Phosphorylation plays a critical role in regulating the structure and function of HISTONE DEACETYLASE15 in Arabidopsis.
Abstract
Mammalian histone deacetylases (HDACs) undergo phosphorylation to regulate their localization, activity, and function. However, little is known about the regulation of plant HDAC function and activity by phosphorylation. Here, we report the crystal structure of the Reduced Potassium Dependency3/Histone Deacetylase1 (RPD3/HDA1) type class II histone deacetylase HDA15 in Arabidopsis (Arabidopsis thaliana). The histone deacetylase domain of HDA15 (HDA15HD) assembles as tetrameric forms with each monomer composed of 12 α-helices and 9 β-sheets. The L1 loop and β2 sheet of HDA15HD are the essential interfaces for the tetramer formation. The N-terminal zinc finger domain enhances HDA15HD dimerization and increases its enzymatic activity. Furthermore, HDA15 can also be phosphorylated at Ser-448 and Ser-452 in etiolated seedlings. The HDA15 phosphorylation status determines its subnuclear localization and oligomerization. Phosphomimetics of HDA15 partially disrupt its oligomerization and cause loss of enzymatic activity and translocation from the nucleolus into nucleoplasm. Together, these data indicate that phosphorylation plays a critical role in regulating the structure and function of HDA15.
In eukaryotes, transcriptional regulation is accompanied with chromatin structure changes, which are strongly influenced by the posttranslational modification of histones, such as acetylation, methylation, phosphorylation, and ubiquitination (Berger, 2002). Histone acetylation is a reversible process, and the acetylation state of chromatin is controlled by histone acetyltransferases and histone deacetylases (HDACs). HDACs can remove acetyl groups from ε-N-acetyl Lys residues (Hollender and Liu, 2008; Liu et al., 2014). Based on phylogenetic and structural conservation, mammalian HDACs can be grouped into four classes (Gregoretti et al., 2004). Class I (HDAC1, HDAC2, HDAC3, and HDAC8), class IIa (HDAC4, HDAC5, HDAC7, and HDAC9), class IIb (HDAC6 and HDAC10), and class IV (HDAC11) are related to the yeast (Saccharomyces cerevisiae) Rpd3, Had1, and Hos3, respectively, and they belong to the zinc-dependent HDACs. Class III (Sirtuins) containing SIRT1 to SIRT7 consists of the NAD+-dependent yeast Sir2 homologs (de Ruijter et al., 2003; Verdin et al., 2003). In Arabidopsis (Arabidopsis thaliana), based on sequence similarity to mammalian HDACs, 12 RPD3/HDA1 type HDACs can be grouped into three classes: class I (HDA6, HDA7, HDA9, HDA10, HDA17, and HDA19), class II (HDA5, HDA8, HDA14, HDA15, and HDA18) and class IV (HDA2; Alinsug et al., 2009). Previous studies indicate that recombinant Arabidopsis HDA19 and HDA5 and the histone deacetylase (HD) domain of HDA18 expressed in Escherichia coli display HDAC deacetylase activity in vitro (Fong et al., 2006; Liu et al., 2013a; Luo et al., 2015). In addition, the amino acids H276, H277, D313, H315, and H316 of HDA15 are necessary for its enzymatic activity and functions in vitro and in vivo (Zhao et al., 2019).
Studies in mammals indicate that HDACs can undergo a variety of posttranslational modifications (Brandl et al., 2009; Eom and Kook, 2014). In particular, phosphorylation is an important regulatory mechanism affecting the function of HDACs. For the human (Homo sapiens) class I HDACs, HDAC1, and HDAC2, each contains homologous phosphorylated Ser residues (Pflum et al., 2001; Segré and Chiocca, 2011). Mutations of these Ser residues to Ala abolish the enzymatic activity of HDAC1/HDAC2 and reduce HDAC-mediated transcriptional repression. Another human class IIb HDAC, HDAC6, can also be phosphorylated on Tyr-570 located at the histone deacetylase domain by epidermal growth factor receptor (Deribe et al., 2009). Phosphorylation of human HDAC6 inhibits the deacetylase activity, resulting in the increased acetylation of ɑ-tubulin after EGF stimulation. Phosphorylation of mammalian HDACs can also affect the subcellular localization of HDACs. Mammalian class I HDACs are localized in the nucleus whereas class IIa HDACs shuttle between the nucleus and cytoplasm (Yang and Grégoire, 2005; Martin et al., 2007). Calcium/calmodulin-dependent protein kinaseI, II, and IV phosphorylate class IIa HDACs at the 14-3-3 binding sites, which promotes nuclear export of these HDACs (McKinsey et al., 2000; Kao et al., 2001; Chawla et al., 2003). Taken together, these studies indicate that phosphorylation regulates the function and enzymatic activity of mammalian HDACs. In Arabidopsis, two phosphorylation sites of the class I HDAC HDA6, Ser-427 and Ser-429, have been identified in the C-terminal region (Yu et al., 2017). Phosphomimetics of HDA6 increases its enzymatic activity. Furthermore, the mutation of Ser-427 of HDA6 to Ala disrupts its interaction with SUVH5 and SUVH6 (Yu et al., 2017). In addition, the MAP kinase MPK3 interacts directly with and phosphorylates the Arabidopsis HD-tuins type HDAC, HD2B, which deacetylates the Lys 9 residue of histone H3 (H3K9; Latrasse et al., 2017).
The first x-ray crystal structure of a HDAC family protein was determined for the histone deacetylase-like protein from the bacterium Aquifex aeolicus (Finnin et al., 1999). Histone deacetylase-like protein contains an α/β-fold and an eight-stranded parallel β-sheet. In human class IIa HDACs, the crystal structures of the HDAC4 and HDAC7 catalytic domains have been identified with high similarity in structure and active sites (Bottomley et al., 2008; Schuetz et al., 2008). Class IIb HDAC6 contains tandem catalytic domains (CD1 and CD2). The crystal structure and enzyme activity of zebrafish (Danio rerio) HDAC6 CD1 and CD2 revealed that CD2 has broad substrate specificity, whereas CD1 is highly specific for the hydrolysis of C-terminal acetyl-Lys substrates (Hai and Christianson, 2016). In addition, the crystal structure of zebrafish HDAC10 with the trifluoromethylketone inhibitor reveals that HDAC10 is a polyamine deacetylase with the active site conferring specificity for N8-acetylspermidine hydrolysis (Hai et al., 2017). Although the crystal structures of several HDACs have been identified in bacteria, yeast, and animals (Finnin et al., 1999; Vannini et al., 2007; Bottomley et al., 2008; Schuetz et al., 2008; Hai and Christianson, 2016; Job et al., 2016; Krämer et al., 2016; Hai et al., 2017), the plant HDAC crystal structures have not been reported yet.
Arabidopsis HDA15 belongs to the RPD3/HDA1 class II HDACs. HDA15 can be recruited by PHYTOCHROME INTERACTING FACTOR3 (PIF3) to repress chlorophyll biosynthetic and photosynthetic genes by histone H4 deacetylation in etiolated seedlings (Liu et al., 2013b). Furthermore, HDA15 can also be recruited by PIF1 to the promoter regions of light-responsive genes involved in multiple hormonal signaling pathways and cellular processes to repress their expression by decreasing the histone H3 acetylation levels in germinating seeds in the dark (Gu et al., 2017). To gain further insight into the function of HDA15, we solved the crystal structure of the HDA15 histone deacetylase domain (HDA15HD). We found that HDA15 is a zinc-dependent HDAC and the HDA15HD assembles as tetrameric forms with each monomer composed of 12 α-helices and nine β-sheets. In addition, the N-terminal zinc finger domain enhances HDA15HD dimerization and enzymatic activity. Furthermore, HDA15 could undergo phosphorylation in etiolated seedlings. Two important phosphorylation sites, Ser-448 and Ser-452, are identified in HDA15. Interestingly, phosphomimetics of HDA15 result in loss of HDAC activity. Moreover, HDA15 phosphomimetics disrupt its oligomerization and result in HDA15 translocation from the nucleolus into the nucleoplasm in vivo.
RESULTS
The Zinc Finger Domain Enhances HDA15 Histone Deacetylase Domain Dimerization with High Catalytic Efficiency in Vitro
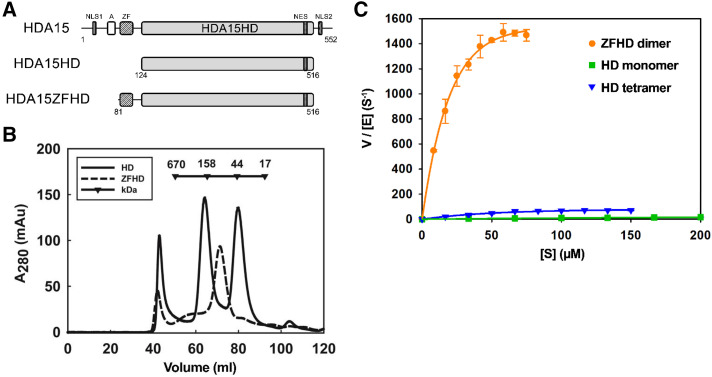
Arabidopsis HDA5, HDA18, HDA14, and HDA15 have high sequence similarities to the human class IIb HDACs, HDAC6, and HDAC10 (Supplemental Fig. S1A; Kumar et al., 2018). To identify the histone deacetylase domain, we performed the sequence alignment of Arabidopsis HDA5, HDA18, and HDA15 with the catalytic domain 2 (CD2) of human HDAC6 and the polyamine deacetylase domain (PDAC) of HDAC10 (Hai and Christianson, 2016; Hai et al., 2017). We identified the region containing amino acid residues 1 to 402 in HDA5, residues 34 to 435 in HDA18, and residues 124 to 516 in HDA15 as the histone deacetylase domain (HD; Fig. 1A; Supplemental Fig. S1B).
Figure 1.
The zinc finger domain enhances HDA15HD dimerization and enzymatic activity in vitro. A, Schematic structure of HDA15 protein domains. A, Asp-rich region; HD, histone deacetylase domain; NLS, nuclear localization signal; ZF, zinc finger domain. B, Size exclusion chromatography of the HDA15HD monomer (42 kD), tetramer (168 kD), and HDA15ZFHD dimer (94 kD). C, Enzyme kinetics of the HDA15HD monomer, tetramer and HDA15ZFHD dimer. The enzyme concentrations [E] of HD monomer, tetramer and HDA15ZFHD dimer were 75, 25, and 2.5 nm, respectively. V indicates the initial velocity (μm s−1). The values represent the mean (± sd) of three replicates.
