Abstract
The small GTPases Rac1 and Cdc42 are key regulators of the morphogenesis of actin-rich dendritic spines in neurons. However, little is known about how activated Rac1/Cdc42 regulates dendritic spines. Insulin receptor substrate 53 (IRSp53), which is highly expressed in the postsynaptic density (PSD), is known to link activated Rac1/Cdc42 to downstream effectors for actin regulation in non-neural cells. Here, we report that IRSp53 interacts with two specific members of the PSD-95 family, PSD-95 and chapsyn-110/PSD-93, in brain. An IRSp53 mutant lacking the C-terminal PSD-95-binding motif shows significant loss of synaptic localization in cultured neurons. Overexpression of IRSp53 in cultured neurons increases the density of dendritic spines but does not affect their length or width. Conversely, short-interfering RNA-mediated knock-down of IRSp53 reduces the density, length, and width of spines. In addition, the density and size of spines are decreased by a dominant-negative IRSp53 with a point mutation in the Src homology 3 (SH3) domain and a dominant-negative proline-rich region of WAVE2 (Wiskott-Aldrich syndrome protein family Verprolin-homologous protein), a downstream effector of IRSp53 that binds to the SH3 domain of IRSp53. These results suggest that PSD-95 interaction is an important determinant of synaptic IRSp53 localization and that the SH3 domain of IRSp53 links activated Rac1/Cdc42 to downstream effectors for the regulation of spine morphogenesis.
Keywords: PSD-95, IRSp53, PDZ, Rac1, Cdc42, spine
Introduction
Dendritic spines are postsynaptic actin-rich protrusions implicated in the regulation of the structure, function, and plasticity of glutamatergic synapses (Harris and Kater, 1994; Matus, 2000; Hering and Sheng, 2001; Yuste and Bonhoeffer, 2001, 2004; Nimchinsky et al., 2002). Synaptic molecules that have received attention for their roles in regulating spine morphogenesis include the Rac1 and Cdc42 Rho family of small GTPases (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Irie and Yamaguchi, 2002; Scott et al., 2003) and their upstream activators, including the Kalirin-7 and p21-activated kinase (PAK) interacting exchange factor (βPIX) guanine nucleotide exchange factors (Penzes et al., 2000, 2001, 2003; Parnas et al., 2001; Park et al., 2003; Zhang et al., 2003). However, the molecular mechanisms linking activated Rac1/Cdc42 to downstream effectors for spine regulation are not well understood.
Insulin receptor substrate 53 (IRSp53) is an effector protein that couples activated Rac1/Cdc42 to actin regulation (Miki et al., 2000; Govind et al., 2001; Krugmann et al., 2001). IRSp53 contains various domains for protein-protein interaction, including an N-terminal Rac binding (RCB) domain, a partial Cdc42 and Rac1-interactive binding (CRIB) motif, an Src homology 3 (SH3) domain, and a C-terminal PDZ [postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1 (ZO-1)]-binding motif. The RCB domain of IRSp53 binds activated Rac1, stimulating formation of lamellipodia and ruffles by mechanisms requiring the interaction of its SH3 domain with WAVE2 (Wiskott-Aldrich syndrom protein family Verprolin-homologous protein), a member of the Wiskott-Aldrich syndrome protein (WASP) family of actin-regulatory proteins (Miki et al., 2000; Miki and Takenawa, 2002). The partial CRIB motif of IRSp53 binds activated Cdc42 and induces the formation of filopodia through the interaction of its SH3 domain with Mena, a member of the Ena/vasodilator-stimulated phosphoprotein family of actin-regulatory proteins (Govind et al., 2001; Krugmann et al., 2001). The SH3 domain of IRSp53 also associates with various polyproline-containing proteins, including the abundant PSD protein Shank/Proline-rich synapse-associated protein (SAP) (Bockmann et al., 2002; Soltau et al., 2002), the actin-bundling protein espin (Sekerkova et al., 2003), and mDia1, a regulator of stress fiber formation and cytokinesis (Fujiwara et al., 2000). The C-terminal PDZ-binding motif binds to the PDZ domain protein MALS (mammalian LIN-seven) (Hori et al., 2003). Recently, the N-terminal domain of IRSp53 (IMD) (overlaps with the RCB domain) was shown to induce filopodia independent of the SH3 domain, which is under the tight control of Rac1/Cdc42 (Yamagishi et al., 2004). These results suggest that IRSp53 is an important Rac1/Cdc42-dependent regulator of the actin cytoskeleton.
In neurons, IRSp53 was initially identified as a component of the PSD (Abbott et al., 1999), but little is known about its neuronal functions. Important steps toward understanding the neuronal functions of IRSp53 would be to determine the molecular mechanism by which IRSp53 is localized to the PSD and whether and how IRSp53 regulates the morphogenesis of dendritic spines. Here, we report that the interaction of IRSp53 with PSD-95, an abundant PSD protein, is important for synaptic IRSp53 localization and that IRSp53, through its SH3 domain, regulates spine morphogenesis.
Materials and Methods
Yeast two-hybrid screen and assay. The PDZ2 domain of PSD-95 (aa 89-299) in the pBHA bait vector was used to screen a human brain yeast two-hybrid cDNA contained in the pACT2 prey vector (Clontech, Palo Alto, CA). IRSp53 (aa 438-521) and a mutant of aa 438-521 in which the last Val residue was changed to Ala were subcloned into the EcoRI-BamHI site of pBHA. PDZ1∼2 (aa 124∼363) and PDZ3 (aa 384∼523) of mouse SAP102 and PDZ1∼2 (aa 1-272) of human ZO-1 were subcloned into the BamHI-EcoRI site of pGAD10. Other PDZ domains in pGAD10 have been described previously (Choi et al., 2002; Mok et al., 2002).
GST pull-down assay. For glutathione S-transferase (GST) fusion proteins, aa 470-521 and 470-517 of IRSp53 and aa 786-899 of Shank1 were subcloned into the BamHI-EcoRI site of pGEX4T-1 (Amersham Biosciences, Arlington, IL). The precipitates were analyzed by immunoblotting with antibodies against Myc (1 μg/ml; for PSD-95 and chapsyn-110), enhanced green fluorescent protein (EGFP) [1 μg/ml; SAP97 and S-SCAM (synaptic scaffolding molecule)], FLAG (1 μg/ml; SAP102), and IRSp53 (1 μg/ml) antibodies.
Antibodies. H6-IRSp53 (human full length) was used as an immunogen to generate rabbit (1452) and guinea pig (1454) polyclonal antibodies. Guinea pig polyclonal PSD-95 (1402) and rabbit polyclonal SAP102 (1445) antibodies were generated using GST-PSD-95 (rat full length) and GST-SAP102 (mouse full length) as immunogens, respectively. Specific antibodies were affinity purified using polyvinylidene difluoride membranes (Amersham Biosciences) or SulfoLink columns (Pierce, Rockford, IL), and the specificity of each antibody for the corresponding PSD-95 family protein was confirmed by immunoblot analysis (data not shown). The following antibodies have been described previously: PSD-95 (HM319) (Kim et al., 1995), PSD-95 (SM55) (Choi et al., 2002), SAP97 (B9591) (Kim et al., 1996), chapsyn-110 (B9594) (Kim et al., 1996), S-SCAM (1146) (Mok et al., 2002), EGFP (1167) (Choi et al., 2002), NMDA receptor subunit 2B (NR2B) (Sheng et al., 1994), Shank (3856) (Naisbitt et al., 1999), and Shank (1123) (Lim et al., 2001). The following antibodies were purchased: synaptophysin, FLAG, β-tubulin, and vinculin hVIN-1 (Sigma, St. Louis, MO); HA mouse monoclonal (Boehringer Mannheim, Philadelphia, PA); and HA and Myc rabbit polyclonal (Santa Cruz Biotechnology, Santa Cruz, CA).
Expression and short-interfering RNA constructs. Human IRSp53 cDNAs (NM_017450; full length aa 1-521 and aa 1-517, which lacks the last four residues) were subcloned into the BamHI-EcoRI site of pcDNA3.1-HA (the HA epitope was inserted into the HindIII-KpnI site of pcDNA3.1, which is located N terminal to the site of IRSp53 insertion) for coimmunoprecipitation in heterologous cells and targeting experiments in neurons) and the BglII-EcoRI site of pIRES2-EGFP (for spine-promoting experiments in neurons; Clontech). Full-length rat IRSp53 (AY037934) was subcloned into the BamHI-EcoRI site of pcDNA3.1-HA [for the analysis of short-interfering RNA (siRNA) constructs]. Mouse SAP102 (full length; aa 1-835) was subcloned into the HindIII-EcoRI site of the pFLAG-CMV2 vector (Sigma). Full-length SAP97 (rat; aa 1-912) and S-SCAM (rat; aa 1-1278) were subcloned into the EcoRI-KpnI site of pEGFP-N1 (Clontech). For siRNA downregulation, pSUPER IRSp53 was generated by annealing oligonucleotides containing nt 522-540 (with the first nucleotide of the start codon as nucleotide 1) of the rat IRSp53 cDNA (GenBank accession number AY037934; critical 19 nucleotide sequence GCTGGAGAACTACGTGTCT) and subcloning into the BglII-HindIII site of pSUPER (OligoEngine Platform, Seattle, WA). For FLAG-WAVE2 Pro, aa 273-402 of WAVE2 was subcloned into the HindIII-KpnI site of p3XFLAG-CMV7.1 (Sigma). IRSp53 constructs with a point mutation in the SH3 domain (I403P) were generated using the QuickChange kit (Stratagene, La Jolla, CA). The following expression constructs have been described previously: GW1-PSD-95-EGFP (Arnold and Clapham, 1999), GW1-Myc-PSD-95 (Hsueh et al., 1997), and GW1-Myc-chapsyn-110 (Hsueh et al., 1997).
