Abstract
Fungal infections, widespread throughout the world, affect a broad range of life forms, including agriculturally relevant plants, humans, and insects. In defending against fungal infections, the fruit fly Drosophila melanogaster employs the Toll pathway to induce a large number of immune peptides. Some have been investigated, such as the antimicrobial peptides (AMPs) and Bomanins (Boms); many, however, remain uncharacterized. Here, we examine the role in innate immunity of two related peptides, Daisho1 and Daisho2 (formerly IM4 and IM14, respectively), found in hemolymph following Toll pathway activation. By generating a CRISPR/Cas9 knockout of both genes, Δdaisho, we find that the Daisho peptides are required for defense against a subset of filamentous fungi, including Fusarium oxysporum, but not other Toll-inducible pathogens, such as Enterococcus faecalis and Candida glabrata. Analysis of null alleles and transgenes revealed that the two daisho genes are each required for defense, although their functions partially overlap. Generating and assaying a genomic epitope-tagged Daisho2 construct, we detected interaction in vitro of Daisho2 peptide in hemolymph with the hyphae of F. oxysporum. Together, these results identify the Daisho peptides as a new class of innate immune effectors with humoral activity against a select set of filamentous fungi.
Keywords: innate immunity, toll, Drosophila, humoral, antifungal
Introduction
Fungal infections have a devastating impact on a wide range of organisms. They are destructive to agricultural plants around the world, including rice, wheat, and tomatoes (1). Additionally, fungi infect more than one million humans annually (2). Existing antifungal treatments are limited, with only one new class of drugs, echinocandins, developed in the past 15 years. Furthermore, extensive usage of limited classes of related antifungals has led to the increasingly frequent appearance of drug-resistant fungi (2). An enhanced understanding of naturally occurring antifungal defenses is thus of tremendous potential benefit.
The fruit fly Drosophila melanogaster is a robust model for fungal infections, replicating many features of murine fungal infections (3, 4). In the wild, flies have been found to be infected with a number of filamentous fungi, including Beauveria, Metarhizium, and Fusarium species (5, 6). In combatting these infections, flies rely on the Toll innate immune pathway (7, 8). Toll provides defense against not only filamentous fungi, but also yeasts and those Gram-positive bacteria that produce a cell wall containing Lys-type peptidoglycan (8–10). A second innate immune pathway, defined by the Imd receptor, provides defense against Gram-negative bacteria and the limited number of Gram-positive bacteria that produce a cell wall containing DAP-type peptidoglycan (11, 12).
Systemic activation of Toll signaling induces a broad set of genes first identified by microarray analysis and mass spectroscopy (13–16). Many of the induced innate immune genes are transcribed in the fly fat body, with the protein products secreted into the hemolymph. These include antimicrobial peptides (AMPs), the Bomanin peptides, and a number of uncharacterized peptides.
Although AMPs, such as the antifungal peptide Drosomycin (Drs) directly kill pathogens in vitro (17, 18) and are immunoprotective when ectopically expressed in vivo (19), recent loss-of-function studies reveal little or no requirement for AMPs in defense against fungi and Gram-positive bacteria (20). In contrast, the Bomanin family of peptides (Boms) are required for defense against both classes of pathogens (21). Boms, which are Drosophila-specific, are readily detected in hemolymph following Toll activation. Here we describe the functional characterization of additional immune effectors, the Daisho peptides, which appear in hemolymph following systemic infection and are required for defense against a subset of filamentous fungi.
Materials and Methods
Fly Husbandry and Strain Generation
Flies were raised at 25°C on cornmeal molasses agar media1. The w1118 strain was used as the wild type. MyD88− flies were MyD88kra1, and imd− flies were imdshadok.
As described in Results, the genes for the immune induced peptides IM4 and IM14 have been given the designations daisho1 and daisho2, respectively. The null allele Δdaisho, deleting both genes, as well as the individual gene deletions, Δdso1 and Δdso2, were generated using CRISPR/Cas9 technology, applying methods described previously (22). Pairs of guide RNAs that targeted Cas9 to delete the region 2R: 20,868,460–20,870,480 for Δdaisho), 2R: 20,868,783–20,869,392 for Δdso1, and 2R: 20,870,332–20,870,728 for Δdso2 were cloned into the pU6-BbsI-chiRNA vector (Addgene plasmid # 45946). Homology arms of ~1 kb were cloned into pHD-DsRed (Addgene plasmid # 51434). Cas9 was provided by plasmid pBS-Hsp70-Cas9 (Addgene plasmid #46294). Constructs were based on target sequences in the w1118 strain and injected into w1118. See Table S1 for primer sequences.
The FLAG epitope tag was cloned between the signal sequence and mature peptide of Dso2 in the context of the pHD-DsRed homologous repair template. This FLAG-Dso2 construct was introduced at the dso2 genomic locus using the Δdso2 guide RNAs.
Plasmids expressing dso1 or dso2 transcripts from the pBomS3 promoter were made using methods previously described (23). Briefly, the BomS3 gene promoter was placed 5′ to the ORF encoding either Dso1 or Dso2. These constructs were then each integrated via ΦC31-mediated transgenesis at an attP landing site located at 86Fb on the D. melanogaster third chromosome (BDSC stock #24749). The transgenes were crossed into the Δdso1 and Δdso2 backgrounds and homozygous stocks were derived. An empty vector control was also introduced at the 86Fb attP landing site.
Microbial Cultures
For survival experiments, microbes were cultured as follows. Enterococcus faecalis NCTC 775 (ATCC 19433) and Enterobacter cloacae were grown overnight at 37°C in LB media and concentrated to an OD600 of 10 in 20% glycerol. Candida glabrata CBS 138 [ATCC 2001] was grown overnight in YPD media at 37°C and concentrated to an OD600 of 100 in PBS, 0.1% Tween. All filamentous fungi were grown on malt extract agar plates at 29°C until sporulation was observed (10–15 days). Fungal material was then strained through glass wool with sterile water to collect spores, which were concentrated in 20% glycerol and stored at −80°C before being used at the following concentrations (in spores/ml): Aspergillus flavus (sequenced strain): 5 × 109; A. fumigatus AF293 (FGSC# A1100): 6 × 109; A. parasiticus Nor-1 mutant (NRRL #6111): 3 × 109; Botrytis cinerea (B05.10): 3 × 109; Fusarium graminearum (NRRL #5883): 8 × 108; F. oxysporum f. sp. lycopersici 4287 (FGSC #9935): 3 × 108; F. verticillioides (FGSC #7415): 3 × 109; Neurospora crassa: 1 × 109.
