Abstract
Background
Essential hypertension (EH) is an inflammatory disease, and endothelial dysfunction induced by chronic inflammation is one of the pathogeneses of EH. The expression of some inflammatory mediators may be regulated by the interaction of circular RNAs (circRNAs) and microRNAs (miRNAs).
Methods
An Agilent human circRNA microarray was used to identify the expression profile of circRNAs in EH. qRT‐PCR was used to evaluate the relative expression of circRNAs in 48 pairs of human whole blood samples (sex and age ± 3 years matched) and endothelial cells. TNF‐α was applied to induce endothelial cells inflammation. CircRNA‐miRNA network was predicted by MiRanda software.
Results
There were 287 circRNAs differentially expressed in the microarray. The top 10 up‐regulated circRNAs in the EH group were hsa_circ_0014243, hsa_circ_0133228, hsa‐circRNA14116‐3, hsa_circ_0079536, hsa‐circRNA13649‐1, hsa_circ_0117886, hsa_circ_0007075, hsa‐circRNA15285‐1, hsa‐circRNA10088‐9, and hsa‐circRNA14119‐10; the top 10 down‐regulated circRNAs were hsa_circ_0100094, hsa_circ_0127342, hsa_circ_0093773, hsa_circ_0096334, hsa_circ_0131618, hsa_circ_0063886, hsa_circ_0097804, hsa_circ_0126640, hsa‐circRNA8935‐1, and hsa_circ_0039978 (fold change in descending order). Hsa_circ_0105015 has two predicted binding sites with hsa‐miR‐637. The relative expression of hsa_circ_0105015 in EH patients was significantly higher than healthy controls (P = .002), and similar results appeared in TNF‐α‐induced endothelial cells. The area under the curve after hsa_circ_0105015 combined with hsa‐miR‐637 was 0.703, P < .001.
Conclusion
Hyperexpression of hsa_circ_0105015 is a significant risk factor of EH and its association with EH involves inflammatory pathways. Hyperexpression of hsa_circ_0105015 combined with hypoexpression of hsa‐miR‐637 indicates vascular inflammation or endothelial dysfunction and has potential as a biomarker for early diagnosis of EH.
Keywords: circRNA, essential hypertension, inflammation, microarray, miRNA
An Agilent human circRNA microarray was used to identify the expression profile of circRNAs in essential hypertension (EH). There were 287 circRNAs differentially expressed in the microarray, and the top 20 circRNAs with up‐regulated and down‐regulated in the EH group were listed. Hsa_circ_0105015 was up‐regulated in the microarray and has two predicted binding sites with hsa‐miR‐637. Hyperexpression of hsa_circ_0105015 combined with hypoexpression of hsa‐miR‐637 indicates vascular inflammation or endothelial dysfunction and has potential as a biomarker for early diagnosis of EH
1. INTRODUCTION
Hypertension is a major risk factor for cardiovascular disease and poses a severe public health burden globally. 1 A multistage study with 98,658 subjects in 31 provinces and municipalities of China indicated that the prevalence of hypertension in China was 33.6% in 2010, equivalent to 335.8 million adult hypertensive patients. 2
Essential hypertension (EH) is a complex multifactorial disease, and vascular endothelial dysfunction induced by chronic inflammation is one of the pathogenesis of EH. 3 Inflammation is the defense response of the vascular system to infection or cell damage. Growing evidence indicates that inflammation is directly involved in EH, 4 , 5 chronic inflammation induces oxidative stress by promoting the production of reactive oxygen species (ROS), resulting in endothelial dysfunction. On the other hand, continuous inflammation inhibits vasodilation by reducing the bioavailability of NO.
The development of chronic inflammation involves abnormal expression of non‐coding RNA (ncRNA). MicroRNAs (miRNAs), as a kind of endogenous ncRNA, are mainly used as post‐transcription inhibitors that specifically bind to the 3’‐UTR of mRNA to inhibit its expression. 6 MiRNAs can regulate vascular inflammation by inhibiting the expression of the inflammatory mediators, such as cell adhesion molecule (CAM), tumor necrosis factor‐α (TNF‐α), and C‐reactive protein (CRP). There is evidence that in endothelial cells, miR‐126 inhibits its expression by binding to the 3'‐UTR of vascular cell adhesion factor (VCAM‐1), and decreased miR‐126 expression promotes the adhesion of leukocytes to endothelial cells. 7
Circular RNAs (circRNAs) have a covalently closed cyclic structure without 5’ or 3’ free terminus to make its structure stable and abundantly express in mammalian cells. 8 , 9 , 10 Based on whether it can be translated, circRNAs can be divided into nc RNA and coding RNA. 11 The proven functions of circRNAs include participation in selective splicing or transcription, regulation of parental gene expression, 12 , 13 , 14 and especially acting as the sponge of miRNAs. 15 Some circRNAs with miRNAs response elements (MREs) can act as competing endogenous RNA (ceRNA) to compete for miRNAs binding sites, thereby releasing the inhibition of miRNAs on downstream mRNAs. 16 Currently, most studies on circRNAs and cardiovascular diseases focus on the function of circRNAs as a miRNAs sponge, which indicates that the circRNA‐miRNA‐mRNA axis is closely related to the pathogenesis of cardiovascular diseases. 17 However, whether the circRNA‐miRNA‐mRNA axis is involved in the pathogenesis of EH remains unclear, and the relationship between circRNAs and EH is worth exploring.
