Abstract
Cryptochromes are evolutionarily related to the light-dependent DNA repair enzyme photolyase, serving as major regulators of circadian rhythms in insects and vertebrate animals. There are two types of cryptochromes in the animal kingdom: Drosophila-like CRYs that act as non-visual photopigments linking circadian rhythms to the environmental light/dark cycle, and vertebrate-like CRYs that do not appear to sense light directly, but control the generation of circadian rhythms by acting as transcriptional repressors. Some animals have both types of CRYs, while others possess only one. Cryptochromes have two domains, the photolyase homology region (PHR) and an extended, intrinsically disordered C-terminus. While all animal CRYs share a high degree of sequence and structural homology in their PHR domains, the C-termini are divergent in both length and sequence identity. Recently, cryptochrome function has been shown to extend beyond its pivotal role in circadian clocks, participating in regulation of the DNA damage response, cancer progression, and glucocorticoid signaling, as well as being implicated as possible magnetoreceptors. In this review, we provide a historical perspective on the discovery of animal cryptochromes, examine similarities and differences of the two types of animal cryptochromes, and explore some of the divergent roles for this class of proteins.
Graphical Abstract
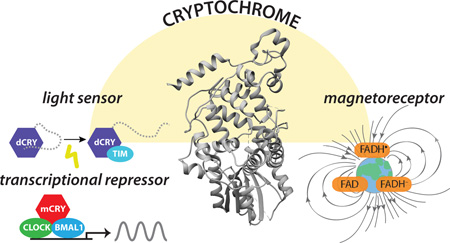
Structurally similar to photolyase, cryptochromes (PDB: 4K0R) regulate circadian rhythms in animals. Left, light absorption leads to a conformational change in Drosophila CRY (dCRY) that regulates its interaction with TIM forcircadian photoentrainment. Mouse cryptochromes (mCRY) repress CLOCK:BMAL1 independently of light to control the circadian transcription-translation feedback loop. Right, CRYs have also been proposed to act as light-dependent magnetoreceptors.
INTRODUCTION
Earth receives almost all of its energy from the Sun’s radiation, which provides the driving force for the photosynthetic production of oxygen, patterns of climate and weather, and the coordination of our behavior and physiology with its daily light-dark cycle. Sunlight also contains harmful ultraviolet (UV) radiation that can induce mutations in DNA to interfere with genetic stability. One of the most elegant solutions for the repair of UV-induced DNA damage occurs by a process termed photoreactivation, in which UV-induced pyrimidine dimers are repaired enzymatically with blue light (1). Photolyase enzymes specifically recognize cyclobutane pyrimidine dimers or pyrimidine-pyrimidone (6–4) photoproducts, both of which accumulate when DNA is exposed to sunlight (2, 3). After recognition of damaged DNA, photolyase captures a photon of blue light with one of its two light-harvesting cofactors, a secondary antenna chromophore (either methenyltetrahydrofolate (MTHF) or the deazaflavin, 8-HDF) or the catalytic chromophore flavin adenine dinucleotide (FAD), which undergoes an electron transfer cycle to split the pyrimidine dimer (4).
Not all organisms possess the capability to photoreactivate; those that don’t have photolyase use mechanisms such as nucleotide excision repair to replace damaged DNA (5). The discovery of photolyase homologs that lack DNA repair activity, known as cryptochromes, were first identified over twenty years ago in plants, followed by insects, vertebrates, and are now found across the domains of life (6). By definition, cryptochromes lack DNA repair activity, but they capitalize on their evolutionary relationship to photolyase to regulate other aspects of our intimate biological relationship with the Sun. Although insect and vertebrate-like cryptochromes have different functions, both remain primarily associated with biological clocks that serve as an interface between host physiology, behavior, and the daily light-dark cycle. However, new roles are being revealed for animal cryptochromes outside circadian clocks, including their involvement in cancer biology and the response to DNA damage, metabolic signaling, and possibly even in magnetoreception. Here, we provide a historical perspective on our understanding of cryptochromes and how our knowledge of photolyase structure and function has informed studies on cryptochrome.
Biological timekeeping by circadian rhythms
Circadian rhythms are nearly-24 hour rhythms of physiology and behavior that are a nearly ubiquitous feature of eukaryotic life. These internal rhythms were first noted by Jean-Jacques de Marain (1678–1771), who observed that the daily opening and closing of the flowers of a Mimosa plant persisted in total darkness. The modern field of circadian rhythms was established by Jürgen Aschoff (1913–1998) and Colin Pittendrigh (1918–1996), who delineated the defining properties of these rhythms: Circadian rhythms are self-sustaining in the absence of external time cues and possess an intrinsic (or free-running) period of approximately 24 hours under constant conditions such as complete darkness. Circadian rhythms are synchronized (or entrained) to the 24-hour solar light-dark cycle and exhibit relatively constant free-running periods with changes in temperature (7).
Under 24-hour light-dark cycles (i.e. 12 hours light and 12 hours dark, or LD 12:12), circadian rhythms demonstrate a period of exactly 24 hours. However, in constant conditions that occur in the absence of environmental cues, these rhythms display their internal free-running periods, which are close to, but usually not exactly, 24 hours. For instance, the human sleep-wake cycle has a free running period of about 24.3 hours, while the mouse has a rest-activity rhythm with a free-running period of about 23.6 hours (8). Light acts as a time cue (or Zeitgeber) to entrain internal rhythms to the exact 24-hour solar day. Light accomplishes this by shifting the phase of free-running rhythms to match the environmental light-dark cycle. Most organisms share a common phase response to light, whereby a short pulse of light during the subjective day (i.e. a time that coincides with daytime based on the free-running clock of an organism in complete darkness) does not shift the clock, but a pulse of light in the early subjective night phase delays the clock, while the same pulse of light given in the late subjective night phase advances the clock. By virtue of being able to shift the phase of internal rhythms with brief pulses of light, circadian clocks provide organisms with the ability to align physiology and behavior to daily environmental cycles for enhanced fitness (9). Remarkably, the spectral character of light that phase shifts the clock is also relatively conserved across a broad range of phylogeny, and is centered in the blue portion of the spectrum.
Circadian rhythms in Drosophila
Colin Pittendrigh was the first to study the circadian rhythms of Drosophila (7). He noted that flies show two separate free-running rhythms, one of eclosion from pupae to adult, and a second in locomotion. Both are entrainable by light. Using Drosophila genetics, Ron Konopka (1947–2015) and Seymour Benzer (1921–2007) performed mutagenesis on flies and identified three mutants with abnormal free-running rhythms (one with a long period, one with a short period, and an arrhythmic mutant) (10). Remarkably, all three mapped to a single gene, dubbed period due its profound effects on the timing, or period, of circadian rhythms (11, 12). Equally remarkably, period mutants affected both eclosion rhythms and locomotor rhythms in the same way. Work over several decades since has demonstrated that the circadian clock mechanism in Drosophila consists of a small number of dedicated clock genes (i.e. period, timeless, clock, cycle) that establish a time-delayed transcription-translation feedback loop controlling circadian expression of a number of genes (13).
Cryptochromes as circadian photoreceptors
The search for a circadian photoreceptor
Drosophila circadian rhythms are entrainable to light-dark cycles. It would be logical to assume that the eyes and visual system mediate this effect. However, studies with eyeless mutants such as norpA and glass demonstrated that blind flies maintained behavioral entrainment to light-dark cycles (14). Further, severe vitamin A depletion that rendered visual opsins non-functional did not block light-induced phase shifting (15). In addition, explanted tissues from Drosophila expressing green fluorescent protein under the control of the period promoter/enhancer elements showed direct photoentrainment in culture (16) despite the absence of opsin expression in these tissues. Collectively, these lines of evidence suggested the presence of a cell autonomous circadian photoreceptor.
