Abstract
α-Neurexins are synaptic organizing molecules implicated in neuropsychiatric disorders. They bind and arrange an array of different partners in the synaptic cleft. The extracellular region of neurexin 1α (n1α) contains six LNS domains (L1-L6) interspersed by three Egf-like repeats. N1α must encode highly evolved structure-function relationships in order to fit into the narrow confines of the synaptic cleft, and also recruit its large, membrane-bound partners. Internal molecular flexibility could provide a solution, however, it is challenging to delineate because currently no structural methods permit high resolution structure determination of large, flexible, multi-domain protein molecules. To investigate the structural plasticity of n1α, in particular the conformation of domains that carry validated binding sites for different protein partners, we used a panel of structural techniques. Individual particle electron tomography (IPET) revealed that the N-terminally and C-terminally tethered domains, L1 and L6, have a surprisingly limited range of conformational freedom with respect to the linear central core containing L2 through L5. A 2.8 Å crystal structure revealed an unexpected arrangement of the L2 and L3 domains. SAXS and ET indicated that incorporation of the alternative splice insert SS6 relieves the restricted conformational freedom between L5 and L6, suggesting that SS6 may work as a molecular toggle. The architecture of n1α thus encodes a combination of rigid and flexibly tethered domains that are uniquely poised to work together to promote its organizing function in the synaptic cleft, and may permit allosterically regulated and/or concerted protein partner binding.
Keywords: synapse, adhesion, neuropsychiatric disorders, protein structure, single-molecule 3D density map
INTRODUCTION
Neurexins form a large portfolio of synaptic adhesion and organizing molecules. They mediate synaptic organization and facilitate synaptic transmission, promoting communication between neurons [1–4]. Neurexins trigger postsynaptic differentiation in contacting dendrites, i.e., the recruitment of a functional postsynaptic signaling machinery [4–6]. Presynaptic α-neurexins use their large extracellular domain to bind and organize an extensive array of proteins in the synaptic cleft; these include postsynaptically tethered partners such as neuroligins (NLGNs), LRRTMs, calsyntenin 3 (CLSTN3), α-dystroglycan, IgSF21, and latrophilin, but also secreted proteins such as neurexophilins, hevin, and cerebellin [4,6–17]. Together with their partners, α-neurexins modulate the number and distribution of synapses, and play distinct roles at excitatory versus inhibitory synapses. α-Neurexins and their partners are implicated in autism spectrum disorder (ASD), schizophrenia (SZ), and mental retardation (MR), and manipulating their levels in animal models replicates behavioral alterations seen in humans with ASD and SZ [4,18–29]. Together, α-neurexins and their partners play a crucial role in mediating connectivities that wire neurons into neural circuits, impacting thereby the communication that traverses these circuits and critical pathways altered in ASD, SZ, and MR.
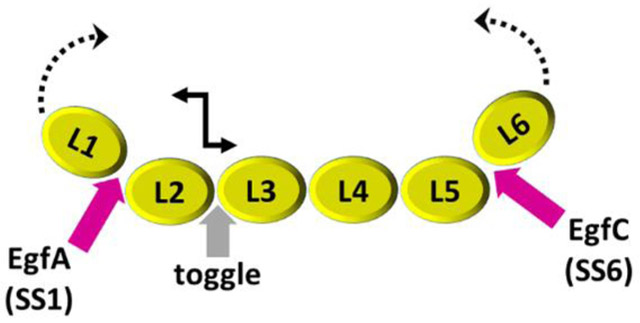
In mammals, there are three neurexin genes (neurexin 1, 2 and 3) and each gene encodes a long α neurexin as well as a short β neurexin [4]. Neurexin 1alpha (n1α) is composed of six LNS domains (L1 through L6) interspersed by three EGF-like repeats (EgfA, EgfB, and EgfC) and it is tethered predominantly to the presynaptic membrane via L6 (Fig. 1a) [4]. The ectodomain has traditionally been divided into three so-called ‘neurexin repeats’ (I, II, and III) consisting of LNS-EGF-LNS. Neurexin mRNA transcripts are diversified through alternative splicing at six sites, SS1 through SS6, generating more than a thousand splice forms (Fig. 1a) [30,31]. Neurexin LNS domains contain a ‘hypervariable surface’ at one edge of their β-sandwich fold formed by loops that host splice inserts and a central Ca2+-binding site [6,32,33]. Most, but not all, neurexin partners bind to these hypervariable surfaces regulated by the presence of splice inserts and/or Ca2+, e.g., neuroligins and LRRTMs [9,34–37].
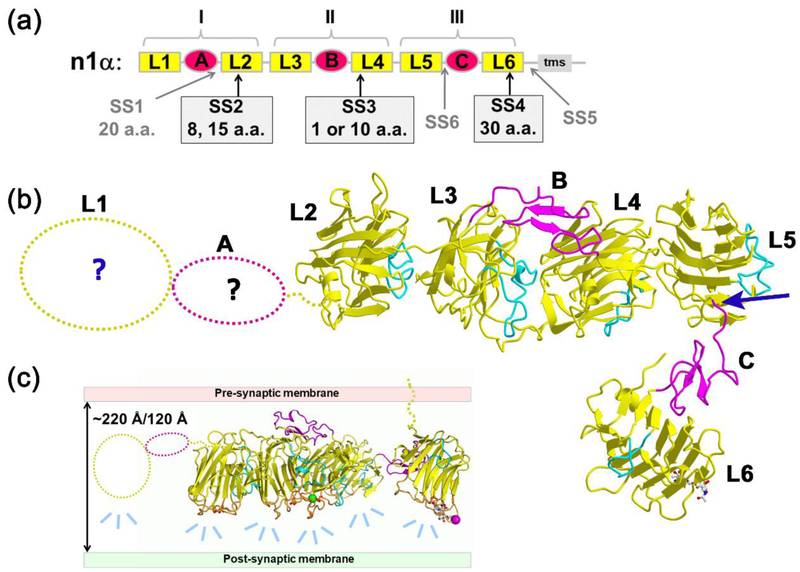
Figure 1: α-Neurexins.
a) Domain structure of n1α. Splice inserts SS1-SS6 are indicated as well as the transmembrane segment (tms).; b) L2 through L6 are ordered in crystal structures (e.g., pdbid:3QCW) while L1-EgfA are disordered (dotted line). A blue arrow indicates a characteristic molecular hinge linking L5 to the EgfC-L6 assembly. The loops β11-β12 that fill the concave side of each β-sandwich are indicated in cyan to help orient the reader.; c) Model depicting how the orientation and architecture of n1α in the synaptic cleft might influence the mode of interaction with postsynaptically-tethered partners via its hyper-variable surfaces (light blue dashes). The hyper-variable surfaces are shown as orange loops; the central Ca2+-binding site at each hyper-variable surface is indicated by a conserved Asp residue that interacts with the Ca2+-ion.
To reveal structure-function relationships, the extracellular domain of n1α has been studied by EM, SAXS and X-ray crystallography. EM analysis of negatively stained n1α L1-L6 particles revealed that five out of six LNS domains arranged in a Y-shape, assigned to L2-L6, while L1-EgfA were too flexible to be visualized [38,39]. The 3D structure of n1α from crystals containing L1-L6 [40] or L2-L6 [41] revealed that L2-L6 are connected into an L-shaped molecule. L2 through L5 adopt a rod-like concatenation attached via a molecular hinge to a ‘foot’ containing EgfC-L6 (Fig. 1b). The three ‘neurexin repeats’ adopted very different domain arrangements with the central neurexin repeat II (L3-EgfB-L4) containing a horse shoe-shaped configuration similar to reelin-repeats, while the other repeats were extended [40]. The placement of the hyper-variable surfaces on one side of n1α led to the suggestion that n1α works as a synaptic organizer by forming a scaffold onto which partners can dock, guiding their arrangement in the synaptic cleft (Fig. 1c). However, how the architecture of n1α controls its organizing ability in the synaptic cleft has remained unclear. In particular, the exact dimensions of n1α have remained uncertain. Also the range of conformations adopted by a population of n1α molecules has remained unknown. Yet these molecular properties are crucial because they determine how n1α fits in the narrow confines of the synaptic cleft and determine how n1α recruits different partners there.
Currently, no single structural technique is well-suited to investigate in 3D at high resolution the conformational preference, flexibility and dynamics within a population of n1α molecules. For example, X-ray crystallography is limited by the need to pack (nearly) identical molecules into a crystal in order to obtain electron density revealing their 3D structure. NMR techniques are still limited by the size of the proteins that can be tackled. SAXS provides low resolution information on the molecular size and shape of an ensemble of molecules that tumble freely in solution averaged over time. Single particle EM analysis requires images of thousands to millions of protein particles that share an identical structure, but in different orientations, that are classified and averaged together to form a limited number of projections (‘class averages’) that are used to computationally produce a 3D reconstruction; however, the portions of each molecule that are conformationally heterogeneous are averaged away revealing only the structurally uniform core. Recently, we reported a method to determine the 3D structure of single protein molecules using individual particle electron tomography (IPET) 3D reconstruction which avoids information loss due to averaging and avoids bias introduced by using models generated from class averages as initial models for 3D reconstruction, though the structures produced are lower resolution because of lower signal-to-noise ratio [11,42–44]. This method has allowed us to analyze the conformational distribution of a broad range of proteins, including large, multi-domain synaptic organizers like Contactin-associated Protein-like 2 (CNTNAP2) and Calsyntenin 3 (CLSTN3), as well as other proteins, e.g., [11,45–49].
Here we used a combination of structural techniques, i.e., IPET, X-ray crystallography and SAXS, to investigate the conformational preference of n1α and identify elements that regulate the flexibility and conformation of two LNS domains, L2 and L6, that contain validated binding sites for protein partners. Using IPET, we determined 3D reconstructions for a total of 110 individual particles of n1α L1-L6 at ~15 Å. Using X-ray crystallography, we determined the structure of n1α L2-L3 to 2.8 Å. Finally using SAXS, we assessed the flexibility of n1α repeat III (L5-EgfC-L6) with and without splice insert SS6. Our studies reveal the range of conformational freedom within a population of n1α molecules, an unexpected possible molecular switch between the L2 and L3 domains that opens and closes the molecule, and the impact of incorporating splice insert SS6. These data extend our understanding of how n1α is poised to recruit and arrange different protein partners in the synaptic cleft, and how its unique architecture provides a platform of structural elements that support possible allosteric control and concerted protein partner binding.
