Abstract
Nitrilases are helical enzymes that convert nitriles to acids and/or amides. All plants have a nitrilase 4 homolog specific for ß-cyanoalanine, while in some plants neofunctionalization has produced nitrilases with altered specificity. Plant nitrilase substrate size and specificity correlate with helical twist, but molecular details of this relationship are lacking. Here we determine, to our knowledge, the first close-to-atomic resolution (3.4 Å) cryo-EM structure of an active helical nitrilase, the nitrilase 4 from Arabidopsis thaliana. We apply site-saturation mutagenesis directed evolution to three residues (R95, S224, and L169) and generate a mutant with an altered helical twist that accepts substrates not catalyzed by known plant nitrilases. We reveal that a loop between α2 and α3 limits the length of the binding pocket and propose that it shifts position as a function of helical twist. These insights will allow us to start designing nitrilases for chemoenzymatic synthesis.
Subject terms: Hydrolases, Protein design, Secondary metabolism, Biocatalysis, Electron microscopy
Andani Mulelu et al. solve the structure of Arabidopsis nitrilase NIT4 using cryo-EM at a 3.4 Å resolution. They show that mutation of three residues alters substrate specificity due to a substantially rearranged supramolecular architecture.
Introduction
Branch 1 nitrilases (EC 3.5.5.1) (NITs) are industrially important1 oligomeric enzymes (forming left-handed twists, spirals, turns or helices) that share a common architecture2–8 and catalyze the hydrolysis of nitriles to carboxylic acids and ammonia and/or amides9. In all plants, the nitrilase 4 (EC 4.2.1.65) (NIT4) group represents the ancestral form, which is highly specific for β-cyano-l-alanine10. The derived NIT1 group, which is restricted to the Brassicaceae family includes NIT1, NIT2, and NIT3, is believed to have arisen from multiple gene duplication events11. NIT1s generally display measurable activity against a large range of aliphatic and aromatic nitriles, but cannot catalyze the hydrolysis of β-cyano-l-alanine12, while NIT4 shows either extremely minor (<0.75%) or no activity for the best substrates of the NIT1 group10. It is thought that the NIT1 group enzymes evolved to catabolise the diverse glucosinolate-derived nitriles found in the Brassicaceae11–13. Interestingly, plant NITs with high specificity against a single substrate or narrow group of substrates have evolved from the ancestral generalist NIT1 group enzymes8,13,14.
We recently discovered that substrate specificity in plant NITs is related to the structure of the NIT helical assembly8. The helical twist (Δϕ) of the NIT filament is correlated with the size of the preferred substrate: NIT4 enzymes have a relatively large helical twist, small substrate size, and high specificity, while NIT1 enzymes have a smaller twist and broader specificity. Specialized NIT1 enzymes that have evolved higher specificity and small substrate size show a larger helical twist8 . Helical twist is not the only determinant of specificity: we have identified plant NITs with similar twists, but different substrate specificities. In this case, exchanging amino acid residues lining the proposed active site pocket led to a change in substrate preference, indicating that these regions play a role in substrate recognition8. The molecular details of this mechanism still remain speculative because only a low-twist helical NIT has been visualized crystallographically: the NIT from Synechocystis sp. PCC6803 (BAA10717.1) (Nit6803) ∆291 (pdb id: 3wuy15) (Δφ = −60°).
Here we present what we believe to be the first close-to-atomic structure of a full-length active helical NIT (EC 3.5.5.1) using cryo-electron microscopy at a resolution of 3.4 Å. We compare the structures of AtNIT4 (with a large twist, high specificity and small substrate) to Nit680315,16 (with small twist, broad specificity and large substrates). We exploit the insights gained in a targeted directed evolution approach17,18 to generate a NIT4 mutant with high activity against substrates not converted by either NIT4 or NIT1 and which shows no activity against the substrate of NIT4. As predicted by our previous work8, this enzyme shows a large decrease in helical twist.
Results
The structure of AtNIT4
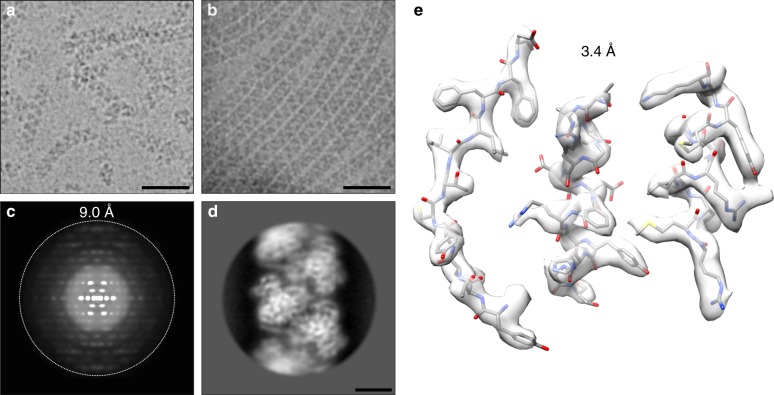
To study the molecular details of helical twist on substrate specificity we reconstructed an active plant NIT by high-resolution cryo-electron microscopy. Preliminary screening of plant NITs revealed a general pattern of heterogeneity with limited helical order. In general, NIT4 produced better filaments than NIT1, but these were still somewhat curved, short and broken (Fig. 1a). We screened buffer conditions and protein concentration in an attempt to improve filament quality without success. Eventually, we unexpectedly discovered that by incubating and blotting freshly prepared Arabidopsis thaliana nitrilase 4 (AtNIT4) (NP_197622.1) before vitrification in a 10% humidity, 25 °C environment, we could improve the quality of the imaged fibers (Fig. 1b). Despite this, we only observed reflections to a resolution of ~9 Å after vertical alignment (Fig. 1c). Clear secondary structure was visible in classified averages, but with a rapid loss of resolution with increasing distance from the center of the images, indicating poor long-range helical order (Fig. 1d). Treating one helical turn (five dimers) as a single particle and selecting a homogeneous dataset through rounds of 2D and 3D classification allowed us to achieve a resolution of 3.4 Å (gold-standard Fourier shell coefficient (FSC) = 0.143) (EMD-0320, pdb id: 6i00) (Fig. 1e and Table 1).
Fig. 1.
Data quality. a In our hands purified recombinant AtNIT4 forms relatively short heterogeneous filaments (scale bar = 40 nm). b The sample after incubating and blotting in a low-humidity environment: the concentrating effect of dehydration resulted in the formation of a monolayer of long, straight filaments stacked up against one another (scale bar = 40 nm). c The filaments showed helical order to a limit of ~9 Å. d Clear secondary structure can be visualized in the classified average, but this deteriorates with increasing distance from the center because of the curved nature of the filaments (scale bar = 5 nm). e By masking out a single helical turn and treating this as a single particle we were able to improve the resolution substantially
Table 1.
