In exo70B1-mediated autoimmunity, TIR-NBS2 associates with CPK5, likely stabilizing the enzyme to maintain kinase activity beyond the initiating calcium-mediated stimulus.
Abstract
Calcium-dependent protein kinases (CPKs) function as calcium sensors and play important roles in plant immunity. Loss of function of the exocyst complex subunit EXO70B1 leads to autoimmunity caused by activation of TN2, a truncated Toll/interleukin-1 receptor-nucleotide binding sequence protein. Here we show, based on a screen for suppressors of exo70B1, that exo70B1-activated autoimmune responses require CPK5. However, the CPK5 homologs CPK4, CPK6, and CPK11, which were previously reported to function redundantly with CPK5 in effector-triggered immunity, did not contribute to exo70B1-associated phenotypes, indicating that CPK5 plays a unique role in plant immunity. Overexpressing CPK5 results in TN2-dependent autoimmunity and enhanced disease resistance, reminiscent of the exo70B1 phenotypes. Ectopic expression of CPK5 in the exo70B1 mutant led to constitutive CPK5 protein kinase activity, which was not detectable in tn2 mutants. Furthermore, TN2 interacts with the CPK5 N-terminal variable and kinase domains, stabilizing CPK5 kinase activity in vitro. This work uncovers a direct functional link between an atypical immune receptor and a crucial component of early immune signaling: increased immunity in exo70B1 depends on TN2 and CPK5 and, in a positive feedback loop, TN2 keeps CPK5 enzymatically active beyond the initiating stimulus.
INTRODUCTION
Plants have evolved a multilayered immune system to battle microbial pathogen attack. Activation of this immune system can induce a series of defense responses, such as increased calcium concentration in the cytoplasm, accumulation of reactive oxygen species (ROS), callose deposition, mitogen-activated protein kinase activation, and pathogenesis-related gene expression (Chisholm et al., 2006; Jones and Dangl, 2006; Dodds and Rathjen, 2010; Dangl et al., 2013). A rapid increase in cytoplasmic calcium concentration is one of the earliest defense responses in pathogen-associated molecular pattern (PAMP)-induced immunity (Kudla et al., 2010; Reddy et al., 2011). Calcium-dependent protein kinases (CDPKs; CPKs in Arabidopsis thaliana) are calcium-sensor proteins that perceive changes in intracellular calcium concentrations and transmit calcium signals via protein phosphorylation to induce downstream signaling responses. CPK family members mediate and transmit defense signals in response to PAMPs as well as pathogen effectors; therefore, CPKs play important roles in plant immunity (Cheng et al., 2002; Boudsocq and Sheen, 2013; Schulz et al., 2013; Romeis and Herde, 2014). For instance, tobacco (Nicotiana tabacum) CDPK2 is activated in Cf-9 tobacco leaves after treatment with the fungal elicitor Avr9 (Romeis et al., 2000, 2001).
Multiple CPKs are activated in Arabidopsis upon elicitation with flg22, a conserved 22-amino acid peptide derived from bacterial flagellin. A specific subgroup of four CPKs, CPK4, 5, 6, and 11, plays critical roles in flg22 signaling (Boudsocq et al., 2010). The cpk4, cpk5, cpk6, and cpk11 mutants do not show defects in the response to Pseudomonas syringae pv tomato (Pto) strain DC3000, but the cpk5 cpk6 double and cpk5 cpk6 cpk11 triple mutants display enhanced susceptibility and reduced flg22-induced ROS production, indicating functional redundancy of closely related CPKs (Boudsocq et al., 2010). Similarly, six CPKs of the same subfamily, CPK1, 2, 4, 5, 6, and 11, contribute to the immune response triggered by the bacterial effectors AvrRpm1, AvrB, and AvrRpt2. In addition, the activation of CPK1 and 2 by effectors controls the onset of cell death, while CPK4, 5, 6, and 11 regulate immune gene expression by directly phosphorylating specific WRKY transcription factors, i.e., WRKY8, 28, and 48 (Gao et al., 2013). Furthermore, CPK5 can directly phosphorylate the NADPH oxidase respiratory burst oxidase homolog D (RBOHD) in vivo, and CPK5 overexpression lines show spontaneous cell death and enhanced disease resistance to bacterial pathogens, as well as increased salicylic acid (SA) accumulation. The activation of CPK5 and RBOHD is critical for rapid defense signal propagation and ROS-mediated cell-to-cell communication (Dubiella et al., 2013).
In contrast to CPK4, 5, 6, and 11, which function as positive regulators in the defense response, CPK28 plays a negative role in immune signaling (Monaghan et al., 2014). Loss of function of CPK28 results in enhanced PAMP-triggered responses, and overexpression of CPK28 inhibits PAMP-triggered immunity. CPK28 phosphorylates the receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1), an important convergent substrate of multiple pattern recognition receptors, and contributes to BIK1 accumulation, suggesting that CPK28 regulates the immunity response by modulating the turnover of RLCKs (Monaghan et al., 2014). Together, these findings indicate that CPKs contribute to plant immunity via different regulatory mechanisms, which are often involved in several distinct aspects of this response, including ROS production, transcriptional reprogramming, and the activation of phytohormone signaling pathways. However, how the primary activation of CPKs is regulated and maintained and how activated CPKs propagate the defense signals that ultimately lead to plant resistance are not well understood.
Powdery mildew fungi are widespread biotrophic pathogens that cause diseases in many important crops, including wheat (Triticum aestivum) and barley (Hordeum vulgare), as well as the model plant Arabidopsis (Adam and Somerville, 1996). Previously, we identified an Arabidopsis exo70B1 allele during a screen for mutants that display enhanced resistance to the powdery mildew pathogen Golovinomyces cichoracearum (Zhao et al., 2015). EXO70B1 encodes a subunit of the exocyst complex, a conserved octameric protein complex that tethers vesicles to the plasma membrane during exocytosis (He and Guo, 2009). In Arabidopsis, the EXO70 protein family has 23 members, some of which are involved in plant immunity (Zárský et al., 2013). For example, EXO70B2 and EXO70H1 are upregulated after treatment with the elicitor elf18, and exo70B2 and exo70H1 mutants are more susceptible to Pseudomonas syringae pv maculicola compared with the wild type (Pecenková et al., 2011). PUB22-mediated ubiquitination and degradation of EXO70B2 contribute to PAMP-triggered responses (Stegmann et al., 2012). EXO70B1 is also implicated in autophagy-related transport to the vacuole (Kulich et al., 2013). Loss of function of EXO70B1 causes reduced numbers of internalized autophagosomes inside the vacuole (Kulich et al., 2013), along with ectopic hypersensitive responses (Kulich et al., 2013; Stegmann et al., 2013; Zhao et al., 2015) and enhanced resistance to several pathogens, including the powdery mildew G. cichoracearum, the bacterial pathogen Pto DC3000, and the oomycete pathogen Hyaloperonospora arabidopsidis Noco2 (Zhao et al., 2015).
The activated immune responses in exo70B1 mutants require the atypical nucleotide binding domain and leucine-rich repeat (NLR) protein TIR-NBS2 (TN2). Moreover, the TN2 transcript accumulates to much higher levels in exo70B1 mutants than in the wild type, and TN2 interacts with EXO70B1, indicating that TN2 is directly involved in exo70B1-activated responses (Zhao et al., 2015). TN2 lacks a leucine-rich repeat (LRR) domain and belongs to the TIR-NBS (TN) family, which has 21 members in Arabidopsis ecotype Col-0 (Meyers et al., 2003). Although the functions of TN proteins are not well understood, studies indicate that they play important roles in plant immunity. For instance, TN1 (also known as CHILLING SENSITIVE1 [CHS1]) contributes to the autoimmunity response at low temperatures, which requires ENHANCED DISEASE SENSITIVITY1 and PHYTOALEXIN DEFICIENT4 (PAD4) (Wang et al., 2013; Zbierzak et al., 2013). In addition, some TN proteins interact with effectors and/or full-length NLR proteins based on yeast two-hybrid (Y2H) analyses (Nandety et al., 2013). However, how TN2 contributes to exo70B1-activated resistance is currently unclear.
Here, by performing a forward genetic screen, we found that CPK5 is required for exo70B1-associated phenotypes, including spontaneous cell death, enhanced resistance to pathogens, increased upregulation of immune-related genes, and increased SA accumulation. Mutants of CPK5 homologs failed to suppress the cell death and resistance to powdery mildew phenotypes of exo70B1, suggesting that CPK5 plays a unique role in plant immunity. The N-terminal domain of CPK5, encompassing the variable and kinase domains, CPK5-VK, interacts directly with TN2, and TN2 is necessary for cell death caused by CPK5 overexpression. Importantly, CPK5 is overactivated in exo70B1 mutants; this is dependent on TN2 function. These findings indicate that TN2 associates with and may stabilize active CPK5 to regulate defense responses, thus uncovering a direct link between the atypical immune receptor TN2 and CPK5, a crucial signaling component involved in early immune responses and the onset of the defense response.
