Abstract
Objective: Previous genome-wide sequencing revealed that Ras-related protein Rab-5B (RAB5B) is a susceptible target in patients with polycystic ovary syndrome (PCOS).
Methods: Direct sequencing was performed to analyze the RAB5B gene rs1045435, rs11550558, rs34962186, rs705700, rs58717357, rs11171718, rs60028217, rs772920 loci genotypes in 300 PCOS patients and 300 healthy controls. The plasma microRNA (miRNA)-24, miR-320 levels were measured by reverse transcription fluorescent quantitative PCR (RT-qPCR).
Results: The risk of PCOS in C allele carriers of RAB5B gene rs1045435 locus was 3.91 times higher than that of G allele. The risk of PCOS in rs11550558 locus G allele was 4.09 times higher than A allele. The risk of PCOS in rs705700 locus C allele was 1.66 times greater than T allele. The risk of PCOS in rs11171718 locus A allele carrier was 3.84 times higher than G allele. The rs11550558 SNP was associated with PCOS risk only in those with age ≥ 31.1 years. And RAB5B gene rs11550558, rs1045435, and rs11171718 SNPs were significantly associated with PCOS risk only in subjects with BMI ≥ 23.8 kg/m2. We also found that the RAB5B gene rs1045435 SNP was associated with plasma miR-24 levels. The RAB5B gene rs11550558, rs705700, rs11171718 SNPs were correlated with plasma miR-230 levels.
Conclusion: The single nucleotide polymorphisms of the rs1045435, rs11550558, rs705700, and rs11171718 loci of the RAB5B gene are associated with PCOS risk. The rs1045435 locus is likely an miR-24 binding site, while rs11550558, rs705700, and rs11171718 loci may be miR-320 binding sites.
Keywords: miR-24, miR-320, polycystic ovary syndrome, Ras-related protein Rab-5B, single nucleotide polymorphism
Introduction
Polycystic ovary syndrome (PCOS) is an endocrine disorder with a complex pathogenesis and a high degree of heterogeneity in clinical manifestations. Signs and symptoms of PCOS include irregular or no menstrual periods, high androgens induced symptoms such as excess hairy and acne, bilateral or unilateral ovarian polycystic changes, difficulty getting pregnant, obesity, insulin resistance and type 2 diabetes [1,2].
PCOS is a common endocrine and metabolic disorder that affects women of childbearing age. Its etiology and pathogenesis are related not only to genetic factors, but also to environmental factors [3–6]. Recent studies have shown that the etiology of PCOS is likely correlated with genes involved in androgen elevated, insulin resistance and insulin-regulating genes, as well as those involved in metabolism, steroid hormones and glandular hormone biosynthesis [7–10].
The Ras-related protein Rab-5B (RAB5B) gene is located on the 12q13.2 chromosome, which encodes a small GTPase involved in cellular phagocytosis, mucosal transport and its receptor expression [11]. These small GTPases mainly affect multiple cellular pathways in the ovarian follicular cells and amplify these signals, which result in corresponding clinical manifestations and endocrine changes in PCOS patients [12,13]. A previous study implicated that the expression of RAB5B gene in skeletal muscle is increased 2–3 times in subjects with insulin resistance [14]. Consistently, RAB5B gene could increase insulin resistance via reducing the function of glucose transporter-4 (GLUT4) on the plasma mucosal surface [15]. Thus, the RAB5B gene might result in the onset of PCOS by affecting insulin function and androgen levels.
In the present study, we aimed to analyze the correlates of the single nucleotide polymorphism of the RAB5B gene on the risk of PCOS. We screened for possible sensitive SNPs from the 3′ URT region of the RAB5B gene. Firstly, eight SNPs were selected by the variation Viewer function in NCBI database with 1000 Genomes MAF above 0.05, which are rs1045435, rs11550558, rs34962186, rs705700, rs58717357, rs11171718, rs60028217, rs772920. We systematically analyzed the correlation between these SNPs and the risk of PCOS in Chinese Han population by a case–control study.
Materials and methods
Ethical statement
The present study was approved by the Institutional Ethics Committees of the following participating institutes: ZheJiang Chinese Medicine and Western Medicine Integrated Hospital/Hangzhou Red Cross Hospital, and First Clinical Medical College at Beijing University of Chinese Medicine. All the participants signed their informed written consent. The present study conforms to the principles outlined in the World Medical Association Declaration of Helsinki.
Study participants
The main study participants comprised 300 cases with PCOS, who were recruited from ZheJiang Chinese Medicine and Western Medicine Integrated Hospital/Hangzhou Red Cross Hospital, and First Clinical Medical College at Beijing University of Chinese Medicine during February 2016 and October 2018. The diagnosis of PCOS was based on two of three criteria described in Rotterdam’s 2003: irregular and/or no ovulation, elevated androgen levels, and ovarian cysts. And subjects with other etiology were excluded from the present study, including congenital adrenal hyperplasia, androgen-secreting tumors, and Cushing’s syndrome related clinical and/or biochemical signs [16]. The irregular or no ovulation usually characterized by oligomenorrhea (<9 cycles per year) or amenorrhea, thus those individuals are under higher risk of infertility. Clinically, hyperandrogenemia is manifested as hirsutism, acne and androgenetic alopecia, and/or total testosterone levels above 55.07 ng/dl (1.91 nmol/l). In the present study, the diagnosis criteria of PCOS is the presence of 12 or more follicles in each ovary, 2 ± 9 mm in diameter, and/or an increase in ovarian volume (>10 ml). PCOS patients with congenital adrenal hyperplasia, Cushing’s syndrome, and androgen-secreting tumors were not included in the study. In addition, patients with diabetes and glucose intolerance as well as PCOS caused by metabolic disorders were also excluded from the present study. Healthy women without any endocrine dysfunction and any other diseases were recruited as controls. All participants in the control group were normal women with normal menstrual cycles, normal ovulation, and no endocrine dysfunction associated with PCOS. The follicular growth of both cases and controls were monitored for 3 months before transvaginal ultrasound detection. All subjects have not received hormonal therapy within 3 months before the present study. We recorded the subject’s age of menarche (AAM) by face-to-face interview and calculated the body mass index (BMI). Peripheral blood was collected from day 3 to day 5 of the menstrual cycle, from all subjects with limosis in the morning. EDTA anticoagulated peripheral blood sample was immediately centrifuged, and then the serum was isolated and stored at −80°C until use. The level of follicle stimulating hormone (FSH), luteinizing hormone (LH), total testosterone, prolactin, thyroid stimulating hormone (TSH), and estradiol (E2) was determined by radioimmunoassay (RIA; Beijing North Institute of Biological Technology of China and the CIS Company of France), the intra- and inter-assay coefficient of variation was controlled within 10%.
