼46% in rcl1hi2452) (Figure 4C; Supplementary Table S5). These results suggest that: (i) the region of 208–218nt region in ITS1 is the primary cleavage site for separating 18S pr-rRNA and 5.8S-28S pre-rRNA, and likely corresponds to site 2 in humans (11,13); and (ii) after cleavage at site 2, further cleavage to separate ITS1 from 18S rRNA is a quick process.
Rcl1 depletion does not halt ITS1 processing after the cleavage at the site 2
We then compared the ITS1 sequence patterns in clones containing -447nt or -97nt or without a 5′ETS (5′ETS-0) between WT and rcl1hi2452, respectively. Strikingly, the majority of clones containing the -447nt 5′ETS harboured the 208-to-218nt ITS1 in both WT (∼74% out of 392 clones) and rcl1hi2452 (∼80% out of 120 clones), irrespective of whether P1, P2 or P3 were used (Figure 4D; Supplementary Table S7). Interestingly, ∼20% of WT and ∼12% of rcl1hi2452 -447nt 5′ETS clones lacked an ITS1 (Figure 4D; Supplementary Table S7). These results strongly suggest that cleavage at site 2 in the ITS1 to separate 18S rRNA from 5.8S–28S rRNAs is the major event prior to cleavage at the A1-site, and that separation of ITS1 from 18S can occur prior to cleavage at the A1-site.
Regarding the clones harbouring the –97nt 5′ETS, only 20 out of 673 WT clones from all three primer pairs were identified, suggesting that cleavage at the A1-site is normally a quick process in the WT. Among the 390 clones harbouring –97nt 5′ETS identified in rcl1hi2452, ∼13% harboured the 208-to-218nt ITS1 and ∼30% lacked an ITS1 (Figure 4D; Supplementary Table S7). These results suggest that 1) cleavage at the A1-site is impaired in rcl1hi2452; and 2) the ITS1 in 18S rRNA intermediates containing the -97nt 5′ETS can be processed further.
Regarding the clones that lacked a 5′ETS (5′ETS-0), the 208-to-218nt ITS1 was found in ∼39% of 254 WT clones, suggesting that cleavage at the A1-site occurred prior to the processing of ITS1 in this case. However, the ratio of the 208-to-218nt ITS1 clones was drastically lower in rcl1hi2452, accounting for only ∼8.3% of 289 clones (Figure 4D; Supplementary Table S7). Interestingly, ∼18% and ∼26% out of 254 WT 5′ETS-0 clones contained the 29-to-33nt and 20-to-26nt sequences, respectively (Figure 4D; Supplementary Table S7). Similarly, a prominent percentage of 29-to-33nt (∼28%) and 20-to-26nt (34%) ITS1 sequences were observed in 289 rcl1hi2452 5′ETS-0 clones (Figure 4D; Supplementary Table S7). These results suggest that (i) the zebrafish 20–33nt ITS1 region might serve as a proximal cleavage site after the cleavage of site 2, and likely corresponds to the E/2a site in humans (11,13); and (ii) Rcl1 depletion does not stop further shortening or processing of the ITS1 towards site E after cleavage at the A1-site and site 2, but somehow delays the cleavage of 20–33nt by exonucleolytic enzymes to produce mature 18S rRNA.
Rcl1 depletion causes an aberrant accumulation of 18S intermediates containing the -97nt 5′ETS
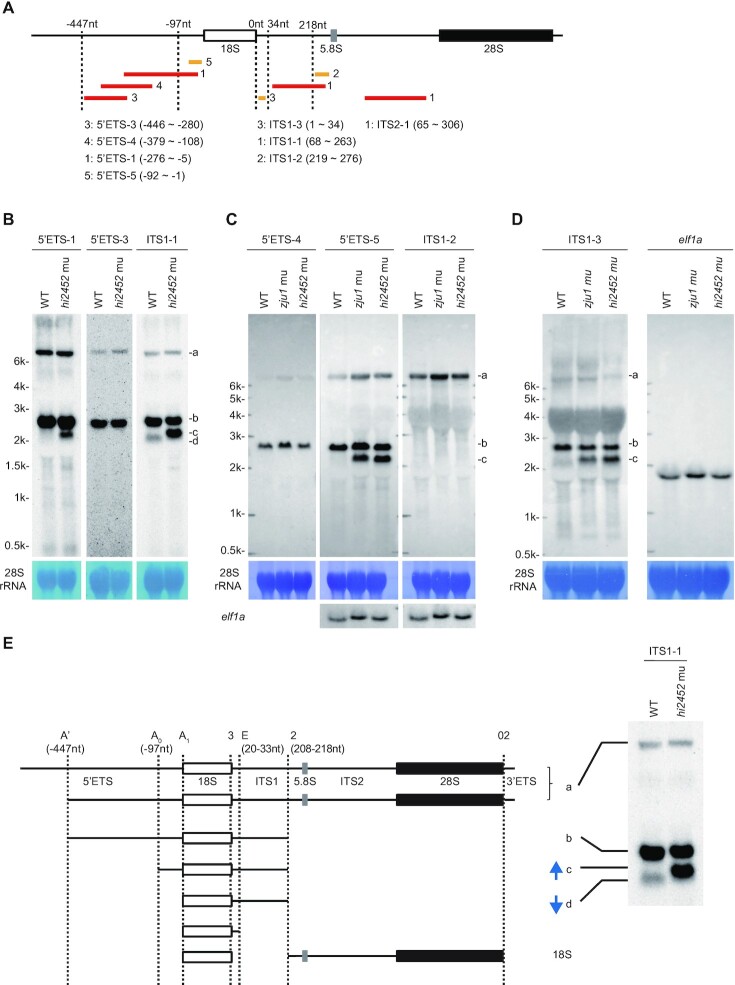
Based on the above, we postulated the identities of the b, c and d bands observed in Figure 3D. The b band is likely the 18S intermediates containing the –447nt 5′ETS and the 208-to-218nt ITS1, the c band likely contains the –97nt 5′ETS and the 208-to-218nt ITS1 and the d band likely contains 5′ETS-0 and the 208-to-218nt ITS1. We designed five additional probes for use in northern blotting to confirm this hypothesis (Figure 5A). The 5′ETS-3 and 5′ETS-4 probes contain the sequences between –280 and –446nt, and between -108 and -379nt upstream of the first base of the mature 18S rRNA, respectively, and are expected to detect the 18S rRNA intermediates harbouring the –447nt 5′ETS but not the –97nt 5′ETS. The 5′ETS-5 probe contains the sequence from -1 to -92nt upstream of the first base of the mature 18S rRNA and is expected to detect 18S rRNA intermediates harbouring either the –447nt or -97nt 5′ETS. The ITS1-3 probe contains the 1–34nt sequence downstream of the last base of the mature 18S rRNA and is expected to detect 18S rRNA intermediates harbouring the 2–33nt ITS1 sequence. The ITS1-2 probe contains the 219–276nt sequence downstream of the last base of the mature 18S rRNA and is expected to detect the 5.8S–28S pre-RNAs (Figure 5A). Northern blot analysis showed that the 5′ETS-3 probe detected bands a and b in WT and rcl1hi2452 (Figure 5B), and that the 5′ETS-4 probe detected bands a and b in the WT and the rcl1hi2452 and rcl1zju1 mutants (Figure 5C), demonstrating that band b contains the -447nt 5′ETS. The 5′ETS-5 probe detected bands a and b in the WT, and bands a, b and c in the rcl1hi2452 and rcl1zju1 mutants (Figure 5C; Supplementary Figure S5B), demonstrating that band c represents the 18S rRNA intermediates containing the -97nt 5′ETS. The ITS1-1 probe (68 to 263nt of ITS1) detected bands a and b in the WT and the rcl1hi2452 and rcl1zju1 mutants (Figures 3D and 5B), suggesting that band b contains both the -447nt 5′ETS and 208-to-218nt ITS1. However, the ITS1-1 probe only detected band d but not c in WT, and only detected band c but not band d in the two mutants (Figures 3D and 5B). These findings suggest that the band d represents the 18S rRNA intermediates containing ITS1, without a 5′ETS, whereas band c represents the 18S rRNA intermediates containing the –97nt 5′ETS and 208–218nt ITS1. Therefore, depletion of Rcl1 impairs the cleavage at the A1-site. Interestingly, ITS1-3 detected bands identical to ITS1-1 (Figure 5D; Supplementary Figure S5C), suggesting that the level of 18S rRNA intermediates containing 5′ETS-0 and 2–33nt ITS1 was very low although such intermediates were detected by IPCR. ITS1-2 only detected band a (Figure 5C; Supplementary Figure S5D), suggesting that the ITS1 sequence is rapidly cleaved in the 5.8S-28S intermediates after cleavage at site 2.
Figure 5.
Defining major processing steps for 18S rRNA maturation in zebrafish. (A) Schematic drawing showing the positions of 8 probes used in northern blotting analysis of different pre-rRNA intermediates. The relative –447nt and -97nt positions in the 5′ETS, 34nt and 218nt positions in the ITS1 are indicated. Red bar: DIG labelled probes; yellow bar: biotin labelled probes. The correspondence between the number after the red or yellow bar and a specific probe is highlighted. (B–D) Northern blot analysis of pre-rRNA intermediates in WT and rcl1hi2452 (B) or in WT, rcl1hi2452 and rcl1zju1 (C and D) at 5dpf using corresponding probes as indicated. The pre-rRNA intermediates are marked to the right as a, b, c and d. Methylene blue staining of 28S rRNA: loading control; elf1a in (C): loading control; elf1a in (D): same samples loaded twice on the same gel for electrophoresis, then the filter was cut and subjected to northern blot analysis using ITS1-3 and elf1a probes, respectively. (E) Diagram illustrates the major processing steps for 18S rRNA maturation in zebrafish. Cleavage sites at A′ (-447nt 5′ETS), A0 (-97nt 5′ETS) and A1 (5′ETS-0) in 5′ETS, at 2 (208–218nt ITS1) and E (20–33nt ITS1) in ITS1 and at 02 in 3′ETS are indicated by vertical dotted lines. The pre-rRNA intermediates a, b, c and d correspond to that detected in northern blots as shown on the right (copied from Figure 5B, panel for the ITS1 probe). Blue arrow indicates the up-regulation of band c and down-regulation of band d in rcl1hi2452 and rcl1zju1 mutants as evident in Figures 3D and 5B-D.
Major steps of 18S rRNA maturation in zebrafish
Based on the above data, we outlined the key steps of 18S rRNA processing in zebrafish (Figure 5E). A full length pre-rRNA transcript has a 631nt 5′ETS and 41nt 3′ETS (Figure 5E, Supplementary Figure S5A). Cleaving the full length pre-rRNA transcript at the A′-site only (producing the –447nt 5′ETS) will yield band a, which can be detected by the 5′ETS-1 to 5′ETS-5 and also the ITS1-1 to ITS1-3 and ITS2 probes (Figures 3D and 5B–E). Cleavage at the A′-site and site 2 two sites will yield band b, which can be detected by 5′ETS-1 to 5′ETS-5, ITS1-1 and ITS1-3, but not by ITS1-2 and ITS2-1 (Figures 3D and 5B–E). Cleavage at the A0-site and site 2 two sites will yield band c, which can be detected by 5′ETS-1 and 5′ETS-5, ITS1-1 and ITS1-3, but not by 5′ETS-3, 5′ETS-4, ITS1-2 and ITS2-1 (Figures 3D and 5B-E). Cleavage at the A1-site and site 2 two sites will yield band d, which can only be detected by the ITS1-1 and ITS1-3 probes (Figures 3D and 5B–E).
Maternal WT rcl1 mRNA is undetectable at 3.5dpf
RNA-seq experiments were planned to study the effect of Rcl1 depletion on expression profiles in the rcl1zju1 mutant. Prior to RNA-seq experiments, we first investigated the fate of maternal WT rcl1 mRNA by obtaining cDNA via RT of total RNA extracted from WT and rcl1zju1 mutant embryos at 3.5dpf, 4.5dpf and 5.5dpf. The purpose of this pilot analysis was to determine the time point for harvesting RNA samples when the effect of the maternal WT Rcl1 on expression profiles in the rcl1zju1 mutant could be exclude. The RT products were used as the template for PCR with a primer pair that amplified the rcl1 cDNA region containing the 16bp insertion in rcl1zju1. The RT-PCR products were cloned into the pGEM-T Easy Vector, and 27–60 single colonies at each time point were picked for sequencing. Sequencing analysis showed that no WT rcl1 transcripts were detected in the rcl1zju1 homozygous mutant embryos at any of the three time points examined (Figure 6A), suggesting that maternal WT rcl1 transcripts were degraded before 3.5dpf. Interestingly, we noticed that in 4.5dpf-old rcl1zju1/+ heterozygotes, the ratio of WT rcl1 clones to rcl1zju1 mutant clones was 30:2, instead of the expected 1:1; this suggests that the rcl1zju1 mutant mRNA is unstable, possibly due to the effect of RNA decay on nonsense mutant mRNA (43,44).