RESULTS
cpk5-2 Suppresses Autoimmune Responses and Powdery Mildew Resistance in exo70B1-3 Mutant
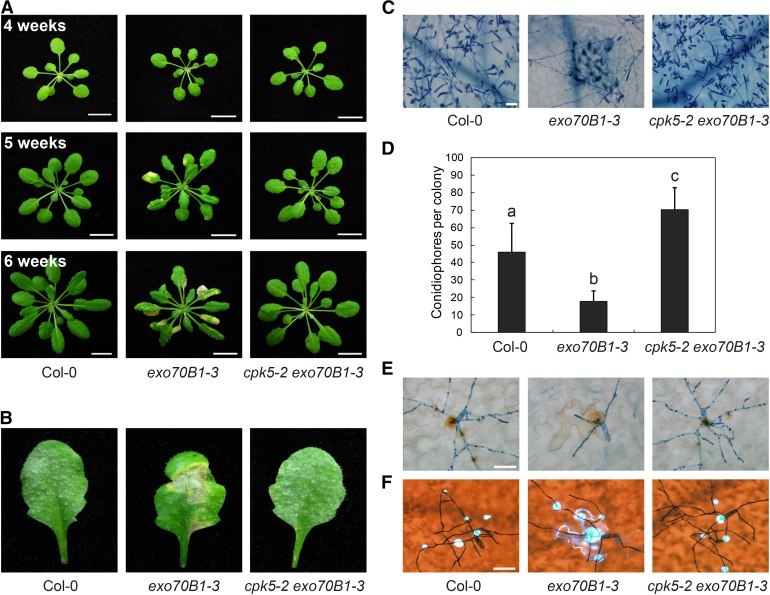
The exo70B1 mutant displays spontaneous cell death and enhanced resistance to the powdery mildew pathogen G.cichoracearum UCSC1 (Kulich et al., 2013; Stegmann et al., 2013; Zhao et al., 2015). To elucidate the underlying molecular mechanism and to identify new components involved in exo70B1-mediated resistance, we performed a forward genetic screen for exo70B1 suppressors. In addition to mutants in TIR-NBS2 (TN2), which we reported previously (Zhao et al., 2015), we identified five cpk5 mutants, which we designated cpk5-2, cpk5-3, cpk5-4, cpk5-5, and cpk5-6, based on subsequent analysis. Since the phenotypes of these exo70B1-3 cpk5 mutants were indistinguishable, we characterized only the cpk5-2 exo70B1-3 mutant in detail in this study. Consistent with previous findings, under short-day conditions, the exo70B1-3 mutant started to display spontaneous cell death in 4-week-old plants, and this cell death was more pronounced in 6-week-old plants (Zhao et al., 2015). By contrast, like the wild type, the cpk5-2 exo70B1-3 mutant did not exhibit cell death (Figure 1A). The exo70B1-3 mutant was resistant to powdery mildew, showing extensive necrotic lesions but no visible powder on leaves at 8 d postinfection (dpi) with G. cichoracearum. By contrast, at 8 dpi, leaves of cpk5-2 exo70B1-3 plants were covered with numerous conidia but lacked visible mildew-induced lesions (Figure 1B). Similarly, trypan blue staining showed that in exo70B1-3, very few fungal hyphae developed and clear cell death occurred. Abundant fungal hyphae and conidiophores were produced in cpk5-2 exo70B1-3 and wild-type plants (Figure 1C). We quantified fungal growth by counting the number of conidiophores per colony in plants at 5 dpi. As shown in Figure 1D, exo70B1-3 mutants had significantly fewer conidiophores than the wild type, but cpk5-2 exo70B1-3 mutants had more conidiophores than the wild type (Figure 1D). These data indicate that the cpk5-2 mutation is fully capable of suppressing cell death and resistance to powdery mildew in the exo70B1-3 mutant.
Figure 1.
cpk5-2 Suppresses exo70B1-3-Mediated Cell Death and Resistance to G. cichoracearum.
(A) Plants were grown under short-day conditions. Uninfected plants of different ages were photographed. The exo70B1-3 plants displayed hypersensitive response-like cell death at 5 weeks, which became more pronounced at 6 weeks, but wild-type Col-0 and cpk5-2 exo70B1-3 plants did not show cell death after 6 weeks. Bar = 1.2 cm.
(B) Four-old-week plants were infected with G. cichoracearum. Representative leaves were detached and photographed at 8 dpi. The exo70B1-3 mutant was more resistant to G. cichoracearum, while the cpk5-2 exo70B1-3 mutant was susceptible, with visible powder produced on the leaves but no cell death, a reaction similar to the wild type.
(C) Infected leaves at 8 dpi were stained with trypan blue to observe fungal structures and dead cells. Few spores were produced in the exo70B1-3 mutant, whereas many fungal spores were produced in both wild-type and cpk5-2 exo70B1-3 plants. Bar = 50 μm.
(D) Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. The exo70B1-3 mutant supported significantly less fungal growth than the wild type, while the cpk5-2 exo70B1-3 mutant supported even higher fungal growth than the wild type. Bars represent mean and sd (n ≥ 14). Lowercase letters indicate statistically significant differences (P < 0.05; one-way ANOVA). The experiment was repeated three times with similar results.
(E) Infected leaves were sequentially stained with 3,3′-diaminobenzidine-HCl and trypan blue at 2 dpi to visualize hydrogen peroxide (brown staining) and fungal structure (blue staining) at the penetration site. Bar = 20 μm.
(F) Infected leaves were sequentially stained with aniline blue and trypan blue at 2 dpi to examine callose deposition (bright blue dots) and fungal structure (dark blue staining) at the penetration site. Bar = 20 μm.
The exo70B1-3 mutant also showed other defense-related phenotypes, including increased H2O2 accumulation and callose deposition, upregulation of pathogenesis-related gene expression, and increased SA accumulation upon powdery mildew infection (Zhao et al., 2015). We investigated the presence of these phenotypes in cpk5-2 exo70B1-3 as well. Consistent with previous findings, exo70B1-3 mutants showed increased H2O2 accumulation and callose deposition at the site of infection at 2 dpi, whereas the increased accumulation of both H2O2 and callose was suppressed by the cpk5-2 mutation (Figures 1E and 1F). Similarly, the cpk5-2 mutation suppressed the mildew-induced upregulation of PR1, PR2, SID2, and PAD4 and accumulation of SA (Supplemental Figures 1 and 2).
In addition to enhanced powdery mildew resistance, the exo70B1-3 mutant shows increased resistance to the bacterial pathogen Pto DC3000 (Zhao et al., 2015). We therefore inoculated wild-type, exo70B1-3, and cpk5-2 exo70B1-3 plants with Pto DC3000. As shown in Supplemental Figure 3, the enhanced resistance to Pto DC3000 in exo70B1-3 was also suppressed by the cpk5-2 mutation. Taken together, these data indicate that CPK5 is required for the activation of defense responses in the exo70B1-3 mutant.
We identified the cpk5-2 mutation using standard map-based cloning (Supplemental Figure 4A). The cpk5-2 mutation (G1522A) is in the end of the second intron of the CPK5 gene, and results in a splicing error that caused an 8-bp deletion in the CPK5 transcript, leading to a premature stop codon (Supplemental Figures 4B and 4C). The CPK5 protein consists of four domains: the N-terminal variable domain, kinase domain, autoinhibitory junction domain, and CaM-like domain (Cheng et al., 2002). The cpk5-2 mutation results in a truncated CPK5 protein with an incomplete kinase domain. In the mutant screen, we identified four additional alleles of CPK5, including three with missense mutations in the kinase domain and one with a missense mutation in the CaM-like domain (Supplemental Figures 4D and 5). The CPK5 genomic clone complemented the cpk5-2 phenotype (Supplemental Figure 6). In addition, cpk5-1, a T-DNA knockout mutation suppressed exo70B1 mutant phenotypes (Figure 2), indicating that the cpk5 alleles identified here are not gain of function or constitutively active mutants.