Cryo-EM data collection, refinement, and validation statistics. AtNIT4: EMD-0320, pdb id: 6i00
| AtNIT4 EMD-0320 PDB 6i00 | |
| Data collection | |
| Microscope | Titan Krios |
| Voltage (kV) | 300 |
| Pixel size (Å) | 0.85 |
| Defocus range (µm) | −0.75 to −2.0 |
| Exposure time (s) | 7 |
| Dose rate (e− pixel−1 s−1) | 6.5 |
| Number of images | 1266 |
| Number of frames per image | 45 |
| Map reconstruction | |
| Initial particle number | 392,263 |
| Final particle number | 133,106 |
| Box size (pixels) | 260 |
| Helical twist (°) | −72.98 |
| Helical rise (Å) | 17.48 |
| Number of monomers per box | 25.3 |
| Box overlap (%) | 85 |
| Resolution (Gold-standard FSC = 0.143) (Å) | 3.4 |
| Map sharpening B factor (Å2) | −124 |
| Model composition | |
| Non-hydrogen atoms (monomer) | 2262 |
| Protein residues (all/built) | 355/289 |
| Ligands | 0 |
| Fit to map | |
| Correlation coefficient (entire box) | 0.759 |
| Correlation coefficient (around atoms) | 0.823 |
| FSC = 0.5 (between model and map) (Å) | 3.49 |
| Protein geometry | |
| Molprobity score | 1.69 |
| All-atom clash score | 2.29 |
| EMRinger score | 3.82 |
| RMSD bonds (Å) | 0.006 |
| RMSD angles (°) | 1.153 |
| Ramachandran favored (%) | 83.92 |
| Ramachandran allowed (%) | 15.30 |
| Ramachandran disallowed (%) | 0.78 |
Helical structure of NIT4
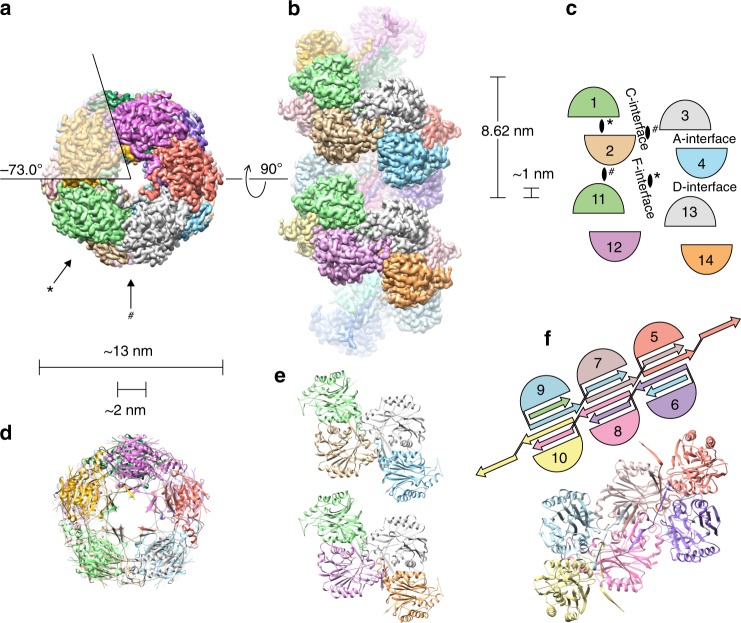
AtNIT4 is a left-handed helical tube with 4.9 sunbunits (dimers) in one helical turn, a pitch of 8.62 nm, an outer diameter of ~13 nm, and a ~2 nm hollow core (Fig. 2a, b). There are two diad axes, which are related to one another by a −5° (negative value: left-handed by convention) rotation and 4.31 nm translation along the helical axis. The first passes through the C- and D-interfaces2 and the second passes between the A- and F-interfaces2,4 (Fig. 2c). A ~1 nm gap (Fig. 2b) across the helical groove shows that contrary to what has been observed previously in negative stain8, this region lacks stabilizing interactions (D- or F-interfaces). Amino acid residues involved in the formation of helical interfaces are shown in Supplementary Fig. 1. Interestingly, the predominant interaction stabilizing the filament is between the C-terminal tails of four adjacent monomers, which all contribute strands that form a row of ß-sheets in the core of the enzyme (Fig. 2d, f).
Fig. 2.
NIT helical structure. a In vitro, AtNIT4 assembles into an extended left-handed helical tube with 4.9 subunits per turn (helical twist of −73.0°) and outer and inner diameters of ~13 nm and ~2 nm, respectively. Arrows indicate the positions of the two diad-axes (#) and (*). b, c The NIT4 filament is comprised of a left-handed helix with a pitch of 8.62 nm. Monomers associate across previously described interfaces2,4; while no interaction was observed across the D- or F-interfaces, they are shown for the sake of completeness. The positions of the two diad-axes (#) and (*) relative to individual monomers are shown (EMD-0320). d About 80% of the model was built with high certainty, 34 N- and 8 C-terminus residues and 24 residues on the outer surface of the filament were disordered and could not be visualized in the map. ß-sheets stabilizing the helical structure can be seen in the core of the enzyme (pdb id: 6i00). e The C-interface is predominantly an interaction across monomers 2 and 3. f The C-terminal tail of each sequential monomer contributes two ß-strands, which form one ß-sheet for every dimer
The substrate-binding pocket
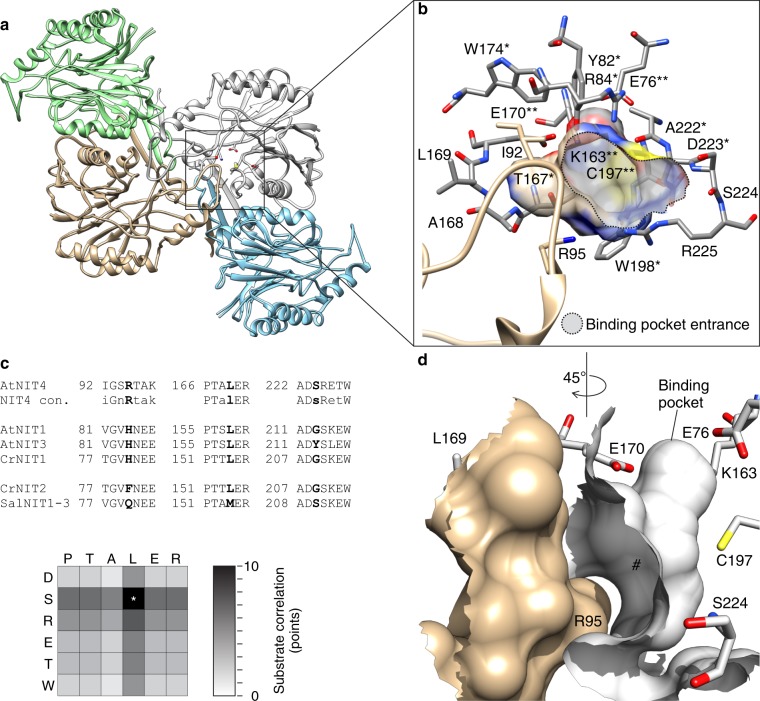
AtNIT4 shows high-structural homology with other members of the NIT superfamily including a conserved αßßα core (Fig. 3a) and CEEK catalytic tetrad (Fig. 3b and Supplementary Fig. 2)19,20. The substrate-binding pocket is a ~7 × 4 Å tunnel lying close to the C-interface (Fig. 3a) that runs almost perpendicular to the helical axis (Fig. 3b). NITs have an insertion between α2 and α3 relative to crystallized NIT superfamily members21, in Nit6803 ∆291 this loop forms a twofold interaction across the C-interface distant from the active-site pocket (pdb id: 3wuy15). Plant NITs have a further five-residue insertion relative to Nit6803 (Supplementary Fig. 1), which conveys this loop across the C-interface, forming a lid over the symmetry-related substrate-binding site (Fig. 3a, b, d).
Fig. 3.
The substrate-binding pocket. a AtNIT4 structure showing two dimers interacting across the C-interface, the position of the catalytic site is indicated by a square. b The active-site pocket, the catalytic residues (E76, K163, E170, and C197) are shown (**), residues conserved in all plant NITs (*). A loop (tan) arising from the adjacent monomer (between α2 and α3) extends over the entrance to the active-site pocket. c As expected, the amino acid residues surrounding the binding pocket correlate with substrate specificity. Three amino acid positions in particular are responsible for the majority of the variation between known plant NITs: 169, 224, and 95 (numbered according to AtNIT4). d A lid (tan), formed by the loop, limits the length of the binding pocket (#). R95 is in the lid loop; S224 is on the outer border of the binding pocket and L169 points toward the light green subunit across the C-interface
The mode of substrate binding
There is good biochemical evidence to suggest that the substrate (or a substrate intermediate) binds covalently to the NIT active-site cysteine10,22. This requires that the nitrile group of the substrate be orientated correctly relative to C197 and imposes a strong constraint on the possible position of the bound substrate. We produced a structural alignment between AtNIT4 and the structures of three NIT superfamily enzymes cocrystallized with substrates/intermediates. The structures used were: a C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase from Agrobacterium sp. KNK712 complexed with N-carbamyl-D-methionine (pdb id: 1uf5)23; an amidase from Pseudomonas aeruginosa with trapped acyl transfer intermediate (pdb id: 2uxy)24 and a C145A mutant of the amidase from Nesterenkonia AN1 complexed with butyramide substrate (pdb id: 4izs)25. The substrates/intermediates localize to the binding pocket and lie in the direction of the lid loop. Their fit is suboptimal though, and the ends of the substrates clash with binding pocket residues on one side, including the lid loop in the case of the rather long substrate N-carbamyl-D-methionine visualized in 1uf523 (Supplementary Fig. 2).
