Abstract
Accurate chromosome segregation in mitosis requires sister kinetochores to bind to microtubules from opposite spindle poles. The stability of kinetochore–microtubule attachments is fine‐tuned to prevent or correct erroneous attachments while preserving amphitelic interactions. Polo kinase has been implicated in both stabilizing and destabilizing kinetochore–microtubule attachments. However, the mechanism underlying Polo–destabilizing activity remains elusive. Here, resorting to an RNAi screen in Drosophila for suppressors of a constitutively active Polo mutant, we identified a strong genetic interaction between Polo and the Rod–ZW10–Zwilch (RZZ) complex, whose kinetochore accumulation has been shown to antagonize microtubule stability. We find that Polo phosphorylates Spindly and impairs its ability to bind to Zwilch. This precludes dynein‐mediated removal of the RZZ from kinetochores and consequently delays the formation of stable end‐on attachments. We propose that high Polo‐kinase activity following mitotic entry directs the RZZ complex to minimize premature stabilization of erroneous attachments, whereas a decrease in active Polo in later mitotic stages allows the formation of stable amphitelic spindle attachments. Our findings demonstrate that Polo tightly regulates the RZZ–Spindly–dynein module during mitosis to ensure the fidelity of chromosome segregation.
Keywords: kinetochore attachments, mitosis, Polo, RZZ, Spindly
Subject Categories: Cell Cycle; Post-translational Modifications, Proteolysis & Proteomics
RNAi screening identifies the RZZ complex as mediator of early mitotic spindle attachment destabilization via high Polo kinase activity in Drosophila.
Introduction
To ensure the fidelity of chromosome segregation, sister kinetochores (KTs) mediate the attachment of chromosomes to microtubules (MTs) of opposite spindle poles (amphitelic attachments). However, the initial contact of KTs with MTs is stochastic and consequently erroneous attachments—syntelic (chromosome bound to MTs from the same spindle pole) or merotelic (same KT bound to MTs from opposite poles)—can be formed during early mitosis (Cimini et al, 2001; Thompson & Compton, 2011; reviewed Foley & Kapoor, 2013). Thus, accurate mitosis requires a tight regulation of KT‐MT turnover so mistakes are prevented or corrected and amphitelic end‐on interactions are stabilized (Bakhoum et al, 2009a,b; Zaytsev & Grishchuk, 2015). This relies heavily on the activity of two conserved mitotic kinases, Aurora B and Polo/Plk1. Aurora B promotes the destabilization of KT‐MT interactions mainly through phosphorylation of proteins of the KMN network (KNL1/Spc105, Mis12 and Ndc80), which decreases their affinity for MTs (Lampson et al, 2004; Cheeseman et al, 2006; Cimini et al, 2006; DeLuca et al, 2006, 2011; Cimini, 2007; Welburn et al, 2010). Interestingly, it has been shown that the RZZ complex (Rod, ZW10 and Zwilch) is able to interact with Ndc80 N‐terminal tail and prevent the adjacent calponin homology (CH) domain from binding to tubulin (Cheerambathur et al, 2013). This Aurora B‐independent destabilizing mechanism is proposed to prevent Ndc80‐mediated binding when KTs are laterally attached, hence reducing the potential for merotely during early mitosis. The RZZ additionally recruits Spindly and the minus end‐directed motor dynein to KTs (Griffis et al, 2007; Gassmann et al, 2008, 2010; Chan et al, 2009; Gama et al, 2017; Mosalaganti et al, 2017), thus providing the means to relieve its inhibitory effect over KT‐MT attachments, as well as to ensure the timely removal of spindle assembly checkpoint (SAC) proteins from KTs (Gassmann et al, 2010; Cheerambathur et al, 2013). However, it remains unclear how RZZ removal by Spindly–dynein is coordinated with end‐on attachment formation.
Polo/Plk1 activity is implicated in both stabilization (Elowe et al, 2007; Matsumura et al, 2007; Liu et al, 2012; Suijkerbuijk et al, 2012; Dumitru et al, 2017) and destabilization (Ahonen et al, 2005; Foley et al, 2011; Moutinho‐Santos et al, 2011; Salimian et al, 2011; Hood et al, 2012; Paschal et al, 2012; Beck et al, 2013) of KT‐MT attachments. While the contribution to the former function has been attributed to PP2A‐B56 phosphatase recruitment through Plk1‐dependent BubR1 phosphorylation (Elowe et al, 2007; Matsumura et al, 2007; Suijkerbuijk et al, 2012), the mechanism underlying Polo/Plk1 destabilizing activity remains unclear. Interestingly, Polo/Plk1 KT localization and activity decrease from early mitosis to metaphase (Ahonen et al, 2005; Conde et al, 2013), concurrent with an increase in KT‐MT stability (Bakhoum et al, 2009a). Moreover, high Plk1 activity at KTs was shown to correlate with decreased stability of KT‐MT attachments during prometaphase (Foley et al, 2011; Paschal et al, 2012; Beck et al, 2013), but the underlying molecular mechanisms have only been marginally addressed (Godek et al, 2015).
Here, we describe the mitotic effect of expressing a constitutively active Polo‐kinase mutant (PoloT182D) in Drosophila neuroblasts and cultured S2 cells. The expression of PoloT182D causes persistent KT‐MT instability and congression defects, extends mitotic timing associated with SAC activation and increases chromosome mis‐segregation. We designed a small‐scale candidate‐based RNAi screen to identify partners/pathways that are affected by constitutive Polo activity in the Drosophila eye epithelium. The screen revealed that downregulation of the RZZ subunit Rod rescues the defects resulting from PoloT182D expression. We show that PoloT182D causes permanent accumulation of the RZZ complex at KTs, which is associated with a delay in achieving stable biorientation. Accordingly, Rod depletion rescues the time required for establishing end‐on KT‐MT attachments and for chromosome congression. We further demonstrate that Polo phosphorylates the dynein‐adaptor Spindly to decrease its affinity for the RZZ. This in turn prevents dynein‐dependent stripping of RZZ from KTs, hence causing a delay in the formation of stable end‐on attachments. Our findings provide a mechanism for the destabilizing action of Polo/Plk1 over KT‐MT attachments. We propose a model in which Polo/Plk1 activity fine‐tunes the RZZ–Spindly–dynein module throughout mitosis to ensure the fidelity of KT‐MT attachments and chromosome segregation.
Results
Constitutively active Polo kinase leads to unstable KT‐MT attachments
Polo/Plk1 has been implicated in both stabilizing and destabilizing KT‐MT interactions. To understand how these apparently opposing actions are coordinated to ensure proper chromosome segregation in mitosis, we first monitored the level of active Polo at KTs during different mitotic stages in Drosophila neuroblasts. Using a phosphospecific antibody for the activating T‐loop phosphorylation (Fig 1A and B), we find that Polo is more active at KTs during prometaphase and its activity markedly decreases at metaphase, when KT‐MT attachments are more stable. Maintaining Plk1 constitutively active in different human cell lines produced conflicting results regarding its effect on KT‐MT attachments and chromosome congression (Liu et al, 2012; Paschal et al, 2012). Thus, we decided to assess the impact of constitutively active Polo on mitotic progression in vivo. The expression of either UASPoloWT (wild‐type) or UASPoloT182D was induced in neuroblasts with the inscuteable‐Gal4 driver (Betschinger et al, 2006) and mitotic progression analysed (Fig 1C; Movie EV1). In contrast to PoloWT, the expression of PoloT182D caused a significant extension of the time from nuclear envelope breakdown to anaphase onset (NEBD‐AO) (Fig 1D). This mitotic delay results mainly from chromosome congression defects, as the time measured from NEBD to alignment of the last chromosome was twice as long in PoloT182D‐expressing neuroblasts (Fig 1E). In vivo measurement of inter‐kinetochore distances revealed that the increased time in prometaphase duration was accompanied by reduced centromeric tension (Fig 1F), indicating that PoloT182D delays the establishment of KT‐MT end‐on attachments. This observation suggests that KT‐MT interactions are more labile in neuroblasts expressing PoloT182D. To confirm this, we monitored the localization of Mad2‐GFP, a SAC protein that decorates unattached KTs. In PoloWT‐expressing neuroblasts, Mad2‐GFP accumulates at KTs during early prometaphase and the signal rapidly fades as stable KT‐MT attachments are established, allowing chromosomes to align at the metaphase plate within a few minutes (Fig 1G and H; Movie EV2). In contrast, Mad2‐GFP persisted for longer periods at KTs of neuroblasts expressing PoloT182D, indicating a reduced MT occupancy on unaligned KTs (Fig 1G and H; Movie EV3). This conclusion is further supported by immunofluorescence analysis showing increased levels of Mad1 at KTs of neuroblasts that express the constitutively active kinase (Appendix Fig S1A and B).
Figure 1. Expression of constitutively active Polo kinase destabilizes kinetochore–microtubule (KT‐MT) interactions.
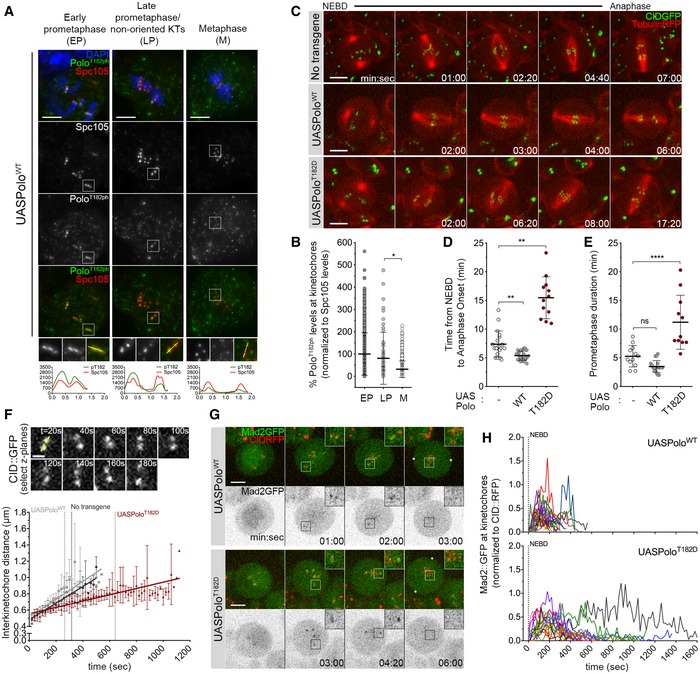
- Representative immunofluorescence images of Polo‐phospho(ph)‐T182 levels at KTs of Drosophila neuroblasts throughout mitotic progression. Insets show magnifications of the outlined regions showing kinetochores (KTs) in early prometaphase (EP), late prometaphase (LP) KTs and aligned KTs in metaphase (M). Plotted profiles of signal intensities of phospho(ph)‐T182 and the KT protein Spc105 are shown for the highlighted KTs.
- Graph represents the levels of Polo phT182 at KTs for neuroblasts shown in (A). phT182 signal was determined relative to Spc105 and all values normalized to the mean value determined for EP, which was set to 100% (n ≥ 67 KTs from at least 19 neuroblasts for each condition, n = 4 independent experiments).
- Selected stills from live imaging analysis of mitotic progression in neuroblasts expressing UASPoloWT or UASPoloT182D under the control of inscuteable‐Gal4 driver. Neuroblasts without Polo overexpression were used as control. Mitotic progression was followed in vivo by direct visualization of tubulin‐RFP and the centromere marker CID‐GFP. Time 0 refers to nuclear envelope breakdown (NEBD).
- Quantification of the mitotic time (from NEBD to anaphase onset) for neuroblasts shown in (C). NEBD was identified as the time tubulin entered the nuclear space and anaphase onset as the time sister KTs separated (n ≥ 12 neuroblasts for each condition, n ≥ 3 independent experiments).
- Quantification of the time spent in prometaphase (from NEBD until last KT alignment at the metaphase plate) for neuroblasts shown in (C) (n ≥ 11 neuroblasts for each condition, n ≥ 3 independent experiments).
- Measurement of inter‐kinetochore distance during mitotic progression for neuroblasts shown in (C). Only KT pairs within the maximum of two consecutive z‐planes were considered eligible for quantification. Images were acquired every 20 s. The graph shows the mean distance between two CID centroids in a KT pair over time (t = 0 is NEBD). A linear regression was applied to the data set. Vertical dashed lines highlight the time at which cells overexpressing PoloWT (grey) or PoloT182D (red) reach the average inter‐kinetochore distance measured in metaphase cells without Polo overexpression (black) (n ≥ 7 neuroblasts for each condition).
- Selected stills from live imaging of neuroblasts expressing Mad2‐GFP to follow KT‐MT attachment status upon expression of PoloWT or PoloT182D. CID‐RFP was used as KT reference. Insets show magnifications of the outlined regions showing single KT pairs that take longer to align at the metaphase plate. Asterisks indicate direction of chromosome segregation (putative spindle pole positions).
- Graph represents the mean fluorescence intensity (MFI) for Mad2‐GFP at KTs measured from NEBD to anaphase onset for neuroblasts shown in (G). Each line represents the average of all KTs measured from a single neuroblast at each time point. Mad2‐GFP MFI was determined relative to CID‐RFP MFI (n ≥ 16 neuroblasts for each condition, n = 5 independent experiments).
To assess in greater detail the configuration of KT‐MT attachments, we turned to cultured S2 cells which also showed increased frequency of unaligned chromosomes when Polo is constitutively active (Fig 2A and B). Detailed analysis of KT‐MT interactions shows that these cells have aligned KTs attached to the side of a MT end, which occurs to a lesser extent in PoloWT‐EGFP expressing cells (Fig 2C and D). Importantly, analysis of neuroblasts expressing PoloT182D allowed us to determine the dynamics and outcome of mitotic progression in a more reproducible manner than cultured cells (Fig 2E–G). Time course analysis shows that these cells undergo a highly asynchronous chromatid migration during anaphase as opposed to the synchrony observed in PoloWT neuroblasts (Fig 2E and G). Moreover, whereas virtually all PoloWT neuroblasts displayed a normal karyotype, we frequently detected aneuploidy in neuroblasts expressing PoloT182D (Fig 2F and G). Collectively, these results demonstrate that constitutive activation of Polo renders KT‐MT attachments unstable and compromises the fidelity of chromosome segregation. These errors are unlikely to result from hyperactivation of Aurora B, as the expression of PoloT182D failed to cause a significant increase in the phosphorylation of Spc105 Ser35 (Appendix Fig S1C and D), a well‐described Aurora B substrate (Welburn et al, 2010; Bajaj et al, 2018).
