Abstract
The oligodendrocyte lineage genes (Olig1/2), encoding basic helix-loop-helix transcription factors, were first identified in screens for master regulators of oligodendrocyte development. OLIG1 is important for differentiation of oligodendrocyte precursors into myelin-forming oligodendrocytes during development and is thought to play a crucial role in remyelination during multiple sclerosis. However, it is still unclear how OLIG1 interacts with its transcriptional cofactors and DNA targets. OLIG1 was reportedly restricted to mammals, but we demonstrate here that zebrafish and other teleosts also possess an OLIG1 homolog. In zebrafish, as in mammals, Olig1 is expressed in the oligodendrocyte lineage. Olig1 associates physically with another myelin-associated transcription factor, Sox10, and the Olig1/Sox10 complex activates mbp (myelin basic protein) transcription via conserved DNA sequence motifs in the mbp promoter region. In contrast, Olig2 does not bind to Sox10 in zebrafish, although both OLIG1 and OLIG2 bind SOX10 in mouse.
Keywords: Olig1, Olig2, Sox10, Mbp, oligodendrocyte, myelin, zebrafish, mouse, evolution, development
Introduction
Myelin, the multilayered glial sheath around axons, is one of the defining features of jawed vertebrates (gnathostomes). It is present in both the central and peripheral nervous systems and facilitates rapid, saltatory conduction of action potentials. In the CNS, oligodendrocytes (OLs) are responsible for myelin synthesis. OLs develop from dedicated precursor cells (OLPs) that are specified in the ventricular zones of the embryonic spinal cord and brain.
Several transcription factors including OLIG1, OLIG2, NKX2.2, SOX10, and OCT6 are known to regulate the development of OLPs (Collarini et al., 1992; Lu et al., 2000; Sun et al., 2001; Stolt et al., 2002; Zhou and Anderson, 2002; Gokhan et al., 2005). In the mouse spinal cord, OLIG2 is necessary for the production of both motor neurons and OLPs (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2002). OLIG1, in contrast, is not required for motor neuron development or the early stages of OLP development but plays a role in OL maturation (Lu et al., 2002). Nkx2.2 directly inhibits transcription from the myelin basic protein (Mbp) promoter (Wei et al., 2005) but promotes transcription from the proteolipid protein (Plp) promoter (Qi et al., 2001). SOX10 also plays a key role in promoting OL terminal differentiation; in the absence of Sox10 activity, OL differentiation is disrupted (Stolt et al., 2002). Moreover, SOX10 directly regulates Mbp transcription (Stolt et al., 2002), and overexpression of SOX10 alone is sufficient to induce myelin gene expression in embryonic chick spinal cord (Liu et al., 2007). OCT6 (also known as TST-1 or SCIP) is expressed in proliferating OLPs in culture and is downregulated when OL differentiation is triggered by the withdrawal of mitogens from the culture medium (Collarini et al., 1992). The regulatory interrelationships among these transcription factors and their DNA targets remain to be worked out.
OLIG1, OLIG2, and another close relative, OLIG3, belong to the large family of basic helix-loop-helix (bHLH) transcription factors that function widely in cellular development and differentiation. The functions of OLIG1 and OLIG2 partly overlap (Lu et al., 2002; Zhou and Anderson, 2002), although OLIG1 plays the key role in OL myelinogenesis (Xin et al., 2005) and in remyelination after experimentally induced CNS demyelination (Arnett et al., 2004). OLIG2 can bind to the E-box (Eb) motif (CANNTG), a DNA binding site that is recognized by several other tissue-specific bHLH factors (Lee et al., 2005). However, OLIG2 is distinct in that it can form homodimers and can function as a transcriptional repressor (Novitch et al., 2001; Lee et al., 2005), whereas most other lineage-restricted bHLH factors act as transcriptional activators. It is reported that the Olig2 gene is structurally conserved from nematode and fly to vertebrates, whereas Olig1 arose as a duplication of Olig2 during later vertebrate evolution and, as a result, occurs only in mammals (Lu et al., 2000). More recently, in silico analysis of Olig genes argues that Olig1 might have appeared first during evolution (Bronchain et al., 2007). OLIG3 is important for specification of class A (dI1–dI3) dorsal spinal cord interneurons and has been implicated in astrocyte development in zebrafish (Filippi et al., 2005; Muller et al., 2005).
Myelin components Protein zero (Mpz), proteolipid protein (Plp1b), and Mbp have been identified in zebrafish (Brosamle and Halpern, 2002). Zebrafish Olig2, like its mammalian counterpart, is required for primary motor neuron and OL development (Park et al., 2002). Sox10, Nkx2.2a, and Oct-6 have all been reported to function in OL differentiation in zebrafish (Levavasseur et al., 1998; Park et al., 2002; Kirby et al., 2006). In this study, we report that zebrafish and other teleosts also possess an olig1 gene. Zebrafish Olig1 is expressed in the OL lineage and can form homodimers as well as heterodimers with Olig2 and the ubiquitous bHLH transcription factor E12. In zebrafish, Olig1 can interact with Sox10 directly, whereas Olig2 cannot. In mouse, both OLIG1 and OLIG2 bind to SOX10. In vitro and in vivo evidence indicates that an Olig1/Sox10 complex can activate mbp transcription by binding to elements in the 5′ region of the mbp gene. These findings cast new light on the role of Olig1 in CNS myelination.
Materials and Methods
Cloning and analysis of the zebrafish olig1 gene.
A tBLASTn (Basic Local Alignment Search Tool) search with the mouse OLIG1 protein sequence (GenBank accession number NP_058664) on the Ensembl zebrafish database (http://www.ensembl.org/Danio_rerio/index.html) identified a zebrafish olig1 homology sequence. Specific primers 1F 5′-GGATCCTCAGAATGCAGGCTGTGTCTGGTG-3′ and 1R 5′-GCGGCCGCTTCGGAAAACGCATGGCTGGATT-3′ were used to amplify olig1 from total RNA of 3 d postfertilization (dpf) zebrafish embryos by reverse transcription (RT)-PCR. The names of zebrafish genes follow the Zebrafish Nomenclature Guidelines (www.zfin.org).
Embryo manipulation.
Zebrafish embryos were raised at 28°C with a 14/10 h light/dark cycle. They were staged according to hours or days after fertilization and morphological criteria. Embryos were fixed and cryoprotected, and sections were cut as described previously (Kazakova et al., 2006). Synthetic mRNA and DNA were injected into embryos at the one-cell stage. For mRNA injection, 3–5 nl of mRNA (100 ng/μl) synthesized by mMESSAGE mMACHINE (Ambion, Austin, TX) was used.
In situ hybridization.
Zebrafish whole-mount in situ hybridization was performed as reported previously (Kazakova et al., 2006). Double-fluorescence in situ hybridization on transverse sections was performed as described previously (Pringle et al., 2003). Digoxigenin and FITC RNA probes were made by in vitro transcription from cloned cDNAs of olig1, olig2, sox10, mbp, plp1b, mpz, and DsRed.