Identifying determinants of specificity
Eleven of the seventeen amino acid residues lining the binding pocket are conserved among all plant NITs regardless of substrate preference (Fig. 3b), many are conserved across all NITs. Of the four loops making up the boundaries of the binding pocket (Supplementary Fig. 1), three show sequence differences between different plant NITs (Fig. 3c). We previously exchanged residues in the (PTSLER) and (ADGSKEW) motifs in a NIT1 and observed changes in substrate preference, while exchanging a single residue in the NIT1 lid loop (VGVHNEE) altered helical twist, substrate size, and degree of specificity8. This residue is a conserved arginine in all known NIT4 enzymes that accept β-cyano-l-alanine as a substrate (EC 4.2.1.65), NIT1s have a histidine in this position and specialized NIT1s show various residues, which are correlated to their specificity and helical twists8. We further refined this analysis to isolate the impact of substrate specificity of individual positions within each of these motifs among the known plant NITs and identified three residue positions with a disproportionate effect on specificity (Fig. 3c and Supplementary Table 1).
We randomized the three residues to alter specificity
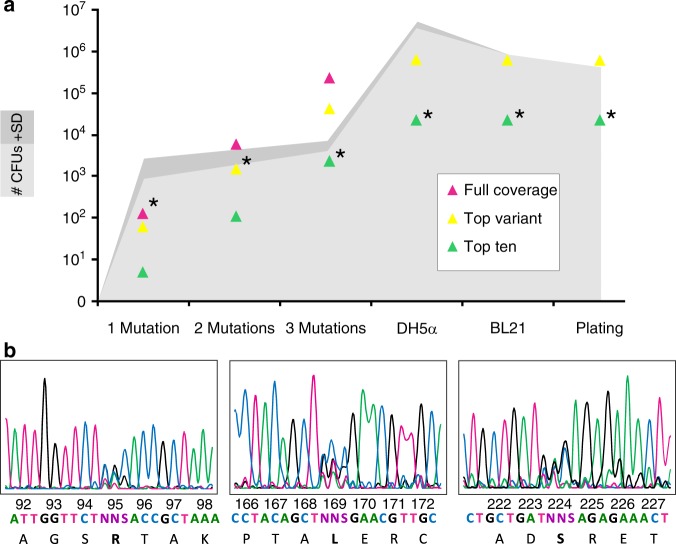
We performed a directed evolution experiment to identify mutants with altered substrate specificity. Target sites (R95, L169, and S224) were randomized (Supplementary Table 2), resulting in a library containing ~5.0 × 103 unique mutants, sufficient to sample at least one of the top ten possible mutants with 95% probability26 (Fig. 4a and Supplementary Table 3). We implemented a strategy of site-saturation mutagenesis of all three sites before selection in order to avoid missing out on synergistic effects between the sites27, which could confound a sequential mutagenesis strategy. To maintain the full diversity of the mutant library in the selection step, 4 × 106 DH5α and 7.7 × 105 BL21 cells were transformed and 4 × 105 cells were plated out to ensure that each unique mutant was present (Fig. 4a). The final library was sequenced as a mix (Fig. 4b) and individual colonies (Table 2) to monitor the successful incorporation of mutations.
Fig. 4.
Generating the library. Mutations were introduced sequentially: R95 (1 mutation), R95 + L169 (2 mutations) and R95 + L169 + S224 (3 mutations). a Graphical representation of the bottlenecks in the experiment. As the library was assembled (one mutation to three mutations), more colony forming units (CFUs) are required to satisfactorily sample the full library diversity. Triangles represent the number of unique mutants present required for 95% probability of full-coverage (all possible protein mutants present), top-mutant (the best amino acid sequence present) and top-ten (one of the 10 best amino acid sequences present) in magenta, yellow, and green, respectively26. The upper (dark gray) and lower (light grey) 95% confidence interval values for number of CFUs show that by the end of library creation at least one of the top ten mutants was present (stars). This diversity was maintained through all cloning and selection steps. b The final library mix was sequenced and showed successful overlapping NNS peaks at amino acid positions 95, 169, and 224 (sample size: 1% of the total CFUs in each sample)
Table 2.
Colonies were sequenced at every stage. No bias toward wild-type sequence or multiple transformants/mixed colonies was observed
| Colony 1 | Colony 2 | Colony 3 | ||
|---|---|---|---|---|
| 1 Mutation | R95 | GAC (Asp) | CGG (Arg) | TAC (Tyr) |
| L169 | CTT (Leu) | CTT (Leu) | CTT (Leu) | |
| S224 | TCA (Ser) | TCA (Ser) | TCA (Ser) | |
| 2 Mutations | R95 | GAC (Asp) | TCC (Ser) | GCC (Ala) |
| L169 | TCG (Ser) | CGC (Arg) | ACC (Thr) | |
| S224 | TCA (Ser) | TCA (Ser) | TCA (Ser) | |
| 3 Mutations | R95 | TAC (Tyr) | GGC (Gly) | CCC (Pro) |
| L169 | GCG (Arg) | CGC (Arg) | AAC (Thr) | |
| S224 | CGG (Arg) | CCC (Pro) | GAC (Asp) |
We identified a NIT4 mutant with broad substrate specificity
In order to select AtNIT4 mutants capable of hydrolyzing a specific nitrile, BL21 Escherichia coli cells harboring mutant enzymes were plated onto minimal media plates containing Isopropyl β-D-1-thiogalactopyranoside (IPTG) and one of 42 different nitriles (Supplementary Table 4). The basis for this assay is (1) nitrile toxicity and (2) the fact that nitrile hydrolysis produces ammonium, which can be used as a nitrogen source by the cell28. Cells expressing NITs capable of hydrolyzing the nitrile present in each plate will therefore have a competitive advantage, form a colony and be identified. After incubation for 13 days, between 1 and 300 colonies were observed on 15 of the nitrile plates (Supplementary Table 1). Growth on the adiponitrile, butanenitrile, 4-cyanopyridine, fluoroacetonitrile plates occurred after 2 days (Fig. 5a–d). Wild-type colonies were observed on the positive-control plate (β-cyano-l-alanine) but not on negative-control plates. Three colonies were randomly picked for sequencing from the most rapidly growing isolated colonies (Fig. 5a–d).
Fig. 5.