Figure 2. Expression of PoloT182D compromises mitotic fidelity.
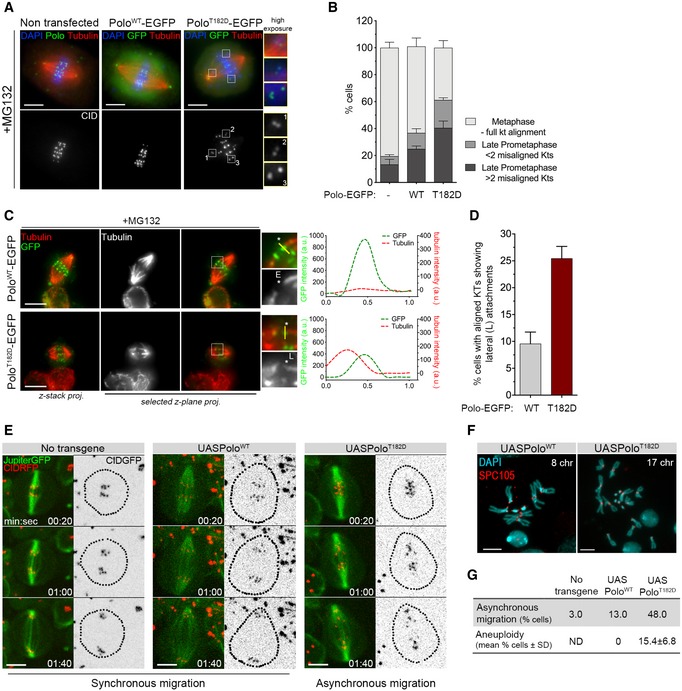
- Representative immunofluorescence images of KT alignment efficiency in mitotic S2 cells expressing PoloWT‐EGFP, PoloT182D‐EGFP or lacking expression of any Polo transgene. Cells were treated with MG132 prior to fixation to prevent cells from exiting mitosis and increase the number of pre‐anaphase figures. Insets display magnifications of the outlined regions, which highlight both low‐tension misaligned (insets 1 and 2) and high‐tension aligned KTs (inset 3).
- Graph represents the percentage of cells in each indicated mitotic state, as shown in (A) (n ≥ 532 cells for each condition, n = 2 independent experiments).
- Representative immunofluorescence images of calcium‐stable KT‐MT attachments in metaphase S2 cells expressing either PoloWT‐EGFP or PoloT182D‐EGFP. Insets display magnifications of the outlined regions. Asterisk highlights either an aligned KT pair attached to MTs in an end‐on fashion (PoloWT‐EGFP) or an aligned KT pair in which a sister KT is laterally attached to the end of a MT fibre (PoloT182D‐EGFP). Plotted profiles show the overlap between Polo‐EGFP and tubulin signals for the highlighted KT.
- Graph represents the percentage of metaphase cells showing at least 1 KT with a lateral interaction, as shown in (C) (asterisk) (n ≥ 58 cells for each condition, n = 2 independent experiments).
- Selected frames from live image analysis of chromosome segregation fidelity in neuroblasts expressing either PoloWT or PoloT182D. Neuroblasts without Polo overexpression were used as control. Jupiter‐GFP was imaged for direct visualization of the mitotic spindle. Time 0 refers to anaphase onset.
- Representative immunofluorescence images of mitotic neuroblasts from squashed brains showing distinct chromosome content. Spc105 was used as reference for KTs. Chromosome content is shown for each neuroblast.
- Table shows the quantification of the percentage of asynchronous migration (n ≥ 23 neuroblasts for each condition, n ≥ 3 independent experiments) and of aneuploid cells shown in (E and F), respectively. For the quantification of aneuploidy, the whole brain was analysed and prometaphase and metaphase neuroblasts were scored for chromosome number. An aneuploid cell was considered when > 8 chromosomes (chr) were visualized (n ≥ 360 neuroblasts from at least seven larvae brains for each condition, n ≥ 3 independent experiments). ND, not determined.
RNAi‐based screen identifies rough deal as a suppressor of PoloT182D expression
To uncover how Polo destabilizes KT‐MT attachments, we designed a small‐scale RNAi screen to identify molecular players mis‐regulated by constitutively active Polo kinase (Fig 3A). We selected 222 candidates (Appendix Tables S1 and S2) biased towards putative Polo interactors annotated on DroID database (http://www.droidb.org) and induced RNAi‐mediated depletion of individual genes in the eye imaginal disc in a PoloT182D background. The expression of PoloT182D with the eye‐specific driver eyeless Gal4 caused abnormal eye development, which ultimately led to a dramatic reduction in the adult structure and a significant reduction in male survival at 25°C (Fig 3B and C). Thus, simply by scoring for the increase in male viability, we identified 24 suppressors of the PoloT182D phenotype (Table EV1). Among these, we identified rough deal, the gene encoding for Rod, a subunit of the RZZ complex (Karess & Glover, 1989). Previous studies in Caenorhabditis elegans showed that Rod interacts with Ndc80 preventing the formation of stable end‐on attachments (Cheerambathur et al, 2013). Thus, we hypothesized that constitutively active Polo might promote KT‐MT instability through inappropriate Rod function.
Figure 3. RNAi‐based screen identifies Rod as a suppressor of the PoloT182D phenotype.
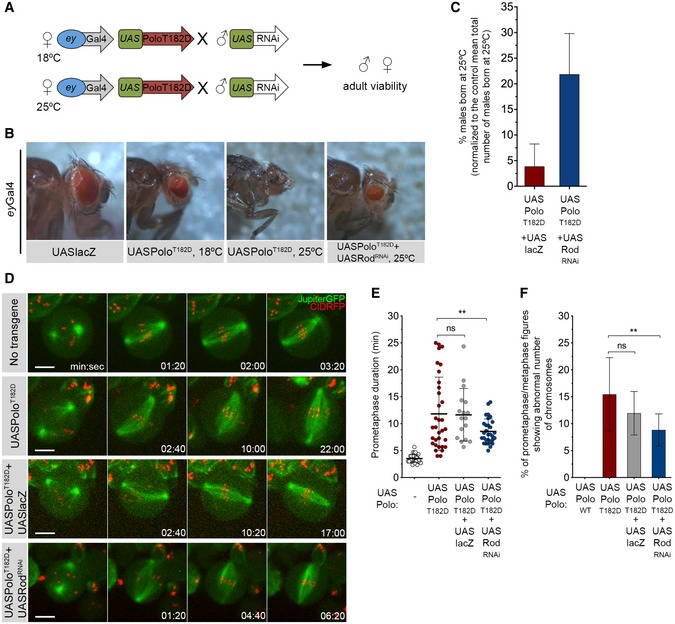
- Schematics of the RNAi screen strategy. Females expressing UASPoloT182D under the control of eyless‐Gal4 promoter were crossed with males carrying a specific UAS‐RNAi transgene. Crosses were left either at 18°C or at 25°C, and adult offspring viability was assessed.
- Representative images of the eye phenotype observed in adult flies that were expressing UASPoloT182D in the eye imaginal disc during development at indicated temperatures. An example of an eye from an adult fly co‐expressing UASPoloT182D with UASRodRNAi is also shown. The expression of UASlacZ transgene in the eye imaginal disc was used as control for overexpression and UAS dilution effect.
- Graph represents the mean percentage of males born at 25°C either expressing eyGal4‐driven UASPoloT182D together with UASlacZ or in combination with UASRodRNAi (n ≥ 3 independent crosses for each condition). The number of males with the genotype of interest born in each cross was normalized to the mean total number of males that are born in a control cross (eyGal4 > UASlacZ, n = 9 independent crosses).
- Selected stills from live‐cell imaging of neuroblasts expressing either in UASPoloT182D alone or in Rod‐depleted background (UASPoloT182D + UASRodRNAi). Neuroblasts without transgene expression were used as control, and neuroblasts expressing UASPoloT182D together with UASlacZ were used as control for UAS dilution effect.
- Quantification of the time spent in prometaphase (from NEBD until last KT alignment at the metaphase plate) for neuroblasts shown in (D) (n ≥ 17 neuroblasts for each condition, n ≥ 4 independent experiments). Statistical analysis was calculated using an one‐way ANOVA test for multiple comparisons. P values: ns, not significant; **< 0.01.
- Quantification of the percentage of prometaphase/metaphase mitotic figures per brain showing abnormal chromosome number (> 8 chromosomes (chr) as shown in Fig 2F and G) (n ≥ 360 neuroblasts from at least seven larvae brains for each condition, n ≥ 3 independent experiments). Statistical analysis was calculated using a Kruskal–Wallis test for multiple comparisons. P values: ns, not significant; **< 0.01.
To evaluate the suppressor effect of Rod depletion in PoloT182D‐expressing neuroblasts, we monitored mitotic progression by live‐cell imaging (Fig 3D and E; Movie EV4). While control neuroblasts efficiently congressed all chromosomes to the metaphase plate within 3.5 min, the expression of PoloT182D significantly compromised this process, extending prometaphase duration up to 13 min. Notably, depletion of Rod partially rescued this delay, with neuroblasts achieving full KT alignment in 8 min (Fig 3E; Movie EV4). Analysis of data dispersion indicates that the absence of Rod particularly improves chromosome congression in PoloT182D neuroblasts (Fig 3E). Importantly, Rod depletion by itself did not significantly affect prometaphase duration, suggesting that SAC mis‐regulation is not responsible for the observed rescue (Fig EV1A–C; Movie EV5). Moreover, depletion of Rod in neuroblasts expressing PoloT182D also rescued the fidelity of chromosome segregation as indicated by the significant decrease in the frequency of aneuploidy (Fig 3F). Rod RNAi led to an equivalent reduction in the accumulation of KT‐associated ZW10 in colchicine‐treated neuroblasts regardless of PoloT182D expression, suggesting that the same level of depletion occurred in both conditions (Fig EV1D and E). Moreover, total levels of PoloT182D were unaffected by Rod RNAi (Fig EV1F and G) and Gal4‐driven expression of UASlacZ failed to suppress the prometaphase delay and aneuploidy caused by PoloT182D (Fig 3D–F). These results exclude titration of the Gal4 transcription factor as the underlying cause for the observed rescue in mitotic fidelity. Taken together, these data strongly suggest that chromosome congression defects and segregation errors observed in PoloT182D neuroblasts are linked to Rod mis‐regulation.
Figure EV1. Absence of the RZZ complex does not impact on the time required for chromosome congression in Drosophila neuroblasts.
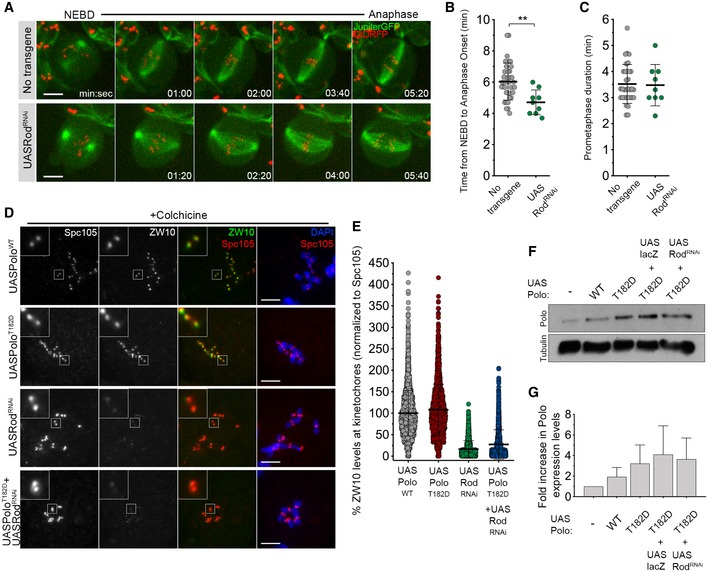
- Selected stills from live imaging analysis of mitotic progression in neuroblasts depleted of Rod (UASRodRNAi). Neuroblasts without transgene expression were used as control.
- Quantification of the mitotic time (time from NEBD to anaphase onset) for neuroblasts shown in (A) (n ≥ 9 neuroblasts for each condition, n = 2 independent experiments).
- Quantification of the time spent in prometaphase (from NEBD until last KT alignment at the metaphase plate) for neuroblasts shown in (A) (n ≥ 9 neuroblasts for each condition, n = 2 independent experiments).
- Representative immunofluorescence images of ZW10 levels in colchicine‐treated neuroblasts expressing UASPoloWT, UASPoloT182D, UASRodRNAi or UASPoloT182D in a Rod‐depleted background (UASPoloT182D + UASRodRNAi). Insets show magnifications of the outlined regions. Neuroblasts expressing UASPoloWT were used as control.
- Graph represents ZW10 levels in neuroblasts for the conditions shown in (D). ZW10 levels were determined relative to Spc105, and all values were normalized to the control mean fluorescence intensity, which are set to 100% (n ≥ 1116 KTs for each condition, n ≥ 2 independent experiments).
- Western blot analysis of UAS dilution effect on Polo overexpression levels in brain extracts from 3rd‐instar larvae expressing UASPoloWT, UASPoloT182D, UASPoloT182D together with UASlacZ or in a Rod‐depleted background (UASPoloT182D + UASRodRNAi). Tubulin was used as a loading control.
- Graph represents the fold increase in Polo protein levels relative to the absence of transgene overexpression. Expected Polo protein levels were determined after normalization to tubulin levels, and fold increase was calculated comparing to the actual Polo levels.
Constitutively active Polo impairs RZZ stripping from late congressing KTs
To determine how Polo activity regulates Rod, we imaged neuroblasts expressing Rod‐GFP under the regulation of its endogenous promoter in the presence of either PoloWT or PoloT182D. In accordance with previous studies (Basto et al, 2004; Siller et al, 2005; Défachelles et al, 2015), Rod‐GFP localized prominently to KTs during prometaphase and was significantly reduced during metaphase in PoloWT neuroblasts (Fig 4A–C). Detailed inspection shows that Rod‐GFP signal starts to decrease as KTs congress and fades when these stably align at the cell equator (Fig 4A and B; Movie EV6). However, in the presence of PoloT182D, Rod‐GFP accumulated at higher levels on late congressing KTs and this correlated with a delay in KT alignment (Fig 4A–C; Movie EV7). A similar increment in KT Rod‐GFP was observed in dynein‐depleted neuroblasts (Fig 4A and C; Movie EV8), where a correlation between Rod‐GFP levels at KTs and delayed biorientation was equally evident (Fig 4A and B). It is interesting to note that the signal of Rod‐GFP does decrease when chromosomes eventually align. Although the mechanism responsible for RZZ removal from dynein‐depleted KTs is still unclear, a similar observation has also been described in human cells (Chan et al, 2009; Gassmann et al, 2010; Raaijmakers et al, 2013).
