This study links HPV-16 and HPV-18 E6 oncoproteins to the modulation of cellular proliferation. The PDZ domain-containing protein NHERF-2 is a tumor suppressor that has been shown to regulate endothelial proliferation; here, we demonstrate that NHERF-2 is targeted by HPV E6 for proteasome-mediated degradation. Interestingly, this indirectly affects p27, cyclin D1, and CDK4 protein levels and, consequently, affects cell proliferation. Hence, this study provides information that will improve our understanding of the molecular basis for HPV E6 function, and it also highlights the importance of the PDZ domain-containing protein NHERF-2 and its tumor-suppressive role in regulating cell proliferation.
KEYWORDS: HPV, E6 oncoprotein, cervical cancer, NHERF-2, p27, cyclin D1, cell proliferation
ABSTRACT
Cancer-causing human papillomavirus (HPV) E6 oncoproteins have a class I PDZ-binding motif (PBM) on their C termini, which play critical roles that are related to the HPV life cycle and HPV-induced malignancies. E6 oncoproteins use these PBMs to interact with, to target for proteasome-mediated degradation, a plethora of cellular substrates that contain PDZ domains and that are involved in the regulation of various cellular pathways. In this study, we show that both HPV-16 and HPV-18 E6 oncoproteins can interact with Na+/H+ exchange regulatory factor 2 (NHERF-2), a PDZ domain-containing protein, which among other cellular functions also behaves as a tumor suppressor regulating endothelial proliferation. The interaction between the E6 oncoproteins and NHERF-2 is PBM dependent and results in proteasome-mediated degradation of NHERF-2. We further confirmed this effect in cells derived from HPV-16- and HPV-18-positive cervical tumors, where we show that NHERF-2 protein turnover is increased in the presence of E6. Finally, our data indicate that E6-mediated NHERF-2 degradation results in p27 downregulation and cyclin D1 upregulation, leading to accelerated cellular proliferation. To our knowledge, this is the first report to demonstrate that E6 oncoproteins can stimulate cell proliferation by indirectly regulating p27 through targeting a PDZ domain-containing protein.
IMPORTANCE This study links HPV-16 and HPV-18 E6 oncoproteins to the modulation of cellular proliferation. The PDZ domain-containing protein NHERF-2 is a tumor suppressor that has been shown to regulate endothelial proliferation; here, we demonstrate that NHERF-2 is targeted by HPV E6 for proteasome-mediated degradation. Interestingly, this indirectly affects p27, cyclin D1, and CDK4 protein levels and, consequently, affects cell proliferation. Hence, this study provides information that will improve our understanding of the molecular basis for HPV E6 function, and it also highlights the importance of the PDZ domain-containing protein NHERF-2 and its tumor-suppressive role in regulating cell proliferation.
INTRODUCTION
Human papillomaviruses (HPVs) are small DNA tumor viruses that have been shown to be the causative agents of cervical cancer, other anogenital cancers, and a number of head and neck cancers (1–3). Of these, cervical cancer is the predominant disease caused by HPVs, with more than 600,000 cancers annually worldwide (4). Approximately 15 mucosotropic HPV types that are associated with human malignancies are referred to as high-risk (HR) types (1). HPV-16 and -18 are the most common HR HPV types and are responsible for approximately 80% of cervical cancers worldwide, while the remaining 20% are caused by the other HR types (5). Numerous studies have shown that the collaborative actions of the two major viral oncoproteins, E6 and E7, are responsible for the development and maintenance of HPV-mediated malignancies (6). These two oncoproteins control various cellular pathways with the aim of maintaining an optimal cellular environment for viral replication. However, in instances where this is perturbed, it can lead to initial changes to the infected cells, which can eventually result in malignant transformation (7). HPV E7 stimulates cell cycle progression by targeting the retinoblastoma tumor suppressor (pRB) and the other two pocket proteins, p107 and p130 (8, 9), while E6 interferes with apoptosis by targeting the tumor suppressor p53 (10). In addition to p53 protein regulation, E6 also regulates p53 gene transactivation by abolishing p53 transcriptional-transactivation activity (5, 6).
Although E6 targeting of p53 is one of the crucial aspects of HPV-induced malignancies, there are also other important functions of E6 that contribute to malignant progression. One of these is the ability of HR HPV E6 oncoproteins to interact with the so-called PDZ domain-containing proteins. The E6 proteins from all of the HR HPV types contain 4 amino acids on their extreme C termini that correspond to a class I (PSD-95/Dlg-1/ZO-1) PDZ-binding motif (PBM). Conversely, this motif is absent from the E6 proteins of low-risk (LR) HPV types, which cause benign warts (11). Multiple studies have shown that the PBM plays critical roles in various E6 functions that are related to the HPV life cycle and malignant transformation. PBM-PDZ interactions lead to increased proliferation of infected cells and are required for optimal amplification and maintenance of viral episomes (12–16). These interactions also play important roles in the process of HPV-induced cellular transformation in tissue culture and in transgenic-mouse models, where they were shown to be required for E6’s ability to induce epithelial tumors in cooperation with E7 (12, 17–20).
HPV E6 oncoproteins interact with a number of PDZ domain-containing proteins that belong to the membrane-associated guanylate kinase (MAGUK) family, and the most extensively studied interacting partners of E6 include the human homologues of the Drosophila disc large protein (hDlg), Scribble (hScrib), and the membrane-associated guanylate kinase with inverted orientation (MAGI) family protein members (11). MAGUK proteins have multiple PDZ domains and, by forming simultaneous interactions with a number of membrane- and cytoplasm-associated cellular proteins, they can serve as scaffolds in forming large complexes. Many of them behave as tumor suppressors and are also involved in the regulation of cell polarity and cell-cell contacts (21, 22). In addition to the MAGUK family proteins, some other PDZ domain-containing proteins involved in cellular signaling and trafficking have also been characterized as E6 substrates (22, 23). One example is a member of the Na+/H+ exchange regulatory factor (NHERF) protein family, NHERF-1, which is involved in a number of important cellular processes, such as signaling and transformation (24). HPV-16 E6 can target NHERF-1 for degradation at the proteasome, leading to activation of the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway, which is an important factor in carcinogenesis (25).
Another member of the NHERF protein family is NHERF-2, which is involved in the regulation of lamellipodium formation and cell migration and which interacts with the N-cadherin/β-catenin (N-Cad/Cat) complex and the platelet-derived growth factor receptor (PDGFR) in epithelial cells (26). NHERF-2 also acts as a scaffold protein for plasma membrane proteins and members of the ezrin/moesin/radixin family, thereby providing a connection between these proteins and the actin cytoskeleton, and controls their surface expression (27). In addition, more recent studies indicate that NHERF-2 is a negative regulator of endothelial proliferation, which is mediated via the cyclin-dependent kinase inhibitor p27 (28).
The fact that NHERF-2 is a PDZ domain-containing protein and is structurally related to NHERF-1, which was previously characterized as an HR HPV-16 E6 oncoprotein substrate, and that it is involved in the regulation of cellular proliferation suggested that NHERF-2 might also be a cellular substrate of the HPV-16 E6 oncoprotein. Here, we report that not only is NHERF-2 a cellular target of the HPV-16 E6 oncoprotein, but also that it binds to other HPV E6 proteins via their PBM motifs. We further report that both HPV-16 and HPV-18 E6 oncoproteins target NHERF-2 for proteasome-mediated degradation. NHERF-2 ablation in the presence of HPV E6 leads to p27 downregulation and, consequently, results in increased cellular proliferation.
RESULTS
E6 oncoproteins from HPV-16, HPV-18, and HPV-33 interact with NHERF-2.
