Routine staining of sputum specimens does not identify acid-fast bacilli as Mycobacterium tuberculosis with utmost precision, limiting its usability as a confirmatory test for pulmonary tuberculosis. We have combined Ziehl-Neelsen staining and fluorescence in situ hybridization (FISH) to detect M. tuberculosis in sputum specimens.
KEYWORDS: Mycobacterium tuberculosis, fluorescence in situ hybridization, tuberculosis, oligonucleotide probe, rpoB
ABSTRACT
Routine staining of sputum specimens does not identify acid-fast bacilli as Mycobacterium tuberculosis with utmost precision, limiting its usability as a confirmatory test for pulmonary tuberculosis. We have combined Ziehl-Neelsen staining and fluorescence in situ hybridization (FISH) to detect M. tuberculosis in sputum specimens. We have developed a new fluorescent oligonucleotide rpoBMTC probe (5′-Alexa-555-AGCGGGGTGATGTCAACCCAG-3′) targeting the M. tuberculosis complex rpoB gene. In silico alignment yielded 100% match for M. tuberculosis complex mycobacteria, 66.6% to 47.6% for other bacteria, and no significant hits for viruses and eukaryotes. Negative binding of rpoBMTC probe to the top six respiratory tract bacterial pathogens and to Mycobacterium abscessus and Mycobacterium avium experimentally confirmed its specificity. As for sensitivity, rpoBMTC-FISH detected 103 CFU/ml M. tuberculosis as confirmed by successful detection of M. tuberculosis in artificially seeded sputum samples. The application of rpoBMTC-FISH to 116 routine sputum specimens yielded a detection of M. tuberculosis in all of the 31 Ziehl-Neelsen-positive and culture-positive specimens, and no detection of M. tuberculosis in the 85 M. tuberculosis-negative specimens. These data established the proof of concept that rpoBMTC-FISH alone or combined with Ziehl-Neelsen staining can specifically “FISH out” M. tuberculosis complex mycobacteria in sputum samples collected from patients suspected of pulmonary mycobacteriosis. We are implementing this probe for the routine and specific detection of M. tuberculosis complex bacteria in sputum exhibiting acid-fast mycobacteria.
INTRODUCTION
Pulmonary tuberculosis (TB) caused by mycobacteria forming the Mycobacterium tuberculosis complex is a deadly infection resulting from lung lesions induced by replicating M. tuberculosis (1). This disease remains in the top three worldwide health priorities in infectious diseases, as a total of 10.4 million new cases and 1.7 million deaths have been notified to the World Health Organization in 2017 (2).
The conclusive detection of M. tuberculosis in a sputum sample remains a priority for the early and accurate diagnosis of TB (3). There are various radiological, molecular, and microscopic techniques available for this purpose (4). However, a limitation in the sensitivity of a single test incorporates the need for more than one confirmatory test for the conclusive detection of M. tuberculosis in the patient samples (4, 5). While the time-consuming culture-based detection of M. tuberculosis from a patient sample still holds its gold standard grade, the initial detection of the tuberculosis bacilli by microscopic observation using appropriate staining, such as Ziehl-Neelsen staining, has a paramount importance in detecting TB in routine diagnostics. It remains the sole diagnostic test that is routinely performed in most of the countries of high endemicity (6).
Ziehl-Neelsen staining of sputum to detect the acid-fast bacilli is practiced worldwide because it is easy and affordable, although this test is highly variable in performance and has 20 to 80% accuracy for the detection of M. tuberculosis in the sputum (4). The use of fluorescence microscopy to scan TB bacilli is an important improvement to the standard Ziehl-Neelsen staining because of its increased sensitivity (4, 7).
Here, we have developed a new protocol for the detection of TB bacilli in sputum samples by combining the standard Ziehl-Neelsen staining with fluorescence in situ hybridization (FISH) with a newly designed oligonucleotide probe, rpoBMTC, targeting the M. tuberculosis complex rpoB gene.
MATERIALS AND METHODS
Specimens and bacterial cultures.