Figure 4. Expression of constitutively active Polo kinase reduces Rod streaming from late congressing KTs.
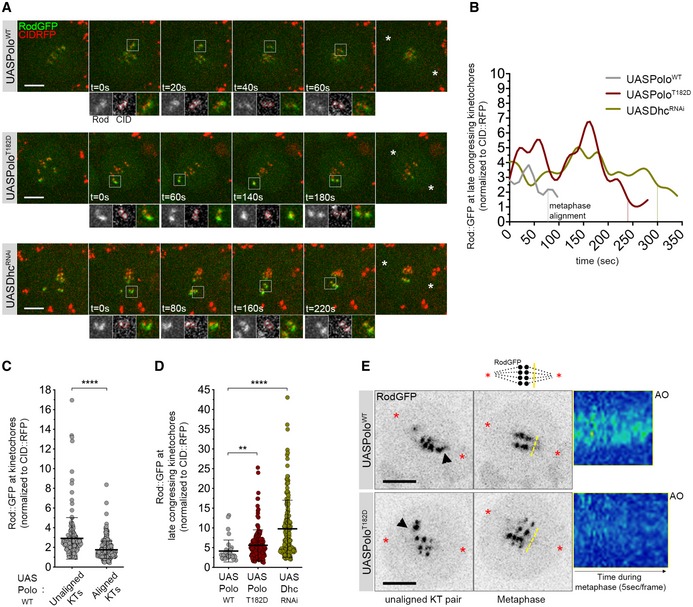
- Selected stills from live‐cell imaging of neuroblasts expressing UASPoloWT, UASPoloT182D or depleted of dynein heavy chain (UASDhcRNAi) and expressing Rod‐GFP under control of its endogenous promoter. CID‐RFP was used as a KT reference. Insets display magnifications of the outlined regions, which highlight late congressing KTs. Asterisks indicate direction of chromosome segregation (putative spindle pole positions).
- Graph represents the mean fluorescence intensity (MFI) for Rod‐GFP determined relative to CID‐RFP MFI for the KT pairs highlighted in (A). Time was measured from the first frame a KT pair moved away from the other congressing KTs, until metaphase alignment (vertical dashed lines).
- Graph represents the levels of KT Rod‐GFP determined relative to CID‐RFP measured throughout mitotic progression for control neuroblasts shown in (A) (UASPoloWT, n = 24 neuroblasts, n = 4 independent experiments).
- Graph represents the levels of Rod‐GFP determined relative to CID‐RFP measured at late congressing KTs (as shown in (A), insets) (n ≥ 15 neuroblasts for each condition, n ≥ 3 independent experiments).
- Selected stills from live imaging analysis of Rod‐GFP streaming from KTs in neuroblasts expressing either PoloWT or PoloT182D. Asterisks indicate direction of chromosome segregation (putative spindle poles positions). Arrowheads highlight streaming of Rod‐GFP from unaligned KTs. Representative kymographs are shown for Rod‐GFP streaming from metaphase until anaphase onset (AO). Frames were acquired every 5 s.
Thus, the expression of PoloT182D mimics the absence of dynein resulting in high levels of RZZ at late congressing KTs (Fig 4D). The role of dynein in RZZ removal from KTs is well established (Howell et al, 2001; Wojcik et al, 2001). Accordingly, we were able to follow dynein‐dependent stripping of Rod‐GFP along spindle MTs in PoloWT neuroblasts (Fig 4E and Appendix Fig S2A and B). Live‐cell imaging with high‐temporal resolution (5 s/frame) shows that Rod‐GFP stripping becomes visible immediately before KTs align at the metaphase plate (Fig 4E, arrowhead; Movie EV9). In contrast, the expression of PoloT182D prevented normal stripping of Rod‐GFP from late congressing KTs (Fig 4E, arrowhead; Movie EV10). The lack of robust stripping becomes even more evident during metaphase, when multiple KTs should contribute to this event, as observed in PoloWT neuroblasts (Fig 4E, kymograph; Appendix Fig S2A and B). These results suggest that constitutive activation of Polo impairs dynein‐mediated removal of Rod from KTs.
The RZZ complex has an inhibitory effect on stable chromosome biorientation
Unaligned KTs in S2 cells expressing PoloT182D‐EGFP often display a tilted configuration relative to the spindle axis (Fig 2A, insets 1 and 2). This arrangement is typical of lateral KT‐MT interactions and is also observed in vivo in late congressing KTs of PoloT182D neuroblasts (Fig 4A, t = 60 s). These KTs accumulate high levels of Rod‐GFP (Fig 4B and D) and recruit Mad2‐GFP (Fig 1E, t = 04:20), thus confirming reduced MT occupancy. We hypothesized that impaired RZZ removal from KTs might be responsible for the delay in the formation of stable end‐on attachments when Polo is constitutively active. To test this, we first prevented the association of dynein with KTs through depletion of dynactin or Spindly, well‐established KT adaptors of dynein (Griffis et al, 2007; Gassmann et al, 2008, 2010; Raaijmakers et al, 2013). Immunofluorescence analysis of ZW10 in these neuroblasts confirmed that in the absence of KT‐associated dynein, the RZZ complex localizes at higher levels on tilted/unaligned KTs (Fig 5A and B). A similar increase in ZW10 accumulation was observed upon expression of PoloT182D (Fig 5A and B), analogous to the pattern described for Rod (Fig 4D).
Figure 5. Expression of constitutively active Polo mimics the depletion of Spindly in the inhibition of stable KT biorientation by the RZZ complex.
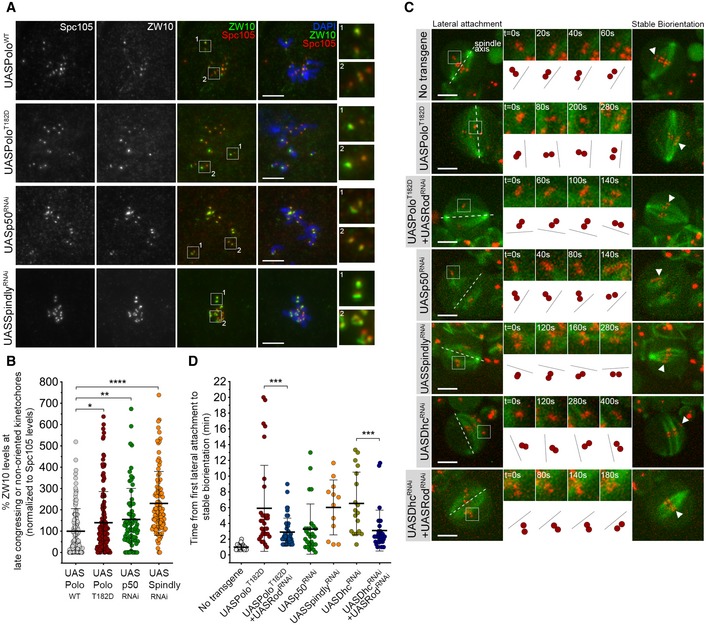
- Representative immunofluorescence images of ZW10 levels at unaligned KTs in neuroblasts expressing UASPoloWT, UASPoloT182D or depleted of dynactin subunit p50 (UASp50RNAi) or Spindly (UASSpindlyRNAi). Spc105 was used as a KT reference. Insets display magnifications of the outlined regions which highlight both late congressing KT (inset 1) and sister KTs that are non‐oriented along the spindle axis (inset 2, except for the UASPoloWT panel which displays congressed KTs). UASPoloWT neuroblasts were used as control.
- Graph represents ZW10 levels at late congressing and non‐oriented KTs for neuroblasts shown in (A). Cells were selected when most KTs were congressed and late congressing KTs were clearly identified. Similarly, non‐oriented KTs were considered when the KT‐KT axis was not parallel to most congressed KTs (or a metaphase plate). ZW10 signal was determined relative to Spc105, and all values were normalized to the control mean fluorescence intensity, which was set to 100% (n ≥ 69 KTs from at least 19 neuroblasts for each condition, n ≥ 3 independent experiments).
- Selected stills from live imaging analysis of KT alignment in neuroblasts expressing UASPoloT182D, either alone or in a Rod‐depleted background (UASPoloT182D + UASRodRNAi), depleted of dynactin (UASp50RNAi), Spindly (UASSpindlyRNAi), dynein (UASDhcRNAi) or Rod in a dynein‐depleted background (UASDhcRNAi + UASRodRNAi). Outlined regions show KTs that are non‐oriented (lateral attachment) relative to spindle axis and that were followed over time until stable biorientation (arrowhead). Cartoon depicts the behaviour of the respective KT pair over successive frames relative to the spindle axis (dashed lines). Neuroblasts without transgene expression were used as control.
- Quantification of the time a KT pair takes from the first contact with MTs until stable biorientation for neuroblasts shown in (C) (n ≥ 12 KTs from at least eight neuroblasts for each condition, n ≥ 5 independent experiments).
To address whether high levels of RZZ delay the formation of stable amphitelic attachments, we followed KT behaviour by time‐lapse imaging and quantified the time from the first lateral contact with the spindle (tilted configuration relative to spindle axis) until biorientation (parallel arrangement relative to spindle axis) (Fig 5C). In control neuroblasts, KTs bioriented efficiently within 1 min; however, the time increased significantly in all conditions where RZZ removal was prevented (Fig 5C and D). Notably, the most pronounced delay in attaining biorientation was observed in neuroblasts expressing PoloT182D or depleted of Spindly/dynein (Fig 5C and D). This defect was efficiently rescued by the depletion of Rod, suggesting that the long‐lived lateral interactions result from the inhibitory action of KT‐associated Rod towards the stability of end‐on attachments (Fig 5C and D). Likewise, preventing KT localization of Rod through depletion of Zw10 was sufficient to restore the accuracy of chromosome congression in S2 cells expressing PoloT182D‐EGFP (Fig EV2A and B; Movie EV11). This improvement in mitotic fidelity is unlikely to result from premature anaphase onset, as loss of SAC function imposed by Mad2 depletion caused PoloT182D‐EGFP cells to transit to anaphase with unaligned KTs, whereas the majority of cells depleted of Zw10 exited mitosis with all KT pairs aligned at the metaphase plate (Fig EV2A and C; Movie EV11).
Figure EV2. Depletion of ZW10 rescues chromosome congression in PoloT182D expressing cells independently of SAC impairment.
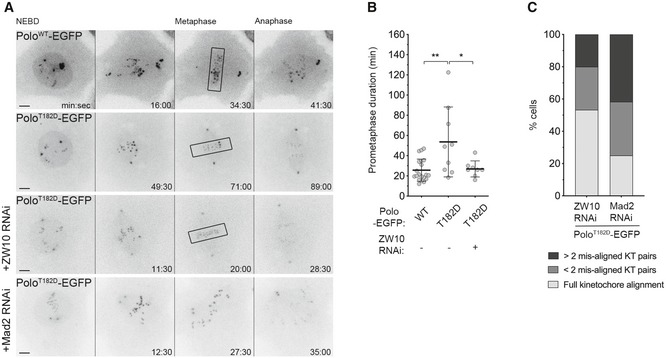
- Selected stills from live imaging analysis of mitotic progression in Drosophila S2 cells expressing PoloWT‐EGFP, PoloT182D‐EGFP and PoloT182D‐EGFP in a ZW10‐ or Mad2‐depleted background. Time 0 refers to nuclear envelope breakdown (NEBD). Black box highlights full KT alignment.
- Quantification of the time spent in prometaphase (from NEBD until last KT alignment at the metaphase plate) for cells shown in A (n ≥ 8 cells for each condition, n ≥ 3 independent experiments). Note that the Mad2‐depleted cell does not achieve full KT alignment.
- Graph represents the percentage of cells that enter anaphase with different levels of KT alignment (n = 15 cells, n ≥ 3 independent experiments).
Interestingly, p50/dynactin and Spindly seem to regulate KT‐MT attachments differently. Depletion of p50/dynactin shows a relatively mild phenotype while the severity in chromosome alignment defects, prometaphase delay and aneuploidy observed upon Spindly depletion resembles the PoloT182D phenotype (Figs 5C and D, and EV3A–D; Movies EV12, EV13, EV14, EV15, EV16 and EV17). Moreover, the expression of PoloT182D or depletion of Spindly in the larval brain affected adult viability, whereas downregulation of p50/dynactin allowed flies to eclose. Collectively, these results strongly suggest that Polo activity affects dynein‐mediated removal of KT RZZ through Spindly.
Figure EV3. Spindly depletion and expression of PoloT182D result in similar mitotic phenotypes.
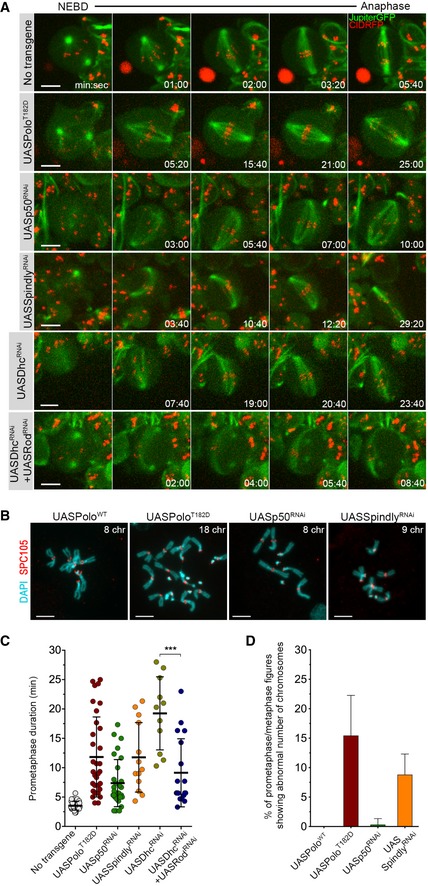
- Selected stills from live imaging analysis of mitotic progression in neuroblasts expressing UASPoloT182D, or depleted of p50 (UASp50RNAi), Spindly (UASSpindlyRNAi), dynein heavy chain (UASDhcRNAi) or Rod in Dhc‐depleted background (UASDhcRNAi + UASRodRNAi).
