Abstract
S100 calcium binding protein A16 (S100A16) is the most recent member of the S100 calcium-binding protein family. The function of S100A16 has been associated with various types of cancer; however, its role in colorectal cancer (CRC) remains unknown. Therefore, the aim of the present study was to investigate the role of S100A16 in CRC progression. The Oncomine dataset used in the current study revealed that the expression of S100A16 was decreased in CRC compared with normal colorectal tissues. Similar results were also determined via immunohistochemistry. In addition, a negative association was identified between S100A16 expression and the prognosis of patients with CRC. Further functional experiments revealed that S100A16 knockdown promoted the proliferation, migration and invasion of HCT116 and SW480 cells, and vice versa in Lovo cells. Epithelial-mesenchymal transition (EMT) was promoted and the JNK/p38 MAPK pathway was activated in HCT116 cells following S100A16 knockdown, as determined via western blotting. Furthermore, S100A16 silencing promoted the migration and invasion of cells. EMT was also reversed when cells were treated with the JNK inhibitor (SP600125) or the p38 inhibitor (SB203580). In summary, the results of the present study demonstrated that S100A16 suppressed the proliferation, migration and invasion of CRC cells partially via the JNK/p38 MAPK signalling pathway and subsequent EMT mediation.
Keywords: S100 calcium binding protein A16, colorectal cancer, tumour metastasis, epithelial-mesenchymal transition
Introduction
Colorectal cancer (CRC) is one of the most common cancer types and is the second cause of cancer-associated mortality worldwide, causing about 600,000 deaths per year (1). Despite progress in diagnostic and therapeutic techniques, the overall survival rate of patients with CRC remains poor, due to diagnosis occurring at a later stage, where metastasis has developed (2,3). Therefore, it is important to gain a greater understanding of the underlying molecular mechanisms involved in CRC, which will contribute to the early diagnosis and effective treatment of the disease.
S100 calcium binding protein A16 (S100A16) is a member of the S100 protein family, which is comprised of acidic proteins with an EF-hand Ca2+ binding motif (4). S100 proteins serve a variety of regulatory functions in cellular processes, such as motility, differentiation, cell proliferation, contraction, transcription, cell cycle processes and secretion (5). The abnormal protein expression of S100 has been exhibited in several types of tumour and is considered to be a potential marker of cancer (6,7). As the most recent member of the S100 family, S100A16 has been implicated in numerous human pathophysiological processes, such as inflammation, adipogenesis, osteoporosis and tumour progression (8–10). In addition, previous studies have reported that the function of S100A16 in cancer is complex (11–14). For instance, S100A16 demonstrates a variable effect in tumour cells of different tissues. Several studies have reported that S100A16 is associated with tumour progression in bladder, lung and breast cancer (11–13). However, an additional study reported the tumour-suppressive function of S100A16 in oral squamous cell carcinoma (14). Moreover, the abnormal expression of S100A16 is involved in CRC progression. It has also been shown that the low membrane expression of S100A16 is associated with a poor prognosis in patients with CRC, potentially serving as a suppressor of cancer (15). However, the specific modulatory role of S100A16 in CRC is yet to be elucidated.
The aim of the present study was to investigate whether S100A16 could inhibit the progression of CRC by regulating JNK/p38 MAPK signalling pathway, which may provide insight into the role of S100A16 in CRC as a potential prognostic marker of patients with CRC.
Materials and methods
Data collection and analysis
The data of 45 colorectal adenocarcinoma and 24 normal colorectal tissues were downloaded from the GSE20916 microarray dataset (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20916) (16). The mRNA expression values of S100A16 in colorectal adenocarcinoma and normal colorectal tissues were analyzed on the Oncomine platform (https://www.oncomine.org/), which collects publicly available cancer microarray data. The term ‘S100A16’ was searched on Oncomine. ‘Cancer type’ and ‘analysis type’ were then defined as ‘colorectal cancer’ and ‘cancer vs. normal analysis’, respectively. The cut-off of P-value and fold change were 0.05 and 2, respectively. The data were also analysed with an unpaired-t test between two groups using GraphPad Prism 6 (GraphPad Software, Inc.).
Tissue samples and microarrays
A total of 10 CRC samples and paired colorectal normal tissues were obtained from patients (age range, 31–71 years; 6 male and 4 female patients) admitted to the Department of Gastrointestinal Surgery, Nanfang Hospital, Southern Medical University between January 2015 and June 2019. All patients provided written informed consent and the study protocol was approved by the Nanfang Hospital Institutional Review Board (approval no. NFEC-2017-147). In addition, microarrays of CRC tissue comprising 100 cases (60 cases of CRC samples with matched, normal colorectal tissue samples; 40 cases were CRC samples alone.) were purchased from Shanghai Outdo Biotech Co., Ltd. All tumours were diagnosed as colorectal adenocarcinoma and all specimens were used for routine pathological processing. Out of all 110 specimens used in the present study, 104 had complete clinicopathological features and complete follow-up data.
Cell culture and treatment
Human CRC cell lines, including SW480, SW620, HCT116 and Lovo, were obtained from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences and identified by the American Type Culture Collection. RKO cells were obtained from the American Type Culture Collection. All cells were cultured in medium containing 90% DMEM (HyClone; Cytiva) and 10% FBS (HyClone; Cytiva) in a 5% CO2 atmosphere at 37°C.
To determine the impact of the JNK or p38 MAPK pathway, 10 µM JNK inhibitor SP600125 (cat. no. 8177; Cell Signaling Technology, Inc.) or 10 µM p38 inhibitor SB203580 (cat. no. 5633; Cell Signaling Technology, Inc.) were applied to HCT116 cells and incubated at 37°C with 5% CO2 for 48 h.