Preparation and immunoblot analysis of PSD and subcellular fractions. Subcellular fractionation of rat brain was performed as described previously (Huttner et al., 1983). PSD fractions were purified as described previously (Cho et al., 1992). Immunoblot analysis of these fractions was performed using IRSp53 (1452; 1 μg/ml), PSD-95 (SM55; 1:500), and synaptophysin (1:1000) antibodies.
Coimmunoprecipitation assay. For coimmunoprecipitation with brain samples, the crude synaptosomal fraction of adult rat brain was solubilized with deoxycholic acid (DOC) buffer (50 mm Tris-HCl, 1% sodium deoxycholate, pH 9.0), dialyzed against binding/dialysis buffer (50 mm Tris-HCl, 0.1% Triton X-100, pH 7.4), and centrifuged. The supernatant was incubated with PSD-95 (1402; 1 μg/ml), IRSp53 (1452; 2 μg/ml), or rabbit/guinea pig IgG (2 μg/ml; negative control) for 2 h and then with protein A-Sepharose (Amersham Biosciences) for 2 h. The precipitates were analyzed by immunoblotting with antibodies against IRSp53 (1452 and 1454; 1 μg/ml), NR2B (1:1000), vinculin (1:1000), PSD-95 (HM319; 1:1000), chapsyn-110 (B9594; 1:1000), SAP97 (B9591; 1:1000), SAP102 (1445; 1:1000), S-SCAM (1146; 1:1000), and Shank (3856; 1:1000).
Immunofluorescence histochemistry on rat brain sections. Rat brain sections (50 μm) were permeabilized by incubation in PBS containing 50% ethanol at room temperature for 30 min. For immunofluorescence staining, brain sections were doubly incubated with IRSp53 (1:500) and synaptophysin (1:1000) antibodies or NeuroTrace fluorescent Nissl stain (1:1000; Molecular Probes) overnight at room temperature, followed by FITC- and CY3-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) for 2 h at room temperature. Images were captured using a Leica (Nussloch, Germany) SP2 confocal laser-scanning microscope.
Immunoelectron microscopy. Rat brain coronal sections (50 μm thick) were cut with a Vibratome and then washed in phosphate buffer (PB); free-floating sections were treated with NaBH4 and locked with normal goat serum (NGS; 10%) in PBS, pH 7.4, followed by incubations in the primary and secondary antibodies. Rabbit anti-IRSp53 antibodies (number 1452) were used at 2 μg/ml diluted in PBS containing 2% NGS. After several washes, sections were treated with a blocking solution containing 5% BSA (Sigma), 0.1% BSA-c (Aurion, Wageningene, The Netherlands), and 5% NGS (Jackson ImmunoResearch), followed by gold-conjugated goat anti-rabbit IgG (Fab coupled to 1 nm gold particles, Electron Microscopy Sciences, Fort Washington, PA; diluted 1:50 in PBS containing 2% NGS). After postfixation with 2% glutaraldehyde, sections were rinsed in 0.1 m sodium acetate, and gold particles were silver-enhanced using IntenseS-EM Silver (Amersham Biosciences). After several washes in sodium acetate and PB, sections were postfixed with 0.5% OsO4, contrasted in 1% uranyl acetate in maleate buffer, dehydrated, and embedded with epoxy resin (Epon/Spurr; Electron Microscopy Sciences). No specific immunoreactivity could be detected when either the primary or the secondary antibody was omitted before silver enhancement. For electron microscopy (EM), ∼70 nm thin sections were cut on a Leica ultra-microtome, mounted on 200 mesh copper grids, contrasted with uranyl acetate and Sato's lead, and then examined in a Philips Tecnai electron microscope at 80 kV. Digital micrographs were acquired with a Gatan (Pleasanton, CA) 12-bit 1024 × 1024 CCD camera. Contrast, density, and sharpness of final images for plates were adjusted using Adobe Photoshop CS (Adobe Systems, San Jose, CA).
Primary neuron culture, transfection, and immunocytochemistry. Cultured hippocampal neurons were prepared from embryonic day 18 (E18) rat brain as described previously (Goslin and Banker, 1991). Cultures were plated on coverslips (Fisher Scientific, Houston, TX) coated with poly-d-lysine (30 μg/ml). Cultures were grown in Neurobasal medium (Invitrogen, San Diego, CA) supplemented with B27 (Invitrogen), 0.5 mm glutamine, and 12.5 μm glutamate. Low-density cultures were used for colocalization studies. At 21 d in vitro (DIV), hippocampal neurons were fixed in PBS containing 4% paraformaldehyde, permeabilized in PBS containing 0.2% Triton X-100, and incubated with primary antibodies against IRSp53 (1452; 2 μg/ml) and PSD-95 (HM319; 3 μg/ml), followed by Cy3- or FITC-conjugated secondary antibodies. To determine the synaptic localization of IRSp53 proteins, neurons at a medium-high density were transfected with HA-IRSp53 [wild type (WT) or mutant] plus PSD-95-EGFP at 17 DIV using the calcium phosphate method and stained at 19 DIV by double immunofluorescence staining with HA monoclonal (for IRSp53; 1 μg/ml) and EGFP (for PSD-95; 1:1000) antibodies. To determine the effects of IRSp53/WAVE2 constructs on dendritic spines, neurons at 13-15 DIV were transfected with the indicated cDNA constructs and visualized at 16-17 DIV by double immunofluorescence staining with IRSp53 (1452; 2 μg/ml), EGFP (1:1000), and Shank (1123; 1:200) antibodies.
Image acquisition and quantification. Fluorescent images were acquired using a confocal microscope (LSM510; Zeiss, Oberkochen, Germany). The same parameter settings were used for all scans. Each experiment was repeated three to five times, and neuronal images for analysis were randomly selected. Morphometric measurements were performed using MetaMorph image analysis software (Universal Imaging, West Chester, CA) and in part by a blind manner. To determine the synaptic localization of IRSp53, the immunofluorescence intensity of a PSD-95-positive spine was compared with that of an adjacent dendritic trunk. The intensity ratios of spine to dendrite were obtained from a total of ∼400 measurements (15-20 neurons and 20 spines/trunks per neuron). For the analysis of spine density, spines were defined as dendritic protrusions of 0.5-3 μm length, with or without a head. Spine density was measured by counting the number of spines on 10-12 neurons (∼100 μm total dendritic length per neuron). For spine dimensions, ∼300-500 spines (10-12 neurons) were measured for each condition. The length of each spine was measured by manually drawing a vertical line from the base of the neck to the furthest point on the spine head. Spine width was measured by drawing a maximal line perpendicular to the length line in the spine head. The density and dimensions of spines from a single neuron were grouped and averaged; means from multiple individual neurons were averaged to obtain a population mean and SEM. Statistical significance was determined by Student's t test.
Results
IRSp53 interacts with PSD-95 family proteins in vitro
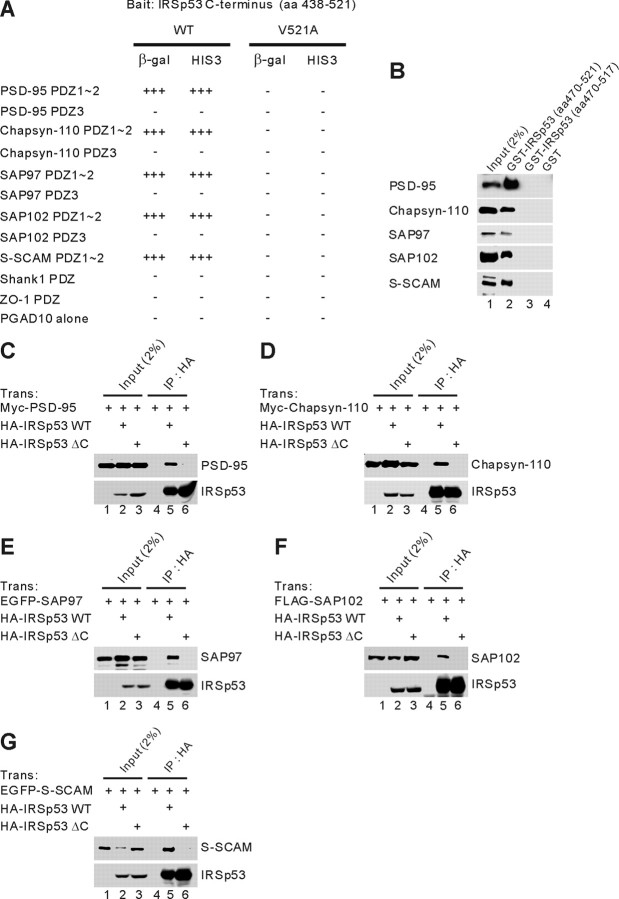
PSD-95 is a key component of the PSD. To identify proteins that contribute to the PSD-95-dependent organization of postsynaptic protein complexes, we searched for binding partners of PSD-95 using the second PDZ domain (aa 89-299) of PSD-95 as bait and a human brain yeast two-hybrid cDNA library. One set of positive clones contained overlapping fragments of IRSp53 that end with Ser-Thr-Val, a potential class I PDZ domain-binding motif (Sheng and Sala, 2001). In the yeast two-hybrid assay, IRSp53 interacted with the first two PDZ domains of all known isoforms of the PSD-95 family (PSD-95/SAP90, SAP97, chapsyn-110/PSD-93, and SAP102) and the S-SCAM multi-PDZ protein but not with the PDZ domains of Shank1 and ZO-1 (Fig. 1A). A point mutation at the C-terminal residue of IRSp53 (Val521 to Ala) eliminated these interactions (Fig. 1A), suggesting that IRSp53 interacts with PSD-95 through the canonical PDZ interaction. In a GST pull-down assay, GST-IRSp53 (aa 470-521), but not GST-IRSp53 lacking the last four residues (aa 470-517) or GST alone, brought down all known PSD-95 family proteins and S-SCAM expressed in heterologous cells (Fig. 1B). In a coimmunoprecipitation analysis in heterologous cells, full-length IRSp53 (IRSp53WT), but not the IRSp53 mutant lacking the last four residues (IRSp53ΔC), formed a complex with all known PSD-95 family proteins and S-SCAM (Fig. 1C-G).
