Abstract
Although originally identified as the components of the complex aiding the cytosolic chaperonin CCT in the folding of actins and tubulins in the cytosol, prefoldins (PFDs) are emerging as novel regulators influencing gene expression in the nucleus. Work conducted mainly in yeast and animals showed that PFDs act as transcriptional regulators and participate in the nuclear proteostasis. To investigate new functions of PFDs, we performed a co-expression analysis in Arabidopsis thaliana. Results revealed co-expression between PFD and the Sm-like (LSM) genes, which encode the LSM2–8 spliceosome core complex, in this model organism. Here, we show that PFDs interact with and are required to maintain adequate levels of the LSM2–8 complex. Our data indicate that levels of the LSM8 protein, which defines and confers the functional specificity of the complex, are reduced in pfd mutants and in response to the Hsp90 inhibitor geldanamycin. We provide biochemical evidence showing that LSM8 is a client of Hsp90 and that PFD4 mediates the interaction between both proteins. Consistent with our results and with the role of the LSM2–8 complex in splicing through the stabilization of the U6 snRNA, pfd mutants showed reduced levels of this snRNA and altered pre-mRNA splicing patterns.
INTRODUCTION
Prefoldins (PFDs) are evolutionarily conserved proteins that were originally identified in humans as the components of a heterohexameric complex, the PFD complex, which acts as a co-chaperone of the chaperonin CCT in the folding of actins and tubulins in the cytosol (1). In addition to this role, increasing evidence indicates that they also perform a role in the regulation of gene expression, either as bona fide transcriptional regulators or through their role in the cellular proteostasis, i.e. folding, assembly or degradation of proteins or protein complexes with diverse roles in gene expression (2). Although there is no indication about the DNA binding ability of PFDs, they participate in the regulation of gene targets in the context of the chromatin. For instance, chromatin immunoprecipitation assays showed that human prefoldin 1 (PFDN1) binds to the transcription start site of the Cyclin A gene to repress its expression (3). We have a better understanding about the mechanism of PFDN5/MM-1 in transcriptional regulation. This PFD regulates c-Myc activity, acting as a bridge protein that recruits a transcriptional co-repressor, TIF1β/KAP1, and the HDAC1 histone deacetylase complex directly to the c-Myc-bound genomic targets to repress their expression (4,5). Furthermore, genetic and molecular analyses in yeast have demonstrated that several PFDs are required for transcription elongation and that are able to bind to actively transcribed gene bodies following the profile of elongating RNA Pol II (6). In particular, PFDs promote histone eviction, thus facilitating the passage of the polymerase through gene bodies during elongation.
PFDs also influence gene expression through their participation in the cellular proteostasis, a role most likely independent of their role as transcriptional regulators. The participation of PFDs in the folding of a transcriptional regulator was recently reported, indeed, interactome analyses in human HeLa cells have related the PFD complex with the activity of the chaperonin CCT in the nucleus, processing the folding of the histone deacetylase HDAC1 prior to its incorporation into transcriptional repressor complexes (7). PFD2, PFD6 and three PFD-like proteins (URI, UXT and PDRG1) form a non-canonical complex, called PFD-like, that is associated to the R2TP complex (8). This complex has been well described in animals and yeast and is formed by RPAP3, PIH1D1 and a heterohexamer of the ATPases RUVBL1 and RUVBL2 (Tah1, Pih1, Rvb1 and Rvb2 in yeast, respectively) (8). This complex acts as a co-chaperone of Hsp90, recruited to the complex through the carboxylate-clamp type TPR domain of RPAP3, in the assembly of several other protein complexes in animals. This includes the nuclear RNA polymerases and the spliceosome U5 small nuclear ribonucleoprotein particle (U5 snRNP) (9–11). Nonetheless, despite proteomic analyses repeatedly identifying the PFD-like complex associated to R2TP (9–13), the actual involvement of any PFD or PFD-like in the complex as co-chaperone of Hsp90 has yet to be demonstrated. PFDs also influence protein stability of transcription factors. In humans, PFDN5/MM-1, in addition to regulate c-Myc activity, promotes its degradation by recruiting an E3 ubiquitin ligase complex (14). Although the mechanism may be different, a role promoting degradation of transcription factors has also been observed in plants. In Arabidopsis, PFDs preferably accumulate in the nucleus upon interaction with the transcriptional regulators DELLA proteins (15). DELLAs are plant-specific proteins whose levels are sensitive to changes in the environment, and that thanks to their ability to interact with numerous transcription factors and transcriptional regulators occupy a key position within the transcriptional network relaying environmental information to the growth and developmental programs (16). When plants are exposed to low temperature, PFD4 moves to the nucleus in a DELLA-dependent manner to promote the ubiquitination and subsequent degradation of the transcription factor HY5, a regulator of the cold acclimation response (17).
Pre-mRNA splicing is a crucial step in gene expression. The alternative processing of many introns and exons not only expands the proteome, as there are multiple mRNA isoforms that give rise in many cases to either slightly different or completely different polypeptides, but also regulates the expression of many genes by producing mRNA species that are not translated (18). Pre-mRNA processing is carried out by the spliceosome, which is formed by the association of several snRNPs, each one including a snRNA (U1, U2, U4, U5 and U6) and numerous associated proteins. Among them, the Sm and Sm-like (LSM) proteins form two core nuclear heteroheptameric complexes, the Sm and the LSM2–8, each one directly binding a snRNA (19). The Sm complex associates to the U1, U2, U4 and U5 snRNAs, while LSM2–8 binds specifically the U6 snRNA. Each complex assures the cellular levels, stability and processing of the corresponding snRNAs, as well as the assembly and subcellular localization of snRNPs.
Here, we have explored new mechanisms by which PFDs would regulate gene expression in Arabidopsis. By harnessing the plethora of transcriptomic data in this species, we identified a striking co-expression between PFD and LSM spliceosome core genes that prompted us to hypothesize that PFDs could contribute to the function of the LSM2–8 complex.
MATERIALS AND METHODS
Plant materials
Arabidopsis thaliana accession Columbia-0 (Col-0) was used as the wild-type (WT). The following mutants have been previously described: pfd4 (17), pfd6-1 (20), and lsm8-1 and lsm8-2 (21). The pfd2 (WiscDsLoxHs096_06D) insertion mutant was obtained from the WiscDsLox (22) T-DNA collection. The transgenic pLSM8:LSM8-GFP (21) Arabidopsis line was introgressed into the pfd2, pfd4, and pfd6-1 mutant backgrounds by crossing. The presence of transgenes in progenies was determined by simultaneous kanamycin and hygromycin resistance. The triple pfd2,4,6 mutant was obtained by genetic crosses. Primers for genotyping all mutant lines are listed in Supplementary Table S1.
Growth conditions
Seeds were stratified for 3–7 days at 4°C, and then exposed to white fluorescent light, either continuously (50 μmol m−2 s−1) or under long photoperiods (16 h of 90 μmol m−2 s−1). Plants were grown at 20°C in pots containing a mixture of organic substrate and vermiculite (3:1) or on Petri dishes containing half MS with 1% (w/v) sucrose and 8 g l−1 agar, pH 5.7 (our control media). Low temperature treatments were performed by transferring 2-week-old plants grown in pots to a growth chamber set to 4°C for different times under long photoperiods (16 h of 40 μmol m−2 s−1).
For experiments including chemicals, seedlings were grown for 7 days in continuous light and then transferred to liquid medium containing 20 μM geldanamycin (GDA; Sigma), 50 μM MG132 (Calbiochem) or both and maintained for 24 h in darkness under mild agitation.
Co-expression analysis
Data of Arabidopsis and mouse gene co-expression were retrieved from ATTED-II database Ath-m c7.1 (23) and COXPRESdb database Mmu-m c4-0 (24), respectively. Graphic co-expressions were represented with Cytoscape (25).
Molecular cloning
pENTR223 clones of PFD2, PFD3, PFD4, PFD6 and LSM8 coding sequences (CDSs) were obtained from the Arabidopsis Biological Resource Center. PFD1 and PFD5 CDSs were amplified from a pool of Arabidopsis cDNA and cloned into the pCR8/GW/TOPO (Invitrogen). The CDSs were transferred to the pYFN43 and pYFC43 destination vectors for Bimolecular Fluorescence Complementation (BiFC) (26), pGADT7 and pGBKT7 destination vectors for yeast two-hybrid assays, or to pEarlyGate104 (27) or pEarlyGate201 to create YFP or HA fusions, respectively, by LR reactions.
The other constructs used for co-IP analysis were obtained using the GoldenBraid 2.0 technology (28): pUPD2 (for PFD2, PFD4, PFD5, PFD6 and LSM8) or pUPD (for PFD1) were used to obtain the constructs containing the CDS pieces lacking stop codons. Since PFD4, PFD5 and PFD6 harbor BsmBI or BsaI recognition sequences, two patches were made by PCR or synthesized (IDT; see Supplementary Table S1). The CDS pieces were combined with pUPD-35Spro, pUPD-35Ster and pUPD-YFP (for PFDs), or pUPD-HA (for LSM8), creating the α constructs. Ω constructs co-expressing two fusion proteins were created from two α constructs. For expression in Arabidopsis, constructs contained also the gene conferring kanamycin resistance.