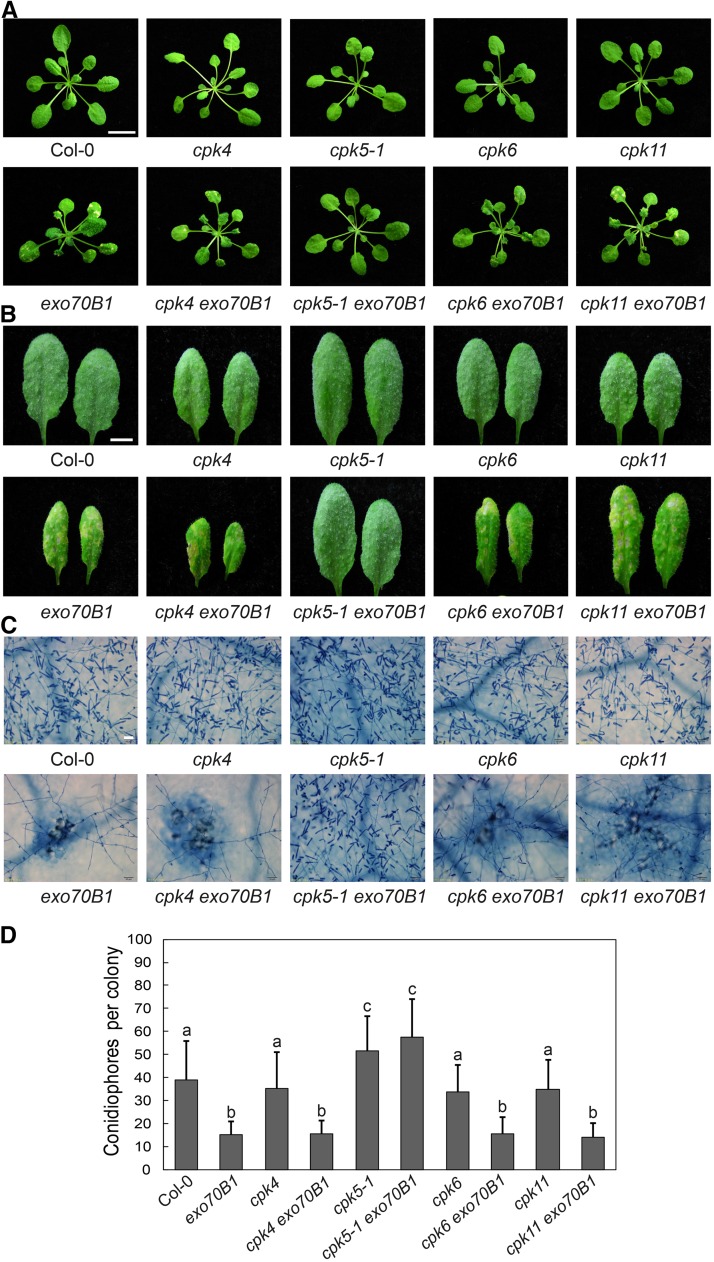
Figure 2.
Crosses with Mutants of CPK4, CPK6, and CPK11 Failed to Suppress exo70B1-3-Mediated Cell Death and Resistance to G. cichoracearum.
(A) Five-week-old plants were photographed under short-day conditions. The cpk4 exo70B1-3, cpk6 exo70B1-3, and cpk11 exo70B1-3 mutants displayed hypersensitive response-like cell death, which was similar to exo70B1-3, but no cell death was observed in the wild type or the cpk5-1 exo70B1-3 mutants. Bar = 1.2 cm.
(B) Four-week-old plants were infected with G. cichoracearum. The leaves were detached and photographed at 8 dpi. The cpk4 exo70B1-3, cpk6 exo70B1-3, and cpk11 exo70B1-3 mutants displayed exo70B1-3-like phenotypes, as they supported much less fungal growth and showed obvious mesophyll cell death compared with cpk5-1 exo70B1-3 and the wild type. Bar = 0.5 cm.
(C) The leaves were stained with trypan blue after infection with G. cichoracearum at 8 dpi. Bar = 50 μm.
(D) Quantification of fungal growth in plants at 5 dpi by counting the number of conidiophores per colony. Bars represent means and sd (n ≥ 20). Lowercase letters indicate statistically significant differences (P < 0.05, one-way ANOVA). The experiment was performed three times with similar results.
“exo70B1” indicates plants carrying the exo70B1-3 allele.
CPK5 Homologs Are Not Required for exo70B1-Activated Resistance
CPK5 belongs to the CDPK gene family; in Arabidopsis, this family has 34 members in four clades, based on phylogenetic analysis. CPK5 belongs to clade I, with CPK4, CPK6, and CPK11 representing the closest homologs of CPK5 (Boudsocq et al., 2010). CPK4, 5, 6, and 11 in this subgroup were proposed to play redundant roles in plant immunity (Boudsocq et al., 2010; Gao et al., 2013). To investigate whether the close homologs of CPK5 contribute to exo70B1-mediated immunity, we generated exo70B1-3 cpk4, exo70B1-3 cpk6, and exo70B1-3 cpk11 double mutants and examined their phenotypes. None of the lines carrying mutations in CPK4, CPK6, or CPK11 showed suppression of exo70B1-mediated spontaneous cell death or powdery mildew resistance (Figures 2A to 2D), indicating that CPK4, CPK6, and CPK11 do not contribute to exo70B1-activated resistance.
The cpk5 Mutant Exhibits Increased Susceptibility to Powdery Mildew
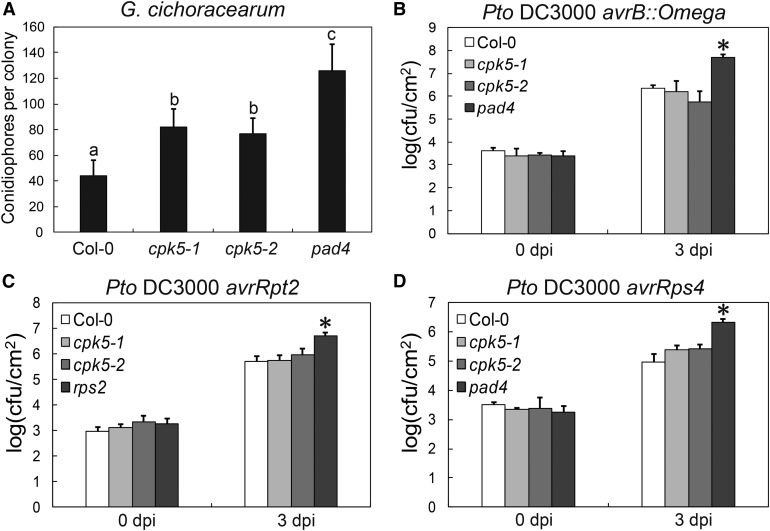
To examine the role of CPK5 and its close homologs in plant immunity, we challenged the cpk4, cpk5, cpk6, and cpk11 single mutants with G. cichoracearum. The cpk5 mutant supported more conidiophore growth than the wild type (Figure 3A), whereas cpk4, cpk6, and cpk11 displayed wild-type-like responses to G. cichoracearum (Figures 2B to 2D), indicating that CPK5 plays an important role in powdery mildew resistance. To further investigate the role of CPK5 in powdery mildew resistance, we crossed cpk5-2 with several well-characterized powdery mildew resistance mutants, including enhanced disease resistance2 (edr2), powdery mildew resistance4 (pmr4-1), and accelerated cell death5 (acd5). As shown in Supplemental Figure 7, cpk5-2 did not affect edr2, pmr4-1, or acd5-mediated resistance to powdery mildew, indicating that the suppression of exo70B1-3 by cpk5 is specific.
Figure 3.
The cpk5 Mutant Displays Enhanced Susceptibility to G. cichoracearum.
(A) Four-week-old plants were infected with G. cichoracearum. Fungal growth was assessed at 5 dpi by counting the number of conidiophores per colony. Bars represent means and sd (n = 14). Lowercase letters indicate statistically significant differences (P < 0.05, one-way ANOVA). The experiment was performed three times with similar results.
(B) to (D) Four-week-old plants were inoculated with different Pto DC3000 strains at OD600 = 0.0005. The number of bacteria was counted at 3 h and 3 dpi. Bars represent mean and sd of three biological samples. cfu, colony-forming units. The asterisk indicates statistically significant difference (P < 0.05, one-way ANOVA). The experiment was performed three times with similar results.
Previously, CPK4, 5, 6, and 11 were proposed to play redundant roles in Pto DC3000 resistance because single cpk mutants are not impaired in the response to Pto DC3000, whereas cpk5 cpk6 double and cpk5 cpk6 cpk11 triple mutants exhibit slightly enhanced susceptibility to Pto DC3000 (Boudsocq et al., 2010). In this study, the cpk5 single mutant displayed enhanced susceptibility to powdery mildew but had wild-type responses to the virulent bacterial pathogen Pto DC3000 and the avirulent Pto DC3000 carrying the effectors avrRpt2 or avrRps4 (Figures 3B to 3D). These results indicate that CPK5 plays a unique role in exo70B1-activated defense and powdery mildew resistance.
