Abstract
Dysregulation of the enzymes involved in the pentose phosphate pathway (PPP) is known to promote tumorigenesis. Our recent study demonstrated that ribose-5-phosphate isomerase (RPIA), a key regulator of the PPP, regulates hepatoma cell proliferation and colony formation. Our studies in zebrafish reveal that RPIA-mediated hepatocarcinogenesis requires extracellular signal-regulated kinase (ERK) and β-catenin signaling. To further investigate RPIA-mediated hepatocarcinogenesis, two independent lines of transgenic zebrafish expressing human RPIA in the liver were generated. These studies reveal that RPIA overexpression triggers lipogenic factor/enzyme expression, steatosis, fibrosis and proliferation of the liver. In addition, the severity of fibrosis and the extent of proliferation are positively correlated with RPIA expression levels. Furthermore, RPIA-mediated induction of hepatocellular carcinoma (HCC) requires the ERK and β-catenin signaling pathway but is not dependent upon transaldolase levels. Our study presents a mechanism for RPIA-mediated hepatocarcinogenesis and suggests that RPIA represents a valuable therapeutic target for the treatment of HCC.
Using a transgenic zebrafish model, we found overexpression RPIA in the liver facilitate the steatosis at 5-months, fibrosis at 7-month, and promote hyperplasia or hepatocellular carcinoma (HCC) at 9 and 11 months. We discovered p-ERK was elevated as early as 5-months, and β-catenin signaling pathway were activated from 7-months revealed by IHC. Using oral gavage to treat 7-months RPIA transgenic fish with the inhibitors for one month, we found blockade ERK pathway diminished the expression of lipogenic factor, while β-catenin inhibitor reverted the upregulation of cell cycle/proliferation markers mediated by RPIA overexpression. The combination of β-catenin and ERK inhibitors synergistically reduced the RPIA-induced cellular proliferation in xenotransplantation assay. Our data suggested activation of ERK pathway induce lipogenesis while activation of β-catenin pathway promoting cancer formation at later stage, inhibition of ERK and β-catenin signaling can significantly reduce HCC progression.
Introduction
Hepatocellular carcinoma (HCC), the predominant form of liver cancer, is one of the most common types of cancers in Eastern Asia and sub-Saharan Africa and is the third most common cause of cancer-related deaths worldwide (1,2). The main causes of liver cancer include alcohol consumption-induced cirrhosis as well as infection with the hepatitis B or hepatitis C viruses. The molecular mechanisms underlying the development of liver cancer are complicated and heterogeneous, and consequently, the therapies to treat late-stage HCC are currently lacking (3). Therefore, it is important to identify precise biomarkers of liver cancer and devise more effective means of diagnosing and treating the disease.
Cancer development is positively correlated with age. Both aging and cancer are considered as metabolic diseases (4) that share some common underlying mechanisms (5). Therefore, understanding the mechanisms by which antiaging genes exert their effects may facilitate the development of new cancer therapies and cures (6). Our previous genetic studies demonstrate that downregulation of RPIA extends lifespan and attenuates neurodegeneration by polyglutamine-induced toxicity in Drosophila (7), ribose-5-phosphate isomerase (RPIA) also plays an important role in the pentose phosphate pathway (PPP), a cellular signaling cascade whose dysregulation can lead to cancer cell survival and proliferation (8). These observations led us to investigate the role of human RPIA in HCC using a transgenic zebrafish model.
The PPP is an important anabolic pathway that participates in cell proliferation. The pathway is critical not only for glucose metabolism but also for nucleotide biosynthesis in tumor cells (9). The PPP reduces NADP+ to nicotine adenine dinucleotide phosphate hydrate (NADPH) through glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD) in its oxidative phase and produces ribose-5-phosphate (R5P) for nucleotide synthesis in its non-oxidative phase (10,11). It has been reported that maintaining the balance between the oxidative and non-oxidative phases of the PPP is critical for cancer cell survival (12) and that highly differentiated cancer cells usually display reduced gluconeogenesis, increased PPP activity and enhanced cell proliferation relative to undifferentiated cancer cells (13).
The PPP plays an important role in cancer development and can induce oncogenesis (14). Previous studies in both humans and transgenic mice have implicated the role of the PPP enzyme transaldolase (TALDO) in cirrhosis and hepatocarcinogenesis (15,16). TALDO deficiency leads to the failure of recycling R5P for the oxidative PPP, a decrease in NADPH, an increase in oxidative stress and activation of β-catenin within the liver. Another key enzyme in the non-oxidative phase of the PPP, transketolase-like 1 (TKTL1), was found to be upregulated in invasive colon carcinoma (17). Moreover, suppression of TKTL1 expression reduced tumor cell growth (18) and decreased liver cancer cell proliferation rates (19). The enzymatic activity of RPIA, a key enzyme in the non-oxidative phase of the PPP, is increased in the serum of most patients with HCC (20), and shRNA-mediated RPIA knockdown inhibits tumor growth in xenotransplantation models (21). These studies indicate that RPIA plays an important role in human cancer development.
Using cell lines and a xenograft mouse model, we demonstrated that RPIA plays a role in liver cancer development (8). In human HCC, activation of the MAPK/MEK/ERK cascade is correlated with tumor growth and progression (22). The serine-threonine protein phosphatase 2A (PP2A) inhibits extracellular signal-regulated kinase (ERK) activation and signaling during cell apoptosis and survival (23,24), functions as a tumor suppressor and is considered a potential target for cancer therapies (25). We demonstrated that RPIA modulates cell proliferation and colony formation by inhibiting PP2A and activating pERK1/2 (8). We also found that in human colorectal cancer (CRC), RPIA stabilizes β-catenin, thereby allowing it to activate its downstream target genes during carcinogenesis of colorectal cancer (26).
The zebrafish (Danio rerio) research model has been used in a variety of biomedical research studies, including angiogenesis studies, cancer studies and drug screenings (27–29). As zebrafish shares up to 87% of their genome with humans, results obtained with zebrafish can often be extrapolated to humans. Moreover, zebrafish reach sexual maturity at ~3 months of age and produce a large number of offspring (100~250 per generation) (30). Another advantage of the zebrafish system is the ability to use Tol2-mediated transgenesis (31), an approach that we have successfully used to create numerous experimental fish lines (32–34). The Tol2 element is a naturally occurring, active transposable element that has a high transposition efficiency and confers the ability to transfer large DNA fragments (35).
To investigate RPIA-mediated hepatocarcinogenesis, we established two independent transgenic zebrafish lines (fabp10a: RPIA; myl7:GFP-Tg1 and fabp10a: RPIA; myl7:GFP-Tg2), in which the liver-specific promoter (fabp10a) drives human RPIA expression and the heart-specific promoter (myl7) expresses GFP as a transgene marker. We demonstrated that human RPIA plays an important role in hepatocarcinogenesis and re-confirmed the involvement of ERK signaling in RPIA-mediated tumorigenesis in HCC. We also found that RPIA-mediated hepatocarcinogenesis requires β-catenin upregulation but is independent of TALDO activation. This study sheds new light on the functions of RPIA as an oncogene and indicates that therapies targeting these molecules may be valuable for the treatment of HCC.
Materials and methods
Transgenic zebrafish lines
Human RPIA cDNA was used to generate the final expression construct, pTol2-fabp10a: RPIA; myl7: EGFP, using the MultiSite Gateway cloning system (Thermo Fisher Scientific). MyI7 is a heart-specific promoter which is placed in front of GFP as an indicator for selecting transgenic fish after microinjection. The coding region of human RPIA (NM_144563.2) was PCR amplified with the attB1-F-RPIA and attB2-R-RPIA primer pair using cDNA from the HEK293 cell line as a template. PCR was performed using a KOD FX (TOYOBO, Japan) and a 994 bp amplicon. The following forward primer was used for Gateway cloning: attB1-F-RPIA (Tm: 59°C): 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTATGCAGCGCCCCGGGCC3′, and the following reverse primer was used for Gateway cloning: attB2-R-RPIA (Tm:58°C):5′GGGGACCACTTTGTACAAGAAAGCTGGGTTCAACAGAAAGGCTTCTCCCTCATG3′. After PCR amplification of RPIA cDNA, MultiSite Gateway Three-Fragment Vector Construction Kit (Thermo Fisher Scientific) was used to generate the final expression construct. Transgenic zebrafish were generated via microinjections of the above constructs and selected as described previously (32). Once the F2 generation was produced, 50 fish each from the RPIA Tg1 and RPIA Tg2 lines as well as 50 EGFP-mCherry transgenic fish (controls) were analyzed in this study. Ten fish each were collected at 3, 5, 7, 9 and 11 months of age, and liver tissue was obtained. One third of tissue was embedded for immunohistochemical (IHC) staining, one-third of the tissue were used to extract RNA and quantitative polymerase chain reaction (QPCR) analysis and remaining one-third of tissue was frozen immediately and stored at −80oC for protein analysis.