Plasmid constructions.
The ORFs of zebrafish olig1, olig2, id1, id2, id3, id4, e12, sox10, ngn1, ngn3, mash1a, and nkx2.2a genes were amplified by RT-PCR and cloned into pCDNA3.1His/V5 (Invitrogen, Paisley, UK). olig1, olig2, and sox10 were then subcloned into the pCDNA4His/Myc (Invitrogen) and pCS2 (olig1 and olig2 with a Myc tag, sox10 with a V5 tag) vectors (University of Michigan, Ann Arbor, MI). In addition, olig1 and olig2 were also subcloned into pACT and pBind vectors of the mammalian two-hybrid system (Promega, Southampton, UK). sox10 was also subcloned into pGEX-4T (GE Healthcare, Bucks, UK). Different fragments of olig1 and sox10 were amplified from cDNAs and cloned into pCDNA4His/myc. A 1.6 kb zebrafish mbp promotor fragment and luciferase ORF were cloned into pIRES-DsRed2 (Clontech Europe, Saint-Germain-en-Laye, France), from which the flanking sequence of the cytomegalovirus promoter had been removed. Different mutations of mbp-reporter constructs were generated with a site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions.
Cell culture and plasmid transfection.
Cos-7 cells were cultured in DMEM supplemented with 10% fetal bovine serum (Invitrogen) at 37°C with 5% CO2. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen). Equal amounts of plasmid DNA were used in cotransfection experiments.
Immunoprecipitation and glutathione S-transferase pull down.
Thirty-six hours after transfection, 107 Cos-7 cells were washed three times in PBS and lysed with 1 ml of lysis buffer [50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% (v/v) Nonidet P-40, 0.5% (v/v) sodium deoxycholate, and 1 tablet of protease inhibitor mixture/50 ml (Roche Diagnostics, Lewes, UK)]. After removing the cell debris by high-speed centrifugation, 500 μl of lysis solution was precleared with 30 μl of protein G beads (GE Healthcare) for 3 h at 4°C on a rotating wheel. After the beads were pelleted, the supernatant was incubated with anti-Myc or anti-V5 rabbit antibody at 4°C for 1 h, mixed with 30 μl of protein G beads, and incubated overnight at 4°C. The beads were washed twice in lysis buffer, twice in high-salt buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 0.1% Nonidet P-40, and 0.05% sodium deoxycholate), and once in low-salt buffer (50 mm Tris-HCl, pH 7.5, 0.1% Nonidet P-40, and 0.05% sodium deoxycholate). The beads were then resuspended in 80 μl of SDS sampling buffer and boiled for 5 min. For protein purification by immunoprecipitation, the antibodies were cross-linked with protein G beads by disuccinimidyl suberate (Pierce, Northumberland, UK), and the proteins were eluted with 80 μl of 0.2 m glycine-HCl, pH 2.8, and neutralized by adding 8 μl of 1 m Tris-HCl, pH 9.5.
For glutathione S-transferase (GST) pull-down assay, GST or GST-sox10 fusion protein was produced in Escherichia coli strain BL21 DE3 (Novagen, Darmstadt, Germany) and purified with glutathione agarose beads following the manufacturer's manual. The immobilized GST fusion protein was then incubated with Cos-7 lysate at 4°C overnight. Bound proteins were washed and eluted.
Luciferase assay.
Luciferase activity was measured using the Dual Luciferase Assay System (Promega). The amount of transfected DNA was calibrated with empty pCDNA vector. The measured firefly luciferase activity was normalized against Renilla luciferase activity. The results were the mean ± SE of three independent transfections. All assays were repeated three times.
Chromatin immunoprecipitation.
Whole zebrafish embryo chromatin immunoprecipitation (ChIP) was performed using a modified protocol (Havis et al., 2006; Wardle et al., 2006). Briefly, 50 embryos at 1 dpf were released from the chorion and fixed in 1.85% (w/v) formaldehyde solution for 15 min at room temperature. The fixation was quenched by glycine for 5 min. The embryos were washed three times with PBS, dissected from the underlying yolk, and homogenized in 500 μl of embryo lysis buffer (10 mm Tris-HCl, pH 7.5, 10 mm NaCl, 0.5% Nonidet P-40, and 1 tablet of protease inhibitor mixture/50 ml). The nuclei were pelleted by centrifugation at 12,000 × g for 30 min at 4°C, resuspended in 100 μl of nuclei lysis buffer [50 mm Tris-HCl, pH 7.5, 10 mm EDTA, 1% (w/v) SDS, and 1 tablet of protease inhibitor mixture/50 ml], sonicated, and diluted by adding 900 μl of lysis buffer. Sheared chromatin solution (500 μl) was used for immunoprecipitation with anti-Myc or anti-V5 antibody. The DNA/protein complex was then eluted with 200 μl of elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, and 1% SDS) at 65°C for 15 min. For PCR, the elute was decross-linked at 65°C for 6 h in 200 mm NaCl. Two primers, MBPF 5′-CCGTTCTTCATGTGCTCTG-3′ and MBPR 5′-TCAGTGGTCTACAGTCTGGAC-3′, were used to amplify a 360 bp mbp promoter fragment.
Western blotting.
Proteins from immunoprecipitation, GST pull down, and ChIP were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Protein bands were visualized by chemiluminescence (ECLplus; GE Healthcare).
Electrophoretic mobility shift assays.
32P-labeled, double-stranded oligonucleotides EbS2 (5′-CAAGTCGGGGCAGATGTGGACTAGAACAATAGCAGCTCC-3′) and S1 (5′-GAGGGGACGACACCTTCAAAGGCCAGCCCTTCGTG-3′) were used as probes (transcription factor binding sites are underlined). Purified protein and 0.1 pmol of radiolabeled probe were incubated at room temperature for 30 min in a 20 μl reaction volume [reaction buffer: 20% (v/v) glycerol, 20 mm HEPES, 100 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, and 0.01 U of poly(dI-dC)]. For competitive binding experiments, 10 pmol of unlabeled EbS2, S1, EbmutS2 (5′-CAAGTCGGGGTTGTAATGGACTAGAACAATAGCAGCTCC-3′), EbS2mut (5′-CAAGTCGGGGCAGATGTGGACTAGGCCGCTAGCAGCTCC-3′), or S1mut (5′-GAGGGGACGACACCGCCTCATGCCAGCCCTTCGTG-3′) was used as competitor (mutant sites underlined). For supershift experiments, 1 μg of mouse anti-Myc or anti-V5 antibody was applied. The mixture was run on a 6% nondenaturing Tris-borate EDTA polyacrylamide Novex minigel (Invitrogen) until the dye migrated off the end of the gel. The gels were dried and visualized by autoradiography.