Genotyping
We collected leukocytes from serum samples of all participants with limosis and extracted genomic DNA using QIAamp DNA Blood Mini Kit 250 (QIAGEN, Germany, Hilden). The experimental procedures were strictly following the standard operating procedures according to the manufacturer, and the extracted genomic DNA was preserved at −80°C until use. Polymerase Chain Reaction (PCR) was performed according to the PCR primer information of SNPs (Table 1). The total volume of the PCR was 50 μl, including 100 ng of extracted genomic DNA, 10 pmol/μl of Forward and Reverse primers, 100 μM of dNTP, and 1 U/μl of Taq DNA polymerase, 2 mM of Mg2+, and add sterile water to the final volume. The PCR conditions were: 94°C, 5 min; 98°C, 10 s; 58°C, 15 s; 72°C, 2 min, a total of 30 cycles followed by a final extension at 72°C for 5 min. All PCR products were sequenced by BioSune Biotechnology Co., Ltd. and sequence data was analyzed using ChromasPro (version 1.42; Technelysium Pty Ltd) software.
Table 1. PCR primer sequences for RAB5B gene SNPs.
| SNPs | PCR sequencing primers (5′–3′) | Product length |
|---|---|---|
| rs1045435 | Forward primer: GGCCTTATGAGGCAGGTGAG; | 112 bp |
| Reverse primer: TAGGAGGTGGACGTCAGAGG | ||
| rs11550558 | Forward primer: AACCTCCATCCCTACCCCTC; | 124 bp |
| Reverse primer: TGGTGTTTGTTGCTGAAGCG | ||
| rs34962186 | Forward primer: GTGTAGGGTATGGTCTGTGGA; | 113 bp |
| Reverse primer: GCCCAAAGATCCACAGACCA | ||
| rs705700 | Forward primer: TGTAGCTTCCACCTCAACGG; | 85 bp |
| Reverse primer: GACTGTGGTGGACTGAAGGG | ||
| rs58717357 | Forward primer: AGGCCTGTCTCTGTCAACTT; | 73 bp |
| Reverse primer: TGAAGAAACGAGGCGGAAGG | ||
| rs11171718 | Forward primer: CCTTCCGCCTCGTTTCTTCA; | 107 bp |
| Reverse primer: AGCCTCCTTCCTAATAGTGTCA | ||
| rs60028217 | Forward primer: AGGTTGGGCATTAGCCTTCT; | 78 bp |
| Reverse primer: GGCTCTTAAGTGTTTGGAAATGGA | ||
| rs772920 | Forward primer: GGGGTGTTAAGCTGCCATTG; | 281 bp |
| Reverse primer: GGGCTAGTCGAATTTCTCCCC |
Abbreviation: SNPs, single nucleotide polymorphisms.
Reverse transcription-polymerase chain reaction for detecting plasma miRNAs levels
Total RNA was extracted using TRIzol reagent (GIBCO BRL, Rockville, MD) according to the manufacturer’s instruction, and finally added in 60 μl of pre-warmed (95°C) nuclease-free water. The cDNA was synthesized by reverse transcription reaction using Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). The plasma levels of miR-24, miR-320 were then determined by the miScript SYBR Green PCR Kit with U6 as a control. The miR-24 primer sequence was: Forward, 5′-GCG CCG GTG ACT CAG TAC AGC-3′; Reverse, 5′-GCG CCG GTG ACT CAG TAC AGC-3′. The miR-320 primer sequence was: Forward, 5′-ACT ATG GAA AAG CTG GGT TGA GAG-3′; Reverse, 5′-ATT CGT TGA GAG ATC ACA AGC GT-3′. The U6 primer sequence was: Forward, 5′-GCC TGC TCC GGC AGC AC-3′; Reverse, 5′-GAG GTA TTC GCA CCA GAG GA-3′. The PCR conditions were: pre-denaturation at 95°C for 60 s, followed by 95°C, 15 s; 60°C, 30 s; 72°C, 30 s; and 40 cycles followed by 95°C, 15 s; 60°C, 30 s; 95°C, 15 s for analyzing the melting curve. Three replicates were prepared for each sample, and the levels of miR-24 and miR-320 relative to the reference U6 were calculated by 2−ΔΔCT.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 (SPSS Inc, Chicago, IL). Continuous variable data were expressed as mean ± SD, categorical variables were expressed as [n (%)]. Fisher’s Exact Test was applied to compare the genotype distribution of RAB5B gene SNPs between PCOS and control groups. To determine the association between RAB5B gene SNPs and PCOS risk, χ2 test was performed to test whether the genotype distribution is consistent with Hardy–Weinberg equilibrium (HWE), based on the distribution of allele frequencies and genetic models (additive, dominant and recessive models), and adjusted for age, BMI. Odds ratio (OR) and 95% confidence interval (CI) were calculated using unconditional logistic regression analysis. The interaction of RAB5B gene SNPs loci was analyzed by a multi-factor dimensionality reduction (MDR) method. All tests were two-tailed, with P < 0.05 was considered as significant differences.
Results
General characteristics of study subjects
The general clinical data of 300 PCOS patients and 300 control subjects are shown in Table 2. There was no significant difference in age, FSH level and AAM between PCOS patients and healthy controls (P>0.05). However, the BMI, LH, total testosterone, Prolactin, TSH and E2 were significantly increased in PCOS patients compared with those in the control group (P < 0.001).
Table 2. General characteristics of PCOS patients and healthy controls.
| Parameters | PCOS (n = 300) | Control (n = 300) | P |
|---|---|---|---|
| Age (years, mean ± SD) | 30.72 ± 6.81 | 31.56 ± 7.04 | 0.11 |
| BMI (kg/m2, mean ± SD) | 25.01 ± 3.70 | 22.64 ± 3.60 | <0.001 |
| FSH (IU/l, mean ± SD) | 7.31 ± 3.19 | 7.35 ± 2.36 | 0.67 |
| LH (IU/l, mean ± SD) | 15.47 ± 5.858 | 4.9 ± 2.15 | <0.001 |
| Total testosterone (ng/ml, mean ± SD) | 0.94 ± 0.42 | 0.52 ± 0.19 | <0.001 |
| Prolactin (ng/ml, mean ± SD) | 17.18 ± 8.87 | 14.65 ± 3.13 | <0.001 |
| TSH (μIU/ml, mean ± SD) | 2.64 ± 0.83 | 1.52 ± 0.42 | <0.001 |
| E2 (pg/ml, mean ± SD) | 222.30 ± 77.41 | 166.56 ± 73.49 | <0.001 |
| AAM (years, mean ± SD) | 14.27 ± 1.52 | 14.21 ± 2.29 | 0.53 |
Abbreviations: AAM, age at menarche; BMI, body mass index; E2, estradiol; FSH, follicle stimulating hormone; LH, luteinizing hormone; PCOS, polycystic ovary syndrome; TSH, thyroid stimulating hormone.
