Abstract
Using a Drosophila model of MECP2 gain-of-function, we identified memory associated KIBRA as a target of MECP2 in regulating dendritic growth. We found that expression of human MECP2 increased kibra expression in Drosophila, and targeted RNAi knockdown of kibra in identified neurons fully rescued dendritic defects as induced by MECP2 gain-of-function. Validation in mouse confirmed that Kibra is similarly regulated by Mecp2 in a mammalian system. We found that Mecp2 gain-of-function in cultured mouse cortical neurons caused dendritic impairments and increased Kibra levels. Accordingly, Mecp2 loss-of-function in vivo led to decreased Kibra levels in hippocampus, cortex, and cerebellum. Together, our results functionally link two neuronal genes of high interest in human health and disease and highlight the translational utility of the Drosophila model for understanding MECP2 function.
Keywords: MECP2, MECP2 duplication syndrome, Dendritic morphology, Drosophila, Disease models
1. Introduction
Methyl-CpG binding protein 2 (MECP2) is a widely abundant, multi-functional regulator of gene expression with highest levels of expression in mature neurons. In humans, both loss- and gain-of-function mutations of MECP2 cause mental retardation and motor dysfunction classified as either Rett Syndrome (RTT, loss-of-function) (Amir et al., 1999) or MECP2 duplication syndrome (MDS, gain-of-function) (Ramocki et al., 2010). In patients and in mouse models both MECP2 loss- and gain- of function can cause changes in dendritic morphology (Armstrong, 2005; Cheng et al., 2014; Wang et al., 2013; Zhou et al., 2006). While there is no MECP2 ortholog in Drosophila, we recently found that expression of human MECP2 (hMECP2) severely reduces the dendritic complexity of identified Drosophila motoneurons while maintaining normal membrane currents (Vonhoff et al., 2012).
Short generation times, facile tools for genetic manipulation (Venken and Bellen, 2005), and a high degree of conservation in fundamental cell biological pathways involved in neuronal development (Degerny et al., 2009; Louvi and Artavanis-Tsakonas, 2006; Rubin et al., 2000) make Drosophila a powerful model to study the cellular and molecular mechanisms by which MECP2 acts to impact dendritic structure. Because the Drosophila genome contains low 5-methylctosine (5′mC) levels (Capuano et al., 2014), it is not expected that this system can be used to elucidate mechanisms associated with the classical 5′mCpG dependent transcription repression function of MECP2 (Nan et al., 1997). However, MECP2 has also been shown to activate transcription (Chahrour et al., 2008) and many additional cellular functions ofMECP2 have been identified which do not require binding to methylated DNA (Cheng et al., 2014; Georgel et al., 2003; Hansen et al., 2010; Skene et al., 2010; Young et al., 2006). Further, other proteins with methyl binding domains (MBD) exist in Drosophila, and many MECP2 interactors, including other parts of the chromatin modifying machinery, have well conserved orthologs (Hendrich and Tweedie, 2003). When expressed in Drosophila, hMECP2 associates with chromatin, can be modified by orthologs of known interacting proteins, and, like in mammals, is phosphorylated at serine 423 (Cukier et al., 2008), and causes specific defects in dendritic morphology (Vonhoff et al., 2012). Furthermore, as observed in humans and mouse models (Collins et al., 2004; Ramocki et al., 2010; Van Esch, 2011), hMECP2 expression in Drosophila causes impairments in motor behavior (Cukier et al., 2008; Vonhoff et al., 2012).
Here, we use RNA-Seq with our Drosophila gain-of-function model to identify KIBRA as a novel gene activated by MECP2 and required for dendritic impairments. Validation in mouse confirms that Mecp2 gain-of-function similarly causes dendritic defects in primary cortical neuron culture coupled with increased Kibra levels. Similarly, we find that loss of Mecp2 decreases Kibra levels in mouse hippocampus, cortex, and cerebellum, thus providing evidence for bidirectional regulation of Kibra by Mecp2 both in vitro and in vivo. We then employ the genetic power of Drosophila to demonstrate a functional consequence of increased kibra with hMECP2 expression, finding that concomitant knockdown of kibra completely rescues the dendritic defects as caused by hMECP2 gain-of-function. Together, these data reveal memory associated KIBRA as a novel target of MECP2 and provide evidence of a role for KIBRA in hMECP2 gain-of-function dendritic growth defects.
2. Materials and methods
2.1. Animals
2.1.1. Drosophila stocks Drosophila melanogaster
were reared in 68 ml vials on a standard yeast corn meal diet at 25 °C and 60% humidity with a 12 h light/dark cycle. UAS-hMECP2 and UAS-hR106W transgenic flies (Cukier et al., 2008; Vonhoff et al., 2012) were kindly provided by Dr. J Botas (Baylor College of Medicine, Houston, Texas). UAS-kibraRNAi on the X chromosome (Genevet et al., 2010) was obtained from the Vienna Drosophila Resource Center. UAS-Dcr2 (Bloomington stock 24650) was used to enhance the efficacy of the RNAi knockdown. Expression of UAS-Dcr2 alone has no effect on MN5 dendritic arborization (Hutchinson et al., 2014). UAS-kibra9 flies were a kind gift of Dr. Hugo Stocker (Institute of Molecular Systems Biology, Zurich, Switzerland). Additional UAS-kibra lines (UAS-kibra18, H. Stocker and UAS-kibra-GFP, Dr. DJ Pan, John Hopkins School of Medicine, Baltimore, Maryland, USA) were used to verify the kibra overexpression phenotype in MN5 (data not shown). ELAV(C155)-GAL4 was used to drive expression of UAS-transgenes pan-neuronally for RNA and/or protein analysis (Lin and Goodman, 1994). For MN5 dendritic analyses, we used C380-GAL4, UAS-mcd8-GFP;;Cha-GAL80, which selectively expresses GFP and other UAS-transgenes in a subset of motoneurons and other unidentified neurons (Sanyal, 2009). The Cha-GAL80 transgene inhibits expression in unidentified cholinergic sensory neurons and interneurons eliminating most known pre-synaptic connections. Thus, effects in MN5 are likely to result from cell autonomous signaling. For all Drosophila experiments, genetic controls were generated by crossing the GAL4 driver line to w1118 flies carrying the same genetic background as transgenic lines.