BiFC, co-immunoprecipitation, and protein analysis
Cells of Agrobacterium tumefaciens strain C58C1 carrying the different constructs were used to infiltrate Nicotiana benthamiana leaves or to transform WT Arabidopsis. Agroinfiltration was performed in leaves from 3- to 4-week-old plants. Leaves were used for analysis 3 days after infiltration.
For BiFC assays, leaf samples were analyzed by confocal microscopy three days after agro-infiltration as described (29).
For co-immunoprecipitations, the ground frozen tissue (occupying a volume of 800 μl) was homogenized in 1 ml of extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% Triton, 2 mM PMSF and 1X protease-inhibitor cocktail [Roche]). Proteins in extracts were quantified using the Bradford assay. Fifty μg of total proteins were set aside to be used as input. One milligram of total proteins was incubated for 2 h at 4°C in a rotating wheel with anti-GFP-coated paramagnetic beads and loaded onto μColumns (Miltenyi). Columns were washed and proteins eluted following manufacturer's instructions (Miltenyi). Samples were analyzed by Western-blot after running two 12% SDS-PAGE in parallel. In one gel, we separated 25 μg of input and 10% of eluted proteins; after wet transfer, the PVDF membrane was incubated with an anti-GFP antibody (JL8, 1:5000; Clontech). In the other gel, we separated 25 μg of input and 90% of eluted proteins and, after transfer, the membrane was incubated with an anti-HA-HRP antibody (3F10, 1:5000; Roche). LSM8-GFP immunoprecipitations in Arabidopsis were carried out following the same procedure, although three times more protein extract (1.5 mg) from the non-transgenic control and the pfd4 mutant were used. To detect the interaction of PFD4-YFP and LSM8-HA with Hsp90 in Arabidopsis double transgenic seedlings, the membrane incubated with anti-HA-HRP was stripped-out and incubated with anti-Hsp90 (1:2500, Agrisera).
For analysis of LSM8-GFP protein levels in Arabidopsis, ground frozen tissue from whole seedlings was homogenized in extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, 0.5% Nonidet P-40, 2 mM PMSF and 1× protease-inhibitor cocktail). Proteins were quantified using the Bradford assay. Total proteins were separated in 12% SDS-PAGE and transferred to a PVDF membrane using a wet transfer system. Membranes were incubated consecutively with anti-GFP antibody (JL8, 1:5000; Clontech) and, after strip-out, with anti-DET3 antibody (1:10000; provided by K. Schumacher, University of Heidelberg, Heidelberg, Germany) as loading control. Protein bands were detected using the LAS-3000 Imaging system (Fujifilm) and quantified using NIH ImageJ software (https://imagej.nih.gov/ij/).
For gel filtration assays, extracts of LSM8-GFP in the WT and pfd4 backgrounds were prepared in extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, 0.5% Nonidet P-40, 2 mM PMSF and 1× protease-inhibitor cocktail). Proteins were quantified using the Bradford assay and loaded in a Superose™ 6 Increase (GE Healthcare) column. The twelve fractions (250 μl each) where LSM8-GFP protein is present are shown in the analysis. Proteins in fractions were precipitated in 10% trichloroacetic acid on ice for 90 min and then washed twice with cold acetone before western-blot analysis. Proteins were separated in 12% SDS-PAGE and transferred to a PVDF membrane using a semi-dry blotting system. Membranes were stained with Ponceau solution and then incubated with anti-GFP antibody (JL8, 1:5000; Clontech).
RNA extraction and quantitative RT-PCR
For quantitative real-time PCR (qPCR) experiments, total RNA was extracted using TRIzol reagent (Life Technologies) and subsequently treated with DNase I (Roche). cDNA was synthesized with the iScript® cDNA Synthesis Kit (Bio-Rad), and used as a template for qPCR assays employing the SsoFast EvaGreen Supermix (Bio-Rad) in an iQ2 thermal cycler machine (Bio-Rad). The relative expression values were calculated using the levels of the At1g13320 (PP2AA3) mRNA as a reference (30), using the ΔΔCT method. Validation of intron retention events was performed by qPCR as previously shown (31). All assays were performed in triplicate with three independent RNA samples. The list of primers and the experimental details for qPCRs (MIQE Guidelines; for primers described for the first time) are in Supplementary Table S1.
For RNA sequencing (RNA-seq) experiments, total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen). The RNA concentration and integrity (RIN) were measured in a RNA nanochip (Bioanalyzer, Agilent Technologies 2100). The TrueSeq stranded mRNA sample preparation kit (Illumina) was used to isolate the poly (A)+ RNA and to prepare libraries, which were sequenced in an Illumina NextSeq™ 500 platform. Library preparation and sequencing were carried out by the Genomics Service of the University of Valencia.
RNA-seq analysis
Approximately, 20 million 75 bp paired-end reads per sample were generated and >90% reads were aligned to the TAIR10 Col-0 reference genome using TopHat v2.1.1 with default parameters. Count tables for the different feature levels were obtained from bam files using custom R scripts.
We used ASpli (32) for the analysis of differential splicing. Briefly, multiexonic genes were partitioned into features defined as ‘bins’ and then classified into exon, intron and annotated alternative splicing bins. These tables were then filtered according to several criteria applied at the gene and bin level. First, bins were considered for differential splicing analysis only if the genes with which they are associated were expressed above a minimum threshold level (>10 reads per gene) in all experimental conditions. Next, bins were considered for differential splicing analysis only if they had >5 reads in at least one experimental condition. After applying these filters, reads summarized at the bin level were normalized to the read counts of their corresponding gene. Differential bins usage was estimated using the edgeR package version 3.14.0 (33), and resulting P-values were adjusted using a false discovery rate (FDR) criterion. We then calculated PSI (percent spliced-in) and PIR (percent intron retention), which were used as a filtering criteria for the splicing analysis combined with FDR. Bins with absolute FC > 1.5, FDR lower than 0.15 and absolute delta PSI/PIR > 5% were considered differential used bins. The events not annotated by ASpli were visualized using Gviz (34) and manually classified. The events annotated by ASpli as ‘Io’ (resulted from the retention/inclusion of two or more bins together) were also plotted using Gviz, classified as IR and included only if they were clear and there were no more IR in these bins. The selected bins considered for differential splicing are included in Supplementary Tables S2–S4. Differential splicing events were used to look for 5′ splice sites as previously described (35,36). The identification of mRNAs with nonsense-mediated decay (NMD) features generated by altered splicing events was done as previously described (31).
Statistical analysis
P values in overlapping splicing events from two particular genotypes were calculated using chi-square tests. Hypergeometric tests were used to calculate P values from representation factors in 5′ splice site sequences. The rest of P values (qPCR, protein abundance and phenotypic analysis) were obtained from one-way ANOVA tests followed by multiple comparison tests.
RESULTS
PFD genes co-express with genes coding for core components of the spliceosome
Given that functionally related genes are usually co-expressed (37,38), we set out to explore new roles of PFDs in Arabidopsis by searching the ATTED-II database (39) for genes correlated with them. We found co-expression between PFD genes and genes encoding the proteins corresponding to LSM2–8 and Sm complexes, which are core components of the spliceosome (Figure 1A and B and Supplementary Table S5) (21,40–44). Interestingly, the Pearson correlation coefficients between PFD and LSM or Sm genes were similar to those between genes of the same complex (Figure 1B and C and Supplementary Table S5), suggesting that the observed inter-complex correlations are indeed significant. Similar co-expression levels were found in mouse (Supplementary Figure S1 and Supplementary Table S5), reinforcing the idea that PFDs and the spliceosome may be functionally related.
Figure 1.
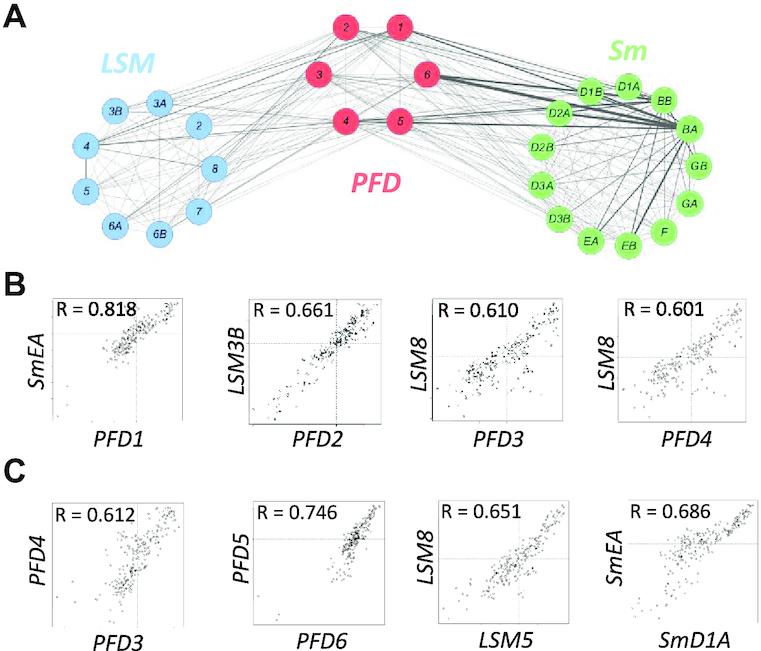
Co-expression and physical interactions between PFDs and core spliceosome proteins. (A) Cytoscape representation of a co-expression network involving LSM, PFD and Sm genes. Co-expression is represented by mutual rank values. This value is directly proportional to the edge thickness. (B, C) Scatter plots comparing the expression level of several Sm or LSM genes with the expression of PFD genes (B) and between genes of the same complex (C) across several tissues and developmental stages. R indicates the Pearson correlation coefficient for each pair of genes.