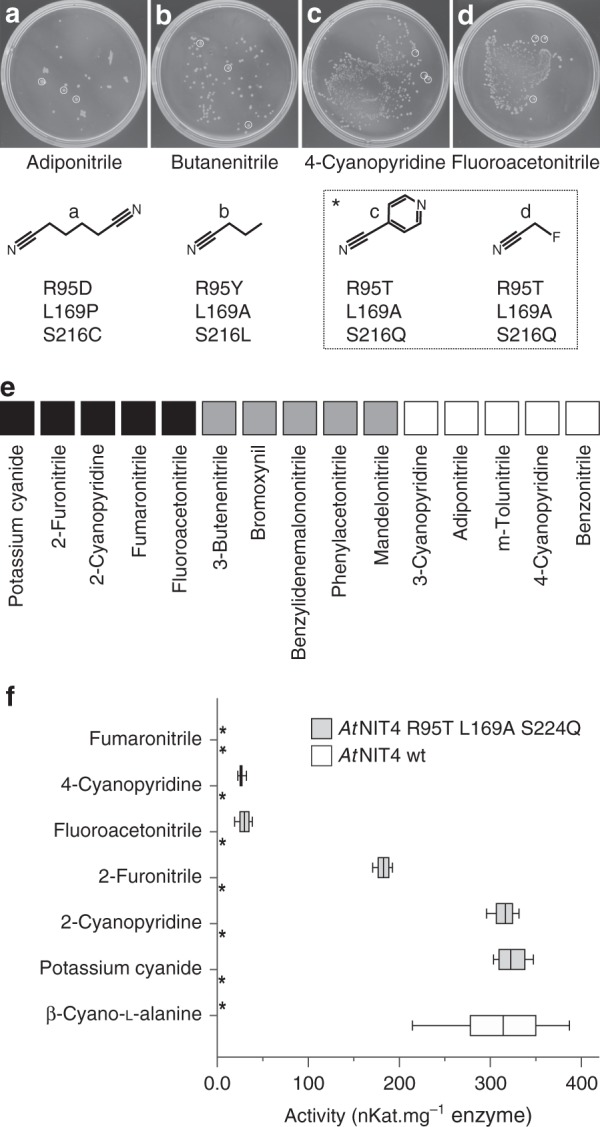
NIT4 mutant selection, screening and characterization. a, b, c, d A variable number of colonies were visible after 2 days of incubation on the selection plates. The plates, substrate structures, and sequencing results are shown. e Screening results indicate measured absorbance after incubating with AtNIT4 wt, AtNIT4 R95T L169A S224Q and an inactivated enzyme control, and assaying with Nessler’s reagent. Black indicates that AtNIT4 R95T L169A S224Q produced more ammonia than AtNIT4 wt. Gray indicates that the substrate reacts to form a high-absorbance product, but after incubating with the mutant, the absorbance was reduced. White indicates a slightly higher AtNIT4 wt activity compared to the mutant. f Enzyme activity results, asterisk indicates no measurable ammonia formation after incubating for 30 min with 5× excess enzyme. AtNIT4 wt showed no nitrilase activity against any of the listed substrates except for β-cyano-l-alanine. The mutant showed high activity against potassium cyanide, 2-cyanopyridine and 2-furonitrile and moderate activity against fluoroacetonitrile and 4-cyanopyridine (1st quartile, median and 2nd quartile ± sd) n = 3 independent activity measurements
At least three different sequences were present on the adiponitrile plate and these were generally mixed, showing that multiple mutants had the capacity to hydrolyze this substrate. Further investigation revealed that very small but visible colonies were present on negative-control adiponitrile selection plates. Purified AtNIT4 enzyme also showed low but measurable activity against adiponitrile (Fig. 5e). These mutants were therefore excluded from further study. All of the colonies picked from the butanenitrile plate showed the same sequence, but attempts to purify this enzyme were unsuccessful and there was no evidence of helical filaments in any of the purification fractions. Interestingly, all three of the colonies from each of the 4-cyanopyridine and fluoroacetonitrile plates gave the same sequence. Since this indicated that the mutant had the potential for broad substrate specificity, we purified this enzyme and performed a substrate screen against our set of 43 nitriles (Supplementary Table 4). This led to the identification of a further four potential substrates: fumaronitrile, potassium cyanide, 2-furonitrile, and 2-cyanopyridine (Fig. 5e). While an excess of purified wt AtNIT4 showed no activity after 30 min incubation with the identified substrates, the mutant showed substantially different substrate specificity with no reduction in activity (Fig. 5f and Supplementary Table 5). We performed activity measurements in triplicate and ensured that at least four data points were observed while the reaction was in the linear range.
AtNIT4 R95T L169A S224Q has a reduced helical twist
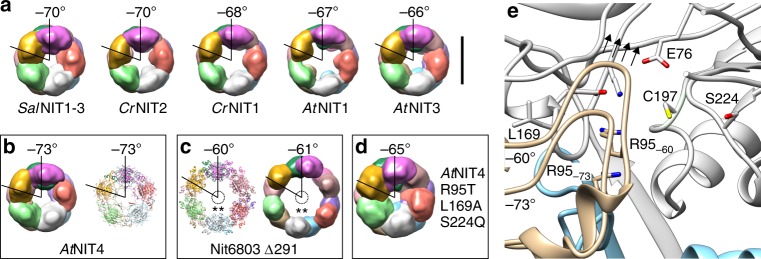
Different plant NITs show a range of helical twists (Fig. 6a): on either side of the two extremes are NITs for which atomic models are available, AtNIT4 (∆φ = −73°) and Nit6803 ∆291, which crystallized in spacegroup P6(5)15 (pdb id: 3wuy), meaning that it has a left-handed helical arrangement with six subunits (dimers) per turn (Δφ = −60°) (Fig. 6b, c). Interestingly, while the diameter of the filament is generally ~constant between the various plant NITs, in Nit6803 ∆291 it is substantially larger. We reconstructed Nit6803 ∆291 by negative-stain electron microscopy (EMD-4407) and found that this was not a crystallization artifact and that the EM and crystal structures largely agree (Fig. 6c). We have previously observed a correlation between helical twist and the length of the preferred substrate as well as the degree of substrate specificity of a given plant NIT8. We therefore hypothesized that AtNIT4 R95T L169A S224Q would show a decrease in helical twist because of its broader substrate range compared to AtNIT4. We reconstructed AtNIT4 R95T L169A S224Q in three dimensions by negative stain electron microscopy (EMD-4406) and found that the helical twist had changed from −73° to −65° (Fig. 6d). Unexpectedly, we also observed that the diameter of this mutant is approximately equal to that of Nit6803 ∆291 (Fig. 6c, d). Note the hollow core lacks density corresponding to the C-terminal tail (Fig. 6c).
Fig. 6.
Comparison of plant NIT helical twists. a A comparison of different plant NITs; each monomer in the filament has been colored differently to illustrate the number of protein subunits in a helical turn (360°), the brown subunit (dimer 6) is more visible with decreasing helical twist. b AtNIT4 negative stain map vs cryo-EM model, AtNIT4 has ~5 dimers per turn. c Nit6803 ∆291 negative stain map vs crystal structure (**) indicates the missing C-terminal residues. d AtNIT4 R95T L169A S224Q negative stain reconstruction, the low helical twist and large diameter show similarities to Nit6803 ∆291, density accounted for by the interacting C-terminal tails can be seen in the enzyme core. e Comparison between the structure of AtNIT4 (labeled −73°) and AtNIT4 with the helical operators of the Nit6803 ∆291 crystal structure applied (labeled −60°). The lid loop shifts in the direction shown by arrows (in the plane of the page), resulting in steric clashes. Geometrically, this movement is greater when the diameter of the fiber remains constant, but the helical twist decreases. Scale bar = 10 nm (applies to all end-on views)
The effect of increasing helical twist
We compared the helical structure of Nit6803 ∆291 with that of AtNIT4 in order to visualize the effect of the differences in helical arrangement between the two filaments. Most notably, in the Nit6803 ∆291 conformation, there is an outward rotation of ~15° about the A- and C-interfaces and a ~3 Å shift in ∆z relative to AtNIT4. Contrary to what we proposed previously8, we observe an overall increase in compression of the monomer with a smaller absolute value of the helical twist. In fact, α1, α2*, α3, α4, α5, and α6 (Supplementary Fig. 1), all lying at the periphery of the enzyme are found closer to the binding pocket in Nit6803 ∆291 than in AtNIT4.
The role of the lid loop
The lid loop limits the length of a substrate bound to C197 (Fig. 3a, b, d). Molecular dynamics in UCSF Chimera29 indicates that it is held in position primarily via a charge–charge interaction between R95 and D317. In silico simulation of R95T allows the lid loop to open outward away from C197 increasing the size of the binding pocket. This explains why mutating R95 affects substrate size and specificity and provides a clue as to why this is correlated with a decrease in helical twist. Applying the symmetry operators of Nit6803 ∆291 to the structure of AtNIT4 (loop in the closed position) results in severe steric clashes (Fig. 6e), moving the loop to the open conformation prevents this clash. This suggests that low helical twist can be used as a sign that the lid loop is in the open conformation. Interestingly, while AtNIT4 L169A S224Q expressed insolubly, AtNIT4 R95T (EMD-4804) showed no change in helical twist compared to the wild type. It also lacked activity for β-cyano-l-alanine or any of the other nitriles in our set (Supplementary table 1), except for some low activity for the rather large substrate, 3-phenylpropanenitrile (8.5 nKat mg−1 enzyme), which neither the wild type nor AtNIT4 R95T L169A S224Q showed activity for.