2.1.2. Drosophila genotypes
Fig. 1 a: C380-GAL4, UAS-mcd8-GFP/Y;;Cha-GAL80 (control). 1b: C380-GAL4, UAS-mcd8-GFP/Y;;Cha-GAL80, UAS-hMECP2. 1c: C380-GAL4, UAS-mcd8-GFP/Y;;Cha-GAL80, UAS-hR106W. 1d–g: ELAV(C155)-GAL4/Y;+; + (control). ELAV(C155)-GAL4/Y;;UAShMECP2. ELAV(C155)-GAL4/Y; +; UAS-hR106W. 3b: C380-GAL4, UAS-mcd8-GFP;;Cha-GAL80 (control). 3c: C380-GAL4, UAS-mcd8-GFP; UAS-hMECP2;Cha-GAL80. 3d: C380-GAL4, UAS-mcd8-GFP, UAS-kibraRNAi;UAS-dcr2, UAS-hMECP2;Cha-GAL80. 3g: C380-GAL4, UAS-mcd8-GFP; UAS-dcr2, UAS-hMECP2;Cha-GAL80. 3h: C380-GAL4, UAS-mcd8-GFP, UAS-kibraRNAi; UAS-dcr2;Cha-GAL80. 3i: C380-GAL4, UAS-mcd8-GFP; UAS-kibra9;Cha-GAL80. S3: ELAV(C155)-GAL4;+; + (control). ELAV(C155)-GAL4, UAS-kibraRNAi;+; + (control).
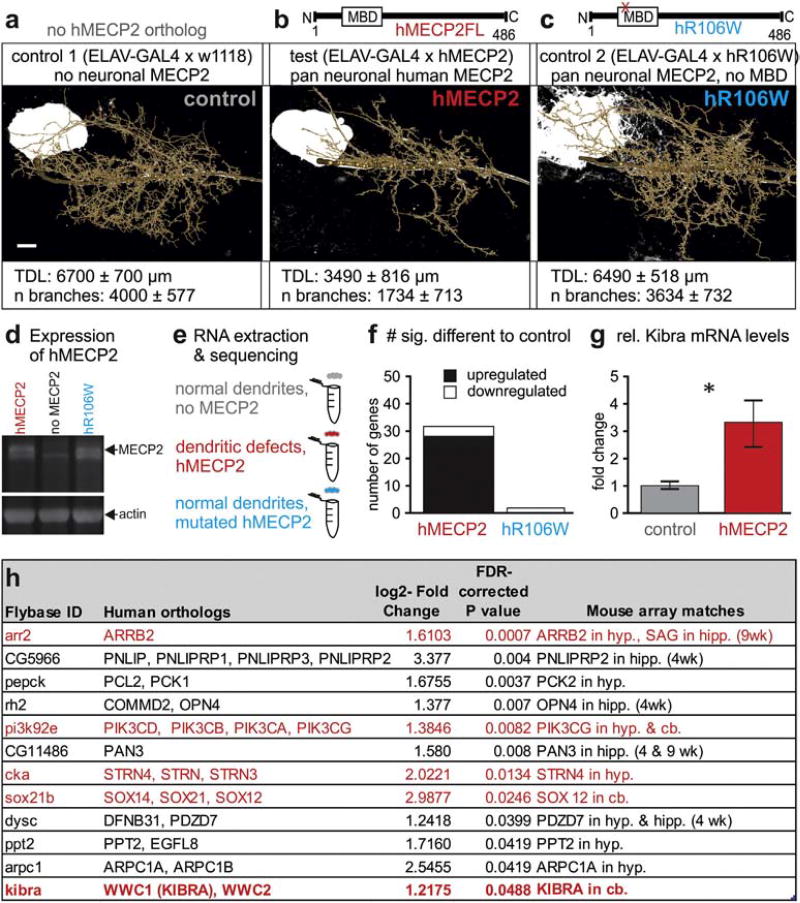
Fig. 1.
Expression of hMECP2 in Drosophila CNS causes MBD dependent changes in gene expression. a–c. Previously reported dendritic phenotypes with targeted hMECP2 expression in the identified Drosophila motoneuron, MN5. Statistically significant reductions in total dendritic length are caused by targeted expression of hMECP2 (b), but not with targeted expression of hMECP2 with a point mutated non-functional MBD, hR106W (c). d. Western blot reveals similar expression levels MECP2 following pan neuronal expression of hMECP2 (left lane) or of hR106W (right lane). The specific MECP2 band (see upper black arrow) is absent from controls (middle lane). Actin was used as loading control (lower black arrow). e. Schematic of RNA-Seq study design following no expression of hMECP2 and pan-neuronal expression of either hMECP2 or hR106W. f. Histogram comparing gene expression differences identified by RNA-Seq. g. qRT-PCR validation of dkibra upregulation with hMECP2 expression (n= 7) versus control (n= 8) (p < 0.05, t13 = 2.56, Welch's corrected two-tailed t-test). h. List of top twelve candidate genes from RNA-Seq. Genes interacting with the Hippo signaling pathway are identified by red text. Hyp.= hypothalamus, cb. = cerebellum, hipp. = hippocampus. Scale bars represent 10 µm.
2.1.3. Mice
Mecp2 null mice (Stock ID 003890) and wild-type colony controls were obtained from The Jackson Laboratory (Bar Harbor, Maine) and were humanely euthanized for experiments at six–seven weeks of age. The use and care of animals complied with institutional guidelines of the Transgenic Facility Mainz at the University Medical Center of the Johannes Gutenberg University Mainz.