RESULTS
Architecture of n1α by OpNS-EM
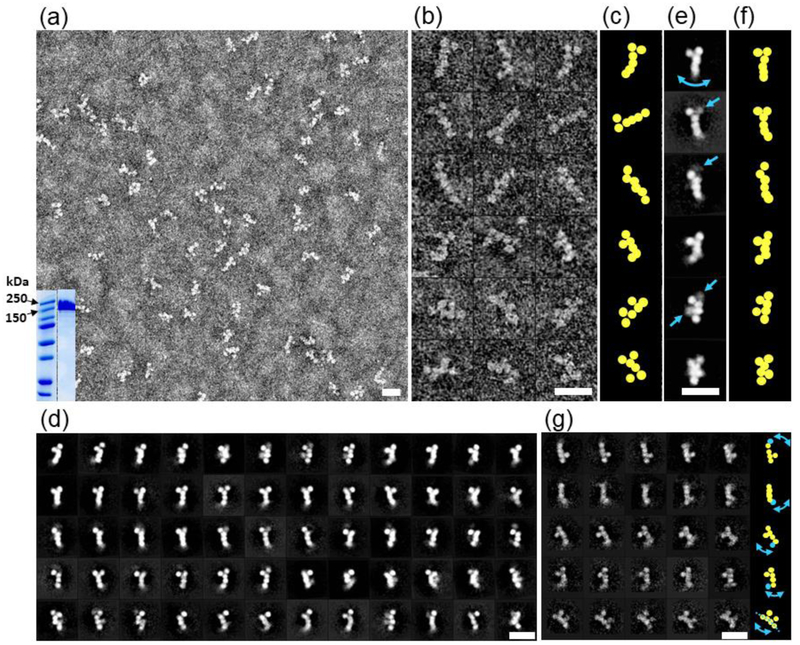
To examine the architecture of n1α L1-L6, we collected EM images of particles using optimized negative-staining (OpNS) and selected reference-free class averages. The survey image (Fig. 2a) showed that n1α particles are monodisperse. All six LNS domains, L1 through L6, were seen clearly, corresponding to round shapes with a diameter ranging from ~35 to 50 Å each, consistent with the size of LNS domains observed in crystal structures [39–41]. However, in addition to the previously reported L-shaped or Y-shaped molecules, additional morphologies were observed including completely linear, F-shaped, π-shaped, and Z-shaped molecules (Fig. 2b; Fig. 2c). To increase the signal-to-noise, approximately 15,000 particles were submitted to reference-free two-dimensional (2D) class averaging using a 100 classes (Fig. 2d). Although six LNS domains could be identified in all the classes, in more than half of the class averages one or two domains were blurred, indicating flexible domains in the protein (see Fig. 2d and arrows in Fig. 2e) which was not alleviated by using more classes (Fig. 2g). This suggested to us that 3D reconstruction of n1α using traditional methods relying on averaging thousands of images from particles was likely obscuring visualization of the distribution of protein conformations. For this reason, we investigated single n1α particles using IPET.
Figure 2: Negative-stain EM images of n1α.
a) Survey view of n1α L1-L6 particles; analysis of n1α L1-L6 by SDS-PAGE with relevant markers indicated in kDa (inset); b) 18 representative raw images of n1α L1-L6 particles.; c) Schematic highlighting the different organizations of domains.; d) 60 representative averaged particles selected from 100 reference-free class averages calculated from 15,402 particles picked from 150 micrographs. Select domains have a blurry appearance because they are averaged away due to conformational heterogeneity.; e) Six representative reference-free class averages. Arrows indicate domains with conformational heterogeneity.; f) Schematic highlighting the organization of domains.; g) 25 representative averaged particles selected from 1,043 reference-free class averages calculated from 15,402 particles showing that even with fewer particles per class there is significant conformational variability within every row (domain with high conformational variability indicated in cyan). Arrows in the schematic on the right panel show the corresponding variation of domains. Scale bars are 200 Å.
Architecture of neurexin 1α by OpNS-IPET
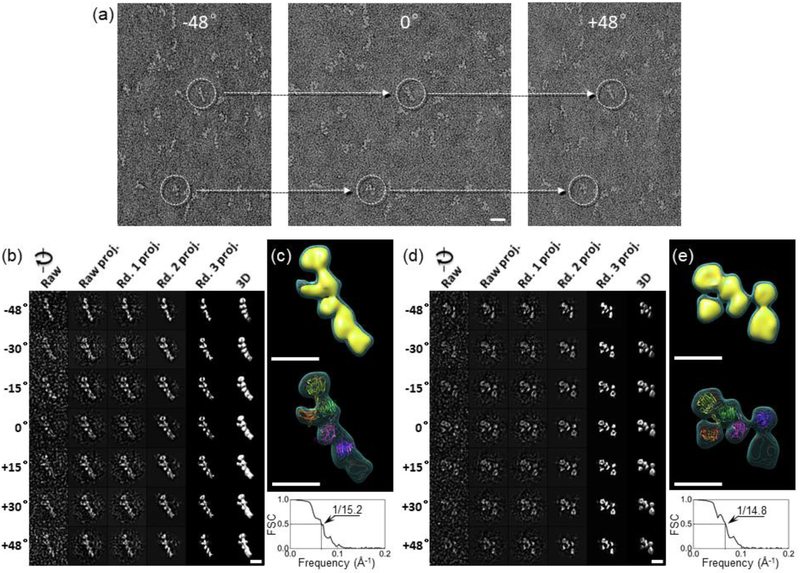
To investigate the conformational variability of n1α, we examined single particles using IPET which entails determining an ab-initio 3D structure of an individual protein particle from a series of tilt images. OpNS-grids containing n1α were used to acquire and align 65 tilting images per particle (Fig. 3a). In total, tilt image series were collected for 110 individual particles. Although the signal-to-noise ratio (SNR) was only ~0.1 to ~0.5 in each tilt image, the overall shape of each n1α particle was still clearly visible, and images could be iteratively aligned to a global center increasing the SNRs in the 3D projections gradually to ~1.8 before achieving a final ab-initio 3D reconstruction (Fig. 3b). As shown for one n1α particle, six globular densities form an overall L-shape or Y-shape at ~15.2 Å resolution that readily accommodates n1α as observed in crystal structures (e.g., pdbid:3QCW; L2 through L6 domains) (Fig. 3c). Demonstrating conformational variability, another representative individual n1α particle was selected and reconstructed using the same IPET procedure, but revealed a very different conformation with a Z-shaped arrangement (Fig. 3d; Fig. 3e). The n1α crystal structure could also be fit into the final 3D EM density map of this particle as well by docking the L3-EgfB-L4-L5 fragment and flexibly docking L2 and the EgfC-L6 entities separately on either side (Fig. 3e).
Figure 3: OpNS-IPET 3D reconstruction of n1α.
a) Two selected individual n1α L1-L6 particles (dashed circles) targeted for a tilt image series. Scale bar is 200 Å.; b) Process to generate representative 3D density maps from an individual n1α L1-L6 particle using IPET.; c) Final 3D density map using the ab initio density map obtained from IPET as an initial model; shown as dual iso-surfaces contoured at volumes corresponding to 1.2 and 2.3 times the molecular mass of ~141 kDa (top). Final 3D density map overlaid with the crystal structure of n1α (pdbid:3QCW) showing L2 (purple), L3 (magenta), L4 (green), L5 (yellow) and L6 (red) (middle). Fourier shell correlation analysis (FSC) (bottom). The structure was oriented in the density map so that L6 formed the foot of the L-shape, in accordance to previous studies.; d) IPET reconstruction of a second individual n1α particle.; e) Final 3D density map for a second particle contoured as in c) (top). Final 3D density map overlaid with the flexibly docked n1α crystal structure (pdbid:3QCW), domain coloring as above in c) (middle). FSC analysis (bottom). The density maps in c) and e) have an effective resolution of ~15.0 Å based on the FSC=0.5 criteria; shown with scale bar 100 Å.
Assessment of the conformation variability of n1α
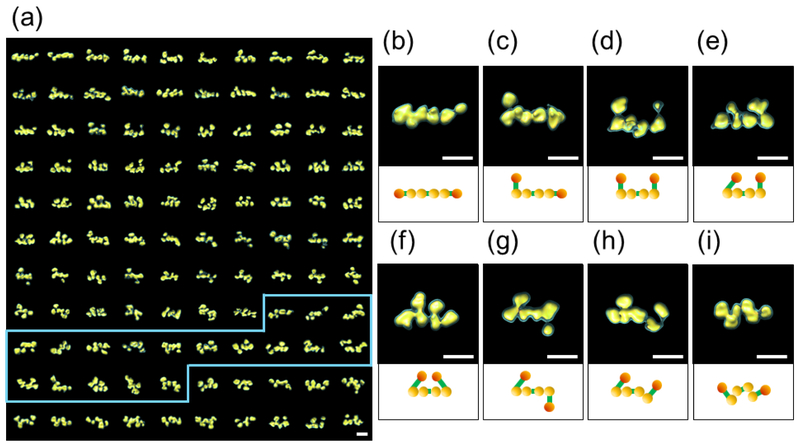
To assess the full range of conformational variability of n1α, we performed IPET 3D reconstructions for a total of 110 individual n1α particles (Fig. 4a). The central core corresponding to domains L2-L3-L4-L5 was observed as a linear array (Fig. 4b through 4g), but the conformation of the outer domains L1 and L6 varied with respect to the central core generating three main groups: i) all six LNS domains in line (Fig. 4b); ii) five LNS domains in line with the sixth deviating (Fig. 4c); iii) four LNS domains in line with the outer L1 and L6 domains deviating and located either on the same side (Fig. 4d, 4e, 4f) or on opposite sides of the central core (Fig. 4g). Strikingly, in a number of cases, the central core was not linear and adopted a variety of bent arrangements that have not been reported before (Fig. 4h, 4i). Taking the variability of the conformations in account, n1α spans 160 - 220 Å in the longest dimension based on 110 IPET reconstructions.
Figure 4: Panel of IPET 3D reconstructions of n1α.
a) IPET 3D structures of 110 individual n1α L1-L6 particles. A subset of the particles (demarcated in blue) appear to have only three domains in the central core, see Discussion.; b) through g) Final 3D density maps for n1α L1-L6 particles exhibiting a linear central core (domains L2-L5); h) and i) Final 3D density maps for n1α L1-L6 particles exhibiting a non-linear central core (domains L2-L5). The IPET reconstructions are contoured at volumes corresponding to 1.2 and 2.3 times the molecular mass of ~141 kDa. Under each IPET reconstruction a schematic of n1α L1-L6 is shown with L1 and L6 in orange and L2 through L5 in yellow. Egf-like repeats are not depicted. Scale bars are 100 Å.
Statistical analysis of the conformational flexibility of n1α.