In a previous study, we revealed the association of hsa_circ_0037909 and hsa‐miR‐637 with EH. 18 In our results, hsa‐miR‐637, originally reported to inhibit the inflammatory mediator CRP, was a protective factor for EH and was down‐regulated in TNF‐α‐induced endothelial cells; hsa_circ_0037909 was a risk factor for EH and was up‐regulated in TNF‐α‐induced endothelial cells. The above evidence indicates that hsa‐miR‐637 regulates vascular inflammation by participating in regulating the expression of inflammatory mediators, and there are certain inflammation‐related circRNAs related to EH. Therefore, it is necessary to further explore the role of inflammation‐related circRNAs in EH through the circRNA‐miRNA network.
The present study aims to predict the circRNAs corresponding to hsa‐miR‐637 through the circRNA‐miRNA network and circRNA microarray, and verify its role in vascular inflammation and EH. The results in the present study may contribute to the exploration of the potential biological mechanism of EH and provide a basis for the early diagnosis for EH. In addition, in order to avoid the bias caused by different samples when evaluating the association between hsa‐miR‐637 and the corresponding circRNA, we replicated the samples and the relative expression results of hsa‐miR‐637 in the previous study. 18
2. MATERIALS AND METHODS
2.1. Microarray processing
The profile of circRNAs expression was analyzed by Agilent human circRNA microarray (CapitalBio Corp) performed with 5 EH and 5 non‐EH whole blood samples (sex and age ± 3 years matched) from Ningbo Seventh Hospital, and 87 935 circRNAs with known sequences were detected, covering 95% of the sequences in circBase. Agilent Feature Extraction software (v10.7) was applied to analyze hybrid images and filter data, and the expression of circRNAs was then determined by fold change using Agilent GeneSpring software. Hierarchical clustering was used to the present differential expression of circRNAs. MiRanda software was used to predict miRNAs corresponding to circRNAs, and circRNA‐miRNA network was drawn by Cytoscape software.
2.2. Population and sample collection
Whole blood samples from 48 EH patients and 48 healthy controls (sex and age ± 3 years matched), aged 35‐75, were obtained from the Affiliated People's Hospital, Ningbo University. All subjects were bled by anterior elbow vein after 12 hours of overnight fasting, and all blood samples were divided into two parts for biochemical analysis and total RNA isolation, loaded into ethylenedia‐minetetraacetic acid (EDTA) anticoagulation tubes and stored at −80℃.
This study was approved by the Ningbo University Medical School Ethics Committee (NBU‐2017‐06). Informed consent was signed by all subjects and assessed by exclusion and inclusion criteria. The criteria for determining hypertension were based on the 2016 guideline for the diagnosis and management of hypertension in adults. 19 In the EH group, the systolic blood pressure (SBP) ≥ 140 mmHg and/or the diastolic blood pressure (DBP) ≥ 90 mmHg. In the control group, blood pressure was lower than 120/80 mmHg. All subjects were included without a history of secondary hypertension, myocardial infarction, stroke, diabetes, renal disease, malignant tumor, mental disorder or substance abuse, and family history of hypertension in the first‐degree relatives.
2.3. Biochemical analysis and RNA extraction
Blood biochemical analysis was measured with Olympus AU5800 automatic biochemical analyzer (Tokyo, Japan). TRIzol kit (TransGen Biotech Corporation) was used to extract total RNA from peripheral blood samples. Full Wavelength Microplate Reader (Thermo Multiskan GO) was used to determine the concentration and purity of extracted RNA. GO ScriptTM Reverse Transcription System (Promega) and miRcute Plus miRNA First‐Strand cDNA Synthesis Kit (TianGen) were applied in the reverse transcription process of circRNAs and miRNAs, respectively.
2.4. Quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR)
The GoTaq® Real‐Time PCR Systems reagent kit (Promega) and the miRcute Plus miRNA qPCR Detection Kit (TianGen) were applied to qRT‐PCR on a Roche LightCycler 480 Real‐Time PCR (Roche Diagnostics Ltd.). We selected glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) gene and cel‐miR‐39 as the external control of circRNAs and miRNAs, respectively. Hsa_circ_0105015 and hsa‐miR‐637 each had three replications of qRT‐PCR.
2.5. Cell culture
Human umbilical vein endothelial cells (HUVECs) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FBS, penicillin (100 μg/mL), and streptomycin (100μg/mL). Human aortic endothelial cells (HAECs) were cultured with RPMI 1640 containing 10% fetal bovine serum (FBS), penicillin (100 μg/mL), streptomycin (100 μg/mL), and 1%2‐[4 ‐ (2‐Hydroxyethyl) −1‐ piperazinyl] ethanesulfonic acid (HEPES). All the cells were cultured in 95% air‐5% carbon dioxide, six‐well plate, 37°C. Each plate was divided into 3 experimental groups and 3 control groups. TNF‐α was used to induce inflammation of HUVECs and HAECs, and the treatment concentration and time were 1 μmol/L 24 h, 1 μmol/L 48 h, 10 μmol/L 24 h, and 10 μmol/L 48 h. The control groups were routinely cultured. The subsequent steps were similar to the previous treatment of whole blood samples, and total RNAs were extracted from HUVECs and HAECs, followed by reverse transcription and qRT‐PCR. Each cell line experiment was repeated at least three times.