PFDs interact with LSM8
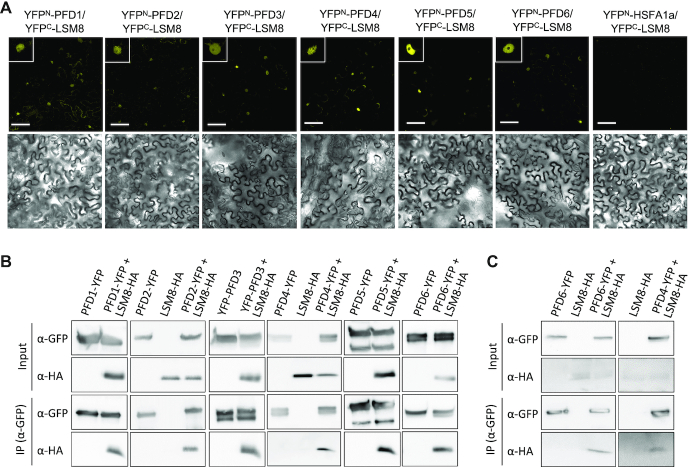
Based on the co-expression data, we hypothesized that PFDs could interact with the nuclear LSM2–8 complex in Arabidopsis. We therefore tested interactions between PFDs and LSM8, the only subunit that is specific to the LSM2–8 complex, as the other subunits are shared with the LSM1–7 complex that regulates mRNA decapping in the cytosol (45), and is required for the complex formation (21). Interactions were assessed after transient expression in leaves of N. benthamiana by BiFC and co-immunoprecipitation assays. The six PFDs were able to interact with LSM8 (Figure 2A and B). Fluorescence from the reconstituted YFP as consequence of interaction was mainly observed in nuclei of epidermal cells in leaves co-expressing YFPC-LSM8 and any YFPN-PFD, whereas no fluorescence was observed when YFPC-LSM8 was co-expressed with YFPN-HSFA1a, an unrelated, nuclear protein (Figure 2A and Supplementary Figure S2). Similarly, LSM8-HA was efficiently pulled-down by anti-GFP antibodies when it was co-expressed with PFDs fused to YFP, but not when it was expressed alone (Figure 2B). Consistent with the interactions observed in N. benthamiana, LSM8-HA was specifically co-immunoprecipitated with anti-GFP antibodies from extracts of available Arabidopsis transgenic plants that also expressed PFD4-YFP or PFD6-YFP (Figure 2C). These results indicated that PFDs form a complex with LSM8, most likely by direct physical interaction.
Figure 2.
PFDs interact with LSM8. (A) BiFC assays in N. benthamiana leaves. PFDs fused to YFPN were co-expressed with LSM8 fused to YFPC. Fluorescence from the reconstituted YFP was detected by confocal microscopy from leaf discs three days after infiltration (upper row). YFPN-HSFA1a, which does not interact with LSM8, was used as negative control. Insets show fluorescence from a representative nucleus. Bright field images are shown in the bottom row. Scale bars: 75 μm. (B) Co-immunoprecipitation assays showing the interaction between PFDs fused to YFP and LSM8-HA in N. benthamiana. Total proteins were immunoprecipitated with anti-GFP antibody-coated paramagnetic beads from extracts of infiltrated N. benthamiana leaves. Proteins were detected with anti-GFP and anti-HA antibodies. (C) Co-immunoprecipitation assays showing the interaction between PFD4 and PFD6 fused to YFP and LSM8-HA in transgenic Arabidopsis. Total proteins were immunoprecipitated with anti-GFP antibody-coated paramagnetic beads from extracts of 7-day-old Arabidopsis seedlings. Proteins were detected with anti-GFP and anti-HA antibodies.
PFDs contribute to maintaining adequate levels of LSM8 protein
Considering the role of PFDs as co-chaperones (8,46) either promoting the folding and stability of client proteins such as tubulins (17,47), or destabilizing protein partners (17), we investigated the LSM8 protein level in several pfd mutants to determine the possible outcome of the interaction. For this purpose, the pLSM8:LSM8-GFP transgene was introgressed by genetic crosses into the pfd4 (17) and pfd6-1 (20) mutant backgrounds. The pfd4 mutation is caused by the insertion of a T-DNA in the second exon of the PFD4 gene that results in undetectable levels of the mRNA (17), whereas the pfd6-1 mutant carries a point mutation that changes the conserved R83 to Q (20). The pLSM8:LSM8-GFP transgene was also introgressed into the newly identified T-DNA insertion mutant pfd2, which showed undetectable levels of the PFD2 full-length transcript (Figure 3A and B) and a smaller size than the WT at adult stage (Figure 3C and D), similar to other pfd mutants (Figure 3C and D) (17,20). LSM8-GFP levels were lower in the pfd4 and pfd6-1 mutants than in the WT in seedlings grown at 20°C, whereas levels were not affected by the pfd2 mutation (Figure 4A and B). As expected, the amounts of LSM8-GFP-containing complexes were reduced in the pfd4 mutant, as deduced from the gel filtration analysis (Figure 4C), suggesting that the amount of the LSM2–8 complex might be also lower in the mutant. Neither the endogenous LSM8 nor the transgenic LSM8-GFP transcript levels were affected by the pfd4 or pfd6-1 mutations (Figure 4D), indicating that PFDs regulate LSM8 at the post-transcriptional level. These results are compatible with PFDs being required to maintain adequate levels of LSM8 protein.
Figure 3.
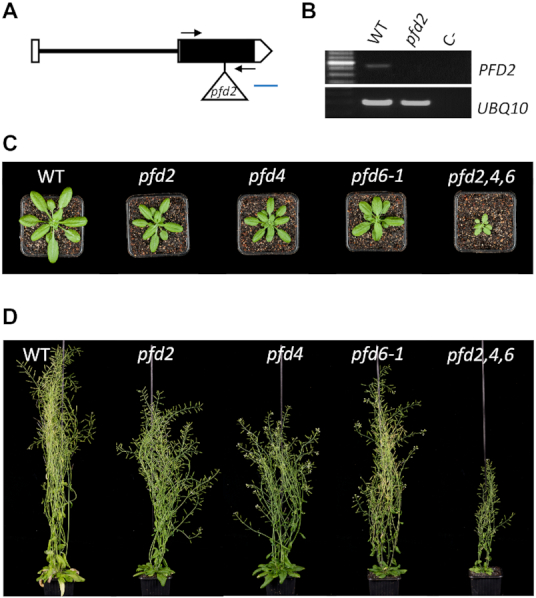
Characterization of novel pfd mutants. (A, B) The novel pfd2 mutant is a null allele. (A) Structure of the PFD2 gene indicating the position of the insertion. Boxes and line indicate exons and intron, respectively. Open boxes correspond to the 5′ and 3′ untranslated regions. The triangle indicates the Ds element insertion. Arrows indicate the position of the primers used for the semi-quantitative RT-PCR. Blue line represents 100 bp. (B) Semi-quantitative RT-PCR analysis of PFD2 expression in WT and mutant plants and the negative control (C-; water). The expression of UBQ10, a housekeeping gene, was used as control. (C) Rosette of 29-d-old plants of the indicated phenotypes at bolting. (D) Global morphological phenotype of 58-day-old plants of the indicated genotypes.
Figure 4.