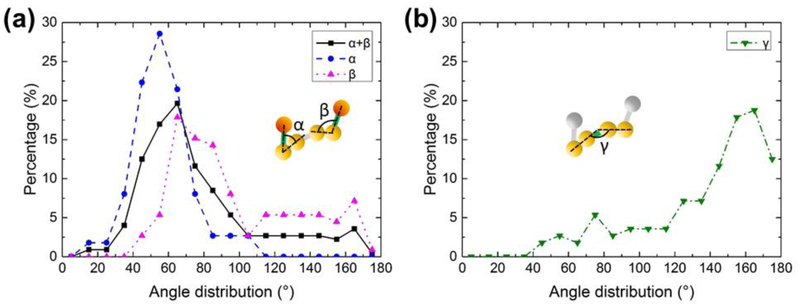
To investigate the conformational preference within a population of n1α particles, we carried out a statistical analysis comparing the 110 IPET 3D reconstructions. We investigated the two outer angles of each molecule, α and β, corresponding to the orientation of the two outer LNS domains with respect to the central core, and the internal torsional angle γ falling within the central core. The angles between domains were calculated by determining the center of mass for the LNS domains obtained from the 110 IPET 3D density maps, and using the coordinates to extract the angles. The molecules were binned according to their α, β, α+β and γ angles generating histograms for their conformational preference (Fig. 5). The histograms revealed a surprising preference for discrete conformations within the molecules. Most n1α particles displayed an α angle in the range 40° to 70° (72%) and an β angle in the range 60° to 90° (47%), considering the three or four most populated bins (Fig. 5a). To circumvent any ambiguity in the assignment of the first and last LNS domain, we examined the distribution of α and β together which still produced a unimodal distribution in the range 40° to 80° (61%) indicating that L1 and L6 adopted a remarkably similar conformations lying close to the central core in most of the molecules. The torsional angle γ, reflecting the internal core of the molecule, adopted a range of 140° to 180° (61%) for the four most populated bins consistent with a largely linear conformation (Fig. 5b). Our analysis suggests that while EgfA and EgfC flexibly tether L1 and L6, respectively, both of these domains have preferred orientations with respect to the central core. The central core featuring EgfB contains LNS domains that arranged in a largely linear array. Strikingly, a small percentage of particles (< 10%) appear to exhibit flexibility within the central core (see Discussion). The conformational flexibility of n1α observed by EM prompted us to investigate the N-terminal and C-terminal regions of n1α more closely using other techniques, focusing on the N-terminal L2 and the C-terminal L6 domains which carry well validated binding sites for post-synaptic partners.
Figure 5: Conformational preference of n1α.
a) Distribution of values for α, β, and α+β derived from 110 n1α L1-L6 IPET 3D reconstructions.; b) Distribution of values for γ derived from 110 n1α IPET 3D reconstructions.
Conformational variability in the N-terminal portion of n1α.
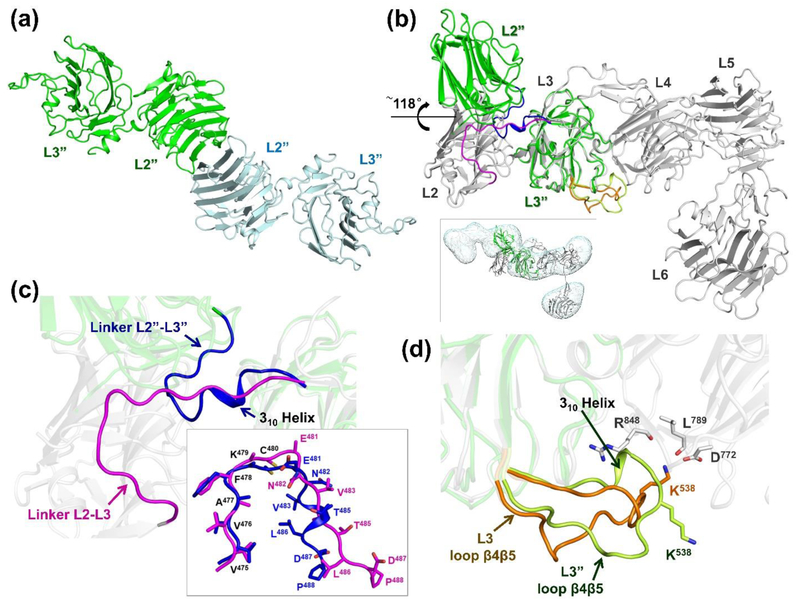
To investigate the conformational variability between domains in the N-terminal portion of n1α, we determined the crystal structure of n1α L2-L3 using X-ray crystallography to a resolution of 2.84 Å (Rwork 22.2 %, Rfree 25.5 %; Table 1). Two independent copies of the tandem were found in the asymmetric unit that were essentially identical (rmsd 0.16 Å for 376 Cα-atoms) (Fig. 6a). However, L2 adopts a dramatically different conformation with respect to L3 compared to that seen in crystal structures containing n1α L1-L6 (pdbid: 3QCW; [40]) or n1α L2-L6 (pdbid:3POY; [41]) (Fig. 6b). The L2 domain has undergone a ~118° rotation swinging away from L3. The crystal structure of n1α L2-L3 is readily docked into an IPET map of a particle with a bent central core (with an associated gamma angle of ~129°; see 3rd map, top row of demarcated particles in Fig. 4a), and it is better accommodated than its counterpart in n1α L2-L6 which houses L2 and L3 in a side-by-side or ‘closed’ conformation (Fig. 6b, inset). We analyzed the ‘open’ and ‘closed’ conformation seen in n1α L2-L3 and n1α L1-L6, respectively, in greater detail to understand the nature of the molecular transition. In the ‘open’ form, the linker between L2 and L3 (residues Gly474-Pro488) has rearranged completely by folding back on itself so that the end-to-end Cα distance is 15.4 Å compared to 24.6 Å in the ‘closed’ form. The N-terminal part of the linker (residues Gly474-Cys480) retains its backbone conformation with an rmsd of 1.0 Å for 7 Cα atoms between forms as these residues are anchored to L2 via a disulfide bond between Cys444 (L2) and Cys480 (linker). However, the C-terminal part of the linker (residues Glu481-Pro488) is dramatically different, converting from an extended conformation to a 310 helical conformation as L2 swings to the ‘open’ form (Fig. 6c). In this ‘open’ conformation, more residues from L2 and L3 interact with the L2-L3 linker, i.e., 20 residues of L2 and 14 residues of L3, compared to in the ‘closed’ conformation with only 18 residues of L2 and 11 residues of L3 (Table 2a). However, in the ‘open’ form, L2 and L3 contact each other directly via fewer residues (7 residues) compared to the ‘closed’ form (24 residues) (Table 2b). Only one residue (Ser289 in L2) mediates direct contact between L2 and L3 in both states Surprisingly, the net chemical interactions at the interface between L2, L3 and their linker do not change very drastically as L2 swings to the ‘open’ conformation, essentially adding only one pair of residues in a salt bridge (Table 2c). Releasing L2 in the ‘open form’ reduces the buried surface by 614 Å2 at the L2-L3 interface (1419 Å2) compared to the ‘closed’ form (2033 Å2; pdbid:3QCW), as calculated by ePISA [50] considering the interface between L2 and linker-L3. Though dramatic changes are seen between their mutual interaction, connecting linker, and orientation, the isolated L2 and L3 domains themselves are very similar, i.e. for L2 an rmsd 0.81 Å for 176 Cα-atoms (residues Lys279-His473) and for L3 an rmsd 0.65 Å for 160 Cα-atoms (residues Ile489-Ala673) excluding the long loop β4-β5 that extends and inserts Lys538 into the Ca2+-binding site of L4 in n1α L1-L6 (rmsd 1.95 Å for 184 Cα-atoms when it is included) (Fig. 6d). The relatively small changes in the buried surface and favorable interactions seen in both the ‘open’ and ‘closed’ forms suggest that residues Glu481-Pro488 of the linker may form a molecular switch that can flip the conformation of L2 with respect to L3 from a ‘closed’ or inline arrangement with the central core to an ‘open’ arrangement repositioning its hyper-variable surface with respect to the rest of the molecule.
Table 1:
Data Collection and Refinement Statistics for n1α L2-L3
| Data collection: | |
| Wavelength (Å) | 1.10208 |
| Space group | P21 |
| Unit cell dimensions | |
| a, b, c (Å) | 87.14, 62.90,113.06 |
| α, β, γ (°) | 90, 97.10, 90 |
| Resolution (Å) | 50.01 - 2.85 (2.95 - 2.84) |
| Observed reflections | 95,955 |
| Unique reflections | 28,540 (2555) |
| Rmerge (%) | 9.1 (52.0) |
| Mean I /σ(I) | 15.2 (2.2) |
| CC1/2 | (0.715) |
| Completeness (%) | 98.4 (89.1) |
| Redundancy | 3.4 (2.4) |
| Refinement: | |
| Resolution (Å) | 50.01 - 2.84 |
| Reflections used | 27112 |
| Rwork / Rfree (%) | 22.2/25.5 |
| Protein (no. residues) | 762 |
| Waters | 12 |
| B factors (Å2) | |
| Protein | 55.4 |
| Waters | 42.1 |
| RMSDs: bond lengths (Å) | 0.013 |
| RMSDs: bond angles (°) | 1.51 |
| Ramachandran plot residues (%) | |
| Favored | 95.4 |
| Allowed | 4.6 |
| Disallowed | 0.0 |
| MolProbity overall score | 2.38 |
Figure 6: Crystal structure of n1α L2-L3.
a) Cartoon diagram of the two n1α L2-L3 tandems in the asymmetric unit.; b) Superposition of n1α L2-L3 (green) on n1α (grey; pdbid:3QCW). Inset shows n1α L2-L3 docked in the IPET map of a n1α L1-L6 particle with a bent central core, shown as dual iso-surfaces contoured at volumes corresponding to 1.2 time the molecular mass of ~141 kDa.; c) Close-up of the interface between L2 and L3 shown in b). The linker between L2 and L3 undergoes a dramatic movement in n1α L2-L3 (blue; linker L2”-L3”) compared to its counterpart in the n1α ectodomain (magenta; linker L2-L3 from pdbid:3QCW). Inset shows the superposition of these linkers; d) Close up of the interface between L3 and L4 shown in b). Loopβ4-β5 in L3 as seen in the n1α L2-L3 fragment (light green) and its counterpart as seen in n1α (orange; pdbid:3QCW). The Ca2+-binding site of L4 is formed by the side chain of D772 and the backbone carbonyls of R848 and L789.