The Membrane Localization and Kinase Activity of CPK5 Are Critical for Its Function in the exo70B1-Mediated Pathway
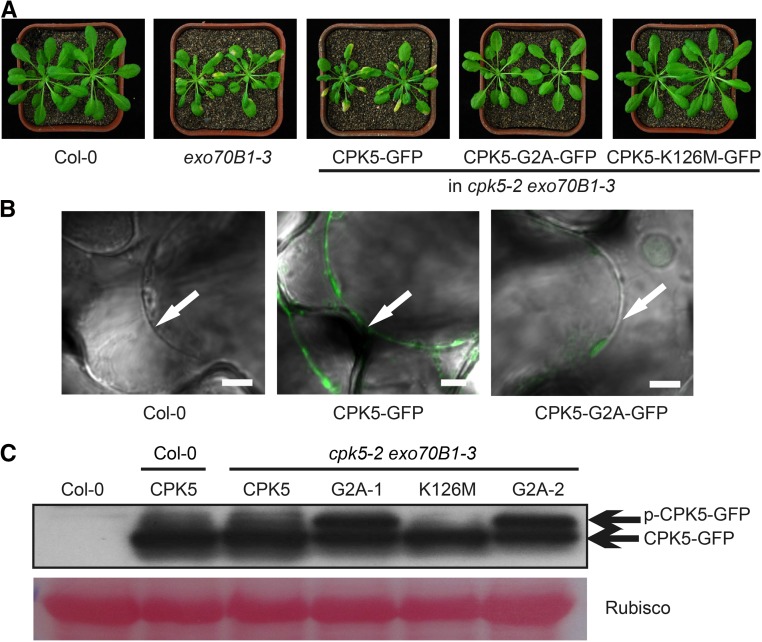
To examine the membrane localization of CPK5, we generated a CPK5-GFP fusion protein driven by the native CPK5 promoter and transformed the resulting ProCPK5:gCPK5-GFP construct into the cpk5-2 exo70B1-3 double mutant. The transgenic plants showed exo70B1-like phenotypes (Figure 4A), indicating that CPK5-GFP has full CPK5 function. CPK5 has a predicted N-myristoylation site, and the second amino acid residue, Gly, is critical for myristoylation, which is necessary for CPK5 plasma membrane localization (Lu and Hrabak, 2013). We therefore generated a CPK5-GFP variant carrying a G2A single amino acid substitution and transformed it into the cpk5-2 exo70B1-3 double mutant. CPK5-G2A-GFP was unable to restore cpk5-2 exo70B1-3 to the exo70B1-3 phenotype (Figure 4A), and CPK5-G2A-GFP no longer localized to the plasma membrane (Figure 4B). These data indicate that CPK5-G2A-GFP is not functional and the membrane localization of CPK5 is necessary for its function.
Figure 4.
Gly, the Second Residue of CPK5, Is Critical for Its Function, and the G2A Mutation Enhances CPK5 Phosphorylation.
(A) Six-week-old plants were photographed under short-day conditions. Cell death was observed in the exo70B1-3 mutant and in cpk5-2 exo70B1-3 transgenic plants carrying CPK5-GFP, but not CPK5-G2A-GFP or CPK5-K126M-GFP.
(B) Four-week-old leaves were soaked in 0.85 M NaCl for 15 min and examined by confocal microscopy. Arrows indicate plasma membrane. Bar = 5 μm.
(C) Various forms of CPK5-GFP protein were examined by immunoblot analysis. CPK5, G2A, and K126M indicate plants carrying CPK5-GFP, CPK5-G2A-GFP, or CPK5-K126M-GFP, respectively. G2A-1 and G2A-2 represent two independent transgenic plants carrying CPK5-G2A-GFP. Ponceau S staining of Rubisco is shown as a protein loading control. Bands corresponding to the phosphorylated or unphosphorylated form of CPK5-GFP are indicated by p-CPK5-GFP or CPK5-GFP, respectively.
When ectopically expressed in leaf mesophyll protoplasts, CPK5 and CPK5-G2A showed protein kinase activities (see below). To investigate whether the kinase activity of CPK5 is required for exo70B1-mediated resistance, we generated a CPK5 kinase-deficient CPK5-K126M-GFP variant by disrupting its ATP binding site and introduced it into cpk5-2 exo70B1-3 plants. We failed to detect the higher molecular mass form of CPK5-K126M-GFP (corresponding to phosphorylated CPK5-GFP) in cpk5-2 exo70B1-3 transgenic plants (Figure 4C), and CPK5-K126M-GFP was unable to restore cpk5-2 exo70B1-3 to the exo70B1-3 phenotype (Figure 4A). These results indicate that CPK5-K126M-GFP is not functional and kinase activity of CPK5 is required for its function in exo70B1-3-mediated resistance phenotypes.
CPK5 Interacts with TN2
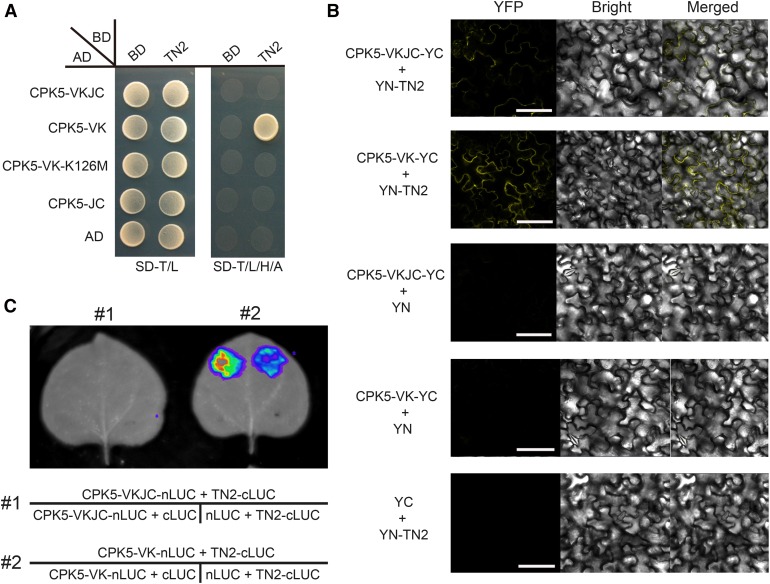
We previously showed that the truncated NLR protein TN2 is required for the phenotypes of exo70B1 and that TN2 associates with EXO70B1. Also, exo70B1 accumulates much higher levels of TN2 transcripts (Zhao et al., 2015). We then measured CPK5 transcript levels in the exo70B1 mutant; however, we did not observe significant differences between the wild type and the exo70B1 mutant (Supplemental Figure 8). Because exo70B1-activated resistance depends on both CPK5 and TN2, we hypothesized that CPK5 and TN2 might function together. To test this hypothesis, we examined the interactions between different domains of CPK5 and TN2 by Y2H. As shown in Figure 5A, CPK5-VK, the truncated variant consisting only of the variable and kinase domains, which displays constitutive, calcium-independent kinase activity, interacted with TN2. However, no interaction was observed with the kinase-deficient form CPK5-VK-K126M or full-length CPK5 and TN2.
Figure 5.
CPK5 Interacts with TN2.
(A) Y2H assay. The coding sequences of CPK5-VKJC, CPK5-VK, CPK5-VK-K126M, and CPK5-JC were fused to the Gal4 transactivation domain (AD). The coding sequence of TN2 was fused to the Gal4 DNA binding domain (BD). Different pairs of constructs were cotransformed into AH109. A 10-μL suspension (OD600 = 0.5) of each cotransformant was dropped onto synthetic dropout (SD) medium lacking Leu and Trp and SD medium lacking Ade, His, Leu, and Trp. Photographs were taken after 5 d of incubation.
(B) BiFC assay. CPK5-VKJC and CPK5-VK were fused to YFPYC, and TN2 was fused to YFPYN. Different pairs of constructs were coexpressed in N. benthamiana. YFP fluorescence was detected by confocal microscopy. Bar = 100 μm.
(C) Firefly LUC complementation imaging assay. N. benthamiana leaves were coinfiltrated with agrobacterial strains containing different pairs of constructs. LUC images were captured using a cooled CCD imaging apparatus.
To confirm the interaction between CPK5-VK and TN2, we performed a bimolecular fluorescence complementation (BiFC) assay. The full-length CPK5-VKJC and N-terminal variant CPK5-VK were fused to the C-terminal fragment of YFP (YFPC) and cotransformed with TN2-YFPN into Nicotiana benthamiana. YFP fluorescence was observed in leaves cotransformed with CPK5-VK-YFPC and TN2-YFPN, whereas weak fluorescence was detected with CPK5-VKJC-YFPC and no fluorescence was detected in the controls (Figure 5B). We next assessed the interaction between CPK5 and TN2 by firefly luciferase complementation imaging (LUC) in N. benthamiana. Again, a strong interaction occurred between TN2-cLUC and N-terminal CPK5-VK-nLUC but not with full-length CPK5-VKJC-nLUC (Figure 5C). Taken together, these results indicate that CPK5 physically associates with TN2 via its N-terminal variable and kinase domains and that CPK5 functions together with TN2 in exo70B1-mediated resistance.
As CPK5 kinase activity is necessary for its function in exo70B1-mediated resistance, we examined whether disrupting the kinase activity of CPK5 would affect its association with TN2. We assessed the interaction between TN2 and two kinase-deficient mutant forms of CPK5, including CPK5-K126M-VK and CPK5-D221A-VK. The amino acids K126 and D221 in CPK5 are critical residues for the ATP binding site and the active site, respectively. Neither the kinase-deficient mutant form CPK5-K126M-VK nor CPK5-D221A-VK interacted with TN2 (Supplemental Figure 9), indicating that the kinase activity of CPK5 is required for its association with TN2.
We previously showed that EXO70B1 interacts with the TIR domain (amino acids 1–219) of TN2 (Zhao et al., 2015). To identify the region of TN2 that interacts with CPK5, we examined the interaction between CPK5-VK and different truncated forms of TN2 by Y2H assay. As shown in Supplemental Figure 10, the presence of the N-terminal amino acids 1 to 160 of TN2 was not sufficient for the interaction with either CPK5-VK or EXO70B1, whereas the remaining amino acids (161–318) of TN2, encompassing the NBS domain, interacted with CPK5-VK but not EXO70B1. Amino acids 1 to 219 of TN2, encompassing the TIR domain, interacted with EXO70B1 but not CPK5-VK. The remaining amino acids (220–318) of TN2 did not interact with CPK5-VK or EXO70B1. These results indicate that CPK5 interacts with the NBS domain of TN2, whereas EXO70B1 associates with the TIR domain of TN2.
