Abstract
The kynurenine pathway is the primary route for l-tryptophan degradation in mammals. Intermediates and side products of this pathway are involved in immune response and neurodegenerative diseases. This makes the study of enzymes, especially those from mammalian sources, of the kynurenine pathway worthwhile. Recent studies on a bacterial version of an enzyme of this pathway, 2-aminomuconate semialdehyde (2-AMS) dehydrogenase (AMSDH), have provided a detailed understanding of the catalytic mechanism and identified residues conserved for muconate semialdehyde recognition and activation. Findings from the bacterial enzyme have prompted the reconsideration of the function of a previously identified human aldehyde dehydrogenase, ALDH8A1 (or ALDH12), which was annotated as a retinal dehydrogenase based on its ability to preferentially oxidize 9-cis-retinal over trans-retinal. Here, we provide compelling bioinformatics and experimental evidence that human ALDH8A1 should be reassigned to the missing 2-AMS dehydrogenase of the kynurenine metabolic pathway. For the first time, the product of the semialdehyde oxidation by AMSDH is also revealed by NMR and high-resolution MS. We found that ALDH8A1 catalyzes the NAD+-dependent oxidation of 2-AMS with a catalytic efficiency equivalent to that of AMSDH from the bacterium Pseudomonas fluorescens. Substitution of active-site residues required for substrate recognition, binding, and isomerization in the bacterial enzyme resulted in human ALDH8A1 variants with 160-fold increased Km or no detectable activity. In conclusion, this molecular study establishes an additional enzymatic step in an important human pathway for tryptophan catabolism.
Keywords: aldehyde dehydrogenase, 2-aminomuconate semialdehyde dehydrogenase, tryptophan catabolism, retinal metabolism, NAD biosynthesis, biodegradation, serotonin, neurological disease, kynurenine pathway, retinal dehydrogenase
Introduction
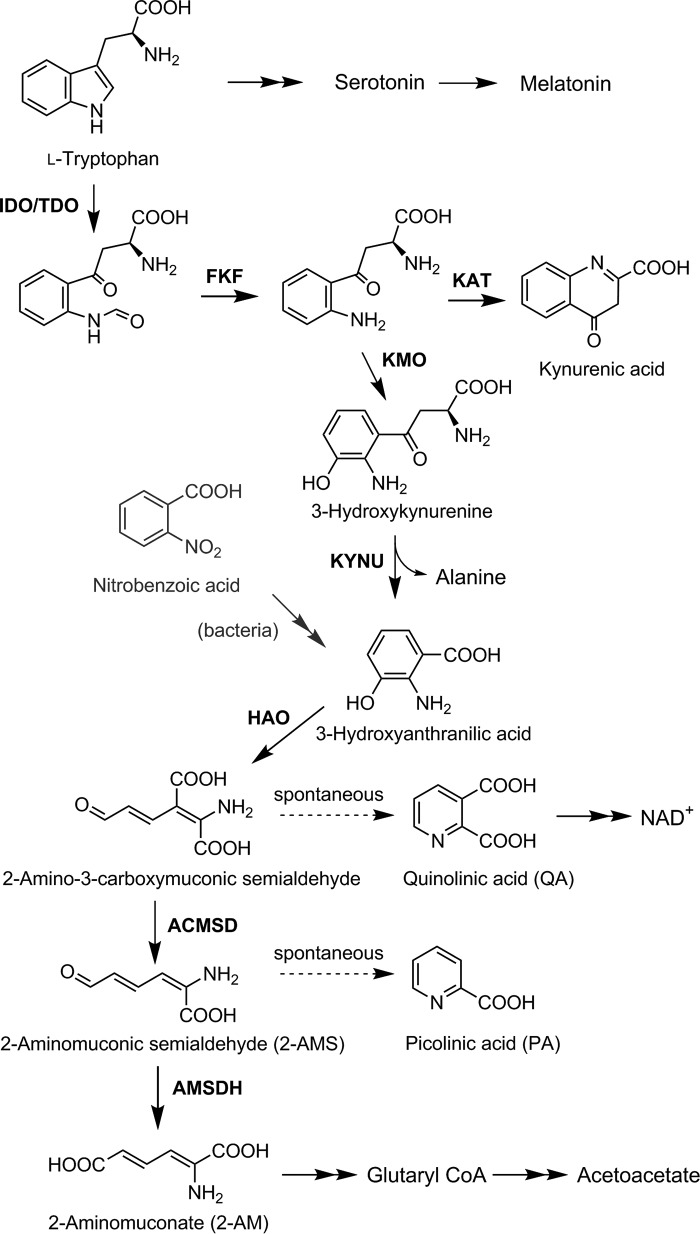
l-Tryptophan, an essential amino acid, has several metabolic fates in mammals: a building block for proteins, the precursor for serotonin and melatonin, and its complete catabolism through the kynurenine pathway (KP)3 to pyruvate via alanine, acetoacetate via glutaryl-CoA (1), NAD/NADH via quinolinic acid (QA) (2), and several neurologically active compounds. Various kynurenine pathway metabolites are linked to the innate immune response and both neuroexcitatory and neurodepressive effects (3–6). Because of its potential medical significance, the KP has received increasing attention. The first and committing enzymes, tryptophan and indolamine dioxygenase, are active drug targets with inhibitors in clinical trials (5, 7, 8). Recently, a downstream enzyme of the KP (Scheme 1), 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase (ACMSD), has received attention as a potential drug target (6). Inhibition of ACMSD has been shown to slow down the reaction competing with QA formation and boost cellular NAD(H) levels (9).
Scheme 1.
The kynurenine pathway. The enzymes identified in the kynurenine pathway are: indoleamine 2,3-dioxygenase (IDO)/tryptophan 2,3-dioxygenase (TDO), N-formyl kynurenine formamidase (FKF), kynurenine 3-monooxygenase (KMO), kynurenine aminotransferase (KAT), kynureninase (KYNU), 3-hydroxyanthranilate-3,4-dioxygenase (HAO), ACMSD, and the proposed enzyme AMSDH.
To date, the KP pathway genes and their corresponding enzymes have not been identified beyond ACMSD (10, 11), although the metabolic pathway was published 53 years ago and has made its way into numerous biochemistry textbooks. One limiting factor for studying KP enzymes is that the identification of their mammalian genes has proved difficult. Initial characterization of the KP enzymes was performed from animal liver extracts (1). Although these studies verified the activities and transformations of the KP, they were unable to provide much insight into the individual enzyme structures and mechanisms. Study of the KP enzymes stagnated until the discovery of an analogous KP in some bacteria (12–17) and that 2-nitrobenzoate biodegradation shares many of the downstream proteins with the eukaryotic kynurenine pathway (18–20). An additional difficulty for studying the KP enzymes, especially downstream proteins, is that several of the metabolic intermediates of the pathway are unstable and commercially unavailable. As shown in Scheme 1, the substrates for ACMSD and its downstream neighbor, 2-aminomuconate semialdehyde dehydrogenase (AMSDH), are both unstable and spontaneously cyclize via a pericyclic reaction to their respective pyridine products, QA and picolinic acid (PA) (2).