Figure 6.
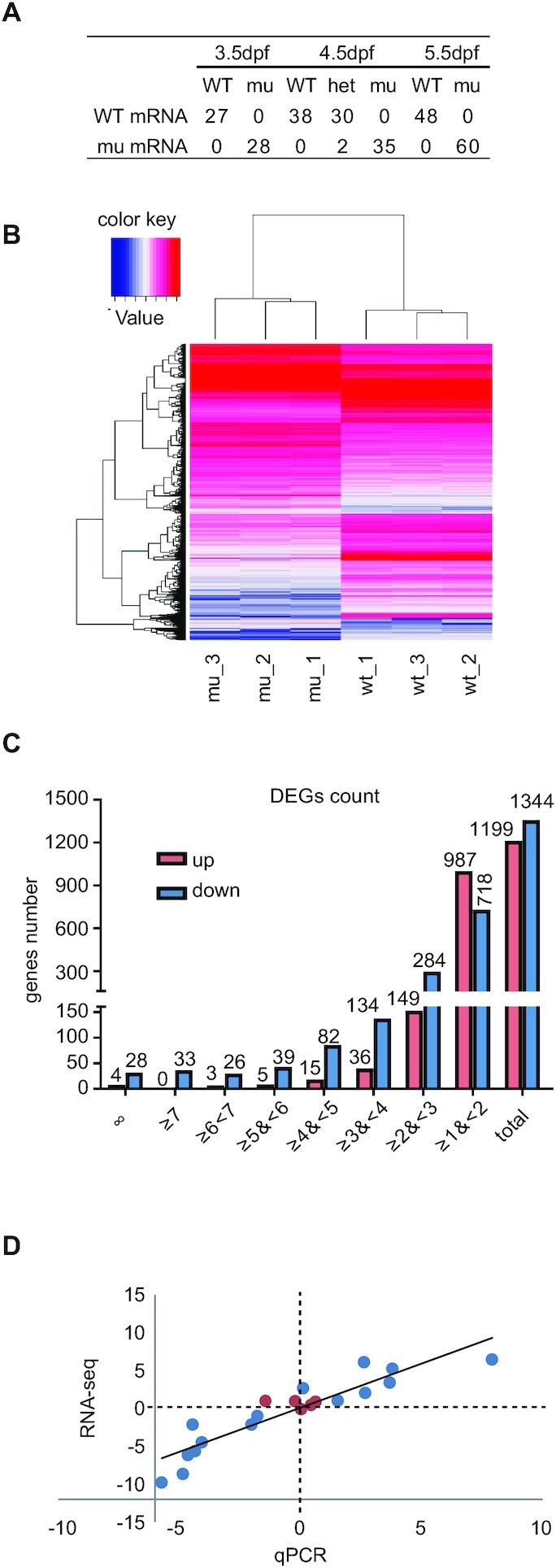
Depletion of Rcl1 causes down-regulation of 1344 genes and up-regulation of 1199 gene. (A) Sequencing analysis of WT rcl1 and mutant rcl1zju1 transcripts in the RT-PCR products obtained from WT, rcl1zju1/+ heterozygous and rcl1zju1 homozygous progenies of the rcl1zju1/+ intercross at 3.5dpf, 4.5dpf and 5.5dpf. (B) Hierarchical Clustering analysis of the RNA-seq data between WT and rcl1zju1 mutant with three biological repeats. (C) The log2 value distributions of the 1,199 significant up-regulated and 1,344 down-regulated genes (cut-off value: log2≧|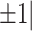 , P <0.05) in rcl1zju1 mutant at 4.5dpf. (D) Confirmation of the RNA-seq data by the qPCR analysis of 20 randomly selected genes in WT and rcl1zju1 mutant. Expression data of genes detected by RNA-seq was plotted against those by qPCR. The reference line indicates the linear correlation between the RNA-seq and qPCR. Blue dot: gene with significant change in RNA-seq; red dot: gene without significant change.
, P <0.05) in rcl1zju1 mutant at 4.5dpf. (D) Confirmation of the RNA-seq data by the qPCR analysis of 20 randomly selected genes in WT and rcl1zju1 mutant. Expression data of genes detected by RNA-seq was plotted against those by qPCR. The reference line indicates the linear correlation between the RNA-seq and qPCR. Blue dot: gene with significant change in RNA-seq; red dot: gene without significant change.
Obtaining high quality RNA-seq data for WT and rcl1zju1 embryos at 4.5dpf
To explore whether and how Rcl1 depletion alters the expression profiles, we performed RNA-seq experiment to compare the transcriptomes between WT and rcl1zju1 mutant embryos at 4.5dpf. We first checked the quality of the RNA-seq data from three independent WT mRNA pools and three rcl1zju1 mutant mRNA pools. Cleans reads from all six samples were exceeded 6GB, and data filtering based on the Clean Q30 Bases Rate program revealed that the Q30 for all six samples was more than 96% (Supplementary Table S8), demonstrating that the RNA-seq data obtained were of high quality. Next, we aligned the clean sequences to the zebrafish genome (Danio_rerio.GRCz10.84). On average, 45,347,516 clean sequences (∼85% of total clean reads) matched a cDNA counterpart in the zebrafish genome (Supplementary Table S9). The expression levels of a zebrafish genes in each sample were calculated based on the RPKM method (Reads per Kilobase Millon Mapped Reads) (45). Clustering analysis using the Hierarchical Cluster showed that the RNA-seq data were highly consistent among the three WT samples and the three mutant samples (Figure 6B). Moreover, a cross comparison of gene expression using the DEseq method (35) identified 1344 down-regulated genes (log2 ≤ –1, P < 0.05) and 1199 up-regulated genes (log2 ≥ 1, P < 0.05) in the rcl1zju1 mutant embryos (Figure 6C; Supplementary Table S10). The log2 values of 1002 out of the 1344 down-regulated genes (∼75%) fell in the range of –1 to –3 (Supplementary Table S11), while among the 1199 up-regulated genes, the log2 values of 1136 (∼95%) genes fell in the range of 1–3 (Supplementary Table S12). The rcl1 transcripts were among the list of the 1344 down-regulated genes (log2 = –2.4, P < 0.05), demonstrating the reliability of the obtained data (Supplementary Table S11). To further evaluate the quality of the data, we analyzed via qPCR the expression of 20 randomly selected genes in WT and rcl1zju1, including seven up-regulated, eight down-regulated and five no-change genes. The data exhibited excellent agreement with the RNA-seq data (Figure 6D; Supplementary Table S13). In addition, a Spearman bivariate correlation analysis showed a highly significant correlation (P < 0.01, coefficient = 0.935) between the results of qPCR and RNA-seq, demonstrating the reliability of our RNA-seq data.