Figure 1.
IRSp53 interacts with all known PSD-95 family proteins and S-SCAM in yeast two-hybrid, GST pull-down, and coimmunoprecipitation assays. A, Interaction of IRSp53 with PSD-95 family proteins and S-SCAM in the yeast two-hybrid assay. PDZ domains from PSD-95 family proteins, S-SCAM, and other PDZ proteins in pGAD10 (prey vector) were tested for binding to IRSp53 (aa 438-521; wild type and the V521A mutant, in which the last Val residue was changed to Ala) in pBHA (bait vector) in the yeast two-hybrid assay. β-Galactosidase (β-gal) activity: +++, <45 min; ++, 45-90 min; +, 90-240 min; -, no significant β-gal activity. HIS3 activity: +++, >60%; ++, 30-60%; +, 10-30%; -, no significant growth. B, Pull down of PSD-95 family proteins and S-SCAM with GST-IRSp53 fusion proteins. GST fusion proteins of IRSp53 (aa 470-521 and aa 470-517, which lacks the last 4 residues) and GST alone were used to bring down the indicated proteins expressed in human embryonic kidney 293T (HEK293T) cells. The precipitates were analyzed by immunoblotting with the indicated antibodies. C, Lysates of HEK293T cells doubly transfected with Myc-PSD-95 plus HA-IRSp53WT or a mutant lacking the last four residues (IRSp53ΔC), or singly with Myc-PSD-95 were immunoprecipitated with HA antibodies and immunoblotted with HA (for IRSp53) and Myc (for PSD-95) antibodies. Trans, Transfection; IP, immunoprecipitation. HA, Myc, FLAG, and EGFP represent epitope tags. D-G, Chapsyn-110 (D), SAP97 (E), SAP102 (F), and S-SCAM (G) were similarly analyzed for coimmunoprecipitation with IRSp53.
Singly transfected PSD-95 family proteins and S-SCAM were not immunoprecipitated in control experiments (Fig. 1C-G, lane 4). The overall coimmunoprecipitation efficiencies, however, were not high, ∼1-2% of the inputs (Fig. 1C-G). Together, these results suggest that IRSp53 associates with all known members of the PSD-95 family and S-SCAM in vitro through the canonical PDZ interaction.
Similar spatiotemporal expression patterns and complex formation of IRSp53 and PSD-95 in brain
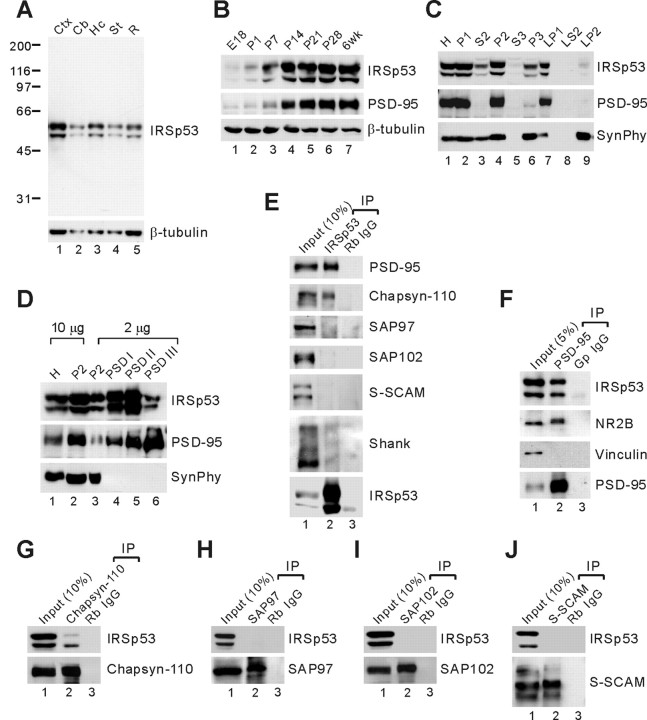
To determine the expression pattern of IRSp53 in brain, we generated polyclonal antibodies against full-length IRSp53 (aa 1-521; rabbit 1452 and guinea pig 1454). These antibodies recognized two previously reported major IRSp53 bands (58 and 53 kDa) in brain samples (Abbott et al., 1999); these may represent splice variants (Abbott et al., 1999; Okamura-Oho et al., 2001; Alvarez et al., 2002). IRSp53 showed widespread expression in various regions of the rat brain, including cortex, cerebellum, hippocampus, and striatum (Fig. 2A), similar to PSD-95 (Cho et al., 1992). During development, the expression levels of IRSp53 and PSD-95 sharply increased together during the second and third postnatal weeks (Fig. 2B). In brain subcellular fractions, IRSp53 and PSD-95 cofractionated into the crude synaptosomal (P2) and synaptic membrane (LP1) fractions (Fig. 2C). In PSD fractions, both IRSp53 and PSD-95 were enriched in all three PSD fractions; IRSp53 was less enriched in the PSD III fraction (Fig. 2D), suggesting that IRSp53 is less tightly associated with the PSD than is PSD-95. Together, these results suggest that IRSp53 and PSD-95 have similar spatiotemporal expression patterns in brain.
Figure 2.
Similar spatiotemporal expression patterns and complex formation of IRSp53 and PSD-95 in the brain. A, Expression of IRSp53 in different brain regions. Membranes from various regions of adult rat brain were immunoblotted for IRSp53 and β-tubulin (control). The widespread expression of IRSp53 is similar to that of PSD-95 (Cho et al., 1992). Cb, Cerebellum; Ctx, cortex; Hc, hippocampus; St, striatum; R, the rest of the brain. B, Similar developmental expression patterns of IRSp53 and PSD-95. Membranes of rat brain at the indicated developmental stages were immunoblotted for IRSp53, PSD-95, and β-tubulin (control). E, Embryonic day; P, postnatal day; wk, week. C, Cofractionation of IRSp53 and PSD-95 in subcellular fractions of adult rat brain. Synaptophysin (SynPhy) was probed as a control. H, Homogenates; P2, crude synaptosomes; S2, supernatant after P2 precipitation; S3, cytosol; P3, light membranes; LP1, synaptosomal membranes; LS2, synaptosomal cytosol; LP2, synaptic vesicle-enriched fraction. D, Coenrichment of IRSp53 and PSD-95 in PSD fractions. PSD fractions of adult rat brain extracted with Triton X-100 once (PSD I), twice (PSD II), or with Triton X-100 and the strong detergent Sarcosyl (PSD III) were immunoblotted with IRSp53, PSD-95, and synaptophysin antibodies. E, DOC extracts of the crude synaptosomal fraction of adult rat brain were immunoprecipitated with IRSp53 antibodies or rabbit IgG (negative control) and immunoblotted with the indicated antibodies. Rb, Rabbit. F-J, The same extracts were immunoprecipitated with PSD-95, chapsyn-110, SAP97, SAP102, and S-SCAM antibodies or control IgGs and immunoblotted with the indicated antibodies. NR2B, NMDA glutamate receptor subunit 2 (positive control); Gp, guinea pig.
To determine the formation of a complex between IRSp53 and PSD-95 in brain, we performed coimmunoprecipitation experiments (Fig. 2E-J). When crude synaptosomal extracts of adult rat brain were immunoprecipitated with IRSp53 antibodies, PSD-95 and chapsyn-110, but not SAP97, SAP102, or S-SCAM, were pulled down (Fig. 2E). IRSp53 antibodies pulled down only a small fraction of Shank (Fig. 2E), a known binding partner of IRSp53 (Bockmann et al., 2002; Soltau et al., 2002). Control rabbit IgG did not bring down any protein tested (Fig. 2E). In the reciprocal coimmunoprecipitation experiments, PSD-95 antibodies brought down IRSp53, along with NR2B (a positive control) (Fig. 2F). Vinculin, a negative control, was not pulled down by PSD-95 antibodies or guinea pig IgG (Fig. 2F). Similarly, chapsyn-110, but not SAP97, SAP102, or S-SCAM antibodies, brought down IRSp53 (Fig. 2G-J). Together, these results suggest that IRSp53 forms complexes with specific members of the PSD-95 family in brain.
Colocalization of IRSp53 with PSD-95 in cultured neurons and synaptic localization of IRSp53 in brain sections
Double immunofluorescence staining of cultured hippocampal neurons revealed that IRSp53 was distributed to dendritic spines, where it colocalized with PSD-95 (Fig. 3A). IRSp53 was also detected in regions outside the synapse, including the cell body and the dendritic trunk (Fig. 3A), consistent with the observed presence of a portion of IRSp53 in the extrasynaptic (S2) fraction (Fig. 2C).
Figure 3.
