Abstract
Regulation of the actin cytoskeleton is crucial for normal development and function of the immune system, as evidenced by the severe immune abnormalities exhibited by patients bearing inactivating mutations in the Wiskott Aldrich Syndrome Protein (WASP), a key regulator of actin dynamics. WASP exerts its effects on actin dynamics through a multi-subunit complex termed Arp2/3. Despite the critical role played by Arp2/3 as an effector of WASP-mediated control over actin polymerization, mutations in protein components of the Arp2/3 complex had not previously been identified as a cause of immunodeficiency. Here, we describe two brothers with hematopoietic and immunologic symptoms reminiscent of Wiskott Aldrich Syndrome (WAS). However, these patients lacked mutations in any of the genes previously associated with WAS. Whole exome sequencing (WES) revealed a homozygous 2 bp deletion, n.c.G623DEL-TC (p.V208fs), in Arp2/3 complex component ARPC1B, that causes a frame shift resulting in premature termination. Modeling of the disease in zebrafish revealed that ARPC1B plays a critical role in supporting T cell and thrombocyte development. Moreover, the defects in development caused by ARPC1B loss could be rescued by the intact human ARPC1B ortholog, but not by the p.V208fs variant identified in the patient. Moreover, we found that the expression of ARPC1B is restricted to hematopoietic cells, potentially explaining why a mutation in ARPC1B has now been observed as a cause of WAS, while mutations in other, more widely expressed, components of the Arp2/3 complex have not been observed.
Introduction
The actin cytoskeleton is a network of actin filaments that are polymerized from actin monomers. A key regulator of this process, in hematopoietic cells, is the WASP protein. WASP regulates actin polymerization by activating the Arp2/3 complex, allowing nucleation of new actin filaments and cross-linking them from end to side-branch (1). The Arp2/3 complex consists of seven subunits: Arp2, Arp3, Arc-p16, Arc-p20, Arc-p21, Arc-p34 and Arc-p41 (2). Among these subunits, Arc-p41, also known as ARPC1B, is proposed to have a regulatory role, facilitating assembly and maintenance of the whole complex.
The precise regulation of actin cytoskeleton dynamics is critical to nearly every stage of the immune response as evidenced by the widespread immunological defects observed upon disruption of this regulation (3). Consequently, it is perhaps not surprising that the molecules regulating this critical process have been linked to the etiology of immunodeficiency. The first described and most extensively studied actin-related protein causing primary immunodeficiency is WASP, resulting in Wiskott-Aldrich syndrome (WAS) (4). WAS is an X-linked immunodeficiency disease with a characteristic clinical phenotype that includes micro-thrombocytopenia, eczema, combined T and B cell immunodeficiency and an increased incidence of autoimmune manifestations and malignancies (5). Mutations in WIP, a known stabilizer of WASP, were also reported to cause a similar clinical phenotype (6). Mutations in the Arp2/3 complex or its activators, have long been sought as a cause of immunodeficiency syndromes with WAS-like pathologies; however, such mutations have not been observed, perhaps because many of these genes are essential for normal development and so the loss of their function would likely result in early lethality (7). In support, the loss of Arp2/3 function in Arc-p21-deficient mice is embryonic lethal (8). Nevertheless, here we report the finding of two brothers with a WAS-like clinical phenotype that harbor a novel mutation in the ARPC1B gene. A distinct mutation in this gene was also recently reported in a patient with defects in platelets and neutrophils (9). We describe here the clinical phenotype of the patients and definitively link their developmental defects in platelets and T cells to the ARPC1B mutation by recapitulating the disease in a zebrafish model.
Materials and Methods
Patients
The patients were diagnosed at the “Edmond and Lily Safra” Children’s Hospital. The Institutional Review Board (Sheba Medical Center, Tel Hashomer) approved this study and a written informed consent was obtained from their parents.
Immunological evaluation
Cells surface markers of peripheral blood mononuclear cells (PBMCs), lymphocyte proliferative responses to mitogens, and the amount of signal joint T-cell receptor excision circles (TRECs) were determined as previously described (10). Serum concentration of immunoglobulins was measured by nephelometry.
ARPC1B immunostaining
Formalin fixed tissues were dehydrated, embedded in paraffin and sectioned at 4 μm. A positive control was added on the right side of the slides. The slides were warmed up to 60°C for 1 hour, followed by fully automated processing. The ARPC1B immunostaining was calibrated on a Benchmark XT staining module (Ventana Medical Systems). Briefly, after sections were dewaxed and rehydrated, a CC1 Standard Benchmark XT pretreatment (Ventana Medical Systems) for antigen retrieval was selected. ARPC1B antibody (Novus Biologicals, USA, NBP1-90114) was diluted 1:100 and incubated 40 minutes at 37°C. Detection and counterstaining were performed with an ultraView detection kit (Ventana Medical Systems) and hematoxylin (Ventana Medical Systems), respectively. At the end of the automated run, slides were dehydrated by passage through increasing concentrations of ethanol. Sections were then cleared in xylene and mounted with Entellan followed by analysis by a pathologist.
IGH and TRG immune repertoire sequencing by NGS
TCR and BCR libraries were generated from at least 150 ng of genomic DNA from the patient’s peripheral blood using primers for conserved regions of V and J genes in the TRG (T cell receptor Gamma) and IGH (immunoglobulin heavy chain) loci respectively, according to manufacturer’s protocol (LymphoTrack, Invivoscribe Technologies, France and USA). Quantified libraries were pooled and sequenced using Mi-Seq Illumina technology (Illumina Inc, USA). For the TRB (T cell receptor Beta) libraries, 2 μg of the same genomic DNA and primers from the V and J genes in the TRB locus were used (Adaptive biotechnologies, USA). The sequences were subjected to bioinformatic analyses (carried out by Invivoscribe Technologies and Adaptive Biotechnology) to generate FASTA sequence files for all the libraries, which then were submitted to the IMGT HighV-QUEST webserver (http://www.imgt.org) and analyzed further for Hierarchical Treemap (Macrofocus Gmbh, Switzerland), Shannon’s H, and Simpson’s D diversity indices and frequency of the different genes. Shannon’s H and Simpson’s D were calculated using the following equations:
R = Total sequences; i = Unique sequences; pi = proportion of the total sequences belonging to the “i”th unique sequence.
Whole exome and Sanger Sequencing
High throughput sequencing was performed for patient 1 and his parents, by Illumina Hiseq2500 using 2×100 bp pair-end. Exome capturing was done using the Illumina Nextera DNA sample preparation kit. Overall ~50M sequence reads were produced for each sample. BWA-mem algorithm(11) for alignment versus the hg19 version of the human genome was applied. Around 40M reads were properly aligned. The median coverage was around 40 reads per base. GATK(12) version 2.4.7 with the UnifiedGenotyper algorithm was applied for variant calling including all steps mentioned in the best practice pipeline. KGG-seq (13) was used for annotation of detected variants and for comparing with allele frequency population databases. In-house scripts were applied for filtering, based on family pedigrees and intersections. The ARPC1B mutation was validated by dideoxy Sanger sequencing in the patients and carriers. PCR amplification products were directly sequenced using BigDye 3.1 Terminator chemistry (Applied Biosystems) and separated on an ABI 3500 genetic analyzer (Applied Biosystems). Data were evaluated using Sequencer v5.0 software (Gene Codes Corporation, Ann Arbor, MI). The resulting data were deposited as a batch deposit of ‘Human; version 1.0’ samples in the NCBI BioSample database (http://www.ncbi.nlm.nih.gov/biosample/; (Submission SUB2910482; Accession STUDY; PRJNA397356, SRP114959).