Association of RAB5B gene SNPs with PCOS risk
The distribution of RAB5B gene SNPs loci genotypes and allelic frequency in the PCOS and control groups is shown in Table 3. The genotype frequencies of the SNPs of the RAB5B gene were consistent with the Hardy–Weinberg equilibrium (P>0.05). The risk of PCOS was significantly increased in heterozygous, homozygous, dominant and recessive models of rs1045435 locus (P < 0.05). And the risk of PCOS in C allele carriers was 3.91 times higher than G allele carriers (95% CI: 2.16–7.17, P<0.001). The risk of PCOS was significantly increased in heterozygous, homozygous, dominant and recessive models of rs11550558 locus (P<0.05), and the risk of PCOS in G allele carriers was 4.09 times higher than A allele carriers (95% CI: 2.22–7.64, P<0.001). While, the genotypes and allele frequencies of rs34962186, rs58717357, rs60028217, and rs772920 were not significantly different between the PCOS and the control groups (P>0.05). The risk of PCOS was higher in the rs705700 locus homozygous, dominant model, and recessive model (P<0.05), and the risk in C allele carrier was 1.66 times higher than those of the T allele (95% CI: 1.28–2.14, P < 0.001). Similarly, the risk of PCOS was significantly increased in heterozygous, homozygous, dominant and recessive models of rs11171718 locus (P<0.05), and the risk in A allele carrier was 3.84 times higher than those of G allele (95% CI: 2.43–6.11, P<0.001).
Table 3. Genotype frequencies of RAB5B gene SNPs loci in PCOS and controls.
| PCOS (n = 300) | Control (n = 300) | HWE P | OR (95%CI)* | P* | |
|---|---|---|---|---|---|
| rs1045435 | |||||
| GG | 255 (85.00%) | 285 (95.00%) | 0.08 | Reference | |
| GC | 32 (10.67%) | 14 (4.67%) | 2.56 (1.28–5.16) | 0.006 | |
| CC | 13 (4.33%) | 1 (0.33%) | 14.53 (1.97–299.74) | 0.001 | |
| Additive model | 1.12 (0.88–1.42) | 0.38 | |||
| Dominant model | 3.35 (1.76–6.46) | <0.001 | |||
| Recessive model | 13.54 (1.84–279.24) | 0.003 | |||
| G | 542 (90.33%) | 584 (97.33%) | Reference | ||
| C | 58 (9.67%) | 16 (2.67%) | 3.91 (2.16–7.17) | <0.001 | |
| rs11550558 | |||||
| AA | 251 (83.67%) | 286 (95.33%) | 0.06 | Reference | |
| AG | 41 (13.67%) | 13 (4.33%) | 3.59 (1.81–7.24) | <0.001 | |
| GG | 8 (2.67%) | 1 (0.33%) | 9.12 (1.15–195.71) | 0.03 | |
| Additive model | 1.14 (0.90–1.45) | 0.30 | |||
| Dominant model | 3.99 (2.08–7.77) | <0.001 | |||
| Recessive model | 8.19 (1.04–175.76) | 0.04 | |||
| A | 543 (90.50%) | 585 (97.50%) | Reference | ||
| G | 57 (9.50%) | 15 (2.50%) | 4.09 (2.22–7.64) | <0.001 | |
| rs34962186 | |||||
| TT | 261 (87.00%) | 266 (88.67%) | 0.06 | Reference | |
| TC | 34 (11.33%) | 31 (10.33%) | 1.12 (0.65–1.93) | 0.77 | |
| CC | 5 (1.67%) | 3 (1.00%) | 1.70 (0.35–9.05) | 0.71 | |
| Additive model | 1.02 (0.80–1.30) | 0.92 | |||
| Dominant model | 1.17 (0.70–1.96) | 0.62 | |||
| Recessive model | 1.68 (0.35–8.93) | 0.72 | |||
| T | 556 (92.67%) | 563 (93.83%) | Reference | ||
| C | 44 (7.33%) | 37 (6.17%) | 1.20 (0.75–1.94) | 0.49 | |
| rs705700 | |||||
| TT | 145 (48.33%) | 173 (57.67%) | 0.08 | Reference | |
| TC | 94 (31.33%) | 102 (34.00%) | 1.10 (0.76–1.60) | 0.67 | |
| CC | 61 (20.33%) | 25 (8.33%) | 2.91 (1.69–5.04) | <0.001 | |
| Additive model | 1.19 (0.90–1.58) | 0.23 | |||
| Dominant model | 1.46 (1.04–2.04) | 0.03 | |||
| Recessive model | 2.80 (1.67–4.76) | <0.001 | |||
| T | 384 (64.00%) | 448 (74.67%) | Reference | ||
| C | 216 (36.00%) | 152 (25.33%) | 1.66 (1.28–2.14) | <0.001 | |
| rs58717357 | |||||
| GG | 234 (78.00%) | 241 (80.33%) | 0.14 | Reference | |
| GC | 61 (20.33%) | 53 (17.67%) | 1.19 (0.77–1.82) | 0.48 | |
| CC | 5 (1.67%) | 6 (2.00%) | 0.86 (0.22–3.22) | 0.80 | |
| Additive model | 1.03 (0.80–1.32) | 0.86 | |||
| Dominant model | 1.15 (0.76–1.74) | 0.55 | |||
| Recessive model | 0.83 (0.22–3.11) | 0.76 | |||
| G | 529 (88.17%) | 535 (89.17%) | Reference | ||
| C | 71 (11.83%) | 65 (10.83%) | 1.11 (0.76–1.60) | 0.65 | |
| rs11171718 | |||||
| GG | 231 (77.00%) | 274 (91.33%) | 0.08 | Reference | |
| GA | 43 (14.33%) | 24 (8.00%) | 2.13 (1.22–3.73) | 0.007 | |
| AA | 26 (8.67%) | 2 (0.67%) | 15.42 (3.51–95.02) | <0.001 | |
| Additive model | 1.19 (0.93–1.52) | 0.18 | |||
| Dominant model | 3.15 (1.89–5.26) | <0.001 | |||
| Recessive model | 14.14 (3.22–87.00) | <0.001 | |||
| G | 505 (84.17%) | 572 (95.33%) | Reference | ||
| A | 95 (15.83%) | 28 (4.67%) | 3.84 (2.43–6.11) | <0.001 | |
| rs60028217 | |||||
| TT | 244 (81.33%) | 251 (83.67%) | 0.07 | Reference | |
| TA | 51 (17.00%) | 44 (14.67%) | 1.19 (0.75–1.90) | 0.50 | |
| AA | 5 (1.67%) | 5 (1.67%) | 1.03 (0.26–4.15) | 0.97 | |
| Additive model | 1.03 (0.81–1.32) | 0.86 | |||
| Dominant model | 1.18 (0.76–1.83) | 0.52 | |||
| Recessive model | \ | \ | |||
| T | 539 (89.83%) | 546 (91.00%) | Reference | ||
| A | 61 (10.17%) | 54 (9.00%) | 1.14 (0.77–1.71) | 0.56 | |
| rs772920 | |||||
| CC | 185 (61.67%) | 177 (59.00%) | 0.06 | Reference | |
| CG | 91 (30.33%) | 99 (33.00%) | 0.88 (0.61–1.27) | 0.53 | |
| GG | 24 (8.00%) | 24 (8.00%) | 0.96 (0.50–1.82) | 0.89 | |
| Additive model | 0.96 (0.73–1.25) | 0.79 | |||
| Dominant model | 0.90 (0.64–1.26) | 0.56 | |||
| Recessive model | \ | \ | |||
| C | 461 (76.83%) | 453 (75.50%) | Reference | ||
| G | 139 (23.17%) | 147 (24.50%) | 0.93 (0.71–1.22) | 0.64 |
Adjusted for age and BMI. Abbreviations: CI, confidence interval; HWE, Hardy–Weinberg equilibrium; OR, odds ratio; PCOS, Polycystic ovary syndrome.