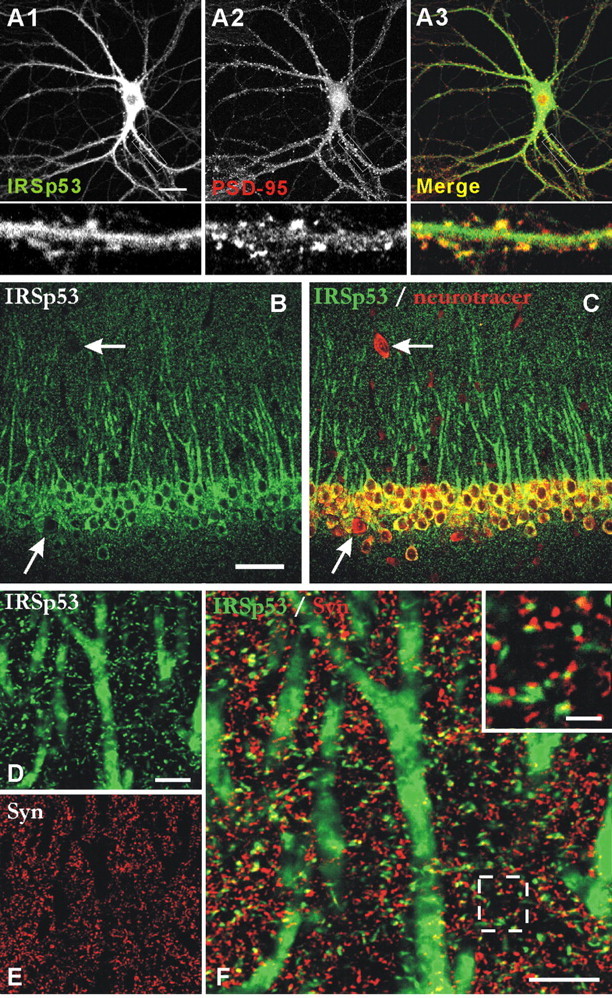
Colocalization of IRSp53 with PSD-95 in cultured neurons and synaptic localization of IRSp53 in brain sections. A, Colocalization of IRSp53 and PSD-95 in cultured neurons. Cultured hippocampal neurons (21 DIV) were doubly stained by immunofluorescence for IRSp53 (A1; green) and PSD-95 (A2; red). The bottom panels are enlarged images of the small white boxes in A1, A2, and A3. B, C, Expression pattern of IRSp53 (B; green) in the CA1 region of rat hippocampus. The Neuro Tracer dye (C; red) was used to visualize all of the neurons. The arrows point to probable GABAergic interneurons. D-F, Colocalization of IRSp53 (D; green) with synaptophysin (E; Syn; red). The inset in the merged image (F) shows the close apposition or partial colocalization between IRSp53 and synaptophysin. Scale bars: A, 20 μm; B, 100 μm; D, F,10 μm; inset, 2 μm.
In rat brain, at light microscopic level, IRSp53 was detected in various brain regions, including cortex, hippocampus, and cerebellum (data not shown), consistent with our immunoblot results (Fig. 2A) and previous reports (Abbott et al., 1999; Bockmann et al., 2002). IRSp53 signals were detected in the cell bodies and apical dendrites of principal neurons (e.g., pyramidal cells in cortex and hippocampus and Purkinje cells in cerebellum) (see Fig. 3, B and C, for images from the CA1 region of hippocampus). In forebrain, IRSp53 was not detected in GABAergic local circuit neurons (data not shown). Numerous puncta were positive in the neuropil (Fig. 3D-F) (CA1 region). Because satisfactory PSD-95 immunostaining of synapses in brain is difficult to achieve (presumably because of poor antibody penetration) unless sections are pretreated with proteases (Fukaya and Watanabe, 2000), we did not try to detect colocalization of IRSp53 and PSD-95. However, double immunofluorescence staining for IRSp53 and the presynaptic marker synaptophysin indicated that most of the IRSp53 puncta were closely apposed to or partially overlapped with synaptophysin puncta (Fig. 3D-F), suggesting that IRSp53 concentrates at synaptic sites in brain. Together, these results suggest that IRSp53 colocalizes with PSD-95 at synaptic sites.
Ultrastructural localization of IRSp53
Despite growing recognition of the importance of IRSp53 in synaptic function, electron microscopic analysis of subcellular distribution at the synapse has not been reported. We used pre-embedding immunogold EM to determine the ultrastructural localization of IRSp53 in rat brain, focusing on the CA1 region of hippocampus (Fig. 4). Silver-enhanced immunogold particles were most numerous at postsynaptic sites, where they labeled both the PSD and spine cytoplasm (Fig. 4A-E). There was no obvious association of immunogold particles with the extrasynaptic spine membrane or intracytoplasmic membranes. Axon terminals also exhibited occasional immunolabeling (Fig. 4B,C,E), but the label was predominantly postsynaptic.
Figure 4.
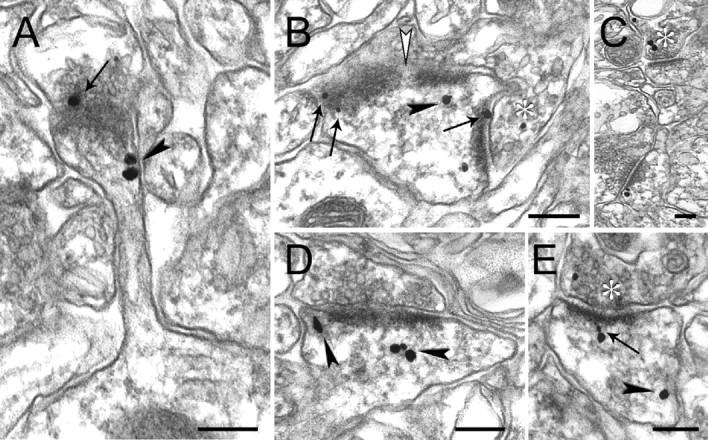
Immunogold-EM localization of IRSp53 in the CA1 stratum radiatum of rat hippocampus. Numerous IRSp53 immunogold particles are associated with postsynaptic structures (A-E). The arrows point to particles that localize within the PSD, and arrowheads point to particles in the spine cytoplasm not associated with the PSD. An example of the association of IRSp53 particles with perforated PSDs is shown (B; white arrowhead). A few particles can be seen at presynaptic sites (B, C, E). The stars indicate immunopositive presynaptic terminals. Scale bars, 200 nm.
Both the C terminus and the SH3 domain are important for the synaptic localization of IRSp53
IRSp53 is less tightly associated with the PSD than is PSD-95 (Fig. 2D), suggesting that the IRSp53-PSD-95 interaction may play a role in the synaptic localization of IRSp53. A previous study has shown that the SH3 domain of IRSp53 binds to a proline-rich region in Shank (Bockmann et al., 2002; Soltau et al., 2002), suggesting that the SH3 domain may be involved in the postsynaptic localization of IRSp53. To test these possibilities, we tested the synaptic localization of the following IRSp53 constructs (Fig. 5): IRSp53 WT, IRSp53 ΔC (a mutant lacking the PSD-95-binding C-terminal residues), IRSp53 SH3 mut (I403P; an SH3 domain mutant incapable of binding Shank) (Fig. 5A), and IRSp53 SH3 mut plus ΔC (double mutant). When expressed in cultured hippocampal neurons at 17 DIV and visualized at 19 DIV, IRSp53 WT showed significant accumulation at PSD-95-positive dendritic spines (Fig. 5B). In contrast, IRSp53 ΔC was mainly diffusely distributed throughout the neuron (Fig. 5C). IRSp53 SH3 mut showed a synaptic distribution between those of IRSp53 WT and IRSp53 ΔC (Fig. 5D). IRSp53 SH3 mut plus ΔC showed the lowest spine localization (Fig. 5E), indicating that the effects of the two mutations are additive. In these targeting experiments, we coexpressed PSD-95, along with the IRSp53 constructs, so that the amount of PSD-95 or PSD-95-associated Shank would not become a limiting factor for synaptic IRSp53 targeting. The expression levels of exogenous IRSp53 and PSD-95 were approximately two to three times higher than those of endogenous proteins (fluorescent intensities in transfected neurons relative to untransfected neurons: IRSp53 WT, 3.41 ± 0.57; IRSp53 SH3 mut, 3.85 ± 0.12; IRSp53 ΔC, 3.13 ± 0.10; IRSp53 SH3 mut plus ΔC; PSD-95, 3.54 ± 0.58; n = 5). In quantitative analysis, IRSp53 ΔC showed a synaptic localization (the ratio of immunofluorescence intensity at a spine over that at an adjacent dendritic trunk, 0.78 ± 0.03; n = 150 spines; *p < 0.001) (Fig. 5F) that is significantly lower than IRSp53WT (2.15 ± 0.02; n = 150) (Fig. 5F). The synaptic accumulation of IRSp53 SH3 mut (1.46 ± 0.02; n = 150; *p < 0.001) (Fig. 5F) was also significantly lower than IRSp53 WT, although higher than that of IRSp53ΔC. Double-mutant IRSp53 SH3 mut plus ΔC displayed the lowest synaptic localization (0.49 ± 0.01; n = 150; *p < 0.001 compared with WT) (Fig. 5F). These results suggest that both the C terminus and the SH3 domain of IRSp53 are important determinants of its synaptic localization.
Figure 5.