2.6. Statistical analyses
Statistical data were analyzed by SPSS 19.0 (IBM Corp) and visualized by GraphPad Prism 6.0 (GraphPad Software). Continuous numeric data were expressed as mean ± SD. Categorical variables were performed by Pearson's χ 2 test. Binary logistic regression was conducted to adjust the confounding factors. The receiver operating characteristic (ROC) curve analysis was used to investigate the diagnostic accuracy. The P‐value < 0.05 was considered to be statistically significant. The ∆Ct method was used to analyze qRT‐PCR data, calculation formula: ΔCt = Ct (target) – Ct (GAPDH/cel‐miR‐39), smaller ∆Ct values imply higher expression.
3. RESULT
3.1. Microarray results and identification of candidate circRNAs
The hierarchical clustering results in Figure 1A indicated the profile of circRNAs in 5 EH patients and 5 healthy controls, red and green represent up‐regulated and down‐regulated, respectively. The volcano plot in Figure 1B showed the results of the differentially expressed circRNAs between the EH group and the non‐EH group. There were 287 differentially expressed circRNAs, of which 209 were up‐regulated and 78 were down‐regulated (fold change > 2 and P < .05). The top 20 circRNAs with up‐regulated and down‐regulated in the EH group were displayed in Table 1.
Figure 1.
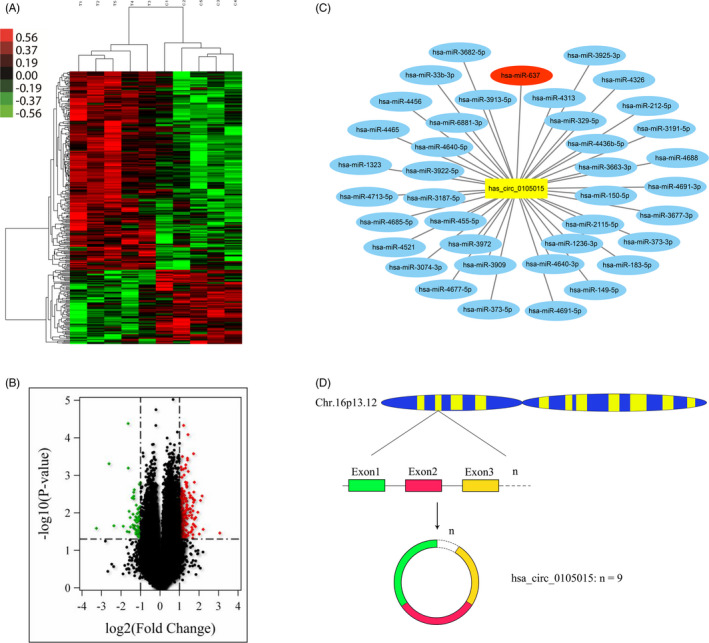
Microarray results and Bioinformatics analysis. (A) The differential expression profile of circRNAs in 5 EH patients and 5 healthy controls. (B) Differences in circRNAs expression between 5 EH patients and 5 healthy controls. There were 287 differentially expressed circRNAs, of which 209 were up‐regulated and 78 were down‐regulated (fold change > 2 and P < .05). (C) MiRanda software predicted miRNAs with potential binding sites for hsa circ 0 105 015, among which hsa‐miR‐637 and hsa_circ_0105015 have two binding sites. (D) The pattern of has_circ_0105015 forming in circular
Table 1.
Top 20 circRNAs with up‐regulated and down‐regulated expressions in EH patients
| The order of fold change | Up‐regulated in EH group | Down‐regulated in EH group | ||||
|---|---|---|---|---|---|---|
| circRNAs ID | Fold change | P‐value | circRNAs ID | Fold change | P‐value | |
| 1 | hsa_circ_0014243 | 8.380 | .035 | hsa_circ_0100094 | 9.616 | .026 |
| 2 | hsa_circ_0133228 | 4.635 | .028 | hsa_circ_0127342 | 6.128 | <.01 |
| 3 | hsa‐circRNA14116‐3* | 4.448 | <.01 | hsa_circ_0093773 | 5.158 | .022 |
| 4 | hsa_circ_0079536 | 4.206 | <.01 | hsa_circ_0096334 | 3.697 | .023 |
| 5 | hsa‐circRNA13649‐1* | 4.031 | .037 | hsa_circ_0131618 | 3.216 | .031 |
| 6 | hsa_circ_0117886 | 3.832 | <.01 | hsa_circ_0063886 | 3.116 | <.01 |
| 7 | hsa_circ_0007075 | 3.561 | .013 | hsa_circ_0097804 | 3.102 | <.01 |
| 8 | hsa‐circRNA15285‐1* | 3.516 | .023 | hsa_circ_0126640 | 3.008 | <.01 |
| 9 | hsa‐circRNA10088‐9* | 3.411 | <.01 | hsa‐circRNA8935‐1* | 2.987 | .032 |
| 10 | hsa‐circRNA14119‐10* | 3.382 | .015 | hsa_circ_0039978 | 2.895 | .013 |
| 11 | hsa‐circRNA15426‐17* | 3.377 | <.01 | hsa_circ_0076255 | 2.855 | .024 |
| 12 | hsa_circ_0136017 | 3.338 | .021 | hsa_circ_0075506 | 2.697 | <.01 |
| 13 | hsa_circ_0112683 | 3.323 | <.01 | hsa_circ_0062990 | 2.626 | .021 |
| 14 | hsa_circ_0112682 | 3.306 | <.01 | hsa_circ_0023966 | 2.605 | .011 |
| 15 | hsa_circ_0117885 | 3.293 | <.01 | hsa_circ_0064250 | 2.559 | .025 |
| 16 | hsa_circ_0070820 | 3.283 | <.01 | hsa_circ_0128635 | 2.555 | <.01 |
| 17 | hsa_circ_0124782 | 3.254 | .020 | hsa_circ_0107472 | 2.541 | <.01 |
| 18 | hsa_circ_0024901 | 3.222 | <.01 | hsa_circ_0013062 | 2.540 | <.01 |
| 19 | hsa_circ_0025088 | 3.221 | .038 | hsa_circ_0062042 | 2.528 | .026 |
| 20 | hsa_circ_0124783 | 3.196 | .048 | hsa_circ_0064890 | 2.500 | <.01 |
CircRNAs that originate from deepBase v2.0 (http://rna.sysu.edu.cn/deepBase/) and cannot be converted into circBase ID are marked with *. The circRNAs without * are from circBase (http://circrna.org/).