Pure cultures of M. tuberculosis H37Rv, Mycobacterium bovis BCG, Mycobacterium canettii, Mycobacterium abscessus, and Mycobacterium avium were grown into a biosafety level 3 laboratory at 37°C under shaking conditions in Middlebrook 7H9 broth with oleic acid-albumin-dextrose-catalase growth supplement and glycerol (as per the manufacturer's instructions; Becton-Dickinson, Le Pont-de-Claix, France). For all mycobacterial cultures, 0.01% Tween was added to the culture medium to prevent clumping of bacterial cells. Clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, Haemophilus influenzae, Escherichia coli, Streptococcus pneumoniae, and Klebsiella pneumoniae were grown in Columbia agar with 5% sheep blood plates (Becton Dickinson), except for Haemophilus influenzae, which was grown on chocolate agar Polyvitex (bioMérieux, La Balme-les-Grottes, France) at 37°C. Ziehl-Neelsen staining was done using a commercially available cold Ziehl-Neelsen staining (Quick-TB kit; RAL Diagnostics, Martillac, France). The identification of all the microorganisms was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), as previously described (8). In order to prepare artificially spiked sputum to set up the FISH protocol, 10 anonymized sputum specimens were collected from 10 unrelated patients without pulmonary tuberculosis. At last, a total of 116 anonymized sputum specimens were prospectively collected to be investigated by FISH. All the sputum specimens were digested by incubating 200 μl sputum with 200 μl of a solution containing 4% sodium hydroxide, 2.9% sodium citrate, and 0.5% N-acetyl-l-cysteine (Sigma-Aldrich, Saint-Quentin-Fancy, France) for 20 min at room temperature. After a 20-min centrifugation at 3,000 × g, the pellet was resuspended into an equal volume of phosphate-buffered saline (PBS), as previously described (9). Specimens were strictly anonymous. This study was approved by the ethics committee of Institut Fédératif de Recherche 48, Marseille, France.
Development of the rpoBMTC probe.
The rpoB sequences of 15 different mycobacteria, including four M. tuberculosis complex mycobacteria and 11 non-M. tuberculosis complex mycobacteria, were retrieved from GenBank in order to design an oligonucleotide probe, rpoBMTC, specifically targeting the M. tuberculosis complex rpoB (GenBank accession no. NC_000962.3). Sequences were aligned using the Mega7 software, and conserved regions specific to M. tuberculosis complex mycobacteria were taken into account. Finally, the Primer3 software (v.0.4.0) was used to design a probe according to length (<22 nucleotides [nt]), GC percent (60% to 65%), and avoidance of internal hairpin structure. The specificity of this probe was assessed in silico using the BLAST algorithm in NCBI database of the Nucleotide collection (nr/nt), with default parameters, except the maximum target sequences were set to 500 bp. The ClustalW pairwise sequence alignment tool (http://www.genome.jp/tools-bin/clustalw) for DNA, with default parameters, was used to determine the sequence similarity of the rpoBMTC probe with the bacterial rpoB sequence available in GenBank.
Development of M. tuberculosis fluorescent in situ hybridization.
The 5′ end of the rpoBMTC probe was labeled with the fluorochrome Alexa-555 (Eurogentec, Angers, France). The universal 16S rRNA bacterial probe EUB-338 (5′-GCTGCCTCCCGTAGGAGT-3′) labeled with Alexa-488 (Eurogentec) was used as a positive control (10), and the nonspecific probe non-EUB (5′-ACTCCTACGGGAGGCAGC-3′) labeled with Alexa-647 (Eurogentec) was used as a negative control.
In a first step, FISH was applied to cultured M. tuberculosis complex mycobacteria. A 100 μl-volume of bacterial suspension in PBS was smeared over a clean glass slide (Fisher Scientific, Illkirch, France) and heat fixed at 90°C for 60 min.