Immunohistochemistry (IHC) staining
Tissue samples were fixed with 4% paraformaldehyde for 2 days at room temperature and embedded in paraffin, after which 4-µm were cut and mounted onto slides. Slides were incubated at 56°C, deparaffinised in xylene and dehydrated in a graded series of alcohol. Heat-induced antigen retrieval was carried out with sodium citrate (pH 6.0) in a microwaving at 100°C for 30 min, IHC staining was performed as previously described (17). Briefly, endogenous peroxidase activity was inhibited using 3% hydrogen peroxide for 10 min at room temperature. After blocking with 5% goat serum (cat. no. BMS0050; Abbkine Scientific Co., Ltd.) diluted with PBS at room temperature for 30 min, sections were incubated with anti-S100A16 antibodies (cat. no. 11456-1-AP; 1:300; ProteinTech Group, Inc.) at 4°C overnight. Samples were then incubated with HRP-conjugated secondary antibodies (cat. no. ab5879; Abcam) at room temperature for 30 min. The resulting protein was presented as a brown pigment using the standard diaminodianiline method (cat. no. SAP 9101; OriGene Technologies, Inc.). Slides were subsequently double-stained with haematoxylin at room temperature for 20 sec, after which immunostaining was assessed using an Olympus BX-53 microscope (Olympus Corporation; magnification, ×100-400). The degree of IHC staining was evaluated as previously described (0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining) (18). Scores of 0–1 were defined as low expression, and scores of 2–3 were defined as high expression. The immunoreactivity score was determined as follows: Immunoreactivity score = the percentage of positive staining × the staining intensity score.
Overexpression and knockdown of S100A16
Based on the S100A16 gene sequence in GenBank (NM_080388; http://www.ncbi.nlm.nih.gov/nuccore), primers specifically matching S100A16 mRNA were constructed, which were amplified, cut by HindIII and BamHI and then sub-cloned into pcDNA3.1 vector (cat. no. P0157; www.miaolingbio.com) or GV287-EGFP lentiviral vector (Shanghai GeneChem Co., Ltd.) to overexpress S100A16 after screening. The empty lentiviral vector was used as a negative control.
To overexpress S100A16, colon cancer cells were transfected with 5 nM S100A16 pcDNA3.1(+) vector using Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Inc.) according to the manufacturers instructions for two days at 37°C. After 48 h of transfection, the expression of S100A16 was determined via western blotting, and the transfected cells were collected for subsequent experiments in vitro.
For lentivirus transduction, 7.5 µg GV287-EGFP-S100A16 (lenti-S100A16) lentiviral vector (Shanghai GeneChem Co., Ltd.), mixed with 5 µg pMD2.G (cat. no. p2062; www.miaolingbio.com) and 10 µg psPAX2 (cat. no. p2061; www.miaolingbio.com), were co-transfected into 293T cells (The Cell Bank of Type Culture Collection of Chinese Academy of Sciences) with Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Inc.) according to the manufacturers instructions at 37°C with 5% CO2 for 48 h. The viral supernatants were collected, centrifuged at 4000 × g for 10 min at 4°C, then filtered through 0.45-µm PVDF membranes. The virus concentration was determined by reverse transcription-quantitative PCR to measure the number of integrated copies of 293T cells. Lentivirus (lentivirus-S100A16) were then used to infect colon cancer cells using 5 µg/ml polybrene (Hanbio Biotechnology Co., Ltd.) with an MOI of 5. The infected cells were cultured with puromycin (5 µg/ml) for 14 days at 37°C to establish stably infected cells for mouse xenografts.
To knockdown S100A16, CRC cells were transfected with 50 nM S100A16 small interfering RNA (siRNA/siS100A16; Shanghai GenePharma Co., Ltd.) using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) according to the manufacturers instructions for 2 days at 37°C. The following sequences were utilized: si-S100A16#1, sense, 5′-GCAUCAGCUUCGAUGAGUATT-3′ and antisense, 5′-UACUCAUCGAAGCUGAUGCTT-3′; si-S100A16#2, sense, 5′-CCAGAACCUGGAUGCCAAUTT-3′ and antisense, 5′-AUUGGCAUCCAGGUUCUGGTT-3′; si-S100A16#3, sense, 5′-GCUGGAGAAGGCAGUCAUUTT-3′ and antisense, 5′-AAUGACUGCCUUCUCCAGCTT-3′; negative control, sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. At 48 h following transfection, the expression of S100A16 was determined via western blotting, and the transfected cells were collected for subsequent experiments.
Cell proliferation assay
After S100A16 siRNA was transfected into SW480 and HCT116 cells or following S100A16 pcDNA3.1(+) vector transfection into Lovo cells, CRC cells (1×103 cells/well) were seeded into 96-well plates. Cells were cultured for 24, 48, 72, 96 and 120 h, after which 10 µl Cell Counting Kit-8 (CCK-8) solution (Dojindo Molecular Technologies, Inc.) was added and incubated in a 5% CO2 atmosphere at 37°C for an additional 4 h. The kit was used according to the manufacturers instructions. Optical density was measured at 450 nm using an automatic enzyme standard.
Cell migration and invasion assays
Cell migration and invasion assays were performed using a Transwell system (Corning, Inc.). For cell invasion, an 8 µm-pore polycarbonate membrane was coated with Matrigel (BD Biosciences) at 37°C for 5 h. Cells at density of 2.5×104 (migration) and 5×104 (invasion) per well were suspended in 500 µl serum-free medium and subsequently added to the upper chamber. A total of 600 µl medium containing 10% FBS was added to the lower chamber. After 24 (migration) or 48 h (invasion) incubation at 37°C, migrated or invaded cells in the lower chambers were stained with 0.1% crystal violet at room temperature for 30 min and counted using light microscopy (magnification, ×100).
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the harvested cells using TRIzol® (Thermo Fisher Scientific, Inc.) according to the manufacturers protocol. First-Strand cDNA Synthesis was carried out by reverse-transcription using PrimeScript™ RT Master Mix (Takara) and qPCR was performed with a LightCycler 480® System (Roche Diagnostics) using SYBR- Green mix (Takara Bio, Inc.), according to the manufacturers instructions. The thermocycling conditions were as follows: i) 94°C for 5 min; ii) 40 amplification cycles at 94°C for 30 sec, 57°C for 30 sec and 72°C for 30 sec; and iii) 72°C for 5 min. The primer sequences used for PCR were as follows: i) S100A16 forward, 5′-TTTTGTCCATCTCCTTTCACCA-3′ and reverse, 5′-CAGGCAAATCAGACTCCCTTC-3′; ii) S100A4 forward, 5′-GTACTCGGGCAAAGAGGGTG-3′ and reverse, 5′-TTGTCCCTGTTGCTGTCCAA-3′; iii) S100A14 forward, 5′-TGCTCTAGAATGGGACAGTGTCGGTCAGCC-3′ and reverse, 5′-CGCGGATCCTCAGTGCCCCCGGA CAGGCCT-3′; and iv) GAPDH (the internal reference) forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′. Relative gene expression was determined using the 2−ΔΔCq method (19).