To test the specificity of the interaction between CPK5 and TN2, we generated CPK4-VK and CPK6-VK variants and subjected them to Y2H and LUC interaction assays with TN2. As shown in Supplemental Figure 11, neither CPK4-VK nor CPK6-VK interacted with TN2. These results suggest that CPK5, but not CPK4 or CPK6, plays a specific role in TN2-dependent exo70B1-mediated resistance.
Overexpression of CPK5 Leads to TN2-Dependent Autoimmune Responses
A CPK5 overexpression line (35S:CPK5-YFP-#7) displayed spontaneous cell death, resistance to Pto DC3000, and increased SA accumulation, which is reminiscent of the exo70B1 mutant phenotype. Plants overexpressing kinase-deficient CPK5 (35S:CPK5m-YFP-#15) showed wild-type-like responses (Dubiella et al., 2013). To further investigate the role of CPK5 in plant immunity, we challenged these two lines with G. cichoracearum. As shown in Supplemental Figure 12, the CPK5 overexpression line displayed mildew-induced cell death and enhanced resistance to powdery mildew, which is reminiscent of the exo70B1 mutant, whereas the kinase-deficient line was indistinguishable from the wild type. Thus, CPK5 overexpression causes enhanced resistance and autoimmune responses, a phenotype similar to that of exo70B1. Indeed, exo70B1-mediated autoimmunity depends on TN2 and TN2 transcript levels are high in exo70B1 mutants (Zhao et al., 2015). To investigate whether CPK5 overexpression leads to higher TN2 expression, we examined TN2 transcript levels in the CPK5 overexpression line. As shown in Supplemental Figure 13, TN2 transcript levels were higher in the CPK5 overexpression line than in the wild type and the CPK5 kinase-deficient line, whereas TN2 transcript levels were similar between the CPK5 overexpression line and exo70B1-3. These data suggest that the autoimmune phenotypes of the CPK5 overexpression line and the exo70B1-3 mutants are caused by a similar mechanism.
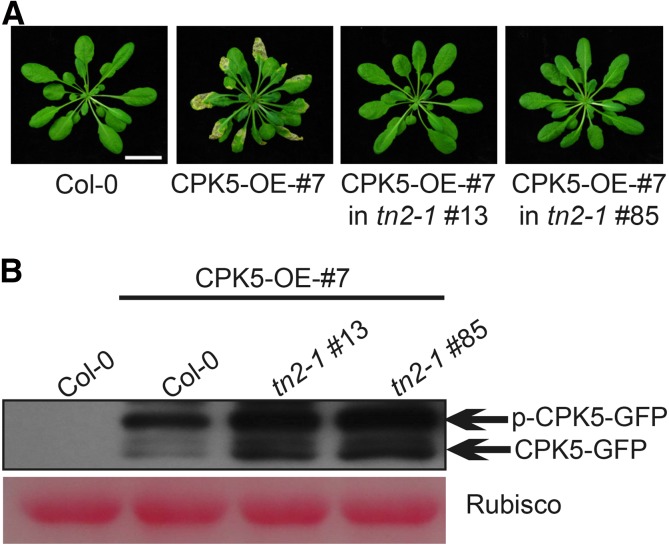
Since exo70B1-mediated resistance requires TN2, and the phenotypes of the CPK5 overexpression line matched those of exo70B1, we hypothesized that the autoimmune response caused by CPK5 overexpression might also depend on TN2. Therefore, we crossed the CPK5 overexpression line with tn2-1 and examined whether the tn2-1 mutation would affect the phenotypes of this line. Interestingly, the tn2-1 mutation suppressed CPK5-dependent cell death in the CPK5 overexpression line (Figure 6A). Immunoblot analysis revealed that CPK5-YFP was still expressed in the tn2-1 background (Figure 6B). These results demonstrate that CPK5 and TN2 function together in exo70B1-mediated resistance, and they suggest the presence of positive feedback between CPK5 and TN2.
Figure 6.
The tn2 Mutation Suppresses Cell Death in the CPK5 Overexpression Line.
(A) Six-week-old plants were photographed under short-day conditions. Bar = 2.4 cm.
(B) The CPK5-YFP fusion protein was examined by immunoblotting. Ponceau S staining of Rubisco is shown as a protein loading control.
CPK5 Is Overactivated in exo70B1-3
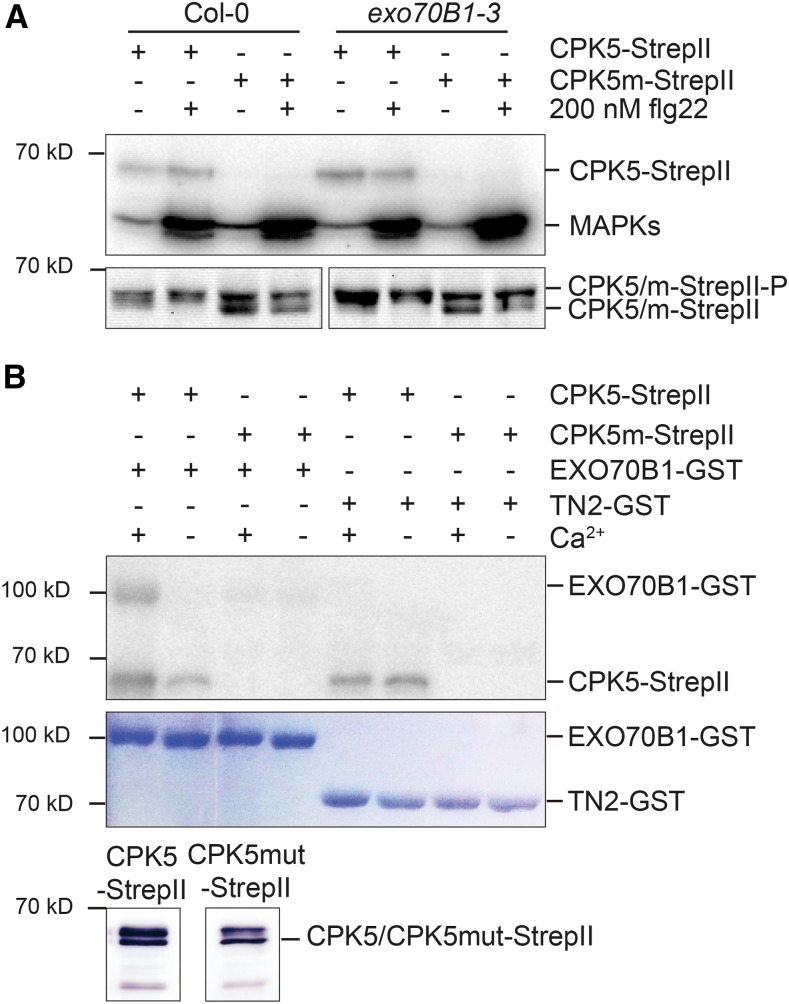
CPK5 functions in innate immune signaling and rapidly becomes biochemically activated after flg22 or elf18 treatment (Dubiella et al., 2013). Because exo70B1-mediated resistance requires CPK5, and the CPK5 overexpression line displayed exo70B1-like phenotypes, we compared CPK5 kinase activity between the wild type and exo70B1. We ectopically expressed StrepII-tagged variants of CPK5 and kinase-deficient CPK5mut in leaf mesophyll protoplasts derived from the respective lines and analyzed in-gel kinase activity in samples taken after 15 min of flg22 exposure (Dubiella et al., 2013). As shown in Figure 7A (upper panel), in exo70B1, constitutive CPK5 kinase activity was already observed at 68 kD in the absence of the flg22 PAMP elicitor, whereas CPK5 kinase activity was inducible in Col-0. No kinase activity was detected when CPK5mut was expressed. Interestingly, flg22-inducible activation of mitogen-activated protein kinase activity at 48 kD was unaltered. Kinase activity correlated with the accumulation of a slower migrating form of CPK5 in immunoblot analysis (lower panel).
Figure 7.
CPK5 Shows Enhanced Biochemical Activity in exo70B1-3 and Can Phosphorylate EXO70B1 but Not TN2.
(A) In-gel kinase assay. CPK5 is active in exo70B1-3 in the absence of flg22 treatment. Protoplasts were isolated from 6-week-old Col-0 and exo70B1-3 plants and transfected with CPK5-StrepII and kinase-deficient CPK5m-StrepII before elicitation with either buffer (−) or 200 nM flg22 (+) for 15 min. Total protein was separated by SDS-PAGE and subjected to an in-gel kinase assay using myelin basic protein as substrate or to immunoblotting. The proteins were visualized by autoradiography (upper panel) or analyzed by immunostaining with Strep-Tactin HRP (lower panel). The experiment was performed three times with similar results.