Depletion of Rcl1 alters the expression of transcription factor and signalling molecule genes associated with the development of digestive organs
Digestive organogenesis involves phases of progenitor specification, cell differentiation, and organ outgrowth. These developmental phases are spatiotemporally governed by specific transcription factors and signalling molecules (46,47). We searched for genes encoding proteins known to regulate the development of digestive organs in the list of down-regulated genes (Supplementary Table S11). We found that hnf4α (gi|34194038, involved in liver development) (48), ptf1a (gi|46518518, involved in exocrine pancreas development) (49,50), and cdx1b (gi|148922965, involved in intestine development) (51,52) were among the down-regulated genes encoding transcription factors. In contrast, p53 is upregulated, as observed in other mutants defective in ribosome biogenesis (30) (Supplementary Table S14). In addition, genes encoding signalling molecules, including Fgf 4B-like (gi|688570986), Fgf 19 (gi|55700026), Bmp 8A (gi|113678863), WNT1 inducible signalling pathway protein 1b (gi|260081535) and TGF-β receptor-associated protein 1-like (gi|688594019), were found in the down-regulated gene panel (Supplementary Table S11). Interestingly, some genes encoding well-known transcription factors involved in the specification of the endoderm and organogenesis of digestive organs were not identified in either the down- or up-regulated gene list (Supplementary Table S14), probably due to the fact that the RNA-seq sample collection time was 4.5dpf, when most of these processes have already been completed. Alterations in the expression of these transcription factors and signalling molecule genes coincided with the hypoplastic digestive organ phenotype exhibited by the rcl1zju1 mutant.
GO analysis of 1,344 down-regulated genes reveals that the metabolic process is the most significantly compromised biological process in rcl1zju1
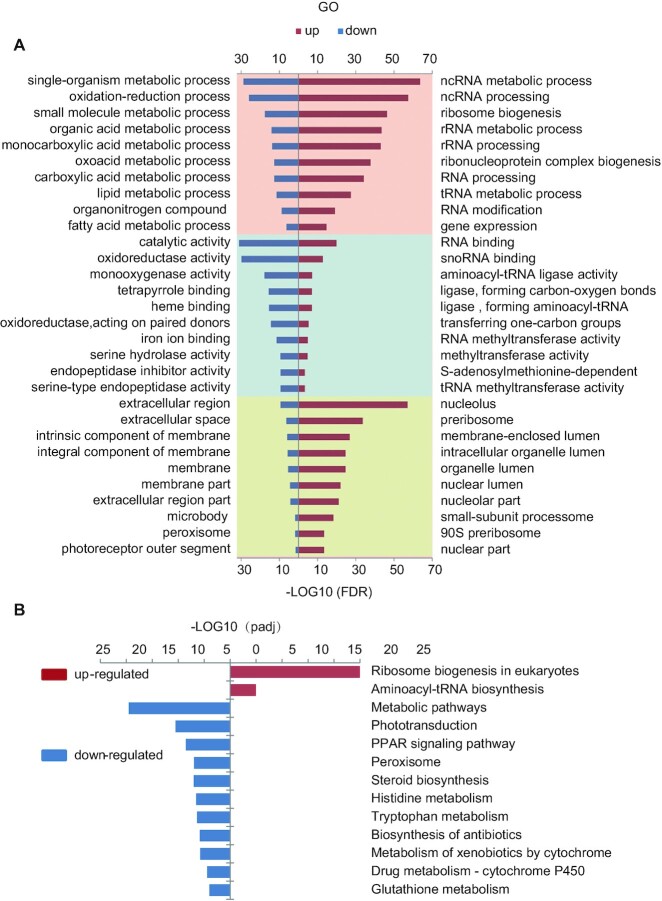
We then adopted the DAVID bioinformatics resources (version 6.8) (53,54) to perform a gene ontology (GO) analysis (36) of the 1,344 down-regulated genes (Supplementary Table S11). The top 10 significantly affected items in both the GO biological process (GO_BP) and molecular function (GO_MF) categories were related to metabolic activities (Figure 7A, left column; Supplementary Table S15). Indeed, all top 10 items in the GO_BP category belonged to metabolic processes, and included genes involved in single-organism metabolic process (233 genes), oxidation-reduction processes (116 genes), small molecule metabolic processes (120 genes), organic acid metabolic processes (74 genes) and lipid metabolic processes (81 genes) (Figure 7A, left column; Supplementary Table S15). The first item in the GO-MF category contained 484 genes that were predicted to encode proteins with catalytic activities (Figure 7A, left column; Supplementary Table S15), followed by genes associated oxidoreductase activity (132 genes), monooxygenase activity (44 genes), tetrapyrrole binding (44 genes), heme binding activity (43 genes) and serine-type endopeptidase activity (Figure 7A, left column; Supplementary Table S15). The down-regulation of genes involved in the metabolic activities in the rcl1zju1 mutant is reflective of the fact that the mutant is severely defective in the development of digestive organs (Figure 2E). Interestingly, in the GO cellular component (GO_CC) category, the top 10 items mainly contained genes associated with the extracellular region/space or the membrane part (Figure 7A; left column; Supplementary Table S15).