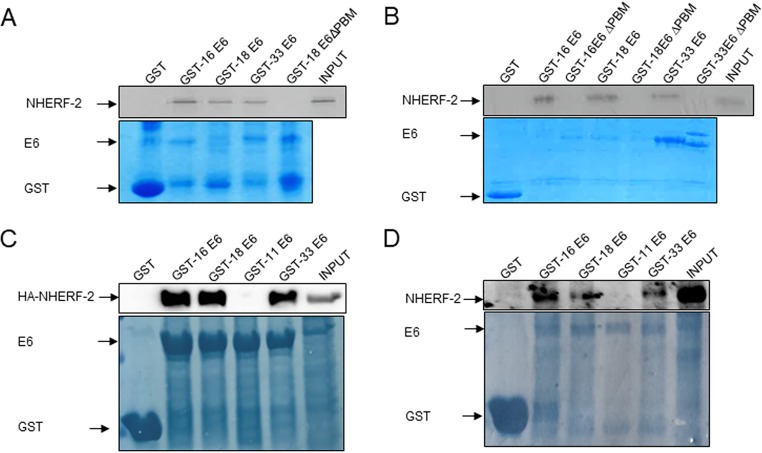
It is well known that the E6 oncoproteins of cancer-causing types of HPV have PBMs through which they can interact with a panel of PDZ domain-containing proteins to elicit a cellular response (11, 21, 22). One of these PDZ domain-containing proteins is NHERF-1, structurally related to NHERF-2, which was previously reported to be bound by HPV-16 E6 and consequently degraded at the proteasome (25). First, therefore, we wanted to investigate whether the PDZ domain-containing NHERF-2 protein could complex with HPV E6 oncoproteins in vitro. A series of glutathione S-transferase (GST) pulldown assays were performed in which in vitro-translated NHERF-2 was incubated with GST-16 E6, GST-18 E6, GST-33 E6, GST-18 E6ΔPBM (E6 protein with the PBM deleted), or GST alone as a control. The results shown in Fig. 1A indicate that HPV-16 E6, HPV-18 E6, and HPV-33 E6 all bind to NHERF-2, and the HPV-16 E6 interaction with NHERF-2 appears to be the strongest, while there is no association between HPV-18 E6ΔPBM and NHERF-2. To confirm that the interaction was PDZ-PBM mediated in each of the HPV types, the assay was repeated, including GST fusion proteins with HPV-16 and HPV-33 E6 proteins with the PBM deleted (HPV-16 E6ΔPBM and HPV-33 E6ΔPBM). The results shown in Fig. 1B indicate that there was no association between NHERF-2 and HPV E6 in the absence of a functional PDZ-binding domain. These results suggest that NHERF-2 can complex with multiple HR E6 proteins and that the interactions are PDZ dependent. We then proceeded to confirm that the interactions between the E6s and NHERF-2 also occur in cultured cells. HEK-293 cells were transfected with hemagglutinin (HA)-tagged NHERF-2; after overnight incubation, the cells were harvested, and proteins were extracted in E1A buffer. The extracts were incubated with GST-16 E6, GST-18 E6, GST-11 E6, GST-33 E6, or GST alone as a control. Bound proteins were detected by SDS-PAGE and Western blotting, and the results are shown in Fig. 1B. In this setting, we observed that NHERF-2 binds with equal strength to HPV-16 E6 and HPV-18 E6, while HPV-33 E6 bound NHERF-2 somewhat weakly. No interaction was detected between the LR HPV-11 E6 and NHERF-2, which was expected, since HPV-11 E6 lacks a PBM. Further, to test whether endogenous NHERF-2 interacts with E6 proteins, we performed GST pulldown assays as previously described, using lysates from C33-A cells. The results shown in Fig. 1C indicate that all the HR E6 oncoproteins tested bind to NHERF-2, with HPV-16 E6 being the strongest interactor, while no interaction with HPV-11 E6 was detected. Together, these results suggest that, although multiple HR E6 oncoproteins bind to NHERF-2, the principal interacting partner is likely to be HPV-16 E6.
FIG 1.
HPV-16 E6, -18 E6, and -33 E6 proteins bind to NHERF-2 in vitro and in vivo. (A) Radiolabeled in vitro-translated NHERF-2 was incubated with GST-16 E6, GST-18 E6, GST-33 E6, and GST-18 E6ΔPBM or GST alone as a control. Bound proteins were assessed by autoradiography, and the input GST fusion proteins were visualized with Coomassie staining (bottom). Input NHERF-2 (20%) is shown. (B) The assay was repeated, including GST-HPV-16 E6ΔPBM and GST-HPV-33 E6ΔPBM. (C) HEK-293 cells were transfected with HA-tagged NHERF-2. After 24 h, the cells were harvested, and the cell lysates were incubated with the indicated GST fusion proteins. GST alone was included as a control. After extensive washing, bound NHERF-2 was detected by Western blotting using anti-HA antibody and compared with the amount of NHERF-2 present in 10% of the input. The lower gel shows the positions of purified GST proteins used in the pulldowns visualized by Coomassie staining. (D) C33-A cell extracts were incubated with the indicated GST fusion proteins. After extensive washing, bound NHERF-2 was detected by Western blotting using anti-NHERF-2 antibody and compared with the amount of NHERF-2 present in 10% of the input. The lower gel shows the positions of purified GST proteins used in the pulldowns visualized by Coomassie staining.
HPV-16 E6, HPV-18 E6, and HPV-33 E6 induce NHERF-2 degradation via the proteasome in a PBM-dependent manner.
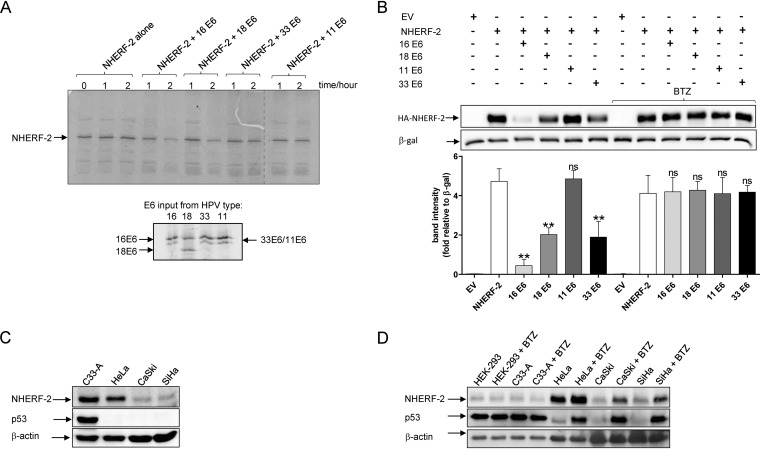
After we had demonstrated that HPV-16 E6, HPV-18 E6, and HPV-33 E6 oncoproteins can interact with NHERF-2, the next obvious question to investigate was the possible consequences of the E6–NHERF-2 interactions. Since substrate degradation is a characteristic of the HPV E6-PDZ interaction (11, 29), we examined whether HPV E6 oncoproteins can likewise direct the degradation of NHERF-2. To do this, HPV-16 E6, -18 E6, -33 E6, and -11 E6 were translated in vitro and coincubated with in vitro-translated NHERF-2 for 1 or 2 h at 30°C. The level of NHERF-2 protein remaining was ascertained by SDS-PAGE and autoradiography. The results shown in Fig. 2A indicate that HPV-16 E6 and HPV-18 E6 were efficient in inducing the degradation of NHERF-2, while HPV-33 E6 induced NHERF-2 degradation less efficiently and HPV-11 E6 did not induce any NHERF-2 degradation. The weaker (HPV-33 E6) or absent (HPV-11 E6) degradative activity is consistent with their lower binding affinity or complete lack of interaction with NHERF-2, respectively.
FIG 2.
A number of HR HPV E6 oncoproteins direct proteasome-mediated degradation of NHERF-2 in vitro and in vivo. (A) NHERF-2 and HPV-16 E6, -18 E6, -33 E6, and -11 E6 were translated and coincubated at 30°C for the times indicated. Residual NHERF-2 was then detected by SDS-PAGE and autoradiography. The E6 inputs are shown below. Note the higher mobility of HPV-18 E6, in agreement with previously published data (36). (B) A plasmid expressing NHERF-2 (HA–NHERF-2) was overexpressed in HEK-293 cells alone or in combination with HPV-16 E6, -18 E6, -11 E6, or -33 E6. Twenty-four hours after transfection, the cells were incubated with or without a proteasome inhibitor (BTZ) for a further 10 h before harvesting. Cell lysates were prepared and analyzed by Western blotting using anti-HA antibody. β-Galactosidase (LacZ) was used as an internal standard to monitor transfection efficiency and as a loading control. Relative densitometry for HA–NHERF-2 under various transfection conditions is shown below. The mean values and SEM of the results of 3 independent experiments are shown. **, P < 0.01; ns, not statistically significant. (C) NHERF-2 protein levels were analyzed by Western blotting in cell lysates from C33-A (HPV-negative), HeLa (HPV-18-positive), and CaSki and SiHa (both HPV-16-positive) cells. (D) The same cell lines were treated with either DMSO or BTZ for 10 h. Cell lysates were then prepared and analyzed by Western blotting using anti-NHERF-2 antibody. (C and D) p53 was used as a control for proteasome inhibition, while β-actin was used as a loading control.
To investigate whether E6 oncoproteins can degrade NHERF-2 in cultured cells, HEK-293 cells were cotransfected with a plasmid expressing HA-tagged NHERF-2 either alone or in combination with HPV-16 E6, HPV-18 E6, HPV-11 E6, or HPV-33 E6 expression plasmids in two sets of experiments. In one set of experiments, the transfected cells were left untreated, while in the other, the transfected cells were treated with the proteasomal inhibitor bortezomib (BTZ). After 24 h, the cells were harvested, and NHERF-2 levels were analyzed by immunoblotting. The Western blotting results, together with quantitative analysis based on band densitometry (Fig. 2B), show that NHERF-2 protein levels were significantly reduced by more than 4-fold in cells expressing HPV-16 E6 and more than 2-fold in those expressing HPV-18 E6 and HPV-33 E6. This result indicates that HPV-16 E6 is the most efficient at inducing NHERF-2 degradation, followed by HPV-18 E6 and HPV-33 E6. In cells expressing the LR HPV-11 E6, there was no significant effect on NHERF-2 levels. Interestingly, when the same transfected cells were treated with BTZ, E6-induced degradation of NHERF-2 was prevented, indicating that it was proteasome mediated.