Zebrafish maintenance
All zebrafish experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the National Health Research Institute (NHRI) and were in accordance with the International Association for the Study of Pain guidelines (protocol number: NHRI-IACUC-104157-A). Zebrafish were maintained at the Taiwan Zebrafish Core Facility at NHRI (TZeHN) under an automated 14-h light, 10-h dark cycle and a constant temperature of 28°C. The TZeNH is a government-funded core facility and has been AAALAC accredited since 2015.
Hematoxylin and eosin (H&E) staining
H&E staining was performed as described previously (32). Briefly, tissue sections were surrounded with a liquid blocker (PapPen) and then incubated with 4% paraformaldehyde (PFA) dissolved in PBS (pH 7.2) for 5 min and then washed with running tap water for 1 min to remove the formaldehyde. The slides were subsequently drained on a tissue paper before being incubated with Mayer’s hematoxylin solution for 5 min to stain the nuclei dark and then washed under warm running tap water for 10 min. Acetic acid (1:100) was then added to a 0.5% eosin solution (Merck 1.09844.1000), which was used to stain the tissues before they were incubated for 10 min. The slides were then placed in a glass chamber and washed thrice with ddH2O for 1 min each to remove the excess eosin. The sections were then dehydrated with 70% ethanol (EtOH) for 1 min, 90% EtOH for 30 s, 100% EtOH for 30 s and xylene for 30 s, after which the slides were mounted with one to two drops of xylene-based mounting media and covered with coverslips in a manner intended to avoid bubble formation. The slides were then pressed under a heavy weight for 10 min at room temperature before being stored at room temperature.
Oil Red O staining
Oil Red O staining was performed as described previously (32). Briefly, tissue specimens were harvested, fixed in 4% PFA/PBS overnight at 4°C and then washed thrice with PBS for 15 min each at 4°C before being transferred to a 10% sucrose/PBS solution for a 3-h incubation at 4°C. The tissues were then transferred to a 30% sucrose/PBS solution and incubated overnight at 4°C. The tissues were subsequently transferred to a 30% sucrose/PBS mixture containing OCT in 1:1 ratio, incubated for 2 h at room temperature then transferred to OCT mounting medium, cut into sections (5 um thick), stored at −80°C and then placed on poly-l-lysine-coated slides. The sections were dried for 30 min at 37°C, washed thrice with PBS for 10 min each, fixed in 4% PFA/PBS for 20 min, washed with PBS for 5 min and washed with 60% isopropanol for 5 min. The sections were then stained with 0.3% Oil Red O in 60% isopropanol for 2 h before being washed with 60% isopropanol for 5 min and tap water for 1 min. The sections were mounted with aqueous mounting media.
Sirius red staining
Sirius red staining was performed as described previously (32). Briefly, tissue sections were deparaffinized using non-xylene, rehydrated in distilled water with EtOH and then treated with 80 µl of solution A for periods of 2 min and treated with 100 µl of solution B before being incubated at 70 rpm for 2 h at room temperature. The sections were subsequently treated with 80 μl of solution C to stop the reaction before being dehydrated and then mounted with a xylene-based paramount solution and examined via polarized light microscopy.
Immunohistochemistry (IHC) staining
IHC staining was performed as described previously (32). The following primary antibodies were used for the immunohistochemistry studies of the zebrafish liver tissue specimens: PCNA mouse mAb (CST#2586, Cell Signaling Technology) and phospho-Erk1/2 (Thr202/Tyr204) rabbit mAb (CST#4370, Cell Signaling), ERK rabbit mAb (CST#4695, Cell Signaling), β-catenin rabbit mAb [E247] (GTX61089, GeneTex), AMPKα mouse mAb (CST#2793, Cell Signaling) and Phospho-AMPKα (Thr172) rabbit mAb (CST#2535, Cell Signaling). The liver tissue sections were deparaffinized using non-xylene and then rehydrated in distilled water with EtOH, after which they were placed in a staining jar containing fresh antigen retrieval buffer (0.05% Tween 20, sodium citrate, 10 mM, pH 6.0) to prevent protein cross-linking that may inhibit antigen binding, heated at 95°C for 20 min and then washed thrice in TBS for 5 min each. The slides were then placed in 3% hydrogen peroxide (H2O2) diluted with 100% methanol for 20 min to minimize background signals and washed thrice with TBS for 5 min each. The sections were subsequently blocked with 5% goat serum (blocking solution) in Tris-buffered saline, 0.1% Tween 20 (TBST) for 1 h to prevent non-specific binding before being incubated with the appropriate primary antibodies overnight at 4°C. The slides were subsequently washed thrice with TBST for 10 min each and then soaked in Tris-buffered saline (TBS), after which they were incubated with the appropriate biotin-conjugated secondary antibodies for 30 min, washed thrice with TBST for 10 min each and soaked in TBS. The sections were subsequently treated with avidin–biotin complex (ABC) reagent (Vectastain Elite ABC Kit, Vector Laboratories) for 30 min, washed thrice with TBST for 10 min each and soaked in TBS. The slides were then stained with 3,3’-Diaminobenzidine (DAB) (Invitrogen Liquid DAB Substrate Kit) and washed with ddH2O after staining brown before being counterstained with hematoxylin for 3 min, centrifuged for 3 min and then washed thrice with ddH2O for 2 min each. The sections were then dehydrated before being mounted using a xylene-based paramount solution and examined via polarized light microscopy.
Immunofluorescent staining
The similar procedures were as described in IHC were used for the liver tissue sections prior to primary antibody staining. The following primary antibodies were used for the immunofluorescence of the zebrafish liver tissue specimens: phospho-Erk1/2 (Thr202/Tyr204) rabbit mAb (CST#4370, Cell Signaling), ERK rabbit mAb (CST#4695, Cell Signaling), β-catenin rabbit mAb [E247] (GTX61089, GeneTex) and Phospho-Histone H3 (Ser10) rabbit mAb (CST#9701, Cell Signaling). The slides were washed thrice with 0.1% TBST for 10 min each and then incubated with the Goat Anti-Rabbit IgG (H&L), DyLight 488 antibody (Thermo Fisher Scientific) in room temperature for 30 min. After wash thrice with 0.1% TBST for 10 min each, sections were counterstained with 1 μg/ml DAPI for 3 min and washed with TBS once for 3 min. The slides were mounted with Aqueous Mount media (REF: RS-C-581, Rapid Science) and examined by fluorescence microscopy.
Liver tissue collection and paraffin section preparation
The fish were killed, and the liver was removed and divided into two portions, which were used for RNA isolation and paraffin section preparation, respectively. Some of the liver tissues were frozen in liquid nitrogen immediately after excision and stored at −80°C until needed for RNA isolation, while the other liver tissues were fixed in 10% formalin solution (Sigma–Aldrich, St. Louis, MO) for histological analysis. The fixed tissues were embedded in paraffin and cut into 5 μm thick sections before being mounted on poly-l-lysine-coated slides and stained with hematoxylin and eosin (H&E staining). This experiment was performed by the pathology facility at the NHRI in Taiwan.
Total RNA isolation, reverse transcription-PCR
Total RNA was isolated from individual liver tissue (~30 mg) using NucleoSpin® RNA Kit (MACHEREY-NAGEL), and complementary DNA (cDNA) was synthesized by a High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s instructions.
Real-time quantitative polymerase-chain-reaction (QPCR)
First-strand cDNA was used as a template for QPCR, which was performed in triplicate using a SYBR Green QPCR Master Mix Kit (Applied Biosystems) and an ABI PRISM 7900 System. After reverse transcription, the cDNA was diluted 100X with RNase-free water. A reaction mixture comprising the following compounds was added to each well of a 384-well QPCR plate: 3.8 μl of cDNA (diluted with RNase-free water), 1.2 μl of 2.5 μM primer mix (forward and reverse) and 5.0 μl of 2x SYBR Green Mix. The plate was then covered with an ‘optical adhesive cover,’ and the bubbles were removed with a ‘sealing comb.’ QPCR was performed on an ABI HT-7900 machine and comprised the following: steps: Stage I: 50°C for 2 min, 95°C for 5 min, followed by 4°C; Stage II: 95°C for 10 min; Stage III (40 cycles): 95°C for 15 s and 60°C for 1 min; and Stage IV: 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s.