Western blotting
Cultured cells were lysed in RIPA buffer containing 1X protease cocktail inhibitor (Sigma-Aldrich; Merck KGaA). Protein concentrations were determined using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). A total of 30 µg proteins from different groups were then separated on 10–12% SDS-polyacrylamide gels for electrophoresis. Separated proteins were transferred to PVDF membranes (EMD Millipore). After being blocked with non-fat milk for 1 h at room temperature, the membranes were incubated with the following primary antibodies at 4°C overnight: anti-rabbit S100A4 (cat. no. 16105-1-AP; 1:500; ProteinTech Group, Inc.), anti-rabbit S100A14 (cat. no. 10489-1-AP; 1:500; ProteinTech Group, Inc.), anti-rabbit S100A16 (cat. no. 11456-1-AP; 1:500; ProteinTech Group, Inc.) anti-phosphorylated (p)-rabbit JNK (cat. no. 9255; Cell Signaling Technology, Inc.), anti-rabbit JNK (cat. no. 9252; Cell Signaling Technology, Inc.), anti-p-rabbit ERK1/2 (cat. no. 4370; Cell Signaling Technology, Inc.), anti-rabbit ERK1/2 (cat. no. 4695; Cell Signaling Technology, Inc.), anti-p-rabbit p38 MAPK (cat. no. 9216; Cell Signaling Technology, Inc.), anti-rabbit p38 MAPK (cat. no. 9212; Cell Signaling Technology, Inc.), anti-rabbit vimentin (cat. no. 12826; Cell Signaling Technology, Inc.), anti-mouse E-cadherin (cat. no. 14472; Cell Signaling Technology, Inc.), anti-mouse N-cadherin (cat. no. 14215; Cell Signaling Technology, Inc.) and anti-mouse GAPDH (cat. no. 51332; Cell Signaling Technology, Inc.). After incubation with horseradish peroxidase-conjugated secondary antibodies (cat. nos. 7074 and 7076; Cell Signaling Technology, Inc.) for 1 h at room temperature, immunoreactive proteins were visualized using chemiluminescence detection reagents (EMD Millipore) using the FluorChem E system (ProteinSimple). Blots were semi-quantified using AlphaView SA software (ProteinSimple; version 3.4.0.0). The grayscale values of protein bands were normalized to that of GAPDH.
Mouse xenografts of CRC cells
A total of 10 BALB/c nude male mice (weight, 12–14 g; age, 4–6 weeks) were purchased from Hunan SJA Laboratory Animal Co., Ltd. The mice were fed with common feed and sterile water ad libitum and housed in 21–23°C with 54–56% humidity and a 12-h light/dark cycle. Lovo cells (2×106) were infected with Lenti-Vector or Lenti-S100A16 (S100A16 overexpression vector) and were injected subcutaneously into the flanks of mice to construct the xenograft model (n=5/group). Tumour volume was measured and recorded every three days. At 21 days after injection, all mice were euthanized via cervical dislocation. The mice were considered to be dead when the heart and breathing stopped. The tumour xenografts were removed to determine their size and weight. The largest tumour diameter was 1.5 cm. All experimental procedures were conducted in accordance with the ethical standards of the Ethical Committee of Nanfang Hospital, Southern Medical University. Study approval was additionally gained from the Institutional Animal Care and Use Committee of Southern Medical University (approval no. L2018160).
Statistical analysis
GraphPad Prism 6 (GraphPad Software, Inc.) and SPSS 22.0 (IBM Corp.) software was used to assess the statistical significance of differences between groups. Data were analysed using an unpaired or a paired two-tailed Students t-test for two groups, or one-way ANOVA followed by Tukeys post hoc test for multiple groups. Two-way ANOVA with Bonferronis correction was used for statistical analyses between different treatments, different cell cohorts or different time points. χ2 or Fishers exact tests were used to compare the differences in age, sex, pathological differentiation, T classification, N classification and American Joint Committee on Cancer (AJCC) stages (Table I). Data are presented as the mean ± SD from a minimum of three experimental repeats. P<0.05 was considered to indicate a statistically significant difference.
Table I.
Association between clinicopathological features and S100A16 expression in 104 patients with colorectal cancer.
| S100A16 expression | |||||
|---|---|---|---|---|---|
| Variables | n | Low (n=71) | High (n=33) | χ2 | P-value |
| Age, years | 0.280 | 0.596 | |||
| ≤50 | 19 | 12 | 7 | ||
| >50 | 85 | 59 | 26 | ||
| Sex | 0.000 | 0.987 | |||
| Female | 44 | 30 | 14 | ||
| Male | 60 | 41 | 19 | ||
| Pathological differentiation | 6.638 | 0.036 | |||
| I (high) | 11 | 7 | 4 | ||
| II (moderate) | 49 | 28 | 21 | ||
| III (poor) | 44 | 36 | 8 | ||
| T classification | 0.133 | 0.715 | |||
| T1 + T2 | 6 | 5 | 1 | ||
| T3 + T4 | 98 | 66 | 32 | ||
| N classification | 4.477 | 0.034 | |||
| N0 | 60 | 36 | 24 | ||
| N1 + N2 | 44 | 35 | 9 | ||
| AJCC stage | 3.311 | 0.069 | |||
| I + II | 59 | 36 | 23 | ||
| III + IV | 45 | 35 | 10 | ||
S100A16, S100 calcium binding protein A16. AJCC, American Joint Committee on Cancer.
