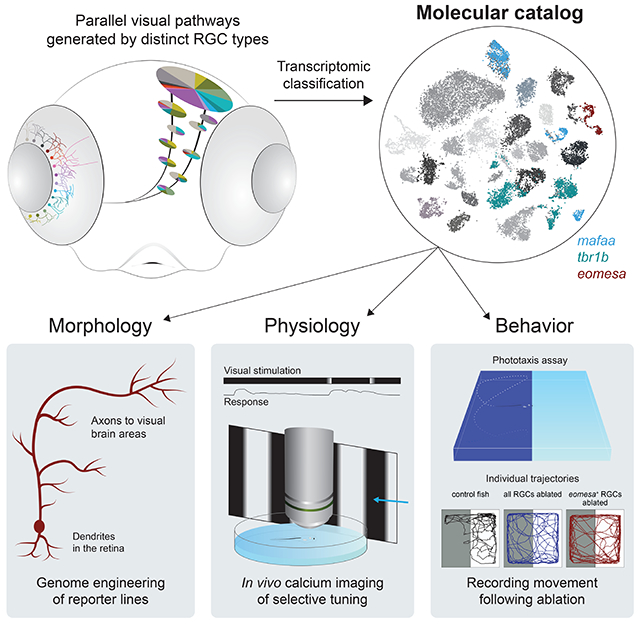
Summary
Retinal ganglion cells (RGCs) form an array of feature detectors, which convey visual information to central brain regions. Characterizing RGC diversity is required to understand the logic of the underlying functional segregation. Using single cell transcriptomics, we systematically classified RGCs in adult and larval zebrafish, thereby identifying marker genes for >30 mature types and several developmental intermediates. We used this dataset to engineer transgenic driver lines, enabling specific experimental access to a subset of RGC types. Expression of one or few transcription factors often predicts dendrite morphologies and axonal projections to specific tectal layers and extratectal targets. In vivo calcium imaging revealed that molecularly defined RGCs exhibit specific functional tuning. Finally, chemogenetic ablation of eomesa+ RGCs, which comprise melanopsin-expressing types with projections to a small subset of central targets, selectively impaired phototaxis. Together, our study establishes a framework for systematically studying the functional architecture of the visual system.
Keywords: single cell transcriptomics, cell atlas, genetic markers, genome engineering, visual pathways, physiology, behavior, eomes, ipRGCs
Graphical Abstract
Introduction
Understanding how the brain regulates behavior requires targeted genetic access to subpopulations of neurons, so they can be characterized and manipulated. Retinal ganglion cells (RGCs) are the sole output neurons of the eye through which all visual information flows as it is transmitted to visual centers of the brain. RGCs can be subdivided into several dozen types based on anatomical, physiological and molecular characteristics (Baden et al., 2016; Bae et al., 2018; Dacey, 1994; Förster et al., 2020; Peng et al., 2019; Robles et al., 2014; Tran et al., 2019; Yan et al., 2020). Their selective tuning to certain visual features, such as luminance transitions, edges, chromaticity or direction of motion, arises from inputs provided by specific subsets of retinal interneurons in the inner plexiform layer (Figure 1A), as well as intrinsic properties (Sanes and Masland, 2015). In many cases, distinct RGC functional types project axons to different brain centers, which in turn are associated with specific perceptual and behavioral functions (Dhande et al., 2015; Kramer et al., 2019; Martersteck et al., 2017; Nikolaou et al., 2012; Robles et al., 2013, 2014; Seabrook et al., 2017; Semmelhack et al., 2014; Temizer et al., 2015). Both the individual synaptic connectivities of RGCs and their biophysical characteristics are determined to a large extent by their cell type-specific gene expression profiles. Owing to the bottleneck role they play in visual processing, RGCs are prime targets for a functional dissection of visually guided behavior.
Figure 1. Single cell transcriptomics defines an adult zebrafish RGC catalog comprising 32 molecularly distinct clusters.
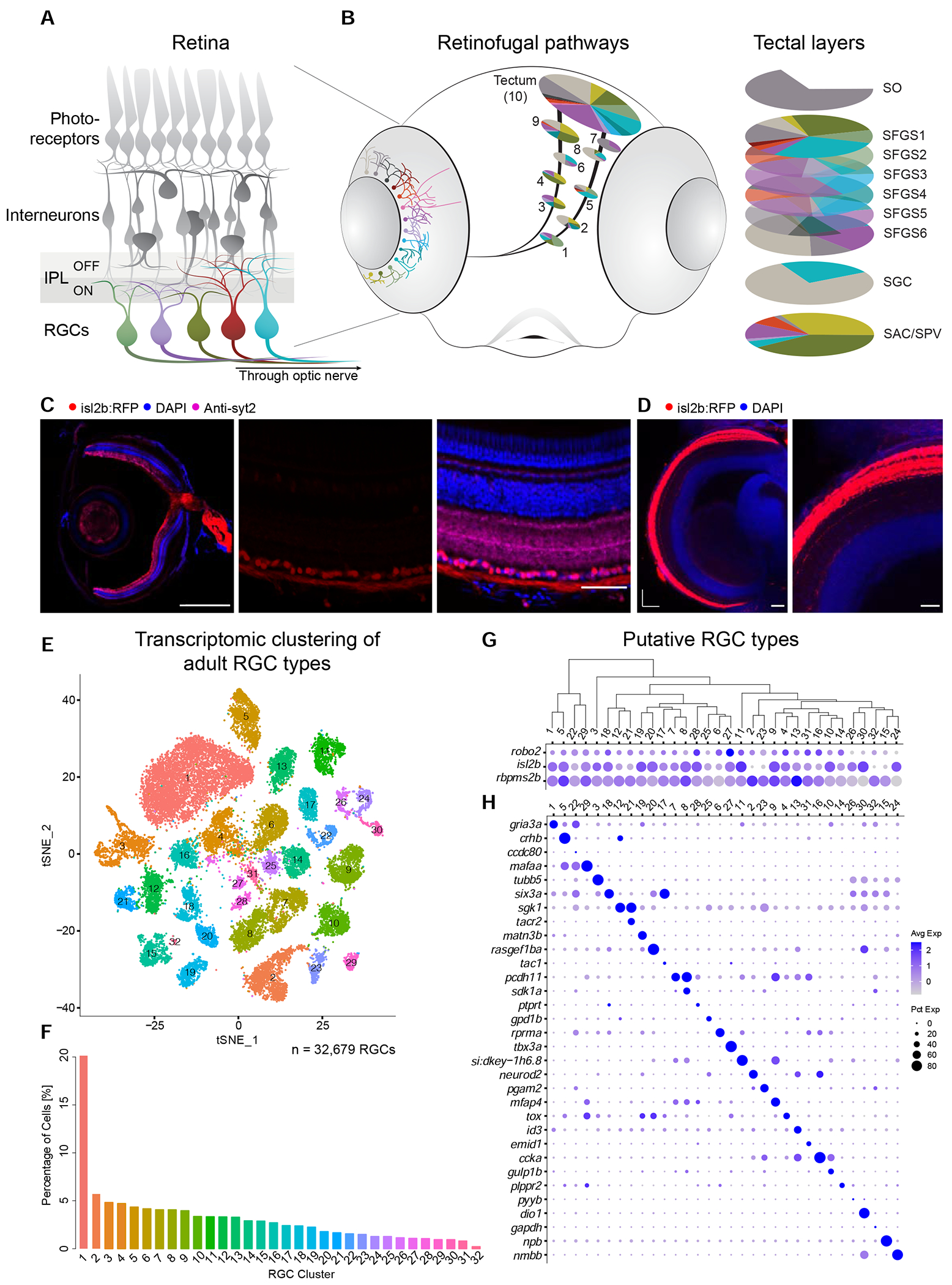
(A) Sketch of the zebrafish retina. RGCs, the innermost retinal neurons, transmit visual information to the rest of the brain through the optic nerve. Unique patterns of dendritic stratification in the inner plexiform layer (IPL), which is divided into two halves that subserve ON and OFF light responses, enables distinct RGC types (colors) to receive presynaptic input from specific interneuron types, rendering individual RGC types sensitive to distinct visual features.
(B) Left: RGC projectome (Robles et al., 2014). RGC types are defined by stereotyped combinations of dendritic stratification patterns in the retina and axonal projections to retinorecipient nuclei, named arborization fields (AFs 1-9) and tectum. Right: Within the tectum, RGC axons project to nine or ten laminae, including SO (stratum opticum), SFGS (stratum fibrosum et griseum superficiale) 1-6, SGC (stratum griseum centrale), and the boundary between SAC/SPV (stratum album centrale/ stratum periventriculare). Each AF or tectal lamina is innervated by a unique set of RGC morphotypes, depicted by colors.
(C) Tg(isl2b:tagRFP) labels RGCs. Left: Section of an adult eye immunostained for RFP, synaptotagmin (syt2) as a neuropil stain, and DAPI counterstain of somata. Middle: Magnified retinal section highlighting RFP-labeled RGCs. Right: Overlay of all markers in the retinal section. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bars, 500 μm (left), 50 μm (middle and right).
(D) Left: Confocal plane covering the RFP-immunostained adult Tg(isl2b:tagRFP) tectum with DAPI counterstain. Right: Magnified area. A, anterior; M, medial. For layer abbreviations, see A. Scale bar, 100 μm.
(E) t-distributed stochastic neighbor embedding (tSNE) visualization of 33 transcriptional clusters (colors) of 32,679 adult zebrafish RGCs (points). Clusters are numbered in the order of decreasing relative frequency.
(F) Relative frequency (y-axis) of adult RGC clusters (x-axis), ordered from highest to lowest. Clusters are colored as in E.
(G) Dotplot showing the expression patterns of three RGC markers (rows) across adult clusters (columns). The area of each circle depicts the percentage of cells expressing the gene, and the color depicts the z-scored expression in cells with nonzero transcripts. Clusters are ordered based on their global transcriptional relatedness depicted using a dendrogram (top), computed using hierarchical clustering.
(H) Dotplot showing expression patterns of markers (rows) that are selectively enriched in adult RGC clusters (columns). Column ordering and expression depiction as in G.
Recent studies have used high-throughput single cell RNA sequencing (scRNA-seq) to generate molecular taxonomies of RGC types in mice (Rheaume et al., 2018; Tran et al., 2019), non-human primates (Peng et al., 2019) and humans (Yan et al., 2020). However, it has remained challenging to associate individual types with their postsynaptic targets and the behaviors their activation elicits. The zebrafish is an attractive model system to address this gap. Despite having diverged from mammals >400 million years ago, retinal architecture and development are conserved in zebrafish (Sanes and Zipursky, 2010). Their visual system develops rapidly and a diverse behavioral repertoire is seen by 5 days post fertilization (dpf) (Fleisch and Neuhauss, 2006; Orger, 2016; Orger and de Polavieja, 2017). Moreover, zebrafish are amenable to rapid and efficient transgenic manipulation and are transparent at larval stages, allowing for imaging of structure and function in vivo (Baier and Scott, 2009).
The diversity of zebrafish RGC types has been documented based on their morphological (Robles et al., 2013, 2014) and functional properties (Gabriel et al., 2012; Nikolaou et al., 2012; Semmelhack et al., 2014; Temizer et al., 2015; Zhou et al., 2020). These studies have revealed that RGCs send their axons to ten different layers in the tectum, as well as nine extratectal arborization fields (AFs), which are numbered according to their position along the optic tract AF1 through 9 (Figure 1B). The AFs are neuropil areas of brain nuclei in the hypothalamus/preoptic area, thalamus and pretectum, which are highly conserved among vertebrates (Burrill and Easter, 1994) and subserve a wide array of visual functions, such as the detection of optic flow, the photo-entrainment of circadian rhythms, prey recognition, visual escape, and phototaxis (Orger, 2016; Portugues and Engert, 2009). Calcium imaging studies have shown that each of the ten tectal layers and the nine AFs receives a distinct combination of feature-selective RGC inputs (e. g., Förster et al., 2020).
Here, we used scRNA-seq to comprehensively classify both larval and adult zebrafish RGCs based on their transcriptomes. Nearly two thirds of larval RGCs exhibited molecular profiles that correspond to their adult counterparts, suggesting that a substantial proportion of the RGC repertoire is established at early larval stages. The remaining third were RGCs largely committed to specific adult fates, but were still in the process of transitioning to their mature state. One small cluster of cells, which persisted into adulthood, comprised postmitotic, yet uncommitted RGCs, reflecting continued neurogenesis and differentiation in the teleost retina. Using CRISPR-Cas9 genome engineering and intersectional strategies, we established transgenic lines to gain exclusive genetic access to several molecularly defined RGC types. We found that molecularly defined types project in stereotyped patterns to visual brain nuclei. A subset of RGCs, which express the T-box transcription factor eomesa, predominantly respond to increases in ambient luminance and express melanopsin-coding genes. Chemogenetic ablation experiments showed that eomesa+ RGCs are required for phototaxis, but dispensable for other visually-guided behaviors. Overall, our study provides an inroad for systematically investigating the development, structure, function, and behavioral contributions of specific cell types in the vertebrate visual system.
Results
Single cell transcriptional profiling generates a molecular taxonomy of RGCs
We isolated RFP-labeled RGCs from adult (4-6 months old) transgenic Tg(isl2b:tagRFP) zebrafish, where nearly all RGCs are labeled with no known bias (Figure 1C–D), and profiled them using droplet-based scRNA-seq (Zheng et al., 2017). Through computational analysis of transcriptional profiles, we separated major cell classes based on the expression of canonical markers. RGCs comprised 67% of cells, with the remainder including rods, Müller glia, amacrine cells, bipolar cells and endothelial cells (Figure S1A). We recovered a total of 32,679 high-quality single RGC transcriptomes with an average of 2,570 transcripts and 1,188 genes per cell. RGC transcriptomes were batch-corrected and analyzed using dimensionality reduction and clustering (STAR Methods), yielding 32 transcriptionally distinct clusters (Figures 1E, S1B–D). Their frequencies ranged from 0.2% to 20.1% likely reflecting the species-specific composition of the RGC complement in zebrafish (Figure 1F). The number, identity, and relative frequency of transcriptional clusters were highly consistent when the analysis was repeated on one of the five biological replicates (Methods). All clusters expressed one or more of the pan-RGC markers rbpms2b, isl2b and robo2 (Figure 1G). Many clusters could be uniquely identified based on selective expression of a single gene, but in a number of cases unique labeling required two genes (Figures 1H, S1E, Table S1). Transcriptomically distinct clusters likely represent individual cell types, or groups of few cell types.
RGC clusters are distinguished by expression of transcription factors and secreted molecules
Transcription factors (TFs) play essential roles in determination of cell identity (Fishell and Heintz, 2013). We therefore surveyed TF expression in our transcriptomic data. From a database of 1,524 TF-encoding genes, we found 186 that were expressed in >30% of cells of at least one RGC cluster (STAR Methods). About 10% (n=17) of these TFs were broadly expressed in most types. The majority (n=169) showed highly variable expression profiles, which were often restricted to one or few clusters (Figures 2A–B, Table S2). Restricted TFs could be further subdivided into two groups based on their degree of cluster-specific expression: 42% (n=71) exhibited multi-type expression in >20% of clusters and included the well-studied RGC-specific POU class 4 homeobox genes pou4f1 and pou4f2, as well as pou6f2, barhl2 and ebf3a. The remaining TFs (n=98) were expressed in 1-6 clusters each and included eomesa (= tbr2), mafaa, tbr1b, foxp2, satbl, satb2 and mef2cb.
Figure 2. Variably expressed transcription factors, cell-surface and secreted molecules, and neuropeptides across adult RGC types.
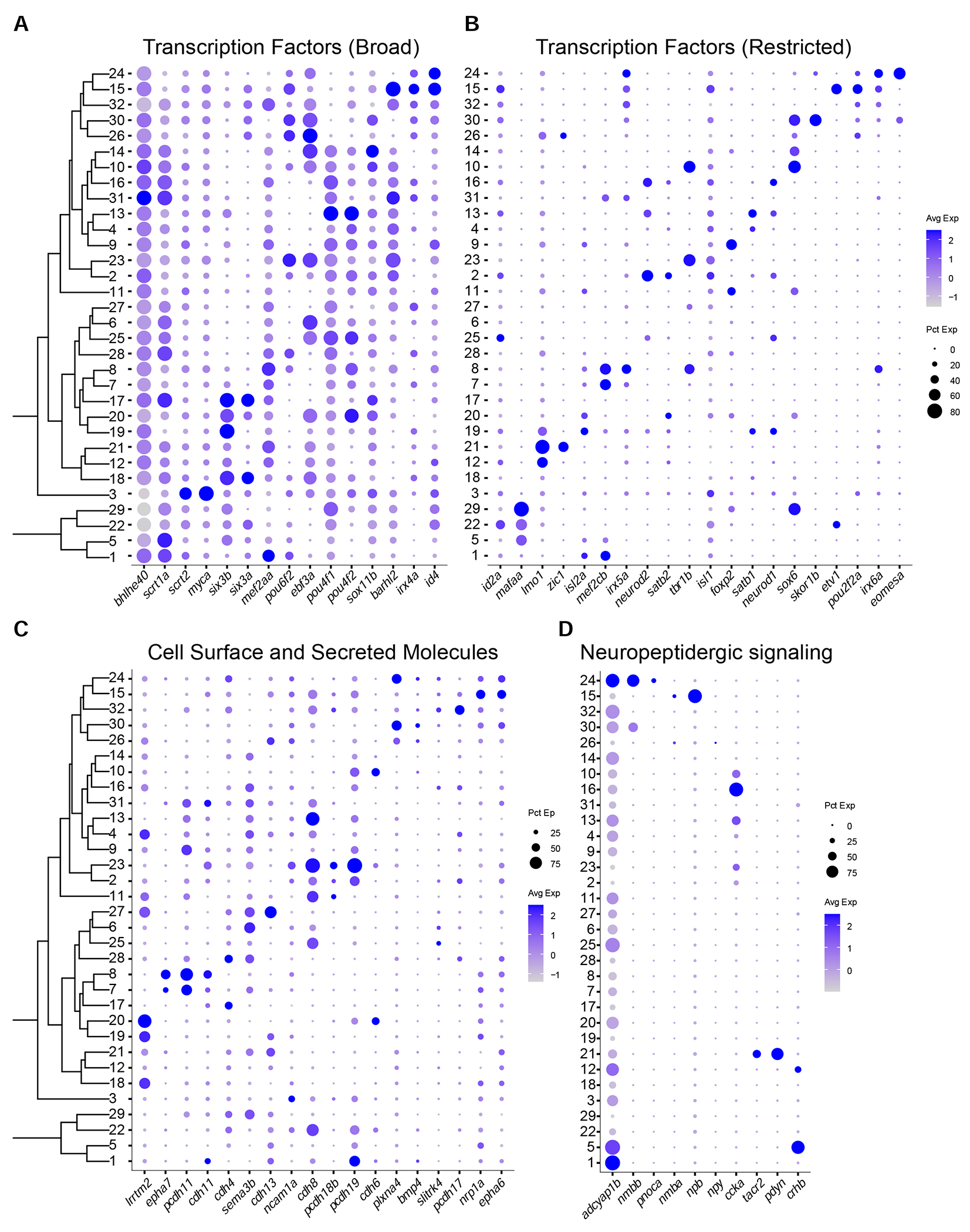
(A)-(B) Dotplots highlighting examples of variably expressed TFs in adult RGC clusters, subdivided into broad (A), and restricted (B) categories. Representation as in Figure 1G. Full list is provided in Table S2.