In a second step, FISH was applied to 10 artificially spiked sputum specimens, as follows: five Mycobacterium-free sputum samples were used to prepare 10-fold serial dilutions of M. tuberculosis mycobacteria (106 to 102 CFU/ml) using only rpoBMTC probe, and five other Mycobacterium-free sputum samples were spiked with 106 M. tuberculosis mycobacteria using both the rpoBMTC and EUB-338 probes. A 10-μl suspension was smeared on a slide and heat fixed as described above. All slides were flooded with 70% ethanol for 30 min and placed on a FISH-hybridizer (Dako; Agilent Technologies, Santa Clara, CA). Slides were flooded for 30 min at 37°C with a 10 mg/ml freshly prepared lysozyme solution (Sigma-Aldrich, Saint-Quentin-Fancy, France) and then with 5 μg/ml proteinase K (Sigma-Aldrich) for 5 min at 37°C (11, 12). After slides were gently washed with distilled water and dried, they were covered with 10 μl of a solution containing 1 μl rpoBMTC probe (10 μmol/liter), 1 μl EUB-338 probe (10 μmol/liter), 1 μl non-EUB probe (10 μmol/liter), 5 μl hybridization buffer (4× saline sodium citrate [SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10% dextran sulfate, 1 mM EDTA, 25% of formamide, 300 ng/ml of sperm of salmon's DNA, and 1× Denhardt solution) (Sigma-Aldrich), 1 μl of 0.1% Triton X-100 (Euromedex, Souffelweyersheim, France), and 1 μl distilled water. Slides were covered with a coverslip, sealed with adhesive (Fixogum; Marabu, Bietigheim-Bissingen, Germany), and incubated for 10 min at 65°C and overnight at 37°C. Further, slides were plunged into a series of baths of SSC buffer of 4×, 2×, 1×, and 0.5× concentrations for 5 min in each bath at room temperature. Air-dried slides were stained with cold Ziehl-Neelsen staining (RAL Diagnostics) and mounted with ProLong Diamond antifade (Fisher Scientific) containing 4′,6-diamidino-2-phenylindole (DAPI). Slides were observed under ×100 magnification with a DMI 6000 fluorescence microscope (Leica Microsystems, Nanterre, France). The fluorescence of the rpoBMTC probe was read with the red filter, the universal probe EUB-338 with the green filter, and the nonspecific non-EUB probe with the far-red filter. Three different microscopic fields were observed in order to measure the percentage of rpoBMTC-positive M. tuberculosis observed among EUB-338-stained bacteria.
Images were registered with a Hamamatsu Orca AG camera (Hamamatsu Photonics, Herrsching-am-Ammersee, Germany) and processed using the Metamorph software (Molecular Devices). The images were registered as black and white, and for publication, false color was applied using Fiji-Image J software. Care was taken not to tamper with the intensity of fluorescence.
In a third step, this FISH protocol was prospectively applied to a series of patients' sputum samples routinely submitted for the diagnosis of pulmonary tuberculosis after sputum digestion, as described above. As for patients' sputum samples, two independent observations were made by two different operators per sample. In order to measure the sensitivity of FISH detection compared to that of Ziehl-Neelsen staining as the gold standard, Ziehl-Neelsen-positive mycobacteria and FISH-positive mycobacteria were counted (10 microscopic fields; magnification, ×100) in a subset of 10 sputum specimens randomly selected among 31 Ziehl-Neelsen-positive and M. tuberculosis-culture-positive sputum samples.
RESULTS
Specificity of the rpoBMTC oligonucleotide probe.
We selected the 21-nucleotide (nt) rpoBMTC oligonucleotide probe (5′-AGCGGGGTGATGTCAACCCAG-3′; 62% GC content), which is close to the 5′ end of the rpoB of M. tuberculosis H37Rv (GenBank accession no. NC_000962.3) with coordinates 760015 to 760035 on the positive strand. Using BLAST (NCBI) with default parameters, we found that hits exhibiting 100% cover and 100% sequence match among the 500 output hits all belonged to M. tuberculosis complex mycobacteria, namely M. tuberculosis, Mycobacterium africanum, Mycobacterium bovis, M. bovis BCG, and Mycobacterium canettii (data not shown). Further checking of the percent similarity of rpoBMTC with the rpoB annotated in M. tuberculosis complex and non-M. tuberculosis complex mycobacteria indicated a 100% match with rpoB of M. tuberculosis, M. africanum, and M. bovis BCG, whereas M. canettii exhibited a single base pair mismatch (Table 1). Among non-M. tuberculosis members, M. abscessus and M. avium, commonly found in respiratory tract samples, exhibited low-percentage matches of 66.6% and 61.9%, respectively (Table 1). Thereafter, we selected rpoB of the six most abundant bacteria in the respiratory tract in our laboratory, namely, S. aureus, P. aeruginosa, H. influenzae, E. coli, S. pneumoniae, and K. pneumoniae, and found very low degrees of similarity of 47.6% to 61.9% with rpoBMTC (Table 1).
