ABSTRACT
Increasing detection of small renal masses by imaging techniques entails the need for accurate discrimination between benign and malignant renal cell tumors (RCTs) as well as among malignant RCTs, owing to differential risk of progression through metastization. Although histone methylation has been implicated in renal tumorigenesis, its potential as biomarker for renal cell carcinoma (RCC) progression remains largely unexplored. Thus, we aimed to characterize the differential expression of histone methyltransferases (HMTs) and histone demethylases (HDMs) in RCTs to assess their potential as metastasis biomarkers. We found that SETDB2 and RIOX2 (encoding for an HMT and an HDM, respectively) expression levels was significantly altered in RCTs; these genes were further selected for validation by quantitative RT-PCR in 160 RCTs. Moreover, SETDB2, RIOX2, and three genes encoding for enzymes involved in histone methylation (NO66, SETD3, and SMYD2), previously reported by our group, were quantified (RT-PCR) in an independent series of 62 clear cell renal cell carcinoma (ccRCC) to assess its potential role in ccRCC metastasis development. Additional validation was performed using TCGA dataset. SETDB2 and RIOX2 transcripts were overexpressed in RCTs compared to renal normal tissues (RNTs) and in oncocytomas vs. RCCs, with ccRCC and papillary renal cell carcinoma (pRCC) displaying the lowest levels. Low SETDB2 expression levels and higher stage independently predicted shorter disease-free survival. In our 62 ccRCC cohort, significantly higher RIOX2, but not SETDB2, expression levels were depicted in cases that developed metastasis during follow-up. These findings were not apparent in TCGA dataset. We concluded that SETDB2 and RIOX2 might be involved in renal tumorigenesis and RCC progression, especially in metastatic spread. Moreover, SETDB2 expression levels might independently discriminate among RCC subgroups with distinct outcome, whereas higher RIOX2 transcript levels might identify ccRCC cases with more propensity to endure metastatic dissemination.
KEYWORDS: biomarker, histone methyltransferase, kidney cancer, metastasis, prognosis, renal cell tumor, renal cell carcinoma, RIOX2, SETDB2
Introduction
Kidney cancer incidence is increasing worldwide, with 62,700 new cases and 14,240 deaths estimated for 2016.1 Increasing incidence has been attributed to the rising number of incidental small renal tumors diagnosed due to widespread use of imaging techniques, as well as to aging, obesity, and smoking, which are known risk factors for the development of kidney cancer.2 Increased detection of small renal masses emphasizes the need for accurate discrimination not only between benign and malignant RCTs, but also among malignant RCTs subtypes. Indeed, renal cell carcinomas (RCCs) that are more likely to behave aggressively and to develop metastases should be clearly distinguished from those that will probably have a more indolent growth and might be managed more conservatively.3,4 Among RCCs, the most frequent are clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (pRCC), and chromophobe renal cell carcinoma (chRCC). Whereas ccRCC is the histotype that more frequently develops metastases, pRCC is more frequently multifocal and chRCC is mostly an indolent cancer that rarely develops metastases, although its differential diagnosis with oncocytoma, a benign tumor, might be challenging.5
Metastasis is the foremost cause of cancer-related mortality, despite improvements in diagnosis, surgical techniques, patient care, and adjuvant therapies. Biologic heterogeneity of tumor cells, as well as differences in metastatic tumor microenvironment at different sites may influence response to therapy.6 Thus, understanding pathogenesis of metastases at cellular and molecular level has become a major goal in cancer research.6,7 Indeed, management of metastatic renal cell carcinoma (mRCC) remains a major clinical challenge. Although median survival of patients with mRCC has been increasing due to therapeutic advances, specifically in antiangiogenic drugs and tyrosine-kinase inhibitors (from approximately 12 months with cytokine therapy to more than 26 months with VEGF inhibitors therapy),8 5-year survival for advanced kidney cancer was only 11.7% in the period 2007–2013.9 Albeit the proportion of patients with mRCC at diagnosis has declined, due to improved imaging techniques as well as more intense screening and incidental case ascertainment, a sizeable number of small RCCs (<4 cm diameter) may present renal capsule invasion, tumor thrombus, or lymphatic and distant metastasis,3,4 and their identification constitutes a major challenge.