After normalization to actin expression levels (an internal control), the expression levels of several genes of interest in the experimental and control groups were calculated using the comparative Ct method. The fold differences in expression between the two groups were calculated based on the △△Ct method using the following formula: △△Ct = (Cttarget − Ctactin)treatment − (Cttarget − Ctactin) control, and fold change = 2−△△Ct. All experiments were performed in triplicate (biologically), and the means of the results of the three experiments are presented. At least three independent samples were used for QPCR, and the standard error was calculated and presented as the median ± standard error.
Cell culture
The hepatoma cell lines Hep3B and PLC5 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Life Technologies) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin and incubated at 37°C and 5% carbon dioxide. Those cell lines were authentically confirmed by Mission Biotech (Taipei, Taiwan) using a Promega GenePrint 10 System. Plasmid DNA transfection for 1 × 106 cells in 10 cm dish was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s manual in Opti-MEM medium.
Fractionation and western blotting analysis
Total protein was extracted from cells using whole-cell extract lysis buffer. Lysates were vibrated for 30 min and centrifuged at 13 200 rpm for 20 min at 4°C. Fractionation protocol and western blotting were performed as described in our previous study (26). Primary antibodies include RPIA (Cat# ab67080, Abcam), β-catenin (Cat# ab22656, Abcam), β-tubulin (Cat# ab52866, Abcam) and lamin A/C (Cat# ab108922, Abcam).
NADP/NADPH quantification
The liver tissues were harvested and washed with 500 μl cold-PBS in the microfuge tube. The samples were prepared and quantified for NADP/NADPH by NADP/NADPH Quantification Colorimetric Kit (Cat # K347-100, BioVision). The tissues were homogenized with 500 μl of NADP/NADPH extraction buffer and then incubated on ice for 10 min. The samples were collected by centrifugation at 11 000 rpm for 10 min at 4°C and the extracted NADP/NADPH solution was transferred into 10 kDa molecular weight cut-off filters (Cat # 1997-25). The extracted NADP/NADPH solution was harvested in a new tube as total NADP/NADPH (NADPt) samples. Fifty miroliter of NADPt and 100 μl of NADP cycling mix (2 μl enzyme + 98 μl buffer) were transferred into a 96-well plate and incubated at room temperature for 5 min to convert NADP to NADPH. Ten microliter NADPH developers were added into each well for reaction 4 h at room temperature. The colorimetric reaction in the plate was read at OD450 nm for analyzing the quantification of NADP/NADPH.
Xenotransplantation
The amount of treated PLC5 was prepared with 9 × 105 cells and re-suspended in 1 ml PBS. Carboxyfluorescein succinimidyl ester (CFSE) with 5 μl was used to stain and monitor the cells. After CFSE incubation, the cells were washed by PBS twice and re-suspended in 20 μl PBS. Injection volume was 4.6 nl containing cells ~200 cells into the 2-day post-fertilization (dpf) embryos yolk. The CFSE-labeled cells were checked 2 h later after transplantation. The fluorescent signal was detected at 1 and 3 day post-injection (dpi). The change in proliferation was determined by the percentage of fluorescent intensity changes between 1 and 3 dpi by dividing the difference of fluorescent intensity by original intensity as the formula: (3 dpi − 1 dpi)/1 dpi.
Statistical analysis
Statistical analysis of the results of the above experiments was performed using two-tailed unpaired t-test with Welch’s correction in Prism graphpad 7. In all statistical analyses, P values of <0.05 were considered statistically significant and were presented as follows: *: 0.01 < P ≤ 0.05; **: 0.001 < P ≤ 0.01; ***: 0.01 <P ≤ 0.001 and ****: 0.001 <P ≤ 0.0001.
Results
RPIA overexpression causes liver steatosis and fibrosis
We demonstrated previously that RPIA plays a crucial role in HCC and that the expression levels of RPIA are correlated with several tumorigenic characteristics (8). To study the underlying mechanisms of hepatocarcinogenesis progression, two independent transgenic zebrafish lines (Tg1 and Tg2) with liver-specific overexpression of RPIA were generated.
To monitor hepatocarcinogenesis in RPIA-transgenic fish, liver samples were collected from zebrafish aged at 3, 5, 7, 9 and 11 months. QPCR analysis confirmed that RPIA expression levels in transgenic fish were significantly upregulated at all time points relative to control fish (fabp10a: EGFP-mCherry) (Supplementary Figure S1A, available at Carcinogenesis Online). In addition, it was observed that the RPIA expression levels were higher in the Tg2 line than in the Tg1 line (Supplementary Figure S1A, available at Carcinogenesis Online).
In humans, fatty liver disease (FLD) is typically the initial step in hepatocarcinogenesis, followed by cirrhosis and HCC. To determine whether RPIA-transgenic zebrafish develops HCC in a similar process, QPCR was used to detect the expression of the lipogenic factors, sterol regulatory element-binding transcription factor 1 (srebp1) and carbohydrate-responsive element-binding protein (chrebp1) as well as the lipid metabolism and synthesis-related enzymes, diglyceride acyltransferase 2 (dgat2) and phosphatidate phosphatase (pap) (Supplementary Figure S1B, available at Carcinogenesis Online). As early as 3 months, the expression levels of chrebp1, dgat2 and pap were upregulated in both transgenic fish lines relative to similarly aged controls. Higher levels of the lipogenic factors srebp1 were detected in the transgenic lines, relative to similarly aged controls, slightly later at 5 months of age (Supplementary Figure S1B, available at Carcinogenesis Online).
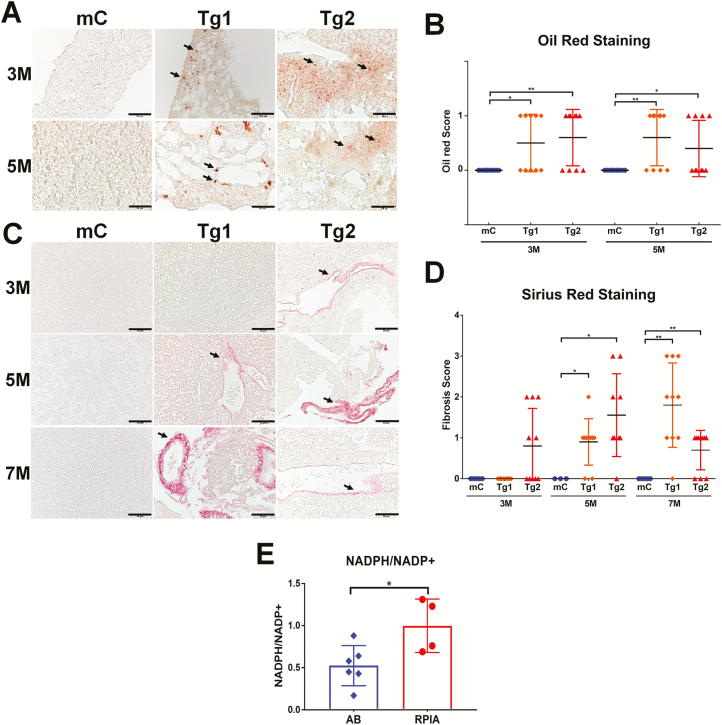
Oil Red O staining was used to stain the neutral triglycerides and lipids on the frozen liver sections of RPIA-transgenic zebrafish, and the results revealed the significant accumulation of lipid droplets indicated as arrow at 3 and 5 months compared with the control (Figure 1A). Statistically, the Tg1 and Tg2 RPIA-transgenic fish both exhibit significantly more lipid accumulation than controls at 3 and 5 months of age (Figure 1B, orange diamond for Tg1, red triangle for Tg2). H&E staining of liver tissue further reveals the presence of clear intracytoplasmic vacuoles indicating that, between 3 and 5 months of age, lipid accumulation happens in the RPIA-transgenic fish (Figure 2). Together these results demonstrate that RPIA-transgenic fish developed steatosis between 3 and 5 months of age, which RPIA overexpression triggers lipogenic factor/enzyme expression and that steatosis is induced at an early stage of development similar to that observed in human hepatocarcinogenesis.
Figure 1.
Steatosis and fibrosis develop in RPIA-transgenic zebrafish. (A) Representative pictures of Oil Red O staining results from liver tissues of 3- and 5-month-old control (fabp10a: EGFP-mCherry, mC) and two Tg (fabp10a: RPIA) lines (Tg1 and Tg2) magnified ×400. Scale bar: 50 μm. (B) Statistical analysis of the Oil Red O staining results for the 3- and 5-month-old RPIA Tg 1 (orange diamond) and Tg2 (red triangle) compared with the control fish (mC, blue). Ten fish from each group were analyzed. (C) Representative image of Sirius Red staining results of the 3- to 7- month-old control (mC) and two RPIA-transgenic fish (Tg1 and Tg2). Control fish exhibited no staining while 7-month-old Tg1 fish and 5-month-old Tg2 fish displayed positive signs of fibrosis, which are shown in red. All of the magnifications were ×400. Scale bar: 50 μm (D) Statistical analysis of the Sirius Red staining results for the 3-, 5- and 7-month-old RPIA Tg 1 (orange diamond) and Tg2 (red triangle) compared with the control fish (mC, blue). (E) The ratio of NADPH/NADP+ was elevated in 5-month-old RPIA-transgenic zebrafish. P < 0.05 was considered to be statistically significant; *: 0.01 < P ≤ 0.05.
