Decoy circadian clock proteins, which bind interacting proteins but prevent their degradation, reveal new functions and interactions of redundant E3 ubiquitin ligases.
Abstract
Eukaryotic circadian clocks utilize the ubiquitin proteasome system to precisely degrade clock proteins. In plants, the F-box-type E3 ubiquitin ligases ZEITLUPE (ZTL), FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), and LOV KELCH PROTEIN2 (LKP2) regulate clock period and couple the clock to photoperiodic flowering in response to end-of-day light conditions. To better understand their functions, we expressed decoy ZTL, FKF1, and LKP2 proteins that associate with target proteins but are unable to ubiquitylate their targets in Arabidopsis (Arabidopsis thaliana). These dominant-negative forms of the proteins inhibit the ubiquitylation of target proteins and allow for the study of ubiquitylation-independent and -dependent functions of ZTL, FKF1, and LKP2. We demonstrate the effects of expressing ZTL, FKF1, and LKP2 decoys on the circadian clock and flowering time. Furthermore, the decoy E3 ligases trap substrate interactions, and using immunoprecipitation-mass spectrometry, we identify interacting partners. We focus studies on the clock transcription factor CCA1 HIKING EXPEDITION (CHE) and show that ZTL interacts directly with CHE and can mediate CHE ubiquitylation. We also demonstrate that CHE protein is degraded in the dark and that degradation is reduced in a ztl mutant plant, showing that CHE is a bona fide ZTL target protein. This work increases our understanding of the genetic and biochemical roles for ZTL, FKF1, and LKP2 and also demonstrates an effective methodology for studying complicated genetic redundancy among E3 ubiquitin ligases.
The circadian clock is an endogenous timekeeper that synchronizes essential biological processes to the outside world. Eukaryotic clocks rely on the ubiquitin proteasome system (UPS) to target core clock factors for degradation, and altering clock protein degradation can change the period length of the clock (Grima et al., 2002; Ko et al., 2002; He et al., 2003; Shirogane et al., 2005; Ito et al., 2012). The UPS is an enzymatic pathway that mediates the covalent attachment of ubiquitin to substrate proteins, which targets them to the proteasome for destruction (Carrano and Bennett, 2013; Kleiger and Mayor, 2014; Hua and Vierstra, 2016). The critical substrate targeting step of this cascade is mediated by E3 ubiquitin ligases, which bridge the interaction between the substrate and activated ubiquitin. Subsequently, ubiquitin is transferred to Lys residues within substrate proteins (Deshaies and Joazeiro, 2009; Hua and Vierstra, 2011).
The SKP1/CULLIN/F-BOX (SCF) is a multisubunit E3 ubiquitin ligase complex that is conserved across eukaryotes (Deshaies and Joazeiro, 2009; Hua and Vierstra, 2011). Within this protein complex, F-box domain-containing proteins act as substrate adaptors using a highly diverse group of protein-protein interaction domains (Gagne et al., 2002; Jin et al., 2004). The F-box domain is an approximately 45-amino acid domain that binds SKP1 (Bai et al., 1996; Deshaies, 1999; Lechner et al., 2006). SKP1 subsequently interacts with the cullin subunit that binds to a RING domain-containing protein, RING-BOX1 (RBX1). RBX1 acts to recruit ubiquitin-charged activated E2 enzymes to facilitate the ubiquitylation of target proteins. Interestingly, all eukaryotic circadian clocks employ SCF-type E3 ligases to mediate the ubiquitylation of clock factors (Grima et al., 2002; Ko et al., 2002; He et al., 2003; Shirogane et al., 2005; Ito et al., 2012).
Identifying the functions of F-box proteins can be challenging for three specific reasons. First, in many species, there is widespread functional redundancy in the F-box family, which makes traditional forward genetic studies difficult or impossible. This problem is greatly exacerbated in plants, as the Arabidopsis (Arabidopsis thaliana) F-box gene family is one of the largest in the genome, containing nearly 700 members (Gagne et al., 2002). Second, the interaction between an F-box protein and its substrate is often difficult to detect because the substrate protein is degraded or dissociates following ubiquitylation. Third, validating putative E3 ubiquitin ligase substrates can be challenging for the same reasons listed above.
These challenges are exemplified by a subfamily of three F-box proteins that regulates the circadian clock and seasonal flowering time in plants. The LOV/F-box/Kelch repeat family of F-boxes has three members: ZEITLUPE (ZTL), LOV KELCH PROTEIN2 (LKP2), and FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1; Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). The members of this F-box subfamily share the same protein domain architecture and have significant overlap in sequence identity. They contain a unique arrangement of protein interaction domains that allow them to communicate light signals to factors controlling the circadian clock and seasonal flowering time (Imaizumi et al., 2003, 2005; Ito et al., 2012; Zoltowski and Imaizumi, 2014; Shim et al., 2017). The N-terminal LIGHT OXYGEN VOLTAGE (LOV) domain is a blue light photoreceptor that interacts with the regulatory protein GIGANTEA (GI) in a light-dependent manner (Fowler et al., 1999; Swarup et al., 1999; Kim et al., 2007; Sawa et al., 2007). It is also predicted that the LOV domains of ZTL, LKP2, and FKF1 interact with the substrate proteins TIMING OF CAB2 EXPRESSION1 (TOC1; sometimes called PRR1) and PSEUDO-RESPONSE REGULATOR5 (PRR5), two homologous transcription factors that regulate the circadian clock (Strayer et al., 2000; Alabadí et al., 2001; Más et al., 2003; Kiba et al., 2007; Kim et al., 2007; Para et al., 2007; Fujiwara et al., 2008; Harmon et al., 2008; Baudry et al., 2010; Gendron et al., 2012). Furthermore, the FKF1 LOV domain interacts with CONSTANS (CO), a critical promoter of flowering (Song et al., 2012, 2014). Interestingly, TOC1 and PRR5 are destabilized through these interactions, while CO is stabilized. The central regions of the ZTL, FKF1, and LKP2 proteins contain the F-box domain, which recruits the ubiquitylation machinery through binding to multiple variants of ARABIDOPSIS SKP1 HOMOLOG1 (ASK1; Zhao et al., 2003; Han et al., 2004; Takahashi et al., 2004; Wang et al., 2006).The C-terminal domain is a typical protein-protein interaction domain composed of Kelch repeats. It also has been shown that both FKF1 and LKP2 can interact with the CYCLING DOF FACTOR (CDF) family of flowering regulators through their Kelch repeat domains and that this interaction can promote the degradation of CDFs by FKF1 (Imaizumi et al., 2005; Fornara et al., 2009, 2015).
Although ZTL, FKF1, and LKP2 play roles in the circadian clock and the regulation of seasonal flowering time, their functions have diverged partially but not entirely, leading to complicated redundancy. For instance, the ztl knockout lengthens clock period by 4 h, while individual lkp2 and fkf1 knockouts have little or no effect on the clock (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). The fkf1 knockout delays flowering time dramatically, while ztl and lkp2 knockouts have minimal effect on flowering (Nelson et al., 2000; Schultz et al., 2001; Somers et al., 2004; Takase et al., 2011). Higher order loss-of-function mutants show synergistic effects. The ztl fkf1 double mutant exhibits a longer period than the ztl mutant alone, and the ztl fkf1 lkp2 triple mutant shows reduced amplitude compared with the ztl fkf1 double mutant. This suggests that all three are necessary for robust 24-h rhythmicity (Baudry et al., 2010). The ztl lkp2 double mutant also shows a slight early-flowering effect (Takase et al., 2011), suggesting that FKF1 plays a specialized role in determining flowering time. Further complexity has been demonstrated by yeast two-hybrid experiments and coimmunoprecipitation in heterologous expression systems. These assays show that the clock factors TOC1 and PRR5 interact with all three family members (Baudry et al., 2010), while the flowering regulator CDF proteins show binding preference for FKF1 and LKP2 (Imaizumi et al., 2005).
A priori knowledge of the substrates’ roles in clock function or shared interacting partners has been required in all work discovering substrates of this subfamily of F-box proteins. Unbiased approaches have been attempted, such as an examination of the FKF1 protein complex by immunoprecipitation followed by mass spectrometry (IP-MS), and the regulatory partners GI and HEAT SHOCK PROTEIN90 (HSP90) were identified (Song et al., 2014). However, the known targets of FKF1 were not identified in these experiments, and to our knowledge, no unbiased approaches have discovered putative substrates of this family of F-box proteins, illustrating the difficulty of de novo substrate identification. Furthermore, genetic techniques have been used to overcome functional redundancy, such as introducing point mutations into the F-box domain of ZTL to create a dominant-negative form (Han et al., 2004). These putative dominant-negatives varied in their effects and, when highly expressed, exhibited arrhythmicity similar to overexpression of the full-length ZTL, suggesting that point mutations in the ZTL F-box domain are not effective at fully preventing interaction with ASK1.
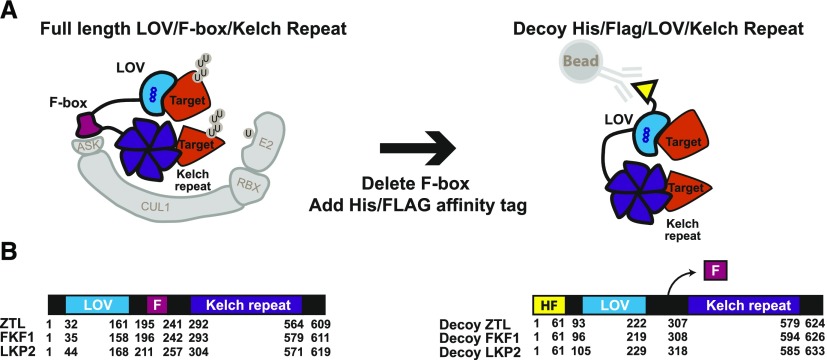
To study plant clock-regulating F-box proteins, we expressed ZTL, LKP2, and FKF1 without their F-box domains. These modified proteins are predicted to maintain the binding of target proteins but prevent ubiquitylation, allowing us to distinguish between ubiquitylation-dependent and -independent functions. Furthermore, we added an affinity tag for biochemical analyses of the stabilized complexes. This strategy allows one to bypass traditional challenges in studying the genetic functions of redundant F-box gene families and also facilitates substrate identification using IP-MS. Using this strategy, we can perform genetic and biochemical experiments in the first generation of transformants, greatly reducing the time it takes to analyze F-box function. We have named this the decoy method because the F-box deletion proteins lure the targets from the endogenous F-box proteins, trapping the interaction and allowing for genetic and biochemical analyses (Fig. 1).
Figure 1.
Schematic of the decoy strategy applied to the LOV-Kelch domain-containing F-box proteins from Arabidopsis. A, F-box proteins bind to substrates (Targets in orange) and mediate the ubiquitylation of targets. Decoy proteins lacking the F-box domain interact with their substrates but cannot target them for ubiquitylation, and the stabilized decoy-substrate complex can be immunoprecipitated via the His-FLAG tag. B, Amino acid numbers are shown for the three domains of ZTL, LKP2, and FKF1 wild-type and decoy proteins used in this study. F, F-box; HF, His-FLAG.
Using this technique, we tested the genetic effects of expressing ZTL, LKP2, and FKF1 decoys on the circadian clock and flowering in Arabidopsis. We show that expressing the ZTL decoy has the greatest effect on the circadian clock, followed by LKP2 and then FKF1. This relationship is inverted with respect to flowering time, as the FKF1 decoy has a strong effect on flowering while LKP2 and ZTL have weaker effects. These genetic results are corroborated by IP-MS studies, showing that FKF1 decoys interact with flowering regulator proteins while LKP2 and ZTL decoys interact with clock proteins. We further employ this unbiased target identification method to identify multiple potential novel targets and regulatory partners of ZTL, including the TCP family transcription factor CCA1 HIKING EXPEDITION (CHE), a critical clock regulator (Pruneda-Paz et al., 2009). We validate CHE as a novel ZTL target by establishing their direct interaction in two heterologous expression systems, reconstituting ZTL ubiquitylation of CHE in mammalian tissue culture cells and demonstrating the requirement of ZTL for maintaining daily rhythms of CHE protein. These results further untangle the complex genetic relationship between the three LOV/F-box/Kelch repeat genes and identify a comprehensive list of potential ZTL, FKF1, and LKP2 interacting proteins. This study also demonstrates an effective methodology for disentangling complicated genetic and biochemical roles of redundant plant F-box proteins.