To further examine the observed E6 effect on NHERF-2, we compared the expression levels of NHERF-2 in HPV-negative C33-A cells, HPV-18-positive HeLa cells, and HPV-16-positive CaSki and SiHa cells by performing Western blot analysis. It is clearly observable in Fig. 2C that endogenous NHERF-2 was abundant in C33-A cells, while a lower level was detected in HeLa cells and it was almost absent in CaSki and SiHa cells. This suggests that the protein turnover rates of endogenous NHERF-2 are increased in the presence of HPV-18 E6 and even further increased in the presence of HPV-16 E6. Furthermore, it implies that the protein turnover of NHERF-2 is more efficiently regulated by HPV-16 E6, which could be attributed to the higher binding capacity of HPV-16 E6 for NHERF-2, as seen in Fig. 1C. To additionally corroborate our initial observations shown in Fig. 2A and B, where we showed that NHERF-2 is targeted for proteasome-mediated degradation by HPV-16 E6 and HPV-18 E6, we cultured the same HPV-positive and HPV-negative cell lines used in Fig. 2C in the presence or absence of the proteasomal inhibitor BTZ. After treatment, cells were harvested, and the levels of NHERF-2 protein were analyzed by Western blotting. In the presence of BTZ, a sharp increase in the levels of NHERF-2 protein was observed in both HPV-16- and HPV-18-positive cell lines compared with the HPV-negative cell lines, where no such increase was observed (Fig. 2D), suggesting that in HPV-positive cell lines derived from cervical tumors, NHERF-2 is subject to proteasome-mediated degradation by E6.
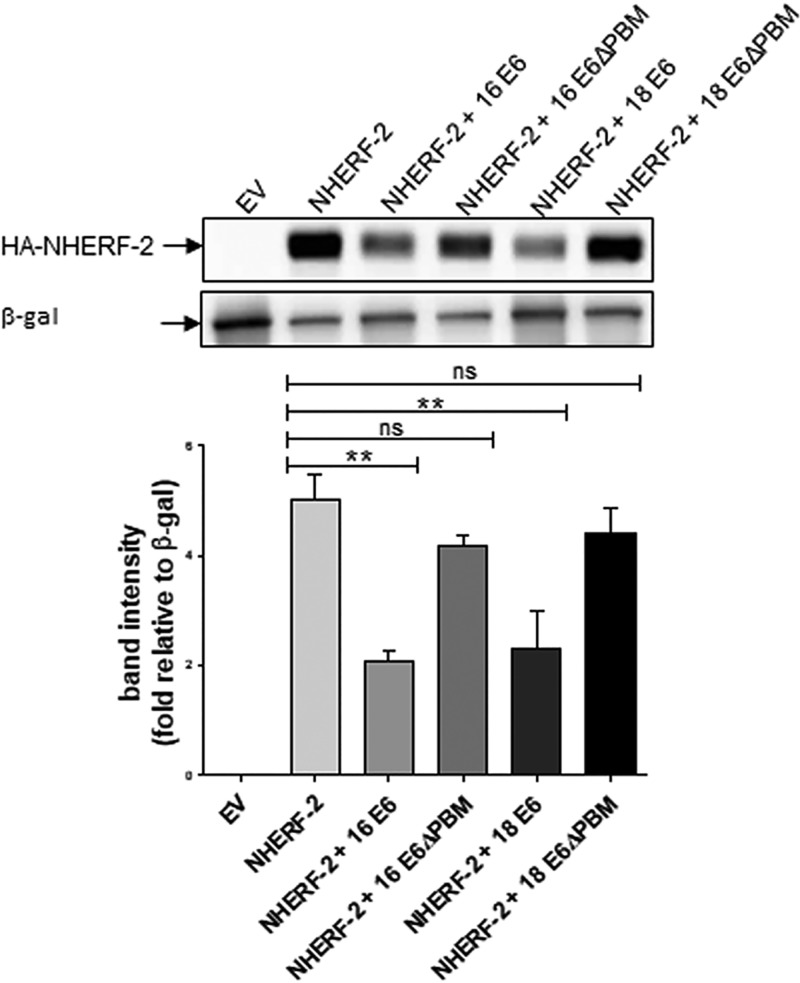
Since cancer-causing HPV E6 proteins have PBMs through which they can interact with a specific panel of proteins (11); including NHERF-2, and then target them for proteasome-mediated degradation, we wondered whether PBM-PDZ interactions are required for NHERF-2 degradation. HEK-293 cells were cotransfected with a plasmid expressing HA-tagged NHERF-2 and plasmids expressing either HPV-16 E6 or HPV-18 E6 or with plasmids expressing the respective mutant E6 proteins, which lack PDZ-binding motifs (16 E6ΔPBM or 18 E6ΔPBM) (30). Again, Western blotting with quantitative analysis based on band densitometry (Fig. 3) showed that NHERF-2 protein levels were significantly downregulated in HEK-293 cells expressing wild-type HPV-16 E6 and HPV-18 E6, but not in those expressing the mutant HPV-16 E6ΔPBM or HPV-18 E6ΔPBM, suggesting that the E6 PBM is required for E6 proteasome-mediated degradation of NHERF-2.
FIG 3.
HPV E6 degradation of NHERF-2 is PBM dependent. HA-tagged NHERF-2 was overexpressed in HEK-293 cells alone or in combination with HPV-16 E6 or -18 E6 or with their respective mutants, HPV-16 E6ΔPBM or -18 E6ΔPBM, as indicated. As a negative control, HEK-293 cells were also transfected with EV. After 24 h of transfection, cells were harvested, and lysates were prepared and analyzed for NHERF-2 protein expression by Western blotting using anti-HA antibody. The expression of β-galactosidase (LacZ) was used as an internal standard to monitor transfection efficiency and loading (lower gel). Relative densitometry for HA–NHERF-2 under various transfection conditions is shown in the graph. The mean values and SEM of the results of 3 independent experiments are shown. **, P < 0.01; ns, not statistically significant.
HPV E6 silencing restores the nuclear pool of NHERF-2.
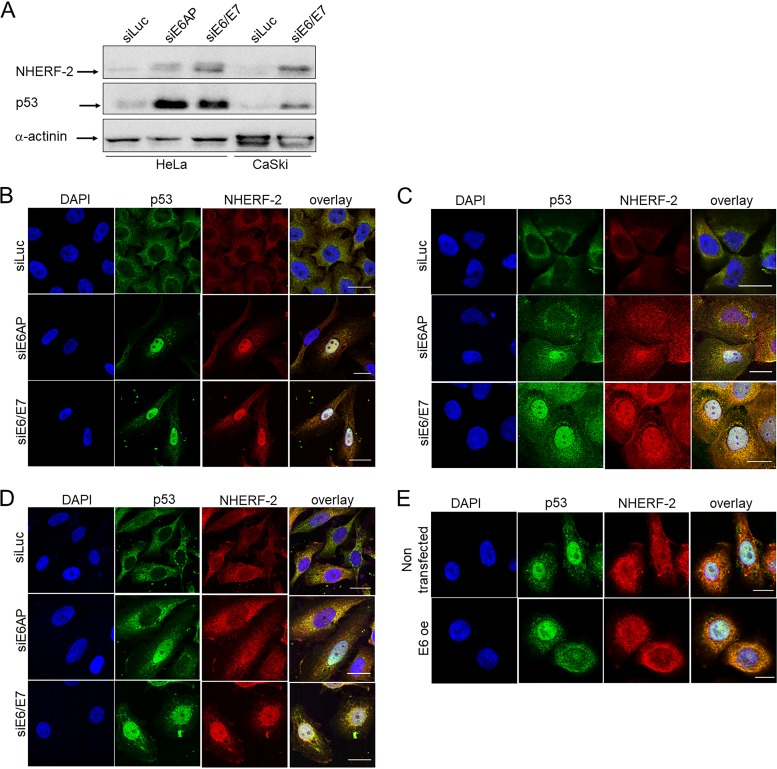
Having found that HPV E6 oncoproteins could degrade NHERF-2, we were next interested in assessing which cellular populations of NHERF-2 were being targeted, as previous studies have indicated both nuclear and cytoplasmic localization of NHERF-2 within the cell (28). In addition, NHERF-1, a protein structurally related to NHERF-2, was previously reported to be detected in the cytoplasm but to be absent from the nucleus in HPV-16 E6/E7-positive primary human foreskin keratinocytes (HFKs) (25). To examine this, we performed small interfering RNA (siRNA) ablation of E6/E7 and also ablated expression of E6AP (E6-associated protein, also known as UBE3A, an E3 ubiquitin protein ligase), as an alternative means of reducing E6 expression levels (31) in HeLa and CaSki cells. After 72 h, the proteins from one set of cells were extracted with E1A buffer, and NHERF-2 protein levels were analyzed by Western blotting; the levels of p53 protein were also analyzed as a control for E6/E7 silencing. The results shown in Fig. 4A indicate NHERF-2 upregulation in both HeLa and CaSki cells upon treatment with siRNA against E6/E7 or E6AP. Simultaneously, cells were fixed and immunolabeled, and the pattern of NHERF-2 localization was monitored by confocal microscopy. Interestingly, E6/E7 downregulation induced the major recovery of NHERF-2 in the nucleus. This pattern was consistent in all HPV-positive cell lines used in the experiment (HeLa, CaSki, and SiHa) (Fig. 4B to D). To further confirm this, we overexpressed HPV-16 E6 in HFKs. After 24 h, the cells were fixed and immunolabeled, and the cellular localization of NHERF-2 was monitored by confocal microscopy. As indicated in Fig. 4E, cells that ectopically expressed HPV-16 E6 exhibited reduced levels of nuclear NHERF-2. Taken together, these results suggest that E6 preferentially targets the nuclear pool of NHERF-2, similarly to NHERF-1 (25).