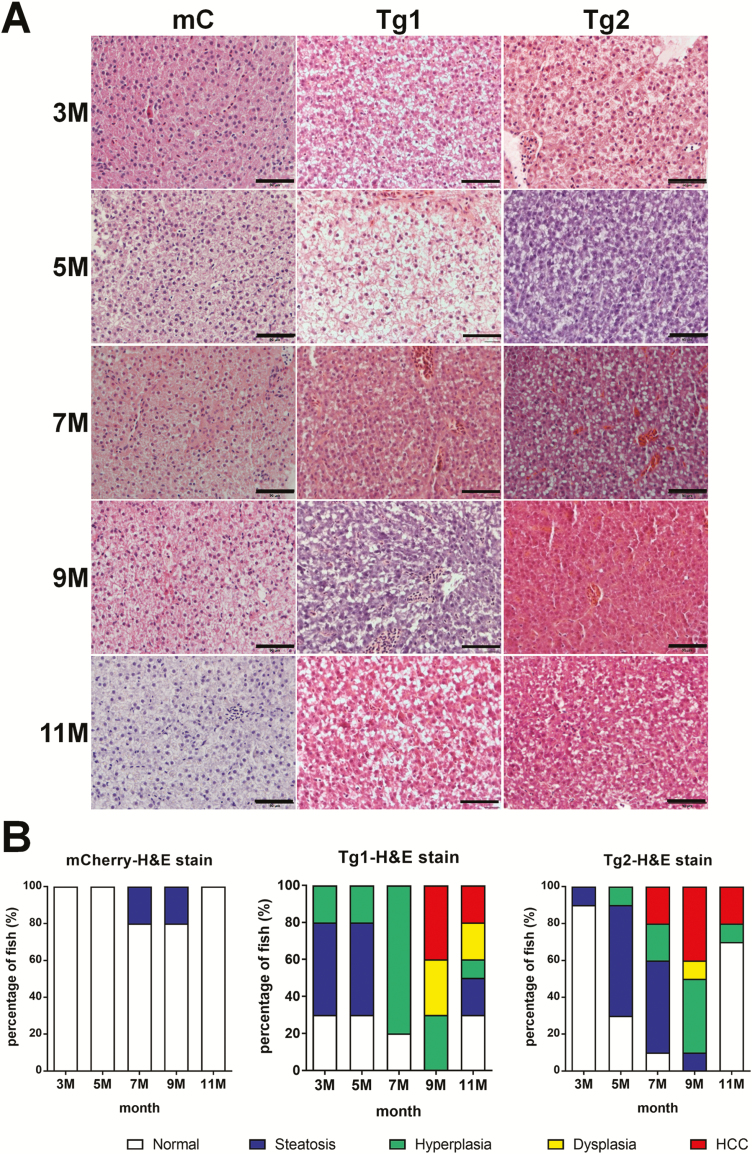
Figure 2.
RPIA-transgenic zebrafish develop steatosis, hyperplasia, dysplasia and HCC at earlier stages than control transgenic zebrafish. (A) Representative images of H&E staining from livers of controls and the two transgenic RPIA lines between 3- and 11-month-old fish. All of the magnifications were ×400. Scale bar: 50 μm. (B) Statistical analysis of H&E stain from 3 to 11 months of control fish (mCherry) and two lines of RPIA-transgenic fish (Tg1 and Tg2). N = 10 for each stage. Tg1 developed HCC as early as 9-month old, while Tg2 developed HCC as early as 7-month old.
In humans, a subgroup of patients with FLD will progress to have liver fibrosis and cirrhosis. Since steatosis occurred between 3 and 5 months of the RPIA-transgenic zebrafish, we next investigated whether RPIA induces liver fibrosis. First, QPCR was utilized to analyze the expression levels of several fibrosis marker genes, including collagen type 1 alpha 1 (col1a1α), connective tissue growth factor a (ctgfa) and heparanase (hpse), in both RPIA-transgenic lines from 3 to 11 months of age (Supplementary Figure S1C, available at Carcinogenesis Online). In the Tg1 line, the induction of fibrosis markers began around 5 months and peaked at 7 months, and expression was maintained until 11 months of age, while similar results were seen with Tg2 although the highest levels of expression were detected at 11 months of age (Supplementary Figure S1C, available at Carcinogenesis Online).
As a further indicator of fibrosis, collagen expression was measured by Sirius Red staining (Figure 1C). Unlike the control line (mC) which was negative for Sirius Red staining indicated as arrow (Figure 1C, left panel), both the Tg1 and Tg2 lines were Sirius Red positive as early as 5 and 3 months with stronger staining at 7 and 5 months, respectively (Figure 1C, middle and right panels). Statistically, the Tg1 RPIA-transgenic fish exhibits significantly more fibrosis than controls at 7 months of age (Figure 1D, orange diamond), while the Tg2 transgenic fish displays significant fibrosis at 5 and 7 months of age (Figure 1D, red triangle). These results suggest that the RPIA overexpression in the liver facilitates the development of fibrosis following steatosis and that RPIA expression levels are positively correlated with the severity of fibrosis. According to our previous studies in the HCC zebrafish models overexpressing other oncogenes, the fibrosis was reduced in later stage and in turn with the development of hyperplasia/dysplasia and HCC (32).
The PPP generates NADPH in the oxidative phase; increased PPP activity enhances cell proliferation. Fatty acid synthesis requires NADPH to supply as reductant. Furthermore, reduction in the cellular NADPH/NADP+ results in significant apoptosis of Sorafenib resistant Huh7 cells (36). To further investigate whether RPIA overexpression in the liver induces PPP flux and increase NADPH production, we measured NADPH/NADP+ ratio in RPIA-transgenic fish and control fish (Figure 1E). Significantly higher NADPH/NADP+ ratio was detected in RPIA-transgenic fish relative to similarly aged WT control. This result may help to explain why RPIA overexpression contributes to fatty liver in the transgenic zebrafish.
RPIA-mediated induction of cirrhosis is TALDO independent
In the transgenic mice, the PPP enzyme TALDO has been shown to play a role in cirrhosis (16). To understand the role of TALDO in RPIA-transgenic fish, TALDO protein levels were examined in the hepatocytes of RPIA-transgenic fish by immunohistochemistry (IHC) analysis. Between 3 and 11 months of age, TALDO proteins levels did not differ between RPIA-transgenic fish (Tg1 and Tg2) and control fish (Supplementary Figure S2, available at Carcinogenesis Online). These results suggest that RPIA overexpression leads to steatosis and cirrhosis independently of TALDO levels.
Examination of histopathological changes in the livers of transgenic RPIA zebrafish by H&E staining
Histopathological examinations of liver tissue taken from control, and RPIA, transgenic fish were assessed for pathology by H&E staining. The criteria for pathology include characteristics specifically generated by three carcinogenesis pathologists, guidelines established by the National Toxicology Program and published literature (32). Liver samples were examined based on these criteria and classified into different histopathological categories including steatosis, hyperplasia, dysplasia and HCC. Steatotic tissues have intracytoplasmic vacuoles, while hyperplastic tissues possess enlarged and mildly irregular nuclei. Dysplastic samples have enlarged nuclei and prominent nucleoli, while HCC samples would have polymorphic nuclei, prominent nucleoli and an increased number of mitotic figures.
We found that the Tg1 RPIA-transgenic fish developed steatosis at 3 months of age, exhibited hyperplasia and dysplasia at between 7 and 9 months of age and developed HCC between 9 and 11 months of age (Figure 2A and B, middle panel). Interestingly, 20% of Tg2 RPIA-transgenic fish developed HCC as early as 7 months (Figure 2A and B, right panel). Some hepatocytes seemed to be normal at 11 months suggesting that zebrafish has the ability to repair liver defects themselves, and similar findings have also been noted in ours and other published papers (32,37). These results indicate that RPIA overexpression in the liver causes hyperplasia, dysplasia and cancer in a progressive fashion consistent with that observed in the progression of human HCC.