Altered epigenetic homeostasis has been implicated in tumorigenesis and epigenetic-based biomarkers and may assist in diagnosis, prognostication, and prediction of response to targeted therapy.10 Histone modifications and chromatin modulators, in particular, have been shown to play an important role in cancer progression.11 In RCC, certain histone modifications associate with progression-free survival and correlate with pathological characteristics of tumors.12 In addition, defects in epigenetic enzymes, involved in chromatin remodeling and packaging, have been implicated in development of RCTs, reflecting the role of these mechanisms in renal tumorigenesis.13 Herein, we aimed to investigate the potential of HMTs and HDMs expression as biomarkers of metastatic progression in RCC, using two independent RCT cohorts, complemented with external validation in TCGA dataset. We selected SETDB2 (an HMT) and RIOX2 (an HDM), based on an extended characterization of histone methyltransferases in RCTs, previously reported by our group.14 Additionally, we also tested SMYD2 and SETD3 (both HMT), as well as NO66 (HDM), previously evaluated in our first RCT cohort and TCGA dataset,14 in the second cohort comprising ccRCC with indolent (non-metastatic) and aggressive (metastatic) disease.
Results
Validation of RIOX2 and SETDB2 expression in RCTs
RIOX2 and SETDB2 expression levels were assessed by quantitative RT-PCR in a series of 160 RCTs and 13 RNTs. The results were fully concordant with those of the TaqMan® Array as both genes were significantly overexpressed in RCTs compared to RNTs (P value <0.0001 for SETDB2 and <0.05 for RIOX2; (Fig. 1A and B). Moreover, RIOX2 and SETDB2 expression levels differed significantly between benign and malignant RCTs (Fig. 1C and D), and among the four RCT subtypes (Table 1). Oncocytomas displayed the highest SETDB2 and RIOX2 expression levels, followed by chRCC (Fig. 1E and F and Table 1). Pairwise comparisons demonstrated that SETDB2 and RIOX2 expression levels significantly differed between chRCC and both pRCC and ccRCC, and between pRCC and both chRCC and oncocytoma. Furthermore, SETDB2 transcript levels differed significantly between chRCCs and oncocytomas (Fig. 1E and F and Table 1).
Figure 1.
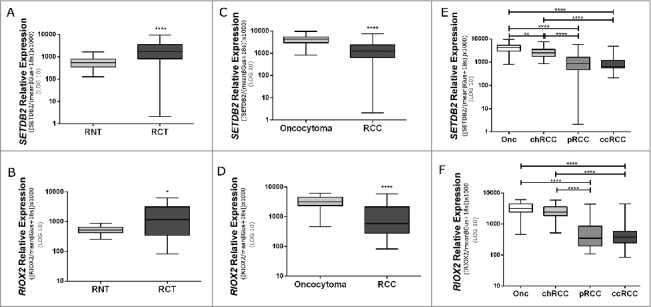
Expression levels of SETDB2 and RIOX2 in cohort #1. A: SETDB2 expression in renal cell tumors (RCTs) and renal normal tissues (RNTs). B: RIOX2 expression in renal cell tumors (RCTs) and renal normal tissues (RNTs). C: SETDB2 expression in benign tumors (oncocytoma) and malignant tumors [renal cell carcinoma (RCCs)]. D: RIOX2 expression in benign tumors (oncocytoma) and malignant tumors (renal cell carcinoma (RCCs)]. E: SETDB2 expression in renal cell tumors subtypes. F: RIOX2 expression in renal cell tumors subtypes. (P values: ****<0.0001; **<0.01; *<0.05).
Table 1.
Comparison of SETDB2 and RIOX2 expression among renal normal tissue (RNT), renal cell tumors (RCT), renal cell carcinoma (RCC), and RCT histotypes. For histotype pairwise comparison, the values were statistically significant when P value <0.0125 (Bonferroni's correction).
| SETDB2 (P value) | RIOX2 (P value) | |
|---|---|---|
| RNT vs. RCT | <0.001 | <0.05 |
| RNT vs. RCC | 0.001 | 0.444 |
| Oncocytoma vs. RCC | <0.001 | <0.001 |
| ccRCC vs. pRCC | 0.392 | 0.658 |
| ccRCC vs. chRCC | <0.001 | <0.001 |
| ccRCC vs. oncocytoma | <0.001 | <0.001 |
| pRCC vs. chRCC | <0.001 | <0.001 |
| pRCC vs. oncocytoma | <0.001 | <0.001 |
| chRCC vs. oncocytoma | <0.001 | 0.131 |
SETDB2 and RIOX2 expression levels and clinicopathological correlates
No significant differences in gender and age were apparent between patients and controls. In RCCs, no statistically significant associations were disclosed between SETDB2 and RIOX2 expression levels and Fuhrman or pathological stage categories. In RCTs, expression levels of both genes were significantly higher in females. Moreover, RIOX2 expression levels significantly associated with patient's age (P value = 0.015). In ccRCCs and pRCCs, SETDB2 expression levels were significantly lower in patients that developed metastases (Fig. 2A and B).
Figure 2.