We generated recombinant proteins of Arabidopsis HDA5, HDA18, and HDA15 histone deacetylase domains (HDA5HD, HDA18HD, and HDA15HD) in E. coli by using size exclusion chromatography. HDA5HD and HDA18HD were in monomer forms (HDA5HD, 45 kD; HDA18HD, 44 kD; Supplemental Fig. S2). Interestingly, HDA15HD contained two forms: the monomer (42 kD) and tetramer (168 kD) forms (Fig. 1B). The aggregation form of HDA15HD was larger than 670 kD. To further understand the catalytic property of these oligomerization forms, we performed enzyme kinetics analysis. The HDA5HD, HDA18HD, and HDA15HD monomers showed a low catalytic efficiency (HDA5HD, kcat/Km = 0.28 μm−1 S−1; HDA18HD, kcat/Km = 0.2 μm−1 S−1, and HDA15HD kcat/Km = 2.22 μm−1 S−1), whereas the HDA15HD tetramer showed a higher catalytic efficiency (kcat/Km = 7.37 μm−1 S−1; Fig. 1C; Table 1). Although the catalytic efficiencies of Arabidopsis HDA5HD, HDA18HD, and HDA15HD were very low, their values were at least 6-fold higher than that of human or zebrafish HDACs (Maltose binding protein[MBP]-hHDAC6 CD2, kcat/Km = 0.00091 μm−1 S−1; zHDAC6 CD2, kcat/Km = 0.031 μm−1 S−1) in vitro (Hai and Christianson, 2016).
Table 1. Arabidopsis class II HDACs oligomerization and enzyme kinetics.
| Proteins | Oligomer State | [E] | Vmax | Km | kcat | kcat/Km |
|---|---|---|---|---|---|---|
| nm | μm s−1 | μm | S−1 | μm−1 S−1 | ||
| Wild type | ||||||
| HDA5HD | Monomer | 25 | 0.65 ± 0.06 | 91.5 ± 26.0 | 26 ± 2.4 | 0.28 |
| HDA18HD | Monomer | 25 | 0.25 ± 0.01 | 47.7 ± 7.63 | 10 ± 0.4 | 0.2 |
| HDA15HD | Monomer | 75 | 2.31 ± 0.85 | 13.95 ± 4.15 | 30.82 ± 11.28 | 2.22 |
| HDA15HD | Tetramer | 25 | 1.02 ± 0.13 | 6.09 ± 1.49 | 40.73 ± 5.32 | 7.37 |
| HDA15ZFHD | Dimer | 2.5 | 3.9 ± 0.49 | 13.75 ± 0.89 | 1558.08 ± 97.71 | 113.78 |
| Interface mutation | ||||||
| HDA15ZFHD E163A | Dimer | 2.5 | 3.01 ± 0.2 | 20.02 ± 4.24 | 1205.83 ± 79.5 | 63.35 |
| HDA15ZFHD E165A | Dimer | 2.5 | 3.42 ± 0.45 | 14.5 ± 1.87 | 1367.99 ± 79.66 | 95.61 |
| HDA15ZFHD Q169A | Dimer | 2.5 | 3.09 ± 0.15 | 19.02 ± 0.99 | 1236.56 ± 58.84 | 65.25 |
| HDA15ZFHD Y228A | Dimer | 2.5 | 3.25 ± 0.14 | 18.59 ± 3.39 | 1299.43 ± 57.21 | 71.83 |
| HDA15ZFHD Y230A | Dimer | 2.5 | 2.41 ± 0.25 | 21.94 ± 3.25 | 964.74 ± 99.34 | 44.45 |
| HDA15ZFHD Y228Y230A | Monomer | 2.5 | 1.93 ± 0.03 | 35.65 ± 4.56 | 772.95 ± 11.58 | 22.02 |
| HDA15ZFHD E173A | Monomer | 25 | 1.49 ± 0.45 | 26.29 ± 6.83 | 59.42 ± 17.85 | 2.3 |
| HDA15ZFHD R177A | Monomer | 625 | 2.39 ± 0.15 | 75.32 ± 24.64 | 3.83 ± 0.24 | 0.06 |
| Phosphorylation mutation | ||||||
| HDA15ZFHD S448D | Monomer | 25 | 4.48 ± 0.14 | 13.35 ± 1.5 | 179.2 ± 5.6 | 13.42 |
| Dimer | 2.5 | 2.58 ± 0.16 | 35.48 ± 5.04 | 1032 ± 64 | 29.08 | |
| HDA15ZFHD S452D | Monomer | 25 | 5.26 ± 0.12 | 11.71 ± 1.0 | 210.4 ± 4.8 | 17.96 |
| Dimer | 2.5 | 3.08 ± 0.08 | 11.65 ± 1.32 | 1232 ± 32 | 105.75 | |
| HDA15ZFHD S448S452D | Monomer | 25 | 3.89 ± 0.17 | 27.39 ± 3.1 | 155.6 ± 6.8 | 5.68 |
| Dimer | 25 | 3.5 ± 0.09 | 21.17 ± 1.69 | 140 ± 3.6 | 6.61 | |
| HDA15ZFHD S448A | Dimer | 2.5 | 4.52 ± 0.17 | 18.12 ± 2.16 | 1808 ± 68 | 99.78 |
| HDA15ZFHD S452A | Dimer | 2.5 | 4.69 ± 0.12 | 12.13 ± 1.27 | 1876 ± 48 | 154.66 |
| HDA15ZFHD S448S452A | Dimer | 2.5 | 4.68 ± 0.2 | 17.58 ± 2.44 | 1872 ± 80 | 106.48 |
HDA15 contains a zinc finger domain of Ran-Binding Protein 2 (RanBP2; Fig. 1A; Singh et al., 1999). Interestingly, the histone deacetylase domain of HDA15 with zinc finger domain (HDA15ZFHD) was in a dimer form (94 kD) and the aggregation form (>670 kD; Fig. 1B). Furthermore, the catalytic efficiency of the HDA15ZFHD dimer (kcat/Km = 113.78 μm−1 S−1) was more than 15-fold higher than that of the HDA15HD tetramer (Fig. 1C; Table 1), suggesting that the zinc finger domain is important for HDA15 enzymatic activity in vitro. The zinc finger domain may stabilize the HDA15HD structure by dimerization to support its high enzyme activity.
Crystal Structure of the HDA15HD Tetramer
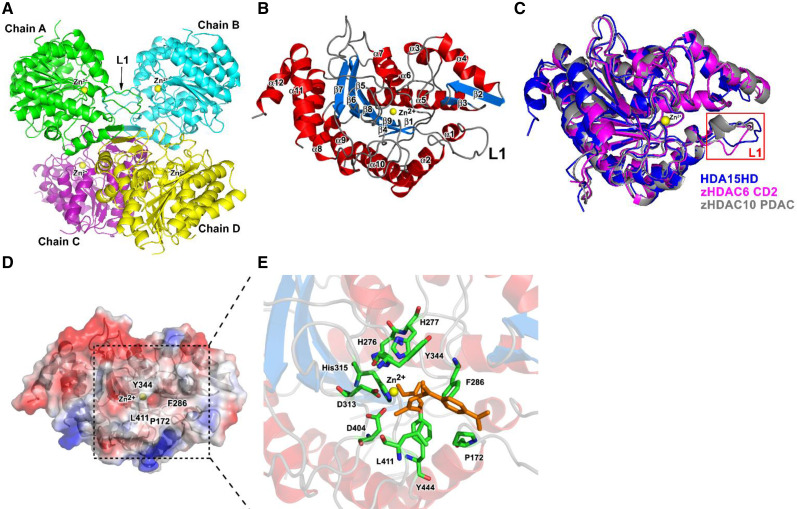
To further investigate the molecular structure of HDA15 in vitro, we determined the crystal structure of the HDA15HD tetramer at 2.36 Å resolution. The crystals of HDA15HD formed a monoclinic space group of P21 with cell dimension a = 60.8 Å, b = 78.8 Å, c = 176.8 Å, α = 90°, β = 94.2°, and γ = 90°. The diffraction data are shown in Table 2. There were four molecules as a tetramer indicated by chain A, B, C, and D in the asymmetric unit (Fig. 2A). The tetrameric quaternary structure significantly influences the accessibility of the active sites. The L1 loops (residues 161–174 of HDA15) of chain A and chain B were very close to the active site of each other, which may affect the activity of chain A and B. This may explain why the catalytic efficiency of the HDA15HD tetramer was lower than that of the HDA15ZFHD dimer. Overall structure of the HDA15HD monomer showed 12 α-helices and 9 β-sheets (Fig. 2B). The enzyme core adopted an open α/β-fold that is characteristic of the HDAC family (Finnin et al., 1999; Somoza et al., 2004).
Table 2. Diffraction and refinement data for the HDA15HD tetramer.
Statistics for the highest-resolution shell are shown in parentheses.
| Crystals | HDA15HD Tetramer |
|---|---|
| Beamline | NSRRC TPS 05A |
| Wavelength (Å) | 1.28198 |
| Data collecting and processing | |
| Space group | P 1 21 1 |
| Cell dimensions (Å and °) | 60.8, 78.8, 176.8 90, 94.2, 90 |
| Resolution (Å; last shell) | 29.55–2.36 (2.44–2.36) |
| Completeness (%; last shell) | 99.1 (97.1) |
| I/σ (last shell) | 13.9 (2.5) |
| Reflections (total/unique) | 274,152/26,756 |
| Redundancy | 3.1 (3.0) |
| Rmerge (%; last shell) | 9.1 (47.3) |
| Refinement | |
| Resolution (Å) | 29.52–2.36 |
| Rwork (%) | 16.7 |
| Rfree (%) | 21.8 |
| No. of nonhydrogen atoms | 11,022 |
| No. of protein atoms | 10,535 |
| No. of water | 439 |
| Protein residues | 1,404 |
| Model quality | |
| R.M.S. deviations | |
| Bond length (Å) | 0.008 |
| Bond angle (°) | 0.93 |
| Average B-factor (Å) | 28.13 |
| B-factor (macromolecules) | 28.07 |
| B-factor (ligands) | 45.62 |
| B-factor (solvent) | 27.53 |
| Ramachandranplot (%) | |
| Most favored | 95.68 |
| Allowed | 3.96 |
| Disallowed | 0.36 |
Figure 2.
Crystal structure of the HDA15HD tetramer and monomer. A, Ribbon diagram representation of the crystal structure of the HDA15HD tetramer in a substrate-free form. Zinc ions are represented by yellow spheres. L1, Loop structure (residues 161 to 174 of HDA15). B, HDA15HD monomer. The α-helices and β-sheets are colored in red and light blue, respectively. C, Superposition of HDA15HD (blue), zHDAC6 CD2 (magenta, PDB 5EEK), and zHDAC10 PDAC (gray, PDB 5TD7). The L1 loop is enclosed by a red frame. D, The active site of the HDA15HD domain is located at the center of the electrostatic surface with four conserved residues, P172, F286, Y344, and L411, around the cavity. E, The model of the HDA15HD–TSA complex showing the binding mode in the active sites with the conserved residues H276, H277, D313, H315, D404, and Y444. TSA is represented by brown.