RESULTS
Genetic and Developmental Analyses of ZTL, FKF1, and LKP2 Decoy Transgenic Lines
ZTL, FKF1, and LKP2 are predicted to regulate clock function and seasonal flowering by mediating the ubiquitylation of critical clock and flowering transcription factors. To investigate the ubiquitylation-dependent and -independent functions of ZTL, FKF1, and LKP2, we expressed decoy versions of each in Arabidopsis. The decoy approach involves expressing the protein without the F-box domain (Fig. 1), allowing it to interact with targets or regulatory partners while preventing recruitment of the machinery that allows for their ubiquitylation function (Banerjee et al., 1995; Margottin et al., 1998; Belaïdouni et al., 2005; Risseeuw et al., 2013; Sharma et al., 2015; Nagels Durand et al., 2016). The effect is that the decoy protein will compete with the endogenous full-length F-box protein and any homologs, thus protecting a portion of the target protein pool from degradation. Genetically, the decoy can reveal dominant-negative (ubiquitylation-dependent) roles, in which phenotypes are similar to knockouts, and dominant-positive (ubiquitylation-independent) roles, in which phenotypes are similar to overexpression of the full-length gene. The main benefit of the decoy dominant-negative approach is the enhanced stability of the target interactions, increasing the chance that IP-MS will identify interactions with target proteins. Thus, we added a dual affinity tag (3XFLAG and 6XHis) to facilitate immunoprecipitation experiments (Fig. 1; Yumimoto et al., 2012).
We constitutively expressed ZTL, LKP2, and FKF1 decoys under the control of a cauliflower mosaic virus 35S promoter in Arabidopsis harboring a circadian clock reporter transgene (CIRCADIAN CLOCK ASSOCIATED1 [CCA1] promoter driving the expression of Luciferase: CCA1p::Luciferase). We entrained T1 generation seedlings in 12-h-light/12-h-dark (LD) conditions for 7 d and then transferred them to constant light (LL) for 1 d prior to imaging in LL (Fig. 2; Supplemental Fig. S1; Supplemental Table S1). We transferred the same plants from the circadian imaging experiment to soil and grew them in inductive long days (16 h of light/8 h of dark) to measure flowering time (Fig. 3). We performed the experiment with dozens of individual T1 transgenic lines to avoid artifacts arising from expression variation from the genomic insertion site. Furthermore, we wished to avoid the pitfalls of following individual transgenic lines that may not show the effects seen in all transgenic lines.
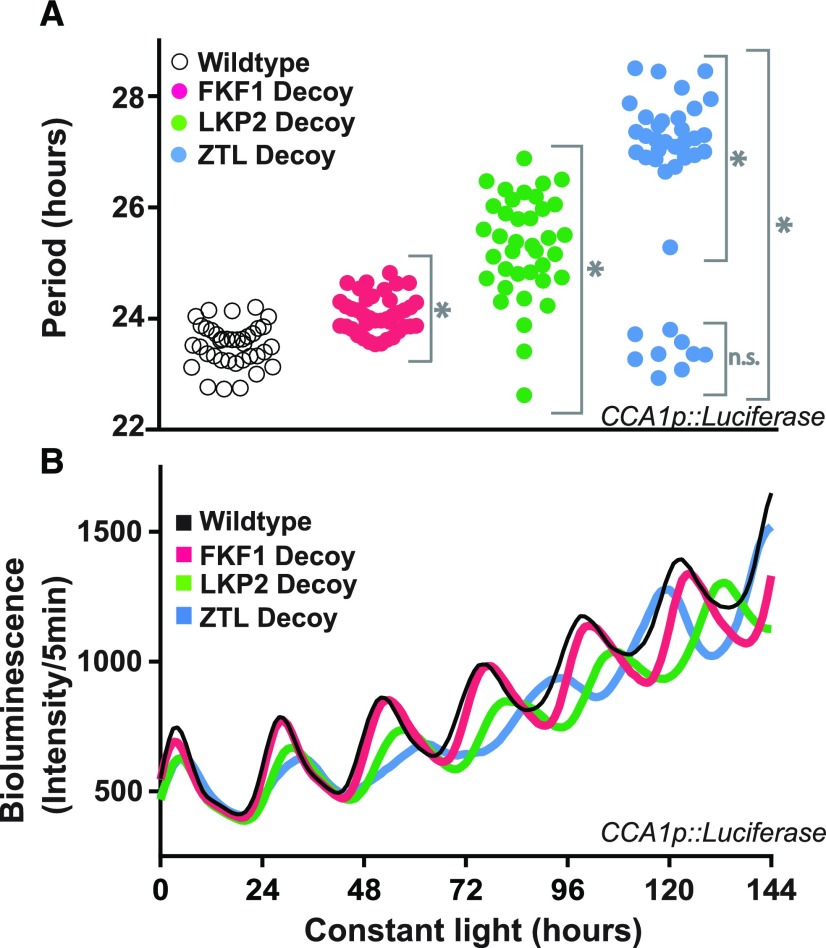
Figure 2.
Decoys reveal the genetic contributions of redundant F-box genes to the circadian clock. A, Period lengths (as measured by CCA1p::Luciferase activity) for T1 transgenic decoys from ZTL (blue; n = 40), LKP2 (green; n = 38), FKF1 (pink; n = 40), and the wild-type parental line CCA1p::Luciferase (white; n = 38). Gray brackets define individual groups used for statistical testing against the wild-type control using a Welch’s t test with a Bonferroni-corrected α of 2 × 10−3. *, P < α; n.s., P > α. B, Average traces from the wild type (parental CCA1p::Luciferase) and the ZTL, LKP2, and FKF1 decoys.
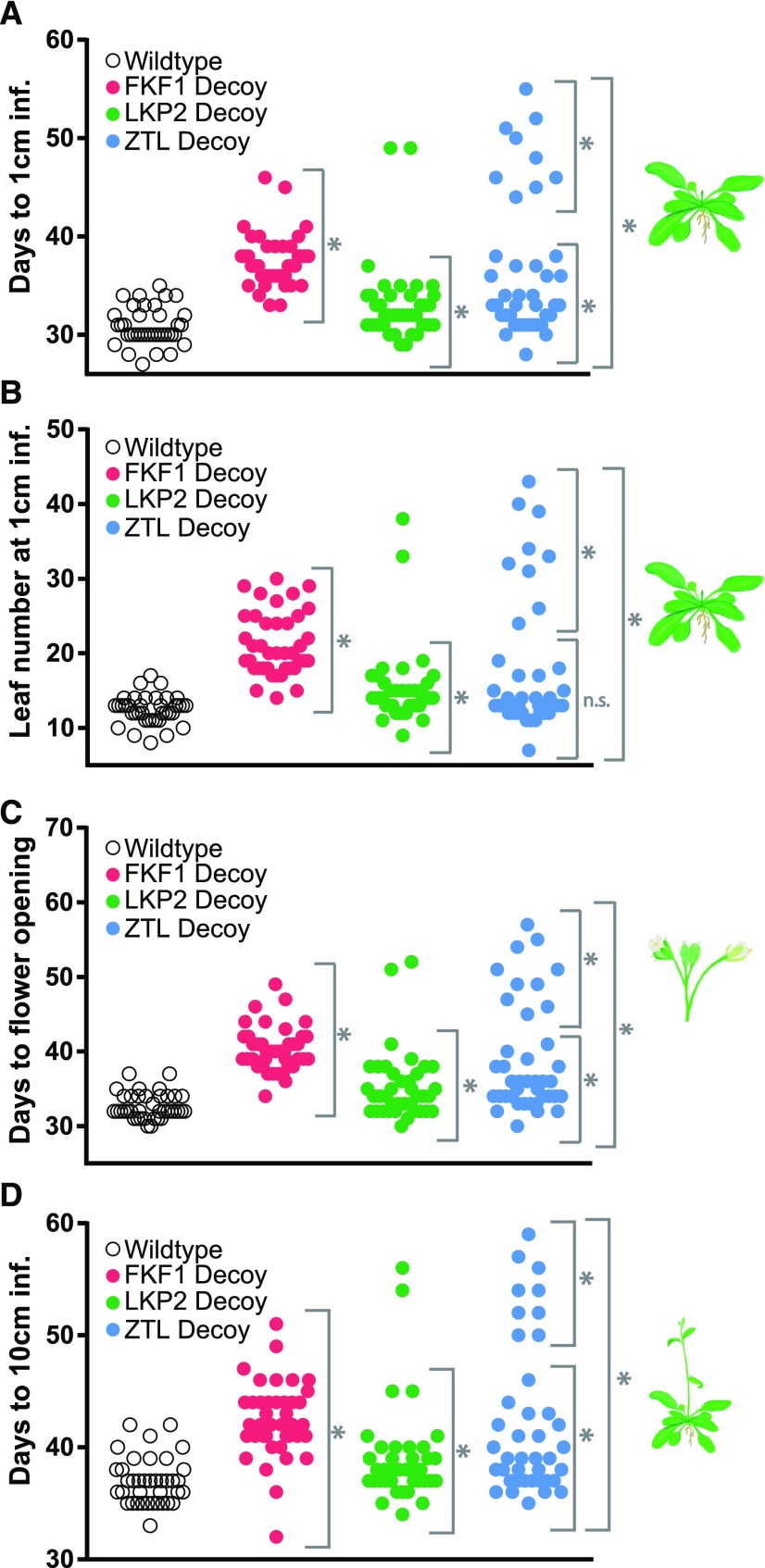
Figure 3.
Decoys reveal the genetic contributions of redundant F-box genes to seasonal flowering time. Flowering time was measured for the wild type and the T1 generation ZTL (n = 40), LKP2 (n = 38), and FKF1 (n = 40) decoys using four parameters: number of days for the inflorescence stem to reach 1 cm (A); number of rosette leaves when the inflorescence stem reaches 1 cm (B); number of days until the first flower opening (C); and number of days for the inflorescence stem to reach 10 cm (D). Gray brackets define individual groups used for statistical testing against the wild-type control (n = 38) using a Welch’s t test with a Bonferroni-corrected α of 2 × 10−3. *, P < α; n.s., P > α. Two lines with a significantly later flowering phenotype than the others were removed from LKP2 statistical analysis and could not be analyzed separately due to the limits of statistical testing on small sample sizes.
Expression of the FKF1 decoy lengthened clock period by approximately 30 min (Fig. 2; Supplemental Fig. S1; Supplemental Table S1, pink symbols) and delayed flowering by around 6.5 d with approximately nine more leaves on average (Fig. 3; Supplemental Table S1, pink symbols). These effects on flowering and circadian period are similar to those of knockout mutants (Nelson et al., 2000), suggesting that these are dominant-negative effects caused by expressing the FKF1 decoy.
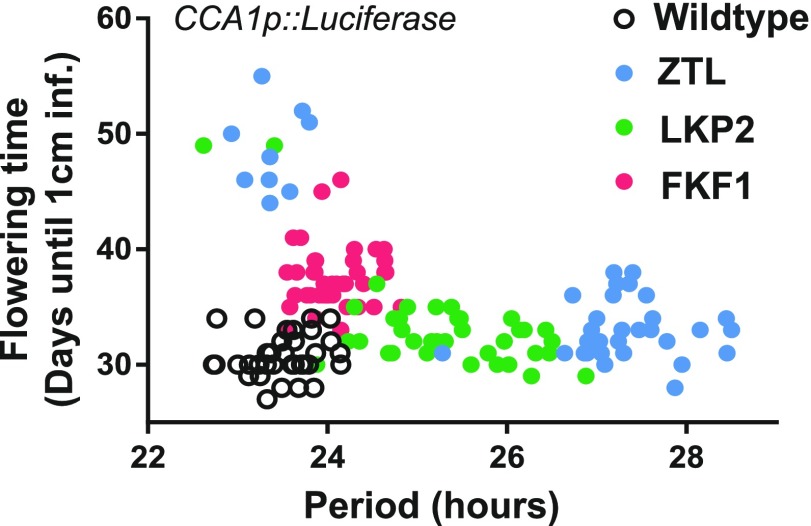
Expression of the ZTL or LKP2 decoys resulted in two subpopulations of transgenic lines with distinct period lengths and flowering times (Figs. 2 and 3; Supplemental Fig. S1; Supplemental Table S1). These subpopulations are best visualized when the flowering time is plotted against the period length (Fig. 4). FKF1 decoys mostly cluster together with a small lengthening of period and delayed flowering. However, 78% of the ZTL decoy lines have lengthened periods near the average of 27.3 h (Fig. 2; Supplemental Table S1, blue symbols) and a small delay in flowering (Fig. 3; Supplemental Table S1), and 95% of the LKP2 decoy lines have lengthened periods near the average of 25.3 h (Fig. 2; Supplemental Table S1, green symbols) and a small delay in flowering (Fig. 3; Supplemental Table S1). These represent the majority populations for each of the decoys. The remaining individuals from the ZTL and LKP2 decoy populations have wild-type period lengths but highly delayed flowering (best seen in Fig. 4 and Supplemental Table S1).
Figure 4.
ZTL and LKP2 decoy transgenic lines fall into separable populations based on flowering and clock phenotypes. Period length data from Figure 2A were plotted against flowering time data from Figure 3A for each individual transgenic line. Seedlings were germinated on one-half-strength Murashige and Skoog (1/2 MS) agar plates under 12-h-light/12-h-dark conditions, transferred to LL conditions for circadian imaging analysis, and then transferred to soil for flowering time analysis.
