Abstract
Cortical pyramidal cells are generated from pallial neuroepithelial precursors, whereas GABAergic interneurons originate in subpallial germinal zones and migrate tangentially to reach the cortex. Using Cre–lox technology in transgenic mice and a series of molecular markers that subdivide the subpallial neuroepithelium into small domains, we fate-map precursor pools and identify interneurons generated from each domain. Cortical interneurons expressing calbindin, parvalbumin, and somatostatin are generated exclusively from Lhx6 (Lim homeobox 6)-expressing precursors in the medial ganglionic eminence (MGE). Martinotti cells that coexpress calretinin and somatostatin are generated from the dorsal region of the MGE neuroepithelium that expresses Nkx6.2 (NK2 transcription factor-related 6.2). Most neuropeptide Y-expressing cells and all bipolar calretinin-expressing interneurons are generated outside the MGE, from the germinal zones of the lateral/caudal ganglionic eminences that express Gsh2 (genomic screened homeobox 2). Our data demonstrate that subpallial neuroepithelial domains defined by expression of genetic determinants generate distinct interneuron subtypes, thereby contributing to the generation of cortical interneuron heterogeneity observed in the adult cortex.
Keywords: cell fate, cortex, Cre-transgenic, development, GABAergic neuron, interneurons
Introduction
GABAergic interneurons represent the minority of neurons in the cortex (∼25%), yet they exhibit remarkable diversity in axonal morphology, electrophysiological properties, and neurochemical content (Markram et al., 2004; Yuste, 2005). The extent to which genetic restriction of neuronal potential at the precursor stage contributes to the generation of interneuron diversity in the adult cortex is still unclear.
Evidence suggests that cortical and hippocampal GABAergic interneurons are not generated from pallial precursors in rodents (Wonders and Anderson, 2006). Instead, they are born in subpallial regions and migrate over long distances to populate the striatum, the cortex, and the hippocampus (Parnavelas, 2000; Marín and Rubenstein, 2001; Marín and Rubenstein, 2003). Pioneering studies had suggested that many cortical GABAergic cells originate in the lateral (LGE) and medial (MGE) ganglionic eminences (Anderson et al., 1997, 2001; Tamamaki et al., 1997; Lavdas et al., 1999; Wichterle et al., 1999; Parnavelas, 2000). Support for migration of cells from these two regions has come from mouse mutants lacking functional germinal zones that show reduced interneuron numbers in the cortex and hippocampus (Anderson et al., 1997; Sussel et al., 1999; Pleasure et al., 2000). Additional attempts to identify subpallial interneurons migrating to the cortex demonstrated migration of cells from the MGE and the caudal ganglionic eminence (CGE) (a caudal extension of the subpallial germinal regions) but not from the LGE, questioning the LGE origin of cortical GABAergic interneurons (Wichterle et al., 2001; Nery et al., 2002; Yozu et al., 2005).
Immature interneurons migrating from the subpallium to the cortex represent a heterogenous population of cells in both neuroepithelial origin and route of migration. Three major groups of interneurons can be identified in the postnatal neocortex on the basis of neurochemical content: parvalbumin (PV)-, calretinin (CR)-, and somatostatin (SST)-expressing cells (Kubota et al., 1994; Gonchar and Burkhalter, 1997). By analogy to the caudal CNS in which distinct interneurons are generated from defined precursor pools, perhaps the precise subpallial origin of cortical interneurons may specify their fate. A series of studies have provided clues into the types of interneurons that are generated from subpallial progenitor domains. The MGE is thought to generate mainly PV-positive (PV+), SST+, and NPY+ cells, as well as a small number of CR+ interneurons (Wichterle et al., 2001; Nery et al., 2002; Valcanis and Tan, 2003; López-Bendito et al., 2004; Xu et al., 2004; Butt et al., 2005). The types of interneurons that are generated from the CGE remain controversial (Nery et al., 2002; Xu et al., 2004; Butt et al., 2005).
We adopted an in vivo Cre–lox-based genetic fate-mapping approach to permanently and reproducibly label the telencephalic neuroepithelium and identify interneuron subtypes generated from different regions. Using molecular markers with distinct and additive expression patterns, we subdivide the neuroepithelium into small domains and examine the spectrum of cortical and hippocampal interneurons that are generated from each region. Our work refines and extends previous studies and provides an accurate genetic map of the origins of pallial interneurons.
Materials and Methods
Transgenic mice. Nkx2.1
(NK2 transcription factor-related 2.1)–Cre, Gsh2 (genomic screened homeobox 2)–Cre, Emx1 (empty spiracles homolog 1)–Cre, and Dbx1 (developing brain homeobox 1)–Cre transgenic mice have been described previously (Fogarty et al., 2005; Kessaris et al., 2006). The Lhx6 (Lim homeobox 6)–Cre transgenic mice will be described elsewhere. Mice expressing Cre under control of the Nkx6.2 gene were generated using bacterial artificial chromosome (BAC) transgenic technology. The codon-improved Cre recombinase (iCre) (Shimshek et al., 2002) containing a nuclear localization signal was fused to the initiation codon of the Nkx6.2 gene using a PCR-based approach. An simian virus 40 polyadenylation signal was inserted at a position immediately after the ATG-encoding exon of Nkx6.2. BAC modification was performed in a bacterial system as described previously (Lee et al., 2001). All mice used in this study were maintained on a C57BL/6/CBA background. Mouse colonies were maintained at the Wolfson Institute for Biomedical Research, University College London, in accordance with United Kingdom legislation.
Two reporter mice have been used in this study: Rosa26R (R26R)–green fluorescent protein (GFP) (Mao et al., 2001) (obtained from The Jackson Laboratory, Bar Harbor, ME) and Rosa26R–yellow fluorescent protein (YFP) (Srinivas et al., 2001). On Cre-mediated recombination, both mice express GFP/YFP under the Rosa26 promoter. However, the efficiency with which recombination takes place and, consequently, the efficiency with which the reporter gene is activated were different between the two mice with the R26R–GFP gene being activated less efficiently than the R26R–YFP. We made use of this delayed activation of the R26R–GFP allele to create sharp borders of reporter activation in Nkx2.1–Cre and Nkx6.2–Cre transgenic mice.
Tissue preparation.
Embryos were fixed overnight in 4% (w/v) paraformaldehyde (PFA) in PBS. Postnatal animals were anesthetized and perfused with 4% (w/v) PFA through the left ventricle of the heart. Adult brains were dissected out, sliced into 2 mm slices using a brain slicer, and immersed in 4% PFA overnight. Fixed samples were cryoprotected overnight by immersion in 20% (w/v) sucrose in PBS. All samples were embedded in Tissue-Tek OCT compound (R. A. Lamb Medical Supplies, Eastbourne, UK), frozen on dry ice, and stored at −80°C.
In situ hybridization.
Solutions used for in situ hybridization were pretreated with 0.1% diethyl pyrocarbonate. The following plasmids were used to generate digoxigenin-labeled probes: mouse full-length Nkx6.2 cDNA and full-length iCre cDNA (kind gift from R. Sprengel, Max-Planck-Institute for Medical Research, Heidelberg, Germany). Details of the in situ hybridization techniques have been described previously (Pringle et al., 1996; Fruttiger et al., 1999).
Immunohistochemistry.