(B) In vitro kinase assay. CPK5 phosphorylates EXO70B1 but not TN2 in vitro. CPK5-StrepII and CPK5m-StrepII were transiently expressed in N. benthamiana leaves, and affinity purification was controlled by immunoblotting (lower panel). The autophosphorylation activity of CPK5-StrepII and CPK5m-StrepII and activity toward recombinant substrate proteins EXO70B1-GST and TN2-GST were assessed in the presence of calcium or EGTA as indicated. Phosphorylation was visualized by autoradiography (upper panel) and the amount of substrate protein was visualized by Coomassie staining (lower panel). The experiment was performed four times with similar results.
Interestingly, the level of CPK5 protein was very low in protoplasts derived from tn2-1 and exo70B1-3 tn2-1 mutant plants compared with those of Col-0 and exo70B1 (Supplemental Figure 14A). To investigate whether the inability to detect high transient expression of CPK5 in the tn2-1 background was related to the function of this enzyme, we assessed the activity of the variant CPK5-G2A in the respective mutant backgrounds. This variant fails to localize to the plasma membrane and therefore cannot complement the exo70B1 cpk5 double mutant phenotype (Figure 4). As shown in Supplemental Figure 14B (upper panel), CPK5-G2A protein accumulated to a much higher level in the exo70B1 mutant background compared with Col-0, whereas only weak expression was detected in tn2-1, and the enzyme displayed flg22-inducible kinase activity (lower panel).
To investigate whether CPK5 can directly phosphorylate either EXO70B1 or TN2, we performed in vitro kinase assays with purified CPK5 and recombinant EXO70B1-GST and TN2-GST, respectively. CPK5 could autophosphorylate (68 kD) and phosphorylate EXO70B1-GST (100 kD) in a calcium-dependent manner (Figure 7B, upper panel). No phosphorylation was detected with TN2-GST at 70 kD. Interestingly, in the presence of TN2, CPK5 was still capable of autophosphorylation, even in the absence of calcium. These results, together with the interaction between TN2 and CPK5-VK but not full-length CPK5, suggest that TN2 binds to and stabilizes the activated enzyme form of CPK5. Thus, the autoimmune responses in exo70B1 appear to be due to the constitutive activation of CPK5 mediated via TN2.
DISCUSSION
EXO70B1 is a subunit of the exocyst, which plays an important role in exocytosis (Zárský et al., 2013). Loss of function of EXO70B1 leads to hypersensitive response-like cell death and activated defense responses (Kulich et al., 2013; Stegmann et al., 2013; Zhao et al., 2015). To elucidate the molecular mechanisms underlying exo70B1-associated resistance, we screened for exo70B1 suppressors and identified tn2 (Zhao et al., 2015) and cpk5 mutants (this study), which fully suppressed the autoimmunity in exo70B1, indicating that TN2 and CPK5 are required for exo70B1-associated resistance.
TN2 is a truncated NLR protein that lacks the LRR domain. Phenotypes associated with exo70B1 are reminiscent of several autoimmune mutants, whose phenotypes result from ectopic activation of NLR immune receptors. For instance, both Arabidopsis accelerated cell death11 and lesion simulating disease1 show autoimmune responses, which are dependent on the TIR-NLR LAZ5 or the coiled-coil-NLRs ADR1-L1 and ADR1-L2 (Palma et al., 2010; Bonardi et al., 2011; Roberts et al., 2013). Based on the potential roles of EXO70B1 in basal defense, we previously proposed that a TN2- or TN2-related immune complex monitors the status of EXO70B1 and that modification of EXO70B1 by pathogen effectors may trigger NLR-mediated defense responses (Zhao et al., 2015). In this model, the exo70B1-associated resistant phenotypes result from effector-triggered immunity (ETI) and EXO70B1 is a potential guardee (guarded effector target) for TN2 or the TN2-associated immune complex, but not a negative regulator of the disease resistance and cell death responses. Consistent with this hypothesis, a recent study showed that the effector AVR-Pii from the rice blast fungus Magnaporthe oryzae directly interacts with OsExo70F3 and that rice (Oryza sativa) NLR protein Pii recognizes AVR-Pii and triggers immunity in an OsExo70F3-dependent manner (Fujisaki et al., 2015). These findings suggest that rice blast fungus targets OsExo70F3 to manipulate plant immunity, while the host plant activates ETI by sensing the modification of OsExo70F3, supporting our hypothesis that EXO70B1 is an effector target. In addition, Phytophthora infestans manipulates plant immunity by targeting the exocyst subunit Sec5 with the RXLR effector AVR1 (Du et al., 2015).
Our finding that CPK5 is required for exo70B1-mediated defense responses provides new insights into not only exo70B1-activated defense, but also the role of CPK5 in TIR-NBS mediated immunity. Interestingly, exo70B1-activated defense depends only on CPK5, not CPK4, 6, and 11, although all of these proteins were previously shown to play redundant roles in immunity (Boudsocq et al., 2010; Gao et al., 2013). Furthermore, only the cpk5 mutant displayed enhanced susceptibility to powdery mildew, while the susceptibility of cpk4, 6, and 11 single mutants was similar to that of the wild type, indicating that CPK5 is unique among CPK5 family members, at least regarding the TN2-activated hypersensitive response-like cell death and disease resistance.
The role of CPK5 in early signaling is well established. CPK5 is transiently biochemically activated during the onset of PAMP immune signaling and plays a dual role in rapid defense signal propagation via ROS and enhanced long-lasting defense, even systemic defense, via transcriptional reprogramming and high SA levels (Dubiella et al., 2013). Overexpressing CPK5 results in constitutive enzyme activity, and CPK5-overexpressing plants show enhanced resistance to Pto DC3000 (Dubiella et al., 2013). Since CPK5 is required for exo70B1-mediated defense, and exo70B1-resistant phenotypes are likely an ETI phenomenon, it will be important to investigate the role of CPK5 in NLR-mediated signaling. We previously showed that EXO70B1 interacts with the TIR domain of TN2 (Zhao et al., 2015). Here, we demonstrated that CPK5 interacts with the NBS domain of TN2. The interaction between CPK5 and TN2 is specific because neither CPK4 nor CPK6 interacted with TN2. Importantly, we showed that CPK5 is overactivated in exo70B1 mutants and that the overactivation of CPK5 requires TN2. CPK5 autophosphorylates, which usually occurs in a calcium-dependent manner. However, intriguingly, in the presence of TN2, CPK5 is autoactive in the presence of little or no calcium. This observation suggests that TN2 positively affects CPK5 activity. Notably, full-length CPK5 (CPK5-VKJC) showed a weaker interaction in both luciferase and BiFC assays (and no interaction in Y2H assay) than the truncated CPK5-VK variant. Moreover, the kinase activity of CPK5 is required for its interaction with TN2 and for CPK5 function. Together, these observations support a model in which TN2 binds to activated, open-folded CPK5 and stabilizes it, in turn resulting in increased and maintained CPK5 activity and, thus, downstream defense signaling. This model is consistent with our interaction data, as full-length CPK5 should more commonly occur in a closed conformation; consequently, the active center would be blocked by the calcium binding domain. By contrast, since CPK5-VK lacks a calcium binding domain, this blockage does not occur, enabling a strong interaction. The tn2 mutant lacks this factor that interacts with CPK5 and stabilizes CPK5 activity. In the exo70B1 mutant, the TN2/CPK5 interaction cannot be released by CPK5-catalyzed phosphorylation of EXO70B1. It is tempting to speculate that effector-modified EXO70B1 is likewise protected from phosphorylation by CPK5. This model predicts an initial priming stimulus resulting in increased cytosolic calcium concentration to induce CPK5 “opening” and activation, e.g., via a PAMP.
Overexpression of CPK5 leads to hypersensitive response-like cell death and enhanced pathogen resistance (Dubiella et al., 2013) (Supplemental Figure 12), a response similar to that of the exo70B1 mutant. Interestingly, overexpression of CPK5 also resulted in high levels of TN2 transcript and the phenotypes associated with CPK5 overexpression require TN2. Therefore, overexpression of CPK5 is not sufficient for the activation of defense responses when TN2 is absent. One possible explanation is that there is positive feedback between TN2 and CPK5. Activation of CPK5 would induce the expression of TN2, and higher levels of TN2 would in turn further activate CPK5. This model is consistent with our hypothesis that TN2 binds to and stabilizes active CPK5, which helps maintain CPK5 activity and downstream immune signaling. Alternatively, a guard model could explain the activation of immunity in the exo70B1 mutant. In this model, CPK5 is guarded by TN2, and loss of EXO70B1 leads to perturbation of CPK5. TN2 senses the change in CPK5 protein and activates downstream defense signaling. Consistent with this model, CPK5 appears to have a very specific role in TN2-mediated immunity, as CPK5 is not required for edr2, pmr4-1, or acd5-mediated resistance to powdery mildew.
TN2 belongs to a family of 21 TIR-NBS proteins in the Arabidopsis ecotype Col-0. Some TN proteins interact with pathogen effectors and the overexpression of some TN genes leads to cell death in tobacco, suggesting that TN proteins are likely involved in plant immunity (Nandety et al., 2013). Among the 21 TIR-NBS proteins, besides TN2, only TN1 (also known as CHS1) has been genetically characterized. TN1 gain-of-function mutation resulted in an autoimmune response at low temperature (Wang et al., 2013; Zbierzak et al., 2013); interestingly, it was recently shown that tn1-associated autoimmunity requires the full-length NLR protein, SOC3 (Zhang et al., 2017). It remains to be determined whether both TN2- and CPK5-activated immunity also requires the function of a full-length NLR.