RPIA-mediated induction of cell cycle/proliferation-related markers and level of PCNA and phospho-histone H3
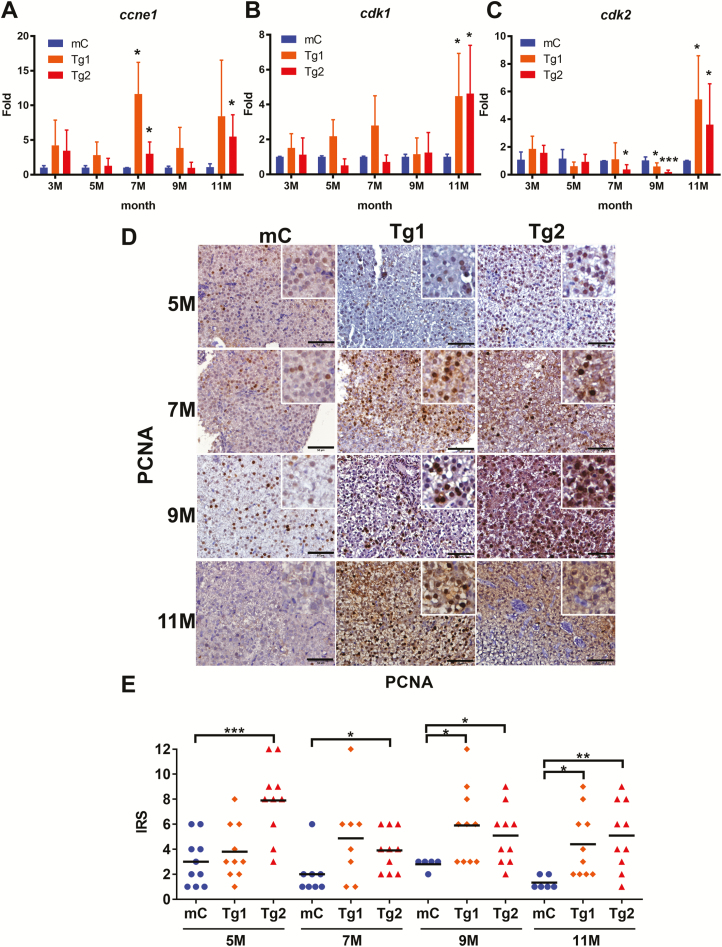
To determine whether RPIA overexpression might induce HCC progression via promoting cellular proliferation, the expression levels of the cell cycle/proliferation-related genes cyclin E1 (ccne1), cyclin-dependent kinase 1 (cdk1) and cyclin-dependent kinase 2 (cdk2) were examined by QPCR (Figure 3A–C). These results showed that ccne1 expression levels were significantly increased between 7 and 11 months of age (Figure 3A), and cdk1 and cdk2 expression levels were significantly upregulated at 11 months of age in both Tg1 and Tg2 RPIA-transgenic fish compared with mCherry control fish (Figure 3B and C).
Figure 3.
RPIA-transgenic fish increase expression of cell cycle/proliferation markers and PCNA nuclear staining. (A–C) QPCR was used to measure the relative mRNA fold induction of the cell cycle/proliferation markers ccne1, cdk1 and cdk2 in two independent lines of RPIA-transgenic fish (Tg1 and Tg2) compared with the control fish (mC) at 3, 5, 7, 9 and 11 months of age. (D) Representative images of PCNA IHC result for the control (mC) and Tg1 and Tg2 fish aged 5–11 months. All of the magnifications were ×400. Scale bar: 50 μm. (E) Statistical analysis of the PCNA IHC staining results from Tg1 and Tg2 compared with control, as determined by the IRS scoring system. Statistical analysis was performed by two-tailed Student’s t-tests. The symbol ‘*’ represents significance between RPIA-transgenic fish and GFP-mCherry control. P < 0.05 was considered to be statistically significant; *: 0.01 < P ≤ 0.05; **: 0.001 < P ≤ 0.01; ***: P ≤ 0.001.
Immunohistochemistry for the proliferation marker, proliferating cell nuclear antigen (PCNA), was performed to detect any differences in PCNA expression between the control and RPIA-transgenic-overexpressing lines (Figure 3D). These results revealed that PCNA was expressed at higher levels in the Tg1 and Tg2 lines relative to controls. PCNA protein levels were then quantified by the immunoreactive score (IRS) which is calculated by multiplying the staining intensity by the proportion of positive cells (38). Statistical analysis revealed that RPIA overexpression in the Tg1 line exhibited significant increase in PCNA expression from 9 to 11 months (Figure 3E, orange diamond), while in Tg2, there is increased PCNA expression between 5 and 11 months of age (Figure 3E, red triangle).
We also examined the phosphorylation status of histone H3 (pH3) at Ser10, which is a hallmark of G2/M phase (Supplementary Figure S3, available at Carcinogenesis Online). Increased levels of pH3 were detected in RPIA-transgenic fish from 5 to 11 months of age compared with control fish. The pH3 was co-localized with DAPI which was considered as metaphase (Supplementary Figure S3, available at Carcinogenesis Online, middle and right panel) and anaphase from 9 to 11 months of age (Supplementary Figure S3, available at Carcinogenesis Online, right panel). These results demonstrate that RPIA overexpression enhances cellular proliferation in the liver.
pERK and pAMPK expression are increased in RPIA-transgenic fish
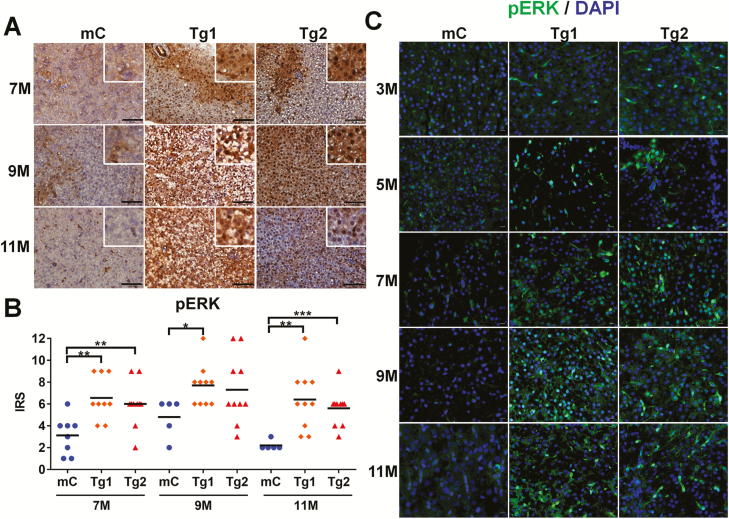
Our previous study showed that ERK signaling participates in RPIA-mediated proliferation and migration in hepatoma cells (8). During activation of the ERK signaling cascade, ERK is phosphorylated (pERK) which induces downstream effects. To investigate the levels of pERK in the livers of RPIA-transgenic fish, IHC staining was performed (Figure 4A). These studies revealed that in both the Tg1 and Tg2 RPIA lines, there are significantly increased pERK staining from 7 to 11 months of age as compared with controls. These results are confirmed in Figure 4B which shows the IRS for the control and transgenic fish between 7 and 11 months of age. In addition, immunofluorescence staining also revealed that the increased expression of pERK occurs as early as 3 months, and pERK is localized in the nucleus in RPIA-transgenic fish (Figure 4C), In contrast, other than at 7 months of age, no differences in total ERK protein levels between RPIA-transgenic fish and controls were detected as measured by IHC and IRS scores (Supplementary Figure S4A and B, available at Carcinogenesis Online); immunofluorescence of ERK also showed no difference among RPIA transgenic and control fish (Supplementary Figure S4C, available at Carcinogenesis Online). These data suggest that activation of ERK signaling is involved in transducing the effects of RPIA-mediated hepatocarcinogenesis in the zebrafish model.
Figure 4.
pERK expression is elevated in RPIA-transgenic fish. (A) Representative images of pERK IHC results for the control (mC), and two RPIA-transgenic fish (Tg1 and Tg2) aged 7–11 months. The control fish displayed only a weak pERK signal. The 7- to 11-month-old Tg1 and Tg2 fish exhibited significant stronger staining. All the magnifications were ×400. Scale bar: 50 μm. (B) Statistical analysis of the IHC results for pERK expression of two independent Tg (fabp10a: RPIA)—Tg1 and Tg2 compared with control (mC) fish. Statistical analysis was performed by unpaired two-tailed t-tests. Asterisks (*) represent the level of significance. *P < 0.05 was considered statistically significant. **P < 0.01; ***P < 0.001. (C) Immunofluorescence staining of pERK in two independent RPIA-transgenic zebrafish (Tg1 and Tg2) compared with control (mC). The images showed that pERK (green) and DAPI (blue) immunofluorescent signals co-localized within nucleus in RPIA-transgenic zebrafish liver. All of the magnifications were ×400. Scale bar: 40 μm.