FIG 4.
HPV-16 E6 and -18 E6 target a nuclear pool of NHERF-2. (A) HeLa and CaSki cells were transfected with the indicated siRNAs. After 72 h, they were harvested and subjected to Western blot analysis using NHERF-2 antibody. p53 served as a control for E6/E7 and E6AP ablation. Overall protein loading was verified using anti-α-actinin antibody. (B to D) HeLa (B), CaSki (C), and SiHa (D) cells were transfected with luciferase siRNA (siLuc), E6/E7 siRNA (siE6/E7), and E6AP siRNA (siE6AP). After 72 h, the cells were fixed and stained for NHERF-2 and for p53, which served as a control for the E6/E7 and E6AP knockdown. (E) HPV-16 E6 was overexpressed (E6 oe) in HFKs, and nontransfected cells were used as a negative control. After 72 h, the cells were fixed and stained for NHERF-2 and for p53, which served as a control for E6 transfection. Scale bars, 20 μm (B, C, and D) and 10 μm (E).
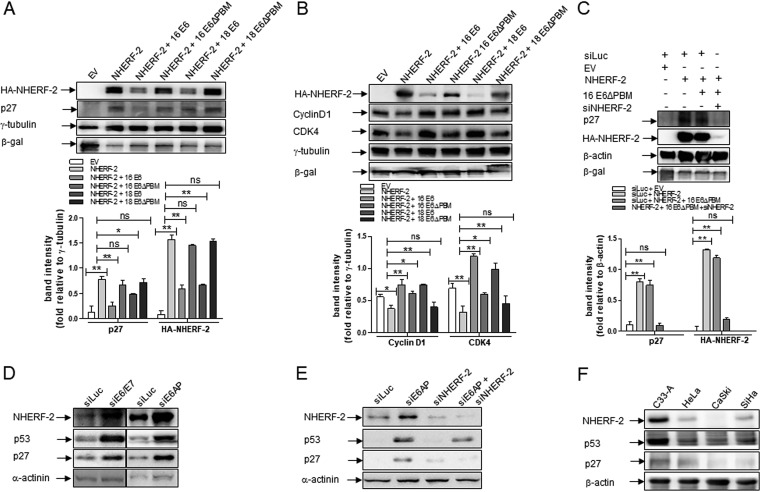
E6 degradation of NHERF-2 regulates the expression of key cell cycle-related proteins.
Cell cycle-related proteins, including cyclins, such as cyclin D; cyclin-dependent kinases, such as CDK2 and CDK4; and cyclin-dependent kinase inhibitors, such as p21 and p27, enable cells to divide (32). For example, p27 is a critical cell cycle regulator, serving as an inhibitor of both CDK2 and CDK4, and its accumulation has been noted to result in cell cycle arrest at the G1/S phase (33, 34). In addition, more recent reports indicate that NHERF-2 has an upregulatory effect on p27 and thus acts as a negative regulator of endothelial cell proliferation (28). Therefore, we asked whether the E6-induced proteasome-mediated degradation of NHERF-2 could have an influence on some of these cell cycle-related proteins, and especially on p27. Hence, we cotransfected HEK-293 cells with plasmids expressing HA–NHERF-2 and those expressing either HPV-16 or HPV-18 E6 wild type or their respective mutated forms, HPV-16 E6ΔPBM or HPV-18 E6ΔPBM. As expected, as shown in Fig. 5A, the protein levels of both p27 and NHERF-2 increased when the cells exogenously expressed NHERF-2 alone but not when NHERF-2 was coexpressed with either HPV-16 E6 or HPV-18 E6 wild types. Coexpression of either of the ΔPBM mutants had no effect on p27 or NHERF-2 levels (relative densitometries are shown in Fig. 5A, bottom).
FIG 5.
HPV regulates p27 protein expression by targeting NHERF-2. (A) HA-tagged NHERF-2 was overexpressed in HEK-293 cells, alone or in combination with HPV-16 E6 or -18 E6 or with their respective mutants, HPV-16 E6ΔPBM or HPV-18 E6ΔPBM, as indicated. As a negative control, HEK-293 cells were transfected with the EV. β-Galactosidase (LacZ) was used as an internal standard to monitor transfection efficiency, and γ-tubulin was used as a loading control. After 24 h of transfection, cells were harvested, and lysates were prepared and analyzed by Western blotting for the expression of p27 and NHERF-2 proteins using anti-p27 and anti-NHERF-2 antibodies. Relative densitometries for p27 and NHERF-2 under various transfection conditions are shown below. (B) Cell lysates from the experiment shown in panel A were used to check the expression levels of HA–NHERF-2, cyclin D1, and CDK4 proteins. Relative densitometries for cyclin D1 and CDK4 under various transfection conditions are shown below. β-Galactosidase (LacZ) was used as an internal standard to monitor transfection efficiency, and γ-tubulin was used as a loading control. (C) HEK-293 cells were cotransfected with the indicated plasmids alone, with the EV, or in combination with the control luciferase siRNA (siLuc) or NHERF-2 siRNA (siNHERF-2). Twenty-four hours after transfection, cells were harvested, and whole-cell lysates were prepared and analyzed by Western blotting using various antibodies as indicated. β-Galactosidase (LacZ) was used as an internal standard to monitor transfection efficiency, while β-actin was used as a loading control. Relative densitometries for p27 and HA–NHERF-2 under various transfection conditions are shown below. One representative of at least 3 independent Western blots is shown. The data are expressed as fold change relative to γ-tubulin (A and B) or to β-actin (C). In each case, the mean values and SEM of the results of 3 independent experiments are shown. *, P < 0.05; **, P < 0.01; ns, not statistically significant. (D and E) HeLa and CaSki cells were transfected with siRNA directed against luciferase (siLuc), E6/E7 (siE6/E7), E6AP (siE6AP), and NHERF-2 (siNHERF-2), alone or in combination. After 72 h, cells were harvested, and the levels of NHERF-2, p53, p27, and the α-actinin loading control were detected by Western blotting. (F) NHERF-2, p53, and p27 protein levels were analyzed by Western blotting in cell lysates from C33-A (HPV-negative), HeLa (HPV-18-positive), and CaSki and SiHa (both HPV-16-positive) cells. β-Actin was used as a loading control, and in each case, one representative of at least three independent Western blots is shown.
In contrast, exogenous expression of NHERF-2 led to decreases in cyclin D1 and CDK4, which was reversed upon coexpression of the wild-type E6, but not the ΔPBM mutants (relative densitometries are shown in Fig. 5B, bottom). These data suggest that the E6-induced degradation of NHERF-2 results in p27 downregulation and upregulation of cyclin D1 and CDK4, which may in turn influence cell proliferation.
Since NHERF-2 overexpression enhances p27 protein levels while E6 degradation of NHERF-2 downregulates it, we wanted to investigate whether the effects of E6 on p27 are exclusively NHERF-2 dependent or if other cellular mechanisms are involved in the process. To do this, HEK-293 cells were cotransfected with plasmids expressing HA-tagged NHERF-2 and HPV-16 E6ΔPBM in the presence or absence of siRNA against NHERF-2 (siNHERF-2). After 72 h, NHERF-2 and p27 levels were analyzed by Western blotting. As shown in Fig. 5C, p27 levels were significantly increased (relative densitometries are shown in Fig. 5C, bottom) in cells ectopically expressing NHERF-2 and remained high in cells coexpressing NHERF-2 and HPV-16 E6ΔPBM, presumably because E6ΔPBM cannot induce NHERF-2 degradation, as shown in Fig. 5A. Interestingly, when NHERF-2 was coexpressed with HPV-16 E6ΔPBM in the presence of siNHERF-2, no significant upregulation of p27 was observed; suggesting that the downregulatory effects of E6 on p27 levels occur exclusively via NHERF-2.