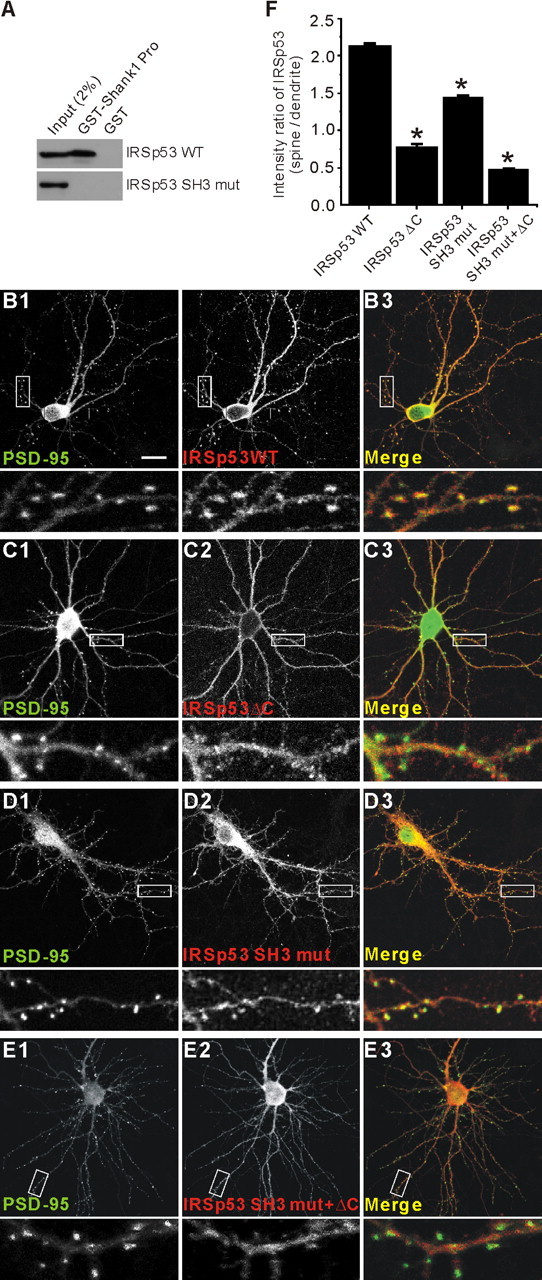
Both the C terminus and the SH3 domain are important for the synaptic localization of IRSp53. A, Loss of Shank binding in an IRSp53 mutant with a point mutation in the SH3 domain (IRSp53 SH3 mut; I403P). HEK293 cell lysates transfected with IRSp53 (WT or SH3 mut) were pulled down by GST-Shank1 Pro (IRSp53-binding proline-rich region in Shank1) and immunoblotted with IRSp53 antibodies. B-E, Cultured hippocampal neurons (17 DIV) were cotransfected with PSD-95-EGFP and HA-IRSp53 constructs; wild-type (IRSp53 WT; B), a PSD-95 binding-defective mutant that lacks the last four residues (IRSp53 ΔC; C), a Shank binding-defective mutant with a point mutation in the SH3 domain (IRSp53 SH3 mut; D), or a double mutant (IRSp53 SH3 mut plus ΔC; E). Transfected neurons were visualized by double immunofluorescence staining for EGFP (for PSD-95) and HA (for IRSp53) at 19 DIV. To measure synaptic localization, the immunofluorescence intensity of IRSp53 at a PSD-95-positive dendritic spine was compared with that in an adjacent dendritic trunk. Scale bar, 20 μm. F, quantitative analysis of the synaptic localization of IRSp53.
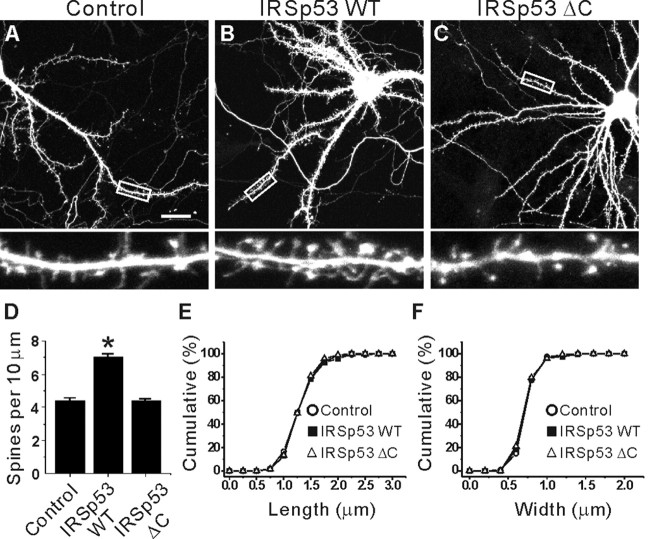
Overexpression of IRSp53 in cultured neurons selectively increases the density of dendritic spines
Rac1/Cdc42 small GTPases have been shown to regulate spine morphogenesis (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000; Irie and Yamaguchi, 2002; Scott et al., 2003). Given that IRSp53 is a downstream effector of Rac1/Cdc42 in non-neural cells, we tested whether IRSp53 is involved in the regulation of dendritic spines (Fig. 6). To this end, cultured neurons at 15 DIV were transfected with IRSp53, and the effects of IRSp53 overexpression on the density and morphology of dendritic spines were analyzed at 22 DIV. Interestingly, IRSp53 overexpression significantly increased the linear density of dendritic spines (7.90 ± 0.12 per 10 μm dendrite; n = 12 neurons; *p < 0.001) (Fig. 6B,D) compared with control neurons transfected with EGFP alone (4.99 ± 0.52 per 10 μm dendrite; n = 14) (Fig. 6A,D). In contrast, IRSp53 ΔC did not increase spine density in transfected neurons (4.39 ± 0.02; n = 10; p = 0.69 compared with EGFP alone controls) (Fig. 6C,D), indicating that synaptic localization of IRSp53 is important for its spine-promoting effects.
Figure 6.
Overexpression of IRSp53 selectively increases the density of dendritic spines in cultured neurons. A-C, Effects of IRSp53 overexpression on dendritic spines. Cultured hippocampal neurons (15 DIV) were transfected with IRSp53 plus EGFP (B), IRSp53 ΔC plus EGFP (C), or EGFP alone (A; control) and visualized at 22 DIV by immunofluorescence staining for EGFP (A-C) and IRSp53 (B, C). Only EGFP images are shown here for comparison. The bottom panels are enlarged images of the small white boxes in the top panels. Scale bar, 20 μm. D, Quantitative analysis of the effects of IRSp53 overexpression on spine density. A significant change is indicated by an asterisk. E, F, The cumulative frequency distribution of dendritic spine length (E) and width (F) in transfected and control neurons.
In contrast, morphometric analysis indicated that the length of dendritic spines on IRSp53-expressing neurons was not significantly different (1.12 ± 0.05 μm; n = 10; p = 0.85) (Fig. 6E) from that of control neurons (EGFP alone; 1.13 ± 0.05 μm; n = 11) (Fig. 6E). Similarly, the width of the dendritic spines in IRSp53-transfected neurons (0.63 ± 0.01 μm; n = 10; p = 0.58) was not significantly different from that of control neurons (0.61 ± 0.04 μm; n = 11) (Fig. 6F). IRSp53 ΔC did not change spine morphology (length, 1.16 ± 0.05; n = 15; p = 0.92 compared with EGFP alone controls; width, 0.77 ± 0.03; n = 15; p = 0.82 compared with EGFP alone controls) (Fig. 6E,F). Together, these results indicate that IRSp53 selectively increases spine density with minimal effects on spine size (length and width).
siRNA downregulation of IRSp53 decreases the density and size of spines
Because IRSp53 overexpression in cultured neurons increases spine density (Fig. 6), we tested whether IRSp53 is required for the maintenance of normal spine density (Fig. 7). To this end, we generated an IRSp53 siRNA construct (pSUPER IRSp53). When coexpressed with IRSp53 in heterologous cells, the expression of IRSp53 was significantly reduced to 13.7 ± 1.2% of control levels (n = 3; data not shown). When this siRNA construct was expressed in cultured neurons for 3 d (from 13-16 DIV), the expression of endogenous IRSp53 was markedly reduced to 10.5 ± 0.5% (n = 7; *p < 0.001) (Fig. 7A,C) of that in control neurons transfected with the empty pSUPER vector (Fig. 7A,B).
Figure 7.
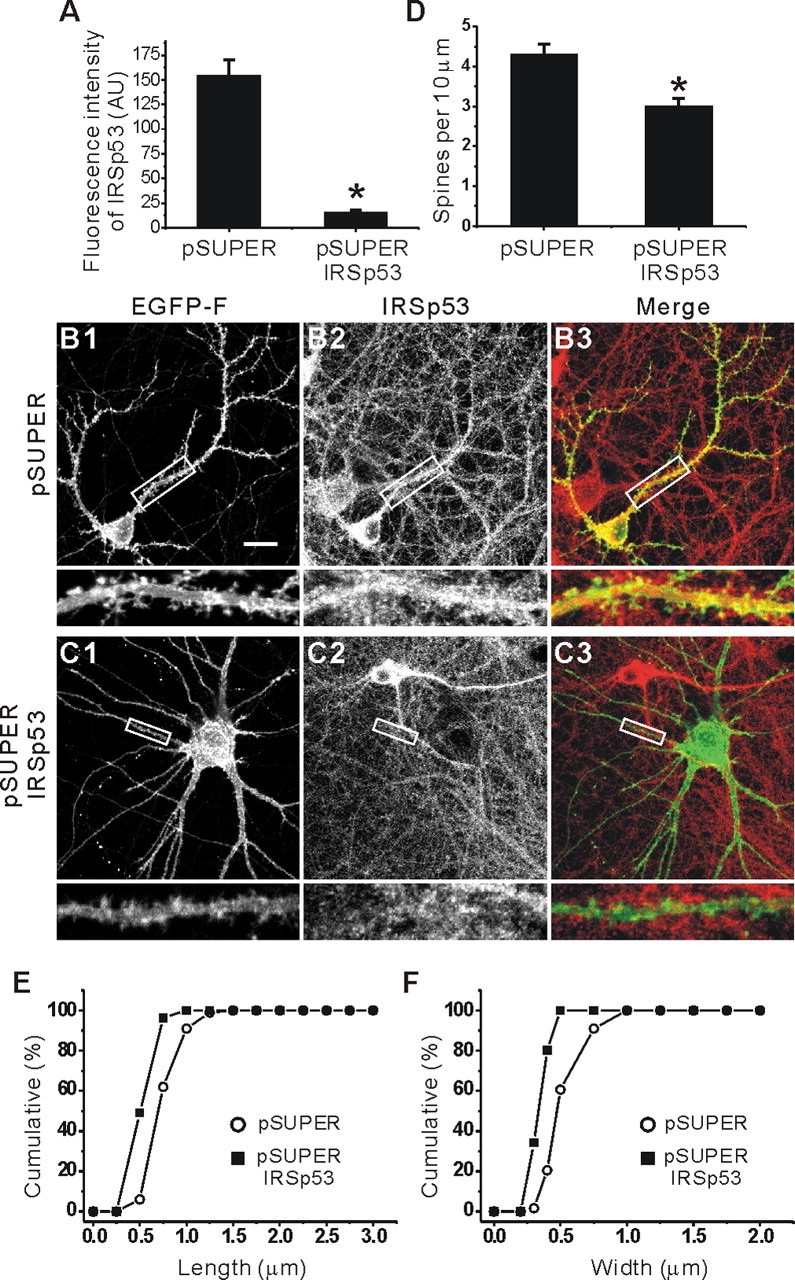
siRNA downregulation of IRSp53 decreases the density and size of spines. A-C, The effects of siRNA IRSp53 downregulation on dendritic spines. Cultured hippocampal neurons (13 DIV) were transfected with pSUPER IRSp53 plus EGFP-F (farnesylated EGFP), or empty pSUPER plus EGFP-F, and visualized at 16 DIV by immunofluorescence staining for EGFP-F and IRSp53. IRSp53 images were used to quantify the siRNA-induced downregulation of endogenous IRSp53 expression (A), and EGFP images were used to measure siRNA-induced changes in dendritic spines. AU, Arbitrary unit. Scale bar, 20 μm. D-F, Quantitative analysis of the effects of siRNA IRSp53 downregulation on the density (D) and size (length and width, E and F, respectively) of dendritic spines.