TABLE 1.
Sequence similarities of rpoBMTC probe with rpoB gene sequences of various species
| Species | rpoB gene NCBI accession no. | % cover | % match of rpoBMTC probe with target gene |
|---|---|---|---|
| Mycobacterium tuberculosis | NC_000962.3 | 100 | 100 |
| Mycobacterium bovis BCG | AM408590.1 | 100 | 100 |
| Mycobacterium africanum | NC_015758.1 | 100 | 100 |
| Mycobacterium canettii | FO203510.1 | 95 | 90.5 |
| Mycobacterium abscessus | AY147164.1 | 67 | 66.6 |
| Mycobacterium avium | NC_008595.1 | 62 | 61.9 |
| Staphylococcus aureus | NC_007795.1 | 52 | 47.6 |
| Pseudomonas aeruginosa | NC_002516.2 | 67 | 52.4 |
| Haemophilus influenzae | NC_000907.1 | 62 | 52.4 |
| Escherichia coli | NC_000913.3 | 62 | 52.4 |
| Streptococcus pneumoniae | NC_003098.1 | 62 | 61.9 |
| Klebsiella pneumoniae | NC_016845.1 | 62 | 61.9 |
Experimental study using FISH confirmed the in silico specificity.
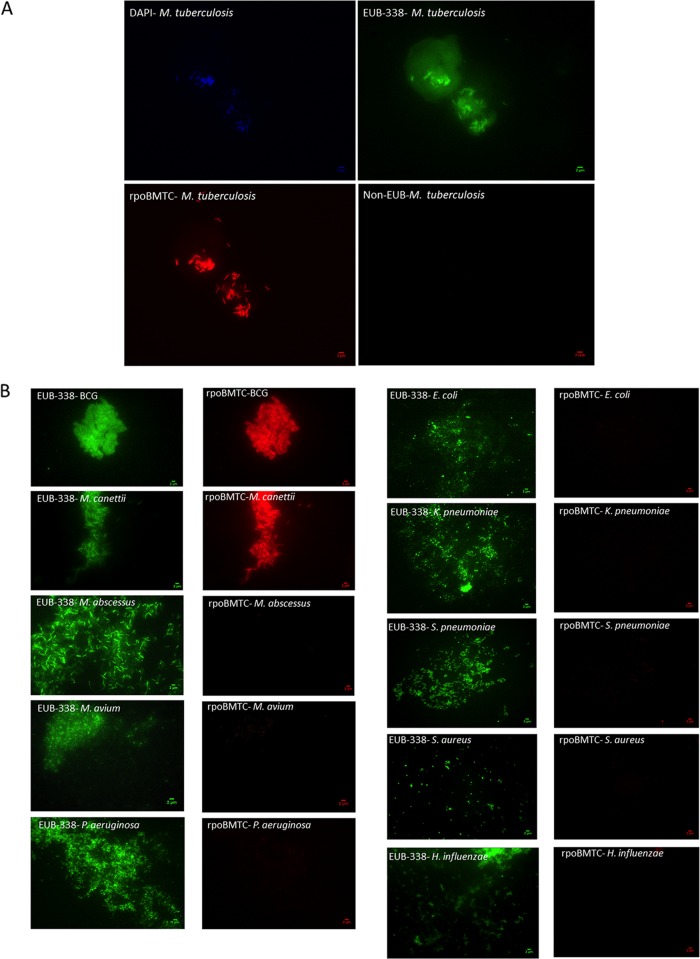
In a first step, our results showed that DAPI and the three oligonucleotide probes used in this study were able to penetrate cultured M. tuberculosis cells (Fig. 1A), as follows: the blue micrograph indicated that cells were stained with DAPI, the green micrograph confirmed EUB-338 binding indicative of bacterial cells, the red micrograph confirmed rpoBMTC binding indicative of M. tuberculosis cells, and negative-control non-EUB probe did not exhibit any binding. Furthermore, FISH yielded rpoBMTC probe detection of M. tuberculosis, M. bovis BCG, and M. canettii, but not M. abscessus, M. avium, S. aureus, P. aeruginosa, H. influenzae, E. coli, S. pneumoniae, or K. pneumoniae, whereas all these bacteria were EUB-338 positive (Fig. 1A and B). We then evaluated the sensitivity of rpoBMTC by determining the minimum number of CFU of bacteria it could detect. We found that rpoBMTC-FISH detected a minimum of 103 CFU/ml for the M. tuberculosis complex mycobacteria, whereas non-MTC mycobacteria up to a concentration of 106 CFU/ml yielded no signal (Table 2).