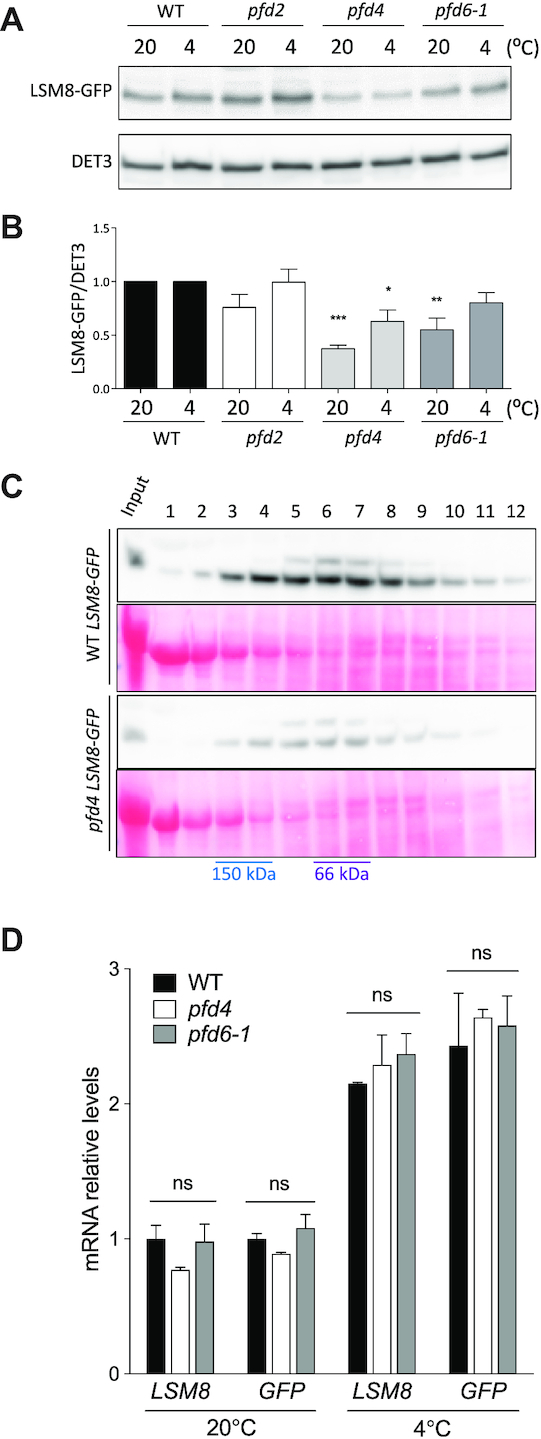
LSM8 protein level depends on PFD activity. (A, B) LSM8-GFP protein levels in the WT and pfd mutants grown at control temperature (20°C) or after being exposed to 4°C for one day. Representative blots are shown in (A). DET3 was used as loading control. (B) Plot showing LSM8-GFP/DET3 ratios. Data are average from three biological replicates. The statistical significance of the difference between mutants and WT is indicated by one, two, and three asterisks, which represent P < 0.01, 0.001 and 0.0001 in Bonferroni's multiple comparison test after an ANOVA test, respectively. (C) Gel filtration fractions were separated in 12% SDS-PAGE and LSM8-GFP detected by western blot with anti-GFP antibodies. (D) RT-qPCR analysis of the LSM8 and LSM8-GFP expression in standard conditions and 24 h after transfer to 4°C. Expression of PP2AA3 gene was used as reference for normalization. Data are average of three biological replicates. Error bars indicate the standard error of the mean. ns, no significant differences.
PFDs contribute to maintaining the activity of the LSM2–8 complex
To establish the relevance of the contribution of PFDs to maintaining LSM8 protein levels, we first investigated to which extent the LSM2–8 function was affected in pfd mutants. In the analysis, we included the pfd2, pfd4 and pfd6-1 mutants, and the triple mutant pfd2,4,6, obtained by genetic crosses. The triple mutant plants are viable, although with marked growth defects (Figure 3C and D). A proxy to estimate LSM2–8 assembly is the analysis of U6 snRNA levels, which is unstable unless incorporated into the U6 snRNP (48). U6 snRNA levels are actually reduced in lsm8 mutants of Arabidopsis (21). In agreement with the requirement of PFDs to maintain LSM2–8 function, we found that levels of U6 snRNA were significantly reduced in pfd6-1 and pfd2,4,6 mutant seedlings, similar to lsm8, and to a lesser extent in pfd4 (Figure 5A). The fact that U6 snRNA levels were reduced to a greater extent in pfd6-1 than in pfd4 mutants, despite the fact that LSM8 protein levels were more reduced in the latter, suggests that other U6 snRNA regulators might be affected by the pfd6-1 mutation. Proteins involved in U6 snRNA biogenesis, for instance, could be among these regulators. In this sense, the U6 BIOGENESIS-LIKE1 protein has been characterized in Arabidopsis and described that it is necessary to achieve adequate levels of U6 snRNA and pre-mRNA splicing (49).
Figure 5.
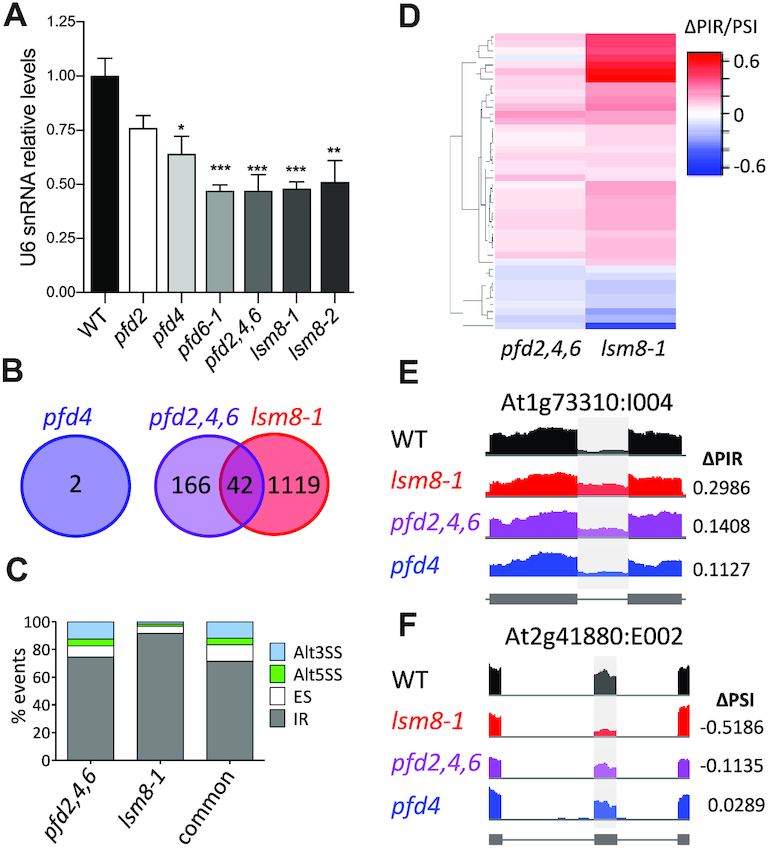
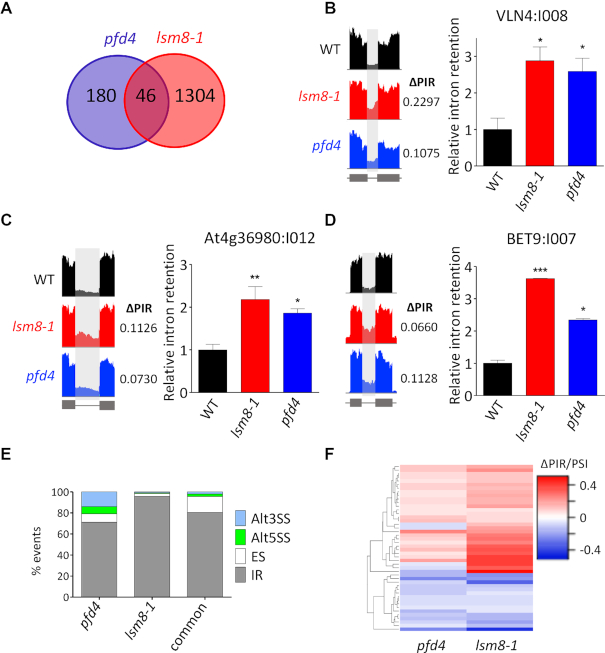
Genome-wide analysis of pre-mRNA splicing in pfd and lsm8-1 mutants by RNA-seq. (A) Relative U6 snRNA levels in pfd and lsm8-1 mutants grown in soil for two weeks. U6 snRNA levels were normalized against PP2AA3 mRNA. Data are average of three biological replicates. One, two, and three asterisks represent P < 0.01, 0.001 and 0.0001 in ANOVA tests, respectively. (B) Venn diagram showing the number of splicing events altered in the mutants. (C) Percentage of the different splicing events affected in each mutant and in both. Alt3SS and Alt5SS, alternative 3′ and 5′ splice site, respectively; ES, exon skipping; IR, intron retention. (D) Hierarchical clustering of common events by ΔPIR/PSI (percent intron retention/percent spliced-in) value. (E, F) IGV plots of altered splicing events: At1g73310:I004 (E) and At2g41880:E002 (F).
The U6 snRNA is functionally related to the U4 snRNA, as both are part of the U4/U6 di-snRNP (50). We therefore investigated whether U4 snRNA levels were also affected in our mutants. U4 snRNA levels were elevated in lsm8 mutants, as observed in yeast (51), and did not change in the pfd mutants (Supplementary Figure S3). The levels of the U5 snRNA, which mainly depends on the Sm proteins, were not affected in either pfd or lsm8 mutants (Supplementary Figure S3).