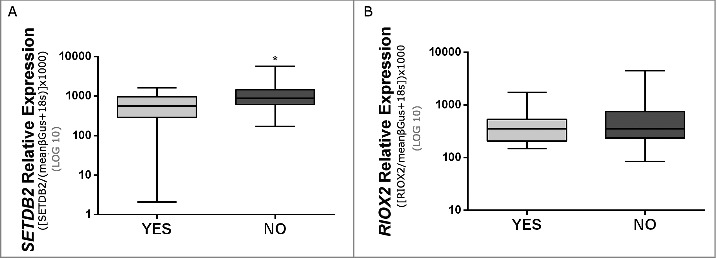
Expression levels of SETDB2 (A) and RIOX2 (B) in clear cell renal cell carcinomas and papillary renal cell carcinomas (cohort #1) with or without metastasis (P value <0.05).
SETDB2 and RIOX2 expression levels as prognostic markers
The median follow-up of RCC patients was 175 months (range: 2–375 months). A total of 15 patients died from RCC during this period. In univariable analysis, higher pathological stage (pT3 or higher) associated with shorter survival, whereas gender, age, histological subtype, and Fuhrman grade did not disclose any prognostic value within the available follow-up time. Disease-specific survival (DSS) analysis showed that low SETDB2 and RIOX2 levels were significantly associated with worse outcome (P value <0.01 and <0.05, respectively; (Fig. 3A and B). Concerning disease-free survival (DFS) analysis, low SETDB2 levels significantly associated with shorter time to disease progression (P value <0.0001; Fig. 3C). The same trend was observed for RIOX2, but statistical significance was not reached (P value = 0.055; Fig. 3D).
Figure 3.
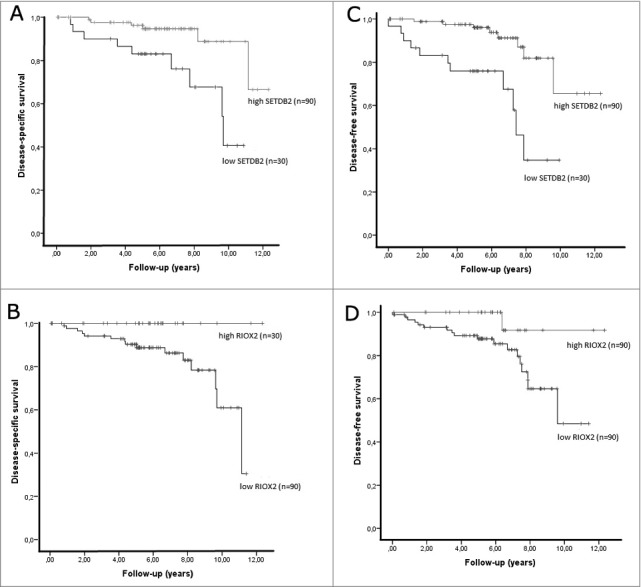
Kaplan-Meier estimated disease-specific survival curves and disease-free survival curves for SETDB2 (respectively A and C) and RIOX2 (respectively B and D).
In this series, only one case with local recurrence presented distant metastasis before local recurrence developed, thus DFS is equivalent to metastasis-free survival in this case. In multivariable analysis, a final model including SETDB2 expression levels and pathological stage was predictive of disease-free survival. Indeed, higher risk of disease progression was depicted for patients with higher pathological stage [HR: 3.03 (1.16-7.80), P value = 0.024] and lower SETDB2 expression levels [HR: 5.11 (1.72-15.24), P value = 0.003].
RIOX2, SETDB2, SETD3, SMYD2, and NO66 expression and risk of metastization in ccRCC
No significant differences were apparent for gender (P value = 0.570) and age (P value = 0.402) between ccRCCs patients that developed metastases and those that did not (cohort #2). Furthermore, no statistically significant associations were disclosed between SETDB2 and RIOX2 expression levels and Fuhrman grade or pathological stage in this cohort. In this cohort, expression levels of SETD3, SMYD2, and NO66, which we have previously found to associate with shorter disease-specific and disease-free survival,14 did not significantly differ between the two groups of ccRCC patients (Fig. 4C and E). Concerning RIOX2 and SETDB2 expression levels, only the former differed significantly between metastasized and non-metastasized ccRCCs (Fig. 4B).
Figure 4.
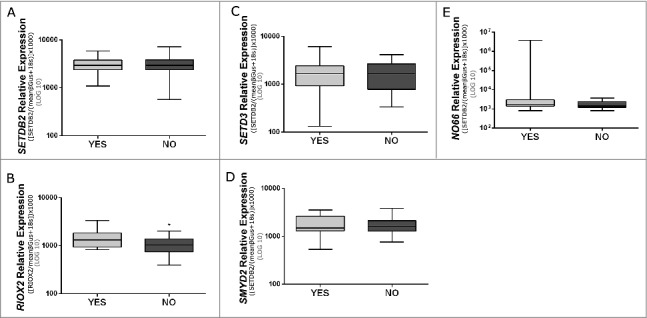
Expression levels of SETDB2 (A), RIOX2 (B), SETD3 (C), SMYD2 (D), and NO66 (E) in clear cell renal cell carcinomas (cohort #2) with or without metastasis (P value <0.05).