Despite the difficulties mentioned above, much progress has been made in understanding the mechanisms of KP enzymes. Recently, AMSDH from Pseudomonas fluorescens (pfAMSDH) identified from the 2-nitrobenzoate biodegradation pathway has been studied at the molecular level. Crystal structures of the resting enzyme, NAD+-bound complex, ternary complex, catalytic thioacyl and thiohemiacetal intermediates, and several mutants have been reported (21). A hidden isomerase activity of AMSDH has also been uncovered (22). Furthermore, the study of pfAMSDH revealed that, in addition to active-site residues that are broadly conserved across all aldehyde dehydrogenases, the hydroxymuconate semialdehyde dehydrogenase (HMSDH) family possesses two conserved arginine residues that are involved in substrate recognition and an isomerization activity (22).
In this work, we have identified a human enzyme annotated as a retinal dehydrogenase (ALDH8A1) that carries the hallmarks of an aminomuconate semialdehyde dehydrogenase. An overexpression system was constructed, and recombinant ALDH8A1 was tested for activity with two muconic semialdehyde substrates. The activity of selected active-site variants was also investigated, and the reaction products were verified with NMR and high-resolution MS. All evidence suggests that ALDH8A1 should be reconsidered as the aldehyde dehydrogenase of the kynurenine pathway of tryptophan catabolism.
Results and discussion
Identification of ALDH8A1 as a potential member of the kynurenine pathway
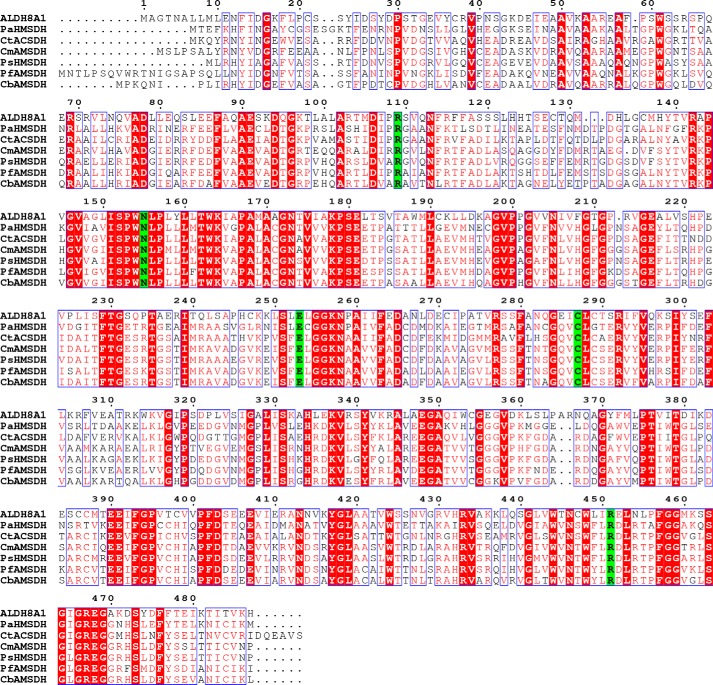
To continue studying the KP pathway at the molecular level, the next pressing step is to identify a mammalian AMSDH, especially the human version. We performed a BLAST search with pfAMSDH as the search sequence. The results revealed a human protein, ALDH8A1 (initially designated ALDH12), with 44% amino acid sequence identity to pfAMSDH. However, ALDH8A1 is currently annotated in the NCBI gene database as a retinal dehydrogenase. It was assigned as cis-retinal dehydrogenase based on its ability to oxidize 9-cis-retinal faster than all-trans-retinal, even though it was most active with benzaldehyde rather than the retinal substrates (23). It was also noted in the original characterization that ALDH8A1 shares the closest nucleotide and protein sequence similarity with AMSDH, but it was not tested for such activity, presumably due to difficulty obtaining 2-aminomuconate semialdehyde (2-AMS). Here, we present evidence that ALDH8A1 may be more appropriately considered human AMSDH of the KP. AMSDH belongs to the HMSDH family. Our alignment of ALDH8A1 against AMSDH and several members of the HMSDH family showed not only high overall conservation but also that, in addition to residues required for aldehyde dehydrogenase activity (Asn-155, Glu-253, and Cys-287), residues responsible for substrate recognition only in HMSDH enzymes are conserved in ALDH8A1, namely Arg-109 and Arg-451 (Fig. 1, generated with ESPript (24)).
Figure 1.
Sequence alignment of ALDH8A1 with HMSDH enzymes. Highly conserved residues are shown with red text and boxed in blue, strictly conserved residues are shown with a red background, and catalytic residues are shown with a green background. The enzymes chosen for alignment are as follows: ALDH8A1, GenBankTM accession number AAI13863; AMSDH from Pseudomonas fluorescens, GenBank accession number BAC65304; HMSDH from Paraglaciecola arctica (Pa), accession number WP_007618756; ACSDH from Comamonas testosterone (Ct), accession number YP_001967696; AMSDH from Cupriavidus metallidurans (Cm), GenBank accession number KWW33428; AMSDH from Cupriavidus basilensis (Cb), GenBank accession number AJG18463; HMSDH from Pseudomonas sp. (Ps) M1, GenBank accession number ETM66811.
A homology structure model of ALDH8A1 was built using the iTASSER server (25). An overlay of the homology model and pfAMSDH is shown in Fig. 2A. The model shows full coverage of the human enzyme sequence, which overlays well with the bacterial enzyme with a root mean square deviation of 1.15 Å for 472 C-α carbons. In addition, all of the catalytically essential active-site residues from pfAMSDH (Arg-109, Asn-155, Glu-253, Cys-287, and Arg-451 by ALDH8A1 numbering) are in the same location in the homology model as in the pfAMSDH structure (Fig. 2B).
Figure 2.
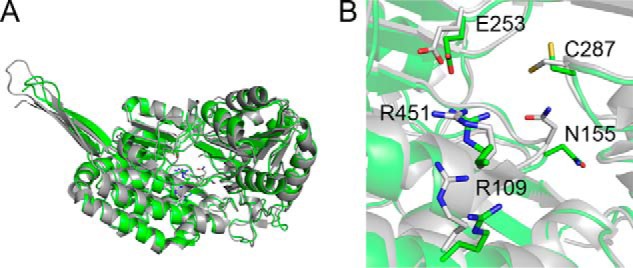
Homology model of ALDH8A1 (green) and crystal structure of pfAMSDH (Protein Data Bank code 4I26) (gray). Shown are an overlay of a single polypeptide (A) and a zoomed-in view of the active site with catalytically relevant residues (B).