Haplotype analysis for RAB5B gene SNPs
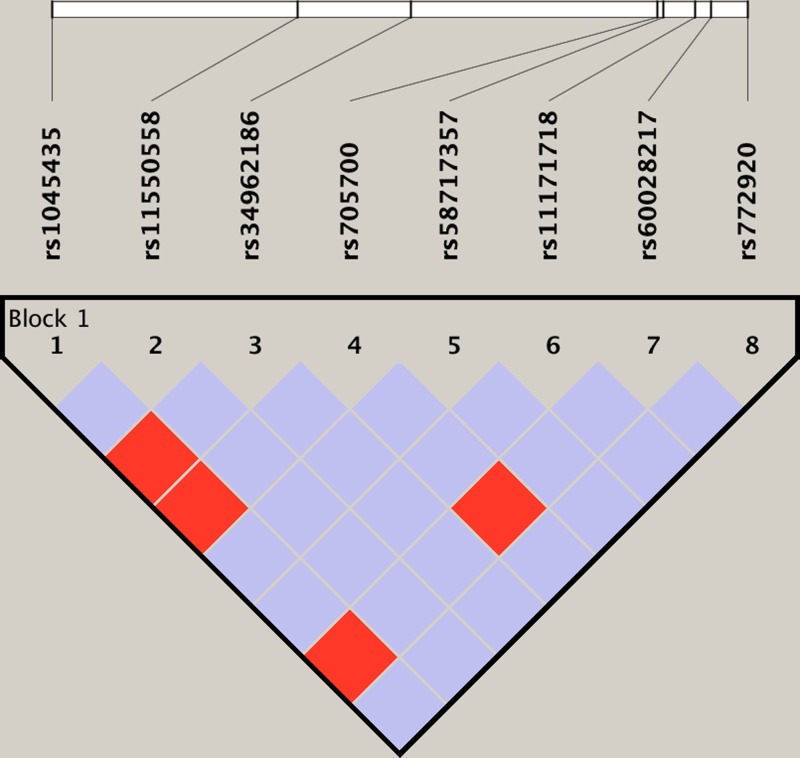
Two haploids were formed in the RAB5B gene rs1045435, rs11550558, rs34962186, rs705700, rs58717357, rs11171718, rs60028217, and rs772920 loci, they are CGCCCAAG and GACCGGTA, respectively. The analysis revealed that CGCCCAAG haplotype is a high risk of PCOS (OR = 3.23, 95% CI: 1.76–5.67, P = 0.005), as shown in Figure 1.
Figure 1. Linkage disequilibrium map of the RAB5B gene SNPs.
Effects of age factor on the relationship between RAB5B gene SNPs and PCOS risk
The mean age of all subjects enrolled in the current study was 31.1 years. We found that the risk of PCOS in subjects with rs1045435 mutation (GC/CC) was significantly higher than wild-type subjects (GG) at ages both <31.1 years and ≥31.1 years (P<0.05) (Table 4). In subjects with age < 31.1 years, there was no statistically significant difference in the risk of PCOS between subjects with rs11550558 mutation (GC/CC) and those with wild-type (GG) (P>0.05). However, in subjects with age > 31.1 years, the risk of PCOS in subjects with rs11550558 mutation (GC/CC) was 9.25 times higher than those of wild-type (GG) (95% CI: 2.97–32.10, P<0.001) (Table 4). Moreover, age factor has no impact in the correlation between rs705700 locus SNPs and PCOS risk (P>0.05) (Table 4). However, in subjects with rs11171718 locus mutation (GA/AA), the risk was significantly higher than those of wild type (GG) at ages both <31.1 and ≥31.1 years (P<0.05) (Table 4).
Table 4. Distribution of genotype frequencies of RAB5B gene SNPs in PCOS and control subjects with different ages.
| PCOS (n = 300) | Control (n = 300) | OR (95% CI)* | P* | |
|---|---|---|---|---|
| <31.1 years | ||||
| rs1045435 | ||||
| GG | 136 (83.44%) | 142 (93.42%) | Reference | |
| GC/CC | 27 (15.56%) | 10 (6.58%) | 2.82 (1.25–6.50) | 0.01 |
| rs11550558 | ||||
| AA | 142 (87.12%) | 142 (93.42%) | Reference | |
| AG/GG | 21 (12.88%) | 10 (6.58%) | 2.10 (0.90–4.98) | 0.09 |
| rs705700 | ||||
| TT | 82 (50.61%) | 92 (60.53%) | Reference | |
| TC/CC | 81 (49.69%) | 60 (39.47%) | 1.52 (0.95–2.43) | 0.09 |
| rs11171718 | ||||
| GG | 120 (73.62%) | 135 (89.47%) | Reference | |
| GA/AA | 43 (26.38%) | 16 (10.53%) | 3.05 (1.57–5.97) | 0.001 |
| ≥31.1 years | ||||
| rs1045435 | ||||
| GG | 119 (86.86%) | 143 (96.62%) | Reference | |
| GC/CC | 18 (13.14%) | 5 (3.38%) | 4.33 (1.46–13.76) | 0.005 |
| rs11550558 | ||||
| AA | 109 (79.56%) | 144 (97.30%) | Reference | |
| AG/GG | 28 (20.44%) | 4 (2.70%) | 9.25 (2.97–32.10) | <0.001 |
| rs705700 | ||||
| TT | 63 (45.99%) | 81 (54.73%) | Reference | |
| TC/CC | 74 (54.01%) | 67 (45.27%) | 1.42 (0.87–2.33) | 0.18 |
| rs11171718 | ||||
| GG | 111 (81.02%) | 138 (93.24%) | Reference | |
| GA/AA | 26 (18.98%) | 10 (6.76%) | 3.23 (1.42–7.53) | 0.003 |
Adjusted for BMI. Abbreviations: CI, confidence interval; OR, odds ratio; PCOS, polycystic ovary syndrome.