The ztl mutants have a lengthened period similar to the majority population of the decoy lines, suggesting that this is a dominant-negative effect. The lkp2 knockout has no effect on clock period (Baudry et al., 2010), and LKP2 overexpression results in arrhythmicity (Schultz et al., 2001). Interestingly, expression of the LKP2 decoy never caused arrhythmicity in our studies (Supplemental Fig. S1), suggesting that the arrhythmicity caused by the overexpression of full-length LKP2 relies on the presence of the F-box domain. The majority populations of the LKP2 or ZTL decoy lines also have a small delay in flowering time of 1.5 or 2.2 d, respectively (Fig. 3; Supplemental Table S1, green and blue symbols). This is opposite to the weak early-flowering effect seen in an lkp2 ztl double knockout (Takase et al., 2011) but similar to the delayed flowering caused by LKP2 or ZTL overexpression (Schultz et al., 2001). This suggests that the role that LKP2 and ZTL play in flowering is not dependent on the F-box domain and that the decoys have a dominant-positive effect in the regulation of flowering.
The minority populations of the ZTL and LKP2 decoy lines have no effect on clock period and a dramatic delay in flowering time (Figs. 3 and 4). These results are consistent with other full-length overexpression studies showing that ZTL or LKP2 overexpression can cause delayed flowering and suggest a dominant-positive effect by the decoys (Kiyosue and Wada, 2000; Schultz et al., 2001; Somers et al., 2004; Kim et al., 2013). In one study, it was also shown that overexpressing ZTL without the Kelch repeat domain will sometimes cause delayed flowering with only minor effects on the clock period (Kim et al., 2013).
These results suggest that the ZTL, FKF1, and LKP2 decoys regulate clock function in a similar manner, albeit with different effectiveness. Conversely, FKF1 regulates flowering in a ubiquitylation-dependent manner, while ZTL and LKP2 regulate flowering in a ubiquitylation-independent manner, a clear divergence in function.
To determine if differences in decoy transgene expression or the expression of endogenous ZTL, FKF1, or LKP2 result in the observed clock and flowering phenotypes, we performed reverse transcription quantitative PCR (RT-qPCR; Supplemental Fig. S2) on a series of decoy transgenic lines. We selected four transgenic lines from each genotype that exhibited the majority phenotype (Supplemental Fig. S2, A and B). The relative expression of the transgenes in these lines was variable, but the phenotypes of the lines are nearly identical. Furthermore, the expression of any decoy results in lower levels of the concomitant endogenous gene, possibly through cosuppression. However, the amount of cosuppression varies while the phenotypes are nearly identical (i.e. ZTL decoy lines 1 and 4 in Supplemental Fig. S2D, LKP2 decoy lines 1 and 3 in Supplemental Fig. S2E, and FKF1 decoy lines 2 and 4 in Supplemental Fig. S2F).
Additionally, a decoy transgene can increase or decrease the expression of the two homologous genes with little effect on phenotype. For instance, expressing the LKP2 decoy can decrease the mRNA expression of ZTL (Supplemental Fig. S2D, LKP2 decoy line 3), but this does not result in a longer period than a line without reduction in ZTL expression (Supplemental Fig. S2D, LKP2 decoy line 1). Furthermore, LKP2 and ZTL decoy-expressing lines can result in higher expression of FKF1 (Supplemental Fig. S2F) but show delayed flowering (Supplemental Fig. S2B), more similar to the fkf1 loss-of-function mutant. These results strengthen the idea that the decoy transgenes act in a dominant fashion irrespective of the expression of the endogenous genes.
We also tested the expression of the transgene and endogenous genes in the late-flowering minority subpopulations of ZTL and LKP2 decoy transgenic lines. Interestingly, both of the ZTL decoy lines and one of the LKP2 decoy lines had the highest expression of the transgene in all tested lines (Supplemental Fig. S2C), suggesting that expression of the ZTL or LKP2 transgene at high levels can lead to late flowering. Furthermore, the expression of FKF1 in one of the ZTL decoy lines and both of the LKP2 decoy lines was higher, suggesting that the late-flowering effect is not due to the suppression of FKF1 expression but rather to high levels of the transgene.
In sum, these results suggest that expression of the decoy is sufficient to cause alterations in phenotype, but if a transgene expression threshold is crossed, variant phenotypes may occur. Furthermore, our data indicate that the expression of the endogenous genes plays little role in the observed phenotypes, as expected when expressing a genetic dominant negative.
Identification of ZTL, LKP2, and FKF1 Interacting Partners
It can be challenging to detect the interactions between full-length F-box proteins and substrates in vivo (for review, see Iconomou and Saunders, 2016). We hypothesize, based on our genetic evidence, that our decoy system traps substrate interactions of ZTL, FKF1, and LKP2 by preventing the substrates from being degraded. Thus, we performed IP-MS on the ZTL, LKP2, and FKF1 decoy transgenic lines using the affinity tag. As FKF1 is known to be involved in flowering, we chose to select flowering inductive conditions for our IP-MS analysis. Additionally, as we hoped to observe the known interactors GI and the CDFs, we selected a time point that was between their daily peaks of expression. We were interested predominantly in the role of ZTL in the circadian clock, so we used day-neutral conditions for this line, but in order to promote comparison between the decoy constructs, we selected the same time point. Thus, tissue was harvested 2 h after dusk in long-day (16 h of light/8 h of dark) conditions for the FKF1 and LKP2 decoy lines and 6 h after dusk in LD (12 h of light/12 h of dark) conditions for the ZTL decoy lines (zeitgeber time 18 [ZT18] in all three cases). For all IP-MS experiments, we used the wild-type parental Arabidopsis (Columbia-0 [Col-0] or Col-0 containing a CCA1p::Luciferase construct) and a 35S::His-FLAG-GFP transgenic line as controls. The wild type acts as a control for proteins that interact with the column, and the 35S::His-FLAG-GFP transgenic line controls for proteins that interact with the affinity tags. After generating a protein list from the mass spectrometry data, we first removed any nonspecific interactors found only in the control samples (Supplemental Table S2). Next, we performed SAINTexpress analysis to identify statistically relevant interactors (Goldfarb et al., 2014; Teo et al., 2014). Proteins with a SAINT score greater than 0.5 were included for downstream analyses (Supplemental Table S3).
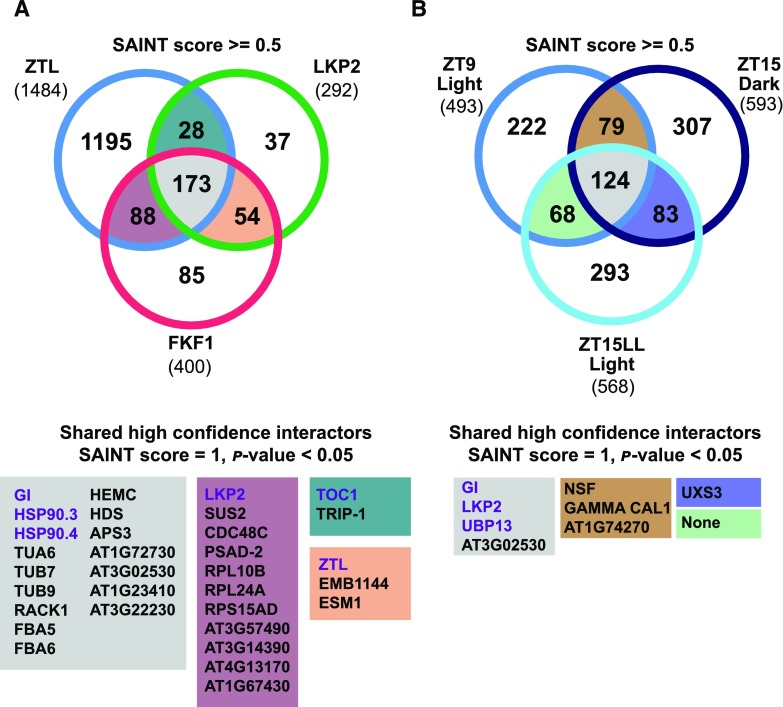
We successfully immunoprecipitated the decoy bait proteins, as demonstrated by the peptide counts of 1,412, 2,855, and 439 for LKP2, FKF1, and ZTL, respectively (Table I; Supplemental Table S3). Subsequently, we compared the common interacting proteins from the three IP-MS experiments (Fig. 5A; Table I; Supplemental Table S4). We found 173 common interactors (SAINT score ≥ 0.5) among the three IP-MS experiments, 16 of which are highly confident, with SAINT score = 1 and P < 0.05 (Fig. 5A; Supplemental Table S4, highlighted in gray). Among these interactors, the top-ranked proteins were GI, HSP90.3, and HSP90.4, which are known regulatory partners of ZTL and FKF1 (Kim et al., 2011; Song et al., 2014; Cha et al., 2017). These results demonstrate the ability of the decoys to form biologically relevant protein complexes in planta.
Table I. Selected IP-MS results from LKP2, FKF1, and ZTL decoys.
At left, the LKP2 and FKF1 decoy immunoprecipitations were harvested at ZT18 under long-day conditions and from the ZTL decoy immunoprecipitation at ZT18 under 12-h-light/12-h-dark conditions. Corresponding full interactor lists are available in Supplemental Table S3. At right, IP-MS results are shown from ZTL decoy immunoprecipitations at 3 h before (ZT9) and after (ZT15) dusk under 12-h-light/12-h-dark diurnal conditions and at 3 h after subjective dusk in continuous light (ZT15-LL). Corresponding full interactor lists are available in Supplemental Table S5.
| Locus | Protein Name | Total Spectral Counts (SAINT Score, SAINT P Value)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LKP2 Decoy ZT18 (LD) | FKF1 Decoy ZT18 (LD) | ZTL Decoy ZT18 (LD) | Wild Typeb | 35S:: FLAG-His-GFPc | ZTL Decoy ZT9 (LD) | ZTL Decoy ZT15 (LD) | ZTL Decoy ZT15 (LL) | Wild Typeb | 35S:: FLAG-His-GFPc | ||
| Bait | |||||||||||
| AT2G18915 | LKP2 | 1,412 | 29 (1*) | 82 (1**) | 0 | 0 | 75 (1**) | 70 (1**) | 74 (1**) | 0 | 0 |
| AT1G68050 | FKF1 | 0 | 2,855 | 0 | 128 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT5G57360 | ZTL | 34 (1*) | 108 (1**) | 439 | 0 | 0 | 466 | 565 | 503 | 0 | 10 |
| Regulatory partners | |||||||||||
| AT1G22770 | GI | 129 (1**) | 265 (1**) | 218 (1**) | 0 | 0 | 250 (1**) | 300 (1**) | 387 (1**) | 0 | 0 |
| AT5G56010 | HSP90.3 | 221 (1**) | 314 (1**) | 466 (1**) | 0 | 0 | 299 (0.5) | 357 (0.5) | 368 (0.5) | 0 | 0 |
| AT5G56000 | HSP90.4 | 192 (1*) | 266 (1*) | 380 (1**) | 6 | 3 | 11 (0.07) | 79 (0.5) | 6 (0) | 18 | 0 |
| Transcription factors | |||||||||||
| AT5G61380 | TOC1 | 74 (1**) | 0 | 28 (1*) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT5G24470 | PRR5 | 55 (1**) | 0 | 0 | 0 | 0 | 0 | 0 | 9 (0.5) | 0 | 0 |
| AT5G39660 | CDF2 | 0 | 6 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT1G72010 | TCP22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 41 (0.5) | 0 | 0 |
| AT5G08330 | CHE (TCP21) | 0 | 0 | 0 | 0 | 0 | 0 | 25 (0.5) | 17 (0.5) | 0 | 0 |
| AT5G52660 | RVE6 | 6 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Transcription regulators | |||||||||||
| AT3G26640 | LWD2 | 0 | 0 | 0 | 0 | 0 | 26 (0.5) | 37 (0.5) | 34 (0.5) | 0 | 0 |
| AT3G20810 | JMJD5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 (0.5) | 0 | 0 |
| AT1G15750 | TPL | 0 | 11 (0.98) | 0 | 9 | 0 | 62 (1**) | 0 | 8 (0.5) | 0 | 0 |
| AT2G19520 | FVE (MSI4) | 0 | 5 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Deubiquitinating enzymes | |||||||||||
| AT5G06600 | UBP12 | 97 (1) | 23 (0) | 162 (1) | 56 | 0 | 156 (0.98*) | 240 (1*) | 193 (0.99*) | 25 | 22 |
| AT3G11910 | UBP13 | 48 (0.99) | 0 | 136 (1) | 34 | 0 | 89 (1*) | 112 (1*) | 89 (1*) | 0 | 0 |
The SAINT scores represent the confidence of protein interactions. The score is from 1 to 0, in which 1 suggests a higher confidence interaction. The SAINT P values are represented by asterisks (*, P ≤ 0.05 and **, P ≤ 0.01). bThe spectral counts of CCA1p:Luciferase and Col-0 were summed to generate the total spectral counts in the Wild Type column. cThe spectral counts from two biological replicates of 35S::FLAG-His-GFP were summed to generate the total spectral counts in the 35S::FLAG-His-GFP column.