FIG 1.
Fluorescence microscopy images of M. tuberculosis to assess the specificity of rpoBMTC against it. (A) Cultured M. tuberculosis cells were targeted with three different FISH probes, EUB-338 (green channel), non-EUB (far-red channel), and rpoBMTC (red channel). The blue channel is for DAPI containing antifade reagent used for the stability of fluorescent probes under laser exposure. (B) Fluorescence microscopy images of various mycobacterial and bacterial species found in human respiratory tract targeted with EUB-338 and rpoBMTC to assess the specificity against M. tuberculosis complex. The green channel is for EUB-338, and the red channel is for rpoBMTC.
TABLE 2.
Sensitivity of rpoBMTC for detection of M. tuberculosis complex bacteria
| Species | Detection by no. of CFU/ml |
||||
|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | |
| M. tuberculosis | + | + | + | + | − |
| M. bovis BCG | + | + | + | + | − |
| M. africanum | + | + | + | + | − |
| M. canettii | + | + | + | + | − |
| M. abscessus | − | − | − | − | − |
| M. avium | − | − | − | − | − |
FISHing M. tuberculosis in sputum samples.
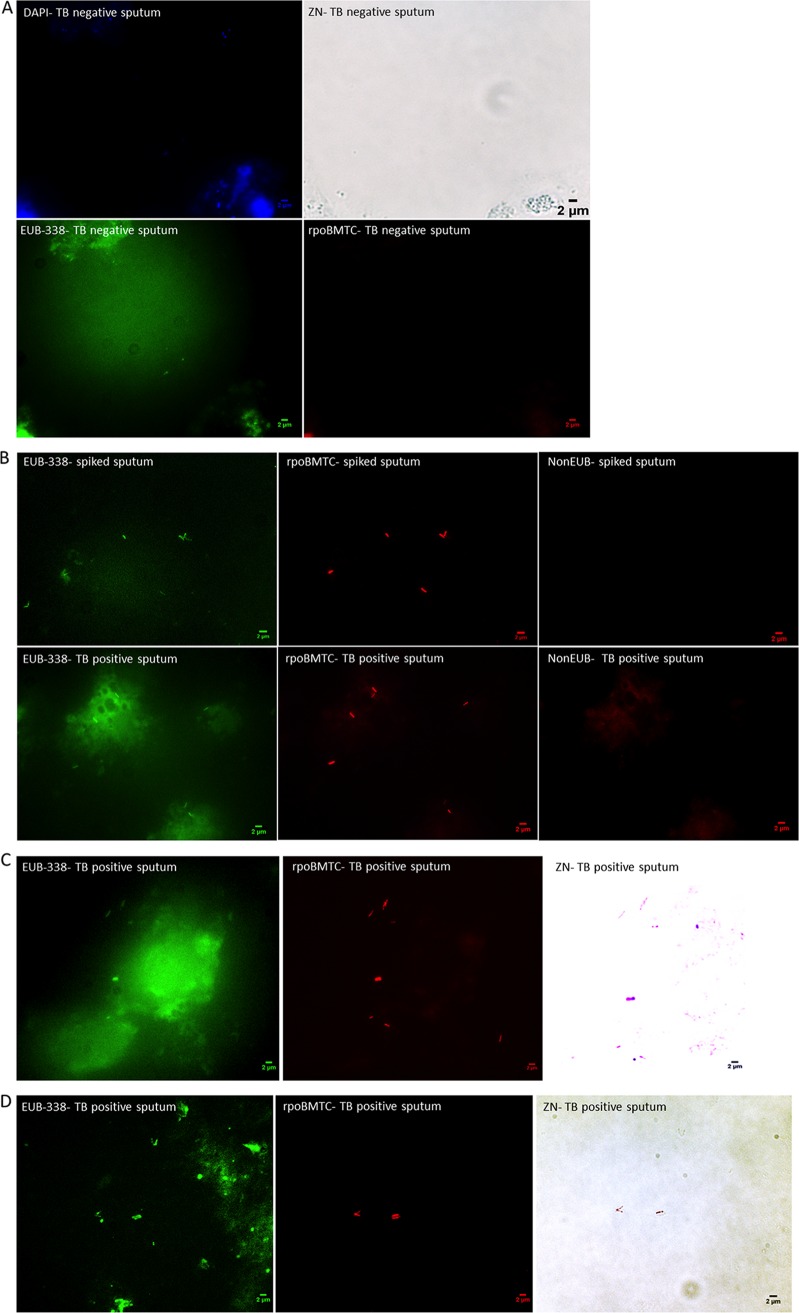
In a second step, the observation of five Ziehl-Neelsen-negative, culture-negative sputum samples spiked with M. tuberculosis mycobacteria indicated that rpoBMTC-positive mycobacteria comprised 36.06% to 43% of the EUB-338-positive bacteria per sample in the presence of negative controls (Fig. 2A and B). To confirm that EUB-338-positive and rpoBMTC-positive bacteria were indeed M. tuberculosis, we combined FISH with cold Ziehl-Neelsen staining. We observed that the red fluorescent bacteria detected by rpoBMTC probe were also Ziehl-Neelsen positive (Fig. 2C). These observations indicated that rpoBMTC-FISH and Ziehl-Neelsen staining could be combined on a single glass slide for detecting M. tuberculosis as red fluorescent and Ziehl-Neelsen-positive cells (Fig. 2C).
FIG 2.