(C) Dotplot highlighting key cell surface and secreted molecules selectively expressed in adult RGC clusters.
(D) Dotplot highlighting neuropeptides selectively expressed in adult RGC clusters.
In addition to TFs, we also identified variably expressed cell-surface recognition molecules, secreted guidance molecules, and neuropeptides (Figures 2C–D, Table S2). These included genes encoding ephrin receptors, epha6 and epha7, which have been implicated in retinotopic axon guidance (Kita et al., 2015), and type II cadherins, known to regulate dendritic targeting in the IPL (Duan et al., 2014; 2018). Among neuropeptides and their receptors, selectively expressed genes included nmbb, npb, ccka and tacr2. Overall, our data indicate that the diverse molecular identities of individual RGC types are reflected in the combinatorial expression patterns of TFs, cell-surface molecules and neuropeptides.
scRNA-seq highlights diversification of RGCs at the larval stage
To survey the molecular diversity of larval RGCs, we profiled RFP-positive cells from 5 dpf Tg(isl2b:tagRFP) fish (Figures S2A–C). We recovered 11,046 RGCs comprising 90.4% of all cells collected (Figure S2D). Our data represent a 10-fold enrichment and 2.5X or 3X coverage over their baseline frequency of 9% assuming ca. 4,000 RGCs (Robles et al., 2014; Zhou et al., 2020; Zimmermann et al., 2018). Larval RGCs were analyzed using the pipeline that had been applied to the adult data, yielding 29 clusters, which could be distinguished using specific markers (STAR Methods, Figures 3A–B and S2E–I). We also observed that a statistically significant proportion of transcription factors (p < 10−132; hypergeometric test), recognition molecules (p < 10−44) and neuropeptides (p < 10−13) expressed in the adult clusters retained their specificity among larval clusters (STAR Methods, Figure S3A–D).
Figure 3. Molecular classification of larval RGCs and their transcriptomic correspondence with adult RGC types.
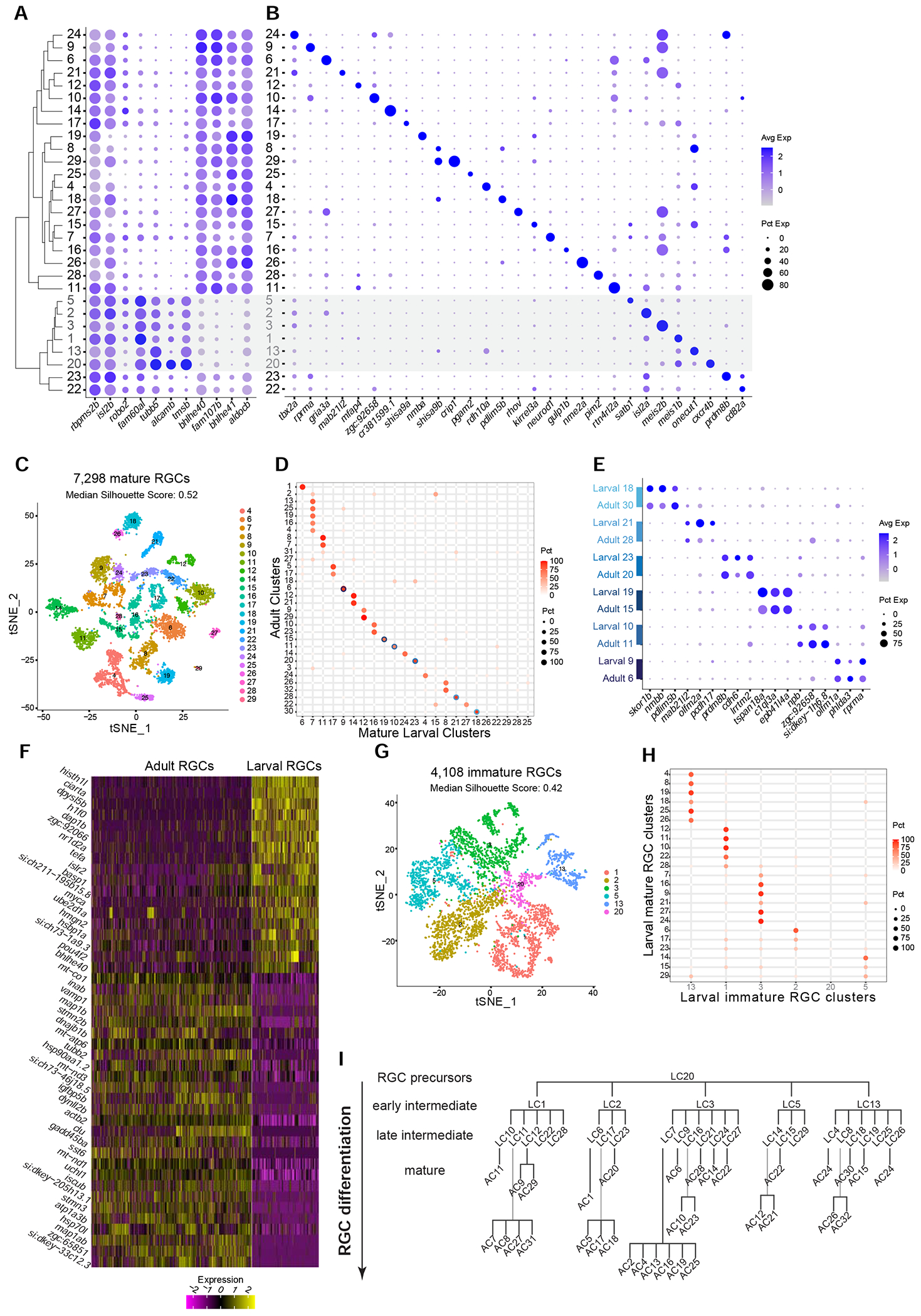
(A) Global transcriptional relatedness (dendrogram, left) of larval RGC clusters (rows) identifies two groups, corresponding to mature and immature RGCs (grey cluster labels and shaded horizontal bar). The dotplot highlights expression of pan-RGC markers rbpms2b, isl2b and robo2, as well as the top differentially expressed genes (n=8) between immature and mature RGC clusters.
(B) Expression patterns of markers (columns) that are selectively enriched in larval RGC clusters (rows), ordered as in panel A.
(C) tSNE visualization of 23 mature RGC clusters (colors) comprising 7,298 cells (points). The median silhouette score was computed for each graph. The score ranges from −1 to 1, with higher values indicating tighter cluster boundaries.
(D) Transcriptional correspondence between adult and mature larval RGC clusters. Circles and colors indicate the proportional representation of adult cluster identities (rows) in mature larval clusters (column) based on a supervised classification analysis using the xgboost algorithm. Each row is normalized to sum to 100%. Blue circles highlight six instances of a 1:1 corresponding pair of adult and larval clusters, which are separately analyzed in panels E-G.
(E) Dotplot showing shared patterns of gene expression between the six 1:1 cluster pairings selected from the classification model (blue circles in panel D). Colored bars (left) indicate matching cluster pairs.
(F) Heatmap showing differentially expressed genes (rows) between adult and larval stages identified from the six 1:1 matching clusters shown in panel D. Columns correspond to individual RGCs grouped by age. Values are row-wise z-scored gene expression values.
(G) tSNE visualization of 6 immature RGC clusters (colors) comprising 4,108 cells (points). The median silhouette score was computed as in C.
(H) Transcriptional correspondence between mature (rows) and immature (columns) larval RGC clusters using an xgboost classifier trained on immature larval RGCs. Representation as in D.
(I) Model of RGC type diversification. Larval clusters (LCs) and adult clusters (ACs) are arranged by their transcriptional correspondences shown in D and H. RGC precursors give rise to immature (early intermediate) RGC clusters and mature (late intermediate) larval clusters, which further diversify into mature adult clusters.
Of the 29 larval clusters (LC1-LC29), 23 shared gene expression features with adult RGCs (Figure 3A, S3E). We refer to these as mature larval RGCs. Through a supervised classification analysis, we assigned adult cluster identities to the mature larval RGCs (Methods), which revealed a highly specific pattern of correspondence between larval and adult clusters (Figures 3D,S3F). Six larval clusters mapped to single adult clusters in a 1:1 fashion, likely representing types whose diversification is complete at the larval stage. We verified these mappings based on the shared expression of cluster-specific markers at both stages (Figure 3E). We found 57 differentially expressed genes between the 1:1 matched larval and adult RGC types associated with global maturational changes (STAR Methods, Figure 3F). The most notable pattern was that genes with sequence-specific DNA binding activity were higher at the larval stage. Examples include histh1l, nr1d2a, h3f3b.1, bhlhe40, and ciarta (Howe et al., 2012). Genes whose expression levels increased with maturation were associated with mature neuronal function, such as snap25a, vamp1, nefma and igfbp5b (Howe et al., 2012). In addition to these global changes, we also found maturational changes that were type-specific (Figure S3G).
An additional fifteen larval clusters mapped to 2 to 4 adult types each. One cluster, LC7, mapped to 6 adult types. Only one cluster, LC28, comprising less than 0.4% of larval RGCs, could not be matched with any adult counterpart, perhaps because it matures into a type too rare to be identified in the adult catalog (Figure S3F). Attempts to resolve substructure in larval clusters that mapped to a multiplicity of adult types were only partially successful. We highlight this using the example of LC7, whose cells showed partial segregation in the 2D embedding when grouped by their adult identity as assigned by the supervised classification (Figure S3H), which is also reflected in transcriptomic differences (Figure S3I). While this observation may reflect insufficient resolution in the larval cells due to low RNA capture and/or a lower sample size, we favor the interpretation that the diversification of these cells is truly incomplete at the molecular level. Thus, our results suggest a substantial level of RGC molecular diversification in larval zebrafish that undergoes further refinement as the fish ages.
Some larval clusters represent immature yet largely committed RGC types
In addition to the 23 larval RGC types with clear relationship to adult types, 6 transcriptionally proximate clusters (36% of larval RGCs) were defined by a distinct gene expression signature (Figure 3A). Notably, these genes were associated with early RGC development, such as the axon outgrowth gene alcamb, known to regulate early RGC axon outgrowth in zebrafish (Diekmann and Stuermer, 2009) as well as tubb5 and tmsb, which are associated with cytoskeletal rearrangement (Ngo et al., 2014; Roth et al., 1999). We therefore reasoned that these clusters may represent immature larval RGCs.
We compared the two groups of clusters, finding three distinctions that support their division into immature and mature larval RGC types. First, genes defining mature larval RGCs (e.g. bhlhe40, bhlhe41 and fam107b) were expressed widely across adult RGC clusters. In contrast, immature larval genes such as alcamb, tubb5 and tmsb were expressed in a single adult RGC cluster (C3), comprising 4.8% of adult RGCs (Figure S3E). Second, clusters of mature larval RGCs were more transcriptionally distinct compared to immature larval RGCs, as judged by their tighter separation in the reduced dimensional space, quantified using the silhouette score (STAR Methods, Figure 3C, G). Third, supervised classification analysis showed that five out of the six immature clusters were transcriptionally related to nonoverlapping sets of 3 to 7 mature larval clusters (Figure 3H). Interestingly, top differentially expressed genes for these immature RGC clusters included satb1, isl2a, meis2b, meis1b and onecut1 (Figure 3B), which are TFs that continue to be selectively expressed among mature larval RGCs and adult RGCs (Figures 2A–B, S3A–B). The observation that the mature types to which immature types mapped were mutually exclusive supports the idea that these five RGC clusters represent committed precursors restricted to specific fates that are gradually refined. Finally, the sixth immature cluster (C20), comprising 2.3% of all larval RGCs, had the highest expression of alcamb, tubb5, and tmsb, and exhibited transcriptional correspondence to adult cluster 3 (data not shown). The unique markers that distinguished this cluster included fgf8, a factor involved in initiation of RGC differentiation (Martinez-Morales et al., 2005), and cxcr4b, a receptor involved in early axon migration (Li, 2005). We propose that C20 might represent uncommitted precursors. Taken together, these results are consistent with a diversification model in which transcriptionally distinct immature RGCs in larvae are specified into adult RGC types in a gradual but possibly asynchronous fashion (Figure 3I).
An intersectional strategy enables genetic access to RGC types
To relate transcriptional clusters to RGC morphology, we chose a set of three TFs with restricted expression to generate transgenic driver lines: mafaa, tbr1b and eomesa. At the larval stage, each TF was expressed in a small number of clusters that formed non-overlapping groups (3 each for mafaa and tbr1b, 5 for eomesa; Figure 4A), together encompassing 37% of larval RGCs. Notably, these TFs maintained a robust cluster-specific expression pattern in adults (Figure 2B). We used a homology-independent target integration method (Suzuki et al., 2016) (STAR Methods, Table S3) to generate driver lines that selectively labeled RGCs expressing mafaa, tbr1b, and eomesa. In this method, a DNA sequence encoding the transcriptional activator QF2 is knocked into the corresponding TF locus. These genes are also expressed outside the retina, often in spatial domains that overlapped with the RGC axonal projection targets (Kunst et al., 2019; https://fishatlas.neuro.mpg.de/; Video S1–4), necessitating an intersectional approach to restrict expression to RGCs (Figure 4B). To this end, we designed an intersectional reporter transgene Tg(QUAS:loxP-GFP-loxP-epNTR-tagRFP), hereafter named Tg(QUAS:switchNTR), containing a conditional cassette that encodes RFP-tagged nitroreductase (NTR). We intersected each driver line with Tg(ath5:Cre) (Förster et al., 2017), wherein Cre recombinase is specifically expressed in RGCs (Kay et al., 2001, 2005) (Figure S4A–B), causing TF-expressing RGCs, to switch from GFP to RFP (Figure 4C, Figure S4C–E).
Figure 4. Molecularly defined RGC clusters exhibit distinct axonal projection patterns.
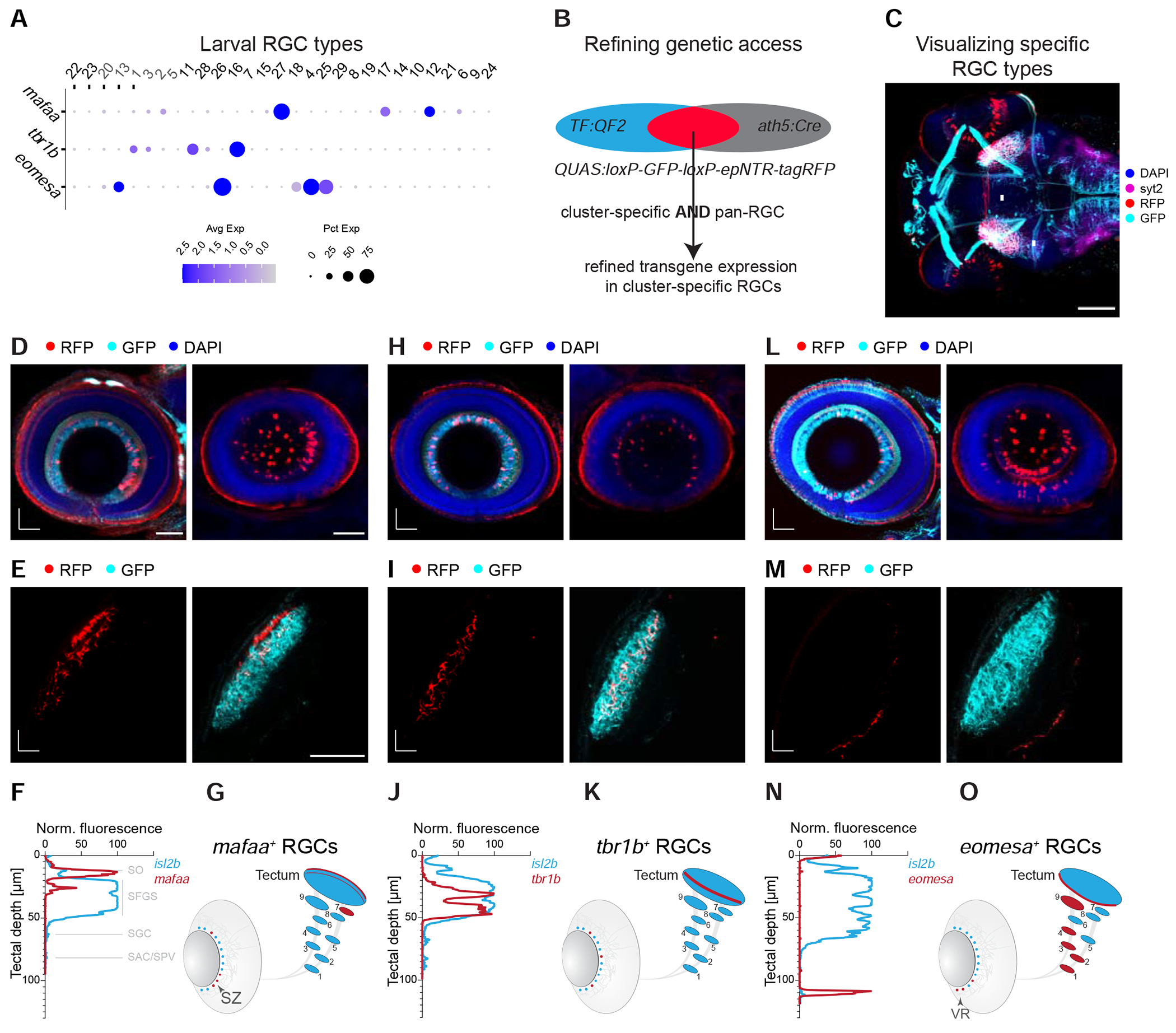
(A) Dotplot showing expression patterns of mafaa, tbr1b, and eomesa (rows) in larval clusters (columns) ordered as in Figure 3A. Cluster numbers (top) correspond to immature (grey) and mature (black) RGC clusters.
