In this study, Robles-Murguia et al. set out to examine whether muscle-secreted hormones regulate feeding. Using Drosophila as a model system combined with several in vivo and in vitro experiments, the authors identify Decapentaplegic (Dpp) as a myokine that can signal from the muscle to the brain to control feeding by altering dopamine synthesis through transcriptional regulation of tyrosine hydroxylase (TH).
Keywords: endocrine signaling, feeding behavior, foraging, Dpp, myokine, skeletal muscle, dopamine, tyrosine hydroxylase, Drosophila
Abstract
In animals, the brain regulates feeding behavior in response to local energy demands of peripheral tissues, which secrete orexigenic and anorexigenic hormones. Although skeletal muscle is a key peripheral tissue, it remains unknown whether muscle-secreted hormones regulate feeding. In Drosophila, we found that decapentaplegic (dpp), the homolog of human bone morphogenetic proteins BMP2 and BMP4, is a muscle-secreted factor (a myokine) that is induced by nutrient sensing and that circulates and signals to the brain. Muscle-restricted dpp RNAi promotes foraging and feeding initiation, whereas dpp overexpression reduces it. This regulation of feeding by muscle-derived Dpp stems from modulation of brain tyrosine hydroxylase (TH) expression and dopamine biosynthesis. Consistently, Dpp receptor signaling in dopaminergic neurons regulates TH expression and feeding initiation via the downstream transcriptional repressor Schnurri. Moreover, pharmacologic modulation of TH activity rescues the changes in feeding initiation due to modulation of dpp expression in muscle. These findings indicate that muscle-to-brain endocrine signaling mediated by the myokine Dpp regulates feeding behavior.
Since their emergence from single cells, multicellular organisms have faced the challenge of monitoring the metabolic status of diverse tissues and organs and of responding to specific local demands by correspondingly adjusting feeding. In animals, peripheral tissues such as the adipose, liver, gut, and pancreas communicate their nutritional status by releasing hormones that signal to the brain, which in turn regulates feeding (Lam 2010; Williams and Elmquist 2012). However, the role of other peripheral tissues in the endocrine control of feeding has not been well explored.
Skeletal muscle comprises ∼40% of the total body mass and is a major determinant of systemic energy expenditure during exercise and in resting conditions. However, its role in the hormonal control of feeding behavior is largely unknown. The observation that muscle contraction influences dietary choices and brain neurotransmitter levels (de Castro and Duncan 1985; Sutoo and Akiyama 2003; Bi et al. 2005; Waters et al. 2013; Liang et al. 2015; Moody et al. 2015; Chen et al. 2017; Friend et al. 2017) suggests that also skeletal muscle regulates feeding behavior by signaling to the brain, as do other peripheral tissues. However, the underlying mechanisms remain largely unknown. In particular, despite the expression of secreted signaling factors (myokines) with endocrine functions by skeletal muscle (Pedersen and Febbraio 2012; Rai and Demontis 2016), it is unknown whether any of such myokines regulates feeding by signaling to the brain.
Studies in the fruit fly Drosophila melanogaster have identified fundamental mechanisms of physiological homeostasis (Sokolowski 2001; Wangler et al. 2015). Many of the neuronal circuits and neurotransmitters that regulate feeding in higher organisms play similar roles in Drosophila (Baker and Thummel 2007; Melcher et al. 2007; Pool and Scott 2014). Moreover, hormones secreted by peripheral tissues act on the brain to regulate metabolic homeostasis also in Drosophila, as exemplified by unpaired 2, a leptin homolog secreted by the Drosophila fat body (Rajan and Perrimon 2012), and by other adipokines such as Stunted and the TNF ligand Eiger (Agrawal et al. 2016; Delanoue et al. 2016). However, as in higher organisms, the role of skeletal muscle in the neuronal control of feeding behavior remains largely unexplored.
Recently, studies in Drosophila have uncovered unexpected endocrine roles of signaling factors previously known only for their local functions during development. For example, the lipophilic morphogen Hedgehog associates with lipoproteins and signals from the gut to the fat body to regulate the storage and release of triacylglycerols during developmental growth in Drosophila (Rodenfels et al. 2014). Moreover, the morphogen decapentaplegic (dpp), homologous to human bone morphogenetic proteins BMP2 and BMP4, has been recently shown to function as an endocrine signal that regulates developmental timing in Drosophila. Specifically, Dpp produced by peripheral tissues, such as wing imaginal discs and the gut, reaches the prothoracic gland, where it inhibits ecdysone biosynthesis (Setiawan et al. 2018; Denton et al. 2019). However, it is unknown whether Dpp functions as an endocrine factor also in the regulation of physiological homeostasis in adults.
In this study, we show that decapentaplegic (dpp) is secreted by skeletal muscle and signals to the brain. Muscle-restricted dpp RNAi promotes food searching and feeding initiation, whereas dpp overexpression reduces it. This feeding response results from modulation of tyrosine hydroxylase (TH) expression and dopamine biosynthesis in the brain by muscle-derived Dpp. Consistently, TH expression and feeding initiation are similarly modulated by cell-autonomous Dpp receptor signaling and TH expression in dopaminergic neurons. In summary, these findings highlight a muscle-to-brain signaling axis that regulates foraging and thus provide evidence for myokine signaling in the endocrine control of feeding.
Results
Muscle-derived Dpp signals to the brain
Skeletal muscle has emerged as an important tissue for regulating many systemic functions via the action of muscle-secreted factors known as myokines (Pedersen and Febbraio 2008; Demontis et al. 2013; Karsenty and Olson 2016). Because dpp is expressed in skeletal muscle (Supplemental Fig. S1) and has been recently identified as an interorgan signal during Drosophila development (Setiawan et al. 2018; Denton et al. 2019), we have examined whether Dpp is an endocrine myokine that signals to distant tissues in adults.
By analyzing the levels of endogenous GFP- and Flag-tagged Dpp by Western blot (Fig. 1), we found that Dpp is efficiently processed via proteolytic cleavage (Kunnapuu et al. 2009) in adults to generate mature Dpp peptides (Fig. 1A).
Figure 1.
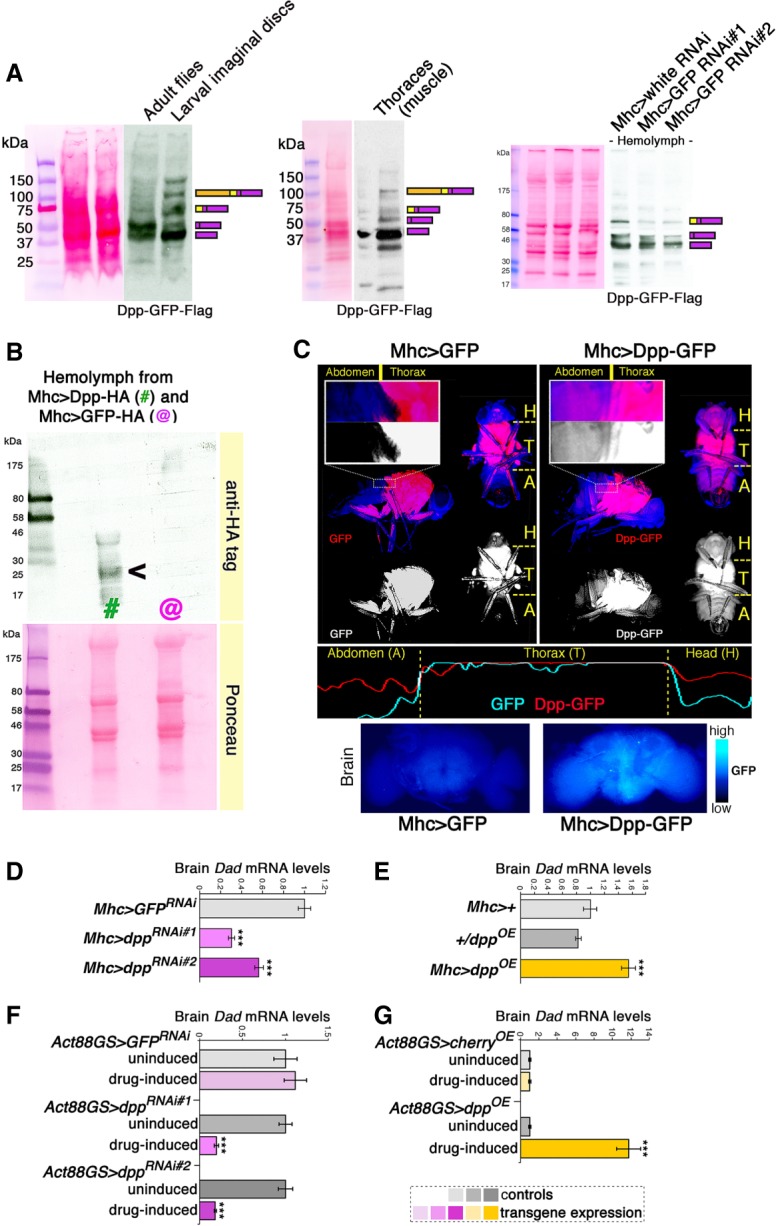
Dpp is an endocrine myokine that signals to the brain. (A) Western blot analyses indicate that endogenous Dpp-GFP-Flag is efficiently cleaved to produce mature Dpp peptides in adult flies and in skeletal muscle (thoraces). Mature Dpp-GFP-Flag is detected in the hemolymph of adult Drosophila, suggesting that it is a circulating factor. Muscle-specific GFP RNAi driven with Mhc-Gal4 leads to a ∼50% reduction in the levels of Dpp-GFP-Flag present in the hemolymph, indicating that skeletal muscle is a major source of circulating Dpp in adults. Note that the inframe fusion with GFP increases the molecular weight of Dpp-GFP-Flag, compared with the Dpp-HA shown in B. (B) Western blot analysis of hemolymph from flies expressing Dpp-HA (#; Mhc > Dpp-HA) and control GFP-HA (@; Mhc > GFP-HA) in skeletal muscle. Muscle-expressed Dpp is found in the fly circulation (hemolymph), suggesting that it is an endocrine myokine. (C) In agreement with this hypothesis, the fluorescence of muscle-produced, GFP-tagged Dpp (red) is detected in tissues of the abdomen (A) and head (H) that are distant from the production site (muscles of the fly thorax [T]). Conversely, the fluorescence of cytosolic GFP is confined to the thorax. Images of fluorescent flies and plots of fluorescence intensity are shown in C, as well as fluorescence of brains from flies with muscle-specific dpp-GFP or GFP overexpression. (D,E) Muscle-specific dpp RNAi reduces brain mRNA levels of Dad, a canonical Dpp/BMP target gene. Converse regulation is seen with muscle-specific dpp overexpression, compared with controls. (***) P < 0.001; n = 4; SEM. (F,G) Drug-induced, muscle-restricted dpp RNAi reduces brain Dad mRNA levels, whereas dpp overexpression increases it, compared with uninduced controls. (***) P < 0.001; n = 4, SEM.
Interestingly, in addition to being detected in skeletal muscle (thoraces) (Fig. 1A), endogenous Dpp-GFP-Flag is found in the hemolymph, suggesting that Dpp is indeed a circulating factor (Fig. 1A). To test whether skeletal muscle is a major source of circulating Dpp, we targeted dpp-GFP-Flag in skeletal muscle via GFP RNAi driven by the skeletal muscle-specific Mhc-Gal4 driver. Interestingly, muscle-specific GFP RNAi led to a ∼50% decrease in the hemolymph levels of Dpp-GFP-Flag. Together, these findings indicate that skeletal muscle is a major source of circulating Dpp.
To further test these findings, we next expressed a Dpp-HA transgene in muscle. Also in this case, muscle-expressed Dpp-HA was detected in the fly circulation (Fig.1B). Moreover, the fluorescence of transgenic Dpp-GFP expressed specifically in skeletal muscle (fly thorax) with Mhc-Gal4 was detected throughout the body, including regions distant from thoracic muscles (e.g., the head), whereas the fluorescence of cytosolic GFP was confined to muscle (Fig. 1C). Consistently, there was higher GFP fluorescence in the brains of flies with muscle-specific dpp-GFP overexpression, compared with GFP overexpression controls (Fig. 1C). These findings suggest that Dpp is a muscle-released factor (a myokine) that may signal to distant tissues including the brain.