For the induction of the Toll response, heat-killed Micrococcus luteus was prepared as previously described (23).
Survival Assays
Groups of 20–25 adult male flies aged 2–7 days were collected and stabbed with a needle dipped in a suspension of bacteria, yeast, or fungal spores. Where needed, MyD88− or BomΔ55C flies were used as controls immunodeficient for the Toll-mediated response. Flies infected with E. faecalis were incubated at 25°C; all other infected flies were incubated at 29°C. Fly deaths were recorded at least twice per day for the duration of each experiment. Any deaths that occurred within the first 6 h were set aside to exclude from the data any deaths due to traumatic injury. The experiment was repeated three times and results combined. Statistical analyses were performed using the Gehan-Breslow-Wilcoxon test.
MALDI-TOF
After Toll induction with heat-killed M. luteus, flies were incubated at 29°C for 24 h, after which hemolymph was collected via capillary as previously described (23). Hemolymph in 0.1% trifluoroacetic acid/50% acetonitrile was mixed 1:1 with Universal Matrix (Sigma-Aldrich). Samples were then dried onto a Bruker MSP 96 ground steel plate. Spectra were collected from 1,500 to 10,000 m/z for linear mode, and 1,000–5,000 m/z for reflectron mode, both with positive polarization. Peptide calibration standard II (Bruker) was used as an external calibration standard. For each genotype, at least five independent samples were collected. Representative spectra are shown. Peaks were identified via corresponding m/z values from previous studies (13, 16). Spectra were visualized using R 3.3.2 and ggplot2 2.2.1 (24, 25).
Quantitation of Pathogen Load
Pathogen load in infected flies was measured by qRT-PCR of fungal RNA (26, 27). Adult male flies, 2–7 days old, were stabbed with a needle dipped in F. verticillioides at 3 × 109 spores/ml. Flies were then incubated at 29°C. Groups of 5–6 flies were collected at the stated times and frozen in liquid nitrogen. Total RNA was isolated with TRIzol (Ambion) and cDNA was made via SuperScript RT II (Invitrogen). EF1A was selected as a proxy gene for fungal load based on its stable expression (28). Measurements by qRT-PCR were performed on the iQ5 cycler (BioRad) with iQ SYBR Green Supermix (BioRad) using the primers listed below. Values were normalized to fly mRNA based on expression of the rp49 gene.
Primers: Fv_EF1A_F1: GGCTTTCACTGACTACCCTCCTCT, Fv_EF1A_R1: ACTTCTCGACGGCCTTGATGACAC, rp49_F1: CAAGGGTATCGACAACAG, rp49_R1: CTTGTTCGATCCGTAACC.
Peptide Gel Electrophoresis and Immunoblotting
Hemolymph samples were collected via the Zymo-Spin IC column method (23) from 30 male flies aged 2–7 days that had been induced with heat-killed M. luteus and incubated for 24 h at 29°C. Samples were run on a SDS-tricine, 18% separating/10% spacer/4% stacking, acrylamide gel2. Protein samples were then transferred to a PDVF membrane, blocked with 5% milk in TBST and stained with primary α-FLAG M2 (Sigma) (1:500) and secondary sheep α-mouse HRP (Amersham Biosciences) (1:1,000). The immunoblot was then treated with West Pico PLUS substrate (Thermo Scientific) and exposed to film.
Peptide Hyphal Binding and Immunofluorescence
The immunostaining protocol was adapted from Luo et al. (29). F. oxysporum was grown in 5 ml malt extract broth from a starting concentration of 2.9 × 105 spores/ml. After overnight shaking at room temperature, fungal hyphae were collected by centrifugation at 1,000 g for 10 min and resuspended in PBS. Hemolymph was collected via the Zymo-Spin IC column method (23) from 420 male flies that had been induced with heat-killed M. luteus 24 h prior and incubated at 29°C, yielding ~35 μl cell-free hemolymph. Next, aliquots of 200 μl hyphae and 35 μl hemolymph were shaken at room temperature for 30 min. The samples were washed three times with PBS before fixation with 4% formaldehyde for 1 h. After washing another three times with PBS, samples were blocked for 1 h with 5% BSA. Samples were then incubated with α-FLAG antibody (1:200) overnight at 4°C. After washing with PBS, samples were stained for 2 h with donkey α-mouse Alexa555 (1:400) and DAPI (1:200) and then washed and mounted on slides. Samples were imaged with a Ti2 Widefield microscope (Nikon) and analyzed with the NIS- elements software and OMERO.
Results
Generation of Flies Null for the daisho Gene Pair
Pioneering mass spectrometry experiments by Bulet et al. identified two dozen peptide IMs (immune-induced molecules) that accumulate in Drosophila hemolymph upon induction of the innate immune response, principally the Toll pathway (13, 16). Among these, the Bomanins have been found to play an essential role against a broad range of pathogens (21, 23) while several, including the 15 aa long IM4 and 24 aa long IM14, have unknown functions. Based on our demonstration of defensive functions for these peptides, we have renamed them Daisho1 and Daisho2, for  daisho, the Japanese term for a matched pair of samurai swords, one short and one long.
daisho, the Japanese term for a matched pair of samurai swords, one short and one long.