ALDH8A1 can perform the NAD+-dependent oxidation of 2-aminomuconic semialdehyde
The next question to arise was whether ALDH8A1 is able to catalyze the NAD+-dependent oxidation of 2-AMS to 2-aminomuconate (2-AM). Pursuant to this end, an overexpression system was generated. The synthesized gene for human ALDH8A1 was ligated into pET-28a(+) vector with a cleavable N-terminal His6 tag and transformed into Escherichia coli BL21(DE3) competent cells. The expressed protein was purified by nickel affinity chromatography (Fig. 3A), and its ability to oxidize 2-AMS in a coupled-enzyme assay with ACMSD was tested. As shown in Fig. 3B, ACMS has a broad absorbance band at 360 nm. Upon ACMSD-catalyzed decarboxylation to 2-AMS, the absorbance maximum red-shifted to 380 nm and then decayed as 2-AMS was nonenzymatically converted to PA. The inclusion of purified ALDH8A1 and 1 mm NAD+ in an otherwise identical assay prevented the red shift, and instead, a broad absorbance band around 350 nm was observed that has been previously assigned as the oxidized product, 2-AM, and NADH (Fig. 3C) according to the reaction scheme shown in Fig. 3D. These results resemble those observed from pfAMSDH and show that ALDH8A1 is able to rapidly oxidize 2-AMS in solution. The presence of the expected product in the coupled-enzyme assay was also verified by NMR spectroscopy. As shown in Fig. 4, the reaction mixture contained resonances consistent with 2-AM and expected cross-peaks in the 1H-1H COSY spectrum to show connectivity. Proton resonances were assigned based on similarity with 2-hydroxymuconic acid, which has been rigorously characterized (26).
Figure 3.
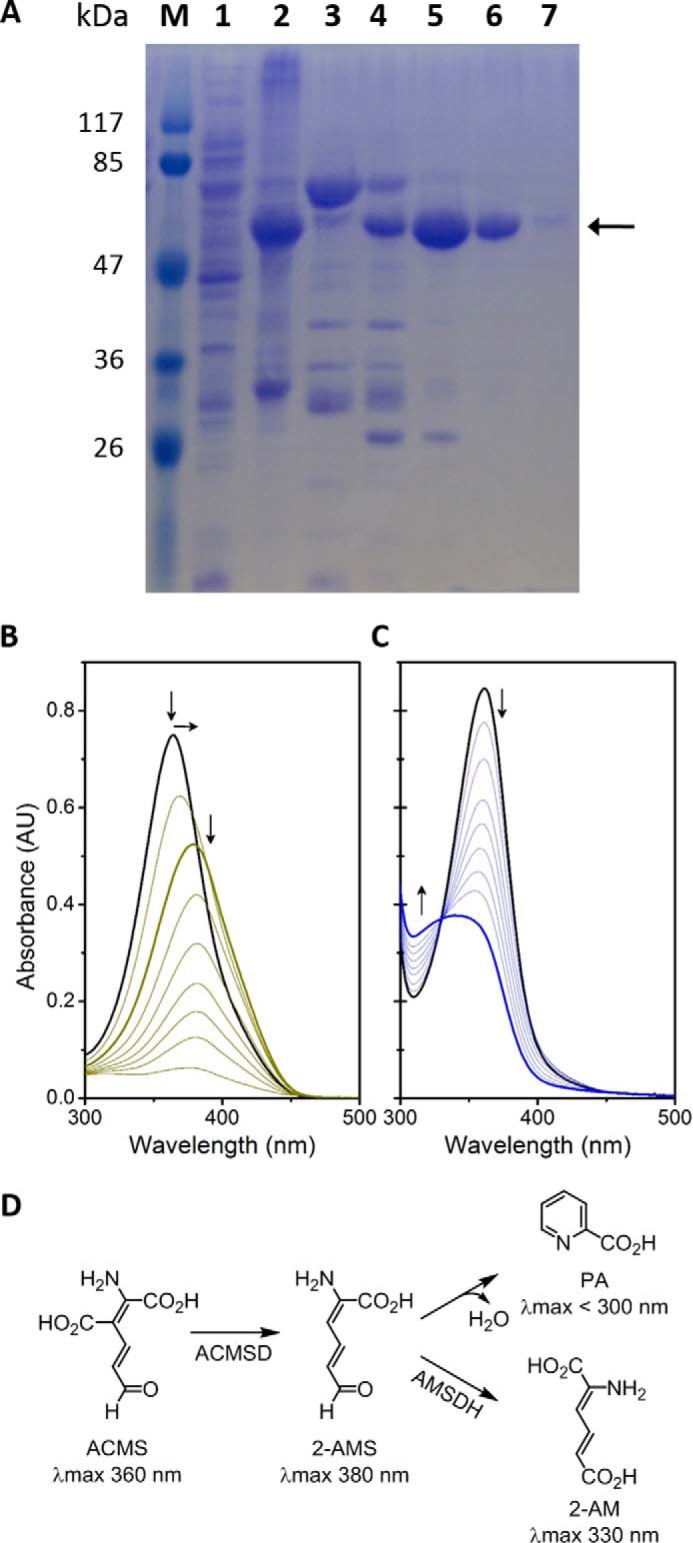
Purification and representative activity assay of ALDH8A1. A, SDS-PAGE of a purification by nickel-nitrilotriacetic acid affinity chromatography. Lane 1 is clarified cell extract, lane 2 is cell pellet, lane 3 is flow-through, and lanes 4–7 are fractions 1–4, respectively. Fractions 2 and 3 were collected for use. B, time course of ACMSD acting on ACMS to produce 2-AMS, which decays to PA. C, coupled-enzyme assay with ACMSD and ALDH8A1 converting ACMS to 2-AM in the presence of NAD+. D, scheme showing the reactions in B and C as the top and bottom branches, respectively. AU, absorbance units.
Figure 4.
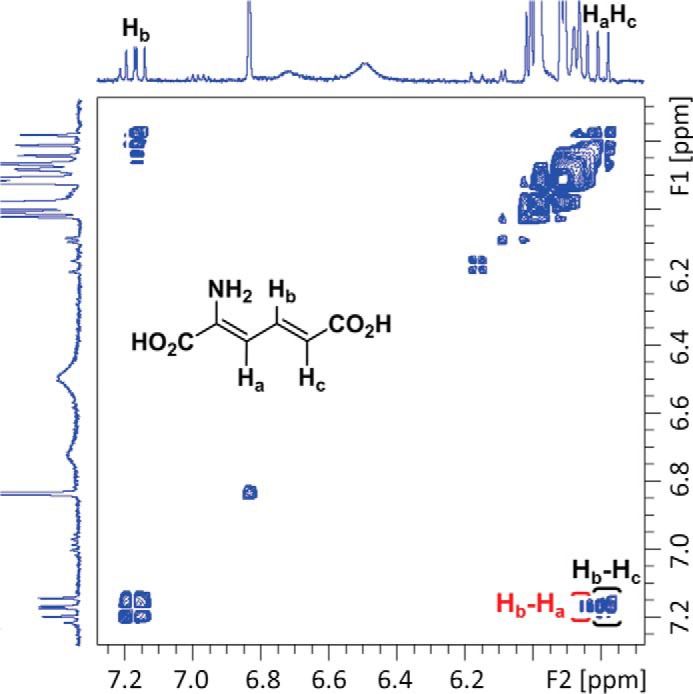
1H-1H NMR COSY spectrum of a coupled-enzyme reaction mixture containing 2-AM showing correlations of Hb with Ha and Hc.