In 1998, Stanewsky et al. conducted a forward mutagenesis screen in Drosophila designed to identify additional clock genes (17). Using a luciferase reporter driven by the period gene, this group found a recessive mutation that blunted circadian gene oscillations. The mutation mapped to the Drosophila cryptochrome (cry) locus, and was named cryb (short for ‘crybaby’). Previously, light was shown to reduce expression of the TIMELESS (TIM) protein, one of the core clock genes in Drosophila; cryb mutant flies no longer exhibited the light-dependent reduction of TIM protein levels. Based on its similarity to photolyase and plant cryptochromes that act as blue light photoreceptors, cry was suspected of encoding a circadian receptor in flies. In addition, cryb flies did not phase shift their rhythms to short, bright pulses of light and overexpression of CRY protein resulted in flies that were super-sensitive to phase shifting by light. Of note, cryb flies still demonstrated free-running behavioral rhythms, and showed some behavioral entrainment to light. However, this entrainment was lost when cryb mutant flies were compounded with the glass mutation (18), which removes all known photoreceptors in the fly eye. Taken together, these results strongly suggested that Drosophila CRY functions as the primary blue light photopigment that mediates circadian photoentrainment.
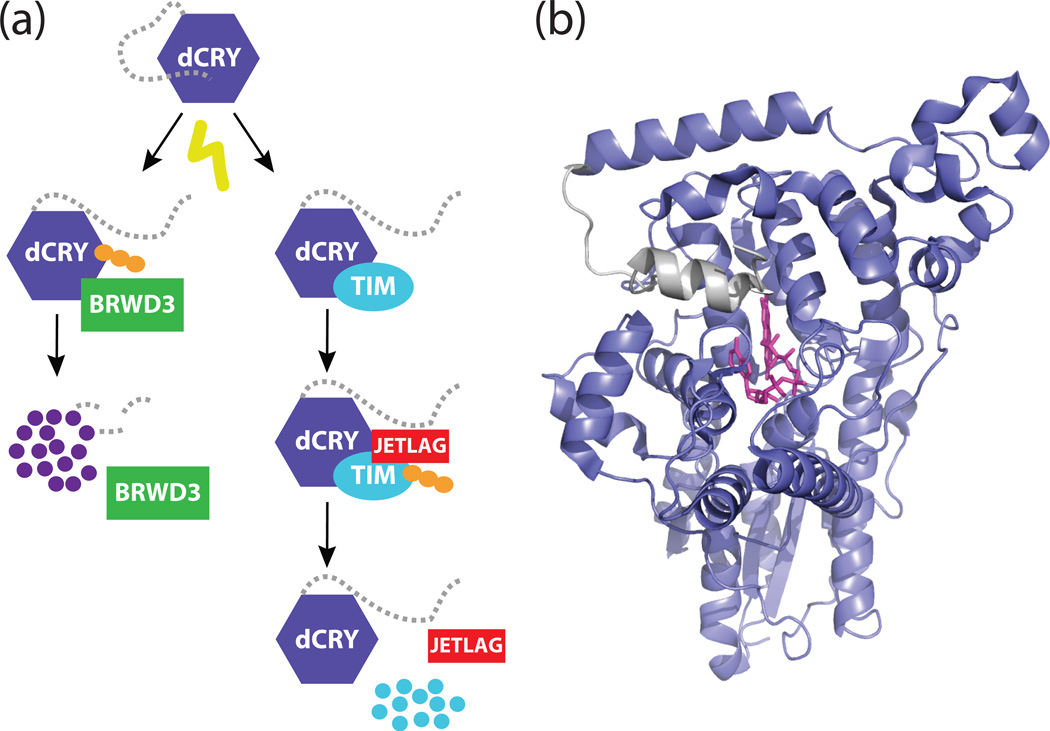
To test this, Emery et al. established an in vitro system to study cryptochrome’s photoreceptive function (19). In cultured Drosophila S2 cells expressing CRY and TIM, exposure to light resulted in the rapid degradation of TIM, followed by the substantially slower degradation of CRY itself. These results suggested that light might induce a structural change in CRY that allows it to interact with TIM, similar to light-dependent changes in conformation observed with Arabidopsis cryptochromes (20, 21). The discovery of two novel alleles of cryptochrome (crym and cryΔ) missing the short, C-terminal extension from the photolyase homology region (PHR) supported this model (19, 22). Wild-type CRY bound TIM only after exposure to light (23, 24), while deletion of the CRY C-terminal extension led to constitutive interactions with TIM that triggered its degradation (19, 25). The E3 ubiquitin ligase encoded by Jetlag was subsequently found to be the protein that recognized TIM after interaction with CRY, marking it for degradation to induce phase shifts in the circadian clock (Fig. 1a) (26–28).
Figure 1. A model for the dCRY C-terminal tail in the Drosophila clock.
(a) Schematic model for the light-responsive degradation of dCRY and TIM. Light stimulates a structural change in dCRY that exposes the binding interface for TIM and JETLAG. This interaction results in the polyubiquitination (depicted as orange circles) and proteasomal degradation of TIM. The same light-induced conformational change in dCRY also renders it sensitive to polyubiquitination by BRWD3. (b) Structure of full-length dCRY (PDB: 4GU5) in the dark-adapted state. A hydrophobic motif in the C-terminal extension (gray) docks onto the PHR domain (purple) in close proximity to the flavin (pink).
Following on these in vitro studies, Chavez et al. developed a cryptochrome-luciferase (CRY-luc) fusion protein to characterize light-mediated CRY degradation in S2 cells (29). They found that a one-hour exposure to light resulted in greater than 80% degradation of the CRY-luc fusion protein. Using this assay, Chavez et al. measured the action spectrum for CRY degradation, demonstrating that maximum degradation of CRY occurred when S2 cells were supplied with near UV light at approximately 380 nm. Light-dependent CRY degradation was blocked by the proteasomal inhibitor MG132, strongly suggesting that CRY, like TIM, is degraded by the proteasome after ubiquitination. However, JETLAG does not regulate CRYs degradation in response to light (30).
In subsequent work, Ozturk et al. (30) knocked down every known F-box E3 ubiquitin ligase present in S2 cells to identify the protein(s) that recognize light-activated cryptochrome in the fly. One E3 ligase, BRWD3 encoded by the ramshackle gene, resulted in loss of light-dependent CRY degradation in S2 cells. CRY and BRWD3 were shown to interact in a light-dependent manner by a yeast two-hybrid assay, and purified CRY and BRWD3 proteins formed complex only after exposure to light to result in the ubiquitination of CRY in vitro. Therefore, Drosophila CRY undergoes a light-dependent change in conformation that regulates its ability to interact with TIM and BRWD3, controlling the stability of both TIM and CRY itself.
Photochemistry and phototransduction of Drosophila CRY
The photocycle of Drosophila cryptochrome was initially assumed to be similar to that of Arabidopsis cryptochromes (31–33). Like their ancestral photolyase homolog, cryptochromes can bind non-covalently to a flavin adenine dinucleotide (FAD) chromophore to absorb blue light. When Drosophila CRY protein was first purified from S2 cells, the catalytic chromophore FAD was found in an oxidized state (FADOX) that has been generally assumed to represent the ground state of cryptochromes. Following exposure to light in vitro, FAD was reduced to the anionic semiquinone form (FAD•−) (34, 35). Hoang et al. demonstrated by fluorescence and paramagnetic spin techniques that both Drosophila and human cryptochromes could be photoreduced by light in Sf21 cell culture (36). Structurally, this electron transfer appears to be dependent on a triad of conserved tryptophan residues (Trp-342, -397, and -420) (37–39). However, mutagenesis of the ‘Trp triad’ as well as a fourth tryptophan (Trp-536) did not affect light-induced proteolysis of CRY or TIM in S2 cells (40, 34). Crane and colleagues then showed that chemical reduction of the dCRY flavin is sufficient to trigger conformational changes in the protein that are similar to those induced by light (41). Therefore, reduction of the flavin by either light excitation or chemical reduction thus appears to be sufficient to cause conformational changes that are important for CRY activation.
Similar to those observed with Arabidopsis CRYs, light-dependent changes in the structure of Drosophila CRY are easily assayed by limited trypsin proteolysis in vitro (27, 20). It was shown that absorption of light efficiently drove changes in both oxidized and reduced CRY in vitro. The structural basis for conformational changes driven by light excitation or chemical reduction of the flavin in Drosophila CRY are suggested by crystal structures of its dark-adapted state, which depict how a hydrophobic motif (“FFW”) in the C-terminal extension docks onto the PHR domain in close proximity to the flavin (Fig. 1b) (37, 38). While some of the mechanistic details of CRY signaling still remain to be determined (42), it appears that light-induced changes in Drosophila CRY structure are relatively long-lasting—on the order of 30 minutes—after even a millisecond pulse of light (43). Further studies on the photocycle of Drosophila CRY and its mechanisms of phototransduction will help inform studies of its biological role in circadian entrainment.