In the tertiary structure, HDA15HD was similar to both zHDAC6 CD2 [PDB 5EEK, 0.87 Å root mean squared deviation (r.m.s.d.) for 323 Cα atoms] and zHDAC10 PDAC (PDB 5TD7, 0.91 Å r.m.s.d. for 323 Cα atoms; Fig. 2C). However, the catalytic efficiency of the HDA15HD monomer was much higher than that of human or zebrafish HDACs, which may be due to their structure difference. The L1 loop of HDA15HD was different from that of zHDAC6 CD2 (4.76 Å r.m.s.d. for 14 Cα atoms) and zHDAC10 PDAC (3.61 Å r.m.s.d. for 14 Cα atoms). One zinc ion located at the active site of HDA15HD was further identified by anomalous scattering. Four residues, P172, F286, Y344, and L411, were located at the opening region of the active site (Fig. 2D). Six inner residues in the active site of HDA15HD were H276, H277, D313, H315, D404, and Y444 (Fig. 2E). These 10 residues at the active sites were largely conserved in sequences and structures within plants and animals (Supplemental Figs. S1B and S3). Our previous study indicates that H276, H277, D313, and H315 are required for the enzymatic activity of HDA15 (Zhao et al., 2019). Furthermore, the HDAC inhibitor Trichostatin A (TSA) was modeled into the active site of HDA15HD (Fig. 2E) based on the structure of the HDAC7–TSA complex (Krissinel and Henrick, 2007). Taken together, our analysis indicated that, similar to human class II HDACs as well as class I and IV HDACs, HDA15 is also a zinc-dependent histone deacetylase.
The Interface of the HDA15ZFHD Dimer Differs from the HDA15HD Tetramer
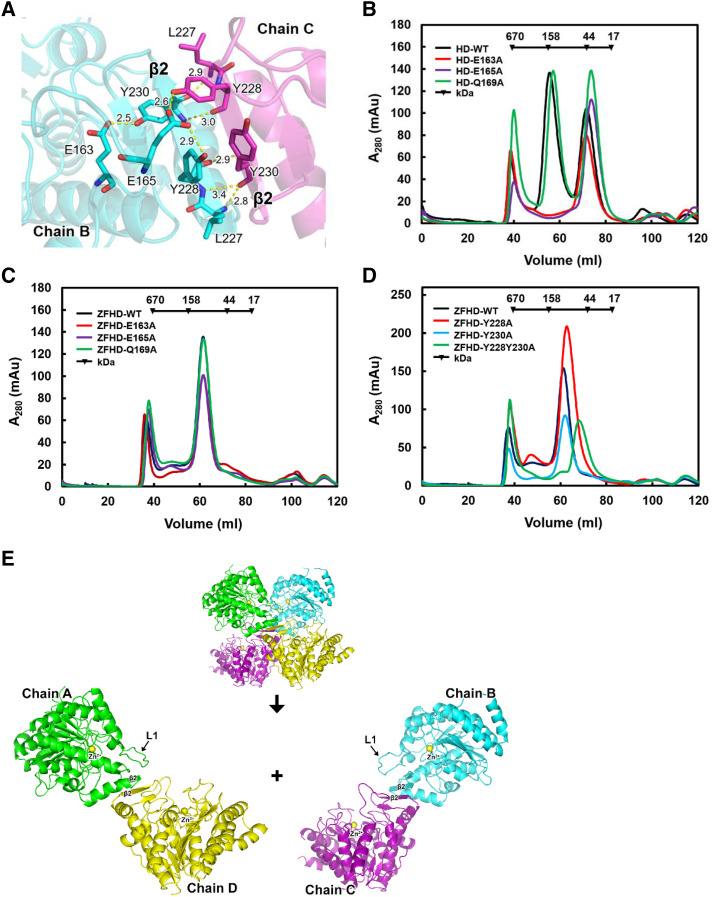
In the interface of the tetramer, two regions were located at the HDA15HD tetramer interface. The first region contained E163 and E165 in the middle of L1 loop. E165 formed hydrogen bonds with Y228 and S233 (Fig. 3A; Supplemental Fig. S4A), whereas E163 formed a hydrogen bond with Y230 (Fig. 3A). The mutations (E163A and E165A) in L1 loop of HDA15HD expressed in E. coli resulted in disruption of HDA15HD tetramers into monomers with low catalytic efficiency (Fig. 3B; Supplemental Table S1). As a control, a mutation in the L1 loop with a nonhydrogen bond amino acid (Q169A) did not affect the HDA15HD tetramer and catalytic efficiency (Supplemental Fig. S4B). However, the catalytic efficiencies of the mutants (E163A, E165A, and Q169A) in the L1 loop of HDA15ZFHD were similar to wild type (ZFHD-WT), suggesting that the HDA15ZFHD dimer is different from the HDA15HD tetramer in the L1 loop (Fig. 3C; Table 1).
Figure 3.
The zinc finger domain enhances HDA15HD dimerization and enzymatic activity in vitro. A, Ribbon diagram representation of the crystal structure of the HDA15HD tetramer interface in L1 (E163 and E165) and β2 (Y228 and Y230). Hydrogen bonds are represented by yellow dotted lines. The number indicates the length of the hydrogen bond. B, Size exclusion chromatography of HDA15HD with mutations in L1 (E163, E165, and Q169). C, Size exclusion chromatography of HDA15ZFHD with mutations in L1 (E163, E165, and Q169). D, Size exclusion chromatography of HDA15ZFHD with mutations in β2 (Y228 and Y230). E, The HDA15HD tetramer is formed by two “head-to-head” dimers (chain A/D × chain B/C). WT, wild type.
The second region of the interface includes Y228 and Y230 at β2 (Fig. 3A). The double mutations Y228A and Y230A resulted in disruption of HDA15ZFHD dimers to monomers with low catalytic efficiency (Fig. 3D; Table 1). From the HDA15HD structure, it appears that Y228 and Y230 not only formed hydrogen bonds by the main chain, the side chains of Y228 and Y230 also formed hydrogen bonds with E163 and E165. In addition, the side chains of Y228 and Y230 also formed π–π stackings (Fig. 3A). These results showed that Y228 and Y230 at β2 are necessary for HDA15ZFHD dimerization. Together, these data indicated that the HDA15HD tetramer is formed by two “head-to-head” dimers (chain A/D × chain B/C; Fig. 3E) mainly mediated by L1 loop and β2 interactions.
Comparative Protein Structure Modeling of HDA15ZFHD
To mimic the structure of the HDA15ZFHD dimer, MODELER was used to predict the three-dimensional structure of the known zinc finger structure with HDA15HD (Webb and Sali, 2016). National Center for Biotechnology Information protein BLAST analysis indicated that the chain B of the human E3 ubiquitin-protein ligase Mdm2 (PDB 4XXB; Supplemental Fig. S5A) was the closest to the HDA15 zinc finger domain (amino acids 86–123) with 30.91% identity. After model building, the zinc finger domains of chain A and B were located in the outer space of the tetramer (Supplemental Fig. S5C). However, the zinc finger domains of chain C and D were inside in the tetramer and extremely close to each other (Supplemental Fig. S5D), which may lead to the inability of HDA15ZFHD to form a tetramer structure.
HDA15HD Forms Tetramers in Vivo
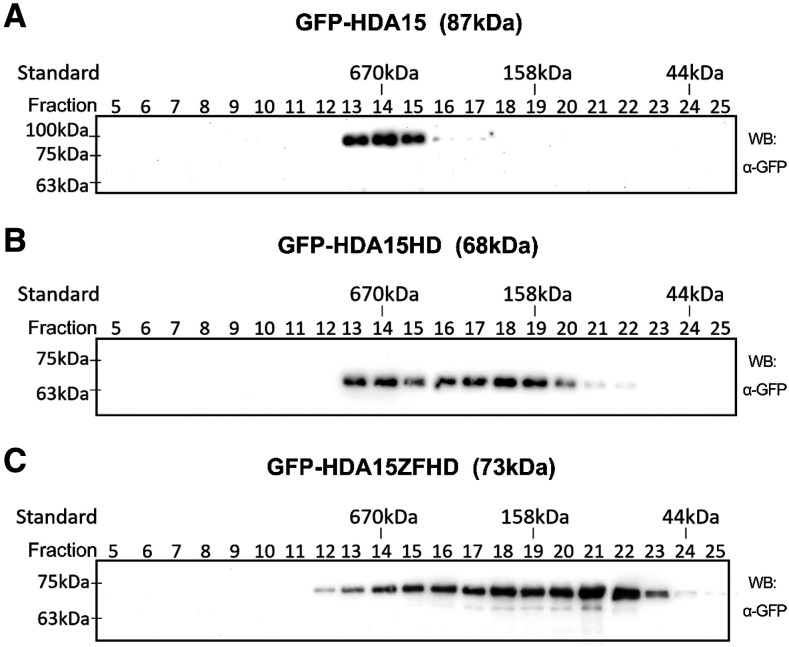
To analyze the possible oligomerization of HDA15 in vivo, HDA15, HDA15HD, and HDA15ZFHD fused with GFP driven by the 35S promoter (35S:GFP-HDA15, 35S:GFP-HDA15HD, and 35S:GFP-HDA15ZFHD) were transformed into hda15-1 mutant plants. Total proteins were extracted from transgenic plants and analyzed by size exclusion chromatography. Proteins immunoprecipitated by GFP-trap beads were further analyzed by immunoblotting using a GFP antibody. As shown in Figure 4A, GFP-HDA15 formed multiprotein complexes or aggregations at a size of ∼670 kD (fraction 13 to 15). However, no monomer (87 kD, fraction 22), dimer (174 kD, fraction 19), or tetramer (348 kD, fraction 17) forms of GFP-HDA15 were identified (Fig. 4A).
Figure 4.
HDA15HD forms tetramers in vivo. Immunoblotting of GFP-HDA15, GFP-HDA15HD, and GFP-HDA15ZFHD from size exclusion chromatography of the total protein extracts. The total protein extracts from 10-d-old seedlings of GFP-HDA15/hda15-1 (A), GFP-HDA15HD/hda15-1 (B), and GFP-HDA15ZFHD/hda15-1 (C) transgenic lines were separated and fractionated. The fractions were immunoprecipitated by GFP-trap beads and immunoblotted with a GFP antibody. WB, Western blot.
Interestingly, GFP-HDA15HD also formed multiprotein complexes with the tetramer form (272 kD, fraction 18) or aggregations (670 kD, fraction 13 and 14), but not the monomer form (68 kD, fraction 23; Fig. 4B). Moreover, GFP-HDA15ZFHD formed a series multiprotein complexes containing both the dimer (146 kD, fraction 20) and monomer (73 kD, fraction 22) forms or aggregations (fraction 12 to 19; Fig. 4C). Taken together, these results indicate that HDA15HD can only form tetramers whereas HDA15ZFHD can form both dimers and monomers in vivo.
The Zinc Finger Domain Is Essential for the Activity and Function of HDA15 in Arabidopsis
To analyze whether the histone deacetylase domain of HDA15 contains enzymatic activity in vivo, we further measured the enzyme activity of GFP-HDA15HD from transgenic plants. As shown in Figure 5A, GFP-HDA15HD did not display HDAC activity, suggesting that other parts of HDA15 are also required for its activity in vivo.
Figure 5.