To test this hypothesis, we examined whether modulation of dpp levels in muscle induces transcriptional changes in the brain. Interestingly, skeletal muscle-specific dpp RNAi and overexpression (Supplemental Figs. S2, S3) induced converse changes in brain mRNA levels of Dad/Smad6 (Fig. 1D–G), a stereotypical target gene of Dpp receptor signaling (Roy et al. 2014). Specifically, muscle-specific dpp RNAi led to a decrease in Dad expression in brains (Fig. 1D,F), whereas muscle-specific dpp overexpression increased it (Fig. 1E,G). Importantly, the Mhc-Gal4 and Act88GS-Gal4 drivers used for these interventions are specific for thoracic skeletal muscle (Schuster et al. 1996; Demontis and Perrimon 2010; Robles-Murguia et al. 2019) and do not drive any transgene expression in the brain (Supplemental Fig. S4). Taken together, these findings indicate that Dpp is an endocrine myokine that signals to the brain.
Muscle-derived Dpp regulates tyrosine hydroxylase expression and dopamine biosynthesis in the brain
The brain is the central regulator of many physiological functions and a proposed target tissue for myokines in Drosophila and mammals (Rai and Demontis 2016; Delezie and Handschin 2018). To determine the outcome of endocrine Dpp signaling, we examined the gene expression changes induced in heads, which consist primarily of brains.
To this purpose, RNA-seq was performed on heads of flies with muscle-specific dpp RNAi, dpp overexpression, and controls. Compared with control flies, several genes were consistently regulated by muscle-specific dpp RNAi (Fig. 2A; Supplemental Table S1). Moreover, a set of genes was regulated conversely by muscle-restricted dpp overexpression compared with dpp RNAi (Fig. 2A,B). Interestingly, these included genes involved in neurotransmission and chemosensory perception, such as TH/ple and Obp57e. Specifically, Obp57e is necessary for sucrose perception by the fly antenna (Galindo and Smith 2001), whereas tyrosine hydroxylase (TH/ple) is the rate-limiting enzyme in the biosynthesis of L-DOPA and dopamine (Friggi-Grelin et al. 2003a; Daubner et al. 2011). Because of the evolutionary conserved importance of dopamine in the regulation of many physiological functions, we next used qPCR to further assess TH/ple expression in brains of flies with muscle-specific dpp RNAi and overexpression. Consistent with the RNA-seq results (Fig. 2A,B), dpp RNAi in muscle increased brain TH/ple mRNA levels, whereas dpp overexpression elicited converse effects compared with controls (Fig. 2C,D). Similar results were obtained by analyzing the expression of ple-RA (Supplemental Fig. S5), a brain-specific ple isoform (Friggi-Grelin et al. 2003b; Kumar et al. 2012). In addition to muscle-derived Dpp, Dpp released by nonmuscle tissues may also contribute to regulate brain TH/ple. However, only a minimal decrease in TH/ple expression is seen in response to dpp overexpression by the fat body (Supplemental Fig. S6).
Figure 2.
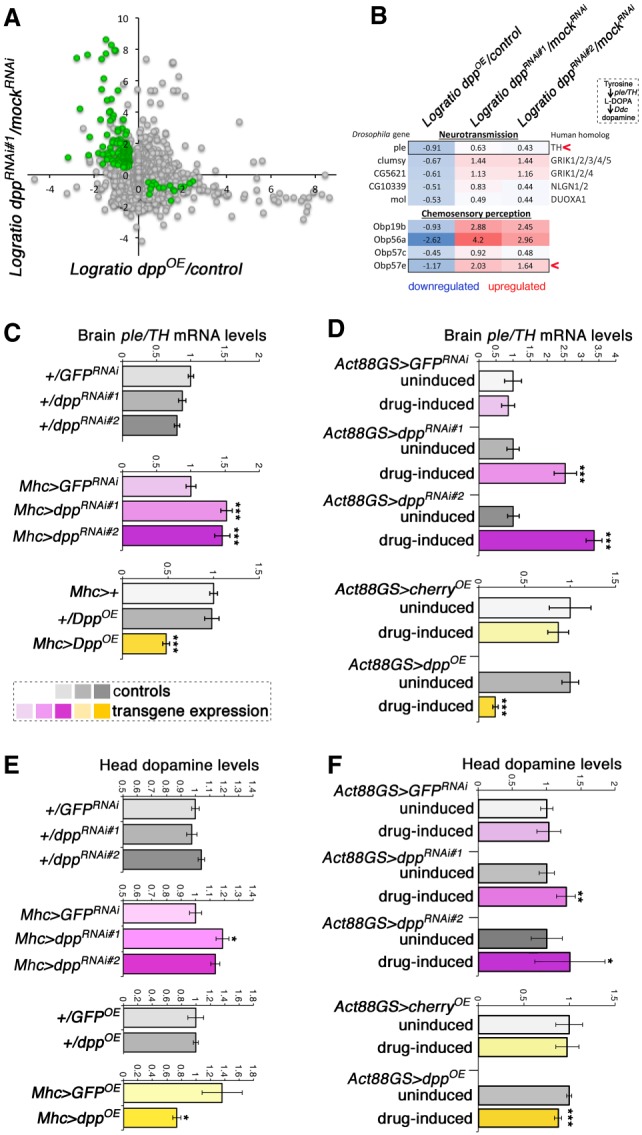
Muscle-derived Dpp regulates brain ple/TH expression and dopamine biosynthesis. (A) RNA-seq from heads of flies with muscle-specific dpp RNAi, dpp overexpression (OE), and controls (n = 3/genotype). The y-axis shows the difference in expression (logratio) of dpp RNAi versus GFP RNAi (mock), and the x-axis shows the logratio of dpp overexpression (Mhc > DppOE) versus control (Mhc/+). Genes that are significantly regulated (P < 0.05) in converse manner by dpp RNAi and dpp overexpression, which include genes involved in neurotransmission and chemosensory perception, are highlighted in green. (B) Among these, ple/TH and Obp57e have been previously implicated in foraging. Ple/TH is the rate-limiting enzyme for L-DOPA and dopamine biosynthesis. (C,D) qRT-PCR from fly brains confirms that muscle-specific dpp RNAi increases ple/TH expression, whereas dpp overexpression has converse effects compared with controls. Similar results are obtained with both Mhc-Gal4 and the drug-inducible Act88-GS-Gal4. (***) P < 0.001; n = 4; SEM. (E,F) UPLC/MS-MS reveals that muscle-specific dpp RNAi increases brain dopamine, whereas dpp overexpression reduces it compared with controls. Similar results are obtained with both Mhc-Gal4 and the drug-inducible Act88-GS-Gal4 driver. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; n[batch of 30 heads] = 5–17. SD.
Because TH is the rate-limiting enzyme in the dopamine biosynthesis pathway (Friggi-Grelin et al. 2003a; Daubner et al. 2011), we next tested whether Dpp-induced changes in TH/ple expression lead to corresponding changes in dopamine levels. To this end, head dopamine levels were measured by UPLC/MS-MS. As expected based on the regulation of TH, dpp overexpression in muscle reduced head dopamine levels by ∼25%, whereas dpp RNAi increased dopamine levels by ∼20% (Fig. 2E,F). Importantly, these changes in head dopamine are similar to those seen in response to exercise (Sutoo and Akiyama 1996, 2003) and other physiological stimuli in mice (Léna et al. 2005; Ferreira et al. 2012; Drgonova et al. 2016) and higher than the circadian variation in dopamine levels seen in Drosophila (Gonzalo-Gomez et al. 2012). Taken together, these findings suggest a role for muscle-derived Dpp in modulating dopaminergic functions.
Modulation of Dpp levels in muscle regulates locomotor activity and foraging
Dopamine levels control spontaneous locomotion in Drosophila and mice (Zhou and Palmiter 1995; Avale et al. 2008; Riemensperger et al. 2011; Sternson et al. 2013), which suggests that modulation of head dopamine biosynthesis by muscle Dpp (Fig. 2) may impact this behavior.
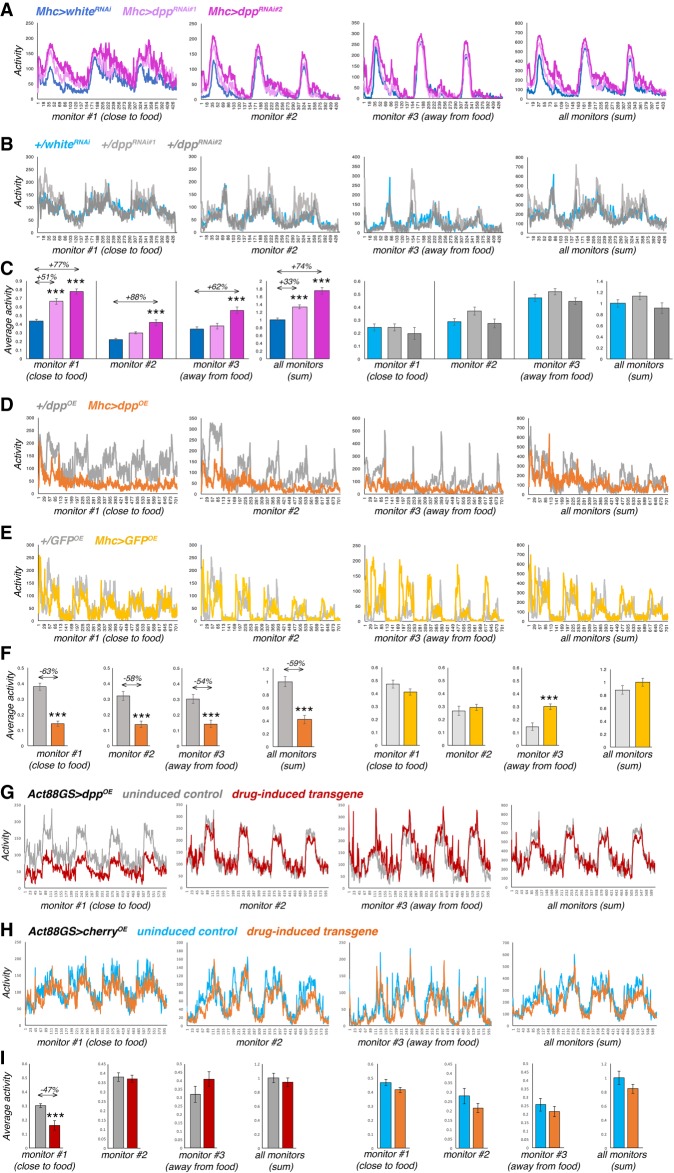
To test this hypothesis, the spontaneous locomotion was monitored via locomotor activity monitors, as done before (Katewa et al. 2012). This system records the spontaneous locomotion of flies as assessed by the interruption of laser beams crossed by flies as they move in a tube with fly food (Pfeiffenberger et al. 2010). As expected based on the regulation of dopamine by muscle-derived Dpp (Fig. 2) and the role of dopamine in spontaneous locomotion (Zhou and Palmiter 1995; Avale et al. 2008; Riemensperger et al. 2011), we found that muscle-specific dpp RNAi increases spontaneous locomotion, whereas muscle-specific dpp overexpression reduces it, compared to controls (Fig. 3). Interestingly, in addition to an overall increase in locomotor activity, dpp RNAi promotes locomotion particularly in proximity to the food (Fig. 3A–C). Conversely, the reduction in activity observed upon dpp overexpression is most prominent in proximity to food (Fig. 3D–I). Altogether, these findings suggest that modulation of spontaneous activity by Dpp is geared toward foraging.
Figure 3.