Figure 5.
Common interacting proteins from IP-MS experiments. Common interactors were identified from the FKF1, LKP2, and ZTL decoy IP-MS (A) and ZTL time-course IP-MS (B). A, The FKF1 and LKP2 decoy samples were harvested at ZT18 under 16 h of light/8 h of dark, and the ZTL decoy sample was harvested at zeitgeber time 18 (ZT18) under 12-h-light/12-h-dark conditions. B, The samples for the ZTL decoy time-course immunoprecipitation were harvested at 3 h before (ZT9) and after (ZT15) dusk under 12-h-light/12-h-dark diurnal conditions and at 3 h after subjective dusk in continuous light (ZT15-LL). Numbers of interacting proteins (SAINT score ≥ 0.5) are presented in the Venn diagrams. The full list of interacting proteins is provided in Supplemental Tables S3 to S6. The highly confident common interactors (SAINT score = 1, P < 0.05) are listed in the matching colored boxes below the Venn diagrams. Known clock regulators are in purple font.
Next, we compared the common interactors in pairwise combinations (Fig. 5A; Supplemental Table S4). There were 28 common interactors between ZTL/LKP2, 54 between LKP2/FKF1, and 88 between ZTL/FKF1 (SAINT score ≥ 0.5). We highlight highly confident common interactors (SAINT score = 1, P < 0.05; Fig. 5A). Interestingly, ZTL and LKP2 both interact with TOC1, a validated target of ZTL and LKP2 (Fig. 5A; Supplemental Table S4, highlighted in teal). This result strengthens the idea that the decoys stabilize target interactions, as expected. Two deubiquitinating enzymes, UBIQUITIN-SPECIFIC PROTEASE12 (UBP12) and UBP13, were identified as common interactors of ZTL and LKP2, with SAINT scores ≥ 0.97 (Supplemental Tables S3 and S4). Mutations in UBP12 or UBP13 cause short-period phenotypes, which is opposite to the long-period phenotype of a ztl mutant (Cui et al., 2013). This may suggest a role for these proteins in ZTL and LKP2 function. We also observed heterodimerization between ZTL, FKF1, and LKP2 (Fig. 5A, highlighted in mauve and peach; Table I; Supplemental Table S4). This result is consistent with previous reports of dimerization demonstrated in yeast and protoplast systems (Yasuhara et al., 2004; Takase et al., 2011). Further validation of the functional significance of these interactions may reveal previously unknown regulatory mechanisms.
We next examined unique interactors for LKP2 and FKF1 with known functions in the clock or flowering time (Fig. 5A; Table I; Supplemental Table S3). In the LKP2 decoy IP-MS, we identified REVELLE6 (RVE6; SAINT score = 1, P = 0.17; Supplemental Table S3), a known regulator of clock function (Hsu et al., 2013; Gray et al., 2017), and PRR5, a well-validated target. PRR5 was missing from the ZTL IP-MS experiment at ZT18, likely because the light conditions were different between the two experiments. The FKF1 decoy shows a divergence in clock function from ZTL and LKP2 (Fig. 2), and in concordance, we did not see interaction with the well-established clock transcription factor targets TOC1 and PRR5. Interestingly, the FKF1 decoy has a strong effect on flowering (Fig. 3) and interacts with a known target involved in flowering, CDF2 (SAINT score = 0.5, P = 0.17; Table I; Supplemental Table S3). In addition, two proteins also involved in flowering time, TOPLESS (TPL; SAINT score = 0.98, P = 0.22; Graeff et al., 2016; Goralogia et al., 2017) and MULTICOPY SUPPRESSOR OF IRA1 4 (FVE/MSI4; SAINT score = 1, P = 0.18; Ausín et al., 2004), interact with the FKF1 decoy but not with the ZTL and LKP2 decoys (Table I; Supplemental Table S3). These results indicate that FKF1 has diverged from ZTL and LKP2 in its genetic and biochemical functions.
To identify a more comprehensive list of ZTL interacting proteins, we performed time-course IP-MS in LD and LL conditions with the ZTL decoy. We entrained T1 ZTL decoy transgenic lines in 12-h-light/12-h-dark growth conditions. We collected tissue in the light (3 h before dusk, ZT9) and dark (3 h after dusk, ZT15), then transferred the plants to LL for 24 h and collected tissue in the first subjective dark period (3 h after subjective dusk, ZT15 in LL conditions [ZT15-LL]). We identified 493, 593, and 568 potential interactors (SAINT score ≥ 0.5) in the ZT9, ZT15, and ZT15-LL IP-MS data sets, respectively (Fig. 5B; Table I; Supplemental Tables S5 and S6). As with the single-time-point IP-MS experiments described earlier, the well-known interactor GI was found in all three conditions (SAINT score = 1, P < 0.01; Fig. 5B; Table I; Supplemental Table S6). In the ZT15-LL IP-MS, we also identified PRR5 (SAINT score = 0.5, P = 0.14). The identification of PRR5 here, along with TOC1 as observed in the ZT18 time point discussed previously, further validates the ability of the ZTL decoy to interact with well-established targets.
The activity and interacting partners of ZTL are regulated by photocycles through conformational changes of the LOV domain between light and dark states (Kim et al., 2007; Pudasaini and Zoltowski, 2013; Zoltowski and Imaizumi, 2014; Pudasaini et al., 2017). We intended to probe the dynamic association of these potential clock regulators with ZTL through comparison of the interactors identified from each condition (Fig. 5B; Supplemental Tables S5 and S6). However, the spectral count ratios of ZTL to GI are similar in all three conditions (466:250 = 1.86 for ZT9, 565:300 = 1.88 for ZT15, and 503:387 = 1.3 for ZT15-LL; Table I), suggesting that the dark-induced dissociation of GI does not occur as expected. Contrary to this, TOC1 and PRR5 only associate with ZTL at later time points, suggesting that some associations remain dynamic with the decoy ZTL. Among 124 common interactors at all three time points, three clock regulators, GI, LKP2, and UBP13, were in the highly confident list (SAINT score = 1, P < 0.05; Fig. 5B; Table I; Supplemental Tables S5 and S6). With a lower stringency cutoff, we found the clock regulators UBP12 (SAINT score ≥ 0.98, P < 0.05) and LIGHT REGULATED WD2 (LWD2; SAINT score = 0.5; Wu et al., 2008, 2016; Wang et al., 2011; Cui et al., 2013). When comparing the IP-MS samples from each time point pairwise, we found few highly confident interacting proteins (SAINT score = 1, P < 0.01; Fig. 5B; Table I; Supplemental Table S6).
We next filtered our list for lower confidence but statistically significant interactors that are known to be involved in clock function. In the late-day experiments (ZT15 and ZT15-LL), we identified CHE/TCP21 (SAINT score = 0.5; Table I; Supplemental Tables S5 and S6), a known clock regulator that interacts with TOC1 (Pruneda-Paz et al., 2009). We also identified JUMONJI DOMAIN CONTAINING5 (JMJD5; SAINT score = 0.5) at ZT15 and TCP22 (SAINT score = 0.5) at ZT15-LL, two additional regulators of clock function (Jones et al., 2010; Jones and Harmer, 2011; Wu et al., 2016).
While we were successful at identifying some of the previously reported targets of ZTL, we did not identify all of the published target proteins (Jarillo et al., 2001; Yasuhara et al., 2004; Fukamatsu et al., 2005; Baudry et al., 2010; Johansson et al., 2011; Takase et al., 2011; Liu et al., 2013; Song et al., 2014; Zhang et al., 2015; Norén et al., 2016; Supplemental Table S7). This is likely due to the age of the tissue, collection times, or tissue types used to perform the IP-MS.
ZTL Interacts with and Ubiquitylates CHE
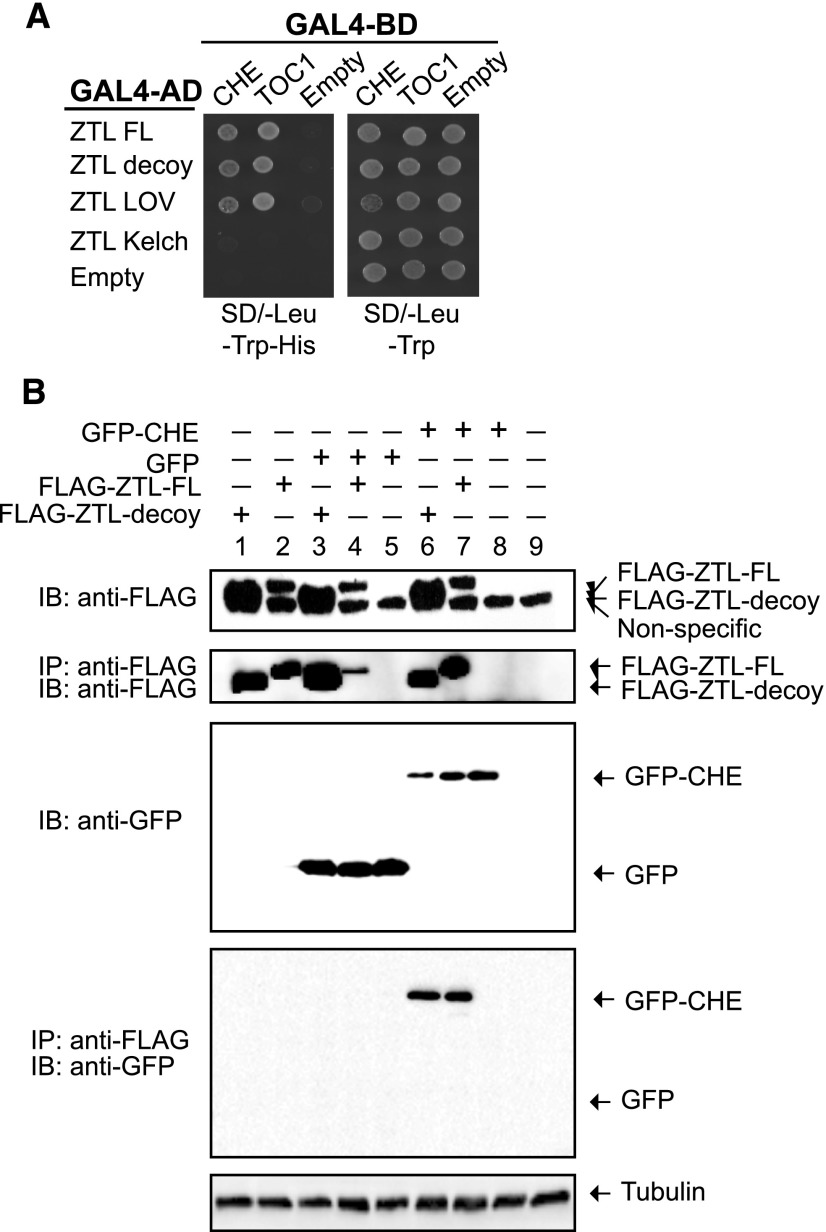
We chose to perform follow-up experiments with CHE because it interacts with TOC1 and regulates CCA1 expression (Pruneda-Paz et al., 2009). As substrate proteins often interact directly with the substrate recognition domain of an E3 ligase, we tested whether CHE interacts directly with ZTL using yeast two-hybrid assays and coimmunoprecipitation in mammalian tissue culture cells. In yeast, CHE interacts directly with the full-length and decoy ZTL (Fig. 6A). Moreover, CHE interacts with the LOV domain of ZTL, similar to the known ZTL target, TOC1 (Fig. 6A; Más et al., 2003).
Figure 6.
ZTL interacts directly with CHE. A, Yeast two-hybrid assays between ZTL and CHE or TOC1. FL, Full length; decoy, translationally fused LOV and Kelch repeat domains; LOV, LOV domain only; Kelch, Kelch repeat domain only. B, Coimmunoprecipitation experiments between CHE and ZTL performed in HEK293T cells. GFP-CHE or GFP alone was coexpressed with full-length or decoy ZTL translationally fused to a FLAG affinity tag. Immunoprecipitation (IP) was performed with the FLAG tag. IB, Immunoblot.
Next, we coexpressed CHE and either full-length or decoy ZTL in HEK293T cells. Figure 6B shows that FLAG-tagged full-length ZTL, FLAG-tagged ZTL decoy, and GFP-tagged CHE can be expressed in HEK293T cells. We immunoprecipitated ZTL using an anti-FLAG antibody and then performed immunoblot analysis with anti-GFP to determine if CHE interacts with ZTL. We detected CHE interaction with the full-length and decoy ZTL (Fig. 6B, lanes 6 and 7), but no interaction occurred in the GFP-only control (Fig. 6B, lanes 3–5) or when CHE-GFP was expressed alone (Fig. 6B, lane 8). This supports the hypothesis that CHE can interact with ZTL in the absence of additional plant proteins, suggesting that the interaction is direct.