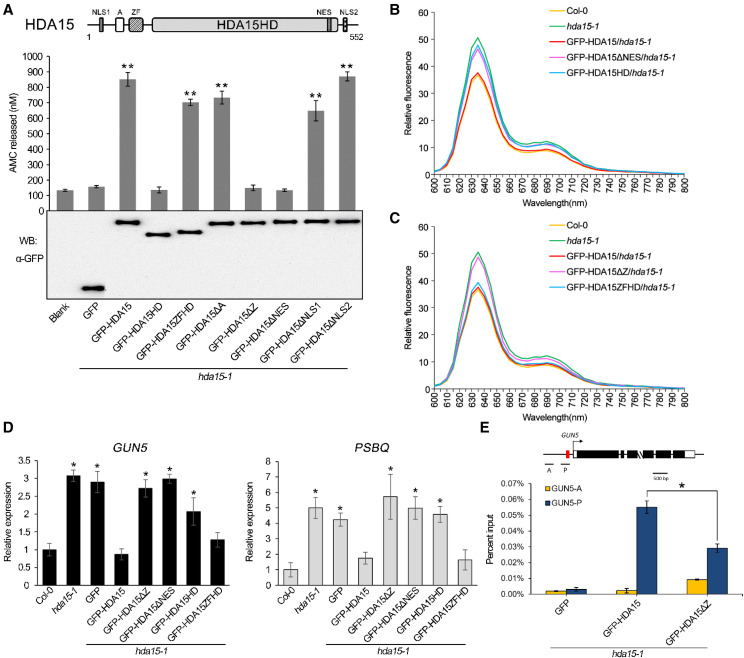
The zinc finger domain is required for the activity and function of HDA15 in Arabidopsis. A, Fluorometric HDAC activity assays of full-length and truncated GFP-HDA15. The schematic structure of HDA15 protein domains is also shown. Proteins were extracted from 10-d-old hda15-1 expressing full-length and truncated GFP-HDA15. Fluorometric HDAC activity assays were performed, and HDAC activity was indicated by AMC released (nanomolar). GFP alone was used as a negative control. Immunoblotting with the GFP antibody indicated the equal protein loading in HDAC activity assays. Data represent the mean (± sd) of three biological replicates compared with GFP alone. Asterisks indicate significant difference using Student’s t test (**P < 0.01). B and C, Relative fluorescence of protochlorophyllide in full emission wavelengths 600 to 800 nm in etiolated transgenic plants expressing full-length and truncated GFP-HDA15. Protochlorophyllide was extracted from 4-d-old etiolated seedlings. D, RT-qPCR analysis of GUN5 and PSBQ expression in 2-d-old etiolated seedlings of Col-0, hda15-1, and transgenic plants expressing full-length and truncated GFP-HDA15. UBQ10 was used as an internal control. E, ChIP-qPCR analysis of the enrichment of GFP-HDA15 and GFP-HDA15ΔZ at P and A regions of GUN5 in etiolated transgenic seedlings. Red box indicates the G-box element (CACGTG). The GFP antibody was used for immunoprecipitation. Data are shown as percentage of input. The values represent the mean (± sd) of three biological replicates. Asterisks indicate significant difference using Student’s t test (*P < 0.05). WB, Western blot.
In addition to the zinc finger domain, HDA15 also contains an Asp-rich region (A), a nuclear export signal (NES) and two nuclear localization signals (Fig. 1A; Alinsug et al., 2009). Different HDA15 deletions fused with GFP were also transformed into hda15-1 mutant plants (Supplemental Fig. S6). Because the NES is located within the histone deacetylase domain, HDA15 without NES (GFP-HDA15ΔNES) showed no enzymatic activity. Interestingly, although GFP-HDA15ZFHD had strong HDAC activity, HDA15 without zinc finger (GFP-HDA15ΔZ) also showed no HDAC activity (Fig. 5A). These results indicate that the zinc finger domain is essentially important for the activity of HDA15 in vivo.
HDA15 negatively regulates chlorophyll biosynthesis in the dark, and etiolated hda15 seedlings have a higher protochlorophyllide content compared to wild type (Liu et al., 2013b). Furthermore, the expression of GUN5 (a gene involved in chlorophyll biosynthesis) and PSBQ (a gene involved in photosynthesis) is also increased in the etiolated hda15 seedlings (Liu et al., 2013b). The protochlorophyllide content and the expression level of GUN5 and PSBQ in etiolated hda15-1 seedlings expressing truncated HDA15 were measured. Consistent with the loss of HDAC activity, GFP-HDA15ΔNES and GFP-HDA15HD could not rescue the hda15 mutant phenotype in the protochlorophyllide content as well as GUN5 and PSBQ expression (Fig. 5, B and D). Similarly, GFP-HDA15ΔZ also could not rescue the hda15 mutant phenotype. On the contrary, GFP-HDA15ZFHD rescued the hda15 phenotype (Fig. 5, C and D).
Previous studies indicate that the N-terminal domain of HDA15 (amino acids 1–146) that contains the zinc finger domain is responsible for its interaction with transcriptional factor PIF3 (Liu et al., 2013b). Moreover, PIF3 recruits HDA15 to the promoters of chlorophyll biosynthetic and photosynthetic genes to repress their transcription by histone deacetylation (Liu et al., 2013b). To further reveal the function of the zinc finger domain of HDA15 in vivo, we used chromatin immunoprecipitation (ChIP) assays to analyze whether the zinc finger domain of HDA15 is important for its binding to the promoter region of GUN5. As shown in Figure 5E, the binding of GFP-HDA15ΔZ to the promoter region of GUN5 decreased compared with GFP-HDA15, indicating that the zinc finger domain is important for the binding of HDA15 to its targets. Taken together, these data indicate that the zinc finger domain is required for the enzymatic activity and function of HDA15 in vivo.
HDA15 can be Phosphorylated in Arabidopsis Etiolated Seedlings
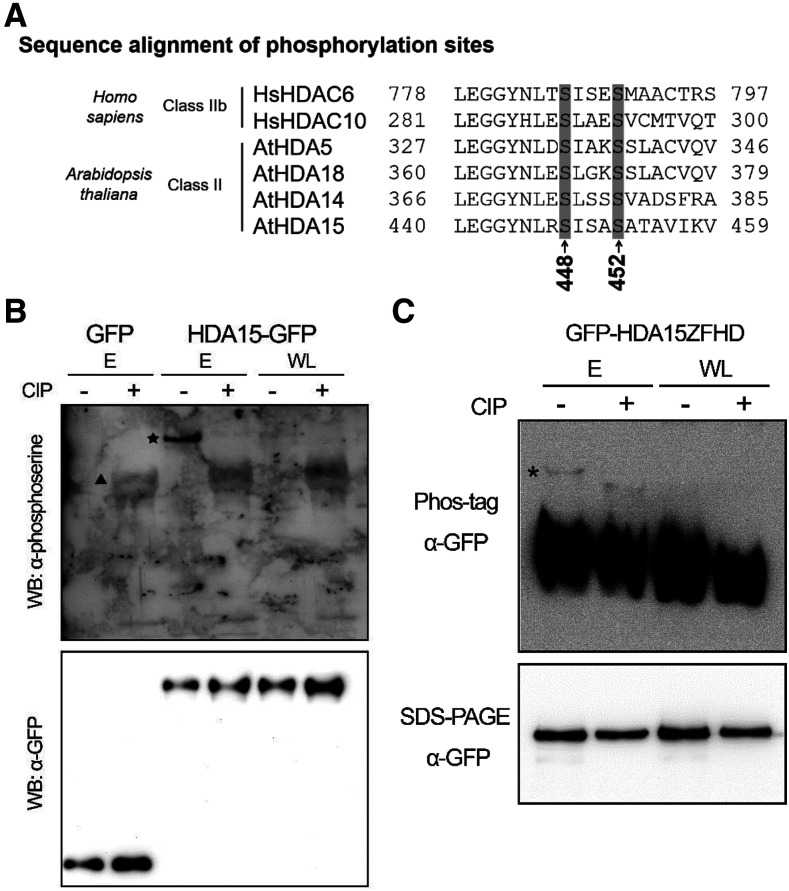
We used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to analyze whether HDA15 contains posttranslational modifications in vivo. For LC-MS/MS analysis, endogenous HDA15 immunoprecipitated by the HDA15 antibody was isolated from SDS-PAGE using etiolated seedlings of Col-0 wild type. After Mascot analysis, 10 phosphorylation sites were identified in the HDA15 protein (Supplemental Fig. S1B; Supplemental Table S2). Interestingly, the most abundant sites were S448, S450, and S452, which were found in all replicates. Moreover, these three phosphorylation sites were all located in the C terminus of the HD domain. Previously, phosphorylation sites were also identified in the HD domain of the human HDACs, including HDAC8, HDAC5, and HDAC6 (Supplemental Fig. S7; Lee et al., 2004; Deribe et al., 2009). Interestingly, sequence alignment of HDA15 with other HDACs revealed that the Arabidopsis class II HDACs, HDA5, HDA18, and HDA14, and human class IIb HDACs, HDAC6, and HDAC10, all contain the two conserved amino acids corresponding to S448 and S452 of HDA15 (Fig. 6A; Supplemental Fig. S1B). In contrast, Arabidopsis class I HDACs HDA6 and HDA19, class II HDAC HDA8, and human class I, class IIa, and class IV HDACs do not have these two conserved serines (Supplemental Fig. S8).
Figure 6.
HDA15 can be phosphorylated in Arabidopsis etiolated seedlings. A, Sequence alignment of the HDA15 phosphorylation sites S448 and S452 with human and Arabidopsis HDACs. B, Immunoblotting of HDA15-GFP with the phospho-Ser antibody. HDA15-GFP immunoprecipitated from PromoterHDA15:HDA15-GFP/hda15-1 transgenic plants in 2-d-old etiolated seedlings (E) and 10-d-old seedlings grown under WL were analyzed by the phospho-Ser antibody after CIP treatment. GFP immunoprecipitated from PromoterHDA15:GFP/hda15-1 transgenic plants in 2-d-old E was used as a negative control. Star indicates the phosphorylated HDA15-GFP. Triangle indicates the phosphatase CIP. Immunoblotting with the GFP antibody indicates the input protein loading. C, Phos-tag gels of GFP-HDA15ZFHD. GFP-HDA15ZFHD immunoprecipitated from 35S:GFP-HDA15ZFHD/hda15-1 transgenic plants in 2-d-old E and 10-d-old seedlings grown under WL were analyzed by the Phos-tag gels or SDS-PAGE after CIP treatment. The asterisks in the Phos-tag gel indicate the phosphorylated GFP-HDA15ZFHD. CIP treatment resulted in GFP-HDA15ZFHD dephosphorylation (the band shifted down in the Phos-tag gels). WB, Western blot.