Characterization of the reaction product of the ALDH8A1-catalyzed reaction
In initial studies of AMSDH, the identity of the product was inferred based on knowledge of the substrate structure and the catalytic cycle of the dehydrogenation reaction. The in crystallo characterization of the AMSDH reaction revealed that, in addition to oxidation of the aldehyde to its corresponding carboxylic acid, pfAMSDH also isomerizes the 2,3-double bond inside the active site prior to substrate oxidation (22). In the process of trying to determine the conformation of the product of the reaction catalyzed by ALDH8A1, we noticed that the product, 2-AM (λmax 330 nm), is unstable and is nonenzymatically bleached in a single kinetic phase with a half-life of 67 min (Fig. 5A). High-resolution mass spectra were acquired of the 2-AM decay product after purification by HPLC. Mass spectra were collected in negative mode, and as shown in Fig. 5B, the parent ion matches tautomerized hydroxymuconate, 2-oxo-3-hexenedioate, with 2.55-ppm mass accuracy. Furthermore, fragment ions from cleaving at either side of the keto group can be observed with nominal masses of 113 and 85 Da.
Figure 5.
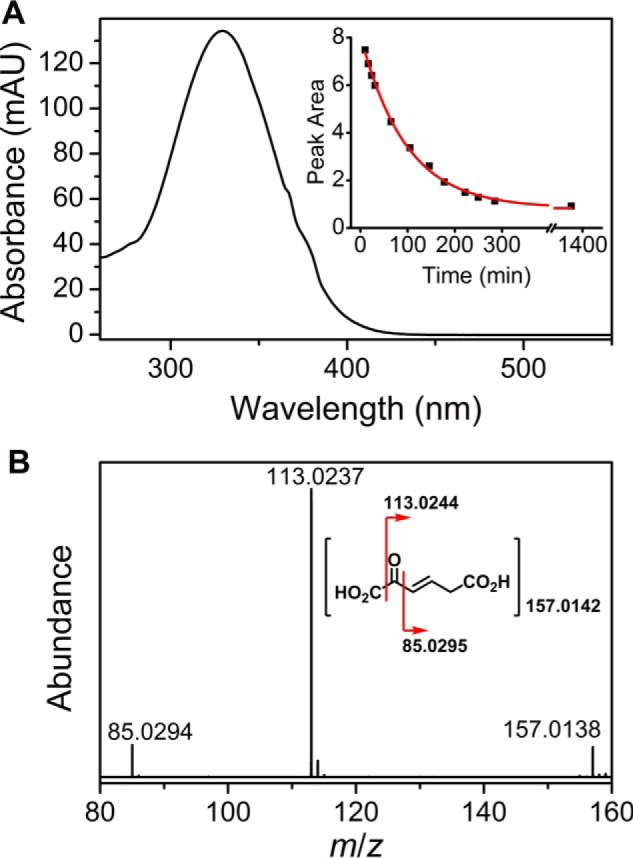
A, UV-visible spectrum of 2-AM and the time course of its nonenzymatic decay (inset). B, MS-MS spectrum of the 2-AM decay product. mAU, milliabsorbance units.
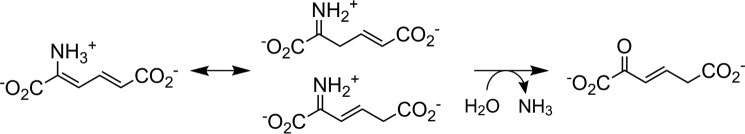
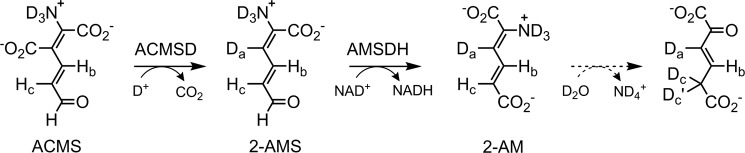
The downstream enzyme of AMSDH performs deamination on 2-AM to produce 2-hydroxymuconate, which is expected to tautomerize to its α,β-unsaturated ketone form, 2-oxo-3-hexenedioate, as shown in Scheme 2. The deamination reaction is not known to be coupled to any other reaction, so it is expected to be thermodynamically preferred. As such, it may proceed nonenzymatically at a slower rate. To lend credence to the proposed nonenzymatic deamination followed by tautomerization, ACMSD–AMSDH coupled-enzyme assays were performed in H2O and D2O in separate experiments, and the reaction products were monitored by NMR spectroscopy (Fig. 6). When performing the reaction in H2O (Fig. 6A), two doublets around 5.8 ppm and a doublet of doublets at 7.15 ppm can be observed to decay while a new doublet at 6.17 ppm and a doublet of triplets at 6.9 ppm arise. These new resonances are consistent with 2-oxohexenedioate observed in the study of 2-hydroxymuconate tautomerization (26). Alternatively, upon enzymatic decarboxylation and oxidation performed in D2O by ACMSD and AMSDH, respectively, the 1H NMR spectrum shows two doublets at 7.1 and 5.8 ppm, corresponding to the protons on carbons 4 and 5 (Hb and Hc) of 2-AM, respectively (Fig. 6B). The proton at the 3-position (Ha) is replaced with deuterium by running the ACMSD reaction in D2O (Scheme 3). The two doublets coalesce into a single resonance at 6.9 ppm over time. The decay of the resonance at 5.8 ppm indicates that the proton at the 5-position can eventually exchange with solvent, and the shift of the doublet at 7.1 to a singlet at 6.9 ppm implies that a chemical change takes place in addition to simple exchanging of protons for deuterons. The most likely candidate for such a chemical change is the replacement of the nitrogen at the 2-position with oxygen derived from water, i.e. deamination of 2-AM. Thus, the NMR spectra of the initial and final products of the ALDH8A1 reaction are consistent with 2-AM being the initially formed product, which then spontaneously deaminates and tautomerizes.
Scheme 2.
Proposed spontaneous decay mechanism for 2-aminomuconate.
Figure 6.
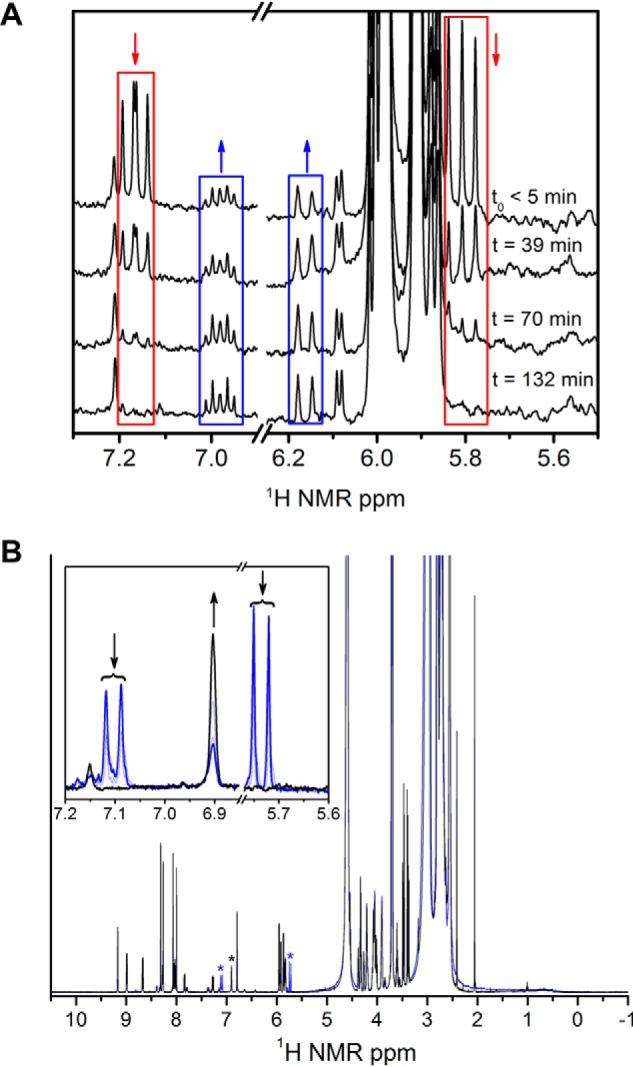
1H-NMR spectra monitoring the decay of 2-AM from a coupled-enzyme assay performed in H2O (A) and D2O (B). 2-AM resonances are highlighted with red boxes, and the decay product, 2-oxo-hexenedioate, is highlighted with blue boxes. A zoomed-in view of the resonances corresponding to 2-AM and its decay product is shown in the inset in B. The initial and final spectra are shown as blue and black, respectively.