2.2. Cell culture
Postnatal day 0 (P0)–P1 mouse cortices were dissociated and transfected by Nucleofection (Lonza) or immediately plated onto polyornithine coated coverslips in 24well plates in Neurobasal medium (Gibco) supplemented with 2% B27 (Gibco), and 1 mM l-glutamine. Mouse MECP2(e2)-GFP, MECP2(e2)-Myc/His, and N3-GFP plasmids were kindly provided by Dr. Vinodh Narayanan (Translational Genomics Research Institute, Phoenix, Arizona). MECP2(e2)-Myc/His was subcloned into pAM/CBA plasmids and used to generate chimeric AAV1/2 vectors as previously described for the control GFP vector (von Jonquieres et al., 2013).
2.3. RNA extraction and sequencing
The brains of 30 individual male flies for each genotype were dissected on ice and flash frozen in liquid nitrogen. RNA was isolated using TRIzol© reagent (Life Technologies) and DNAse treated with the TURBO DNA-free™ Kit (Life Technologies). 50 ng of pooled RNA quantified via Ribogreen (Invitrogen) was reversed transcribed and linearly amplified using Ovation RNA-Seq System V2 (NuGEN). 1 µg of cDNA was fragmented using focused-ultrasonication (Covaris) and libraries were subsequently prepared using Encore SP Rapid Library Systems (Nugen). Libraries were validated on a 2100 Bioanalyzer (Agilent Technologies) and quantified by qPCR with the KAPA Library Quantification Kit (KAPA Biosystems). Whole transcriptome pairedend data was generated with the HiSeq 2000 (Illumina) with samples multiplexed across three sequencing lanes. 150–180 million paired-end reads (90mers) were generated for each sample. Reads were aligned to D. melanogaster BDGP5 v67 with TopHat (version 2.0.2, bowtie version 0.12.7) using default parameters. Differential expression was analyzed using Cufflinks (version 0.9.3). The cutoff for determining significant differences between experimental groups (hMECP2 and hR106W) and control was a log scale fold difference of 0.2 and a false discovery rate (FDR) of 0 < .05 for the T-statistic corresponding to each comparison.
Mouse array matches were determined by cross comparison with published array data from Mecp2 TG (gain-of-function mouse) and Mecp2−/Y (null mouse) in hypothalamus (Chahrour et al., 2008), cerebellum (Ben-Shachar et al., 2009), striatum (null only)(Zhao et al., 2013), hippocampus (null only) (Baker et al., 2013) and amygdala (Mecp2 TG only) (Samaco et al., 2012a). Human orthologs were obtained using the DRSC Integrative Ortholog Prediction Tool (Hu et al., 2011), and only genes with scores ≥2.0 are listed.
2.4. Quantitative real-time PCR
Brain dissection and RNA extraction was completed as described above, with each sample consisting of five pooled male brains. cDNA was synthesized with Superscript III Reverse Transcriptase (Life Technologies) and treated with RNAse H (New England Biolabs). qRT-PCR was performed with the Applied Biosystems 7900HT Fast PCR system and KAPA SYBR® FAST qPCR Kit Master Mix (KAPA Biosystems). Samples were run in triplicate and were normalized to αTub84B using the comparative Ct method. Samples with high variability within replicates (standard deviation > 1.5 cycles) were removed from analysis. Primers used were as follows:
dkibra rev: 5′-CGTTTTCAATAGCCCAGGCG-3′.
dkibra fwd: 5′-GCCCAAGTCAGTCACAAAACT-3′.
dαTub84B fwd: 5′-CTTGTCGCGTGTGAAACACT-3′.
dαTub84 rev: 5′-AGCAGTAGAGCTCCCAGCAG-3′.
2.5. Western blotting
For cell culture experiments, cells were harvested by scraping in cold lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton-X) containing protease and phosphatase inhibitors (Roche). Mouse brains were dissected and distributed directly into lysis buffer. Whole adult Drosophila brains (10/sample) were dissected directly into cold lysis buffer. Proteins were separated by SDS-PAGE and transferred onto PVDF (Immobilion-P, Millipore) or nitrocellulose (Karl Roth) membranes. Membranes were blocked in 4% milk in TBST (0.05 M Tris, 0.15M NaCl, pH 7.2, 0.1% (v/v) Tween20) for 30 min at room temperature and primary antibodies were applied overnight at 4 °C in blocking medium. Secondary antibodies were applied for one hour at room temperature. Band intensities were measured using ImageJ. Relative Kibra levels were normalized to the mean control value for each blot.
2.6. Intracellular staining, image acquisition, and geometric reconstructions
Adult Drosophila (1–2 days old) were dissected and dye filled with Neurobiotin solution (7% Neurobiotin (Vector Labs) and lysine fixable rhodamine-dextran 3000 (Life Technologies) in 2Mpotassium acetate) using sharp electrodes as described previously (Vonhoff and Duch, 2010; Vonhoff et al., 2012). After staining, ganglia were fixed in 4% paraformaldehyde (PFA) in PBS for 1 h and washed in PBS. Tissue was permeated with 0.5% Triton X in PBS (6 × 30 min washes), and primary antibodies and secondary antibodies were sequentially applied overnight at 4 °C in 0.3% Triton X in PBS or pure PBS, respectively. Preparations were washed in PBS, progressively dehydrated in an ascending ethanol series, and mounted in methyl salicylate.
For MN5 dendritic morphometric analysis, stacks of 0.3 µm optical sections (Drosophila whole mount) with 1024 × 1024 resolution were acquired on a Leica TCS SP8 laser-scanning confocal microscope with a 40× oil-immersion, 1.2 NA lens. Image stacks were further processed with AMIRA 4.1.1 software (TGS) and custom AMIRA plug-ins were used for geometric dendrite reconstructions as previously published (Evers et al., 2005).