To further confirm the underlying mechanisms, HeLa cells were transfected with siRNA against E6/E7 and E6AP, since loss of E6AP can destabilize E6 (31); siRNA against luciferase (siLuc) was used as a control. After 72 h, proteins from the cellular lysates were analyzed by Western blotting for NHERF-2, p27, and α-actinin. The results shown in Fig. 5D indicate that ablation of E6 by using either siRNAE6/E7 or siE6AP leads to upregulation of NHERF-2 and p27. Secondly, we transfected CaSki cells with siRNA directed against E6AP and NHERF-2, in combination or separately, using siRNA against luciferase as a control. In this setting, we also analyzed the effect on p27 levels of a double knockdown of both E6AP and NHERF-2. After 72 h, the cellular lysates were analyzed by Western blotting for NHERF-2, p27, and α-actinin. The results shown in Fig. 5E confirm that downregulation of E6 (through siE6AP) leads to upregulation of NHERF-2 and p27, and interestingly, in the cells that were doubly knocked down for E6AP and NHERF-2, there was little or no upregulation of p27. We further explored the endogenous p27 protein levels in relation to those of NHERF-2 and p53 in HPV-negative C33-A cells, HPV-18-positive HeLa cells, and HPV-16-positive CaSki and SiHa cells by performing Western blot analysis. As shown in Fig. 5F, endogenous NHERF-2 protein levels were again abundant in C33-A cells compared with HeLa, CaSki, and SiHa cells. Interestingly, a similar trend was also observed for the endogenous protein levels of both p27 and p53, where p27 and p53 protein levels were abundant in C33-A cells compared with HeLa, CaSki, and SiHa cells. Taken together, these results additionally support the notion that the effects of E6 on p27 expression levels are primarily dependent on the E6–NHERF-2 interaction.
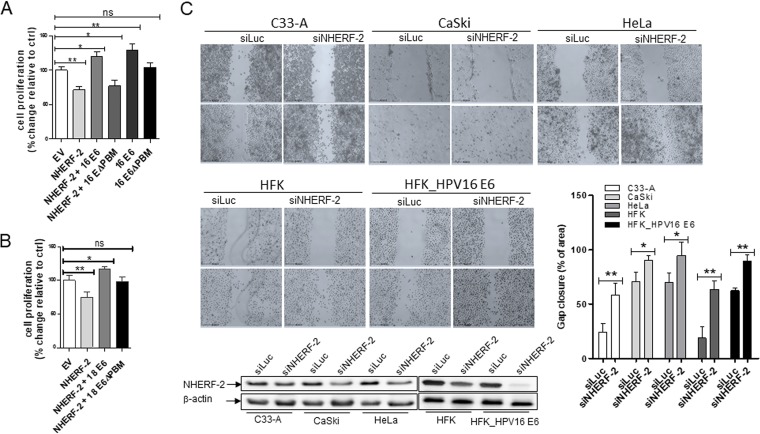
HPV E6 increases cellular proliferative capacity by degrading NHERF-2.
Having shown that NHERF-2 overexpression can decrease cyclin D1 and CDK4 protein levels while increasing p27 protein levels, we questioned whether this effect might affect cell proliferation and, if so, what influence E6-induced degradation of NHERF-2 might also have. NHERF-2 has been reported to negatively regulate endothelial cell proliferation (28), and studies suggest that p27 accumulation can inhibit the cyclin D1-CDK4 complex, leading to cell cycle arrest at G1/S (32), all of which makes these questions compelling. To answer them, we transfected HEK-293 cells with empty vector (EV) or with vectors expressing HA–NHERF-2, HPV-16 E6, HPV-18 E6, HPV-16 E6ΔPBM, and HPV-18 E6ΔPBM, alone or in combination, as indicated (Fig. 6). Cell proliferation was then evaluated with the Uptiblue cell proliferation assay, and the results are shown in Fig. 6. Exogenous expression of NHERF-2 alone significantly decreased proliferation compared with EV-transfected cells. However, proliferation significantly increased in cells coexpressing NHERF-2 with HPV-16 or HPV-18 E6 wild type, but not with the E6 ΔPBM mutants (Fig. 6A and B), indicating that the E6ΔPBM mutants were not able to target NHERF-2 like the wild-type E6s but were still able to stimulate cell proliferation via other mechanisms (35). When cells were transfected with plasmids expressing HPV-16 E6 alone, a significant increase in cell proliferation was observed compared with EV-transfected cells, while transfection with the HPV-16 E6ΔPBM mutant alone showed no significant change in cell proliferation (Fig. 6A). Furthermore, in cells ectopically expressing HPV-16 E6 or HPV-16 E6ΔPBM alone, cellular proliferation was stimulated more strongly than in cells coexpressing NHERF-2 (Fig. 6A), suggesting that the difference in cell proliferation might be due to the antiproliferative effects of NHERF-2. To confirm this, expression of NHERF-2 was modulated in HPV-negative C33-A cells and HFK cells, HFK cells containing the HPV-16 E6 genome (HFK_HPV16 E6), HPV-18-positive HeLa cells, and HPV-16-positive CaSki and SiHa cells. Each cell line was transfected with a control siRNA (siLuc) or NHERF-2 siRNA (siNHERF-2) as indicated. After 48 h, a scratch wound was generated in the confluent cells and immediately photographed. The cells were photographed again 24 h later, and the gap closure, which represents wound healing, was calculated. Figure 6C shows a representative assay, together with Western blot analysis and a histogram of the collated results of at least three assays. Compared with the control siRNA (siLuc) groups, ablation of NHERF-2 using siRNA not only caused a decrease in the endogenous protein levels of NHERF-2, but also significantly increased wound healing in both HPV-negative and HPV-positive cell lines, thereby confirming the antiproliferative effects of NHERF-2 in the different cell lines. Taken together, these data suggest that NHERF-2 downregulation can increase cell proliferation, while its overexpression can decrease cell proliferation through upregulation of p27 and inhibition of cyclin D1 and CDK4. Moreover, HPV E6-mediated NHERF-2 degradation can lead to an increase in cellular proliferation. This is of obvious importance in inducing a cellular state permissive for viral DNA replication but can also contribute to the ability of HR HPV types to cause malignancy.
FIG 6.
HPV E6 increases cellular proliferative capacity by degrading NHERF-2. (A) HEK-293 cells were transfected with the EV or with plasmids expressing HA-tagged NHERF-2 and HPV-16 E6 and -16 E6ΔPBM, alone or in combination as indicated. (B) HEK-293 cells were transfected with the EV or with plasmids expressing HA-tagged NHERF-2 and HPV-18 E6 and -18 E6ΔPBM, alone or in combination as indicated. After 48 h of transfection, cell proliferation was analyzed as described in Materials and Methods. (A and B) For all experiments, the data are expressed as percent change relative to EV-transfected cells, which was normalized to 100%. In each case, the mean values and SEM of the results of 3 independent experiments are shown. *, P < 0.05; **, P < 0.01; ns, not statistically significant. (C) Confluent cells (C33-A, CaSki, and HeLa cells; HFKs; and HFKs containing the HPV-16 E6 genome [HFK_HPV16 E6]) were scratched with a plastic pipette tip 48 h after being transfected with either siLuc or siNHERF-2. The cells were photographed to capture gaps immediately postscratch (0 h) and after 24 h. The bar chart shows areas of gap closure (percent) at 24 h. The same cells were then harvested and lysed, and NHERF-2 protein levels were analyzed by Western blotting. β-Actin was used as a loading control. The data are presented as means and standard deviations (SD) of the results from three independent experiments. *, P < 0.05; **, P < 0.01 versus the control (siLuc).
DISCUSSION
Numerous studies have demonstrated that HR HPV E6 oncoproteins bind and degrade various PDZ domain-containing proteins (11), and so far, the majority of the identified PDZ targets of E6 belong to the MAGUK protein family. Two of these MAGUK family members, hScrib and MAGI-I, are preferentially targeted by HPV-16 E6 (36) and HPV-18 E6 (37), respectively. Interestingly, it has been shown that hDIg, a third member of the MAGUK protein family, can be bound by E6 oncoproteins from a wide range of HR HPV types, indicating the evolutionary conservation and importance of proteins involved in various E6 functions (38). Furthermore, it was reported that HPV-16 E6 can also bind and degrade NHERF-1, a PDZ domain-containing protein and a member of the NHERF protein family. HPV-16 E6 degradation of NHERF-1 results in the activation of the PI3K/AKT signaling pathway, which plays a crucial role in carcinogenesis (25). Most LR HPV E6 oncoproteins do not have a PBM, while all of the HR types contain a class I PBM, implying that this HR hallmark is a key feature in HPV-mediated carcinogenesis. This is further supported by tissue culture and in vivo animal model studies, which showed that the interactions between HPV E6 and PDZ domain substrates play a major role in cellular transformation, in cooperation with E7, and in the induction of epithelial tumors (12, 17–20). So far, however, little is known about the effect of HPV E6 oncoproteins on the PDZ domain-containing protein NHERF-2, even though NHERF-2 is structurally related to NHERF-1, which was previously characterized as an HR HPV-16 E6 oncoprotein substrate (25). We therefore speculated that NHERF-2, which, like NHERF-1, is involved in various cellular processes, such as signaling and proliferation control, is also likely to be a cellular substrate of some of the HPV E6 oncoproteins.