Results
S100A16 is decreased in human CRC tissues
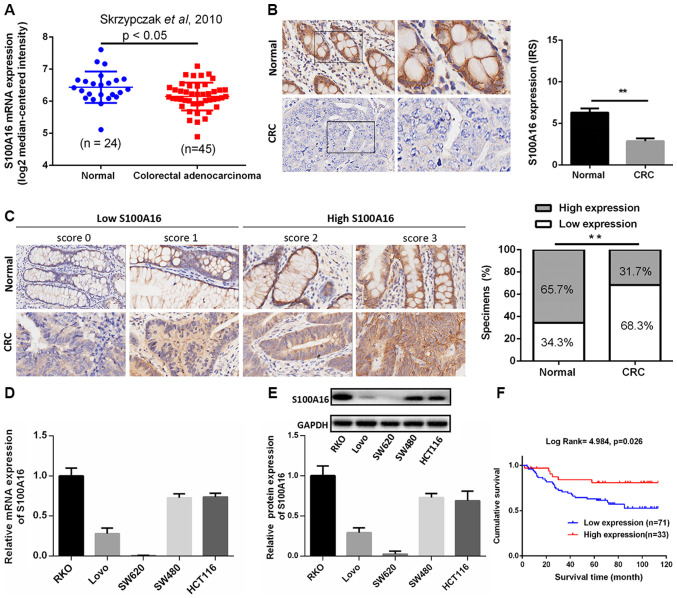
To identify the potential role of S100A16 in CRC, gene expression profiles were analysed from Oncomine microarray datasets (16). The results revealed that S100A16 mRNA was significantly downregulated in colorectal adenocarcinoma compared with normal tissues (Fig. 1A). The expression of S100A16 was subsequently determined in 70 CRC samples (clinical samples from Nanfang Hospital, n=10; paired samples in CRC tissue microarrays purchased from Shanghai Outdo Biotech Co., Ltd., n=60) and paired normal adjacent tissues. As presented in Fig. 1B, IHC analysis demonstrated that S100A16 exhibited weaker staining in CRC tumour tissues compared with normal tissues. In addition, S100A16 protein was localized to the cell membrane and cytoplasm of CRC cells. To confirm the expression of S100A16 in CRC, CRC microarrays were screened alongside the CRC samples obtained in the present study. The results demonstrated that S100A16 was decreased in CRC tissues compared with normal colon tissues (Fig. 1C).
Figure 1.
S100A16 is decreased in human colorectal cancer. (A)Analysis of S100A16 expression in normal colorectal tissues and colorectal adenocarcinoma tissues in the Skrzypczak in Oncomine microarray dataset, as assessed using an unpaired-t test. P<0.05. (B) Representative images of S100A16 protein expression in CRC tumour tissues and paired normal adjacent tissues, as assessed via immunohistochemistry. Magnification, ×400. **P<0.01, paired Students t-test. (C) Representative images of difference scores indicating S100A16 protein expression in CRC tumour tissues and paired normal adjacent tissues (magnification, ×400). The proportion of S100A16-expressing in CRC tissue samples was also analysed. **P<0.01, χ2 test. S100A16 (D) mRNA and (E) protein expression in CRC cell lines, as determined via reverse transcription-quantitative PCR and western blotting, respectively. (F) Kaplan-Meier analysis of the survival rates in patients with CRC with S100A16 low or high expression. CRC, colorectal cancer; S100A16, S100 calcium binding protein A16, IRS, immunoreactive score.
The expression levels of S100A16 mRNA and protein in five CRC cell lines were examined via RT-qPCR and western blotting, respectively. The results indicated that S100A16 mRNA and protein expression levels were markedly decreased in Lovo and SW620 CRC cell lines (Fig. 1D and E). Among the CRC cell lines assessed in the present study, HCT116 and SW480 cells exhibited higher expression levels of S100A16, and as such, were selected to perform loss-of-function experiments. Lovo cells exhibiting lower expression levels of S100A16 were selected for gain-of-function experiments. As SW620 cells did not express an appropriate level of S100A16, they were not selected for further study.
S100A16 is negatively associated with CRC progression and positively associated with patient survival
To investigate the clinical importance of S100A16 in CRC, the relationship between patient clinicopathological features and S100A16 expression was analysed. Patients with CRC were classified into low and high S100A16 expression groups based on immunostaining scores (16). The results suggested that the expression of S100A16 was closely associated with pathological differentiation (P=0.036) and N classification (P=0.034). However, no association was identified between age, sex, T classification and AJCC stage (Table I). Furthermore, patients in the low S100A16 expression group demonstrated poor prognosis, with a lower survival time (Fig. 1F). The results indicated that S100A16 may be involved in CRC development and could serve as a potential therapeutic target for CRC.