(B) Marker intersection refines genetic access to TF+ RGC types. In a TF:QF2 driver line, TF+ cells activate expression of GFP through a QUAS:switchNTR reporter. Combination with the pan-RGC Tg(ath5:Cre) line results in TF+ RGCs switching to RFP expression, while TF+ non-RGCs continue to express GFP.
(C) Visualization of RGC types, shown here for mafaa+ RGCs (arrows, red labeling), by immunostaining in a triple-transgenic Tg(TF:QF2, QUAS:switchNTR, ath5:Cre) larva. Scale bar, 100 μm.
(D-O) Anatomical characterization of RGC types labeled by mafaa (D-G), tbr1b (H-K) and eomesa (L-O) using quadruple-transgenic Tg(TF:QF2, QUAS:switchNTR, ath5:Cre, isl2b:GFP) larvae. In each case Tg(isl2b:GFP) serves as a label for landmarks of RGC projections. Confocal visualizations showing a single plane (left) and RGC soma distribution (maximum z projection, right) in en face views of the immunostained retina (D, H, L), in vivo images of axonal projections in the tectum (E, I, M), fluorescence profile across retinotectal laminae measured from the pan-RGC reporter isl2b and marker-specific RGC axons (F, J, N) as well as a schematic representation of the soma distribution in the retina and the projection pattern indicating TF+ RGCs in red against all RGCs in blue (G, K, O). Asterisks (*) in (E, I, M) denote layers innervated by TF+ RGCs. D, dorsal; T, temporal; A, anterior; M, medial. SZ, strike zone enrichment. VR, ventral retina enrichment. Scale bar in D for D, H, L, 50 μm. Scale bar in E for E, I, M, 50 μm.
Molecularly defined RGC subclasses exhibit distinct retinal asymmetries and projection patterns
In zebrafish, RGC axons project to ten tectal laminae and nine extra-tectal arborization fields (AF1-9) (Figure 1B). We previously classified zebrafish RGCs at single cell resolution based on stereotyped combinations of dendritic morphologies and axonal projections (Robles et al., 2014). Of note, most (97%) RGCs project axons to the tectum, forming arborizations restricted to a single tectal layer (for nomenclature of the layers, see Figure 1B). In addition, about half of the RGCs form en route axon collaterals to one or several extratectal targets AF1-AF9. Three percent of RGCs do not reach the tectum, but extend their most distal axon arbor to AF9, the neuropil of the periventricular pretectal nucleus (H. Baier and M. Wullimann, manuscript in preparation). To ask if axonal patterns of molecularly defined types match individual projection classes described previously (Robles et al., 2014), we crossed the mafaa, tbr1b and eomesa intersectional lines to Tg(isl2b:GFP), in which all RGCs express GFP. The GFP signal allowed us to compare the intraretinal distribution and axonal projection patterns of RGCs that express each of these three TFs to those that do not.
mafaa+ RGCs were distributed asymmetrically in the retina with an enrichment in the previously described “strike zone” (SZ), a zone for high acuity vision in the ventrotemporal retina, which serves as a fovea-like specialization for the capture of small prey items (Bianco et al., 2011; Gahtan et al., 2005; Mearns et al., 2020; Zhou et al., 2020; Zimmermann et al., 2018) (Figure 4D, single plane, left, and z projection, right). mafaa+ RGCs showed two projection patterns: They either terminated in the tectal SO, with axon collaterals in AF7, a projection type ascribed to prey-tuned RGCs (Semmelhack et al., 2014); or they arborized exclusively in SFGS2, with no extratectal collaterals, which corresponds to projection class 5 (Robles et al., 2014) (Figure 4E–G, S4F). Notably, the innervation of AF7 was heavily biased to the ventral side, suggesting a functional subdivision of this neuropil.
In contrast, tbr1b+ RGCs were distributed throughout the retina (Figure 4H, single plane, left, and z projection, right). Axons of tbr1b+ RGCs directly innervated intermediate tectal layers (SFGS3 and SFGS5) with no axon collaterals in extratectal AFs, corresponding to projection classes 6 and 8, respectively (Robles et al., 2014) (Figure 4I–K).
Somata of eomesa+ RGCs were enriched in the ventral retina (VR; Figure 4L, single plane, left, and z projection, right) and were found to innervate multiple extratectal areas in preoptic area/hypothalamus, thalamus and pretectum (AF1, AF2, AF3, AF4 and AF9) (Figure S4G). In the tectum, axons terminated exclusively in the SAC/SPV layer (Figure 4M–O), matching projection classes 15 to 20 (Robles et al., 2014). Within the pretectal AF9 neuropil, eomesa+ axon collaterals were restricted to the dorsal half, supporting the previously described subdivision between ventral and dorsal AF9 (Robles et al., 2014).
RGC types within a subclass exhibit distinct morphologies
We next sought to additionally resolve individual RGC types within the groups defined by tbr1b, eomesa and mafaa. The transcription factor tbx3a, which labels a single tbr1b+ adult type (C27 in Figure 1H), was specifically expressed by one of the tbr1b+ larval types albeit at low expression levels (Figure 5A). We therefore used the intersectional genome engineering approach to generate a line in which tbx3a+ RGCs were labeled (Figure S5A). The line sparsely labeled tbx3a+ RGCs, which demonstrated diffuse dendrites corresponding to the D2 type (Robles et al., 2014) (Figure 5B). The morphological features of tbx3a+ RGCs were consistent with them being a subset of tbr1b+ RGCs. tbx3a+ axons terminated exclusively in SFGS5 (Figures 5C–D), which is one of the tectal layers to which tbr1b+ RGCs project (Figures 4I–K).
Figure 5. Morphological types within RGC subclasses.
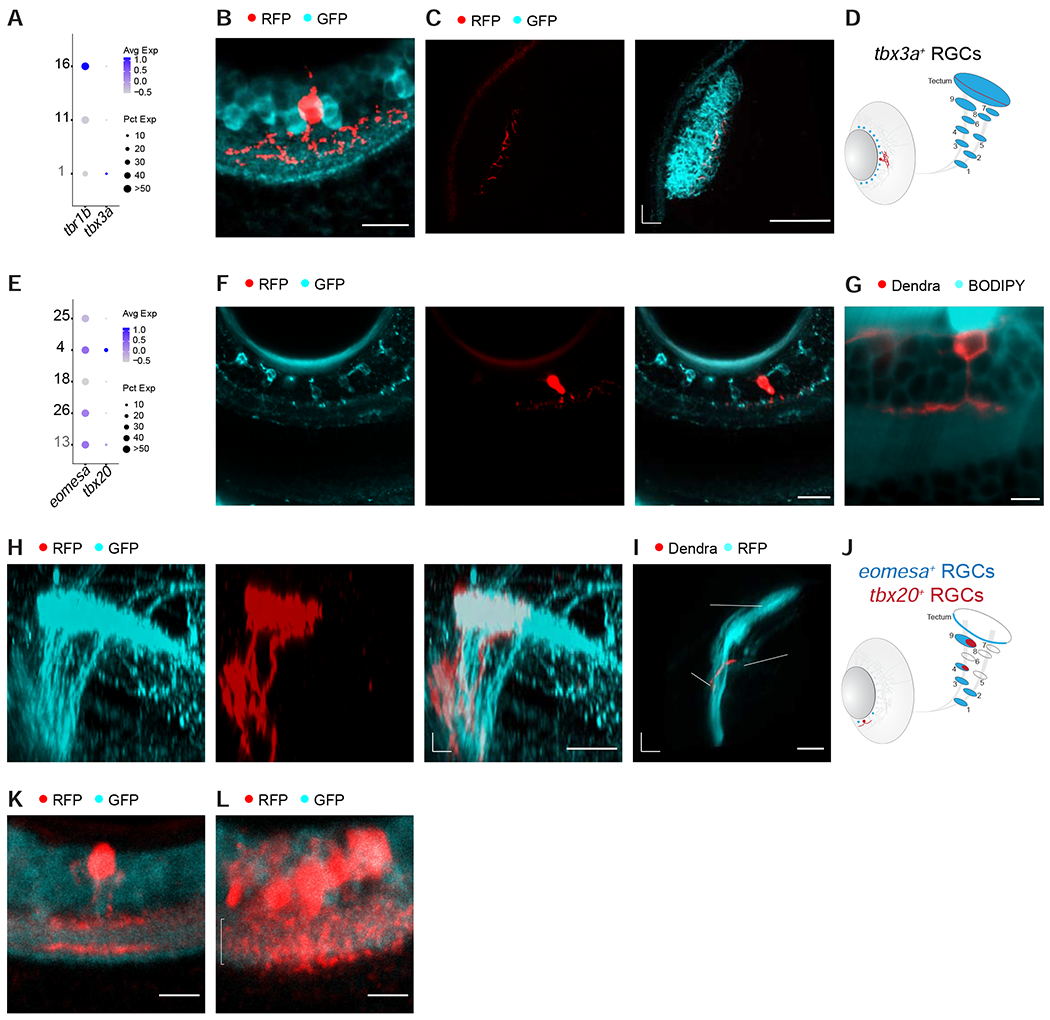
(A) Dotplot showing selective co-expression of tbx3a in larval tbr1b+ cluster 1.
(B) Immunostained retina of a Tg(tbx3a:QF2, QUAS:switchNTR, ath5:Cre, isl2b:GFP) larva shows diffuse dendrites of tbx3a+ RGCs in the IPL. GCL, ganglion cell layer; IPL, inner plexiform layer. Scale bar, 10 μm.
(C) Confocal plane of a live Tg(tbx3a:QF2, QUAS:switchNTR, ath5:Cre, isl2b:GFP) larva shows tbx3a+ RGC axons terminating in a deep SFGS layer. A, anterior; M, medial. Scale bar, 50 μm.
(D) Schematic representation of the soma distribution and projection patterns of the RGC type labeled by tbx3a (red) against all RGCs (blue).
(E) Dotplot showing specific expression of tbx20 in larval eomesa+ cluster 4.
(F) Immunostained retinal section of a quadruple-transgenic Tg(eomesa:QF2, QUAS:GFP, tbx20:Gal4, UAS:NTR-mCherry) larva showing GFP-labeled eomesa+ RGCs (left), one of which also expresses tbx20+ based on RFP-staining (right, star indicates the co-labeled cell). Scale bar, 20 μm.
(G) Confocal plane of a live Tg(tbx20:Gal4, UAS:Dendra) larval retina with a BODIPY neuropil counterstain shows tbx20+ RGCs exhibiting monostratified dendrites in the ON sublayer of the IPL. Scale bar, 5 μm.
(H) Confocal image of GFP-immunostained eomesa+ RGC axons and RFP-immunostained eomesa+tbx20+ RGC axons, innervating AF4 and AF9. D, dorsal; P, posterior. Scale bar, 20 μm.
(I) 3D side view of the optic tract imaged in a live Tg(tbx20:Gal4, UAS:Dendra, isl2b:tagRFP) larva, showing that tbx20+ RGC axons innervate AF4 and terminate in AF9. D, dorsal; P, posterior. Scale bar, 50 μm.
(J) Schematic of soma distribution and axon projections of the RGC type labeled by tbx20 (red) against all eomesa+ RGCs (blue).
(K) Single mafaa+ RGC in the periphery of the immunostained retina with bistratified dendrite (arrowheads) in the IPL. This stratification pattern matches the B2 morphology (Robles et al., 2014).
(L) Dense cluster of immunostained mafaa+ RGCs in the SZ with dendritic arborizations that extend throughout the width of the IPL (indicated by bracket). Some mafaa+ RGCs appear to have D1 morphology (Robles et al., 2014).
One of the five eomesa+ RGC types specifically expressed the transcription factor tbx20 (Figure 5E, Table S1). We labeled the eomesa+tbx20+ RGCs by crossing Tg(eomesa:QF2, QUAS:switchNTR) to Tg(tbx20:Gal4, UAS:NTR-mCherry) (Förster et al., 2017) (Figure S5B). The subset of eomesa+ RGCs that was tbx20+ had monostratified dendrites in the ON layer (Figure 5F, G) and axons that extended collaterals into AF4 and terminated in AF9, corresponding to projection class 15 (Robles et al., 2014) (Figure 5H–J). In contrast, eomesa+tbx20− RGCs projected further to the tectal SAC/SPV layer.
Individual RGCs that project to AF7 and tectal SO were either of a bistratified, B2, or a narrow diffuse, D1, dendritic type (Robles et al., 2014; Semmelhack et al., 2014). We asked if the stratification patterns of single RGCs in the mafaa+ line, matched any or both of these two morphologies. Indeed, solitary mafaa+ RGCs outside of the SZ often exhibited a bistratified dendritic arbor, resembling the B2 type (Figure 5K). mafaa+ RGCs in the SZ were densely clustered and could therefore not be imaged individually. However, many of their dendrites were bushy, with branches extending through the depth of the IPL, corresponding to a diffuse type (Figure 5L). Together, inspection of individual RGCs in our reporter lines revealed that a molecularly defined subclass can comprise multiple types with distinct, position-dependent dendritic stratification and axonal projection patterns.
Molecularly defined RGC types form visual feature-specific channels
To ask whether molecularly distinct RGC types also exhibit distinct functional properties, we harnessed eomesa and tbx20 as markers for an RGC subclass and a unique type within that subclass, respectively. We expressed the cytosolic calcium sensor GCaMP6s in RGCs and recorded axonal calcium transients in response to a battery of visual stimuli, which were displayed on a screen in front of the immobilized fish larva. The stimulus set was similar to the one reported previously for recording responses in tectal cells (Förster et al., 2020) and included luminance changes (bright, ‘ON’, and dark, ‘OFF’ flashes), as well as more complex stimuli such as moving black and white stripes (gratings), small moving dots (resembling prey) and rapidly expanding disks (resembling an approaching predator or an object on a collision course) (STAR Methods, Figure 6A). Because tbx20+ RGCs do not innervate the tectum, we focused our initial functional analysis on RGC axons in pretectal AF9 (Figure 6B–D).
Figure 6. RGC types exhibit specific physiological profiles.