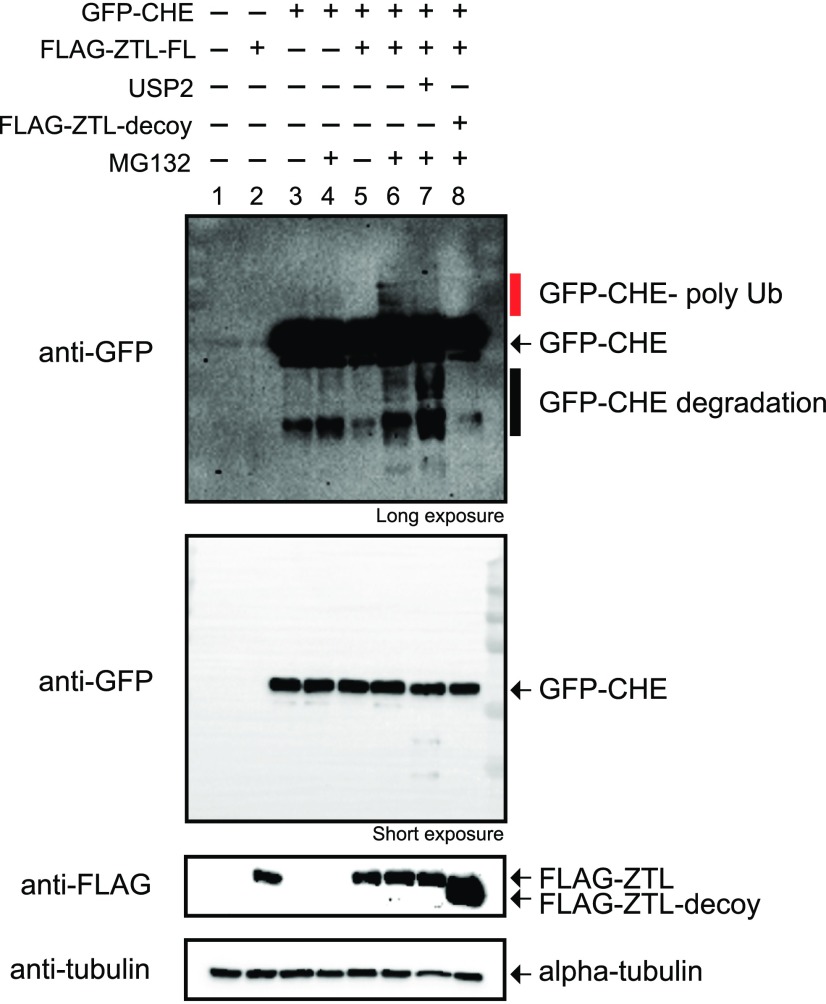
Ubiquitylation studies are often technically difficult and time consuming to perform and interpret in plant model systems. In order to determine whether ZTL mediates the ubiquitylation of CHE, we used mammalian tissue culture cells as a heterologous ubiquitylation system. Mammalian tissue culture has three distinct advantages for testing the relationship between plant F-box proteins and potential targets: (1) the mammalian UPS supports ubiquitylation by plant F-box proteins; (2) redundant plant proteins do not complicate the interpretation of results; and (3) there are a wealth of reagents available for protein expression and ubiquitylation assays in mammalian cells. We coexpressed GFP-CHE and FLAG-ZTL in the presence or absence of the proteasome inhibitor MG132 (Fig. 7). When full-length ZTL is coexpressed with CHE in the presence of a proteasome inhibitor, a higher Mr laddering of the CHE protein occurs (Fig. 7, lane 6). This laddering diminishes upon addition of the catalytic domain of a general deubiquitylating enzyme, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE2 (USP2cc), to the lysate (Fig. 7, lane 7), demonstrating that these bands are ubiquitylated forms of CHE. Furthermore, the ZTL-dependent laddering of CHE is blocked by the ZTL decoy (Fig. 7, lane 8), demonstrating that our decoy inhibits the function of the full-length protein, as predicted.
Figure 7.
ZTL promotes CHE ubiquitylation. GFP-CHE was coexpressed with full-length (FL) or decoy ZTL in HEK293T cells. They were expressed in the presence or absence of 30 µm MG132 or 2 µg of USP2cc. The red bar denotes polyubiquitylated forms of CHE.
CHE Is Less Stable in the Dark
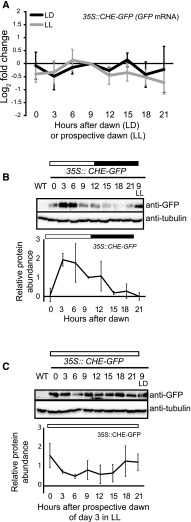
Based on our interaction and ubiquitylation studies, we hypothesized that CHE protein would be destabilized in the dark when ZTL was actively degrading target proteins (Más et al., 2003; Kiba et al., 2007). This would result in the cycling of CHE protein levels in LD conditions even in the absence of transcriptional changes. To test this, we constitutively expressed CHE-GFP under the control of a 35S promoter (35S::CHE-GFP) in Arabidopsis (Fig. 8A) and performed time-course immunoblotting in LD and LL conditions (Fig. 8, B and C). In order to cross-compare protein levels from the two time-course experiments, we loaded the sample from 9 h after lights on (LD) or subjective lights on in LL conditions in the immunoblots of the other time course (Fig. 8, B and C, lanes LL or LD). In LD conditions, CHE protein levels cycled robustly, peaking in the light (Fig. 8B). In LL, CHE protein levels did not show robust 24-h cycles but remained at a constant high level (Fig. 8C). This demonstrates that CHE protein levels are controlled by light cycles, which also control ZTL activity. This suggests that, although CHE mRNA expression is controlled by the circadian clock (Pruneda-Paz et al., 2009), CHE protein stability is controlled by LD cycles. This was shown previously for PRR5, where extending the length of the day decoupled the phase of PRR5 protein from the phase of mRNA expression (Kiba et al., 2007).
Figure 8.
CHE protein cycles over daily time courses. A, CHE mRNA expression was measured using RT-qPCR in a 35S::CHE-GFP transgenic line in LD (12 h of light/12 h of dark) and LL growth conditions. B and C, CHE protein levels were measured using immunoblotting in a 35S::CHE-GFP transgenic line in LD (B) and LL (C). All plants were grown in LD conditions for 12 d and then transferred to the aforementioned light conditions for 48 h prior to the start of the time course. Sampling began at dawn or prospective dawn of day 14. ZT9 samples are presented for cross-comparison of relative protein levels between LD and LL time courses. All quantifications are averages of three biological replicates, with error bars showing sd. WT, Wild type.
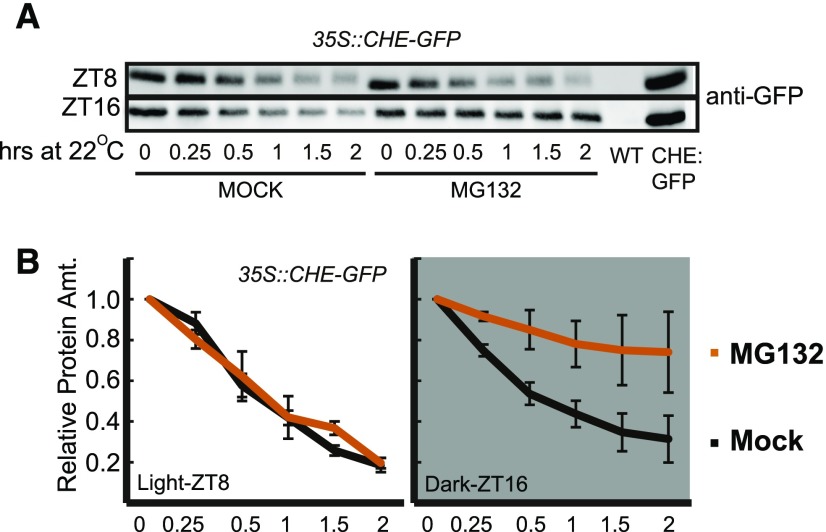
It is possible that CHE is stabilized in the light rather than destabilized in the dark. To determine whether CHE is degraded by the proteasome in the dark, we performed a cell-free degradation assay on 35S::CHE-GFP tissue collected 4 h before and after dusk (ZT8 and ZT16) in an LD (12 h of light/12 h of dark) time course. We separated the tissue into a mock- and MG132-containing sample buffer and measured protein levels at various time points over 2 h via immunoblotting (Fig. 9). MG132 stabilized CHE-GFP in the ZT16 samples but not in the ZT8 samples, demonstrating that the ubiquitin proteasome system acts on CHE stability at night during ZTL’s peak activity. This is similar to what has been demonstrated for known ZTL targets, such as TOC1 (Más et al., 2003).
Figure 9.
CHE protein stability is controlled by the proteasome. A cell-free degradation assay was performed on 35S::CHE-GFP grown in LD cycles. Samples were collected at ZT8 (light) or ZT16 (dark), and protein was extracted at 4°C and split into tubes at room temperature for mock treatment or treatment with 1 µm MG132 for the indicated times. Samples were normalized to time 0 for quantification. Error bars represent sd of three biological replicates. WT, Wild type.
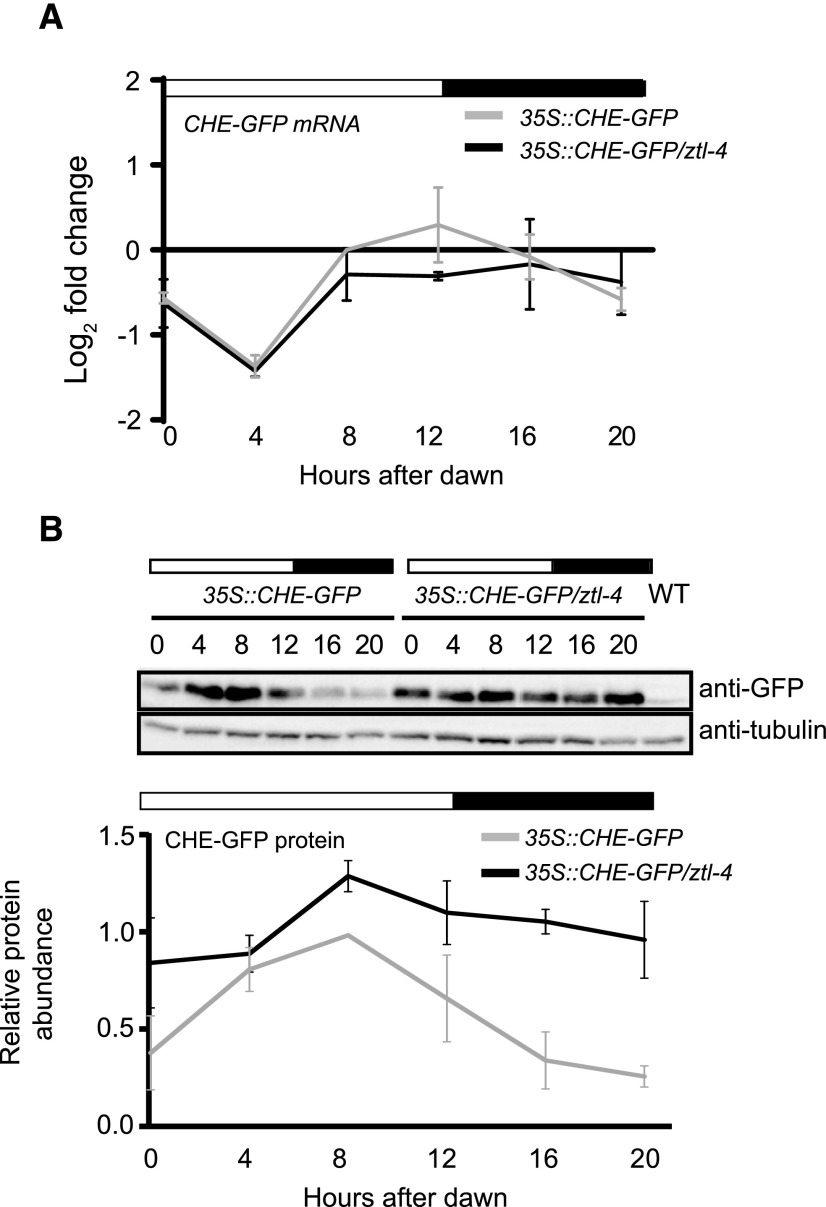
ZTL Controls the Stability of CHE Protein
To determine if ZTL is responsible for the cycling levels of CHE protein across the day, we crossed the 35S::CHE-GFP transgenic line into the ztl-4 mutant (Salomé and McClung, 2005). We used RT-qPCR to show that the levels of CHE-GFP mRNA were mostly constant across the day in the wild type and the ztl-4 mutant (Fig. 10A). We then used immunoblotting to measure CHE-GFP protein (Fig. 10B). As expected for a ZTL target protein, the levels of CHE are higher in the ztl-4 mutant compared with the wild type. Most notably, the CHE-GFP protein levels remain high in the dark, with only a very slight reduction. Together with the interacting and ubiquitination assays, this evidence demonstrates that CHE is a bona fide target of ZTL.