S100A16 inhibits CRC cell proliferation, migration and invasion
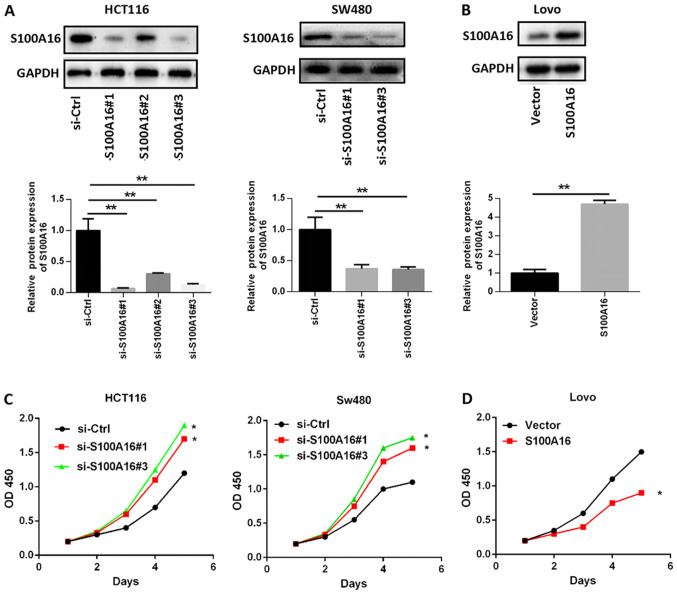
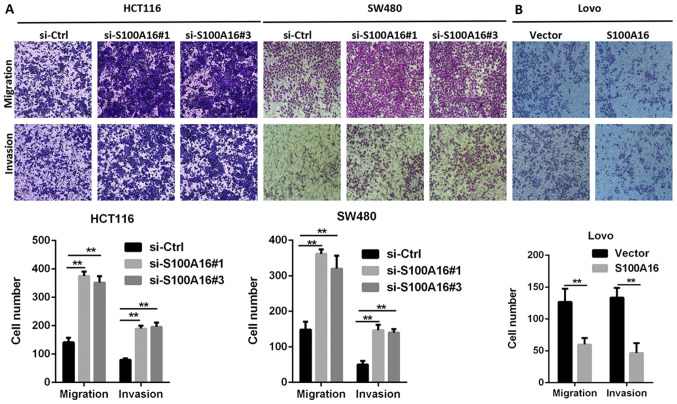
To determine whether S100A16 affects CRC cell proliferation, migration and invasion, siRNAs targeting S100A16 were transfected into HCT116 and SW480 cells. The results demonstrated that S100A16 protein expression was significantly decreased following siRNA transfection. si-S100A16#1, si-S100A16#2, and si-S100A16#3 were all efficiently in silencing S100A16; thus, only two siRNAs were arbitrarily selected for subsequent experiments (Fig. 2A). However, no significant differences were identified in cells transfected with S100A4 and S100A14 siRNA, indicating that S100A16 knockdown was specific to S100A16 (Fig. S1A and B). Subsequently, the S100A16 overexpression vector was transfected into Lovo cells for S100A16 upregulation (Fig. 2B). CCK-8 assays were then performed to determine cell proliferation. The results indicated that proliferation was increased in S100A16-silenced HCT116 and SW480 cells, but decreased in S100A16-overexpressing Lovo cells, compared with the corresponding control groups (Fig. 2C and D). The results of Transwell assays demonstrated that S100A16 knockdown promoted the migration and invasion of HCT116 and SW480 cells, whereas S100A16 overexpression had the opposite effect in Lovo cells (Fig. 3A and B).
Figure 2.
S100A16 inhibits the proliferation of CRC cells. (A) Western blot analysis of S100A16 protein expression in CRC cells transfected with S100A16 siRNAs compared with si-Ctrl. Data were analysed using a one-way ANOVA followed by Tukeys post hoc test. **P<0.01. (B) Western blot analysis of S100A16 protein expression in CRC cells transfected with overexpression plasmids compared vectors, as assessed using an unpaired Students t-test. **P<0.01. (C) S100A16 knockdown promoted the proliferation of HCT116 and SW480 cells. *P<0.05, vs. si-Ctrl. (D) S100A16 overexpression suppressed the proliferation of Lovo cells. *P<0.05. Cell proliferation was examined by performing Cell Counting Kit-8 assays and analysed using a two-way ANOVA with Bonferronis correction. CRC, colorectal cancer; siRNA/si, small interfering RNA; Ctrl, control; S100A16, S100 calcium binding protein A16; OD, optical density.
Figure 3.
S100A16 inhibits CRC cell migration and invasion. (A) S100A16 knockdown promoted the migration and invasion of the two CRC cell lines, which was analysed using a one-way ANOVA followed by Tukeys post hoc test. **P<0.01. (B) S100A16 overexpression suppressed the migration and invasion of Lovo cells, which was analysed using an unpaired Students t-test. **P<0.01. Cell migration and invasion were determined by performing Transwell assays. CRC, colorectal cancer; S100A16, S100 calcium binding protein A16; siRNA/si, small interfering RNA; Ctrl, control.
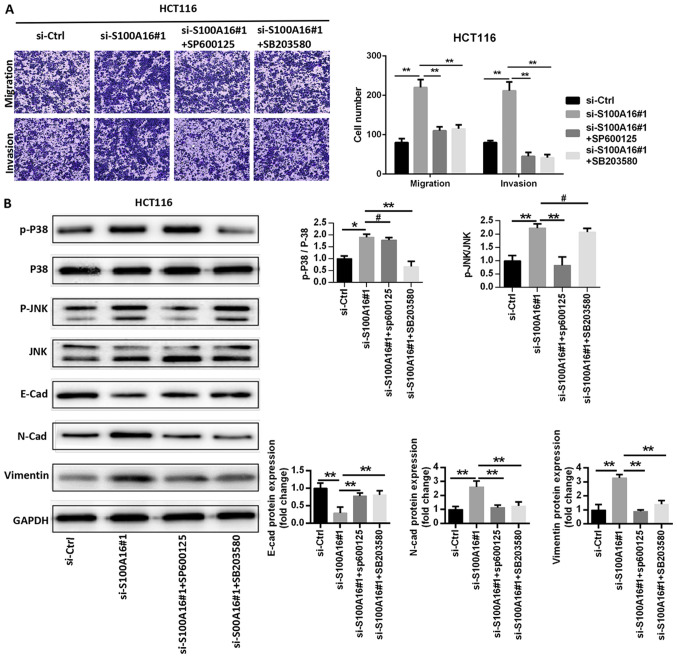
S100A16 knockdown activates the JNK/p38 MAPK signalling pathway to promote epithelial-mesenchymal transition (EMT)
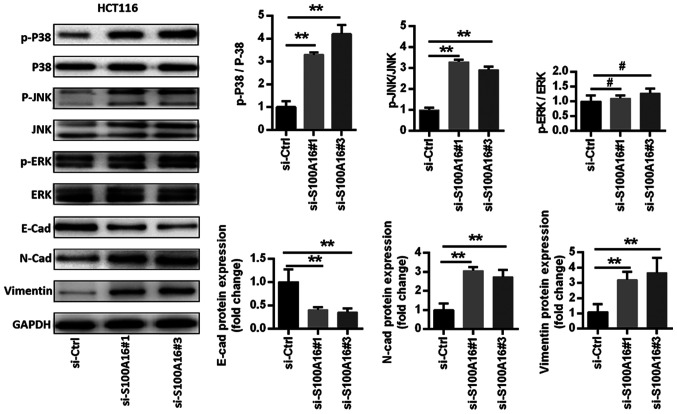
The signalling pathways associated with the S100A16-mediated suppression of CRC cell proliferation, migration and invasion were assessed in the current study. Previous reports have revealed that S100A16 affects the JNK/p38 MAPK signalling pathway (9,20), which serves an important role in CRC (21,22). Thus, western blotting was performed to assess the factors associated with the MAPK signalling pathway in S100A16-inhibited HCT116 cells. The results identified that p38, ERK and JNK phosphorylation levels were increased in HCT116 cells following S100A16 knockdown. However, ERK phosphorylation levels were not as high as those observed for p38 and JNK (Fig. 4). Additionally, S100A16 knockdown increased the expression levels of the mesenchymal markers N-cadherin and vimentin, as well as decreased the expression of the epithelial marker E-cadherin (Fig. 4).