Table 2:
Impact of the molecular switch between L2 and L3. a) Comparison of the residues in L2 and L3 that contact the L2L3 linker (a.a. 474-488) within 5 Å. Differences between n1α L2-L3 and n1α L1-L6 are in bold.; b) Comparison of the residues in L2 and L3 that form interdomain contacts (within 5 Å) independent of the L2L3 linker. Differences between n1α L2-L3 and n1α L1-L6 are in bold.; c) Interactions at the interface between L2 and L3 that are selectively found in the ‘open’ and ‘closed’ form (L2 domain: a.a. 279-473; L2L3 linker: a.a. 474-488; L3 domain: a.a. 489-673).
| (a) | (b) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n1α L2-L3 | n1α L1-L6 (pdbid:3QCW) |
n1α L2-L3 | n1α L1-L6 (pdbid:3QCW) |
|||||||||
| L2 domain | L2 domain | |||||||||||
| ILE283 | ALA284 | THR285 | ILE283 | ALA284 | THR285 | LYS287 | SER289 | GLU290 | GLY288 | SER289 | GLN316 | |
| PHE286 | LYS287 | SER289 | LYS287 | SER289 | ASN337 | PRO427 | SER428 | |||||
| GLU290 | PHE292 | GLU290 | TYR291 | THR429 | ALA430 | ASP431 | ||||||
| LYS313 | THR314 | LEU315 | LYS313 | LEU315 | SER438 | ASN439 | ASN440 | |||||
| ASN365 | TRP367 | MET442 | ASN365 | TRP367 | MET442 | MET442 | ||||||
| GLY443 | CYS444 | CYS444 | Ser460 | |||||||||
| ALA463 | LYS464 | ILE472 | ALA463 | LYS464 | ILE472 | |||||||
| HIS473 | HIS473 | |||||||||||
| L3 domain | L3 domain | |||||||||||
| ILE489 | THR490 | ASP514 | ILE489 | THR490 | ASP514 | TRP582 | ARG652 | THR508 | GLY509 | SER510 | ||
| TRP582 | HIS584 | CYS650 | TRP582 | ASP653 | SER512 | GLN588 | THR600 | |||||
| ILE651 | ARG652 | ASP653 | ILE651 | ARG652 | ASP653 | PHE655 | ASP657 | GLY658 | ||||
| PHE655 | Gly658 | GLN659 | SER660 | |||||||||
| SER660 | LYS661 | ASP662 | SER660 | LYS661 | ASP662 | |||||||
| Ile663 | Arg664 | |||||||||||
| (c) | |||||
|---|---|---|---|---|---|
| n1α L2-L3 | n1α L1-L6 (pdbid:3QCW) | ||||
| Interaction L2 with L2L3 linker | |||||
| Hydrogen bonds: | Hydrogen bonds: | ||||
| L2 | [Å] | L2L3 linker | L2 | [Å] | L2L3 linker |
| GLU290[OE2] | 2.9 | VAL475[ N ] | |||
| ILE472[ O ] | 3.1 | GLY474[ N ] | |||
| Interaction L3 with L2L3 linker | |||||
| Hydrogen bonds: | |||||
| L3 | [Å] | L2L3 linker | |||
| ASP653[ N ] | 3.2 | THR485[ O ] | |||
| ASP653[ Oδ1 ] | 2.6 | THR485[ Oγ1 ] | |||
| Interaction L2 and L3 | |||||
| Hydrogen bonds: | Hydrogen bonds: | ||||
| L2 | [Å] | L3 | L2 | [Å] | L3 |
| LYS287[ O ] | 3.4 | ARG652[ NH1 ] | |||
| ASN440[ N ] | 3.1 | GLY658 [ O ] | |||
| ASN440[ Nδ2 ] | 2.9 | GLY658 [ O ] | |||
| ASP431[ Oδ2 ] | 2.7 | THR508[ Oγ1 ] | |||
| Salt bridges: | |||||
| L2 | [Å] | L3 | |||
| LYS287[ Nζ ] | 2.9 | ASP514[ Oδ2 ] | |||
| LYS287[ Nζ ] | 3.2 | ASP653[ Oδ2 ] | |||
Conformational variability in the C-terminal portion of n1α.
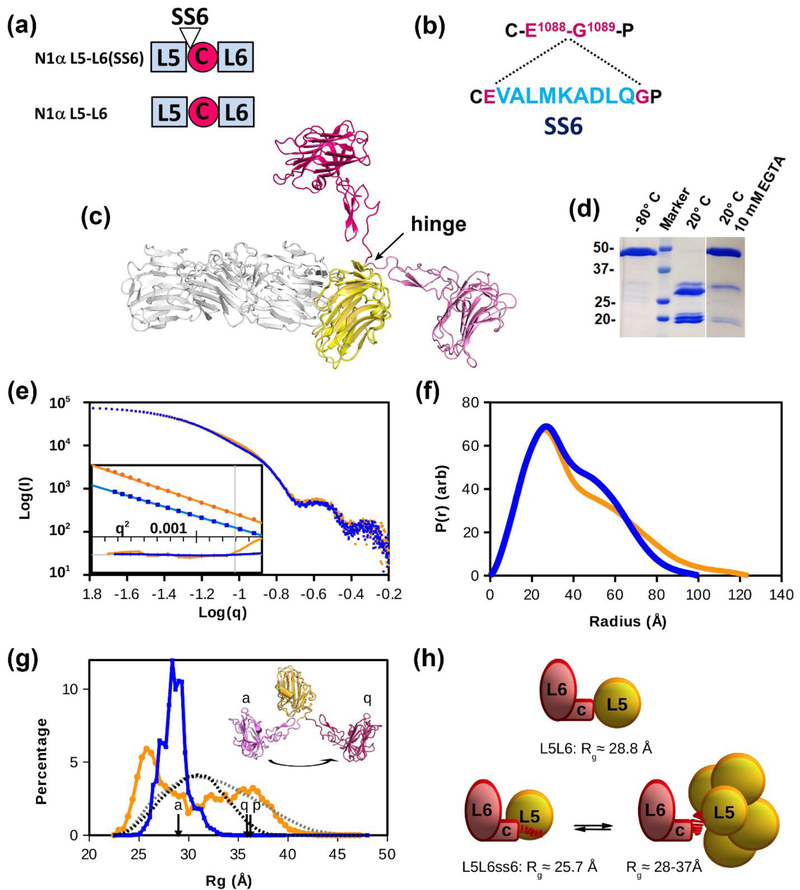
To investigate the conformational variability between domains in the C-terminal portion of n1α, we assessed the conformation of n1α L5-L6 in solution using small angle solution scattering (SAXS) (Fig. 7a, Fig. 7b). SS6 inserts at a molecular hinge between L5 and EgfC (7c; [31,39–41]. Incorporation of SS6 rendered n1α L5-L6 susceptible to proteolysis and cleavage between the residues Asp and Leu of the splice insert (E1088 V A L M K A D ↓ L Q G1089) as determined by N-terminal sequencing. However, addition of EGTA or EDTA prevented proteolysis suggesting that SS6 is a substrate for metalloproteases (Fig. 7d). For this reason, samples were recovered and analyzed by SDS-PAGE after each SAXS experiment to confirm that proteolysis had not taken place during data collection. Scattering data were collected for a dilution series of n1α L5-L6 and n1α L5-L6(SS6) generating SAXS scattering curves (Fig. 7e). The Guinier plots revealed roughly parallel lines suggesting that the radius of gyration (Rg), i.e. the average root mean square distance of all atoms to the center of mass, was very similar between n1α L5-L6 and n1α L5L(SS6) with Rg values of 30.2 Å and 32.4 Å, respectively (Fig. 7e). The pair-distance distribution function plot P(r) which reveals the distribution of distances between all pairs of atoms within a scattering macromolecule shows a similar major peak for n1α L5-L6 and n1α L5-L6(SS6) consistent with putative inter-atomic distances found within neurexin LNS domains (Fig. 7f). Shoulder peaks in the P(r) plots for n1α L5-L6 and n1α L5-L6(SS6) mapped to large interatomic distances (> 40 Å) as would be expected between the L5 and EgfC-L6 entities, i.e., two large moieties arranged in sequence (Fig. 7f). However, the shoulder peak for n1α L5-L6(SS6) was markedly different and broader than that for n1α L5-L6, suggesting that SS6 enables the moieties to separate further apart on average with a range of relative motions between the domains (Fig. 7f; Table 3). This was further exemplified by estimating the maximum length (Dmax) from the P(r) distance distribution plot revealing that it was larger for n1α L5-L6(SS6) (~ 124 Å) than for n1α L5-L6 (~100 Å), suggesting that SS6 enables L5 and L6 to move farther apart and works as a spacer to generate a more rod-shaped assembly.
Figure 7. Comparison of n1α L5-L6 and n1α L5-L6(SS6) by SAXS.
a) N1α L5-L6 and n1α L5-L6(SS6) constructs used.; b) Location of splice insert SS6 (light blue) in the amino acid sequence of n1α at the molecular hinge E1088-G1089.; c) Superposition of n1α L5-L6 from the crystal structure of the isolated fragment (pink/yellow; pdbid:3ASI) and n1α (magenta/yellow/grey; pdbid:3QCW). The hinge incorporating SS6 is indicated.; d) N1α L5-L6(SS6) is proteolytically cleaved at 20°C and proteolysis is inhibited by 10 mM EGTA as shown by SDS-PAGE (markers in kDA).; e) SAXS data Log-Log plots for n1α L5-L6 (◾) and n1α L5-L6(SS6) (●). Inset shows the Guinier fits (Log(I) vs. q2), offset for clarity, with their normalized residuals shown below (▬ n1α L5-L6, ▬ n1α L5-L6(SS6)). The vertical grey line marks the maximum value, q~1.3/RG, used for fitting. The residual range is ± 3 sigma. f) The pair-distance distribution function P(r) for n1α L5-L6 (▬) and n1α L5-L6(SS6) (▬) were calculated from the inverse Fourier transform of the scattering intensity using GNOM. P(r) is expressed in arbitrary units (‘arb’).; g) Conformational ensemble modeling of the SAXS data for n1α L5-L6 (χ2=1.1) and n1α L5-L6(SS6) (χ2=1.3). Rg distributions are shown for n1α L5-L6 (▬), and n1α L5-L6(SS6) (▬), as well as the model-pool distributions n1α L5-L6 (▪▪▪) and n1α L5-L6(SS6) (▪▪▪). The n1α L5-L6 fragment extracted from crystallographic models (PDBID: 3ASI, 3QCW, and 3POY) are indicated with ‘a’, ‘q’, and ‘p’, respectively, and shown for ‘a’ and ‘q’, see also c).; h) Schematic of the conformations of n1α L5-L6 (top) and n1α L5-L6(SS6) (bottom) deduced from the SAXS data. N1α L5-L6(SS6) appears in equilibrium between a compact form (left) and extended conformations (right). The SS6 insert is shown as a dashed line (▮ ▮ ▮ ) between the L5 and EgfC-L6 entities.