METHODS
Plant Materials and Growth Conditions
The cpk5-2 exo70B1-3 mutant was identified from an EMS exo70B1-3 population. The cpk5-2 single mutant was identified from the progeny of a cross between cpk5-2 exo70B1-3 and Col-0. The other mutants and transgenic plants used in this study include cpk4 (SALK-025708C), cpk5-1 (SAIL_657C06) (Boudsocq et al., 2010), cpk6 (SALK-025460C) (Boudsocq et al., 2010), cpk11 (SALK_054495) (Boudsocq et al., 2010), tn2-1 (Zhao et al., 2015), sid2 (Wildermuth et al., 2001), pad4 (Glazebrook et al., 1997), rps2 (Mindrinos et al., 1994), edr2 (Tang et al., 2005), pmr4-1 (Vogel and Somerville, 2000), acd5 (Greenberg et al., 2000), 35S:CPK5-YFP-#7, and 35S:CPK5m-YFP-#15 (Dubiella et al., 2013). All mutants were in the Col-0 background. Double mutants were obtained by genetic crosses and identified by PCR. Arabidopsis thaliana plants were grown in the growth room at 20 to 22°C and ∼60% relative humidity with 16/8-h day/night photoperiod for seed setting and 9/15-h day/night photoperiod for phenotyping, with light intensity of 7000 to 8000 lux. Nicotiana benthamiana plants for transient expression were grown under the same short-day conditions as Arabidopsis (Wu et al., 2015).
Pathogen Infection
Powdery mildew strain Golovinomyces cichoracearum UCSC1 was used to infect Arabidopsis plants (Adam and Somerville, 1996). To observe hyphal growth and lesion formation, plants were inoculated at a high spore density. To quantify the number of conidiophores per colony, the plants were inoculated at a lower spore density, and conidiophores were counted at 5 dpi (Wang et al., 2011). The infected leaves were stained with trypan blue at 8 dpi to visualize hyphae and dead cells (Frye and Innes, 1998). The infected leaves were stained with 3,3′-diaminobenzidine hydrochloride to visualize H2O2 accumulation and aniline blue to visualize callose deposition, followed by trypan blue staining at 2 dpi (Wang et al., 2009). Pto DC3000 infection was performed by leaf infiltration as described (Chen et al., 2000).
Map-Based Cloning
The exo70B1-3 mutation was introgressed into the Landsberg erecta (Ler) background by genetic crossing. To obtain the mapping population, cpk5-2 exo70B1-3 was crossed with the derived exo70B1-3 (Ler) plants. Using ∼3000 F2 plants, the mutation was narrowed down to a 28-kb region on BAC-F23E12 on chromosome 4. The mutation in CPK5 was subsequently identified by sequencing all genes in this region.
Vector Construction
The CPK5 genomic sequence, including the 1985-bp promoter and 2187-bp genomic sequence without the stop codon, was cloned into the pEGAD vector to create ProCPK5:gCPK5-GFP-pEGAD by digestion using SacI and AgeI restriction sites. The same CPK5 genomic sequence was amplified by PCR from ProCPK5:gCPK5-GFP-pEGAD and inserted into Gateway vector pDONR207 using a BP Clonase Kit (Invitrogen). Simultaneously, two mutant forms, CPK5-G2A and CPK5-K126M, were created from ProCPK5:gCPK5-GFP-pDONR207 using site-directed mutagenesis (Stratagene). The inserts were then cloned into vector pMDC107 using an LP Clonase Kit (Invitrogen).
RT-qPCR
Total RNA extraction and RT-qPCR were performed as described previously (Shi et al., 2013).
SA Quantification
SA extraction and measurement were performed as described previously (Li et al., 1999; Gou et al., 2009).
Protein Extraction and Immunoblotting
The proteins were extracted and analyzed by immunoblotting as described previously (Shi et al., 2013). The primary anti-GFP antibody was obtained from Roche (1:1000), and the secondary antibody was anti-mouse (1:10,000) HRP-linked. Detection was performed with a chemiluminescent HRP substrate kit (Millipore).
Plasmolysis
The plasmolysis process was performed as described previously (Underwood and Somerville, 2013). Four-week-old leaves were soaked in 0.85 M NaCl for 15 min and observed by confocal microscopy.
Y2H Assay
The Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was used for Y2H assay. To examine the interactions between CPKs and TN2, CPK5-VKJC, CPK5-VK, CPK5-JC, CPK5-VK-K126M, CPK5-VK-D221A, CPK4-VK, and CPK6-VK were cloned into pGADT-7. The full-length TN2 and different truncated fragments of TN2 were cloned into pGBKT-7. Different pairs of constructs were cotransformed into AH109, and the cells were grown on SD-Trp/-Leu medium. Three single clones from SD-Trp/-Leu plates were transferred to SD-Trp/-Leu/-His/-Ade plates with or without X-α-Gal. A single clone was incubated in SD-Trp/-Leu liquid medium, and 10-μL suspensions (diluted to OD600 = 0.5) of the overnight cultures were dropped onto different plates to test the interactions of the proteins.
BiFC Analysis
CPK5-VKJC and CPK5-VK were cloned into vector pSY738 by enzyme digestion and ligation. CPK5-VKJC-YFPC and CPK5-VK-YFPC were then cloned into pDONR207 and finally into pMDC32 using the Gateway system. TN2-YFPN was described previously (Zhao et al., 2015). Different pairs of constructs were cotransformed into N. benthamiana, and YFP fluorescence was observed by confocal microscopy after 2 d.
LUC Analysis
Two vectors, pCAMBIA1300-nLUC and pCAMBIA1300-cLUC, were used for LUC analysis. The following constructs were first obtained by enzyme digestion and ligation: CPK5-VKJC/CPK5-VK/CPK4-VK/CPK6-VK-nLUC and TN2-cLUC. Different pairs of constructs were cotransformed into N. benthamiana. After 2 d, 1 mM precooled luciferin was sprayed onto the leaves, and the samples were incubated in the dark for 5 to 10 min. LUC images were captured using a cooled CCD imaging apparatus (Chen et al., 2008).
Transient Protoplast Expression Assay
Protoplast isolation and transient expression of CPK5-StrepII were conducted using the Tape-Arabidopsis Sandwich method (Wu et al., 2009) as described by Yoo et al. (2007). At 15 h after transfection, flg22 was applied to the samples. Expression of CPK5-StrepII protein was monitored by immunoblot analysis (Strep-Tactin HRP conjugate, 1:5000; IBA), and protein kinase activity was determined using an in-gel kinase assay.
In-Gel Kinase Assay
Protoplasts were harvested by centrifugation and frozen in liquid nitrogen. Protoplast pellets were resuspended by vortexing in 20 μL extraction buffer (100 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM DTT, 10 mM NaF, 10 mM Na3VO4, 10 mM β-glycerol-phosphate, 0.5 mM AEBSF, 100 μg/mL avidin, and 1× protease inhibitor mixture [Sigma-Aldrich]) and incubated on ice for 10 min. The samples were heated for 3 min at 70°C, and proteins were separated by SDS-PAGE in a gel containing 0.25 mg/mL myelin basic protein (Life Technologies). The gel was washed three times for 1 h per wash in wash buffer (25 mM Tris-HCl, pH 7.5, 0.5 mM DTT, 5 mM NaF, 0.1 mM Na3VO4, 0.5 mg/mL BSA, and 0.1% Triton X-100). To renature the proteins, the gel was incubated in renaturation buffer (25 mM Tris-HCl, pH 7.5, 0.5 mM DTT, 5 mM NaF, and 0.1 mM Na3VO4) for 1 h at room temperature, overnight at 4°C and 1 h at room temperature, with several buffer changes. After equilibration of the gel for 1 h in reaction buffer (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 0.1 mM Na3VO4, 12 mM MgCl2, and 1 mM CaCl2), a kinase assay was performed for 1.5 h using reaction buffer containing 1 nM ATP and 50 µCi [γ-32P]ATP. The reaction was stopped by washing six to eight times with 5% trichloroacetic acid and 1% phosphoric acid for a total of 3 to 4 h. The gel was dried, and phosphorylated proteins were visualized by autoradiography.
In Vitro Protein Kinase Assay
CPK5-StrepII and kinase-deficient CPK5m-StrepII were transiently expressed in N. benthamiana leaves. First, 0.5 g leaf material was ground in liquid nitrogen and homogenized in 1 mL plant extraction buffer. The supernatant was then incubated with 40 μL Strep-Tactin Macroprep (50% slurry; IBA) in a rotation wheel for 2 h at 4°C. The samples were washed five times with wash buffer (100 mM Tris, pH 8, 0.5 mM EDTA, 150 mM NaCl, and 0.05% Triton X-100) and stored overnight on ice in 500 μL wash buffer.