Figure 7.
GO and KEGG enrichment analysis of the DEGs reveled an up-regulation of a cohort of genes related to ribosome biogenesis. (A) Top 10 significantly enrichment GO analysis for up-regulated (right panel, red bar) and down-regulate (left panel, blue bar) DEGs in BP, MF and CC items. BP: biological process (pink panel, top); MF: molecular function (light blue panel, middle); CC: cellular component (yellow panel, bottom). (B) Significantly enriched KEGG pathway for up-regulated (right panel, red bar) and down-regulate (left panel, blue bar) DEGs.
GO analysis of 1,199 up-regulated genes reveals that a cohort of genes involved in rRNA maturation and ribosome biogenesis are up-regulated in rcl1zju1
Next, the 1,199 up-regulated genes were subjected to a GO analysis. Strikingly, the GO_BP analysis of the up-regulated genes revealed that among the top 10 significantly affected items, 9 were related to RNA regulation, including non-coding RNA (ncRNA) metabolic process (113 genes), ncRNA processing (95 genes), ribosome biogenesis (79 genes), rRNA metabolic process (65 genes) and rRNA biogenesis (64 genes) (Figure 7A, right column; Supplementary Table S16), which suggests abnormally high rRNA processing and ribosome biogenesis activity in the rcl1zju1 mutant. This observation was further confirmed by the cellular component (GO_CC) analysis, which showed that 5 out of the top 10 items were involved in nucleolar function, including items of the nucleolus (83 genes), preribosome (43 genes), nucleolar part (28 genes), SSU processome (23 genes) and 90S preribosome (18 genes) (Figure 7A, right column; Supplementary Table S16). Surprisingly, none of the most significantly affected top 10 items of the 1344 down-regulated genes in either the GO-BP or GO-CC category were reflected in the regulation of rRNA processing or ribosome biogenesis (Figure 7A, left column; Supplementary Table S15). Therefore, the depletion of Rcl1 up-regulated the expression of a cohort of genes involved in rRNA processing and ribosome biogenesis (Supplementary Figure S6A and B).
Depletion of Rcl1 causes up-regulation of a cohort of genes involved in the tRNA metabolic process
In addition to the genes involved in rRNA production, we found that the tRNA metabolic process (53 genes) was among the top 10 items in the GO_BP analysis of the up-regulated genes (Figure 7A, right column; Supplementary Table S16). In the GO_MF analysis, 2 of the top 10 items involved genes encoding ligase activity for the formation of aminoacyl-tRNA and related compounds (17 genes) and those encoding tRNA methyltransferase activity (13 genes) (Figure 7A, right column; Supplementary Table S16). Moreover, 17 of the 45 genes associated with ligase activity for the formation of aminoacyl-tRNA and related compounds genes were significantly up-regulated. The remaining genes, which are located in the mitochondria, maintained a stable expression level under Rcl1 depletion compared with WT, with the exception of si:ch211-256m1.8, which was significantly decreased (Supplementary Figure S6C).
Consistency between KEGG and GO analysis
We also used DAVID (53,54) to perform a Kyoto Encyclopedia of Genes and Genomes (KEGG) (55) analysis to systematically analyze the down- and up-regulated genes in the rcl1zju1 mutant. A KEGG analysis of the 1,344 down-regulated genes identified that metabolic pathways were the most significantly affected (149 genes, P < 2.4e−22) (Figure 7B, Supplementary Table S17). Of the 1,199 up-regulated genes, KEGG analysis revealed that the ribosome biogenesis (37 genes) and aminoacyl-tRNA biosynthesis (16 genes) were the two most significantly affected pathways (Figure 7B, Supplementary Table S17). Importantly, all of the above genes are among the corresponding list of genes in the corresponding items in the GO_BP category (Supplementary Table S16). Therefore, both the KEGG and GO analysis suggests that a cohort of genes involved in ribosome biogenesis and tRNA metabolic process is significantly up-regulated in the rcl1zju1 mutant.
DISCUSSION
The processes of 18S rRNA maturation and ribosome biogenesis in yeast, mouse and human cells have been defined previously (1,8–12). However, little is known about the steps of 18S rRNA maturation in other higher eukaryotes, especially in multicellular animals at the whole organismal level. In this report, we used four pairs of primers to perform the inverted RT-PCR, using the circularized RNA from zebrafish as the substrate. The P1, P2 and P3 three primer pairs were designed to amplify the precursor containing 5′ETS and ITS1 whereas the P0 pair (18S-R and 28S-F) for the precursor containing the 5′ETS and 3′ETS. Sequencing individual clones containing the inverted RT-PCR product allowed us to identify different intermediates during pre-rRNA processing. Based on the sequence data obtained in this study and the known human rRNA processing pathway, we propose for the first time the cleavage steps during18S rRNA maturation in zebrafish and the related sites within the 5′ETS and ITS1 (Figure 5E) (1,56).
Separation of the 18S precursor from the 5.8S–28S rRNA precursors by cleavages within the ITS1 is a key step in 18S maturation. In yeast, cleavage at the proximal site A2, rather than the distal site A3, is usually the initial step to separate the pre-40S and pre-60S processing pathways. In contrast, in human cells, cleavage at the distal site 2 is the predominant cleavage that separates pre-40S and pre-60S processing (8–13). In both humans and yeast, RNase MRP has been identified as the endonuclease that cleaves at the distal ITS1 site (i.e. human site 2 and yeast site A3), indicating that the importance of MRP mediated cleavage is different between human and yeast (10,13). Although the function of Rcl1 as an endonuclease for the A2 (yeast) and E/2a (humans) cleavage has not been fully established (13,16,17), it is known that depletion of yeast Rcl1 or knockdown of human Rcl1 both did not obviously affect the cleavage at site A3 (yeast) or site 2 (humans), however, did cause accumulation of aberrant precursors without cleavage at site A2 or E/2a (13,15), demonstrating that the RNase MRP cleavage is independent of Rcl1 function. Our data strongly suggest that the cleavage at the distal site (208–218nt) in the zebrafish ITS1 is the primary cleavage responsible for separation of the 18S pre-rRNA and 5.8S–28S pre-rRNA. Furthermore, cleavage at this site appears to be independent of Rcl1 function, given that the 18S rRNA intermediates in both the WT and rcl1−/− mutant harbour an identical ITS1 sequence of the length 208-to-218nt ITS1 sequence. Therefore, it appears that the process of zebrafish 18S rRNA maturation resembles that in humans. Future work is required to determine whether RNase MRP is responsible for the site 2 (208-to-218nt) cleavage in zebrafish.