Figure 10.
ZTL controls rhythmic CHE protein levels in diurnal conditions. A, The mRNA expression of 35S::CHE-GFP in the Col-0 or ztl-4 background in LD (12 h of light/12 h of dark) conditions was measured with RT-qPCR. B, The CHE-GFP protein from the same time points was detected with anti-GFP antibody. Immunoblots shown here are representative of three biological repeats. Quantifications are averages of three biological replicates, with error bars showing sd. WT, Wild type.
DISCUSSION
Decoy Genetic Studies
ZTL, LKP2, and FKF1 have a complicated genetic relationship demonstrated in the multiple studies exploring the effects of mutations and overexpression on flowering time and circadian clock function (Baudry et al., 2010; Ito et al., 2012). This study aimed to address their ubiquitylation-independent and -dependent roles by expressing the genes in the absence of the F-box domain, which is necessary to recruit activated ubiquitin for substrate ubiquitylation. The benefit of this approach is that the F-box-less decoys act dominantly to their endogenous counterparts, bypassing issues of functional redundancy (Iconomou and Saunders, 2016). The decoy experiment further demonstrates that FKF1 plays a specialized role in controlling flowering time that is distinct from ZTL or LKP2 (Figs. 2 and 3). This was confirmed in our interaction studies that showed that FKF1 alone interacts with CDF2 and other flowering proteins in vivo (Table I). Conversely, ZTL and LKP2 decoys have strong effects on circadian clock period and only mild effects on flowering time (Figs. 2 and 3). This is supported by interaction studies showing that ZTL and LKP2 interact with core clock transcription factors from the PRR/TOC1 family of transcriptional regulators but not with the CDFs (Table I).
Our results support the idea that ZTL, LKP2, and FKF1 have diverged into specialized roles in the circadian clock and flowering. LKP2 is only partially redundant with ZTL or FKF1 in Arabidopsis, but, intriguingly, Brassica rapa has three copies of LKP2 and no ZTL or FKF1 (Lou et al., 2012), suggesting that LKP2 is sufficient to drive seasonal flowering and maintain circadian period in other plant species. Our results suggest that LKP2 has the biochemical potential to play a role in regulating clock period length, and previous studies support the notion that it can function similar to ZTL but is restricted in function by its low expression level (Baudry et al., 2010). Our data also support the idea put forth that expression is not the only limiting factor in LKP2 function, as expressing the LKP2 decoy has a weaker effect on the circadian clock than the ZTL decoy, despite similar levels of expression. The detection of PRR5 and TOC1 in the LKP2 decoy IP-MS suggests that LKP2 has the potential to mediate the ubiquitylation of clock factors in planta and is consistent with previous results that LKP2 is involved in PRR5 turnover (Wang et al., 2010). We also detect endogenous LKP2 interacting with the ZTL decoy in IP-MS, showing that the endogenous protein is expressed despite predicted low relative mRNA expression.
CHE Is a Bona Fide Target of ZTL
Our decoy approach coupled to a series of validation experiments demonstrates that CHE is a previously unknown target of ZTL. CHE is a TCP-type transcription factor that interacts with TOC1 to control CCA1 expression, but it has no apparent sequence similarity with the PRRs (Pruneda-Paz et al., 2009). This suggests that ZTL can regulate diverse targets, and ZTL may be a master regulator of nighttime repressors of CCA1. The CHE protein is stabilized in the ztl-4 mutant (Fig. 10), providing genetic evidence that ZTL degrades CHE in a light-dependent manner in vivo in a fashion analogous to the previously known ZTL substrates, TOC1 and PRR5. As with other oscillatory biological processes, E3 ubiquitin ligases with multiple substrates can target them sequentially (Rape et al., 2006; Song and Rape, 2011; Mocciaro and Rape, 2012). It will be interesting to use our in vivo and heterologous expression systems to test the order of degradation of ZTL targets during the day. Furthermore, as with other ZTL substrates, it will be important to determine whether CHE can be ubiquitylated by LKP2 or FKF1.
Potential New Regulatory Mechanisms
The association of UBP12 and UBP13 with ZTL (Table I) is intriguing and requires further investigation. Biochemically, ZTL is an E3 ligase that targets substrates for ubiquitylation, whereas UBP12 and UBP13 are active deubiquitinating enzymes, cleaving ubiquitin from ubiquitylated proteins (Cui et al., 2013). Genetically, ztl-4 has a long-period clock phenotype, while the ubp12-2w mutant shows the opposite short-period phenotype. UBP12 and UBP13 also were found in a time-course IP-MS experiment with GI (Krahmer et al., 2017), although it is not clear if their presence is dependent on direct interaction with GI or through a secondary interaction with ZTL, FKF1, or LKP2. These findings suggest a plausible model in which UBP12 and UBP13 can counteract the ubiquitin ligase activity of ZTL to stabilize ZTL targets, such as TOC1, PRR5, and CHE.
In accordance with previous studies, we identified heterodimerization between ZTL, LKP2, and FKF1 in our IP-MS studies (Han et al., 2004; Yasuhara et al., 2004; Takase et al., 2011). Our work suggests that they may form higher order complexes in vivo, possibly to regulate target stability or to regulate their own stability. It is likely that at least part of the effect of the decoys is caused by disrupting endogenous homodimers or heterodimers between the three E3 ligases. Future studies will likely tease apart the functional significance of these putative dimerization events, and the decoy strategy provides a unique opportunity to study this in vivo.
Mammalian Tissue Culture for Plant Protein Complex Studies
We demonstrate the ability to reconstitute CHE ubiquitylation by ZTL in HEK293T cells, a mammalian tissue culture system (Fig. 7). One of the drawbacks of IP-MS approaches is the prevalence of false positives that cloud the interpretation of results. Mammalian tissue culture is a rapid eukaryotic protein expression system that can be used to demonstrate the ubiquitylation of a substrate by a plant F-box protein. This allows for rapid screening of potential F-box targets to quickly generate hypotheses that can then be tested in plant systems (as in Figs. 8–10), which are traditionally time consuming. This method also reduces the possibility of indirect interactions through interacting partners of the F-box protein or target of interest, which would be present in an in vivo assay. The combined decoy and heterologous ubiquitylation assays is a powerful method that can be used to understand the role of any E3 ubiquitin ligase from plants.
The Decoy Method
Three challenges inhibit progress in studying plant F-box proteins: genetic redundancy, transient interactions with substrate proteins, and validation of substrates. The decoy method presented here inverts the function of an E3 ubiquitin ligase, overcoming all three issues by stabilizing substrate proteins even in the presence of the endogenous F-box or functionally redundant F-box proteins. This creates a dominant-negative genetic effect that can be used to determine the functions of any F-box protein family, even in the presence of functional redundancy. This dominant-negative feature of our technique is especially useful in plant species and other species with genome duplications, where redundancy is prevalent. Furthermore, this platform presents a means for rapid identification of F-box substrate proteins using IP-MS and a subsequent method for the validation of substrate ubiquitylation by the assayed F-box protein.
The decoy method can be used for high-throughput reverse genetic screening of F-box-containing genes. The results presented here demonstrate the effectiveness of using the decoy approach to break apart redundancy within a gene family. There are additional benefits of this approach that make it amenable as a screening platform. First, nearly all F-box proteins have the F-box domain in the N terminus, providing a simple two-primer PCR strategy for high-throughput cloning of F-box decoys. Second, a screen can be done in the T1 generation because of the dominant genetic effects of decoys (Figs. 2–4; Supplemental Fig. S1). Additionally, all members of a family need not be identified, as would be the case in a mutant or knockout screen, as the dominant-negative effects overcome redundancy. Third, T1 lines of varying expression can be assayed for phenotypes, reducing the danger of expression artifacts. Furthermore, due to the dominant-negative nature of decoys, the expression of the transgene can vary but still have the same effect (Supplemental Fig. S2). Finally, genetic screening can be followed by rapid IP-MS experiments to determine the interacting proteins that may be stabilized by the decoy F-box. Such screening would be difficult and potentially fruitless in the case of a full-length overexpression, as the native forms of these proteins would cause lower substrate protein levels, inhibiting identification.
The decoy strategy employed here is a streamlined method that should be used in combination with knockout and overexpression studies to untangle the complex genetic and biochemical functions of E3 ubiquitin ligases. Our results illustrate the promise of this technique, which we believe can be applied effectively to many systems and can be particularly potent when studying genetically intractable plant species or experimental systems with high levels of genetic redundancy.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The decoy constructs for ZTL (AT5G57360), LKP2 (AT2G18915), and FKF1 (AT1G68050) were constructed by fusion of the LOV and Kelch domains using overlap extension PCR with the primers listed in Supplemental Table S8. The design, including amino acid numbers for ZTL, LKP2, and FKF1 decoys, is shown in Figure 1. The PCR products were cloned into pENTR/D-TOPO vectors (Invitrogen, catalog no. K240020). The decoys were then fused to FLAG and His tags at the N terminus and under the control of a cauliflower mosaic virus 35S promoter by recombination into the plant binary pDEST vector pB7-HFN (Huang et al., 2016a,b) using LR recombination. The decoy constructs were transformed into Arabidopsis (Arabidopsis thaliana) Col-0 expressing the circadian reporter CCA1p::Luciferase (Pruneda-Paz et al., 2009) or Col-0 by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101. The 35S::CHE-GFP transgenic lines were generated in the Col-0 background as described previously (Pruneda-Paz et al., 2009) and crossed into ztl-4 (SALK_012440; Salomé and McClung, 2005).
For the growth of Arabidopsis seedlings, Arabidopsis seeds were surface sterilized in 70% (v/v) ethanol and 0.01% (v/v) Triton X-100, sown on 1/2 MS plates (2.15 g L−1 Murashige and Skoog medium, pH 5.7 [Caisson Laboratories, catalog no. MSP01] and 0.8% [w/v] bacteriological agar [AmericanBio, catalog no. AB01185]), and stratified at 4°C for 2 d. Following stratification, seeds were grown at 22°C in a 12-h-light/12-h-dark cycle at a fluence rate of 130 μmol m−2 s−1, unless specified otherwise. For soil-grown Arabidopsis, Arabidopsis seedlings were geminated on 1/2 MS plates, and 14-d-old seedlings were transferred to soil (Fafard II) and grown at 22°C in a 16-h-light/8-h-dark cycle with a light fluence rate of 135 μmol m−2 s−1, unless specified otherwise.
Measurement of Circadian Rhythms and Flowering Time
The decoy and control CCA1p::Luciferase seeds were grown on 1/2 MS plates with or without 15 µg mL−1 ammonium glufosinate (Santa Cruz Biotechnology, catalog no. 77182-82-2). Seven-day-old T1 transgenic seedlings were arrayed on 1/2 MS in a 10 × 10 grid on a 100-mm square dish and then treated with 5 mm d-luciferin (Cayman Chemical, catalog no. 115144-35-9) dissolved in 0.01% (v/v) Triton X-100. Seedlings were imaged at 22°C under constant white light provided by two LED light panels (Heliospectra, model no. L1) with a light fluence rate of 21 μmol m−2 s−1. The imaging regime was as follows: each hour, lights were turned off for 2 min, then an image was collected using a 5-min exposure on an Andor iKon-M CCD camera; lights remained off for 1 min after the exposure was completed, and then lights returned to the normal lighting regime. The CCD camera was controlled using Micromanager (Edelstein et al., 2014) using the following settings: binning of 2, preamp gain of 2, and a 0.05-MHz readout mode. Using this setup, 400 seedlings were imaged simultaneously across four plates. Images were acquired each 1 h for approximately 6.5 d. Data collected between the first dawn of LL and the dawn of day 6 were used for analyses.
The mean intensity of each seedling at each time point was calculated using ImageJ (Schneider et al., 2012). The calculated values were imported into the Biological Rhythms Analysis Software System for analysis. The Fast Fourier Transform Nonlinear Least Squares algorithm (Moore et al., 2014) was used to calculate period and relative amplitude of the rhythms from each individual seedling.
Following circadian analysis, seedlings were transferred to soil and grown in inductive conditions: 22°C in a 16-h-light/8-h-dark cycle with a light fluence rate of 135 μmol m−2 s−1. Plants were monitored daily for flowering status, and the dates when each individual reached 1-cm inflorescence height, 10-cm inflorescence height, and showed the first open flower bud were recorded. Additionally, the leaf number for each plant was counted when the inflorescence reached 1 cm.
The entire experiment was performed three times with similar results. The data presented are from one representative experiment.