Immunohistochemical detection of calbindin (CB), CR, PV, SST, and NPY was performed on 30 μm floating cryosections of brains from adult mice (postnatal day 45 or older). Sections from embryonic brains (15 μm thickness) were collected directly on Superfrost plus slides (BDH Laboratory Supplies, Poole, UK). All sections were blocked in PBS containing 10% heat-inactivated sheep serum (Sigma, St. Louis, MO) and 0.1% Triton X-100 (Sigma) at room temperature for a minimum of 1 h. For immunodetection of GFP using the rat anti-GFP antibody, sections were not allowed to dry out at any stage before application of the primary antibody. Primary antibodies were applied overnight at 4°C. Secondary antibodies were applied for >60 min at room temperature. All antibodies were diluted in block solution. The Lhx6 and Cre primary antibodies were detected using a biotin-conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch, West Grove, PA), followed by 40 min incubation with a preformed avidin–biotin HRP complex (Vectastain ABC kit; Vector Laboratories, Burlingame, CA). Cyanine 3 tyramide (TSA Plus Fluorescence Systems technology; PerkinElmer, Wellesley, MA) was applied at 1:100 dilution, and the signal was developed for 5 min. After antibody treatment, sections were treated with Hoescht 33258 (1:104; Sigma) to detect cell nuclei and washed with PBS. Floating sections were transferred onto Superfrost plus slides (BDH Laboratory Supplies) and air dried. All sections were mounted in Citifluor (City University, UK) anti-fade reagent. Primary antibodies used were the following: rabbit anti-GFP (1:6000; Abcam, Cambridge, UK); rat anti-GFP IgG2a (1:1000; Nacalai Tesque, Kyoto, Japan); rabbit anti-Cre (1:1000; Novagen, San Diego, CA); rabbit anti-Gsh2 (1:2000; kind gift from K. Campbell, University of Cincinnati College of Medicine, Cincinnati, OH); rabbit anti-Nkx2.1 (1:500; Biopat Immunotechnologies, Caserta, Italy); rabbit anti-calbindin (1:2000; Swant, Bellizona, Switzerland); rabbit anti-calretinin (1:2000; Swant); mouse anti-calretinin (1:1000; Swant); rabbit anti-parvalbumin (1:2000; Chemicon, Temecula, CA); rabbit anti-NPY (1:2000; ImmunoStar, Hudson, WI), and rabbit anti-somatostatin (1:200; Peninsula Laboratories, Belmont, CA). The rabbit anti-Lhx6 (1:500) antibody used in this study has been described previously (Lavdas et al., 1999). The antibody had been raised against a mouse Lhx6 peptide spanning part of the second LIM domain and the spacer region between the LIM and homeobox domains. Control experiments demonstrated lack of cross-reactivity with other LIM homeodomain proteins, including LHX7 (our unpublished observations). Secondary antibodies used were AlexaFluor 488-conjugated, AlexaFluor 568-conjugated, and AlexaFluor 647-conjugated goat anti-rabbit IgG or goat anti-rat IgG or goat anti-mouse IgG (all used at 1:750; Invitrogen, Carlsbad, CA).
Quantification.
The extent of colocalization between GFP/YFP and the interneuron markers CB, CR, PV, SST, and NPY in each of the transgenic mice (Nkx2.1–Cre, Nkx6.2–Cre/Nkx2.1–Cre, Lhx6–Cre, Gsh2–Cre/Nkx2.1–Cre, and Emx1–Cre/Dbx1–Cre) carrying a Cre reporter allele (R26R–GFP or R26R–YFP) were determined as follows. Briefly, 30 μm coronal sections immunostained for the presence of GFP and one of the interneuron markers were photographed and assembled into montages of 850 × 1250 μm2 spanning the entire pial–ependymal extent of the motor or somatosensory cortex using Adobe Photoshop (Adobe Systems, San Jose, CA). At least three montages from sections >300 μm apart were taken from cortical regions between bregma levels 0.14 and −1.06 mm (approximately from the beginning of the hippocampus to the level of the anterior commissure). At least three animals older than 45 d were used for each genotype. Data are presented either as percentage GFP (in cases in which GFP was only expressed in interneuron populations) or as percentage interneuron marker (average ± SD).
Results
“Nkx2.1”-expressing precursors in the central and ventral MGE generate mainly calbindin-, parvalbumin-, and somatostatin-expressing interneurons
The homeobox transcription factor Nkx2.1 (also known as Ttf1 or T/ebp) is one of the key regulators that specify the MGE (Kimura et al., 1996; Sussel et al., 1999). We took advantage of a P1-derived artificial chromosome (PAC) transgenic mouse line that expresses Cre recombinase under transcriptional control of the Nkx2.1 gene to lineally label MGE precursors and identify interneurons that are generated from this region (Kessaris et al., 2006). When crossed to the Cre reporter mouse R26R–GFP (Mao et al., 2001), there is near complete recombination and activation of GFP in the neuroepithelium of the MGE and the anterior endopenducular area (AEP) (Fig. 1 A–D). Unlike the endogenous Nkx2.1 gene, however, there is no expression of the GFP reporter gene in the most dorsal region of the MGE (dMGE) (Fig. 1 E–G), most likely attributable to failure of the specific PAC to support strong expression of Cre in this region (Fig. 1 H–J). In addition, unlike the endogenous gene, Cre expression remained on at high levels in migrating interneurons until early postnatal stages (Fig. 1 I and data not shown). The transcription factor Lhx6, which identifies postmitotic neurons generated in the MGE and is required for migration to the cortex (Grigoriou et al., 1998; Lavdas et al., 1999; Sussel et al., 1999; Marín et al., 2000; Alifragis et al., 2004; Choi et al., 2005; Liodis et al., 2007), is abundantly expressed throughout this region in Nkx2.1–Cre/R26R–GFP embryos (Fig. 1 K). At early stages, before extensive migration of MGE cells occurs, all Lhx6+ cells express GFP in the pallidal mantle region. However, there are Lhx6+ cells near the dMGE that are negative for GFP, confirming the poor recombination of the R26R–GFP allele in this region of the ventral zone (Fig. 1 K–O). Within the cortex, the majority of GFP+ cells coexpress Lhx6. GFP+Lhx6-negative (Lhx6−) cells are likely to represent early oligodendrocyte progenitors that emigrate to the cortex from subpallial regions (Wichterle et al., 2001; Kessaris et al., 2006).
Figure 1.
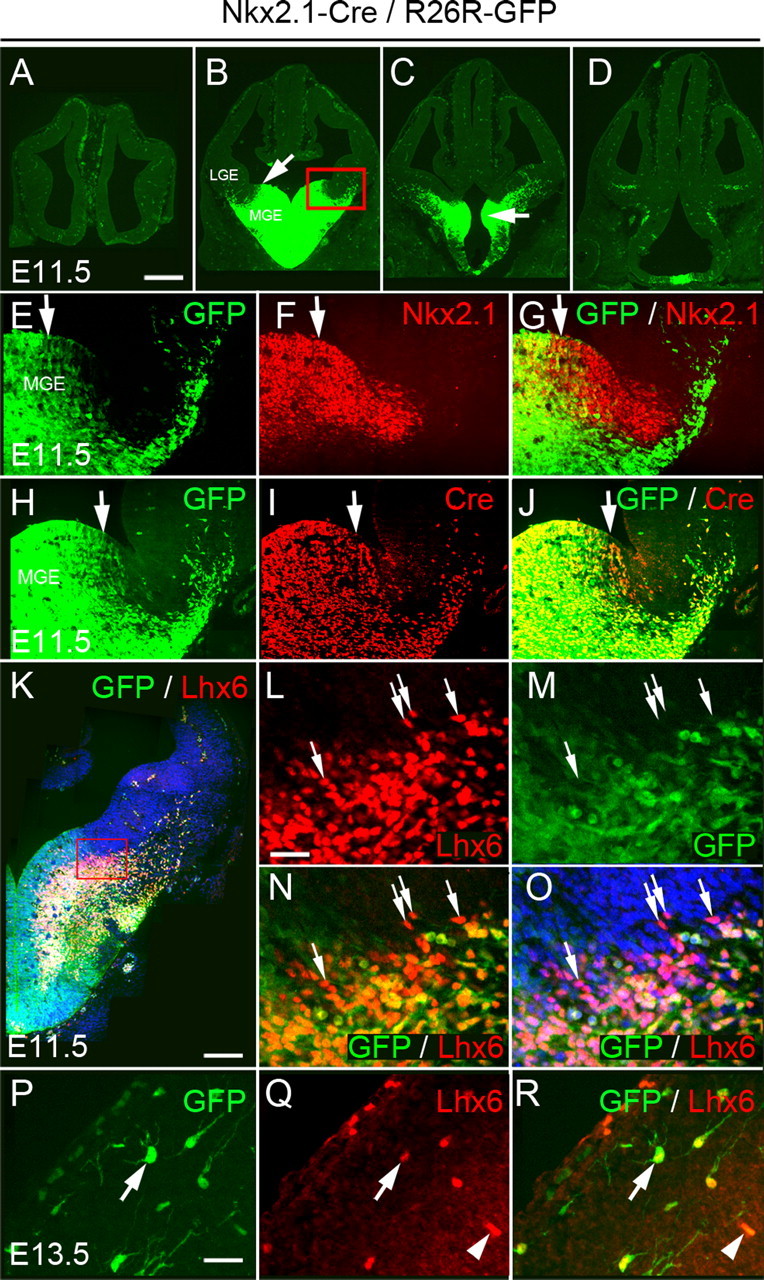
Activity of Cre recombinase in Nkx2.1–Cre/R26R–GFP transgenic embryos. A–D, Expression of GFP at four rostrocaudal levels in embryonic day 11.5 (E11.5) transgenic embryos. Arrow in B indicates the dorsal limit of GFP expression. Arrow in C shows expression in the AEP. The red box in B indicates the area shown in E–J. E–G, Comparison between the expression of the endogenous Nkx2.1 gene and GFP at E11.5. Nkx2.1 expression extends farther dorsally than GFP. H–J, Expression of Cre and GFP at E11.5. K–O, Expression of Lhx6 in E11.5 Nkx2.1–Cre/R26R–GFP transgenic embryos. Arrows indicate Lhx6+GFP− cells in the dMGE (L–O). P–Q, GFP+ cells in the cortex express Lhx6 (arrows). Arrowheads indicate blood vessels. Scale bars: A–D, 350 μm; E–J, 60 μm; K, 1250 μm; L–O, 25 μm; P–R, 15 μm.