Regulation of locomotor activity and foraging by muscle-derived Dpp. (A,B) Muscle-specific dpp RNAi increases spontaneous locomotion (A), compared with controls (B). (C) In addition to an overall increase in spontaneous activity (sum of all monitors), dpp RNAi increases activity particularly in proximity to the food (monitor #1), compared with whiteRNAi and transgene-alone controls. Mean values are shown in A and B, and the corresponding bar graphs with mean ± SEM are shown in C, with n[Mhc > whiteRNAi] = 12, n[Mhc > dppRNAi] = 10, n[+/whiteRNAi] = 12, and n[+/dppRNAi] = 10 batches of 10 flies each. (D,E) Muscle-specific dpp overexpression reduces spontaneous locomotion (D), compared with controls (E). Mean values are shown in D and E, and the corresponding bar graphs with mean ± SEM are shown in F, with n = 8 batches each consisting of 10 flies for each genotype. (G,H) Muscle-specific dpp overexpression with Act88GS reduces spontaneous locomotion primarily in proximity to food (monitor #1) (G), compared with uninduced controls and cherry overexpression (H). Mean values are shown in G and H, and the corresponding bar graphs with mean ± SEM are shown in I, with n = 16 batches each consisting of 10 flies for each condition. Overall, the reduction in activity observed upon dpp overexpression is most prominent in proximity to food (F,I). (***) P < 0.001.
Muscle-derived Dpp regulates feeding initiation
Because muscle-derived Dpp regulates head dopamine biosynthesis (Fig. 2) and foraging (Fig. 3) and because of the known roles of dopaminergic neurons in feeding initiation in Drosophila (Inagaki et al. 2012; Marella et al. 2012; Kain and Dahanukar 2015; Landayan et al. 2018), we next asked whether muscle-derived Dpp regulates feeding. To this purpose, PER assays were used, whereby flies are stimulated on their labellum (a primary taste organ near the mouth) with a sucrose solution and the occurrence of a proboscis extension response (PER) is scored. PER reflects the gustatory perception and acceptance of dietary sugar by the neuronal circuits that modulate feeding (Shiraiwa and Carlson 2007; Marella et al. 2012; Itskov and Ribeiro 2013): A higher chance of PER is typically observed in flies that are starved or when flies are presented with increasingly higher sucrose concentrations, whereas the chance of PER is lower in optimally fed flies and in mutants that are defective in sugar chemosensation (Galindo and Smith 2001; Swarup et al. 2014).
In agreement with a role of muscle-derived Dpp in feeding initiation, we found that muscle-restricted dpp RNAi increases the probability of PER (Fig. 4A), whereas muscle-restricted dpp overexpression reduces it, compared with controls (Fig. 4B).
Figure 4.
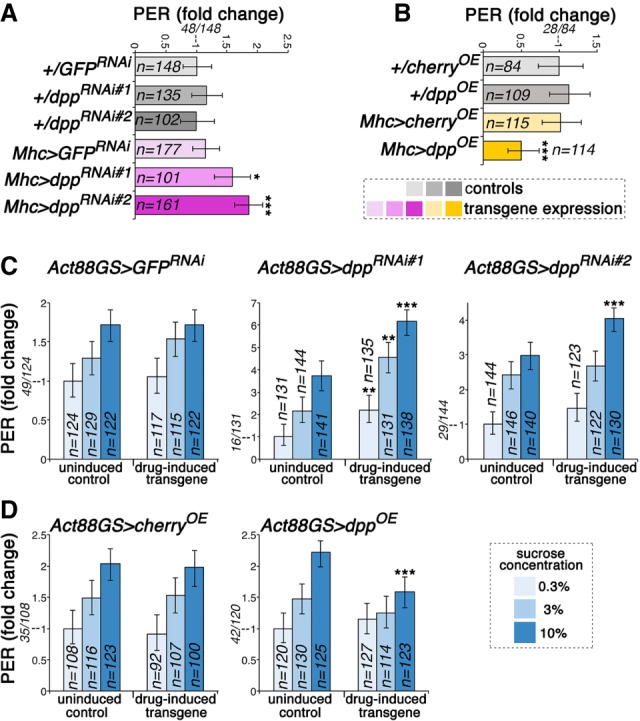
Muscle-derived Dpp regulates feeding initiation. (A) PER assays indicate that dpp RNAi (Mhc > dppRNAi#1 and RNAi#2) increases the propensity of flies to initiate feeding compared with control GFP RNAi (Mhc > GFPRNAi) and transgene-alone controls (+/dppRNAi#1; +/dppRNAi#2; +/GFPRNAi). (*) P < 0.05; (***) P < 0.001; n = 101–177. (B) Conversely, dpp overexpression reduces the chance of PER, compared with controls (+/cherryOE; +/dppOE; Mhc > cherryOE). (*) P < 0.05; n = 84–114. (C) PER assays done at different sucrose concentrations (0.3%, 3%, 10%). Drug-induced muscle-specific dpp RNAi increases the chance of PER even at low sucrose concentrations, compared with uninduced controls and to GFP RNAi. (D) Conversely, the chance of PER is low in flies with muscle-specific dpp overexpression, even at high sucrose concentrations. (**) P < 0.01; (***) P < 0.001; n = 100–141. In A–D the proportion of flies with PER versus the total is indicated for the control condition in each panel.
Previous studies have shown that initiation of feeding on sucrose is strongly influenced by its concentration (Galindo and Smith 2001; Marella et al. 2012). Therefore, we next tested whether muscle-derived Dpp affects the chance of PER in flies that are given increasing sucrose concentrations. For these studies, the drug-inducible Act88GS-Gal4 driver (Robles-Murguia et al. 2019) was used to drive transgene expression in the indirect flight muscles of adult flies in response to the drug RU486. Importantly, drug-induced expression of a control GFP RNAi in muscle did not significantly affect PER compared with the uninduced control, indicating that the RU486 drug used to induce transgene expression is not responsible for changes in PER (Fig. 4C).
Interestingly, the chance of PER was higher in drug-induced dpp RNAi expression than in uninduced controls, even at low sucrose concentrations, whereas GFP RNAi did not significantly induce PER (Fig. 4C). Conversely, whereas an increase in the chance of PER was typically observed when increasing the sucrose concentrations, flies with drug-induced dpp overexpression in muscle displayed little increase in PER (Fig. 4D). Taken together, these findings indicate that the myokine Dpp regulates feeding initiation in a manner that is consistent with its modulation of dopamine biosynthesis (Fig. 2) and foraging (Fig. 3).
Up-regulation of TH/ple expression in different dopaminergic neuronal subsets exerts distinct effects on foraging and feeding initiation
In Drosophila, several dopaminergic neuronal subsets have been implicated in feeding initiation, as inferred from the experimental modulation of their activity (Inagaki et al. 2012; Marella et al. 2012; Kain and Dahanukar 2015; Landayan et al. 2018). However, it is unknown whether an increase in TH/ple levels in some of these neuronal subsets would similarly regulate foraging and feeding initiation.
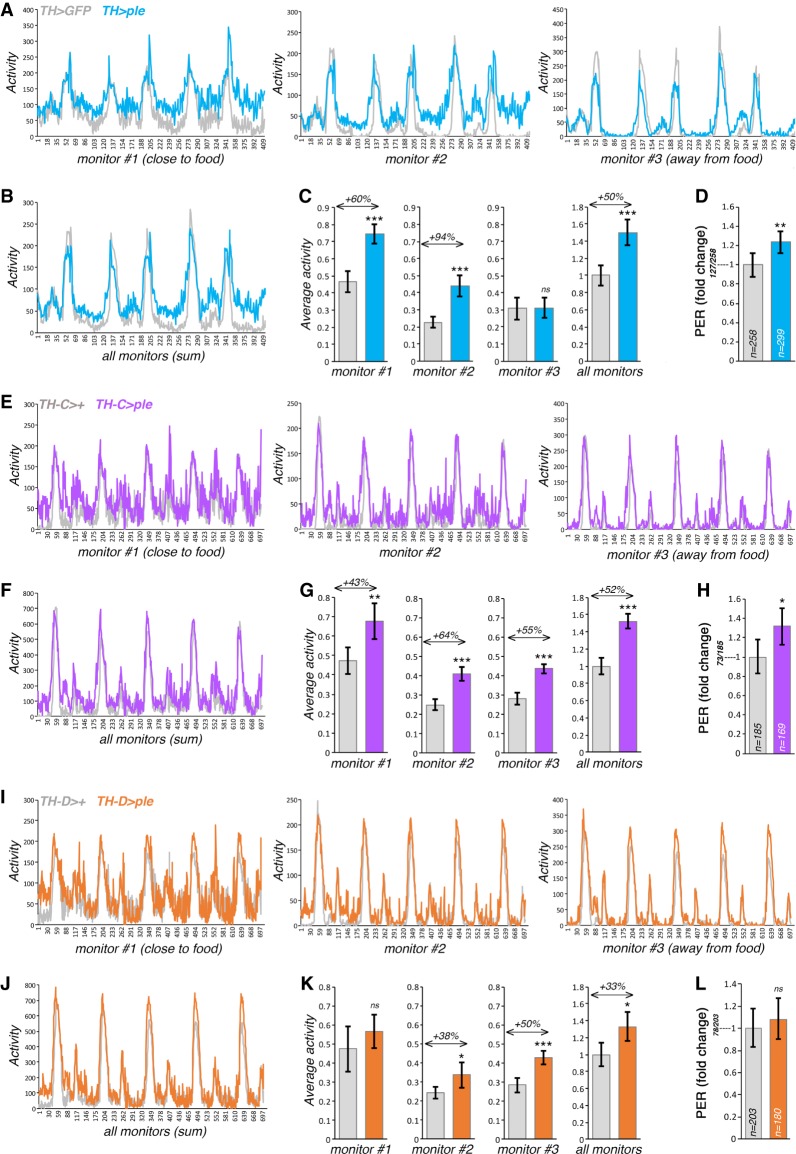
To address this question, we used a neuronal-specific TH/ple transgene (UAS-DTH1m) (Cichewicz et al. 2017) and general and subset-specific TH-Gal4 drivers (Friggi-Grelin et al. 2003a; Xie et al. 2018). Consistent with the hypothesis that muscle-derived Dpp regulates foraging and feeding initiation via brain TH/ple, we found that a general increase of TH/ple expression in dopaminergic neurons promotes overall spontaneous activity and locomotion in proximity to the food (Fig. 5A–C). Consistently, measurement of feeding initiation via PER assays reveals that increased expression of TH/ple leads to higher chance of feeding initiation, compared with controls (Fig. 5D).
Figure 5.
Increase in ple/TH expression in dopaminergic neurons promotes foraging. (A–C) Monitoring of the spontaneous locomotion of flies with general ple/TH overexpression in dopaminergic neurons. Three monitors are located at different distance from the food. In addition to increasing overall activity (B,C), overexpression of ple/TH with TH-Gal4 promotes locomotion nearby the food (monitor #1) (A,C) compared with control GFP overexpression. Mean values are shown in A and B, and the corresponding bar graphs with mean ± SD are shown in C, with n[TH > GFP] = 6 and n[TH > ple] = 6 batches of 15 flies each. (D) ple/TH overexpression with TH-Gal4 promotes feeding initiation, as assessed with PER assays, with n[TH > GFP] = 258 and n[TH > ple] = 299; P < 0.05, 95% CI. (E–G) ple/TH overexpression in subsets of dopaminergic neurons with TH-C-Gal4 promotes spontaneous locomotion in proximity of food (monitor #1) (E,G), whereas this does not occur with TH-D-Gal4 (I,K), which drives expression in a different subset of dopaminergic neurons. (F,G,J,K) ple/TH overexpression with both TH-C-Gal4 and TH-D-Gal4 increases overall activity. (H,L) ple/TH overexpression with TH-C-Gal4 but not with TH-D-Gal4 promotes feeding initiation, as estimated with PER assays, with n[TH-C > GFP] = 185, n[TH-C > ple] = 169, n[TH-D > GFP] = 203, and n[TH-D > ple] = 180. P < 0.05; 95% CI. Mean values are shown in E, F, I, and J), and the corresponding bar graphs with mean ± SD are shown in G and K, with n[TH-C > GFP] = 5, n[TH-C > ple] = 4, n[TH-D > GFP] = 5, and n[TH-D > ple] = 4 batches of 15 flies each.