Discussion
NITs are attractive biocatalysts for the synthesis of amides and carboxylic acids for use in the manufacture of drugs and fine chemicals30. Novel specificities are generally discovered by environmental sampling31 or sequence data mining32,33. In some cases, activity against a particular substrate or substrate class has been increased by up to ~3–8-fold by rational design15 or directed evolution34. We recently saw a ~500-fold improvement in activity for a substrate after a single amino acid exchange and discovered that this correlated with a change in the helical twist of the NIT filament8. At the time, the molecular basis for this change was unclear, because we did not have an atomic resolution structure of an active helical NIT. We speculated that the change in substrate size preference resulted from compression of the active-site pocket at the helical interface. This is noteworthy because one of the distinguishing features of branch 1 NITs (EC 3.5.5.1) is that they form helical-, turn-, or spiral-quaternary structures, the basic architecture of these assemblies is conserved in phylogenetically diverse NITs2–8,35,36 and appear necessary for activity21,37.
Crystal structures of members of the NIT superfamily have been available for almost 20 years38,39 and today there >20 crystal structures of various members deposited in the Protein Data Bank (https://www.rcsb.org/). Extensive attempts to crystallize a helical NIT by ourselves and others failed and Thuku et al.21 attributed this to the irregular/ ~5-fold symmetry of the NIT helix, which presumably interferes with crystallization. This limits our structural understanding of the active NIT oligomer to ~20 Å electron microscopy information because of the heterogeneous nature of the filaments, until the publication in 2014 by Zhang et al.15 of the first helical NIT crystal structure. Interestingly, they observed that 55 C-terminal amino acids were cleaved off before crystallization. This fortuitously led to the formation of filaments with a helical twist of −61° with sufficient flexibility to allow crystallization, but unfortunately also altered the oligomeric structure of the protein and abolished activity (our unpublished results).
Here, we describe the first active NIT filament structure interpretable to atomic resolution. The structure shares conserved features with other members of the superfamily, such as an αßßα core (Figs. 2 and 3a) arranged into dimers across the A-interface2,21 and CEEK catalytic tetrad (Fig. 3b)19,20. Dimers interact across the C-interface forming left-handed helices with ~5 dimers per turn (Fig. 2). The extended C-terminal tail of each monomer forms two ß-strands, which assemble, along with strands from monomers across the A- and C-interfaces, into a row of 4-stranded ß-sheets in the interior of the tubular filament (Fig. 2). These interactions stabilize the C- and A-interfaces, explaining why C-terminal truncation decreases thermostability40 and why the C-terminal tail is an important factor in oligomerization3,41,42. Interactions across the helical groove (D- or F-interfaces) were not observed (Fig. 2) and this could explain the plasticity of NIT helical twist8.
The architecture of the substrate-binding pocket came as a surprise to us and provided an interesting insight into the structural mechanism of substrate size selection as well as the substrate altering/inducing effect of heterocomplex formation in some plant NITs8,43. The NIT superfamily binding pocket has been described23,24, but in addition to these shared elements, in AtNIT4 a loop from the adjacent monomer extends over the C-interface in line with C197 (Fig. 3a, b). We superimposed bound substrates and reaction intermediates from NIT superfamily members23–25 on our structure (Supplementary Fig. 2) and propose that this loop places a limit on the maximum bound substrate length (Fig. 3d). Crucially, we observed via molecular dynamics, that this loop is held in position by a charge–charge interaction between R95 and D317. On this basis we refer to the loop as a lid, R95 as a lock, and the flexible ends of the loop as a hinge, which open or close, shifting the lid relative to C197. We previously noted that all ß-cyano-l-alanine converting NIT4s have an arginine in this position, while NIT1s have a histidine and specialist NIT1s have various residues, which correlate with substrate specificity8. In addition to this, D137 and surrounding motif: KFDFD is conserved in all known NIT4s, while NIT1s have: KLYFD in this position (Supplementary Fig. 1). This may explain why we did not observe any mutations similar to those in NIT1 in our screen, even though many NIT1 substrates were present. This could be tested in the future by mutating residue 317 in addition to the three residues targeted here.
A major challenge in protein engineering is the generation of enzymes with novel substrate specificities. The typical starting point is an enzyme with at least measureable promiscuous activity for the targeted substrate(s)27. Rounds of directed evolution or rational engineering are then used to move the protein toward a point of higher activity in the fitness landscape44. An analogous process is believed to be responsible for the enormous diversity of modern proteins seen today. Following gene duplication, new enzyme copies, no longer constrained by their original physiological function(s) can diverge by selective pressure on existing promiscuous reactions45. However, in nature, broadly specific NIT1 enzymes presumably evolved from a highly specific ancestral NIT411, with no or very low activity for NIT1 substrates. To explore this process we used a focused-library approach, targeting three amino acid residues within the binding pocket that influence substrate specificity disproportionately27. We selected altered mutants over one round of directed evolution with a small library of only ~5000 mutants, but which nevertheless sampled at least one of the top ten mutants with 95% probability (Fig. 4). This strategy was effective, generating a mutant (AtNIT4 R95T L169A S224Q) with altered substrate specificity and no activity against the wild-type substrate.
We observed in our initial screen that some substrates formed a high-absorbance product with the assay reagents, which reduced after incubation with the enzyme, but with no corresponding increase in ammonia (Fig. 5e). We interpret this as evidence of possible amide formation because amide formation accounts for ~50% of the total activity of wild-type AtNIT410, but we did not explore this phenomenon further. In some cases mutants identified on one substrate showed activity against other substrates when tested using purified enzyme, but showed no growth on these conditions. This phenomenon may result from certain substrates or products being more toxic than others, decomposition of the substrates during incubation or low substrate solubility. In fact this is a key weakness of this strategy and in some cases may mean that interesting variants are lost during selection.
We reconstructed AtNIT4 R95T L169A S224Q filaments in three dimensions by negative stain EM and found that it has a helical twist of −65°, very similar to AtNIT3 (Δφ = −66°) (Fig. 6a, c), which has a broad substrate specificity and a preference for large substrates8. In contrast to AtNIT3, the mutant has a larger diameter, more similar to Nit6803 ∆291 (Fig. 6c, d), Nit6803 also shows a very broad substrate specificity16. This was fortuitous, because in effect, the two available atomic resolution NIT structures: AtNIT4 (Δφ = −73°) and Nit6803 ∆291 (Δφ = −60°) represent the helical structure extremes. This allowed us to visualize both structural states, which suggested a mechanism for the observed changes in substrate specificity (Fig. 6e). We propose: R95T eliminates the positive charge at position 95, which disrupts the interaction with D317 and allows the lid loop to open and shift away from C197 (Fig. 3b). In NIT4 R95T this allows 3-phenylpropanenitrile to bind to C197 by opening up the binding pocket, but does not result in a change in helical twist. Introduction of the other two mutations (AtNIT4 R95T L169A S224Q) results in a large-scale change in helical twist (Fig. 6b, d).
The S224Q exchange on the peripheral border of the binding pocket might have a direct effect on the substrate, as we previously observed for glycine to tryptophan mutants in this position8 potentially accounting for the particular combination of substrates observed here. The close proximity of R95 to the catalytic residues may mean that there is direct interaction with or selection for the substrate by a residue in this position. This would account for cases where there is an almost identical helical twist, but mutating this residue still increases activity against a particular substrate8. In the case of heterocomplex formation8,43, the loop of one subunit and active-site pocket of another subunit interact to produce the observed activity.