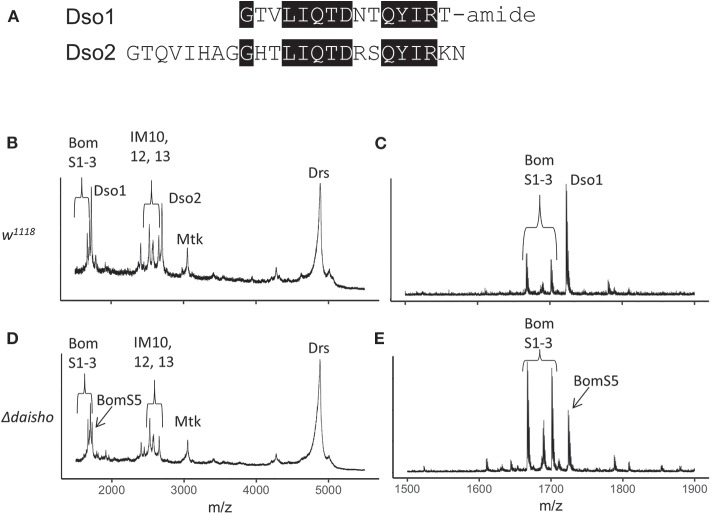
The Daisho peptides are closely related to one another and occupy adjacent positions in the genome, where they are divergently transcribed (Figure S1). As shown in Figure 1A, the sequence of amidated mature Daisho1 (Dso1) has 67% identity with the corresponding region of the mature Daisho2 (Dso2) peptide. Like the Bomanins, the daisho genes are widespread among the Drosophila genus, but not identified elsewhere. To investigate the potential role of the daisho genes in innate immunity, we used CRISPR/Cas9 technology to delete both genes. The 2.0 kb deleted region includes the entire dso1 gene, the upstream region for both genes, and the first exon of dso2 (including the start codon). Flies homozygous for the Δdaisho1,2 deletion, hereafter Δdaisho, were viable and fertile.
Figure 1.
Deletion of Drosophila daisho1 and daisho2 gene pair. (A) Alignment of mature Daisho1 and Daisho2 peptide sequences. Identical residues are highlighted. (B–E) Mass spectrometry analysis of Toll-induced hemolymph in linear (B,C) and reflectron (D,E) mode, illustrating loss of Daisho1 (Dso1, formerly IM4) and Daisho2 (Dso2, formerly IM14) signal in Δdaisho deletion mutant. The Dso1 signal overlaps with the BomS5 signal, which is readily apparent in the Δdaisho mutant analyzed in reflectron mode. Mtk, Metchnikowin; Drs, Drosomycin.
With the Δdaisho stock in hand, we carried out MALDI-TOF studies of hemolymph (Figures 1B–E). As described above, following Toll activation wild-type hemolymph displays robust expression of immune peptides, including the Daisho peptides, Bomanins, and AMPs. The signals from Dso1 and Dso2 were ablated in Δdaisho, as evidenced by the loss of signal at 1,722 mass/charge (m/z) (Dso1) and 2,694 m/z (Dso2). Furthermore, the spectra of induced Δdaisho hemolymph was wild-type for all previously identified peaks other than Dso1 and Dso2, including the Bomanins and AMPs, Metchnikowin (Mtk), and Drosomycin (Drs). The absence of Dso1 and Dso2 thus did not detectably alter the accumulation or modification of other Toll-induced peptides in the hemolymph.
In addition to previously identified peaks, Δdaisho hemolymph contained one previously unseen signal. The 1,724 m/z signal of this peak, readily apparent in reflectron mode, is identical to that predicted for the BomS5 amidated peptide, previously known as CG15065 (Figure 1E). This signal had not been detected previously because in the wild type it lies in the shoulder of the robust Dso1(IM4) peak. Its existence in Toll-induced hemolymph was expected, however, on the basis of microarray and RNAseq data demonstrating strong Toll-activated induction of the BomS5 locus (15, 30).
The daisho Genes Are Specifically Required for Defense Against F. oxysporum
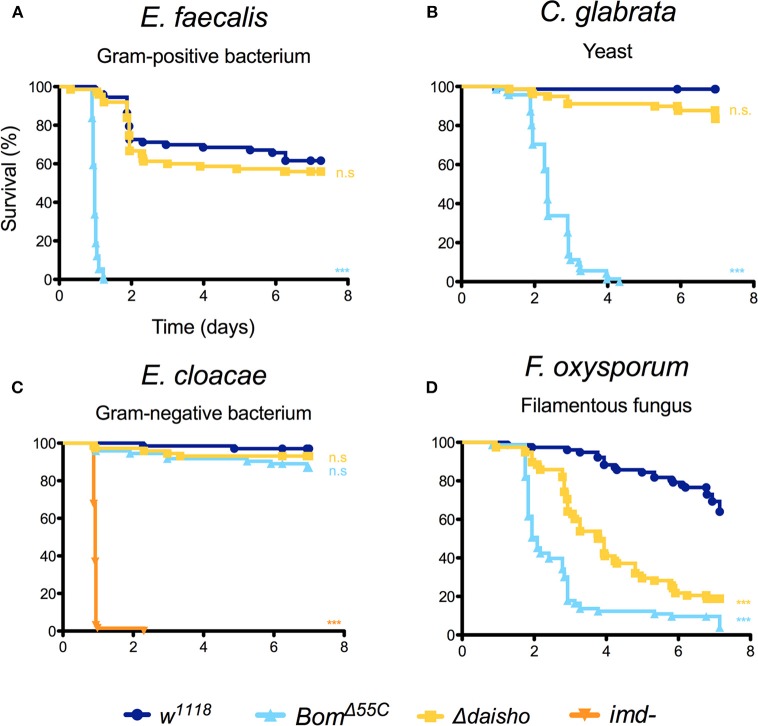
We next turned to a functional assay to determine whether the absence of the Daisho peptides impaired survival following systemic infection. Because the Toll pathway responds to and protects against infection by many Gram-positive bacteria and fungi, we focused on these classes of pathogens. We stabbed adult flies with a needle dipped in a suspension of bacteria, yeast, or fungal spores and then monitored survival. We used w1118 flies as our wild-type, i.e., immunocompetent, control and BomΔ55C flies, which lack the 10-gene Bom cluster, as an immunodeficient control (21).
For a number of the pathogens tested, Δdaisho flies behaved identically to the wild type. Roughly 50% of both wild-type and Δdaisho flies survived 6 or more days following infection with the Gram-positive bacteria Enterococcus faecalis, whereas 100% of BomΔ55C flies died within 2 days (Figure 2A). Likewise, wild-type and Δdaisho flies survived a week or longer after infection with the yeast Candida glabrata, whereas BomΔ55C flies died in 4 days or fewer (Figure 2B). We also found no effect of Δdaisho on immune defenses mediated by the Imd pathway: wild-type, Δdaisho, and BomΔ55C flies all survived infection with the Gram-negative bacteria Enterobacter cloacae, whereas, control imd- flies died within 1 day (Figure 2C).
Figure 2.