RIOX2 and SETDB2 expression in RCC Patients from TCGA Dataset
In TCGA dataset, significantly lower RIOX2 expression levels were found in RCC compared to RNT, contrarily to our results. Nevertheless, among RCCs, pairwise comparisons showed that RIOX2 expression levels were significantly higher in chRCCs compared to ccRCCs and pRCCs, paralleling our findings. In ccRCCs from TCGA database, no statistically significant difference was disclosed for RIOX2 expression levels between the group of patients that developed metastases and those that did not.
Concerning SETDB2, lower expression levels were depicted in RCC compared to RNT, as well. In line with our results, however, pairwise comparisons demonstrated that SETDB2 expression levels were significantly higher in chRCCs compared to ccRCCs and pRCCs, and expression levels significantly differed among subtypes. Considering only ccRCCs from TCGA database, no statistically significant difference was apparent for SETDB2 expression levels between the group of patients that developed metastases and those that did not, paralleling our results.
Discussion
Due to the widespread use of imaging tests, the frequency of incidentally detected RCTs has significantly increased, consisting mainly of small and early stage tumors. However, as lymph node and distant metastases may occur even in small RCCs,15 and the latter constitute the main cause RCC-related mortality,16 there is an unmet need for biomarkers capable of accurately discriminate tumors that will metastasize from those that will not, especially among pT1, which are the most amenable to nephron-sparing surgery. Because epigenetic-based biomarkers may assist in diagnosis, prognostication and prediction of response to targeted therapy,10 we hypothesized that histone modifications and chromatin modulators11 might aid in the identification of RCCs more prone to recur and metastasize. Indeed, previous reports have shown that, in RCC, histone modifications are associated with pathological features and DFS,12 and defects in chromatin remodelers and chromatin packaging are implicated in RCT development.13
In this study, we focused mostly on RIOX2 and SETDB2 expression levels as candidate biomarkers for RCC prognostication. Whereas RIOX2 encodes for an HDM, displaying high transcript or protein levels in RCC and several other cancers, associating with poor prognosis,17–24 SETDB2 encodes for an HMT involved in leukemogenesis,15 but was not previously associated with solid tumors. These two genes were selected based on previously published array data from our team,14 which were confirmed through analysis of cohort #1 tissue samples. Indeed, both RIOX 2 and SETDB2 were found overexpressed in RCTs compared to non-paired normal renal tissues, suggesting that their deregulation is associated with neoplastic transformation of renal parenchymal cells. Interestingly, renal oncocytomas displayed the highest RIOX2 and SETDB2 expression levels, significantly differing from RCCs. This result might be of practical value for distinction between oncocytoma and chRCC, especially the eosinophilic variant, as both histotypes display variable degree of morphological overlap, rendering differential diagnosis problematic, particularly in small core biopsies.25
Unexpectedly, when TCGA dataset was interrogated for RIOX2 and SETDB2 expression in renal tissues, RCCs displayed significantly lower expression levels than normal renal tissues, contrarily to our findings in cohort #1. Nonetheless, the expression ranking among RCCs in TCGA paralleled our findings, with chRCC displaying the highest expression levels, compared to ccRCC and pRCC. This discrepancy might be related with the origin of the “normal renal tissue” analyzed. Indeed, whereas in our study RNT samples derived from kidneys not harboring RCC, those of TCGA dataset were collected from macroscopically normal looking areas of organs involved in RCC. As we have previously shown, these “paired” normal renal tissue samples disclose significant epigenetic alterations that may precede neoplastic transformation.26 Thus, the results from cohort #1 and TCGA dataset analysis may not be directly comparable.
A major goal of this study was to ascertain the prognostic value of RIOX2 and SETDB2 expression levels in RCCs. Whereas, in univariable analysis, lower RIOX2 and SETDB2 expression levels associated with worse DSS, only lower SETDB2 levels associated with worse DFS. Interestingly, among standard clinicopathological parameters, only stage reached statistical significance. In multivariable analysis, however, only low SETDB2 expression and stage retained statistical significance, suggesting that assessment of SETDB2 expression might add relevant prognostic information for the management of RCC patients. Although a previous report has associated higher RIOX2 immunoexpression with shorter DSS in RCC,17 these results are not directly comparable with ours as we did not assess protein expression and the proportion of RCC subtypes also differed. Indeed, in our series, survival analysis results were mostly influenced by ccRCC and pRCC, which displayed the lowest expression levels among RCCs. Moreover, RIOX2 overexpression has been associated with worse prognosis in esophageal cancer24 but with favorable outcome in lung cancer,27 emphasizing that the biological and clinical impact of RIOX2 expression is strongly dependent on the primary location and the specific cancer type.