We next determined whether an increase in TH/ple levels restricted to some dopaminergic neuronal subsets similarly modulates foraging. For these studies, we utilized TH-C-Gal4 and TH-D-Gal4, which drive expression in different combinations of dopaminergic neurons (Cording et al. 2017; Xie et al. 2018). Specifically, TH-C-Gal4 drives transgene expression in PAL, PPM1, PPM2, PPL2ab, PPL2c, and VUM dopaminergic neurons, whereas TH-D-Gal4 is specific for PPL1, PPM1, PPM2, and PPM3 dopaminergic neurons (Cording et al. 2017; Xie et al. 2018). In response to TH/ple overexpression, there was an overall increase in spontaneous locomotion with both drivers (Fig. 5E–G,I–K). However, ple/TH overexpression with TH-C-Gal4 increases activity in proximity to food (Fig. 5G) and the chance of PER (Fig. 5H), whereas this does not occur with TH-D-Gal4 (Fig. 5K,L), suggesting that subsets of dopaminergic neurons modulated by TH-C-Gal4 but not by TH-D-Gal4 are responsible for food-seeking in response to an increase in TH/ple levels. Specifically, TH-C-Gal4 drives transgene expression in the dopaminergic TH-VUM neuron, which had been already implicated in feeding initiation in Drosophila (Inagaki et al. 2012; Marella et al. 2012), whereas TH-D-Gal4 does not (Cording et al. 2017; Xie et al. 2018). On this basis, modulation of TH/ple levels in the TH-VUM neuron may contribute to feeding initiation in response to muscle-derived Dpp.
Dpp receptor signal transduction in dopaminergic neurons regulates TH expression and feeding initiation via the downstream transcriptional repressor Schnurri
Dpp signaling is initiated by binding of Dpp to its receptor, which consists of a dimer formed by the type I receptor Thickveins (Tkv, which confers ligand-specificity) and the type II receptor Punt, which also transduces signaling from other BMP and TGF-β ligands (Miyazono et al. 2010; Hamaratoglu et al. 2014).
To test whether Dpp receptor signaling in dopaminergic neurons elicits similar effects as modulation of Dpp levels in muscle, we first determined the expression pattern of tkv in the brain. To this end, brains from tkv > GFP flies were immunostained with antibodies for GFP and TH, which marks dopaminergic neurons. Interestingly, tkv-Gal4 induced GFP expression in brain cells that were TH positive, suggesting that Tkv is preferentially enriched in subsets of dopaminergic neurons (Fig. 6A; Supplemental Fig. S7), including the TH-VUM dopaminergic neuron previously implicated in foraging and feeding initiation (Fig. 5; Inagaki et al. 2012; Marella et al. 2012; Landayan et al. 2018). Moreover, compared with control GFP overexpression, muscle-specific dpp overexpression led to a preferential increase of phospho-Smad3 levels (indicative of Dpp signaling) in dopaminergic neurons versus nearby cells (Fig. 6B). Together, these findings suggest that dopaminergic neurons may be particularly competent in responding to changes in Dpp levels.
Figure 6.
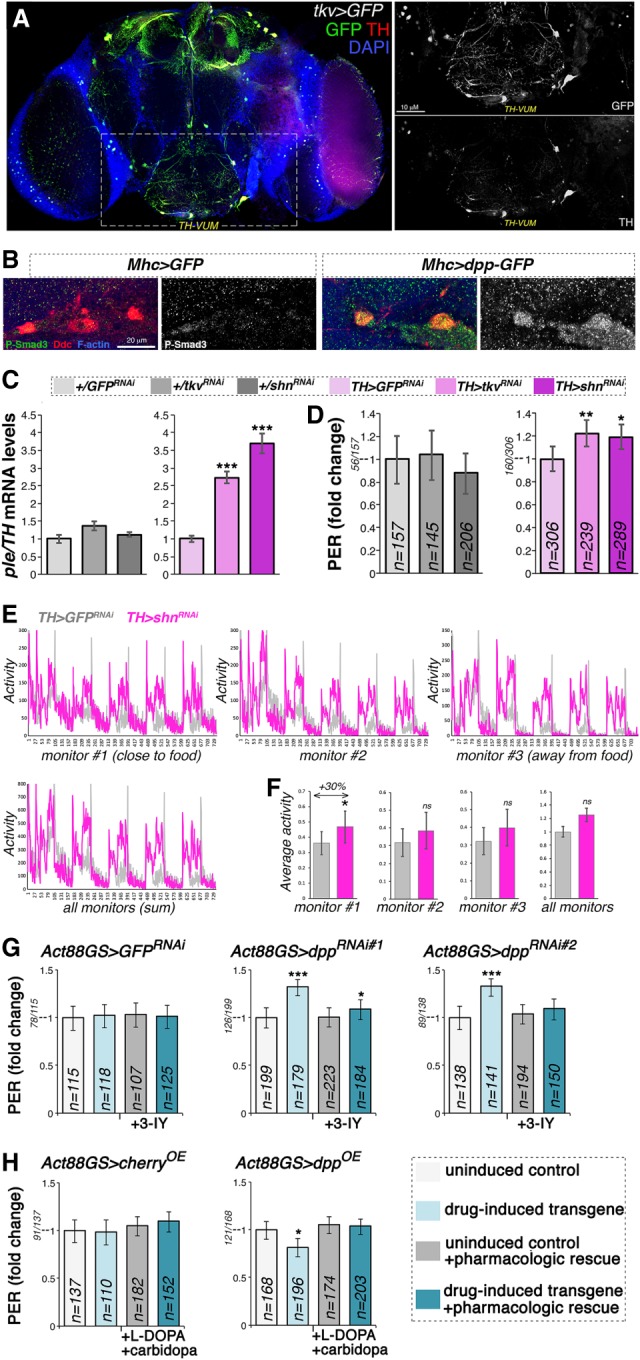
Cell-autonomous modulation of Dpp receptor signaling in dopaminergic neurons regulates ple/TH expression and the propensity to feed on sucrose. (A) Immunostaining of brains from flies that express GFP under control of the Tkv promoter (Tkv > GFP). Many neurons that express TH (red) are also positive for the Dpp receptor Tkv (green), including the TH-VUM neuron previously implicated in the PER. (B) Immunostaining of brains from flies that express GFP and dpp-GFP in skeletal muscle (Mhc > GFP vs. Mhc > dpp-GFP). Overall brain P-Smad3 immunoreactivity (indicative of BMP signaling) increases in response to muscle overexpression of dpp-GFP, and this change is particularly prominent in dopaminergic neurons. (C) RNAi for the Dpp receptor Tkv in dopaminergic neurons increases brain ple/TH levels, similar to RNAi for the transcriptional repressor Schnurri (Shn), which acts downstream from Tkv. (***) P < 0.001; n = 4; SEM. (D) RNAi for Tkv and Shn in dopaminergic neurons promotes PER. (*) P < 0.05; (**) P < 0.01; n = 145–306; 95% CI. (E) RNAi for Shn in dopaminergic neurons promotes spontaneous activity, in particular in proximity to the food (F), compared with control GFP RNAi. (*) P < 0.05; with n = 7 tubes [TH > GFPRNAi] and n = 9 tubes [TH > shnRNAi], each with 15 flies. Error bars indicate the SD. (G) The TH inhibitor 3-IY blunts PER induction by drug-induced dpp RNAi in muscle, indicating that TH is key for this adaptive response. n = 107–223; 95% CI. (H) Decrease in PER due to drug-induced, muscle-specific dpp overexpression is compensated for by the dietary administration of L-DOPA, the precursor of dopamine produced by TH. Carbidopa is coadministered to ensure preferential delivery of L-DOPA to the brain. n = 110–196; 95% CI.
To test whether Dpp receptor signal transduction regulates TH/ple expression in a cell-autonomous manner, RNAi for the Dpp receptor thickveins was driven in dopaminergic neurons by TH-Gal4. Interestingly, tkv RNAi promoted TH/ple mRNA expression (Fig. 6C), similar to the effect seen with muscle-restricted dpp RNAi (Fig. 2). These findings indicate that TH/ple expression is modulated by Dpp receptor signaling in dopaminergic neurons.
The observation that inhibition of Dpp receptor signaling increases TH/ple expression suggests the involvement of a downstream transcriptional repressor. Typically, Dpp receptor signaling induces phosphorylation and activation of the transcription factor Mad, which works in complex with Medea, similar to the homologous Smad1/5–Smad4 complex regulated by BMP signaling in vertebrates (Miyazono et al. 2010; Hamaratoglu et al. 2014). Although Mad/Medea activates the transcription of target genes, its association with Schnurri, an evolutionary conserved transcriptional repressor, inhibits target gene expression (Grieder et al. 1995; Marty et al. 2000; Pyrowolakis et al. 2004). To test whether Schnurri is a transcriptional repressor that inhibits ple/TH expression, shn RNAi was driven in dopaminergic neurons. Similar to tkv RNAi, shn RNAi promoted ple/TH expression, compared with GFP RNAi (Fig. 6C). Moreover, also shnurri appeared to be preferentially expressed in subsets of dopaminergic neurons, as shown by immunostaining of brains from shn > GFP flies (Supplemental Fig. S7).
To investigate the underlying molecular mechanisms, we searched publicly available Drosophila ChIP-seq data (Van Bortle et al. 2015) for the presence of Shn peaks, which were detected in the promoter of the canonical target gene brinker but not in that of ple/TH (data not shown). However, several Shn peaks were found in the promoter of dikar (Supplemental Fig. S8A), a gene located next to ple/TH. Analysis of high-throughput chromosome conformation capture (Hi-C) data (Sexton et al. 2012) revealed that the dikar promoter region characterized by Shn peaks is in the same topologically associated domain (TAD) and has high frequency of interactions with the ple promoter region (Supplemental Fig. S8A). To directly assess the putative silencer activity of the dikar promoter region, a ∼500-bp sequence with the strongest Shn binding was cloned upstream of a Renilla luciferase reporter driven by a minimal promoter. Luciferase assays in S2R+ cells revealed that this region reduced transcription in response to treatment with recombinant Dpp or in the presence of a constitutively active form of Tkv (TkvQ253D), compared with control GFP expression (Supplemental Fig. S8D). Conversely, RNAi for Tkv and Shn increased luciferase activity (Supplemental Fig. S8E). These findings suggest that Shn represses TH/ple expression through long-range interactions of a Shn-bound silencer with the proximal ple/TH promoter (Supplemental Fig. S8).
Considering that TH expression levels and dopaminergic neurons modulate the PER (Fig. 5), and that Dpp receptor signaling in dopaminergic neurons regulates TH/ple expression (Fig. 6A,C), which is key for dopaminergic functions (Friggi-Grelin et al. 2003a; Daubner et al. 2011), we next tested whether Dpp receptor signaling in dopaminergic neurons regulates the chance of PER. In agreement with this hypothesis, tkv RNAi and shn RNAi significantly increased the chance of PER compared with control GFP RNAi (Fig. 6D). In summary, cell-autonomous reduction of Dpp receptor signaling in dopaminergic neurons increases TH/ple expression and the chance of PER. Together, these findings suggest that muscle-derived Dpp may regulate brain TH levels and feeding initiation by modulating Dpp receptor signal transduction in dopaminergic neurons.
Having found that shn RNAi promotes TH/ple expression and the chance of PER (Fig. 6C,D), we next assessed whether it also modulates foraging. For these studies, shn RNAi was driven in dopaminergic neurons with TH-Gal4 in the presence of tubulin-Gal80ts (a similar analysis was not possible for TH > tkv RNAi due to its semilethality and hence impossibility of recording foraging over several days).
Consistent with the hypothesis that Dpp regulates foraging and feeding initiation by modulating brain TH/ple expression via shn, we found that shn RNAi promotes spontaneous activity in particular in proximity to the food (monitor #1) compared with control GFP RNAi (Fig. 6E,F).