We took advantage of the reduced expression of Nkx2.1–Cre in the dMGE to identify interneurons generated in the central and ventral neuroepithelium of the MGE. We immunostained forebrain sections from adult Nkx2.1–Cre/R26R–GFP transgenic mice for GFP and a panel of markers thought to collectively identify the vast majority of GABAergic interneurons present in the mouse cortex (Gonchar and Burkhalter, 1997). These markers included the calcium binding proteins CB, CR, and PV and the neuropeptides SST and NPY. Unlike CR, PV, SST, and NPY, which are expressed predominantly in GABAergic interneurons, the CB+ population represents a heterogenous mixture of GABAergic and non-GABAergic cortical cells (Gonchar and Burkhalter, 1997). Non-GABAergic CB+ cells are present in superficial layers, show weak expression of CB, and were excluded from this study. GFP colocalized with all five interneuron markers examined (Fig. 2 A–O), but the extent of colocalization varied among different interneuron subtypes. PV- and SST-expressing cells, which are thought to represent primarily nonoverlapping populations of interneurons in the cortex, were the most abundant interneuron subtypes generated from Nkx2.1-expressing precursors (∼45 and ∼35%, respectively, of the GFP+ populations) (Fig. 2 R). The majority of PV+ and SST+ cells present in the adult motor and somatosensory cortex were derived from Nkx2.1-expressing precursors (70–80%) (Fig. 2 P). CB+ cells represented ∼30% of the GFP+ cells in the two cortical areas examined (motor and somatosensory), and CB+GFP+ cells represented ∼80% of the total CB+ cells in the cortex (Fig. 2 P–R). Finally, a small number of Nkx2.1-derived GFP+ cells coexpressed CR or NPY (∼5%) (Fig. 2 R) and contributed ∼15–20% to the total number of CR+ and NPY+ interneuron populations in the cortex (Fig. 2 P). Virtually all CR+GFP+ cells in the Nkx2.1–Cre/R26R–GFP transgenic mice had a stellate morphology, in contrast to the CR+GFP− neurons, which had a characteristic bipolar morphology, and did not express GFP (Fig. 2 B).
Figure 2.
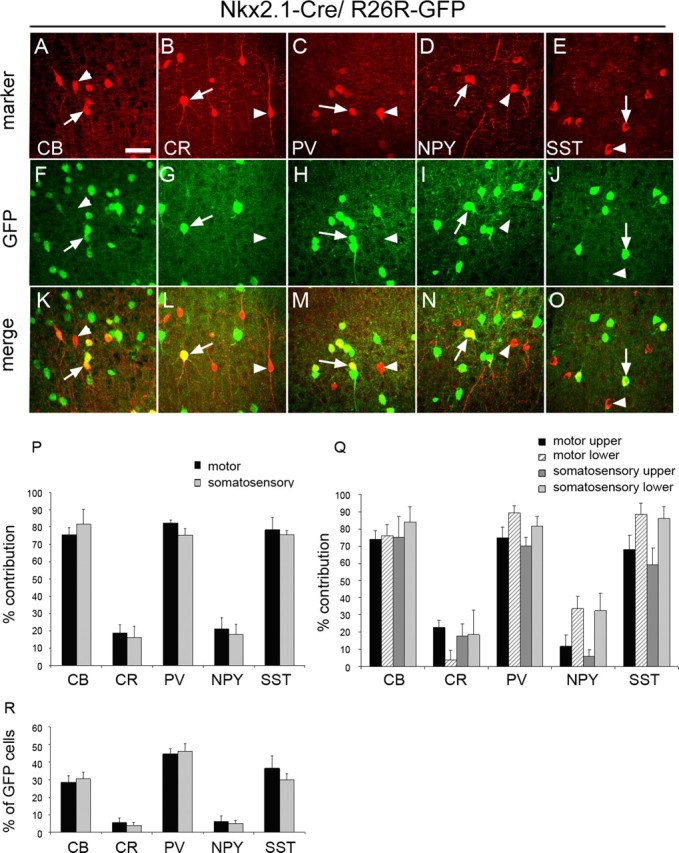
Cortical interneurons derived from Nkx2.1-expressing precursors. A–O, Coexpression of GFP with CB, CR, PV, NPY, and SST. Arrows indicate interneurons coexpressing GFP. Arrowheads point to interneurons that are negative for GFP. P, The contribution of Nkx2.1-expressing precursors to interneuron populations expressing CB, CR, PV, NPY, or SST in the motor and somatosensory cortex. The extent of colocalization between GFP and each of the five markers was quantified, and the data are presented as percentage of the total number of cells expressing each of the five markers. Q, The contribution of Nkx2.1-expressing precursors to interneuron populations in the upper and lower layers of the motor and somatosensory cortex. R, The number of Nkx2.1-derived cells coexpressing GFP and one of the markers CB, CR, PV, NPY, or SST are presented as a percentage of the total number of GFP+ cells. Scale bar: (in A) A–O, 20 μm.