Recombinant EXO70B1-GST and TN2-GST were expressed in Escherichia coli BL21 using autoinduction medium (Novagen). Harvested cells were suspended in 1 mL GST extraction buffer (50 mM Tris, pH 8, 250 mM NaCl, 1 mM EDTA, 0.2% [v/v] Triton X-100, 1 mM DTT, and 1 mM AEBSF) and sonicated, followed by incubation of the cell lysate in 100 μL GST matrix (50% slurry) for 2 h at 4°C in a rotation wheel. The samples were washed five times with GST wash buffer (100 mM Tris, pH 8, 150 mM NaCl, 0.2% [v/v] Nonidet-P40, and 1 mM AEBSF) and eluted three times for 10 min at room temperature and 750 rpm with 1:1 elution buffer (100 mM Tris, pH 8.0, and 20 mM reduced GSH).
For the in vitro CPK phosphorylation assay, 0.2 µg of purified EXO70B1-GST or TN2-GST in GST elution buffer was incubated in a 16 μL slurry of immobilized CPK5-StrepII and CPK5m-StrepII, respectively, in buffer E (50 mM HEPES, pH 7.4, 2 mM DTT, and 0.1 mM EDTA) and 6 μL reaction mix (60 mM MgCl2, 60 µM CaCl2, 6 µM ATP, and 18 µCi [γ- 32P]ATP). To test for calcium-independent phosphorylation, calcium was replaced with 12 mM EGTA in the reaction mix. After 30 min incubation at 25°C and 650 rpm, the samples were centrifuged and the supernatant was mixed with 5× SDS loading buffer. The samples were heated for 5 min at 95°C and separated by SDS-PAGE. The gel was stained with Coomassie Brilliant Blue and dried, and protein phosphorylation was visualized by autoradiography.
Oligonucleotide Sequences
The primers used in this study are listed in Supplemental Table 1.
Statistical Analyses
The ANOVA tables for the statistical analyses are listed in Supplemental Table 2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis EXO70B1 (At5g58430), CPK4 (At4g09570), CPK5 (At4g35310), CPK6 (At2g17290), CPK11 (At1g35670), TN2 (At1g17615), PMR4 (AT4g03550), ACD5 (AT5g51290), EDR2 (At4g19040), PR1 (At2g14610), PR2 (AT3g57260), SID2 (AT1g74710), PAD4 (At3g52430), RPS2 (At4g26090), and ACTIN2 (At3g18780).
Supplemental Data
Supplemental Figure 1. cpk5-2 suppresses the expression of defense-related genes in exo70B1-3.
Supplemental Figure 2. cpk5-2 suppresses powdery mildew-induced salicylic acid accumulation in exo70B1-3.
Supplemental Figure 3. cpk5-2 suppresses exo70B1-3-mediated resistance to Pto DC3000.
Supplemental Figure 4. Map-based cloning of CPK5.
Supplemental Figure 5. The cpk5 alleles suppress cell death and resistance to G. cichoracearum and PR1 accumulation in exo70B1-3.
Supplemental Figure 6. Complementation of the cpk5-2 mutation.
Supplemental Figure 7. cpk5-2 does not suppress edr2-, pmr4-, or acd5-mediated resistance to G. cichoracearum.
Supplemental Figure 8. CPK5 transcript level was not significantly changed in exo70B1-3.
Supplemental Figure 9. The K126 and D221 residues of CPK5 are critical for the interaction between CPK5-VK and TN2.
Supplemental Figure 10. CPK5-VK interacts with the TN2-NBS domain, but not the TIR domain.
Supplemental Figure 11. CPK4-VK and CPK6-VK do not interact with TN2.
Supplemental Figure 12. CPK5-OE transgenic plants display cell death and resistance to G. cichoracearum.
Supplemental Figure 13. TN2 transcript accumulates in the CPK5 overexpression line.
Supplemental Figure 14. CPK5 and CPK5-G2A accumulate after ectopic expression in exo70B1-3 but not in tn2-1.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. ANOVA tables for statistical analyses.
Supplementary Material
Acknowledgments
We thank Jeff Dangl for reading the manuscript and the ABRC for providing T-DNA insertion lines. The work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11020100), National Science Fund for Distinguished Young Scholars of China (31525019), and the Ministry of Science and Technology of China (2015CB910200 and 2014DFA31540) to D.T., and by SFB973 (project C2) of the German Science Foundation DFG to T.R.
AUTHOR CONTRIBUTIONS
D.T. and N.L. conceived and initiated the research. D.T., N.L., and T.R. designed the experiments. N.L., K.H., W.W., and T.Z. performed the experiments. N.L., K.H., W.W., T.Z., T.R., and D.T. analyzed the data. D.T. and N.L. wrote the article. D.T. and T.R. contributed to significant discussion and revision.
Glossary
- ROS
reactive oxygen species
- PAMP
pathogen-associated molecular pattern
- SA
salicylic acid
- Y2H
yeast two-hybrid
- dpi
days postinfection
- BiFC
bimolecular fluorescence complementation
- ETI
effector-triggered immunity
References
- Adam L., Somerville S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9: 341–356. [DOI] [PubMed] [Google Scholar]
- Bonardi V., Tang S., Stallmann A., Roberts M., Cherkis K., Dangl J.L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. USA 108: 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Sheen J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.-H., Sheen J. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Kloek A.P., Boch J., Katagiri F., Kunkel B.N. (2000). The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol. Plant Microbe Interact. 13: 1312–1321. [DOI] [PubMed] [Google Scholar]
- Cheng S.-H., Willmann M.R., Chen H.-C., Sheen J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129: 469–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Horvath D.M., Staskawicz B.J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Du Y., Mpina M.H., Birch P.R., Bouwmeester K., Govers F. (2015). Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169: 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.P., Schulze W.X., Romeis T. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C.A., Innes R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki K., Abe Y., Ito A., Saitoh H., Yoshida K., Kanzaki H., Kanzaki E., Utsushi H., Yamashita T., Kamoun S., Terauchi R. (2015). Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 83: 875–887. [DOI] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Zook M., Mert F., Kagan I., Rogers E.E., Crute I.R., Holub E.B., Hammerschmidt R., Ausubel F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Su N., Zheng J., Huai J., Wu G., Zhao J., He J., Tang D., Yang S., Wang G. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60: 757–770. [DOI] [PubMed] [Google Scholar]
- Greenberg J.T., Silverman F.P., Liang H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Guo W. (2009). The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 21: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kudla J., Batistic O., Hashimoto K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich I., Pečenková T., Sekereš J., Smetana O., Fendrych M., Foissner I., Höftberger M., Zárský V. (2013). Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14: 1155–1165. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Y., Clarke J.D., Li Y., Dong X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339. [DOI] [PubMed] [Google Scholar]
- Lu S.X., Hrabak E.M. (2013). The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol. Biol. 82: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M., Katagiri F., Yu G.L., Ausubel F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Monaghan J., Matschi S., Shorinola O., Rovenich H., Matei A., Segonzac C., Malinovsky F.G., Rathjen J.P., MacLean D., Romeis T., Zipfel C. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615. [DOI] [PubMed] [Google Scholar]
- Nandety R.S., Caplan J.L., Cavanaugh K., Perroud B., Wroblewski T., Michelmore R.W., Meyers B.C. (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 162: 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6: e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecenková T., Hála M., Kulich I., Kocourková D., Drdová E., Fendrych M., Toupalová H., Zársky V. (2011). The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 62: 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S.N., Ali G.S., Celesnik H., Day I.S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., Tang S., Stallmann A., Dangl J.L., Bonardi V. (2013). Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 9: e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Herde M. (2014). From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 20: 1–10. [DOI] [PubMed] [Google Scholar]
- Romeis T., Piedras P., Jones J.D. (2000). Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12: 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Ludwig A.A., Martin R., Jones J.D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20: 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz P., Herde M., Romeis T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 163: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Ichimura K., Pecenkova T., Reuter P., Žársky V., McDowell J.M., Shirasu K., Trujillo M. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Westphal L., Rosahl S., McDowell J.M., Trujillo M. (2013). The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal. Behav. 8: e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2005). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W., Somerville S.C. (2013). Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. USA 110: 12492–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Somerville S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97: 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wen Y., Berkey R., Xiao S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21: 2898–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68: 74–87. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Wang Z., Zhang X., Yang S. (2013). A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J. 75: 553–565. [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Liu S., Zhao Y., Wang W., Kong Z., Tang D. (2015). ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. Plant Cell 27: 857–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zárský V., Kulich I., Fendrych M., Pečenková T. (2013). Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 16: 726–733. [DOI] [PubMed] [Google Scholar]
- Zbierzak A.M., Porfirova S., Griebel T., Melzer M., Parker J.E., Dörmann P. (2013). A TIR-NBS protein encoded by Arabidopsis Chilling Sensitive 1 (CHS1) limits chloroplast damage and cell death at low temperature. Plant J. 75: 539–552. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Liu J., Ding Y., Wang S., Zhang X., Liu Y., Yang S. (2017). Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3. New Phytol. 213: 1330–1345. [DOI] [PubMed] [Google Scholar]
- Zhao T., Rui L., Li J., Nishimura M.T., Vogel J.P., Liu N., Liu S., Zhao Y., Dangl J.L., Tang D. (2015). A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 11: e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.