In summary, these findings indicate that Dpp receptor signal transduction in dopaminergic neurons regulates TH/ple expression and feeding initiation via the downstream transcriptional repressor Schnurri.
Rescue of Dpp-induced changes in feeding behavior by pharmacologic modulation of TH
In addition to TH/ple, muscle-derived Dpp nonautonomously regulates the expression of Obp57e (Fig. 2B), which has been implicated in sucrose perception and PER (Galindo and Smith 2001). Furthermore, Dpp also induces additional gene expression changes (Fig. 2A,B) that may contribute to feeding behavior.
To test whether modulation of TH/ple expression is a prominent mechanism by which muscle-derived Dpp regulates PER, pharmacologic treatments were done to inhibit TH and, conversely, to compensate for the lack of TH activity. In the first set of experiments, the TH inhibitor 3-iodotyrosine (3-IY) was administered to flies having muscle-specific dpp RNAi driven by Act88-GS-Gal4. Consistent with a key role for TH downstream from Dpp signaling, pharmacologic inhibition of TH greatly reduced the changes in PER due to muscle-specific dpp RNAi (Fig. 6D).
A second set of experiments consisted of pharmacologic rescues with L-DOPA, the rate-limiting dopamine precursor synthesized by TH from L-tyrosine. To improve its delivery to the brain, L-DOPA is typically administered with carbidopa, a DOPA decarboxylase inhibitor that prevents L-DOPA utilization by peripheral tissues (Maas et al. 2017; Poewe et al. 2017). This strategy has been effective in Drosophila, whereby administration of L-DOPA with carbidopa rescues TH/ple mutants (Nall et al. 2016; Cichewicz et al. 2017). Therefore, we tested whether dietary supplementation of L-DOPA and carbidopa rescues PER defects seen in flies with muscle-specific dpp overexpression. This was found to be the case (Fig. 6E), which is consistent with the hypothesis that defects in TH can be bypassed by providing sufficient amounts of L-DOPA to restore its levels to those required for normal dopamine synthesis. In summary, pharmacologic rescue experiments indicate that TH/ple is key for modulating PER by muscle-derived Dpp (Fig. 7).
Figure 7.
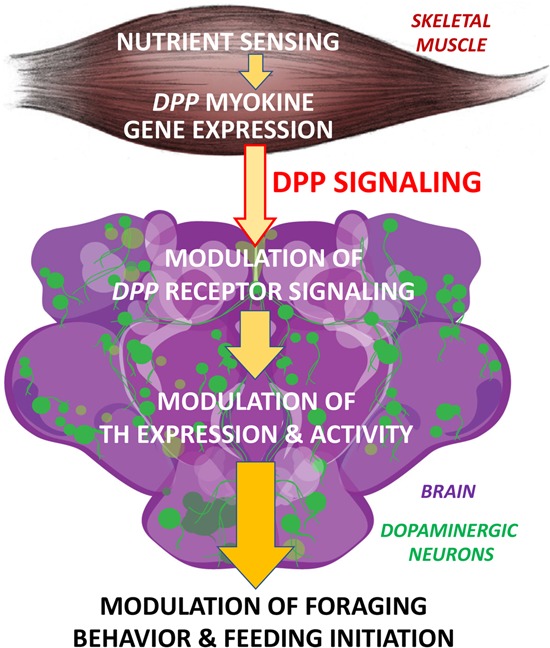
Endocrine regulation of feeding behavior by myokine signaling. Dpp is an endocrine myokine that signals to the brain. In skeletal muscle, dpp expression is induced by nutrient sensing (mTOR signaling). Consistently, muscle-restricted dpp RNAi leads to higher pale/tyrosine hydroxylase (TH) expression in the brain and higher chance of foraging and feeding initiation, whereas muscle-restricted dpp overexpression elicits converse effects. Cell-autonomous decrease of Dpp receptor signaling in dopaminergic neurons leads to higher ple/TH expression and higher propensity for feeding initiation. These results highlight a muscle-to-brain signaling axis that regulates feeding behavior.
Discussion
Since its discovery in 1982 (Spencer et al. 1982), Dpp has been primarily characterized as a morphogen during development. We have found that muscle-derived Dpp is an endocrine factor that signals to the brain, in line with increasing evidence for a role of Dpp in intertissue signaling (Li et al. 2013; Kux and Pitsouli 2014) and more recently for its capacity to act as an endocrine factor in Drosophila (Setiawan et al. 2018; Denton et al. 2019). These roles of Dpp in intertissue communication are in line with the increasing knowledge about BMPs in mammals (Wagner et al. 2010) and the observation that BMP2 and BMP4 (the human homologs of Dpp) are detected in the bloodstream (Son et al. 2011; Umemoto et al. 2011; van Baardewijk et al. 2013; Zhang et al. 2015; Eslaminejad et al. 2019).
A number of different factors, such as posttranslational modifications, could be involved in determining the range of Dpp signaling. In particular, because Dpp proprotein processing has been shown to be key for the range of Dpp signaling during development (Kunnapuu et al. 2009), it is possible that it may also influence the capacity of Dpp to function as an endocrine factor. Specifically, three furin sites are present in the Dpp proprotein and cleavage at the first site has been shown to be critical and sufficient for long-range signaling within wing imaginal discs (Kunnapuu et al. 2009). However, cleavage at the other sites may modulate the interaction of Dpp with the extracellular matrix and with Dpp-binding proteins (Umulis et al. 2009), and thus influence the capacity of Dpp to signal in a paracrine versus endocrine manner. Consistent with this hypothesis, there is a difference in the relative abundance of Dpp variants derived from proprotein processing in adults versus larval imaginal discs (Fig. 1A).
Although the functions of circulating BMP2 and BMP4 are not known (Son et al. 2011; Umemoto et al. 2011; van Baardewijk et al. 2013; Zhang et al. 2015; Eslaminejad et al. 2019), many circulating factors contribute to the endocrine control of important physiological body functions by signaling to the brain. For example, several neural circuits in the brain regulate feeding by integrating environmental cues and signaling by hormones released by peripheral tissues such as the gut, pancreas, liver, and adipose (Williams and Elmquist 2012). Recently, a broader contribution of peripheral tissues to the endocrine control of feeding has emerged, as exemplified by an unanticipated role of bone in this process (Mosialou et al. 2017). However, the contribution of many peripheral tissues remains largely unexplored. In particular, although skeletal muscle is the most abundant tissue and a major site of nutrient uptake, how the brain senses changes in the metabolic status of the muscle still remains unknown.
Myokines are muscle-secreted growth factors and cytokines that circulate in the bloodstream (Pedersen and Febbraio 2012) and that might mediate such muscle-to-brain signaling (Rai and Demontis 2016; Delezie and Handschin 2018). However, thus far there has been no evidence for myokine signaling to the brain. In this study we report that muscle-secreted Dpp regulates the expression of brain TH (Fig. 2), the rate-limiting enzyme for dopamine synthesis. Furthermore, cell-autonomous modulation of Dpp receptor signaling in dopaminergic neurons recapitulates the effects of muscle-derived Dpp, supporting the model that modulation of TH expression results from muscle-to-brain signaling mediated by the myokine Dpp.
Although our study provides evidence for modulation of TH/ple expression in the brain by muscle-specific Dpp (Fig. 2), several distinct mechanisms may be responsible for such regulation. One possibility is that muscle-derived Dpp regulates TH/ple expression in dopaminergic neurons indirectly via a relay tissue. For example, muscle-derived Dpp may signal to the glia (which is part of the blood brain barrier), which, in turn, may cross-talk with dopaminergic neurons via a relay signal (Kanai et al. 2018; Maksoud et al. 2019). Alternatively, Dpp may cross the blood-brain barrier and directly engage Dpp receptors in dopaminergic neurons, as observed for other circulating factors that regulate feeding behavior by directly acting on orexigenic and anorexigenic neurons (Lam 2010; Williams and Elmquist 2012). Altogether, the myokine Dpp may signal directly to dopaminergic neurons or indirectly via an intermediate signal, such as a BMP ligand produced by a relay tissue.
Dopaminergic neurons have long been implicated in promoting feeding initiation in Drosophila (Inagaki et al. 2012; Marella et al. 2012; Kain and Dahanukar 2015; Landayan et al. 2018). Similarly, changes in dopamine levels regulate feeding behavior in Drosophila and mice (Zhou and Palmiter 1995; Szczypka et al. 1999, 2000, 2001; Sotak et al. 2005; Palmiter 2007; Avale et al. 2008; Riemensperger et al. 2011; Pristerà et al. 2015; Doan et al. 2016; Chen et al. 2017). Our findings that muscle-derived Dpp regulates feeding initiation via the modulation of dopamine biosynthesis therefore suggest that this evolutionary conserved myokine signaling pathway may play a role in the integrated control of feeding behavior also in higher organisms. Specifically, BMP2 and BMP4, which are known to be expressed by skeletal muscle in mammals (Umemoto et al. 2011), may signal to the brain to regulate dopamine biosynthesis and feeding behavior also in humans.
Consistent with a role for muscle-derived Dpp in the endocrine control of foraging and feeding initiation, we found that dpp expression in skeletal muscle is modulated by nutrient sensing (Supplemental Fig. S9), as observed for other orexigenic and anorexigenic hormones secreted by peripheral tissues (Williams and Elmquist 2012). Specifically, activation of the target of rapamycin (mTOR) nutrient sensing pathway promotes dpp transcription, whereas mTOR inhibition via the Tsc1/2 tuberous sclerosis complex represses dpp expression via the transcription factor Mnt (Supplemental Fig. S9). Altogether, these findings indicate that reduced mTOR signaling in muscle reduces Dpp levels. Thus, the mTOR/Mnt-dependent expression of dpp may contribute to its role in muscle-to-brain systemic signaling, and explain how the brain perceives nutritional scarcity within skeletal muscle.
In addition to key roles in locomotor activity and feeding, TH activity and the dopaminergic system are key for normal brain function and are altered in human diseases such as Parkinson's disease, schizophrenia, and addiction. On this basis, our findings that muscle-derived Dpp cell nonautonomously regulates brain TH activity and dopamine biosynthesis suggest that skeletal muscle may influence via myokine signaling several brain functions and disease processes that depend on dopamine. Endocrine signaling by myokines may thus constitute an additional layer of regulation of TH activity and dopaminergic functions in physiologic and pathologic conditions.
Materials and methods
Fly stocks
The Gal4 drivers used in this study were TH-Gal4 (Friggi-Grelin et al. 2003a), TH-C-Gal4, and TH-D-Gal4 (Xie et al. 2018), Mhc-Gal4 (Schuster et al. 1996; Demontis and Perrimon 2010), Act88GS-Gal4 (Act88F-GeneSwitch-Gal4) (Robles-Murguia et al. 2019), tkv-Gal4 (BL#63958), shn-Gal4 (VDRC#205415), and lipophorin-Gal4 (Rajan and Perrimon 2012). The following Gal4-responsive UAS stocks were used: UAS-cherry (cherryOE; BL#35787), UAS-Dpp-GFP (DppOE) (Entchev et al. 2000), UAS-Dpp-HA (Kunnapuu et al. 2009), UAS-mito-GFP-HA (GFP-HA; BL#8442), UAS-GFP (Shen et al. 2009), UAS-GFPRNAi (BL#41552 as control RNAi; BL#41554 and BL#41556 to knock down dpp-GFP-Flag, driven at 29°C for a week), UAS-cherryRNAi (BL#35785), UAS-dppRNAi#1 (BL#25782), UAS-dppRNAi#2 (BL#33618), UAS-tkvRNAi (NIG-Fly#14026R-1) (previously used and validated in Li et al. 2013), UAS-shnRNAi (VDRC#105643) (previously used and validated in Peterson and O'Connor 2013), and UAS-DTH1m (Cichewicz et al. 2017). The following additional fly stocks were used: dpp-GFP-Flag (VDRC#318414) and shn-GFP (BL#42671). UAS-Tsc1 + Tsc2, UAS-Mnt, UAS-Rheb (BL#9688), and Mnt null flies were previously described (Loo et al. 2005; Demontis and Perrimon 2009).