Nkx6.2-expressing precursors in the dMGE generate distinct interneuron subtypes that migrate to the cortex
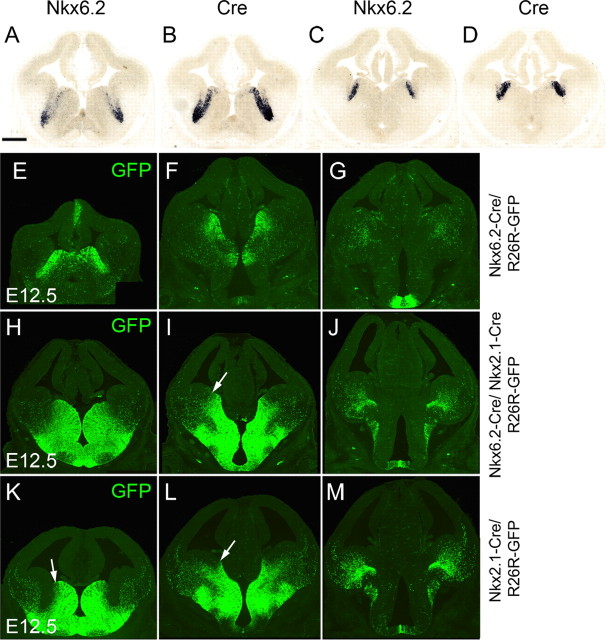
To examine the contribution of the dMGE to interneuron populations in the neocortex, we took advantage of the expression of the homeobox-encoding gene Nkx6.2. Nkx6.2 transcripts can be observed at the sulcus between the MGE and LGE, with a strong expression in the dMGE (Stenman et al., 2003) (Fig. 3 A–D) (supplemental Fig. 1A–D, available at www.jneurosci.org as supplemental material). We generated a transgenic mouse line expressing Cre under control of the Nkx6.2 gene using BAC recombineering and transgenic mouse technology (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Expression of Cre in Nkx6.2–Cre/R26R–GFP transgenic embryos recapitulated the endogenous Nkx6.2 pattern (Fig. 3 A–D) (supplemental Fig. 1A–D, available at www.jneurosci.org as supplemental material) and activation of GFP in Nkx6.2–Cre/R26R–GFP transgenic embryos was observed predominantly in the dMGE (Fig. 3 E–G). However, GFP was also observed in scattered neuroepithelial cells located more ventrally in the central and ventral MGE (Fig. 3 E–G) in which neither Nkx6.2 nor Cre could be detected (Fig. 3 A–D). The extent of GFP expression in the ventral MGE was dependent on the Cre reporter allele used, with R26R–YFP showing a wider activation in this region (supplemental Fig. 1E, available at www.jneurosci.org as supplemental material). The scattered GFP/YFP expression in the central and ventral MGE presumably reflects an early expression of the gene. It is unclear whether it results from an early ectopic expression of the Nkx6.2–Cre transgene or a transient expression of Nkx6.2. The latter possibility would be analogous to the spinal cord in which early expression of Nkx6.2 occurs in a wide area of the ventral neuroepithelium and later narrows to a small domain (Vallstedt et al., 2001). Nevertheless, because of the scattered activation of GFP in the entire MGE, we combined the activities of Nkx2.1–Cre and Nkx6.2–Cre to determine the additive contribution of these regions to cortical interneuron populations. Activation of GFP in these double-transgenic mice can be observed throughout the entire MGE and AEP (Fig. 3 H–J). Expression was undetectable in the LGE and CGE in all anteroposterior regions. This pattern of GFP activation is clearly wider than that of Nkx2.1–Cre alone, confirming that the two transgenic mice have additive expression domains (Fig. 3, compare H–J with K–M).
Figure 3.
Activity of Cre recombinase in Nkx6.2–Cre/R26R–GFP and compound Nkx6.2–Cre/Nkx2.1–Cre/R26R–GFP transgenic embryos. A–D, Expression of Nkx6.2 and Cre transcripts in E12.5 Nkx6.2–Cre transgenic embryos. E–G, Expression of GFP at three rostrocaudal levels in E12.5 Nkx6.2–Cre/R26R–GFP transgenic embryos. H–J, Expression of GFP at three rostrocaudal levels in E12.5 Nkx6.2–Cre/Nkx2.1–Cre/R26R–GFP transgenic embryos and comparison with equivalent sections from Nkx2.1–Cre/R26R–GFP transgenic embryos (K–M). Scale bar: (in A) A–M, 300 μm.
We made use of single and compound transgenic mice expressing Cre in the dMGE (Nkx6.2–Cre), the entire MGE (Nkx6.2–Cre/Nkx2.1–Cre) or the central and ventral MGE (Nkx2.1–Cre; these mice show some labeling in the dMGE) to examine the contribution of subdivisions of the MGE germinal region to cortical interneuron populations in the adult cortex. Whereas in single Nkx6.2–Cre/R26R–GFP transgenic mice only small subsets of interneurons coexpressed GFP and one of the interneuron markers examined (CB, CR, PV, NPY, or SST), in Nkx6.2–Cre/Nkx2.1–Cre/R26R–GFP double transgenic mice, the majority of CB+, PV+, and SST+ cells were labeled with GFP (Fig. 4 A). This suggests that the bulk of CB+, PV+, and SST+ interneurons are generated in the central and ventral MGE. However, the extent of colocalization between CR and GFP or NPY and GFP was remarkably similar between single Nkx6.2–Cre/R26R–GFP and double Nkx6.2–Cre/Nkx2.1–Cre/R26R–GFP transgenic mice (Student's t test, p < 0.99 for CR and p < 0.03 for NPY in the motor cortex) (Fig. 4 A), suggesting that most, if not all, of the MGE-derived CR+ and NPY+ interneurons originate in the dorsal region of the MGE that expresses Nkx6.2. A comparison between Nkx2.1–Cre +/Nkx6.2–Cre + precursors versus Nkx2.1–Cre+ precursors revealed significant differences in the contribution of the respective precursor pools to CR+, NPY+, and SST+ interneuron populations in the cortex (Fig. 4 B). The most significant differences were observed in the extent of colocalization between GFP and the interneuron markers CR and SST in the upper layers of the motor cortex (Student's t test, p < 0.0007 and p < 0.0003, respectively) (Fig. 4 C) and NPY in the lower layers of the motor and somatosensory cortex (Student's t test, p < 0.0007 and p < 0.0006) (Fig. 4 D). Smaller differences were observed in CR+, PV+, and SST+ cells in the upper layers of the somatosensory cortex (Fig. 4 C) (Student's t test, p < 0.0027, p < 0.0024, and p < 0.0015, respectively) and CR+ cells in the lower levels of the motor cortex (p < 0.0036) (Fig. 4 D). Collectively, the data suggest that Nkx6.2-expressing precursors in the dMGE contribute mainly CR-, SST-, and NPY-expressing interneurons to the neocortex. In addition to interneurons, Nkx6.2–Cre/R26R–GFP and Nkx2.1–Cre/Nkx6.2–Cre/R26R–GFP transgenic mice showed extensive colocalization between GFP and the oligodendrocyte lineage marker Sox10 (sex-determining region Y-box containing gene 10) (data not shown). This recapitulates the expression of the endogenous Nkx6.2 gene in oligodendrocytes undergoing maturation (Awatramani et al., 1997).
Figure 4.
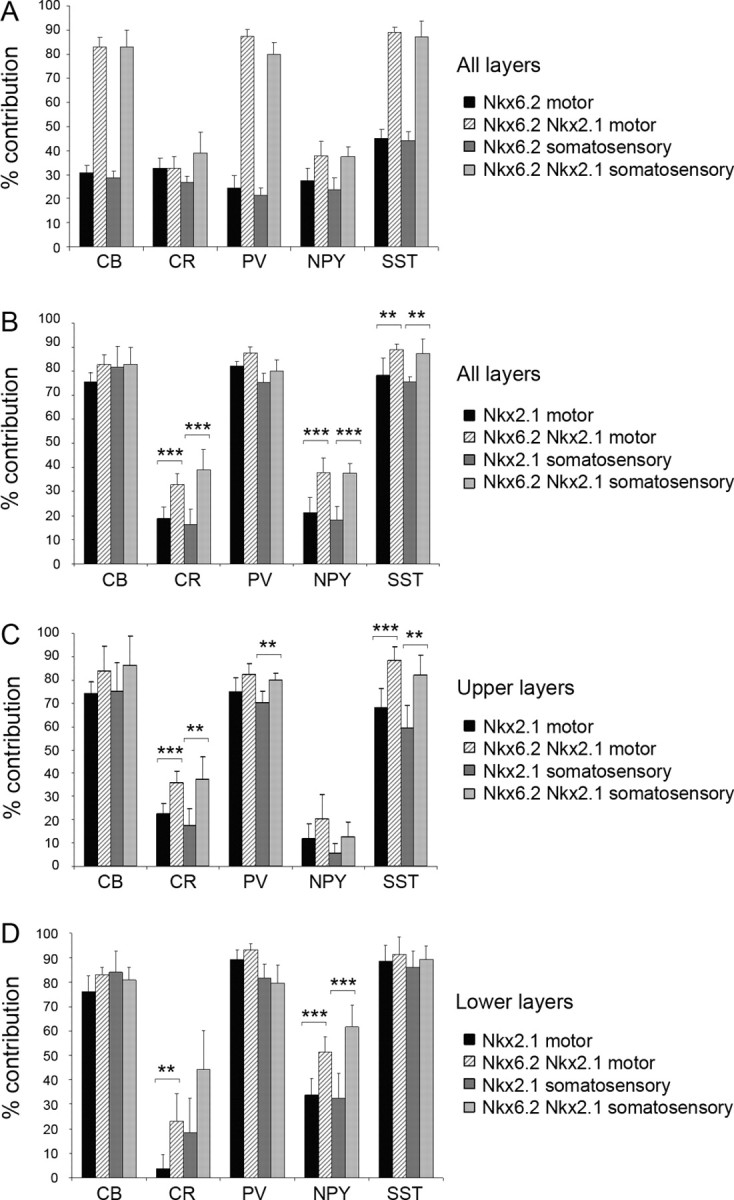
Cortical interneurons derived from Nkx6.2-, Nkx2.1/Nkx6.2-, and Nkx2.1-expressing precursors. A, The coexpression between GFP and one of the markers CB, CR, PV, NPY, and SST in Nkx6.2–Cre/R26R–GFP and Nkx6.2–Cre/Nkx2.1–Cre/R26R–GFP transgenic mice was quantified and presented as percentage of the total number of cells expressing the markers in the motor and somatosensory cortex. B–D, The contribution of Nkx2.1–Cre-expressing precursors to interneuron populations expressing CB, CR, PV, NPY, and SST was compared with that of Nkx2.1-Cre/Nkx6.2-Cre-expressing precursors (B) in upper (C) and lower (D) levels of the motor and somatosensory cortex. **p < 0.004; ***p < 0.001.