In this study, we report that the E6 oncoproteins of HPV-16, HPV-18, and HPV-33 can interact with NHERF-2. Our data indicate that the E6–NHERF-2 interaction is PDZ-PBM mediated and that the binding with HPV-16 E6 is the strongest, less strong with HPV-18 E6, and rather weak with HPV-33 E6, while the LR HPV-11 E6, on the other hand, does not bind NHERF-2. The interactions of HPV-16, HPV-18, and HPV-33 E6s with NHERF-2 lead to its proteasome-mediated degradation both in vitro and in vivo. Of the E6 oncoproteins examined, HPV-16 E6 is the most efficient inducer of NHERF-2 degradation, while HPV-33 E6 is the least efficient, directly correlating with the intensities of their NHERF-2 binding. Previous studies have shown that NHERF-1 interacts exclusively with HPV-16 E6 (25). Interestingly, despite their structural similarities, this is not the case with NHERF-2, which can interact with multiple E6 proteins. Although the two NHERF family proteins are similar, it is likely that variations within their PDZ domains influence the selection of their interacting partners (39, 40). Namely, it is well known that even a single amino acid change in the PBM of HPV E6 protein can alter the preferred target selection. In addition, it has been shown that other amino acids upstream of the canonical PDZ recognition motif in E6 can influence the PBM-PDZ interactions, and even minor changes in these amino acids can also have an effect on the strength of interaction (36, 40). All of these facts can explain the differences in the strengths of interactions between the different E6 proteins and NHERF-2, as well as the corresponding differences in their degradative capabilities.
In agreement with the overexpression assays, NHERF-2 turnover is also regulated by E6 and the proteasome in HPV-positive cells. Although the endogenous protein levels of NHERF-2 are significantly lower in HPV-16-positive CaSki and SiHa cells than in HPV-18-positive HeLa cells, it appears that NHERF-2 protein turnover is regulated via the proteasome in all the HPV-positive cell lines tested, since NHERF-2 levels were stabilized in the presence of proteasome inhibitors. Remarkably, however, even though the interaction between HPV-18 E6 and NHERF-2 is weaker than that of HPV-16 E6, it appears to be sufficient to induce proteasome-mediated NHERF-2 degradation. This finding is further supported by the restoration of the nuclear pool of NHERF-2 following ablation of E6 in HeLa, CaSki, and SiHa cells. Conversely, this is not the case in the HPV-negative C33-A cell line, where in the presence of proteasome inhibitors, there is no significant increase in the expression levels of endogenous NHERF-2, indicating the importance of NHERF-2 regulation in the HPV life cycle and HPV-mediated malignancies.
Previous studies revealed that E6 can induce cellular proliferation by deregulating the G1/S transition, which is thought to be mainly an E7-controlled function (41). Our results provide new insights into the mechanisms used by HPV-16 E6 and HPV-18 E6 in involving p27 to induce cellular proliferation in epithelial cells. Intriguingly, NHERF-2 can behave as a tumor suppressor, since it negatively regulates endothelial proliferation primarily by upregulating the expression of p27 protein (28). We present new evidence for a direct role of E6 in manipulating NHERF-2 regulation of the cell proliferation mechanism in epithelial cells, where by targeting NHERF-2, E6 downregulates p27 and increases the expression of cyclin D1 and CDK4 proteins, which ultimately results in increased cell proliferation. This is the first report showing an indirect effect of E6 on p27, which, as a consequence, enhances cell proliferation. Previous studies have shown the importance of HPV E7 and p27 interactions for HPV-driven malignancies, indicating that HPV-16 and HPV-18 E7 proteins enhance cytoplasmic retention of p27, which results in increased cellular proliferation; this p27 localization to the cytoplasm was also revealed to be a marker of poor prognosis for several cancer types (42, 43). Hence, it appears that HR HPV-16 and -18 have developed two autonomous mechanisms for targeting the p27 cellular pathway. In one of them, E7 inactivates p27 by preserving it in the cytoplasm, resulting in increased cellular proliferation (42, 43), while in the other, HPV E6 targets the PDZ domain-containing protein NHERF-2 for proteasomal degradation, leading to the downregulation of p27, thereby promoting cellular proliferation. These two mechanisms emphasize the relevance of the p27 pathway, both for the HPV life cycle and for the development of HPV-induced malignancies.
MATERIALS AND METHODS
Cell culture and transfections.
HFKs and HFKs containing the HPV-16 genome (HFK_HPV16 E6) were cultured in keratinocyte serum-free medium (K-SFM) (Gibco) and penicillin/streptomycin. All other cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin/streptomycin (Gibco). The cells were cultured at 37°C in an atmosphere enriched with 10% CO2. HFKs, HFK_HPV16 E6, HEK-293 cells (human embryonic kidney cells), C33-A cells (cervical carcinoma cells, HPV negative), HeLa cells (HPV-18-positive cervical carcinoma cells), CaSki cells (HPV-16-positive cervical carcinoma cells), and SiHa cells (HPV-16-positive cervical carcinoma cells) were transfected using calcium phosphate precipitation (44), Lipofectamine RNAimax (Invitrogen), or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Plasmids.
Wild-type HA-tagged HPV-16 E6, HA-tagged HPV-18 E6, HA-tagged HPV-33 E6, HA-tagged HPV-11 E6, HA-tagged HPV-16 E6ΔPBM, HA-tagged HPV-18 E6ΔPBM, and HA-tagged NHERF-2, which have all been described previously (29, 30, 45, 46), were used. The GST fusion proteins GST-16 E6, GST-18 E6, GST-33 E6, GST-11 E6, and GST-18 E6ΔPBM have also been previously described (31, 47, 48).
Antibodies.
The following antibodies were used: anti-NHERF-2, anti-p53 (DO-1), anti-α-actinin, anti-p21, anti-cyclin D1, and anti-CDK4, which were all purchased from Santa Cruz Biotechnology; anti-HA-peroxidase (clone HA-7) (Sigma); β-galactosidase (LacZ; Promega); and mouse and rabbit secondary antibodies conjugated to horseradish peroxidase (HRP) (Dako) and rhodamine or Alexa Fluor (Invitrogen).
Inhibitors.
The following inhibitors were dissolved in dimethyl sulfoxide (DMSO) and used at the indicated concentrations: the proteasome inhibitor Z-Leu-Leu-Leu-al (CBZ [MG-132]; Sigma; 50 μM) and the proteasome inhibitor BTZ (Sigma; 10 μM). Cocktail set I protease inhibitors (Calbiochem) were dissolved in water.
Fusion protein purification and in vitro binding assays.
GST fusion protein synthesis in Escherichia coli DH5α competent cells and protein purification were performed as previously described (49). Proteins were translated in vitro using a Promega TNT kit and radiolabeled with [35S]cysteine or [35S]methionine (Perkin Elmer). Equal amounts of in vitro-translated proteins were added to the GST fusion proteins bound to glutathione agarose (Sigma) and incubated for 1 h at 4°C. After extensive washing with phosphate-buffered saline (PBS) containing 0.25% NP-40, the bound proteins were analyzed by SDS-PAGE and autoradiography.
GST pulldowns using cellular extracts were performed by incubating GST fusion proteins immobilized on glutathione agarose with cells extracted in E1A buffer (25 mM HEPES [pH 7.0], 0.1% NP-40, 150 mM NaCl, plus protease inhibitor cocktail set I [Calbiochem]) for 1 h at 4°C on a rotating wheel. After extensive washing, the bound proteins were detected using SDS-PAGE and Western blotting.
Immunofluorescence assays.