Survival of Δdaisho against E. faecalis (A), C. glabrata (B), E. cloacae (C), and F. oxysporum (D) infection. Shown is the combination of three independent experiments for each pathogen with 20-25 flies per genotype per experiment. Survival curves were compared using the Gehan-Breslow-Wilcoxon test. Significance is shown relative to w1118 (***p < 0.0001; n.s., not significant; p > 0.01).
For one pathogen in the initial test set, the filamentous fungus Fusarium oxysporum, deletion of the daisho genes had a marked effect on survival (Figure 2D): 50% of flies homozygous for Δdaisho died within 4 days of infection. In contrast, <70% of wild-type flies survived 7 or more days post-infection. Thus, loss of the Daisho peptides disrupts defense against F. oxysporum, but not other tested pathogens. Interestingly, loss of the Daisho peptides did not impact survival peptides did not impact survival as severely as did loss of the Boms, which led to 50% death after 2 days, very similar to complete loss of Toll signaling (21).
Δdaisho Flies Are Susceptible to Some but Not All Filamentous Fungi
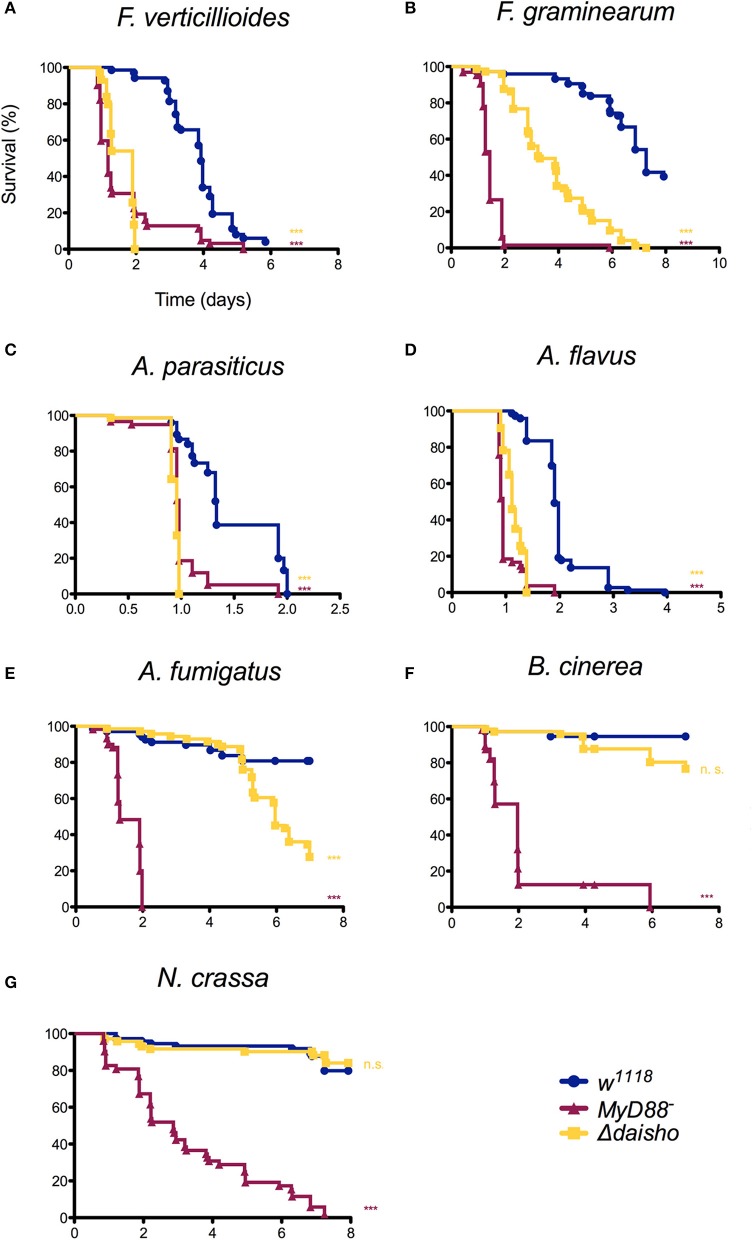
We next investigated whether the susceptibility of Δdaisho flies to F. oxysporum reflected a general susceptibility to filamentous fungi. For these studies, we focused on filamentous fungi for which flies deficient for Toll signaling, and thus for induction of Daisho1, Daisho2, and other Toll effectors, exhibit a significantly decreased survival relative to wild type (Figure 3). The control fly strains in each case were w1118 (wild type) and kra-1 (MyD88−), a loss-of-function allele for an essential mediator of Toll signaling (31).
Figure 3.
Survival of Δdaisho against F. verticillioides, (A), F. graminearum (B), A. parasiticus (C), A. flavus (D), A. fumigatus (E), B. cinerea (F), and N. crassa (G). The combination of three independent experiments for each pathogen with 20-25 flies per genotype per experiment is shown. Survival curves were compared using the Gehan-Breslow-Wilcoxon test. Significance is shown relative to w1118 (***p > 0.0001; n.s., not significant; p > 0.01).
As shown in Figure 3, susceptibility of Δdaisho flies to the filamentous fungi species varied. Survival was significantly less than wild-type for F. verticillioides and F. graminearum (panels A, B), two Fusarium species closely related to F. oxysporum. In the case of F. graminearum, survival of Δdaisho flies was intermediate between that of wild-type and MyD88− flies, a pattern very similar to that observed with F. oxysporum, where Δdaisho survival falls between wild type and BomΔ55C, which behaves similarly to MyD88− (21). In contrast, Δdaisho flies displayed a much greater immune impairment upon infection with F. verticillioides than with F. oxysporum, dying to a comparable extent and at a similar rate as the MyD88− control (compare Figures 2D, 3A).
Variation in survival was also observed among Aspergillus species. The survival curves of Δdaisho infected with either A. parasiticus or A. flavus largely tracked with MyD88− (panels C, D). Upon A. fumigatus infection, however, Δdaisho flies survived at least twice as long as MyD88− flies (Figure 3E).