Primary neuron cultures were fixed for 10 min in 4%PFA on cover-slips in 24 well plates, and washed in PBS. Cells were permeated with 0.1% Triton X in PBS for two minutes, washed in PBS and incubated with primary antibodies in PBS for 1–2 h at room temperature or overnight at 4 °C. Secondary antibodies were applied for 30–60 min and coverslips were mounted onto glass slides with Flouromount™ (Sigma-Aldrich). Images were acquired and processed as described above using a 20× oil-immersion (0.75 NA lens) to capture 1 µmoptical sections. mMecp2 or control neurons were identified by positive GFP or myc labeling and neuronal identity was confirmed with B-III tubulin and NeuN co-labels. Positively transfected neurons were randomly selected and compared to the closest non-transfected neighbor within an individual coverslip to control for variability between cultures. At least ten cells per group were quantified and averaged for each transfection.
2.7. Antibodies
2.7.1. Western blot
The mouse Kibra (raised in rabbit) antibody was a kind gift from Dr. Richard Huganir (The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA) and was used at 1:1000 for Western blots (WB). GAPDH (rabbit; Bethyl Laboratories A300–641 A) and MECP2 (rabbit; Thermo Scientific PA1–887) antibodies were both used at 1:5000 for WB. The Drosophila kibra (raised in rabbit) antibody was a kind gift from Dr. Nicolas Tapon (London Research Institute, London, UK) and used at 1:1000 for WB. Anti-HSP90 (rabbit, Cell Signaling Technology #4874) or anti-Actin (mouse, Developmental Studies Hybridoma Bank JLA20) were used as loading controls at 1:1000 and 1:10,000, respectively, for Drosophila blots. Anti-rabbit horseradish peroxidase (HRP) (Dianova #111-035-144) or anti-mouse HRP (Dianova #115-035-062) were used as secondary antibodies at 1:10,000.
2.7.2. Immunohistochemistry
The following primary antibodies were used for cell culture and/or whole mount stainings: B-III tubulin (chicken, abcam® ab107216) (1:1000), NeuN (rabbit, Merck Millipore #ABN78) (1:1000), GFP (mouse, Life Technologies A11121) (1:200), c-myc (mouse, Developmental Studies Hybridoma Bank 9E 10) (1:200), and MECP2 (mouse, abcam® ab50005) (1:1,000). Cy3-streptavidin (Jackson ImmunoResearch 016-160-084) (1:400) was used to visualize neurobiotin filled neurons. Additional secondary antibodies, including goat anti-mouse Alexa 488 (Life Technologies A-11001), donkey anti-mouse Alexa 647 (Jackson Immuno Research 715-605-150), anti-rabbit Cy3 (Life Technologies A10042), and donkey anti-chicken Cy5 (Jackson ImmunoResearch 703-175-155) were applied at a concentration of 1:400.
2.8. Statistical analysis
Statistical analyses were conducted using GraphPad Prism 6.0 or R Statistical Software. Non parametric statistics were used in the event a data set did not meet the assumptions of normality or equal variance between groups. Graphical representations were prepared using GraphPad Prism 6.07 and CorelDraw × 7. Error bars are mean ± SEM in all figures unless otherwise noted.
3. Results
3.1. Expression of hMECP2 in Drosophila causes MBD dependent changes in gene expression
As in many neurodevelopmental diseases (Kaufmann and Moser, 2000; Kulkarni and Firestein, 2012) misregulation of MECP2 causes dendritic defects across many model systems, but the underlying mechanisms are poorly understood. In Drosophila, hMECP2 gain-of-function dendritic defects are dependent on an intact MBD, as expression of an MBD point mutated human variant R106W (hR106W) has no effect on dendritic morphology (Vonhoff et al., 2012) (Fig. 1a–c). Expression of either hR106W or hMECP2 under the control of the pan neuronal GAL4 driver Elav yields similar MECP2 protein levels (Fig. 1d). To identify genes that specifically regulate dendrite development downstream of MECP2, we compared the transcriptomes of Drosophila brains with pan neuronal expression of hMECP2, hR106W, or without any MECP2 isoform (Fig. 1a–e). This differential RNA-Seq revealed 32 genes that were potentially associated with dendritic defects because their expression was specifically altered with expression of hMECP2 but not hR106W (Fig. 1f). Consistent with array data from Mecp2 gain- and loss- of function mouse models (Ben-Shachar et al., 2009; Chahrour et al., 2008; Zhao et al., 2013), most genes were activated instead of repressed by hMECP2 (Fig. 1f). Of the 32 genes uniquely modified by hMECP2 and not hR106W, we narrowed our candidate genes to the 20 with known human orthologs, twelve of which have been identified to be regulated by MECP2 in the same direction in comparative gene expression analyses from MECP2 gain- or loss- of function mouse models (Baker et al., 2013; Ben-Shachar et al., 2009; Chahrour et al., 2008) (Supplementary Table 1). This provided a select subset of genes modified by MECP2 in both mouse and in Drosophila (Fig.1h). Notably, five of the twelve top candidate genes identified in the RNA-Seq screen have direct links to the Hippo signaling cascade, a pathway involved in regulation of dendritic morphology and cell size (Emoto et al., 2006; Yu et al., 2010) (Supplementary Fig. 1/Fig. 1h), two key cellular properties disrupted by MECP2 gain- and loss- of function (Jentarra et al., 2010).
3.2. KIBRA is bidirectionally regulated by MECP2
Of these five genes, we selected KIBRA (HGNC Gene ID:WWC1) as a primary candidate gene for further analysis due to its position as an activator of this signaling cascade (Genevet et al., 2010) and used quantitative RT-PCR (qRT-PCR) to validate upregulation of Drosophila kibra (dkibra) with hMECP2 expression (Fig.1g). KIBRA was additionally of high interest due to its association with learning and memory in humans and mouse models (Makuch et al., 2011; Papassotiropoulos et al., 2006). While KIBRA has not been directly linked to intellectual disability or cognitive impairments, its postsynaptic localization and role in synaptic plasticity could provide a direct link to shared symptoms of RTT, MDS, and other autism spectrum disorders (Zoghbi and Bear, 2012).