To further investigate the phosphorylation of HDA15, we generated the PromoterHDA15:HDA15-GFP/hda15-1 transgenic plants, in which HDA15-GFP driven by the HDA15 native promoter was expressed in hda15-1 plants. HDA15-GFP was immunoprecipitated from PromoterHDA15:HDA15-GFP/hda15-1 transgenic plants and analyzed by the phospho-Ser antibody after in vitro phosphatase treatment. The phosphorylated HDA15-GFP could be observed through the phospho-Ser antibody in mock etiolated seedlings (no phosphatase) and the nonspecific phosphatase, calf intestinal phosphatase (CIP), completely dephosphorylated HDA15-GFP (Fig. 6B). However, the phosphorylated HDA15-GFP could not be detected in the 10-d-old seedlings grown under white light (WL) conditions.
Phos-tag gels were performed to analyze the phosphorylation status of GFP-HDA15ZFHD after in vitro phosphatase treatment. One band representing phosphorylated GFP-HDA15ZFHD could be observed in etiolated seedlings (Fig. 6C), which shifted down when incubated with CIP (indicated by an asterisk in Fig. 6C). However, no band shift was found when plants were grown in the WL conditions, indicating that HDA15 could be phosphorylated in etiolated seedlings but not in the 10-d-old seedlings grown under WL conditions. Taken together, these results indicate that HDA15 can be phosphorylated in vivo and its phosphorylation status can be regulated upon different developmental stages or in different environmental conditions.
Ser-448 and Ser-452 Phosphorylation Affects the Enzymatic Activity and Oligomerization of HDA15
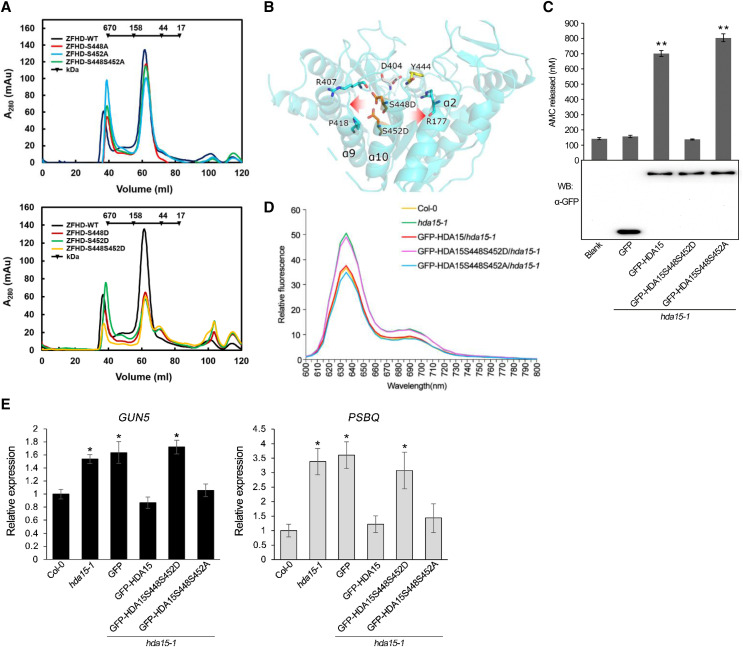
To reveal whether phosphorylation affects histone deacetylase activity of HDA15, we generated the HDA15ZFHD mutations S448D and S452D (to mimic phosphorylation) or S448A and S452A (nonphosphorylated amino acid) and expressed these in E. coli. Size exclusion chromatography analysis indicated that HDA15ZFHD S448A, S452A, and S448S452A were in dimer forms with strong catalytic efficiency (Fig. 7A; Table 1). Although phosphomimetics of HDA15ZFHD (S448D, S452D, and S448S452D) still occurred in dimer forms, a small portion of them also occurred in monomer forms (Fig. 7A). However, with the exception of the HDA15ZFHD S452D dimer, the catalytic efficiencies of other phosphomimetic HDA15ZFHD dimers and monomers were very low, which was similar to the HDA15HD tetramer (Table 1). These data indicated that S448 and S452 are two phosphorylation sites important for enzymatic activity and oligomerization in vitro. The decreased enzyme activity of the phosphomimetics of HDA15ZFHD may result from the dimer structure change and the transition of the dimer into the monomer.
Figure 7.
Phosphorylation of HDA15 on Ser-448 and Ser-452 is critical for enzymatic activity and oligomerization. A, Size exclusion chromatography of the phosphorylation site mutants of HDA15ZFHD. B, The ribbon diagram model showing how phosphomimetics of HDA15 on S448 and S452 may regulate its enzymatic activity and oligomerization. C, Fluorometric HDAC activity assays of the phosphorylation site mutants of GFP-HDA15 in Arabidopsis. Proteins were extracted from 10-d-old etiolated hda15-1 seedlings expressing truncated HDA15. Immunoblotting with the GFP antibody indicated the protein loading in HDAC activity assays. Data represent the mean (± sd) of three biological replicates. Asterisks indicate significant difference using Student’s t test (**P < 0.01). D, Relative fluorescence of protochlorophyllide in full emission wavelengths 600 to 800 nm in etiolated transgenic plants expressing phosphorylation site mutants of GFP-HDA15. Protochlorophyllide was extracted from 4-d-old etiolated seedlings. E, RT-qPCR analysis of GUN5 and PSBQ expression in 2-d-old etiolated transgenic seedlings expressing phosphorylation site mutants of GFP-HDA15. UBQ10 was used as an internal control. Data represent the mean (± sd) of three biological replicates. Asterisks indicate significant difference using Student’s t test (*P < 0.05). WT, wild type.
According to the HDA15HD tetramer crystal structure, the side chain lengths of S448 and S452 were 2.4 Å (Supplemental Fig. S9A). In the simulation diagram, the side chain lengths of the phosphomimetics on S448 and S452 (S448D and S452D) were 3.6 and 3.7 Å, respectively (Supplemental Fig. S9B). Moreover, the side chain lengths of the phosphorylation on S448 and S452 were speculated to be 4.5 to 4.9 Å (Supplemental Fig. S9C). These data suggest that the side chain length of S448 and S452 may increase when being phosphorylated. The increased side chain might push the neighbor helix structures of α9 (near active site D404 and Y444) and α2 (near active site R177), which may affect enzyme activity and oligomerization (Fig. 7B).
Phosphomimetics of HDA15 on Ser-448 and Ser-452 Result in a Loss of HDAC Activity and Functions in Arabidopsis
To further identify the function of HDA15 phosphorylation in vivo, we generated 35S:GFP-HDA15S448S452D and 35S:GFP-HDA15S448S452A transgenic plants, in which S448 and S452 were mutated to D and A, respectively, in the hda15-1 mutant background. Similar with the in vitro data, GFP-HDA15S448S452D had no HDAC activity, whereas GFP-HDA15S448S452A retained HDAC activity (Fig. 7C). Furthermore, GFP-HDA15S448S452D could not rescue the hda15 phenotype in both protochlorophyllide biosynthesis and the expression of GUN5 and PSBQ (Fig. 7, D and E). In contrast, GFP-HDA15S448S452A rescued the hda15 mutant phenotype. Taken together, these data support that the phosphomimetics of HDA15 on S448 and S452 result in a loss of HDAC activity and functions in Arabidopsis.
Phosphomimetics of HDA15 Disrupt its Oligomerization and Result in Translocation from the Nucleolus into Nucleoplasm
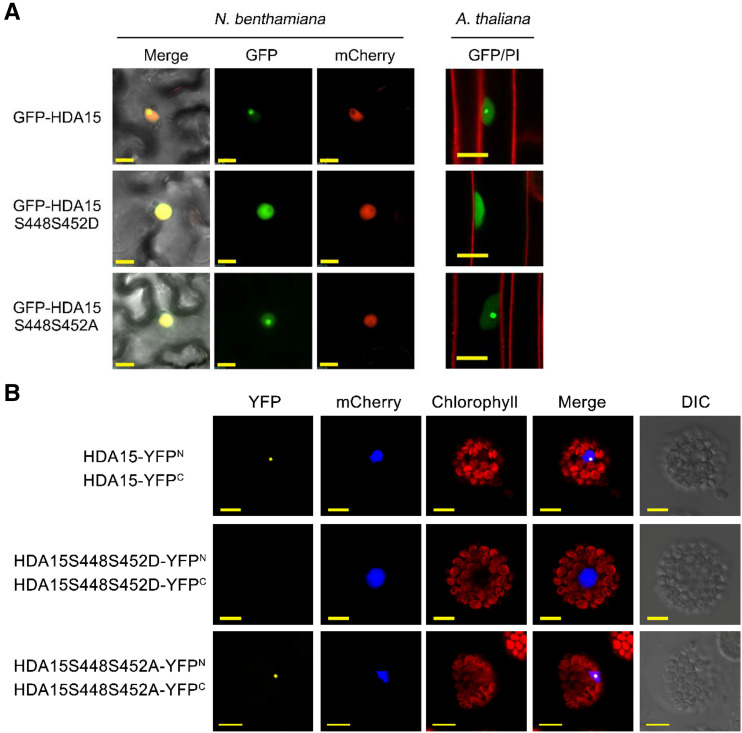
Mammalian class IIa HDACs shuttle between the nucleus and cytosol relying on the phosphorylation status (Ha et al., 2010; Mihaylova and Shaw, 2013). We further investigated whether HDA15 phosphorylation affects its subcellular localization in Nicotiana benthamiana and transgenic Arabidopsis. HDA15 localized in the nucleus with a strong signal in the nucleolus (Fig. 8A). Interestingly, mutations of S448 and S452 into phosphomimetics, S448S452D, resulted in the translocation of HDA15 from the nucleolus to the nucleoplasm. In comparison, mutations to Ala did not affect the nucleolus localization of HDA15. These results suggest that phosphomimetics of HDA15 in S448 and S452 contribute to the translocation of HDA15 from the nucleolus to the nucleoplasm.
Figure 8.
Subcellular localization and oligomerization of HDA15 phosphorylation status. A, Subcellular localization of GFP-HDA15 and mutants with S448 and S452 changed into Asp (D) or Ala (A) in Nicotiana benthamiana (scale bar = 15 μm) and Arabidopsis (scale bar = 10 μm). mCherry carrying a nuclear localization signal was used as the nuclear marker. Cell walls were stained by propidium iodide. B, bimolecular fluorescence complementation (BiFC) assays of Arabidopsis protoplasts showing the oligomerization of HDA15 and the mutants with S448 and S452 changed into Asp (S448S452D) or Ala (S448S452A). Wild-type (WT) and mutated HDA15 fused with the C terminus (YFPC) or the N terminus (YFPN) of YFP were cotransfected into protoplasts and visualized by confocal microscopy. Scale bar = 10 μm. DIC, Differential interference contrast.
HDA15ZFHD is in a dimer form and phosphomimetics of HDA15ZFHD partially turn it into the monomer form in vitro. We further analyzed the relationship between phosphorylation, oligomerization, and subcellular localization by using bimolecular fluorescence complementation assays in Arabidopsis protoplasts. Fluorescence signals were observed in the nucleolus from wild-type HDA15 and S448S452A mutants but not from phosphomimetics HDA15 (S448S452D; Fig. 8B). Taken together, these data indicate that HDA15 could form oligomers in the nucleolus. Phosphomimetics in HDA15 may affect its oligomerization and result in its translocation from the nucleolus into nucleoplasm.