As we previously demonstrated in hepatoma cell lines, the increase of pERK level was due to the reduced PP2A activity by RPIA overexpression (8). PP2A also can dephosphorylate the Thr172 of phospho-AMP-activated protein kinase α (pAMPKα) (39). pAMPK plays an oncogenic role for Myc-induced HCC progression (40) and is essential for maintaining NADPH (41). Immunohistochemistry using antibodies detecting AMPKα and pAMPKα (Thr172) was performed to examine the protein levels in RPIA transgenic and control fish. The results showed that pAMPKα (Thr172) levels were elevated in RPIA-transgenic liver in 3 and 5 months of age but no significant differences in AMPKα levels between RPIA transgenic and control fish (Supplementary Figure S5, available at Carcinogenesis Online). It suggests that pAMPK may also be involved in RPIA-mediated HCC progression.
β-catenin is activated in RPIA-transgenic fish
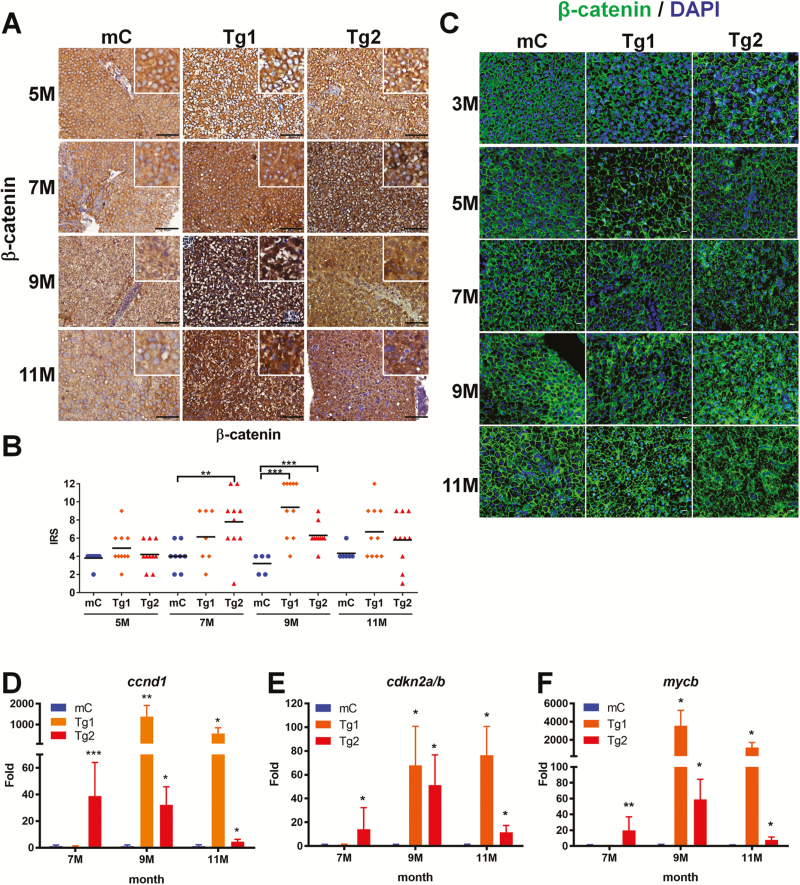
Our previous study identified a new role for RPIA in the promotion of colorectal cancer formation via the stabilization and activation of β-catenin via a unique C-terminal domain (26). Thus, we hypothesized that RPIA may use similar mechanisms to mediate liver cancer as well as colorectal cancer. IHC staining was used to examine β-catenin protein levels. There was a strong β-catenin protein staining in RPIA-transgenic fish relative to controls (Figure 5A). Furthermore, the IRS revealed that β-catenin protein level was significantly higher at 9 months (in Tg1) and 7 months (in Tg2) relative to the transgenic control line (Figure 5B). Using immunofluorescent assay, the localization of β-catenin (green) with DAPI (blue) was observed in both Tg1 and Tg2 (Figure 5C, middle and right panel). In control fish, β-catenin was completely localized at the adherent junction; however, the membrane junctional pattern was disrupted in RPIA-transgenic fish, and nuclear localization was observed in 11-month Tg1 and 9-month Tg2 of RPIA-transgenic fish (Figure 5C). These data suggest RPIA overexpression induces the transcriptional function of β-catenin rather than cellular junctional function.
Figure 5.
The expression of β-catenin and downstream target genes was upregulated in RPIA-transgenic fish aged 7–11 months. (A) Representative images of β-catenin IHC staining from 5 to 11 months of control (mC), Tg1- and Tg2-RPIA-transgenic fish. A weaker β-catenin signal was detected in transgenic, control tissues (mC) relative to tissues from the RPIA-transgenic fish, and staining in the control fish revealed expression in the area surrounding the nucleus. In the 9 and 11 months of Tg (fabp10a: RPIA) fish lines, the amount of β-catenin in nucleus was upregulated. All of the magnifications were ×400. Scale bar: 50 μm. (B) Statistical analysis of β-catenin immunostaining in 5- to 11-month-old Tg1- and Tg2-RPIA-transgenic fish compared with control. Statistical analysis was performed by unpaired two-tailed t-tests. Asterisks (*) represent the level of significance. *P < 0.05 was considered statistically significant. **P < 0.01; ***P < 0.001. N = 10 each stage. (C) Immunofluorescence staining of β-catenin in Tg1- and Tg2-RPIA-transgenic zebrafish compared with control (mC). The images showed that β-catenin (green) lost the expression for cell junction and co-localized with DAPI (blue) within nucleus in RPIA-transgenic zebrafish liver. All of the magnifications were ×400. Scale bar: 40 μm. (D–F) QPCR was used to measure the induction of the β-catenin target genes ccnd1 (D), cdkn2a/b (E), and mycb (F) in two independent lines of Tg (fabp10a: RPIA), which compared with control fish (mC) aged 3, 5, 7, 9 and 11 months. A significant increase in the expression levels of β-catenin target genes was found in 9- to 11-month-old Tg1 and 7- to 9-month-old Tg2 transgenic fish as compared with the transgenic control fish (mCherry). Statistical analysis was performed by unpaired two-tailed t-tests. Asterisks (*) represent the level of significance. *P < 0.05 was considered statistically significant. **P < 0.01; ***P < 0.001. N = 10 each stage.
To further verify that mentioned above, QPCR was utilized to analyze the expression levels of β-catenin downstream target genes cyclin D1 (ccnd1) (42,43), cyclin-dependent kinase inhibitor 2A/2B (cdkn2a/b) (43) and MYC proto-oncogene (mycb) (42,43). The expression of the β-catenin downstream target genes was elevated at 9–11 months in Tg1 RPIA-transgenic fish and increased at 7 months in Tg2 RPIA-transgenic fish (Figure 5D–F) as compared with the transgenic controls. These results suggest that RPIA may mediate its effects in liver cancer in a mechanism similar to that involved in RPIA-mediated colorectal cancer.
We also examined the β-catenin protein levels in Hep3B and PLC5, two hepatoma cell lines, upon knockdown of RPIA. These experiments revealed that there was decreased β-catenin protein expression in the nucleus after siRNA-mediated knockdown of RPIA (Supplementary Figure S6A and B, available at Carcinogenesis Online); however, RPIA was expressed in the cytoplasm exclusively in the hepatoma cell. We suspect that RPIA activates β-catenin during hepatocarcinogenesis in a manner similar to that seen in CRC, mediated by the stabilization of β-catenin so that it can enter the nucleus. Nevertheless, these results demonstrate that β-catenin signaling is also involved in transducing the effects of RPIA-mediated hepatocarcinogenesis in zebrafish and that RPIA expression levels are positively correlated with β-catenin activation.