A more direct conclusive measure of LSM2–8 function is the analysis of pre-mRNA splicing. Thus, we performed a RNA-seq analysis of three biological replicates of 14-day-old WT, pfd4, pfd2,4,6 and lsm8-1 seedlings grown under long days (16 h light:8 h darkness) at 20°C (our standard conditions; Supplementary Table S2). Importantly, a total of 208 splicing events corresponding to 195 different genes were altered in the triple pfd2,4,6 mutant compared to the WT, while only two events corresponding to one gene were affected by the single pfd4 mutation (Figure 5B and Table 1). The lack of splicing defects in pfd4 seedlings suggests, therefore, that the low levels of both LSM8 protein (Figure 4A–C) and U6 snRNA (Figure 5A) observed in the mutant should be above the minimum functional threshold. It is worth to mention that 25 out of the 195 different genes that showed altered splicing events in the pfd2,4,6 mutant, for instance DEK1, RCD1, YAB3, TTG1 or BPS1, have been implicated in regulating different developmental processes (52–56) (Supplementary Table S2) and, therefore, could account for the growth defects exhibited by this mutant (Figure 3C and D). Interestingly, 20% (P < 0.0001, chi-square test) of the altered splicing events detected in pfd2,4,6 seedlings were also detected in the lsm8-1 mutant (Figure 5B-F, Supplementary Figure S4 and Table 1). The most abundant splicing event affected was intron retention (IR) (Figure 5C and Table 1). It reached 74.5% in pfd2,4,6, 91.5% in lsm8-1 and 71.4% among the events affected in both. The levels of IR in lsm8-1 seedlings were in line with previous results (31). Importantly, the hierarchical cluster analysis indicated that, despite the range of ΔPIR/PSI values in the pfd2,4,6 mutant was smaller compared to lsm8-1, the majority of common events showed a similar trend in each mutant (Figure 5D), as expected if PFDs and LSM2–8 act in the same pathway to process these introns. For instance, Figure 5E and F show IGV images of one IR and one exon skipping affected similarly in pfd2,4,6 and lsm8-1 mutant seedlings. Together, these results indicate that PFD function is required for pre-mRNA splicing of a particular set of genes and that part of this action is likely mediated by its effect on LSM8 protein levels.
Table 1.
Classification of splicing events found in this work
| Type of events (n) | Type of events (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Condition | Genotype | Events | Genes with events | IR | ES | Alt5SS | Alt3SS | IR | ES | Alt5SS | Alt3SS |
| Standard (20°C) | pfd4 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 100 | 0 | 0 |
| pfd2,4,6 | 208 | 195 | 155 | 17 | 10 | 26 | 74.52 | 8.17 | 4.81 | 12.5 | |
| lsm8-1 | 1161 | 898 | 1063 | 61 | 16 | 21 | 91.56 | 5.25 | 1.38 | 1.81 | |
| pfd2,4,6 and lsm8-1 | 42 | 39 | 30 | 5 | 2 | 5 | 71.43 | 11.9 | 4.76 | 11.9 | |
| Cold (4°C) | pfd4 | 226 | 216 | 161 | 18 | 15 | 32 | 71.24 | 7.96 | 6.64 | 14.16 |
| lsm8-1 | 1350 | 1031 | 1292 | 36 | 9 | 13 | 95.7 | 2.67 | 0.67 | 0.96 | |
| pfd4 and lsm8-1 | 46 | 46 | 37 | 7 | 1 | 1 | 80.43 | 15.22 | 2.17 | 2.17 | |
| NaCl (150 mM) | lsm8-1 | 1764 | 1449 | 1723 | 14 | 13 | 14 | 97.68 | 0.79 | 0.74 | 0.79 |
Altered splicing events usually produce mRNA variants that carry features associated to NMD (57). Notably, the great majority of splicing events altered in the pfd2,4,6 mutant, in the lsm8-1 mutant or in both, were IR that contain at least one feature potentially triggering NMD (Table 2) (31). The proportion of transcripts with IR and NMD features that are translated and, therefore, susceptible of entering the decay pathway is particularly small in plants (35). Interestingly, it has been demonstrated that the IR-containing, NMD-resistant splice variants of two Arabidopsis genes are retained in the nucleus (58), which opens the possibility that this mechanism extends to other alternatively spliced transcripts. Therefore, PFDs might contribute to regulate gene expression post-transcriptionally through its action on pre-mRNA splicing.
Table 2.
Genes containing NMD signatures in the IR events found in pfd4, pfd2,4,6 and lsm8-1 mutants
| Genes with NMD-features | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genes with events | Genes analyzed | PTCa | 3′UTR > 350ntb | SC→intron > 55ntc | 5′uORFd | uORF overlape | NMD genes (%)f | Non-NMD genes (%)g | |
| pfd2,4,6 at 20°C | 195 | 183 | 102 | 124 | 141 | 65 | 16 | 173 (94.5) | 10 (5.5) |
| lsm8-1 at 20°C | 898 | 842 | 531 | 583 | 761 | 229 | 80 | 816 (96.9) | 26 (3.1) |
| pfd4 at 4°C | 216 | 201 | 60 | 127 | 92 | 67 | 21 | 172 (85.6) | 29 (14.4) |
| lsm8-1 at 4°C | 1031 | 970 | 628 | 678 | 877 | 250 | 84 | 941 (97.0) | 29 (3.0) |
aPremature termination codon (PTC) and a 3′UTR longer than 350nt.
b3′UTR longer than 350nt.
cMore than 55 nt between stop codon (SC) and a downstream intron.
dUpstream open reading frame (uORF) longer than 35 amino acids.
euORF overlapping with the start codon of the main ORF.
fTotal number of genes containing at least one retained intron causing NMD features.
gTotal number of genes containing retained introns that do not generate NMD features.
PFD4 is required for adequate LSM2–8 function depending on the environmental conditions
The lack of splicing defects in the pfd4 mutant prompted us to hypothesize that PFD4 function might be particularly relevant under specific environmental conditions. PFD4 and LSM8 mRNA levels increase 24 h after low temperature exposure, while the accumulation of the corresponding proteins is evident after 48 h (17,31). It is important to note that the activity of both proteins is necessary to attenuate the cold acclimation response (17,31). As expected, PFD4 mRNA levels were upregulated in response to cold in our transcriptomic analysis (Supplementary Figure S5A). Therefore, we reasoned that PFD4 activity might be important to ensure cellular levels of LSM8 under cold conditions. LSM8-GFP protein levels were specifically reduced in the pfd4 mutant compared to the WT at 4°C (Figure 4A and B), while transcript levels were not affected (Figure 4D), suggesting that post-transcriptional regulation of LSM8 by PFDs also operates at low temperature. Indeed, the upregulation of U6 snRNA at 4°C (31) is attenuated in the pfd4 mutant, similarly to lsm8-1 (Supplementary Figure S6). Then, we investigated the pre-mRNA splicing in three independent biological replicates of WT, pfd4 and lsm8-1 mutant seedlings grown under long days at 20°C and transferred to 4°C for 24 h (Supplementary Table S3). In sharp contrast to the lack of splicing defects in the pfd4 mutant grown at 20°C (Figure 5B), we observed 226 altered splicing events corresponding to 216 different genes after cold exposure, with 46 of them (20%; P < 0.0001, chi-square test) being also detected in the lsm8-1 mutant (Figure 6A, Supplementary Figure S7 and Table 1). The retention of three introns detected in the transcriptomic analysis that belong to three common genes was confirmed by RT-qPCR using independent RNA samples, validating the RNA-seq data (Figure 6B-D). Interestingly, 92 out of the 216 genes (42.6%) showing altered splicing events in the pfd4 mutant under cold conditions, for instance VLN4, BET9, CHIL, CBL10 or At4g36980, have been described to be regulated in response to low temperature (59) (Supplementary Table S3). The majority of events altered in both mutants at 4°C were IR containing NMD features (Figure 6E and Tables 1 and 2), suggesting that PFD4 action on pre-mRNA splicing might impact gene expression post-transcriptionally under this environmental condition as well. The hierarchical clustering of the common events indicated a positive correlation in both mutants, albeit changes in the pfd4 mutant were of lower magnitude (Figure 6F). These results indicate that PFD4 is required for proper LSM2–8 function at low temperature, most likely by ensuring adequate levels of the LSM2–8 complex.
Figure 6.
Analysis of pre-mRNA splicing by RNA-seq in pfd4 and lsm8-1 mutants in response to cold treatment. (A) Venn diagram showing the number of splicing events altered in the pfd4 and lsm8-1 mutants. (B–D) Examples of common IR highlighted in IGV plots (left) and validated by RT-qPCR (right); VLN4:I008 (B), At4g36980:I012 (C), and BET9:I007 (D). (E) Percentage of the different splicing events affected in each mutant and in both. Alt3SS and Alt5SS, alternative 3′ and 5′ splice site, respectively; ES, exon skipping; IR, intron retention. (F) Hierarchical clustering of common events by ΔPIR/PSI (percent intron retention/percent spliced-in) value.