Because DFS was analogous to metastasis-free survival in cohort #1, we looked for differences in SETDB2 expression levels among tumors with and without metastasis. Interestingly, we found significantly lower SETDB2 expression in ccRCC and pRCC (the more clinically aggressive histotypes) with metastasis. However, when we attempted to validate these findings is an independent cohort comprising ccRCC with and without metastasis (cohort #2) and in ccRCCs from TCGA dataset, no significant differences were disclosed. Nonetheless, significantly higher RIOX2 transcript levels were depicted in ccRCC that developed metastasis during follow-up compared to matched ccRCC without metastasis, suggesting that RIOX2 expression might be a biomarker of progression (metastization) in ccRCC. Interestingly, high RIOX2 expression levels have been associated with development of lymphatic or distant metastasis in bile duct, gastric and pancreatic carcinomas.22,23,28 In TCGA dataset, however, no differences in RIOX2 expression levels were apparent between ccRCCs that developed metastases and those that did not. These discrepancies might be due to differences in follow-up time and enrolment criteria, as we excluded from analysis cases that presented metastasis at diagnosis and only analyzed cases in which metastases developed after curative-intent surgical treatment. Although we further evaluated three genes encoding for histone modifying enzymes (SMYD2, SETD3, and NO66) previously shown to be associated with worse prognosis in RCC14 in cohort #2, their expression levels did not significantly differ between ccRCC with and without metastasis.
Although our findings might be limited by the sample size, it should be emphasized that the most frequent histotypes are represented, whereas in many studies only ccRCC cases have been included. Moreover, survival analysis is based on long-term follow-up data, including two patient cohorts and TCGA dataset. Finally, although candidate biomarkers were previously identified in array-based analysis, validation in independent patient's series was undertaken, whereas several previous studies proposing array-based biomarkers for RCC, have not validated them or have just performed validation in limited series of patients,29–32 precluding the assessment of its clinical usefulness.
In conclusion, our results suggest that SETDB2 and RIOX2 might be involved in renal tumorigenesis and RCC progression, especially in metastatic spread. Moreover, SETDB2 expression levels might independently discriminate among RCC patient subgroups with distinct outcome, whereas higher RIOX2 transcript levels might identify ccRCC cases with more propensity to endure metastatic dissemination.
Materials and Methods
Patients and sample collection
A series of 160 RCTs (cohort #1) comprising 40 cases of each subtype (ccRCC, pRCC, chRCC, and oncocytoma) was prospectively collected from patients consecutively diagnosed and submitted to nephrectomy at the Portuguese Oncology Institute of Porto (IPO Porto). As controls, 13 renal normal tissue (RNT) samples were procured from patients submitted to nephro-ureterectomy due to upper urinary tract urothelial carcinoma, not involving the renal parenchyma. All tissues were immediately frozen and stored at −80°C. Sampling of more than 70% of malignant cells was confirmed using two slides stained with hematoxylin and eosin (H&E) taken before and after frozen section collection for RNA extraction. Relevant clinical data was retrieved from clinical charts.
An independent series of 62 ccRCCc (cohort #2) comprising 31 ccRCCs that have developed metastasis and 31 ccRCCs that did not progress, matched for gender, age, tumor size, grade, and stage at diagnosis were also retrieved from the archives. Tissue samples were prepared as described above.
This study was approved by the ethics committee of IPO Porto (CES-IPOPFG-EPE 518/10).
RNA extraction
For RNA extraction, samples were suspended in TRIzol® reagent (Invitrogen™, Carlsbad, CA, USA; Cat. #15596018) and chloroform (Merck Millipore, Darmstadt, Germany; Cat. #MCX10601) was added after the cells were lysed. RNA concentrations and purity ratios were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Samples were then stored at −80°c.
HMT and HDM selection
Selection of candidate HMTs and HDMs was based on the results of previously reported custom made TaqMan® Array 96-Well expression plates (Applied Biosystems®, Foster City, CA, USA; Cat. #4391528).14 SMYD2, SETD3, and NO66 had been previously validated and found to be associated with shorter disease-specific and disease-free survival.14 Additionally, SETDB2 and RIOX2 expression also displayed high fold variation between RCTs and RCCs, as well as between chRCC and oncocytomas, and were thus selected for further validation. Both presented higher expression in RCT compared to RNT, as well as in oncocytomas than in chRCC.