Scheme 3.
Coupled ACMSD–AMSDH assay performed in D2O.
Determination of the kinetic parameters of ALDH8A1 and selected site-directed mutants
Although the coupled-enzyme assays presented above show that 2-AMS can serve as a substrate for ALDH8A1, such experiments are not amenable to extraction of enzymatic kinetic parameters. Instead, a substrate analog, 2-hydroxymuconate semialdehyde (2-HMS), in which the amino group of 2-AMS has been replaced with a hydroxyl group to prevent the nonenzymatic cyclization reaction, is used to determine kinetic parameters. The 1H NMR spectrum of 2-HMS can be found in Fig. 7 along with corresponding 1D NOESY spectra, which show not only through-space interactions between protons but also in-phase resonances for the enol tautomer, which was previously implicated as an intermediate in the 2,3-bond isomerization reaction. The observation of resonances consistent with the enol form of 2-HMS in solution lends credence to the previously proposed tautomerization mechanism in pfAMSDH by showing that the enol form is energetically accessible. ALDH8A1 exhibits typical steady-state kinetics when acting on 2-HMS (Fig. 8A). The data were fitted with the Michaelis–Menten equation to provide a kcat, Km, and kcat/Km of 0.42 s−1, 590 nm, and 7.1 × 105 m−1 s−1, respectively. A submicromolar Km is at the lower end for the KP enzymes; however, such high commitment may be necessary to efficiently compete with the rapid decay of its substrate to PA.
Figure 7.
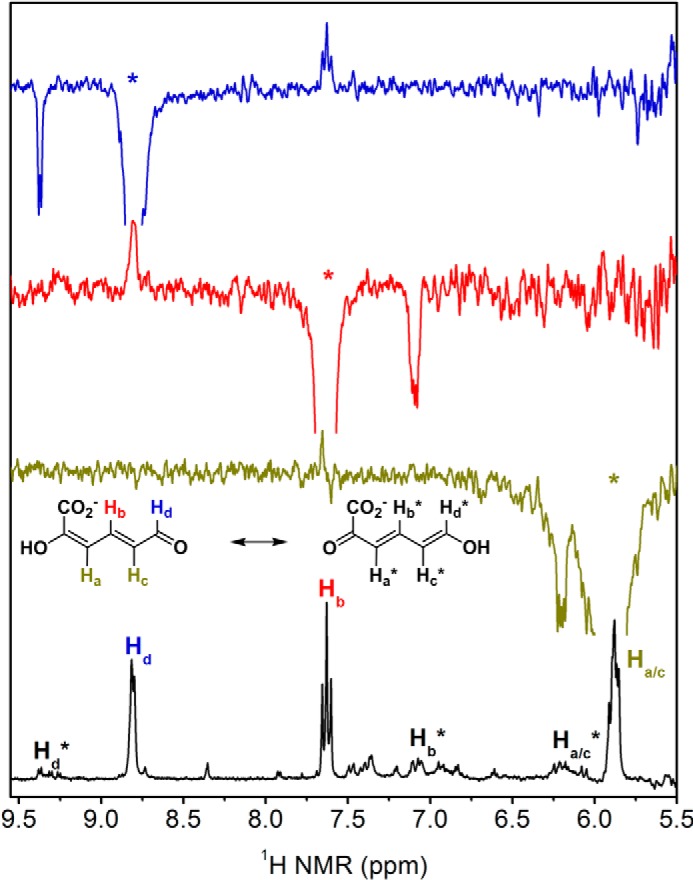
1H NMR spectrum of 2-HMS (bottom) and 1D NOESY spectra show enol tautomer. NOESY spectra were acquired by irradiating at the resonance marked with an asterisk corresponding to the color-coded proton (Hd as blue, Hb as red, and Ha/c as mustard). Out-of-phase, positive resonances show through-space interactions between protons, and in-phase, negative resonances show the same proton in the enol tautomer as indicated by the isomerization shown.
Figure 8.
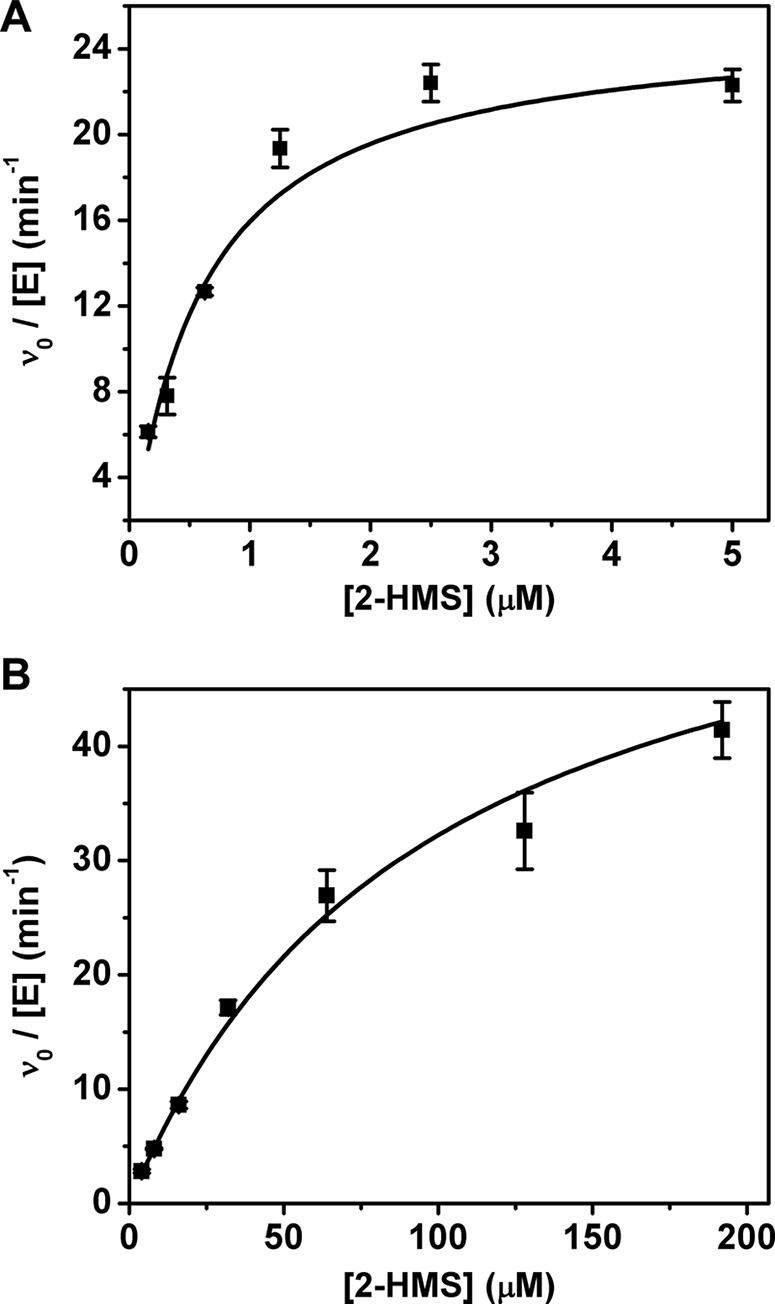
Determination of Michaelis–Menten parameters of ALDH8A1 (A) and the R109A variant (B) for 2-HMS. Reactions were monitored by the decrease in absorbance at 375 nm. Error bars represent S.D.