The LSM2–8 complex and PFD3 and 5 operate as positive regulators of Arabidopsis tolerance to salt stress (31,47,60). We observed that pfd4 seedlings were hypersensitive to high salt treatment, indicating that PFD4 activity is also required by the plant to properly respond to this stress (Supplementary Figure S8). Therefore, we decided to investigate if PFD4 is required for the function of the LSM2–8 complex under high salt conditions. Interestingly, our RNA-seq analysis on high salt-treated samples only identified a single splicing event altered in the pfd4 mutant compared to the WT, while 1764 events corresponding to 1449 genes were altered in the lsm8-1 mutant (Supplementary Figure S9A and Supplementary Table S4), being the great majority IR (Table 1 and Supplementary Figure S9B-E), in line with previous results (31). Contrarily to the upregulation of PFD4 gene in response to cold, its expression was no altered by salt treatment (Supplementary Figure S5B). These data indicate that, in contrast to its role under cold conditions, PFD4 activity does not seem to be required for adequate LSM2–8 function in response to salt stress. PFD4, therefore, seems to constitute a regulatory element of the complex, depending on the environmental conditions.
PFD4 acts as co-chaperone of Hsp90 to control levels of LSM8
Having shown that PFD activity is required to maintain cellular levels of LSM8 protein thus ensuring proper pre-mRNA splicing, we investigated the molecular mechanism underlying the LSM8 accumulation. PFDs are co-chaperones of two major cellular chaperones, the chaperonin CCT and Hsp90 (8,46). We focused on Hsp90 as the levels of its client proteins are usually reduced when the activity of the chaperone is compromised (61). To investigate if LSM8 is a client of Hsp90, we treated pLSM8:LSM8-GFP seedlings with the Hsp90 inhibitor GDA (62) and analyzed LSM8-GFP levels by Western-blot. GDA is commonly used to impair Hsp90 activity in Arabidopsis (63). LSM8-GFP levels were reduced by nearly 50% in response to either short (Figure 7A and B) or long (Supplementary Figure S10A and B) GDA-treatments and, consistent with the 26S-proteasome being involved in the destabilization (61), LSM8-GFP levels were restored to control ones by simultaneous treatment with GDA and the 26S-proteasome inhibitor MG132 (Figure 7A and B and Supplementary Figure S10A and B). These results suggested that Hsp90 activity contributes to attain adequate levels of LSM8. We reasoned that this could be the result of PFDs acting as co-chaperones of Hsp90. In agreement with this idea, Hsp90 and LSM8-HA were co-immunoprecipitated with anti-GFP antibodies from extracts of plants co-expressing LSM8-HA and either PFD4 or PFD6 fused to YFP (Figure 7C), suggesting the presence of LSM8-PFD4-Hsp90 and LSM8-PFD6-Hsp90 complexes in the cells. The interaction of Hsp90 with LSM8 and PFD4 was also observed in seedlings expressing either LSM8-GFP or PFD4-GFP under the control of their own promoters (Figure 7D). Importantly, the interaction between Hsp90 and LSM8-GFP is primarily dependent on PFD4, as we were unable to detect co-immunoprecipitated Hsp90 after LSM8-GFP was pulled-down from pfd4 extracts (Figure 7D). Reaching adequate LSM8 levels has consequences in the assembly of the LSM2–8 complex, as U6 snRNA levels were reduced when Hsp90 activity was compromised by GDA-treatment, while the reduction was reverted by simultaneous treatment with GDA and MG132; the MG132-treatment alone did not have any significant effect on U6 snRNA levels (Figure 7E).
Figure 7.
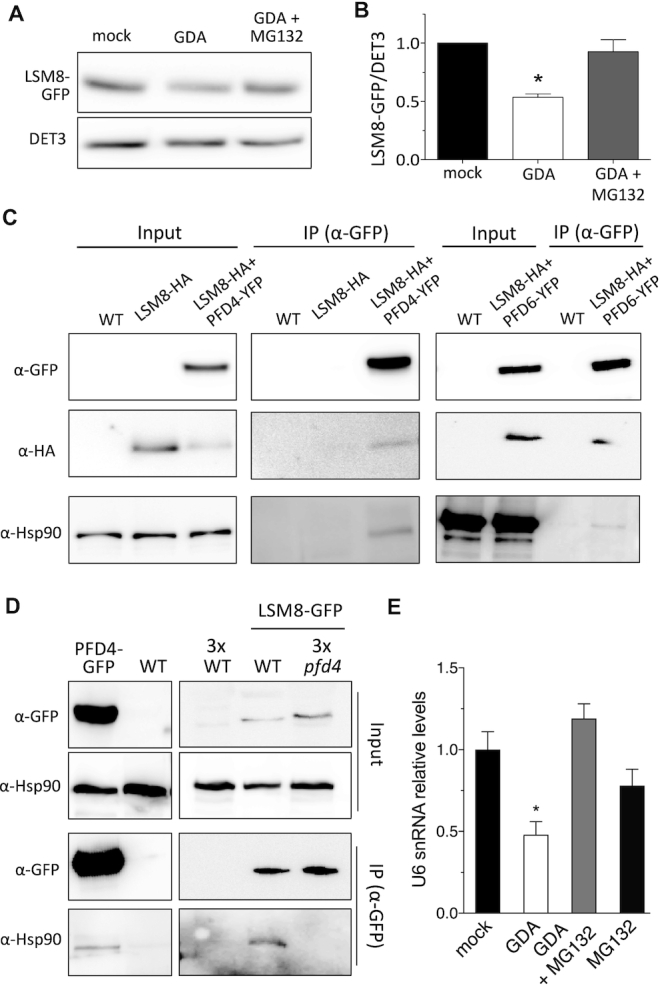
Hsp90 stabilizes LSM8 protein. (A, B) LSM8-GFP protein levels in response to 24 h treatments with 20 μM GDA or with 20 μM GDA + 50 μM MG132. A representative blot is shown in (A). (B) Plot showing the LSM8-GFP levels after normalization to DET3 that is used as loading control. Data are average from two biological replicates. One asterisk represents P < 0.01 in an ANOVA test. (C) Co-immunoprecipitation assay showing the interaction of PFD4-YFP or PFD6-YFP with LSM8-HA and Hsp90. Total proteins were immunoprecipitated with anti-GFP antibody-coated paramagnetic beads from extracts of 7-day-old seedlings. Proteins were detected with anti-GFP, anti-HA and anti-Hsp90 antibodies. (D) Co-immunoprecipitation assay showing that PFD4 mediates the interaction of Hsp90 with LSM8-GFP in Arabidopsis. Total proteins were immunoprecipitated with anti-GFP antibody-coated paramagnetic beads from extracts of 7-day-old seedlings. Proteins were detected with anti-GFP and anti-Hsp90 antibodies. Note that we used three times the amount of protein extract from non-transgenic control and pfd4 seedlings for the immunoprecipitation. (E) Relative U6 snRNA levels in response to treatments with 20 μM GDA, with 20 μM GDA + 50 μM MG132 or with 50 μM MG132. U6 snRNA levels were normalized against PP2AA3 mRNA. Data are average of three biological replicates. Error bars indicate standard error of mean.
DISCUSSION
Despite the activity of the LSM and Sm complexes is key for pre-mRNA splicing (44), our current knowledge about mechanisms regulating their activity is scarce. For instance, the LSM2–8 complex is regulated by interacting with Pat1b, a protein formerly identified as interactor of the LSM1–7 complex in the cytoplasm and that is also required for proper splicing in animals and algae (64,65). Furthermore, some LSM and Sm proteins are di-methylated in animals (66) and plants (67), where this modification is required for proper assembly of the snRNPs (68). Here, we show that an extra layer of post-translational control operates at the core of the spliceosome. The results presented here, led us to propose a model in which the level of the protein LSM8 is dependent on the major chaperone Hsp90 and that PFDs act as co-chaperones.
Hsp90 acts when its clients are close to their native conformation and is assisted by numerous co-chaperones that either drive the conformational changes associated to the chaperone activity cycle, act as adaptors to recruit client proteins, or both (69). We propose that PFD4 would act as an adaptor for the recruitment of LSM8 to the chaperone. The interaction between PFD4 and LSM8 is likely direct. However, it is unclear at present whether PFD4 interacts directly with Hsp90 or if it requires another co-chaperone. TPR domain-containing co-chaperones that bind directly to the Hsp90 and have been related to PFDs in the context of the R2TP/prefoldin-like complex (8) are good candidates.