- Representative immunofluorescence images of mitotic neuroblasts from squashed brains showing distinct chromosome content. Spc105 was used as a KT reference. Neuroblasts were identified as the larger cells. Chromosome content is shown for each neuroblast.
- Quantification of the time spent in prometaphase (from NEBD until last KT alignment at the metaphase plate) for neuroblasts shown in (A) (n ≥ 11 neuroblasts for each condition, n ≥ 6 independent experiments).
- Graph represents the % of aneuploidy among mitotic figures for conditions shown in (C). For the quantification of aneuploidy, the whole brain was analysed and prometaphase and metaphase mitotic neuroblasts were scored for chromosome number (aneuploid cell when > 8 chromosomes (chr) were visualized). PoloWT neuroblasts were used as control (n ≥ 305 neuroblasts from at least six larvae brains for each condition, n ≥ 3 independent experiments).
Constitutively active Polo impairs proper Spindly retention at unaligned KTs
To address whether Polo regulates Spindly, we started by examining Spindly localization in S2 cells expressing PoloWT‐EGFP or PoloT182D‐EGFP transgenes (Fig 6A and B; Appendix Fig S3A). Immunofluorescence analysis of PoloWT cells revealed that Spindly is absent from metaphase‐aligned KTs and accumulates prominently at late congressing KTs (Fig 6A and B). However, the expression of PoloT182D severely reduced Spindly association with late congressing KTs (Fig 6A and B). To examine Spindly KT behaviour with adequate temporal resolution, we monitored S2 cells expressing Spindly–EGFP by time‐lapse imaging (Fig 6C). In PoloWT cells, Spindly–EGFP signal peaks at unaligned KTs and sharply decreases as KTs congress, reaching residual values immediately before KTs become correctly aligned at the metaphase plate (Fig 6C, inset; D; Movie EV18). However, Spindly–EGFP accumulation on unaligned KTs is significantly delayed when PoloT182D is expressed, thus confirming the localization pattern observed in fixed cells (Fig 6C, inset; D; Movie EV19).
Figure 6. Constitutively active Polo delays Spindly accumulation at congressing KTs.
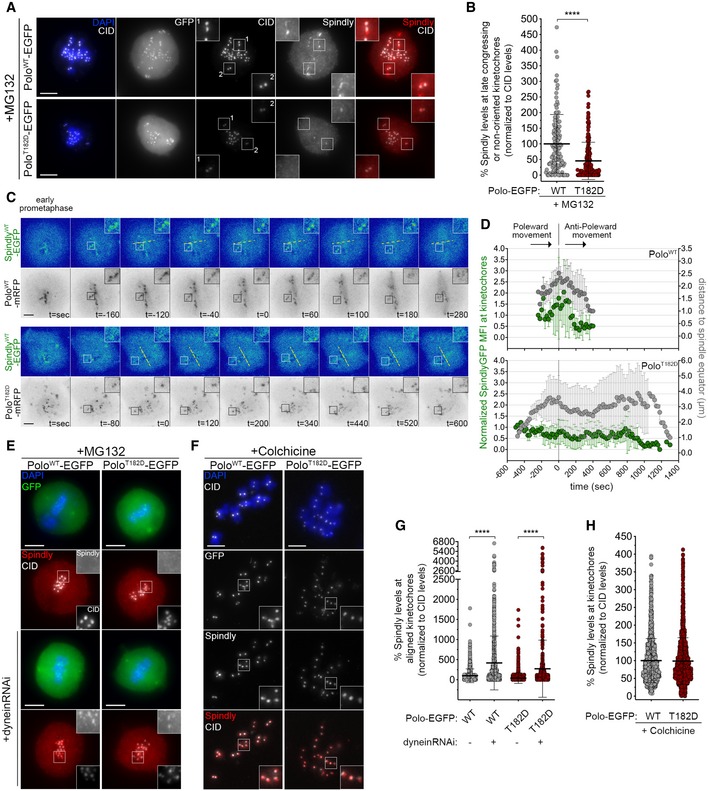
- Representative immunofluorescence images of Spindly localization to unaligned KTs in Drosophila S2 cells expressing either PoloWT‐EGFP or PoloT182D‐EGFP. Insets display magnifications of the outlined regions. Cells were treated with MG132 prior to fixation to increase the number of late prometaphase figures and allow better identification of late congressing KTs. CID was used as a KT reference.
- Graph represents the percentage of Spindly levels at late congressing and non‐oriented (not oriented parallel to the spindle axis) KTs for cells shown in (A). Spindly levels were determined relative to CID, and all values were normalized to the mean fluorescence intensity quantified in cells expressing PoloWT‐EGFP, which was set to 100% (n ≥ 121 KTs from at least 39 cells for each condition, n = 3 independent experiments).
- Selected frames from live imaging analysis of SpindlyWT–EGFP behaviour during KT alignment in Drosophila S2 cells expressing either PoloWT‐mRFP or PoloT182D‐mRFP. Insets show magnifications of the outlined regions that highlight KTs with delayed congression to the spindle equator (yellow dashed line). Time 0 is the first shift in KT direction from poleward to anti‐poleward movement.
- Graph represents the variation in SpindlyWT–EGFP levels at KTs highlighted in (C) and its correlation with efficiency of alignment. Spindly–EGFP mean fluorescence intensity (MFI) levels were corrected for photobleaching during acquisition and normalized to the first frame of analysis to account for differences in expression levels. KTs were tracked during the process of congression (using Polo‐mRFP signal as a KT reference), and the distance from the unaligned KT pair to the spindle equator was measured. All values were normalized to the last frame of analysis (stable integration in metaphase plate). Time 0 is the first shift in KT direction from poleward to anti‐poleward movement (n ≥ 9 KTs from at least five cells for each condition, n ≥ 4 independent experiments).
- Representative immunofluorescence images of Spindly accumulation at aligned KTs in Drosophila S2 cells expressing either PoloWT‐EGFP or PoloT182D‐EGFP, in control or upon depletion of dynein heavy chain. Insets display magnifications of the outlined regions. CID was used as a KT reference.
- Representative immunofluorescence images of Spindly accumulation at unattached KTs in Drosophila S2 cells expressing either PoloWT‐EGFP or PoloT182D‐EGFP. Insets display magnifications of the outlined regions. Cells were treated with colchicine prior to fixation to generate unattached KTs. CID was used as a KT reference.
- Graph represents Spindly levels at aligned KTs for cells shown in (E). Spindly levels were determined relative to CID, and all values were normalized to the mean fluorescence intensity quantified in control PoloWT‐EGFP expressing cells, which was set to 100% (n ≥ 633 KTs from at least 35 cells for each condition, n = 2 independent experiments).
- Graph represents Spindly levels at unattached KTs for cells shown in (F). Spindly levels were determined relative to CID, and all values were normalized to the mean fluorescence intensity quantified in PoloWT‐EGFP expressing cells, which was set to 100% (n ≥ 1,282 KTs from at least 59 cells for each condition, n = 2 independent experiments).
Dynein mediates Spindly–RZZ removal from KTs along spindle MTs (Griffis et al, 2007; Gassmann et al, 2010; Sacristan et al, 2018). Accordingly, dynein depletion in S2 cells expressing PoloWT prevented stripping of Spindly causing its accumulation at aligned KTs (Fig 6E and G). Notably, a similar result was observed when dynein was depleted from cells expressing PoloT182D, hence suggesting that constitutively active Polo prevents proper accumulation of Spindly at late congressing KTs through dynein‐dependent activity (Fig 6E and G). Consistent with this, we found that colchicine‐induced depolymerization of MTs restored the KT localization of Spindly in PoloT182D cells to similar levels as in cells expressing PoloWT (Fig 6F and H). Together, these results show that reduced accumulation of Spindly at late congressing KTs of PoloT182D cells is not caused by defects in Spindly recruitment but rather by its premature dynein‐mediated removal along MTs.
Polo phosphorylates Spindly at unattached and unaligned KTs
To test whether Polo directly regulates Spindly, we performed in vitro kinase assays. We found that Polo phosphorylates MBP‐Spindly (Fig 7A) and mass‐spectrometry analysis identified Ser499 as a phosphorylated residue (Fig 7B) within a canonical Polo‐phosphorylation consensus signature (Santamaria et al, 2011). Interestingly, Ser499 lies within a motif that is conserved among different dynein‐adaptors, but whose molecular function remains uncharacterized (Fig 7C).
Figure 7. Polo phosphorylates Spindly at unattached and unaligned KTs.
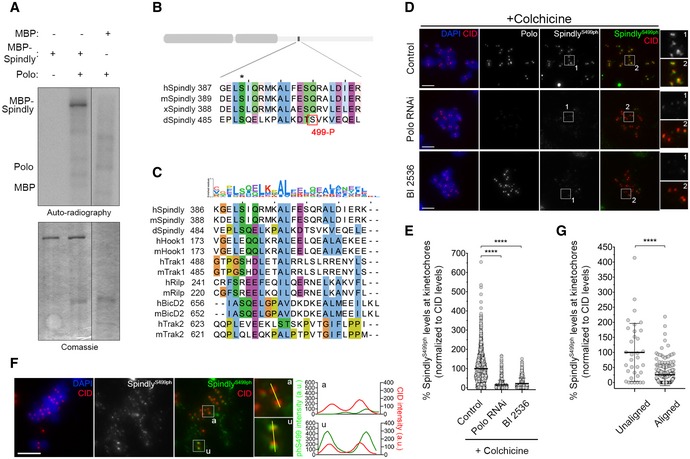
- In vitro kinase assays with the indicated recombinant protein in the presence of [γ‐32P]‐ATP for 30 min. Phosphorylation was detected by autoradiography, and levels of proteins were visualized by Coomassie Blue staining.
- ClustalW sequence alignment for Spindly orthologues. The region encompasses the Polo‐dependent phosphorylation site identified in Drosophila Spindly (red box). Asterisk highlights a conserved residue identified in silico by the Eukaryotic Linear Motif (ELM) resource as a putative Polo‐dependent phosphorylation site in human Spindly. Sequence alignment was coloured according to clustal colour scheme in Jalview (http://www.jalview.org, v2.10.5). Abbreviations: h, Homo sapiens; m, Mus musculus; x, Xenopus laevis; d, Drosophila melanogaster.
- Sequence alignment showing that the Polo‐dependent phosphorylation site in Drosophila Spindly lies within a conserved region found in different dynein‐adaptors. Abbreviations: h, Homo sapiens; m, Mus musculus; d, Drosophila melanogaster. Sequence alignment was coloured according to clustal colour scheme in Jalview (http://www.jalview.org, v2.10.5). The binomial log10 sequence logo was created using the PSSMSearch software (Krystkowiak et al, 2018).
- Representative immunofluorescence images of Spindly‐phospho(ph)Ser499 levels at unattached KTs in control Drosophila S2 cells and in Polo‐depleted or BI2536‐treated cells. Insets display magnifications of the outlined regions. Cells were treated with colchicine prior to fixation to generate unattached KTs. CID was used as a KT reference.
- Graph represents the levels of Spindly phS499 at unattached KTs for cells shown in (D). Spindly phS499 levels were determined relative to CID, and all values were normalized to the control mean fluorescence intensity, which was set to 100% (n ≥ 1246 KTs from at least 63 cells for each condition, n = 2 independent experiments). Statistical analysis was calculated using a Kruskal–Wallis test for multiple comparisons. P values: ****< 0.0001.
- Representative immunofluorescence images of Spindly phS499 levels at unaligned and aligned KTs in a PoloWT‐EGFP expressing Drosophila S2 cell. Insets show magnifications of the outlined regions, which highlight either a unaligned (u) KT or an aligned (a) KT. CID was used as a KT reference. Plotted profiles of signal intensities of phS499 and CID are shown for the highlighted KTs.
- Graph represents the levels of Spindly phS499 at unaligned KTs versus aligned KTs for cells shown in (F). Spindly phS499 levels were determined relative to CID, and all values were normalized to the mean fluorescence intensity of unaligned KTs, which was set to 100% (n ≥ 34 KTs from 14 cells, n = 2 experiments). Statistical analysis was calculated using an unpaired t‐test (Mann–Whitney). P values: ****< 0.0001.
To establish whether Polo phosphorylates Spindly Ser499 in cells, we generated a polyclonal antibody against Ser499 phosphopeptides and performed immunofluorescence analysis in S2 cells. Phosphorylation of Spindly on Ser499 (Ser499Ph) was uniformly detected at unattached KTs of colchicine‐treated cells (Fig 7D and E). However, KT levels of Ser499Ph were severely reduced following RNAi‐mediated depletion of Polo or inhibition of its kinase activity with BI2356 (Fig 7D and E). Depletion of Spindly or replacing the endogenous protein with a phosphodefective mutant for Ser499 (SpindlyS499A–EGFP) also compromised Ser499Ph staining, which further validates the specificity of the phosphoantibody (Fig EV4A–D). Collectively, these results strongly support the notion that Polo phosphorylates Spindly Ser499 in a cellular context. We then assessed the phosphorylation of Spindly Ser499 in unperturbed mitosis of asynchronous cultured S2 cells. The KT signal of SerS499Ph was reduced on KT pairs that had congressed to the cell equator but remained enriched at unaligned KTs (Fig 7F and G). This pattern correlates both with the activation status of Polo and with the KT levels of Spindly observed during chromosome congression (Figs 1A and B, and 6C and D). Thus, we conclude that Polo preferentially phosphorylates Spindly Ser499 when KTs lack stable end‐on attachments.
Figure EV4. Phospho(ph)Ser499 antibody recognizes a specific epitope on Spindly.
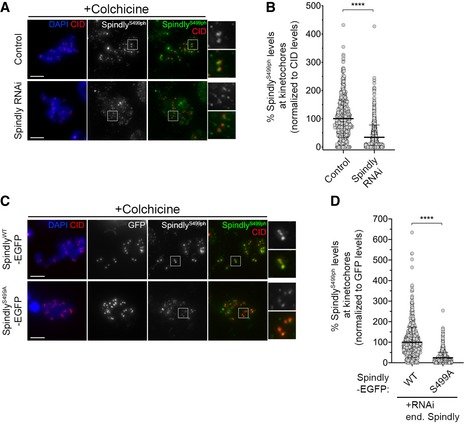
- Representative immunofluorescence images of Spindly‐phospho(ph)Ser499 levels at unattached KTs in control Drosophila S2 cells and in Spindly‐depleted cells. Insets display magnifications of the outlined regions. Cells were treated with colchicine prior to fixation to generate unattached KTs. CID was used as a KT reference.