Drosophila CRY as a magnetoreceptor
Remarkably, in addition to light sensing, it appears that cryptochromes may also mediate magnetoreception in Drosophila. In a binary-choice behavioral assay for magnetosensitivity, wild-type flies showed significant naïve and trained responses to a magnetic field under full-spectrum light, but could not respond to a magnetic field when short wavelengths were blocked (44). Moreover, cryb flies did not exhibit responses to a magnetic field under full-spectrum light, nor did the cryΔ mutant lacking the C-terminal extension (22). Theoretical studies have suggested models explaining how light absorption could induce a paramagnetic triplet excited state in cryptochromes that allows for light-dependent magnetoreception, but it remains to be demonstrated that this is essential for magnetoreception (45). Moreover, CRY was also recently shown to interact with Drosophila CG8198 (named MagR for Magnetic Receptor) to yield a protein complex that exhibits light-dependent responses to an induced magnetic field in vitro (46). Genetic studies substituting Drosophila CRY with cryptochromes from vertebrates suggest that the capability to sense both light and magnetic field may be conserved in animals, while others have provided evidence of light-dependent conformational changes in some vertebrate cryptochromes that are expressed in the retina (47, 43, 48).
Circadian photoreception in vertebrates
In the late 1990s, two cryptochrome homologs (Cry1 and Cry2) were identified in mice (49, 50). Initially, a role for mammalian CRYs in circadian photoreception was suggested by photoentrainment experiments that mirrored early work done in flies. Mice blind from degeneration of the outer retina due to the rd1 mutation continued to show circadian entrainment to light, even to light as dim as 1–2 lux (51, 52). Unlike the fly, which utilizes a cell autonomous mechanism for circadian entrainment, mice that lacked eyes (through enucleation) or optic nerves did not exhibit circadian photoentrainment, demonstrating that mammals require retinal phototransduction to entrain to a light/dark cycle (53). Cryptochromes seemed to be viable candidates for non-visual phototransduction based on their expression in retinal ganglion cells (54) and similarity to Drosophila and Arabidopsis cryptochromes. Upon genetic deletion of both mouse cryptochromes, several lines of evidence suggested that the function of vertebrate cryptochromes might be quite different from Drosophila cryptochrome. While flies lacking cryptochrome continued to show behavioral rhythmicity, Cry1−/−;Cry2−/− mice no longer consolidated behavioral rhythms into circadian patterns (55, 56). Subsequent work demonstrated that mammalian cryptochromes had taken over a role similar to TIM in the fly clock as the major binding partner of PER in the negative transcriptional complex of the core clock mechanism (57, 58).
Although the finding that cryptochromes function as essential transcriptional regulators in the mammalian clock mechanism complicated studies of entrainment, it did not formally eliminate the possibility that they might also function as circadian photopigments. Several lines of evidence initially supported this possibility. First, mice with severe vitamin A depletion (due to dietary vitamin A starvation in a retinol binding protein knockout background) continued to show light-dependent activation of gene expression in the suprachiasmatic nucleus of the hypothalamus, the site of the light-responsive ‘master clock’ in mammals (59, 60). Second, while Cry1−/−;Cry2−/− animals continued to show masking-type effects in a light-dark cycle, the Cry1−/−;Cry2−/−;rd1/rd1 triple mutant resulted in substantially reduced rhythmicity and reduced c-fos activation in the suprachiasmatic nucleus in response to light (61).
Melanopsin as a circadian photoreceptor
The discovery of mammalian melanopsin, a rhabdomeric opsin photopigment expressed in a small number of retinal ganglion cells, in 2000 soon led to its rise as the most likely candidate for a dedicated mammalian circadian photoreceptor (62). Although originally discovered in the photosensitive dermal melanocytes of Xenopus (63), melanopsin was soon found to form a network of intrinsically photosensitive retinal ganglion cells in mice (64–67). While knockouts of melanopsin (Opn4) by itself had little effect on circadian entrainment, combining the rd1/rd1 mutation with deletion of melanopsin had a stronger phenotype than seen in mice with the rd1 mutation that also lacked both cryptochromes. Specifically, rd1/rd1;Opn4−/− animals had free-running rhythms that never entrained to external light-dark cycles, and had no pupillary light responses under any light intensities (68), while rd1/rd1;Cry1−/−;Cry2−/− mutants showed pupillary light responses, albeit with markedly decreased sensitivity (69).
It appears that a combination of the unique signaling properties of melanopsin and effects due to the loss of circadian rhythms lead to the misattribution of cryptochrome as a circadian photopigment in mammals. Vitamin A depletion renders mice visually blind, as the ciliary opsins (rod and cone opsins) release their chromophore with each photocycle and are thus highly dependent on new, vitamin-A-derived photopigment for restoring function. Melanopsin, by contrast, is a rhabdomeric opsin that appears to bind its chromophore irreversibly, using a combination of second photon absorption and thermal relaxation to regenerate its chromophore (70). Subsequent work has shown that melanopsin is remarkably resistant to photic bleaching, and can function in the absence of retinal pigment epithelium and other enzymes of the visual photocycle (62, 71). Thus, earlier studies with vitamin A depletion likely did not result in loss of melanopsin function.
How to explain the reduced pupillary light responses in rd1/rd1;Cry1−/−;Cry2−/− animals? Several lines of evidence suggest that Cry-dependent disruption of the retinal circadian clock reduces melanopsin-dependent pupillary light responses. Identical phenotypes are seen with deletion of other core clock genes that also disrupt circadian rhythms, notably in Bmal1−/−;Opn4−/− and Per1−/−; Per2−/−;Opn4−/− mice (72). Taken together, these results confirm that there is a circadian rhythm of retinal function; in the absence of a circadian clock, reduced retinal sensitivity results in decreased photosignaling to the SCN and pupillary light response centers.
Thus, it appears that the role of cryptochromes is significantly different between insects and vertebrates. Whereas CRY is primarily a blue light photoreceptor that entrains circadian rhythmicity in Drosophila and other insects, cryptochromes in mammals and other vertebrates have evolved to become light-independent transcriptional regulators within the core clock mechanism. Such results, however, must be reconciled with the finding that some vertebrate cryptochromes can bind flavin when purified from eukaryotic expression systems (73, 74, 36) and appear capable of substituting for Drosophila CRY in magnetoreception assays in vivo (75). However, if vertebrate cryptochromes retain the ability to signal in response to light, the physiology behind these influences remains to be discovered.
Commonalities and differences in animal cryptochromes
Evolutionary diversity of animal cryptochromes
Phylogenetic analysis of the photolyase/cryptochrome superfamily distributes the group into seven broad subfamilies: cyclobutane pyrimidine dimer (CPD) photolyase classes I–III, (6–4) photolyase, plant cryptochromes, animal cryptochromes and CRY-DASH, which selectively repairs UV-induced damage in single-stranded DNA and RNA (76–78, 6). Animal cryptochromes are more similar to (6–4) photolyase than photolyases that repair cyclobutane pyrimidine dimers. The animal cryptochrome subfamily can be further divided into two groups: Type I cryptochromes, which are light-responsive circadian photoreceptors found only in Drosophila and other insects; and Type II cryptochromes in vertebrates (and some insects) that act as transcriptional repressors. Notably, one feature that appears to distinguish the molecular architecture of Drosophila and vertebrate clocks is the functional role of cryptochromes—does CRY act as a non-visual photopigment for entrainment (Drosophila) or does it act as a transcriptional repressor (vertebrates)? However, even this distinction may not be clear-cut, as some studies show that Drosophila CRY may act as a transcriptional repressor in some tissues (13). Furthermore, some species of insects have both types of animal cryptochromes (monarch butterflies and mosquitos), while others have only a vertebrate-like Type II cryptochrome (honeybees and ants) (79–81). In yet another interesting twist, some vertebrates (chickens and zebrafish) possess yet another type of cryptochrome (Type IV) that is also sensitive to light, although functional roles for these cryptochromes have yet to be firmly established (6, 43, 48).