Depletion of yeast Rcl1 and knockdown of human Rcl1 are known to affect the cleavage at the A1-site (yeast) and site 1 (human) (13,15). We found that that the 18S rRNA intermediates harbouring the -97nt 5′ETS sequence were accumulated in the zebrafish rcl1−/− mutant, strongly suggesting that depletion of Rcl1 impairs the cleavage at the A1-site. Therefore, the function of Rcl1 in cleavage at the A1-site is conserved among these species.
In both yeast and human, it is clear that Rcl1 is needed for both A0 and A2 cleavage (yeast) and A0 and E/2a cleavage (human). The loss of Rcl1 causes the ITS1 spacer region to be cleaved by RNase MRP at the distal site (i.e. human site 2 or yeast site A3) only, leading to the accumulation of precursors with a 3′-end at this site (13,15). Surprisingly, no obvious accumulation of pre-rRNA intermediates uncleaved at these two sites was observed in rcl1−/− mutants. In contrast, depletion of Rcl1 in zebrafish appears to increase the ratio of intermediates containing the 20–33nt ITS1 region after the cleavage at the A1 site. One explanation for this difference is that in zebrafish, unlike in yeast, Rcl1 is the key endonuclease responsible for cleavage at the A1-site rather than the site E. Alternatively, Rcl1 may cleave at the site E, and the functions of other endonucleases (such as Utp24) compensates for the depletion of Rcl1 (13). Due to a lack of concrete evidence to establish vertebrate Rcl1 as an endonuclease, it is also possible that depletion of Rcl1 simply alters the tertiary structure of the 90S pre-ribosome, which hinders the access of the endonuclease responsible for cleaving the A1-site. However, we cannot exclude the possibility that after cleavage of the A1-site and site 2, Rcl1-independent exonucleases such as the RNA exosome further process the intermediates from the 3′-end towards the site E; in this case, cleavage at the proximal site E/2a might not be essential for 18S maturation (10,11,13). Further experiments are needed to clarify this point.
As Rcl1 interacts with Bms1l to form a sub-complex in the SSU processome (5,6,20,21), we considered whether the bms1l−/− genetic mutant would also exhibit similar 18S rRNA processing defects. The results of northern blotting result showed that no aberrant -97nt 5′ETS products were detected when using the 5′ETS and ITS1 probes (39). Other zebrafish SSU processome component knockout mutants, such as sas10, mpp10, def/utp25 and bap28/utp10, also show different processing defect patterns from the rcl1 mutant (30,40,41). Therefore, it appears that Rcl1 has a unique function in RNA processing, compared with other SSU processome proteins in zebrafish.
Ribosome biogenesis is an extremely complex process that requires the fine-tuned coordination of hundreds of nucleolar proteins and RNA molecules to produce 18S, 5.8S and 28S rRNA (2). An intriguing question is whether the genes encoding these factors are expressed in a coordinative way, and if yes, how is this coordination achieved? We found some clues suggesting an answer to this question in our RNA-seq data. It is somewhat expected that many genes related to metabolic activity would be down-regulated in the rcl−/− mutant, given that the mutant embryos suffered from hypoplasia of the entire digestive system. Additionally, we found that the expression of genes encoding transcription factors responsible for the differentiated functions of digestive organs, for example, hnf4α in hepatocytes, ptf1a in the exocrine pancreas and cdx1b in the intestinal tube, were down-regulated in the rcl−/− mutant. Surprisingly, we noticed that depletion of Rcl1 up-regulated the expression of a cohort of genes related to rRNA processing and ribosome biogenesis, which were among the top 10 items in the GO analysis. Moreover, a cohort of genes related to tRNA function and maturation were also found to be up-regulated. EXOSC8 and EXOSC9 are components of the RNA exosome, which is essential for ribosome biogenesis (57,58). We noted that a remarkable ratio of genes related to ribosome biogenesis (63 out of the 70 transcripts) were upregulated in the exosc8 and exosc9 zebrafish mutants (58). Despite reviewing the RNAseq data obtained from morphants, including RPS19, RPS24, RPL5 and RPL11, after gene knockdown using specific morpholinos (MO) (59–62), we failed to identify uniform gene signatures among these data. One explanation for this could be that the genes encode ribosomal proteins whose roles in rRNA processing are negligible. Alternatively, the sampling stages or MO injection might make the data non-comparable. Nevertheless, certain ribosome biogenesis genes were upregulated in these cases (62). These results suggest that cells might have evolved a common mechanism to coordinate the expression of genes involved in rRNA transcription, processing, maturation and ribosome biogenesis and those encoding the components of protein translation in response to defective rRNA processing and ribosome biogenesis. Considering the diverse biochemical nature of the different factors involved in rRNA processing and ribosome biogenesis, the abnormal intermediates of pre-18S rRNA may serve as guide RNA to activate the gene compensation system and thus up-regulate a cohort of genes that share a common regulatory cis-element responsive to the gene compensation pathway. It would be of great interest to explore the mechanism behind these findings in the future.
Defective in proteins or protein function involved in ribosome biogenesis and function has been reported to lead to ribosomopathies (63). A subset of ribosomopathies are related to abnormalities in neural crest development, which will give rise to a diverse cell lineage, including melanocytes, smooth muscle cells, craniofacial cartilage and bone, peripheral and enteric neurons, and glia (64). The rcl−/− mutant exhibits abnormal craniofacial and muscle development (data not shown) in addition to digestive organ defects, suggesting that depletion of Rcl1, which is highly expressed in the brain region at 3dpf, also influences the craniofacial development. Human RCL1 is highly expressed in a subclass of astrocytes in the brain, and a rare missense in the rcl1 gene appears to be associated with depression across multiple generations in extended families (18). Based on the rcl−/− mutant phenotypes and protein similarity with human RCL1, it would be interesting to exploit the zebrafish rcl+/− mutant to study depression.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Drs Yong Wang, Ce Gao, Yihong Guan, Shuyi Zhao, Enhui Shen and all members in the labs of JRP and JC for their valuable suggestions. The authors are grateful to Zhengxin Xu and Xiangfeng Shen for their technical support with animal studies.