Figure 4.
S100A16 knockdown activates the JNK/p38 MAPK signalling pathway and promotes EMT. MAPK pathway-associated and EMT-associated proteins were assessed via western blotting in HCT116 cells. Data were analysed with a one-way ANOVA followed by Tukeys post hoc test. **P<0.01; #P>0.05 (not significant). EMT, epithelial-mesenchymal transition; S100A16, S100 calcium binding protein A16; p-, phosphorylated; E-cad, E-cadherin; siRNA/si, small interfering RNA; Ctrl, control; N-cad, N-cadherin.
To confirm that the MAPK signalling pathway served a role in S100A16-modulated CRC cellular activity, the JNK inhibitor SP600125 and the p38 inhibitor SB203580 were administered to HCT116 cells. SP600125 and SB203580 significantly repressed the increase in HCT116 cell migration and invasion caused by S100A16 silencing (Fig. 5A). The results of western blotting suggested that SP600125 and SB203580 treatments significantly inhibited the S100A16 knockdown-mediated activation of JNK and p38 in HCT116 cells. SP600125 and SB203580 also suppressed the augmented protein expression levels of N-cadherin and vimentin, and reversed the effect of S100A16 silencing on the protein expression of E-cadherin in HCT116 cells (Fig. 5B). The results suggested that S100A16 suppressed EMT and that S100A16 may exert its effects via the JNK/p38 MAPK signalling pathway.
Figure 5.
JNK/p38 MAPK signalling pathway inactivation is required for S100A16 knockdown-mediated HCT116 cellular effects. (A) Representative images of Transwell assays after treatment with the p38 inhibitor (SB203580) or the JNK inhibitor (SP600125) following western blotting in S100A16-silenced HCT116 cells. (B) MAPK and epithelial-mesenchymal transition markers were examined via western blotting after treatment with the p38 inhibitor (SB203580) or the JNK inhibitor (SP600125) in S100A16-silenced HCT116 cells. Data were analysed with a one-way ANOVA followed by Tukeys post hoc test. **P<0.01 and *P<0.05; #P>0.05. S100A16, S100 calcium binding protein A16; p-, phosphorylated; E-cad, E-cadherin; siRNA/si, small interfering RNA; Ctrl, control; N-cad, N-cadherin.
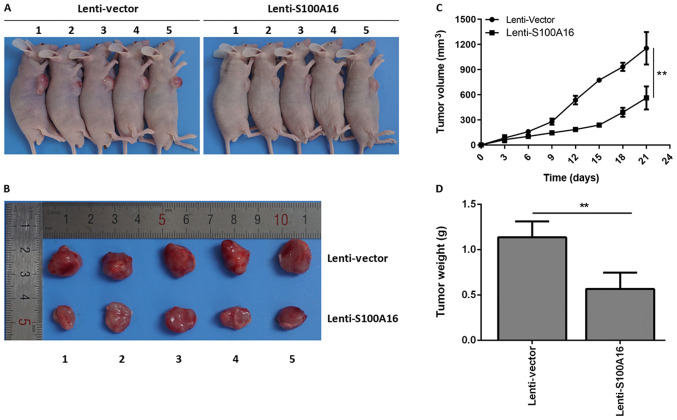
S100A16 elevation suppresses tumour growth in vivo
To further determine the effects of S100A16 on colorectal cancer in vivo, tumour-bearing mice were constructed. Lenti-Vector and Lenti-S100A16 were transfected into Lovo cells to elevate S100A6 expression (Fig. S1C). The largest tumour diameter observed was 1.5 cm. It was determined that the size of tumours in mice of the S100A16 overexpression group were markedly decreased compared with the control group (Fig. 6A and B). Additionally, S100A16 overexpression mice exhibited significantly decreased tumour volumes and weights relative to the control mice (Fig. 6C and D). The results indicated that S100A16 overexpression inhibited CRC tumours in vivo.
Figure 6.
Overexpression of S100A16 inhibits tumour growth in vivo. (A) Images of tumour xenografts in mice of the vector and S100A16 groups. (B) Images of the isolated tumours. (C) Tumour volume and (D) weight were separately compared between the two groups. **P<0.01. S100A16, S100 calcium binding protein A16.
Discussion
The S100 family comprises 21 identified Ca2+ binding proteins, several of which are upregulated in various tumours, serving vital roles in tumour progression (4,23). Among these proteins, S100A6 promotes the proliferation and migration of CRC cells (24). Additionally, S100A11 promotes CRC aggressiveness by modulating the TGFβ/Smad signalling pathway (25). Previous studies have reported that several S100 members are upregulated in CRC, including S100A2, S100A3, S100A4 and S100A8, and function as tumour promoters (26–29). In addition, low and high expression levels of S100A14 and S100A4, respectively, are associated with increased CRC metastasis (30). S100A14 interacts with S100A16 and regulates its expression in human cancer cells (31). Furthermore, S100A16 expression is decreased in CRC, which is associated with shorter overall survival times in patients with CRC (15). However, the role of S100A16 in CRC is yet to be fully elucidated. Therefore, the aim of the present study was to determine whether S100A16 serves a functional role in CRC progression.
The present study analysed the expression of S100A16 in the Oncomine microarray dataset, which revealed that S100A16 was downregulated in CRC. The expression of S100A16 was further examined in CRC tissue samples and paired adjacent normal tissues via IHC analysis. The results indicated that S100A16 expression was significantly decreased in CRC tumour tissues. The association between patient clinicopathological characteristics and S100A16 expression was subsequently determined. The results demonstrated that S100A16 expression was significantly associated with pathological differentiation and N classification. Furthermore, decreased S100A16 expression levels were significantly associated with a poor patient prognosis, which is in line with the results of a previous study (15).