The dependence of LSM8 levels on PFDs/Hsp90 contributes to the activity of the spliceosome, based on the splicing defects observed in mutants impaired in PFD activity. The selection of the right splice sites in a particular pre-mRNA is key for the accuracy of the spliceosome activity. Interestingly, this is strongly influenced by the levels of spliceosome core proteins (70). For instance, downregulation of LSM or Sm proteins results in the use of alternative splicing sites in animals (71) and in plants (31,40–43,72). Thus, the splice site selection can be adjusted by controlling the level of these proteins, being some splice sites more sensitive to the concentration of a particular core protein than to others when its level is limiting (70,72). The positive effect of PFDs on LSM8 levels provides, therefore, a likely mechanism to explain the selection of splicing events that require the concerted action of both proteins, i.e. the splicing of these events is most sensitive to the level of LSM8. It is tempting to speculate that PFDs act here as regulatory proteins that affect the extent to which a particular intron is alternatively spliced, which might be particularly relevant under challenging environmental conditions. The cold-specific requirement of PFD4 activity for pre-mRNA splicing of a set of genes, together with the fact that the expression of the PFD4 gene is upregulated in response to cold (Supplementary Figure S5A) (17), indicate that it might well represent an entry point to feed the spliceosome with temperature information by controlling the level of LSM8. The differential behavior of the pfd4 mutant in response to the two environmental challenges is striking. We understand that the differential regulation of the PFD4 expression under the two conditions, i.e. upregulation at low temperature and no change under salt stress (Supplementary Figure S5A and B), might be at the base of the differential response. Nonetheless, a contribution of particular biophysical features of PFD4 that allow it to efficiently perform at low temperature, but not under salt stress, cannot be discarded.
Three aspects of the analysis of pre-mRNA splicing in the pfd mutants drew particularly our attention. First, LSM8 levels are less affected by the loss of PFD4 at 4°C than 20°C, while defects in splicing occur only at the low temperature in the mutant (Figures 4–6). Although these results seem incompatible with the requirement of PFD4 to maintain adequate levels of LSM8 contributing to splicing at 4°C, several pieces of evidence support the idea that the plant requires more LSM8 and, therefore, is more sensitive to its changes under low temperature conditions: (i) LSM8 accumulates at 4°C (31); (ii) PFD4 accumulates and moves into the nucleus at 4°C (17); (iii) U6 snRNA levels increase at 4°C in a PFD4- and LSM8-dependent manner (Supplementary Figure S6); and (iv) lsm8 and pfd4 mutants showed altered cold acclimation (17,31). Second, the small overlap of altered events between pfd2,4,6 and lsm8-1 under standard conditions and between pfd4 and lsm8-1 in response to low temperature (Figures 5B and 6A). This overlap would reflect a partial contribution of PFD/Hsp90 to maintain adequate LSM8 levels. And third, the majority of events altered in pfd mutants are not affected by the lsm8-1 mutation (Figures 5B and 6A and Supplementary Figures S11 and S12), which indicates that the effect of PFDs on LSM8 protein levels represents only a partial explanation for the effect of PFDs in pre-mRNA splicing and that they should act through LSM8-independent pathways as well. This idea is supported by three additional evidences: (i) the splicing profiles of pfd and lsm8-1 mutants are different despite the U6 snRNA levels are reduced to a similar extent (Figure 5A); (ii) the percentages of the various types of splicing events affected in pfd and lsm8-1 mutants are different (Figures 5C and 6E), which might be considered as a molecular signature of partially overlapping pathways; and (iii) the -1 and -2 positions of the 5′ splice junction of all introns showing altered splicing in pfd2,4,6 and pfd4 mutants deviate from the consensus, i.e. are weak sites, whereas they are mostly conserved in lsm8-1 mutants (Supplementary Figures S13 and S14). We suggest that PFDs might also affect pre-mRNA splicing through the Sm complex, which participates in the maturation of many pre-mRNAs (72) showing low sensitivity to diminished LSM2–8 activity in Arabidopsis. The differential sensitivity of particular splicing events to levels of different core components has been previously demonstrated in animals (73,74). Consistent with this assumption, some PFD genes are co-expressed with Sm genes, both in Arabidopsis and in mouse (Figure 1A, Supplementary Figure S1 and Supplementary Table S5), and PFD3 and PFD6 interact physically with SmD1A and SmD2A (Supplementary Figure S15). PFDs, therefore, might also participate in controlling adequate levels of Sm proteins.
In summary, our results indicate that PFD activity is required to maintain cellular levels of the LSM2–8 spliceosome core complex by mediating the interaction of LSM8 with the Hsp90 chaperone. The action of PFDs/Hsp90 represents, therefore, a novel layer of regulation for the LSM2–8 complex that might operate in other eukaryotes as well, given the elevated conservation of PFDs and spliceosome components. Indeed, the strong genetic interaction between the PFD4 mutant gim3 and lsm8 in Saccharomyces cerevisiae (75) supports this possibility.
DATA AVAILABILITY
The data supporting the findings of this study are available from the corresponding authors upon request. Raw sequences (fastq files) and count tables at gene, exon, intron, AS bin and junction levels used in this study have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE124812).
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124812.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Chávez and J. Forment for helpful comments on the manuscript and bioinformatics support, respectively, and K. Schumacher (University of Heidelberg, Heidelberg, Germany) for the anti-DET3 antibody. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness and ‘Agencia Española de Investigación’/FEDER/European Union [BIO2016-79133-P to D.A., BIO2016-79187-R to J.S.]; European Union Research and Innovation Staff Exchange [H2020-MSCA-RISE-2014-644435 to M.A.B., D.A. and M.J.Y]; N.B.-T. was recipient of a ‘Formación de Personal Investigador’ predoctoral fellowship from the Spanish Ministry of Economy and Competitiveness. Funding for open access charge: Spanish Ministry of Economy and Competitiveness [BIO2016-79133-P].
Conflict of interest statement. None declared.
REFERENCES
- 1. Vainberg I.E., Lewis S.A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H.L., Cowan N.J.. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998; 93:863–873. [DOI] [PubMed] [Google Scholar]
- 2. Payán-Bravo L., Peñate X., Chávez S.. Functional contributions of prefoldin to gene expression. Adv Exp Med Biol. 2018; 1106:1–10. [DOI] [PubMed] [Google Scholar]
- 3. Wang D., Shi W., Tang Y., Liu Y., He K., Hu Y., Li J., Yang Y., Song J.. Prefoldin 1 promotes EMT and lung cancer progression by suppressing cyclin A expression. Oncogene. 2017; 36:885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Satou A., Taira T., Iguchi-Ariga S.M., Ariga H.. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 2001; 276:46562–46567. [DOI] [PubMed] [Google Scholar]
- 5. Mori K., Maeda Y., Kitaura H., Taira T., Iguchi-Ariga S.M., Ariga H.. MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J. Biol. Chem. 1998; 273:29794–29800. [DOI] [PubMed] [Google Scholar]
- 6. Millán-Zambrano G., Rodríguez-Gil A., Peñate X., de Miguel-Jiménez L., Morillo-Huesca M., Krogan N., Chávez S.. The prefoldin complex regulates chromatin dynamics during transcription elongation. PLoS Genet. 2013; 9:e1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banks C.A.S., Miah S., Adams M.K., Eubanks C.G., Thornton J.L., Florens L., Washburn M.P.. Differential HDAC1/2 network analysis reveals a role for prefoldin/CCT in HDAC1/2 complex assembly. Sci. Rep. 2018; 8:13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houry W.A., Bertrand E., Coulombe B.. The PAQosome, an R2TP-based chaperone for quaternary structure formation. Trends Biochem Sci. 2018; 43:4–9. [DOI] [PubMed] [Google Scholar]
- 9. Malinova A., Cvackova Z., Mateju D., Horejsi Z., Abeza C., Vandermoere F., Bertrand E., Stanek D., Verheggen C.. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017; 216:1579–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson E., Bouchard A., Faubert D., Chabot B., Coulombe B.. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat Commun. 2017; 8:15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M.C., Ahmad Y., Neel H. et al.. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell. 2010; 39:912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zur Lage P., Stefanopoulou P., Styczynska-Soczka K., Quinn N., Mali G., von Kriegsheim A., Mill P., Jarman A.P.. Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J. Cell Biol. 2018; 217:2583–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mita P., Savas J.N., Ha S., Djouder N., Yates J.R. 3rd, Logan S.K.. Analysis of URI nuclear interaction with RPB5 and components of the R2TP/prefoldin-like complex. PLoS One. 2013; 8:e63879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kimura Y., Nagao A., Fujioka Y., Satou A., Taira T., Iguchi-Ariga S.M., Ariga H.. MM-1 facilitates degradation of c-Myc by recruiting proteasome and a novel ubiquitin E3 ligase. Int. J. Oncol. 2007; 31:829–836. [PubMed] [Google Scholar]
- 15. Locascio A., Blázquez M.A., Alabadí D.. Dynamic regulation of cortical microtubule organization through prefoldin-DELLA Interaction. Curr Biol. 2013; 23:804–809. [DOI] [PubMed] [Google Scholar]
- 16. Claeys H., De Bodt S., Inze D.. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2013; 19:231–239. [DOI] [PubMed] [Google Scholar]
- 17. Perea-Resa C., Rodríguez-Milla M.A., Iniesto E., Rubio V., Salinas J.. Prefoldins negatively regulate cold acclimation in arabidopsis thaliana by promoting nuclear proteasome-mediated HY5 Degradation. Mol. Plant. 2017; 10:791–804. [DOI] [PubMed] [Google Scholar]