After prediction by MiRanda software, among 287 differentially expressed circRNAs, there were two predicted binding sites between hsa_circ_0105015 (fold change = 2.381, P = .003) and hsa‐miR‐637. The network in Figure 1C showed the relationship between hsa_circ_0105015 and its corresponding miRNA. The gene of hsa_circ_0105015 is located in the 11979037bp ‐ 11991892bp region of chromosome 16 and has a length of 346 nucleotides (nt) splicing sequence including 9 exons (Figure 1D). Therefore, we take hsa_circ_0105015 as the candidate circRNA for this study.
3.2. Basic demographic and clinical characteristics of the subjects
In this case‐control study, 96 participants aged 35‐75 were divided equally into the EH group and the non‐EH group (sex and age ± 3 years matched). In Table 2, the clinical and demographic characteristics of all participants are within the normal range, and there was no significant difference in age and sex between the EH and the non‐EH group.
Table 2.
Basic demographic and clinical characteristics of the subjects
| Characteristics | Non‐EH (n = 48) | EH (n = 48) | χ 2/t | P‐value |
|---|---|---|---|---|
| Drinking (Y/N) | 6/42 | 10/38 | 1.20 | .273 |
| Smoking (Y/N) | 7/41 | 11/37 | 1.09 | .296 |
| Sex (F/M) | 30/18 | 30/18 | 0 | 1 |
| Age (year) | 55.31 ± 9.76 | 55.58 ± 9.67 | 0.14 | .892 |
| BMI (Kg/m2) | 23.03 ± 2.68 | 23.89 ± 2.69 | 1.55 | .125 |
| ALT (IU/L) | 23.10 ± 14.12 | 24.34 ± 16.08 | 0.40 | .691 |
| AST (IU/L) | 23.88 ± 6.38 | 23.98 ± 7.44 | 0.07 | .942 |
| Scr (μmol/L) | 78.88 ± 16.52 | 76.02 ± 21.31 | −7.28 | .467 |
| BUN (μmol/L) | 5.28 ± 1.10 | 5.22 ± 1.10 | −0.27 | .790 |
| BUA (μmol/L) | 356.29 ± 66.08 | 351.02 ± 79.83 | −0.35 | .726 |
| Glu (mmol/L) | 5.06 ± 0.71 | 5.25 ± 0.75 | 1.27 | .207 |
| RBC | 4.59 ± 0.40 | 4.65 ± 0.49 | 0.640 | .527 |
| Lym | 2.17 ± 0.58 | 2.13 ± 0.56 | −0.33 | .746 |
| Mono | 0.36 ± 0.15 | 0.37 ± 0.15 | 0.42 | .676 |
| Neut | 3.52 ± 1.16 | 3.72 ± 1.39 | 0.80 | .428 |
| WBC | 6.09 ± 1.52 | 6.36 ± 1.63 | 0.84 | .402 |
| LDL (mmol/L) | 2.88 ± 0.85 | 2.77 ± 0.64 | −0.69 | .490 |
| TG (mmol/L) | 1.65 ± 0.73 | 1.45 ± 0.55 | −1.52 | .132 |
| HDL (mmol/L) | 1.41 ± 0.31 | 1.31 ± 0.27 | −1.75 | .084 |
| TC (mmol/L) | 5.29 ± 0.90 | 5.18 ± 0.75 | −0.67 | .507 |
P‐value less than 0.05 is marked with *.
Abbreviations: ALT, alanine aminotransferase; AST, glutamic oxalacetic transaminase; BMI, body mass index; BUA, blood uric acid; BUN, blood urea nitrogen; Glu, blood glucose; HDL, high‐density lipoprotein; LDL, low density lipoprotein; Lym, lymphocyte ratio; Monor, monocyte ratio; Neut, neutrophil ratio; RBC, Red blood cell; Scr, serum creatinine; TC, total cholesterol; TG, triglycerdes; WBC, White blood cell.
3.3. Verification of relative expression of candidate circRNA and miRNA
The ∆Ct of hsa_circ_0105015 in the EH group and the non‐EH group was significantly different (t = 3.21, P = .002) in Table 3 and Figure 2A. The relative expression of hsa_circ_0105015 was up‐regulated in the EH group compared with the non‐EH group. Meanwhile, in the previous study, under the same group of subjects, the ∆Ct of hsa‐miR‐637 in the EH group was significantly different from that in the non‐EH group (t = −2.10, P = .039), 18 and hsa‐miR‐637 was down‐regulated in the EH group compared with the non‐EH group.
Table 3.