Cells were stained and fixed for immunofluorescence assay as previously described (48). In brief, HeLa, CaSki, and SiHa cells were each grown overnight on glass coverslips before transfection with siRNA against luciferase, E6AP, or E6/E7 as indicated for 72 h and then fixed with 4% paraformaldehyde for 10 min at room temperature, followed by permeabilization in PBS containing 0.1% Triton X-100. Immunostaining was performed by incubating the coverslips in PBS containing antibodies against p53 (Santa Cruz Biotechnology) or NHERF-2 (Santa Cruz Biotechnology) as indicated overnight in a humidified chamber at 4°C. Secondary anti-rabbit or anti-mouse antibody conjugated with Alexa Fluor or rhodamine was used as appropriate (Invitrogen). Nuclei were labeled with DAPI (4′,6-diamidino-2-phenylindole). The coverslips were slide mounted using Fluoroshield mounting medium with DAPI (GR271388-1; Cambridge, United Kingdom). Confocal fluorescence microscopy was performed using a Leica TCS SP8 X laser scanning microscope equipped with an nHC PL APO CS2 63×/1.40 oil objective, a 405-nm-diode laser, and argon and supercontinuum excitation lasers (Leica Microsystems). Images were acquired by sequential scanning with excitation at 405 nm for DAPI, 488 nm for Alexa 488, and 570 nm for rhodamine red. Detection ranges were 413 to 460 nm for DAPI, 496 to 559 nm for Alexa 488, and 578 to 650 nm for rhodamine red.
In vitro degradation assays.
Proteins were transcribed and translated in vitro in rabbit reticulocyte lysate using the Promega TNT system according to the manufacturer’s instructions. The HPV-16 E6, -18 E6, -33 E6, and -11 E6 proteins were radiolabeled with [35S]cysteine, while NHERF-2 was labeled with [35S]methionine. Degradation assays were performed as previously described (50). Briefly, radiolabeled proteins were mixed and incubated for the indicated times at 30°C. Volumes were adjusted using water-primed lysate. The remaining NHERF-2 was analyzed by SDS-PAGE and autoradiography.
In vivo degradation assays.
Transfected or nontransfected cells (3.5 × 105) seeded on 60-mm dishes were treated either with DMSO alone as a control or with the proteasome inhibitor MG-132 or BTZ, both of which were dissolved in DMSO. Cells were harvested 24 h after treatment, and cellular proteins were extracted for analysis by Western blotting.
Cell proliferation assay.
Cells (3.5 × 105) were seeded into a 60-mm dish in a total volume of 2.5 ml of cell culture medium. The cells were cultured overnight and were then transfected with either the Lipofectamine 2000 (according to the manufacturer’s protocol) or calcium phosphate (44) method, using a total of 1.0 μg of plasmid DNA. The cells were transfected with plasmids expressing NHERF-2, HPV-16 E6, HPV-16 E6ΔPBM, HPV-18 E6, or HPV-18 E6ΔPBM, alone or in combination as indicated in the figures and/or corresponding figure legends. In order to monitor transfection efficiency, cells were cotransfected with β-galactosidase (LacZ) and checked by Western blotting using the appropriate antibody; 16 h after transfection, the medium was aspirated, and the cells were washed with sterile PBS, counted, and seeded at 0.3 × 104 cells per well to a final volume of 100 μl in a 96-well plate and incubated for a further 10 h for the cells to attach. Cell proliferation was monitored using Uptiblue reagent (Interchim) as previously described (51). Uptiblue reagent (5% [vol/vol]) was added to the culture medium, and fluorescence was measured (excitation, 540 nm; emission, 590 nm) on a Tecan fluorescence multiwell plate reader (Tecan Group Ltd., Männedorf, Switzerland) after 48 h. The results are expressed as a percentage of the number of untransfected cells or of the EV with the standard error of the mean (SEM) versus transfected cells or untransfected cells.
Wound healing/scratch assay.
A monolayer scratch/wound healing assay was employed as previously described (38). Briefly, C33-A, CaSki, and HeLa cells; HFKs; and HFKs containing the HPV 16 genome (HFK_HPV16 E6) were transfected with a control siRNA (siLuc) or NHERF-2 siRNA (siNHERF-2) as indicated. After 48 h, a scratch wound was generated in the confluent cells with a sterile Artline p2 pipette tip (Thermo Scientific). The wounds were immediately photographed under a microscope using a Dino-Eye digital eyepiece camera [AM7023(R4); IDCP B.V., Naarden, The Netherlands] that was connected to a computer and DinoCapture 2.0 microscope imaging software. After a further 24 h, the wounds were photographed again, and wound closure was calculated; the images were saved in TIFF format, and gap areas were measured using the MRI wound healing tool macro for ImageJ software (NIH) (http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool ). The cells were then harvested in RIPA lysis buffer, and NHERF-2 protein levels were analyzed by Western blotting. β-Actin was used as a loading control.
Western blotting.
Extraction of cellular proteins was performed as previously described (31). In brief, following incubation of cells with the proteasome inhibitors, cells were collected in cold PBS, pH 7.4, and centrifuged together with the cell culture medium at 4°C and 250 × g for 4 min. After two washing steps with cold PBS, the cells were lysed with 100 μl of RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% sodium desoxycholate, 1% Triton X-100, 0.1% SDS) supplemented with the cocktail set I protease inhibitors (Calbiochem) according to the manufacturer’s instructions. The cell lysate was left on ice for 15 min and subjected to sonification (3 times for 1 min each time) at 4°C, and then cell debris was removed by centrifugation at 16,250 × g at 4°C for 10 min. The protein content of the supernatant was determined according to the Bradford method using the Bio-Rad protein assay reagent (Bio-Rad).
Proteins were separated on either a 10 or 12% sodium dodecyl sulfate-polyacrylamide gel and transferred onto a nitrocellulose membrane by tank blotting. The membrane was blocked with 5% dry milk in PBS containing 0.1% Tween 20 for 1 h at room temperature and then incubated with the specific antibody, which was diluted in PBS with 0.1% Tween 20 containing 1% dry milk powder. The membrane was washed with PBS-Tween 20 containing 1% skim milk (3 times for 10 min each time) before being incubated with a peroxidase-coupled secondary antibody (anti-rabbit or anti-mouse; 1:1,000) for 1 h at room temperature. The membrane was washed again in PBS-Tween 20 (3 times for 10 min each time). Signals were developed, visualized, and quantified using the Uvitec Cambridge-Alliance 4.7 imaging system (Cleaver Scientific, Rugby, Warwickshire, United Kingdom).
Statistical analysis.
GraphPad Prism (GraphPad Inc.) software was used to analyze the data. All the values are averages of the results of at least 3 independent experiments done in triplicate except where specified. The error bars shown in the figures represent SEM, and all results were expressed as arithmetic means with SEM. Differences between the experimental groups were analyzed using one-way analysis of variance (ANOVA) or Student's t test (two-tailed; unpaired). Differences with P values of ≤0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the Croatian Science Foundation (grant no. 2246), by an ICGEB Early Career Return Grant (grant no. CRP/16/018), and by a Ruđer Bošković Institute Support Grant to V.T. L.B. gratefully acknowledges research support from the Associazione Italiana per la Ricerca sul Cancro (grant no. 18578).