Identification of zebrafish arpc1b ortholog
The zebrafish ortholog (NM_213156) of human ARPC1B was identified using the reciprocal ‘best-hits’ BLAST search strategy (14). Then multiple alignments of amino acid sequences for human, mouse and zebrafish ARPC1B were obtained by using the Clustal Omega algorithm (http://www.ebi.ac.uk/Tools/msa/clustalo/). Synteny analysis for zebrafish arpc1b was performed as described previously (15).
Zebrafish experiments and Morpholino knockdown
Wild-type zebrafish (AB), Tg(cd41:EGFP)la2 and Tg(lck:EGFP)cz2 transgenic lines were maintained under standard conditions as previously described (16). Two different morpholino oligonucleotides (MO) against arpc1b were designed and purchased from Gene Tools (Gene Tools, LLC, Philomath, OR, USA). The sequences of the arpc1b-ATG and splice-site MO are as follows: MO: arpc1b-ATG, 5′-ATGGTCGCTTTTCCCAGGAAACCGT-3′); and arpc1b exon4 (arpc1b-i3e4) MO, 5′-AATGCCTTGGTGTTGATGAGAAAAC-3′. All MO were injected at the 1-cell stage. The efficacy of ATG-MO for translational silencing was tested by EGFP reporter assay at 6hpf and 24hpf (17). The knockdown effectiveness of splicing MO was validated by RT-PCR at 26hpf. The RT-PCR primers were: Forward:5′-AACAAAATCCACGTGCTGAAGG -3′, Reverse:5′-ACCCATCCTGAAGTACCTGTTG -3′.
In Situ hybridization
Whole mount in situ hybridization (WISH) for arpc1b was performed as previously described (18). Primers used to clone the arpc1b probe sequence into pCS2+ vector were: Forward: 5′-TACGAATTCGTGCGTGACTTTCATCACTGAG-3′; Reverse:5′-TACCTCGAGAAAAGCAATCTATGACCAGCATT-3′.
Primers for cloning the arpc1a WISH probe into pCS2+ vector were: Forward: 5′-ATAGGATCCCCAGTGGAAACAGATTGGCTTG -3′; Reverse:5′-ATACTCGAGAGTGTCACAACAGTTCACACCA-3′. The embryos were mounted in 3% methylcellulose and images were taken from the Nikon SMZ1500 stereomicroscope equipped with DS-Fi1 digital camera and Nikon Ar imaging software.
Plasmid constructs and Rescue experiments in zebrafish
For rescue experiments in zebrafish, the human WT ARPC1B coding sequence was PCR-amplified using cDNA from human Jurkat cells and then cloned into pCS2+ vector. The PCR primers used were: Forward:5′-CGCGATCGATATGGCCTACCACAGCTTCCT -3′; Reverse:5′-CGCGCTCGAGTCATTTGATCTTGAGGTCCTT-3′. Site-directed mutagenesis was subsequently performed to recreate the patient allele of ARPC1B (c.G623DEL-TC; P.V208fs) according to manufacturer’s instruction (Invitrogen, Geneart kit). Capped mRNAs were made from these pCS2+ plasmids using the mMessage mMachine kit (Ambion) and then co-injected with arpc1b-i3e4-MO into 1-cell stage Tg(cd41:EGFP)la2 embryos for rescue analysis. For heat shock mediated inducible expression of human WT and mutant ARPC1B, their cDNAs were subcloned into the heat-inducible pSGH2 vector (19). Plasmids were mixed with 0.5×I-SceI buffer, and 0.5 units/l I-SceI meganuclease (New England Biolabs) and microinjected (150pg) into 1-cell stage embryos. Injected embryos were heat shocked at 37° for 1hr at 20hpf, following which GFP+ embryos were analyzed by WISH at 5dpf.
Results
Clinical characteristics
The index patient (p1) was a male child of consanguineous Jewish parents. Family history was negative for immunodeficiency or thrombocytopenia. He was initially evaluated during infancy for colitis, an eczematous rash, papulovesicular lesions on the scalp and severe thrombocytopenia. No bleeding tendency was noted. Later, he developed severe life-threatening infections including peri-anal abscess, lymphadenitis, recurrent chest infections and CMV pneumonitis. He was placed on IVIG treatment and prophylactic antibiotics. At the age of 5 years, while awaiting bone marrow transplantation, the patient had a severe adenovirus infection, which resulted in multi-organ failure and death. His younger brother (p2) was evaluated at age of 7 weeks because of fever, severe eczema, purulent otitis media, bloody mucous stools and thrombocytopenia. Blood cultures were positive for pneumococcus pneumonia and enterococcus. This patient unfortunately succumbed to severe veno-occlusive disease (VOD) after bone marrow transplantation from a matched related donor. Initial laboratory values of both brothers during infancy are shown in Table 1. In both patients, similar immunologic abnormalities were demonstrated including reduced thymic output, increased B lymphocytes and high IgG and IgE levels. Analysis of peripheral blood obtained from patient 1 at year of age (Table 1) revealed reduced percentages and numbers of CD3+ cells, with CD8+ cells more affected than CD4+ cells. The reduction in CD8+ T cells persisted until 4 years of age (Table 1). The number of B cells was elevated, whereas the percentage and number of CD56+ NK cells were normal. Serum immunoglobulins increased remarkably from birth to age 4. The response to PHA mitogenic stimulation was normal in pt1 but reduced in pt2 (Table 1). Importantly, the response to anti-CD3 mitogenic stimulation was profoundly reduced in both patients, as has been observed previously in patients with WAS (20). Thymus output at 4 years of age, as determined by T cell receptor excision circles (TREC) in peripheral blood (10), was undetectable. Similar immunologic abnormalities were found in p2, p2 including reduced TREC and an impaired response to anti-CD3 mitogenic stimulation. P1’s bone marrow was hyper-cellular with megakaryocytic hyperplasia (Data not shown). The red cell lines showed relatively numerous immature cells. Eosinophil numbers were mildly increased.
TABLE 1.