The ΔCt of target circRNA‐miRNA between EH and non‐EH group
| Characteristics | Non‐EH (n = 48) | EH (n = 48) | t‐value | P‐value |
|---|---|---|---|---|
| hsa‐miR‐637 (ΔCt) | 1.33 ± 1.14 | 1.91 ± 1.51 | −2.095 | .039* |
| hsa_circ_0105015 (ΔCt) | 12.65 ± 1.235 | 11.98 ± 0.76 | 3.205 | .002* |
P‐value less than 0.05 is marked with *.
Figure 2.
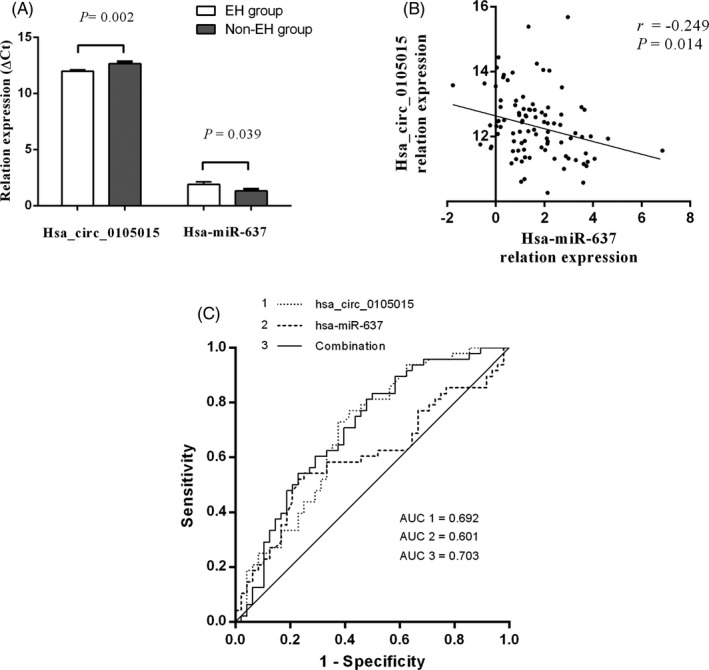
Association between candidate RNAs and EH (A) The ΔCt of hsa_circ_0105015 and hsa‐miR‐637 in the EH group and the non‐EH group. (B) Correlation between the relative expression of hsa_circ_0105015 and hsa‐miR‐637. (C) The ROC analysis of hsa_circ_0105015, hsa‐miR‐637, and the combination
After adjusting for confounding factors by binary logistic regression (Table 4), hyperexpression of hsa_circ_0105015 presented as a risk factor for EH, OR (95%CI) = 0.489(0.285, 0.838), P = .009; hyperexpression of hsa‐miR‐637 presented as a protective factor of EH, OR (95%CI) = 1.415(1.013, 1.977), P = .042. After adjusting for both hsa_circ_0105015 and hsa‐miR‐637 as well as other confounders, hsa_circ_0105015 remained a risk factor for EH, OR (95%CI) = 0.549(0.316, 0.953), P = .033, and there was no statistical difference in the results of hsa‐miR‐637, OR (95%CI) = 1.255(0.878, 1.795), P = .213.
Table 4.
Logistic regression of hsa‐miR‐637 and hsa_circ_0105015 as well as other confounders
| circRNA | miRNA | circRNA‐miRNA | ||||
|---|---|---|---|---|---|---|
| OR (95%CI) | P‐value | OR (95%CI) | P‐value | OR (95%CI) | P‐value | |
| hsa‐miR‐637(ΔCt) | ‐ | ‐ | 1.415 (1.013, 1.977) | .042* | 1.255 (0.878, 1.795) | .213 |
| hsa_circ_0105015(ΔCt) | 0.489 (0.285, 0.838) | .009* | ‐ | ‐ | 0.549 (0.316, 0.953) | .033* |
| Age (year) | 1.019 (0.969, 1.070) | .469 | 1.023 (0.974, 1.074) | .366 | 1.021 (0.971, 1.074) | .418 |
| Smoking | 0.618 (0.124, 3.081) | .557 | 0.842 (0.166, 4.284) | .836 | 1.341 (0.258, 6.971) | .728 |
| Drinking | 0.781 (0.150, 4.075) | .770 | 0.524 (0.101, 2.701) | .439 | 1.478 (0.274, 7.964) | .649 |
| Scr (μmol/L) | 0.995 (0.965, 1.027) | .762 | 0.987 (0.957, 1.017) | .383 | 0.994 (0.963, 1.026) | .706 |
| LDL (mmol/L) | 1.152 (0.378, 3.513) | .804 | 0.945 (0.312, 2.858) | .920 | 1.078 (0.348, 3.342) | .896 |
| TG (mmol/L) | 0.495 (0.220, 1.115) | .090 | 0.560 (0.270, 1.162) | .119 | 0.495 (0.218, 1.120) | .091 |
| CHO (mmol/L) | 0.495 (0.974, 1.055) | .946 | 0.861 (0.318, 2.327) | .768 | 0.998 (0.355, 2.811) | .997 |
P‐value less than 0.05 is marked with *.
3.4. Association analysis between the relative expression of candidate circRNA and miRNA
The correlation between the relative expression (∆Ct) of hsa_circ_0105015 and hsa‐miR‐637 was analyzed by Person Correlation. In Figure 2B, there was a negative correlation between the relative expression of hsa_circ_0105015 and hsa‐miR‐637, r = −.249, P = .014.
ROC results were shown in Figure 2C, and the area under the curve (AUC) of hsa_circ_0105015 was 0.692, P < .001; the AUC of hsa‐miR‐637 was 0.601, P = .087. After the combination of hsa_circ_0105015 and hsa‐miR‐637, the AUC increased to 0.703, P < .001.