Cryptochromes share a high degree of structural homology within their PHR domain both to each other and to all photolyases (82, 37, 39). Despite this overarching similarity, variation in the PHR domains of animal cryptochromes may play a role in their profoundly different functions. Ning Zheng and colleagues first showed that mouse CRY2, a representative vertebrate cryptochrome, appears to have decreased affinity for FAD due to the presence of a much shallower FAD-binding pocket, further supporting their light-insensitive roles as transcriptional repressors (74). Although the pockets in vertebrate CRYs may not be dedicated to chromophore binding, the conservation of these pockets might point to new roles for repressor-type CRYs.
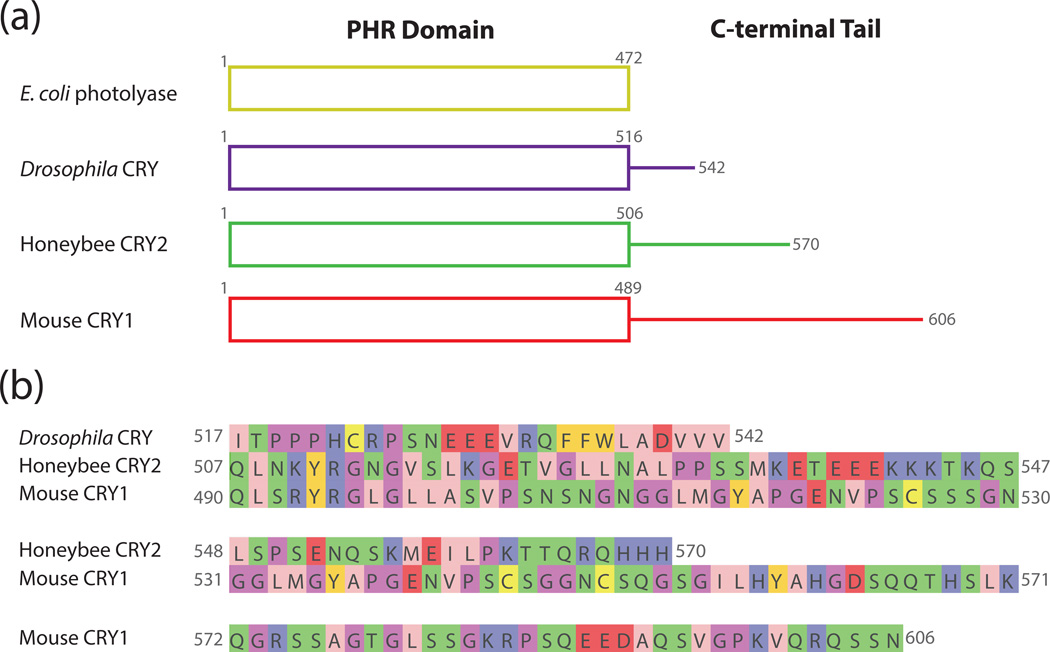
There is also some indication that variation in the sequence composition and length of Type I and Type II cryptochromes may help to explain their diverse functions (Fig. 2a). For example, mouse CRY1 and Drosophila CRY are only 43% identical, whereas honeybee CRY2 and mouse CRY1, both Type II cryptochromes, are 69% identical. A great deal of the sequence variation in animal cryptochromes falls outside of the PHR domain in their extended and intrinsically unstructured C-termini (Fig. 2b). While the role of the Drosophila CRY C-terminus in regulating light-dependent interactions with target proteins is well established (19, 25, 24), the role of C-termini in vertebrate-like cryptochrome function is less resolved. Studies of light-dependent Type IV cryptochromes suggest that their C-termini act similarly to Drosophila cryptochromes, exhibiting changes in conformation and/or interaction with the PHR domain in response to light (48). The C-termini of Type II CRYs are not essential for circadian transcriptional repression, but their deletion led to changes in circadian period and the amplitude of cycling in genetic reconstitution assays (83–85). Several groups have shown that phosphorylation of the C-termini of CRY1 and CRY2 regulates their stability, suggesting one mechanism by which the C-termini could affect circadian rhythms (86, 87). The C-terminus of human CRY2 was shown to bind the PHR domain and adopt a proteolytically stable structure (20); however, no crystal structures of vertebrate cryptochromes thus far have managed to capture the PHR domain bound to its disordered C-terminus, so a complete reckoning of the vertebrate cryptochrome structure still remains to be determined (37, 74).
Figure 2. Comparison of C-terminal tail extensions in representative photolyase and cryptochrome proteins.
(a) Schematic representation of the domains present in E. coli photolyase, Drosophila CRY, honeybee CRY2, and mouse CRY1. The overall structure and organization of the PHR domain remains relatively unchanged between the different proteins, but the C-terminal tails vary in length. (b) Sequences of the unstructured C-terminal extensions of Drosophila CRY, honeybee CRY2, and mouse CRY1. Amino acids are colored according to their physicochemical properties using the Jalview Zappo coloring scheme (138): pink, aliphatic/hydrophobic; gold, aromatic; purple, positive; red, negative; green, hydrophilic; light purple, conformationally special; yellow, cysteine.
Cryptochromes as transcriptional repressors
Circadian rhythms in vertebrates
Circadian rhythms in vertebrates and most insects are driven by a time-delayed transcription-translation feedback loop that shares similarity with the Drosophila clock mechanism through moderate conservation of most of the core clock genes. In vertebrates, the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) domain-containing transcription factor complex CLOCK:BMAL1 binds to conserved E-box sequences in the promoters of clock-controlled genes to activate their transcription on a daily basis. Of the approximately 40% of the genome that is rhythmically driven by the circadian clock (88), a small subset of these target genes (Per1, Per2, Cry1, Cry2) make up the core negative component of this feedback loop. Additionally, the nuclear receptors ROR and REV-ERB make up an additional feedback loop that interfaces with the primary loop by controlling the expression of a subset of clock genes, including Bmal1 (89). Large, heteromultimeric PER:CRY complexes slowly enter the nucleus, where they repress CLOCK:BMAL1 activity (90, 91). Recent data demonstrate the presence of at least two distinct phases of repression: an early repressive PER:CRY complex and a late complex where CRY1 directly inhibits CLOCK:BMAL1-driven transcription independently of PER (92–94). Ultimately, the regulated degradation of PER and CRY proteins alleviates repression of CLOCK:BMAL1 activity to allow this cyclical process to begin again. Precise control of protein levels, cellular location and specific complex formation all play an important role in allowing this cycle to occur with ~24 hour periodicity.
Control of cryptochrome expression
As mentioned above, proper regulation of CRY abundance and subcellular localization is critical for accurate clock timing. At the transcriptional level, specific regulatory mechanisms result in the postponement of Cry1 transcription in relation to Cry2; this phase delay in Cry1 expression is required for 24 hour cycling (95). Post-translational regulation of CRY stability also helps to create necessary delays in abundance that contribute to the 24-hour period of the clock. In the nucleus, CRY1 and CRY2 stability is regulated by the Skp1-Cul1-F-box protein FBXL3, which ubiquitinates cryptochromes to induce proteasomal degradation (96–98). The related protein FBXL21 was shown to ubiquitinate CRY1 and CRY2 in the cytoplasm (99). Interestingly, FBXL21 activity was found to antagonize the more efficient activity of the nuclear E3 ligase FBXL3 to fine-tune CRY protein levels. The small molecule KL001 was shown to bind directly to CRYs and prevent this ubiquitin-dependent degradation, demonstrating for the first time that CRYs could be valuable targets for clock-based therapeutics. KL001 binds in the FAD binding pocket to compete with FBXL3 binding, resulting in the stabilization of CRY protein levels and lengthening of the circadian period (100, 101). Clearly, the ability to fine-tune CRY protein levels is important for an accurate and robust cellular clock, and also represents a powerful example of a pharmacological strategy to manipulate clock function.