Fluorescence microscopic images of patients' sputum samples combining FISH with Ziehl-Neelsen (ZN) staining. (A) An M. tuberculosis-negative-control sputum sample exhibited bacteria stained in a green (EUB-338) channel, whereas the red (rpoBMTC) channel remained negative, and the DAPI channel was positive for all cells. (B) An M. tuberculosis-positive sputum sample exhibited green (EUB-338) channel-positive bacteria also stained in red (rpoBMTC) channel (for M. tuberculosis [TB]), while the negative-control far-red (non-EUB) channel has no fluorescence. (C) To confirm that rpoBMTC specifically bound M. tuberculosis, Ziehl-Neelsen staining was combined with FISH on the very same microscopic field of one patient's sputum. (D) In some specimens, FISH detected M. tuberculosis mycobacteria that were not detected by Ziehl-Neelsen staining. Light microscopic images of Ziehl-Neelsen staining were made using a DFC425 C digital microscope camera (Leica Microsystems); fluorescence microscopic images of FISH were made using an Orca AG camera (Hamamatsu Photonics). The two cameras operated at the same ×100 magnification but yielded slightly different output formats.
We then applied this protocol on 31 Ziehl-Neelsen-positive and M. tuberculosis-culture-positive sputum samples and on 85 Ziehl-Neelsen-negative and M. tuberculosis-culture-negative sputum samples. In all 31 Ziehl-Neelsen- and culture-positive sputum samples, rpoBMTC detected TB bacilli, whereas no detection was visible for any of the 85 TB-negative sputum samples. Counting mycobacteria in a subset of 10 sputum specimens, found 1.87 ± 1.85 Ziehl-Neelsen-stained mycobacteria per microscopic field and 2.14 ± 1.62 FISH-stained mycobacteria per microscopic field. Indeed, in nine sputum specimens, all mycobacteria detected by Ziehl-Neelsen staining were also detected by FISH staining, while some FISH-stained mycobacteria were not stained by Ziehl-Neelsen (Fig. 2D). These results confirmed the efficiency of rpoBMTC to detect M. tuberculosis or a member of the M. tuberculosis complex (MTBC) in sputum samples.