These insights have placed us in a position where we can semi-rationally design plant NITs in a very successful way. In fact, the mutant that we generated shows wild-type levels of activity (~300 nKat.mg−1 enzyme) for substrates that the wild type is inactive against. This is made possible by the structural flexibility of the lid loop system and explains how by shifting this loop and making a small number of other changes in the binding pocket, a variety of novel substrate specificities evolved in plants from NIT4, a highly specific precursor. More work is needed to further explore this relationship. Certainly the near-atomic resolution structure of a plant NIT1 will allow us to observe the effect of the low-twist helical state on the position of the lid loop. It would also be interesting to target NIT1 sites to determine if the substrate specificity of NIT1 can be further broadened to generate economically interesting enzymes. We also cannot currently explain why AtNIT4 R95T L169A S224Q shows activity against cyanide, the smallest nitrile (Fig. 5e, f), further work will need to be done to explain this.
Methods
NIT expression and purification
His-tagged NITs in pET-21b(+) (Novagen) were expressed at 37 °C in E. coli BL21-CodonPlus (DE3)-RIL (Agilent Technologies) for 4 h in LB containing 100 μg ml−1 ampicillin and 1 mM IPTG. The cells were pelleted at 4 °C and resuspended in 50 mM Tris–Hcl pH 8.0, 100 mM NaCl with cOmpleteTM protease inhibitor (Roche) and sonicated on ice for 4 min (at 15 s intervals) (Sonicator® 3000, Misonix). The clarified supernatant was loaded onto a Ni2+-NTA-agarose (Qiagen) column and washed with 50 mM Tris–Hcl pH 8.0, 500 mM NaCl, 20 mM imidazole and eluted over a gradient up to 500 mM imidazole. The NIT-containing peak was pooled and loaded onto a TSKgel PWXL4000 HPLC size exclusion column (Tosoh Corp), equilibrated with 50 mM Tris–Hcl pH 8.0, 100 mM NaCl and the leading edge of the NIT-containing peak was collected. Cryo-EM grids were immediately prepared using this material and aliquots destined for long-term storage were flash frozen in liquid nitrogen and stored at −80 °C after addition of 1 mM DTT. Activity assays were performed after buffer exchange into 50 mM potassium phosphate pH 8.0, 1 mM DTT.
Negative-stain electron microscopy and image processing
NIT filaments were negatively stained according to standard practices (e.g., Booth et al.46). Briefly, 3μl of NIT solution was incubated for 30 s on glow-discharged carbon-coated copper grids before being washed three times with distilled water and stained with two exchanges of 2% uranyl acetate. The grids were loaded into a FEI/Tecnai F20 FEGTEM equipped with a 4k × 4k CCD camera (GATAN US4000 Ultrascan, CA, USA) and imaged at a sampling of 2.11 Å pixel−1 at 200 kV with a defocus of −1.0 µm under standard low-dose conditions. Helical segments were extracted using Eman47 Boxer with 90% overlap and reconstructed using the iterative helical real-space reconstruction48 algorithm using SPIDER49 using a featureless cylinder as a starting model as described8.
Cryo-electron microscopy and image processing
Purified AtNIT4 filaments (2.5 μl at ~0.15 mg ml−1) were applied to glow-discharged 1.2/1.3 gold grids (Quantifoil, Micro Tools), incubated for ~30 s, blotted from both sides for ~3.5 s and vitrified using a Vitrobot (FEI) with humidifier turned off (~10% humidity). The grids were imaged on a Titan Krios microscope (FEI) with a K2 Summit direct detector camera (Gatan) using EPU (FEI) at 300 kV. A total of 1266 movies, consisting of 45 frames per movie with a dose rate of 6.5 e− pixel−1 s−1 over 7 s, were collected over a defocus range of −0.75 to −2.0 µm with a physical pixel size of 0.85 Å pixel−1.
Image processing
RELION50 was used for image processing and reconstruction. MotionCor251 was used to correct beam-induced motion and Gctf 1.0652 was used to determine parameters for CTF correction. ~3000 particles from straight helical filaments were picked manually from 50 micrographs and used to generate 2D classified averages. The best classes were used as auto-picking templates resulting in ~392,000 particles, poor particles were eliminated by two rounds of 2D classification. Helical reconstruction53 was initiated with the known helical parameters of AtNIT48 using a negative stain reconstruction, which was low-pass filtered to a resolution of 60 Å as a starting model. The best ~133,000 particles were masked, 3D classified, and subjected to 3D autorefinement and postprocessing to yield a map with an overall resolution of 3.4 Å (gold-standard FSC = 0.143 criterion with a soft mask applied to the two half-maps).
Model building, refinement, and validation
De novo polypeptide chain fitting into the map was performed using Buccaneer54 within the CCPEM software suite55. Obvious errors in connectivity, unbuilt residues, and incorrect rotamers were corrected against the EM map in Coot56 and refined using Phenix Real Space Refine57,58. After symmetrization in UCSF Chimera29 the complete helical assembly was refined using Phenix. Additional rounds of manual fitting and refinement with secondary structure imposed resulted in a final model-to-map correlation of 0.8.
Data visualization
All molecular visualization and high quality image rendering were performed using UCSF Chimera29.
Molecular dynamics
The molecular dynamics simulation tool using default parameters was used in UCSF Chimera29.
Substrate correlation value
Plant NITs for which substrate specificity data is available were used: A. thaliana NIT1, NIT312, and NIT410 (AtNIT1: NP_851011.1; AtNIT3: NP_190018.1; AtNIT4: NP_197622.1); Salix alba NIT1-314 (SalNIT1-3: MH567030); Capsella rubella NIT1 and NIT28 (CrNIT1: XP_006291436.1; CrNIT2: XM_006284056.1). Sequences were allocated to NIT4, NIT1, and specialist NIT1 groups and compared in pairs: amino acid positions were allocated one point when a residue differed between the groups and the substrate differed, or when the residue remained the same when the substrate remained the same. Points were subtracted when this was not the case.
Library construction
The A. thaliana nitrilase 4 (AT5G22300) pET-21b(+) vector was a gift from Markus Piotrowski (Ruhr-Universität Bochum). Sequential randomization of the chosen sites was achieved with NNS (N = A/T/C/G; S = G/C) degenerate codons applying a modified QuikChange® PCR protocol59 using HiFi Hotstart Readymix (Kappa Biosystems). PCR reactions (50 μl) with 1–2 ng of template DNA were run under the following conditions: 95 °C for 3 min (1 cycle); 98 °C for 20 s, 62 °C for 15 s, 72 °C for 4 min (20–30 cycles); and 72 °C for 10 min (1 cycle) and subsequently digested with DpnI at 37 °C for 1 h. Primers are listed in Supplementary Table 2. Sufficient DNA for large-scale transformation was obtained by pooling several rounds of PCR product and gel purifying (Zyppy™ Gel Purification Kit, Zymo Research). Transformations were completed by electroporation in 400 μl electroporation cuvettes (Eppendorf). DNA sequencing was performed by the Central DNA Sequencing Facility (University of Stellenbosch, South Africa).
Statistics and reproducibility
The theoretical library size required for site-saturation mutagenesis on three amino acid residues was estimated using the TopLib calculator26 (http://stat.haifa.ac.il/~yuval/toplib/); 100% yield was assumed. A sample consisting of 1% of the volume of the recovered cells was plated out to estimate the total number of cells transformed. The resulting colonies were counted and a confidence interval was derived using: (n ± z*√n) * N, where n is the number of colonies counted, z* are the values from a standard normal distribution for the appropriate probability level, and N is the sampling60. Oversampling requirements were calculated using the formula: P(Missing 0 mutants) = e−λ where λ = n(1 – 1·n−1)m for n unique mutants in a library of size m61. Activity assays and selection experiments were performed at least in triplicate; quartiles, medians and standard deviations were calculated in Microsoft Excel.