Mechanism of transcriptional repression by cryptochrome
Over the last decade, studies have begun to provide some insight into how cryptochromes inhibit CLOCK:BMAL1-driven transcription. Genetic screens performed concurrently by several groups led to the identification of mutations on both CLOCK and BMAL1 that disrupted the ability of CRY to repress transcription (102, 103); a similar study by the Green lab also identified critical residues on cryptochromes (104). Further investigations into cryptochrome function demonstrated distinct functions of CRY1 and CRY2 that exist at the biochemical level, showing that only CRY1 is capable of generating cell-autonomous circadian rhythms in fibroblasts and in tissues outside the SCN, while CRY2 cannot (85, 105). The molecular basis by which a few modest changes in amino acid identity between mammalian CRY1 and CRY2 confers the ability to cycle is not currently understood.
In the late phase of repression, cryptochromes interact directly with CLOCK:BMAL1 on DNA. Aziz Sancar’s group first showed this with purified clock proteins, demonstrating their ability to interact both on and off E-box elements in DNA in vitro (94). A genetic screen in mammalian cells identified that mutations in the PAS-B domain of CLOCK and C-terminal transactivation domain of BMAL1 are important for CRY repression (102). Subsequent biophysical studies showed that transcriptional repression by CRY1 occurs through competition with transcriptional coactivators for binding the unstructured C-terminal transactivation domain of BMAL1; notably, mutations that altered the balance of repressor and activator binding on BMAL1 were critical for establishing proper period length (106). In this way, cryptochromes achieve repression of CLOCK:BMAL1 by sequestering the transactivation domain that is needed for activity (103). Cryptochromes utilize multivalent interactions with both CLOCK and BMAL1 to make stable complexes that allow them to repress activity efficiently when expressed to near-stoichiometric levels with the transcription factor (107). Notably, introducing only a few point mutations on CLOCK PAS-B as well as the BMAL1 transactivation domain eliminated the ability of CRY1 to repress CLOCK:BMAL1 activity (106).
Transcriptional regulation by CRYs
While our understanding of circadian regulation of transcription has steadily grown over the last fifteen years, until recently, it has not been clear whether the activity of circadian transcriptional regulators was strictly limited to regulation of CLOCK:BMAL1, or whether their influence reaches beyond this complex. Koike et al. addressed this with a large-scale chromatin immunoprecipitation sequencing (ChIP-seq) study that examined patterns of recruitment for the six core clock proteins genome-wide in mouse liver over the course of an entire day (92). This study cataloged daily patterns in recognition and circadian transcriptional control, not only for the core clock proteins, but also for Pol II and other epigenetic markers, to clearly demonstrate genome-wide control of the transcriptional landscape by the clock. In addition to providing valuable information about the phase distribution of DNA binding by circadian regulators, this study also identified a large number of sites that are uniquely bound by each of the clock proteins analyzed. For example, CRY1 was found at over 16,000 sites throughout the genome, nearly one-third of which were unique to CRY1 (amongst all the clock proteins), and CRY2 was found at over 10,000 sites (one-fifth of which were unique to CRY2) (92). These data strongly suggest that cryptochromes also work outside the core clock transcription-translation loop to regulate transcription throughout the genome. It will be exciting to see how cryptochromes provide temporal regulation to pathways traditionally considered outside the purview of clock-mediated transcription.
New roles for cryptochromes
Communication of temporal information by cryptochromes
While it is well established that cryptochromes are essential for circadian timekeeping, the involvement of CRYs in numerous additional signaling pathways is just beginning to be elucidated (108, 55). As core clock genes, the expression of cryptochromes follows a rhythmic, circadian pattern of accumulation in nearly every tissue. This universal and time-dependent accumulation of CRY has the potential to impart circadian control on the regulation of downstream pathways throughout the body. In this way, CRY proteins act as a type of time-telling second messenger between the core clock and other cellular processes in which they participate. Many of the signaling pathways that CRYs are known to regulate aside from CLOCK:BMAL1 play key roles in the maintenance of cellular homeostasis such as metabolism, inflammation and DNA damage (109–112). These pathways help to maintain cellular and genomic integrity by sensing the current status of the cell (i.e. metabolic, genotoxic stress) and responding to maintain homeostasis (Fig. 3). In some interesting cases, the stability of CRY proteins is conversely regulated by metabolic and DNA damage signals, helping to form feedback loops that mediate crosstalk between systems (110, 113). Eliminating cryptochrome-mediated crosstalk through knockdown or genetic ablation of CRYs has been shown in some cases to have deleterious effects (114). However, the mechanisms by which cryptochromes directly regulate pathways outside of the clock are often not fully understood, especially in cancer (115). Here, we discuss a few known regulatory roles of cryptochromes outside of the regulation of CLOCK:BMAL1 activity.
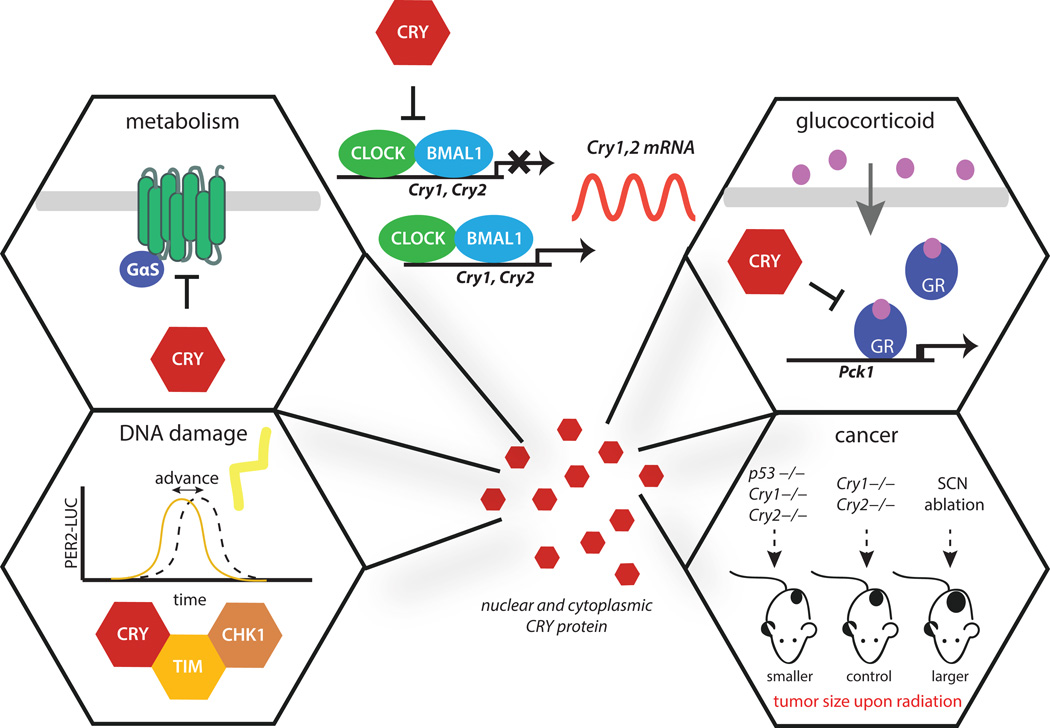
Figure 3. Roles of CRY outside of CLOCK:BMAL1 regulation.
In mammals, CRYs negatively regulate CLOCK:BMAL1 activity to generate a ~24 hour clock that regulates ~40% of the genome (88). CRY is also reported to regulate GPCR signaling and downstream metabolism through interaction with the Gsα subunit to block glucagon-stimulated increases in intracellular cAMP (top left). CRY negatively regulates the glucocorticoid receptor to maintain glucose homeostasis, partly through regulation of Pck1 expression (top right). Interaction of CRY with components of the ATR-mediated DNA damage checkpoint control phase shifting of the clock in response to DNA damage (bottom left). While ablation of the SCN increases tumor formation in mouse models, deletion of cryptochromes extends lifespan after ionizing radiation in a p53 null background (bottom right).