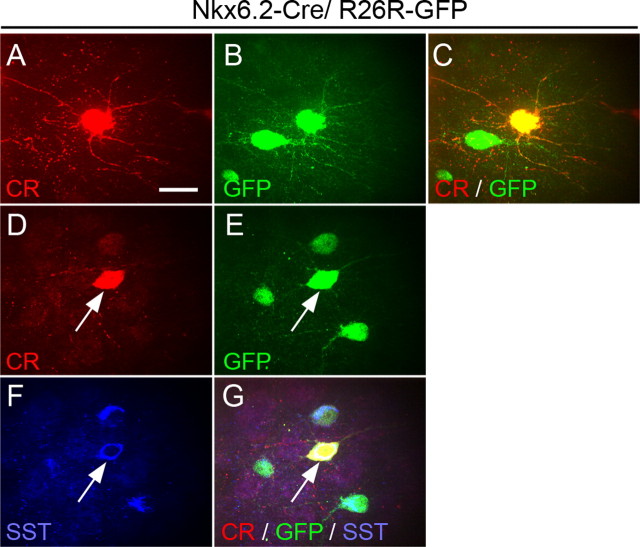
Recent work has identified a subpopulation of CR+ cells in the mouse cortex that coexpress SST and have morphological and physiological characteristics of Martinotti cells (Xu et al., 2006). They represent 33% of the total CR+ population in the cortical areas examined (Xu et al., 2006). This is comparable with the contribution of Nkx6.2-expressing precursors to the CR+ cortical interneuron population. We therefore examined the characteristics of CR+ cells in Nkx6.2–Cre/R26R–GFP transgenic mice. We find that CR+GFP+ interneurons have a stellate morphology (Fig. 5 A–C). Bipolar CR+ interneurons did not express GFP, indicating that they are generated elsewhere. We characterized and quantified the colocalization of CR, SST, and GFP in brain sections of adult Nkx6.2–Cre/R26R–GFP mice (Fig. 5 D–G). We find that 82.5 ± 2.6% of CR+SST+ cells present in the motor and somatosensory cortex coexpress GFP, and 93.3 ± 3.3% of CR+GFP+ cells coexpress SST. These data indicate that CR+ interneurons generated from Nkx6.2-expressing precursors in the dMGE correspond to putative CR+ SST+ Martinotti cells and that these cells are generated from Nkx6.2-expressing precursors in the dMGE.
Figure 5.
Coexpression of CR, SST, and GFP in cortical interneurons of Nkx6.2–Cre/R26R–GFP transgenic mice. A–C, CR+GFP+ cells have a stellate morphology. D–G, Coexpression of SST and GFP in all CR+ cortical interneurons derived from Nkx6.2-expressing precursors. Scale bar, 10 μm.
Cortical interneurons generated from Lhx6-expressing precursors
Our data so far showed that the majority of CB+, SST+, and PV+ interneurons are indeed generated from Nkx2.1–Cre + and Nkx6.2–Cre + pallidal precursors. However, there were small numbers of these cells (10–20% in Nkx2.1–Cre/Nkx6.2–Cre/R26R–GFP transgenic mice) that remained unaccounted for, suggesting that they may be generated outside the MGE. Alternatively, the absence of GFP expression in these cells may be attributable to inefficient recombination in the MGE of our Cre transgenic mice. To distinguish between these two possibilities, we generated BAC transgenic mice that express Cre under transcriptional control of Lhx6. Lhx6 is thought to be expressed in all MGE-derived neurons as they emerge from the neuroepithelium, and expression persists in PV+ and SST+ interneurons in the adult cortex (Cobos et al., 2005, 2006; Liodis et al., 2007).
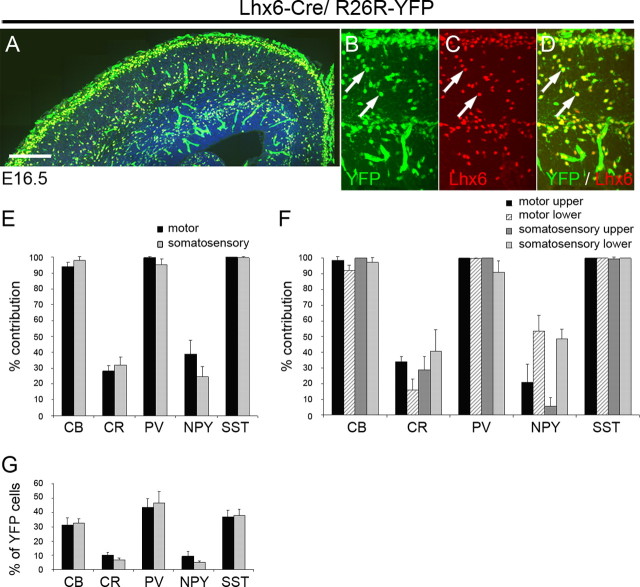
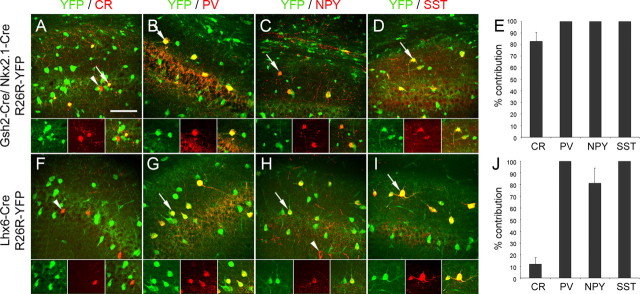
When crossed to the R26R–YFP reporter mice, activation of YFP in Lhx6–Cre/R26R–YFP transgenic mice was observed in all Lhx6 + cells, indicating that expression of the transgene recapitulates the endogenous pattern (Fig. 6 A–D). Activation of YFP was also observed in blood vessels scattered throughout the brain. This may reflect an early expression of Lhx6 or the Cre transgene in the developing vasculature. To identify interneuron subtypes that are generated from Lhx6 + cells, we immunolabeled sections from adult Lhx6–Cre/R26R–YFP brains for YFP and the interneuron markers described in previous sections. We observed a striking 100% contribution to the SST+ population and a near 100% contribution to PV+ and CB+ cells in both motor and somatosensory cortex (Fig. 6 E), indicating that these two interneuron populations are generated exclusively from Lhx6 + precursors. Interneurons expressing CB, PV, or SST were the predominant interneuron subtypes generated from Lhx6-expressing precursors (Fig. 6 G). The contribution of Lhx6 + cells to CR+ and NPY+ populations was comparable with that of Nkx6.2 + precursors (compare Figs. 4 A,B, 6 E). In the motor cortex, Lhx6-derived CR+ interneurons had a stellate morphology and were located mainly in the upper layers of the cortex, whereas NPY+YFP+ interneurons were concentrated in lower layers (Fig. 6 F). Immunolabeling for CR, SST, and YFP showed that 100% of CR+SST+ cells in the motor and somatosensory cortex coexpress YFP and 100% of CR+GFP+ cells coexpress SST. These results together with the Nkx6.2–Cre data indicate that cortical CR+ SST+ Martinotti cells are generated from Nkx6.2/Lhx6-expressing precursors.