Cell lines
Drosophila S2R+ cells/cm2 were cultured in Schneider's medium containing 10% FCS and penicillin-streptomycin (Gibco).
Drosophila husbandry and GeneSwitch-mediated transgenic expression
All experiments were done with 10- to 14-day-old male flies kept (25–30 flies/tube) at 25°C, 60% humidity, and a 12-h/12-h light-dark cycle. For experiments with the GeneSwitch system, flies were raised for 4 d on normal food posteclosion and then kept for 10 d on food supplemented with 200 µM RU486 (mifepristone; Calbiochem #475838) dissolved in ethanol or with ethanol alone (control). Each RU486-treated genotype was compared with the corresponding ethanol-treated control.
Monitoring of spontaneous activity and foraging
The Drosophila locomotor activity monitor (LAM; TriKinetics #LAM25H-3) was used to record the spontaneous locomotion of flies (Katewa et al. 2012), as assessed based on the interruption of laser beams crossed by flies as they move in a tube with fly food (Pfeiffenberger et al. 2010). For these studies, 10 or 15 male flies/tube (as indicated) were monitored over >2 d under a 12:12 h light/dark cycle at 25°C and 60% humidity. Three monitors were used to record every 10 min the activity of flies in three different areas of the tube, i.e., close to the food (monitor #1), away from the food (monitor #3), and at an intermediate position (monitor #2). To avoid a specific effects of RU486 on spontaneous locomotion in experiments with Act88GS flies (Fig. 3G–I), flies were treated for 10 d with 20 µM RU486, left for 3 d on plain food, and then assessed for spontaneous activity. All genotypes and corresponding control conditions were assessed at the same time.
Proboscis extension response (PER) assays
PER assays were performed as previously described (Galindo and Smith 2001; Shiraiwa and Carlson 2007; Marella et al. 2012; Steck et al. 2018), but without any prior starvation of flies. Briefly, flies were glued on their back to a plastic surface and after recovery were allowed to drink distilled water ad libitum that was delivered via a fine-pointed mop made from a kimwipe. Subsequently, a cone-like shaped wick made from a kimwipe was dipped into a 10% sucrose solution and presented to the labellum by gentle contact, and flies were scored on the basis of whether a proboscis extension occurred. For the assays reported in Figure 4, C and D, flies were sequentially given 0.3%, 3%, and 10% sucrose solutions. PER assays were done under constant light conditions between ZT4 and ZT10.
Hemolymph preparation and Western blots
Hemolymph was prepared from >30 flies, using standard procedures (Géminard et al. 2009). Specifically, flies were decapitated, placed in a 0.5-mL tube, centrifuged at 1500g for 6 min, and the hemolymph collected in an underlying tube at 4°C. Equal volumes of hemolymph from all conditions were mixed with SDS-containing blue loading buffer with protease inhibitors, denatured by incubation for 5 min at 95°C, and probed with SDS-PAGE and Western blots with anti-HA (Roche #12CA5) or anti-Flag (Sigma #F3165) antibodies, as indicated, followed by Ponceau S staining. Note that the presence of an in-frame GFP fusion results in an ∼25-kDa band shift when comparing the MW of Dpp-GFP-Flag (Fig. 1A) versus Dpp-HA (Fig. 1B). All membranes were blocked with 10% BSA.
Western blots for whole flies, thoraces, and imaginal discs were processed as described above, with the difference that the tissues were first homogenized in RIPA buffer with protease inhibitors by using a bullet blender (Next Advance), followed by mixing with SDS containing blue loading buffer, denaturation at 95°C, and SDS-PAGE. Imaginal discs were dissected as done before (Demontis and Dahmann 2007).
qRT-PCR
qRT-PCR was performed as previously described (Demontis et al. 2014). Total RNA was extracted with the TRIzol reagent (Life Technologies) from Drosophila thoraces, heads, or brains (as indicated), followed by reverse transcription with the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed with SYBR Green and a CFX96 apparatus (Bio-Rad). Alpha-tubulin 84B was used as a normalization reference. pale-RA expression was monitored as described before (Kumar et al. 2012). The comparative CT method was used for relative quantitation of mRNA levels.
RNA sequencing
Total RNA was extracted as described above from heads. Three biological replicates for each sample were prepared for RNA-seq with the TruSeq stranded mRNA library preparation kit (Illumina) and sequenced on the Illumina HiSeq 2000 platform. FASTQ sequences derived from mRNA paired-end 100-bp sequences were mapped to the Drosophila melanogaster genome (BDGP release 5) with the STAR aligner (version 2.5.3a). Transcript level data were counted by using HTSeq (version 0.6.1p1) based on the BDGP5 gtf release 75. For the analyses in Figure 2, the TMM method was used to calculate the normalization factors. Then linear modeling was carried out on the log-CPM (count per million) values, which are assumed to be normally distributed, and the mean-variance relationship was accommodated using precision weights calculated by the voom function. The lmFit and contrasts.fit functions were used for the linear modelling. All analyses were done by limma package in R 3.2.3. The RNA-seq data are available in Supplemental Table S1 and at the Gene Expression Omnibus with accession number GSE140391.
Dopamine measurement with UPLC-MS/MS
Groups of 30 heads were analyzed by ultraperformance liquid chromatography tandem mass spectrometry (UPLC-MS/MS), following derivatization with benzoyl chloride (BzCl) (Wong et al. 2016). Briefly, frozen fly heads were suspended in buffer (100 µL of 0.1 M TCA with 10−2 M sodium acetate, 10−4 M EDTA, and 10.5% methanol pH 3.8) and homogenized with a handheld sonicating probe. Then, 20 µL of the homogenate was used for protein determination with the BCA Protein Assay Kit (Thermo Scientific). Samples were then centrifuged, and 5 µL of the supernatant was diluted in 20 µL of ice-cold acetonitrile. Subsequently, 10 µL of 50 mM aqueous sodium carbonate was added, followed by 10 µL of 2% BzCl solution in acetonitrile. Samples were vortexed and neutralized by adding 20 µL of 13C-BzCl internal standard solution (20% acetonitrile in water with 3% sulfuric acid) and 40 µL of water, and analyzed by UPLC-MS/MS. Dopamine concentrations were quantified by comparing peak areas of samples with those of dopamine standards (Wong et al. 2016).
Brain immunostaining and confocal microscopy
Brains were dissected, fixed, and immunostained as described previously (Demontis and Perrimon 2010) with rabbit anti-TH (Millipore #AB152), chicken anti-GFP (Abcam #13970), rabbit anti-P-Smad3 (S423 + S425, Abcam #52903; previously used to monitor BMP receptor signaling in Drosophila) (Akiyama et al. 2012), and/or rat anti-Ddc (gift of Dr. Jay Hirsh) (Riemensperger et al. 2011). DAPI (1:1000) was used to stain nuclei, and Alexa635-phalloidin or anti-Bruchpilot antibodies (DSHB, #nc82) were used to visualize the overall brain architecture.
Pharmacologic rescue experiments
For pharmacologic inhibition of TH, 1.25 mg/mL of 3-iodotyrosine (3-IY; Sigma #I8250) was added to the fly food, as described previously in flies (Cichewicz et al. 2017). For pharmacologic rescue of TH defects, 3 mg/mL of L-DOPA and 0.0125 mg/mL of carbidopa (Abcam #ab120573 and #ab144725) were mixed with the fly food as previously done (Nall et al. 2016; Cichewicz et al. 2017). A similar L-DOPA/carbidopa regimen was previously shown to rescue defects in dopamine levels of TH/ple mutant flies but not to increase dopamine abundance beyond normal levels (Cichewicz et al. 2017).
For pharmacologic rescue experiments, flies were fed for 10 d, with regular changes of food every 2–3 d, before PER assays.
Fluorescence distribution
Epifluorescence microscopy was performed with a Zeiss microscope and the Axiovision software. NIH ImageJ was used to plot the GFP fluorescence intensity across the fly body.
ChIP-seq and HiC analysis
ChIP-seq data were downloaded from GSE68654 (Van Bortle et al. 2015) and mapped to the Drosophila genome (BDGP release 5). Data quality control was performed and bigwig tracks for visualization (normalized to 15 M unique mapped reads) were generated as described previously (Aldiri et al. 2017). Hi-C processed data from GSE34453 (Sexton et al. 2012) were downloaded from Juicebox (Durand et al. 2016) and visualized with ProteinPaint (Zhou et al. 2016).
ChIP-qPCR
Heads from shn-GFP flies were collected by vortexing (Jensen et al. 2013) and maintained in a frozen state using liquid nitrogen while being ground into powder with a pestle and mortar. Samples were immediately fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated by adding lysis buffer, followed by sonication using the EpiShear Probe Sonicator (Active Motif #53051) with an EpiShear Cooled Sonication Platform (Active Motif #53080), and the DNA was sheared to an average length of 300–500 bp. Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase, proteinase K, and heat for de-crosslinking, followed by ethanol precipitation. Pellets were resuspended, and the resulting DNA was quantified on a CLARIOstar spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield.
Aliquots of chromatin (5 µg) were precleared with protein A agarose beads (Invitrogen). Genomic DNA regions bound by Shn-GFP were isolated by using a ChIP-grade antibody against GFP (Abcam #ab290) that is routinely used by the ModENCODE project (Mod et al. 2010). Complexes were washed, eluted from the beads with SDS buffer, and treated with RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation.
The qPCR reactions were carried out in triplicate using SYBR Green Supermix (Bio-Rad #170-8882) on a CFX Connect Real Time PCR detection system. One negative control region and eight sites of interest were queried. Resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using input DNA (pooled unprecipitated genomic DNA from each sample).
Cloning and luciferase assays
To generate the Silencer #A-Renilla luciferase reporter, a region containing the strongest Shn-bound ChIP-seq peak (3L: 6743499..6743994) nearby dikar was synthetically synthesized and cloned into the pRL-null vector (Promega) with EcoRI-HF and SpeI-HF.
Luciferase assays were done as before (Demontis and Perrimon 2009; Zhou et al. 2013). Briefly, 15 × 104 S2R+ cells/cm2 were seeded in Schneider's medium containing 10% FCS and penicillin-streptomycin (Gibco), and transfected 1 d later, using the Effectene Transfection Kit (Qiagen). Cells in each well of a 96-well plate (0.32 cm2) were transfected with a mixture of 60 ng of the Silencer #A-Renilla luciferase reporter, 2 ng of the polIII-firefly luciferase reporter, 20 ng of Actin-Gal4, 20 ng of UAS-RNAi, 10 ng of Act-GFP, and/or 10 ng of Act-TkvQ253D DNA plasmids. The Act-GFP and Act-TkvQ253D DNA plasmids have been previously described (Pyrowolakis et al. 2004). The UAS-tkvRNAi and UAS-shnRNAi DNA plasmids were obtained from the VDRC (#dna2549 and #dna1644) and the pUAST plasmid was used as empty vector (EV) control. Other plasmids were described before (Demontis and Perrimon 2009, 2010; Demontis et al. 2014). Four days after transfection, recombinant Drosophila Dpp (R&D #159-DP-020) was added to the culture medium at a final concentration of 4.1 µg/mL. The following day, the luciferase assay was performed with the Dual-Glo Luciferase Assay (Promega) according to manufacturer's instructions. The polIII-firefly luciferase reporter was cotransfected as normalization control, and luciferase activity represents the ratio of Renilla to firefly luciferase activity.
Statistical analyses
The unpaired two-tailed Student's t-test was used to compare the means of two independent groups to each other. One-way ANOVA with Tukey's post hoc test was used for multiple comparisons of more than two groups of normally distributed data. PER data were analyzed by the Fisher's exact test (two-tailed), and 95% confidence intervals (CI) were obtained by the modified Wald method. The “n” for each experiment can be found in the figures or legends and represents independently generated samples consisting of cell populations/wells for luciferase reporter assays, and individual flies, batches of flies, or batches of fly tissues for in vivo experiments. Bar graphs represent the mean ± SD, ±SEM, or ± CI as indicated in the figure legends. Throughout the figures, asterisks indicate the significance of the P value: P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***). A significant result was defined as P < 0.05. Statistical analyses were done with Excel and GraphPad Prism.