When the effects of pSUPER IRSp53 were quantified, the linear density of dendritic spines on siRNA-transfected neurons was significantly reduced (3.0 ± 0.09 per 10 μm dendrite; n = 13; *p < 0.001) (Fig. 7C,D) compared with that of control neurons transfected with the empty pSUPER vector (4.30 ± 0.14 per 10 μm dendrite; n = 12) (Fig. 7B,D). Intriguingly, the length and width of the dendritic spines on siRNA-transfected neurons were also significantly reduced (length, 0.43 ± 0.02 μm, n = 10, *p < 0.001; width, 0.24 ± 0.01 μm, n = 10, *p < 0.001) (Fig. 7E,F) compared with those of control neurons (length, 0.88 ± 0.03 μm, n = 10; width, 0.58 ± 0.02 μm, n = 10) (Fig. 7 E, F). These results suggest that IRSp53 is required for the maintenance of both spine density and size.
A dominant-negative IRSp53 SH3 mutant decreases the density and size of spines
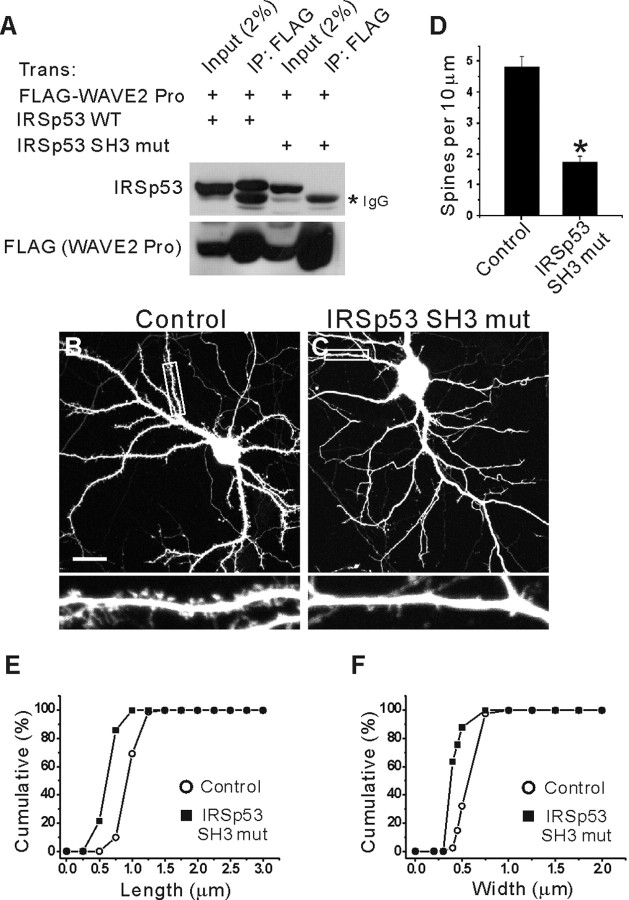
The present results suggest that IRSp53 is important for spine regulation (Figs. 6, 7), which prompts the question of just how IRSp53 exerts its spine-promoting effects. One possibility is that IRSp53, through its SH3 domain, binds to various actin-modulating proteins, including WAVE2, Mena, and Shank. To test this hypothesis, we used dominant-negative IRSp53 SH3 mutants that cannot link activated Rac1/Cdc42 to SH3-binding downstream effectors. When the IRSp53 SH3 mut (I403P) (incapable of binding Shank, as described above) was further tested for WAVE2 binding, there was no detectable interaction with WAVE2, as inferred from the lack of coimmunoprecipitation with the SH3-binding proline-rich region of WAVE2 in heterologous cells (Fig. 8A). This suggests that the IRSp53 SH3 mut (I403P) is likely to block the coupling of activated Rac1/Cdc42 with various downstream SH3-binding proteins in a dominant-negative manner.
Figure 8.
A dominant-negative IRSp53 SH3 mutant decreases the density and size of spines. A, Loss of WAVE2 interaction in IRSp53 SH3 mutant (I403P). Lysates of HEK293T cells transfected with HA-IRSp53 (WT or SH3 mut) plus FLAG-WAVE2 Pro (proline-rich region; aa273-402) were immunoprecipitated with FLAG antibodies and immunoblotted with IRSp53 and FLAG (for WAVE2 Pro) antibodies. B, C, Cultured hippocampal neurons (15 DIV) were transfected with IRSp53 SH3 mut plus EGFP (C), or EGFP alone (B), and visualized at 17 DIV by double immunofluorescence staining for EGFP and IRSp53. Only EGFP images are shown for comparison. Scale bar, 20 μm. D-F, Quantitative analysis of the effects of IRSp53 SH3 mut expression on the density (D) and size (length and width, E and F, respectively) of spines.
When IRSp53 SH3 mut was expressed in cultured neurons for 2 d (15-17 DIV), the density of dendritic spines was significantly decreased (1.76 ± 0.18 per 10 μm dendrite; n = 11; *p < 0.001) (Fig. 8C,D) compared with control neurons (4.82 ± 0.36 per 10 μm dendrite; n = 12) (Fig. 8B,D). In addition, the length and width of the dendritic spines on IRSp53 SH3 mut-transfected neurons were significantly reduced (length, 0.63 ± 0.02 μm, n = 9, *p < 0.001; width, 0.38 ± 0.01 μm, n = 9, *p < 0.001) (Fig. 8E,F) compared with those of control neurons (length, 0.95 ± 0.02 μm, n = 10; width, 0.59 ± 0.01 μm, n = 10) (Fig. 8E,F). Notably, the expression of IRSp53 SH3 mut significantly reduced the synaptic localization of Shank, one of the IRSp53 SH3-interacting proteins, as determined by the spine/dendrite immunofluorescence ratio (1.45 ± 0.03 in IRSp53 SH3 mut-expressing neurons, n = 120, vs 2.63 ± 0.05 in EGFP alone-expressing control neurons, n = 120; *p < 0.001). A possible explanation for this is that Shank may depend on IRSp53 interaction for its synaptic localization. Alternatively, this effect may be caused by the general shrinkage of spines associated with spine density reduction. Together, these results suggest that the SH3 domain plays an important role in the IRSp53-dependent regulation of dendritic spines.
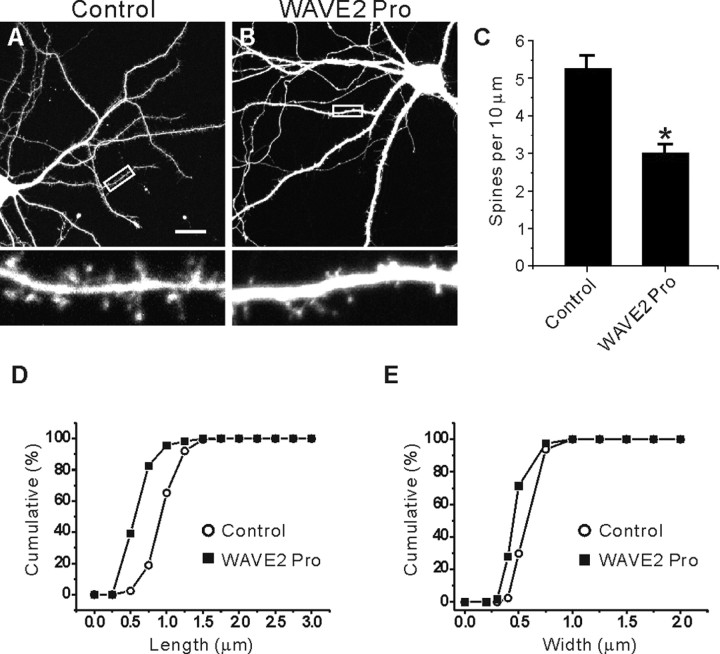
A dominant-negative WAVE2 proline-rich region decreases spine density and size
WAVE2, which binds to the IRSp53 SH3 via its proline-rich region, links activated Rac1 to the Arp2/3 actin nucleation complex-dependent formation of lamellipodia in non-neural cells (Miki et al., 2000; Miki and Takenawa, 2002). A useful dominant negative that disrupts the functional connection between IRSp53 and WAVE2 is a construct containing the proline-rich region of WAVE2 (Miki et al., 2000). When this dominant-negative construct (termed WAVE2 Pro) was expressed in cultured neurons for 3 d (14-17 DIV), spine density was significantly decreased (3.01 ± 0.10; n = 11; *p < 0.001) (Fig. 9B,C) compared with control neurons transfected with EGFP alone (5.06 ± 0.15; n = 10) (Fig. 9A,C). In addition, the length and width of dendritic spines on WAVE2 Pro-transfected neurons were significantly reduced (length, 0.60 ± 0.04, n = 9, *p < 0.001; width, 0.46 ± 0.01, n = 9, *p < 0.001) (Fig. 9D,E), compared with those of control neurons (length, 0.96 ± 0.03, n = 9; width, 0.55 ± 0.01, n = 9) (Fig. 9D,E). WAVE2 Pro expression did not significantly change the synaptic localization of IRSp53, as determined by the spine/dendrite immunofluorescence ratio (1.28 ± 0.03 in WAVE2 Pro-expressing neurons, n = 100, vs 1.31 ± 0.02 in EGFP alone-expressing control neurons, n = 100; p = 0.81). A possible explanation for this is that IRSp53, although its SH3 domain is mainly blocked by WAVE2 Pro, can still be targeted to synapses by the interaction of its C terminus with PSD-95. Together, these results suggest that WAVE2 may be an important mediator of IRSp53-dependent spine regulation.