To further investigate the specificity of ALDH8A1 for α-substituted muconate semialdehydes, several active-site mutants were constructed. The two strictly conserved residues among the HMSDH family previously shown to be responsible for substrate recognition and binding, Arg-109, and Arg-451, were mutated to alanine, and their kinetic parameters for 2-HMS were determined. As summarized in Table 1, deletion of Arg-109 by mutation to alanine generated a variant with a similar turnover number but approximately 160-fold increased Km as compared with WT (Fig. 8B). No detectable activity could be measured for the R451A variant. Additionally, the active-site asparagine (Asn-155) responsible for stabilizing tetrahedral, oxyanion intermediates in general and involved in substrate isomerization in AMSDH was mutated to alanine, aspartic acid, and glutamine. The activity of the mutants was too low to determine kinetic parameters; however, specific activities for N155A, N155D, and N155Q were 7.4 ± 0.1, 20 ± 1, and 0.31 ± 0.03 nmol/mg/min, respectively. Of the same mutants in pfAMSDH, N169D also showed the highest activity (22). In other aldehyde dehydrogenases, mutation of the corresponding asparagine to alanine or aspartic acid reduced the activity by approximately 1,000-fold or below detectable limits (27, 28).
Table 1.
Kinetic parameters of ALDH8A1 and mutants for 2-HMS
| kcat | Km | kcat/Km | |
|---|---|---|---|
| s−1 | μm | s−1 m−1 | |
| ALDH8A1 | 0.42 ± 0.03 | 0.59 ± 0.10 | 7.1 × 105 |
| R109A | 1.06 ± 0.12 | 97 ± 13 | 1.1 × 104 |
| R451A | NDa | ND | ND |
| N169A/D/Q | <0.02 | ND | ND |
a ND, not determined.
Conclusion
The human enzyme ALDH8A1 (ALDH12) was shown to catalyze the NAD+-dependent oxidation of 2-AMS with catalytic efficiency comparable with pfAMSDH. Mutation of the active-site residues, which were shown to be heavily involved with substrate recognition, binding, and isomerization in the bacterial enzyme, resulted in variants with 100-fold increased Km or no detectable activity. As such, the ALDH8A1 enzyme, which was previously assigned as a cis-retinal dehydrogenase, should be reassigned as human AMSDH. It was also shown that the reaction product, 2-AM, can spontaneously deaminate in solution, ultimately forming 2-oxo-3-hexenedioate. This work thus establishes that the aldehyde dehydrogenase of the kynurenine pathway, first discovered 53 years ago from liver extracts (1), is ALDH8A1 (ALDH12). The kynurenine pathway of the tryptophan catabolic pathway in humans is therefore extended to AMSDH.
Experimental procedures
Cloning and site-directed mutagenesis
A DNA sequence that codes for human ALDH8A1 (accession number AF303134) was purchased from DNASU (Arizona State University) and ligated into pET28a(+) vector with NheI and HindIII restriction sites, creating an N-terminal His6-tagged construct. The resultant plasmid was transformed into E. coli cell line BL21 (DE3), which was stored at −80 °C as a 20% (v/v) glycerol stock. Overexpression systems for R109A, R451A, N155A, N155Q, and N155D were constructed by PCR overlap extension using the WT as the starting template. The forward primers were 5′-CCATGGACATTCCCGCGTCTGTGCAGAA and 5′-CTGCTGGCTCATCGCGGAGCTGAACCTT for R109A and R451A, respectively, and 5′-GCTGGTCTGATCAGCCCCTGGGCTTTGCCACTCTACTTGCTGACC, 5′-GCTGGTCTGATCAGCCCCTGGCAGTTGCCACTCTACTTGCTGACC, and 5′-GCTGGTCTGATCAGCCCCTGGGACTTGCCACTCTACTTGCTGACC for N155A, N155Q, and N155D, respectively. Nucleotides corresponding to the mutated residues are underlined.
Protein preparation
For all cultures, antibiotic selection under kanamycin was used. Cultures were started by streaking the appropriate glycerol stock onto an LB-agar plate, which was incubated overnight at 37 °C. A single colony was selected for further incubation in 15 ml of LB-Miller broth at 37 °C with 220 rpm shaking until an A600 of approximately 0.6 was achieved. Then 50 ml of LB-Miller broth was inoculated to an A600 of 0.0002 and incubated at 37 °C with 220 rpm shaking. Finally, once the 50-ml flask reached an A600 of approximately 0.6, it was used to inoculate 6 liters of LB-Miller broth in 12 2-liter baffled flasks to an A600 of 0.0002. The flasks were incubated at 37 °C with 220 rpm shaking. Upon reaching an A600 of 0.5, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 800 μm to induce protein expression, the temperature was lowered to 16 °C, and the culture was incubated for an additional 12 h. Cells were harvested by centrifugation at 8,000 × g and resuspended in 50 mm KPi, 150 mm NaCl buffered to pH 8.0 with 0.1% (v/v) β-mercaptoethanol. Protein was released by cell disruption (LS-20, Microfluidics), and the cell debris was removed by centrifugation at 27,000 × g.
The protein of interest was purified by nickel affinity chromatography. Clarified, cell-free extract was applied to a nickel-nitrilotriacetic acid column and eluted with an imidazole gradient. The running and elution buffers were 50 mm KPi, 150 mm NaCl buffered to pH 8.0 with 5 mm 1,4-DTT with the elution buffer also containing 500 mm imidazole. The purified protein was then desalted to 25 mm HEPES buffer, pH 7.5, 5 mm 1,4-DTT, 1 mm NAD+, 5% glycerol (w/v); concentrated to approximately 1 mm by 30-kDa centrifugal filters; flash frozen in liquid nitrogen; and stored at −80 °C until use. ACMSD used for coupled-enzyme assays was prepared as reported previously (11, 29–33).