Table 3:
SAXS analysis of n1α L5L6 and n1α L5L6 (SS6)
| L5-L6 | L5-L6 | L5-L6 | L5-L6 | L5-L6 (SS6) |
L5-L6 (SS6) |
L5-L6 (SS6) |
L5-L6 (SS6) |
|
|---|---|---|---|---|---|---|---|---|
| Conc. (mg/ml) | 4 | 3 | 2 | m | 4 | 3 | 2 | m |
| Exposure (H) | 10 | 12 | 14 | * | 10 | 12 | 14 | * |
| Q-range (Å−1) | 0.015 - 0.50 | 0.015 - 0.50 | 0.014 - 0.50 | 0.016 - 0.50 | 0.011 - 0.50 | 0.011 - 0.50 | 0.011 - 0.50 | 0.018 - 0.50 |
| Rg (Å) | 30.4 (2) | 29.7 (4) | 30.1 (3) | 30.2 (1) | 32 (2) | 31.8 (7) | 31 (2) | 32.4 (9) |
| Dmax (Å) | 99 | 101 | 107 | 100 | 117 | 113 | 115 | 124 |
| Shanum (Å−1) | 0.685 | 0.582 | 0.522 | 0.570 | 0.583 | 0.582 | 0.553 | 0.553 |
| MW (kDa) | 44.5 | 44.5 | 44.5 | 44.5 | 45.5 | 45.5 | 45.5 | 45.5 |
| MW(Io) (kDa) | 40.6 | 40.9 | 39.0 | 39.6 | 48.8 | 46.5 | 44.2 | 45.1 |
|
MWRAMBO (kDa) |
40.0 | 40.5 | 40.0 | 39.8 | 38.2 | 35.2 | 34.7 | 33.4 |
| 3ASI χ2 | 4.0 | 3.9 | 3.1 | 6.2 | 31 | 27 | 14 | 65 |
| CORAL χ2 | ND | ND | ND | 1.3 | 3.2 | 2.0 | 1.6 | 3.2 |
|
EOM Rg1 (Å) (Occupancy) |
ND | ND | 28.5 (94%) |
28.5 (100%) |
ND | 26.3 (46%) |
26.4 (48%) |
25.7 (38%) |
|
EOM Rg2 (Å) (Occupancy) |
31.2 (6%) |
− (0%) |
34.9 (54%) |
35.5 (52%) |
28–37 (72%) |
To further assess the impact of SS6 and its impact on the conformation of L5 versus L6, we performed a Polydispersity/Conformational Ensemble analysis whereby a pool of 10,000 randomly oriented L5 and EgfC-L6 moieties was created that met the criteria of 1) a specified linker length, and 2) no steric clashe (see Methods); a separate pool with free L5 and EgfC-L6 moieties was created as a test for proteolysis. The selection of models that fit the solution scattering data was then assessed using EOM, a tool which uses an ensemble representation of atomic models and assesses their fit to experimental SAXS data (Fig. 7g). Absence of proteolysis in the samples was supported by EOM Polydispersity analysis of the SAXS data because the selected models did not cluster at the lowest values of Rg (~ 22.5 Å; Fig. 7g), nor were models selected from the pools of free L5 or free EgfC-L6 (not shown); furthermore, samples after SAXS analysis showed no signs of proteolysis by SDS-PAGE. The distribution of the selected models revealed that n1α L5-L6 maintained a distinct state with an Rg=28.5 Å and a narrow distribution of <1.2 Å width (Fig. 7g). On the other hand, the ensemble of models for n1α L5-L6(SS6) demonstrated a broad multi-state distribution featuring a compact peak shifted to Rg ~25.7 Å as well as a broad continuum of expanded states, that although extended, did not reach the maximum permitted by the pool of 10,000 random models (i.e., the model with the largest Rg of ~ 47 Å) suggesting these species are not due to aggregation (Fig. 7g). Although these analyses were performed on scattering data merged from the different protein concentrations, similar analyses using data from each individual concentration produced similar results (Table 3). Because crystal structures containing n1α L5-L6 are known, in isolation as a fragment (pdbid:3ASI) and in the context of the almost complete ectodomain (pdbid:3QCW and the very similar 3POY) revealing a dramatic molecular motion (Fig. 6c), we assessed the fit between the experimental SAXS data and the calculated scattering curves derived from the crystal structure counterparts. The SAXS scattering curve for n1α L5-L6 is poorly modeled by 3ASI (χ2 = 6.2; Table 3), though some of the error may be due to C-terminal residues in the protein used for the SAXS experiments that are missing in the crystal structure (residues E1336, V1337, P1338, S1339, and a 10 a.a. affinity tag). However, the SAXS scattering curve for n1α L5-L6(SS6) could not be modeled by 3ASI at all (χ2 = 65), indicating a significant change in the shape of the molecule when the SS6 insert is present (Table 3). Likewise, isolated L5-L6 fragments extracted from the n1α ectodomain structures (pdbid: 3QCW and 3POY) also gave very poor fits with the SAXS data for both n1α L5-L6 and n1α L5-L6(SS6) (χ2> 100). The fit of the calculated scattering factors from the crystal structures could be greatly improved through a rigid body analysis that modeled missing residues and incorporated flexibility between the L5 and EgfC-L6 domains for n1α L5-L6 (χ2= 1.3) and n1α L5-L6(SS6) (χ2= 3.2) to produce an average conformation of the ensembles entailing moieties that are tethered, but not otherwise interacting.
Taken together, direct interpretation of the SAXS data as well as ensemble modeling analyses indicate that n1α L5-L6 maintains a remarkable preference for discrete conformations in solution. The insert SS6 imparts flexibility to n1α L5-L6(SS6) permitting two discrete states: a compact state resembling n1α L5-L6 and a range of more extended states (Fig. 7h).
DISCUSSION
The studies presented here indicate that α-neurexins contain unique structural features in their architecture that form a dynamic platform to support their role as synaptic organizers. To circumvent limitations of current structural approaches, we used a combination of individual particle electron tomography (IPET), X-ray crystallography, and small angle solution scattering (SAXS) to delineate different conformations of n1α. First, we show that n1α adopts several discrete conformations including novel linear, F-, and Z-shaped molecules, in addition to the previously observed Y-/L- architecture. Taking these different architectures into account, n1α can span 160 Å to 220 Å in length, and is ~96 Å wide. Next, we show through analysis of more than a 100 individual single particles that the outer L1 and L6 domains are tethered with similar angles with respect to the central core of the molecule (L2-L3-L4-L5) via the two main hinges within n1α. Strikingly, however, the linker between L2 and L3 also permits a large conformational rearrangement, enabling L2 (and the attached L1-EgfA) to alter between a ‘closed’ and ‘open’ state with respect to the rest of the ectodomain. Finally, we show that splice insert SS6 incorporated at the molecular hinge between L5 and L6 enables these domains to alter between compact and extended conformations. Though previous structural studies have focused on the invariant nature of the rod-shaped n1α, our results collectively indicate that α-neurexins contain very specific structural elements that regulate their architecture, setting the stage for these molecules to work as dynamically regulated synaptic organizers.
Conformation of n1α L1-L6
The dimensions of n1α are important because they dictate how n1α fits in the synaptic cleft and orients its binding sites towards postsynaptically tethered or secreted protein partners. The excitatory synaptic cleft is estimated to span ~200-240 Å [51–53] and the inhibitory synaptic cleft ~120 Å [54], though narrower dimensions were recently proposed (~160 Å for excitatory and 100 Å for inhibitory clefts) [54]. Crystal structures of n1α spanning L2 through L6 were estimated to be ~130 × ~100 × ~60 Å [40,41]. Analysis of n1α by SAXS, revealed a molecular length (Dmax) of 145 Å for n1α L2-L6 and a Dmax of 170 Å for the full-length n1α L1-L6 [38]. However, in crystal structures, electron density is only seen for those regions of a molecule that are the same in crystallographically related copies, which can generate the impression that a protein has a particular, uniform architecture and SAXS data contain information on molecules as they rotate in solution averaged over time. So, while previous structural studies have been enormously valuable to gain insight into the overall architecture of n1α and the fold of the individual domains, an orthogonal technique was needed to reveal the conformation of individual molecules, as well as the conformational preference within a population. We turned to IPET to obtain structural information on a population of individual n1α molecules and reveal that n1α has a range of distances spanning 160 Å - 220 Å. The majority of the molecules adopt a rod-like shape (68 %) for the core domains L2-L3-L4-L5, while the N-terminal L1 and C-terminal L6 domains veer away with a surprisingly uniform angle of 50 – 70°. Thus, the dimensions of n1α support that it fits in the synaptic cleft parallel to the membranes while L1 and L6 have limited freedom to orientate themselves with respect to the central core.
Plasticity at the n1α L2-L3 interface
The exact arrangement of domains within the N-terminus of α-neurexins is important for their function, because L1 in neurexin 2α binds IgSF21 [13], while L2 binds α-dystroglycans [12] and neurexophilins [15]. The crystal structure of n1α L2-L3 presented here reveals two key architectural properties. First, L2 and L3 can reside side-by-side each other in a ‘closed’ conformation or in an ‘open’ conformation in n1α. In the crystal structures containing the ‘open’ and ‘closed’ forms (PDBIDs 6CW1, 3QCW), the L2 and L3 domains are held in place by significant molecular contacts (> 3000 Å2) making it difficult to assess which molecular conformation would be more likely in solution. Analysis of the interface between L2 and L3 suggest that both conformations are similarly favorable. This suggests that the long 15 residues linker (Gly474-Pro488) between L2 and L3 may govern the conformational preference of L2 with respect to L3. The linker between L2 and L3 might be influenced by the binding of protein partners or even serve as their binding site. Such a situation is observed with synaptic organizers from the LAR-RPTP family where the linker between Ig2 and Ig3 (encoded by alternative splice insert MeB) is critical for establishing the binding site for different partners in the synaptic cleft such as Slitrk1, Slitrk2, IL1RacP, and IL1RAPL1, as well as governing the orientation of Ig2 versus Ig3 [55–57]. Second, the 18 residue loop β4-β5 (Gly529-Asp546) in L3 adopts a very different, well ordered conformation in the crystal structure of the n1α L2-L3 fragment that no longer reaches across to insert Lys538 into the Ca2+-binding site found at the ‘hyper-variable’ surface of L4, an interaction that would putatively stabilize the horseshoe-shaped L3-EgfB-L4 repeat (refer back to Fig. 6). It is possible that loop β4-β5 in L3 may release from L4 upon protein partner or metal-ion binding. Many of the n1α particles with a bent central core (Z-shaped molecules; 15-20% of the total particles analyzed by IPET, see subset of n1α particles demarcated in blue in Fig. 4a) are consistent with a LNS domain being freed from the linear central core leaving just three left in a linear array as would be expected for example from the ‘open’ conformation of n1α L2-L3. Therefore, the linker between L2 and L3, and the loop β4-β5 in L3 may be able to transmit allosteric changes that ripple through the n1α architecture upon protein partner binding, affecting the recruitment of other partners.