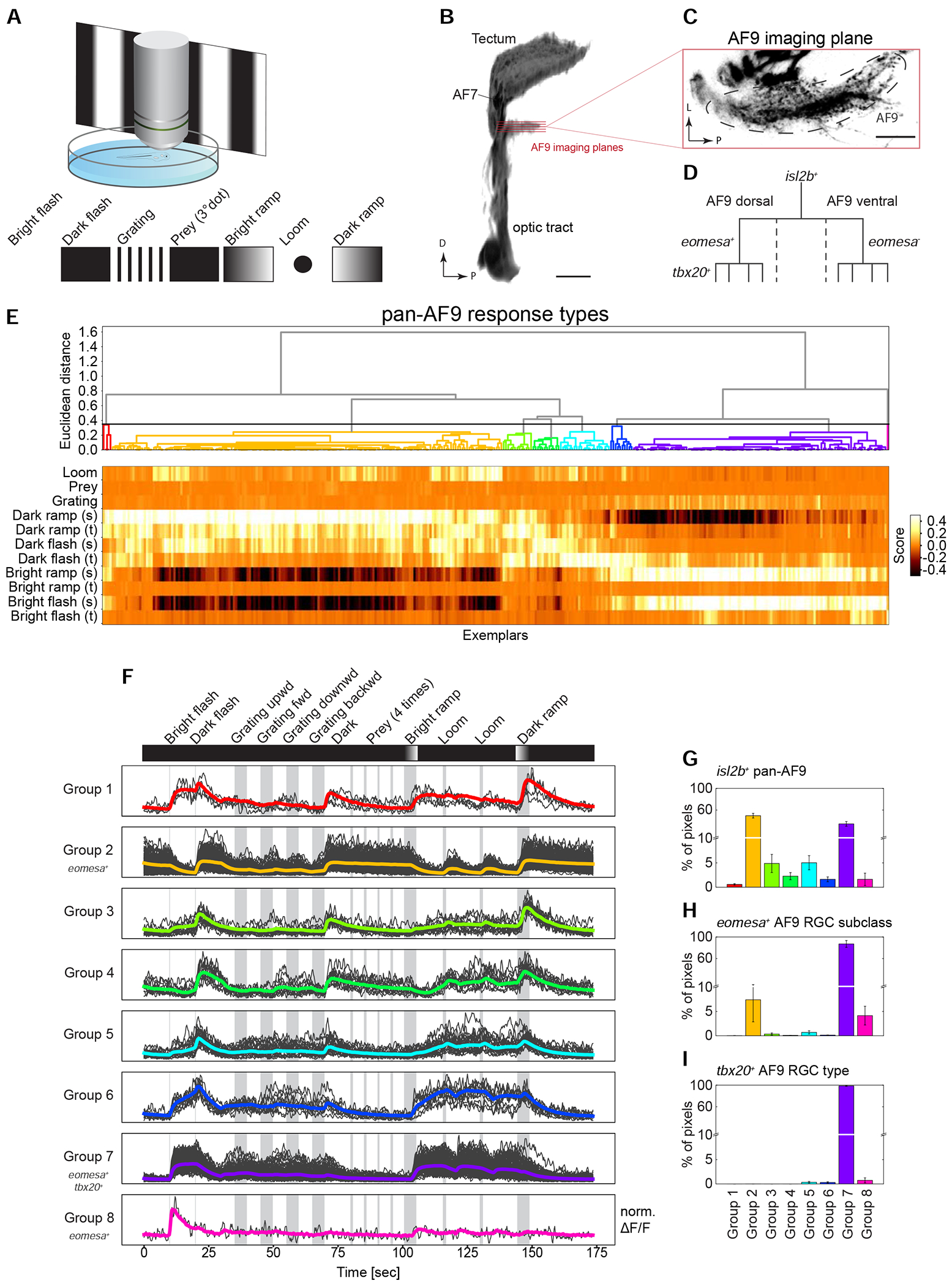
(A) Functional imaging of RGC types. Neuronal activity was recorded using two-photon calcium imaging from immobilized larvae expressing GCaMP6s in RGC axon terminals during presentation of a battery of visual stimuli displayed on a projection screen.
(B) 3D projection of the optic tract indicating the imaging planes in both ventral and dorsal subdivisions of AF9. D, dorsal; P, posterior. Scale bar, 50 μm.
(C) Single imaging plane in AF9. L, lateral; P, posterior. Scale bar, 20 μm.
(D) Hierarchical relationship of RGC subpopulations used for functional imaging: isl2b labels all RGCs, eomesa marks a subclass in dorsal AF9, wherein tbx20 is expressed by a single type among eomesa+ RGCs.
(E) Diversity of isl2b+ RGC responses to visual stimuli in AF9-projecting axons. Neural activity recordings derived from single pixels were clustered using affinity propagation to reduce noise, resulting in 345 clusters represented by exemplars (STAR Methods). Hierarchical clustering divided exemplar activity into eight major response groups (dendrogram, top). Heatmap (bottom) depicts calculated score of exemplars (columns) to each component of visual stimulus (rows). Sustained (s) and transient (t) activity was observed in responses to changing luminance levels.
(F) Activity traces of eight classified response groups shown in E aligned with the visual stimulus sequence. Shown are the normalized averaged activity traces (colored lines) and all representing exemplars that fall into the group (grey lines) over time. Response groups encompass different numbers of exemplars depending on their abundance.
(G-I) Relative frequencies of the eight response groups in isl2b+ RGCs (G), eomesa+ RGCs (H) and tbx20+ RGCs (I).
We began by characterizing the baseline diversity responses in AF9 by measuring activity from axons in the Tg(isl2b:Gal4, UAS:GCamP6s) line, which labels all RGCs (Figures 1G, 3A). Combined regression and clustering analysis classified RGC responses based on their activity to the stimulus components into 8 distinct pretectal groups (STAR Methods, Figure 6E). This analysis revealed that AF9-projecting RGCs responded robustly to broad ON or OFF stimuli, i. e., increments or decrements of ambient light (Figure 6F). By visual inspection, we categorized these groups as “sustained ON” (group 7), “transient ON” (group 8), “sustained OFF” (groups 2, 4) or “sustained ON – transient OFF” (groups 1, 3, 5, 6). The relative distribution of these responses varied, with sustained ON and OFF types (groups 2 and 7) predominating (Figure 6G and S6A). AF9-projecting RGC terminals were not reliably activated by patterned stimuli in our set, such as drifting gratings, motile dots or looming disks (Figure 6F).
We next recorded from eomesa+ RGCs, a subpopulation of isl2b+ RGCs. 87% of eomesa+ RGCs mapped to response group 7 (“sustained ON”; STAR Methods, Figure 6H and S6B). The remaining eomesa+ RGCs, mapped to response groups 2 (7%) and 8 (4%). Finally, eomesa+tbx20+ RGCs, a single type within the eomesa+ RGC subclass, mapped almost exclusively to response group 7 (Figure 6I and S6C), suggesting that this group of RGCs encodes ambient luminance increments. Thus, the observed functional specification aligns to our molecular and morphological definitions, with tbx20+ RGCs representing a unique type of eomesa+ RGCs, which in turn form a subset of isl2b+ RGCs.
To ask whether the diversity of responses in AF9 represent the full range of response types, we recorded from the retinotectal laminae of Tg(isl2b:Gal4, UAS:GCaMP6s) larvae. Using the computational pipeline applied to AF9, we observed 14 response groups. The retinotectal RGCs also responded to the presentation of visual objects and patterns, such as prey-like stimuli or direction of movement, features that are invisible to AF9-projecting RGCs (Figure S6D–F). Interestingly, tectal laminae received input from distinct response groups, which is attributable to dedicated innervation by unique sets of RGC types (Figure S6G). These results demonstrate that specific visual representations are relayed to distinct retinorecipient brain areas, with AF9 receiving a small subset of the overall visual information. Together, our findings demonstrate a tight correspondence between molecular, morphological and physiological properties within identified cell types.
eomesa+ RGCs express melanopsin
In mammals, Eomes (also known as Tbr2), the ortholog of eomesa, is expressed selectively, although not exclusively, by intrinsically photosensitive RGCs (ipRGCs; Mao et al., 2014; Peng et al., 2019; Sweeney et al., 2014; Tran et al., 2019), raising the possibility that at least some eomesa+ RGCs in zebrafish are ipRGCs. The canonical marker of ipRGCs is the photopigment melanopsin (Opn4) (Berson et al., 2002; Do, 2019; Gooley et al., 2001). We therefore assessed expression of opn4 in our dataset. While mammals have a single melanopsin gene (Opn4), zebrafish contain five melanopsin homologs, three of which (opn4.1, opn4a and opn4b) are related to mammalian Opn4 and two (opn4xa and opn4xb) are more closely related to the Xenopus gene Opn4x (Bellingham et al., 2006; Matos-Cruz et al., 2011). Previous studies found zebrafish melanopsin genes to be expressed across multiple cell classes in the larval retina, but only opn4xa was expressed in RGCs (Matos-Cruz et al., 2011; Zhang et al., 2017). Surprisingly, many RNA-seq reads within these four genes mapped to short sequences within presumptive introns (Figure S7A, B). These reads might represent unspliced precursor mRNA or unannotated exons (see Zhang et al., 2020). In fact, in each case, the sequence encoded at least one open reading frame, but the predicted amino acid sequence displayed no significant homology to opsins in any species (data not shown). Nonetheless, because numbers of intronic and exonic reads were correlated among cells and types, we combined them to assess cell-type specific expression patterns (La Manno et al., 2018) (STAR Methods). This analysis suggested that while opn4.1, opn4a and opn4xb were largely expressed in non-RGCs (Figure S7C), opn4xa and opn4b were robustly enriched in RGCs at both the larval and adult stages. Strikingly, within RGCs, their expression was restricted to eomesa+ RGC types and an additional type (Figure 7A, S7D). Taken together, these results provide evidence that the eomesa+ subclass contains ipRGC types.
Figure 7. eomesa+ RGCs regulate phototaxis.
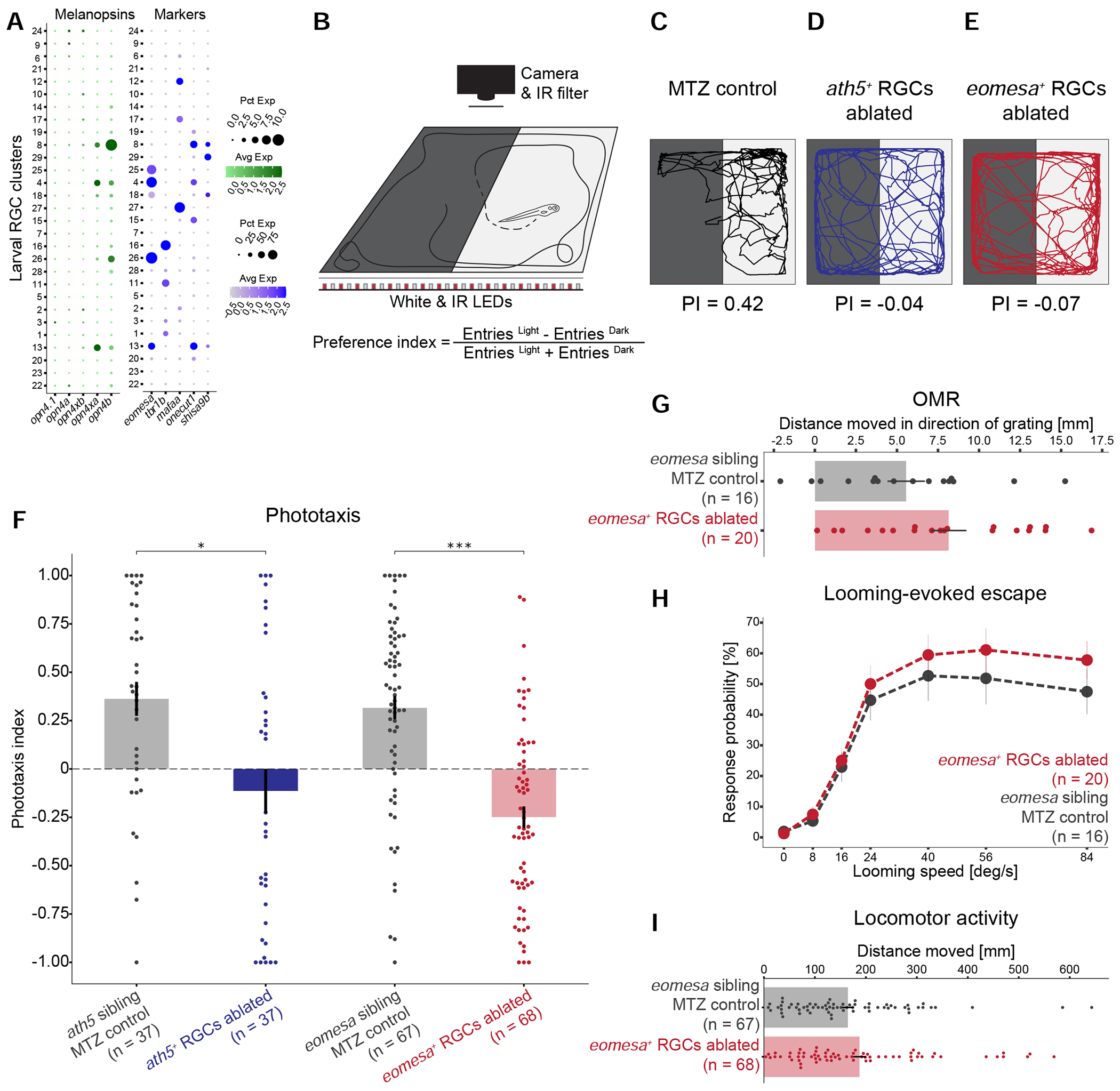
(A) Dotplots showing type-specific expression of melanopsin in larval RGCs. Left: Of five melanopsin homologs (columns), only opn4xa and opn4b have discernible expression in specific larval RGC clusters (rows). Right: opn4xa+ and opn4b+ clusters include eomesa+ RGC types, but not mafaa+ or tbr1b+ types. An opn+eomesa− RGC type is marked by the co-expression of onecut1 and shisa9b. Larval clusters are ordered as in Figure 3A. Expression patterns are conserved in adult RGC types (Figure S7D).
(B) Phototaxis assay. Larvae are placed in a light-dark choice arena, and their positions are tracked over time. A phototaxis index (PI) quantifies attraction towards the light source.
(C-E) Representative traces and PI values of an MTZ-treated control larva (C), ath5+ RGC-ablated blind larva (D), and eomesa+ RGC-ablated larva (E).
(F) PI values for NTR− Tg(ath5:QF2) and Tg(eomesa:QF2) control siblings as well as ath5+ RGC ablated blind fish, and eomesa+ RGC-ablated larvae. Shown is a barplot with a superimposed dotplot, where each dot represents one fish. Error bars represent SEM. * p<0.05, *** p<0.001 (Dunn post-hoc test).
(G) Quantification of optomotor response in MTZ-treated controls and eomesa+ RGC-ablated larvae, plotted as in F.
(H) Escape probability of MTZ-treated controls and eomesa+ RGC-ablated larvae to a looming disc. Each dot represents the mean value at a given stimulus expansion rate. Error bars represent SEM.
(I) Quantification of locomotor activity of MTZ-treated controls and eomesa+ RGC-ablated larvae, plotted as in F.
eomesa+ RGCs are required for phototaxis
To test if eomesa+ RGCs subserve a specific behavior, we selectively ablated them and tested fish in a battery of behavioral assays: (a) phototaxis, the tendency to move towards a light source; (b) the optomotor response (OMR), a reflexive behavior that stabilizes the animal’s position in response to optic flow; (c) escape behavior evoked by a looming stimulus; and (d) overall locomotor activity. In each case, we used a cross of Tg(eomesa:QF2, QUAS:switchNTR) and Tg(ath5:Cre) fish. In the resulting triple-transgenic fish, nitroreductase (NTR) is specifically expressed by eomesa+ RGCs. NTR converts the substrate metronidazole (MTZ) into a cytotoxic compound (Curado et al., 2008; Tabor et al., 2014). Administration of MTZ to these larvae effectively and selectively ablated eomesa+ RGCs (STAR Methods, Figure S7E).
To examine phototaxis, we tracked the position of larvae in a dark-light choice arena and calculated a preference index (PI) (STAR Methods, Figure 7B). Control larvae preferred the illuminated half of the arena, quantified as positive phototaxis (Figures 7C, 7F; PI = 0.36 ± 0.08 for ath5:QF2 clutch siblings, PI = 0.32 ± 0.06 for eomesa:QF2 clutch siblings, mean ± SEM). As expected, phototaxis was severely disrupted upon NTR activation in a pan-RGC line, a treatment that lesioned all RGCs. (Figures 7D, 7F; PI = −0.11 ± 0.11, p=0.013, Dunn’s test with Bonferroni correction). Phototaxis was similarly abolished upon selective ablation of eomesa+ RGCs (Figure 7E, F and S7F; PI = −0.25 ± 0.06, p=1. 176 x 10−7, Dunn’s test with Bonferroni correction). In contrast, we found no significant effect of removing eomesa+ RGCs on OMR, escape behavior or overall locomotor activity (STAR Methods, Figure 7G–I). In conclusion, this defined RGC subclass is required specifically for phototaxis.
Discussion
Using single-cell transcriptomics, we assembled a molecular catalog of RGC types in adult and larval zebrafish. We identified transcription factors (TFs), cell recognition molecules, and neuropeptides that were expressed in one or few of the RGC clusters, serving as single or combinatorial markers for individual cell types. We note that orthologs of variably expressed TFs in zebrafish often also label RGC types in mouse, macaque and human (e.g. Pou4f3, Eomes, Tbr1, Irx3, Mafa, Satb1, Satb2, Foxp2) (Liu et al., 2018; Peng et al., 2019, 2017; Rousso et al., 2016; Tran et al., 2019; Yan et al., 2020). This observation highlights the conservation of TF expression patterns and potentially cell type identities across the vertebrate lineage. We chose four type-specific TFs to genome-engineer driver lines and used them to investigate the morphological and physiological properties of RGC types. Finally, we exploited this genetic access to causally associate a molecularly, anatomically and functionally defined retinofugal pathway with a specific visual behavior. Together, our work provides a comprehensive survey of RGC diversity as a resource for future studies and paves the way to explore the development, structure and function of the vertebrate visual system.
Diversity of zebrafish RGCs
We identified 32 transcriptionally distinct RGC types in adult zebrafish. The number and frequency distribution of cell types resembles those seen in other vertebrates like mice (~40-46 types: Rheaume et al., 2018; Tran et al., 2019), Peromyscus and chick (Sanes and co-workers, unpublished). On the other hand, the primate and human visual system contains only 15 to 18 molecular RGC types (Peng et al., 2019; Yan et al., 2020) with the four most frequent types, the so called ON and OFF midget and parasol RGCs, accounting for ~85% of all RGCs. In contrast, the four most frequent RGC types in zebrafish, mice, chick and Peromyscus account for <30% of all RGCs.