3.5. Relative expression of candidate circRNA and miRNA in endothelial cells
The verification of endothelial cells in vitro is a further exploration of human whole blood samples. qRT‐PCR analysis results were shown in Figure 3. In Figure 3A and Figure 3C (HAEC and HUVEC), the relative expression of hsa_circ_0105015 was all significantly different between the four different treatment groups and the control groups, and the four treatments were as follows (1) TNF‐α 1μmol/L, 24h vs. control; (2) TNF‐α 1μmol/L, 48h vs. control; (3) TNF‐α 10μmol/L, 24h vs. control (4) TNF‐α 10 μmol/L, 48 h vs. control. Similar to the results in whole blood samples, compared with the control groups, hsa_circ_0105015 was up‐regulated in TNF‐α‐treated HAEC and HUVEC, respectively. In contrast, in Figure 3B and Figure 3D, hsa‐miR‐637 was down‐regulated in TNF‐α‐treated HAEC and HUVEC compared to the control groups. 18 The above evidence confirms the role of hsa_circ_0105015 and hsa‐miR‐637 in vascular inflammation. Under continuous inflammation conditions, hsa_circ_0105015 was up‐regulated and hsa‐miR‐637 was down‐regulated, which was consistent with the results in the whole blood samples of EH patients.
Figure 3.
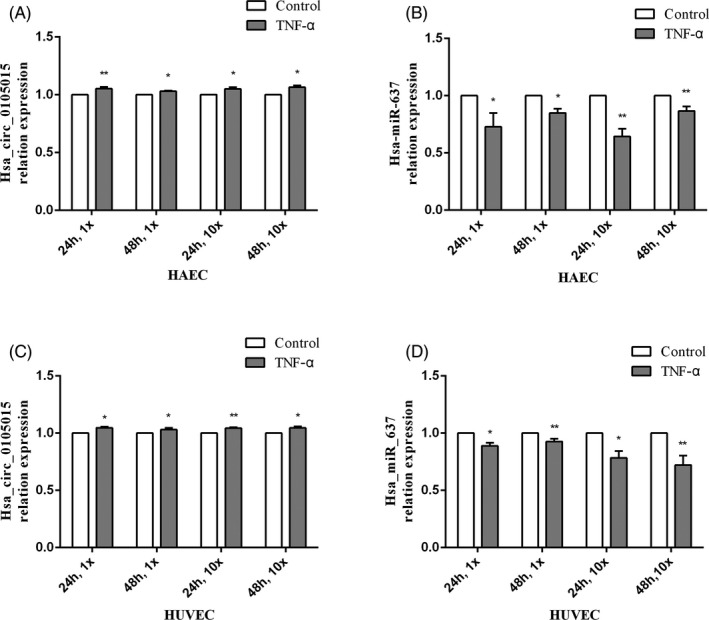
Relative expression of candidate circRNA and miRNA in endothelial cells with four treatment: (1) TNF‐α 1μmol/L, 24h vs. control; (2) TNF‐α 1μmol/L, 48h vs. control; (3) TNF‐α 10μmol/L, 24h vs. control (4) TNF‐α 10μmol/L, 48h vs. control. (A) The relative expression of hsa_circ_0105015 in HAEC. (B) The relative expression of hsa‐miR‐637 in HAEC. (C) The relative expression of hsa_circ_0105015 in HUVEC. (D) The relative expression of hsa‐miR‐637 in HUVEC. * P < .05; ** P < .01. Each cell line experiment was repeated at least three times, and the result was represented by mean ± SD, n = 3
4. DISCUSSION
Herein, we explored the association between circRNAs and EH. Our study is the first to reveal the role of hsa_circ_0105015 in EH and provides new insights into the pathogenesis of EH from the perspective of inflammation and endothelial dysfunction, which might be a new field of EH research. Moreover, our finding suggests the possibility of circRNA‐miRNA interaction in EH and may provide a basis for the early diagnosis of EH.
Dysfunctional endothelial cells are involved in the pathogenesis of hypertension through multiple pathways, including reduced bioavailability of NO, impaired vasodilation, and release of inflammatory mediators. More and more evidence shows that the dysregulated expression of miRNAs has an effect on the expression of inflammatory mediators, which leads to endothelial dysfunction. As an inflammatory mediator, CRP is a biomarker of cardiovascular diseases related to inflammation, the increase of oxidative stress in hypertensive patients is related to the level of CRP. 20 Hsa‐miR‐637 was confirmed to be related to CRP. Kim et al revealed that hsa‐miR‐637 can effectively inhibit the expression of CRP in the competition of HuR, which implies that hsa‐miR‐637 may play a protective role in EH. 21 Similarly, in our results, hsa‐miR‐637 was a protective factor for EH (Table 4), which was down‐regulated in the EH group (Table 3). In addition, compared with the control groups, hsa‐miR‐637 was down‐regulated in TNF‐α‐treated endothelial cells (Figure 3, B, and D), suggesting that hsa‐miR‐637 is associated with inflammatory mediators. To sum up, the process of vascular inflammation is accompanied by dysregulation of hsa‐miR‐637 expression, which is closely related to the occurrence and development of EH.