Validation of selected enzymes
RIOX2 and SEDTB2 mRNA expression levels were firstly evaluated in cohort #1. Subsequently, RIOX2, SETDB2, SMYD2, NO66, and SETD3 expression was assessed in cohort #2.
For validation in cohort #1, 300 ng of mRNA was reverse transcribed and amplified using TransPlex®Whole Transcriptome Amplification Kit (Sigma-Aldrich®, St. Louis, MO, USA) with subsequent purification using QIAquick PCR Purification Kit (QIAGEN, Germany), according to manufacturer's instructions. RIOX2 and SETDB2 mRNA levels were evaluated using TaqMan® Gene Expression Assays [Applied Biosystems®, Foster City, CA, USA; Hs99999908 m1 (GUSβ), Hs99999901 s1 (18S), Hs01126272 m1 (SETDB2), Hs00262155 m1 (RIOX2)] according to manufacturer's instructions. For each sample, expression levels were normalized using two internal reference gene, GUSβ and 18S, according to the formula: target gene relative expression = target gene expression level / ((GUSβ expression level + 18S expression level) / 2). Each plate included multiple non-template controls and serial dilutions of a cDNA Human Reference Total RNA (Agilent Technologies, La Jolla, CA, USA; Cat. #750500) to construct a standard curve.
For validation in cohort #2, 1 μg of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit according to the manufacturer's instructions. RIOX2, SETDB2, SMYD2, NO66, and SETD3 mRNA levels were evaluated using TaqMan® Gene Expression Assays [Applied Biosystems®, Foster City, CA, USA; Cat. #4331182 Hs00220210 m1 (SMYD2), Hs00260120 m1 (SETD3), Hs02743012 s1 (NO66), Hs99999908 m1 (GUSβ), Hs99999901 s1 (18S), Hs01126272 m1 (SETDB2), Hs00262155 m1 (RIOX2)] according to manufacturer's instructions. For each sample, expression levels were normalized using two internal reference gene, GUSβ and 18S, according to the formula: target gene relative expression = target gene expression level / ((GUSβ expression level + 18S expression level) / 2). Each plate included multiple non-template controls and serial dilutions of a cDNA Human Reference Total RNA (Agilent Technologies, La Jolla, CA, USA; Cat. #750500) in order to construct a standard curve.
TCGA dataset analysis in pRCC, chRCC, and ccRCCs patients
The Cancer Genome Atlas (TCGA) dataset was interrogated for data on RIOX2 and SETDB2 expression and clinical information, when available, from ccRCCs, pRCCs, and ccRCCs patients. All expression data from samples hybridized at the University of North Carolina, Lineberger Comprehensive Cancer Center, using Illumina HiSeq 2000 RNA Sequencing version 2 analysis, were downloaded from TCGA data matrix (http://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). This dataset included 533 ccRCC, 290 pRCC, and 66 chRCC. The provided value was pre-processed and normalized according to “level 3” specifications of TCGA (see http://cancergenome.nih.gov/dataportal/ for details). Biospecimen Core Resources (BCRs) provided the clinical data of each patient. This data is available for download through TCGA data matrix (http://tcga-data.nci.nih.gov/tcga/dataAccesMatrix.htm).
Statistical analysis
Non-parametric tests were used to ascertain statistical significance of comparisons among groups. Kruskal-Wallis test (KW) was used for comparisons among multiple groups and Mann-Whitney U test (MW) was used for pairwise comparisons. The prognostic significance of clinicopathological variables (age, gender, histological subtype, pathological stage, Fuhrman grade) and HMTs and HDMs expression levels was assessed by constructing disease-specific and disease-free survival curves using the Kaplan-Meier method with log-rank test (univariable test). The expression levels of SETDB2 and RIOX2 were classified as low or high based on the cutoff value of 25th percentile for SETDB2 expression and 75th percentile for RIOX2. A Cox-regression model comprising the different variables (multivariable test) was also constructed. For this analysis, the 120 RCC patients from cohort #1 were included.
Statistical significance was set at P value <0.05. Bonferroni correction was applied for pairwise comparisons following multiple groups' analyses. Statistical analysis was performed using SPSS software for Windows, version 22.0 (IBM-SPSS Inc., Chicago, IL, USA), and graphs were built using GraphPad Prism 6.0 software for Windows (GraphPad Software Inc., La Jolla, CA, USA).