DISCUSSION
Here, we have developed a new protocol combining Ziehl-Neelsen staining and FISH for the microscopic detection of TB bacilli in sputum smear. The results reported here were obtained in the presence of appropriate negative controls and were reproducible.
Penetration of oligonucleotide probes into TB bacilli has been reported as inefficient due to the thick and complex nature of the mycobacterial cell wall (13). To circumvent this limitation, oligonucleotide probes have been replaced with peptide nucleic acid probes, which were found to be more penetrable and have better detection capacity for TB bacilli than oligonucleotide probes (13, 14). The results reported here indicate that oligonucleotide probes can indeed be a suitable alternative to peptide nucleic acid probes to FISH-out TB bacilli, depending on the setup of a suitable protocol. The one here reported combines a consecutive treatment of mycobacterial cells with lysozyme, proteinase K, and Triton X.
So far, probes targeting the 16S rRNA have been devised and used to detect TB bacilli in different patient samples. The 16S rRNA probes, however, have been found to be less specific toward some closely related mycobacterial species which could be also encountered in respiratory tract specimens (15). Moreover, RNA is an unstable molecule, and 16S rRNA-based FISH could be negatively influenced by the dormant state or inside dead mycobacteria, such as under antituberculosis treatment (16). We have previously demonstrated that the rpoB gene is a useful tool for the detection and identification of mycobacteria (15, 17). Therefore, here we have developed a rpoB gene-based FISH protocol using oligonucleotide probes. For this purpose, a new oligonucleotide-probe named rpoBMTC was designed against the species-specific region of the rpoB gene of M. tuberculosis complex bacteria (17). This probe, rpoBMTC, targeting the rpoB DNA and not the encoded mRNA, circumvents the problem of unstable RNA inside bacterial cells. The results reported here clearly demonstrate the specificity of the rpoBMTC probe for detecting M. tuberculosis, along with other important M. tuberculosis complex bacteria, including M. bovis BCG, M. africanum, and M. canettii (18) while differentiating these M. tuberculosis complex mycobacteria from non-M. tuberculosis complex mycobacteria and the other bacteria most frequently encountered in respiratory tract samples.
One step forward, we developed a novel methodology combining FISH with standard Ziehl-Neelsen-staining to confirm the specificity of rpoBMTC and consequently improve the specificity of the Ziehl-Neelsen staining for the patient sputum samples. This approach led to the observation that FISH was more sensitive than the gold standard Ziehl-Neelsen staining, as in the majority of sputum specimens, FISH-positive mycobacteria were Ziehl-Neelsen “ghosts.” Indeed, we evaluated a sensitivity of 103 CFU for FISH, one log of magnitude lower than what is commonly reported for Ziehl-Neelsen staining (19). Ziehl-Neelsen-staining is in use in spite of its well-known vulnerability (6), as it is easy to perform and cost-effective. Our protocol is thus easily adaptable with the routine diagnostic procedure, as we have performed our protocol directly on liquefied sputum smears. However, the necessity of fluorescence microscopy may limit the spread of this protocol in some laboratories in some resource-limited countries. Accordingly, cost-effectiveness studies will have to be done in order to determine in which settings FISH could be reasonably implemented.
Our results indicate the specificity of this approach and suggest that it could be implemented in the routine practice as an additional laboratory tool for the one-shot microscopic detection of TB bacilli. Accordingly, these proof-of-concept data sets based our decision to ongoing implementation of the rpoBMTC-FISH into the routine detection of M. tuberculosis complex mycobacteria in the Ziehl-Neelsen-positive sputum samples collected from patients suspected of pulmonary tuberculosis.