Effects of BMI on the relationship between RAB5B gene SNPs and PCOS risk
The mean BMI for all subjects enrolled in the current study was 23.8 kg/m2. We observed that in subjects with BMI <23.8 kg/m2, the risk of PCOS in subjects with rs1045435, rs11550558, rs705700, and rs11171718 mutations was not significant difference from those of wild-type (P>0.05) (Table 5). Among subjects with BMI≥23.8 kg/m2, the risk of PCOS in those with rs11550558, rs1045435 and rs11171718 mutations was significantly higher than that in wild type (P<0.05), however, there was no significant difference in PCOS risk between rs705700 locus mutation and wild-type subjects (P>0.05) (Table 5).
Table 5. Distribution of genotype frequencies of RAB5B gene SNPs in PCOS and control subjects with different BMI.
| PCOS (n = 300) | Control (n = 300) | OR (95% CI)* | P* | |
|---|---|---|---|---|
| <23.8 kg/m2 | ||||
| rs1045435 | ||||
| GG | 68 (98.55%) | 136 (96.45%) | Reference | |
| GC/CC | 1 (1.45%) | 5 (3.55%) | 0.40 (0.02–3.63) | 0.68 |
| rs11550558 | ||||
| AA | 66 (95.65%) | 134 (95.04%) | Reference | |
| AG/GG | 3 (4.35%) | 7 (4.96%) | 0.87 (0.17–3.90) | 0.84 |
| rs705700 | ||||
| TT | 32 (46.38%) | 82 (58.16%) | Reference | |
| TC/CC | 37 (53.62%) | 59 (41.84%) | 1.61 (0.87–2.99) | 0.14 |
| rs11171718 | ||||
| GG | 64 (92.75%) | 127 (90.07%) | Reference | |
| GA/AA | 5 (7.25%) | 14 (9.93%) | 0.71 (0.21–2.23) | 0.70 |
| ≥23.8 kg/m2 | ||||
| rs1045435 | ||||
| GG | 187 (80.95%) | 149 (93.71%) | Reference | |
| GC/CC | 44 (19.05%) | 10 (6.29%) | 3.51 (1.64–7.71) | 0.001 |
| rs11550558 | ||||
| AA | 185 (80.09%) | 152 (95.60%) | Reference | |
| AG/GG | 46 (19.91%) | 7 (4.40%) | 5.40 (2.26–13.51) | <0.001 |
| rs705700 | ||||
| TT | 113 (48.92%) | 91 (57.23%) | Reference | |
| TC/CC | 118 (51.08%) | 68 (42.77%) | 1.40 (0.91–2.14) | 0.13 |
| rs11171718 | ||||
| GG | 167 (72.29%) | 147 (92.45%) | Reference | |
| GA/AA | 64 (27.71%) | 12 (7.55%) | 4.70 (2.35–9.57) | <0.001 |
Adjusted for age. Abbreviations: CI, confidence interval; OR, odds ratio; PCOS, polycystic ovary syndrome.
MDR analysis of RAB5B gene SNPs loci gene–gene interaction
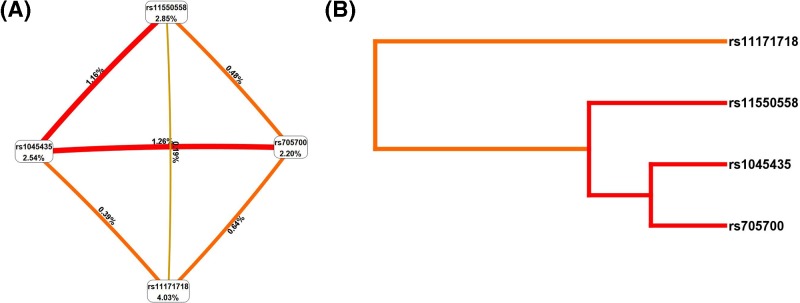
We performed multi-factor dimensionality reduction (MDR) to analyze the interaction of RAB5B gene SNPS rs1045435, rs11550558, rs705700, rs11171718 with the risk of PCOS, and adjusted for age and BMI factors by logistic regression analysis. We found that the interaction between rs1045435 and rs705700 was the best model with consistency of 10/10 (Table 6), and the interaction between rs1045435 and rs705700 loci was the most robust (Figure 2). Subjects with both rs1045435 CC genotype and rs705700 CC genotype had a higher risk of PCOS (OR = 4.12, 95% CI: 2.23–7.31, P<0.001).
Table 6. RAB5B gene rs1045435, rs11550558, rs705700, rs11171718 loci gene–gene interaction.
| Model | Bal. Acc. CV testing | CV consistency | P |
|---|---|---|---|
| rs11171718 | 0.54 | 5/10 | 0.65 |
| rs1045435, rs705700 | 0.65 | 10/10 | <0.001 |
| rs1045435, rs11550558, rs705700 | 0.58 | 9/10 | 0.06 |
| rs1045435, rs11550558, rs705700, rs11171718 | 0.53 | 7/10 | 0.08 |
Abbreviations: Bal. Acc., balanced accuracy; CV, cross-validation.
Figure 2. MDR analysis of RAB5B gene rs1045435, rs11550558, rs705700, rs11171718 loci gene–gene interaction.
(A) Circle graph, the percentage at the bottom of each polymorphism represented entropy of it, and the percentage on each line represented in interaction percentage of entropy between two polymorphisms. (B) Dendogram, red lines represent synergistic interactions, with darker colors representing stronger interactions and orange colors representing weaker interactions.
MDR analysis of the interaction between RAB5B gene SNP loci gene and environmental factors
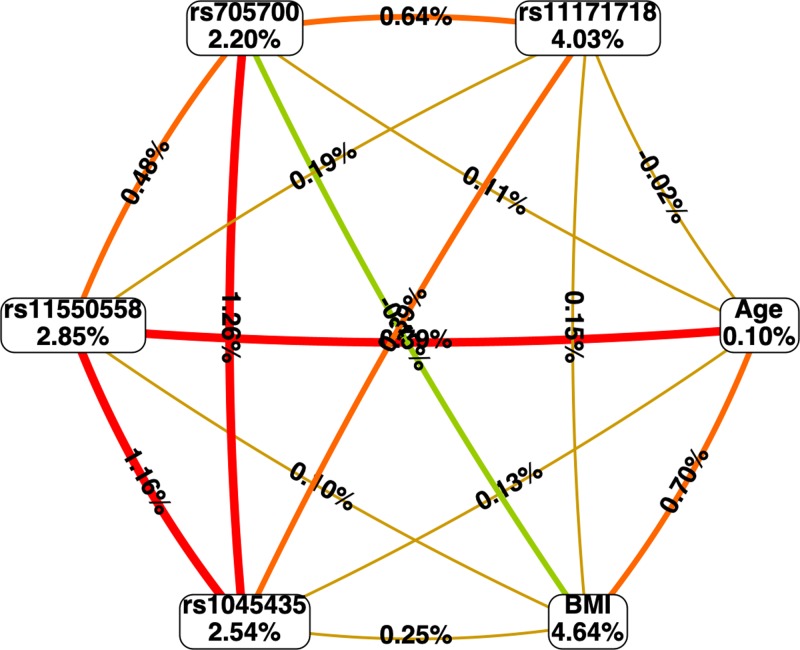
We used MDR to analyze the association between RAB5B gene rs1045435, rs11550558, rs705700, rs11171718 loci and age, BMI with PCOS. The results showed that there was a strong interaction between rs11550558 and BMI (Figure 3), while subjects carrying rs11550558 GG genotype and a BMI more than 23.8 kg/m2 had a higher risk of PCOS (OR = 2.24, 95% CI: 1.13–4.16, P = 0.005).