Laboratory characteristics of patients during the first year of life (patient 1 and 2) and at age 4 (patient 1). Age-matched reference ranges (in square brackets) are provided for both patients
| Value | Patient 1 | Patient 2 | Patient 1 |
|---|---|---|---|
| Age when blood tests were performed (months) | 12 | 2 | 48 |
| Complete blood count | |||
| WBC (K/micl) | 33.7 [6.1–13] | 13.8 [7.2–18] | 15.3 [5.2–11] |
| Hemoglobin (g/dl) | 9.5 [10.5–14] | 11.3 [10.5–14] | 13.5 [11–14.5] |
| Platelets (K/micl) | 46 [150–400] | 76 [150–400] | 42 [150–400] |
| Lymphocyte Subsetsa (cells per microliter ×10−3) | |||
| Lymphocytes | 11.6 [3.6–8.9] | 6.1 [3.4–7.6] | 7.1 [2.3–5.4] |
| T (CD3+) | 1.29 [2.1–6.2] | 2.6 [2.5–5.5] | 1.77 [1.4–3.7] |
| T helper (CD4+) | 1.15 [1.3–3.4] | 1.7 [1.6–4] | 1.56 [0.7–2.2] |
| T cytotoxic (CD8+) | 0.54 [0.6–2] | 0.5 [0.6–1.7] | 0.21 [0.49–1.3] |
| CD3+αβ+ | ND | ND | 28% [>20%] |
| CD3+γδ+ | ND | ND | 9% [<5%] |
| CD20+ | 7.57 [0.7–2.6] | 3.02 [0.3–2] | 4.54 [0.4–1.4] |
| CD20+ (%) | 70% [16–35%] | 44% [6–32%] | 52% [5–25%] |
| CD56+ | 0.216 [0.2–0.9] | 0.36 [0.17–1.1] | 0.55 [0.15–0.5] |
| T cell proliferation (cpmb) | |||
| PHA 6 μg/ml | ND | 26300 [56000] | 81000 [84000] |
| PHA 25 μg/ml | ND | 59040 [100000] | 128000] 120000] |
| Anti-CD3 | ND | 4200 [13000] | 37000] 10400] |
| TREC | ND | 258 [> 400] | 0 [> 400] |
| Serum Ig | |||
| IgM (mg/dl) | 103 [40–170] | 97 [17–105] | 742 [40–200] |
| IgA (mg/dl) | 761 [10–100] | 86 [2–50] | 625 [33–230] |
| IgG (mg/dl) | 1750 [170–1000] | 930 [250–900] | 3100 [600–1500] |
| IgE (IU/ml) | 3590 [0–100] | 1790 [0–100] | ND |
CPM - Counts Per Minute; PHA – phytohaemagglutinin. ND –not done
control numbers represents 50–100 healthy donors, ages 1–2 years, with percentages/counts presented as median (10th and 90th percentiles) (depicted from ref 33)
H3-thymidine uptake in response to mitogens.
Differential restriction and clonal expansion of patients’ T cell receptor, but not immunoglobulin, repertoire
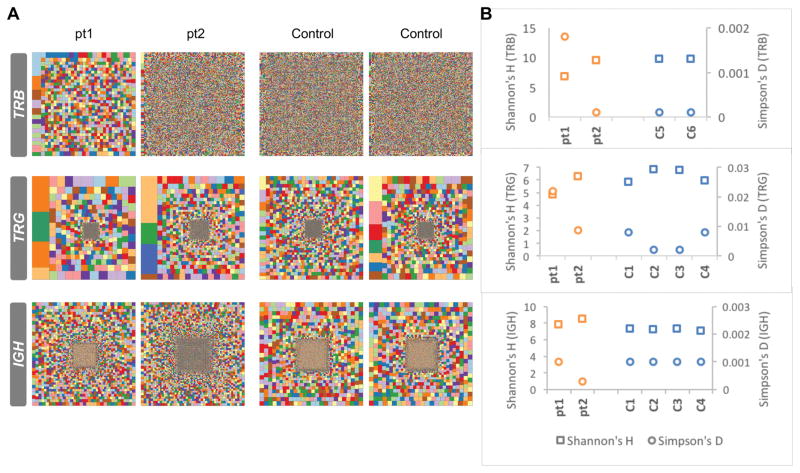
The patients displayed clinical symptoms involving both T and B lymphocyte immunity; therefore, their T cell receptor (TCR) and B cell receptor (BCR) repertoires were investigated in-depth. TCR and BCR repertoires were determined by specifically sequencing the variable regions of the TCR beta (TRB) and gamma (TRG) and immunoglobulin heavy chain (IGH). The complexity of the repertoires was represented graphically by Treemap, where each square represents a unique CDR3 (based on amino acids) and the size of the square represents the frequency. The Treemaps illustrate that the TRB and TRG repertoires of pt1 are restricted and clonally expanded, whereas those of pt2 are not restricted and contain only few expanded clones (Fig. 1A). The diversity of the patients’ IGH repertoire is indistinguishable from the normal control (Fig. 1A). In order to quantify the overall diversity of the repertoires, we used the Shannon’s H diversity measure, which estimates diversity by taking into account the number of unique CDR3 reads relative the total number of sequences obtained from an individual. Thus, we see that relative to controls, pt1’s Shannon’s H index of diversity was slightly decreased and the Simpson index of unevenness was markedly increased for both the TRB and TRG repertoires (Fig. 1B). Interestingly, this is not observed in pt2, where there is no clear difference from the controls for either the TRB and TRG repertoires for pt2 (Fig. 1B). The Shannon’s H and Simpson’s D diversity indices for the IGH repertoires of both patients are comparable to controls (Fig. 1B). Nevertheless, there were slight alterations in the Shannon’s H and Simpson’s D values for pt2, perhaps attributable to the increased B cell numbers (Fig. 1B). Taken together, both graphical and quantitative measures of the diversity of the repertoire suggest that the TRB and TRG repertoire for pt1, but not for pt2, are restricted and clonally expanded, whereas the IGH repertoires of both patients are largely unaffected.
FIGURE 1. Analysis of the patients’ TRB, TRG and IGH repertoires.
Hierarchical Treemaps (A), Shannon’s H, and Simpson’s D diversity indices (B) were generated for TRB, TRG and IGH repertoires by analysis of NGS of samples from two patients and two healthy donor controls for TRB repertoire and four healthy donor controls for TRG and IGH repertoires.