Funding Statement
This study was funded by research grants from Research Center of Portuguese Oncology Institute - Porto (CI-IPOP 4-2012 and CI-IPOP 27) and from Associação Portuguesa de Urologia (APU-2010). ASP-L was supported by FCT-Fundação para a Ciência e a Tecnologia fellowship (SFRH/SINTD/94217/2013). CSG is supported by FCT- Fundação para a Ciência e Tecnologia PhD fellowships (SFRH/BD/92786/2013) and BMC is funded by FCT-Fundação para a Ciência e a Tecnologia (IF/00601/2012).
Disclosure statement
None of the authors have any conflict of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67:519–30. doi: 10.1016/j.eururo.2014.10.002. PMID:25449206 [DOI] [PubMed] [Google Scholar]
- 3.Guethmundsson E, Hellborg H, Lundstam S, Erikson S, Ljungberg B, Swedish Kidney Cancer Quality Register G . Metastatic potential in renal cell carcinomas </ = 7 cm: Swedish Kidney Cancer Quality Register data. Eur Urol. 2011;60:975–82. doi: 10.1016/j.eururo.2011.06.029. PMID:21741160 [DOI] [PubMed] [Google Scholar]
- 4.Jewett MA, Mattar K, Basiuk J, Morash CG, Pautler SE, Siemens DR, Tanguay S, Rendon RA, Gleave ME, Drachenberg DE, et al.. Active surveillance of small renal masses: progression patterns of early stage kidney cancer. Eur Urol. 2011;60:39–44. doi: 10.1016/j.eururo.2011.03.030. PMID:21477920 [DOI] [PubMed] [Google Scholar]
- 5.Moch H, Humphrey PA, Ulbright TM, Reuter VE. WHO Classification of Tumours of the Urinary System and Male Genital Organs. Lyon, France: IARC Press, 2016. [DOI] [PubMed] [Google Scholar]
- 6.Sleeman JP. The metastatic niche and stromal progression. Cancer Metastasis Rev. 2012;31:429–40. doi: 10.1007/s10555-012-9373-9. PMID:22699312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. PMID:17110329 [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Vida A, Hutson TE, Bellmunt J, Strijbos MH. New treatment options for metastatic renal cell carcinoma. ESMO Open. 2017;2:e000185. doi: 10.1136/esmoopen-2017-000185. PMID:28761748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.SEER cancer stat facts: kidney and renal pelvis cancer. Bethesda M, National Cancer Institute. [Google Scholar]
- 10.Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, Pickering MA, O'Loughlin MA, Ryan NK, Raymond WA, et al.. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–22. doi: 10.1158/1055-9965.EPI-10-0555. PMID:20841388 [DOI] [PubMed] [Google Scholar]
- 11.Chi P, Allis CD, Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nature reviews Cancer 2010; 10:457–69. doi: 10.1038/nrc2876. PMID:20574448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramakrishnan S, Ellis L, Pili R. Histone modifications: implications in renal cell carcinoma. Epigenomics 2013; 5:453–62. doi: 10.2217/epi.13.40. PMID:23895657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J, Goh XY, Vetter M, Pickering L, Swanton C. Epigenetic regulation in RCC: opportunities for therapeutic intervention? Nat Rev Urol. 2012;9:147–55. doi: 10.1038/nrurol.2011.236. PMID:22249190 [DOI] [PubMed] [Google Scholar]
- 14.Pires-Luis AS, Vieira-Coimbra M, Vieira FQ, Costa-Pinheiro P, Silva-Santos R, Dias PC, Antunes L, Lobo F, Oliveira J, Goncalves CS, et al.. Expression of histone methyltransferases as novel biomarkers for renal cell tumor diagnosis and prognostication. Epigenetics. 2015;10:1033–43. doi: 10.1080/15592294.2015.1103578. PMID:26488939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, Kipps TJ, Bullrich F, Croce CM. Cloning and characterization of CLLD6, CLLD7, and CLLD8, novel candidate genes for leukemogenesis at chromosome 13q14, a region commonly deleted in B-cell chronic lymphocytic leukemia. Cancer Res. 2001;61:2870–7. PMID:11306461 [PubMed] [Google Scholar]
- 16.Chin AI, Lam JS, Figlin RA, Belldegrun AS. Surveillance strategies for renal cell carcinoma patients following nephrectomy. Rev Urol. 2006;8:1–7. PMID:16985554 [PMC free article] [PubMed] [Google Scholar]