Results
Zebrafish olig1 is expressed in the OL lineage
Zebrafish olig1 transcripts were first detected in the diencephalon at 48 h postfertilization (hpf) by in situ hybridization (Fig. 1A), whereas olig2 and olig3 appeared earlier (Park et al., 2002; Filippi et al., 2005). Subsequently, the olig1-expressing domain extended caudally and eventually spread through the entire brain and spinal cord by 72 hpf (Fig. 1B,C).
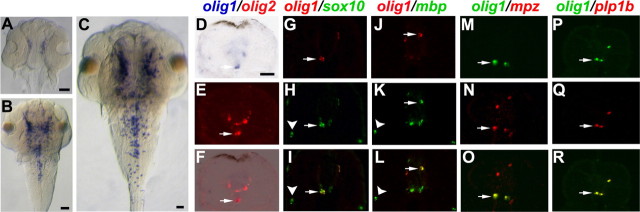
Figure 1.
Zebrafish olig1 is expressed in the OL lineage. olig1 expression was revealed by in situ hybridization. A–C, Whole-mount dorsal view; anterior is to the top. Some cells in the diencephalon began to express olig1 at 48 hpf (A), and then olig1 expression extended posteriorly at 60 hpf (B). At 72 hpf, the expression domain expanded through the entire brain and spinal cord (C). D–R, Transverse sections through trunk; dorsal is to the top. In the spinal cord at 72 hpf, olig1 partly colocalized with olig2 in white matter (D–F, arrows) and partly with other OL lineage markers sox10 (G–I), mbp (J–L), mpz (M–O), and plp1b (P–R). sox10 and mbp were also expressed in the PNS (arrowheads). Scale bars: 20 μm.
To identify the olig1 mRNA-expressing cell lineage, we performed double in situ hybridization on transverse sections of trunk spinal cord. In spinal cord, olig1 expression began at ∼72 hpf. olig1 was expressed only in the circumferential white matter where it colocalized with markers of OLPs (olig2, sox10) as well as markers of differentiating OLs (sox10, mpz, plp1b, mbp) (Fig. 1D–R).
Olig1 and Olig2 bind to different transcriptional partners in zebrafish
The sequence relatedness between Olig1 and Olig2 is less than that between Olig2 and Olig3 (Bronchain et al., 2007). Moreover, as stated above, the temporal and spatial expression of olig1 and olig2 is different. These data suggest that Olig1 and Olig2 might have different functions. To investigate their functions further, we transfected expression vectors encoding Myc or V5 epitope-tagged zebrafish Olig1 or Olig2 into cultured Cos-7 cells together with vectors encoding a range of potential transcriptional cofactors (also epitope tagged). We then performed coimmunoprecipitation assays on cell lysates with anti-Myc and anti-V5 to test for physical associations (Fig. 2). As in mammals, zebrafish Olig1 and Olig2 could both form homodimers as well as heterodimers with universal bHLH transcription factor E12 (Fig. 2A,B). Moreover, Olig1 and Olig2 could form heterodimers with each other. The formation of Olig1/2 homodimers and heterodimers was further confirmed by mammalian two-hybrid assay. Association of Olig1/Olig1, Olig1/Olig2, or Olig2/Olig2 all increased luciferase activity approximately eightfold (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). However, in contrast to mouse OLIG1 and OLIG2, both of which can bind to bHLH proteins ID2 and ID4 (Samanta and Kessler, 2004), zebrafish Olig1/2 could not interact with Id1, Id2, Id3, or Id4 (Fig. 2A,B). All of these Id proteins could interact with zebrafish E12, however (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). Previous reports indicate that mouse OLIG2 physically interacts with NGN2 and NKX2.2 to regulate OL and motor neuron development (Sun et al., 2003; Lee et al., 2005), which is consistent with our finding that in zebrafish, which lacks an NGN2 homolog, Olig2 can interact with Ngn3 instead.
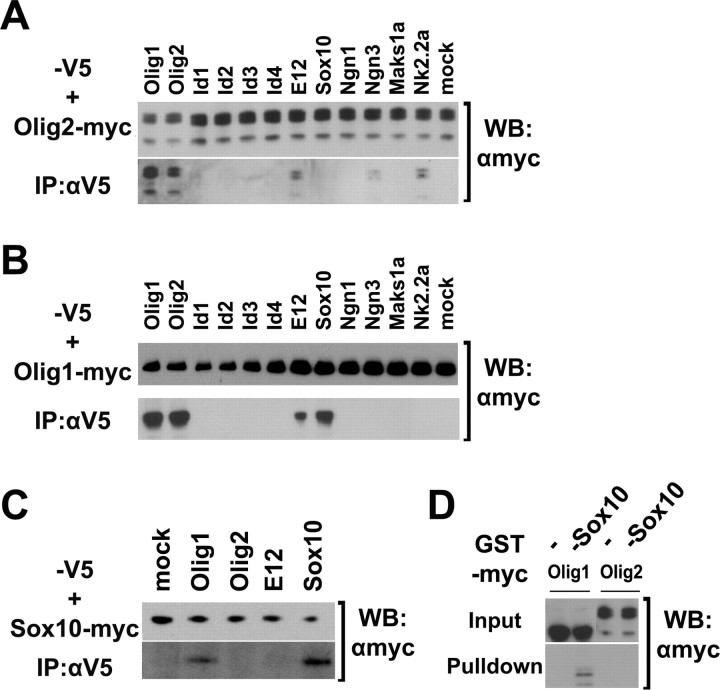
Figure 2.
Olig1 and Olig2 bind to different partners. A, B, Cos-7 cells were cotransfected with Myc-tagged olig2 (A) or olig1 (B) and V5-tagged olig1, olig2, id1, id2, id3, id4, e12, sox10, ngn1, ngn3, mash1a, or nkx2.2a. Cell lysates were used for immunoprecipitation with rabbit anti-V5 antibody, followed by Western blotting using mouse anti-Myc antibody. Empty V5 vector was used as a negative control. One-twentieth of the cell lysate was subjected directly to Western blotting with anti-Myc. C, Cell lysates from cotransfection of Myc-tagged sox10 and V5-tagged olig1, olig2, e12, or sox10 were immunoprecipitated with rabbit anti-V5 antibody, followed by Western blotting with mouse anti-Myc antibody. D, GST pull-down assay. Lysates from Cos-7 cells transfected with Myc-tagged olig1 or olig2 were incubated with GST or GST-Sox10 fusion protein. The GST fusion protein-bound products were separated by SDS-PAGE and detected with anti-Myc antibody. IP, Immunoprecipitation; WB, Western blot.