KIBRA was previously identified as one of hundreds of genes downregulated in the cerebellum of a mouse loss-of-function model (Mecp2tm1 · 1Bird or Mecp2−/y) (Guy et al., 2001) (Fig. 1h), but was not significantly altered on similar arrays of striatum, amygdala, hippocampus, or hypothalamus (Baker et al., 2013; Ben-Shachar et al., 2009; Chahrour et al., 2008; Samaco et al., 2012b; Zhao et al., 2013). However, of the thousands of genes differentially expressed with Mecp2 loss-and/or gain- of function, very few are consistently altered in all brain regions with previously published microarray data (Zhao et al., 2013). Furthermore, in mouse models, MECP2 dendritic impairments are variable across cell types, MECP2 mutation, and age, but have primarily been identified in pyramidal neurons of the cortex or hippocampus, regions with the highest basal Kibra expression (Lein et al., 2007; Wang et al., 2013; Zhou et al., 2006). We therefore sought to verify the regulation of Kibra by mouse MECP2 (mMECP2) in cortex and hippocampus, where gain- or loss- of MECP2 function is associated with dendritic impairments (Armstrong, 2005; Cheng et al., 2014; Wang et al., 2013; Zhou et al., 2006).
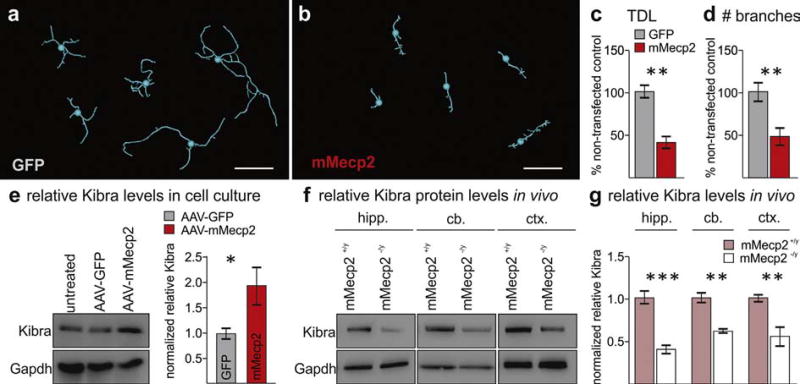
While reduced dendritic complexity has not been observed with mMecp2 gain-of-function in vivo, overexpression in primary neuron culture and in slice culture consistently impairs dendritic growth (Cheng et al., 2014; Zhou et al., 2006). To test if Kibra is upregulated with mMecp2 gain-of-function in cultured neurons, we used both electroporation (0 DIV) and an adeno associated virus (AAV) (1 DIV) to overexpress mMecp2 in cortical neurons. Three days after mMecp2 delivery we found a 50% reduction in both total dendritic length and branching compared to controls (Fig. 2a–d; Supplementary Fig. 2). Immunoblots for mouse Kibra (mKibra) following AAV-mMecp2 transduction revealed a nearly two-fold increase in total mKibra protein levels (Fig. 2e), suggesting thatMECP2 gain-of-function inmouse affects KIBRA similarly to hMECP2 gain-of-function in Drosophila.
Fig. 2.
MECP2 disrupts dendritic function and bidirectionally regulates Kibra levels in mouse brain. a,b. Representative traces of transfected primary cortical neurons from each replicate experiment (n = 5). c,d. Quantification of total dendritic length (TDL) and number of branches following mMECP2 overexpression (**p < 0.005 Tukey post-hoc following two-way ANOVA, F1,16 = 35.94, p < 0.0001). Error bars are mean ± SEM for replicate experiments. e. Representative Western blot and quantification of relative mKibra levels following AAV mediated overexpression of mMecp2 in primary cortical neurons (*p < 0.05, U = 4, Mann-Whitney U test, n = 6 independent assays per group). f,g. Representative Western blots of mKibra from hippocampus (hipp.), cerebellum (cb.), and cortex (ctx) of Mecp2−/y mice and Mecp2+/y controls (**p < 0.01, *p < 0.05 Tukey post-hoc following two-way ANOVA, F1,18 = 62.28, p < 0.0001, n = 4 per genotype). Scale bar represents 50 µm.
We additionally assessed mKibra levels in Mecp2−/y mice to determine whether mMecp2 loss-of-function also regulates Kibra in vivo in brain regions with associated dendritic impairments. We confirmed the decrease in mKibra in Mecp2−/y cerebella compared to with-in strain age matched controls, and additionally found decreased mKibra in both cortex and hippocampus (Fig. 2f–g). Together, these results confirm that MECP2 bidirectionally regulates KIBRA in brain regions of model systems where it also impairs dendrite development.