Supplementary Material
Acknowledgments
Fly stocks, antibodies, and DNA plasmids were generously provided by Dr. Osamu Shimmi, Dr. Giorgos Dr. Pyrowolakis, Dr. Christian Dahmann, Dr. Mark Wu, and Dr. Jay Hirsh and the VDRC, NIG-Fly, and Bloomington stock centers. We thank Paris Hazelton-Cavill for technical help, Dr. Richard Smeyne and Amar Pani for initial HPLC measurements of catecholamine levels, and the Vanderbilt University Neurochemistry Core, which is supported by the Vanderbilt Brain Institute and the Vanderbilt Kennedy Center (U54HD083211). We also thank the Light Microscopy facility and the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. F.D. is supported by research grants from The American Parkinson Disease Association, The Glenn Foundation for Medical Research, The Ellison Medical Foundation, The Hartwell Foundation, The American Federation for Aging Research, and the National Institute on Aging of the National Institutes of Health (R01AG055532 and R56AG063806). Research at St. Jude Children's Research Hospital is supported by the ALSAC.
Author contributions: M.R-M., D.R., and F.D. performed the experiments. D.F., B.X., and Y.F. did computational analyses. F.D. designed the project, analyzed the data, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.329110.119.
References
- Agrawal N, Delanoue R, Mauri A, Basco D, Pasco M, Thorens B, Léopold P. 2016. The Drosophila TNF eiger is an adipokine that acts on insulin-producing cells to mediate nutrient response. Cell Metab 23: 675–684. 10.1016/j.cmet.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Akiyama T, Marques G, Wharton KA. 2012. A large bioactive BMP ligand with distinct signaling properties is produced by alternative proconvertase processing. Sci Signal 5: ra28 10.1126/scisignal.2002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I, Xu B, Wang L, Chen X, Hiler D, Griffiths L, Valentine M, Shirinifard A, Thiagarajan S, Sablauer A, et al. 2017. The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron 94: 550–568.e10. 10.1016/j.neuron.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avale ME, Faure P, Pons S, Robledo P, Deltheil T, David DJ, Gardier AM, Maldonado R, Granon S, Changeux JP, et al. 2008. Interplay of β2* nicotinic receptors and dopamine pathways in the control of spontaneous locomotion. Proc Natl Acad Sci 105: 15991–15996. 10.1073/pnas.0807635105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. 2007. Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab 6: 257–266. 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi S, Scott KA, Hyun J, Ladenheim EE, Moran TH. 2005. Running wheel activity prevents hyperphagia and obesity in Otsuka long-evans Tokushima fatty rats: role of hypothalamic signaling. Endocrinology 146: 1676–1685. 10.1210/en.2004-1441 [DOI] [PubMed] [Google Scholar]
- Chen W, Wang HJ, Shang NN, Liu J, Li J, Tang DH, Li Q. 2017. Moderate intensity treadmill exercise alters food preference via dopaminergic plasticity of ventral tegmental area-nucleus accumbens in obese mice. Neurosci Lett 641: 56–61. 10.1016/j.neulet.2017.01.055 [DOI] [PubMed] [Google Scholar]
- Cichewicz K, Garren EJ, Adiele C, Aso Y, Wang Z, Wu M, Birman S, Rubin GM, Hirsh J. 2017. A new brain dopamine-deficient Drosophila and its pharmacological and genetic rescue. Genes Brain Behav 16: 394–403. 10.1111/gbb.12353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cording AC, Shiaelis N, Petridi S, Middleton CA, Wilson LG, Elliott CJH. 2017. Targeted kinase inhibition relieves slowness and tremor in a Drosophila model of LRRK2 Parkinson's disease. NPJ Parkinsons Dis 3: 34 10.1038/s41531-017-0036-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner SC, Le T, Wang S. 2011. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch Biochem Biophys 508: 1–12. 10.1016/j.abb.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro JM, Duncan G. 1985. Operantly conditioned running: effects on brain catecholamine concentrations and receptor densities in the rat. Pharmacol Biochem Behav 23: 495–500. 10.1016/0091-3057(85)90407-1 [DOI] [PubMed] [Google Scholar]
- Delanoue R, Meschi E, Agrawal N, Mauri A, Tsatskis Y, McNeill H, Leopold P. 2016. Drosophila insulin release is triggered by adipose Stunted ligand to brain Methuselah receptor. Science 353: 1553–1556. 10.1126/science.aaf8430 [DOI] [PubMed] [Google Scholar]
- Delezie J, Handschin C. 2018. Endocrine crosstalk between skeletal muscle and the brain. Front Neurol 9: 698 10.3389/fneur.2018.00698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Dahmann C. 2007. Apical and lateral cell protrusions interconnect epithelial cells in live Drosophila wing imaginal discs. Dev Dyn 236: 3408–3418. 10.1002/dvdy.21324 [DOI] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. 2009. Integration of insulin receptor/foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136: 983–993. 10.1242/dev.027466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143: 813–825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Piccirillo R, Goldberg AL, Perrimon N. 2013. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell 12: 943–949. 10.1111/acel.12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Patel VK, Swindell WR, Perrimon N. 2014. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep 7: 1481–1494. 10.1016/j.celrep.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Xu T, Dayan S, Nicolson S, Kumar S. 2019. Dpp regulates autophagy-dependent midgut removal and signals to block ecdysone production. Cell Death Differ 26: 763–778. 10.1038/s41418-018-0154-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan KV, Kinyua AW, Yang DJ, Ko CM, Moh SH, Shong KE, Kim H, Park SK, Kim DH, Kim I, et al. 2016. FoxO1 in dopaminergic neurons regulates energy homeostasis and targets tyrosine hydroxylase. Nat Commun 7: 12733 10.1038/ncomms12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonova J, Walther D, Hartstein GL, Bukhari MO, Baumann MH, Katz J, Hall FS, Arnold ER, Flax S, Riley A, et al. 2016. Cadherin 13: human cis-regulation and selectively altered addiction phenotypes and cerebral cortical dopamine in knockout mice. Mol Med 22: 537–547. 10.2119/molmed.2015.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3: 99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev EV, Schwabedissen A, González-Gaitán M. 2000. Gradient formation of the TGF-β homolog Dpp. Cell 103: 981–992. 10.1016/S0092-8674(00)00200-2 [DOI] [PubMed] [Google Scholar]
- Eslaminejad F, Mashayekhi F, Osalou MA, Sasani ST, Salehi Z. 2019. BMP4 circulating levels and promoter (rs17563) polymorphism in risk prediction of idiopathic male infertility. Br J Biomed Sci 76: 98–100. 10.1080/09674845.2018.1564419 [DOI] [PubMed] [Google Scholar]
- Ferreira JG, Tellez LA, Ren X, Yeckel CW, de Araujo IE. 2012. Regulation of fat intake in the absence of flavour signalling. J Physiol 590: 953–972. 10.1113/jphysiol.2011.218289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend DM, Devarakonda K, O'Neal TJ, Skirzewski M, Papazoglou I, Kaplan AR, Liow J-S, Guo J, Rane SG, Rubinstein M, et al. 2017. Basal ganglia dysfunction contributes to physical inactivity in obesity. Cell Metab 25: 312–321. 10.1016/j.cmet.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. 2003a. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol 54: 618–627. 10.1002/neu.10185 [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Iché M, Birman S. 2003b. Tissue-specific developmental requirements of Drosophila tyrosine hydroxylase isoforms. Genesis 35: 175–184. 10.1002/gene.10178 [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. 2001. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159: 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géminard C, Rulifson EJ, Léopold P. 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab 10: 199–207. 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Gonzalo-Gomez A, Turiegano E, León Y, Molina I, Torroja L, Canal I. 2012. Ih current is necessary to maintain normal dopamine fluctuations and sleep consolidation in Drosophila. PLoS One 7: e36477 10.1371/journal.pone.0036477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder NC, Nellen D, Burke R, Basler K, Affolter M. 1995. Schnurri is required for Drosophila Dpp signaling and encodes a zinc finger protein similar to the mammalian transcription factor PRDII-BF1. Cell 81: 791–800. 10.1016/0092-8674(95)90540-5 [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Affolter M, Pyrowolakis G. 2014. Dpp/BMP signaling in flies: from molecules to biology. Semin Cell Dev Biol 32: 128–136. 10.1016/j.semcdb.2014.04.036 [DOI] [PubMed] [Google Scholar]
- Inagaki HK, Ben-Tabou de-Leon S, Wong AM, Jagadish S, Ishimoto H, Barnea G, Kitamoto T, Axel R, Anderson DJ. 2012. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell 148: 583–595. 10.1016/j.cell.2011.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskov PM, Ribeiro C. 2013. The dilemmas of the gourmet fly: the molecular and neuronal mechanisms of feeding and nutrient decision making in Drosophila. Front Neurosci 7: 12 10.3389/fnins.2013.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Sanchez-Garcia J, Williams C, Khare S, Mathur K, Graze RM, Hahn DA, McIntyre LM, Rincon-Limas DE, Fernandez-Funez P. 2013. Purification of transcripts and metabolites from Drosophila heads. J Vis Exp 2013: e50245 10.3791/50245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain P, Dahanukar A. 2015. Secondary taste neurons that convey sweet taste and starvation in the Drosophila brain. Neuron 85: 819–832. 10.1016/j.neuron.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Kanai MI, Kim MJ, Akiyama T, Takemura M, Wharton K, O'Connor MB, Nakato H. 2018. Regulation of neuroblast proliferation by surface glia in the Drosophila larval brain. Sci Rep 8: 3730 10.1038/s41598-018-22028-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, Olson EN. 2016. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell 164: 1248–1256. 10.1016/j.cell.2016.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. 2012. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab 16: 97–103. 10.1016/j.cmet.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Chen D, Sehgal A. 2012. Dopamine acts through Cryptochrome to promote acute arousal in Drosophila. Genes Dev 26: 1224–1234. 10.1101/gad.186338.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnapuu J, Bjorkgren I, Shimmi O. 2009. The Drosophila DPP signal is produced by cleavage of its proprotein at evolutionary diversified furin-recognition sites. Proc Natl Acad Sci 106: 8501–8506. 10.1073/pnas.0809885106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kux K, Pitsouli C. 2014. Tissue communication in regenerative inflammatory signaling: lessons from the fly gut. Front Cell Infect Microbiol 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TK. 2010. Neuronal regulation of homeostasis by nutrient sensing. Nat Med 16: 392–395. 10.1038/nm0410-392 [DOI] [PubMed] [Google Scholar]
- Landayan D, Feldman DS, Wolf FW. 2018. Satiation state-dependent dopaminergic control of foraging in Drosophila. Sci Rep 8: 5777 10.1038/s41598-018-24217-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léna I, Parrot S, Deschaux O, Muffat-Joly S, Sauvinet V, Renaud B, Suaud-Chagny MF, Gottesmann C. 2005. Variations in extracellular levels of dopamine, noradrenaline, glutamate, and aspartate across the sleep–wake cycle in the medial prefrontal cortex and nucleus accumbens of freely moving rats. J Neurosci Res 81: 891–899. 10.1002/jnr.20602 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Han L, Shi L, Lin X. 2013. Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev Cell 24: 133–143. 10.1016/j.devcel.2012.12.010 [DOI] [PubMed] [Google Scholar]