Interestingly, we observed that zebrafish Olig1 could complex with Sox10, whereas Olig2 could not (Fig. 2A,B). To explore the binding specificity of Sox10 further, we transfected epitope-tagged Sox10 expression vectors into Cos-7 cells along with vectors encoding a range of potential binding partners and performed coimmunoprecipitation assays as before. We found that Sox10 can form homodimers and can also bind to Olig1 but fails to bind to Olig2 or E12. To further confirm the specificity of binding between Olig1 and Sox10, we performed GST pull-down assays. GST-Sox10 fusion protein was expressed in E. coli and purified on an affinity column. The fusion protein could bind to Olig1 in cell lysate of olig1-transfected Cos-7 cells but failed to bind to Olig2 under analogous conditions (Fig. 2D). These results reveal that in zebrafish, Olig1 but not Olig2 can form a protein complex with Sox10. This contrasts with mouse, in which OLIG2 can bind to SOX10 in vitro (Wissmuller et al., 2006). OLIG1 was not examined in the latter study.
We looked into the reason for the species difference between mouse and zebrafish and found that mouse SOX10 can interact not only with mouse OLIG1 and OLIG2 but also zebrafish Olig1 and Olig2, whereas zebrafish Sox10 interacts with zebrafish Olig1 and mouse OLIG1 but not with Olig2 from either species (supplemental Fig. 1C,D, available at www.jneurosci.org as supplemental material). Comparison of the predicted sequences of mouse and zebrafish Sox10 revealed several amino acid differences between mouse SOX10 and zebrafish Sox10, specifically in the H3 and C-terminal region of their high-mobility group (HMG) domains (supplemental Fig. 1E, available at www.jneurosci.org as supplemental material), which have been shown to play a crucial role in the interaction between SOX proteins and their partners (Remenyi et al., 2003; Wissmuller et al., 2006).
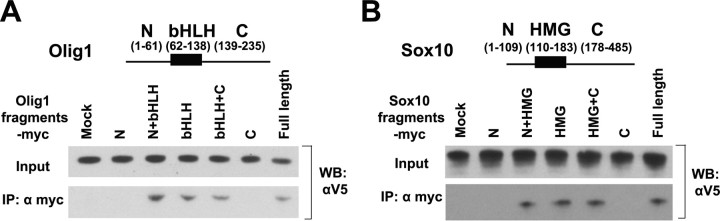
To map the interacting domains of Olig1 and Sox10, we made a series of constructs expressing different subdomains of Olig1 and Sox10 and performed coimmunoprecipitation assays in transfected Cos-7 cells. The bHLH domain of Olig1 was sufficient to coprecipitate Sox10, and conversely, the HMG domain of Sox10 was sufficient to pull down Olig1 (Fig. 3). Therefore, the physical interaction between Olig1 and Sox10 is mediated through their bHLH and HMG domains, respectively. The bHLH motif was previously known to be involved in DNA binding and interactions with other transcription factors (Sun et al., 2003; Wissmuller et al., 2006).
Figure 3.
Interaction of Olig1 and Sox10 is mediated by their respective bHLH and HMG domains. A, B, Cos-7 cells were cotransfected with V5-tagged, full-length Sox10 and Myc-tagged subdomains of Olig1 (A) or V5-tagged, full-length Olig1 and Myc-tagged subdomains of Sox10 (B). Cell lysates were subjected to immunoprecipitation with rabbit anti-Myc, followed by Western blot using mouse anti-V5. Empty Myc-vector was used as the mock control. One-twentieth of the aliquot of the cell lysate was subjected directly to Western blotting with anti-V5. N, N terminus; C, C terminus; IP, Immunoprecipitation; WB, Western blot.
Olig1 and Sox10 coregulate mbp transcription in vitro
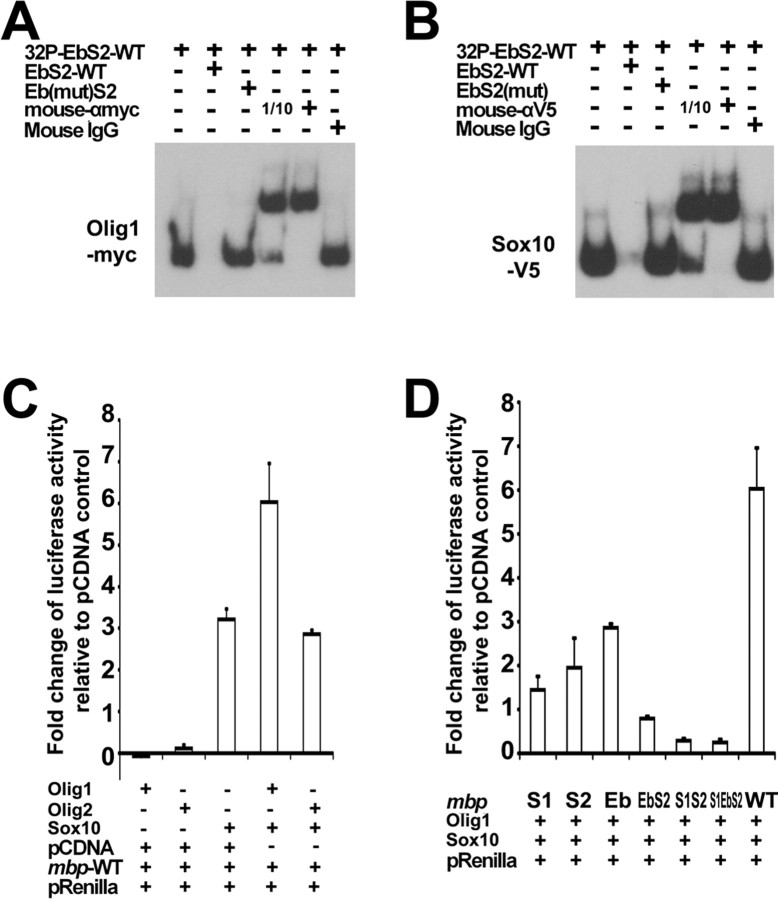
Previous studies have shown that, in mice, SOX10 can activate the Mbp and P0 gene promoters (Peirano and Wegner, 2000; Wei et al., 2004) and that OLIG1 can upregulate Mbp and Plp (Xin et al., 2005). Because zebrafish Olig1 can physically interact with Sox10, we investigated the potential collaborative role of Olig1 and Sox10 in mbp transcription. In the mbp upstream sequence, we found two Sox binding sites (named S1 and S2) that are completely conserved among human, mouse, rat, chick, and zebrafish (Fig. 4A). Just 8 bp upstream of site S2, there is a conserved bHLH binding site (Eb). To test whether these DNA motifs mediate the direct binding of Olig1 and/or Sox10 to the mbp gene, we performed electrophoretic mobility shift assays (EMSAs). A 39 bp double-stranded oligonucleotide containing Eb and S2 sites (EbS2) was used as a probe. The mobility of the 32P-labeled probe could be altered by immunopurified Olig1 or Sox10 proteins (epitope tagged) (Fig. 5A,B). Binding to the radiolabeled probe was inhibited by an excess of unlabeled wild-type EbS2 but not by mutated EbS2, confirming the specificity of protein–DNA binding (Fig. 5A,B). The shifted bands could also be supershifted with anti-Myc or anti-V5 antibodies (Fig. 5A,B). We found that an analogous oligonucleotide probe, EbS1, could also be shifted by Sox10 (data not shown).