Inhibition of ERK and β-catenin can disrupt HCC progression in zebrafish
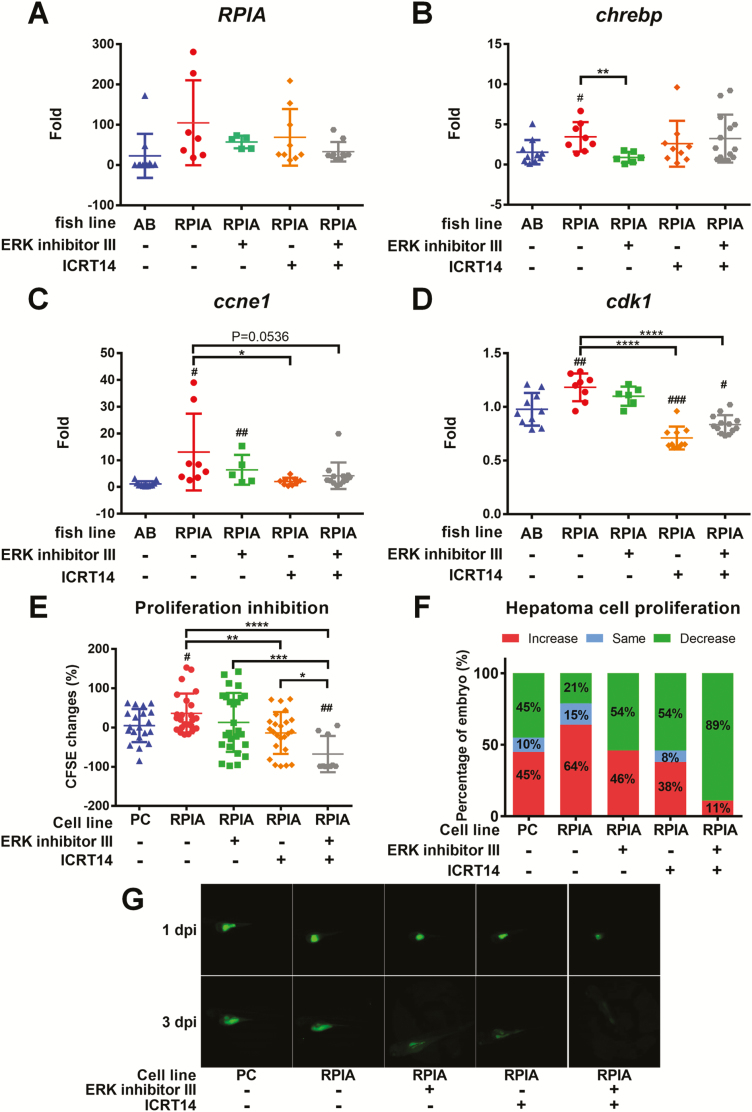
To explore whether blockade of ERK and β-catenin signaling can prevent HCC progression, we used oral gavage to treat RPIA-transgenic fish of 7 months of age with ERK inhibitor (ERK inhibitor III), β-catenin inhibitor (ICRT14) or mixture of these two inhibitors for 1 month. The expression of the related genes was verified by QPCR. As predicted, the expression level of RPIA transgene was not affected by the inhibitors (Figure 6A). However, ERK inhibitor III treatment diminished the expression of lipogenic factor, chrebp, in RPIA-transgenic fish (Figure 6B). Intriguingly, β-catenin inhibitor-ICRT14 reverted the upregulation of cell cycle/proliferation markers, ccne1 and cdk1, mediated by RPIA overexpression (Figure 6C and D).
Figure 6.
ERK inhibition diminished lipogenesis while β-Catenin inhibition reverted cellular proliferation in zebrafish. (A–D) QPCR was used to measure the relative mRNA fold induction of RPIA, lipogenic factor (chrebp) and cell cycle/proliferation markers (ccne1 and cdk1) in two independent lines of Tg (fabp10a: RPIA) compared with the control fish (mC) after ERK inhibitor III and β-catenin inhibitor (ICRT14) treatment by oral feeding. (E) PLC5 with RPIA-overexpressed cells proliferation ability was examined by xenotransplantation assay. The CFSE‐labeled cells were injected to 2-day-old embryos. PLC5 was pretreated with ERK inhibitor and β-catenin inhibitor or mixture of these two inhibitors for 24 h before injection. The cell proliferation was analyzed between 1 and 3 dpi. RPIA-overexpressed PLC5 (RPIA) has higher proliferation rate than the empty vector control (pcDNA3; PC). β-catenin inhibition (ICRT14) significantly reduced RPIA-expressed cell proliferation ability. Statistical analysis was performed by unpaired two-tailed t-tests with Welch’s correction. P < 0.05 was considered to be statistically significant. Hashtag (#) represented the comparison between RPIA-transgenic lines and AB; Asterisk (*) represented the comparison between RPIA Tg lines with and without inhibitor treatment. #0.01 < P ≤ 0.05; ###P ≤ 0.001; **0.001 < P ≤ 0.01; ****P ≤ 0.0001. (F) The percentage of embryos with increased, the same or decreased CFSE‐labeled cells compared between 3 and 1 dpi. β-catenin inhibition decreased the proportion of fish with RPIA-induced proliferative cells. Combination of β-catenin and ERK inhibitors revealed synergistic inhibition on RPIA-induced cell proliferation. (G) Representative fluorescence images of xenotransplanted zebrafish at 1 and 3 dpi for each treatment.
Similar results were also revealed by xenotransplantation analysis. CFSE-labeled PLC5 hepatoma cells were injected to 2-day-old zebrafish embryos. Inhibition of β-catenin by ICRT14 significantly reduced RPIA-mediated cell proliferation ability (Figure 6E), but the blockade of ERK alone did not have significant effect. The combination of β-catenin and ERK inhibitors synergistically reduced the RPIA-induced cellular proliferation. Besides, the proportion of the fish with RPIA-induced proliferative cells was also decreased with the β-catenin inhibitor treatment (Figure 6F). Together, these results suggest that ERK contributes to induce lipogenesis, while β-catenin mediates in HCC progression by promoting cellular proliferation in zebrafish, and inhibition of ERK and β-catenin signaling can significantly reduce HCC progression.
Discussion
The PPP is essential in the production of NAPDH, pentoses and ribose-5-phosphate as a precursor for nucleotide synthesis. NADPH is required for fatty acid synthesis. Alterations in the PPP play an important role in the metabolism and survival of cancer cells (44). The formation of HCC involves the dysregulation of PPP enzymes such as G6PD (45), TALDO (15,16) and TKTL1 protein (19). RPIA is also important in the PPP and regulates the transition between the oxidative and non-oxidative phases of PPP. In this study, we demonstrated that overexpression of RPIA in transgenic zebrafish promotes hepatocarcinogenesis via the activation of both the ERK and β-catenin signaling pathways.
In addition to the activation of signaling pathways, RPIA overexpression causes the PPP to transition to a non-oxidative phase, and reductions in NADPH levels may result in increases in intracellular reactive oxygen species levels, potentially resulting in cell death (44). While we did not detect differences in TALDO levels between RPIA transgenic and control fish, we observed the increased levels of NAPDH in RPIA-transgenic fish, opposite to Sorafenib resistant Huh7 cells with reduction in the cellular NADPH/NADP+ (36). To gain a comprehensive view of how RPIA overexpression contributes to inflammation, fatty liver and cancer formation, we plan to measure the reduced glutathione (GSH), oxidized glutathione (GSSG) and the expression of glutathione-related genes (46) using high-performance liquid chromatography. These studies have been used to detect oxidative stress in zebrafish (47) as well as effects on liver function. Blood samples from adult zebrafish will be collected to examine alanine aminotransferase levels, a marker for human liver damage markers (48). Those results shall provide more insight into the role of the PPP enzyme RPIA in cirrhosis and hepatocarcinogenesis.
Liver plays a major role in the homeostasis of lipid and glucose metabolism, and dysregulation of the PPP coupled with lipogenesis promotes non-alcoholic FLD (NAFLD), which may lead to cirrhosis and HCC (49). As RPIA overexpression in transgenic zebrafish liver induces hepatocyte steatosis at earlier ages, followed by fibrosis and eventual development of HCC, we suspect dramatic upregulation of the non-oxidative phase of the PPP can produce glyceraldehyde-3-phosphate, which can be converted to pyruvate. We will further determine the physiological relevance and pathophysiological connection between metabolites produced during hepatocarcinogenesis in RPIA-transgenic fish and further develop novel therapeutics for treatment of NAFLD in humans.
This study demonstrates that the hepatocarcinogenesis induced by RPIA overexpression in zebrafish is similar to that seen in human HCC patients. In addition, RPIA overexpression in the liver induces ERK phosphorylation, activates the β-catenin signaling pathway and promotes liver cancer cell proliferation. The information of this study will facilitate the development of treatments for HCC using transgenic zebrafish as a drug screening platform, to identify small molecular inhibitors or other interventions that may be useful for treating cancer in humans.
Funding
This study was supported by Taiwan’s Ministry of Health and Welfare (MOHW), Advance Medical Plan (107-0324-01-19-03), MOST (105-2314-B-400-031-MY2) and NHRI (MG-105-PP-06) to Dr. Chiou-Hwa Yuh, as well as the Veterans General Hospitals University System of Taiwan (VGHUST 104-G6-1-1) and MOST (102-2311-B-007-008-MY3; 106-2311-B-007-006-MY2) to Dr. Horng-Dar Wang.