DISCUSSION
HDA15 Is a Zinc-Dependent Enzyme and its Histone Deacetylase Domain Assembles as Tetrameric Forms
To date, the crystal structures of several HDACs have been reported in bacteria, yeast, and animals (Vannini et al., 2007; Schuetz et al., 2008; Hai and Christianson, 2016; Job et al., 2016; Krämer et al., 2016; Hai et al., 2017). Structural comparisons among different mammalian class I, II, IV HDACs, as well as HDAC homologs from other species, indicate that they are all zinc-dependent and share significant sequence similarity, suggesting that these HDACs contain a conserved group of active sites and a common mechanism for acetylating substrates (Seto and Yoshida, 2014; Yoshida et al., 2017). D198, D291, and H200 of Arabidopsis HDA5 are the conserved residues responsible for chelating zinc ions, and mutations of these conserved residues to Ala result in loss of histone deacetylase activity, indicating that HDA5 is a zinc-dependent enzyme (Luo et al., 2015). In this study, we solved the x-ray crystal structure of the histone deacetylase domain of the Arabidopsis RPD3/HDA1 type class II HDAC, HDA15, which assembles as tetrameric forms. Four residues (P172, F286, Y344, and L411) located at the opening region of the active site of HDA15 and six inner residues (H276, H277, D313, H315, D404, and Y444) in the active site of HDA15 are largely conserved in plant and animal HDACs. One zinc ion located at the active site was identified, suggesting that HDA15 is also a zinc-dependent histone deacetylase. Interestingly, the L1 loop and β2 sheet are essential interfaces for the tetramer formation, and mutations of the critical residues in the loop disrupt the HDA15 tetramer to the monomer. Because the amino acid sequence and structure of the L1 loop in HDA15 are different from zebrafish HDAC6 and HDAC10, the formation of tetramers may be a unique structure feature for HDA15. The crystal structure of PA3774, a histone deacetylase homolog (HDAH) from Pseudomonas aeruginosa, was also identified as a tetramer composed of “head-to-head” dimers (Krämer et al., 2016). The L1 loop of PA3774 is necessary for the formation of the tetramer structure. However, both the L1 loop and β2 sheet are essential for the tetramer formation of HDA15HD. The tetrameric quaternary structure significantly influences the accessibility of the active site and may represent a crucial determinant of molecular recognition and substrate selectivity. This indicates that the evolutionary trends of HDACs between species tend to use oligomers to modulate biological functions.
The Zinc Finger Domain Is Important for HDA15 Dimerization, Enzymatic Activity, and Function
Human HDAC1 and HDAC2 could form homo- and heterodimers with themselves or with each other, and dimer formation is required for their HDAC activity (Taplick et al., 2001; Brunmeir et al., 2009). The N terminus of human HDAC1 is important for the formation of homodimers and heterodimers with HDAC2 and HDAC3 and for catalytic activity of the enzyme (Taplick et al., 2001). Moreover, HDAC3 can also form homodimers and homotrimers in vitro and in vivo (Yang et al., 2002). Previous studies indicated that several types of zinc-finger domains are required for protein dimerization (Payre et al., 1997; Wang and Pabo, 1999). In human HDACs, only HDAC6 contains a zinc finger ubiquitin binding domain (ZnF-UBD), which is necessary for recruiting polyubiquitinated protein aggregation and loading misfolded proteins to the lysosome (Hook et al., 2002; Kawaguchi et al., 2003). Truncating ZnF-UBD from HDAC6 enhances its enzymatic activity (Miyake et al., 2016). Arabidopsis HDA15 contains a zinc finger domain of RanBP2 (Singh et al., 1999). Interestingly, the RanBP2 type zinc finger domain of HDA15 is important for its dimerization and enzymatic activity. Deletion of the zinc finger in HDA15 results in loss of enzymatic activity and decreased binding ability to target genes. The zinc finger domain in HDA15 may stabilize its structure by dimerization to increase the enzymatic activity and function.
Phosphorylation on Ser-448 and Ser-452 of HDA15 Is Conserved and Functionally Important
Studies from mammals indicate that HDACs undergo various posttranslational modifications to regulate HDAC activity, stability, localization, and protein-protein interactions (Sengupta and Seto, 2004; Dokmanovic et al., 2007). The human class I HDACs, HDAC1 and HDAC2, are phosphorylated at multiple serines in the C-terminal region (Pflum et al., 2001; Tsai and Seto, 2002; Sun et al., 2007). In this study, we found that Arabidopsis HDA15 could be phosphorylated in etiolated seedlings and contains two important phosphorylation sites at Ser-448 and Ser-452. Phosphomimetics of HDA15 (S448D and S452D) cannot rescue the hda15 mutant phenotype due to the loss of enzymatic activity, indicating that the phosphorylation of HDA15 is important for its function.
Similar to Arabidopsis HDA15, phosphomimetics of human HDAC8 and HDAC6 also inhibit deacetylase activity (Lee et al., 2004; Deribe et al., 2009). The phosphorylation sites of human HDAC8 and HDAC6 as well as Arabidopsis HDA15 are all located in the HD domain. Therefore, phosphorylation in the HD domain in these HDACs may affect their structure and/or substrate binding. The crystal structure of HDAC8 reveals that Ser-39 phosphorylation of HDAC8 may induce a conformation change preventing the substrate binding (Somoza et al., 2004). The crystal structure of HDA15 also suggests that phosphorylation of S448 and S452 might enhance the side chain length to push the neighbor α9 (near active site D404 and Y444) and α2 (near active site R177) into the active site and interface, which disrupts enzyme activity and dimerization. The human class IIa HDAC5 also contains a phosphorylation site (Ser-755) in the HD domain. However, the mutation of this Ser does not affect HDAC activity because Ser-755 is located distal to the catalytic pocket of HDAC5 (Greco et al., 2011).
Several phosphorylation sites are highly conserved in HDACs. The human class I HDACs, HDAC1 and HDAC2, possess extensive amino acid sequence homology and can be phosphorylated in Ser-421 and Ser-423 for HDAC1 and Ser-422 and Ser-424 for HDAC2 (Pflum et al., 2001; Tsai and Seto, 2002; Segré and Chiocca, 2011). Interestingly, the conserved phosphorylation motif of human HDAC1 and HDAC2 is also found in the Arabidopsis RPD3/HDA1 class I HDAC, HDA6 (Yu et al., 2017). Phosphorylation in Ser-427 and Ser-429 of HDA6 promotes its enzyme activity and affects its interaction with other proteins (Yu et al., 2017). The N-terminal domain on the human class IIa HDACs also contains conserved Ser residues that could be phosphorylated (Mihaylova and Shaw, 2013; Eom and Kook, 2014). Moreover, the Arabidopsis HD-tuins type HDAC, HD2B, can be phosphorylated by the MAP kinase MPK3 in plant innate immunity (Latrasse et al., 2017). In this study, we find that the Arabidopsis RPD3/HDA1 class II HDAC, HDA15, contains two phosphorylation residues at Ser-448 and Ser-452, which are also conserved in the Arabidopsis class II HDACs, HDA5, HDA18, and HDA14 as well as the human class IIb HDACs, HDAC6, and HDAC10. It remains to be determined whether these conserved serines in human class IIb HDACs can also be phosphorylated and are functionally important.
Phosphomimetics of HDA15 Regulate its Subcellular Localization
Human class IIa HDACs can shuttle from the nucleus to the cytosol, depending on the phosphorylation of the conserved Ser residues in the N-terminal region (Mihaylova and Shaw, 2013). Actually, phosphorylation on the conserved Ser residues within the N-terminal region promotes the binding of the class IIa HDACs to 14-3-3 proteins, which triggers HDACs export from the nucleus (Grozinger and Schreiber, 2000; McKinsey et al., 2000). For instance, activated protein kinase D or calcium/calmodulin-dependent protein kinase phosphorylates HDAC5 at the 14-3-3 binding sites Ser-259 and Ser-498, resulting in translocation of HDAC5 to the cytoplasm through 14-3-3 proteins. The disassociation of HDAC5 from transcription factors permits transcriptional expression of the downstream genes (Grozinger and Schreiber, 2000; McKinsey et al., 2000). Phosphorylation of HDAC1 and HDAC2 enhances their enzymatic activities, whereas nonphosphorylated HDAC1/2 displays low activity. Moreover, the phosphorylation of HDAC2 mediated by CK2 results in the dissociation of the HDAC1/2 heterodimer during the mitosis stage (Khan et al., 2013). In Arabidopsis, MAMP-triggered MPK3 phosphorylation of HD2B induces the nuclear relocalization of HD2B from the nucleolus to the nucleoplasm (Latrasse et al., 2017). In this study, we found that unphosphorylated HDA15 forms homodimers in the nucleolus whereas phosphomimetics of HDA15 at Ser-448 and Ser-452 disrupt the homodimer formation and result in translocation from the nucleolus to the nucleoplasm, indicating that the subcellular localization of RPD3/HDA1 class II HDACs is regulated by phosphorylation in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The HDA15 T-DNA insertion mutant, hda15-1 (SALK_004027), was described previously (Liu et al., 2013b). Arabidopsis (Arabidopsis thaliana) plants were grown under long-day conditions (16-h light/8-h dark cycle) at 23°C after a 3-d stratification period. For measurement of morphogenetic phenotypes, seeds were plated on one-half strength Murashige-Skoog medium agar plates containing 0.3% (w/v) Suc and imbibed for 3 d at 4°C in the dark. After germination was induced under WL for 6 h, the seedlings were grown in the dark for 2 or 4 d at 23°C (Shin et al., 2009).
Plasmid Constructs
The full-length complementary DNA (cDNA), deletion, and the phosphorylation site mutants of HDA15 were amplified by PCR and subsequently cloned into the entry vector, pENTR/SD/D-TOPO, or pCR8/GW-TOPO (Invitrogen). To generate the PromoterHDA15:GFP construct, the 2k-bp promoter of HDA15 was amplified by PCR and subsequently cloned into the pCambia 1300sGFP plasmid. To generate the PromoterHDA15:HDA15-GFP construct, the full-length cDNA of HDA15 was amplified by PCR and subsequently cloned into the plasmid containing PromoterHDA15:GFP. The LR clonase enzyme mix (Invitrogen) was used to transfer the insert to the destination vector, pK7WGF2 (Karimi et al., 2005), with the GFP tag positioned at the N terminus of the insert. The primers using for PCR to generate constructs are listed in Supplemental Table S3. The Arabidopsis transgenic plants were generated using the floral dip method (Clough and Bent, 1998).