CircRNAs are considered to be related to human diseases, and microarray is an effective and vital tool for circRNAs profile. Li et al integrated 87 935 circRNAs sequences in circBase to design a microarray probe for each circRNA back‐splice site and found that about 80 000 circRNAs were expressed in cervix tumors and normal tissues, of which about 25 000 differential expressions. 22 Li et al concluded that circRNA microarrays are more effective than RNA‐seq. In the present study, we integrated 87 935 circRNA sequences in the circRNAs microarray, and the results showed that there were 287 circRNAs differential expressions in human whole blood samples, of which hsa_circ_0105015 was up‐regulated in the EH group. In the subsequent verification of human whole blood samples and endothelial cells, the results were consistent with the microarray analysis results. Hsa_circ_0105015 was up‐regulated in the EH group (Table 3) and TNF‐α‐treated group (Figure 3A and C), respectively.
Growing evidence suggests that circRNAs can act as miRNAs sponges to inhibit their effects on mRNAs. In cancer, Xia et al revealed that circSMC3 reduced miR‐4720‐3p expression by acting as a miRNA sponge, thereby promoting the expression of the tight junction protein 1 (TJP1), the target of miR‐4720‐3p. The silence of circSMC3 inhibits the proliferation of gastric cancer cells. 23 For cardiovascular disease, circRNA‐miRNA‐mRNA axis is present in diseases such as MI, 24 cardiac fibrosis, 25 atherosclerosis, 26 and myocardial hypertrophy. 27 In our study, the results of bioinformatics analysis predicted that hsa_circ_0105015 and hsa‐miR‐637 had two potential binding sites, and the relative expression of hsa_circ_0105015 and hsa‐miR‐637 was negatively correlated (Figure 2B), which provides evidence for the interaction of hsa_circ_0105015 and hsa‐miR‐637. In addition, in logistic regression, after adding both hsa_circ_0105015 and hsa‐miR‐637 into the equation, hsa‐miR‐637 was no longer a protective factor for EH, which may be caused by the correlation between hsa_circ_0105015 and hsa‐miR‐637, the existence of hsa_circ_0105015 limits the protective effect of hsa‐miR‐637 on EH.
As an important proinflammatory factor, TNF‐α can induce the adhesion of monocytes to endothelial cells, activate the NF‐κB pathway, and promote the occurrence and development of inflammation. Jang et al concluded that quinic acid (QA) inhibits the MAP kinase and NK‐κB signaling pathway, thereby reducing the VCAM‐1 in TNF‐α‐stimulated vascular smooth muscle cell (VSMC) and ultimately inhibiting inflammation. 28 Han et al induced the inflammation of SH‐Sy5y cells by TNF‐α and found that MI‐related transcript 2 (Mirt2) inhibits the expression of miR‐101 to block the NF‐κB/p38MAPK pathway triggered by TNF‐α. 29 Moreover, the association between TNF‐α and EH has been reported. Bautista et al concluded from a case‐control study that TNF‐α and IL‐6 could be independent risk factors of EH. 30 In our study, HUVECs and HAECs were treated with TNF‐α to induce vascular inflammation. In both the EH group and TNF‐α treatment group, the relative expression of hsa‐miR‐637 and hsa_circ_0105015 was down‐regulated and up‐regulated, respectively (Table 3 and Figure 3). This indicates that vascular inflammation as an early lesion of EH is accompanied by dysregulation of the expression of hsa‐miR‐637 and hsa_circ_0105015, both of which have the potential to be biomarkers for early diagnosis of EH. Combining the correlation between hsa_circ_0105015 and hsa‐miR‐637, we speculate that hsa_circ_0105015 may activate inflammation by targeting hsa‐miR‐637 to promote EH.
The stable expression of circRNAs in organisms makes it more suitable as a biomarker for human disease than long‐chain non‐coding RNA (lncRNA). In our results, the combined AUC of hsa_circ_0105015 and hsa‐miR‐637 was 0.703 (Figure 2C). Hyperexpression of hsa_circ_0105015 combined with hypoexpression of hsa‐miR‐637 in human whole blood may predict the occurrence of vascular inflammation and endothelial dysfunction, and the combination of the two has potential as a biomarker for early diagnosis and intervention of EH.
EH is a systemic disease and there is no specific target organ, which increases the difficulty of exploring its pathogenesis at the molecular level. To our knowledge, this study is the first to explore the association between hsa_circ_0105015 and EH risk, provides new evidence for the pathogenesis of EH, and lays the foundation for subsequent mechanism research. However, there are still some limitations in this study, one is that whether hsa_circ_0105015 can act as a sponge for hsa‐miR‐637 in EH remains to be further explored. Another limitation is that this study only considers the effect of hsa_circ_0105015 on EH in terms of vascular inflammation and endothelial dysfunction, and hsa_circ_0105015 may also be associated with EH through other pathways such as binding to other miRNAs or proteins. Therefore, in the future, large‐scale research should focus on elucidating the causal mechanism between circRNA‐miRNA interaction and EH.
In conclusion, hyperexpression of hsa_circ_0105015 is a significant risk factor of EH, and the process of vascular inflammation is accompanied by a dysregulated expression of hsa_circ_0105015. Hyperexpression of hsa_circ_0105015 combined with hypoexpression of hsa‐miR‐637 indicates vascular inflammation or endothelial dysfunction and has potential as a biomarker for early diagnosis of EH. We speculate that hsa_circ_0105015 may promote EH by targeting hsa‐miR‐637 to activate the inflammatory pathway.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest regarding the publication of this article.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant No. 81773528); Ningbo Scientific Innovation Team for Environmental Hazardous Factor Control and Prevention (Grant No. 2016C51001); KC Wong Magna Fund in Ningbo University; Natural Science Foundation of Ningbo (NO:2018A610406). Scientific Research Foundation of Graduate School of Ningbo University (G19129). The Student Research and Innovation Program of Ningbo University (2019SRIP1923)
He X, Bao X, Tao Z, et al. The microarray identification circular RNA hsa_circ_0105015 up‐regulated involving inflammation pathway in essential hypertension. J Clin Lab Anal. 2021;35:e23603 10.1002/jcla.23603
Xin He and Xingjie Bao authors contributed equally to this work.