For some filamentous fungi, loss of Daisho1 and Daisho2 did not affect survival. For example, 80% of wild-type and Δdaisho flies survived for at least 7 days after infection with Botrytis cinerea, whereas >50% of MyD88− flies died after 2 days (Figure 3F). Likewise, wild-type and Δdaisho flies survived Neurospora crassa infection for 6 days or more, but 50% of MyD88− flies died after 3 days (Figure 3G). Overall, we find that the Daisho peptides play a vital role in survival after infection with certain species of filamentous fungi, but are not important for infections with others.
daisho1 and daisho2 Are Each Required for Defense
Daisho1 and Daisho2 are highly similar in sequence and expression pattern. Are they functionally redundant? To address this question, we explored the function of each individual locus. We again used CRISPR/Cas9, generating deletions that removed the entire coding sequence for either daisho1 or daisho2. The 5' endpoints of each deletion were chosen to lie within 100 bp of the transcriptional start site, minimizing potential disruption of elements in the regulatory region separating the two genes (Figure S1). For both deletions, MALDI-TOF analysis of induced hemolymph confirmed loss of the deleted gene product but no other peptides, indicating that either Daisho1 or Daisho2 can be stably expressed in the absence of the other (Figure 4).
Figure 4.
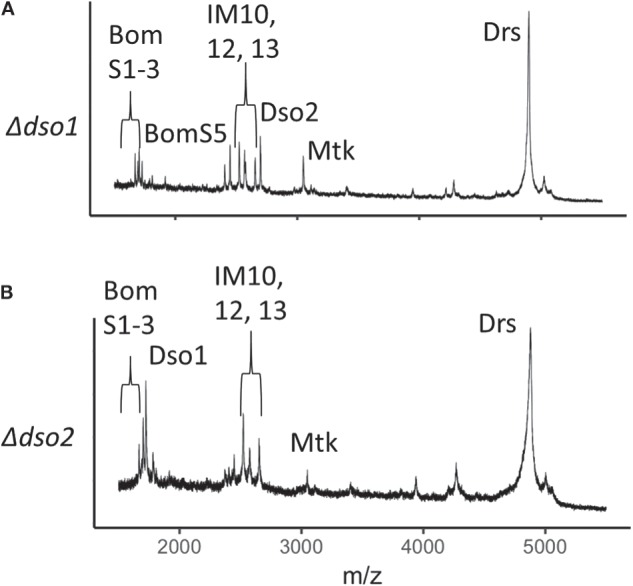
MALDI-TOF spectra for Δdso1 and Δdso2 hemolymph. (A,B) Mass spectrometry analysis of Toll-induced hemolymph in linear mode, highlighting loss of Dso1 (A) and Dso2 (B) in deletion mutants.
To test the effect on defense of deleting dso1 or dso2, we stabbed adults with F. verticillioides spores, for which Δdaisho flies have a reduced survival. Deleting either the dso1 or dso2 gene resulted in susceptibility to F. verticillioides markedly different from wild-type and comparable to that of the double deletion (Figure 5). Thus, Daisho1 and Daisho2 each act in defense against F. verticillioides infection.
Figure 5.
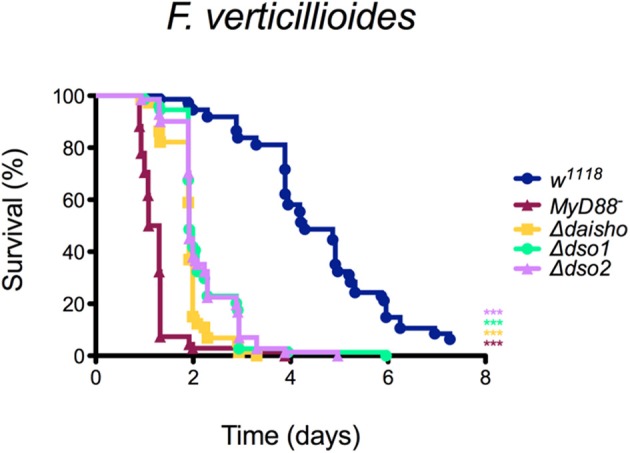
Survival of Δdso1 and Δdso2 against F. verticillioides. Shown is the combination of three independent experiments with 20–25 flies per genotype per experiment. Survival curves were compared using the Gehan-Breslow-Wilcoxon test. Significance is shown relative to w1118 (***p < 0.0001).
Since deletion of either dso1 or dso2 had as severe an effect on survival as the double mutant, it was possible that each gene has a specific and distinct function in antifungal defense. Alternatively, survival might depend only on total dosage for the two genes, with loss of either dropping expression below the threshold required. To distinguish between these models, we generated transgenes placing each ORF under control of pBomS3, shown previously to be strongly Toll-responsive promoter (23), and then assayed the transgenes for rescue of Δdso1 or Δdso2. As shown in Table 1, pBomS3-driven dso1 rescued Δdso1, improving the median survival from 46 to 93 h (p < 0.0001). The same was true of pBomS3-driven dso2 in the Δdso2 background (p < 0.0001) (see Figure S2 for full survival curves). Flies expressing the empty vector construct at the same chromosomal location did not show any increase in survival (Figure S3).
Table 1.
Median survival in hours of dso1 and dso2 deletion mutations rescued by homotypic and heterotypic transgenes.
| No transgene | pBomS3-dso1 | pBomS3-dso2 | |
|---|---|---|---|
| MyD88− | 29 | n.a. | n.a. |
| Δdaisho | 46 | n.a. | n.a. |
| Δdso1 | 46 | 93 | 78 |
| Δdso2 | 46 | 55 | 93 |
| w1118 | 103 | n.a. | n.a. |
Data derived from Figure S2. n.a., not applicable.
Having confirmed the activity of the two constructs, we expressed each in a background deficient for the other. dso2 expression significantly improved survival of Δdso1 flies, increasing median survival from 46 to 78 h (p < 0.0001). Similarly, dso1 expressed in a Δdso2 background improved median survival from 46 to 55 h (p = 0.0005). Nevertheless, rescue was incomplete. The median survival of dso2 expressed in Δdso1 background (78 h) did not reach median survival of Δdso1 rescued with dso1 (93 h) (n.s., p = 0.09). Furthermore, dso1 did not rescue survival of Δdso2 (55 h) to the same level as dso2 (93 h) (p < 0.0001). The data thus indicate that the two loci encode functions that are neither fully distinct nor fully redundant.