3.3. Increased kibra is necessary, but not sufficient, to cause dendritic defects
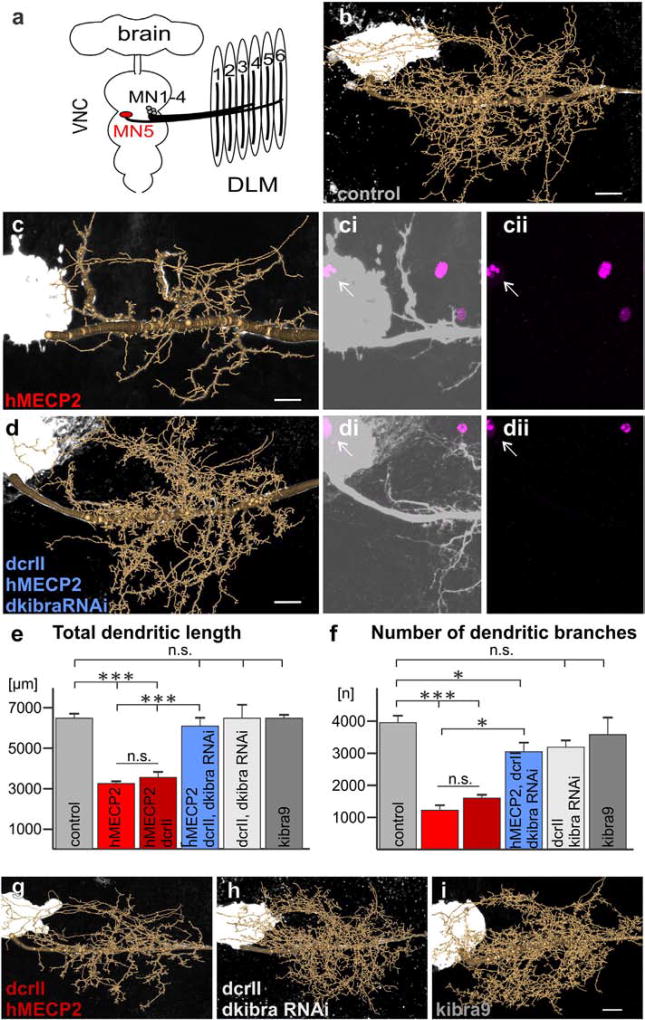
To test whether manipulation of KIBRA could work as a useful tool to rescue MECP2 gain-of-function dendritic growth impairments, we returned to the Drosophila model. We expressed hMECP2 in an identified motoneuron (MN5), which displays a stereotyped and well described dendritic morphology, thus allowing for quantitative in vivo dendritic structure analysis following genetic interaction experiments (Duch et al., 2008; Hutchinson et al., 2014; Vonhoff and Duch, 2010; Vonhoff et al., 2013; Vonhoff et al., 2012). As previously reported, expression of full length hMECP2 in MN5 reduced total dendritic length and the number of dendritic branches by 50% compared to controls (Fig. 3b–c, e–f). RNAi mediated knockdown of dKibra with hMECP2 expression (dkibraRNAi;hMECP2), however, completely rescued total dendritic length to that of controls (Fig. 3d–e, Supplementary Fig. 4). By contrast, the number of branches was only partially rescued (Fig. 3f). Sholl analysis revealed that this was due to fewer branches closer to the primary neurite (between 5 and 20 µm) in dkibraRNAi;hMECP2 flies, an effect that was also observed with dkibraRNAi alone (Supplementary Fig. 3). Despite subtle changes in branch patterning, dkibraRNAi alone had no significant effect on total dendritic length or branch number (Fig. 3h, e–f). Moreover, while overexpression of dkibra alone (kibra9, Fig. 3i) also caused subtle changes in branch patterning (Supplementary Fig. 3), it had no effect on total dendritic length or branch number (Fig. 3, e–f). This suggests that the increase in dkibra with hMECP2 expression is necessary to disrupt dendritic morphology, but altered dkibra levels alone are not sufficient to cause defects. This finding was verified with three independent UAS-dkibra lines (data not shown) and indicates that the major effects of kibra on dendritic growth must therefore be dependent on additional hMECP2 gain-of-function induced changes (see discussion).
Fig. 3.
induced dendritic defects in Drosophila MN5. a. Schematic of Drosophila CNS and dorsal longitudinal flight depressor muscle innervation by MNs 1–5. b–d, g–i. Representative geometric reconstructions of MN5 dendritic trees superimposed onto representative projection views for each labeled group. ci–ii, di–ii. hMECP2 (magenta) is expressed and localizes to the nucleus of MN5 (white arrows) with (di–ii) and without (ci–ii) knockdown of dkibra. e,f. Quantification of total dendritic length (e) and number of branches (f) for all experimental and control genotypes (***p < 0.0001, *p < 0.05 Tukey post-hoc following two-way ANOVA, F5,46 = 56.30, p < 0.0001, n = 3–7 cells per group). All constructs were expressed using the C380-GAL4, cha-GAL80 driver line (see Online Methods for complete list of genotypes). Scale bars represent 10 µm.
4. Discussion
While the Drosophila genetic model is increasingly utilized for analyzing specific cellular and genetic components of neurodevelopmental disease, data obtained from this model requires careful interpretation. Due to the lack of a fly MECP2 ortholog, the gain-of-function model presented here relies on heterologous expression of a human MECP2 allele. Unlike mammalian models systems with endogenous MECP2, Drosophila cannot be used to precisely model the genetic mutations or copy number variations that cause RTT or MDS in humans. Thus, there is always the possibility that results obtained with the Drosophila model system could be attributed to nonspecific effects resulting from heterologous expression and not a direct function ofMECP2. The external validity of the model is dependent on validation in mammalian model systems with MECP2. Nonetheless, quick generation times and facile genetic tools render the fly useful to test novel hypotheses on MECP2 function which can then be validated in mouse models and ultimately in human brain.
Our study identifies KIBRA as a novel player in MECP2 related dendritic pathology. While genetic and behavioral proof of principle for using Drosophila to study hMECP2 gain-of-function has been well established (Cukier et al., 2008; Vonhoff et al., 2012), we now utilize this system to identify new MECP2 targets. We then focus on KIBRA as one such target, confirm it in the mouse model with endogenous MECP2, and employ the genetic power of Drosophila to rescue MECP2 induced cellular defects by concomitant reduction of KIBRA.
In addition to KIBRA, we have identified eleven genes modified by MECP2 in both Drosophila and in mouse models (see Fig. 1g) that may be involved in MECP2 dependent dendritic defects. Interestingly, four of these genes have direct links to the Hippo kinase signaling network (Fig. S1), which is known to regulate dendrite development (Emoto, 2011; Ultanir et al., 2012; Yang et al., 2011) but has not previously been associated with MECP2 induced neurodevelopmental defects. Our data indicate a potential role of the Hippo signaling network in Rett/MDS pathology. Disruption of this pathway has been associated with both cancer and other neurological diseases, including amyotrophic lateral sclerosis, Alzheimer's disease, and schizophrenia (Lee et al., 2013; Melka et al., 2015; Orcholski et al., 2011; Plouffe et al., 2015). However, additional work will be needed to determine whetherMECP2 disruption of Hippo signaling is relevant to the cellular and behavioral phenotypes of Rett and MDS.