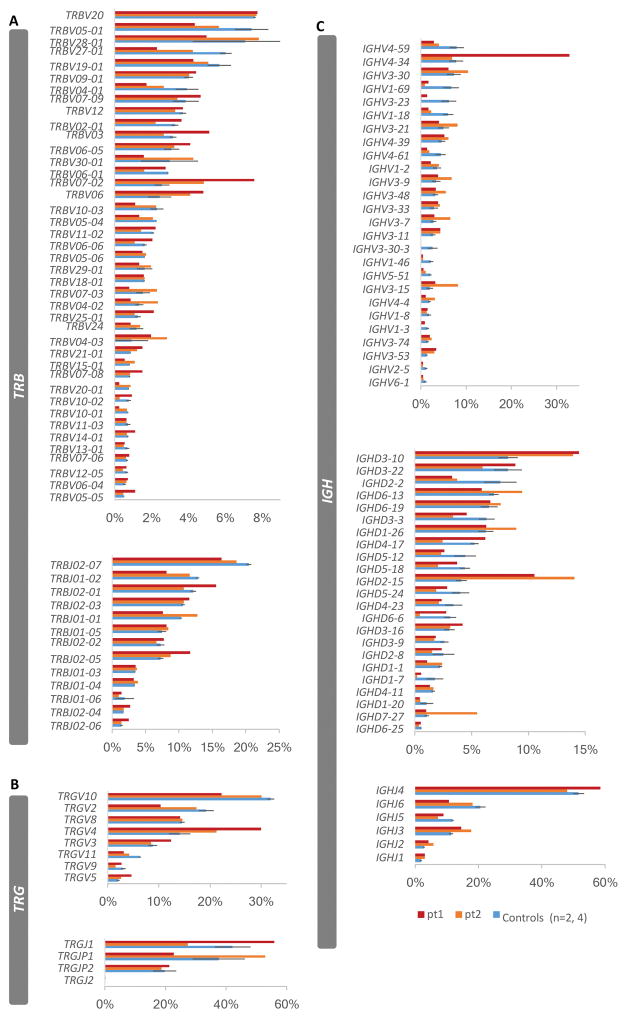
Differential V(D)J gene usages in the TRB, TRG, and IGH repertoires of the patients
We determined whether, as for the patient’s CDR3 sequences, we would observe alterations in V(D)J gene usage. Therefore, we first calculated the frequencies of the V(D)J gene usage for the TRB, TRG and IGH repertoires based on the total sequences. The gene usages of TRBV7-2, TRBV20-1 and TRBJ2-5 of the TRB repertoire of both patients showed two standard errors above the mean of the controls (Fig. 2A and Supplementary Table I). As for the TRG repertoire, TRGV4 and TRGV5 genes were preferentially utilized, with two standard errors above the mean of the controls only for pt1 (Fig. 2B). Although the overall diversity of the IGH repertoire of the patients was comparable to controls, IGHV3-11, IGHV3-53, IGHD3-10, IGHD2-15, IGHJ3, IGHJ2 and IGHJ1 are preferentially utilized in both patients, with two standard errors above the mean of the controls (Fig. 2C). In addition, two standard errors above the mean of the controls are observed for genes TRBV20, TRBV3, TRBV6-5, TRBV11-2, TRBV6-6, TRBV25-1, TRBV7-8, TRBV14-1, TRBV7-6, TRBV6-4, TRBV5-5, TRBJ2-1, TRBJ2-3, TRBJ2-4, TRBJ2-6, TRGV3, TRGV5, IGHV4-34, IGHV3-66, IGHV3-43 and IGHD3-16, IGHD1-14 and IGHJ4 in pt1, and observed for the genes TRBV5-6, TRBV4-2, TRBV4-3, TRBV15-1, TRBV20-1, TRBJ1-1, TRBJ1-3, TRBJ1-4, IGHV3-30, IGHV3-21, IGHV3-9, IGHV3-48, IGHV3-7, IGHV3-15, IGHV4-4, IGHV3-74, IGHV3-13, IGHD6-13, IGHD1-26 and IGHD7-27 in pt2 (Fig. 2 and Supplementary Table I). In addition, we have also calculated the V(D)J gene usages based on unique sequences. Overall the gene usage profiles analyzed on unique sequences (Supplementary Fig 1) were similar to the analysis of done on total sequences, except for IGHV3-11, IGHJ1 and IGHJ2, which were no longer exhibited a significant difference in representation relative to that in controls (Supplementary Table 1). Moreover, the representation of TRBJ1-3, TRBJ2-4, TRGV5, IGHV3-73, IGHD1-26 and IGHD2-15 was only significantly greater than that in controls when analyzed relative to unique sequences (Supplementary Table 1). Thus, the V gene repertoires are altered in these patients, with some genes being preferentially used in both patients while others are more highly represented in only one of the patients compared to the controls. This is largely the case irrespective of comparing representation to the total number of sequences or to the number of unique sequences. Taken together, our data demonstrates that indeed there is differential usage of multiple V(D)J genes in the T cell population as well as in the B cell population. The alteration in B cell V(D)J usage may be a consequence of the inability of T cells to productively support B cell activation.
FIGURE 2. Differential VDJ gene usage relative to total sequences in the that are patients’ TRB, TRG and IGH repertoires.
The frequencies of gene usages based on total sequences for the patients for TRBV and TRBJ genes (A), TRGV and TRGJ genes (B) and IGHV, IGHD and IGHJ genes (C), were compared to the frequencies (average ± SEM) of gene usage of two controls in TRB repertoire and four controls in TRG and IGH repertoires.
Genetic evaluation
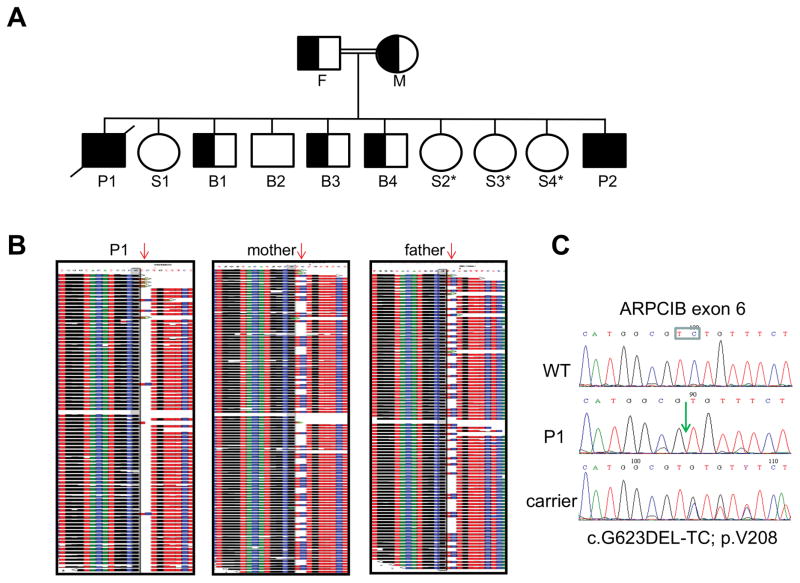
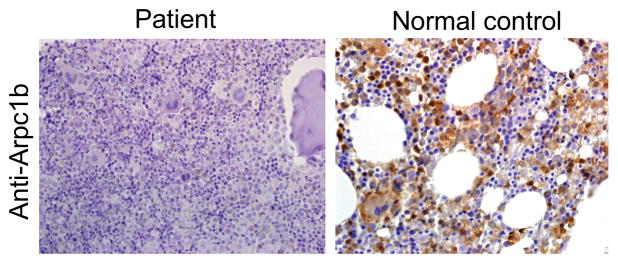
The consanguinity of the patient’s parents suggested autosomal recessive inheritance (Fig. 3). The clinical constellation and initial workup of the patient were suggestive of a WAS like syndrome. Nevertheless, direct dideoxy-sequencing of the WAS, WASL and WIP coding regions revealed no abnormalities. Consequently, whole exome sequencing (WES) was performed for patient 1 and his parents (trio). Bioinformatic analysis filtered out variants present in ≥0.01 of our in-house exomes (n=700) and variants with a minor allele frequency (MAF) of ≥ 0.01 in the 1000 Genomes Project (1KG; http://browser.1000genomes.org/index.html), the dbSNP 135 database, or the NHLBI Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/). Recessive analysis reduced the variants list of the patient to 22 candidate genes (Table 2). The most attractive variant among these candidates was ARPC1B, with a homozygous 2 bp deletion, n.c.G623DEL-TC; p.V208fs, causing a putative frame shift resulting in premature termination (Fig. 3A,B). This gene was pursued because of its known interaction with WASP. Dideoxy Sanger sequencing confirmed the presence of this mutation in the patient, which fully segregated within the family (Fig. 3C). Pt2 was genetically diagnosed based on his similar clinical and laboratory characteristics and was determined to have the same 2 bp deletion by direct sequencing of the ARPC1B gene. The 2bp deletion led to loss of ARPB1C expression, since immune staining of bone marrow from the patient with anti-ARPB1C antibody, failed to detect protein, which was readily detected in the cytoplasm of bone marrow cells from a normal healthy control (Fig. 4). This pathology finding indicates that full protein was not produced. Since the anti-ARPB1C antibody used does not recognize the ARPC1B N-terminal, the possible presence of a truncated protein cannot be excluded. Nevertheless, the genetic analysis suggested that the 2bp deletion in ARPB1C was responsible for the immune deficiency in these patients, as well as for their other clinical manifestations. In-depth analysis of the remaining candidates (Table 2) failed to explain the immunodeficiency in our patients.