The number of molecular types identified in this study (32) is substantially lower than the ~75 types described previously based on dendritic stratification and axonal projection patterns (Robles et al., 2014). As morphological validation of all transcriptional clusters remains to be completed, one can only speculate on the reasons for this discrepancy. Our transcriptomic catalog may underestimate the true diversity for the following reasons: (a) Despite broad RFP expression, it is possible that some types are not captured due to incomplete labeling by the isl2b promoter used to purify RGCs for sequencing (Zhou et al., 2020). (b) Some types may have been undersampled because of biases in expression of the transgene or selective vulnerability in the purification process. (c) Some infrequent types might be unresolved, because the computational power to resolve types into separate clusters depends on transcriptional separation and the number of cells sequenced (Shekhar et al., 2016). (d) Distinctive transcriptional signatures may exist only during very early development and are downregulated at the stages we sampled, as observed in previous studies (Li et al., 2017; Tran et al., 2019). Using more sensitive RNA sequencing methods or measuring other modalities (e.g. proteome or epigenome) might potentially resolve these “hidden” types. On the other hand, the projectome study may have overestimated the cell type diversity: (a) Some morphologies might be developmentally transient. (b) Some morphologically sampled cells may be developmental errors that are pruned or error-corrected at later stages. (c) Some cells viewed as morphologically distinct may have been variants of a single molecular type. Taken together, it seems plausible that the true number of RGC types in zebrafish is greater than 32 and fewer than 75. Continued efforts to map the transcriptional profiles of specific types to their morphology, function and distribution in the retina, at different stages of development, will settle this issue and produce a definitive account of RGC diversity.
Progressive diversification of RGCs
Unlike in mammals and birds, the teleost retina grows throughout life, constantly adding new RGCs at its margins, although expansion and neurogenesis rates drop in the adult (Marcus et al., 1999). Against this backdrop of continued growth and rewiring (Hu and Easter, 1999; Kay et al., 2005), the zebrafish visual system supports a variety of behaviors already at early larval stages. We therefore expected to find a mix of fully differentiated and immature RGCs, especially in our larval dataset. Because most larval behaviors persist into adulthood, we also hypothesized that many larval RGC types would have matching counterparts in the adult. Comparison of the larval and adult RGC clusters confirmed both of these hypotheses. Twenty-three mature larval clusters, containing two-thirds of the RGCs at this stage, could be successfully mapped onto one or very few adult clusters each. Six of the 29 larval clusters, displayed signatures of ongoing differentiation, including genes associated with cytoskeletal reorganization and axon guidance. Five of these immature clusters exhibited transcriptomic signatures that mapped onto distinct subsets of mature larval RGC types, indicating that they are cells in transition to maturity. The sixth cluster may represent postmitotic RGC precursors that are apparently not yet committed to specific fates. These immature RGCs persist into adulthood (adult cluster 3). However, such immature RGCs were not detected in mouse, primate or human retinas, which do not add new neurons at adult stages (Macosko et al., 2015; Peng et al., 2019; Rheaume et al., 2018; Shekhar et al., 2016; Tran et al., 2019; Yan et al., 2020).
These results suggest a model in which RGC types arise by progressive diversification, proceeding from a precursor, through immature and incompletely specified larval types that finally mature into specific adult types. A corollary is that not all RGC types are completely specified in larvae. Those that do form later may support behaviors that emerge at juvenile stages, such as shoaling (Larsch and Baier, 2018) or recognition of geometric visual cues (Yashina et al., 2019). This diversification proceeds in parallel with global as well as type-specific gene expression changes even in the stable clusters. Nonetheless, we find potential larval counterparts of all adult types, suggesting a progressive addition of new types by splitting of already existing ones.
Common molecular, physiological and morphological properties of single RGC types
One of the goals in classifying neuronal cell types is to harmonize multiple aspects of cell identity (Regev et al., 2017; Sanes and Masland, 2015; Vlasits et al., 2019; Zeng and Sanes, 2017). Recent studies in mice have demonstrated congruence between molecularly, physiologically and morphologically defined RGC types (Baden et al., 2016; Bae et al., 2018; Franke et al., 2017; Tran et al., 2019), and efforts to catalog all properties are underway (rgctypes.org). In larval zebrafish, we were able to assign molecular identities to RGC subpopulations with shared structural, developmental and functional properties. Our study, together with a recent comprehensive survey of RGC feature selectivities (Förster et al., 2020), has thus laid the foundation for a systematic annotation of all RGC types in a vertebrate.
By exploiting selectively expressed TFs, we engineered reporter lines that target either an individual or a small group of closely related RGC types. In each case, we uncovered an anatomically distinct visual pathway with dedicated projection targets. Intriguingly, RGCs resembling the mafaa+ projection pattern and the corresponding two dendrite morphologies have previously been implicated in the recognition of small, motile prey (Semmelhack et al., 2014). Molecular and morphological characteristics also mapped to discrete physiological tuning profiles. For example, tbx20+ RGCs, which constitute a rare RGC type that projects to two pretectal nuclei (AF4 and AF9), but does not extend to the tectum, possess transcriptomic, morphological and activity patterns that are consistent with being a single type within the group of eomesa+ RGCs. We speculate that eomesa+ RGC-specific expression of the secreted morphogen bmp4 and axon guidance receptor plxna4 contribute to their asymmetrical position within the retina and their axonal projection patterns within the brain, respectively. In addition, this group of RGCs may exert neuropeptidergic functions via their synthesis of the neuropeptide nmbb. These hypotheses can now be tested, using the genetic access provided by our work.
Specific role of eomesa+ RGCs in phototaxis
In mammals, melanopsin confers the ability to sense ambient light stimuli (Berson et al., 2002; Do, 2019; Fu et al., 2005; Hattar, 2002; Schmidt et al., 2011). Tbr2, the mammalian homolog of eomesa, is expressed in melanopsin-expressing ipRGCs and is essential for their establishment and maintenance (Mao et al., 2014). In zebrafish, eomesa+ RGCs co-expressed not only the melanopsin gene opn4xa as previously suggested (Matos-Cruz et al., 2011), but also opn4b, consistent with the idea that they are ipRGCs. Functional imaging during presentation with a battery of visual stimuli revealed that eomesa+ RGCs indeed encode ambient luminance levels. Zebrafish melanopsin-expressing RGCs have previously been linked to phototaxis (Zhang et al., 2017), but lack of precise genetic access has precluded a direct test of this connection. By selectively ablating eomesa+ RGCs, we showed that they are required for phototaxis, but are dispensable for several other visually guided behaviors. These results add to the growing consensus (see Baden et al., 2016; Chen et al., 2011; Dhande et al., 2013; Sanes and Masland, 2015) that genetically defined and anatomically separable axonal pathways convey specific visual features to downstream processing centers for initiation of appropriate behavioral responses.
In conclusion, we have shown that eomesa+ RGCs form a specific functional pathway and suggest that the same may be true for other types. Neural circuits downstream of the tectum and the other retinorecipient nuclei in the hypothalamus, thalamus and pretectum may be organized in a similar ‘labeled line’ fashion. For example, the tectal motor map was recently shown to channel commands to hindbrain circuits via two parallel pathways, each dedicated to a specific behavioral response (Helmbrecht et al., 2018), and a small cluster of pretectal neurons, which receive input from direction-selective RGCs, was shown to be necessary and sufficient to drive optokinetic responses (Wu et al., 2020). Our scRNA-seq approach to RGC diversity is expected to serve as a blueprint for the molecular dissection of other parts of the central nervous system.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Herwig Baier (hbaier@neuro.mpg.de).
Materials Availability
Unique materials such as plasmids and transgenic lines generated in this study will be made available upon request without any restrictions.
Data and Code Availability
Computational scripts detailing scRNA-seq analysis reported in this paper are available at https://github.com/shekharlab/ZebrafishRGC. We have also provided R markdown (Rmd) files that show step-by-step reproduction of the key results. All raw and processed scRNA-seq datasets reported in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under the accession number GSE152842. Visualization of the zebrafish RGC atlas is available at https://portals.broadinstitute.org/single_cell (Study ID: SCP992). All data and custom software for functional imaging analysis and behavioral tests will be made available upon request.
Experimental model and subject details
Zebrafish
Adult and larval zebrafish were maintained on a 14:10 hour light:dark cycle at 28°C. Embryos were bred in Danieau’s solution (17 mM NaCl, 2 mM KCl, 0.12 mM MgSO4, 1.8 mM Ca(NO3)2, 1.5 mM HEPES). All animal procedures conformed to the institutional guidelines set by the Max Planck Society, with an animal protocol approved by the regional government (Regierung von Oberbayern) as well as by the Harvard University/Faculty of Arts & Sciences Standing Committee on the Use of Animals in Research and Teaching (IACUC). All animals used were anesthetized in a lethal overdose of tricaine and rapidly euthanized by immersion in ice water for 10 min. Zebrafish larvae used in this study were between 5 and 7 days post fertilization. For single cell transcriptomic profiling, 5 dpf larval and 4–6 months old female and male adult Tg(isl2b:tagRFP) zebrafish were used for retina dissection, tissue dissociation and cell purification.
To establish intersectional transgenic tools, the Tg(ath5:Cre) transgene was characterized using Tg(actb2:loxP-eGFP-loxP-lynTagRFPT) fish.
For morphological analysis of specific RGC types, the pan-RGC transgenes Tg(isl2b:tagRFP) or Tg(isl2b:GFP) served as a visual landmark of target brain nuclei. We generated the RGC type-specific transgenic lines Tg(eomesa:QF2), Tg(mafaa:QF2), Tg(tbr1b:QF2), Tg(tbx3a:QF2) and combined them with Tg(QUAS:switchNTR) or Tg(QUAS:switchG6s) reporters. In addition, we used Tg(tbx20:Gal4) in combination with Tg(UAS:NTR-mCherry) or Tg(UAS:Dendra). Larvae were bred in 0.003% PTU-Danieau’s to suppress pigmentation prior to staining.
Functional imaging data were obtained from mitfa−/− larvae expressing GCaMP6s in RGCs by crossing Tg(isl2b:Gal4) or Tg(tbx20:Gal4) to Tg(UAS:GCaMP6s) or generating triple-transgenic Tg(eomesa:QF2, QUAS:switchG6s, ath5:Cre) fish. Targeted cell ablation and subsequent behavioral experiments were performed using Tg(ath5:QF2, QUAS:epNTR-tagRFP) and triple-transgenic Tg(eomesa:QF2, QUAS:switchNTR, ath5:Cre) larvae.
METHOD DETAILS
RGC purification and droplet based single cell RNA sequencing
RGCs were labeled using transgenic Tg(isl2b:tagRFP) zebrafish that express RFP in all RGCs (Mumm et al., 2006; Pittman et al., 2008). Retinas from larval or adult fish were dissected in oxygenated (ox) Ames and transferred into ox Ames on ice until tissue collection was completed. Retinas were digested in papain 20U/ml, DNAseI 80U/ml, L-cysteine 1.5mM in ox Ames at 28°C for 30 (larval retinas) or 45 minutes (adult retinas). To stop the digestion, the papain solution was replaced by papain inhibitor solution containing ovomucoid 15mg/ml and BSA 15mg/ml. Tissue was gently dissociated by trituration using a flamed glass pipette in papain inhibitor solution. To wash the cell suspension, cells were pelleted at 250g for 8 minutes and resuspended in ox. Ames containing 0.4% BSA. The cell suspension was filtered through a 30μm strainer prior to fluorescence-activated cell sorting (FACS) purification. Non-transgenic wildtype retinas were used to determine background fluorescence levels and adjust sorting gates. Calcein blue was added to distinguish live RFP+ RGCs. Cells were washed and resuspended in PBS 0.04% BSA and loaded onto the microfluidic device within ~45 minutes after FACS enrichment. Droplet RNA sequencing experiments using the 10X chromium platform were performed according to the manufacturer’s instructions with no modifications.
For the larval dataset, 200 manually dissected retinas were dissociated in one experiment and single cell profiles were collected across 3 replicates. For the adult dataset, about 20 retinas per batch were dissected and dissociated and droplet RNA sequencing was performed collecting a total of 15 replicates across 5 experiments. The cDNA libraries were sequenced on the Illumina HiSeq 2500 to a depth of ~30,000 reads per cell.
Computational analysis of single cell transcriptomics data
Alignment and quantification of gene expression
Initial preprocessing was performed using the cellranger software suite (version 2.1.0, 10X Genomics), following steps described previously (Pandey et al., 2018). Briefly, sequencing reads were demultiplexed using “cellranger mkfastq” to obtain a separate set of fastq.gz files for each of 15 adult and 3 larval samples. Reads for each channel were aligned to the zebrafish reference transcriptome (ENSEMBL zv10, release 82) using “cellranger count” with default parameters to obtain a digital gene expression (DGE) matrix (genes x cells) summarizing transcript counts. For each of the adult and larval experiments, we combined the DGEs from different channels and analyzed them, as described below using the Seurat R package (Satija et al., 2015). We used the default parameter values for all Seurat functions, unless stated otherwise. In the context of tasks such as clustering, we also verified the robustness of the results to variations in select parameters. We have documented the full details of our analyses in markdown scripts available at https://github.com/shekharlab/ZebrafishRGC.
Adult RGC catalog
Preprocessing and batch integration:
The adult DGE matrix was filtered to remove genes expressed in fewer than 25 cells, and cells expressing fewer than 450 genes resulting in 24,105 genes and 48,551 cells. To align the five biological replicates, we used the Canonical Correlation Analysis (CCA) based integration framework in Seurat to embed the cells in a shared, reduced dimensional gene expression space. Briefly, each cell was normalized to a total library size of 10,000 and the normalized counts were log-transformed (X ← log(X + 1)) using the function Seurat::NormalizeData. We used Seurat::FindVariableFetures with option selection.method = “vst” to identify the top 2000 highly variable genes (HVGs) (Hafemeister and Satija, 2019) in each batch. Next, we used Seurat::FindIntegrationAnchors and Seurat::IntegrateData, both with options “dims=1:40” to perform Canonical Correlation Analysis (CCA)-based batch correction on the reduced expression matrix consisting of the HVGs. Seurat::LocalStruct was used to estimate how well the structure of each batch was conserved after integration. For each batch, LocalStruct computes the top nearest neighbors in PCA and corrected PCA space and returns the size of the intersection. The median score was 0.83, 0.77, 0.77, 0.83, and 0.77 for batches 1 through 5, respectively, indicating that the local structure of the individual datasets is not substantially impacted by the integration. The “integrated” expression values were combined across batches, and used for dimensionality reduction and clustering.
Dimensionality Reduction, Clustering and Visualization:
To remove scale disparities between genes arising from differences in average expression levels, the integrated expression values for each HVG were z-scored across the cells using Seurat::ScaleData. Next, we performed Principal Component Analysis (PCA) on the scaled matrix, and used Seurat::ElbowPlot to select 40 principal components (PCs). In this reduced dimensional space of 40 PCs, we built a k-nearest neighbor graph using Seurat::FindNeighbors and identified transcriptionally distinct clusters using Seurat::FindClusters, using a resolution parameter of 0.5. We then varied the resolution parameter from 0.3 to 1.5 and found the cluster definitions were robust in this range as assessed by the adjusted rand index (ARI), a measure of the similarity of data clusterings ranging from 0 (poor correspondence) to 1 (full correspondence).
The ARI values were higher than 0.8 throughout this range, indicating a high correspondence. The number of clusters increased from 32 clusters at resolution = 0.5 to 37 clusters at resolution = 1.0. The new clusters nominated were splits of older clusters, but post hoc analysis did not reveal genuine differentially expressed genes (data not shown). We therefore elected to report our results at resolution = 0.5.
Using the top 40 PCs, we also embedded the cells onto a 2D map using t-distributed stochastic neighbor embedding (tSNE) (Linderman et al., 2019; Maaten and Hinton, 2008). These embeddings were used downstream to visualize gene expression patterns as well as the distribution of various metadata (batch ID, cluster ID, cell quality, etc.).
Identification of RGCs and filtering contaminant classes:
RGC clusters were identified based on expression of the pan-RGC markers rbpms2b (Hörnberg et al., 2013) and isl2b (Pittman et al., 2008). Clusters were removed if they contained an abnormally low number of average genes per cell, did not express rbpms2b, or expressed genes present in contaminant cell types. Examples of such genes include rlbp1a and apoeb for Muller glia (Bernardos et al., 2007), vsx1 for bipolar cells (Vitorino et al., 2009), gad1 and gad2 for amacrine cells (Sandell et al., 1994), pde6 for photoreceptors (Abalo et al., 2020), and cldn19 for endothelial cells (Kolosov et al., 2013). A total of 15,909 cells corresponding to these cell classes were removed. This contamination most likely arises from transgenic labeling of other retinal cells by Tg(isl2b:tagRFP) that fall into the same FACS gate as RGCs. Interestingly, we found a much lower proportion of contaminants at the larval stage (see below), which suggests that promiscuous expression of the transgene may increase with age. The RGCs were separated and analyzed beginning from raw counts, with integration, PCA, clustering, and visualization performed in the same way detailed above.
We also estimated the relative expression of mitochondrial genes in clusters (Methods Figure 1). Further, we found that a more stringent filtering of the data via removal of cells that contained >10% mitochondrial origin transcripts did not impact cluster assignments (Methods Figure 2).