Figure 4.
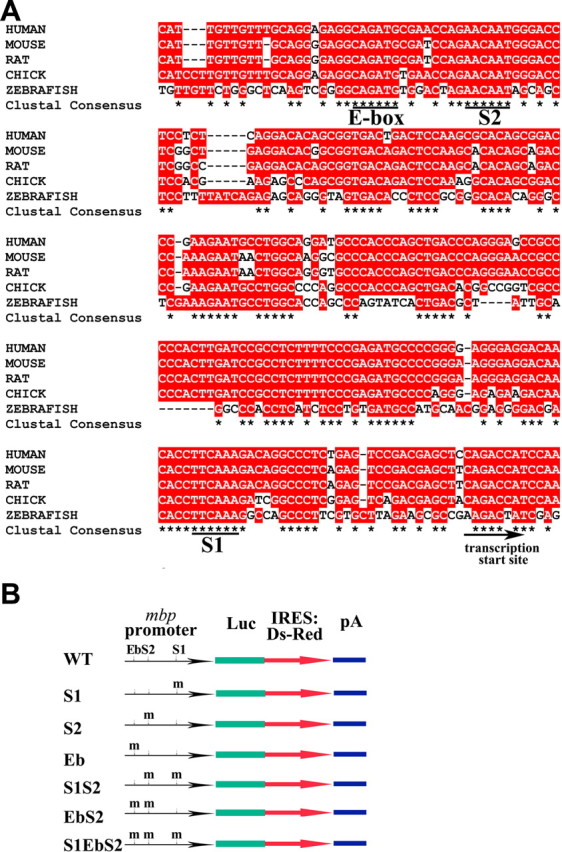
Transcription factor binding sites in the mbp promoter region. A, Alignment of the mbp promoter sequences from human, mouse, rat, chick, and zebrafish. Conserved nucleotides are in red, and clustal consensus sequences are indicated by asterisks. Conserved Sox10 and Olig1 binding sites are underlined. B, Schematic maps of a series of mbp reporter constructs. A 1.6 kbp mbp promoter was used to direct luciferase and IRES-DsRED expression. S1, S2, and Eb sites were mutated, respectively. WT, Wild type.
Figure 5.
Olig1 and Sox10 regulate mbp transcription in vitro. A, B, Analysis of Olig1 and Sox10 DNA binding activities by EMSA. Purified Olig1 and Sox10 could shift a 32P-labeled 39 bp double-stranded oligonucleotide containing Eb and S2 sites. A 100-fold excess of unlabeled wild-type oligonucleotide, mutated Eb (for Olig1), or mutated S2 (for Sox10) oligonucleotide was used as the competitor. Mouse IgG, mouse anti-Myc IgG (for Olig1), or mouse anti-V5 IgG (for Sox10) was used for supershift. Parallel lanes contained 10-fold different amounts of supershifting antibodies (labeled 1/10 or +). C, D, Luciferase assay was performed with cell lysates from transfected Cos-7 cells. pCDNA control vector was used to normalize the amount of transfected DNA. The firefly luciferase activity was standardized by reference to the Renilla luciferase activity. The results are displayed as fold increase of luciferase activity compared with control transfection of pRenilla and pCDNA only. Results are the mean ± SE of three independent experiments. WT, Wild type.
To further study the function of Olig1/Sox10 in regulating mbp transcription, we made a series of luciferase reporter constructs controlled by a 1.6 kbp zebrafish mbp promoter fragment (Fig. 4B). In Cos-7 cells cotransfected with the wild-type mbp-luciferase reporter and the Sox10 expression vector, there was a more than threefold increase in luciferase activity compared with reporter plus the pCDNA vector-only control (Fig. 5C). In parallel experiments, neither Olig1 nor Olig2 alone induced an obvious change in luciferase activity (Fig. 5C). However, when Olig1 and Sox10 were cotransfected along with the luciferase reporter, there was an additional twofold increase in reporter activity compared with reporter-plus-Sox10-alone transfection (Fig. 5C). Cotransfection of Olig2 and Sox10 did not have this augmenting effect (Fig. 5C).
To characterize Olig1/Sox10-specific DNA binding sites on the mbp promoter, the S1, S2, or Eb site in the 1.6 kbp mbp promoter-luciferase vector was mutated by PCR-based site-directed mutagenesis (Fig. 4B). The mutated luciferase constructs were then cotransfected with either Olig1 or Sox10 expression vectors. Mutation of Sox10 binding site S1 or S2 decreased luciferase activity more than threefold compared with the nonmutated construct (Fig. 5D). Mutation of Olig1 binding site Eb also decreased luciferase activity, although the effect was less marked (Fig. 5D). Double mutation of S2 and Eb further decreased luciferase activity, and double mutation of S2 and S1 or triple mutation of S1, S2, and Eb brought the luciferase activity down to background levels (Fig. 5D). Together, these in vitro results suggest that Sox10 alone is capable of activating mbp transcription and that Olig1, but not Olig2, can enhance Sox10 activity.
The Olig1/Sox10 complex regulates mbp transcription in vivo
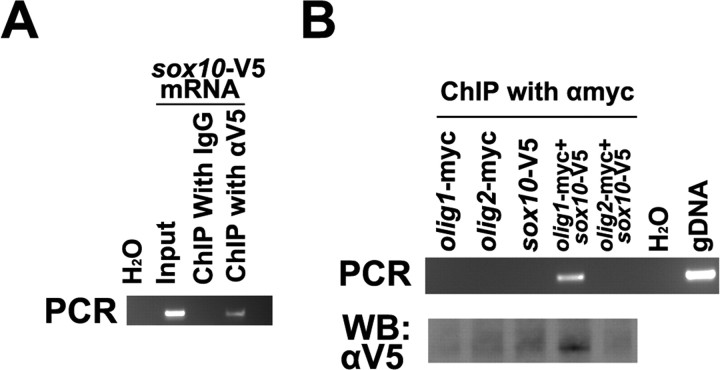
ChIP assays were performed to monitor DNA and protein interactions in zebrafish embryos in vivo. In vitro transcripts of PCS-olig1-myc, PCS-olig2-myc, and/or PCS-sox10-V5 were injected on their own into zebrafish one-cell-stage embryos. After 24 h, the embryos were collected and homogenized. Chromatin was cross-linked, fragmented by sonication, and immunoprecipitated with anti-Myc or anti-V5 antibodies. The presence or absence of the mbp promoter sequence was then detected in the immunoprecipitate by PCR. Sox10 could bind to the mbp promoter on its own (Fig. 6A), whereas Olig1 or Olig2 failed to bind (Fig. 6B). However, the mbp promoter could be precipitated by anti-Myc antibodies after coinjection of olig1-myc and sox10-V5 mRNAs. In contrast, a negative result was obtained with anti-Myc when olig2-myc and sox10-V5 mRNAs were coinjected under the same conditions (Fig. 6B). These data indicate that Olig1 can form an mbp binding complex with Sox10 but that Olig2 cannot. To test whether the Olig1/Sox10 complex can activate mbp transcription, we injected olig1-myc mRNA and/or sox10-V5 mRNA into one-cell-stage zebrafish embryos and collected embryos at 3 dpf to detect mbp mRNA expression by in situ hybridization on transverse spinal cord sections. Compared with olig1 or sox10 single injections, olig1/sox10 double injection gave rise to a significant increase in the number and intensity of mbp-expressing cells in the spinal cord (Fig. 7A). In addition, some ectopically expressed mbp signal was detected in somites after olig1/sox10 double injection.