Previous studies have revealed that high levels of S100A16 are associated with increased proliferation, migration and invasion (13,23,32). Additionally, S100A16 has been found to affect metastasis (20). However, the effect of S100A16 on the biological activity of CRC cells remains undetermined. The present study therefore investigated the proliferation, migration and invasion of CRC cells in vitro. The results demonstrated that S100A16 knockdown promoted the proliferation, migration and invasion of HCT116 and SW480 cells. By contrast, following S100A16 overexpression, the proliferation, migration and invasion of Lovo cells were decreased. These results indicated that S100A16 may serve as a tumour suppressor in CRC.
Since it has been reported that S100A16 interacts with the MAPK signalling pathway (9,20), which regulates cell proliferation and migration, it was hypothesized that the MAPK signalling pathway may be involved in S100A16-mediated CRC. In the present study, markers of the MAPK signalling pathway were assessed following S100A16 knockdown. It was identified that S100A16 knockdown activated the JNK/p38 MAPK signalling pathway. EMT is a physiological process that increases the migration and invasion of cells, as well as induces tumour metastasis and development (1,33,34). Additionally, EMT is regulated by S100A16 in breast cancer (35). Therefore, the present study assessed EMT markers to investigate the underlying mechanism of CRC progression using MAPK inhibitors following S100A16 knockdown. The results indicated that S100A16 knockdown promoted CRC progression partially via the JNK/P38 MAPK pathway and its subsequent EMT. However, the detailed mechanism via which S100A16 affects the JNK/P38 MAPK pathway was not fully assessed in the present study and may involve additional factors, such as transcription target genes or downstream effectors.
In conclusion, the present study investigated the potential function of S100A16 in CRC, the results of which provided insights into the tumour-suppressive role of S100A16 in CRC cells. S100A16 may serve as a prognostic indicator of CRC, potentially providing a reference for the early diagnosis of CRC, as well as a novel therapeutic target. However, the current study had several limitations. Additional experiments are therefore required to reveal the detailed underlying mechanisms of S100A16 in CRC regulation and should be the focus of future research.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81970451, 81500398 and 81470790), the Natural Science Foundation of Guangdong Province (grant nos. 2019A1515010667, 2015A030310480 and 2015A030313295) and the Guangdong Gastrointestinal Disease Research Center (grant no. 2017B02029003).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors contributions
SO conceived and designed the experiments, performed the experiments, analysed the data, contributed reagents/materials/analytical tools, wrote the manuscript, prepared figures and tables and reviewed the manuscript. YL, JS, JT, YY, FW, WW, JF and FX performed the experiments, analysed the data, contributed reagents/materials/analysis tools and prepared figures and tables. LB conceived and designed the experiments, as well as drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent for publication
All patients provided written informed consent and the study protocol for the use of clinical samples was approved by The Nanfang Hospital Institutional Review Board (approval no. NFEC-2017-147). Approval for animal studies was additionally gained from The Institutional Animal Care and Use Committee of Southern Medical University (approval no. L2018160).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Keane MG, Johnson GJ. Early diagnosis improves survival in colorectal cancer. Practitioner. 2012;256:15–18. 2. [PubMed] [Google Scholar]
- 4.Sturchler E, Cox JA, Durussel I, Weibel M, Heizmann CW. S100A16, a novel calcium-binding protein of the EF-hand superfamily. J Biol Chem. 2006;281:38905–38917. doi: 10.1074/jbc.M605798200. [DOI] [PubMed] [Google Scholar]
- 5.Donato R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/S1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 6.Sedaghat F, Notopoulos A. S100 protein family and its application in clinical practice. Hippokratia. 2008;12:198–204. [PMC free article] [PubMed] [Google Scholar]
- 7.Heizmann CW. The multifunctional S100 protein family. Methods Mol Biol. 2002;172:69–80. doi: 10.1385/1-59259-183-3:069. [DOI] [PubMed] [Google Scholar]
- 8.Domínguez B, Pardo BG, Noia M, Millán A, Gómez-Tato A, Martínez P, Leiro J, Lamas J. Microarray analysis of the inflammatory and immune responses in head kidney turbot leucocytes treated with resveratrol. Int Immunopharmacol. 2013;15:588–596. doi: 10.1016/j.intimp.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Zhang R, Zhu W, Xue Y, Zhang Y, Huang Q, Liu M, Liu Y. S100A16 inhibits osteogenesis but stimulates adipogenesis. Mol Biol Rep. 2013;40:3465–3473. doi: 10.1007/s11033-012-2413-2. [DOI] [PubMed] [Google Scholar]