Figure 3. Circle graph showing the MDR analysis of interactions between RAB5B gene rs1045435, rs11550558, rs705700, rs11171718 loci gene–environmental interaction.
The percentage at the bottom of each polymorphism represented entropy of it, and the percentage on each line represented in interaction percentage of entropy between SNP and environment factors.
The levels of plasma miR-24 and miR-320 in PCOS patients and controls
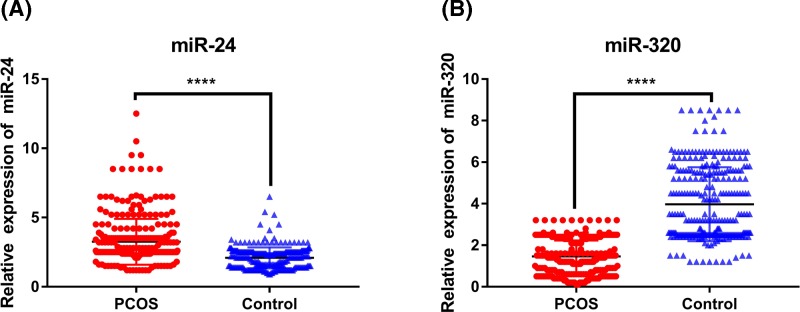
The levels of miR-24 and miR-320 in plasma of PCOS patients and control subjects were determined by RT-PCR. We found that the level of miR-24 in plasma of PCOS patients was significantly higher than that of controls, while the level of miR-320 was significantly lower than that of controls, with both differences were statistically significant (P < 0.001) (Figure 4).
Figure 4. The plasma miR-24 and miR-320 levels in PCOS and controls.
****P<0.001.
Correlations of RAB5B gene SNPs with levels of plasma miR-24, miR-230
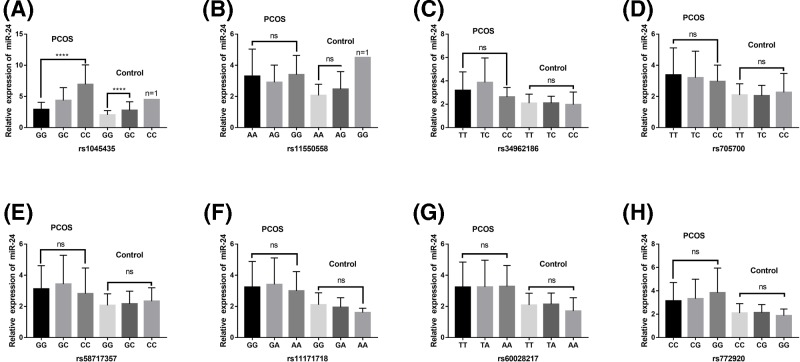
The plasma miR-24 level of RAB5B gene rs1045435 mutation in PCOS patients was significantly higher than that of wild type (P<0.001). Consistently, in controls there is only one case of homozygotes, and the plasma miR-24 levels were significantly higher in heterozygous than wild type (P < 0.001). In both PCOS and control groups, the plasma level of miR-24 was not significant difference in different genotypes of rs11550558, rs34962186, rs705700, rs58717357, rs11171718, rs60028217, rs772920 loci (P>0.05) (Figure 5).
Figure 5. Associations of plasma miR-24 level with RAB5B gene 3′UTR region SNPs in both PCOS and controls.
Plasma miR-24 levels in subjects with different genotypes at (A) rs1045435; (B) rs11550558; (C) rs34962186; (D) rs705700; (E) rs58717357; (F) rs11171718; (G) rs60028217; (H) rs772920. ****P<0.001. Abbreviations: ns, no statistical difference; PCOS, polycystic ovary syndrome.
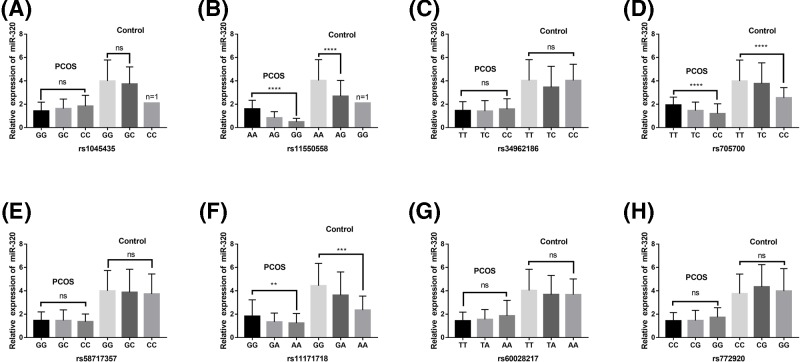
In contrast, in both PCOS and control groups, the plasma miR-230 levels of RAB5B gene rs11550558, rs705700, rs11171718 mutants were significantly lower than those in the wild type (P<0.05). However, there were no significant difference in plasma miR-230 levels between subjects with different genotypes of rs1045435, rs34962186, rs58717357, rs60028217, and rs772920 (P>0.05) (Figure 6).
Figure 6. Associations of plasma miR-320 level with RAB5B gene 3′UTR region SNPs in both PCOS and controls.
Plasma miR-320 levels in subjects with different genotypes at (A) rs1045435; (B) rs11550558; (C) rs34962186; (D) rs705700; (E) rs58717357; (F) rs11171718; (G) rs60028217; (H) rs772920. ****P<0.0001. ***P<0.001. **P<0.01. Abbreviations: ns, no statistical difference; PCOS, polycystic ovary syndrome.
Discussion
PCOS is a highly heterogeneous and multi-gene mediated complex disease, it has been drawn a considerable attention by multiple studies, however, the precise underlying mechanisms remain elusive [1,17,18]. A growing body of evidence suggests that PCOS is affected by a multi-gene interactions together with environmental factors [19,20]. In the last decades, genome-wide association research (GWAS) has become a new approach to analyze the influence of genetic factors on the pathogenesis of PCOS. Using GWAS method for the first time, previous studies have reported three susceptibility genes in the case–control study of Chinese Han PCOS patients [21]. Further, also by GWAS, Shi et al. [22] revealed some new susceptibility genes after expanding the sample size. Although some genes were verified in different populations, the exact functions of these genes were still unclear and controversial. It is obviously necessary to investigate the function of a single gene. Therefore, in the present study, we selected one of the RAB5B genes as a target, aiming to explore its role in the pathogenesis of PCOS through a case–control study.