Figure 6.
Olig1 and Sox10 bind synergistically to the mbp promotor in vivo. ChIP assays were performed with 24 hpf, mRNA-injected embryos, fixed with formaldehyde. A, B, Mouse IgG and mouse anti-V5 IgG (A) or mouse anti-Myc IgG (B) were used for immunoprecipitation. PCR was used to detect the mbp promotor region. This fragment was also amplified from the lysate (input), genomic DNA (gDNA), and water control (H2O). The immunoprecipitated DNA/protein complex was also detected by Western blotting (WB; B).
Figure 7.
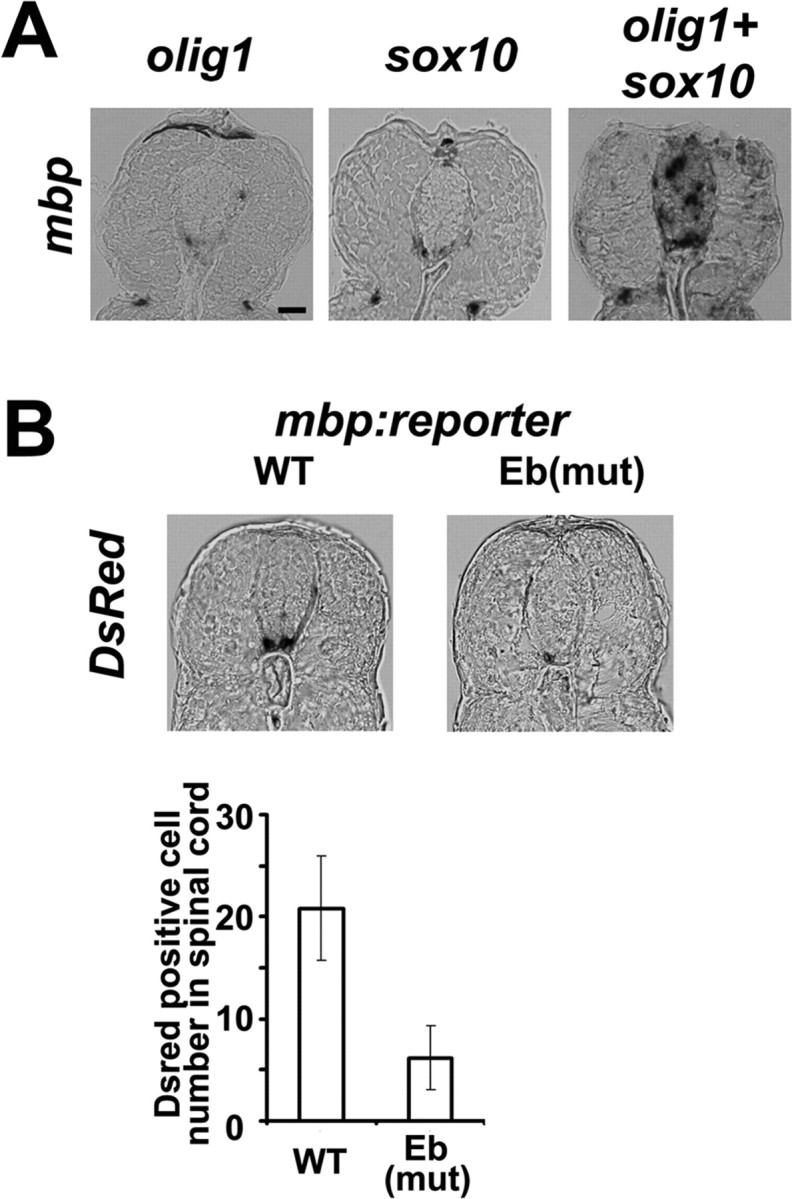
Olig1 and Sox10 regulate mbp transcription in vivo. Transverse sections through trunk; dorsal is to the top. A, The mbp expression in olig1/sox10 mRNA-injected embryos was revealed by in situ hybridization at 3 dpf. B, Different mbp reporter constructs were injected into one-cell-stage embryos. mbp transcription was evaluated by in situ hybridization at 3 dpf with a DsRed-specific probe. No DsRed signal was detected in embryos injected with S1, S2, Eb+S2, S1+S1, or S1+Eb+S2 mutated constructs (data not shown). Quantification of DsRed-positive cells in spinal cord (10 sections) showed that there were many more DsRed-expressing cells in spinal cord of embryos injected with the wild-type (WT) construct than in embryos injected with the Eb mutated (mut) construct. Error bars indicate SEM. Scale bar, 20 μm.
The series of mbp-luciferase reporters referred to above also contain a downstream IRES-DsRed sequence, so that that they are, in effect, double reporters of luciferase and DsRed. We injected these reporter constructs into one-cell-stage embryos and subsequently performed in situ hybridization with a DsRed probe (Fig. 7B). There were many more DsRed-expressing cells in embryos injected with wild-type mbp-DsRed reporter DNA (five fish, 10 sections per fish) than in embryos injected with the reporter containing a mutated Eb (five fish, 10 sections per fish) (Fig. 7B). In embryos injected with mbp-DsRed reporter containing mutated S1, S2, Eb+S2, S1+S1, or S1+Eb+S2 binding sites, no DsRed signal whatsoever was detected (data not shown). Overall, our data provide strong evidence that a protein complex of Olig1 and Sox10 directly binds to and activates transcription from the mbp promoter.