Methods Figure 1:
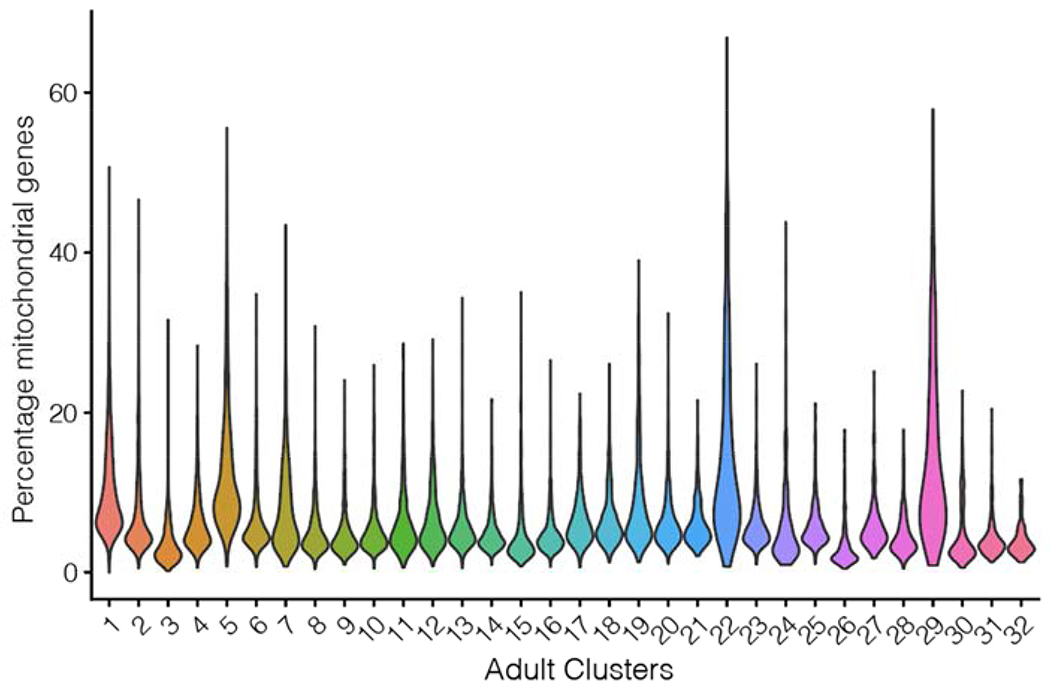
Percentage of mitochondrial genes in adult clusters.
Methods Figure 2:
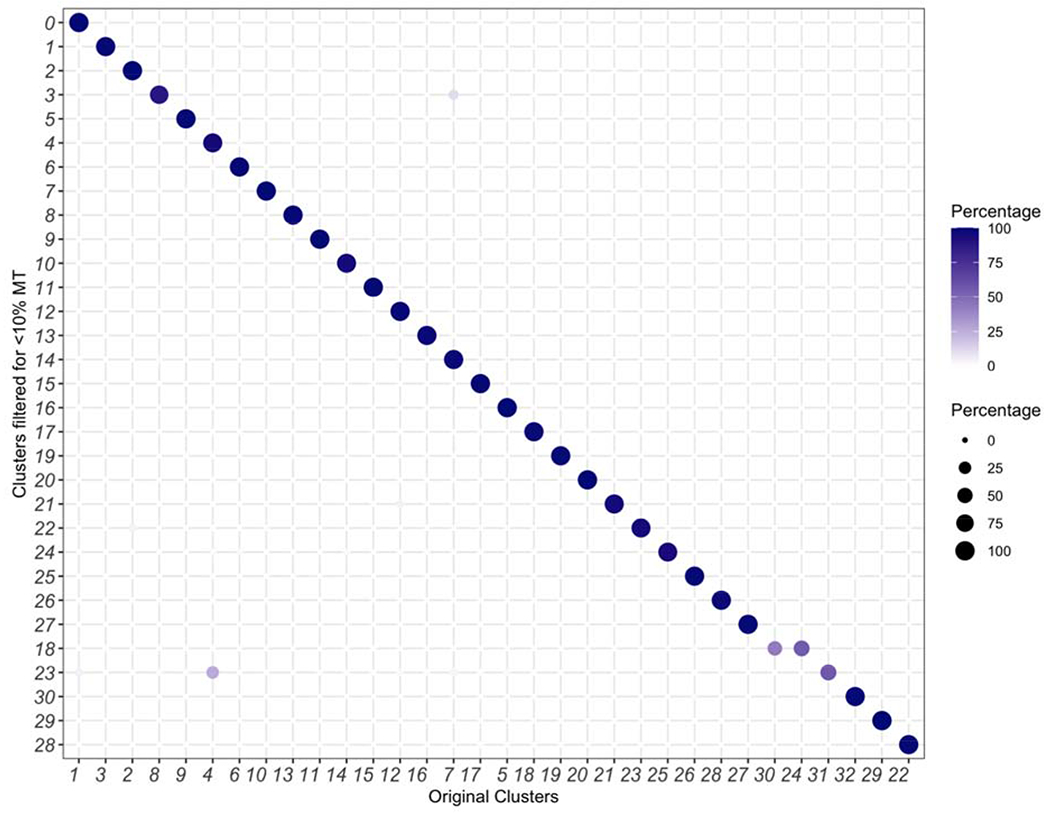
Removing cells with a mitochondrial percentage greater than 10% does not significantly impact cluster discovery.
Differential expression analysis and hierarchical clustering:
We used Seurat::FindMarkers with options test.use =“MAST”, max.cells.per.ident = 1000 to identify differentially expressed genes in each RGC cluster. To identify transcriptional relationships between RGC clusters, we used Seurat::FindVariableFeatures to recalculate the top 500 most variable genes. The average expression values of genes in each cluster were used as input for hierarchical clustering, performed using Seurat::BuildClusterTree. The resulting output was visualized as a dendrogram.
Larval RGC catalog
The larval DGE was analyzed by following the steps entitled “Preprocessing”, “Dimensionality Reduction”, “Clustering” and “2D Visualization” described above. Filtering genes expressed in fewer than 25 cells, and cells expressing fewer than 450 genes resulted in 24,105 genes in 12,698 cells. Each cell was normalized and log-transformed, and the top 2000 HVGs were identified in each batch as before. z-scored expression values along these HVGs were used to calculate PCs, and the top 30 PCs were used to define clusters as well as embed cells on a tSNE map. As in the case of the adult, we found that the number and identity of clusters were quite robust when we varied the resolution parameter in the range 0.3 to 1.5 (not shown). We choose to report results at resolution = 0.5.
We annotated clusters based on their expression of cell-class specific markers as before, and removed non-RGC clusters, which comprised ~9.3% of the data. The relative expression levels of mitochondrially derived transcripts across larval clusters was low (Methods Figure 3).
Methods Figure 3:
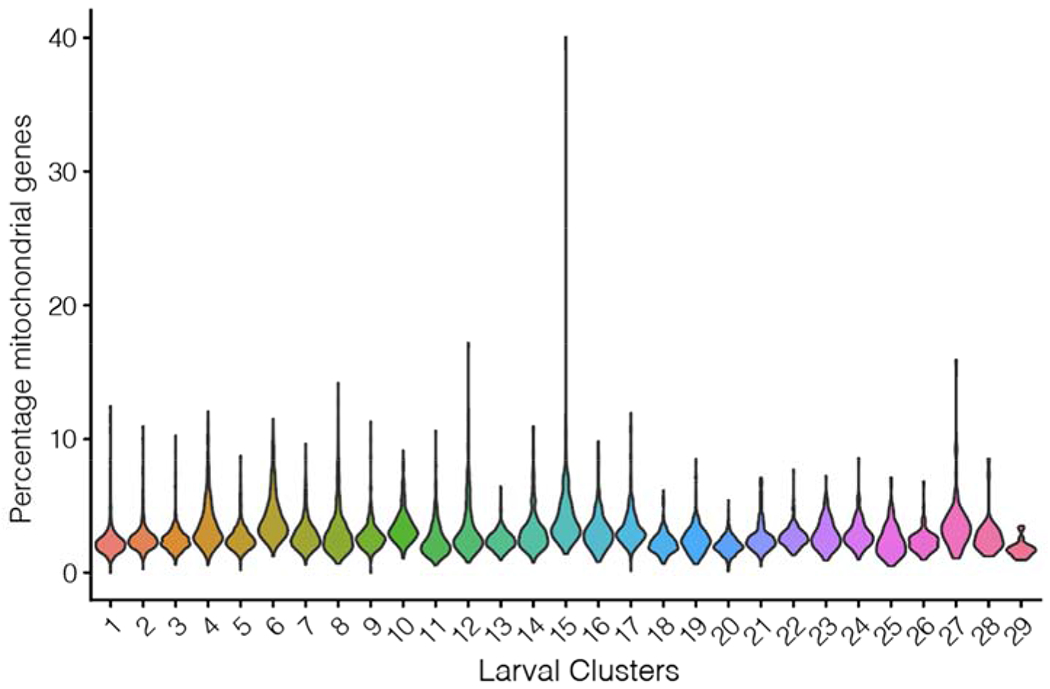
Percentage of mitochondrial genes in larval clusters.
We performed differential expression analysis among the RGC clusters (all of which robustly expressed rbpms2b and isl2b) to define cluster-specific markers. The transcriptional interrelatedness between the larval RGC clusters was visualized on a dendrogram built using hierarchical clustering. Six clusters (1, 2, 3, 5, 13, 20) were identified as “immature” based on shared expression of alcamb, tmsb, and tubb5, while the remaining 23 clusters were labeled as “mature”. Immature and mature larval RGCs were separately visualized on tSNE maps. To compare the clustering quality between the two subsets, we computed the silhouette score for each cell within each subset. For each point, the silhouette score is defined as a(i) – b(i) / max{a(i),b(i)}, where a(i) is the mean distance between point i and all other points in the cluster containing i and b(i) is the minimum mean distance of point i to all points in any cluster not containing i. Using the silhouette function of the R package cluster with the tSNE embeddings and cluster labels as inputs, the median silhouette score was computed for each subset.
Surveying expression of cluster-enriched transcription factors, neuropeptides, and recognition molecules
Initial databases of 1,524 transcription factors, 158 neuropeptides, and 387 candidates involved in axon guidance were assembled from the Zebrafish Information Network website (zfin.org) by selecting genes with search terms “transcription factor”, “neuropeptide” or “axon guidance”. The recognition molecule library was expanded from the axon guidance list to 515 genes by searching the larval and adult DGEs for genes that began with cntn (Contactins), eph (ephrin receptors), efn (Ephrin proteins), robo (Roundabout family of guidance molecules), slit (Slit guidance ligands and receptors), sema (Semaphorins), plx (Plexins), nrp (Neuropilins), cdh (Cadherins), pcdh (Protocadherins), ncam (Neuronal cell adhesion molecules), cadm (Cell adhesion molecule genes), and lrrtm (Leucine-rich repeat transmembrane proteins). These databases were filtered to include only genes expressed in >30% of cells in at least one cluster within the catalog (adult and larval).
From the database of 1,524 transcription factors, 184 and 186 transcription factors were expressed in the larval and adult RGC catalog, respectively. Of these, 147 candidates were expressed in both the larval and adult datasets, which represents a highly significant overlap unlikely to occur by chance (p < 10−132, hypergeometric test). Out of 158 candidate neuropeptides, 10 were expressed in the larva and 11 were expressed in the adult (N=8 shared). Among 515 candidate cell surface and secreted molecules, 67 and 76 were expressed in the larva and adult respectively (N=57 shared). These overlaps were also highly significant based on the hypergeometric test (p < 10−13 and p < 10−44 respectively).
Supervised classification analysis of transcriptional correspondence between adult RGC catalog and larval RGC types
Feature Selection:
We first assembled a list of cluster specific markers in both larval and adult RGCs as features for training a multi-class classifier. We applied Seurat::FindAllMarkers with arguments “only.pos = TRUE, test.use = ‘wilcox’, min.pct = 0.25, logfc.threshold = 0.25” separately to the adult and larval catalogs to identify genes differentially expressed (DE) within each adult or larval cluster. 641 genes expressed with a significance level of p<10−10 in at least one adult or larval cluster were selected as features.
Assigning adult identities to larval RGCs:
Using the 641 DE genes as features, we trained gradient boosted decision trees on the 32 adult clusters. This was implemented using the R package xgboost (Chen and Guestrin, 2016), as in our previous publications (Peng et al., 2019; Tran et al., 2019). For training, expression values along each feature were z-scored to remove scale disparities. We split the adult RGCs 60%/40% into training and validation sets, respectively. To avoid overrepresentation of the largest clusters, we capped the representation of each cluster to a maximum of 400 cells. We used the “held-out” labels of the validation set to assess the performance of the classifier, which was found to have an average error rate of 8.7% (min 2.8%, max 29%) for each of the 32 clusters.
The adult RGC-trained classifier was used to assign an adult identity to each mature larval RGC based on its expression values along the 641 features. For consistency, the larval RGC expression matrix was z-scored along each of these features. The results were summarized as a confusion matrix, which plots the relative proportion of cells in each larval cluster (column) that map to each adult cluster (rows). Importantly, we note that information regarding the larval clustering was not used in either the training or classification steps.
Assigning mature larval identities to immature RGCs:
We followed a procedure analogous to the one outlined above, with the exception that the classifier was trained and validated on the 6 immature clusters, and applied to each mature cell.
Maturational changes
A mapping was considered 1:1 if more than 60% of the cells within an adult cluster mapped to a single larval cluster, and that larval cluster received no more than 25% of its mappings from any other adult cluster. Six 1:1 mappings were found in the model. Seurat::FindMarkers was used to determine differentially expressed genes between all six adult and larval clusters. A gene was considered to be associated with global maturational changes if the magnitude of its average log fold change (logFC) was greater than 1 and expressed in at least 50% of all larval or adult cells. Seurat:FindMarkers was then used to determine differentially expressed genes between each larval and adult cluster that mapped to each other. A gene was considered to be associated with type specific maturational changes if the magnitude of its average logFC was greater than 1, the gene was expressed in at least 50% of either the larval or adult cells, and it was not associated with global maturational changes.
Combining intronic and exonic reads to elucidate RGC type-specific expression of melanopsin genes
To include intron aligned reads in the quantification of gene expression, we employed velocyto (La Manno et al., 2018), which uses cellranger-generated binary alignment map (BAM) files to calculate separate DGE matrices corresponding to “spliced” and “unspliced” transcripts by distinguishing between intron-aligned and exon-aligned reads. We ran velocyto on each of the adult and larval samples’ BAM files individually using the following general command line invocation,
velocyto run -b barcodes.tsv -o /output/path -m mask.gtf bamfile.bam genes.gtf
The transcriptome annotation (genes.gtf) file used here was also used for alignment by cellranger. “barcodes.tsv” refers to the list of valid cell barcodes in the sample, an output from the cellranger pipeline. The masking file for suppressing alignment to repetitive elements was downloaded from the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables) Sep. 2014 (GRCz10/danRer10) assembly. Velocyto output loom files were processed using in house scripts to compute the spliced and unspliced DGE matrices (DGEspliced and DGEunspliced), which were summed into a consolidated expression matrix DGEtot = DGEspliced+DGEunspliced. We used DGEtot for examining the expression patterns of melanopsin genes in RGC types.
Establishment of Q-system intersectional transgenic tools
To generate intersectional QUAS plasmids, a Tol2-QUAS; cmlc2:mCherry plasmid (Fernandes et al., 2019) was linearized to insert effector genes. For the QUAS:switchNTR construct, a loxP-GFPcaax-loxP fragment (Förster et al., 2017) and an epNTR-tagRFP fragment (Tabor et al., 2014) were inserted by In-Fusion cloning (Takara, Cat# 638909). Similarly, for the QUAS:switchG6s construct, a loxP-tdTomatocaax-loxP fragment (Förster et al., 2017) and a GCaMP6s fragment (Thiele et al., 2014) were inserted. QUAS reporter lines were generated by Tol2-transgenesis as described previously (Suster et al., 2011).
Locus-specific transgenesis using CRISPR-Cas9
gRNA target sequences were selected using the CCTop tool. Donor plasmids were cloned using the GoldenGATEway strategy (Kirchmaier et al., 2013) recombining entry vectors carrying fragments with sequences for GBait (pGGEV_-1), target gRNA (pGGEV_2 was mimicked by annealed oligonucleotides), basal promoter e1b (pGGEV_3), QF2-polyA (pGGEV_4), and polyA (pGGEV_5’) into a pGGDestSC_-ATG vector.
CRISPR-Cas9 RNP complex was prepared at a concentration of 1.5 μM as described before (Essner, 2016). Briefly, gRNA was produced by annealing customized crRNA (IDT, Alt-R® CRISPR-Cas9 crRNA) with tracrRNA (IDT, CAT# 1072533) in buffer (IDT, CAT# 11-05-01-12). gRNA was incubated with Cas9 protein (IDT, CAT# 1081060) for 15 minutes at 37°C and donor plasmid was added to a final concentration of 25 ng/μl. The freshly prepared CRISPR-Cas9 cocktail was injected into the cell of transgenic Tg(QUAS:switchNTR, ath5:Cre) zygotes. Transient expressors were raised and screened for germline transmission.
Histological methods
Immunohistochemical staining on whole fish larvae or dissected adult brains was performed following PACT tissue clearing as described previously (Kunst et al., 2019; Treweek et al., 2015). In brief, larvae were fixed in PACT hydrogel monomer solution, deoxygenated and polymerized. Samples were cleared for several days and washed in PBT prior to staining. For adult brains, fixation and clearing time was adapted to 48 hours and two weeks, respectively. After clearing, samples were permeabilized and blocked. Primary antibody incubation on larval fish took place for 7 days while incubation for adult brains was prolonged to 14 days. Following thorough washing, Alexa-conjugated secondary antibodies were incubated for 3 and 7 days for larval and adult brain samples, respectively. Samples were washed, post-fixed in paraformaldehyde and stored in 87% glycerol. Imaging was performed at a Leica SP8 confocal microscope. Prior to image acquisition, samples were mounted in 2% low melting point agarose in 87% glycerol on bridge slides and coverslipped.