We studied eight SNPs in the 3′UTR region of RAB5B gene, which are rs1045435, rs11550558, rs34962186, rs705700, rs58717357, rs11171718, rs60028217, rs772920. We found that the risk of PCOS was increased significantly in subjects carrying the C allele at the rs1045435 locus of RAB5B gene, exclusion the interferences of age and BMI factors. In order to assess the effects of age and BMI, we further performed multiple analyses to age and BMI factors. We grouped all data by the median of age and the median of BMI, and found that only in subjects with age ≥31.1 years, the risk of PCOS was associated with rs11550558 SNP. And only in subjects with BMI≥23.8 kg/m2, the risk of PCOS was associated with rs11550558, rs1045435 and rs11171718 loci SNPs. These results indicate that for older female subjects, the risk of PCOS is higher in rs11550558 G allele carriers, and for obese (BMI ≥ 23.8 kg/m2) Chinese Han women, the risk of PCOS is higher in rs11550558 G allele carriers, the rs1045435 C allele carriers or the rs11171718 locus A allele carriers. Hence, it is necessary for these patients to take precautions in advance. Using MDR analysis, we revealed that rs1045435 had robust interaction with rs705700, suggesting that a higher risk of PCOS might we exist for those carrying both rs1045435 C allele and rs705700 C allele. These results also provided clinical significance for PCOS prevention.
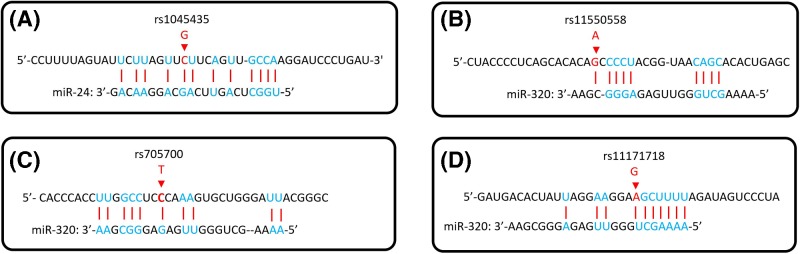
The 3′UTR region contains binding sites for microRNAs (miRNAs). To further investigate whether miRNAs play a major role in the correlation between SNPs in the 3′UTR region of RAB5B gene and PCOS risk, we conducted TargetScan 3.1 analysis (http://www.targetscan.org/mamm_31/), and found two miRNAs, miR-24 and miR-320. The predicted results showed that the binding site of miR-24 to the 3′UTR region of RAB5B gene was located at rs1045435 (Figure 7A), while miR-320 binding site to the 3′UTR region of the RAB5B gene was located at the rs11550558 (Figure 7B), rs705700 (Figure 7C), and rs11171718 loci (Figure 7D). The Smads family proteins play a key role in the transduction of transforming growth factor beta (TGF-β) signaling from cell surface to the nucleus, and different Smads proteins mediate signal transduction by different TGF-β family members [23,24]. Previous studies have shown that miR-24 might inhibit the production of estradiol by suppressing the expression of Smad protein, which causes inhibition of the TGF-β signaling pathway [25]. In the present study, we found that the level of miR-24 in plasma of PCOS patients was significantly higher than that of the control group. However, Sorensen et al. [26] reported that the level of miR-24-3p in follicular fluid of PCOS patients was lower than that of the control group, which is inconsistent with the results of the present study. We considered it may be related to ethnic differences. Further, the regulation of miRNAs expression in vivo is very complicated, and the levels of miRNA might be affected by multiple factors. Meanwhile, we found that the plasma miR-24 level was associated with rs1045435 SNP, and the C allele carrier had both higher risk for PCOS and plasma miR-24 levels. For the underlying mechanism of the relationship between plasma miR-24 levels and rs1045435 SNPs observed in the current study, we speculate that the rs1045435 SNP is associated with the risk of PCOS, while the plasma miR-24 levels are higher in patients with PCOS; however, the higher levels of plasma miR-24 in PCOS patients are not determined by the rs1045435 SNP.
Figure 7. Analyses of miR-24, miR-230 binding sites in RAB5B gene 3′UTR region.
The red arrow indicates the base of the SNPs loci mutations in the 3′UTR region of RAB5B gene.
Furthermore, in consistent with Sorensen et al. [27] and Sang et al. [28], we found that the level of miR-320 in patients with PCOS was significantly lower than that in the control group. Our further studies showed that the level of miR-320 in plasma was correlated with s11550558, rs705700, and rs11171718 SNPs. Therefore, we speculated that the diverse alleles of SNPs may have different binding efficiency to miR-320, which lead to different regulation efficiency of RAB5B gene expression miR-320, and finally cause the difference in the risk of POCS in participants. However, there is no direct evidence in the current study to support this speculation, and further research is needed.
There were several limitations in the present study. Firstly, it is difficult to recruit large samples of PCOS patients. Therefore, the sample size is fairly small for the rare mutant patients, which may affect the objectivity of our results. Secondly, we did not have ex vivo study to verify the precise mechanism underlying regulation of RAB5B gene expression by miR-24 and miR-320, which is one of the difficulties. In addition, we considered that other related genes may also have certain effects on the etiology of PCOS, such as genes involved in androgen elevation, insulin resistance and insulin-regulation, metabolism, steroid hormones, and genes that promote glandular hormone biosynthesis. It is valuable to analyze the interaction between multiple genes, which may represent one of the possible directions for future study. Another limitation of the current study is that we did not analyze the association between these SNPs and clinical symptoms.
Conclusions
Here, we report that RAB5B gene rs1045435, rs11550558, rs705700 and rs11171718 SNPs were associated with PCOS risk, rs1045435 SNP was associated with plasma miR-24, and rs11550558, rs705700, rs11171718 SNPs were correlated to plasma level of miR-320. It is speculated that rs1045435 may be a binding site for miR-24, while rs11550558, rs705700 and rs11171718 may be binding sites for miR-320, which needs further studies to support.
Abbreviations
- BMI
body mass index
- CI
95% confidence interval
- FSH
follicle stimulating hormone
- GWAS
genome-wide association research
- HWE
Hardy–Weinberg equilibrium
- LH
luteinizing hormone
- miRNA
microRNA
- OR
odds ratio
- PCOS
polycystic ovary syndrome
- PCR
polymerase chain reaction
- RAB5B
Ras-related protein Rab-5B
- RT-qPCR
reverse transcription fluorescent quantitative PCR
- TGF-β
transforming growth factor beta
- TSH
thyroid stimulating hormone
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province [grant number LQ19L27005].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
C.W. conceived and designed the experiments. J.Y., C.D., and S.G. performed the experiments. J.Y. and C.D. analyzed the data. J.Y. and C.W. wrote the paper.