Protein Expression and Purification
Wild-type and mutated HDA5HD, HDA18HD, HDA15HD, and HDA15ZFHD were constructed into the pET-28a vector. All target proteins were produced in Escherichia coli-BL21 codon plus (DE3)-RIL strain growing in Luria-Bertani broth upon OD600 0.6 to 1 and induced with 0.1 mm isopropylthio-β-galactoside overnight at 4°C. Bacteria were harvested by centrifugation and resuspended in a lysis buffer of 25 mm HEPES pH 7.5, 150 mm KCl, 30% (v/v) glycerol, 1% (v/v) Triton X-100, and 1 mm Tris[2-carboxyethyl]phosphine (TCEP). Bacteria were lysed by Sonicator-3000 (Misonix), and the lysate was centrifuged at 12,500 r.p.m. for 25 min at 4°C. The supernatant was diluted 2-fold with 25 mm HEPES, 150 mm KCl, and 1 mm TCEP. The his tag-only fusion protein was purified by HisTrap FF (GE healthcare); nontarget proteins were eluted by elution buffer A (25 mm HEPES pH 7.5, 150 mm KCl, 5% [v/v] glycerol, 1 mm TCEP, and 50 mm imidazole), and the target protein was further eluted by elution buffer B (25 mM HEPES pH 7.5, 150 mm KCl, 5% [v/v] glycerol, 1 mm TCEP, and 300 mm imidazole) to obtain the purified target protein. The his tag was removed by Tobacco etch virus protease and the different oligomeric states of the target protein were separated by size exclusion chromatography (Superdex-200, 160 mL; GE healthcare). The target protein with dual tags was purified by sequential affinity step purification by His-trap FF and MBP-trap HP (GE healthcare). The target protein was further bound on to the MBP column with MBP elution buffer (25 mm HEPES pH 7.5, 150 mm KCl, 5% [v/v] glycerol, 1 mm TCEP, and 10 mm maltose). The purified target protein was subjected to size exclusion chromatography, and its oligomeric state was analyzed. Before size exclusion chromatography, all protein buffers were exchanged in the gel filtration (GF) buffer (25 mm HEPES pH 7.5, 150 mm KCl, and 5% [v/v] glycerol). Size exclusion chromatography was performed in 1 mL/ min−1 with GF buffer and using protein standard markers (Biorad). All procedures were operated on AKTA Prime (GE healthcare).
Size Exclusion Chromatography and Immunoprecipitation for Arabidopsis Transgenic Plants
Total proteins were extracted from 2 g of 10-d-old seedings grown under WL using 2 mL cold protein extraction buffer (50 mm Tris-HCl pH 7.5, 150 mm NaCl, 1 mm PMSF, 10% [v/v] glycerol, and 1% [v/v] CA-630) containing the protease inhibitor cocktail (Roche). The homogenates were microcentrifuged twice for 15 min, and the supernatants were filtered through 0.2 μm filters. Before size exclusion chromatography, all protein buffers were exchanged in elution buffer (50 mm Tris-HCl pH 7.5, 150 mm NaCl, 1 mm PMSF, and 10% [v/v] glycerol). Size exclusion chromatography (Superdex-200, 160 mL; GE healthcare) was performed in 1 mL min−1 with elution buffer and using protein standard markers (Biorad Inc). Each fraction was collected at 3 mL. The fractions were immunoprecipitated by 5 μL GFP-trap beads (Chromotek) and immunoblotted with a GFP antibody (Abcam, ab290).
HDAC Enzymatic Activity Assays
For Arabidopsis transgenic plants, total proteins were extracted from 1 g of 2-d-old etiolated seedlings using 2 mL cold protein extraction buffer containing the protease inhibitor. Before HDAC enzymatic activity was measured, protein extracts were incubated with 5 μL GFP-trap beads (Chromotek) to immunoprecipitate GFP-tagged HDA15 overnight at 4°C and washed by the wash buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 10% [v/v] glycerol, and 1% [v/v] CA-630). Immunoblotting was carried out to identify the immunoprecipitation efficiency of GFP-trap beads using the GFP antibody (Abcam, ab290).
HDAC enzymatic activity assays were performed using the Fluorometric HDAC Activity Assay Kit (BioVision) following the manufacturer’s instructions. For Arabidopsis transgenic plants, 103 μL double distilled water, 12 μL 10× HDAC assay buffer, and 5 μL substrate [0.02 mm Boc-Lys(Ac)-7-amino-4-methylcoumarin (AMC)] were used. For kinetics assays, different substrate concentrations were used for the HDA15HD monomer, tetramer, and HDA15ZFHD dimer. The 75 nm protein of monomeric HDA15HD and 25 nm protein of tetrameric HDA15HD were reacted with the substrate, and 2.5 nm protein of dimeric HDA15ZFHD was used. The total reaction volume was 120 µL, and the deacetylation reaction was incubated at 37°C for 30 min. The reaction was stopped by adding 80 μl Lys developer and incubated at 37°C for 30 min.
The HDAC activity was then measured using the RFU (Relative fluorescence unit) by ELISA reader (TECAN Infinite M200, Ex/Em = 350/440 nm) on a 96-well black plate. A standard curve was prepared using the AMC Standard (range from 0 to 2 μm). AMC released was used to stand for HDAC enzymatic activity. For kinetics assays, the results were further transformed to molecular concentration for enzyme kinetics calculations.
Crystallization of Tetrameric HDA15HD
After size exclusion chromatography, the HDA15HD tetramer was further collected for protein crystallization. Tetrameric HDA15HD was concentrated to 9 mg mL−1 and stored in the GF buffer. Tetrameric HDA15HD was mixed with the crystallization buffer (0.1 m Tris-HCl pH 8.0 and 0.8 m LiSO4) in 0.5 µL: 0.5 µL in sitting drop at 22°C. Protein crystals were observed after 4 d.
Data Collection, Structure Determination, and Refinement
X-ray diffraction data were collected at the BL13B1 beam line at the National Synchrotron Radiation Research Center (Hsinchu, Taiwan) from single crystals flash-frozen in liquid nitrogen. Data were indexed and integrated with HKL2000 (Otwinowski and Minor, 1997). The Phenix program suite (Adams et al., 2010) was used for further data processing and analysis. The structure of HDA15HD was solved by molecular replacement with phaser (McCoy, 2007), and the HDAC7 structure (PDB ID: 3C10) was trimmed by the phenix program sculptor (Bunkóczi and Read, 2011) as the searching template. Manual building and refinement used phenix refine (Adams et al., 2010) and Coot (Emsley et al., 2010), respectively. All structural images were generated by PyMOL (DeLano Scientific). Atomic coordinate and structure factors for the reported crystal structure have been deposited in the Protein Data Bank with accession number 6J6T.
Protochlorophyllide Determination
Protochlorophyllide contents were measured as described (Shin et al., 2009). Ten frozen 4-d-old etiolated seedlings were ground into powder in opaque tubes, and pigments were extracted by adding 1 mL of cold 80% acetone for 1 h on a rotator in the dark at 4°C. After centrifugation at 4°C for 10 min, the supernatant was collected and the relative fluorescence was measured with a fluorescence spectrophotometer. The excitation wavelength was 440 nm, and the fluorescence emission spectra were recorded between 600 and 800 nm with a bandwidth of 5 nm.
RNA Extraction and RT-qPCR
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. One microgram of total RNA was used to synthesize cDNA. RT-qPCR was performed using iQ SYBR Green Supermix (Bio-Rad) and the CFX96 real-time PCR system (Bio-Rad). The gene-specific primers used for real-time PCR are listed in Supplemental Table S3. Each sample was quantified at least in triplicate and normalized using Ubiquitin10 (UBQ10) as an internal control.
LC-MS/MS
The HDA15 protein extracted from 15 g of 2-d-old etiolated seedlings of Col-0 wild type was immunoprecipitated by 500 μg HDA15 antibody (Liu et al., 2013b) and isolated from SDS-PAGE in 4 biological replicates. After trypsin digestion and desalting, peptides were used to perform LC-MS/MS by Thermo Orbitrap Elite Mass Spectrometer and for Mascot analysis.
In Vitro Phosphatase Treatment by using the Phospho-Ser Antibody and Phos-Tag Polyacrylamide Gels
To harvest phosphorylated HDA15 for in vitro phosphatase treatment, total proteins were extracted from 2-d-old etiolated seedlings and 10-d-old seedings grown under WL using the cold protein extraction buffer containing the protease inhibitor and PhosSTOP phosphatase inhibitor (Roche). The protein extracts were incubated with 15 μL GFP-trap beads to immunoprecipitate GFP-tagged HDA15 at 4°C for 3 h and washed by the wash buffer (100 mm NaCl, 10 mm Tris-HCl pH 7.5, 0.5% [v/v] CA-630, 10% [v/v] glycerol, and protease inhibitor) and the phosphatase treatment buffer (10 mm Tris-HCl pH 7.5, 10% [v/v] glycerol, and protease inhibitor). The GFP-trap beads and bound proteins were treated with 1 μL calf intestinal alkaline phosphatase (CIP, NEB) in 1× CutSmart buffer at 37°C for 30 min. The reaction was stopped by adding the loading buffer for 10 min on boiling water. For the phosphorylated HDA15-GFP, immunoblotting was carried out by using the SuperBlock T20 (TBS) blocking buffer (Thermo) and the phospho-Ser antibody (Thermo, MA1-90649). The GFP antibody (Abcam, ab290) was used to indicate the input protein loading.
For GFP-HDA15ZFHD, Zn2+-Phos-Tag polyacrylamide gels were performed as described previously (Bekešová et al., 2015) with some modification. Phos-Tag gel was prepared using 6% acrylamide and 25 μm Phos-tag for the resolving gel (6% [w/v] acrylamide/bis 37.5:1, 350 mm Bis-Tris/HCl pH 6.8, 25 μm Phos-tagTM AAL [Wako], 50 μm ZnSO4, 0.05% [v/v] TEMED [N,N,N′,N′-tetramethylethylenediamine], and 0.025% [w/v] ammonium persulfate) and 4% acrylamide for the stacking gel (4% acrylamide/bis 37.5:1, 350 mm Bis-Tris/HCl pH 6.8, 0.1% [v/v] TEMED and 0.05% [w/v] APS). The gel was run at 10 mA for 16 h at 4°C using 1× running buffer (100 mm Tris base, 100 mm MOPS, 0.1% [w/v] SDS, and 5 mm NaHSO3). The resolving gel was then soaked twice with the transfer buffer (192 mm Gly, 25 mm Tris-base, and 20% [v/v] methanol) containing 10 mm EDTA for 15 min each time and then washed once for 15 min in the transfer buffer without EDTA. Proteins were blotted onto polyvinylidene difluoride membranes by the wet tank procedure at 30 V for 16 h at 4°C. Immunoblotting was carried out by using the GFP antibody (Abcam, ab290).
ChIP Assays
ChIP assays were performed as described (Gendrel et al., 2005; Liu et al., 2013b). Chromatin was extracted from 2-d-old etiolated seedlings. After fixation with 1% formaldehyde, the chromatin was sheared to an average length of 500 bp by sonication and then immunoprecipitated with the GFP antibody (Abcam, ab290). The cross-linking was then reversed, and the amount of each precipitated DNA fragment was determined by qPCR using specific primers in Supplemental Table S3. Three biological replicates were performed, and three technical repeats were carried out for each biological replicate. Representative results from one biological replicate were shown.