Figure 9.
A dominant-negative WAVE2 proline-rich region decreases spine density and size. A, B, Cultured hippocampal neurons (14 DIV) were transfected with a WAVE2 dominant-negative construct containing the proline-rich region (WAVE2 Pro) plus EGFP (B), or EGFP alone (A), and visualized at 17 DIV for IRSp53 and EGFP. Scale bar, 20 μm. C-E, Quantitative analysis of the effects of WAVE2 Pro expression on the density (C) and size (D, E) of spines.
Discussion
Role of the IRSp53-PSD-95 interaction in the synaptic localization of IRSp53
In the present study, we demonstrated that IRSp53 interacts with PSD-95. This corroborates the recent finding by Soltau et al. (2004) that IRSp53 associates with PSD-95. In addition, we found that IRSp53 associates with specific members of the PSD-95 family (PSD-95 and chapsyn-110/PSD-93) in brain (Fig. 2E-J). This in vivo specificity could be attributable to differences in the spatiotemporal distribution patterns of PSD-95 family proteins; PSD-95 and chapsyn-110 are generally synaptic, whereas SAP97 and SAP102 are more widespread and implicated in traf-ficking (Kim and Sheng, 2004). Previous work showed that a member of the transmembrane AMPA receptor regulatory protein/stargazin family interacts specifically with PSD-95 and chapsyn-110/PSD-93 (Dakoji et al., 2003). In addition, preferential associations have been demonstrated for PSD-95 and NR2A, SAP102 and NR2B, and SAP97 and glutamate receptor 1 (Leonard et al., 1998; Sans et al., 2000; Townsend et al., 2003). Thus, our results contribute to the emerging notion that PSD-95 family proteins have differential functions.
Our data indicate that both PSD-95 and Shank interactions are required for synaptic IRSp53 localization (Fig. 5). Direct comparison of the relative contribution of PSD-95 and Shank to synaptic IRSp53 localization is difficult, because the relative amounts of PSD-95, Shank, and IRSp53 in transfected neurons, in addition to IRSp53 mutation/deletion, may alter the experimental outcomes. However, in vivo coimmunoprecipitation results indicate that a much smaller portion of Shank, compared with PSD-95, is in complex with IRSp53 (Fig. 2E). Because PSD-95 is several fold more abundant than Shank1, as demonstrated by a quantitative mass spectrometric analysis (Peng et al., 2004), this result suggests that the majority of IRSp53 is associated with PSD-95 or that IRSp53 has a higher affinity for PSD-95 relative to Shank. Thus, it is likely that, in the PSD, the C-terminal motif of IRSp53 is occupied by PSD-95, whereas the SH3 domain may be free for other functions. The binding partners of this SH3 domain vary depending on the nature of the associated small GTPases (Rac1 vs Cdc42). Thus, it would be advantageous for the C terminus to engage in synaptic anchoring through the generally strong PDZ interaction, whereas the SH3 domain serves as a versatile Rac1/Cdc42-dependent switch that associates with various target proteins for dynamic regulation of spine morphogenesis.
Role of IRSp53 in the regulation of dendritic spines
Our results indicate that IRSp53 overexpression selectively increases spine density (Fig. 6). Conversely, siRNA-mediated knock-down of IRSp53 reduces spine density and size (Fig. 7). IRSp53 SH3 mut and WAVE2 Pro reduce spine density and size (Figs. 8, 9). Because IRSp53 links activated Rac1/Cdc42 to the regulation of the actin cytoskeleton in non-neural cells (Miki et al., 2000), these results suggest that IRSp53 may be an important regulator of spine morphogenesis that acts downstream of Rac1/Cdc42 in neurons, along with PAK and N-WASP, which have been suggested to be downstream effectors of Rac1/Cdc42 in EphB receptor-dependent spine morphogenesis (Irie and Yamaguchi, 2002; Penzes et al., 2003).
The SH3 domain appears to be important in the spine-promoting effects of IRSp53. In support of this, IRSp53 SH3 mut and WAVE2 Pro reduced the spine density and size (Figs. 8, 9). In addition, these dominant-negative effects resembled those seen following siRNA-mediated knock-down of IRSp53, suggesting that the IRSp53 effects are, in large part, mediated by the SH3 domain. Moreover, IRSp53 SH3 mut, despite containing the N-terminal IMD domain that regulates the actin cytoskeleton independent of the SH3 domain (Yamagishi et al., 2004), did not have any spine-promoting effects (Fig. 8), suggesting that the SH3 domain is more important. Together, these results strongly suggest that IRSp53 links activated Rac1/Cdc42 to downstream effectors through its SH3 domain.
The above results raise the question of how the IRSp53SH3 mediates the spine-promoting effects of IRSp53. One possibility is that the IRSp53 SH3 domain interacts with actin regulating downstream effectors. Known binding partners of the IRSp53 SH3 that are implicated in actin regulation include WAVE2, Mena, and Shank (Miki et al., 2000; Krugmann et al., 2001; Bockmann et al., 2002; Soltau et al., 2002). We have shown here that WAVE2 Pro reduces spine density and size (Fig. 9), suggesting that the effects of IRSp53 may be mediated by WAVE2. WAVE2 is expressed in the brain at both the mRNA and protein levels (Suetsugu et al., 1999) (J. Choi and E. Kim, unpublished results), although its neuronal functions remain to be determined. The IRSp53-induced increase in spine density may be attributable to the association of WAVE2 with the Arp2/3 actin nucleation complex (Miki et al., 2000), which adds new branches to the sides of existing actin filaments, causing dynamic extension of actin-based structures. Conversely, the siRNA-induced reduction in spine density and size may be attributable to spine shrinkage caused by a lack of basal level activation of the Arp2/3 machinery. However, it should be noted that IRSp53 SH3 mut (I403P) disrupts the interaction of IRSp53 with both Shank and WAVE2 (Figs. 5A, 8A), suggesting that overexpression of WAVE2 Pro dominant negative may block SH3 binding of WAVE2 as well as other proteins. Another candidate that may mediate the IRSp53 effects is Mena, which is known to link Cdc42 activation to filopodia formation in non-neural cells (Govind et al., 2001; Krugmann et al., 2001) and is widely expressed in both developing and, to a lesser degree, adult brain (Lanier et al., 1999). Mena is required for the formation of growth cone filopodia and is implicated in axon outgrowth and guidance (Lebrand et al., 2004), but its role in spine morphogenesis remains an open question.
Shank may be the best characterized protein among the known binding partners of the IRSp53 SH3. Shank has been shown to regulate spine morphogenesis (Sala et al., 2001). In particular, Shank acts in the maturation of spines, causing the width and length of spines to increase without significantly affecting spine density (Sala et al., 2001), which contrast to the selective effects of IRSp53 on spine density in this study. Cortactin, a Shank-associated protein that binds to F-actin and the Arp2/3 actin nucleation complex (Weed et al., 2000; Uruno et al., 2001), has been shown to induce the formation of protrusions in aspiny neurons and to be required for the maintenance of normal spine density, as demonstrated by siRNA downregulation (Hering and Sheng, 2003). However, the main effect of cortactin overexpression is to increase the length of spines but not their width and density (Hering and Sheng, 2003), which again differs from the spine density-specific effects of IRSp53 (Fig. 6). Another Shank-associated protein that may contribute to the IRSp53-dependent spine regulation is Abp1, an F-actin binding protein that is enriched in the PSD (Qualmann et al., 2004). Notably, Abp1 is recruited to spines by synaptic stimulation (Qualmann et al., 2004), but the specific role for Abp1 in spine regulation is not known. Thus, it remains to be determined whether Shank, cortactin, and Abp1 mediate the effects of IRSp53.
Possible role of PSD-95 as an organizer of the Rac1 signaling pathway in spines
Our data show that PSD-95 is important for the synaptic localization of IRSp53, a downstream effector of activated Rac1/Cdc42. PSD-95 is also required for the synaptic localization of Kalirin-7 [Rac1 guanine nucleotide exchange factor (GEF)] (Penzes et al., 2001). These results suggest that PSD-95, which can form multimers (Kim et al., 1996; Hsueh et al., 1997; Christopherson et al., 2003), may bring Kalirin-7 and IRSp53 close together and mediate their possible functional coupling (activation of IRSp53 by Kalirin-7-activated Rac1). In addition, Shank, which is linked to PSD-95 through guanylate kinase-associated protein (GKAP) (Sheng and Kim, 2000), might interact with βPIX (Rac1/Cdc42 GEF) (Park et al., 2003) to bring βPIX close to IRSp53 for their possible functional coupling (activation of IRSp53 by βPIX-activated Rac1/Cdc42). Together, it appears that the well-known PSD-95-GKAP-Shank axis may bring together various Rac1/Cdc42-related signaling molecules (Kalirin-7, βPIX, and IRSp53) and coordinate their functional coupling to regulate spine morphogenesis.