- Graph represents the levels of Spindly phS499 at unattached KTs for cells shown in (A). Spindly phS499 levels were determined relative to CID, and all values were normalized to the control mean fluorescence intensity, which was set to 100% (n ≥ 701 KTs from at least 35 cells for each condition, n = 2 independent experiments).
- Representative immunofluorescence images of Spindly phS499 levels at unattached KTs in Drosophila S2 cells expressing either SpindlyWT–EGFP or SpindlyS499A–EGFP and depleted of endogenous Spindly. Insets display magnifications of the outlined regions. Cells were treated with colchicine prior to fixation to generate unattached KTs. CID was used as a KT reference.
- Graph represents the levels of Spindly phS499 at unattached KTs for cells shown in (C). Spindly phS499 levels were determined relative to GFP, and all values were normalized to the mean fluorescence intensity levels quantified in SpindlyWT–EGFP expressing cells, which was set to 100% (n ≥ 824 KTs from at least 42 cells for each condition, n = 2 independent experiments).
Polo‐mediated phosphorylation of Spindly decreases its binding to Zwilch and delays its accumulation at KTs
Human Spindly has been shown to interact with the RZZ complex through Rod in a farnesylation‐dependent manner (Mosalaganti et al, 2017). Drosophila Spindly lacks a farnesylation motif, hence resembling the C. elegans orthologue, which instead binds to the RZZ through the Zwilch subunit (Gama et al, 2017). Thus, we sought to evaluate whether Drosophila Spindly also binds to Zwilch and the impact that Polo‐mediated Ser499 phosphorylation might have on this interaction. Immobilized MBP‐ZwilchFL/WT and MBP‐SpindlyFL/WT were able to, respectively, pull‐down 6xHis‐SpindlyFL/WT and 6xHis‐ZwilchFL/WT, thereby confirming a direct interaction between Spindly and Zwilch fly orthologues (Fig 8A and Appendix Fig S4A). Strikingly, a full‐length version of phosphomimetic Spindly for Ser499 (6xHis‐SpindlyFL/S499D) failed to bind MBP‐ZwilchFL/WT (Fig 8A and C). However, removing the last 270 amino acids from Spindly C‐terminus enabled the truncated protein to efficiently associate with MBP‐ZwilchFL/WT regardless of S499D phosphomimetic mutation (6xHis‐Spindly1−510/S499D; Fig 8B and C). These results suggest that phosphorylation of Spindly Ser499 by Polo promotes a negative auto‐regulatory action of Spindly C‐terminal region over the N‐terminus, thereby decreasing its interaction with Zwilch.
Figure 8. Polo‐mediated phosphorylation of Spindly on Ser499 impairs its binding to the RZZ and delays KT accumulation during chromosome congression.
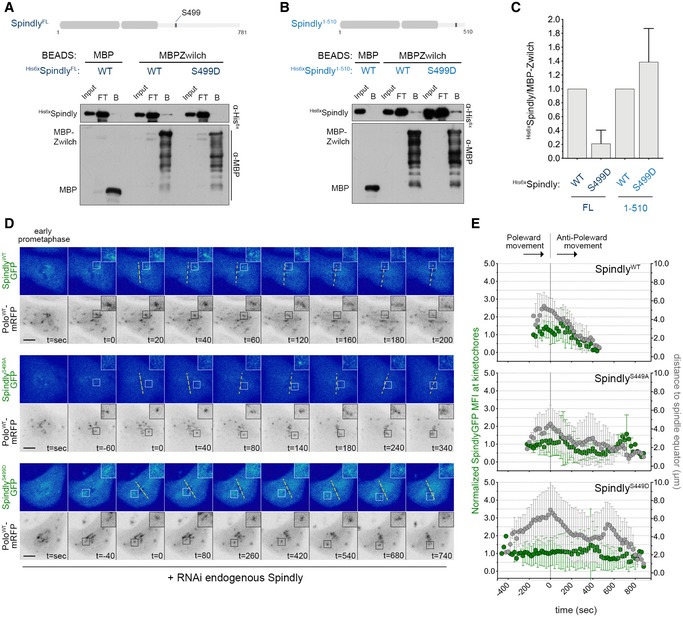
- Western blot analysis of MBP‐Zwilch pull downs of full‐length (FL) his6x‐SpindlyWT and phosphomimetic mutant his6x‐SpindlyS499D. Immobilized MBP was used as negative control. B: beads, FT: flow‐through.
- Western blot analysis of MBP‐Zwilch pull downs of his6x‐SpindlyWT or his6x‐SpindlyS499D fragments (aa1–510). Immobilized MBP was used as negative control. B: beads, FT: flow‐through.
- Graph represents the levels of his6x‐Spindly relative to MBP‐Zwilch from at least three independent pull‐down experiments. The values obtained for his6x‐SpindlyWT were set to 1.
- Selected frames from live imaging analysis of SpindlyS499A–EGFP or SpindlyS499D–EGFP behaviour during KT alignment in Drosophila S2 cells expressing PoloWT‐mRFP. SpindlyWT–EGFP expressing cells were used as control. All Spindly–EGFP constructs were resistant to the RNAi targeted against endogenous Spindly. Insets show magnifications of the outlined regions that highlight KTs that delay in congressing to the spindle equator (yellow dashed line). Time 0 is the first shift in KT direction from poleward to anti‐poleward movement.
- Graph represents the variation in SpindlyWT–EGFP, SpindlyS499A–EGFP and SpindlyS499D–EGFP levels at KTs highlighted in (D) and its correlation with efficiency of alignment. All values were determined as described in Fig 6D. Time 0 is the first shift in KT direction from poleward to anti‐poleward movement (n ≥ 5 KTs from at least four cells for each condition, n ≥ 2 independent experiments).
To dissect the impact of Ser499 phosphorylation in a cellular context, we started by monitoring the localization pattern of SpindlyWT–EGFP, SpindlyS499A–EGFP and SpindlyS499D–EGFP by live‐cell imaging. All transgenes were rendered resistant against RNAi used to deplete endogenous Spindly (Appendix Fig S3B), and the expression of PoloWT‐mRFP was used to trace KTs. As shown before, SpindlyWT–EGFP levels peak at late congressing KTs just before stable congression to the cell equator is initiated (Fig 8D, inset; E; Movie EV20). A similar behaviour was observed for SpindlyS499A–EGFP (Fig 8D, inset; Fig 8E; Movie EV21). Conversely, SpindlyS499D–EGFP accumulation on late congressing KTs is significantly delayed, which is concurrent with an increase in the time required for KTs to align at the metaphase plate (Fig 8D, inset; Fig 8E; Movie EV22). These results demonstrate that Polo‐mediated phosphorylation of Spindly on Ser499 decreases its ability to interact with Zwilch and to timely accumulate on KTs at levels required for efficient chromosome congression.
Polo‐mediated phosphorylation of Spindly prevents RZZ removal from KTs and delays the conversion of lateral KT‐MT interactions into stable end‐on attachments
The expression of SpindlyS499D–EGFP caused a significant increase in the levels of Zw10 at late congressing KTs, when compared to the expression of wild‐type or phosphodefective (SpindlyS499A–EGFP) transgenes (Fig 9A and B). SpindlyS499D–EGFP cells further resembled cells expressing constitutively active Polo with respect to the chromosome congression defects exhibited. Congressing KTs of SpindlyWT–EGFP and SpindlyS499A–EGFP cells displayed stable directional movement towards the equatorial plane and became correctly aligned, on average, within 16 min (Fig 9C and D; [Link], [Link]). However, cells expressing SpindlyS499D–EGFP required significantly longer time (~ 23 min) to align all KT pairs at the metaphase plate (Fig 9C and D; Movie EV25). Furthermore, these KTs failed to exhibit a stable alignment behaviour, presenting instead a tilted configuration typical of lateral attachments (Fig 8D, inset) and similar to that observed in PoloT182D cells. Strikingly, depletion of ZW10 restored normal chromosome congression (Fig 9C and D; Movie EV26), thus suggesting that the alignment defects in SpindlyS499D–EGFP cells are caused by high levels of RZZ at KTs. Conversely, preventing Spindly Ser499 phosphorylation facilitates RZZ removal from KTs (Fig 10A–C). In metaphase cells expressing SpindlyWT–EGFP, ZW10 was frequently observed as puncta over the spindle region (Fig 10A and B). This pattern resembles poleward streaming along microtubules and, accordingly, was completely abrogated by depletion of dynein (Fig 10A). Notably, Zw10 streaming was consistently more robust in cells expressing SpindlyS499A–EGFP, where a “fibre‐like” pattern of Zw10 could be observed as opposed to the puncta detected in control cells (Fig 10A–C). Together, these data suggest that phosphorylation of Spindly on Ser499 controls the strength by which the dynein–Spindly complex binds to KT‐associated RZZ, and the ability to stream it along spindle MTs.
Figure 9. Phosphorylation of Spindly Ser499 imposes an RZZ‐dependent delay on chromosome congression.
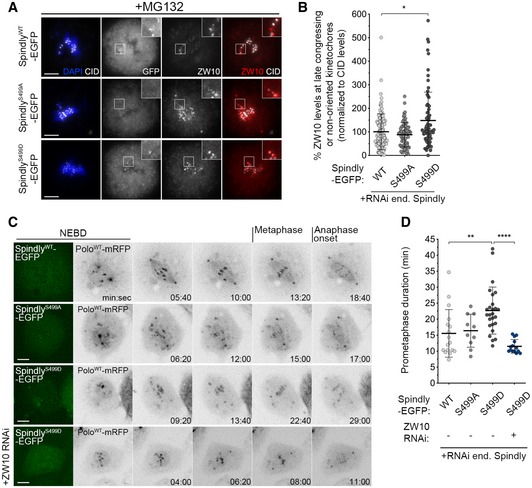
- Representative immunofluorescence images of ZW10 localization to unaligned KTs in Drosophila S2 cells expressing either SpindlyWT‐, SpindlyS499A‐ or SpindlyS499D–EGFP. Insets display magnifications of the outlined regions, which highlight late congressing KTs. CID was used as a KT reference.
- Graph represents the percentage of ZW10 levels at late congressing and non‐oriented (not oriented parallel to the spindle axis) KTs for cells shown in (A). ZW10 levels were determined relative to CID, and all values were normalized to the control mean fluorescence intensity, which was set to 100% (n ≥ 72 KTs from at least 26 cells for each condition, n = 4 independent experiments).
- Selected frames from live imaging analysis of chromosome congression in Drosophila S2 cells expressing SpindlyWT–EGFP, SpindlyS499A–EGFP, SpindlyS499D–EGFP or SpindlyS499D–EGFP in a ZW10‐depleted background. The first frame shows EGFP‐positive cells. Chromosome alignment was followed in vivo by direct visualization of KT‐associated PoloWT‐mRFP signal. Time 0 refers to nuclear envelope breakdown (NEBD), metaphase refers to full KT alignment, and anaphase onset refers to onset of chromosome segregation.
- Quantification of the time spent in prometaphase (from NEBD until last KT alignment at the metaphase plate) for cells shown in (C) (n ≥ 10 cells for each condition, n ≥ 2 independent experiments).
Figure 10. Polo activity fine‐tunes the RZZ–Spindly–dynein module to ensure the accuracy of KT‐MT attachments.
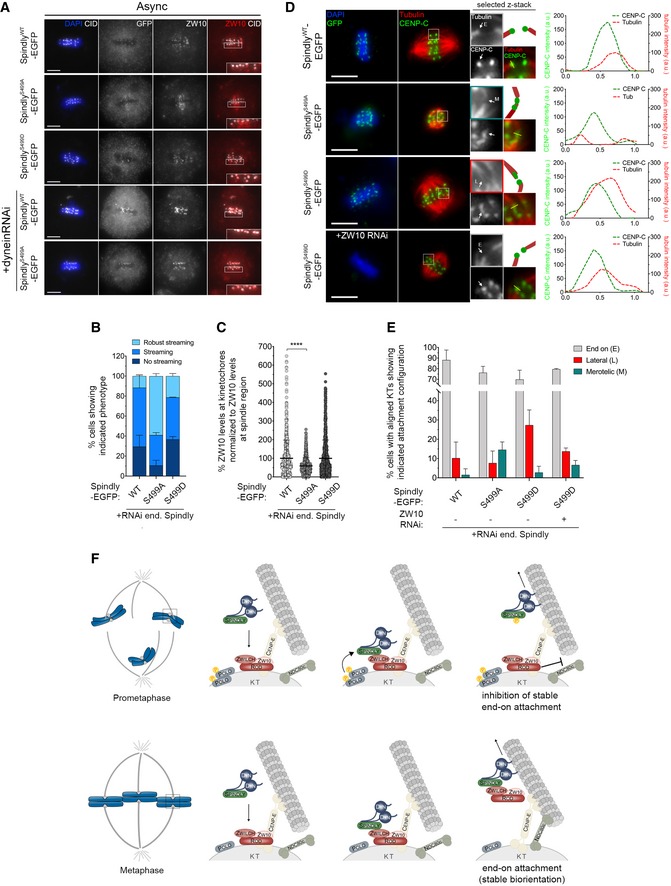
- Representative immunofluorescence images of ZW10 streaming from KTs in Drosophila S2 cells in metaphase expressing either SpindlyWT‐, SpindlyS499A‐ or SpindlyS499D–EGFP. ZW10 streaming is also shown in S2 cells expressing SpindlyWT– or SpindlyS499A–EGFP in a dynein‐depleted background. Insets display magnifications of the outlined regions, which highlight streaming robustness.
- Graph represents the percentage of cells in metaphase showing different levels of ZW10 streaming (n ≥ 35 cells for each condition, n = 2 independent experiments).
- Graph represents the percentage of ZW10 levels at aligned KTs normalized to ZW10 levels at the spindle region. All values were normalized to the mean fluorescence intensity quantified in SpindlyWT–EGFP expressing cells, which was set to 100% (n ≥ 470 KTs from at least 35 cells for each condition, n = 2 independent experiments).