Contributor Information
Fade Zhong, Email: zhanglina@nbu.edu.cn, Email: zfdyf@163.com.
Lina Zhang, Email: zhanglina@nbu.edu.cn.
REFERENCES
- 1. Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2224‐2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bundy JD, He J. Hypertension and related cardiovascular disease burden in China. Ann Glob Health. 2016;82(2):227‐233. [DOI] [PubMed] [Google Scholar]
- 3. Agita A, Alsagaff MT. Inflammation, Immunity, and Hypertension. Acta Med Indones. 2017;49(2):158‐165. [PubMed] [Google Scholar]
- 4. Androulakis ES, Tousoulis D, Papageorgiou N, Tsioufis C, Kallikazaros I, Stefanadis C. Essential hypertension: is there a role for inflammatory mechanisms? Cardiol Rev. 2009;17(5):216‐221. [DOI] [PubMed] [Google Scholar]
- 5. Hage FG. C‐reactive protein and hypertension. J Hum Hypertens. 2014;28(7):410‐415. [DOI] [PubMed] [Google Scholar]
- 6. Davis‐Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148(4):381‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA‐126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105(5):1516‐1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141‐148. [DOI] [PubMed] [Google Scholar]
- 9. Salzman J, Circular RNA. Expression: its potential regulation and function. Trends Genet. 2016;32(5):309‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de la Pena M. Circular RNAs biogenesis in eukaryotes through self‐cleaving hammerhead ribozymes. Adv Exp Med Biol. 2018;1087:53‐63. [DOI] [PubMed] [Google Scholar]
- 11. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor‐suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630‐3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ashwal‐Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell. 2014;56(1):55‐66. [DOI] [PubMed] [Google Scholar]
- 13. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42‐51. [DOI] [PubMed] [Google Scholar]
- 14. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256‐264. [DOI] [PubMed] [Google Scholar]
- 15. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384‐388. [DOI] [PubMed] [Google Scholar]
- 17. Lim TB, Lavenniah A, Foo RS. Circles in the heart and cardiovascular system. Cardiovasc Res. 2020;116(2):269‐278. [DOI] [PubMed] [Google Scholar]
- 18. Bao X, He X, Zheng S, et al. Up‐regulation of circular RNA hsa_circ_0037909 promotes essential hypertension. J Clin Lab Anal. 2019;33(4):e22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gabb GM, Mangoni AA, Arnolda L. Guideline for the diagnosis and management of hypertension in adults ‐ 2016. Med J Aust. 2017;206(3):141. [DOI] [PubMed] [Google Scholar]
- 20. Yasunari K, Maeda K, Nakamura M, Yoshikawa J. Oxidative stress in leukocytes is a possible link between blood pressure, blood glucose, and C‐reacting protein. Hypertension. 2002;39(3):777‐780. [DOI] [PubMed] [Google Scholar]
- 21. Kim Y, Noren Hooten N, Dluzen DF, Martindale JL, Gorospe M, Evans MK. Posttranscriptional Regulation Of The Inflammatory marker C‐reactive protein by the RNA‐binding protein HuR and MicroRNA 637. Mol Cell Biol. 2015;35(24):4212‐4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20(4):1420‐1433. [DOI] [PubMed] [Google Scholar]
- 23. Xia T, Pan Z, Zhang J. CircSMC3 regulates gastric cancer tumorigenesis by targeting miR‐4720‐3p/TJP1 axis. Cancer Med. 2020;9(12):4299‐4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang K, Gan TY, Li N, et al. Circular RNA mediates cardiomyocyte death via miRNA‐dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24(6):1111‐1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu Y, Pan W, Yang T, et al. Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR‐433. Front Genet. 2019;10:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun Y, Yang Z, Zheng B, et al. A novel regulatory mechanism of smooth muscle alpha‐actin expression by NRG‐1/circACTA2/miR‐548f‐5p Axis. Circ Res. 2017;121(6):628‐635. [DOI] [PubMed] [Google Scholar]
- 27. Lim TB, Aliwarga E, Luu TDA, et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res. 2019;115(14):1998‐2007. [DOI] [PubMed] [Google Scholar]
- 28. Jang SA, Park DW, Kwon JE, et al. Quinic acid inhibits vascular inflammation in TNF‐alpha‐stimulated vascular smooth muscle cells. Biomed Pharmacother. 2017;96:563‐571. [DOI] [PubMed] [Google Scholar]
- 29. Han Y, Kang C, Kang M, Quan W, Gao H, Zhong Z. Long non‐coding RNA Mirt2 prevents TNF‐alpha‐triggered inflammation via the repression of microRNA‐101. Int Immunopharmacol. 2019;76:105878. [DOI] [PubMed] [Google Scholar]
- 30. Bautista LE, Vera LM, Arenas IA, Gamarra G. Independent association between inflammatory markers (C‐reactive protein, interleukin‐6, and TNF‐alpha) and essential hypertension. J Hum Hypertens. 2005;19(2):149‐154. [DOI] [PubMed] [Google Scholar]