Selection
Minimal media agar plates without nitrogen were made according to an adapted protocol62. M9 minimal salts with agar (bacteriological agar, Merck) was supplemented with 0.4% glucose, 0.1 mM CaCl2, 2 mM MgSO4, 1 mM IPTG, and ampicillin (100 μg ml−1) and one of a set of 42 nitriles (10 mM) (Supplementary Table 1) added to the warm agar. Nitrile stocks (250 mM) were made up by vortexing in distilled water at 37 °C where possible. Hydrophobic nitriles were dissolved in a gradually increasing volume of methanol at 37 °C while vortexing and made up to the correct volume with warm water. In some cases with highly water insoluble nitriles small particles remained after vortexing in 100% methanol, these were used as is. Library plasmid DNA was transformed into BL21 E. coli according to calculated oversampling requirements. The cells were pelleted and washed with ice-cold 1× minimal salts to remove any source of nitrogen; ~4 × 105 transformants were plated on each nitrile plate. E. coli transformed with AtNIT4 was grown on selection plates containing β-CN(Ala) as a positive control, while AtNIT4 was grown on 4-hydroxyphenyacetonitrile, 6-heptenenitrile, mandelonitrile, butanenitrile, 4-cyanopyridine, fluoroacetonitrile, fumaronitrile, potassium cyanide, 2-furonitrile, and 2-cyanopyridine as negative controls. Untransformed E. coli on plates lacking nitriles did not form observable colonies. The plates were grown over several days at 37 °C.
Activity assays
Specific activity measurements were conducted using the indophenol blue assay43 or Nessler reagent assay63 with substrates purchased from Sigma-Aldrich. Briefly, 2.5 mM substrate, 1 mM DTT and between 0.5 and 10 μg of protein were incubated in 100 μl reaction tubes containing phosphate buffer at 37 °C in intervals of between 5 and 30 min. Initial substrate screening was performed as above, except that 2 μg of protein and 10 mM substrate were incubated for 2 h. Nessler assay: 100 μl of 100 mM KI, 27 mM HgCl2 and 330 mM NaOH was added immediately following incubation and left at room temperature for 10 min before measuring the absorbance at 420 nm. Indophenol blue assay: reactions were terminated by the addition of 4 μl of 100 mM phenol (in ethanol), followed by the addition of 4 μl of 19 mM sodium nitroprusside and 10 μl of oxidizing solution (1 part of 470 mM NaClO and 4 parts of 775 mM Na3C6H5O7 and 256 mM NaOH) in distilled water and boiled for 4 min, 1 ml distilled water was added before measuring absorbance at 640 nm. These procedures were conducted in triplicate. Controls consisted of identical reaction mixtures containing enzyme inactivated by boiling for 5 min prior to addition.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We acknowledge Diamond for access and support of the Cryo-EM facilities at the UK national electron bio-imaging centre (eBIC), (proposal EM20975-1), funded by the Wellcome Trust, MRC, and BBSRC. T. Sewell and M. Piotrowski for their mentorship and Y. Qayiya, N. Arendse, M. Jaffer, B. Weber, and S. Neumann for technical assistance. This work was funded by a Research Career Advancement Fellowship from the National Research Foundation of South Africa (92556) and GCRF-START (UK Science and Technology Facilities Council grant ref. ST/R002754/1).
Author contributions
JDW designed and supervised the study, wrote the manuscript and performed model building, refinement and validation. AEM and JDW reconstructed the nitrilase helices. AEM performed electron microscopy and carried out substrate-preference determinations. AMK constructed and screened the mutant library and performed statistical analyses.
Data availability
Expression plasmids generated in this study are available from the corresponding author upon reasonable request. Supplementary Tables 1–5 summarize the data underlying Figs. 3–5. The NIT maps and coordinates have been deposited in the Electron Microscopy Data Bank (http://www.ebi.ac.uk/pdbe/emdb/) and Protein Data Bank (https://www.rcsb.org/): AtNIT4 (Δϕ = −73.0°) EMD-0320, pdb id: 6i00; AtNIT4 R95T L169A S224Q EMD-4406; AtNIT4 R95T (Δϕ = −73.3°) EMD-4804, Nit6803 Δ291 EMD-4407. Protein sequences are available from Genbank: AtNIT1 NP_851011.1; AtNIT3 NP_190018.1; AtNIT4 NP_197622.1; CrNIT1 XP_006291436.1; CrNIT2 XP_006284056.1; SalNIT1-3 MH567030; Nit6803 BAA10717.1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0505-4.
References
- 1.Martínková, L., Rucká, L., Nešvera, J. & Pátek, M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J. Microbiol. Biotechnol. 33, 8 (2017). [DOI] [PubMed]
- 2.Sewell BT, Berman MN, Meyers PR, Jandhyala D, Benedik MJ. The cyanide degrading nitrilase from Pseudomonas stutzeri AK61 is a two-fold symmetric, 14-subunit spiral. Structure. 2003;11:1413–1422. doi: 10.1016/j.str.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Thuku RN, Weber BW, Varsani A, Sewell BT. Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J. 2007;274:2099–2108. doi: 10.1111/j.1742-4658.2007.05752.x. [DOI] [PubMed] [Google Scholar]
- 4.Woodward JD, et al. Helical structure of unidirectionally shadowed metal replicas of cyanide hydratase from Gloeocercospora sorghi. J. Struct. Biol. 2008;161:111–119. doi: 10.1016/j.jsb.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Dent KC, Weber BW, Benedik MJ, Sewell BT. The cyanide hydratase from Neurospora crassa forms a helix which has a dimeric repeat. Appl. Microbiol. Biotechnol. 2009;82:271–278. doi: 10.1007/s00253-008-1735-4. [DOI] [PubMed] [Google Scholar]
- 6.Williamson DS, et al. Structural and biochemical characterization of a nitrilase from the thermophilic bacterium, Geobacillus pallidus RAPc8. Appl. Microbiol. Biotechnol. 2010;88:143–153. doi: 10.1007/s00253-010-2734-9. [DOI] [PubMed] [Google Scholar]
- 7.Doskočilová A, et al. NITRILASE1 regulates the exit from proliferation, genome stability and plant development. New Phytol. 2013;198:685–698. doi: 10.1111/nph.12185. [DOI] [PubMed] [Google Scholar]
- 8.Woodward JD, Trompetter I, Sewell BT, Piotrowski M. Substrate specificity of plant nitrilase complexes is affected by their helical twist. Commun. Biol. 2018;1:10. doi: 10.1038/s42003-018-0186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pace HC, Brenner C. The nitrilase superfamily: classification, structure and function. Genome Biol. 2001;2:1–9. doi: 10.1186/gb-2001-2-1-reviews0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piotrowski M, Schönfelder S, Weiler EW. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode??-Cyano-L-alanine hydratase/nitrilase. J. Biol. Chem. 2001;276:2616–2621. doi: 10.1074/jbc.M007890200. [DOI] [PubMed] [Google Scholar]
- 11.Piotrowski M. Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry. 2008;69:2655–2667. doi: 10.1016/j.phytochem.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Vorwerk S, et al. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta. 2001;212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- 13.Janowitz T, Trompetter I, Piotrowski M. Evolution of nitrilases in glucosinolate-containing plants. Phytochemistry. 2009;70:1680–1686. doi: 10.1016/j.phytochem.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 14.Agerbirk N, Warwick SI, Hansen PR, Olsen CE. Sinapis phylogeny and evolution of glucosinolates and specific nitrile degrading enzymes. Phytochemistry. 2008;69:2937–2949. doi: 10.1016/j.phytochem.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, et al. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803. J. Struct. Biol. 2014;188:93–101. doi: 10.1016/j.jsb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Heinemann U., Engels D., Burger S., Kiziak C., Mattes R., Stolz A. Cloning of a Nitrilase Gene from the Cyanobacterium Synechocystis sp. Strain PCC6803 and Heterologous Expression and Characterization of the Encoded Protein. Applied and Environmental Microbiology. 2003;69(8):4359–4366. doi: 10.1128/AEM.69.8.4359-4366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005;16:378–384. doi: 10.1016/j.copbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Lutz S. Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol. 2011;21:734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber BW, et al. The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning. J. Biol. Chem. 2013;288:28514–28523. doi: 10.1074/jbc.M113.503284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer, D., Lohkamp, B., Krumpel, M. & Widersten, M. Crystal structure and pH-dependent allosteric regulation of human β -ureidopropionase, an enzyme involved in anticancer drug metabolism. Biochem. J. 475, 2395–2416 (2018). [DOI] [PubMed]
- 21.Thuku RN, Brady D, Benedik MJ, Sewell BT. Microbial nitrilases: versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 2009;106:703–727. doi: 10.1111/j.1365-2672.2008.03941.x. [DOI] [PubMed] [Google Scholar]
- 22.Stevenson DE, Feng R, Storer AC. Detection of covalent enzyme-substrate complexes of nitrilase by ion-spray mass spectroscopy. FEBS Lett. 1990;277:112–114. doi: 10.1016/0014-5793(90)80821-Y. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto, H. et al. Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase. RCSB Protein Data Bank. 10.2210/pdb1uf5/pdb (2003)
- 24.Andrade, J., Karmali, A., CarrondoM. A. & FrazãoC. Structure of amidase from Pseudomonas aeruginosa showing a trapped acyl transfer reaction intermediate state. J. Biol. Chem. 282, 19598–19605 (2007). [DOI] [PubMed]
- 25.Kimani, S. W., Hunter, R., Vlok, M., Watermeyer, J. & Sewell, B. T. Covalent modifications of the active site cysteine occur as a result of mutating the glutamate of the catalytic triad in the amidase from Nesterenkonia sp. RCSB Protein Data Bank. 10.2210/pdb4izs/pdb (2013).