Deleting daisho1 and daisho2 Results in an Elevated Pathogen Load in Infected Flies
To investigate whether Daisho1 and Daisho2 affect pathogen growth during infection, we measured fungal load after infection in Δdaisho and wild-type flies. After stabbing adult males with F. verticillioides, groups of 5–6 infected flies were collected and RNA was extracted. Fungal EF1A transcript levels were measured as a proxy for pathogen load and normalized to the fly reference gene rp49. Directly after infection (2 h), there was no significant difference between Δdaisho and wild-type flies by Mann-Whitney U test (Figure S4). By the next day, however, Δdaisho flies had a pathogen load roughly 10-fold greater than wild-type (p = 0.0317).
FLAG-Dso2 Binds to F. oxysporum Hyphae
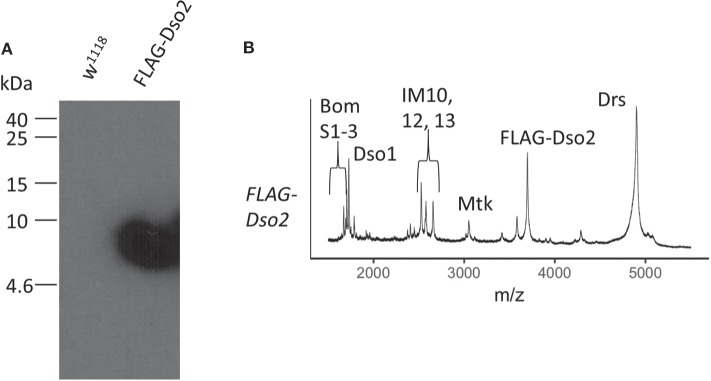
We next tagged Dso2, the larger of the two peptides, with the FLAG epitope, using CRISPR/Cas9 to introduce the tag at the amino-terminus of the endogenously expressed mature peptide. Immunoblot analysis of induced hemolymph from FLAG-Dso2 flies revealed a single band detectable with α-FLAG antibody (Figure 6A). MALDI-TOF analysis of hemolymph confirmed the loss of the Dso2 peak at 2,694 m/z and the appearance of a peak with an m/z ratio of 3,689, the value expected for FLAG-Dso2 (Figure 6B).
Figure 6.
Characterization of FLAG-Dso2 gene product. (A) Immunoblot stained with mouse α-FLAG M2 (1:500) and sheep α-mouse HRP (1:1,000). Two μl of Toll-induced hemolymph was loaded per lane. (B) MALDI-TOF analysis of FLAG-Dso2 Toll-induced hemolymph in linear mode.
Having confirmed that FLAG-Dso2 peptide is stably expressed, we next assayed its activity in providing antifungal defense. Specifically, flies homozygous for FLAG-dso2 at the dso2 locus were infected with F. verticillioides and their survival was compared to both wild-type flies and Δdso2 flies. Survival of FLAG-Dso2 flies was not wild-type, but was significantly better than that of flies lacking the dso2 gene (Figure S5). We conclude that the FLAG-Dso2 peptide is active in providing defense against F. verticillioides infection.
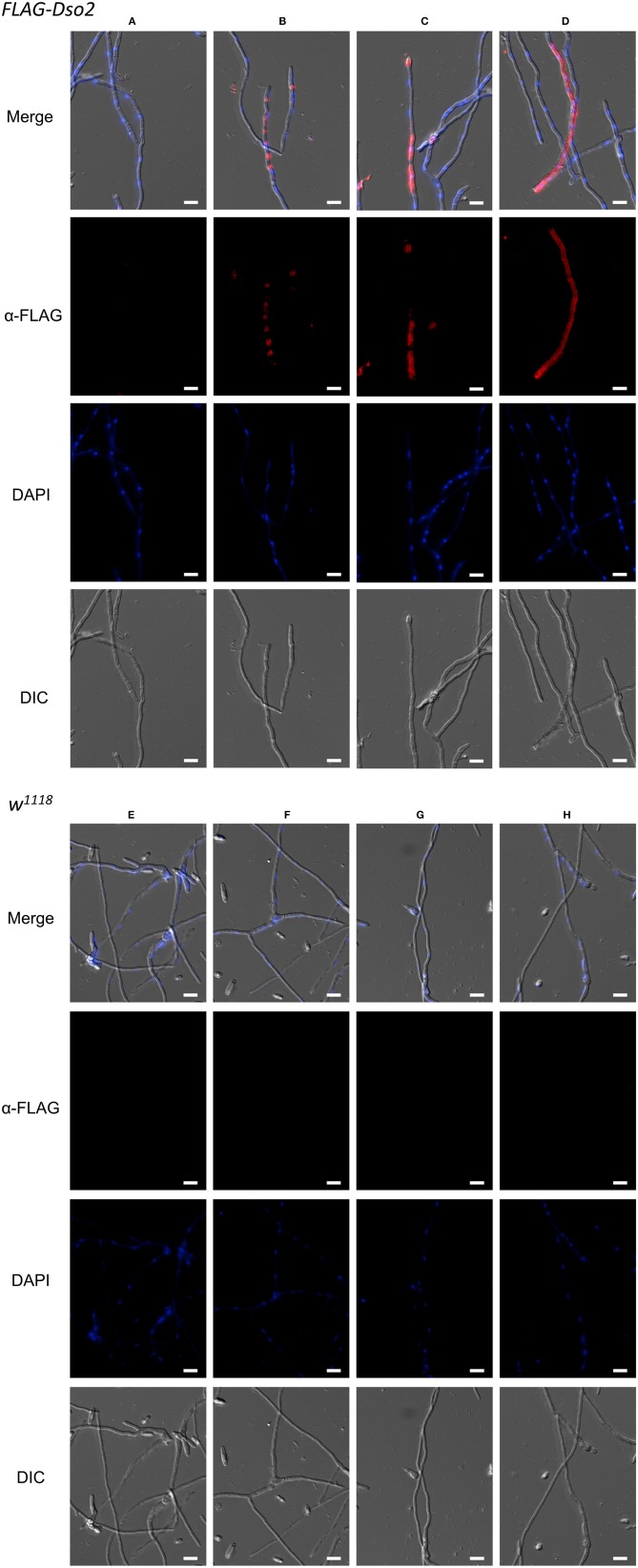
Next, we assayed FLAG-tagged Dso2 peptide in hemolymph for its ability to bind fungus. We collected hemolymph from Toll induced flies, incubated it with hyphae from F. oxysporum, and fixed samples. The majority (>80%) of F. oxysporum hyphae had no visible signal when stained with α-FLAG antibody (Figure 7A). Among the remaining hyphae, we observed a variety of staining patterns, including, but not limited to, signals concentrated in the regions between nuclei (Figure 7B), extending across greater fractions of hyphae (Figure 7C) or spanning the length of hyphae (Figure 7D). In parallel experiments with untagged wild-type hemolymph, no signal was detected (Figures 7E–H). We conclude that Daisho2 peptide in hemolymph can bind to F. oxysporum hyphae.