The mechanism by which hMECP2 acts to modify expression of kibra and the other genes identified through the RNA-Seq screen remains unknown. Because the Drosophila genome is sparsely methylated (Capuano et al., 2014), it is unlikely that hMECP2 acts by binding to 5′ mCpG and repressing transcription (Nan et al., 1997). Accordingly, all twelve of the genes modified in both Drosophila and mouse models are activated by MECP2 in both model systems. Overall, we found far more genes activated by hMECP2 in Drosophila CNS than repressed. This is consistent with array data from mouse hypothalamus and cerebellum that suggestsMECP2may be more of an activator than a repressor (Chahrour et al., 2008). HowMECP2 acts to activate genes is unclear in any system, but in mouse it has been found to associate with the major transcriptional activator CREB1 in the promoter region of activated genes (Chahrour et al., 2008). In one such example, the binding of MECP2 and CREB1 to the promoter region of transcriptional target Glut3 is dependent on 5′mCpG binding, but MECP2 still activates Glut3 expression in the absence of DNA methylation (Chen et al., 2013). Thus, it is possible that hMECP2 can activate many of the same genes in Drosophila without direct 5′mCpG binding, potentially by binding histone tails (Fuks et al., 2003). MECP2 can directly bind methylated histones (Fuks et al., 2003) and influence local gene expression both in the presence and absence of genomic DNA methylation (Zhao et al., 2005). Comparative analysis of patterning and regulatory association of DNA methylation and histone modifications in insects has revealed that active histone methylation in Drosophila is localized in genomic regions with conserved DNA methylation in both fire ants and honey bees, insects with higher overall levels of genomic methylation (Hunt et al., 2013). Thus, histone modifications may be in some degree redundant to other forms of epigenetic information and MBD dependent binding of hMECP2 to methylated histones in Drosophila could influence the same genes as when bound to methylated DNA in other systems. In Drosophila, an intact MBD is critical for the effects of hMECP2 gain-of-function on dendritic morphology (Vonhoff et al., 2012) and gene expression (Fig. 1); thus, it is likely that the genes identified through the RNA-Seq screen are direct transcriptional targets influenced by local histone modifications. Alternatively, hMECP2 may also act in the fly by one of its many identified functions not dependent on DNA methylation, such as RNA splicing or suppression of microRNA processing, the latter of which has a demonstrated role in MECP2 dendritic growth defects (Cheng et al., 2014; Young et al., 2006).
The primary motivation to screen for MECP2 targets is to identify potential future treatment targets to ameliorate cellular defects as induced by mis-regulation of MECP2. We show here that, at least in Drosophila, knockdown of dkibra with hMECP2 expression can rescue the severe dendritic impairments observed with hMECP2 alone, but manipulation of dkibra alone is not sufficient to cause similar impairments. This is consistent with the fact that both MECP2 gain- and loss-of function impair dendritic growth but have opposing effects on KIBRA levels. The role of KIBRA in Hippo signaling could potentially provide an explanation on how this could occur. In Drosophila it is known that dkibra activates the Hippo signaling pathway through interactions with Mer and Ex, which ultimately inhibits the activation of the transcription factor Yki (Yu et al., 2010). Kibra itself is also a Yki target, and therefore can regulate its own expression through a Hippo mediated negative feedback loop (Genevet et al., 2010). This may also provide an explanation as to how we found a large difference in mKibra at the protein level in hippocampus with mMecp2 loss-of-function, while mkibra transcript levels have previously been reported unchanged at similar ages (Baker et al., 2013). Furthermore, overexpression and knockdown manipulations may have only subtle effects on overall KIBRA levels, while the activation and/or inhibition of Hippo signaling via KIBRA and additional regulators (see Fig. S1) may result in more dramatic effects. Accordingly, while the Drosophila kibraRNAi line used for kibra knockdown produced robust eye and wing growth phenotypes in an RNAi screen strong enough to identify a novel function of KIBRA (Genevet et al., 2010), our Western blot analysis from brains collected following pan-neuronal dkibra knockdown revealed a significant but modest 30% decrease in total dkibra levels (see Fig. S4). Considering that this subtle reduction in dkibra leads to a near complete reversal of thehMECP2 dendritic phenotype but has minimal impact on dendritic arborization alone, targeting KIBRA and/or Hippo signaling could be an exciting option for the development of potential future therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Dr. Juan Botas, Dr. D.J. Pan, & Dr. Hugo Stocker for UAS-MeCP2 and UAS-kibra flies, Dr. Richard Huganir for the mouse Kibra antibody, Dr. Nic Tapon for the Drosophila kibra antibody, and Dr. Vinodh Narayanan for mouse MECP2 plasmids. The authors acknowledge the excellent technical writing support of Matt De Both from the Neurogenomics Division at the Translational Genomics Research Institute, and additional technical support from Veronika Prantl, Simone Dahms-Praetorius and Nicole Knauer at Johannes Gutenberg University Mainz. We gratefully acknowledge support from the German Science Foundation (DFG Du331/9-1 to CD) and the Natural and Medical Sciences Research Center of the University of Medicine Mainz (NMFZ to CD and RW). AAW was supported under National Institute of Health R01NS072128 and fellowship funding from Science Foundation Arizona.
Abbreviations
- MECP2
Methyl-CpG-binding protein 2
- RTT
Rett syndrome
- MDS
MECP2 duplication syndrome
- hMECP2
human MECP2
- 5′Mc
5-mehtylcytosine
- MBD
methyl binding domain
- hR106W
human MECP2 variant R106W
- qRT-PCR
quantitative real-time polymerase chain reaction
- Mecp2−/Y
Mecp2 null mouse model
- mMecp2
mouse Mecp2
- mKibra
mouse Kibra
- dkibra
Drosophila kibra.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nbd.2016.03.019.
