Hu et al. show that sumoylation of the viral RNA sensors RIG-I and MDA5 by TRIM38 in uninfected and early-infected cells, and their desumoylation by SENP2 in the late phase of infection, ensure efficient innate immune responses to RNA viruses and their timely termination upon resolution of infection.
Abstract
Sensing of viral RNA by the cytosolic receptors RIG-I and melanoma differentiation-associated gene 5 (MDA5) leads to innate antiviral response. How RIG-I and MDA5 are dynamically regulated in innate antiviral response is not well understood. Here, we show that TRIM38 positively regulates MDA5- and RIG-I–mediated induction of downstream genes and acts as a SUMO E3 ligase for their dynamic sumoylation at K43/K865 and K96/K888, respectively, before and after viral infection. The sumoylation of MDA5 and RIG-I suppresses their K48-linked polyubiquitination and degradation in uninfected or early-infected cells. Sumoylation of the caspase recruitment domains of MDA5 and RIG-I is also required for their dephosphorylation by PP1 and activation upon viral infection. At the late phase of viral infection, both MDA5 and RIG-I are desumoylated by SENP2, resulting in their K48-linked polyubiquitination and degradation. These findings suggest that dynamic sumoylation and desumoylation of MDA5 and RIG-I modulate efficient innate immunity to RNA virus and its timely termination.
Introduction
The innate immune system is the first line of host defense against pathogen invasion. After detection of structurally conserved pathogen–associated molecular patterns (PAMPs) via germline-encoded pathogen recognition receptors (PRRs), the host cells initiate a series of signaling cascades which ultimately induce the expression of downstream antiviral genes, such as type I IFNs and inflammatory cytokines, to inhibit replication of pathogens, clear pathogen-infected cells, and facilitate adaptive immune response (Akira et al., 2006; Hiscott, 2007; Gürtler and Bowie, 2013; Carpenter et al., 2014).
Innate immune response to cytosolic viral RNA is mediated by RIG-I-like receptors (RLRs) including retinoic acid-inducible gene 1 protein (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which contain two N-terminal tandem caspase recruitment domain (CARDs), a helicase domain, and a C-terminal domain (CTD) and recognize different types of RNA viruses (Yoneyama and Fujita, 2008). In the absence of viral infection, RIG-I and MDA5 are phosphorylated in their respective CARDs to suppress their activation in resting cells (Gack et al., 2010; Nistal-Villán et al., 2010; Wies et al., 2013). Additionally, RIG-I but not MDA5 exhibit an autoinhibition state through the intramolecular interaction of its CARDs and CTD in uninfected cells (Saito et al., 2007). After recognition of cytosolic viral RNA, RIG-I and MDA5 undergo conformational changes and recruit PP1 for their dephosphorylation (Wies et al., 2013), followed by their K63-linked polyubiquitination (Gack et al., 2007; Zeng et al., 2010; Yan et al., 2014) and translocation to the outer membrane of mitochondria on which they further recruit and activate the central adaptor virus-induced signaling adaptor (VISA, also known as MAVS, CARDIF, and IPS-1; Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). VISA in turn recruits TNF receptor–associated factor 2/6 (TRAF2/6) and mitochondrial mediator of IFN regulatory factor 3 (IRF3) activation (MITA, also known as STING) to activate the GSK3β-TBK1 and IKK complexes, which then phosphorylate the transcriptional factors IRF3 and NF-κB, respectively, leading to the ultimate induction of downstream antiviral genes (Zhong et al., 2008; Lei et al., 2010; Hou et al., 2011; Liu et al., 2013). In addition, at the late phase of viral infection, RIG-I and MDA5 are regulated by K48-linked polyubiquitination and degradation to avoid their sustained activation (Arimoto et al., 2007; Chen et al., 2013; Hao et al., 2015). However, how RIG-I and MDA5 are optimally activated in early-infected cells, and then timely turned-off at the late phase of viral infection, is still enigmatic.
In this study, we report that RIG-I and MDA5 are dynamically sumoylated by tripartite motif-containing protein 38 (TRIM38) in uninfected or early-infected cells to ensure their optimal activation, and then undergo desumoylation by sentrin/sumo-specific protease 2 (SENP2) and degradation at the late phase of viral infection to turn off the sustained induction of downstream antiviral genes. Our study provides exciting insights into the mechanisms on how innate immune response to RNA virus is efficiently mounted upon infection and terminated in a timely manner at the late phase of infection to avoid excessive and harmful immune damage to the host.