ACKNOWLEDGMENTS
This study was supported by IHU Méditerranée Infection, Marseille, France, and by the French Government under the “Investissements d'Avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research) (reference Méditerranée Infection 10-IAHU-03). This work was supported by Région Provence Alpes Côte d'Azur and European funding FEDER PRIMI. A.L. benefits from a Ph.D. grant from Aix-Marseille Université.
We acknowledge Olga Cusack for her expert assistance in editing the manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Cambau E, Drancourt M. 2014. Steps towards the discovery of Mycobacterium tuberculosis by Robert Koch, 1882. Clin Microbiol Infect 20:196–201. doi: 10.1111/1469-0691.12555. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2017. Global tuberculosis report. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf. [Google Scholar]
- 3.Asmar S, Drancourt M. 2015. Rapid culture-based diagnosis of pulmonary tuberculosis in developed and developing countries. Front Microbiol 6:1184. doi: 10.3389/fmicb.2015.01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu YJ. 2015. Diagnosis of pulmonary tuberculosis: recent advances and diagnostic algorithms. Tuberc Respir Dis (Seoul) 78:64–71. doi: 10.4046/trd.2015.78.2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clifford V, Tebruegge M, Curtis N. 2015. Limitations of current tuberculosis screening tests in immunosuppressed patients. BMJ 350:h2226. doi: 10.1136/bmj.h2226. [DOI] [PubMed] [Google Scholar]
- 6.Parsons LM, Somoskovi A, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, Spector S, Roscigno G, Nkengasong J. 2011. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev 24:314–350. doi: 10.1128/CMR.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steingart KR, Ramsay A, Pai M. 2007. Optimizing sputum smear microscopy for the diagnosis of pulmonary tuberculosis. Expert Rev Anti Infect Ther 5:327–331. doi: 10.1586/14787210.5.3.327. [DOI] [PubMed] [Google Scholar]
- 8.Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D. 2013. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent PT, Kubica GP. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 10.St. Amand AL, Frank DN, De Groote MA, Basaraba RJ, Orme IM, Pace NR. 2005. Use of specific rRNA oligonucleotide probes for microscopic detection of Mycobacterium tuberculosis in culture and tissue specimens. J Clin Microbiol 43:5369–5371. doi: 10.1128/JCM.43.10.5369-5371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimino M, Alamo L, Salazar L. 2006. Permeabilization of the mycobacterial envelope for protein cytolocalization studies by immunofluorescence microscopy. BMC Microbiol 6:35. doi: 10.1186/1471-2180-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Nuñez J, Avelar FJ, Marquez F, Rivas-Santiago B, Quinones C, Guerrero-Barrera AL. 2012. Mycobacterium tuberculosis complex detected by modified fluorescent in situ hybridization in lymph nodes of clinical samples. J Infect Dev Ctries 6:58–66. [DOI] [PubMed] [Google Scholar]
- 13.Lefmann M, Schweickert B, Buchholz P, Gobel UB, Ulrichs T, Seiler P, Theegarten D, Moter A. 2006. Evaluation of peptide nucleic acid-fluorescence in situ hybridization for identification of clinically relevant mycobacteria in clinical specimens and tissue sections. J Clin Microbiol 44:3760–3767. doi: 10.1128/JCM.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stender H, Mollerup TA, Lund K, Petersen KH, Hongmanee P, Godtfredsen SE. 1999. Direct detection and identification of Mycobacterium tuberculosis in smear-positive sputum samples by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes. Int J Tuberc Lung Dis 3:830–837. [PubMed] [Google Scholar]
- 15.Adekambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol 41:5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutscher MP. 2003. Degradation of stable RNA in bacteria. J Biol Chem 278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 17.Adekambi T, Drancourt M, Raoult D. 2009. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol 17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Aboubaker Osman D, Bouzid F, Canaan S, Drancourt M. 2015. Smooth tubercle bacilli: neglected opportunistic tropical pathogens. Front Public Health 3:283. doi: 10.1016/j.hjdsi.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiruviluamala P, Reichman LB. 2002. Tuberculosis. Annu Rev Public Health 23:403–426. doi: 10.1146/annurev.publhealth.23.100901.140519. [DOI] [PubMed] [Google Scholar]