Discussion
olig1 is expressed by OL lineage cells during zebrafish CNS development
Zebrafish olig1 and olig2, like their mammalian counterparts, were expressed strictly in the CNS (Fig. 1). In mouse spinal cord, both OLIG1 and OLIG2 are expressed in the pMN (progenitors of motor neurons) domain in the ventral neuroepithelium, which is known to give rise to both motor neurons and OLs (Lu et al., 2000; Zhou et al., 2000). OLIG1 and OLIG2 are rapidly downregulated in postmitotic motor neurons but continue to be expressed in OL lineage cells throughout development and in the adult. In zebrafish, olig2 is expressed in a pMN-like region of neural plate cells at 9.5 hpf (Park et al., 2002) where olig1 is not expressed (data not shown). After motor neuronogenesis, expression of olig2 remains on in OLPs but is downregulated in differentiating OLs and motor neurons. olig1 first emerges in zebrafish spinal cord at 72 hpf (Fig. 1C) and partly overlaps with olig2 in white matter (Fig. 1D–F), suggesting that some migrating OLPs express olig1. Unlike olig2, olig1 partly colocalizes with differentiated OL markers mbp, plp1b, and mpz (Fig. 1J–R) at 72 hpf, continuing into adulthood (data not shown). The differences between the expression patterns of zebrafish and mammalian olig1 and between zebrafish olig1 and olig2 imply some functional differences between Olig1 and Olig2 and perhaps between Olig1 in fish versus mammals.
Functional differences between Olig1 and Olig2
As described originally, Olig1 knock-out mice (with a PGKneo cassette at the Olig1 locus) have delayed OL development but are otherwise normal (Lu et al., 2002). However, it was subsequently reported that after removal of the PGKneo cassette, the mice were dysmyelinating (they failed to activate myelin gene expression, developed tremor and seizures, and died in the third postnatal week) (Xin et al., 2005). This implied that the active PGKneo cassette might cause cis upregulation of the adjacent Olig2 gene and hence rescue the “true” Olig1 null phenotype. We performed olig1 knock-down experiments with specific antisense morpholinos but did not observe any reduction in mbp expression. This might not be surprising given that olig1 is not expressed until 72 hpf, which is close to the limit of the effectiveness of morpholinos. However, coinjection of olig1and sox10 mRNAs strongly increased mbp expression in fish embryos compared with injection of sox10 or olig1 alone.
OLIG2 is necessary for motor neuron and OL development both in rodents (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2002) and zebrafish (Park et al., 2002). OLIG2 is reported to interact with a range of different transcription factors including E12, IDs (Samanta and Kessler, 2004), NKX2.2 (Sun et al., 2003), and NGN2 (Lee et al., 2005), whereas the interacting partners of OLIG1 have not yet been established. We analyzed binding partners of Olig1 and Olig2 in zebrafish by coimmunoprecipitation. Zebrafish Olig2, like mouse OLIG2, can form homodimers and can also bind to E12, Nkx2.2a, and Ngn3. Zebrafish Olig1, in contrast, can form homodimers and heterodimers with Olig2 or E12 but fails to interact with Nkx2.2a or Ngn3.
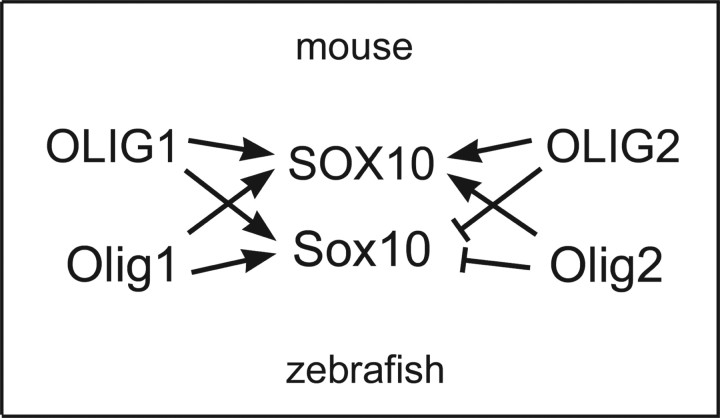
Another difference between zebrafish Olig1 and Olig2 is that Olig1 can bind to Sox10 whereas Olig2 cannot. This seemed to contradict the previous report that OLIG2 can bind to SOX10 in mouse (Wissmuller et al., 2006). However, we have shown that this is a true species difference: OLIG2/Olig2 from mice or zebrafish can both bind mouse SOX10 but not zebrafish Sox10. This probably reflects several key amino acid differences between the HMG domains of mouse SOX10 and zebrafish Sox10 (supplemental Fig. 1E, available at www.jneurosci.org as supplemental material), specifically in the H3 and C-terminal regions, which are known to be important for protein–protein interactions of SOX proteins (Remenyi et al., 2003; Wissmuller et al., 2006). In addition to the species divergence between SOX10 and Sox10, there is also divergence between the binding properties of OLIG1 and OLIG2 (and between Olig1 and Olig2), illustrated by the fact that OLIG1/Olig1 but not OLIG2/Olig2 can bind zebrafish Sox10 (summarized in Fig. 8). There is 22% amino acid sequence divergence between the bHLH domains of OLIG1 and OLIG2 (36% divergence between bHLH domains of zebrafish Olig1 and Olig2), but it is not yet possible to pinpoint which amino acids are responsible for their different Sox10 binding activities because the crystal structure of OLIG proteins has not yet been determined. It is also not known whether, or how, the different structural properties of OLIG1/Olig1 and OLIG2/Olig2 might be reflected in their interactions with other transcriptional cofactors in mice or fish.
Figure 8.
Summary of the interactions among SOX10, OLIG1, and OLIG2 from mouse and zebrafish. Arrows indicate binding, and “T” symbols indicate lack of binding.
The Olig1/Sox10 complex directly activates mbp transcription
SOX10 is a key mediator of OL terminal differentiation (Stolt et al., 2002). In Sox10 null mouse embryos, expression of myelin genes including Mbp, Plp, and Mag is lost (Stolt et al., 2002). SOX10 can bind to multiple sites in the proximal region of the Mbp promoter to activate its transcription (Stolt et al., 2002, 2006; Wei et al., 2004). SOX10 can also stimulate expression of P0 in the CNS (Brosamle and Halpern, 2002; Schweitzer et al., 2003) as well as in Schwann cells in the peripheral nervous system (Peirano and Wegner, 2000; Peirano et al., 2000). OLIG1 is crucial for OL myelinogenesis in brain and is required for transcription of Mbp, Plp, and Mag (Xin et al., 2005). Here, we uncovered a functional relationship between Olig1, Sox10, and mbp. There are two evolutionarily conserved Sox10 binding sites (S1 and S2) in the proximal region of the mbp promoter as well as an Eb that lies 8 bp upstream of S2 (Fig. 4A). Sox10 can bind to S1 and S2, whereas both Olig1 and Olig2 can bind to Eb (Fig. 5A,B) (and data not shown). Although these results suggest that Sox10 is not absolutely required for the binding of Olig1 to the mbp promoter in vitro, ChIP with mRNA-injected embryos indicated that Sox10 is needed for Olig1 binding to the endogenous mbp promoter in vivo (Fig. 6B).