Results
TRIM38 positively regulates RIG-I– and MDA5-mediated signaling
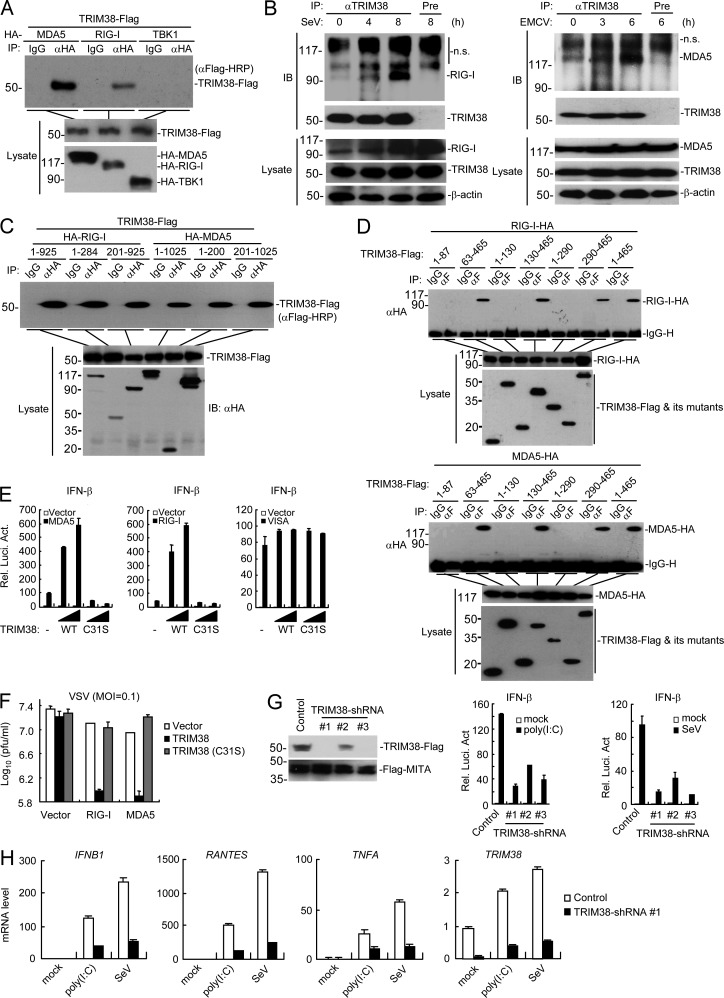
Using a two-step immunoaffinity purification and “shot-gun” mass spectrometry analysis, we identified TRIM38 as a candidate protein associated with MDA5. As it has been shown that certain TRIM family members are involved in regulation of innate immune responses (Versteeg et al., 2013), we investigated whether TRIM38 is involved in MDA5-mediated signaling. Coimmunoprecipitation experiments indicated that TRIM38 interacted with MDA5 as well as RIG-I in mammalian overexpression system (Fig. 1 A). Endogenous TRIM38 constitutively interacted with RIG-I and MDA5 in uninfected cells, and their interactions were increased after infection with the RNA viruses Sendai virus (SeV) and encephalomyocarditis virus (EMCV; Fig. 1 B), which have been shown to be sensed by RIG-I and MDA, respectively (Loo and Gale, 2011). Domain mapping experiments indicated that TRIM38 interacted with both the N-terminal CARD-containing (aa 1–284 of RIG-I or aa 1–200 of MDA5) and the C-terminal Helicase-containing (aa 201-925 of RIG-I or aa 201-1025 of MDA5) domains of RIG-I and MDA5 via its PRY-SPRY (aa 290-465) domain (Fig. 1, C and D). Interestingly, unlike the negatively regulatory roles of TRIM38 in TLR3/4-mediated or TNF/IL-1-triggered signaling (Hu et al., 2014, 2015), TRIM38 but not its enzymatic-inactive mutant TRIM38(C31S) dramatically potentiated RIG-I– and MDA5-, but not VISA-mediated activation of the IFN-β promoter in reporter assays (Fig. 1 E). Consistently, TRIM38 but not TRIM38(C31S) markedly potentiated RIG-I– and MDA5-mediated cellular antiviral responses (Fig. 1 F). Knockdown of TRIM38 markedly inhibited SeV and transfected polyinosine-polycytidylic acid (poly(I:C))–triggered activation of the IFN-β promoter in reporter assays (Fig. 1 G) and transcription of downstream antiviral genes, including IFNB1, CXCL10, and TNFA (Fig. 1 H). These results suggest that TRIM38 positively regulates RIG-I– and MDA5-mediated induction of downstream antiviral genes.