- Representative immunofluorescence images of calcium‐stable KT‐MT attachments in metaphase S2 cells expressing SpindlyWT‐, SpindlyS499A‐, SpindlyS499D‐ or SpindlyS499D–EGFP in a ZW10‐depleted background. Insets display magnifications of the outlined regions which highlight different attachment configurations (E—end on; L—lateral; M—merotelic). Cartoon depicts the attachment configuration of the respective KT pair. Asterisk highlights an aligned KT pair in which a sister KT appears to be laterally attached to the end of a MT fibre. Plotted profiles show the overlap between CENP‐C and tubulin signals for the highlighted KT. CENP‐C was used as a KT reference.
- Graph represents the percentage of metaphase cells showing only end‐on attachments or at least one KT with lateral or merotelic attachment, as shown in (D) (n ≥ 44 cells for each condition, n ≥ 2 independent experiments).
- Proposed model for Polo‐mediated regulation of RZZ‐Spindly and its impact on KT‐MT attachment. High levels of active Polo during early mitosis render Spindly phosphorylated on Ser499, thus weakening its interaction with the RZZ complex at KTs. On laterally attached KTs, the presence of minus end‐directed motor dynein allows the removal of phosphorylated Spindly, while the RZZ complex is retained at high levels to inhibit premature formation of stable end‐on attachments and therefore avoid merotely. As protein phosphatases are recruited to KTs, Polo activation status declines and Spindly is eventually dephosphorylated. Spindly is now able to bind Zwilch, and the Spindly–RZZ complex is stripped from KTs through dynein, which enables the stable conversion from lateral to end‐on attachments and SAC silencing.
To establish whether mis‐regulated phosphorylation of Ser499 impacts on KT‐MT interactions, cells expressing different Spindly–EGFP versions were pre‐treated with calcium to depolymerize non‐KT microtubules and the nature of attachments in metaphase cells examined. As expected, in the vast majority of cells expressing SpindlyWT–EGFP, KTs were amphitelically attached to spindle MTs with an end‐on configuration (Fig 10D and E). However, aligned KTs of SpindlyS499D–EGFP cells frequently interacted with MTs in a lateral fashion (Fig 10D and E), similarly to cells expressing constitutively active Polo (Fig 2C and D). Depletion of Zw10 restored the accuracy of KT‐MT attachments, thus indicating that failure in end‐on conversion when Ser499 is constitutively phosphorylated is largely due to elevated levels of the RZZ complex at late congressing KTs. Importantly, an increase in the frequency of merotelic attachments was observed in cells expressing SpindlyS499A–EGFP. This is in agreement with a more robust streaming of RZZ in this mutant, which allows a premature stabilization of KT‐MT interactions (Fig 10D and E).
Together with the biochemical data from Fig 8, these findings strongly support a model in which Polo‐mediated phosphorylation of Spindly decreases its affinity for Zwilch, hence preventing RZZ streaming by dynein. This transiently ensures elevated levels of RZZ at congressing KTs to prevent premature conversion of lateral into end‐on MT attachments and thereby minimizing the potential for merotelic attachments.
Discussion
KT‐MT attachments at metaphase must be sufficiently stable to satisfy the spindle assembly checkpoint and sustain chromatid segregation during anaphase (Bakhoum et al, 2009b; Bakhoum & Compton, 2012; Musacchio and Salmon,2007; McEwen & Dong, 2009). On the other hand, during prometaphase, MTs must be able to rapidly detach from KTs to allow efficient correction of erroneous attachments (Maiato et al, 2004; Bakhoum et al, 2009a). How KTs regulate the balance of MTs stabilizing and destabilizing forces during successive mitotic stages has remained unclear. Here, we show that Polo kinase plays a critical role in this process through control of the RZZ–Spindly–dynein module at KTs (Fig 10F). Polo‐mediated phosphorylation of Spindly on Ser499 results in a transient increase in RZZ accumulation at KTs, which inhibits stable end‐on attachments and likely minimizes merotely in early mitosis (Cheerambathur et al, 2013). However, permanent Spindly Ser499 phosphorylation is deleterious for mitotic fidelity since it prevents stable KT biorientation and timely chromosome congression.
Polo/Plk1 has been implicated in the stabilization of KT‐MT attachments (Elowe et al, 2007; Matsumura et al, 2007; Liu et al, 2012; Suijkerbuijk et al, 2012; Dumitru et al, 2017). Intriguingly, however, attachments are most stable during metaphase, when Polo/Plk1 activity is reduced. We show that maintaining Polo active in Drosophila larval neuroblasts markedly decreases the stability of KT‐MT interactions, which is in line with previous observations in RPE‐cultured cells (Paschal et al, 2012). It is important to mention that insc‐GaL4‐driven expression of PoloWT and PoloT182D consistently yielded higher levels of the latter protein (Fig EV1F and G). This, however, unlikely explains the different phenotypic consequences observed in these neuroblasts, as analogous experiments with S2 cells expressing equivalent levels of PoloWT and PoloT182D (Appendix Fig S3A) mimic the decrease in the efficiency of chromosome congression and KT‐MT stability when constitutively active Polo is expressed (Fig 2C and D). Moreover, we previously showed that Drosophila S2 cells depleted of Polo accumulated hyperstable attachments and that this phenotype was not exclusively attributed to reduced Aurora B activity (Moutinho‐Santos et al, 2011). A requirement for Polo in fine‐tuning the RZZ‐Spindly–dynein axis offers a mechanistic explanation for these observations. During early mitosis, high levels of active Polo at KTs ensure that as soon as Spindly is recruited to RZZ, it is efficiently phosphorylated on Ser499. This promptly reduces Spindly affinity towards Zwilch, sensitizing RZZ‐uncoupled Spindly for dynein‐mediated transport away from KTs. As a result, the RZZ complex is retained at KTs to levels that normally inhibit the formation of stable end‐on attachments by the Ndc80 complex (Cheerambathur et al, 2013) and maintain the SAC signalling active (Buffin et al, 2005; Kops et al, 2005). Rod interacts with the basic tail of Ndc80 and, in this way, precludes binding of MTs to the calponin homology domain of Ndc80 (Cheerambathur et al, 2013). Thus, the conversion of lateral attachments preferentially formed at early stages of mitosis into stable amphitelic interactions that are essential for faithful chromosome segregation requires the relief of this Rod‐mediated inhibitory mechanism. We provide evidence that a decrease in Polo activity and, consequently, in Spindly phosphorylation, is critical for this transition by allowing the RZZ to fully engage with the Spindly–dynein complex and to be stripped from KTs. This raises the question of how and when Polo activity and Ser499 phosphorylation are antagonized to allow timely formation of stable end‐on attachments. PP2A‐B56 phosphatase may have a role in this process, since impairing its association with BubR1 was recently shown to dramatically increase the frequency of laterally attached KTs in human cells (Shrestha et al, 2017). However, because BubR1‐PP2A‐B56 is already present at high levels on early mitotic KTs, we reason that additional mechanisms must operate to prevent premature end‐on conversion (Moura & Conde, 2019). It is plausible that the switch is determined by the levels of cyclin A, which have been shown to function as a timer in prometaphase to destabilize attachments and facilitate error correction (Kabeche & Compton, 2013). Since Cdk1/CycA is able to phosphorylate human Spindly in vitro (Barisic et al, 2010), we hypothesize that this phosphorylation primes Spindly for Polo binding and increases Ser499 phosphorylation to levels that surpass the opposing phosphatase activity. As mitosis progresses, degradation of Cyclin A tips the balance towards Ser499 dephosphorylation, hence favouring stabilization of end‐on attachments. This concurs with an increase in tension across KTs that allows the recruitment of PP1, whose role in Polo T‐loop dephosphorylation has been described (Yamashiro et al, 2008; Dumitru et al, 2017).
Although the Polo‐phosphorylation site in Drosophila Spindly is not conserved in vertebrates, additional residues conforming to Polo/Plk1 consensus signature (Santamaria et al, 2011) are present within the same domain, hinting that an analogous regulatory mechanism may take place in these organisms. Interestingly, Ser499 lies within motif that is conserved among different dynein‐adaptors. Two other conserved domains have also been recently described for a number of adaptors and shown to act as regulatory modules involved in the interaction with dynein (Gama et al, 2017; Sacristan et al, 2018). Thus, we envision that the motif identified here might provide an additional level of regulation in controlling dynein‐adaptor complex formation.
Our results suggest that Polo‐mediated phosphorylation of Spindly on Ser499 uncouples dynein‐mediated transport of the RZZ complex from Spindly. Moreover, we propose that phosphorylation of Ser499 causes Spindly C‐terminal domain to elicit a negative regulatory action over the N‐terminus Zwilch binding domain. In line with these results, it has been recently shown that intramolecular interactions occur within Spindly, causing it to fold on itself at different regions (Sacristan et al, 2018). Spindly C‐terminal region could be involved in facilitating these interactions since it is thought to be of disordered nature (Sacristan et al, 2018). This structural organization resembles that of BicD/BicD2, a dynein‐adaptor which is predicted to share with Spindly a similar mechanism of interaction with dynein (Gama et al, 2017; Mosalaganti et al, 2017). It is therefore noteworthy that Polo has been shown to activate BicD–dynein transport during oogenesis (Mirouse et al, 2006). Furthermore, several point mutations in BicD/BicD2 were shown to hyperactivate dynein for cargo transport (Liu et al, 2013; Huynh & Vale, 2017). It will be interesting to establish whether Spindly Ser499 phosphorylation could also impact on dynein complex motility/processivity.
Long‐lasting Polo activation or permanent Spindly Ser499 phosphorylation stalls KTs in labile interactions with MTs. Our data confirm a destabilizing role for Polo in KT‐MT attachments which has also been shown to operate through the control the kinase exerts over the recruitment and activation of Aurora B and the MT depolymerizing motor Kif2b (Hood et al, 2012; Conde et al, 2013; Carmena et al, 2014). Hence, high levels of active Polo in early mitosis ensure efficient correction of merotelic and syntelic attachments, errors that typically occur upon nuclear envelope breakdown as a result of stochastic interactions between KTs and spindle MTs (Paul et al, 2009; Zaytsev & Grishchuk, 2015). Paradoxically, Plk1 activity has also been implicated in stabilization of KT‐MT attachments through phosphorylation of BubR1 (Suijkerbuijk et al, 2012). In light of ours and previous findings, we envisage a model where these apparently antagonistic Polo‐directed inputs are not mutually exclusive but rather cooperate to establish proper attachments. Phosphorylation of BubR1 by Polo/Plk1 in prometaphase promotes the accumulation of PP2A‐B56, which opposes Aurora B destabilizing phosphorylations on Ndc80. This is important to allow binding of MTs to the Ndc80 complex during the end‐on conversion process, tipping the balance against the KT‐MT destabilizing environment, particularly when Cyclin A levels drop. The observation that disrupting Plk1 activity rescues the attachment defects otherwise generated by depletion of PP2A‐B56 (Foley et al, 2011) strongly argues in favour of this integrated model for Polo‐regulated stabilizing and destabilizing forces.
In summary, our findings demonstrate that the RZZ–Spindly–dynein module is tightly regulated by Polo kinase to ensure accurate chromosome segregation. Spindly phosphorylation by Polo on early mitotic KTs ensures RZZ‐mediated inhibition of end‐on interactions, hence preventing premature stabilization of erroneous attachments. As mitosis progresses, decreased Polo‐kinase activity and concurrent Spindly dephosphorylation render the RZZ prone for removal from KTs by Spindly–dynein. This alleviates RZZ antagonism of MT binding by the Ndc80 complex, thus allowing timely conversion of labile lateral interactions into stable amphitelic attachments ensuring proper sister chromatid segregation.
Materials and Methods
Fly stocks
All RNAi lines used for the screen were obtained from VDRC (Vienna Drosophila Resource Center, VDRC, Vienna, Austria) and are listed in Appendix Table S1. We used the following stocks (described in FlyBase, unless stated otherwise): eyelessGal4 (Bloomington Stock Center, BDSC, IN, USA), inscuteableGal4 (BDSC), UASlacZ (#8529, BDSC), UASSpindlyRNAi (#34933, BDSC), UAS‐p50RNAi (#28596, BDSC), UAS‐Dhc64CRNAi (#28054, VDRC), UASRodRNAi (#19152, VDRC), Mad2‐GFP (Buffin et al, 2005), Rod‐GFP (Basto et al, 2004), CID‐RFP (Oliveira et al, 2007), α‐Tubulin‐RFP (Mathieu et al, 2011), CID‐GFP (Schuh et al, 2007), Jupiter‐GFP (Karpova et al, 2006). Stocks expressing pUASp‐PoloWT (Mirouse et al, 2006; Martins et al, 2009) or pExPress‐UAS‐PoloT182D (#8434, BDSC) was used in this study.
S2 cell culture, RNAi treatment and cell lysates
Drosophila S2 cells were cultured, and dsRNA incubation was performed as previously described (Maiato et al, 2003). For the depletion of endogenous Spindly, a set of primers was selected from the DRSC/TRiP Functional Genomics Resources (#DRSC31559). A dsRNA‐resistant cassette was synthetized (GeneArt, Thermo Fisher Scientific, Waltham, MA, USA) and replaced a region encompassing the DRSC31559‐targeted sequence by cloning it into EcoNI/BstEII sites on exogenous SpindlyWT–EGFP constructs. The dsRNA‐resistant cassette had almost every third base of a codon on the DRSC31559‐targeted sequence mutated without altering the coding sequence. High depletion efficiency of endogenous Spindly was achieved at 120 h post‐dsRNA incubation. The expression of RNAi‐resistant SpindlyWT–, SpindlyS499A– and SpindlyS499D–EGFP constructs was induced at 96 h post‐dsRNA incubation.
For the preparation of S2 cell lysates to measure protein depletion levels, cells were collected by centrifugation at 10,621 g for 10 min at 4°C. After washing the pellet in PBS 1× supplemented with 1× protease inhibitor cocktail (Roche, Basel, Switzerland), cells were pelleted by centrifugation at 452 g for 5 min at 4°C. The pellet was resuspended in lysis buffer (150 mM KCl, 75 mM HEPES, pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 15% glycerol, 0.1% NP‐40, 1× protease inhibitors cocktail (Roche) and 1× phosphatase inhibitors cocktail 3 (Sigma‐Aldrich, St. Louis, MO, USA) before immersion in liquid nitrogen for cell disruption. Protein extracts were recovered by centrifugation at 10,621 g for 10 min at 4°C and quantified by Bradford protein assay (Bio‐Rad, Hercules, CA, USA). For the preparation of drosophila brain lysates, a minimum amount of 10 3rd‐instar larvae brains was dissected in PBS 1× an immediately transferred to a volume of lysis buffer. The lysates were sonicated and clarified by centrifugation at 4°C.