FIGURE 3. Genetic and genomic analysis of the patient’s inheritance of the ARPB1C mutation.
A, Family pedigree. Solid symbols represent the affected subjects P1 (diagonal line, deceased) and P2. Half solid symbols represent unaffected relatives, which are carriers for the mutation. Open symbols represent unaffected relatives, including three sisters, marked by asterisks, that did not undergo genetic analysis for the familial mutation but clinically are unaffected. F=father; M=mother; P=patient; S=sister; B=brother. B, Whole exome sequencing of patient 1 and his parents (trio). Deep sequences around the 2 bp deletion (red arrow) are shown, demonstrating the homozygosity of the patient and the heterozygosity of his parents. C, Dideoxy Sanger sequencing of the different ARPC1B genotypes detected in the studied pedigree. The deleted TC nucleotides are boxed and their position in the patient’s sequence is marked by a green arrow.
Table 2.
Whole exome sequencing analysis, filtered for de-novo variants, revealing 22 candidate genes including ARPC1B*
| Chr | Pos | Gene | Description |
|---|---|---|---|
| 19 | 56561923 | NLRP5 | NLR FAMILY, PYRIN DOMAIN CONTAINING 5 (APPROVED) |
| 19 | 51848635 | ETFB | ELECTRON-TRANSFER-FLAVOPROTEIN, BETA POLYPEPTIDE (APPROVED) |
| 7 | 98988637 | ARPC1B | ACTIN RELATED PROTEIN 2/3 COMPLEX, SUBUNIT 1B, 41KDA (APPROVED) |
| 3 | 195508503 | MUC4 | MUCIN 4, CELL SURFACE ASSOCIATED (APPROVED) |
| 3 | 195508498 | MUC4 | MUCIN 4, CELL SURFACE ASSOCIATED (APPROVED) |
| 10 | 129055687 | DOCK1 | DEDICATOR OF CYTOKINESIS 1 (APPROVED) |
| 2 | 120776692 | EPB41L5 | ERYTHROCYTE MEMBRANE PROTEIN BAND 4.1 LIKE 5 (APPROVED) |
| 1 | 175372381 | TNR | TENASCIN R (APPROVED) |
| 10 | 120801964 | EIF3A | EUKARYOTIC TRANSLATION INITIATION FACTOR 3, SUBUNIT A (APPROVED) |
| 19 | 52091623 | ZNF175 | ZINC FINGER PROTEIN 175 (APPROVED) |
| X | 15721061 | CA5BP1 | CARBONIC ANHYDRASE VB PSEUDOGENE 1 (APPROVED) |
| 19 | 54760648 | LILRB5 | LEUKOCYTE IMMUNOGLOBULIN-LIKE RECEPTOR, SUBFAMILY B (WITH TM AND ITIM DOMAINS), MEMBER 5 (APPROVED) |
| 15 | 101606082 | LRRK1 | LEUCINE-RICH REPEAT KINASE 1 (APPROVED) |
| 7 | 122027137 | CADPS2 | CA++-DEPENDENT SECRETION ACTIVATOR 2 (APPROVED) |
| 2 | 131521888 | AMER3 | APC MEMBRANE RECRUITMENT PROTEIN 3 (APPROVED) |
| 20 | 31685511 | BPIFB4 | BPI FOLD CONTAINING FAMILY B, MEMBER 4 (APPROVED) |
| 7 | 122635687 | TAS2R16 | TASTE RECEPTOR, TYPE 2, MEMBER 16 (APPROVED) |
| 1 | 169337581 | BLZF1 | BASIC LEUCINE ZIPPER NUCLEAR FACTOR 1 (APPROVED) |
| 4 | 70146635 | UGT2B28 | UDP GLUCURONOSYLTRANSFERASE 2 FAMILY, POLYPEPTIDE B28 (APPROVED) |
| 7 | 100637805 | MUC12 | MUCIN 12, CELL SURFACE ASSOCIATED (APPROVED) |
| 7 | 100682216 | MUC17 | MUCIN 17, CELL SURFACE ASSOCIATED (APPROVED) |
| 19 | 55972877 | ISOC2 | ISOCHORISMATASE DOMAIN CONTAINING 2 (APPROVED) |
Table 2 summarizes the chromosome number (Chr), chromosomal position (Pos), and description of the gene function for each of the 22 candidate variant genes identified by whole exome sequencing analysis of patient 1.
FIGURE 4. Expression of ARPB1C protein in the patient’s bone marrow cells.
Bone marrow was immunostained with anti-ARPC1B, which revealed that immunoreactivity to ARPB1C was absent from the patient’s bone marrow cells (left panel), but was abundant in the normal control (right panel). The magnification of the images in 400X.
Identification of the zebrafish ortholog of human ARPC1B
In order to test the role of ARPC1B function in hematopoietic development, we turned to the zebrafish, as human hematopoietic processes are effectively modeled in zebrafish (21, 22). Using both synteny and homology searching in the zebrafish genome database, we identified the zebrafish ortholog of ARPC1B (Supplementary Fig. 2A,B). The zebrafish arpc1b cDNA (NM_213156) encodes a protein of 369 amino acids (~41kDa) that is 81% identical with its human ortholog. Using whole mount in situ hybridization (WISH), we determined that arpc1b expression is largely confined to the hematopoietic system, including the aorta-gonad-mesonephros (AGM) and posterior blood island (PBI) at 26 hours post-fertilization (hpf) (Supplementary Fig. 2C). Interestingly, the expression of arpc1a, another member of the ARP2/3 complex, was not detected in hematopoietic tissues (Supplementary Fig. 2D). The relative restriction of arpc1b expression to the hematopoietic system, may explain why mutation in ARPC1B observed in the patient was not lethal, as has been observed upon genetic ablation of other Arp2/3 complex components in mice.