References
- 1.Rosenfield R.L. and Ehrmann D.A. (2016) The pathogenesis of polycystic ovary syndrome (PCOS): the hypothesis of PCOS as functional ovarian hyperandrogenism revisited. Endocr. Rev. 37, 467–520 10.1210/er.2015-1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumesic D.A., Oberfield S.E., Stener-Victorin E., Marshall J.C., Laven J.S. and Legro R.S. (2015) Scientific statement on the diagnostic criteria, epidemiology, pathophysiology, and molecular genetics of polycystic ovary syndrome. Endocr. Rev. 36, 487–525 10.1210/er.2015-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dadachanji R., Shaikh N. and Mukherjee S. (2018) Genetic variants associated with hyperandrogenemia in PCOS pathophysiology. Genet Res. Int. 2018, 7624932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosova G. and Urbanek M. (2013) Genetics of the polycystic ovary syndrome. Mol. Cell. Endocrinol. 373, 29–38 10.1016/j.mce.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rojas J., Chavez M., Olivar L., Rojas M., Morillo J., Mejias J.. et al. (2014) Polycystic ovary syndrome, insulin resistance, and obesity: navigating the pathophysiologic labyrinth. Int. J. Reprod. Med. 2014, 719050 10.1155/2014/719050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldani D.P., Skrgatic L. and Ougouag R. (2015) Polycystic ovary syndrome: important underrecognised cardiometabolic risk factor in reproductive-age women. Int. J. Endocrinol. 2015, 786362 10.1155/2015/786362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X.H., Kou L.C., Wang H.M., Bo C.M. and Song X.C. (2019) Genetic polymorphisms of melatonin receptors 1A and 1B may result in disordered lipid metabolism in obese patients with polycystic ovary syndrome. Mol. Med. Rep. 19, 2220–2230 10.3892/mmr.2019.9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crespo R.P., Bachega T., Mendonca B.B. and Gomes L.G. (2018) An update of genetic basis of PCOS pathogenesis. Arch. Endocrinol. Metab. 62, 352–361 10.20945/2359-3997000000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legro R.S., Driscoll D., Strauss J.F. 3rd, Fox J. and Dunaif A. (1998) Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc. Natl. Acad. Sci. U.S.A. 95, 14956–14960 10.1073/pnas.95.25.14956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamanti-Kandarakis E., Kandarakis H. and Legro R.S. (2006) The role of genes and environment in the etiology of PCOS. Endocrine 30, 19–26 10.1385/ENDO:30:1:19 [DOI] [PubMed] [Google Scholar]
- 11.Jean S. and Kiger A.A. (2012) Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat. Rev. Mol. Cell Biol. 13, 463–470 10.1038/nrm3379 [DOI] [PubMed] [Google Scholar]
- 12.Chi X.X., Zhang T., Chu X.L., Zhen J.L. and Zhang D.J. (2018) The regulatory effect of Genistein on granulosa cell in ovary of rat with PCOS through Bcl-2 and Bax signaling pathways. J. Vet. Med. Sci. 80, 1348–1355 10.1292/jvms.17-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornes R., Ormazabal P., Rosas C., Gabler F., Vantman D., Romero C.. et al. (2010) Changes in the expression of insulin signaling pathway molecules in endometria from polycystic ovary syndrome women with or without hyperinsulinemia. Mol. Med. 16, 129–136 10.2119/molmed.2009.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao S., Zhu J. and Garvey W.T. (1998) Cloning of Rab GTPases expressed in human skeletal muscle: studies in insulin-resistant subjects. Horm. Metab. Res. 30, 656–662 10.1055/s-2007-978953 [DOI] [PubMed] [Google Scholar]
- 15.Tessneer K.L., Jackson R.M., Griesel B.A. and Olson A.L. (2014) Rab5 activity regulates GLUT4 sorting into insulin-responsive and non-insulin-responsive endosomal compartments: a potential mechanism for development of insulin resistance. Endocrinology 155, 3315–3328 10.1210/en.2013-2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayasena C.N. and Franks S. (2014) The management of patients with polycystic ovary syndrome. Nat. Rev. Endocrinol. 10, 624–636 10.1038/nrendo.2014.102 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y.D., Li Y., Feng S.X., Ye D.S., Chen X., Zhou X.Y.. et al. (2017) Long noncoding RNAs: potential regulators involved in the pathogenesis of polycystic ovary syndrome. Endocrinology 158, 3890–3899 10.1210/en.2017-00605 [DOI] [PubMed] [Google Scholar]
- 18.de Melo A.S., Dias S.V., Cavalli Rde C., Cardoso V.C., Bettiol H., Barbieri M.A.. et al. (2015) Pathogenesis of polycystic ovary syndrome: multifactorial assessment from the foetal stage to menopause. Reproduction 150, R11–24 10.1530/REP-14-0499 [DOI] [PubMed] [Google Scholar]
- 19.Diamanti-Kandarakis E., Piperi C., Spina J., Argyrakopoulou G., Papanastasiou L., Bergiele A.. et al. (2006) Polycystic ovary syndrome: the influence of environmental and genetic factors. Hormones (Athens) 5, 17–34 10.14310/horm.2002.11165 [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta S. and Reddy B.M. (2013) The role of epistasis in the etiology of Polycystic Ovary Syndrome among Indian women: SNP-SNP and SNP-environment interactions. Ann. Hum. Genet. 77, 288–298 10.1111/ahg.12020 [DOI] [PubMed] [Google Scholar]
- 21.Chen Z.J., Zhao H., He L., Shi Y., Qin Y., Shi Y.. et al. (2011) Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat. Genet. 43, 55–59 10.1038/ng.732 [DOI] [PubMed] [Google Scholar]
- 22.Shi Y., Zhao H., Shi Y., Cao Y., Yang D., Li Z.. et al. (2012) Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 44, 1020–1025 10.1038/ng.2384 [DOI] [PubMed] [Google Scholar]
- 23.Xu F., Liu C., Zhou D. and Zhang L. (2016) TGF-beta/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 64, 157–167 10.1369/0022155415627681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derynck R. and Zhang Y.E. (2003) Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577–584 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- 25.Chan M.C., Hilyard A.C., Wu C., Davis B.N., Hill N.S., Lal A.. et al. (2010) Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 29, 559–573 10.1038/emboj.2009.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen A.E., Wissing M.L., Englund A.L. and Dalgaard L.T. (2016) MicroRNA species in follicular fluid associating with polycystic ovary syndrome and related intermediary phenotypes. J. Clin. Endocrinol. Metab. 101, 1579–1589 10.1210/jc.2015-3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen A.E., Wissing M.L., Salo S., Englund A.L. and Dalgaard L.T. (2014) MicroRNAs related to polycystic ovary syndrome (PCOS). Genes (Basel) 5, 684–708 10.3390/genes5030684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sang Q., Yao Z., Wang H., Feng R., Wang H., Zhao X.. et al. (2013) Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J. Clin. Endocrinol. Metab. 98, 3068–3079 10.1210/jc.2013-1715 [DOI] [PubMed] [Google Scholar]