Contributor Information
Qinfang Zhu, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
Boxiang Tao, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
Hong Chen, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
Hui Shi, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
Ling Huang, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
Jun Chen, College of Life Sciences, Zhejiang University, Hangzhou 310058, China.
Minjie Hu, Department of Embryology, Carnegie Institution for Science, Baltimore, MD 21218, USA.
Li Jan Lo, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
Jinrong Peng, MOE Key Laboratory for Molecular Animal Nutrition, College of Animal Sciences, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (http://www.nsfc.gov.cn/); ‘973 Program’ of the Ministry of Science and Technology of China [31771596, 31830113, 31571495, 2017YFA0504501]. Funding for open access charge: National Natural Science Foundation of China [31571495].
Conflict of interest statement. None declared.
REFERENCES
- 1. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2015; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Woolford J.J., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fromont-Racine M., Senger B., Saveanu C., Fasiolo F.. Ribosome assembly in eukaryotes. Gene. 2003; 313:17–42. [DOI] [PubMed] [Google Scholar]
- 4. Tao B., Lo L.J., Peng J., He J.. rDNA subtypes and their transcriptional expression in zebrafish at different developmental stages. Biochem. Biophys. Res. Commun. 2020; 529:819–825. [DOI] [PubMed] [Google Scholar]
- 5. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Kos-Braun I., Kos M., Thoms M., Berninghausen O., Beckmann R.et al.. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell. 2016; 166:380–393. [DOI] [PubMed] [Google Scholar]
- 6. Chaker-Margot M., Barandun J., Hunziker M., Klinge S.. Architecture of the yeast small subunit processome. Science. 2017; 355:eaal1880. [DOI] [PubMed] [Google Scholar]
- 7. Barandun J., Hunziker M., Klinge S.. Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 2018; 49:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charette J.M., Baserga S.J.. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA. 2010; 16:2156–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mullineux S.T., Lafontaine D.L.. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand. Biochimie. 2012; 94:1521–1532. [DOI] [PubMed] [Google Scholar]
- 10. Cerezo E., Plisson-Chastang C., Henras A.K., Lebaron S., Gleizes P.E., O’Donohue M.F., Romeo Y., Henry Y.. Maturation of pre-40S particles in yeast and humans. Wiley Interdiscip. Rev. RNA. 2019; 10:e1516. [DOI] [PubMed] [Google Scholar]
- 11. Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J., Watkins N.J.. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013; 200:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang M., Anikin L., Pestov D.G.. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res. 2014; 42:11180–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wells G.R., Weichmann F., Colvin D., Sloan K.E., Kudla G., Tollervey D., Watkins N.J., Schneider C.. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 2016; 44:5399–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Billy E., Hess D., Hofsteenge J., Filipowicz W.. Characterization of the adenylation site in the RNA 3'-terminal phosphate cyclase from Escherichia coli. J. Biol. Chem. 1999; 274:34955–34960. [DOI] [PubMed] [Google Scholar]
- 15. Billy E., Wegierski T., Nasr F., Filipowicz W.. Rcl1p, the yeast protein similar to the RNA 3'-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000; 19:2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horn D.M., Mason S.L., Karbstein K.. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 2011; 286:34082–34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka N., Smith P., Shuman S.. Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis. RNA. 2011; 17:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amin N., de Vrij F., Baghdadi M., Brouwer R., van Rooij J., Jovanova O., Uitterlinden A.G., Hofman A., Janssen H., Darwish M.S.et al.. A rare missense variant in RCL1 segregates with depression in extended families. Mol. Psychiatry. 2018; 23:1120–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karbstein K., Jonas S., Doudna J.A.. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005; 20:633–643. [DOI] [PubMed] [Google Scholar]
- 20. Delprato A., Al K.Y., Perebaskine N., Monfoulet C., Henry Y., Henras A.K., Fribourg S.. Crucial role of the Rcl1p-Bms1p interaction for yeast pre-ribosomal RNA processing. Nucleic Acids Res. 2014; 42:10161–10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Zhu Q., Huang L., Zhu Y., Chen J., Peng J., Lo L.J.. Interaction between Bms1 and Rcl1, two ribosome biogenesis factors, is evolutionally conserved in zebrafish and human. J Genet Genomics. 2016; 43:467–469. [DOI] [PubMed] [Google Scholar]
- 22. Cheng J., Kellner N., Berninghausen O., Hurt E., Beckmann R.. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017; 24:954–964. [DOI] [PubMed] [Google Scholar]
- 23. Marneros A.G. BMS1 is mutated in aplasia cutis congenita. PLos Genet. 2013; 9:e1003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S., Hopkins N.. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.W., Xi J.J.. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013; 23:465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao C., Huang W., Gao Y., Lo L.J., Luo L., Huang H., Chen J., Peng J.. Zebrafish hhex-null mutant develops an intrahepatic intestinal tube due to de-repression of cdx1b and pdx1. J. Mol. Cell Biol. 2019; 11:448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo J., Lee S., Xu M., Liu F., Ruan H., Eun A., He Y., Ma W., Wang W., Wen Z.et al.. 15000 unique zebrafish EST clusters and their future use in microarray for profiling gene expression patterns during embryogenesis. Genome Res. 2003; 13:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Q., Mao Z., Li S., Hu J., Zhu Y.. A non-radioactive method for small RNA detection by northern blotting. Rice (N Y). 2014; 7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guan Y., Huang D., Chen F., Gao C., Tao T., Shi H., Zhao S., Liao Z., Lo L.J., Wang Y.et al.. Phosphorylation of Def regulates nucleolar p53 turnover and cell cycle progression through Def recruitment of Calpain3. PLoS Biol. 2016; 14:e1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao S., Chen Y., Chen F., Huang D., Shi H., Lo L.J., Chen J., Peng J.. Sas10 controls ribosome biogenesis by stabilizing Mpp10 and delivering the Mpp10-Imp3-Imp4 complex to nucleolus. Nucleic Acids Res. 2019; 47:2996–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J., Ruan H., Ng S.M., Gao C., Soo H.M., Wu W., Zhang Z., Wen Z., Lane D.P., Peng J.. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005; 19:2900–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rene O., Alix J.H.. Late steps of ribosome assembly in E. coli are sensitive to a severe heat stress but are assisted by the HSP70 chaperone machine. Nucleic Acids Res. 2011; 39:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L., Feng Z., Wang X., Wang X., Zhang X.. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010; 26:136–138. [DOI] [PubMed] [Google Scholar]