Assessment of the requirement of Arpc1b in zebrafish hematopoiesis
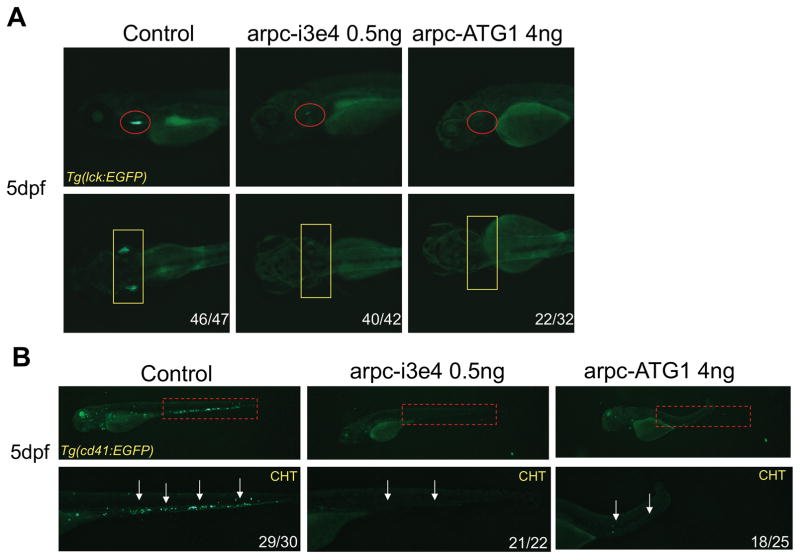
To determine if the ARPC1B mutation was responsible for the immune anomalies exhibited by the patient, we performed functional analysis in zebrafish. In doing so, we designed morpholino (MO) oligonucleotides that attenuate Arpc1b expression in zebrafish, either by interfering with the splicing of arpc1b mRNA or its translation into protein (Supplementary Fig. 3A–C). Treatment of zebrafish embryos with these MO blocked the development of T cells, as indicated by the depletion at 5dpf of T lineage progenitors marked by an lck:EGFP transgene (Fig. 5A; red circles and yellow rectangles). Moreover, development of thrombocytes was also impaired, since arpc1b MO blocked the development of CD41:EGFP labeled cells in caudal hematopoietic tissue (CHT) at 3.5dpf and 5dpf (Fig. 5B and Supplementary Fig. 3D, white arrows). These data indicate that arpc1b plays a critical role in supporting the development of both T lymphocytes and thrombocytes.
FIGURE 5. Impairment of T cell and thrombocyte development upon knockdown of arpc1b in zebrafish.
A, T cell development was disrupted after injection of MO, which disrupt arpc1b splicing (arpc-i3e4) or suppress translation of arpc1b mRNA (arpc-ATG1). T cell development was assessed at 5dpf using Tg(lck:EGFP) transgenic embryos to mark T cell progenitors (lateral view, red circles; dorsal view, yellow rectangles). B, Development of CD41+ thrombocytes was impaired following arpc1b knockdown. The effect of MO knockdown (as above) on thrombocyte development was assessed at 5dpf (red rectangles and white arrows). The embryos were photographed from lateral views. CHT: caudal hematopoietic tissue. The numbers on the images refer to the fraction of embryos with the depicted phenotype.
Functional analysis of the mutant ARPC1B allele
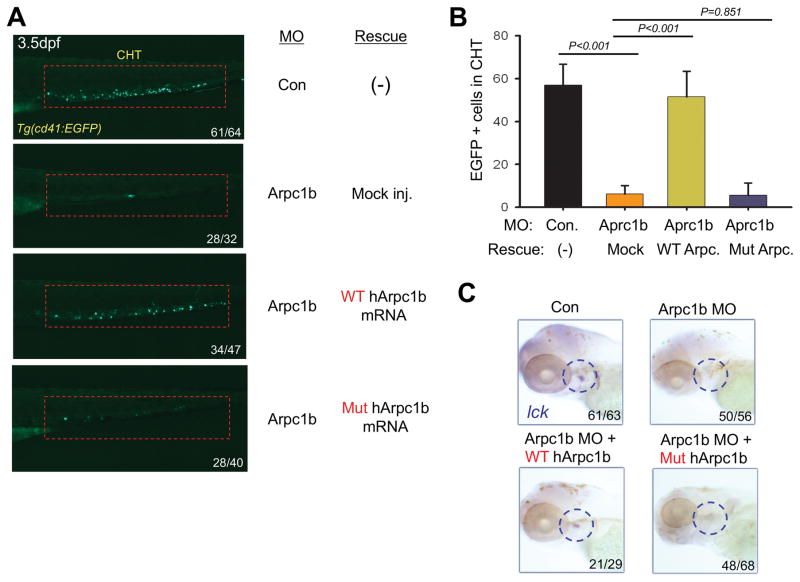
To determine if the ARPC1B mutation observed in the patient attenuated ARPC1B function, we re-expressed the wild-type and mutant human orthologs in arpc1b morphant zebrafish. Interestingly, injection of mRNA encoding intact, but not mutant, ARPC1B rescued the development of CD41:EGFP labeled thrombocyte precursors in arpc1b morphant zebrafish at 3.5dpf (Fig. 6A,B). The presence of mRNA encoding both wild-type and mutant ARPC1B in the injected embryos was confirmed by RT-PCR (Supplementary Figure 3E). Moreover, using a heat-inducible expression system, we observed that re-expression of intact, but not mutant, ARPC1B protein restored the development of T lymphocytes in the thymus, which were identified by performing WISH staining with a probe for lck mRNA (Fig. 6C). Taken together, these data conclusively link the mutant ARPC1B allele observed in the patients to the observed constellation of hematopoietic anomalies.
FIGURE 6. Patient mutation in ARPC1B fails to rescue developmental anomalies resulting from Arpc1b knockdown.
A, Overexpression of human WT but not mutant ARPC1B mRNA (150pg) partially rescued the thrombocyte development in 3.5dpf Tg(CD41:EGFP) embryos (lateral view, red rectangles) treated with arpc1b-i3e4 MO (Arpc1b). The numbers refer to the fractions of embryos exhibiting the depicted phenotypes. B, Quantitation of EGFP+ cell numbers in the CHT region as depicted in the red boxes of Fig. 6A as the mean of five embryos of each phenotype. T bar indicates the standard deviation. C, Overexpression ARPC1B in arpc1b morphant embryos using heat-shock inducible plasmids (150pg) encoding WT or mutant human ARPC1B. Developing T cells were identified by performing WISH analysis at 5dpf for using a probe for lck (blue circles, lateral view). The numbers refer to the fractions of embryos exhibiting the depicted phenotypes.
Discussions
Actin and proteins that regulate its dynamics or interactions have well-established roles in the cytoplasm where they function as key components of the cytoskeleton to control diverse processes of the immune system, including cellular infrastructure, cellular motility, cell signaling, and vesicle transport (23). Consequently, defects in this machinery result in severe immunodeficiency and even lethality, as is observed in WAS patients. Here we identify two brothers whose immunodeficiency results from a homozygous 2 bp deletion in the ARPC1B gene, a member of the Arp2/3 complex through which WAS controls actin cytoskeletal dynamics. As has been observed previously in WAS, these patients displayed immunodeficiency, thrombocytopenia, colitis and eczema. We demonstrated that ARPC1B plays a key role in the development of T cells and thrombocytes by showing that its knockdown in zebrafish disrupts these developmental processes. Moreover, the anomalies caused by loss of zebrafish Arpb1c were restored by re-expression of the intact human ARPB1C protein but not by the mutant allele identified in the patient, thus definitively identifying a novel ARPC1B mutation as the cause of the disease in these patients. Accordingly, we identified a cause for immunodeficiency resulting from a defect in a component of the Arp2/3 complex.