Cryptochromes in metabolism
Epidemiological studies of shift workers show that disruption of circadian rhythms is correlated with increased incidence of metabolic disturbances (116). In rodents, disruption of the molecular circadian clock led to increased insulin resistance and obesity, emphasizing the intricate link between the circadian clock and metabolism (117). Increasing evidence shows that cryptochromes play a direct role in glucose homeostasis through mechanisms that are often independent of CLOCK:BMAL1-regulated transcription. Both CRY1 and CRY2 interact with the glucocorticoid receptor (GR) to repress glucocorticoid-stimulated changes in transcription (Fig. 3) (110). The importance of glucocorticoid signaling to synchronize circadian clocks in peripheral tissues has long been recognized, and recent studies have revealed that CRYs rhythmically antagonize this pathway (118). Interestingly, CRYs modulate only a subset of GR transcriptional targets; the basis of this selectivity is not yet understood. One of the genes negatively regulated by the GR-CRY1 interaction is the rate-limiting gluconeogenic enzyme, phosphoenolpyruvate carboxykinase 1 (Pck1). By directly controlling the expression of this essential regulatory enzyme, CRYs globally regulate metabolism in response to circulating factors and presumably function to limit glucocorticoid-induced hyperglycemia. Other studies have also identified that cryptochromes interact with the heterotrimeric G protein subunit Gsα to modulate the cAMP pathway and CREB activity downstream of G-protein coupled receptors during fasting (Fig. 3) (114). Consistent with these collective findings, glucose homeostasis is severely disrupted in Cry-deficient mice, further highlighting the importance of cryptochromes in metabolic disease.
DNA damage response
As mentioned previously in this review, CRYs share evolutionary conservation and structural similarity to the DNA damage repair enzyme photolyase. By definition, cryptochromes do not directly repair DNA lesions like photolyase, but they do interface with pathways that modulate the cellular responses to DNA damage (119, 120). In mammals, CRY1 modulates the ATR-mediated DNA damage checkpoint by interacting with the cell cycle protein TIMELESS (TIM) (121). There is a substantial degree of sequence homology between mammalian TIM and its counterpart in Drosophila; however, they appear to have evolved different functions in these parallel, yet distinct, mechanisms of timekeeping. Drosophila contains two Tim paralogs, Tim1 (Timeless) and Tim2 (Timeout). While Drosophila TIMELESS is a core clock component that works with PER and CRY to control clock timing in flies (122), TIMEOUT acts outside of the central clock to regulate light entrainment of the adult circadian clock and is essential for DNA metabolism and chromosome integrity (123). Mammalian TIMELESS is more similar to Drosophila TIMEOUT. Consistent with this, ablation of TIM in mammals resulted in embryonic lethality and thus, it has been difficult to study behavioral rhythmicity and classify it as an essential clock gene (124). To date, mammalian TIM is not considered a central clock component. Instead, TIM mediates DNA damage signaling in the ATR-Chk1 pathway to control cell cycle checkpoints (125–127). In mammals, cryptochromes maintain their ability to interact with TIM, where it competes with ATR-Chk1 for a binding site on the N-terminus of TIM (109, 128). Knockdown of TIM attenuates the canonical phase advance of the circadian system upon DNA damage insults. Mechanistic studies show that TIM interacts with the CC helix (α22) of CRY1 (109). This helical region of CRY1 is a hotspot for regulation by other proteins, including PER2, FBXL3 and BMAL1 (129, 130, 74, 106). Indeed, Tamanini and colleagues found that co-expression of PER2 abolishes formation of a TIM:CRY1 complex, presumably through competition at the CC helix (109). This competitive mechanism for interactions with CRY1 between TIM and other clock proteins that regulate CRY1 stability could represent one manner in which circadian phase is altered in response to genotoxic stress.
Cryptochromes in cancer
Both epidemiological and animal studies show that disruption of circadian rhythms through environmental stimuli (light at night or shift work) or genetic means can lead to an increase incidence of cancer; however, this is confounded by genetic deletion of various clock components and the subsequent array of cancer phenotypes (131, 132). Circadian disruption appears to play a role in the deregulation of cellular homeostasis and cell cycle control, but the mechanisms by which this occur are still being identified (133). Considering that cryptochromes are necessary for circadian timing and regulate vital metabolic processes as well as the UV-induced DNA damage response, deletion of cryptochromes was expected to increase the risk of developing cancer. Furthermore, Cry1−/−;Cry2−/− mice show elevated Wee1 levels in the liver and impaired regeneration, linking cryptochromes and the clock to cell cycle control (134). Surprisingly, in vivo studies on Cry1−/−;Cry2−/− mice performed by Sancar and colleagues found that these knockout mice do not show an increase in cancer rate compared to wild-type mice, even after exposure to ionizing radiation (135). Further pursuit of these confounding results found that CRY proteins are actually protective using a p53 mutant background that are more prone to cancer. Deletion of cryptochromes extends the median lifespan of p53−/−;Cry1−/−;Cry2−/− mice by 1.5-fold compared to p53−/− mice (136). Mechanistic studies in these triple knockout fibroblasts demonstrated that they were more susceptible than p53−/− knockout cells to UV-induced apoptosis, implicating cryptochromes in the transmission of p53-independent apoptotic signals in response to DNA damage (136). Furthermore, downregulation of CRYs was found to modulate levels of inflammatory cytokines and the NF-κβ-stimulated transcriptional response that sensitizes cells to apoptosis, linking CRYs to the inflammatory response (111). These studies revealed that reprogrammed regulatory networks involving CRYs in cancer that provide potential for tailored chrono- and chemotherapeutic approaches in p53 null tumors.
While deletion of both cryptochromes is required to render mice arrhythmic due to intercellular coupling the SCN, differences in their clock phenotypes has long suggested that Cry1 and Cry2 likely have different functions. This was demonstrated in recent study that CRY1 and CRY2 have divergent functions in response to DNA damage (113). Lamia and co-workers showed that CRY1 was stabilized by the ubiquitin-specific protease HAUSP following DNA damage insults, while CRY2 was simultaneously destabilized through preferential interaction with its ubiquitin ligase, FBXL3. Accordingly, Cry1−/− cells exhibited an increased response to genotoxic stress, while Cry2−/− cells exhibited a decreased response. These opposing phenotypes suggest that future mechanistic studies are needed to examine independent roles that CRY1 and CRY2 may play, especially as they relate to cancer. Understanding the diverse mechanisms by which cryptochromes work, both inside and outside of the circadian clock, will strengthen our understanding of connection between circadian disruption and disease.
SUMMARY AND PERSPECTIVES
The very name of cryptochromes appropriately hints at our cryptic understanding of their many biological functions (137). Since their discovery over twenty years ago in plants, a large part of our understanding of cryptochrome function has been drawn from our knowledge of photolyase structure and function (4). However, it is clear that cryptochromes are a family of functionally diverse proteins in their own right. Some cryptochromes act as photopigments to control circadian photoentrainment and possibly even magnetoreception, while others have apparently lost the ability to sense light and have been co-opted as transcriptional regulators and participants in intracellular signaling cascades that control circadian timing, metabolism, and cellular responses to DNA damage. As more insight is brought to light about cryptochrome structure and function, we hope to fully realize their far-reaching influence over many aspects of animal behavior and physiology.
Acknowledgments
We would like to thank Aziz Sancar for being a valued collaborator, mentor and friend. This work has been supported in part by funds from NIH GM107069 (C.L.P.) and an unrestricted grant from Research to Prevent Blindness (R.N.V.G.). A.K.M was supported by NIH fellowship F31 CA189660.
Footnotes
This article is part of the Special Issue highlighting Dr. Aziz Sancar's outstanding contributions to various aspects of the repair of DNA photodamage in honor of his recent Nobel Prize in Chemistry.