- 36. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tao T., Shi H., Guan Y., Huang D., Chen Y., Lane D.P., Chen J., Peng J.. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013; 23:620–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tao T., Shi H., Huang D., Peng J.. Def functions as a cell autonomous factor in organogenesis of digestive organs in zebrafish. PLoS One. 2013; 8:e58858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Luo Y., Hong Y., Peng J., Lo L.. Ribosome biogenesis factor Bms1-like is essential for liver development in zebrafish. J Genet Genomics. 2012; 39:451–462. [DOI] [PubMed] [Google Scholar]
- 40. Azuma M., Toyama R., Laver E., Dawid I.B.. Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J. Biol. Chem. 2006; 281:13309–13316. [DOI] [PubMed] [Google Scholar]
- 41. Tao T., Sondalle S.B., Shi H., Zhu S., Perez-Atayde A.R., Peng J., Baserga S.J., Look A.T.. The pre-rRNA processing factor DEF is rate limiting for the pathogenesis of MYCN-driven neuroblastoma. Oncogene. 2017; 36:3852–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boglev Y., Badrock A.P., Trotter A.J., Du Q., Richardson E.J., Parslow A.C., Markmiller S.J., Hall N.E., de Jong-Curtain T.A., Ng A.Y.et al.. Autophagy induction is a Tor- and Tp53-independent cell survival response in a zebrafish model of disrupted ribosome biogenesis. PLos Genet. 2013; 9:e1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baker K.E., Parker R.. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 2004; 16:293–299. [DOI] [PubMed] [Google Scholar]
- 44. Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J.. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019; 568:259–263. [DOI] [PubMed] [Google Scholar]
- 45. Wagner G.P., Kin K., Lynch V.J.. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012; 131:281–285. [DOI] [PubMed] [Google Scholar]
- 46. Wilkins B.J., Pack M.. Zebrafish models of human liver development and disease. Compr Physiol. 2013; 3:1213–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S., Miller S.R., Ober E.A., Sadler K.C.. Making it new again: insight into liver development, regeneration, and disease from zebrafish research. Curr. Top. Dev. Biol. 2017; 124:161–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Field H.A., Ober E.A., Roeser T., Stainier D.Y.. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 2003; 253:279–290. [DOI] [PubMed] [Google Scholar]
- 49. Lin J.W., Biankin A.V., Horb M.E., Ghosh B., Prasad N.B., Yee N.S., Pack M.A., Leach S.D.. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev. Biol. 2004; 274:491–503. [DOI] [PubMed] [Google Scholar]
- 50. Zecchin E., Mavropoulos A., Devos N., Filippi A., Tiso N., Meyer D., Peers B., Bortolussi M., Argenton F.. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev. Biol. 2004; 268:174–184. [DOI] [PubMed] [Google Scholar]
- 51. Chen Y.H., Lu Y.F., Ko T.Y., Tsai M.Y., Lin C.Y., Lin C.C., Hwang S.P.. Zebrafish cdx1b regulates differentiation of various intestinal cell lineages. Dev. Dyn. 2009; 238:1021–1032. [DOI] [PubMed] [Google Scholar]
- 52. Flores M.V., Hall C.J., Davidson A.J., Singh P.P., Mahagaonkar A.A., Zon L.I., Crosier K.E., Crosier P.S.. Intestinal differentiation in zebrafish requires Cdx1b, a functional equivalent of mammalian Cdx2. Gastroenterology. 2008; 135:1665–1675. [DOI] [PubMed] [Google Scholar]
- 53. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 54. Huang D.W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoshikawa H., Ishikawa H., Izumikawa K., Miura Y., Hayano T., Isobe T., Simpson R.J., Takahashi N.. Human nucleolar protein Nop52 (RRP1/NNP-1) is involved in site 2 cleavage in internal transcribed spacer 1 of pre-rRNAs at early stages of ribosome biogenesis. Nucleic Acids Res. 2015; 43:5524–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu Q., Greimann J.C., Lima C.D.. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006; 127:1223–1237. [DOI] [PubMed] [Google Scholar]
- 58. Muller J.S., Burns D.T., Griffin H., Wells G.R., Zendah R.A., Munro B., Schneider C., Horvath R.. RNA exosome mutations in pontocerebellar hypoplasia alter ribosome biogenesis and p53 levels. Life Sci. Alliance. 2020; 3:e202000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jia Q., Zhang Q., Zhang Z., Wang Y., Zhang W., Zhou Y., Wan Y., Cheng T., Zhu X., Fang X.et al.. Transcriptome analysis of the zebrafish model of Diamond-Blackfan anemia from RPS19 deficiency via p53-dependent and -independent pathways. PLoS One. 2013; 8:e71782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Z., Jia H., Zhang Q., Wan Y., Zhou Y., Jia Q., Zhang W., Yuan W., Cheng T., Zhu X.et al.. Assessment of hematopoietic failure due to Rpl11 deficiency in a zebrafish model of Diamond-Blackfan anemia by deep sequencing. BMC Genomics. 2013; 14:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song B., Zhang Q., Zhang Z., Wan Y., Jia Q., Wang X., Zhu X., Leung A.Y., Cheng T., Fang X.et al.. Systematic transcriptome analysis of the zebrafish model of diamond-blackfan anemia induced by RPS24 deficiency. BMC Genomics. 2014; 15:759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wan Y., Zhang Q., Zhang Z., Song B., Wang X., Zhang Y., Jia Q., Cheng T., Zhu X., Leung A.Y.et al.. Transcriptome analysis reveals a ribosome constituents disorder involved in the RPL5 downregulated zebrafish model of Diamond-Blackfan anemia. Bmc. Med. Genomics. 2016; 9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mills E.W., Green R.. Ribosomopathies: there's strength in numbers. Science. 2017; 358:eaan2755. [DOI] [PubMed] [Google Scholar]
- 64. Ross A.P., Zarbalis K.S.. The emerging roles of ribosome biogenesis in craniofacial development. Front. Physiol. 2014; 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.