Other mutations in the ARPC1B gene have also been reported in patients with WAS-like features. Kuijpers et al, reported a homozygous complex frameshift mutation (c.491_495TCAAGdelCCTGCCCins) in a 7 year-old male patient with reduced platelets and impaired neutrophil function who had experienced recurrent infections, allergic reactions, vasculitis, and mild bleeding (9). Kahr at al. reported a child with a homozygous frameshift mutation (p.Val91Trpfs*30) in ARPC1B who presented with microthrombocytopenia, eosinophilia, and inflammatory bowel disease (24). Both of the patients in our study exhibited impaired T cell function, while one also manifested reductions in T cell numbers. This was not reported for the patients above. There are two potential explanations for the distinct constellation of symptoms observed in the different patients. It is possible that the distinct mutations in those patients impair ARPC1B function in a different manner. Alternatively, it is possible that the phenotypic differences result from variable expressivity. Variable expressivity, which is widely observed in genetic disorders, is the term applied to situations in which distinct individuals with identical mutations manifest marked differences in disease severity (25–27). Variable expressivity is thought to result either from environmental influences or from the action of different constellations of mutations in modifier genes. Interestingly, there is an element of variable expressivity in the siblings under study here. Indeed, while pt1 exhibited a profound alteration in the TCR V gene usage, this was not observed in his sibling. The basis for this difference remains unclear.
We have expanded our understanding of how inactivation of ARPB1C perturbs the development of immune cells by characterizing T and B cell receptor repertoires in patient’s cells. The TRB, TRG and IGH repertoires of our patients were evaluated by NGS, which revealed that there is no apparent defect in the initial generation of the repertoire, as can be seen by the constant Shannon’s H diversity index among the healthy donor controls and patients for the IGH repertoire. Despite the fact that B and T cells share a common VDJ recombination processes, the reduction in repertoire diversity was restricted to the TCR repertoires of pt1, which is likely to be a secondary consequence of altered selection during T cell development, differential survival in the periphery and/or altered immune responses caused by the loss of ARPC1B function. The TRB and IGH repertoires in WAS patients were not restricted; however, they were reported to exhibit substantial increases in clonal expansions in both the TRB and IGH repertoires (28). In support, patients with ARPC1B mutations are probably on the same spectrum of clinical presentation of WAS patients. Both WASP, WIP (29, 30) and probably ARPC1B (31) are involved in nucleation of branched actin filaments, which is critical in the formation of the immune synapse (IS) in T cells, and in the endocytosis and phagocytosis of antigen presenting cells. Defects in IS formation would be expected to interfere with proper immune responses in both T and B cells and so could explain perturbation of the repertoires.
Although the diversity of the patient’s IGH repertoire was comparable to the controls, IGHV gene usage was altered in our patients. The alteration in IGH usage may be impacted by altered selection during development; however, a more likely explanation is that it is a secondary consequence of attenuated T cell function, which blocks the provision of adequate survival signals and B lymphoid help during the response to recurrent infections experienced by the patient. Interestingly, we do observe a pronounced increase in the IGHV4-34 gene for Pt1, which is highly associated with patients with Systemic lupus erythematosus (SLE), infections (32) and other autoimmune disorders (33) and was observed previously in another WAS patient (28). The increased utilization of TRGV4 and TRGV5 in the peripheral blood repertoire of pt1 and TRGV4 in pt2 may be due to the involvement of specific γδ T cells that express TRGV4 and TRGV5 in the eczematous rash and papulovesicular lesions on the scalp that were observed in the patients. While murine Trgv4 and Trgv5 bear no relationship with those V regions in humans, these murine Trgv4 and Trgv5 are associated with localization of γδ T cells to the dermis and epidermis, respectively (34). Together, these findings raise the possibility that the alterations in the IGH and TRG immune repertoire may contribute in some way to aspects of the constellation of clinical manifestations observed in the patient.
Interestingly, Arpc1b mutant mice have been established (Arpc1btm1a(EUCOMM)Wtsi) (9); however, the development of T cells and thrombocytes in those animals is normal. The only significant immunological phenotypes observed are an increase in IgE levels and vasculitis (9). It is unclear why the inactivation of Arpb1c in these mice does not produce a constellation of defects similar to those in our patients or those observed upon Arpc1b loss in zebrafish. Nevertheless, there are several explanations for this apparent discrepancy. First, the mutation generated in that report is not a complete Arpc1b knockout and so the Arpb1c mutant protein may be a hypomorph that retains some function. Second, it is possible that Arpc1b function differs slightly in the mouse, providing an alternative explanation for the incomplete penetrance and phenotypic variability. Here, we exploited the zebrafish, which enables rapid and effective modeling of human disease (35). Using this model, we demonstrate that Arpc1b plays essential roles in supporting the development of both T cells and thrombocytes, in agreement with the constellation of hematopoietic and immune phenotypes observed our patients. Because the defects in T cell and thrombocyte production can be rescued by intact, but not mutant ARPC1B, we were able to definitively demonstrate that the human p.V208fs mutation is responsible for the hematopoietic and immune phenotypes exhibited by the patients in our study. It is also interesting that our WISH analysis in zebrafish embryos revealed that arpc1b is the only actin complex-related gene with expression that is largely restricted to hematopoietic cells (Supplementary Fig. 2C), while other components (e.g arp2, arp3, arpc1a, arpc2, arpc3, arpc4, arpc4l, arpc5a and arpc5b) are ubiquitously expressed in zebrafish embryos (https://zfin.org/). This distinct expression pattern of arpc1b provides a plausible explanation for why mutations in ARPC1B are not lethal in humans. Indeed, through expression data mining, we also confirmed that ARPC1B has very strong expression in human bone marrow (https://www.ebi.ac.uk/gxa/home). Interestingly, we find that Arpc1b is predominantly expressed in the central nervous system in mouse (https://www.ebi.ac.uk/gxa/home; http://www.informatics.jax.org/gxd), rather than in the hematopoietic and immune systems, as in zebrafish and humans. Together, these data raise the possibility that the differences in anomalies observed upon Arpb1c loss in mouse relative to human and zebrafish may reflect the differences in Arpb1c expression observed in these different species.
In conclusion, our results demonstrate the important role that ARPC1B plays in T cell and thrombocyte development in both humans and zebrafish, and provides a potential explanation for why, to date, ARPC1B is unique among the members of the Arp2/3 complex in being linked to immunodeficiency. Nevertheless, the precise manner in which ARPC1B and its p.V208fs mutation regulates the development of T lymphocytes and thrombocytes remains to be established.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants P30 CA006927 and R21 AI111208 (DW), an appropriation from the Commonwealth of Pennsylvania (DW) and the Bishop Fund (DW), the M.D. Anderson Cancer Center. YZ was supported by the Leukemia SPORE grant CA100632 and the institutional postdoctoral training grant T32 CA009035.
We thank the following Fox Chase Cancer Center core facilities for their vital service: Genomics, DNA Sequencing, Imaging, and Laboratory Animal/Zebrafish. We thank the Jeffrey Modell Foundation (JMF) for their consistent support of Raz Somech and Sheba Medical Center.