REFERENCES
- 1.Sancar GB. Enzymatic photoreactivation: 50 years and counting. Mutat Res. 2000;451:25–37. doi: 10.1016/s0027-5107(00)00038-5. [DOI] [PubMed] [Google Scholar]
- 2.Brash DE, Franklin WA, Sancar GB, Sancar A, Haseltine WA. Escherichia coli DNA photolyase reverses cyclobutane pyrimidine dimers but not pyrimidine-pyrimidone (6-4) photoproducts. J Biol Chem. 1985;260:11438–11441. [PubMed] [Google Scholar]
- 3.Todo T, Takemori H, Ryo H, Ihara M, Matsunaga T, Nikaido O, Sato K, Nomura T. A new photoreactivating enzyme that specifically repairs ultraviolet light-induced (6-4)photoproducts. Nature. 1993;361:371–374. doi: 10.1038/361371a0. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 5.Sancar A, Reardon JT. Nucleotide excision repair in E. coli and man. Adv Protein Chem. 2004;69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]
- 6.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 7.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 8.Czeisler CA, Duffy JF, Shanahan TL, Brown EN, Mitchell JF, Rimmer DW, Ronda JM, Silva EJ, Allan JS, Emens JS, Dijk DJ, Kronauer RE. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science. 1999;284:2177–2181. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 9.Emerson KJ, Bradshaw WE, Holzapfel CM. Concordance of the circadian clock with the environment is necessary to maximize fitness in natural populations. Evolution. 2008;62:979–983. doi: 10.1111/j.1558-5646.2008.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 12.Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M, Hall JC. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984;39:369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]
- 13.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Advances in genetics. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helfrich-Forster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerman WF, Goldsmith TH. Photosensitivity of the circadian rhythm and of visual receptors in carotenoid-depleted Drosophila. Science. 1971;171:1167–1169. doi: 10.1126/science.171.3976.1167. [DOI] [PubMed] [Google Scholar]
- 16.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 17.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 18.Mealey-Ferrara ML, Montalvo AG, Hall JC. Effects of combining a cryptochrome mutation with other visual-system variants on entrainment of locomotor and adult-emergence rhythms in Drosophila. J Neurogenet. 2003;17:171–221. [PubMed] [Google Scholar]
- 19.Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 20.Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- 21.Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 22.Fedele G, Green EW, Rosato E, Kyriacou CP. An electromagnetic field disrupts negative geotaxis in Drosophila via a CRY-dependent pathway. Nat Commun. 2014;5:4391. doi: 10.1038/ncomms5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 24.Rosato E, Codd V, Mazzotta G, Piccin A, Zordan M, Costa R, Kyriacou CP. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr Biol. 2001;11:909–917. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- 25.Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- 26.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction mechanism of Drosophila cryptochrome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peschel N, Veleri S, Stanewsky R. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila's circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17313–17318. doi: 10.1073/pnas.0606675103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanVickle-Chavez SJ, Van Gelder RN. Action spectrum of Drosophila cryptochrome. J Biol Chem. 2007;282:10561–10566. doi: 10.1074/jbc.M609314200. [DOI] [PubMed] [Google Scholar]
- 30.Ozturk N, VanVickle-Chavez SJ, Akileswaran L, Van Gelder RN, Sancar A. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1-CUL4-ROC1 E3 ligase complex. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4980–4985. doi: 10.1073/pnas.1303234110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 32.Deisenhofer J. DNA photolyases and cryptochromes. Mutat Res. 2000;460:143–149. doi: 10.1016/s0921-8777(00)00023-9. [DOI] [PubMed] [Google Scholar]
- 33.Sancar A. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. [DOI] [PubMed] [Google Scholar]
- 34.Ozturk N, Song SH, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 35.Song SH, Ozturk N, Denaro TR, Arat NO, Kao YT, Zhu H, Zhong D, Reppert SM, Sancar A. Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J Biol Chem. 2007;282:17608–17612. doi: 10.1074/jbc.M702874200. [DOI] [PubMed] [Google Scholar]
- 36.Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, Wu W, Berndt A, Wolf E, Bittl R, Ahmad M. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS biology. 2008;6:e160. doi: 10.1371/journal.pbio.0060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czarna A, Berndt A, Singh HR, Grudziecki A, Ladurner AG, Timinszky G, Kramer A, Wolf E. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013;153:1394–1405. doi: 10.1016/j.cell.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Levy C, Zoltowski BD, Jones AR, Vaidya AT, Top D, Widom J, Young MW, Scrutton NS, Crane BR, Leys D. Updated structure of Drosophila cryptochrome. Nature. 2013;495:E3–E4. doi: 10.1038/nature11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoltowski BD, Vaidya AT, Top D, Widom J, Young MW, Crane BR. Structure of full-length Drosophila cryptochrome. Nature. 2011;480:396–399. doi: 10.1038/nature10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozturk N, Selby CP, Zhong D, Sancar A. Mechanism of photosignaling by Drosophila cryptochrome: role of the redox status of the flavin chromophore. J Biol Chem. 2014;289:4634–4642. doi: 10.1074/jbc.M113.542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaidya AT, Top D, Manahan CC, Tokuda JM, Zhang S, Pollack L, Young MW, Crane BR. Flavin reduction activates Drosophila cryptochrome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20455–20460. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nature chemical biology. 2014;10:801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozturk N, Selby CP, Song SH, Ye R, Tan C, Kao YT, Zhong D, Sancar A. Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry. 2009;48:8585–8593. doi: 10.1021/bi901043s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gegear RJ, Casselman A, Waddell S, Reppert SM. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature. 2008;454:1014–1018. doi: 10.1038/nature07183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller M, Ahmad P. Light-activated cryptochrome reacts with molecular oxygen to form a flavin-superoxide radical pair consistent with magnetoreception. J Biol Chem. 2011;286:21033–21040. doi: 10.1074/jbc.M111.228940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin S, Yin H, Yang C, Dou Y, Liu Z, Zhang P, Yu H, Huang Y, Feng J, Hao J, Hao J, Deng L, Yan X, Dong X, Zhao Z, Jiang T, Wang HW, Luo SJ, Xie C. A magnetic protein biocompass. Nat Mater. 2016;15:217–226. doi: 10.1038/nmat4484. [DOI] [PubMed] [Google Scholar]
- 47.Niessner C, Denzau S, Malkemper EP, Gross JC, Burda H, Winklhofer M, Peichl L. Cryptochrome 1 in Retinal Cone Photoreceptors Suggests a Novel Functional Role in Mammals. Sci Rep. 2016;6:21848. doi: 10.1038/srep21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watari R, Yamaguchi C, Zemba W, Kubo Y, Okano K, Okano T. Light-dependent structural change of chicken retinal Cryptochrome4. J Biol Chem. 2012;287:42634–42641. doi: 10.1074/jbc.M112.395731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, Todo T, Wei YF, Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 50.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Similarity among the Drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 51.Foster RG, Provencio I, Hudson D, Fiske S, De Grip W, Menaker M. Circadian photoreception in the retinally degenerate mouse (rd/rd) J Comp Physiol A. 1991;169:39–50. doi: 10.1007/BF00198171. [DOI] [PubMed] [Google Scholar]
- 52.Freedman MS, Lucas RJ, Soni B, von Schantz M, Munoz M, David-Gray Z, Foster R. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:502–504. doi: 10.1126/science.284.5413.502. [DOI] [PubMed] [Google Scholar]
- 53.Wee R, Castrucci AM, Provencio I, Gan L, Van Gelder RN. Loss of photic entrainment and altered free-running circadian rhythms in math5−/− mice. J Neurosci. 2002;22:10427–10433. doi: 10.1523/JNEUROSCI.22-23-10427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 56.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirayama J, Nakamura H, Ishikawa T, Kobayashi Y, Todo T. Functional and structural analyses of cryptochrome. Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J Biol Chem. 2003;278:35620–35628. doi: 10.1074/jbc.M305028200. [DOI] [PubMed] [Google Scholar]
- 58.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 59.Thompson CL, Blaner WS, Van Gelder RN, Lai K, Quadro L, Colantuoni V, Gottesman ME, Sancar A. Preservation of light signaling to the suprachiasmatic nucleus in vitamin A-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11708–11713. doi: 10.1073/pnas.201301498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson CL, Selby CP, Van Gelder RN, Blaner WS, Lee J, Quadro L, Lai K, Gottesman ME, Sancar A. Effect of vitamin A depletion on nonvisual phototransduction pathways in cryptochromeless mice. Journal of biological rhythms. 2004;19:504–517. doi: 10.1177/0748730404270519. [DOI] [PubMed] [Google Scholar]
- 61.Selby CP, Thompson C, Schmitz TM, Van Gelder RN, Sancar A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14697–14702. doi: 10.1073/pnas.260498597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sexton T, Buhr E, Van Gelder RN. Melanopsin and mechanisms of non-visual ocular photoreception. J Biol Chem. 2012;287:1649–1656. doi: 10.1074/jbc.R111.301226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: An opsin in melanophores, brain, and eye. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- 65.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science. 2002;298:2213–2216. doi: 10.1126/science.1076848. [DOI] [PubMed] [Google Scholar]