Figure 1.
TRIM38 positively regulates RIG-I– and MDA5-mediated signaling. (A) Coimmunoprecipitation of TRIM38 with MDA5 and RIG-I in mammalian overexpression system. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by coimmunoprecipitation experiments and immunoblotting analysis. (B) Endogenous association of TRIM38 with RIG-I and MDA5. THP-1 cells were left uninfected or infected with SeV (top) or EMCV (bottom) for the indicated times followed by coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. (C and D) Domain mapping of TRIM38 with RIG-I and MDA5. Experiments were performed as in B, except for the different transfected plasmids. (E) Effects of TRIM38 or TRIM38(C31S) on RIG-I– and MDA5-mediated activation of the IFN-β promoter. HEK293 cells were transfected with the indicated plasmids for 24 h before luciferase assays. (F) Effects of TRIM38 or TRIM38(C31S) on RIG-I– and MDA5-mediated cellular antiviral response. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by VSV (MOI = 0.1) infection for 24 h, and then the supernatants were collected for plaque assays to determine the viral titers. (G) Effects of knockdown of TRIM38 on SeV- or poly(I:C)-induced activation of the IFN-β promoter. (left) HEK293 cells were transfected with the indicated plasmids for 24 h before immunoblot analysis. (right) HEK293 cells were transfected with the indicated plasmids for 36 h, and then transfected with infected with SeV or poly(I:C) for 18 h for 10 h before luciferase assays. (H) Effects of TRIM38 knockdown on SeV- or poly(I:C)-induced transcription of downstream antiviral genes. HEK293 cells were transfected and treated as in (I) before qPCR analysis. Data are from one representative experiment with four (E and G) or three (H) technical replicates. The error bars are mean ± SD in E, G, and H. All experiments were repeated twice.
TRIM38 is required for RLR-mediated innate immune response to RNA virus
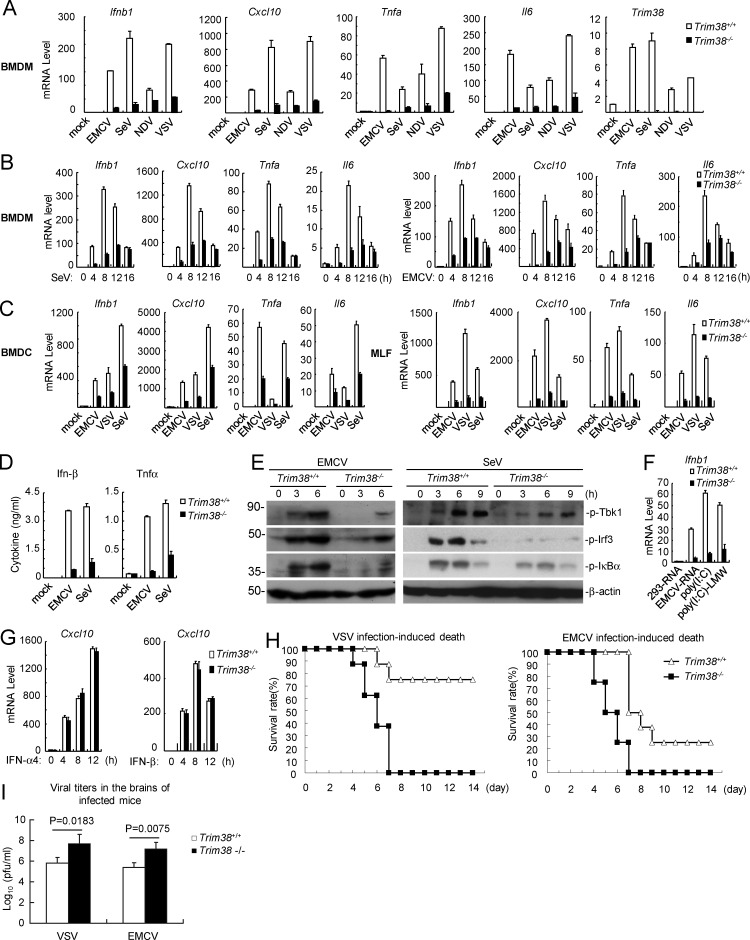
To further confirm a role of TRIM38 in RLR-mediated innate immune response to RNA virus, we infected Trim38+/+ and Trim38−/− BM-derived macrophages (BMDMs) with different types of RNA viruses, including MDA5-sensed EMCV, and RIG-I–sensed SeV, Newcastle disease virus (NDV), and vesicular stomatitis virus (VSV). The results indicated that Trim38 (referred as the murine orthologue of human TRIM38) deficiency significantly impaired transcription of downstream antiviral genes including Ifnb1, Cxcl10, Tnfa, and Il6 induced by these RNA viruses (Fig. 2 A), especially at the early phase (4–12 h) but not late phase (16 h) after viral infection (Fig. 2 B). Trim38 deficiency also dramatically inhibited transcription of downstream antiviral genes induced by EMCV, VSV, and SeV in mouse BMDCs or mouse lung fibroblasts (MLFs; Fig. 2 C). ELISAs further confirmed that secretion of Ifn-β and Tnf induced by EMCV and SeV was significantly inhibited in Trim38-deficient BMDMs (Fig. 2 D). Consistently, EMCV- and SeV-induced phosphorylation of Tbk1, Irf3, and IκBα, which are