Footnotes
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Pizarro-Cerda J, Chorev DS, Geiger B, Cossart P. The Diverse Family of Arp2/3 Complexes. Trends Cell Biol. 2017;27:93–100. doi: 10.1016/j.tcb.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyler JJ, Allwood EG, Ayscough KR. WASP family proteins, more than Arp2/3 activators. Biochem Soc Trans. 2016;44:1339–1345. doi: 10.1042/BST20160176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaney KF, Li R. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr Opin Cell Biol. 2016;42:63–72. doi: 10.1016/j.ceb.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 5.Chandra S, Bronicki L, Nagaraj CB, Zhang K. WAS-Related Disorders. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1993. [PubMed] [Google Scholar]
- 6.Lanzi G, Moratto D, Vairo D, Masneri S, Delmonte O, Paganini T, Parolini S, Tabellini G, Mazza C, Savoldi G, Montin D, Martino S, Tovo P, Pessach IM, Massaad MJ, Ramesh N, Porta F, Plebani A, Notarangelo LD, Geha RS, Giliani S. A novel primary human immunodeficiency due to deficiency in the WASP-interacting protein WIP. J Exp Med. 2012;209:29–34. doi: 10.1084/jem.20110896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulding DA, Record J, Malinova D, Thrasher AJ. Actin cytoskeletal defects in immunodeficiency. Immunol Rev. 2013;256:282–299. doi: 10.1111/imr.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yae K, V, Keng W, Koike M, Yusa K, Kouno M, Uno Y, Kondoh G, Gotow T, Uchiyama Y, Horie K, Takeda J. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol Cell Biol. 2006;26:6185–6196. doi: 10.1128/MCB.00018-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuijpers TW, Tool AT, van der Bijl I, de Boer M, van Houdt M, de Cuyper IM, Roos D, van Alphen F, van Leeuwen K, Cambridge EL, Arends MJ, Dougan G, Clare S, Ramirez-Solis R, Pals ST, Adams DJ, Meijer AB, van den Berg TK. Combined immunodeficiency with severe inflammation and allergy caused by ARPC1B deficiency. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.09.061. [DOI] [PubMed] [Google Scholar]
- 10.Amariglio N, Lev A, Simon A, Rosenthal E, Spirer Z, Efrati O, Broides A, Rechavi G, Somech R. Molecular assessment of thymus capabilities in the evaluation of T-cell immunodeficiency. Pediatr Res. 2010;67:211–216. doi: 10.1203/PDR.0b013e3181c6e554. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li MX, Gui HS, Kwan JS, Bao SY, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic acids research. 2012;40:e53. doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang HT, Kathrein KL, Barton A, Gitlin Z, Huang YH, Ward TP, Hofmann O, Dibiase A, Song A, Tyekucheva S, Hide W, Zhou Y, Zon LI. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat Cell Biol. 2013;15:1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Duc AC, Rao S, Sun XL, Bilbee AN, Rhodes M, Li Q, Kappes DJ, Rhodes J, Wiest DL. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell. 2013;24:411–425. doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, Zon LI, Look AT, Trede NS. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A. 2004;101:7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, O’Leary MN, Peri S, Wang M, Zha J, Melov S, Kappes DJ, Feng Q, Rhodes J, Amieux PS, Morris DR, Kennedy BK, Wiest DL. Ribosomal Proteins Rpl22 and Rpl22l1 Control Morphogenesis by Regulating Pre-mRNA Splicing. Cell Rep. 2017;18:545–556. doi: 10.1016/j.celrep.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 19.Bajoghli B, Aghaallaei N, Heimbucher T, Czerny T. An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev Biol. 2004;271:416–430. doi: 10.1016/j.ydbio.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Molina IJ, Sancho J, Terhorst C, Rosen FS, Remold-O’Donnell E. T cells of patients with the Wiskott-Aldrich syndrome have a restricted defect in proliferative responses. J Immunol. 1993;151:4383–4390. [PubMed] [Google Scholar]
- 21.de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 22.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verboon JM, Sugumar B, Parkhurst SM. Wiskott-Aldrich syndrome proteins in the nucleus: aWASH with possibilities. Nucleus. 2015;6:349–359. doi: 10.1080/19491034.2015.1086051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahr WH, Pluthero FG, Elkadri A, Warner N, Drobac M, Chen CH, Lo RW, Li L, Li R, Li Q, Thoeni C, Pan J, Leung G, Lara-Corrales I, Murchie R, Cutz E, Laxer RM, Upton J, Roifman CM, Yeung RS, Brumell JH, Muise AM. Loss of the Arp2/3 complex component ARPC1B causes platelet abnormalities and predisposes to inflammatory disease. Nat Commun. 2017;8:14816. doi: 10.1038/ncomms14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonafe L, Dermitzakis ET, Unger S, Greenberg CR, Campos-Xavier BA, Zankl A, Ucla C, Antonarakis SE, Superti-Furga A, Reymond A. Evolutionary comparison provides evidence for pathogenicity of RMRP mutations. PLoS Genet. 2005;1:e47. doi: 10.1371/journal.pgen.0010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A. 2015;112:E7128–7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wegman-Ostrosky T, Savage SA. The genomics of inherited bone marrow failure: from mechanism to the clinic. British journal of haematology. 2017;177:526–542. doi: 10.1111/bjh.14535. [DOI] [PubMed] [Google Scholar]
- 28.O’Connell AE, Volpi S, Dobbs K, Fiorini C, Tsitsikov E, de Boer H, Barlan IB, Despotovic JM, Espinosa-Rosales FJ, Hanson IC, Kanariou MG, Martinez-Beckerat R, Mayorga-Sirera A, Mejia-Carvajal C, Radwan N, Weiss AR, Pai SY, Lee YN, Notarangelo LD. Next generation sequencing reveals skewing of the T and B cell receptor repertoires in patients with wiskott-Aldrich syndrome. Front Immunol. 2014;5:340. doi: 10.3389/fimmu.2014.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodnick-Smith M, Luan Q, Liu SL, Nolen BJ. Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc Natl Acad Sci U S A. 2016;113:E3834–3843. doi: 10.1073/pnas.1517798113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasahara Y. WASP-WIP complex in the molecular pathogenesis of Wiskott-Aldrich syndrome. Pediatr Int. 2016;58:4–7. doi: 10.1111/ped.12819. [DOI] [PubMed] [Google Scholar]
- 31.Abella JV, Galloni C, Pernier J, Barry DJ, Kjaer S, Carlier MF, Way M. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2016;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- 32.Mockridge CI, Rahman A, Buchan S, Hamblin T, Isenberg DA, Stevenson FK, Potter KN. Common patterns of B cell perturbation and expanded V4–34 immunoglobulin gene usage in autoimmunity and infection. Autoimmunity. 2004;37:9–15. doi: 10.1080/08916930310001624656. [DOI] [PubMed] [Google Scholar]
- 33.Doorenspleet ME, Klarenbeek PL, de Hair MJ, van Schaik BD, Esveldt RE, van Kampen AH, Gerlag DM, Musters A, Baas F, Tak PP, de Vries N. Rheumatoid arthritis synovial tissue harbours dominant B-cell and plasma-cell clones associated with autoreactivity. Ann Rheum Dis. 2014;73:756–762. doi: 10.1136/annrheumdis-2012-202861. [DOI] [PubMed] [Google Scholar]
- 34.Turchinovich G, Pennington DJ. T cell receptor signalling in gammadelta cell development: strength isn’t everything. Trends Immunol. 2011;32:567–573. doi: 10.1016/j.it.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.