Figure 6.
Activity of Cre recombinase in Lhx6–Cre/R26R–YFP transgenic embryos and contribution of Lhx6-expressing precursors to cortical interneuron populations in the adult mouse. A–D, Expression of GFP and Lhx6 in E16.6 transgenic embryos. E, F, The contribution of Lhx6-expressing precursors to interneuron populations expressing CB, CR, PV, NPY, or SST in the motor and somatosensory cortex. The extent of colocalization between YFP and each of the five markers was quantified, and the data are presented as percentage of the total number of cells expressing each of the five markers. G, The number of Lhx6-derived cells coexpressing YFP and one of the markers CB, CR, PV, NPY, or SST are presented as a percentage of the total number of YFP+ cells. Scale bar: A, 100 μm; B–D, 20 μm.
Subpallial origin of calretinin- and neuropeptide Y-expressing neocortical interneurons
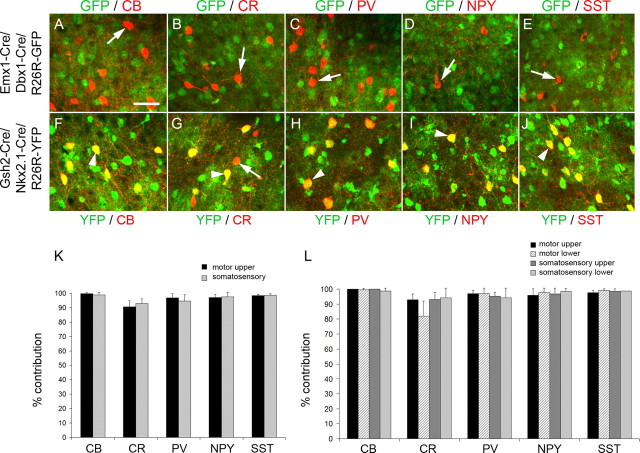
To determine the embryonic origin of CR+ and NPY+ cortical interneurons that remained unlabeled in the Cre transgenic mice we described thus far, we fate mapped the remaining pallial and subpallial neuroepithelial regions. To identify interneurons originating in the pallium, we used mice expressing Cre under transcriptional control of the homeobox-encoding genes Emx1 and Dbx1 to mark pallial precursors (supplemental Fig. 3, available at www.jneurosci.org as supplemental material) (Fogarty et al., 2005; Kessaris et al., 2006). Expression of Emx1 is restricted to the dorsal pallium, whereas expression of Dbx1 is restricted to the pallial–subpallial boundary and identifies the lateral and ventral pallial regions (Simeone et al., 1992; Lu et al., 1994). There was no colocalization between GFP/YFP and any of the interneuron markers examined in cortical sections of adult Emx1–Cre/Dbx1–Cre/R26R–GFP mice in which the pallium and all its cell derivatives express GFP (Fig. 7 A–E), reinforcing the view that the pallium does not generate interneurons at any stage in vivo (Iwasato et al., 2000; Gorski et al., 2002). To identify interneurons originating in all subpallial regions, we used another transgenic mouse that expresses Cre recombinase under transcriptional control of the Gsh2 gene (Kessaris et al., 2006). Although expression of Gsh2 is observed throughout the subpallium (Hsieh-Li et al., 1995), in our transgenic mice, expression of Cre and activation of the YFP reporter was poor in the MGE (Kessaris et al., 2006). Indeed, in single transgenic mice (Gsh2–Cre/R26R–YFP), the contribution of Gsh2–Cre + precursors to MGE-derived interneuron populations such as CB+, PV+, and SST+ interneurons was significantly lower than the contribution of precursors expressing Cre in the MGE (Nkx2.1–Cre or Nkx2.1–Cre/Nkx6.2–Cre, or Lhx6–Cre), confirming that Cre expression and activation of the reporter gene in this region is inefficient (supplemental Fig. 4 and see supplemental Fig. 5 for Cre expression in the LGE/CGE, available at www.jneurosci.org as supplemental material). We therefore used compound Gsh2–Cre/Nkx2.1–Cre/R26R–YFP transgenic mice to label the majority of the subpallial neuroepithelium and identify emerging interneurons (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). In Nkx2.1–Cre/Gsh2–Cre/R26R–YFP transgenic mice, nearly all interneurons identified in the cortex coexpressed YFP (Fig. 7 F–L). The complete colocalization of all interneuron markers with YFP in the Gsh2–Cre/Nkx2.1–Cre/R26R–YFP transgenic mice and the partial colocalization of NPY and CR with YFP in Lhx6–Cre/R26R–YFP transgenic mice indicate that the majority of CR- and NPY-expressing interneurons in the neocortex are generated outside the MGE, possibly from precursors residing within the LGE/CGE (compare Figs. 6 E, 7 K). These included all bipolar CR+ interneurons.
Figure 7.
Cortical interneurons derived from Emx1/Dbx1- and Gsh2/Nkx2.1-expressing precursors. A–J, Colocalization between GFP/YFP and the interneuron markers CB, CR, PV, NPY, and SST in adult Emx1–Cre/Dbx1–Cre/R26R–GFP transgenic mice (A–E) and Gsh2–Cre/Nkx2.1–Cre/R26R–YFP transgenic mice (F–J). Arrowheads indicate colocalization between GFP/YFP and the interneuron marker. Arrows indicate interneurons that do not express GFP/YFP. K–L, The contribution of Gsh2/Nkx2.1-expressing precursors to interneuron populations expressing CB, CR, PV, NPY, or SST in the motor and somatosensory cortex. The colocalization between GFP and each of the five markers was quantified, and the data are presented as percentage of the total number of cells expressing each of the five markers. Scale bar: (in A) A–J, 20 μm.
The embryonic origin of hippocampal interneurons
Previous studies had suggested that hippocampal interneurons generated during embryonic development are derived predominantly from subpallial regions and migrate tangentially into the cortex (Pleasure et al., 2000; Wichterle et al., 2001). Using our panel of transgenic mice, we examined the contribution of the pallium (Emx1 +/Dbx1 +) and the subpallium (Gsh2 +/Nkx2.1 +) as well as the contribution of the MGE alone (Lhx6+) to interneurons in the CA1 area of the hippocampus. Pallial precursors did not generate any interneurons in CA1 identified by immunohistochemical staining for GFP and CR, PV, SST, or NPY (data not shown). The majority of the CR+ neurons in the dentate gyrus of the hippocampus were positive for GFP in the Emx1–Cre/Dbx1–Cre/R26R–GFP transgenic mice, indicating a pallial origin. These CR+ cells represent a transient stage of adult newly born granule neurons (data not shown) (Brandt et al., 2003). Gsh2 +/Nkx2.1 + precursors generated nearly all of the CR+, PV+, NPY+, and SST+ interneurons in the CA1 area of the hippocampus (Fig. 8 A–E), confirming the exclusive subpallial origin of hippocampal interneurons. The contribution of Lhx6 + precursor populations to hippocampal interneurons was similar to that of the neocortex. All SST+ and PV+ interneurons were positive for YFP in Lhx6–Cre/R26R–YFP transgenic mice, indicating an MGE origin for these cells (Fig. 8 F–J). Contribution to the CR+ populations of the hippocampus was also comparable with that of the neocortex with only a small number of these cells being generated in the MGE. Unlike the neocortex, however, Lhx6 + precursors generated most of the NPY+ interneurons found in the hippocampal CA1 area in Lhx6–Cre/R26R–YFP transgenic mice (Fig. 8 F–J).
Figure 8.