To better characterize labeling in the retina, retinal tissue was sectioned on a cryostat. Transgenic larvae or dissected adult eyes were fixed in 4% PFA in PBT at 4°C overnight, washed and incubated in 35% sucrose in PBST for cryoprotection. Tissue was embedded in TissueTek (Sakura), sectioned at 30μm, washed in PBT and blocked in 5% goat serum, 1% BSA, and 1% DMSO in PBT. Staining occurred by incubation with primary antibody for 4 days and secondary antibody for 2 days. Following washing and post-fixation, sections were coverslipped for imaging.
Images were processed using the FIJI software.
Functional imaging and computational methods for characterization of RGC responses
For functional characterization of RGCs, we immobilized transgenic zebrafish larvae expressing GCaMP6s in 2% low melting point agarose. In vivo calcium imaging was performed on a two-photon microscope (Femtonics 3DRC, Femtonics, Tuzlo, Hungary) equipped with a water-immersion objective (Olympus 20X, 1.0 NA); a Ti:Sapphire laser source (Chameleon Ultra II, Coherent) tuned to 920 nm; using the green detection channel (Semrock Brightline 525/50). A stimulus sequence described below was projected onto a white diffusive screen (Rosco, 10cm wide, 6cm high) using a LED projector (LG, Model No. PA72G) with the addition of a 561nm longpass filter, using only the red LED channel which emits 600–635 nm (peak emission at around 615 nm). At maximum intensity the filtered red channel, as measured just before the diffusive screen, was 52.9 μW/cm2 (105.6 lux), with a contrast ratio of 41:1. The projection was presented monocularly from the animal’s left side and covered 120° of the larva’s field of view. Visual stimulation was designed using PsychoPy2 as follows: dark screen (10 sec), bright flash (10 sec), dark flash (10 sec), grating moving in four main cardinal directions (10° black bars interspaced at 30°, 5 sec stationary then 5 sec moving at 20°/sec), dark screen (10 sec), prey-like stimulus (4 repetitions of a 3° bright dot sweeping across the black screen at 90°/sec, 20 seconds), bright ramp (dark to bright red in 5 seconds), bright screen (10 seconds), loom (2 repetitions of a black disc expanding at 30°/second on a white screen, 30 seconds), dark ramp (bright red to dark in 5 seconds), dark screen (10 sec). The total stimulus duration was 160 seconds. The frame size for AF9 was 178 x 380 pixels and the recording frame rate was ~3.6 Hz. For tectal recordings frame size was 345 x 345 pixels and the recording frame rate was ~2 Hz. The pixel size was .25 um by .25 um.
Recorded imaging data were pre-processed as described previously (Helmbrecht et al., 2018). In brief, images were motion-corrected using the CaImAn package, uniform filtered over 3 frames and the dF/F was calculated using the 5th percentile of the traces. In total, 11 regressors for all stimulus components were created and convolved with a GCaMP6s kernel. Neuronal activity was analyzed pixel-wise by calculating a score of all regressors to the calcium responses of each pixel using a linear regression model of the selected response window with the regressor (Python scikit-learn). For the score, the coefficient of the regression (corresponding to the dF/F) was multiplied by the R2.
To determine overall response types, the scores were normalized per fish to the 99th percentile of all pixels recorded. For the functional clustering of the responsive pixels, 2 Tg(isl2b:Gal4; UAS:GCaMP6s) fish (6 planes each, 380 x 178 pixels) were analyzed by first removing pixels with maximum scores smaller than 0.3 (49823 pix). Next, to reduce noise, affinity propagation clustering (scikit learn – preference: median of similarities) was performed. Keeping clusters with at least 50 pixels (0.1 percent of all pixels) yielded in total 345 clusters with chosen exemplars. To extract the global cluster structure, these 345 exemplars were further clustered using hierarchical clustering (scipy.cluster) using correlation as distance metric. Setting a distance threshold of 0.35 classified a total of 42,444 pixels into 8 distinct clusters, each comprising more than 100 pixels. The activity profile of these 8 clusters, referred to as groups in the text, is shown with an individual min/max normalization in Figure 6 and S6. Classification of response types in the tectum presented in Figure S6 was performed similarly and yielded 14 final clusters.
To map response types of genetically-defined RGC populations, pixels from eomesa and tbx20 recordings from five fish each, were first analyzed to calculate the scores to each regressor as described above. Likewise, pixels with maximum scores smaller than 0.3 were removed. A k-nearest neighbor classifier was trained on the isl2b clustered pixel (42,444 pix with cluster labels, k = 100) and the scores of every mapped fish were assigned to the clusters of the isl2b dataset using either predicted labels for the pixel distribution or probability estimates for the population distributions.
Cell ablation
Larvae expressing the enhanced potency nitroreductase enzyme in RGCs as judged by RFP presence in RGC axons were sorted at 3 dpf. Cell ablation was induced at 4 dpf by bathing larvae in 7.5mM metronidazole for 24 hours, followed by continued treatment in 5mM metronidazole for 12 hours. Healthy, well developed larvae showing normal body posture and locomotive activity were then given a recovery period for a minimum of 24 hours and behavioral experiments were carried out at 7 dpf. Successful ablation was confirmed by confocal imaging before and after treatment with metronidazole using randomly selected clutch mates.
Phototaxis assay
All tests were performed between 9AM and 5PM. Light-preference behavior was assayed by methods modified from (Zhang et al., 2017). Larvae were tested, six animals at a time, in custom-made square chambers (3 × 3 cm each chamber), which was placed inside the ZebraBox, a device for automated observation and tracking of zebrafish behavior. White light and infrared light were projected from the bottom. Half of a chamber was covered with two dichroic Film Polarizers stacked on top of each other creating a gradient of light intensity near the boundary. The lux intensity of the dark and the light side was around 50 lux and 180 lux, respectively. Animals were allowed to adapt to the arena and light conditions for at least 30 minutes prior to behavioral testing. Calculation of the distance travelled and number of entries to each field was performed by the ZebraBox software. Independent validation of tracking of animals was performed with the TheRealFishTracker and results were plotted with custom-written python scripts (Python 3.7) for examples shown on Figure 7. The duration Preference Index was calculated as the difference between the duration (every 10 seconds) in the lit side and the dim side divided by the total duration of the experiment (30 minutes per recording session). The entries PI was calculated as the difference between the number of counts in the lit side and the dim side divided by the total number of counts. Positive PI values indicate light preference. Plots and statistical analysis were performed with custom-written python scripts.
Looming-evoked escape and optomotor response assay
We adapted previously established methods (Fernandes et al., 2019; Larsch and Baier, 2018) to test optomotor and escape responses. In brief, larvae are placed in glass dishes of 10 cm diameter separated by walls to prevent visual contact. Nine animals could be tested at a time. Diffuse Infrared illumination was provided from below to record animal behavior at 30 fps using a camera equipped with an IR bandpass filter. Visual stimuli were projected onto the projection film from underneath via a cold mirror. Image processing and stimulus generation were performed with Bonsai (Lopes et al., 2015). Before behavioral tests, animals were kept for at least 30 minutes in a petri dish floating above a fully lit portion of the projection screen to allow habituation to light and temperature conditions of the experiment.
Looming stimuli were presented as stationary dots expanding for half a second (15 frames) with a linear increase in diameter. Stimuli were presented 1 cm from the fish at angles of 90° or 270° relative to the animals’ center of mass and orientation at the beginning of the stimulus. Looming stimuli were presented once every 90 seconds.
The order of different stimuli (looming expansion rates used: 8, 16, 24, 40, 56 and 84 deg/s, either left or right of the animal) was randomized for each group of animals. To drive larvae towards the center of the dishes using their optomotor response (OMR), a moving grating was presented for 20 seconds ending 10 seconds before the presentation of the next looming stimulus. Distance travelled towards the center during grating presentation was used to measure performance of OMR.
Supplementary Material
Table S1. Summary of differentially expressed genes in larval and adult RGC clusters computed using MAST, Related to Figure 1 and Figure 3.
Table S2. Summary of expression of transcription factors, neural recognition molecules, and neuropeptides expressed selectively in larval and adult RGC clusters, Related to Figure 2.
Table S3. gRNA target sequences for CRISPR/Cas9-mediated transgenesis, Related to Figure 4.
Video S1. Expression pattern of the mafaa transgenic line, Related to Figure 4.
Video S2. Expression pattern of the tbr1b transgenic line, Related to Figure 4.
Video S3. Expression pattern of the eomesa transgenic line, Related to Figure 4.
Video S4. Expression pattern of the tbx3a transgenic line, Related to Figure 4.
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Invitrogen | CAT# A10262 |
| Rabbit polyclonal anti-tagRFP | Invitrogen | CAT# R10367 |
| Mouse monoclonal anti-synaptotagmin 2 (syt2) | ZIRC | AB_10013783 |
| Goat anti-chicken A488 | Invitrogen | CAT# A32931 |
| Goat anti-rabbit A555 | Invitrogen | CAT# A32732 |
| Goat anti-mouse A647 | Invitrogen | CAT# A32728 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AMES’ Medium | Sigma | CAT# A1420 |
| BSA (Bovine Serum Albumin) | Sigma | CAT# A9418 |
| Calcein Blue | ThermoFisher | CAT# C1429 |
| DAPI | Invitrogen | CAT# D1306 |
| DNAseI | Sigma | CAT# D4527 |
| L-cysteine | Sigma | CAT# C1276 |
| Low melting point agarose | Invitrogen | CAT# 10143954 |
| Metronidazole | Sigma | CAT# M1547 |
| Ovomucoid | Worthington | CAT# LS003087 |
| Papain | Worthington | CAT# LS003126 |
| Paraformaldehyde | Alfa Aesar | CAT# 43368 |
| PBS (for single cell transcriptomics) | Gibco | CAT# 10010001 |
| PTU (Phenylthiourea) | Sigma | CAT# P7629 |
| TissueTek | Sakura | CAT# 4583 |
| Tricaine – MS222 | Sigma | CAT# E10521 |
| Critical Commercial Assays | ||
| Chromium Single Cell 30 Library & Gel Bead Kit v2 | 10X Genomics | CAT# 120237 |
| Chromium Single Cell A Chip Kit | 10X Genomics | CAT# 1000009 |
| Chromium i7 Multiplex Kit | 10X Genomics | CAT# 120262 |
| Deposited Data | ||
| Raw data files for RNA-sequencing | GEO | GSE152842 |
| Visualized zebrafish RGC atlas | https://portals.broadinstitute.org/single_cell | SCP992 |
| Guide for reproduction of key results | https://github.com/shekharlab/ZebrafishRGC | |
| Experimental Models: Organisms/Strains | ||
| isl2b:tagRFP | Chien lab (Poulain and Chien, 2013) | zc80Tg |
| isl2b:GFP | Chien lab (Pittman et al., 2008) | zc7Tg |
| isl2b:Gal4 | Bonkowsky lab (Fujimoto et al., 2011) | zc65Tg |
| tbx20:Gal4 | Baier lab (Förster et al., 2017) | mpn220 |
| ath5:Cre | Baier lab (Förster et al., 2017) | mpn221 |
| UAS:GCaMP6s | Baier lab (Thiele et al., 2014) | mpn101 |
| UAS:NTR-mCherry | Halpern lab (Davison et al., 2007) | c264Tg |
| UAS:Dendra | Baier lab (Arrenberg et al., 2009) | s1998 |
| ath5:QF2 | Baier lab (Fernandes et al., 2019) | mpn405 |
| eomesa:QF2 | This study | mpn406 |
| mafaa:QF2 | This study | mpn408 |
| tbr1b:QF2 | This study | mpn409 |
| tbx3a:QF2 | This study | mpn410 |
| QUAS:epNTR-tagRFP | Baier lab (Fernandes et al., 2019) | mpn165 |
| QUAS:switchNTR (QUAS:loxP-GFPcaax-loxP-epNTR-tagRFP) | This study | mpn168 |
| QUAS:switchG6s (QUAS:loxP-tdTomatocaax-loxP-GCamP6s) | This study | mpn169 |
| actb2:loxP-eGFP-loxP-lynTagRFPT | Burgess lab (Marquart et al., 2015) | y272 |
| CRISPR-Cas9 reagents & gRNA’s | ||
| Alt-R® S.p. HiFi Cas9 Nuclease V3 | IDT | CAT# 1081060 |
| Alt-R® CRISPR-Cas9 tracrRNA | IDT | CAT# 1072533 |
| Alt-R® CRISPR-Cas9 crRNA | IDT | customized |
| Nuclease-free Duplex Buffer | IDT | CAT# 11-05-01-12 |
| Recombinant DNA | ||
| pTol2 - QUAS:loxP-GFPcaax-loxP-epNTR-tagRFP | This paper | |
| pTol2 - QUAS:loxP-tdTomatocaax-loxP-GCaMP6s | This paper | |
| pGGEV-1 | This paper | |
| pGGEV-2 (locus-specific gRNA target sequence) | This paper | |
| pGGEV-3 e1b | This paper | |
| pGGEV-4 QF2-polyA | This paper | |
| pGGEV-5’ polyA | This paper | |
| Software and Algorithms | ||
| Cell Ranger v2.1.0 | 10X Genomics | www.support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| FIJI | (Schindelin et al., 2012) | www.fiji.sc |
| Zen | Zeiss | www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| LAS X | Leica | www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Psychopy2 | (Peirce et al., 2019) | www.psychopy.org |
| R | R version 3.6 | www.r-project.org/ |
| Python 2.7.12 | Anaconda2 | www.anaconda.com |
| Python 3.7. | Anaconda3 | www.anaconda.com |
| CCTop | (Stemmer et al., 2015) | https://crispr.cos.uni-heidelberg.de |
| The Real Fish Tracker | N/A | http://www.dgp.toronto.edu/~mccrae/projects/FishTracker/ |
| Other | ||
| Camera | IDS | UI-3370CP-NIR |
| Chromium Controller | 10X Genomics | N/A |
| Film Polarizer | Thorlabs | LPVISE2X2 |
| HiSeq 2500 System | Illumina | N/A |
| Leica SP8 confocal microscope | Leica | N/A |
| Lens | Edmund Optics | 86-572 |
| ZebraBox | ViewPoint | N/A |
Acknowledgements
Funding for this study was provided by the Max Planck Society (YK, TOH, MS, AMF, SL, EL, IAA, HB), the DFG-SPP1926 (HB), National Institute of Health grants EY022073 and NS029169 (JRS) and R00EY028625 (KS), and startup funding from UC Berkeley (KS, JH, SB). We acknowledge travel grant support from the Graduate School of Systemic Neurosciences, GSN-LMU Munich. The authors thank all members of our labs, in particular Johannes Larsch, Greg Marquart, Joe Donovan, Inbal Shainer and Duncan Mearns for comments and discussions. Thanks to Julia Kuhl for creating scientific illustrations, Robert Kasper for imaging support, and Irene Whitney, Yirong Peng, Emily Martersteck, Mallory Laboulaye and Dustin Herrmann for experimental assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Abalo XM, Lagman D, Heras G, Eggert J, and Larhammar D (2020). Circadian regulation of phosphodiesterase 6 genes in zebrafish differs between cones and rods: Implications for photopic and scotopic vision. Vision Res. 166, 43–51. [DOI] [PubMed] [Google Scholar]
- Arrenberg AB, Del Bene F, and Baier H (2009). Optical control of zebrafish behavior with halorhodopsin. Proc. Natl. Acad. Sci 106, 17968–17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, and Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JA, Mu S, Kim JS, Briggman KL, Seung HS, Bae JA, Mu S, Kim JS, Turner NL, Tartavull I, et al. (2018). Digital Museum of Retinal Ganglion Cells with Dense Anatomy and Physiology. Cell 173, 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier H, and Scott EK (2009). Genetic and optical targeting of neural circuits and behavior — zebrafish in the spotlight. Curr. Opin. Neurobiol 19, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingham J, Chaurasia SS, Melyan Z, Liu C, Cameron MA, Tarttelin EE, luvone PM, Hankins MW, Tosini G, and Lucas RJ (2006). Evolution of Melanopsin Photoreceptors: Discovery and Characterization of a New Melanopsin in Nonmammalian Vertebrates. PLoS Biol. 4, e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, and Raymond PA (2007). Late-Stage Neuronal Progenitors in the Retina Are Radial Müller Glia That Function as Retinal Stem Cells. J. Neurosci 27, 7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn F. a, and Takao M (2002). Phototransduction by retinal ganglion cells that set the circadian clock. Science (80-.). 295, 1070–1073. [DOI] [PubMed] [Google Scholar]
- Bianco IH, Kampff AR, and Engert F (2011). Prey Capture Behavior Evoked by Simple Visual Stimuli in Larval Zebrafish. Front. Syst. Neurosci 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill JD, and Easter SS (1994). Development of the retinofugal projections in the embryonic and larval zebrafish (Brachydanio rerio). J. Comp. Neurol 346, 583–600. [DOI] [PubMed] [Google Scholar]
- Chen T, and Guestrin C (2016). XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794. [Google Scholar]
- Chen S-K, Badea TC, and Hattar S (2011). Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S, Stainier DYR, and Anderson RM (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: A spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc 3, 948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacey DM (1994). Physiology, Morphology and Spatial Densities of Identified Ganglion Cell Types in Primate Retina. In Novartis Foundation Symposia, pp. 12–34. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, and Parsons MJ (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol 304, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Estevez ME, Quattrochi LE, El-Danaf RN, Nguyen PL, Berson DM, and Huberman AD (2013). Genetic Dissection of Retinal Inputs to Brainstem Nuclei Controlling Image Stabilization. J. Neurosci 33, 17797–17813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Stafford BK, Lim JA, and Huberman AD (2015). Contributions of Retinal Ganglion Cells to Subcortical Visual Processing and Behaviors. Annu. Rev. Vis. Sci 1, 291–328. [DOI] [PubMed] [Google Scholar]
- Diekmann H, and Stuermer CAO (2009). Zebrafish neurolin-a and -b, orthologs of ALCAM, are involved in retinal ganglion cell differentiation and retinal axon pathfinding. J. Comp. Neurol 513, 38–50. [DOI] [PubMed] [Google Scholar]