- 26.Nov Y. When second best is good enough: nother probabilistic look at saturation mutagenesis. Appl. Environ. Microbiol. 2012;78:258–262. doi: 10.1128/AEM.06265-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeymer, C. & Hilvert, D. Directed evolution of protein catalysts. Annu. Rev. Biochem.87, 131–157 (2018). [DOI] [PubMed]
- 28.WARE GC, PAINTER HA. Bacterial utilization of cyanide. Nature. 1955;175:900. doi: 10.1038/175900a0. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.DeSantis, G. & DiCosimo, R. Applications of nitrile hydratases and nitrilases. In Biocatalysis for the Pharmaceutical Industry (eds Tao, J.A., Lin, G.-Q. & Liese, A.) (John Wiley & Sons, Singapore, 2009).
- 31.Stalker DM, McBride KE, Malyj LD. Herbicide resistance in transgenic plants expressing a bacterial detoxification gene. Science. 1988;242:419–423. doi: 10.1126/science.242.4877.419. [DOI] [PubMed] [Google Scholar]
- 32.Seffernick JL, Samanta SK, Louie TM, Wackett LP, Subramanian M. Investigative mining of sequence data for novel enzymes: a case study with nitrilases. J. Biotechnol. 2009;143:17–26. doi: 10.1016/j.jbiotec.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan O, et al. Genome mining for the discovery of new nitrilases in filamentous fungi. Biotechnol. Lett. 2011;33:309–312. doi: 10.1007/s10529-010-0421-7. [DOI] [PubMed] [Google Scholar]
- 34.Schreiner U, et al. Directed evolution of Alcaligenes faecalis nitrilase. Enzym. Microb. Technol. 2010;47:140–146. doi: 10.1016/j.enzmictec.2010.05.012. [DOI] [Google Scholar]
- 35.Meyers PR, Rawlings DE, Woods DR, Lindsey GG. Isolation and characterization of a cyanide dihydratase from Bacillus pumilus C1. J. Bacteriol. 1993;175:6105–6112. doi: 10.1128/jb.175.19.6105-6112.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jandhyala D, et al. CynD, the cyanide dihydratase from Bacillus pumilus: gene cloning and structural. Studies. 2003;69:4794–4805. doi: 10.1128/AEM.69.8.4794-4805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JM, Mulelu A, Sewell BT, Benedik MJ. Probing an interfacial surface in the cyanide dihydratase from Bacillus pumilus, a spiral forming nitrilase. Front. Microbiol. 2016;6:1479. doi: 10.3389/fmicb.2015.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pace HC, et al. Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers. Curr. Biol. 2000;10:907–917. doi: 10.1016/S0960-9822(00)00621-7. [DOI] [PubMed] [Google Scholar]
- 39.Nakai T, et al. Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure. 2000;8:729–737. doi: 10.1016/S0969-2126(00)00160-X. [DOI] [PubMed] [Google Scholar]
- 40.Crum MAN, Park JM, Mulelu AE, Sewell BT, Benedik MJ. Probing C-terminal interactions of the Pseudomonas stutzeri cyanide-degrading CynD protein. Appl. Microbiol. Biotechnol. 2015;99:3093–3102. doi: 10.1007/s00253-014-6335-x. [DOI] [PubMed] [Google Scholar]
- 41.Kiziak C, Klein J, Stolz A. Influence of different carboxy-terminal mutations on the substrate-, reaction- and enantiospecificity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Protein Eng. Des. Sel. 2007;20:385–396. doi: 10.1093/protein/gzm032. [DOI] [PubMed] [Google Scholar]
- 42.Crum MA, Park JM, Sewell BT, Benedik MJ. C-terminal hybrid mutant of Bacillus pumilus cyanide dihydratase dramatically enhances thermal stability and pH tolerance by reinforcing oligomerization. J. Appl. Microbiol. 2015;118:881–889. doi: 10.1111/jam.12754. [DOI] [PubMed] [Google Scholar]
- 43.Jenrich R, et al. Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc. Natl Acad. Sci. USA. 2007;104:18848–18853. doi: 10.1073/pnas.0709315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkisyan KS, et al. Local fitness landscape of the green fluorescent protein. Nature. 2016;533:397–401. doi: 10.1038/nature17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen RA. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 46.Booth, D. S., Avila-Sakar, A. & Cheng, Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. 58, 3227. 10.3791/3227 (2011) [DOI] [PMC free article] [PubMed]
- 47.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 48.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/S0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 49.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 50.Scheres, S. H. W. Processing of structurally heterogeneous Cryo-EM data in RELION. Methods Enzymol.579, 125–157 (2016). [DOI] [PubMed]
- 51.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He, S. & Scheres, S. H. W. Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed]
- 54.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. Sect. D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 55.Burnley T, Palmer CM, Winn M. Recent developments in the CCP-EM software suite. Acta Crystallogr. Sect. D Struct. Biol. 2017;73:469–477. doi: 10.1107/S2059798317007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution; pp. 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr. Sect. D. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115–e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abbott JH, Rosenblatt JI. Two stage estimation with one observation on the first stage. Ann. Inst. Stat. Math. 1962;14:229–235. doi: 10.1007/BF02868644. [DOI] [Google Scholar]
- 61.Denault, M. & Pelletier, J. N. Protein Library Design and Screening.In Protein Engineering Protocols. (Humana Press, New York, 2007).
- 62.Maniatis, T., Fritsch, E. F. & Sambrook, J. Molecular Cloning. A Laboratory Manual. (Cold Spring Harbor Laboratory, New York, 1982).
- 63.Nichols ML, Willits CO. Reactions of Nessler’s solution. J. Am. Chem. Soc. 1934;56:769–774. doi: 10.1021/ja01319a003. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expression plasmids generated in this study are available from the corresponding author upon reasonable request. Supplementary Tables 1–5 summarize the data underlying Figs. 3–5. The NIT maps and coordinates have been deposited in the Electron Microscopy Data Bank (http://www.ebi.ac.uk/pdbe/emdb/) and Protein Data Bank (https://www.rcsb.org/): AtNIT4 (Δϕ = −73.0°) EMD-0320, pdb id: 6i00; AtNIT4 R95T L169A S224Q EMD-4406; AtNIT4 R95T (Δϕ = −73.3°) EMD-4804, Nit6803 Δ291 EMD-4407. Protein sequences are available from Genbank: AtNIT1 NP_851011.1; AtNIT3 NP_190018.1; AtNIT4 NP_197622.1; CrNIT1 XP_006291436.1; CrNIT2 XP_006284056.1; SalNIT1-3 MH567030; Nit6803 BAA10717.1.