References
- Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Armstrong Can we relate MeCP2 deficiency to the structural and chemical abnormalities in the Rett brain? Brain Dev. 2005;27:S72–S76. doi: 10.1016/j.braindev.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Baker Steven A, et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, et al. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum. Mol. Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano F, et al. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal. Chem. 2014;86:3697–3702. doi: 10.1021/ac500447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, et al. Creb1-Mecp2-mCpG complex transactivates postnatal murine neuronal glucose transporter isoform 3 expression. Endocrinology. 2013;154:1598–1611. doi: 10.1210/en.2012-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TL, et al. MeCP2 suppresses nuclear MicroRNA processing and dendritic growth by regulating the DGCR8/drosha complex. Dev. Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- Collins AL, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Cukier HN, et al. Genetic modifiers of MeCP2 function in Drosophila. Plos Genetics. 2008;4 doi: 10.1371/journal.pgen.1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degerny C, et al. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 2009;87:77+. doi: 10.1139/O08-114. [DOI] [PubMed] [Google Scholar]
- Duch, et al. Dendrite elongation and dendritic branching are affected separately by different forms of intrinsic motoneuron excitability. J. Neurophysiol. 2008;100:2525–2536. doi: 10.1152/jn.90758.2008. [DOI] [PubMed] [Google Scholar]
- Emoto K. The growing role of the Hippo–NDR kinase signalling in neuronal development and disease. J. Biochem. 2011;150:133–141. doi: 10.1093/jb/mvr080. [DOI] [PubMed] [Google Scholar]
- Emoto, et al. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Evers JF, et al. Progress in functional neuroanatomy: precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. J. Neurophysiol. 2005;93:2331–2342. doi: 10.1152/jn.00761.2004. [DOI] [PubMed] [Google Scholar]
- Fuks F, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Genevet A, et al. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel PT, et al. Chromatin compaction by human MeCP2: assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- Guy J, et al. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hansen JC, et al. Binding of the Rett syndrome protein, MeCP2, tomethylated and unmethylated DNA and chromatin. IUBMB Life. 2010;62:732–738. doi: 10.1002/iub.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- Hu Y, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, et al. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol. Evol. 2013;5:591–598. doi: 10.1093/gbe/evt030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson KM, et al. Dscam1 Is required for normal dendrite growth and branching but not for dendritic spacing in Drosophila motoneurons. J. Neurosci. 2014;34:1924–1931. doi: 10.1523/JNEUROSCI.3448-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentarra GM, et al. Abnormalities of cell packing density and dendritic complexity in the MeCP2 A140V mouse model of Rett syndrome/X-linked mental retardation. BMC Neurosci. 2010;11 doi: 10.1186/1471-2202-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kulkarni VA, Firestein BL. The dendritic tree and brain disorders. Mol. Cell. Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Lee JK, et al. MST1 functions as a key modulator of neurodegeneration in amouse model of ALS. Proc. Natl. Acad. Sci. 2013;110:12066–12071. doi: 10.1073/pnas.1300894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of fasciclin-Ii alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Makuch L, et al. Regulation of AMPA receptor function by the human memory-associated gene KIBRA. Neuron. 2011;71:1022–1029. doi: 10.1016/j.neuron.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melka MG, et al. Insights into the origin of DNA methylation differences between monozygotic twins discordant for schizophrenia. J. Mol. Psychiatry. 2015;3:7. doi: 10.1186/s40303-015-0013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, et al. MeCP2 Is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Orcholski ME, et al. Signaling via amyloid precursor-like proteins APLP1 and APLP2. J. Alzheimers Di. 2011;23:689–699. doi: 10.3233/JAD-2010-101470. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, et al. Common KIBRA alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Plouffe SW, et al. Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 2015;21:212–222. doi: 10.1016/j.molmed.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki, et al. The MECP2 duplication syndrome. Am. J. Med. Genet. A. 2010;152a:1079–1088. doi: 10.1002/ajmg.a.33184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, et al. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat. Genet. 2012a;44:206–211. doi: 10.1038/ng.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, et al. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat. Genet. 2012b;44:206–211. doi: 10.1038/ng.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S. Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr. Patterns. 2009;9:371–380. doi: 10.1016/j.gep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultanir SK, et al. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 uncovers their roles in dendrite arborization and spine development. Neuron. 2012;73:1127–1142. doi: 10.1016/j.neuron.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esch H. MECP2 duplication syndrome. Mol. Syndr. 2011;2:128–136. doi: 10.1159/000329580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 2005;6:167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- von Jonquieres G, et al. Glial promoter selectivity following AAV-delivery to the immature brain. PLoS One. 2013;8:e65646. doi: 10.1371/journal.pone.0065646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonhoff F, Duch C. Tiling among stereotyped dendritic branches in an identified Drosophila motoneuron. J. Comp. Neurol. 2010;518:2169–2185. doi: 10.1002/cne.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonhoff F, et al. Drosophila as amodel forMECP2 gain of function in neurons. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonhoff F, et al. Temporal coherency between receptor expression, neural activity and AP-1-dependent transcription regulates Drosophila motoneuron dendrite development. Development. 2013;140:606–616. doi: 10.1242/dev.089235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ITJ, et al. Neuronal morphology in MeCP2 mouse models is intrinsically variable and depends on age, cell type, and Mecp2 mutation. Neurobiol. Dis. 2013;58:3–12. doi: 10.1016/j.nbd.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WK, et al. Nak regulates localization of clathrin sites in higher-order dendrites to promote local dendrite growth. Neuron. 2011;72:285–299. doi: 10.1016/j.neuron.2011.08.028. [DOI] [PubMed] [Google Scholar]
- Young, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2 (vol 102, pg 17551, 2005) Proc. Natl. Acad. Sci. U. S. A. 2006;103:1656. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JZ, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, et al. The essential role of histone H3 Lys9 di-methylation and MeCP2 binding in MGMT silencing with poor DNA methylation of the promoter CpG Island. J. Biochem. 2005;137:431–440. doi: 10.1093/jb/mvi048. [DOI] [PubMed] [Google Scholar]
- Zhao, et al. Loss of MeCP2 function is associated with distinct gene expression changes in the striatum. Neurobiol. Dis. 2013;59:257–266. doi: 10.1016/j.nbd.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZL, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.