Bimolecular Fluorescence Complementation Assays and Microscopes
Wild-type and mutant HDA15 were subcloned into the pCR8/GW/TOPO vector and then recombined into the YFPN (pEarleyGate201-YFPN) and YPFC (pEarleyGate202-YFPC) vectors (Lu et al., 2010). Constructed vectors were transiently transformed into Arabidopsis protoplasts and the florescence was observed using a Zeiss LSM 780 Confocal microscope. Leaves of 3- to 4-week-old Nicotiana benthamiana were infiltrated with Agrobacterium tumefaciens strains (GV3101) containing the YFPN and YFPC construct pairs. YFP florescence of epidermal cell layers were examined 3 to 4 d after infiltration using Delta Vision Core.
Homozygous transgenic plants were used for confocal imaging. Seeds were plated on half-strength Murashige-Skoog medium agar plates for 4 d. Roots were counterstained with 10 μg mL−1 PI (Sigma-Aldrich), washed once in distilled water, and mounted in water for Zeiss LSM 780 Confocal microscopy (Ex/Em = 493/636 nm).
Accession Numbers
Sequence data from this article can be found in the EMBL/National Center for Biotechnology Information/GenBank data libraries under the following accession numbers: HDA15 (AT3G18520); HDA5 (AT5G61060); HDA18 (T5G61070); HDA14 (AT4G33470); HDA8 (AT1G08460); HDA6 (AT5G63110); HDA19 (AT4G38130); HsHDAC1 (BAG70111.1); HsHDAC4 (NP_001365345.1); HsHDAC5 (NP_001015053.1); HsHDAC6 (AAH69243.1); HsHDAC7 (NP_001091886.1); HsHDAC8 (NP_060956.1); HsHDAC9 (NP_001191073.1); HsHDAC10 (NP_001152758.1); HsHDAC11 (NP_001129513.1); and Clr3 (NP_595104.1).
Supplemental Data
The following materials are available as supplemental data.
Supplemental Figure S1. Phylogenetic tree and sequence alignment of human and Arabidopsis class II HDACs.
Supplemental Figure S2. HDA5HD and HDA18HD proteins expressed in E. coli.
Supplemental Figure S3. Sequence alignment of Arabidopsis class II HDACs with human class IIa and yeast HDACs.
Supplemental Figure S4. The interface hydrogen bonds of the HDA15HD tetramer.
Supplemental Figure S5. Comparative protein structure modeling of HDA15ZFHD.
Supplemental Figure S6. Schematic structures of HDA15 protein domains and deletion constructs.
Supplemental Figure S7. Phosphorylation sites in the HD domain of human HDACs and Arabidopsis HDA15.
Supplemental Figure S8. Sequence alignment of HDA15 phosphorylation sites S448 and S452 with human and Arabidopsis HDACs.
Supplemental Figure S9. The side chain length of S448 and S452 in wild-type and phosphomimetic HDA15.
Supplemental Table S1. Mutant HDA15HD oligomerization and enzyme kinetics.
Supplemental Table S2. LC-MS/MS identification of the HDA15 phosphorylation sites in etiolated seedlings.
Supplemental Table S3. List of primers used in this study.
Acknowledgments
We are grateful to the Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Bio-Rad real-time PCR system, the confocal spectral microscope Zeiss LSM 780, and Delta Vision systems. We are thankful for the technical supports provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan.
Footnotes
This work was supported by the Ministry of Science and Technology, Taiwan (grant nos. 108–2311–B–002–013–MY3 and 108–2311–B–002–001 to K.W., and 107–2311–B–002–010 and 107–2313–B–002–003 to Y.-S.C.) and National Taiwan University (grant nos. NTU–AS–108L104310 to K.W. and 108L893105 to Y.-S.C.).
Articles can be viewed without a subscription.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alinsug MV, Yu CW, Wu K(2009) Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekešová S, Komis G, Křenek P, Vyplelová P, Ovečka M, Luptovčiak I, Illés P, Kuchařová A, Šamaj J(2015) Monitoring protein phosphorylation by acrylamide pendant Phos-TagTM in various plants. Front Plant Sci 6: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL.(2002) Histone modifications in transcriptional regulation. Curr Opin Genet Dev 12: 142–148 [DOI] [PubMed] [Google Scholar]
- Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, Jones P, Neddermann P, De Francesco R, Steinkühler C, et al. (2008) Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem 283: 26694–26704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl A, Heinzel T, Krämer OH(2009) Histone deacetylases: Salesmen and customers in the post-translational modification market. Biol Cell 101: 193–205 [DOI] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Seiser C(2009) Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol 53: 275–289 [DOI] [PubMed] [Google Scholar]
- Bunkóczi G, Read RJ(2011) Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr 67: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJ, Huang CLH, Bading H(2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem 85: 151–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB(2003) Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J 370: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe YL, Wild P, Chandrashaker A, Curak J, Schmidt MHH, Kalaidzidis Y, Milutinovic N, Kratchmarova I, Buerkle L, Fetchko MJ, et al. (2009) Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci Signal 2: ra84-ra84 [DOI] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA(2007) Histone deacetylase inhibitors: Overview and perspectives. Mol Cancer Res 5: 981–989 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K(2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom GH, Kook H(2014) Posttranslational modifications of histone deacetylases: Implications for cardiovascular diseases. Pharmacol Ther 143: 168–180 [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP(1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193 [DOI] [PubMed] [Google Scholar]
- Fong PM, Tian L, Chen ZJ(2006) Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro. Cell Res 16: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A-V, Lippman Z, Martienssen R, Colot V(2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Greco TM, Yu F, Guise AJ, Cristea IM(2011) Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics 10: M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV(2004) Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol 338: 17–31 [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL(2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci USA 97: 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D, Chen CY, Zhao M, Zhao L, Duan X, Duan J, Wu K, Liu X(2017) Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res 45: 7137–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CH, Kim JY, Zhao J, Wang W, Jhun BS, Wong C, Jin ZG(2010) PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. Proc Natl Acad Sci USA 107: 15467–15472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai Y, Christianson DW(2016) Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol 12: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai Y, Shinsky SA, Porter NJ, Christianson DW(2017) Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat Commun 8: 15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C, Liu Z(2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50: 875–885 [DOI] [PubMed] [Google Scholar]
- Hook SS, Orian A, Cowley SM, Eisenman RN(2002) Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc Natl Acad Sci USA 99: 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job G, Brugger C, Xu T, Lowe BR, Pfister Y, Qu C, Shanker S, Baños Sanz JI, Partridge JF, Schalch T(2016) SHREC silences heterochromatin via distinct remodeling and deacetylation modules. Mol Cell 62: 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S(2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem 276: 47496–47507 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P(2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP(2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Khan DH, He S, Yu J, Winter S, Cao W, Seiser C, Davie JR(2013) Protein kinase CK2 regulates the dimerization of histone deacetylase 1 (HDAC1) and HDAC2 during mitosis. J Biol Chem 288: 16518–16528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Wagner T, Yildiz Ö, Meyer-Almes FJ(2016) Crystal structure of a histone deacetylase homologue from Pseudomonas aeruginosa. Biochemistry 55: 6858–6868 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K(2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K(2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Jégu T, Li H, de Zelicourt A, Raynaud C, Legras S, Gust A, Samajova O, Veluchamy A, Rayapuram N, et al. (2017) MAPK-triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Biol 18: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Rezai-Zadeh N, Seto E(2004) Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol 24: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li LC, Chen WQ, Chen X, Xu ZH, Bai SN(2013a) HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 25: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen CY, Wang KC, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, et al. (2013b) PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Tai R, Wu K(2014) Transcriptional repression by histone deacetylases in plants. Mol Plant 7: 764–772 [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Luo M, Tai R, Yu CW, Yang S, Chen CY, Lin WD, Schmidt W, Wu K(2015) Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J 82: 925–936 [DOI] [PubMed] [Google Scholar]
- Martin M, Kettmann R, Dequiedt F(2007) Class IIa histone deacetylases: Regulating the regulators. Oncogene 26: 5450–5467 [DOI] [PubMed] [Google Scholar]
- McCoy AJ.(2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN(2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ(2013) Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol Metab 24: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Keusch JJ, Wang L, Saito M, Hess D, Wang X, Melancon BJ, Helquist P, Gut H, Matthias P(2016) Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat Chem Biol 12: 748–754 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W(1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Payre F, Buono P, Vanzo N, Vincent A(1997) Two types of zinc fingers are required for dimerization of the serendipity delta transcriptional activator. Mol Cell Biol 17: 3137–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum MKH, Tong JK, Lane WS, Schreiber SL(2001) Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem 276: 47733–47741 [DOI] [PubMed] [Google Scholar]
- Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, et al. (2008) Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem 283: 11355–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segré CV, Chiocca S(2011) Regulating the regulators: The post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol 2011: 690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E(2004) Regulation of histone deacetylase activities. J Cell Biochem 93: 57–67 [DOI] [PubMed] [Google Scholar]
- Seto E, Yoshida M(2014) Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G(2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA(1999) The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem 274: 37370–37378 [DOI] [PubMed] [Google Scholar]
- Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, et al. (2004) Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Sun JM, Chen HY, Davie JR(2007) Differential distribution of unmodified and phosphorylated histone deacetylase 2 in chromatin. J Biol Chem 282: 33227–33236 [DOI] [PubMed] [Google Scholar]
- Taplick J, Kurtev V, Kroboth K, Posch M, Lechner T, Seiser C(2001) Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J Mol Biol 308: 27–38 [DOI] [PubMed] [Google Scholar]
- Tsai SC, Seto E(2002) Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem 277: 31826–31833 [DOI] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfí A, De Francesco R, Steinkühler C, Di Marco S(2007) Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep 8: 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG(2003) Class II histone deacetylases: Versatile regulators. Trends Genet 19: 286–293 [DOI] [PubMed] [Google Scholar]
- Wang BS, Pabo CO(1999) Dimerization of zinc fingers mediated by peptides evolved in vitro from random sequences. Proc Natl Acad Sci USA 96: 9568–9573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A(2016) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54: 5.6.1-5.6.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Tsai SC, Wen YD, Fejer G, Seto E(2002) Functional domains of histone deacetylase-3. J Biol Chem 277: 9447–9454 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Grégoire S(2005) Class II histone deacetylases: From sequence to function, regulation, and clinical implication. Mol Cell Biol 25: 2873–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kudo N, Kosono S, Ito A(2017) Chemical and structural biology of protein lysine deacetylases. Proc Jpn Acad, Ser B, Phys Biol Sci 93: 297–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CW, Tai R, Wang SC, Yang P, Luo M, Yang S, Cheng K, Wang WC, Cheng YS, Wu K(2017) HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 29: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Peng T, Chen CY, Ji R, Gu D, Li T, Zhang D, Tu YT, Wu K, Liu X(2019) HY5 interacts with the histone deacetylase HDA15 to repress hypocotyl cell elongation in photomorphogenesis. Plant Physiol 180: 1450–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]