- 17.Ishizaki H, Yano H, Tsuneoka M, Ogasawara S, Akiba J, Nishida N, Kojiro S, Fukahori S, Moriya F, Matsuoka K, et al.. Overexpression of the myc target gene Mina53 in advanced renal cell carcinoma. Pathol Int. 2007;57:672–80. doi: 10.1111/j.1440-1827.2007.02156.x. PMID:17803656 [DOI] [PubMed] [Google Scholar]
- 18.Teye K, Tsuneoka M, Arima N, Koda Y, Nakamura Y, Ueta Y, Shirouzu K, Kimura H. Increased expression of a Myc target gene Mina53 in human colon cancer. Am J Pathol. 2004;164:205–16. doi: 10.1016/S0002-9440(10)63111-2. PMID:14695334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogasawara S, Komuta M, Nakashima O, Akiba J, Tsuneoka M, Yano H. Accelerated expression of a Myc target gene Mina53 in aggressive hepatocellular carcinoma. Hepatol Res. 2010;40:330–6. doi: 10.1111/j.1872-034X.2009.00604.x. PMID:20070393 [DOI] [PubMed] [Google Scholar]
- 20.Komiya K, Sueoka-Aragane N, Sato A, Hisatomi T, Sakuragi T, Mitsuoka M, Sato T, Hayashi S, Izumi H, Tsuneoka M, et al.. Mina53, a novel c-Myc target gene, is frequently expressed in lung cancers and exerts oncogenic property in NIH/3T3 cells. J Cancer Res Clin Oncol. 2010;136:465–73. doi: 10.1007/s00432-009-0679-0. PMID:19756735 [DOI] [PubMed] [Google Scholar]
- 21.Teye K, Arima N, Nakamura Y, Sakamoto K, Sueoka E, Kimura H, Tsuneoka M. Expression of Myc target gene mina53 in subtypes of human lymphoma. Oncol Reports. 2007;18:841–8. [PubMed] [Google Scholar]
- 22.Tan XP, Zhang Q, Dong WG, Lei XW, Yang ZR. Upregulated expression of Mina53 in cholangiocarcinoma and its clinical significance. Oncol Letters. 2012;3:1037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan XP, Dong WG, Zhang Q, Yang ZR, Lei XF, Ai MH. Potential effects of Mina53 on tumor growth in human pancreatic cancer. Cell Biochem Biophys. 2014;69:619–25. doi: 10.1007/s12013-014-9841-7. PMID:24522517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuneoka M, Fujita H, Arima N, Teye K, Okamura T, Inutsuka H, Koda Y, Shirouzu K, Kimura H. Mina53 as a potential prognostic factor for esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:7347–56. doi: 10.1158/1078-0432.CCR-03-0543. PMID:15534111 [DOI] [PubMed] [Google Scholar]
- 25.Volpe A, Finelli A, Gill IS, Jewett MA, Martignoni G, Polascik TJ, Remzi M, Uzzo RG. Rationale for percutaneous biopsy and histologic characterisation of renal tumours. Eur Urol. 2012;62:491–504. doi: 10.1016/j.eururo.2012.05.009. PMID:22633318 [DOI] [PubMed] [Google Scholar]
- 26.Costa VL, Henrique R, Ribeiro FR, Pinto M, Oliveira J, Lobo F, Teixeira MR, Jeronimo C. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer. 2007;7:133. doi: 10.1186/1471-2407-7-133. PMID:17645803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komiya K, Sueoka-Aragane N, Sato A, Hisatomi T, Sakuragi T, Mitsuoka M, Sato T, Hayashi S, Izumi H, Tsuneoka M, et al.. Expression of Mina53, a novel c-Myc target gene, is a favorable prognostic marker in early stage lung cancer. Lung Cancer. 2010;69:232–8. doi: 10.1016/j.lungcan.2009.10.010. PMID:19914733 [DOI] [PubMed] [Google Scholar]
- 28.Xing J, Wang K, Liu PW, Miao Q, Chen XY. Mina53, a novel molecular marker for the diagnosis and prognosis of gastric adenocarcinoma. Oncol Reports. 2014;31:634–40. doi: 10.3892/or.2013.2918. [DOI] [PubMed] [Google Scholar]
- 29.Jeronimo C, Henrique R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer Lett. 2014;342:264–74. doi: 10.1016/j.canlet.2011.12.026. PMID:22198482 [DOI] [PubMed] [Google Scholar]
- 30.Junker K, Hindermann W, von Eggeling F, Diegmann J, Haessler K, Schubert J. CD70: a new tumor specific biomarker for renal cell carcinoma. J Urol. 2005;173:2150–3. doi: 10.1097/01.ju.0000158121.49085.ba. PMID:15879877 [DOI] [PubMed] [Google Scholar]
- 31.Meyer HA, Tolle A, Jung M, Fritzsche FR, Haendler B, Kristiansen I, Gaspert A, Johannsen M, Jung K, Kristiansen G. Identification of stanniocalcin 2 as prognostic marker in renal cell carcinoma. Eur Urol. 2009;55:669–78. doi: 10.1016/j.eururo.2008.04.001. PMID:18450365 [DOI] [PubMed] [Google Scholar]
- 32.Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R, Slaby O. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Trans Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]