Plasticity at the n1α L5-L6 interface
The exact arrangement of domains within the C-terminus of n1α is important as well. In particular, control of the orientation of L6 is important because it binds to the majority of the partners identified for α-neurexins, including postsynaptically tethered partners in the synaptic cleft such as neuroligins, LRRTMs, calsyntenin 3, α-dystroglycan, cerebellins, and latrophilins [4]. Crystal structures have shown that L6 packs against EgfC with an extensive interface of ~470 Å2 (pdbid:3QCW) producing a relatively autonomous entity [39–41]. The EgfC-L6 unit is connected to L5 via a characteristic hinge formed by Glu1088-Gly1089. The L5 and EgfC-L6 moieties adopt an extended conformation with an elbow angle of ~139° between L5 and L6 (pdbid: 3QCW; 3POY) but L5 can also pivot moving towards L6 generating an elbow angle of ~90° between L5 and L6 (pdbid: 3ASI). For this reason, the recently identified splice insert SS6 which is specifically expressed in brain and maps exactly to this molecular hinge is particularly interesting [31,58]. Its location suggests that the physiological role could be to add strategic flexibility to n1α by altering the orientation of EgfC-L6 with respect to the rest of the molecule, impacting its function. Our SAXS data show that in absence of SS6, L5-L6 adopts a primarily compact conformation that corresponds well with the L5 and L6 domains pivoting towards each other; a result that is consistent with the n1α populations observed by IPET where L6 bends towards the central core. In contrast, L5-L6(SS6) adopts a bimodal distribution of molecular states containing a population of compact conformation(s), as well as a population of extended architectures. This is a surprising result, because if SS6 were to work solely as a tether, a purely unimodal distribution of increasingly extended states would be expected. Incorporation of SS6 may thus add a molecular toggle to n1α, positioning protein partner binding surfaces within the synaptic cleft, and/or modifying actual binding sites or the accessibility to them. Alternatively, the major function of SS6 may be to render the n1α molecule sensitive to proteolysis by enabling the region L1-L5 to be shed, reducing n1α to a molecule similar to neurexin 1β (which contains a single LNS domain with identical sequence to L6). The splice insert SS1 that tethers L1-EgfA to L2 was reported to be proteolytically sensitive as well, shearing L1-EgfA from n1α [39]. Thus, hinges linking L1 and L6 and their bordering EGF domains, EgfA and EgfC, may regulate the function of n1α structurally and/or its susceptibility to proteolysis may be a mechanism to prevent n1α from recruiting protein partners that selectively bind n1α specific domains.
Conclusion
By using a combination of different structural techniques, i.e., IPET, X-ray crystallography and SAXS, our data establish that n1α, a large synaptic organizer containing nine domains, possesses strategic hinges with unexpected flexibility as well as conformational restraint (Fig. 8). More work is needed to determine whether these hinges mediate solely select end-states and work as true switches or toggles, or whether they enable a larger continuum of conformations. Regardless, our results suggest that α-neurexins contain structural elements in their architecture that provide a platform that may support allosteric control or concerted protein partner binding, dynamically impacting how α-neurexins recruit and arrange protein networks in the synaptic cleft.
Figure 8: Possible molecular hinges and toggles in n1α.
The architecture of n1α reveals unexpected conformational freedom between L2 and L3, and unexpected conformational restraint between the central core and both L1 and L6.
MATERIALS AND METHODS
Protein expression and purification
The following constructs were made: bovine neurexin 1 alpha ectodomain n1α L1-L6 (here referred to as n1α) (nm_174404; Met1-Ser1339 carrying the endogenous signal peptide but no splice inserts, and accommodating SS1, SS2, SS3, and SS4 but not SS6 in the numbering scheme), n1α L5-L6 (residues Ala911-Ser1339), and n1α L5-L6(SS6) (residues Ala911- Ser1339, carrying SS6, i.e., VALMKADLQ, between Glu1088 and Gly1089). Each construct was designed with a C-terminal tag ASTSHHHHHH, produced using baculo-virus mediated overexpression in HighFive cells, and purified as described in [40]. Briefly, medium containing the secreted proteins was concentrated, dialyzed, and purified using the following columns: Ni-NTA (Qiagen; in 25 mM sodium phosphate pH 8, 500 mM NaCl, eluted with an imidazole gradient 0-250 mM), Mono Q (GE Healthcare, in 25 mM Tris pH 8, 50 mM NaCl, eluted with increasing NaCl), and Superdex-200 (GE Healthcare, in 25 mM Tris pH 8, 100 mM NaCl). The purified proteins were stored in 25 mM Tris pH 8, 100 mM NaCl in flash-frozen aliquots. Bovine neurexin n1α L2-L3 (residues Glu258–Gly674 which includes SS#1: E258DNNVEGLAHLMMGDQGKSK277) was expressed as a thrombin-cleavable GST-fusion protein in E. coli BL21(DE3) and purified using glutathione-agarose beads, ion-exchange and gel filtration as previously described [32,59]; purified proteins were stored in 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA in flash-frozen aliquots. The intact nature of full length n1α L1-L6 (calculated molecular weight 137 kDa) was confirmed by SDS-PAGE (Fig. 2a) and mass spectrometry (141 kDa).
Negative staining EM specimen preparation
EM grids of n1α were prepared using the OpNS method [44,60,61], except skipped the water washing step. Our OpNS procedure was refined starting from a conventional protocol and entailed the following modifications: i) use of 1% (w/v) uranyl formate (UF) as the negative stain; ii) 0.02 μm filtering the stain right before use; iii) use of DPBS as the sample dilution buffer; iv) staining the grids in the dark to avoid light-induced precipitation of UF; and v) drying the grids under N2 gas to avoid the potential oxidation of samples during the drying process. In brief, n1α (1.0 mg/ml) was diluted to 0.005 mg/ml with 25 mM Tris pH 8, 100 mM NaCl, 3 mM CaCl2 and a 4 μl aliquot placed on an ultrathin-carbon-coated 200 mesh copper grid (CF200-Cu-UL, EMS, Hatfield, PA, USA; Cu-200CN, Pacific Grid-Tech, San Francisco, CA, USA) that had been glow-discharged for 15 s. After ~1 min incubation, excess solution was blotted with filter paper, and the grid was stained for ~15 s by sequential submersion in two drops of 1% (w/v) uranyl formate (UF) (~35 µl; 0.02 µm filtered) on parafilm within a dark box before being nitrogen-gas-dried at room temperature. Insertion of SS6 rendered n1α proteolytically sensitive in our hands, therefore we did not perform EM studies on n1α carrying SS6.
Electron microscopy data acquisition and IPET 3D reconstruction
NS (negative stain)-EM micrographs were acquired at 80,000× magnification on a Zeiss Libra 120 transmission electron microscope (Carl Zeiss NTS, Overkochen, Germany) operating at 120 kV, with a Gatan UltraScan 4K×4K CCD. Each pixel of the micrographs corresponded to 1.48 Å. For 2D analysis, a total of 150 focus pairs of untilted micrographs were acquired under near Scherzer focus (0.1 µm) and defocus of 0.6 µm. For 2D reference-free class averaging analysis, 150 untilted micrographs under near Scherzer focus were low-pass filtered to 15 Å and high-pass filtered to 600 Å after X-ray speckles were removed. A total of 15,402 particles were windowed and selected by EMAN [62], and masked by SPIDER [63]. These particles were aligned and averaged by either 100 or 1,043 classes, respectively, to assess the conformational heterogeneity of the sample [62]. For IPET 3D analysis, a total of five tilt series were collected from −60° to +60° at 1.5° increments under defocus of ~0.6 μm, using Gatan tomography software and in-house developed fully mechanically controlled automated ET software [64]. The electron dose per tilt series is ~3,240 e−/Å2. Each set contained 81 tilt images/micrographs, in which CTF corrected by TOMOCTF [65]. Two sets were initially aligned by IMOD, and used for 3D reconstructions. In each tilt series, there were roughly 100 particles, in which ~80 particles that were not overlapped to others or missed their tilt images were selected for 3D reconstructions by IPET [42]. The tilt series of each targeted particle was submitted for IPET 3D reconstruction. During the last step of this process, data from −48° to +48° were used to produce the final IPET 3D reconstruction applying a missing-wedge computational correction to counter potential artifacts, e.g., elongation, and blurring as a result of the limited tilt angle range [49]. The resolution of each IPET 3D reconstructed density map was analyzed by Fourier shell correlation (FSC) as described before [42,47,48,66]. In brief, the center refined raw ET images (after CTF correction) were split into two groups according to their even- or odd-numbered index in the tilting angle series. Each group was used to generate an independent IPET 3D reconstruction; the two IPET 3D reconstructions were then used to compute the FSC curve over their corresponding spatial frequency shells in Fourier space. The frequency at which the FSC curve falls to a value of 0.5 was used to assess the resolution of the final IPET 3D density map. To estimate the signal of each protein particle, the SNR was calculated using the equation SNR = (Is - Ib)/Nb, where Is is the average density of the particle, Ib is the average density outside the particle, and Nb is the standard deviation of the noise that was calculated from the standard deviation of the background outside the particle area [47]. The particle area was defined using a particle-shaped mask generated from the IPET final 3D reconstruction that was low-pass filtered to ~25-30 Å and the volume was set to 3 times the molecular weight of the protein, using the volume command in EMAN, which assumes a density of 1.35 g/ml (0.81 Da/A3)[62]. A similar method was used to calculate the 2D SNR, except that the 2D mask was generated from the 3D projection at each tilt angle. This method provides us with a conservative estimate of the signal-to-noise for each particle. We used 110 particles for 3D reconstruction out of a total of 400 particles targeted and imaged, given that 110 maps were sufficient to demonstrate novel conformations of n1α L1-L6 and to carry out statistical analyses.
Crystal structure docking
The crystal structure of n1α (pdbid:3QCW) was used for docking studies in 3D EM density maps using Chimera. The remaining unoccupied density corresponded to the n1α L1 domain.