- 10.Babini E, Bertini I, Borsi V, Calderone V, Hu X, Luchinat C, Parigi G. Structural characterization of human S100A16, a low-affinity calcium binder. J Biol Inorg Chem. 2011;16:243–256. doi: 10.1007/s00775-010-0721-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Zhu X, Li A, Yang S, Qiao R, Zhang J. S100A16 regulated by Snail promotes the chemoresistance of nonmuscle invasive bladder cancer through the AKT/Bcl-2 pathway. Cancer Manag Res. 2019;11:2449–2456. doi: 10.2147/CMAR.S196450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Luo L, Liang C. Aberrant S100A16 expression might be an independent prognostic indicator of unfavorable survival in non-small cell lung adenocarcinoma. PLoS One. 2018;13:e197402. doi: 10.1371/journal.pone.0197402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W, Pan H, Xia T, Xue J, Cheng L, Fan P, Zhang Y, Zhu W, Xue Y, Liu X, et al. Up-regulation of S100A16 expression promotes epithelial-mesenchymal transition via Notch1 pathway in breast cancer. J Biomed Sci. 2014;21:97. doi: 10.1186/s12929-014-0097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapkota D, Bruland O, Parajuli H, Osman TA, Teh MT, Johannessen AC, Costea DE. S100A16 promotes differentiation and contributes to a less aggressive tumor phenotype in oral squamous cell carcinoma. BMC Cancer. 2015;15:631. doi: 10.1186/s12885-015-1622-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Wang T, Zhang C, Ning K, Guan ZR, Chen SX, Hong TT, Hua D. S100A16 is a prognostic marker for colorectal cancer. J Surg Oncol. 2018;117:275–283. doi: 10.1002/jso.24822. [DOI] [PubMed] [Google Scholar]
- 16.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:5. doi: 10.1371/annotation/8c585739-a354-4fc9-a7d0-d5ae26fa06ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi J, Sun S, Liao Y, Tang J, Xu X, Qin B, Qin C, Peng L, Luo M, Bai L, et al. Advanced oxidation protein products induce G1 phase arrest in intestinal epithelial cells via a RAGE/CD36-JNK-p27kip1 mediated pathway. Redox Biol. 2019;25:101196. doi: 10.1016/j.redox.2019.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang J, Liao Y, He S, Shi J, Peng L, Xu X, Xie F, Diao N, Huang J, Xie Q, et al. Autocrine parathyroid hormone-like hormone promotes intrahepatic cholangiocarcinoma cell proliferation via increased ERK/JNK-ATF2-cyclinD1 signaling. J Transl Med. 2017;15:238. doi: 10.1186/s12967-017-1342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Zhu W, Xue Y, Liang C, Zhang R, Zhang Z, Li H, Su D, Liang X, Zhang Y, Huang Q, et al. S100A16 promotes cell proliferation and metastasis via AKT and ERK cell signaling pathways in human prostate cancer. Tumour Biol. 2016;37:12241–12250. doi: 10.1007/s13277-016-5096-9. [DOI] [PubMed] [Google Scholar]
- 21.Yang MH, Zhao L, Wang L, Ou-Yang W, Hu SS, Li WL, Ai ML, Wang YQ, Han Y, Li TT, et al. Nuclear lncRNA HOXD-AS1 suppresses colorectal carcinoma growth and metastasis via inhibiting HOXD3-induced integrin β3 transcriptional activating and MAPK/AKT signalling. Mol Cancer. 2019;18:31. doi: 10.1186/s12943-019-0955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroyer AL, Stimes NW, Abi Saab WF, Chadee DN. MLK3 phosphorylation by ERK1/2 is required for oxidative stress-induced invasion of colorectal cancer cells. Oncogene. 2018;37:1031–1040. doi: 10.1038/onc.2017.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marenholz I, Heizmann CW. S100A16, a ubiquitously expressed EF-hand protein which is up-regulated in tumors. Biochem Biophys Res Commun. 2004;313:237–244. doi: 10.1016/j.bbrc.2003.11.115. [DOI] [PubMed] [Google Scholar]
- 24.Duan L, Wu R, Zou Z, Wang H, Ye L, Li H, Yuan S, Li X, Zha H, Sun H, et al. S100A6 stimulates proliferation and migration of colorectal carcinoma cells through activation of the MAPK pathways. Int J Oncol. 2014;44:781–790. doi: 10.3892/ijo.2013.2231. [DOI] [PubMed] [Google Scholar]
- 25.Niu Y, Shao Z, Wang H, Yang J, Zhang F, Luo Y, Xu L, Ding Y, Zhao L. LASP1-S100A11 axis promotes colorectal cancer aggressiveness by modulating TGFβ/Smad signaling. Sci Rep. 2016;6:26112. doi: 10.1038/srep26112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda T, Ishikawa T, Mogushi K, Okazaki S, Ishiguro M, Iida S, Mizushima H, Tanaka H, Uetake H, Sugihara K. Overexpression of the S100A2 protein as a prognostic marker for patients with stage II and III colorectal cancer. Int J Oncol. 2016;48:975–982. doi: 10.3892/ijo.2016.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Sun WY, Zhi CY, Lu TC, Gao HM, Zhou JH, Yan WQ, Gao HC. Role of S100A3 in human colorectal cancer and the anticancer effect of cantharidinate. Exp Ther Med. 2013;6:1499–1503. doi: 10.3892/etm.2013.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlmann M, Okhrimenko A, Marcinkowski P, Osterland M, Herrmann P, Smith J, Heizmann CW, Schlag PM, Stein U. RAGE mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognostic biomarker for human colorectal cancer metastasis. Oncotarget. 2014;5:3220–3233. doi: 10.18632/oncotarget.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zha H, Sun H, Li X, Duan L, Li A, Gu Y, Zeng Z, Zhao J, Xie J, Yuan S, et al. S100A8 facilitates the migration of colorectal cancer cells through regulating macrophages in the inflammatory microenvironment. Oncol Rep. 2016;36:279–290. doi: 10.3892/or.2016.4790. [DOI] [PubMed] [Google Scholar]
- 30.Wang HY, Zhang JY, Cui JT, Tan XH, Li WM, Gu J, Lu YY. Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol Rep. 2010;23:45–52. [PubMed] [Google Scholar]
- 31.Sapkota D, Costea DE, Ibrahim SO, Johannessen AC, Bruland O. S100A14 interacts with S100A16 and regulates its expression in human cancer cells. PLoS One. 2013;8:e76058. doi: 10.1371/journal.pone.0076058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi M, Nagashio R, Saito K, Aguilar-Bonavides C, Ryuge S, Katono K, Igawa S, Tsuchiya B, Jiang SX, Ichinoe M, et al. Prognostic significance of S100A16 subcellular localization in lung adenocarcinoma. Hum Pathol. 2018;74:148–155. doi: 10.1016/j.humpath.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 34.Zeindl-Eberhart E, Brandl L, Liebmann S, Ormanns S, Scheel SK, Brabletz T, Kirchner T, Jung A. Epithelial-mesenchymal transition induces endoplasmic-reticulum-stress response in human colorectal tumor cells. PLoS One. 2014;9:e87386. doi: 10.1371/journal.pone.0087386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka M, Ichikawa-Tomikawa N, Shishito N, Nishiura K, Miura T, Hozumi A, Chiba H, Yoshida S, Ohtake T, Sugino T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer. 2015;15:53. doi: 10.1186/s12885-015-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.