According to previous reports, OLIG1/2 are transcriptional repressors (Novitch et al., 2001; Lee et al., 2005). However, through binding to different partners or by phosphorylation at different sites, the properties of a transcription factor can change (Ju and Rosenfeld, 2006). By in vitro luciferase assay, we found that Sox10 might be primarily responsible for mbp transcription but that Olig1 can enhance transcription approximately twofold. This stimulating activity disappeared after mutagenesis of Olig1 and/or Sox10 binding sites. The functional synergy between Olig1 and Sox10 was further verified in vivo. Coinjection of olig1 and sox10 mRNA into zebrafish one-cell embryos strongly enhanced mbp expression, whereas injection of either mRNA individually had little effect (Fig. 7A). Moreover, Olig1/Sox10 complex formation could not enhance plp1b or mpz expression in the spinal cord (data not shown), consistent with the observation that there are no conserved bHLH/HMG binding motifs in the upstream regions of the plp1b or mpz genes.
Liu et al. (2007) showed that Sox10 alone could activate mbp expression when a Sox10-expressing retrovirus was electroporated into the embryonic chick spinal cord. Our failure to observe a similar effect of Sox10 alone after mRNA injection into fish embryos could simply reflect mRNA decay over the time course of our experiment, resulting in low levels of Sox10 expression. However, this does not affect our conclusion that Sox10 and Olig1 cooperate to elevate mbp expression both in vitro and in vivo.
Mouse OLIG2 has been shown to bind SOX10 in vitro (Wissmuller et al., 2006), but it remains to be shown whether OLIG2 binds to SOX10 in vivo, or whether OLIG2 can cooperate with SOX10 to drive Mbp transcription in vivo.
Evolution of CNS myelin
Compact myelin is a defining characteristic of gnathostomes (vertebrates with jaws) and must have played a crucial role in vertebrate evolution. By permitting much faster conduction of action potentials in both the CNS and PNS, the presence of compact myelin would have allowed more rapid sensory and motor reflexes and would presumably have conferred a strong selective advantage for rapid locomotion, favoring predatory behavior and/or escape from predators, for example. Other benefits would undoubtedly have included more central processing power (evolution of a complex, portable brain) and better central control over distal appendages, thereby allowing body size to increase. Given the crucial influence of OLIG2 on OL development, exploring the molecular evolution of olig genes might yield useful clues to myelin evolution.
There is only one member of the olig gene family in invertebrates (Lowe et al., 2006; Bronchain et al., 2007). So far, there is no information about olig genes in agnathans (lampreys and hagfish, which lack myelin), but in the genome of a myelinated cartilaginous fish (elephant shark or chimera, Callorhinchus milii) we identified orthologs of olig2 and olig3 but not olig1 (data not shown). This finding suggests that olig2 and olig3 were originally derived by duplication of a single ancestral olig gene, perhaps during the whole genome duplication that is known to have occurred early in the vertebrate lineage, and that olig1 appeared later in bony fish through local duplication at the olig2 locus. This interpretation contrasts with the conclusion of a recent in silico analysis that olig1 is the founder member of the gene family (Bronchain et al., 2007). Distinguishing between these models requires further comparative analysis, in particular whether the lack of olig1 is a general property of cartilaginous fish.
Why did OLIG2 acquire the SOX10 binding properties of OLIG1 after the separation of fish and mammalian lineages? We suggest that Olig1 first appeared in teleost fish, increasing myelin gene expression and augmenting the rate or extent of myelination. Subsequently, in mammals, OLIG2 acquired the ability to bind SOX10, perhaps further enhancing myelin gene transcription and myelination. Testing this model will require further comparison of OLIG1 and OLIG2 activities in vitro and in vivo.
Footnotes
This work was supported by grants from the Wellcome Trust and the United Kingdom Medical Research Council (W.D.R., H.K.S.). We thank our colleagues in the Wolfson Institute for Biomedical Research for helpful discussions. We also thank Steve Wilson and members of his research group for invaluable help and advice, Carole Wilson for fish stock maintenance, and B. Venkatesh for helping us to analyze the elephant shark genomic database.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Bronchain OJ, Pollet N, Ymlahi-Ouazzani Q, Dhorne-Pollet S, Helbling JC, Lecarpentier JE, Percheron K, Wegnez M. The olig family: phylogenetic analysis and early gene expression in Xenopus tropicalis. Dev Genes Evol. 2007;217:485–497. doi: 10.1007/s00427-007-0158-z. [DOI] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Collarini EJ, Kuhn R, Marshall CJ, Monuki ES, Lemke G, Richardson WD. Down-regulation of the POU transcription factor SCIP is an early event in oligodendrocyte differentiation in vitro. Development. 1992;116:193–200. doi: 10.1242/dev.116.1.193. [DOI] [PubMed] [Google Scholar]
- Filippi A, Tiso N, Deflorian G, Zecchin E, Bortolussi M, Argenton F. The basic helix-loop-helix olig3 establishes the neural plate boundary of the trunk and is necessary for development of the dorsal spinal cord. Proc Natl Acad Sci USA. 2005;102:4377–4382. doi: 10.1073/pnas.0407284102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havis E, Anselme I, Schneider-Maunoury S. Whole embryo chromatin immunoprecipitation protocol for the in vivo study of zebrafish development. Biotechniques. 2006;40:34, 36, 38. doi: 10.2144/000112098. [DOI] [PubMed] [Google Scholar]
- Ju BG, Rosenfeld MG. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle. 2006;5:2557–2560. doi: 10.4161/cc.5.22.3497. [DOI] [PubMed] [Google Scholar]
- Kazakova N, Li H, Mora A, Jessen KR, Mirsky R, Richardson WD, Smith HK. A screen for mutations in zebrafish that affect myelin gene expression in Schwann cells and oligodendrocytes. Dev Biol. 2006;297:1–13. doi: 10.1016/j.ydbio.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levavasseur F, Mandemakers W, Visser P, Broos L, Grosveld F, Zivkovic D, Meijer D. Comparison of sequence and function of the Oct-6 genes in zebrafish, chicken and mouse. Mech Dev. 1998;74:89–98. doi: 10.1016/s0925-4773(98)00067-7. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, Smith M, Kirschner M, Gerhart J. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Muller T, Anlag K, Wildner H, Britsch S, Treier M, Birchmeier C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005;19:733–743. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Peirano RI, Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol Cell Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, Richardson WD. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003;130:93–102. doi: 10.1242/dev.00184. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Becker T, Becker CG, Schachner M. Expression of protein zero is increased in lesioned axon pathways in the central nervous system of adult zebrafish. Glia. 2003;41:301–317. doi: 10.1002/glia.10192. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk DI, Kaing S, Stiles CD, Rowitch DH. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11:1413–1420. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23:9547–9556. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol. 2006;7:R71. doi: 10.1186/gb-2006-7-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Sox10 acts as a tissue-specific transcription factor enhancing activation of the myelin basic protein gene promoter by p27Kip1 and Sp1. J Neurosci Res. 2004;78:796–802. doi: 10.1002/jnr.20342. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J Biol Chem. 2005;280:16284–16294. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]