Statistical analysis of molecular angles within n1α
To implement an unbiased approach, we assigned generic positions (P1-P6) to the LNS domains within each n1α 3D map as follows. First, the central core containing four LNS domains, corresponding to P2-P3-P4-P5, was assigned in each particle. For most particles, the connectivity was clear for the central core, otherwise we chose four LNS domains in a linear arrangement. The P1 and P6 positions were chosen to coincide with the two outer LNS domains farthest away from the rigid core. We assigned the outer domain with the smaller angle with respect to the central core as P1 (angle α) and the outer domain with the larger angle as P6 (angle β). The coordinates corresponding to the center of mass of the individual LNS domains were obtained from the IPET reconstructions of 110 n1α particles with Chimera. The angles α (P1-P2-P3) and β (P4-P5-P6), and the angle γ (calculated between the two vectors and ) were then calculated using these coordinates. The distribution of angles was plotted as histograms. We investigated the two outer angles of each molecule, α and β, as well as their combined distribution (α and β) to control for bias in assigning P1 and P6, and the internal torsional angle γ, corresponding to the central core of each molecule. This approach enabled us to investigate the molecular angles irrespective of the exact identity of the N-terminal versus C-terminal ends of each molecule, with the assumption that the central core contains P2-P3-P4-P5.
Crystallization and structure determination of n1α L2-L3
Crystals of n1α L2-L3 were grown by hanging drop vapor diffusion at 21°C in 0.9 M NaCitrate, 0.1 M Tris pH 8, 5 mM CaCl2. Prior to data collection, crystals were cryo-protected in 1 M NaCitrate, 25 mM Tris pH 8, 5 mM CaCl2, 30% glycerol for 5 min at room temperature and flash-cooled in liquid nitrogen. The crystals have the symmetry of space group P21 with cell dimensions a=87.144 Å, b=62.901 Å, c=113.061 Å, α= 90.0°, β=97.1°, γ=90.0° and contain 2 molecules per asymmetric unit. Diffraction data were collected at LS-CAT 21-ID-D at 1.10208 Å on a MarMosaic CCD 300 detector. The data were integrated and scaled with HKL2000 [67]. The structure was solved by molecular replacement using PHASER [68] in the CCP4 suite [69] searching for two separate L2 domains and two separate L3 domains from n1α (pdbid:3QCW; [40]. Model building was iteratively carried out with the program Coot [70] interspersed with refinement using Phenix [71] and Refmac [72]. The refined model consists of 762 residues (Lys279/G278 – Ala673/Gly674) with good geometry, 95.4 % in the preferred region (723 residues), 4.6 % in the allowed region (35 residues) and no outliers of the Ramachandran plot and twelve water molecules. Data collection and refinement statistics are summarized in Table 1. Figures were generated using Pymol.
Small angle X-ray scattering (SAXS)
SAXS data were collected using a Rigaku BioSAXS-1000 camera on a FR-E++ Cu X-ray source. After purification, and prior to buffer equilibration, samples of n1α L5-L6 and n1α L5-L6(SS6) were treated with 20 mM EDTA to remove unwanted metal ions which might cause aggregation. Each sample was then buffer exchanged into 20 mM HEPES pH 8, 150 mM NaCl, 0.5mM CaCl2 and concentrated. The flow-through from the micro-concentrators was used in the scattering experiments as the matching buffer for the buffer subtraction. SAXS data were collected for each protein sample from a series of protein concentrations, i.e., 2.0, 3.0, and 4.0 mg/ml. For each protein concentration, 70 μl of sample and its matching buffer were loaded into an aligned quartz flow-cell mounted in the BioSAXS camera under vacuum using an ASC-96 Automatic Sample Changer. For each sample, a series of one hour exposures was collected at 10°C spanning a total of 10 to 16 hours and averaged in SAXLab to produce separate sample and buffer curves (Table 3). Data were collected in the range 0.008 Å−1 < q < 0.68 Å−1, and the analysis used all significant data to 0.50 Å−1. No radiation-induced or time-dependent changes were observed. Buffer subtraction, absorption correction, and molecular weight (MW) calibration were performed using the SAXNS-ES server (http://xray.utmb.edu/SAXNS), which also uses the concentration and intensity independent method of Rambo and Tainer [73] to determine the MW of the proteins. Data analysis, including the merging of curves, was performed with the Primus program and the P(r) was calculated using DATGNOM from the ATSAS suite [74,75]. The ab initio molecular shape was generated from an average of 15 DAMMIF runs [76], using the saxns_dammif utility. The dilution series for n1α L5-L6 and n1α L5-L6(SS6) displayed similar mild q-dependencies, indicative of molecular crowding at the highest concentrations. A polydispersity/conformational ensemble analysis was performed using the pdbid:3ASI model in EOM [77] to create 10,000 possible combinations for each construct (L5-L6, and L5-L6(SS6)). The missing C-terminal residues (E1336, V1337, P1338, S1339, and 10 a.a. affinity tag) and the flexible hinge region between the domains L5 and L6 (C1043EGPST1048) for n1α L5-L6 and 14 residues (C1043EVALMKADLQGPST1048) for n1α L5-L6(SS6), respectively, were modeled as a flexible Cα chain in order to tether the L5 and L6 domains to each other in a physiologically meaningful way. The same models and flexible regions were also used for rigid-body modeling in CORAL [74]. This Polydispersity/Conformational Ensemble analysis is a tool to assess the kinds of conformations that fit the experimental SAXS data. Dimerization of n1α L2-L3 during the course of 8-hour SAXS experiments, even at low protein concentration (0.7 mg/ml), precluded straightforward use of this technique to analyze the conformation of n1α L2-L3 in solution
ACCESSION NUMBERS
The coordinates for n1α L2-L3 have been deposited in the Protein Data Bank with accession number 6CW1. 3D maps of n1α IPET reconstruction have been deposited in the EM Data Bank with the following accession codes: EMD-7639, EMD-7659 through EMD-7719, EMD-7722 through EMD-7768. SAXS data have been deposited at SASBDB with accession codes SASDD95 and SASDDA5.
HIGHLIGHTS.
Neurexin 1alpha (n1α) organizes protein networks in the synaptic cleft.
A panel of structural techniques reveals strategic conformational freedom in n1α.
Novel molecular switches are identified between domains.
The architecture of n1α suggests allosteric control and concerted partner binding.
ACKNOWLEDGEMENTS
This work was funded by NIMH (R01MH077303) with additional support provided by the Sealy Center for Structural Biology and Molecular Biophysics at UTMB and a UT BRAIN Award. The Advanced Photon Source (LS-CAT, SBC-CAT and IMCA-CAT) are thanked for access to synchrotron radiation. Work at the Molecular Foundry (J. Liu and G. Ren) was supported by the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 and the National Heart, Lung, and Blood Institute of the National Institutes of Health (R01HL115153). Prashant Rajbhandari is thanked for preliminary studies.
ABBREVIATIONS:
- 3D
three dimensional
- EGF
epidermal growth factor
- EM
electron microscopy
- ET
electron tomography
- FSC
Fourier Shell Coefficient
- IPET
individual particle electron tomography
- LAR-RPTP
leukocyte common antigen-related receptor protein tyrosine phosphatase
- LNS
laminin, neurexin, sex hormone-binding globulin
- rmsd
root mean square deviation
- SAXS
small angle X-ray scattering
Footnotes
Declaration of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- [1].Chen LY, Jiang M, Zhang B, Gokce O, Südhof TC, Conditional Deletion of All Neurexins Defines Diversity of Essential Synaptic Organizer Functions for Neurexins, Neuron. 94 (2017) 611–625.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fuccillo MV, Földy C, Gökce Ö, Rothwell PE, Sun GL, Malenka RC, Südhof TC, Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms, Neuron. 87 (2015) 326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, Yi F, Wernig M, Südhof TC, Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1, Cell Stem Cell. 17 (2015) 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Südhof TC, Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits, Cell. 171 (2017) 745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Siddiqui TJ, Craig AM, Synaptic organizing complexes, Curr. Opin. Neurobiol. 21 (2011) 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Reissner C, Runkel F, Missler M, Neurexins, Genome Biol 14 (2013) 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ko J, Fuccillo MV, Malenka RC, Südhof TC, LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation, Neuron. 64 (2009) 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].De Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR III, Comoletti D, Taylor P, Ghosh A, LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation, Neuron. 64 (2009) 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM, LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development, J. Neurosci 30 (2010) 7495–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pettem KL, Yokomaku D, Takahashi H, Ge Y, Craig AM, Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development, J Cell Biol. (2013) jcb–201206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu Z, Wang Y, Chen F, Tong H, Reddy MS, Luo L, Seshadrinathan S, Zhang L, Holthauzen LMF, Craig AM, Calsyntenin-3 molecular architecture and interaction with neurexin 1α, J. Biol. Chem. 289 (2014) 34530–34542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sugita S, Saito F, Tang J, Satz J, Campbell K, Südhof TC, A stoichiometric complex of neurexins and dystroglycan in brain, J Cell Biol. 154 (2001) 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tanabe Y, Naito Y, Vasuta C, Lee AK, Soumounou Y, Linhoff MW, Takahashi H, IgSF21 promotes differentiation of inhibitory synapses via binding to neurexin2α, Nat. Commun 8 (2017) 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Boucard AA, Ko J, Südhof TC, High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex, J. Biol. Chem. 287 (2012) 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Born G, Breuer D, Wang S, Rohlmann A, Coulon P, Vakili P, Reissner C, Kiefer F, Heine M, Pape H-C, Modulation of synaptic function through the α-neurexin–specific ligand neurexophilin-1, Proc. Natl. Acad. Sci. 111 (2014) E1274–E1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz L-J, Kim IH, Manhaes AC, Rodrigues WS Jr, Pamukcu A, Astrocytes assemble thalamocortical synapses by bridging NRX1α and NL1 via hevin, Cell. 164 (2016) 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Uemura T, Lee S-J, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M, Trans-synaptic interaction of GluRδ2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum, Cell. 141 (2010) 1068–1079. [DOI] [PubMed] [Google Scholar]
- [18].Francks C, Maegawa S, Laurén J, Abrahams BS, Velayos-Baeza A, Medland SE, Colella S, Groszer M, McAuley EZ, Caffrey TM, LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia, Mol. Psychiatry. 12 (2007) 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hines RM, Wu L, Hines DJ, Steenland H, Mansour S, Dahlhaus R, Singaraja RR, Cao X, Sammler E, Hormuzdi SG, Synaptic imbalance, stereotypies, and impaired social interactions in mice with altered neuroligin 2 expression, J. Neurosci. 28 (2008) 6055–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yan J, Noltner K, Feng J, Li W, Schroer R, Skinner C, Zeng W, Schwartz CE, Sommer SS, Neurexin 1α structural variants associated with autism, Neurosci. Lett. 438 (2008) 368–370. [DOI] [PubMed] [Google Scholar]