- Liang NC, Bello NT, Moran TH. 2015. Wheel running reduces high-fat diet intake, preference and mu-opioid agonist stimulated intake. Behav Brain Res 284: 1–10. 10.1016/j.bbr.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LW, Secombe J, Little JT, Carlos LS, Yost C, Cheng PF, Flynn EM, Edgar BA, Eisenman RN. 2005. The transcriptional repressor dMnt is a regulator of growth in Drosophila melanogaster. Mol Cell Biol 25: 7078–7091. 10.1128/MCB.25.16.7078-7091.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas RPPWM, Wassenberg T, Lin JP, van de Warrenburg BPC, Willemsen MAAP. 2017. l-Dopa in dystonia: a modern perspective. Neurology 88: 1865–1871. 10.1212/WNL.0000000000003897 [DOI] [PubMed] [Google Scholar]
- Maksoud E, Liao EH, Haghighi AP. 2019. A neuron-glial trans-signaling cascade mediates LRRK2-induced neurodegeneration. Cell Rep 26: 1774–1786.e4. 10.1016/j.celrep.2019.01.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella S, Mann K, Scott K. 2012. Dopaminergic modulation of sucrose acceptance behavior in Drosophila. Neuron 73: 941–950. 10.1016/j.neuron.2011.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty T, Müller B, Basler K, Affolter M. 2000. Schnurri mediates Dpp-dependent repression of brinker transcription. Nat Cell Biol 2: 745–749. 10.1038/35036383 [DOI] [PubMed] [Google Scholar]
- Melcher C, Bader R, Pankratz MJ. 2007. Amino acids, taste circuits, and feeding behavior in Drosophila: towards understanding the psychology of feeding in flies and man. J Endocrinol 192: 467–472. 10.1677/JOE-06-0066 [DOI] [PubMed] [Google Scholar]
- Miyazono K, Kamiya Y, Morikawa M. 2010. Bone morphogenetic protein receptors and signal transduction. J Biochem 147: 35–51. 10.1093/jb/mvp148 [DOI] [PubMed] [Google Scholar]
- Mod EC, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody L, Liang J, Choi PP, Moran TH, Liang NC. 2015. Wheel running decreases palatable diet preference in Sprague-Dawley rats. Physiol Behav 150: 53–63. 10.1016/j.physbeh.2015.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialou I, Shikhel S, Liu JM, Maurizi A, Luo N, He Z, Huang Y, Zong H, Friedman RA, Barasch J, et al. 2017. MC4R-dependent suppression of appetite by bone-derived lipocalin 2. Nature 543: 385–390. 10.1038/nature21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall AH, Shakhmantsir I, Cichewicz K, Birman S, Hirsh J, Sehgal A. 2016. Caffeine promotes wakefulness via dopamine signaling in Drosophila. Sci Rep 6: 20938 10.1038/srep20938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD. 2007. Is dopamine a physiologically relevant mediator of feeding behavior? Trends Neurosci 30: 375–381. 10.1016/j.tins.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Febbraio MA. 2008. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol Rev 88: 1379–1406. 10.1152/physrev.90100.2007 [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Febbraio MA. 2012. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol 8: 457–465. 10.1038/nrendo.2012.49 [DOI] [PubMed] [Google Scholar]
- Peterson AJ, O'Connor MB. 2013. Activin receptor inhibition by Smad2 regulates Drosophila wing disc patterning through BMP-response elements. Development 140: 649–659. 10.1242/dev.085605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Lear BC, Keegan KP, Allada R. 2010. Locomotor activity level monitoring using the Drosophila activity monitoring (DAM) system. Cold Spring Harb Protoc 2010: pdb prot5518. [DOI] [PubMed] [Google Scholar]
- Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, Schrag AE, Lang AE. 2017. Parkinson disease. Nat Rev Dis Primers 3: 17013 10.1038/nrdp.2017.13 [DOI] [PubMed] [Google Scholar]
- Pool AH, Scott K. 2014. Feeding regulation in Drosophila. Curr Opin Neurobiol 29: 57–63. 10.1016/j.conb.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pristerà A, Lin W, Kaufmann AK, Brimblecombe KR, Threlfell S, Dodson PD, Magill PJ, Fernandes C, Cragg SJ, Ang SL. 2015. Transcription factors FOXA1 and FOXA2 maintain dopaminergic neuronal properties and control feeding behavior in adult mice. Proc Natl Acad Sci 112: E4929–E4938. 10.1073/pnas.1503911112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Müller B, Basler K, Affolter M. 2004. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell 7: 229–240. 10.1016/j.devcel.2004.07.008 [DOI] [PubMed] [Google Scholar]
- Rai M, Demontis F. 2016. Systemic nutrient and stress signaling via myokines and myometabolites. Annu Rev Physiol 78: 85–107. 10.1146/annurev-physiol-021115-105305 [DOI] [PubMed] [Google Scholar]
- Rajan A, Perrimon N. 2012. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151: 123–137. 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Isabel G, Coulom H, Neuser K, Seugnet L, Kume K, Iche-Torres M, Cassar M, Strauss R, Preat T, et al. 2011. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci 108: 834–839. 10.1073/pnas.1010930108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Murguia M, Hunt LC, Finkelstein D, Fan Y, Demontis F. 2019. Tissue-specific alteration of gene expression and function by RU486 and the GeneSwitch system. NPJ Aging Mech Dis 5: 6 10.1038/s41514-019-0036-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenfels J, Lavrynenko O, Ayciriex S, Sampaio JL, Carvalho M, Shevchenko A, Eaton S. 2014. Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev 28: 2636–2651. 10.1101/gad.249763.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB. 2014. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science 343: 1244624 10.1126/science.1244624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. 1996. Genetic dissection of structural and functional components of synaptic plasticity. II. fasciclin II controls presynaptic structural plasticity. Neuron 17: 655–667. 10.1016/S0896-6273(00)80198-1 [DOI] [PubMed] [Google Scholar]
- Setiawan L, Pan X, Woods AL, O'Connor MB, Hariharan IK. 2018. The BMP2/4 ortholog Dpp can function as an inter-organ signal that regulates developmental timing. Life Sci Alliance 1: e201800216 10.26508/lsa.201800216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shen J, Curtis C, Tavaré S, Tower J. 2009. A screen of apoptosis and senescence regulatory genes for life span effects when over-expressed in Drosophila. Aging (Albany NY) 1: 191–211. 10.18632/aging.100018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraiwa T, Carlson JR. 2007. Proboscis extension response (PER) assay in Drosophila. J Vis Exp 3: 193 10.3791/193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski MB. 2001. Drosophila: genetics meets behaviour. Nat Rev Genet 2: 879–890. 10.1038/35098592 [DOI] [PubMed] [Google Scholar]
- Son JW, Jang EH, Kim MK, Baek KH, Song KH, Yoon KH, Cha BY, Son HY, Lee KW, Jo H, et al. 2011. Serum BMP-4 levels in relation to arterial stiffness and carotid atherosclerosis in patients with Type 2 diabetes. Biomark Med 5: 827–835. 10.2217/bmm.11.81 [DOI] [PubMed] [Google Scholar]
- Sotak BN, Hnasko TS, Robinson S, Kremer EJ, Palmiter RD. 2005. Dysregulation of dopamine signaling in the dorsal striatum inhibits feeding. Brain Res 1061: 88–96. 10.1016/j.brainres.2005.08.053 [DOI] [PubMed] [Google Scholar]
- Spencer FA, Hoffmann FM, Gelbart WM. 1982. Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell 28: 451–461. 10.1016/0092-8674(82)90199-4 [DOI] [PubMed] [Google Scholar]
- Steck K, Walker SJ, Itskov PM, Baltazar C, Moreira JM, Ribeiro C. 2018. Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. Elife 7: e31625 10.7554/eLife.31625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternson SM, Nicholas Betley J, Cao ZF. 2013. Neural circuits and motivational processes for hunger. Curr Opin Neurobiol 23: 353–360. 10.1016/j.conb.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoo DE, Akiyama K. 1996. The mechanism by which exercise modifies brain function. Physiol Behav 60: 177–181. 10.1016/0031-9384(96)00011-X [DOI] [PubMed] [Google Scholar]
- Sutoo D, Akiyama K. 2003. Regulation of brain function by exercise. Neurobiol Dis 13: 1–14. 10.1016/S0969-9961(03)00030-5 [DOI] [PubMed] [Google Scholar]
- Swarup S, Morozova TV, Sridhar S, Nokes M, Anholt RR. 2014. Modulation of feeding behavior by odorant-binding proteins in Drosophila melanogaster. Chem Senses 39: 125–132. 10.1093/chemse/bjt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Rainey MA, Kim DS, Alaynick WA, Marck BT, Matsumoto AM, Palmiter RD. 1999. Feeding behavior in dopamine-deficient mice. Proc Natl Acad Sci 96: 12138–12143. 10.1073/pnas.96.21.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Rainey MA, Palmiter RD. 2000. Dopamine is required for hyperphagia in Lepob/ob mice. Nat Genet 25: 102–104. 10.1038/75484 [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Kwok K, Brot MD, Marck BT, Matsumoto AM, Donahue BA, Palmiter RD. 2001. Dopamine production in the caudate putamen restores feeding in dopamine-deficient mice. Neuron 30: 819–828. 10.1016/S0896-6273(01)00319-1 [DOI] [PubMed] [Google Scholar]
- Umemoto T, Furutani Y, Murakami M, Matsui T, Funaba M. 2011. Endogenous Bmp4 in myoblasts is required for myotube formation in C2C12 cells. Biochim Biophys Acta 1810: 1127–1135. 10.1016/j.bbagen.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Umulis D, O'Connor MB, Blair SS. 2009. The extracellular regulation of bone morphogenetic protein signaling. Development 136: 3715–3728. 10.1242/dev.031534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baardewijk LJ, van der Ende J, Lissenberg-Thunnissen S, Romijn LM, Hawinkels LJ, Sier CF, Schipper IB. 2013. Circulating bone morphogenetic protein levels and delayed fracture healing. Int Orthop 37: 523–527. 10.1007/s00264-012-1750-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Peterson AJ, Takenaka N, O'Connor MB, Corces VG. 2015. CTCF-dependent co-localization of canonical Smad signaling factors at architectural protein binding sites in D. melanogaster. Cell Cycle 14: 2677–2687. 10.1080/15384101.2015.1053670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DO, Sieber C, Bhushan R, Borgermann JH, Graf D, Knaus P. 2010. BMPs: from bone to body morphogenetic proteins. Sci Signal 3: mr1. [DOI] [PubMed] [Google Scholar]
- Wangler MF, Yamamoto S, Bellen HJ. 2015. Fruit flies in biomedical research. Genetics 199: 639–653. 10.1534/genetics.114.171785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters RP, Pringle RB, Forster GL, Renner KJ, Malisch JL, Garland T Jr, Swallow JG. 2013. Selection for increased voluntary wheel-running affects behavior and brain monoamines in mice. Brain Res 1508: 9–22. 10.1016/j.brainres.2013.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KW, Elmquist JK. 2012. From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat Neurosci 15: 1350–1355. 10.1038/nn.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Malec PA, Mabrouk OS, Ro J, Dus M, Kennedy RT. 2016. Benzoyl chloride derivatization with liquid chromatography-mass spectrometry for targeted metabolomics of neurochemicals in biological samples. J Chromatogr A 1446: 78–90. 10.1016/j.chroma.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Ho MCW, Liu Q, Horiuchi W, Lin CC, Task D, Luan H, White BH, Potter CJ, Wu MN. 2018. A genetic toolkit for dissecting dopamine circuit function in Drosophila. Cell Rep 23: 652–665. 10.1016/j.celrep.2018.03.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Sara JD, Wang FL, Liu LP, Su LX, Zhe J, Wu X, Liu JH. 2015. Increased plasma BMP-2 levels are associated with atherosclerosis burden and coronary calcification in type 2 diabetic patients. Cardiovasc Diabetol 14: 64 10.1186/s12933-015-0214-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Palmiter RD. 1995. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell 83: 1197–1209. 10.1016/0092-8674(95)90145-0 [DOI] [PubMed] [Google Scholar]
- Zhou R, Mohr S, Hannon GJ, Perrimon N. 2013. Inducing RNAi in Drosophila cells by transfection with dsRNA. Cold Spring Harb Protoc 2013: 461–463. 10.1101/pdb.prot074351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Edmonson MN, Wilkinson MR, Patel A, Wu G, Liu Y, Li Y, Zhang Z, Rusch MC, Parker M, et al. 2016. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet 48: 4–6. 10.1038/ng.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.