Constructs and S2 cell transfection
PoloWT‐EGFP and PoloT182D‐EGFP constructs were previously characterized (Conde et al, 2013). For the generation of the expression vectors PoloWT‐mRFP and PoloT182D‐mRFP, available pENTR‐PoloWT and pENTR‐PoloT182D entry clones were used for Gateway recombination (Thermo Fischer Scientific) together with pHWR destination vectors (heat‐shock inducible promoter, C‐terminal mRFP tag). Spindly–EGFP (under the regulation of a metallothionein promoter) was a gift from Eric Griffis (Griffis et al, 2007). RNAi‐resistant SpindlyWT–EGFP was generated as described above. Both SpindlyS499A– and SpindlyS499D–EGFP constructs were generated by site‐directed mutagenesis with a primer harbouring the desired mutation. The PCR was performed with Phusion polymerase (New England Biolabs, Ipswich, MA, USA) using RNAi‐resistant SpindlyWT–EGFP as template. Stable transfection of indicated vectors into S2 cells was performed using Effectene Transfection Reagent (Qiagen, Hilden, Germany), according to the manufacturer's instructions. To induce Spindly–EGFP expression, S2 cells were incubated overnight with 20 μM CuSO4 at 25°C. For the co‐expression of PoloWT‐ or PoloT182D‐mRFP, cells were heat‐shocked for 45 min at 37°C prior to incubation with CuSO4. To induce PoloWT‐ or PoloT182D‐EGFP expression, cells were heat‐shocked for 45 min at 37°C and allowed to rest for at least 6 h before being processed for immunofluorescence.
Live imaging
For time‐lapse recording of Drosophila neuroblasts, brains from 3rd‐instar larvae were dissected in PBS1× and placed on a drop of PBS1× in a coverslip. The preparation was gently squashed using a smaller coverslip, and excess media was removed. The sample was sealed with Halocarbon oil 700 (Sigma‐Aldrich). Images were obtained at 20‐s intervals unless stated otherwise. For live imaging of S2 cells, cells were plated onto dishes with coverslip bottoms (MatTek Corporation, Ashland, MA, USA) previously treated with 0.2 mg/ml concanavalin A (Sigma‐Aldrich). Images were obtained at 20‐s intervals unless stated otherwise. All images were acquired at 25°C with a spinning disc confocal system (Revolution; Andor) equipped with an electron multiplying charge‐coupled device camera (iXonEM+; Andor) and a CSU‐22 unit (Yokogawa) based on an inverted microscope (IX81; Olympus). The microscope is served with two laser lines, 488 and 561 nm, for the excitation of EGFP and mCherry/mRFP, respectively. The system was driven by iQ software (Andor). Time‐lapse imaging of z stacks with 0.5 μm steps covering the entire volume of the cell was collected, and image sequence analysis and video assembly were done with Fiji software (https://fiji.sc/). For the quantification of Mad2‐GFP and Rod‐GFP at kinetochores (KTs) in neuroblasts, the mean fluorescence intensity was measured within a specific predefined region of interest (ROI) where KT pairs could fit. KTs identified based on the constitutive KT marker CID‐RFP. After background subtraction, identified as a region in the cell with no KTs, the intensities of Rod‐GFP or Mad2‐GFP were determined relative to CID‐RFP. A similar approach was employed for the quantification of Spindly–EGFP in S2 cells, where Polo‐mRFP signal was used as a KT reference. The changes in Spindly–EGFP fluorescence intensities with time were plotted as normalized signal relative to the signal at the first frame a chromosome moved away from the majority of congressing chromosomes. For the quantification of Rod‐GFP signal along microtubules (MTs) during metaphase, the mean fluorescence intensity was measured within a specific predefined region of interest (ROI) where the entire width of both half spindles could fit (a and b boxes in Appendix Fig S2). After background subtraction, measured in a region outside the spindle region, the intensity measured for Rod‐GFP was determined relative to Tubulin‐RFP to account for slight changes in the focal plane.
Immunofluorescence analysis
For immunofluorescence analysis of Drosophila neuroblasts, third‐instar larval brains were dissected in PBS and processed for fixation in 1.8% formaldehyde (Sigma‐Aldrich) and 45% glacial acetic acid for 5 min. When required, brains were incubated in 50 μM Colchicine (Sigma‐Aldrich) for 1.5 h prior to fixation. After fixation, brains were squashed between a coverslip and a slide and immersed in liquid nitrogen. Then, the coverslip was flipped off and the slides were incubated in cold ethanol for 10 min. The slides were further washed in 0.1% Triton X‐100 in PBS1× for 10 min for permeabilization. The slides were processed for immunostaining as previously described (Moura et al, 2017). Neuroblasts were identified as the larger cells based on the background signal from the different antibodies used that made the cell shape visible. For immunofluorescence analysis of chromosome alignment efficiency and KT‐MT attachment status in S2 cells, 105 cells were seeded onto coverslips previously treated with 0.2 mg/ml concanavalin (Sigma‐Aldrich). Two hours before fixation, cells were treated with 20 μM MG132 (Calbiochem, San Diego, CA, USA). For detection of calcium‐stable k‐fibres, cells were processed for fixation and immunostaining as previously described (Kapoor et al, 2000). For the scoring of chromosome alignment, cells were processed for fixation on 3.7% formaldehyde (Sigma‐Aldrich) in PHEM (60 mM PIPES, 25 mM HEPES, pH 7.0, 10 mM EGTA, 4 mM MgSO4) for 12 min and then detergent extracted with 0.5% Triton X‐100 in PBS1× three times for 5 min each. For immunofluorescence analysis of MT‐free mitotic S2 cells, cells were incubated with 30 μM Colchicine (Sigma‐Aldrich) for 2 h before fixation. When required, 100 nM BI2536 (Boehringer Ingelheim) was added to the media 1 h before fixation. Then, 105 cells were centrifuged onto slides for 5 min, at 239 g, and processed for simultaneous fixation and extraction in 3.7% formaldehyde, 0.5% Triton X‐100 in PBS1× for 10 min. The slides were washed three times in 0.05% Tween 20 in PBS1× for 5 min each. Immunostaining was performed as previously described (Conde et al, 2013). Images were acquired in Zeiss Axio Imager Z1 microscope (Carl Zeiss, Germany) using an AxioCam MR ver.3.0 (Carl Zeiss). The system was driven by AxioVision software (Carl Zeiss). All images were analysed using the Fiji software (https://fiji.sc/). For immunofluorescence quantification, the mean fluorescence intensity of KT proteins was measured within a specific predefined region of interest (ROI) where individual KTs could fit. The KT was identified based on the KT constitutive markers CID or Spc105. After background subtraction, identified as a region in the cell with no KTs, the intensities of KT proteins of interest were determined relative to CID or Spc105. Control values were averaged and used for normalization of values determined in the different biological conditions tested.
Antibodies
Rabbit polyclonal anti‐Spindly‐phospho(ph)Ser499 was raised against a phosphorylated peptide (493‐PALKDT‐Sp‐VKVEQE‐505, generated by Eurogentec, Seraing, Belgium). The primary antibodies used for immunofluorescence were rabbit anti‐phosphorylated Ser499‐Spindly used at 1:50; mouse anti‐α‐tubulin B512 (Sigma‐Aldrich) used at 1:5,000; rat anti‐CID (Rat4) used at 1:500; chicken anti‐GFP (#ab13970, Abcam, Cambridge, UK) used at 1:2,000; rabbit ZW10 (Rb85, Williams et al, 1992), used at 1:1,000; rabbit anti‐Mad1 (Rb1) used at 1:1,000; rat anti‐Spc105 used at 1:500 (Conde et al, 2013); rabbit anti‐Spindly (gift from Eric Griffis) used at 1:1,000; mouse anti‐phosphorylated Thr210‐Polo (#ab39068, Abcam) used at 1:1,500; rabbit anti‐phosphorylated Ser60‐AuroraB (gift from Iain Cheeseman) used at 1:1,500; rabbit anti‐CENP‐C (Rb1, Heeger et al, 2005) used at 1:1,000; and mouse anti‐Polo (MA294) used at 1:100. The primary antibodies used for Western blotting analysis were mouse anti‐Polo (MA294) used at 1:30; mouse anti‐α‐tubulin DM1A (Sigma‐Aldrich) used at 1:10,000; mouse anti‐MBP (New England Biolabs) used at 1:5,000; mouse anti‐His6x (#HisH8, Millipore, Burlington, MA, USA) used at 1:2,500; and rabbit anti‐Spindly (gift from Eric Griffis) used at 1:2,000.
RNAi screen
The list of Polo interactors was obtained from the Drosophila interactions database (DroID, http://www.droidb.org). The search retrieved 222 candidate genes for which we ordered RNAi lines (Appendix Table S1). The rational for the screen was as follows: test whether downregulation of a particular gene could rescue the lethality associated with the expression of UASPoloT182D in the eye imaginal discs. We first tested whether the expression of individual RNAi lines in the eye imaginal discs was lethal on its own (Appendix Table S1, RNAi lines highlighted in red). For the screen, with the exception of six RNAi lines that were not tested (Appendix Table S1, RNAi lines highlighted in yellow), we performed one round of crosses (done in replicates, same parents) both at 25°C and at 18°C. Most crosses (160) were repeated at least once (in replicates, same parents) at 25°C. We found more unambiguous to score any suppression of the phenotype based on the rescue in adult lethality rather than on eye morphology. The number of males and females with the genotype of interest (both PoloT182D and UAS‐RNAi transgenes) that was born was scored for the period estimated until the eclosion of flies from second generation to account for late eclosing flies. Although females showed defects in eye morphology, adult viability was not significantly affected by PoloT182D expression in the eye disc and was only consider for exclusion of conflicting results. The number of males was then normalized to the mean total number of males that are born in a control cross. The percentages were plotted and compared using one‐way ANOVA without correcting for multiple comparisons (P < 0.05). Although this approach increases the chance of making a type I error, any false‐positive finding could be detected in subsequent cellular characterizations. From the 26 genes found to be suppressors of the PoloT182D phenotype, two were excluded due to conflicting results obtained at 18 and 25°C. From the final hit list, Rod was selected based on recent evidences for the involvement of RZZ in the regulation of KT‐MT interactions and on the fact that the RNAi line is described as having no off‐targets.
Expression and purification of recombinant proteins
Full‐length Zwilch and SpindlyWT were either tagged with six histidines (His6x) at the N‐terminus or fused C‐terminal to MBP. SpindlyS499D was tagged with His6x. Spindly fragments encompassing the first 510 amino acids (WT or harbouring S499D mutation) were also tagged with His6x. For His6x‐tag, PCR products harbouring the coding sequences of interest were cloned into SalI/EcoRI sites of pET30a (+) expression vector (Novagen, Darmstadt, Germany). Recombinant vectors were subsequently transformed into BL21‐star competent cells, and expression was induced overnight at 15°C by the addition of 0.05 mM IPTG. Glucose was added to the media to control for the stress of the expression system. Lysates were sonicated and clarified by centrifugation at 4°C. The recombinant proteins were purified using the Ni‐NTA purification system (Qiagen) and eluted with 250 mM Imidazole, 300 mM NaCl, 50 mM NaH2PO4, pH 8.0. For MBP fusion, PCR products harbouring Zwilch or Spindly coding sequences were cloned into EcoRI/XbaI sites of pMal‐c2 expression vector (New England Biolabs). Recombinant vectors were subsequently transformed into BL21‐star competent cells, and expression was induced as for His6x‐tag recombinant constructs. Lysates were sonicated and clarified by centrifugation at 4°C. The recombinant proteins were then incubated with MBP amylose magnetic beads (New England Biolabs) for 1 h at 4°C and washed with 200 mM NaCl, 20 mM Tris–HCl, 1 mM EDTA, 1 mM DTT, pH 7.4, without protein elution. The same protocol was followed for expression and purification of MBP protein alone.
In vitro kinase assay and mass‐spectrometry analysis
For in vitro kinase assay, recombinant MBP or MBP‐SpindlyWT (full length) was incubated with 0.2 μg of active recombinant Polo kinase (gift from Bill Sulivan) for 45 min at 25°C in kinase reaction buffer (5 mM MOPS, pH 7.2, 2.5 mM β‐glycerol‐phosphate, 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, 0.25 mM DTT, 200 μM ATP and 1 × phosphatase inhibitors cocktail 3, supplemented with 10 μCi [γ‐32P] ATP [3,000 Ci/mmol, 10 mCi/ml). The reaction was stopped by the addition of Laemmli sample buffer (4% SDS, 10% mercaptoethanol, 0.125 M Tris–HCl, 20% glycerol, 0.004% bromophenol blue) and heated for 5 min at 95°C. Samples were resolved in a SDS–PAGE, and the gel was subsequently allowed to dry. The gel was then exposed to an X‐ray film (Fuji Medical X‐Ray Film) for detection of 32P incorporation. For the identification of phosphorylation sites on Spindly by Polo, the same protocol was followed, except no [γ‐32P] ATP was included in the reaction mix. Furthermore, the reaction was stopped by addition of 6 M urea and samples containing either MBP or MBP‐SpindlyWT were analysed by liquid chromatography directly coupled with mass spectrometry. Samples were digested with LysC/Trypsin and/or GluC and prepared for LC‐MS/MS analysis as previously described (Rappsilber et al, 2007). Peptides (100 ng) were separated on a Thermo ScientificTM EASY‐nLC 1000 HPLC system (Thermo Fisher ScientificTM) for 1 h from 5 to 60% acetonitrile with 0.1% formic acid and directly sprayed via a nano‐electrospray source in a quadrupole‐Orbitrap mass spectrometer (Q ExactiveTM, Thermo Fisher ScientificTM) (Michalski et al, 2011). The Q ExactiveTM was operated in data‐dependent mode acquiring one survey scan and subsequently ten MS/MS scans (Olsen et al, 2007). Resulting raw files were processed with the MaxQuant software (version 1.5.2.18) using a reduced database containing only the proteins of interest and giving phosphorylation on serine, threonine and tyrosine as variable modification (Cox & Mann, 2008). A false discovery rate cut‐off of 1% was applied at the peptide and protein levels and the phosphorylation site decoy fraction.