- [21].Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism, Proc. Natl. Acad. Sci. 105 (2008) 1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, Südhof TC, Powell CM, Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2, Genes Brain Behav. 8 (2009) 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Etherton MR, Blaiss CA, Powell CM, Südhof TC, Mouse neurexin-1α deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments, Proc. Natl. Acad. Sci. 106 (2009) 17998–18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guilmatre A, Dubourg C, Mosca A-L, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation, Arch. Gen. Psychiatry. 66 (2009) 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun C, Cheng M-C, Qin R, Liao D-L, Chen T-T, Koong F-J, Chen G, Chen C-H, Identification and functional characterization of rare mutations of the neuroligin-2 gene (NLGN2) associated with schizophrenia, Hum. Mol. Genet. 20 (2011) 3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafrenière RG, Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia, Hum. Genet. 130 (2011) 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dachtler J, Ivorra JL, Rowland TE, Lever C, Rodgers RJ, Clapcote SJ, Heterozygous deletion of α-neurexin I or α-neurexin II results in behaviors relevant to autism and schizophrenia, Behav. Neurosci. 129 (2015) 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liang J, Xu W, Hsu YT, Yee AX, Chen L, Südhof TC, Conditional neuroligin-2 knockout in adult medial prefrontal cortex links chronic changes in synaptic inhibition to cognitive impairments, Mol. Psychiatry. 20 (2015) 850. [DOI] [PubMed] [Google Scholar]
- [29].Babaev O, Botta P, Meyer E, Müller C, Ehrenreich H, Brose N, Lüthi A, Krueger-Burg D, Neuroligin 2 deletion alters inhibitory synapse function and anxiety-associated neuronal activation in the amygdala, Neuropharmacology. 100 (2016) 56–65. [DOI] [PubMed] [Google Scholar]
- [30].Schreiner D, Nguyen T-M, Russo G, Heber S, Patrignani A, Ahrné E, Scheiffele P, Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins, Neuron. 84 (2014) 386–398. [DOI] [PubMed] [Google Scholar]
- [31].Treutlein B, Gokce O, Quake SR, Südhof TC, Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing, Proc. Natl. Acad. Sci. 111 (2014) E1291–E1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rudenko G, Nguyen T, Chelliah Y, Sudhof TC, Deisenhofer J, The structure of the ligand-binding domain of neurexin Ib: regulation of LNS domain function by alternative splicing, Cell. 99 (1999) 93–102. [DOI] [PubMed] [Google Scholar]
- [33].Shen KC, Kuczynska DA, Wu IJ, Murray BH, Sheckler LR, Rudenko G, Regulation of neurexin 1β tertiary structure and ligand binding through alternative splicing, Structure. 16 (2008) 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Araç D, Boucard AA, Özkan E, Strop P, Newell E, Südhof TC, Brunger AT, Structures of neuroligin-1 and the neuroligin-1/neurexin-1β complex reveal specific protein-protein and protein-Ca2+ interactions, Neuron. 56 (2007) 992–1003. [DOI] [PubMed] [Google Scholar]
- [35].Graf ER, Kang Y, Hauner AM, Craig AM, Structure function and splice site analysis of the synaptogenic activity of the neurexin-1β LNS domain, J. Neurosci. 26 (2006) 4256–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P, Structural analysis of the synaptic protein neuroligin and its β-neurexin complex: determinants for folding and cell adhesion, Neuron. 56 (2007) 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen X, Liu H, Shim AH, Focia PJ, He X, Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions, Nat. Struct. Mol. Biol. 15 (2008) 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Comoletti D, Miller MT, Jeffries CM, Wilson J, Demeler B, Taylor P, Trewhella J, Nakagawa T, The macromolecular architecture of extracellular domain of αNRXN1: domain organization, flexibility, and insights into trans-synaptic disposition, Structure. 18 (2010) 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tanaka H, Nogi T, Yasui N, Iwasaki K, Takagi J, Structural basis for variant-specific neuroligin-binding by α-neurexin, PloS One. 6 (2011) e19411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen F, Venugopal V, Murray B, Rudenko G, The structure of neurexin 1α reveals features promoting a role as synaptic organizer, Structure. 19 (2011) 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Miller MT, Mileni M, Comoletti D, Stevens RC, Harel M, Taylor P, The crystal structure of the α-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function, Structure. 19 (2011) 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang L, Ren G, IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure, PloS One. 7 (2012) e30249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tong H, Zhang L, Kaspar A, Rames MJ, Huang L, Woodnutt G, Ren G, Peptide-conjugation induced conformational changes in human IgG1 observed by optimized negative-staining and individual-particle electron tomography, Sci. Rep 3 (2013) 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang L, Tong H, Garewal M, Ren G, Optimized negative-staining electron microscopy for lipoprotein studies, Biochim. Biophys. Acta BBA-Gen. Subj. 1830 (2013) 2150–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lu Z, Reddy MS, Liu J, Kalichava A, Liu J, Zhang L, Chen F, Wang Y, Holthauzen LMF, White MA, Molecular architecture of contactin-associated protein-like 2 (CNTNAP2) and its interaction with contactin 2 (CNTN2), J. Biol. Chem 291 (2016) 24133–24147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang L, Yan F, Zhang S, Lei D, Charles MA, Cavigiolio G, Oda M, Krauss RM, Weisgraber KH, Rye K-A, Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein, Nat. Chem. Biol. 8 (2012) 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang X, Zhang L, Tong H, Peng B, Rames MJ, Zhang S, Ren G, 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography, Sci. Rep. 5 (2015) 9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang L, Lei D, Smith JM, Zhang M, Tong H, Zhang X, Lu Z, Liu J, Alivisatos AP, Ren G, Three-dimensional structural dynamics and fluctuations of DNA-nanogold conjugates by individual-particle electron tomography, Nat. Commun. 7 (2016) 11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lei D, Marras AE, Liu J, Huang C-M, Zhou L, Castro CE, Su H-J, Ren G, Three-dimensional structural dynamics of DNA origami Bennett linkages using individual-particle electron tomography, Nat. Commun 9 (2018) 592. doi: 10.1038/s41467-018-03018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state, J. Mol. Biol. 372 (2007) 774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- [51].Harris KM, Weinberg RJ, Ultrastructure of synapses in the mammalian brain, Cold Spring Harb. Perspect. Biol 4 (2012) a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lucˇic V, Yang T, Schweikert G, Förster F, Baumeister W, Morphological characterization of molecular complexes present in the synaptic cleft, Structure. 13 (2005) 423–434. [DOI] [PubMed] [Google Scholar]
- [53].Zuber B, Nikonenko I, Klauser P, Muller D, Dubochet J, The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes, Proc. Natl. Acad. Sci. U. S. A. 102 (2005) 19192–19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].High B, Cole AA, Chen X, Reese TS, Electron microscopic tomography reveals discrete transcleft elements at excitatory and inhibitory synapses, Front. Synaptic Neurosci. 7 (2015) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Um JW, Kim KH, Park BS, Choi Y, Kim D, Kim CY, Kim SJ, Kim M, Ko JS, Lee SG, Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion, Nat. Commun. 5 (2014) 5423. [DOI] [PubMed] [Google Scholar]
- [56].Yamagata A, Sato Y, Goto-Ito S, Structure of Slitrk2-PTP complex reveals mechanisms for splicing-dependent trans-synaptic adhesion, Sci. Rep 5 (2015) 9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yamagata A, Yoshida T, Sato Y, Goto-Ito S, Uemura T, Maeda A, Shiroshima T, Iwasawa-Okamoto S, Mori H, Mishina M, Fukai S, Mechanisms of splicing-dependent trans-synaptic adhesion by PTPδ-IL1RAPL1/IL-1RAcP for synaptic differentiation, Nat. Commun 6 (2015) 6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Serova OV, Radionov NV, Shayahmetova DM, Deyev IE, Petrenko AG, Structural and functional analyses of the sixth site of neurexin alternative splicing, Dokl. Biochem. Biophys. 463 (2015) 239–242. [DOI] [PubMed] [Google Scholar]
- [59].Sheckler LR, Henry L, Sugita S, Südhof TC, Rudenko G, Crystal Structure of the Second LNS/LG Domain from Neurexin 1α Ca2+ binding and the effects of alternative splicing, J. Biol. Chem. 281 (2006) 22896–22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rames M, Yu Y, Ren G, Optimized negative staining: a high-throughput protocol for examining small and asymmetric protein structure by electron microscopy, J. Vis. Exp. JoVE (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang L, Song J, Newhouse Y, Zhang S, Weisgraber KH, Ren G, An optimized negative-staining protocol of electron microscopy for apoE4• POPC lipoprotein, J. Lipid Res. 51 (2010) 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ludtke SJ, Baldwin PR, Chiu W, EMAN: semiautomated software for high-resolution single-particle reconstructions, J. Struct. Biol. 128 (1999) 82–97. [DOI] [PubMed] [Google Scholar]
- [63].Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A, SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields, J. Struct. Biol. 116 (1996) 190–199. [DOI] [PubMed] [Google Scholar]
- [64].Liu J, Li H, Zhang L, Rames M, Zhang M, Yu Y, Peng B, Celis CD, Xu A, Zou Q, Yang X, Chen X, Ren G, Fully Mechanically Controlled Automated Electron Microscopic Tomography, Sci. Rep 6 (2016) 29231. doi: 10.1038/srep29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fernandez JJ, Li S, Crowther RA, CTF determination and correction in electron cryotomography, Ultramicroscopy. 106 (2006) 587–596. [DOI] [PubMed] [Google Scholar]
- [66].Yu Y, Kuang Y-L, Lei D, Zhai X, Zhang M, Krauss RM, Ren G, Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo-electron tomography1, J. Lipid Res. 57 (2016) 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr A 276: 307–326, 1997. [DOI] [PubMed] [Google Scholar]
- [68].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J. Appl. Crystallogr 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, Overview of the CCP4 suite and current developments, Acta Crystallogr. D Biol. Crystallogr 67 (2011) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kovalevskiy O, Nicholls RA, Long F, Carlon A, Murshudov GN, Overview of refinement procedures within REFMAC5: utilizing data from different sources, Acta Crystallogr. Sect. Struct. Biol. 74 (2018) 215–227. doi: 10.1107/S2059798318000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rambo RP, Tainer JA, Accurate assessment of mass, models and resolution by small-angle scattering, Nature. 496 (2013) 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI, New developments in the ATSAS program package for small-angle scattering data analysis, J. Appl. Crystallogr. 45 (2012) 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions, J. Appl. Crystallogr. 50 (2017) 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Franke D, Svergun DI, DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering, J. Appl. Crystallogr. 42 (2009) 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI, Structural characterization of flexible proteins using small-angle X-ray scattering, J. Am. Chem. Soc. 129 (2007) 5656–5664. [DOI] [PubMed] [Google Scholar]