- 18. Mockenhaupt S., Makeyev E.V.. Non-coding functions of alternative pre-mRNA splicing in development. Semin. Cell Dev. Biol. 2015; 47-48:32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Catalá R., Carrasco-López C., Perea-Resa C., Hernández-Verdeja T., Salinas J.. Emerging roles of LSM complexes in posttranscriptional regulation of plant response to abiotic Stress. Front Plant Sci. 2019; 10:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Y., Deng Z., Paredez A.R., DeBolt S., Wang Z.Y., Somerville C.. Prefoldin 6 is required for normal microtubule dynamics and organization in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 2008; 105:18064–18069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perea-Resa C., Hernández-Verdeja T., López-Cobollo R., del Mar Castellano M., Salinas J.. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell. 2012; 24:4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J.. The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 2007; 120:157–165. [DOI] [PubMed] [Google Scholar]
- 23. Obayashi T., Aoki Y., Tadaka S., Kagaya Y., Kinoshita K.. ATTED-II in 2018: A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 2018; 59:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Obayashi T., Kagaya Y., Aoki Y., Tadaka S., Kinoshita K.. COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019; 47:D55–D62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Belda-Palazón B., Ruíz L., Martí E., Tárraga S., Tiburcio A.F., Culiáñez F., Farràs R., Carrasco P., Ferrando A.. Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One. 2012; 7:e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S.. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006; 45:616–629. [DOI] [PubMed] [Google Scholar]
- 28. Sarrión-Perdigones A., Palaci J., Granell A., Orzáez D.. Design and construction of multigenic constructs for plant biotechnology using the GoldenBraid cloning strategy. Methods Mol Biol. 2014; 1116:133–151. [DOI] [PubMed] [Google Scholar]
- 29. Olate E., Jiménez-Gomez J.M., Holuigue L., Salinas J.. NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat Plants. 2018; 4:811–823. [DOI] [PubMed] [Google Scholar]
- 30. Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R.. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005; 139:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carrasco-López C., Hernández-Verdeja T., Perea-Resa C., Abia D., Catalá R., Salinas J.. Environment-dependent regulation of spliceosome activity by the LSM2–8 complex in Arabidopsis. Nucleic Acids Res. 2017; 45:7416–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mancini E., Iserte J., Yanovsky M., Chernomoretz A.. ASpli: Analysis of alternative splicing using RNA-Seq. R package version 1.6.0. 2017; [Google Scholar]
- 33. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hahne F., Ivanek R.. Visualizing genomic data using gviz and bioconductor. Methods Mol Biol. 2016; 1418:335–351. [DOI] [PubMed] [Google Scholar]
- 35. Yu H., Tian C., Yu Y., Jiao Y.. Transcriptome survey of the contribution of alternative splicing to proteome diversity in arabidopsis thaliana. Mol Plant. 2016; 9:749–752. [DOI] [PubMed] [Google Scholar]
- 36. Hernando C.E., Sánchez S.E., Mancini E., Yanovsky M.J.. Genome wide comparative analysis of the effects of PRMT5 and PRMT4/CARM1 arginine methyltransferases on the Arabidopsis thaliana transcriptome. BMC Genomics. 2015; 16:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eisen M.B., Spellman P.T., Brown P.O., Botstein D.. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. U.S.A. 1998; 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee H.K., Hsu A.K., Sajdak J., Qin J., Pavlidis P.. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004; 14:1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H.. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007; 35:D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pérez-Santángelo S., Mancini E., Francey L.J., Schlaen R.G., Chernomoretz A., Hogenesch J.B., Yanovsky M.J.. A role for LSM genes in the regulation of circadian rhythms. Proc. Natl Acad. Sci. U.S.A. 2014; 111:15166–15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui P., Zhang S., Ding F., Ali S., Xiong L.. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 2014; 15:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Golisz A., Sikorski P.J., Kruszka K., Kufel J.. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013; 41:6232–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elvira-Matelot E., Bardou F., Ariel F., Jauvion V., Bouteiller N., Le Masson I., Cao J., Crespi M.D., Vaucheret H.. The nuclear ribonucleoprotein SmD1 interplays with splicing, RNA quality control, and posttranscriptional gene silencing in arabidopsis. Plant Cell. 2016; 28:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell Mol. Biol. 2009; 272:149–189. [DOI] [PubMed] [Google Scholar]
- 46. Sahlan M., Zako T., Yohda M.. Prefoldin, a jellyfish-like molecular chaperone: functional cooperation with a group II chaperonin and beyond. Biophys. Rev. 2018; 10:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodríguez-Milla M.A., Salinas J.. Prefoldins 3 and 5 play an essential role in Arabidopsis tolerance to salt stress. Mol. Plant. 2009; 2:526–534. [DOI] [PubMed] [Google Scholar]
- 48. Pannone B.K., Xue D., Wolin S.L.. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998; 17:7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li J., Fu J., Chen Y., Fan K., He C., Zhang Z., Li L., Liu Y., Zheng J., Ren D. et al.. The U6 biogenesis-like 1 plays an important role in maize kernel and seedling development by Affecting the 3′ end processing of U6 snRNA. Mol. Plant. 2017; 10:470–482. [DOI] [PubMed] [Google Scholar]
- 50. Kambach C., Walke S., Nagai K.. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 1999; 9:222–230. [DOI] [PubMed] [Google Scholar]
- 51. Reijns M.A., Auchynnikava T., Beggs J.D.. Analysis of Lsm1p and Lsm8p domains in the cellular localization of Lsm complexes in budding yeast. FEBS J. 2009; 276:3602–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galletti R., Johnson K.L., Scofield S., San-Bento R., Watt A.M., Murray J.A., Ingram G.C.. DEFECTIVE KERNEL 1 promotes and maintains plant epidermal differentiation. Development. 2015; 142:1978–1983. [DOI] [PubMed] [Google Scholar]
- 53. Teotia S., Lamb R.S.. The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 2009; 151:180–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F.. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell. 2009; 21:3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C.. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999; 11:1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van Norman J.M., Frederick R.L., Sieburth L.E.. BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol. 2004; 14:1739–1746. [DOI] [PubMed] [Google Scholar]
- 57. Kalyna M., Simpson C.G., Syed N.H., Lewandowska D., Marquez Y., Kusenda B., Marshall J., Fuller J., Cardle L., McNicol J et al.. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012; 40:2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gohring J., Jacak J., Barta A.. Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis. Plant Cell. 2014; 26:754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K.. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007; 50:347–363. [DOI] [PubMed] [Google Scholar]
- 60. Xiong L., Gong Z., Rock C.D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J.K.. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell. 2001; 1:771–781. [DOI] [PubMed] [Google Scholar]
- 61. Theodoraki M.A., Caplan A.J.. Quality control and fate determination of Hsp90 client proteins. Biochim. Biophys. Acta. 2012; 1823:683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Whitesell L., Mimnaugh E.G., De Costa B., Myers C.E., Neckers L.M.. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:8324–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Queitsch C., Sangster T.A., Lindquist S.. Hsp90 as a capacitor of phenotypic variation. Nature. 2002; 417:618–624. [DOI] [PubMed] [Google Scholar]
- 64. Reimer K.A., Stark M.R., Aguilar L.C., Stark S.R., Burke R.D., Moore J., Fahlman R.P., Yip C.K., Kuroiwa H., Oeffinger M. et al.. The sole LSm complex in Cyanidioschyzon merolae associates with pre-mRNA splicing and mRNA degradation factors. RNA. 2017; 23:952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vindry C., Marnef A., Broomhead H., Twyffels L., Ozgur S., Stoecklin G., Llorian M., Smith C.W., Mata J., Weil D. et al.. Dual RNA processing roles of pat1b via cytoplasmic Lsm1-7 and nuclear Lsm2–8 complexes. Cell Rep. 2017; 20:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brahms H., Meheus L., de Brabandere V., Fischer U., Luhrmann R.. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001; 7:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Z., Zhang S., Zhang Y., Wang X., Li D., Li Q., Yue M., Li Q., Zhang Y.E., Xu Y. et al.. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell. 2011; 23:396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chari A., Golas M.M., Klingenhager M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U.. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008; 135:497–509. [DOI] [PubMed] [Google Scholar]
- 69. Schopf F.H., Biebl M.M., Buchner J.. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017; 18:345–360. [DOI] [PubMed] [Google Scholar]
- 70. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saltzman A.L., Pan Q., Blencowe B.J.. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011; 25:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Huertas R., Catala R., Jimenez-Gomez J., Castellano M.M., Crevillen P., Pineiro M., Jarillo J.A., Salinas J.. Arabidopsis SME1 regulates plant development and response to abiotic stress by determining spliceosome activity specificity. Plant Cell. 2019; 31:537–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Corrionero A., Minana B., Valcarcel J.. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011; 25:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Papasaikas P., Tejedor J.R., Vigevani L., Valcarcel J.. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell. 2015; 57:7–22. [DOI] [PubMed] [Google Scholar]
- 75. Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D. et al.. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016; 353:aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon request. Raw sequences (fastq files) and count tables at gene, exon, intron, AS bin and junction levels used in this study have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE124812).
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124812.