A correlation between PSD-95 assembly and spine morphogenesis has been demonstrated in several studies. Time-lapse imaging revealed that local assembly of PSD-95 spatiotemporally correlates with spine formation (Okabe et al., 2001). PSD-95 overexpression in cultured neurons increases the density and size of dendritic spines (El-Husseini et al., 2000). The degradation of PSD-95 triggered by an inducible kinase results in the loss of dendritic spines (Pak and Sheng, 2003). Considering the important role of PSD-95 in synaptic IRSp53 localization and the role of IRSp53 in spine regulation, dynamic changes in the synaptic content of PSD-95 during development and plasticity may be linked via concomitant changes in synaptic IRSp53 levels to the regulation of spine morphogenesis.
In conclusion, our results indicate that IRSp53 interacts with two specific members of the PSD-95 family, PSD-95 and chapsyn-110/PSD-93, and that this interaction is important for synaptic IRSp53 localization. In addition, we have demonstrated that IRSp53, a downstream effector of Rac1/Cdc42, regulates spine morphogenesis through its SH3 domain. The next step is to identify the molecular mechanisms acting downstream of IRSp53 and genetically confirm the roles for IRSp53 in the regulation of dendritic spines.
Footnotes
This work was supported by the National Creative Research Initiative Program of the Korean Ministry of Science and Technology (E.K.) and by National Institutes of Health Grant NS-39444 (R.J.W.).
Correspondence should be addressed to Eunjoon Kim, Department of Biological Sciences, Korea Advanced Institute of Science and Technology, Daejeon 305-701, Korea. E-mail: kime@kaist.ac.kr.
Copyright © 2005 Society for Neuroscience 0270-6474/05/250869-11$15.00/0
References
- Abbott MA, Wells DG, Fallon JR (1999) The insulin receptor tyrosine kinase substrate p58/53 and the insulin receptor are components of CNS synapses. J Neurosci 19: 7300-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez CE, Sutcliffe JG, Thomas EA (2002) Novel isoform of insulin receptor substrate p53/p58 is generated by alternative splicing in the CRIB/SH3-binding region. J Biol Chem 277: 24728-24734. [DOI] [PubMed] [Google Scholar]
- Arnold DB, Clapham DE (1999) Molecular determinants for subcellular localization of PSD-95 with an interacting K+ channel. Neuron 23: 149-157. [DOI] [PubMed] [Google Scholar]
- Bockmann J, Kreutz MR, Gundelfinger ED, Bockers TM (2002) ProSAP/Shank postsynaptic density proteins interact with insulin receptor tyrosine kinase substrate IRSp53. J Neurochem 83: 1013-1017. [DOI] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB (1992) The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9: 929-942. [DOI] [PubMed] [Google Scholar]
- Choi J, Ko J, Park E, Lee JR, Yoon J, Lim S, Kim E (2002) Phosphorylation of stargazin by protein kinase A regulates its interaction with PSD-95. J Biol Chem 277: 12359-12363. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Sweeney NT, Craven SE, Kang R, El-Husseini Ael D, Bredt DS (2003) Lipid- and protein-mediated multimerization of PSD-95: implications for receptor clustering and assembly of synaptic protein networks. J Cell Sci 116: 3213-3219. [DOI] [PubMed] [Google Scholar]
- Dakoji S, Tomita S, Karimzadegan S, Nicoll RA, Bredt DS (2003) Interaction of transmembrane AMPA receptor regulatory proteins with multiple membrane associated guanylate kinases. Neuropharmacology 45: 849-856. [DOI] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science 290: 1364-1368. [PubMed] [Google Scholar]
- Fujiwara T, Mammoto A, Kim Y, Takai Y (2000) Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem Biophys Res Commun 271: 626-629. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Watanabe M (2000) Improved immunohistochemical detection of postsynaptically located PSD-95/SAP90 protein family by protease section pretreatment: a study in the adult mouse brain. J Comp Neurol 426: 572-586. [PubMed] [Google Scholar]
- Goslin K, Banker G (1991) Rat hippocampal neurons in low-density culture. In: Culturing nerve cells (Banker G, Goslin K, eds), pp 337-370. Cambridge, MA: MIT.
- Govind S, Kozma R, Monfries C, Lim L, Ahmed S (2001) Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kD insulin receptor substrate to filamentous actin. J Cell Biol 152: 579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Kater SB (1994) Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu Rev Neurosci 17: 341-371. [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M (2001) Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci 2: 880-888. [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M (2003) Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci 23: 11759-11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Konno D, Maruoka H, Sobue K (2003) MALS is a binding partner of IRSp53 at cell-cell contacts. FEBS Lett 554: 30-34. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Kim E, Sheng M (1997) Disulfide-linked head-to-head multimerization in the mechanism of ion channel clustering by PSD-95. Neuron 18: 803-814. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Schiebler W, Greengard P, De Camilli P (1983) Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol 96: 1374-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y (2002) EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci 5: 1117-1118. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M (2004) PDZ domain proteins of synapses. Nat Rev Neurosci 5: 771-781. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M (1995) Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378: 85-88. [DOI] [PubMed] [Google Scholar]
- Kim E, Cho KO, Rothschild A, Sheng M (1996) Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron 17: 103-113. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A (2001) Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol 11: 1645-1655. [DOI] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB (1999) Mena is required for neurulation and commissure formation. Neuron 22: 313-325. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB (2004) Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron 42: 37-49. [DOI] [PubMed] [Google Scholar]
- Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW (1998) SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem 273: 19518-19524. [DOI] [PubMed] [Google Scholar]
- Lim S, Sala C, Yoon J, Park S, Kuroda S, Sheng M, Kim E (2001) Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci 17: 385-397. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN (1996) Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379: 837-840. [DOI] [PubMed] [Google Scholar]
- Matus A (2000) Actin-based plasticity in dendritic spines. Science 290: 754-758. [DOI] [PubMed] [Google Scholar]
- Miki H, Takenawa T (2002) WAVE2 serves a functional partner of IRSp53 by regulating its interaction with Rac. Biochem Biophys Res Commun 293: 93-99. [DOI] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T (2000) IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408: 732-735. [DOI] [PubMed] [Google Scholar]
- Mok H, Shin H, Kim S, Lee JR, Yoon J, Kim E (2002) Association of the kinesin superfamily motor protein KIF1Bα with postsynaptic density-95 (PSD-95), synapse-associated protein-97, and synaptic scaffolding molecule PSD-95/discs large/zona occludens-1 proteins. J Neurosci 22: 5253-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M (1999) Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23: 569-582. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L (2000) Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci 20: 5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K (2002) Structure and function of dendritic spines. Annu Rev Physiol 64: 313-353. [DOI] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H (2001) Spine formation and correlated assembly of presynaptic and postsynaptic molecules. J Neurosci 21: 6105-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura-Oho Y, Miyashita T, Yamada M (2001) Distinctive tissue distribution and phosphorylation of IRSp53 isoforms. Biochem Biophys Res Commun 289: 957-960. [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M (2003) Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302: 1368-1373. [DOI] [PubMed] [Google Scholar]
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E (2003) The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. [DOI] [PubMed]
- Parnas D, Haghighi AP, Fetter RD, Kim SW, Goodman CS (2001) Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron 32: 415-424. [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M (2004) Semi-quantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem 279: 21003-21011. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Alam MR, Kambampati V, Mains RE, Eipper BA (2000) An isoform of kalirin, a brain-specific GDP/GTP exchange factor, is enriched in the postsynaptic density fraction. J Biol Chem 275: 6395-6403. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA (2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29: 229-242. [DOI] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL (2003) Rapid induction of dendritic spine morphogenesis by transsynaptic EphrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron 37: 263-274. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Boeckers TM, Jeromin M, Gundelfinger ED, Kessels MM (2004) Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J Neurosci 24: 2481-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M (2001) Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115-130. [DOI] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos Jr J, Hell JW, Wenthold RJ (2000) A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci 20: 1260-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EK, Reuter JE, Luo L (2003) Small GTPase Cdc42 is required for multiple aspects of dendritic morphogenesis. J Neurosci 23: 3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekerkova G, Loomis PA, Changyaleket B, Zheng L, Eytan R, Chen B, Mugnaini E, Bartles JR (2003) Novel espin actin-bundling proteins are localized to Purkinje cell dendritic spines and bind the Src homology 3 adapter protein insulin receptor substrate p53. J Neurosci 23: 1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E (2000) The shank family of scaffold proteins. J Cell Sci 113: 1851-1856. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24: 1-29. [DOI] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY (1994) Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368: 144-147. [DOI] [PubMed] [Google Scholar]
- Soltau M, Richter D, Kreienkamp HJ (2002) The insulin receptor substrate IRSp53 links postsynaptic shank1 to the small G-protein cdc42. Mol Cell Neurosci 21: 575-583. [DOI] [PubMed] [Google Scholar]
- Soltau M, Berhorster K, Kindler S, Buck F, Richter D, Kreienkamp HJ (2004) Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J Neurochem 90: 659-665. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T (1999) Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun 260: 296-302. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R (2000) Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex 10: 927-938. [DOI] [PubMed] [Google Scholar]
- Townsend M, Yoshii A, Mishina M, Constantine-Paton M (2003) Developmental loss of miniature N-methyl-d-aspartate receptor currents in NR2A knockout mice. Proc Natl Acad Sci USA 100: 1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X (2001) Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol 3: 259-266. [DOI] [PubMed] [Google Scholar]
- Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT (2000) Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol 151: 29-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N (2004) A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem 279: 14929-14936. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2001) Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci 24: 1071-1089. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5: 24-34. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Horwitz AF (2003) Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol 161: 131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]