Kinetic assays
The substrate analog 2-hydroxymuconic semialdehyde was prepared as reported previously (21). Briefly, 3-hydroxyanthralinic acid was converted to 2-amino-3-carboxymuconic semialdehyde by purified 3-hydroxyanthranilic acid dioxygenase. 2-Amino-3-carboxymuconic semialdehyde was then nonenzymatically converted to 2-hydroxymuconic semialdehyde by lowering the pH below 2. After conversion, the solution was neutralized, and excess enzyme was removed by filtration.
The coupled-enzyme assays were initiated by addition of excess ACMSD (1 μm) to rapidly convert all ACMS to 2-AMS, which is in turn was converted to 2-aminomuconic acid by ALDH8A1. In the absence of ALDH8A1 and 1 mm NAD+, the 2-AMS nonenzymatically decays to picolinic acid. Catalytic parameters were obtained using 2-HMS as the substrate. The decrease in absorbance as 2-HMS (λmax at 375 nm, ϵ375 of 43,000 m−1 cm−1) and NAD+ are converted to 2-hydroxymuconic acid and NADH (ϵ375 of 1,900 m−1 cm−1) was measured with an Agilent 8453 diode-array spectrometer. The reaction rate was calculated as the change in absorbance divided by the sum of the extinction coefficients of 2-HMS and NADH. Initial rates versus substrate concentration were fitted with the Michaelis–Menten equation.
| (1) |
Nonlinear least squares regression was performed with OriginPro 8.5.
NMR spectroscopy
All NMR spectra were recorded on a Bruker (Billerica, MA) Avance III HD 500-MHz spectrometer equipped with a CryoProdigy Probe at 300 K running TopSpin 3.5pl6. Spectra were recorded in D2O or 90% H2O, 10% D2O and referenced to residual solvent (1H, 4.70 ppm). One-dimensional 1H spectra (pulse sequence, zg30) were recorded with 1-s relaxation delays, 65,536 data points, and multiplied with an exponential function for a line broadening of 0.3 Hz before Fourier transformation. Double quantum–filtered COSY (pulse sequence, cosygpmfppqf) spectra were acquired with spectral widths of 13.0 ppm with 2,048 × 128 data points and a relaxation delay of 2.0 s. 1D gradient-selected NOESY (pulse sequence, selnogp) spectra were recorded with a mixing time of 0.3 s and a 2-s relaxation delay and multiplied with an exponential function for a line broadening of 3 Hz before Fourier transformation. All NMR data were processed using MestReNova NMR version 11.0.3 software.
Mass spectrometry
The AMSDH reaction product, 2-aminomuconate, was isolated for mass spectrometric characterization by reverse-phase HPLC with an InertSustain C18 column (5-μm particle size, 4.6-mm inner diameter × 100 mm; GL Sciences Inc.) on a Dionex Ultimate 3000 HPLC equipped with a diode-array detector (Sunnyvale, CA). The crude reaction mixture was ultrafiltered (10-kDa cutoff) to remove protein, and separation was achieved using isocratic elution with 95% H2O, 5% acetonitrile, and 0.5% formic acid. Mass spectra were collected on a maXis plus quadrupole-TOF mass spectrometer equipped with an electrospray ionization source (Bruker Daltonics). The instrument was operated in the negative ionization mode in the range 50 ≤ m/z ≤ 1,500 and calibrated using ESI-L Low Concentration Tuning Mix (Agilent Technologies). Samples were introduced via syringe pump at a constant flow rate of 3 μl/min. Relevant source parameters are summarized as follows: capillary voltage, 3500 V with a set end plate offset of −500 V; nebulizer gas pressure, 0.4 bar; dry gas flow rate, 4.0 liter/min; source temperature, 200 °C. Mass spectra were averages of 1 min of scans collected at a rate of 1 scan per second. Collision-induced dissociation was achieved using a set collision energy of −20 eV. OtofControl software version 6.3 was used for data acquisition, and Compass Data Analysis software version 4.3 (Bruker Daltonics) was used to process all mass spectra. mMass software version 5.5.0 was used for all exact mass calculations (34).
Author contributions
I. D. and A. L. conceptualization; I. D., Y. Y., and D. W. data curation; I. D., Y. Y., and D. W. formal analysis; I. D. and Y. Y. investigation; I. D. writing-original draft; I. D., D. W., and A. L. writing-review and editing; D. W. and A. L. resources; D. W. methodology; A. L. supervision; A. L. funding acquisition; A. L. validation; A. L. project administration.
Acknowledgments
We thank Dr. Wendell Griffith for assisting with MS analysis. The mass spectrometry facility was sponsored by National Institutes of Health Grant G12MD007591. The NMR spectrometer is a shared instrument sponsored by the National Science Foundation under Award 1625963.
This work was supported by National Science Foundation Grant CHE-1623856; National Institutes of Health Grants GM107529, GM108988, and MH107985; and the Lutcher Brown Distinguished Chair Endowment fund (to A. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- KP
- kynurenine pathway
- 2-AMS
- 2-aminomuconate semialdehyde
- AMSDH
- 2-aminomuconate semialdehyde dehydrogenase
- QA
- quinolinic acid
- ACMSD
- 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase
- PA
- picolinic acid
- pfAMSDH
- AMSDH from P. fluorescens
- HMSDH
- hydroxymuconate semialdehyde dehydrogenase
- 2-AM
- 2-aminomuconate
- 2-HMS
- 2-hydroxymuconate semialdehyde
- ACMS
- 2-amino-3-carboxymuconate 6-semialdehyde.