Supplementary Material
Acknowledgements
The authors thank Drs Micheline Laurent and Theodore Brummel for critically editing the manuscript. We would like to thank the Taiwan Zebrafish Core Facility at NHRI (TZeNH) for providing the fish lines and resources used herein. TZeNH is supported by a grant from MOST (105-2319-B-400-001). The fellowships from both NHRI and Liver Disease Prevention & Treatment Research Foundation (2018) to Dr Y.-T. C. is acknowledged.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CFSE
carboxyfluorescein succinimidyl ester
- dpi
day post-injection
- ERK
extracellular signal-regulated kinase
- EtOH
ethanol
- FLD
fatty liver disease
- H&E
hematoxylin and eosin
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- NADPH
nicotine adenine dinucleotide phosphate hydrate
- PCNA
proliferating cell nuclear antigen
- PP2A
protein phosphatase 2A
- PPP
pentose phosphate pathway
- QPCR
quantitative polymerase chain reaction
- RPIA
ribose-5-phosphate isomerase
- TALDO
transaldolase
References
- 1. El-Serag H.B., et al. (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology, 132, 2557–2576. [DOI] [PubMed] [Google Scholar]
- 2. Nordenstedt H., et al. (2010) The changing pattern of epidemiology in hepatocellular carcinoma. Dig. Liver Dis., 42 (suppl. 3), S206–S214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farazi P.A., et al. (2006) Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer, 6, 674–687. [DOI] [PubMed] [Google Scholar]
- 4. Seyfried T.N. (2015) Cancer as a mitochondrial metabolic disease. Front. Cell Dev. Biol., 3, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pawelec G., et al. (2008) Are cancer and ageing different sides of the same coin? Conference on cancer and ageing. EMBO Rep., 9, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Longo V.D., et al. (2008) Turning anti-ageing genes against cancer. Nat. Rev. Mol. Cell Biol., 9, 903–910. [DOI] [PubMed] [Google Scholar]
- 7. Wang C.T., et al. (2012) Reduced neuronal expression of ribose-5-phosphate isomerase enhances tolerance to oxidative stress, extends lifespan, and attenuates polyglutamine toxicity in Drosophila. Aging Cell, 11, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ciou S.C., et al. (2015) Ribose-5-phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signaling. Int. J. Cancer, 137, 104–115. [DOI] [PubMed] [Google Scholar]
- 9. Liang Y., et al. (2013) The regulation of cellular metabolism by tumor suppressor p53. Cell Biosci., 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jain M., et al. (2003) Glucose-6-phosphate dehydrogenase modulates cytosolic redox status and contractile phenotype in adult cardiomyocytes. Circ. Res., 93, e9–16. [DOI] [PubMed] [Google Scholar]
- 11. Hove-Jensen B. (1988) Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J. Bacteriol., 170, 1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramos-Montoya A., et al. (2006) Pentose phosphate cycle oxidative and nonoxidative balance: a new vulnerable target for overcoming drug resistance in cancer. Int. J. Cancer, 119, 2733–2741. [DOI] [PubMed] [Google Scholar]
- 13. Bannasch P., et al. (1997) Early bioenergetic changes in hepatocarcinogenesis: preneoplastic phenotypes mimic responses to insulin and thyroid hormone. J. Bioenerg. Biomembr., 29, 303–313. [DOI] [PubMed] [Google Scholar]
- 14. Riganti C., et al. (2012) The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Radic. Biol. Med., 53, 421–436. [DOI] [PubMed] [Google Scholar]
- 15. Hanczko R., et al. (2009) Prevention of hepatocarcinogenesis and increased susceptibility to acetaminophen-induced liver failure in transaldolase-deficient mice by N-acetylcysteine. J. Clin. Invest., 119, 1546–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perl A., et al. (2011) Oxidative stress, inflammation and carcinogenesis are controlled through the pentose phosphate pathway by transaldolase. Trends Mol. Med., 17, 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langbein S., et al. (2006) Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br. J. Cancer, 94, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu X., et al. (2009) Transketolase-like protein 1 (TKTL1) is required for rapid cell growth and full viability of human tumor cells. Int. J. Cancer, 124, 1330–1337. [DOI] [PubMed] [Google Scholar]
- 19. Zhang S., et al. (2007) Gene silencing of TKTL1 by RNAi inhibits cell proliferation in human hepatoma cells. Cancer Lett., 253, 108–114. [DOI] [PubMed] [Google Scholar]
- 20. Mitsuyama T. (1979) [Experimental and clinical studies on ribosephosphate isomerase (author’s transl)]. Hokkaido Igaku Zasshi., 54, 387–400. [PubMed] [Google Scholar]
- 21. Ying H., et al. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell, 149, 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt C.M., et al. (1997) Increased MAPK expression and activity in primary human hepatocellular carcinoma. Biochem. Biophys. Res. Commun., 236, 54–58. [DOI] [PubMed] [Google Scholar]
- 23. Junttila M.R., et al. (2007) CIP2A inhibits PP2A in human malignancies. Cell, 130, 51–62. [DOI] [PubMed] [Google Scholar]
- 24. Rao R.K., et al. (2002) Regulation of protein phosphatase 2A by hydrogen peroxide and glutathionylation. Biochem. Biophys. Res. Commun., 293, 610–616. [DOI] [PubMed] [Google Scholar]
- 25. Kalev P., et al. (2011) Protein phosphatase 2A as a potential target for anticancer therapy. Anticancer. Agents Med. Chem., 11, 38–46. [DOI] [PubMed] [Google Scholar]
- 26. Chou Y.T., et al. (2018) Identification of a noncanonical function for ribose-5-phosphate isomerase A promotes colorectal cancer formation by stabilizing and activating β-catenin via a novel C-terminal domain. PLoS Biol., 16, e2003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spitsbergen J.M., et al. (2003) The state of the art of the zebrafish model for toxicology and toxicologic pathology research–advantages and current limitations. Toxicol. Pathol., 31 (suppl), 62–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zon L.I., et al. (2005) In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov., 4, 35–44. [DOI] [PubMed] [Google Scholar]
- 29. Lieschke G.J., et al. (2007) Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet., 8, 353–367. [DOI] [PubMed] [Google Scholar]
- 30. Feitsma H., et al. (2008) Zebrafish as a cancer model. Mol. Cancer Res., 6, 685–694. [DOI] [PubMed] [Google Scholar]
- 31. Suster M.L., et al. (2009) Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol., 561, 41–63. [DOI] [PubMed] [Google Scholar]
- 32. Lu J.W., et al. (2013) Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS One, 8, e76951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu J.W., et al. (2014) Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS One, 9, e85318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu J.W., et al. (2013) Hepatitis B virus X antigen and aflatoxin B1 synergistically cause hepatitis, steatosis and liver hyperplasia in transgenic zebrafish. Acta Histochem., 115, 728–739. [DOI] [PubMed] [Google Scholar]
- 35. Urasaki A., et al. (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics, 174, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim M.J., et al. (2017) PPARδ reprograms glutamine metabolism in sorafenib-resistant HCC. Mol. Cancer Res., 15, 1230–1242. [DOI] [PubMed] [Google Scholar]
- 37. Mudbhary R., et al. (2014) UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell, 25, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fedchenko N., et al. (2014) Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue—a review. Diagn. Pathol., 9, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Y., et al. (2007) Activation of protein phosphatase 2A by palmitate inhibits AMP-activated protein kinase. J. Biol. Chem., 282, 9777–9788. [DOI] [PubMed] [Google Scholar]
- 40. Liu L., et al. (2012) Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature, 483, 608–612. [DOI] [PubMed] [Google Scholar]
- 41. Jeon S.M., et al. (2012) AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature, 485, 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Röhrs S., et al. (2009) Chronological expression of Wnt target genes Ccnd1, Myc, Cdkn1a, Tfrc, Plf1 and Ramp3. Cell Biol. Int., 33, 501–508. [DOI] [PubMed] [Google Scholar]
- 43. Herbst A., et al. (2014) Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics, 15, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patra K.C., et al. (2014) The pentose phosphate pathway and cancer. Trends Biochem. Sci., 39, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kowalik M.A., et al. (2017) Emerging role of the pentose phosphate pathway in hepatocellular carcinoma. Front. Oncol., 7, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Timme-Laragy A.R., et al. (2013) Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic. Biol. Med., 65, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mugoni V., et al. (2014) Analysis of oxidative stress in zebrafish embryos. J. Vis. Exp., 89, 51328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zang L., et al. (2015) Repeated blood collection for blood tests in adult zebrafish. J. Vis. Exp., 102, e53272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bechmann L.P., et al. (2012) The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol., 56, 952–964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.