- 67.Ruby NF, Brennan TJ, Xie X, Cao V, Franken P, Heller HC, O'Hara BF. Role of melanopsin in circadian responses to light. Science. 2002;298:2211–2213. doi: 10.1126/science.1076701. [DOI] [PubMed] [Google Scholar]
- 68.Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, Pletcher MT, Sato TK, Wiltshire T, Andahazy M, Kay SA, Van Gelder RN, Hogenesch JB. Melanopsin is required for non-image-forming photic responses in blind mice. Science. 2003;301:525–527. doi: 10.1126/science.1086179. [DOI] [PubMed] [Google Scholar]
- 69.Van Gelder RN, Wee R, Lee JA, Tu DC. Reduced pupillary light responses in mice lacking cryptochromes. Science. 2003;299:222. doi: 10.1126/science.1079536. [DOI] [PubMed] [Google Scholar]
- 70.Emanuel AJ, Do MT. Melanopsin tristability for sustained and broadband phototransduction. Neuron. 2015;85:1043–1055. doi: 10.1016/j.neuron.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sexton TJ, Golczak M, Palczewski K, Van Gelder RN. Melanopsin is highly resistant to light and chemical bleaching in vivo. J Biol Chem. 2012;287:20888–20897. doi: 10.1074/jbc.M111.325969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Owens L, Buhr E, Tu DC, Lamprecht TL, Lee J, Van Gelder RN. Effect of circadian clock gene mutations on nonvisual photoreception in the mouse. Invest Ophthalmol Vis Sci. 2012;53:454–460. doi: 10.1167/iovs.11-8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozgur S, Sancar A. Purification and properties of human blue-light photoreceptor cryptochrome 2. Biochemistry. 2003;42:2926–2932. doi: 10.1021/bi026963n. [DOI] [PubMed] [Google Scholar]
- 74.Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, Zheng N. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Foley LE, Gegear RJ, Reppert SM. Human cryptochrome exhibits light-dependent magnetosensitivity. Nat Commun. 2011;2:356. doi: 10.1038/ncomms1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Partch CL, Sancar A. Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 78.Yu X, Liu H, Klejnot J, Lin C. The Cryptochrome Blue Light Receptors. Arabidopsis Book. 2010;8:e0135. doi: 10.1199/tab.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubin EB, Shemesh Y, Cohen M, Elgavish S, Robertson HM, Bloch G. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 2006;16:1352–1365. doi: 10.1101/gr.5094806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 81.Ingram KK, Kutowoi A, Wurm Y, Shoemaker D, Meier R, Bloch G. The molecular clockwork of the fire ant Solenopsis invicta. PloS one. 2012;7:e45715. doi: 10.1371/journal.pone.0045715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park HW, Kim ST, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 83.Chaves I, Yagita K, Barnhoorn S, Okamura H, van der Horst GT, Tamanini F. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol Cell Biol. 2006;26:1743–1753. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Schalie EA, Conte FE, Marz KE, Green CB. Structure/function analysis of Xenopus cryptochromes 1 and 2 reveals differential nuclear localization mechanisms and functional domains important for interaction with and repression of CLOCK-BMAL1. Mol Cell Biol. 2007;27:2120–2129. doi: 10.1128/MCB.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khan SK, Xu H, Ukai-Tadenuma M, Burton B, Wang Y, Ueda HR, Liu AC. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem. 2012;287:25917–25926. doi: 10.1074/jbc.M112.368001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 87.Gao P, Yoo SH, Lee KJ, Rosensweig C, Takahashi JS, Chen BP, Green CB. Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J Biol Chem. 2013;288:35277–35286. doi: 10.1074/jbc.M113.509604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gustafson CL, Partch CL. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2015;54:134–149. doi: 10.1021/bi500731f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 91.Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol. 2014;21:126–132. doi: 10.1038/nsmb.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stratmann M, Stadler F, Tamanini F, van der Horst GT, Ripperger JA. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev. 2010;24:1317–1328. doi: 10.1101/gad.578810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. 2011;286:25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 96.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 97.Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O'Neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 98.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, Nussbaum J, Liu X, Chen Z, Chen ZJ, Green CB, Takahashi JS. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, Doyle FJ, 3rd, Schultz PG, Kay SA. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nangle S, Xing W, Zheng N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 2013;23:1417–1419. doi: 10.1038/cr.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kiyohara YB, Tagao S, Tamanini F, Morita A, Sugisawa Y, Yasuda M, Yamanaka I, Ueda HR, van der Horst GT, Kondo T, Yagita K. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCarthy EV, Baggs JE, Geskes JM, Hogenesch JB, Green CB. Generation of a novel allelic series of cryptochrome mutants via mutagenesis reveals residues involved in protein-protein interaction and CRY2-specific repression. Mol Cell Biol. 2009;29:5465–5476. doi: 10.1128/MCB.00641-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Evans JA, Pan H, Liu AC, Welsh DK. Cry1−/− circadian rhythmicity depends on SCN intercellular coupling. Journal of biological rhythms. 2012;27:443–452. doi: 10.1177/0748730412461246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee HW, Liu AC, Partch CL. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol. 2015;22:476–484. doi: 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 109.Engelen E, Janssens RC, Yagita K, Smits VA, van der Horst GT, Tamanini F. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PloS one. 2013;8:e56623. doi: 10.1371/journal.pone.0056623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee JH, Sancar A. Circadian clock disruption improves the efficacy of chemotherapy through p73-mediated apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10668–10672. doi: 10.1073/pnas.1106284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12662–12667. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Papp SJ, Huber AL, Jordan SD, Kriebs A, Nguyen M, Moresco JJ, Yates JR, Lamia KA. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. Elife. 2015;4 doi: 10.7554/eLife.04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sancar A, Lindsey-Boltz LA, Kang TH, Reardon JT, Lee JH, Ozturk N. Circadian clock control of the cellular response to DNA damage. FEBS Lett. 2010;584:2618–2625. doi: 10.1016/j.febslet.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang XS, Armstrong ME, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med (Lond) 2011;61:78–89. doi: 10.1093/occmed/kqr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Current biology : CB. 2013;23:372–381. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reddy AB, Maywood ES, Karp NA, King VM, Inoue Y, Gonzalez FJ, Lilley KS, Kyriacou CP, Hastings MH. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007;45:1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- 119.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665–1667. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 121.Lee TH, Park JM, Leem SH, Kang TH. Coordinated regulation of XPA stability by ATR and HERC2 during nucleotide excision repair. Oncogene. 2014;33:19–25. doi: 10.1038/onc.2012.539. [DOI] [PubMed] [Google Scholar]
- 122.Rosato E, Tauber E, Kyriacou CP. Molecular genetics of the fruit-fly circadian clock. Eur J Hum Genet. 2006;14:729–738. doi: 10.1038/sj.ejhg.5201547. [DOI] [PubMed] [Google Scholar]
- 123.Benna C, Bonaccorsi S, Wulbeck C, Helfrich-Forster C, Gatti M, Kyriacou CP, Costa R, Sandrelli F. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Current biology : CB. 2010;20:346–352. doi: 10.1016/j.cub.2009.12.048. [DOI] [PubMed] [Google Scholar]
- 124.Gotter AL, Manganaro T, Weaver DR, Kolakowski LF, Jr, Possidente B, Sriram S, MacLaughlin DT, Reppert SM. A time-less function for mouse timeless. Nat Neurosci. 2000;3:755–756. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- 125.Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 128.Kang TH, Leem SH. Modulation of ATR-mediated DNA damage checkpoint response by cryptochrome 1. Nucleic Acids Res. 2014;42:4427–4434. doi: 10.1093/nar/gku094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, Zheng N. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife. 2014;3:e03674. doi: 10.7554/eLife.03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, Benda C, Kramer A, Wolf E. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell. 2014;157:1203–1215. doi: 10.1016/j.cell.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 131.Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden MG, Jacks T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016 doi: 10.1016/j.cmet.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Davis S, Mirick DK. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer Causes Control. 2006;17:539–545. doi: 10.1007/s10552-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 133.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 135.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 136.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.J G. Blue light of photoreception. Photochemistry and Photobiology. 1979;30:749–754. [Google Scholar]
- 138.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]