Quantification of mRNA Expression in Decoy Transgenic Lines
To determine the relative mRNA expression levels of decoy transgenic lines, leaf tissue from T1 decoy lines was harvested at ZT14 directly following the completion of flowering time experiments (after the inflorescence stem reached 10 cm). ZT14 was chosen because it is likely that, at this time, all genes would be expressed to measurable levels. The tissue was frozen and ground in liquid nitrogen. Total RNA was extracted using the RNeasy Plant Mini Kit and treated with RNase-Free DNase (Qiagen, catalog nos. 74904 and 79254) following the manufacturer’s protocols. cDNA was prepared from 100 ng of total RNA using iScript Reverse Transcription Supermix (Bio-Rad, catalog no. 1708841), diluted 10-fold, and then used directly as the template for the PCR. RT-qPCR was performed with 4 µL of diluted cDNA and 500 nm primers listed in Supplemental Table S8 using iTaq Universal SYBR Green Supermix (Bio-Rad, catalog no. 1725121) with the CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The RT-qPCR started with a denaturation step at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 10 s, primer annealing at 55°C for 10 s, and primer extension at 72°C for 30 s. ISOPENTENYL PYROPHOSPHATE:DIMETHYLALLYL PYROPHOSPHATE ISOMERASE2 (IPP2; AT3G02780) was used as an internal control. Relative expression represents means of log2(2-ΔCT) from three technical replicates, in which ΔCT = (CT of gene of interest − CT of IPP2).
IP-MS Analysis
For the IP-MS of the ZTL decoy (ZT9, ZT15, ZT18, and ZT15-LL), approximately 20 T1 ZTL decoy transgenic plants (Col-0 background) along with Col-0 and 35S::His-FLAG-GFP controls were grown on 1/2 MS plates with or without 15 mg mL−1 ammonium glufosinate (Santa Cruz Biotechnology). Seven-day-old T1 transgenic lines were transferred to soil and grown under 16-h-light/8-h-dark conditions at 22°C for 3 weeks. Prior to harvest, plants were entrained in 12 h of light/12 h of dark with a light fluence rate of 165 μmol m−2 s−1 at 22°C for 1 week. Samples were harvested at 6 h after dusk (ZT18), 3 h prior to dusk (ZT9; collected in light), and 3 h after dusk (ZT15; collected in dark) in LD (12 h of light/12 h of dark) conditions. At dawn (ZT0), the plants were transferred to continuous light conditions, and leaf tissue was harvested at 3 h after subjective dusk (ZT15 in LL conditions). One or two leaves were harvested from each individual plant, and approximately 20 mature leaves from the ZTL decoy lines were harvested at each time point to compare the interacting dynamics of ZTL. For the controls, an equal number of leaves were harvested from the two control lines at four time points under appropriate light conditions and combined prior to further processing.
Tissue samples were ground in liquid nitrogen using the Mixer Mill MM400 system (Retsch). The immunoprecipitation was done as described previously (Huang et al., 2016a,b) with modifications for one-step immunoprecipitation. Briefly, protein from 2 mL of tissue powder was extracted with sonication in SII buffer (100 mm sodium phosphate, pH 8, 150 mm NaCl, 5 mm EDTA, and 0.1% [v/v] Triton X-100) with cOmplete EDTA-free Protease Inhibitor Cocktail (Roche, catalog no. 11873580001), 1 mm phenylmethylsulfonyl fluoride, and a PhosSTOP tablet (Roche, catalog no. 04906845001). The anti-FLAG antibodies were cross-linked to Dynabeads M-270 Epoxy (Thermo Fisher Scientific, catalog no. 14311D) for immunoprecipitation. Immunoprecipitation was performed by incubation with beads at 4°C for 2 h on a tube rocker. The beads were then washed with SII buffer three times, 25 mm ammonium bicarbonate three times, and then 10 mm ammonium bicarbonate twice before being subjected to trypsin digestion (0.5 µg; Promega, catalog no. V5113) at 37°C overnight. We vacuum dried the digested peptides using a SpeedVac and then dissolved them in 5% (v/v) formic acid/0.1% (v/v) trifluoroacetic acid. The protein concentration was determined by Nanodrop measurement (A260/A280; Thermo Scientific Nanodrop 2000 UV-Vis Spectrophotometer). An aliquot of each sample was then further diluted with 0.1% (v/v) trifluoroacetic acid to 0.1 µg µL−1. A total of 0.5 µg (5 µL) was injected for liquid chromatography tandem-mass spectrometry (LC-MS/MS) analysis at the Keck MS & Proteomics Resource Laboratory at Yale University.
LC-MS/MS analysis was performed on a Thermo Scientific Orbitrap Elite mass spectrometer equipped with a Waters nanoAcquity ultra-performance liquid chromatography system utilizing a binary solvent system (buffer A, 0.1) (v/v) formic acid; buffer B, 0.1% (v/v) formic acid in acetonitrile). Trapping was performed at 5 µL min−1, 97% (v/v) buffer A for 3 min using a Waters Symmetry C18 180-µm × 20-mm trap column. Peptides were separated using an ACQUITY UPLC PST (BEH) C18 nanoACQUITY Column, 1.7 µm, 75 µm × 250 mm (37°C), and eluted at 300 nL min−1 with the following gradient: 3% (v/v) buffer B at initial conditions; 5% (v/v) B at 3 min; 35% (v/v) B at 140 min; 50% (v/v) B at 155 min; 85% (v/v) B at 160 to 165 min; then returned to initial conditions at 166 min. Mass spectra were acquired in the Orbitrap in profile mode over the 300 to 1,700 m/z range using one microscan, 30,000 resolution, AGC target of 1E6, and a full maximum ion time of 50 ms. Up to 15 MS/MS scans were collected per MS scan using collision-induced dissociation on species with an intensity threshold of 5,000 and charge states 2 and above. Data-dependent MS/MS scans were acquired in centroid mode in the ion trap using one microscan, AGC target of 2E4, full maximum ion time of 100 ms, 2 m/z isolation window, and normalized collision energy of 35. Dynamic exclusion was enabled with a repeat count of one, repeat duration of 30 s, exclusion list size of 500, and exclusion duration of 60 s.
After using the Mascot Distiller program to generate Mascot-compatible files, the MS/MS spectra were searched in house using the Mascot algorithm (version 2.4.0; Perkins et al., 1999). The data were searched against the Protein SwissProt_2016_05.fasta Arabidopsis database and the significance threshold P < 0.5, allowing for oxidation (M), phosphorylation (STY), and ubiquitination (diGLY-K) as variable modifications. Peptide mass tolerance was set to 10 ppm, the MS/MS fragment tolerance was set to 0.5 D, and the maximum number of missed cleavages by trypsin was set to two. Normal and decoy database searches were run to determine the false discovery rates. The confidence level was set to 95%. The raw data files and the mapped peptide information will be deposited to proteomexchange (http://www.proteomexchange.org/submission/index.html). As for the complexity of the protein identified by IP-MS, we filtered the protein list by first removing the proteins present only in the parental lines or GFP controls. Then, we applied SAINTexpress analysis (Goldfarb et al., 2014) to evaluate the confidence of interactors.
We repeated the ZTL decoy time-course experiments with long-period ZTL decoy T1 lines in the CCA1p::Luciferase background along with CCA1p::Luciferase and 35S::His-FLAG-GFP controls. Briefly, 7-d-old T1 ZTL decoy seedlings selected on 1/2 MS plates were transferred to new 1/2 MS plates without selection for circadian period analysis, as described above. Then, ZTL decoy plants with lengthened circadian periods (approximately 28 h) were grown and entrained under the same condition as described above and harvested at ZT9, ZT15, and ZT15-LL. The control samples were harvested at the same time points and pooled before immunoprecipitation. The sample processing, immunoprecipitation, mass spectrometry, and data analyses were done as described above (Huang et al., 2016a,b).
For the IP-MS experiments of the FKF1 and LKP2 decoys, T2 decoy transgenic lines along with parental CCA1p::Luciferase lines and 35S::His-FLAG-GFP plants were grown on soil in inductive conditions (16 h of light/8 h of dark at 22°C) for 3 weeks prior to collection at 2 h after dusk in long-day conditions (ZT18). Approximately 20 individual rosettes were collected for each genotype. The immunoprecipitation was done as described previously (Huang et al., 2016a,b). The immunoprecipitated proteins were digested with trypsin, and the samples were analyzed by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific) as described previously (Perkins et al., 1999; Gendron et al., 2016). The mass spectrometry data were processed as described above, except that the search parameters were optimized to the mass tolerance of fragment ions with 50 ppm for precursor ions and 0.01 D for fragments to fit the sensitivity of the Q-Exactive mass spectrometer.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed according to the Yeast Protocol Handbook (Clontech, catalog no. P3024). Briefly, CHE and TOC1 coding sequences in pENTR/D-TOPO vectors were recombined into the pGBKT7-GW destination vector (Gateway-compatible pGBKT7 vector). This resulted in a translational fusion of CHE and TOC1 to the GAL4 DNA-binding domain (Lu et al., 2010). These constructs were transformed into the yeast (Saccharomyces cerevisiae) Y187 strain. The full-length, LOV domain (amino acids 1–196), Kelch domain (amino acids 246–609), and decoy (the LOV domain fused to the Kelch domain) ZTL coding sequences in pENTR/D-TOPO vectors were recombined into the pGADT7-GW vector (Gateway-compatible pGADT7 vector), resulting in a translational fusion to the GAL4 activation domain (Lu et al., 2010). These were transformed into the yeast AH109 strain. To test protein-protein interactions, diploid yeast was generated by yeast mating of Y187 and AH109 strains bearing pGBKT7 and pGADT7 vectors, respectively, and tested on synthetic dropout/-Leu-Trp and synthetic dropout/-Leu-Trp-His plates. The empty pGBKT7-GW and pGADT7-GW vectors were included as negative controls.
Coimmunoprecipitation and in Vivo Ubiquitylation Assays Using HEK293T Cells
The full-length (FL) or decoy ZTL (in the pENTR/D-TOPO vectors) was recombined into a pEZYflag destination vector to generate translational fusions to the FLAG affinity tag. CHE (in the pENTR/D-TOPO vectors) was recombined into the pEZYegfp destination vector to generate a CHE translational fusion to GFP (Guo et al., 2008). Approximately 2 × 106 HEK293T cells in 60-mm petri dishes were transfected with pEZYflag-ZTL-FL, pEZYflag-ZTL-decoy, pEZYegfp-CHE, and/or pEZYegfp vectors using Lipofectamine 2000 (Thermo Fisher Scientific, catalog no. 11668027) for 24 h according to the manufacturer’s protocol. Prior to harvest, the cells were treated with 30 µm MG132 (Peptide Institute, catalog no. 3175-v) for 6 h. The cells were lysed by sonication in RIPA buffer (Sigma-Aldrich, catalog no. R0278) with cOmplete EDTA-free Protease Inhibitor Cocktail. Coimmunoprecipitation was performed by incubation of cell lysate with anti-FLAG antibody conjugated to SureBeads Protein G Magnetic Beads (Bio-Rad, catalog no. 161-4023) at 4°C for 1 h. The protein on the beads was washed with SII buffer three times and eluted by boiling in Laemmli sample buffer for subsequent immunoblot analyses.
For the heterologous ubiquitination assays in mammalian cells, approximately 2 × 106 HEK293T cells in 60-mm petri dishes were transfected with pEZYegfp-CHE, pEZYegfp, pEZYflag-ZTL-FL, and/or pEZYflag-ZTL-decoy vectors using Lipofectamine 2000 (Thermo Fisher Scientific, catalog no. 11668027) for 24 h and then treated with 30 µm MG132 (Peptide Institute, catalog no. 3175-v) or DMSO vehicle for 6 h. The cells were lysed in urea buffer (8 m urea, 50 mm sodium phosphate, pH 8, 150 mm NaCl, 1 mm DTT, and cOmplete EDTA-free Protease Inhibitor Cocktail) for immunoblot analyses. For the deubiquitination of GFP-CHE with USP2cc, the cell lysate from pEZYegfp-CHE and pEZYflag-ZTL-FL transfected and MG132-treated cells was used. The cell lysate was diluted 3-fold with 50 mm sodium phosphate and incubated with 2 µg of USP2cc (Sigma-Aldrich, catalog no. U6653) at 37°C for 2 h. The mouse anti-FLAG and rabbit anti-GFP (Abcam, catalog no. Ab-290) antibodies were used to detect FLAG- or GFP-fused proteins, respectively. Experiments were repeated three times, and one representative immunoblot is shown.
Measurement of mRNA and Protein Levels in CHE-GFP Lines
35S::CHE-GFP lines were grown on 1/2 MS plates and entrained in 12 h of light followed by 12 h of dark with a light fluence rate of 165 μmol m−2 s−1 at 22°C for 12 d and then shifted to LL or kept in LD for 48 h prior to the beginning of the time course. 35S::CHE-GFP/ztl-4 lines were grown and approximately 15 seedlings were harvested as described above, but only in the LD condition. For immunoblotting, the protein was extracted from ground seedlings in 8 m urea buffer (8 m urea, 50 mm Tris, pH 8.2, 75 mm NaCl, and cOmplete EDTA-free Protease Inhibitor Cocktail [Roche, catalog no. 11836170001]) and quantified with a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, catalog no. 23225). Approximately 40 µg of total protein was separated on 10% (w/v) SDS-PAGE for immunoblot analyses. CHE-GFP protein levels were detected with anti-GFP antibody, and tubulin was detected with anti-α-tubulin antibody (Sigma-Aldrich, catalog no. T9026). Quantification of signal intensity was performed with the Gel Doc XR+ System (Bio-Rad) and analyzed with Image Lab software (Bio-Rad). To cross-compare the protein levels among different blots, the ZT9 samples from LL and LD were included on each blot. The relative protein levels were calculated by the average of log2[(CHE-GFP/α-tubulin)time point A, condition X/(CHE-GFP/α-tubulin)ZT9, LD] from three biological replicates.