We are most grateful to Magdalena Grce for valuable comments on the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. 2012. Biological agents, vol 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum 100:1–441. [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V, WHO International Agency for Research on Cancer Monograph Working Group . 2009. A review of human carcinogens. Part B: biological agents. Lancet Oncol 10:321–322. doi: 10.1016/S1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 3.Zur Hausen H. 2002. Papillomaviruses, and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM, Bray F. 2006. Chapter 2: the burden of HPV-related cancers. Vaccine 24(Suppl 3):11–25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 5.Tomaić V. 2016. Functional roles of E6 and E7 oncoproteins in HPV-induced malignancies at diverse anatomical sites. Cancers 8:95. doi: 10.3390/cancers8100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 7.Mittal S, Banks L. 2017. Molecular mechanisms underlying human papillomavirus E6 and E7 oncoprotein-induced cell transformation. Mutat Res Rev Mutat Res 772:23–35. doi: 10.1016/j.mrrev.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Münger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J 8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helt A-M, Galloway DA. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24:159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- 10.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 11.Thomas M, Narayan N, Pim D, Tomaić V, Massimi P, Nagasaka K, Kranjec C, Gammoh N, Banks L. 2008. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27:7018–7030. doi: 10.1038/onc.2008.351. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen ML, Nguyen MM, Lee D, Griep AE, Lambert PF. 2003. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6’s induction of epithelial hyperplasia in vivo. J Virol 77:6957–6964. doi: 10.1128/jvi.77.12.6957-6964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C, Laimins LA. 2004. Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31. J Virol 78:12366–12377. doi: 10.1128/JVI.78.22.12366-12377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolaides L, Davy C, Raj K, Kranjec C, Banks L, Doorbar J. 2011. Stabilization of HPV16 E6 protein by PDZ proteins, and potential implications for genome maintenance. Virology 414:137–145. doi: 10.1016/j.virol.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Delury CP, Marsh EK, James CD, Boon SS, Banks L, Knight GL, Roberts S. 2013. The role of protein kinase A regulation of the E6 PDZ-binding domain during the differentiation-dependent life cycle of human papillomavirus type 18. J Virol 87:9463–9472. doi: 10.1128/JVI.01234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brimer N, Vande Pol SB. 2014. Papillomavirus E6 PDZ interactions can be replaced by repression of p53 to promote episomal human papillomavirus genome maintenance. J Virol 88:3027–3030. doi: 10.1128/JVI.02360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci U S A 94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley RR, Duensing S, Brake T, Münger K, Lambert PF, Arbeit JM. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res 63:4862–4871. [PubMed] [Google Scholar]
- 19.Watson RA, Thomas M, Banks L, Roberts S. 2003. Activity of the human papillomavirus E6 PDZ-binding motif correlates with an enhanced morphological transformation of immortalized human keratinocytes. J Cell Sci 116:4925–4934. doi: 10.1242/jcs.00809. [DOI] [PubMed] [Google Scholar]
- 20.Shai A, Pitot HC, Lambert PF. 2010. E6-associated protein is required for human papillomavirus type 16 E6 to cause cervical cancer in mice. Cancer Res 70:5064–5073. doi: 10.1158/0008-5472.CAN-09-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganti K, Broniarczyk J, Manoubi W, Massimi P, Mittal S, Pim D, Szalmas A, Thatte J, Thomas M, Tomaić V, Banks L. 2015. The human papillomavirus E6 PDZ binding motif: from life cycle to malignancy. Viruses 7:3530–3551. doi: 10.3390/v7072785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pim D, Bergant M, Boon SS, Ganti K, Kranjec C, Massimi P, Subbaiah VK, Thomas M, Tomaić V, Banks L. 2012. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J 279:3530–3537. doi: 10.1111/j.1742-4658.2012.08709.x. [DOI] [PubMed] [Google Scholar]
- 23.James CD, Roberts S. 2016. Viral interactions with PDZ domain-containing proteins—an oncogenic trait? Pathogens 5:8. doi: 10.3390/pathogens5010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaquero J, Nguyen Ho-Bouldoires TH, Clapéron A, Fouassier L. 2017. Role of the PDZ-scaffold protein NHERF1/EBP50 in cancer biology: from signaling regulation to clinical relevance. Oncogene 36:3067–3079. doi: 10.1038/onc.2016.462. [DOI] [PubMed] [Google Scholar]
- 25.Accardi R, Rubino R, Scalise M, Gheit T, Shahzad N, Thomas M, Banks L, Indiveri C, Sylla BS, Cardone RA, Reshkin SJ, Tommasino M. 2011. E6 and E7 from human papillomavirus type 16 cooperate to target the PDZ protein Na/H exchange regulatory factor 1. J Virol 85:8208–8216. doi: 10.1128/JVI.00114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theisen CS, Wahl JK, Johnson KR, Wheelock MJ. 2007. NHERF links the N-cadherin/catenin complex to the platelet-derived growth factor receptor to modulate the actin cytoskeleton and regulate cell motility. Mol Biol Cell 18:1220–1232. doi: 10.1091/mbc.e06-10-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinman EJ, Hall RA, Friedman PA, Liu-Chen L-Y, Shenolikar S. 2006. The association of NHERF adaptor proteins with g protein-coupled receptors and receptor tyrosine kinases. Annu Rev Physiol 68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya R, Wang E, Dutta SK, Vohra PK, E G, Prakash YS, Mukhopadhyay D. 2012. NHERF-2 maintains endothelial homeostasis. Blood 119:4798–4806. doi: 10.1182/blood-2011-11-392563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomaić V, Gardiol D, Massimi P, Ozbun M, Myers M, Banks L. 2009. Human and primate tumour viruses use PDZ binding as an evolutionarily conserved mechanism of targeting cell polarity regulators. Oncogene 28:1–8. doi: 10.1038/onc.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas M, Dasgupta J, Zhang Y, Chen X, Banks L. 2008. Analysis of specificity determinants in the interactions of different HPV E6 proteins with their PDZ domain-containing substrates. Virology 376:371–378. doi: 10.1016/j.virol.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 31.Tomaić V, Pim D, Banks L. 2009. The stability of the human papillomavirus E6 oncoprotein is E6AP dependent. Virology 393:7–10. doi: 10.1016/j.virol.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 32.Satyanarayana A, Kaldis P. 2009. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 33.Vermeulen K, Van Bockstaele DR, Berneman ZN. 2003. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif 36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolfe M, Chiu MI, Pagano M. 1997. The ubiquitin-mediated proteolytic pathway as a therapeutic area. J Mol Med 75:5–17. doi: 10.1007/s001090050081. [DOI] [PubMed] [Google Scholar]
- 35.DeFilippis RA, Goodwin EC, Wu L, DiMaio D. 2003. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J Virol 77:1551–1563. doi: 10.1128/jvi.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas M, Massimi P, Navarro C, Borg J-P, Banks L. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222–6230. doi: 10.1038/sj.onc.1208757. [DOI] [PubMed] [Google Scholar]
- 37.Kranjec C, Massimi P, Banks L. 2014. Restoration of MAGI-1 expression in human papillomavirus-positive tumor cells induces cell growth arrest and apoptosis. J Virol 88:7155–7169. doi: 10.1128/JVI.03247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas M, Myers MP, Massimi P, Guarnaccia C, Banks L. 2016. Analysis of multiple HPV E6 PDZ interactions defines type-specific PDZ fingerprints that predict oncogenic potential. PLoS Pathog 12:e1005766. doi: 10.1371/journal.ppat.1005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham R, Biswas R, Steplock D, Shenolikar S, Weinman E. 2010. Role of NHERF and scaffolding proteins in proximal tubule transport. Urol Res 38:257–262. doi: 10.1007/s00240-010-0294-1. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Dasgupta J, Ma RZ, Banks L, Thomas M, Chen XS. 2007. Structures of a human papillomavirus (HPV) E6 polypeptide bound to MAGUK proteins: mechanisms of targeting tumor suppressors by a high-risk HPV oncoprotein. J Virol 81:3618–3626. doi: 10.1128/JVI.02044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malanchi I, Caldeira S, Krützfeldt M, Giarre M, Alunni-Fabbroni M, Tommasino M. 2002. Identification of a novel activity of human papillomavirus type 16 E6 protein in deregulating the G1/S transition. Oncogene 21:5665–5672. doi: 10.1038/sj.onc.1205617. [DOI] [PubMed] [Google Scholar]
- 42.Charette ST, McCance DJ. 2007. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 26:7386–7390. doi: 10.1038/sj.onc.1210541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan X, Shah W, Jing L, Chen H, Wang Y. 2010. High-risk human papillomavirus type 18 E7 caused p27 elevation and cytoplasmic localization. Cancer Biol Ther 9:728–735. doi: 10.4161/cbt.9.9.11442. [DOI] [PubMed] [Google Scholar]
- 44.Matlashewski G, Schneider J, Banks L, Jones N, Murray A, Crawford L. 1987. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J 6:1741–1746. doi: 10.1002/j.1460-2075.1987.tb02426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau AG, Hall RA. 2001. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry 40:8572–8580. doi: 10.1021/bi0103516. [DOI] [PubMed] [Google Scholar]
- 46.Tomaic V, Pim D, Thomas M, Massimi P, Myers MP, Banks L. 2011. Regulation of the human papillomavirus type 18 E6/E6AP ubiquitin ligase complex by the HECT domain-containing protein EDD. J Virol 85:3120–3127. doi: 10.1128/JVI.02004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pim D, Tomaic V, Banks L. 2009. The human papillomavirus (HPV) E6* proteins from high-risk, mucosal HPVs can direct degradation of cellular proteins in the absence of full-length E6 protein. J Virol 83:9863–9874. doi: 10.1128/JVI.00539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganti K, Massimi P, Manzo-Merino J, Tomaić V, Pim D, Playford MP, Lizano M, Roberts S, Kranjec C, Doorbar J, Banks L. 2016. Interaction of the human papillomavirus E6 oncoprotein with sorting Nexin 27 modulates endocytic cargo transport pathways. PLoS Pathog 12:e1005854. doi: 10.1371/journal.ppat.1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szalmás A, Tomaić V, Basukala O, Massimi P, Mittal S, Kónya J, Banks L. 2017. The PTPN14 tumor suppressor is a degradation target of human papillomavirus E7. J Virol 91:e00057-17. doi: 10.1128/JVI.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas M, Glaunsinger B, Pim D, Javier R, Banks L. 2001. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20:5431–5439. doi: 10.1038/sj.onc.1204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett Saidu NE, Bretagne M, Mansuet AL, Just P-A, Leroy K, Cerles O, Chouzenoux S, Nicco C, Damotte D, Alifano M, Borghese B, Goldwasser F, Batteux F, Alexandre J. 2018. Dimethyl fumarate is highly cytotoxic in KRAS mutated cancer cells but spares non-tumorigenic cells. Oncotarget 9:9088–9099. doi: 10.18632/oncotarget.24144. [DOI] [PMC free article] [PubMed] [Google Scholar]