Figure 7.
Immunofluorescence staining of F. oxysporum hyphae. Images showing various staining patterns of hyphae incubated with FLAG-Dso2 (A–D) or w1118 hemolymph (E–H) and then stained with mouse α-FLAG M2 (1:200) and donkey α-mouse Alexa 555 (1:400). DAPI marks fungal DNA. Scale bar is 10 μm. Images were generated as focused images from Z-stacks.
In summary, our results demonstrate that the pair of immune-induced peptides, Daisho1 and Daisho2, mediate Toll-induced defense against specific filamentous fungi, most likely via a humoral effect on fungal hyphae.
Discussion
Role of the Daisho Peptides in Antifungal Defense
In this study we found that the related peptides Daisho1 and Daisho2 are required in D. melanogaster for defense against a subset of filamentous fungi. We have also demonstrated that the two peptides have partially overlapping functions. Survival data reveal a dependence on the overall level of Dso1 and Dso2, with each peptide able to partially compensate for the absence of the other. Furthermore, each peptide accumulates in the absence of the other.
The Daisho peptides lack known motifs of defined function. As noted previously (21), there is a similarity in size and sequence between Dso1 and Dso2 and the Bomanin peptides. There are, however noteworthy differences, including the presence of a CxxC motif in the Bomanins and the broader requirement for the Bomanins in Toll-mediated defense.
Among those fungi for which deleting dso1 and dso2 decreases survival, Δdaisho flies nevertheless often exhibit significantly greater survival than do MyD88− or BomΔ55C flies (see e.g., F. oxysporum and F. graminearum). Thus, in contrast to the Bom effectors, which are strictly required for Toll defenses against a broad range of pathogens, the Daisho peptides appear to be required for some, but not all Toll functions and to be active against only a select group of pathogens against which Toll mounts defense.
Like the Bomanins, dso1 and dso2 are found only within the Drosophila genus. Taxonomically-restricted genes (TRGs), while often studied only sparingly, represent 10–20% of most genomes and frequently have essential functions (32). TRGs have been identified in the immune pathways of many invertebrates, including flies, mosquitoes, and cnidarians. Within immune systems they are abundant among effectors, but rare among signal transduction factors (33, 34).
Specificity of daisho Genes in Antifungal Defense
In tracking survival following systemic infection, we find considerable variability with regard to which pathogens exhibit increased virulence toward D. melanogaster in the absence of both daisho genes. Categorizing the fungi against which the daisho genes provide defense, we detect no simple relationship to fungal phylogeny. For example, the daisho genes are required to defend against all the Fusarium species tested and some of the Aspergillus species, but not Neurospora crassa. Yet Fusarium and Neurospora are both members of the class Sordariomycetes, whereas Aspergillus is part of the less closely related Eurotiomycetes class (35, 36). Furthermore, Δdaisho flies exhibit differential susceptibility to fungi within a single genus: the Δdaisho deletion substantially decreases survival against A. flavus and A. parasiticus, but has a much smaller effect on survival following A. fumigatus infection.
Although susceptibility of Δdaisho flies does not track simply with fungal phylogeny, susceptibility does appear to be closely related to fungal pathogenicity. Consider four filamentous fungi that are particularly pathogenic for wild-type flies: F. verticillioides, F. graminearum, A. flavus, and A. parasiticus. Infection with any of these four pathogens kills >50% of wild-type flies within 7 days. For each of these four, Δdaisho greatly decreases survival. By comparison, consider filamentous fungi with low pathogenicity, e.g., A. fumigatus, N. crassa, and B. cinerea. For each, >80% of wild-type and Δdaisho flies survive for 7 or more days post-infection. Note that we observe this association of susceptibility with pathogenicity only among filamentous fungi: for the strongly pathogenic Gram-positive bacterium E. faecalis, the Δdaisho deletion had no effect on survival.
Although the Bomanins are strictly required for Toll humoral defenses, we have found a correlation between pathogenicity and the level of Bomanin function required to confer resistance (21). It thus appears that for both Bomanins and the daisho genes, pathogenicity tracks with the strength of effector function required for defense.
Activity of Daisho Peptides
How do the Daisho peptides provide defense against filamentous fungi? One mechanism could be directly binding and killing the pathogens. Consistent with this idea, we find a modest but significant increase in pathogen load in Δdaisho flies. In addition, our immunofluorescence data demonstrate that Daisho2 can interact in vitro with at least one filamentous fungus that it targets. Antifungal peptides, such as mammalian LL-37 and plant defensin NaD1, also bind hyphae of fungal pathogens against which they are active (29, 37). The Daisho2 peptide's ability to bind fungal hyphae could indicate an antimicrobial function. Given that our assay was carried out with crude hemolymph, we cannot state whether the observed interaction of Daisho2 with hyphae is direct or is mediated by one or more unidentified hemolymph components.
The Daisho peptides might themselves interfere with pathogen growth, survival, or proliferation, or they might enable the fungicidal activity of other factors. The same is true of the Bomanins, which are required for hemolymph mediated killing of C. glabrata, but for which fungicidal activity of synthetic peptides has not been observed (23). Given that daisho genes are required for defense against only a subset of Toll and Bomanin targets, the function of Daisho1 and Daisho2 may be to meet a specific challenge posed by certain fungi to the entry or activity of antimicrobial factors. Further investigation of the Daisho peptides, as well as other hemolymph immune effectors, is likely to be informative in this regard.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
LC and SW conceived the project and wrote the paper. LC, SAL, YX, and SJHL performed and analyzed the experiments.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Bo Zhou, Beatriz Pinatti, and André Soares for the generation of the pU6-chiRNA constructs. We would also like to thank Philipp Weckwerth and Alisa Huffaker for the generous gift of F. graminearum, F. verticillioides, A. flavus, A. parasiticus, and B. cinerea. We thank Andrés Mauricio Caraballo Rodríguez and the Dorrestein lab for access to and assistance with MALDI-TOF. We also thank the McGinnis, Gantz, Posakony, and Bier labs for access to equipment. We thank Anita Hermann for help with immunofluorescence and imaging and Eric Griffis and Daphne Bindels from the Nikon Imaging Center at UC San Diego for microscopy expertise. We thank Matt Daugherty, Bill McGinnis, and Kirthi Reddy for thoughtful reading of the manuscript and helpful comments.