Pull‐down assay and western blotting
Coomassie staining of proteins resolved by SDS–PAGE was first conducted to determine comparable amounts of recombinant protein in solution or bound to amylose magnetic beads to be used in the pull‐down assays. Recombinant protein bound to beads were washed two times in column buffer A (200 mM NaCl, 20 mM Tris–HCl, 1 mM EDTA, 1 mM DTT, 0.05% Tween 20, pH 7.4). Similar amounts of His6x‐tagged recombinant proteins were prepared in column buffer B (200 mM NaCl, 20 mM Tris–HCl, 1 mM EDTA, 1 mM DTT, pH 7.4) and incubated with MBP fusion proteins for 1.5 h at 4°C, under constant rotation. The flow‐through was collected, and the fraction bound to beads was further washed three times in column buffer A. Magnetic beads and bound protein were resuspended in Laemmli sample buffer. Samples were heated at 95°C for 5 min before being resolved in SDS–PAGE and probed for proteins of interest through Western blotting. For Western blotting analysis, resolved proteins were transferred to a nitrocellulose membrane, using the iBlot Dry Blotting System (Thermo Fisher Scientific) according to the manufacturer's instructions. Membranes were incubated for at least 1 h at room temperature in blocking solution (5% powder milk in PBS1×, 0.05% Tween 20). All primary and secondary antibodies were diluted in the blocking solution. Membranes were incubated with primary antibody solutions overnight at 4°C under constant stirring and washed three times in PBS1×, 0.05% Tween 20 for 10 min each. Then, membranes were incubated with secondary antibody solutions for 1 h at room temperature under constant stirring. Secondary antibodies were conjugated to horseradish peroxidase (GE Healthcare, Little Chalfont, UK). The blots were visualized by ECL detection and exposure to X‐ray films.
Statistical analysis
All statistical analysis was performed with GraphPad Prism V8.0 (GraphPad Software, Inc.). Values were considered statistically different whenever P < 0.05.
Author contributions
CC and CS conceived the project. JB, TM, LT and TB performed the experiments. JB and CC analysed the data. JB and CC wrote the paper and CS revised it.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
Movie EV11
Movie EV12
Movie EV13
Movie EV14
Movie EV15
Movie EV16
Movie EV17
Movie EV18
Movie EV19
Movie EV20
Movie EV21
Movie EV22
Movie EV23
Movie EV24
Movie EV25
Movie EV26
Review Process File
Acknowledgements
We thank Eric Griffis (University of Dundee, Scotland, UK), for the Spindly antibody and SpindlyWT–EGFP construct, Roger Karess (IJM, Paris, France) for fly stocks, Bill Sulivan (University of California, Santa Cruz, USA), for recombinant Polo kinase. This article is a result of the project Norte‐01‐0145‐FEDER‐000029—Advancing Cancer Research: from basic knowledge to application, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER). JB is supported by an FCT PhD grant SFRH/BD/87871/2012. CC is supported by an FCT investigator position and funding (IF/01755/2014).
The EMBO Journal (2020) 39: e100789
Contributor Information
Carlos Conde, Email: cconde@ibmc.up.pt.
Claudio Sunkel, Email: cesunkel@ibmc.up.pt.
References
- Ahonen LJ, Kallio MJ, Daum JR, Bolton M, Manke IA, Yaffe MB, Stukenberg PT, Gorbsky GJ (2005) Polo‐like kinase 1 creates the tension‐sensing 3F3/2 phosphoepitope and modulates the association of spindle‐checkpoint proteins at kinetochores. Curr Biol 15: 1078–1089 [DOI] [PubMed] [Google Scholar]
- Bajaj R, Bollen M, Peti W, Page R (2018) KNL1 binding to PP1 and microtubules is mutually exclusive. Structure 26: 1327–1336.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA (2009a) Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol 19: 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA (2009b) Genome stability is ensured by temporal control of kinetochore‐microtubule dynamics. Nat Cell Biol 11: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Compton DA (2012) Kinetochores and disease: keeping microtubule dynamics in check!. Curr Opin Cell Biol 24: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M, Sohm B, Mikolcevic P, Wandke C, Rauch V, Ringer T, Hess M, Bonn G, Geley S (2010) Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell 21: 1968–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Scaerou F, Mische S, Wojcik E, Lefebvre C, Gomes R, Hays T, Karess R (2004) In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr Biol 14: 56–61 [DOI] [PubMed] [Google Scholar]
- Beck J, Maerki S, Posch M, Metzger T, Persaud A, Scheel H, Hofmann K, Rotin D, Pedrioli P, Swedlow JR et al (2013) Ubiquitylation‐dependent localization of PLK1 in mitosis. Nat Cell Biol 15: 430–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA (2006) Asymmetric segregation of the tumor suppressor brat regulates self‐renewal in Drosophila neural stem cells. Cell 124: 1241–1253 [DOI] [PubMed] [Google Scholar]
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15: 856–861 [DOI] [PubMed] [Google Scholar]
- Carmena M, Lombardia MO, Ogawa H, Earnshaw WC (2014) Polo kinase regulates the localization and activity of the chromosomal passenger complex in meiosis and mitosis in Drosophila melanogaster . Open Biol 4: 140162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Fava LL, Uldschmid A, Schmitz MH, Gerlich DW, Nigg EA, Santamaria A (2009) Mitotic control of kinetochore‐associated dynein and spindle orientation by human Spindly. J Cell Biol 185: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A (2013) Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 342: 1239–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson‐Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule‐binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED (2001) Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED (2006) Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol 16: 1711–1718 [DOI] [PubMed] [Google Scholar]
- Cimini D (2007) Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle 6: 1558–1564 [DOI] [PubMed] [Google Scholar]
- Conde C, Osswald M, Barbosa J, Moutinho‐Santos T, Pinheiro D, Guimarães S, Matos I, Maiato H, Sunkel CE (2013) Drosophila polo regulates the spindle assembly checkpoint through Mps1‐dependent BubR1 phosphorylation. EMBO J 32: 1761–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Défachelles L, Raich N, Terracol R, Baudin X, Williams B, Goldberg M, Karess RE (2015) RZZ and Mad1 dynamics in Drosophila mitosis. Chromosome Res 23: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- DeLuca KF, Lens SM, DeLuca JG (2011) Temporal changes in Hec1 phosphorylation control kinetochore‐microtubule attachment stability during mitosis. J Cell Sci 124: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru AMG, Rusin SF, Clark AEM, Kettenbach AN, Compton DA (2017) Cyclin A/Cdk1 modulates Plk1 activity in prometaphase to regulate kinetochore‐microtubule attachment stability. Elife 6: e29303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S, Hümmer S, Uldschmid A, Li X, Nigg EA (2007) Tension‐sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev 21: 2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM (2011) Formation of stable attachments between kinetochores and microtubules depends on the B56‐PP2A phosphatase. Nat Cell Biol 13: 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM (2013) Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama JB, Pereira C, Simões PA, Celestino R, Reis RM, Barbosa DJ, Pires HR, Carvalho C, Amorim J, Carvalho AX et al (2017) Molecular mechanism of dynein recruitment to kinetochores by the Rod‐Zw10‐Zwilch complex and Spindly. J Cell Biol 216: 943–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Essex A, Hu JS, Maddox PS, Motegi F, Sugimoto A, O'Rourke SM, Bowerman B, McLeod I, Yates JR III et al (2008) A new mechanism controlling kinetochore‐microtubule interactions revealed by comparison of two dynein‐targeting components: SPDL‐1 and the Rod/Zwilch/Zw10 complex. Genes Dev 22: 2385–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A (2010) Removal of Spindly from microtubule‐attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev 24: 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godek KM, Kabeche L, Compton DA (2015) Regulation of kinetochore‐microtubule attachments through homeostatic control during mitosis. Nat Rev Mol Cell Biol 16: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, Lehner CF (2005) Genetic interactions of separase regulatory subunits reveal the diverged Drosophila CENP‐C homolog. Genes Dev 19: 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EA, Kettenbach AN, Gerber SA, Compton DA (2012) Plk1 regulates the kinesin‐13 protein Kif2b to promote faithful chromosome segregation. Mol Biol Cell 23: 2264–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh W, Vale RD (2017) Disease‐associated mutations in human BICD2 hyperactivate motility of dynein‐dynactin. J Cell Biol 216: 3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA (2013) Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature 502: 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ (2000) Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150: 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R, Glover D (1989) rough deal: a gene required for proper mitotic segregation in Drosophila . J Cell Biol 109: 2951–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A (2006) Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskeleton 63: 301–312 [DOI] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR III, Tagaya M, Cleveland DW (2005) ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 169: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystkowiak I, Manguy J, Davey NE (2018) PSSMSearch: a server for modeling, visualization, proteome‐wide discovery and annotation of protein motif specificity determinants. Nucleic Acids Res 46: W235–W241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM (2004) Correcting improper chromosome‐spindle attachments during cell division. Nat Cell Biol 6: 232–237 [DOI] [PubMed] [Google Scholar]
- Liu D, Davydenko O, Lampson MA (2012) Polo‐like kinase‐1 regulates kinetochore‐microtubule dynamics and spindle checkpoint silencing. J Cell Biol 198: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, Lukavsky PJ, Walshaw J, Bullock SL (2013) Bicaudal‐D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev 27: 1233–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore‐microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Maiato H, Sunkel CE, Earnshaw WC (2003) Dissecting mitosis by RNAi in Drosophila tissue culture cells. Biol Proced Online 5: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T, Maia AF, Steffensen S, Sunkel CE (2009) Sgt1, a co‐chaperone of Hsp90 stabilizes Polo and is required for centrosome organization. EMBO J 28: 234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Cauvin C, Moch C, Radford SJ, Sampaio P, Perdigoto C, Schweisguth F, Bardin A, Sunkel CE, McKim K et al (2011) Aurora B and Cyclin B have opposite effect on the timing of cytokinesis abscission in Drosophila germ cells and in vertebrate somatic cells. Dev Cell 26: 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S, Toyoshima F, Nishida E (2007) Polo‐like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J Biol Chem 282: 15217–15227 [DOI] [PubMed] [Google Scholar]
- McEwen BF, Dong Y (2009) Releasing the spindle assembly checkpoint without tension. J Cell Biol 184: 355–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S (2011) Mass spectrometry‐based proteomics using Q Exactive, a high‐performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics 10: M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouse V, Formstecher E, Couderc JL (2006) Interaction between Polo and BicD proteins links oocyte determination and meiosis control in Drosophila . Development 133: 4005–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosalaganti S, Keller J, Altenfeld A, Winzker M, Rombaut P, Saur M, Petrovic A, Wehenkel A, Wohlgemuth S, Müller F et al (2017) Structure of the RZZ complex and molecular basis of its interaction with Spindly. J Cell Biol 216: 961–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura M, Osswald M, Leça N, Barbosa J, Pereira AJ, Maiato H, Sunkel CE, Conde C (2017) Protein phosphatase 1 inactivates Mps1 to ensure efficient Spindle Assembly Checkpoint silencing. Elife 6: e25366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura M, Conde C (2019) Phosphatases in mitosis: roles and regulation. Biomolecules 9: E55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho‐Santos T, Conde C, Sunkel CE (2011) POLO ensures chromosome bi‐orientation by preventing and correcting erroneous chromosome‐spindle attachments. J Cell Sci 125: 576–583 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle‐assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Heidmann S, Sunkel CE (2007) Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma 116: 259–274 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M (2007) Higher‐energy C‐trap dissociation for peptide modification analysis. Nat Methods 4: 709–712 [DOI] [PubMed] [Google Scholar]
- Paschal CR, Maciejowski J, Jallepalli PV (2012) A stringent requirement for Plk1 T210 phosphorylation during K‐fiber assembly and chromosome congression. Chromosoma 121: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner A (2009) Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc Natl Acad Sci USA 106: 15708–15713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH (2013) Systematic dissection of dynein regulators in mitosis. J Cell Biol 201: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Sacristan C, Ahmad MUD, Keller J, Fermie J, Groenewold V, Tromer E, Fish A, Melero R, Carazo JM, Klumperman J et al (2018) Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat Cell Biol 20: 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, Black BE (2011) Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol 21: 1158–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Silljé HH, Körner R, Nigg EA (2011) The Plk1‐dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics 10: M110.004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S (2007) Incorporation of Drosophila CID/CENP‐A and CENP‐C into centromeres during early embryonic anaphase. Curr Biol 17: 237–243 [DOI] [PubMed] [Google Scholar]
- Shrestha RL, Conti D, Tamura N, Braun D, Ramalingam RA, Cieslinski K, Ries J, Draviam VM (2017) Aurora‐B kinase pathway controls the lateral to end‐on conversion of kinetochore‐microtubule attachments in human cells. Nat Commun 8: 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Serr M, Steward R, Hays TS, Doe CQ (2005) Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell 16: 5127–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ (2012) Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore‐microtubule attachments. Dev Cell 23: 745–755 [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA (2011) Chromosome missegregation in human cells arises through specific types of kinetochore‐microtubule attachment errors. Proc Natl Acad Sci USA 108: 17974–17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR III, Lampson MA, Fukagawa T, Cheeseman IM (2010) Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore‐microtubule interface. Mol Cell 38: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Karr TL, Montgomery JM, Goldberg ML (1992) The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J Cell Biol 118: 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E, Basto R, Serr M, Scaërou F, Karess R, Hays T (2001) Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol 3: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, Hartshorne DJ, Matsumura F (2008) Myosin phosphatase‐targeting subunit 1 regulates mitosis by antagonizing polo‐like kinase 1. Dev Cell 14: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Grishchuk EL (2015) Basic mechanism for biorientation of mitotic chromosomes is provided by the kinetochore geometry and indiscriminate turnover of kinetochore microtubules. Mol Biol Cell 26: 3985–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
Movie EV11
Movie EV12
Movie EV13
Movie EV14
Movie EV15
Movie EV16
Movie EV17
Movie EV18
Movie EV19
Movie EV20
Movie EV21
Movie EV22
Movie EV23
Movie EV24
Movie EV25
Movie EV26
Review Process File