Interneurons in the hippocampus derived from Gsh2/Nkx2.1 and Lhx6-expressing precursors. A–D, F–I, Colocalization between YFP and the interneuron markers CR, PV, NPY, and SST in CA1 region of the hippocampus in adult Gsh2–Cre/Nkx2.1–Cre/R26R–YFP transgenic mice (A–D) and adult Lhx6–Cre/R26R–YFP transgenic mice (F–I). Arrows indicate colocalization between GFP/YFP and the interneuron marker. Arrowheads indicate interneurons that do not express GFP/YFP. Cells indicated by arrows/arrowheads are shown in the insets. E, J, Quantification of the contribution of Gsh2/Nkx2.1-expressing precursors (E) and Lhx6-expressing precursors (J) to various interneuron populations in the CA1 area of the hippocampus. Data are presented as percentage of the total number of cells expressing each of the four markers. Scale bar: (in A) A–D, F–I, 40 μm.
Discussion
Using Cre–lox fate mapping in transgenic mice, we demonstrate that subpallial precursor domains identified by expression of molecular markers generate neurochemically distinct interneuron subtypes and thus contribute to the generation of interneuron diversity in the adult cortex.
(1) SST+, PV+, and CB+ interneurons present in the adult cortex are generated from Nkx2.1/Lhx6-expressing precursors in the MGE. (2) All CR+SST+ Martinotti cells are generated from dMGE precursors that express Nkx6.2 and Lhx6. (3) Small but significant numbers of NPY+ interneurons are generated from the dMGE, and these are distributed in lower cortical layers. (4) The majority of CR+ and NPY+ interneurons are generated outside the pallidal neuroepithelium from precursors expressing Gsh2. The data are summarized in Table 1.
Table 1.
The origin of interneuron subtypes from subpallial neuroepithelial domains identified by genetic fate mapping
| Subpallial germinal zone | Cre mice used for in vivo fate mapping (level of Cre activity) | Interneuron subtype |
|---|---|---|
| Central and ventral MGE | Nkx2.1–Cre (high) | SST+CR− |
| Lhx6–Cre (high) | PV+ | |
| Gsh2–Cre (low) | CB+ | |
| Nkx6.2–Cre (low) | ||
| Dorsal MGE | Nkx6.2–Cre (high) | CR+SST+ (Martinotti) |
| Lhx6–Cre (high) | NPY+ | |
| Nkx2.1–Cre (low) | ||
| Gsh2–Cre (low) | ||
| LGE/dorsal CGE | Gsh2–Cre (high) | CR+SST− (bipolar) |
| NPY+ |
Transgenic mice expressing Cre recombinase in distinct and overlapping domains were used to fate map neuroepithelial stem cell zones in vivo. Neurochemically distinct interneuron subtypes originate from discrete neuroepithelial domains identified by expression of molecular markers.
Neuroepithelial origin of cortical interneurons and generation of diversity in the adult cortex
The origin of pallial interneurons has been controversial. Previous work had suggested that the majority of GABAergic interneurons in humans are generated from cortical precursors (Letinic et al., 2002), and in vitro studies in rodents have shown a capacity for cortical neuroepithelial cells to generate GABAergic interneurons (Gotz et al., 1995; He et al., 2001; Bellion et al., 2003; Gulacsi and Lillien, 2003). However, the bulk of data support a major subpallial origin for cortical interneurons in rodents (for review, see Wonders and Anderson, 2006). In our studies, the absence of colocalization between GFP and any of the interneuron markers examined in R26R–GFP mice expressing both Emx1–Cre and Dbx1–Cre, and the near complete colocalization of all interneuron markers with YFP in R26R–YFP mice expressing Cre in the subpallium support the notion of an exclusive subpallial origin.
Cortical interneurons are heterogenous with respect to morphology, electrophysiology, and neurochemical properties. It is unknown how this diversity arises. One possibility is that diversity is generated through regional genetic patterning of the subpallial neuroepithelium, as is well established for more caudal regions of the CNS, such as the brainstem and spinal cord. Indeed, morphologically distinct subdivisions of the subpallial neuroepithelium generate different subsets of interneurons. However, this cannot account for the large spectrum of interneuron subtypes observed in the adult cortex. The demonstration that cortical Martinotti cells that coexpress CR and SST are generated from a small dorsal subdomain of the MGE neuroepithelium (identified by expression of Nkx6.2) suggests that genetic patterning may parcellate the neuroepithelium into smaller precursor pools, restricting their neurogenic potential and contributing toward larger precursor heterogeneity. Recent evidence has shown that there are also temporal shifts in interneuron subtype generation from embryonic telencephalic precursors (Miyoshi et al., 2007). Together, the spatial and temporal control of neuronal subtype specification from embryonic precursor cells may account for the large interneuron diversity observed in the adult cortex. The extent to which interneurons diversify further at postnatal stages remains unclear.
The embryonic origin of PV+, SST+, and CB+ GABAergic interneurons
PV+ and SST+ interneurons represent two nonoverlapping populations in the rodent cortex (Kubota et al., 1994; Gonchar and Burkhalter, 1997), whereas expression of CB in nonpyramidal cells is thought to be associated mainly with the SST-expressing group (Gonchar and Burkhalter, 1997). Several studies have shown that PV+ interneurons are generated in the MGE and represent the majority of interneurons emerging from this region [data range from 30% (Nery et al., 2002), 57% (Butt et al., 2005), 67% (Valcanis and Tan, 2003), and 70% (Wichterle et al., 2001)]. Transplantation studies examining the origins of SST+ and CB+ cells in the telencephalon had shown that 26–35% of MGE-derived interneurons express SST (Wichterle et al., 2001; Nery et al., 2002; Butt et al., 2005) and 13% express CB (Nery et al., 2002). Our data agree partly with these findings. PV+ and SST+ cells represent ∼45 and 35%, respectively, of the interneurons generated from Nkx2.1 + or Lhx6 + cells. CB+ cells show very similar distribution with PV- and SST-expressing interneurons and may well correspond to either or both of those interneuron populations. More importantly, however, we demonstrate that all PV+, CB+, and SST+ interneurons are generated from Lhx6-expressing precursors. In vivo transplantation studies had previously identified a small percentage of CGE-derived cells expressing PV (2.8%) (Nery et al., 2002), 27–35% expressing SST (Nery et al., 2002; Butt et al., 2005), and 17% expressing CB. These cells may correspond to MGE-derived interneurons migrating caudally into the cortex. Alternatively, they may originate from Lhx6 + cells present in the ventral CGE, which is a caudal extension of the MGE. We cannot distinguish between these two possibilities. Nevertheless, our data together with data from Nkx2.1 null mutant mice, which fail to develop PV+ or SST+ cells in vitro (Xu et al., 2004), and Lhx6 null mice, which show a dramatic reduction of PV+ and SST+ cells (Liodis et al., 2007), demonstrate that these two interneuron populations of the cortex originate entirely from Nkx2.1- and Lhx6-expressing cells in the developing subpallium. Although expression of Nkx2.1 is abolished soon after interneurons emerge from the MGE (Sussel et al., 1999), expression of Lhx6 persists in most PV- and SST-expressing cells in the adult cortex (Cobos et al., 2005, 2006; Liodis et al., 2007). Thus, expression of Lhx6 identifies predominantly PV- and SST-expressing interneurons at all embryonic and adult stages.
The embryonic origin of NPY+ GABAergic interneurons
Expression of NPY in the cortex can be highly dynamic: it can easily be induced by neuronal excitation or epileptiform activity and is transiently activated by cortical pyramidal cells at early embryonic stages (Gall et al., 1990; Schwarzer et al., 1996). In addition to this contingent expression of the neurotransmitter in cortical neurons, constitutive expression can also be observed in certain interneuron populations of the cortex. In the rat cortex, NPY primarily colocalizes with SST (Kawaguchi and Kubota, 1997). In Nkx2.1 null mutant mice in which the MGE is respecified into LGE, there is a lack NPY+ cells at birth (Anderson et al., 2001), and telencephalic cultures from these mice fail to develop NPY+ interneurons (Xu et al., 2004). However, our in vivo lineage tracing work demonstrates that, compared with SST, only a small number of cells generated from the MGE express NPY. These originate mainly in the dMGE, in the region of the neuroepithelium that expresses Nkx6.2. They represent a significant proportion of NPY+ cells in the cortex and are localized mainly in deep cortical layers. The majority of NPY+ interneurons are generated outside the MGE, presumably from the LGE and/or CGE. Our data agree with transplantation studies that showed a much higher frequency of NPY+ cells emerging from the CGE rather than the MGE (Butt et al., 2005).