Funding. This work was supported by National Institutes of Health (NIH) grant R01 GM050545 (to SW).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00009/full#supplementary-material
References
- 1.Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. (2012) 13:414–30. 10.1111/j.1364-3703.2012.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janbon G, Quintin J, Lanternier F, d'Enfert C. Studying fungal pathogens of humans and fungal infections: fungal diversity and diversity of approaches. Genes Immun. (2019) 20:403–14. 10.1038/s41435-019-0071-2 [DOI] [PubMed] [Google Scholar]
- 3.Brunke S, Quintin J, Kasper L, Jacobsen ID, Richter ME, Hiller E, et al. Of mice, flies–and men? Comparing fungal infection models for large-scale screening efforts. Dis Model Mech. (2015) 8:473–86. 10.1242/dmm.019901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dionne MS, Schneider DS. Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis Model Mech. (2008) 1:43–9. 10.1242/dmm.000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma L, Marques G. Fusarium, an entomopathogen—a myth or reality? Pathogens. (2018) 7:E93 10.3390/pathogens7040093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuthbertson AGS, Audsley N. Further screening of entomopathogenic fungi and nematodes as control agents for drosophila suzukii. Insects. (2016) 7:E24. 10.3390/insects7020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imler JL. Overview of drosophila immunity: a historical perspective. Dev Comp Immunol. (2014) 42:3–15. 10.1016/j.dci.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 8.Valanne S, Wang JH, Rämet M. The Drosophila toll signaling pathway. J Immunol. (2011) 186:649–56. 10.4049/jimmunol.1002302 [DOI] [PubMed] [Google Scholar]
- 9.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- 10.Lindsay SA, Wasserman SA. Conventional and non-conventional Drosophila Toll signaling. Dev Comp Immunol. (2014) 42:16–24. 10.1016/j.dci.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. (2001) 1:503–14. 10.1016/S1534-5807(01)00059-4 [DOI] [PubMed] [Google Scholar]
- 12.Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat Rev Immunol. (2014) 14:796–810. 10.1038/nri3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci USA. (1998) 95:11342–7. 10.1073/pnas.95.19.11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. (2001) 98:12590–5. 10.1073/pnas.221458698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. (2002) 21:2568–79. 10.1093/emboj/21.11.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy F, Rabel D, Charlet M, Bulet P, Hoffmann JA, Ehret-Sabatier L. Peptidomic and proteomic analyses of the systemic immune response of Drosophila. Biochimie. (2004) 86:607–16. 10.1016/j.biochi.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 17.Levashina EA, Ohresser S, Bulet P, Reichhart J, Hetru C, Hoffmann JA. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur J Biochem. (1995) 233:694–700. 10.1111/j.1432-1033.1995.694_2.x [DOI] [PubMed] [Google Scholar]
- 18.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert WF, Hetru C, et al. Insect immunity: Septic injury of drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. (1994) 269:33159–63. [PubMed] [Google Scholar]
- 19.Tzou P, Reichhart JM, Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc Natl Acad Sci USA. (2002) 99:2152–7. 10.1073/pnas.042411999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife. (2019) 8:e44341 10.7554/eLife.44341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for Drosophila Toll-mediated immunity. PLoS Pathog. (2015) 11:e1004876. 10.1371/journal.ppat.1004876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. (2014) 196:961–71. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsay SA, Lin SJH, Wasserman SA. Short-Form bomanins mediate humoral immunity in Drosophila. J Innate Immun. (2018) 10:306–14. 10.1159/000489831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; (2016). [Google Scholar]
- 25.Wickham H. ggplot2: Elegant Graphics for Data Analysis. (2016). [Google Scholar]
- 26.Troha K, Buchon N. Methods for the study of innate immunity in Drosophila melanogaster. Wiley Interdiscip Rev Dev Biol. (2019) 8:e344. 10.1002/wdev.344 [DOI] [PubMed] [Google Scholar]
- 27.Dostálová A, Rommelaere S, Poidevin M, Lemaitre B. Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Biol. (2017) 15:79. 10.1186/s12915-017-0408-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HK, Yun SH. Evaluation of potential reference genes for quantitative RTPCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol J. (2011). Available online at: https://peerj.com/preprints/27537.pdf (accessed August 8, 2019).
- 29.Luo XL, Li JX, Huang HR, Duan JL, Dai RX, Tao RJ, et al. LL37 inhibits aspergillus fumigatus infection via directly binding to the fungus and preventing excessive inflammation. Front Immunol. (2019) 10:283. 10.3389/fimmu.2019.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valanne S, Salminen TS, Järvelä-Stölting M, Vesala L, Rämet M. Immune-inducible non-coding RNA molecule lincRNA-IBIN connects immunity and metabolism in Drosophila melanogaster. PLoS Pathog. (2019) 15:e1007504 10.1371/journal.ppat.1007504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. (2002) 3:91–7. 10.1038/ni747 [DOI] [PubMed] [Google Scholar]
- 32.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. (2009) 25:404–13. 10.1016/j.tig.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 33.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. (2007) 316:1738–43. 10.1126/science.1139862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. (2007) 39:1461–8. 10.1038/ng.2007.60 [DOI] [PubMed] [Google Scholar]
- 35.Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia. (2006) 98:1076–87. 10.3852/mycologia.98.6.1076 [DOI] [PubMed] [Google Scholar]
- 36.Schoch CL, Sung GH, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, et al. The ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. (2009) 58:224–39. 10.1093/sysbio/syp020 [DOI] [PubMed] [Google Scholar]
- 37.van der Weerden NL, Lay FT, Anderson MA. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem. (2008) 283:14445–52. 10.1074/jbc.M709867200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.