- Do MTH (2019). Melanopsin and the Intrinsically Photosensitive Retinal Ganglion Cells: Biophysics to Behavior. Neuron 104, 205–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, and Sanes JR (2014). Type II Cadherins Guide Assembly of a Direction-Selective Retinal Circuit. Cell 158, 793–807. [DOI] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, Laboulaye MA, Liu J, Peng Y, Yamagata M, Toma K, and Sanes JR (2018). Cadherin Combinations Recruit Dendrites of Distinct Retinal Neurons to a Shared Interneuronal Scaffold. Neuron 99, 1145–1154.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner J (2016). Zebrafish embryo microinjection. User Methods, IDT Inc 9–10. [Google Scholar]
- Fernandes AM, Larsch J, Donovan JC, Helmbrecht TO, Mearns D, Kölsch Y, Maschio MD, and Baier H (2019). Neuronal circuitry for stimulus selection in the visual system. BioRxiv 598383. [DOI] [PubMed] [Google Scholar]
- Fishell G, and Heintz N (2013). The Neuron Identity Problem: Form Meets Function. Neuron 80, 602–612. [DOI] [PubMed] [Google Scholar]
- Fleisch VC, and Neuhauss SCF (2006). Visual Behavior in Zebrafish. Zebrafish 3, 191–201. [DOI] [PubMed] [Google Scholar]
- Förster D, Arnold-Ammer I, Laurell E, Barker AJ, Fernandes AM, Finger-Baier K, Filosa A, Helmbrecht TO, Kölsch Y, Kühn E, et al. (2017). Genetic targeting and anatomical registration of neuronal populations in the zebrafish brain with a new set of BAC transgenic tools. Sci. Rep 7, 5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster D, Helmbrecht T, Mearns DS, Jordan L, Mokayes N, and Baier H (2020). Retinotectal circuitry of larval zebrafish is adapted to detection and pursuit of prey. “in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Berens P, Schubert T, Bethge M, Euler T, and Baden T (2017). Inhibition decorrelates visual feature representations in the inner retina. Nature 542, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman MS (1999). Regulation of Mammalian Circadian Behavior by Non-rod, Non-cone, Ocular Photoreceptors. Science (80-.). 284, 502–504. [DOI] [PubMed] [Google Scholar]
- Fu Y, Liao H-W, Do MTH, and Yau K-W (2005). Non-image-forming ocular photoreception in vertebrates. Curr. Opin. Neurobiol 15, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto E, Gaynes B, Brimley CJ, Chien C-B, and Bonkowsky JL (2011). Gal80 intersectional regulation of cell-type specific expression in vertebrates. Dev. Dyn 240, 2324–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel JP, Trivedi CA, Maurer CM, Ryu S, and Bollmann JH (2012). Layer-Specific Targeting of Direction-Selective Neurons in the Zebrafish Optic Tectum. Neuron 76, 1147–1160. [DOI] [PubMed] [Google Scholar]
- Gahtan E (2005). Visual Prey Capture in Larval Zebrafish Is Controlled by Identified Reticulospinal Neurons Downstream of the Tectum. J. Neurosci 25, 9294–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley JJ, Lu J, Chou TC, Scammell TE, and Saper CB (2001). Melanopsin in cells of origin of the retinohypothalamic tract. Nat. Neurosci 4, 1165–1165. [DOI] [PubMed] [Google Scholar]
- Güler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao H-W, Barnard AR, Cahill H, Badea TC, Zhao H, et al. (2008). Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S (2002). Melanopsin-Containing Retinal Ganglion Cells: Architecture, Projections, and Intrinsic Photosensitivity. Science 295, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Lucas RJ, Mrosovsky N, Thompson S, Douglas RH, Hankins MW, Lem J, Biel M, Hofmann F, Foster RG, et al. (2003). Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature 424, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbrecht TO, dal Maschio M, Donovan JC, Koutsouli S, and Baier H (2018). Topography of a Visuomotor Transformation. Neuron 100, 1429–1445.e4. [DOI] [PubMed] [Google Scholar]
- Hörnberg H, Wollerton-van Horck F, Maurus D, Zwart M, Svoboda H, Harris WA, and Holt CE (2013). RNA-Binding Protein Hermes/RBPMS Inversely Affects Synapse Density and Axon Arbor Formation in Retinal Ganglion Cells In Vivo. J. Neurosci 33, 10384–10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SAT, et al. (2012). ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 41, D854–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, and Easter SS (1999). Retinal Neurogenesis: The Formation of the Initial Central Patch of Postmitotic Cells. Dev. Biol 207, 309–321. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, and Baier H (2001). Retinal Ganglion Cell Genesis Requires lakritz, a Zebrafish atonal Homolog. Neuron 30, 725–736. [DOI] [PubMed] [Google Scholar]
- Kay JN, Link B, and Baier H (2005). Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development 132, 2573–2585. [DOI] [PubMed] [Google Scholar]
- Kirchmaier S, Lust K, and Wittbrodt J (2013). Golden GATEway Cloning – A Combinatorial Approach to Generate Fusion and Recombination Constructs. PLoS One 8, e76117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita EM, Scott EK, and Goodhill GJ (2015). Topographic wiring of the retinotectal connection in zebrafish. Dev. Neurobiol 75, 542–556. [DOI] [PubMed] [Google Scholar]
- Kolosov D, Bui P, Chasiotis H, and Kelly SP (2013). Claudins in teleost fishes. Tissue Barriers 1, e25391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Wu Y, Baier H, and Kubo F (2019). Neuronal Architecture of a Visual Center that Processes Optic Flow. Neuron 103, 118–132.e7. [DOI] [PubMed] [Google Scholar]
- Kunst M, Laurell E, Mokayes N, Kramer A, Kubo F, Fernandes AM, Förster D, Dal Maschio M, and Baier H (2019). A Cellular-Resolution Atlas of the Larval Zebrafish Brain. Neuron 103, 21–38.e5. [DOI] [PubMed] [Google Scholar]
- Larsch J, and Baier H (2018). Biological Motion as an Innate Perceptual Mechanism Driving Social Affiliation. Curr. Biol 28, 3523–3532.e4. [DOI] [PubMed] [Google Scholar]
- Li Q (2005). Chemokine Signaling Guides Axons within the Retina in Zebrafish. J. Neurosci 25, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Horns F, Wu B, Xie Q, Li J, Li T, Luginbuhl DJ, Quake SR, and Luo L (2017). Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman GC, Rachh M, Hoskins JG, Steinerberger S, and Kluger Y (2019). Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat. Methods 16, 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Reggiani JDS, Laboulaye MA, Pandey S, Chen B, Rubenstein JLR, Krishnaswamy A, and Sanes JR (2018). Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nat. Neurosci 21, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes G, Bonacchi N, Frazão J, Neto JP, Atallah BV, Soares S, Moreira L, Matias S, Itskov PM, Correia PA, et al. (2015). Bonsai: an event-based framework for processing and controlling data streams. Front. Neuroinform 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten L Van Der, and Hinton G (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C-A, Li H, Zhang Z, Kiyama T, Panda S, Hattar S, Ribelayga CP, Mills SL, and Wang SW (2014). T-box Transcription Regulator Tbr2 Is Essential for the Formation and Maintenance of Opn4/Melanopsin-Expressing Intrinsically Photosensitive Retinal Ganglion Cells. J. Neurosci 34, 13083–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Delaney CL, and Easter SS (1999). Neurogenesis in the visual system of embryonic and adult zebrafish ( Danio rerio ). Vis. Neurosci 16, 417–424. [DOI] [PubMed] [Google Scholar]
- Marquart GD, Tabor KM, Brown M, Strykowski JL, Varshney GK, LaFave MC, Mueller T, Burgess SM, Higashijima S, and Burgess HA (2015). A 3D Searchable Database of Transgenic Zebrafish Gal4 and Cre Lines for Functional Neuroanatomy Studies. Front. Neural Circuits 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, et al. (2017). Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Rep. 18, 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Morales J-R, Del Bene F, Nica G, Hammerschmidt M, Bovolenta P, and Wittbrodt J (2005). Differentiation of the Vertebrate Retina Is Coordinated by an FGF Signaling Center. Dev. Cell 8, 565–574. [DOI] [PubMed] [Google Scholar]
- Matos-Cruz V, Blasic J, Nickle B, Robinson PR, Hattar S, and Halpern ME (2011). Unexpected Diversity and Photoperiod Dependence of the Zebrafish Melanopsin System. PLoS One 6, e25111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mearns DS, Donovan JC, Fernandes AM, Semmelhack JL, and Baier H (2020). Deconstructing Hunting Behavior Reveals a Tightly Coupled Stimulus-Response Loop. Curr. Biol 30, 54–69.e9. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Williams PR, Godinho L, Koerber A, Pittman AJ, Roeser T, Chien C-B, Baier H, and Wong ROL (2006). In Vivo Imaging Reveals Dendritic Targeting of Laminated Afferents by Zebrafish Retinal Ganglion Cells. Neuron 52, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo L, Haas M, Qu Z, Li SS, Zenker J, Teng KSL, Gunnersen JM, Breuss M, Habgood M, Keays DA, et al. (2014). TUBB5 and its disease-associated mutations influence the terminal differentiation and dendritic spine densities of cerebral cortical neurons. Hum. Mol. Genet 23, 5147–5158. [DOI] [PubMed] [Google Scholar]
- Nikolaou N, Lowe AS, Walker AS, Abbas F, Hunter PR, Thompson ID, and Meyer MP (2012). Parametric Functional Maps of Visual Inputs to the Tectum. Neuron 76, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orger MB (2016). The Cellular Organization of Zebrafish Visuomotor Circuits. Curr. Biol 26, R377–R385. [DOI] [PubMed] [Google Scholar]
- Orger MB, and de Polavieja GG (2017). Zebrafish Behavior: Opportunities and Challenges. Annu. Rev. Neurosci 40, 125–147. [DOI] [PubMed] [Google Scholar]
- Pandey S, Shekhar K, Regev A, and Schier AF (2018). Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA-Seq. Curr. Biol 28, 1052–1065.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce J, Gray JR, Simpson S, MacAskill M, Höchenberger R, Sogo H, Kastman E, and Lindeløv JK (2019). PsychoPy2: Experiments in behavior made easy. Behav. Res. Methods 51, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y-R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Tran NM, Krishnaswamy A, Kostadinov D, Martersteck EM, and Sanes JR (2017). Satb1 Regulates Contactin 5 to Pattern Dendrites of a Mammalian Retinal Ganglion Cell. Neuron 95, 869–883.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo DM, El-Danaf RN, Huberman AD, and Niell CM (2013). Diverse Visual Features Encoded in Mouse Lateral Geniculate Nucleus. J. Neurosci 33, 4642–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman AJ, Law M-Y, and Chien C-B (2008). Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development 135, 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugues R, and Engert F (2009). The neural basis of visual behaviors in the larval zebrafish. Curr. Opin. Neurobiol 19, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain FE, and Chien C-B (2013). Proteoglycan-Mediated Axon Degeneration Corrects Pretarget Topographic Sorting Errors. Neuron 78, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. (2017). The Human Cell Atlas. Elife 6, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles E, Filosa A, and Baier H (2013). Precise Lamination of Retinal Axons Generates Multiple Parallel Input Pathways in the Tectum. J. Neurosci 33, 5027–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles E, Laurell E, and Baier H (2014). The Retinal Projectome Reveals Brain-Area-Specific Visual Representations Generated by Ganglion Cell Diversity. Curr. Biol 24, 2085–2096. [DOI] [PubMed] [Google Scholar]
- Roth LWA, Bormann P, Bonnet A, and Reinhard E (1999). Beta-Thymosin is required for axonal tract formation in developing zebrafish brain. Development 126, 1365–1374. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Qiao M, Kagan RD, Yamagata M, Palmiter RD, and Sanes JR (2016). Two Pairs of ON and OFF Retinal Ganglion Cells Are Defined by Intersectional Patterns of Transcription Factor Expression. Cell Rep. 15, 1930–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell JH, Martin SC, and Heinrich G (1994). The development of GABA immunoreactivity in the retina of the zebrafish (brachydanio rerio). J. Comp. Neurol 345, 596–601. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Masland RH (2015). The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annu. Rev. Neurosci 38, 221–246. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Zipursky SL (2010). Design Principles of Insect and Vertebrate Visual Systems. Neuron 66, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TM, Chen S-K, and Hattar S (2011). Intrinsically photosensitive retinal ganglion cells: many subtypes, diverse functions. Trends Neurosci. 34, 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabrook TA, Burbridge TJ, Crair MC, and Huberman AD (2017). Architecture, Function, and Assembly of the Mouse Visual System. Annu. Rev. Neurosci 40, 499–538. [DOI] [PubMed] [Google Scholar]
- Semmelhack JL, Donovan JC, Thiele TR, Kuehn E, Laurell E, and Baier H (2014). A dedicated visual pathway for prey detection in larval zebrafish. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, and Mateo JL (2015). CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS One 10, e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, and Kawakami K (2011). Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc 6, 1998–2021. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, and Feldheim DA (2014). Tbr2 Is Required to Generate a Neural Circuit Mediating the Pupillary Light Reflex. J. Neurosci 34, 5447–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor KM, Bergeron SA, Horstick EJ, Jordan DC, Aho V, Porkka-Heiskanen T, Haspel G, and Burgess HA (2014). Direct activation of the Mauthner cell by electric field pulses drives ultrarapid escape responses. J. Neurophysiol 112, 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temizer I, Donovan JC, Baier H, and Semmelhack JL (2015). A Visual Pathway for Looming-Evoked Escape in Larval Zebrafish. Curr. Biol 25, 1823–1834. [DOI] [PubMed] [Google Scholar]
- Thiele TR, Donovan JC, and Baier H (2014). Descending Control of Swim Posture by a Midbrain Nucleus in Zebrafish. Neuron 83, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, et al. (2019). Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 104, 1039–1055.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, et al. (2015). Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc 10, 1860–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitorino M, Jusuf PR, Maurus D, Kimura Y, Higashijima S, and Harris WA (2009). Vsx2 in the zebrafish retina: restricted lineages through derepression. Neural Dev. 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasits AL, Euler T, and Franke K (2019). Function first: classifying cell types and circuits of the retina. Curr. Opin. Neurobiol 56, 8–15. [DOI] [PubMed] [Google Scholar]
- Wu Y, dal Maschio M, Kubo F, and Baier H (2020). An Optical Illusion Pinpoints an Essential Circuit Node for Global Motion Processing. Neuron. [DOI] [PubMed] [Google Scholar]
- Yan W, Peng Y, van Zyl T, Regev A, Shekhar K, Juric D, and Sanes JR (2020). Cell Atlas Of The Human Fovea And Peripheral Retina. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashina K, Tejero-Cantero Á, Herz A, and Baier H (2019). Zebrafish Exploit Visual Cues and Geometric Relationships to Form a Spatial Memory. IScience 19, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, and Sanes JR (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci 18, 530–546. [DOI] [PubMed] [Google Scholar]
- Zhang B, Yao Y, Zhang H, Kawakami K, and Du J (2017). Left Habenula Mediates Light-Preference Behavior in Zebrafish via an Asymmetrical Visual Pathway. Neuron 93, 914–928.e4. [DOI] [PubMed] [Google Scholar]
- Zhang D, Guelfi S, Garcia-Ruiz S, Costa B, Reynolds RH, D’Sa K, Liu W, Courtin T, Peterson A, Jaffe AE, et al. (2020). Incomplete annotation has a disproportionate impact on our understanding of Mendelian and complex neurogenetic disorders. Sci. Adv 6, eaay8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Bear J, Roberts PA, Janiak FK, Semmelhack J, Yoshimatsu T, and Baden T (2020). Zebrafish Retinal Ganglion Cells Asymmetrically Encode Spectral and Temporal Information across Visual Space. Curr. Biol 30, 2927–2942.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann MJY, Nevala NE, Yoshimatsu T, Osorio D, Nilsson D-E, Berens P, and Baden T (2018). Zebrafish Differentially Process Color across Visual Space to Match Natural Scenes. Curr. Biol 28, 2018–2032.e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of differentially expressed genes in larval and adult RGC clusters computed using MAST, Related to Figure 1 and Figure 3.
Table S2. Summary of expression of transcription factors, neural recognition molecules, and neuropeptides expressed selectively in larval and adult RGC clusters, Related to Figure 2.
Table S3. gRNA target sequences for CRISPR/Cas9-mediated transgenesis, Related to Figure 4.
Video S1. Expression pattern of the mafaa transgenic line, Related to Figure 4.
Video S2. Expression pattern of the tbr1b transgenic line, Related to Figure 4.
Video S3. Expression pattern of the eomesa transgenic line, Related to Figure 4.
Video S4. Expression pattern of the tbx3a transgenic line, Related to Figure 4.
Data Availability Statement
Computational scripts detailing scRNA-seq analysis reported in this paper are available at https://github.com/shekharlab/ZebrafishRGC. We have also provided R markdown (Rmd) files that show step-by-step reproduction of the key results. All raw and processed scRNA-seq datasets reported in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under the accession number GSE152842. Visualization of the zebrafish RGC atlas is available at https://portals.broadinstitute.org/single_cell (Study ID: SCP992). All data and custom software for functional imaging analysis and behavioral tests will be made available upon request.