References
- 1. Nishizuka Y., Ichiyama A., Gholson R. K., and Hayaishi O. (1965) Studies on the metabolism of the benzene ring of tryptophan in mammalian tissues. I. Enzymatic formation of glutaric acid from 3-hydroxyanthranilic acid. J. Biol. Chem. 240, 733–739 [PubMed] [Google Scholar]
- 2. Colabroy K. L., and Begley T. P. (2005) The pyridine ring of NAD is formed by a nonenzymatic pericyclic reaction. J. Am. Chem. Soc. 127, 840–841 10.1021/ja0446395 [DOI] [PubMed] [Google Scholar]
- 3. Stone T. W., and Darlington L. G. (2002) Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug Discov. 1, 609–620 10.1038/nrd870 [DOI] [PubMed] [Google Scholar]
- 4. Schwarcz R. (2004) The kynurenine pathway of tryptophan degradation as a drug target. Curr. Opin. Pharmacol. 4, 12–17 10.1016/j.coph.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 5. Cervenka I., Agudelo L. Z., and Ruas J. L. (2017) Kynurenines: tryptophan's metabolites in exercise, inflammation, and mental health. Science 357, eaaf9794 10.1126/science.aaf9794 [DOI] [PubMed] [Google Scholar]
- 6. Davis I., and Liu A. (2015) What is the tryptophan kynurenine pathway and why is it important to neurotherapeutics? Expert Rev. Neurother. 15, 719–721 10.1586/14737175.2015.1049999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ananieva E. (2015) Targeting amino acid metabolism in cancer growth and anti-tumor immune response. World J. Biol. Chem. 6, 281–289 10.4331/wjbc.v6.i4.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pilotte L., Larrieu P., Stroobant V., Colau D., Dolusic E., Frédérick R., De Plaen E., Uyttenhove C., Wouters J., Masereel B., and Van den Eynde B. J. (2012) Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 109, 2497–2502 10.1073/pnas.1113873109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pellicciari R., Liscio P., Giacchè N., De Franco F., Carotti A., Robertson J., Cialabrini L., Katsyuba E., Raffaelli N., and Auwerx J. (2018) α-Amino-β-carboxymuconate-ϵ-semialdehyde Decarboxylase (ACMSD) inhibitors as novel modulators of de novo nicotinamide adenine dinucleotide (NAD+) biosynthesis. J. Med. Chem. 61, 745–759 10.1021/acs.jmedchem.7b01254 [DOI] [PubMed] [Google Scholar]
- 10. Fukuoka S., Ishiguro K., Yanagihara K., Tanabe A., Egashira Y., Sanada H., and Shibata K. (2002) Identification and expression of a cDNA encoding human α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD). A key enzyme for the tryptophan-niacine pathway and “quinolinate hypothesis.” J. Biol. Chem. 277, 35162–35167 10.1074/jbc.M200819200 [DOI] [PubMed] [Google Scholar]
- 11. Huo L., Liu F., Iwaki H., Li T., Hasegawa Y., and Liu A. (2015) Human α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase (ACMSD): a structural and mechanistic unveiling. Proteins 83, 178–187 10.1002/prot.24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurnasov O., Goral V., Colabroy K., Gerdes S., Anantha S., Osterman A., and Begley T. P. (2003) NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10, 1195–1204 10.1016/j.chembiol.2003.11.011 [DOI] [PubMed] [Google Scholar]
- 13. Colabroy K. L., and Begley T. P. (2005) Tryptophan catabolism: identification and characterization of a new degradative pathway. J. Bacteriol. 187, 7866–7869 10.1128/JB.187.22.7866-7869.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lima W. C., Varani A. M., and Menck C. F. (2009) NAD biosynthesis evolution in bacteria: lateral gene transfer of kynurenine pathway in xanthomonadales and flavobacteriales. Mol. Biol. Evol. 26, 399–406 10.1093/molbev/msn261 [DOI] [PubMed] [Google Scholar]
- 15. Phillips R. S. (2014) Structure and mechanism of kynureninase. Arch. Biochem. Biophys. 544, 69–74 10.1016/j.abb.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 16. Wogulis M., Chew E. R., Donohoue P. D., and Wilson D. K. (2008) Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry 47, 1608–1621 10.1021/bi701172v [DOI] [PubMed] [Google Scholar]
- 17. Phillips R. S., Anderson A. D., Gentry H. G., Güner O. F., and Bowen J. P. (2017) Substrate and inhibitor specificity of kynurenine monooxygenase from Cytophaga hutchinsonii. Bioorg. Med. Chem. Lett. 27, 1705–1708 10.1016/j.bmcl.2017.02.080 [DOI] [PubMed] [Google Scholar]
- 18. Muraki T., Taki M., Hasegawa Y., Iwaki H., and Lau P. C. (2003) Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate 3,4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 69, 1564–1572 10.1128/AEM.69.3.1564-1572.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishino S. F., and Spain J. C. (1993) Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59, 2520–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He Z., Davis J. K., and Spain J. C. (1998) Purification, characterization, and sequence analysis of 2-aminomuconic 6-semialdehyde dehydrogenase from Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 180, 4591–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huo L., Davis I., Liu F., Andi B., Esaki S., Iwaki H., Hasegawa Y., Orville A. M., and Liu A. (2015) Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action. Nat. Commun. 6, 5935 10.1038/ncomms6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Y., Davis I., Ha U., Wang Y., Shin I., and Liu A. (2016) A pitcher-and-catcher mechanism drives endogenous substrate isomerization by a dehydrogenase in kynurenine metabolism. J. Biol. Chem. 291, 26252–26261 10.1074/jbc.M116.759712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin M., and Napoli J. L. (2000) cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and identification of a rat ortholog, ALDH12. J. Biol. Chem. 275, 40106–40112 10.1074/jbc.M008027200 [DOI] [PubMed] [Google Scholar]
- 24. Gouet P., Robert X., and Courcelle E. (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 10.1093/nar/gkg556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitman C. P., Aird B. A., Gillespie W. R., and Stolowich N. J. (1991) Chemical and enzymic ketonization of 2-hydroxymuconate, a conjugated enol. J. Am. Chem. Soc. 113, 3154–3162 10.1021/ja00008a052 [DOI] [Google Scholar]
- 27. Cobessi D., Tête-Favier F., Marchal S., Branlant G., and Aubry A. (2000) Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J. Mol. Biol. 300, 141–152 10.1006/jmbi.2000.3824 [DOI] [PubMed] [Google Scholar]
- 28. Park J., and Rhee S. (2013) Structural basis for a cofactor-dependent oxidation protection and catalysis of cyanobacterial succinic semialdehyde dehydrogenase. J. Biol. Chem. 288, 15760–15770 10.1074/jbc.M113.460428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li T., Walker A. L., Iwaki H., Hasegawa Y., and Liu A. (2005) Kinetic and spectroscopic characterization of ACMSD from Pseudomonas fluorescens reveals a pentacoordinate mononuclear metallocofactor. J. Am. Chem. Soc. 127, 12282–12290 10.1021/ja0532234 [DOI] [PubMed] [Google Scholar]
- 30. Li T., Ma J. K., Hosler J. P., Davidson V. L., and Liu A. (2007) Detection of transient intermediates in the metal-dependent non-oxidative decarboxylation catalyzed by α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase. J. Am. Chem. Soc. 129, 9278–9279 10.1021/ja073648l [DOI] [PubMed] [Google Scholar]
- 31. Li T., Iwaki H., Fu R., Hasegawa Y., Zhang H., and Liu A. (2006) α-Amino-β-carboxymuconic-ϵ-semialdehyde decarboxylase (ACMSD) is a new member of the amidohydrolase superfamily. Biochemistry 45, 6628–6634 10.1021/bi060108c [DOI] [PubMed] [Google Scholar]
- 32. Huo L., Fielding A. J., Chen Y., Li T., Iwaki H., Hosler J. P., Chen L., Hasegawa Y., Que L. Jr., and Liu A. (2012) Evidence for a dual role of an active site histidine in α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase. Biochemistry 51, 5811–5821 10.1021/bi300635b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huo L., Davis I., Chen L., and Liu A. (2013) The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in α-amino-β-carboxymuconate-ϵ-semialdehyde decarboxylase. J. Biol. Chem. 288, 30862–30871 10.1074/jbc.M113.496869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strohalm M., Hassman M., Kosata B., and Kodícek M. (2008) mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom. 22, 905–908 10.1002/rcm.3444 [DOI] [PubMed] [Google Scholar]