For RT-qPCR, total RNA was extracted and RT-qPCR was performed as described above for the quantification of mRNA expression in decoy transgenic lines. The relative expression of CHE-GFP represents means of log2(2-ΔΔCT) from three biological replicates, in which ΔΔCT = (CT of IPP2 − CT of CHE-GFP)time point − (CT of IPP2 − CT of CHE-GFP)ZT9.
Cell-Free Degradation Assay
A cell-free degradation assay was conducted as described previously (Más et al., 2003) with modifications as noted below. Frozen 14-d-old seedlings, approximately 20 per sample, grown on 1/2 MS plates under a 12-h-light/12-h-dark cycle with a light fluence rate of 165 μmol m−2 s−1 at 22°C, were collected at 4 h before and after dusk (ZT8 and ZT16) and ground in liquid nitrogen. Powder was resuspended with ice-cold assay buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 10 mm MgCl2, 5 mm DTT, 5 mm ATP, and 0.5% [v/v] Triton X-100). The plant slurry was split into two tubes and incubated at room temperature with 50 µm MG132 or DMSO vehicle. An equal volume of sample was taken out at designated time points and mixed with Laemmli sample buffer to stop the reactions. All assay steps were carried out in the dark. After boiling for 10 min, the samples were clarified by centrifugation for 10 min at 16,000g. SDS-PAGE was performed on clarified samples for immunoblotting. CHE-GFP and tubulin were detected with anti-GFP antibody and anti-α-tubulin antibody, respectively, as described above. Triplicate biological repeats were performed for each time point.
Accession Numbers
The Arabidopsis Genome Initiative numbers of featured genes are as follows: ZTL (AT5G57360), FKF1 (AT1G68050), LKP2 (AT2G18915), GI (AT1G22770), TOC1 (AT5G61380), PRR5 (AT5G24470), CHE (AT5G08330), CCA1 (AT2G46830), CDF2 (AT5G39660), RVE6 (AT5G52660), LWD2 (AT3G26640), JMJD5 (AT3G20810), TPL (AT1G15750), FVE (AT2G19520), UBP12 (AT5G06600), UBP13 (AT3G11910), TCP22 (AT1G72010), HSP90.3 (AT5G56010), and HSP90.4 (AT5G56000).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Decoy transgenic lines have rhythmic circadian clocks.
Supplemental Figure S2. RT-qPCR analysis of transgene and endogenous gene expression levels.
Supplemental Table S1. Statistical analyses of the period and flowering time effects of the ZTL, LKP2, and FKF1 decoy lines from Figures 2A and 3.
Supplemental Table S2. Proteins identified by IP-MS in the wild type or 35S::His-FLAG-GFP controls.
Supplemental Table S3. Proteins identified by IP-MS in decoy lines.
Supplemental Table S4. Common interactors of FKF1, LKP2, and ZTL decoys as identified by IP-MS.
Supplemental Table S5. Proteins identified by IP-MS in the ZTL decoy time-course experiment.
Supplemental Table S6. Common interactors of the ZTL decoy time-course experiment.
Supplemental Table S7. Peptide counts for selected published interactors in the ZTL decoy time-course experiment.
Supplemental Table S8. Primers used for cloning decoy constructs and RT-qPCR experiments.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Dmitri Nusinow and the Keck Proteomics Facility at Yale for materials and assistance with proteomics. We also thank Elan Sherazee, Suyuna Eng Ren, Marshall Deline, Denise George, Catherine Chamberlin, and Antonette Lestelle for technical and administrative support. We thank Dr. Wei Liu, Dr. Vivian Irish, and Dr. Adam Saffer for helpful comments on the article.
Footnotes
This work was supported by the National Science Foundation (EAGER #1548538) to J.M.G.; by a Rudolph J. Anderson Fund Fellowship and Forest B.H. and Elizabeth D.W. Brown Fund Fellowship to C.-M.L.; by the National Institutes of Health (T32 GM007499), the Gruber Foundation, and the National Science Foundation (GRFP DGE-1122492) to A.F.; by the National Institutes of Health (R01 GM056006, R01 GM067837, and RC2 GM092412) to S.A.K.; by the Ellison Medical Foundation (New Scholar Award) and a Hellman Fellowship to E.J.B.; and by the National Institutes of Health (R01 GM056006) to J.P.-P.
Articles can be viewed without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Banerjee A, Deshaies RJ, Chau V (1995) Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J Biol Chem 270: 26209–26215 [DOI] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaïdouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guérin C (2005) Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene 24: 2271–2276 [DOI] [PubMed] [Google Scholar]
- Carrano AC, Bennett EJ (2013) Using the ubiquitin-modified proteome to monitor protein homeostasis function. Mol Cell Proteomics 12: 3521–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JY, Kim J, Kim TS, Zeng Q, Wang L, Lee SY, Kim WY, Somers DE (2017) GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat Commun 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui X, Lu F, Li Y, Xue Y, Kang Y, Zhang S, Qiu Q, Cui X, Zheng S, Liu B, et al. (2013) Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol 162: 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N (2014) Advanced methods of microscope control using μManager software. J Biol Methods 1: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Ver Loren van Themaat E, Huettel B, Davis SJ, Coupland G (2015) The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Fukamatsu Y, Mitsui S, Yasuhara M, Tokioka Y, Ihara N, Fujita S, Kiyosue T (2005) Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol 46: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Webb K, Yang B, Rising L, Zuzow N, Bennett EJ (2016) Using the ubiquitin-modified proteome to monitor distinct and spatially restricted protein homeostasis dysfunction. Mol Cell Proteomics 15: 2576–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb D, Hast BE, Wang W, Major MB (2014) Spotlite: web application and augmented algorithms for predicting co-complexed proteins from affinity purification-mass spectrometry data. J Proteome Res 13: 5944–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralogia GS, Liu TK, Zhao L, Panipinto PM, Groover ED, Bains YS, Imaizumi T (2017) CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J 92: 244–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S (2016) MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet 12: e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Shalit-Kaneh A, Chu DN, Hsu PY, Harmer SL (2017) The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol 173: 2308–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, Rouyer F (2002) The F-box protein slimb controls the levels of clock proteins Period and Timeless. Nature 420: 178–182 [DOI] [PubMed] [Google Scholar]
- Guo F, Chiang MY, Wang Y, Zhang YZ (2008) An in vitro recombination method to convert restriction- and ligation-independent expression vectors. Biotechnol J 3: 370–377 [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE (2004) Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Harmon F, Imaizumi T, Gray WM (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, He Q, Yu H, Liu Y (2003) FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J 22: 4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD (2016) Ubiquitin goes green. Trends Cell Biol 26: 3–5 [DOI] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA (2016a) Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics 15: 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Nusinow DA (2016b) Data on the identification of protein interactors with the Evening Complex and PCH1 in Arabidopsis using tandem affinity purification and mass spectrometry (TAP-MS). Data Brief 8: 56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iconomou M, Saunders DN (2016) Systematic approaches to identify E3 ligase substrates. Biochem J 473: 4083–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Imaizumi T (2012) LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18: 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, McWatters HG, Bakó L, Takata N, Gyula P, Hall A, Somers DE, Millar AJ, Eriksson ME (2011) Partners in time: EARLY BIRD associates with ZEITLUPE and regulates the speed of the Arabidopsis clock. Plant Physiol 155: 2108–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Harmer S (2011) JMJD5 functions in concert with TOC1 in the Arabidopsis circadian system. Plant Signal Behav 6: 445–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL (2010) Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA 107: 21623–21628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Geng R, Gallenstein RA, Somers DE (2013) The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140: 4060–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Kim WY, Fujiwara S, Kim J, Cha JY, Park JH, Lee SY, Somers DE (2011) HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc Natl Acad Sci USA 108: 16843–16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Wada M (2000) LKP1 (LOV kelch protein 1): a factor involved in the regulation of flowering time in Arabidopsis. Plant J 23: 807–815 [DOI] [PubMed] [Google Scholar]
- Kleiger G, Mayor T (2014) Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol 24: 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I (2002) Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678 [DOI] [PubMed] [Google Scholar]
- Krahmer J, Goralogia G, Kubota A, Johnson RS, Song YH, Halliday K, MacCoss MJ, LeBihan T, Imaizumi T, Millar A (2017) Time-resolved interaction proteomics of the putative scaffold protein GIGANTEA in Arabidopsis thaliana. bioRxiv doi/https://doi.org/10.1101/162271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9: 631–638 [DOI] [PubMed] [Google Scholar]
- Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM, Lin C (2013) Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci USA 110: 17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou P, Wu J, Cheng F, Cressman LG, Wang X, McClung CR (2012) Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 24: 2415–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R (1998) A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell 1: 565–574 [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Mocciaro A, Rape M (2012) Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. J Cell Sci 125: 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A, Zielinski T, Millar AJ (2014) Online period estimation and determination of rhythmicity in circadian data, using the BioDare data infrastructure. Methods Mol Biol 1158: 13–44 [DOI] [PubMed] [Google Scholar]
- Nagels Durand A, Iñigo S, Ritter A, Iniesto E, De Clercq R, Staes A, Van Leene J, Rubio V, Gevaert K, De Jaeger G, et al. (2016) The Arabidopsis iron-sulfur protein GRXS17 is a target of the ubiquitin E3 ligases RGLG3 and RGLG4. Plant Cell Physiol 57: 1801–1813 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Norén L, Kindgren P, Stachula P, Rühl M, Eriksson ME, Hurry V, Strand Å (2016) Circadian and plastid signaling pathways are integrated to ensure correct expression of the CBF and COR genes during photoperiodic growth. Plant Physiol 171: 1392–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudasaini A, Zoltowski BD (2013) Zeitlupe senses blue-light fluence to mediate circadian timing in Arabidopsis thaliana. Biochemistry 52: 7150–7158 [DOI] [PubMed] [Google Scholar]
- Pudasaini A, Shim JS, Song YH, Shi H, Kiba T, Somers DE, Imaizumi T, Zoltowski BD (2017) Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. eLife 6: e21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW (2006) The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell 124: 89–103 [DOI] [PubMed] [Google Scholar]
- Risseeuw E, Venglat P, Xiang D, Komendant K, Daskalchuk T, Babic V, Crosby W, Datla R (2013) An activated form of UFO alters leaf development and produces ectopic floral and inflorescence meristems. PLoS ONE 8: e83807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR (2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Williams PJ, Gupta A, McCluskey B, Bhaskaran S, Muñoz S, Oyajobi BO (2015) A dominant-negative F-box deleted mutant of E3 ubiquitin ligase, β-TrCP1/FWD1, markedly reduces myeloma cell growth and survival in mice. Oncotarget 6: 21589–21602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane T, Jin J, Ang XL, Harper JW (2005) SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem 280: 26863–26872 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Rape M (2011) Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle 10: 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T (2014) Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci USA 111: 17672–17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67–77 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kuroda H, Kuromori T, Hirayama T, Seki M, Shinozaki K, Shimada H, Matsui M (2004) Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol 45: 83–91 [DOI] [PubMed] [Google Scholar]
- Takase T, Nishiyama Y, Tanihigashi H, Ogura Y, Miyazaki Y, Yamada Y, Kiyosue T (2011) LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J 67: 608–621 [DOI] [PubMed] [Google Scholar]
- Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H (2014) SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics 100: 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fujiwara S, Somers DE (2010) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ni W, Ge X, Zhang J, Ma H, Cao K (2006) Proteomic identification of potential target proteins regulated by an ASK1-mediated proteolysis pathway. Cell Res 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH (2011) LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JF, Wang Y, Wu SH (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol 148: 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JF, Tsai HL, Joanito I, Wu YC, Chang CW, Li YH, Wang Y, Hong JC, Chu JW, Hsu CP, et al. (2016) LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun 7: 13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T (2004) Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot 55: 2015–2027 [DOI] [PubMed] [Google Scholar]
- Yumimoto K, Matsumoto M, Oyamada K, Moroishi T, Nakayama KI (2012) Comprehensive identification of substrates for F-box proteins by differential proteomics analysis. J Proteome Res 11: 3175–3185 [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ni W, Feng B, Han T, Petrasek MG, Ma H (2003) Members of the Arabidopsis-SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol 133: 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Imaizumi T (2014) Structure and function of the ZTL/FKF1/LKP2 group proteins in Arabidopsis. Enzymes 35: 213–239 [DOI] [PMC free article] [PubMed] [Google Scholar]