The embryonic origin of CR+ GABAergic interneurons
Similar to NPY-expressing interneurons, the majority of CR+ cells are generated outside the MGE, possibly within the CGE (Xu et al., 2004; Butt et al., 2005). Most CR+ interneurons have bipolar morphology and reside in the outer layers of the cortex. They were not labeled with GFP/YFP in Nkx2.1–Cre/R26R–GFP, Nkx6.2–Cre/Nkx2.1–Cre/R26R–GFP, or Lhx6–Cre/R26R–YFP transgenic mice.
A small but significant population of CR+ interneurons are derived from Nkx6.2-expressing precursors. These cells coexpress SST and have a stellate morphology thus correspond to the recently identified subpopulation of Martinotti cells (Xu et al., 2006). They are derived entirely from Nkx6.2-expressing subpallial precursors most likely in the dMGE. It is conceivable, however, that CR+ or NPY+ cells may arise from Nkx6.2-expressing precursors in the preoptic area that do not express the Nkx2.1–Cre transgene (Fig. 1 C). Although migration of interneurons from this region to the cortex has not been demonstrated, additional research is required to examine this possibility.
The origin of hippocampal interneurons in rodents
Interneurons of the hippocampus are generated during embryogenesis. They are born earlier than pyramidal cells (Soriano et al., 1989a,b), and evidence shows that they originate from different precursors. Projection neurons are generated from hippocampal neuroepithelial cells, whereas interneurons are generated in the subpallium and migrate tangentially to reach the cortex (Pleasure et al., 2000; Wichterle et al., 2001). We confirm that all CR+, PV+, NPY+, and SST+ interneurons in the CA1 area are generated from the subpallial neuroepithelium. PV+, NPY+, and SST+ cells are born from Lhx6-expressing cells in the MGE. CR+ interneurons are born outside the MGE. The pallial neuroepithelium gave rise to all CR+ granule neurons in the dentate gyrus that are generated at postnatal stages (Brandt et al., 2003).
In conclusion, our work has shown that genetic restriction of neuronal potential at the precursor stage contributes toward the extensive pallial interneuron diversity observed in the adult cortex. Identification of additional genetic subdivisions of the subpallial neuroepithelium may not only help us understand how this diversity arises but will also provide us with a handle on specific interneuron populations, allowing us to manipulate their properties and dissect their functions in vivo.
Footnotes
This work was supported by a United Kingdom Medical Research council (MRC) New Investigator Award (N.K.).
References
- Alifragis P, Liapi A, Parnavelas JG. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J Neurosci. 2004;24:5643–5648. doi: 10.1523/JNEUROSCI.1245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Awatramani R, Scherer S, Grinspan J, Collarini E, Skoff R, O'Hagan D, Garbern J, Kamholz J. Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination. J Neurosci. 1997;17:6657–6668. doi: 10.1523/JNEUROSCI.17-17-06657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion A, Wassef M, Metin C. Early differences in axonal outgrowth cell migration GABAergic differentiation properties between the dorsal lateral cortex. Cereb Cortex. 2003;13:203–214. doi: 10.1093/cercor/13.2.203. [DOI] [PubMed] [Google Scholar]
- Brandt MD, Jessberger S, Steiner B, Kronenberg G, Reuter K, Bick-Sander A, von der BW, Kempermann G. Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Mol Cell Neurosci. 2003;24:603–613. doi: 10.1016/s1044-7431(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Choi GB, Dong HW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Swanson LW, Anderson DJ. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46:647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;1(Suppl 16):i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- Fruttiger M, Karlsson L, Hall AC, Abramsson A, Calver AR, Bostrom H, Willetts K, Bertold CH, Heath JK, Betsholtz C, Richardson WD. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development. 1999;126:457–467. doi: 10.1242/dev.126.3.457. [DOI] [PubMed] [Google Scholar]
- Gall C, Lauterborn J, Isackson P, White J. Seizures, neuropeptide regulation, and mRNA expression in the hippocampus. Prog Brain Res. 1990;83:371–390. doi: 10.1016/s0079-6123(08)61263-7. [DOI] [PubMed] [Google Scholar]
- Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex. 1997;7:347–358. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Williams BP, Bolz J, Price J. The specification of neuronal fate: a common precursor for neurotransmitter subtypes in the rat cerebral cortex in vitro. Eur J Neurosci. 1995;7:889–898. doi: 10.1111/j.1460-9568.1995.tb01076.x. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125:2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Gulacsi A, Lillien L. Sonic hedgehog and bone morphogenetic protein regulate interneuron development from dorsal telencephalic progenitors in vitro . J Neurosci. 2003;23:9862–9872. doi: 10.1523/JNEUROSCI.23-30-09862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Ingraham C, Rising L, Goderie S, Temple S. Multipotent stem cells from the mouse basal forebrain contribute GABAergic neurons and oligodendrocytes to the cerebral cortex during embryogenesis. J Neurosci. 2001;21:8854–8862. doi: 10.1523/JNEUROSCI.21-22-08854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Witte DP, Szucsik JC, Weinstein M, Li H, Potter SS. Gsh-2, a murine homeobox gene expressed in the developing brain. Mech Dev. 1995;50:177–186. doi: 10.1016/0925-4773(94)00334-j. [DOI] [PubMed] [Google Scholar]
- Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, Knopfel T, Erzurumlu RS, Itohara S. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Hattori R, Yui Y. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain Res. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez d V, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–1133. doi: 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- Lu S, Wise TL, Ruddle FH. Mouse homeobox gene Dbx: sequence, gene structure and expression pattern during mid-gestation. Mech Dev. 1994;47:187–195. doi: 10.1016/0925-4773(94)90089-2. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Marín O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- Marín O, Anderson SA, Rubenstein JLR. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Parnavelas JG. The origin and migration of cortical neurones: new vistas. Trends Neurosci. 2000;23:126–131. doi: 10.1016/s0166-2236(00)01553-8. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marín O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Yu WP, Guthrie S, Roelink H, Lumsden A, Peterson AC, Richardson WD. Determination of neuroepithelial cell fate: induction of the oligodendrocyte lineage by ventral midline cells and sonic hedgehog. Dev Biol. 1996;177:30–42. doi: 10.1006/dbio.1996.0142. [DOI] [PubMed] [Google Scholar]
- Schwarzer C, Sperk G, Samanin R, Rizzi M, Gariboldi M, Vezzani A. Neuropeptides-immunoreactivity and their mRNA expression in kindling: functional implications for limbic epileptogenesis. Brain Res Brain Res Rev. 1996;22:27–50. [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Simeone A, Gulisano M, Acampora D, Stornaiuolo A, Rambaldi M, Boncinelli E. Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. EMBO J. 1992;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano E, Cobas A, Fairen A. Neurogenesis of glutamic acid decarboxylase immunoreactive cells in the hippocampus of the mouse. I. Regio superior and regio inferior. J Comp Neurol. 1989a;281:586–602. doi: 10.1002/cne.902810408. [DOI] [PubMed] [Google Scholar]
- Soriano E, Cobas A, Fairen A. Neurogenesis of glutamic acid decarboxylase immunoreactive cells in the hippocampus of the mouse. II. Area dentata. J Comp Neurol. 1989b;281:603–611. doi: 10.1002/cne.902810409. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marín O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Fujimori KE, Takauji R. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J Neurosci. 1997;17:8313–8323. doi: 10.1523/JNEUROSCI.17-21-08313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23:5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31:743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;2:461–466. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, de la CE, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Yozu M, Tabata H, Nakajima K. The caudal migratory stream: a novel migratory stream of interneurons derived from the caudal ganglionic eminence in the developing mouse forebrain. J Neurosci. 2005;25:7268–7277. doi: 10.1523/JNEUROSCI.2072-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Origin and classification of neocortical interneurons. Neuron. 48:524–527. doi: 10.1016/j.neuron.2005.11.012. [DOI] [PubMed] [Google Scholar]