hallmarks of activation of downstream signaling components, was markedly decreased in Trim38−/− compared with Trim38+/+ BMDMs (Fig. 2 E). Additionally, transcription of downstream genes induced by transfected EMCV RNA and synthetic RNA, including high molecular weight poly(I:C) (recognized by MDA5) and low molecular weight poly(I:C) (poly(I:C)-LMW, recognized by RIG-I), was impaired in Trim38−/− BMDMs (Fig. 2 F). In similar experiments, transcription of Cxcl10 induced by Ifn-α4 and Ifn-β was fully comparable between Trim38−/− and Trim38+/+ BMDMs (Fig. 2 G). Collectively, these results suggest that Trim38 plays a specific and essential role in RLR-mediated induction of downstream antiviral genes in various cell types.
Figure 2.
TRIM38 is required for RLR-mediated innate immune response. (A and B) Effects of Trim38 deficiency on RNA virus–induced transcription of downstream antiviral genes in BMDMs. The indicated cells were left uninfected or infected with the indicated viruses for 6 h (A) or infected with SeV or EMCV for the indicated times (B) before qPCR analysis. (C) Effects of Trim38 deficiency on RNA virus–induced transcription of downstream antiviral genes in BMDCs or MLFs. Trim38+/+ and Trim38−/− BMDCs (A) or MLFs (B) were left uninfected or infected with EMCV, VSV, or SeV for 6 h before qPCR analysis. (D) Effects of Trim38 deficiency on EMCV- and SeV-induced secretion of Ifn-β and Tnfα cytokines in BMDMs. The indicated cells were left uninfected or infected with EMCV or SeV for 18 h before ELISA with the culture medium. (E) Effects of Trim38 deficiency on EMCV- or SeV-induced phosphorylation of Tbk1, Irf3, and IκBα in BMDMs. The indicated cells were left uninfected or infected with EMCV or SeV for the indicated times, followed by immunoblotting analysis. (F) Effects of Trim38 deficiency on transcription of antiviral genes induced by transfected nucleic acids in MLFs. The indicated cells were transfected with the indicated nucleic acids for 6 h before qPCR analysis. (G) Effects of Trim38 deficiency on IFN-α4- or IFN-β–induced transcription of Cxcl10 in BMDMs. The indicated cells were left untreated or treated with IFN-α4 or IFN-β for the indicated times for the indicated times before qPCR analysis. (H) Effects of Trim38 deficiency on VSV- or EMCV-induced death of mice. Trim38+/+ and Trim38−/− mice (n = 16) were intranasally infected with VSV at 108 PFU per mouse or EMCV at 105 PFU per mouse, and the survival rates of mice were observed and recorded for two weeks. (I) Measurement of viral titers in the brain of infected mice. Trim38+/+ and Trim38−/− mice (n = 3) were intranasally infected with VSV at 108 PFU per mouse or EMCV at 105 PFU per mouse. 2 d later, the brains of the infected mice were extracted for measurement of viral titers. The P-values were calculated using the Student’s t test. Data in A, B, and C are from four biological replicates. Data in F and G are from one representative experiment with three technical replicates. The error bars are mean ± SD in A–D, F, G, and I. Experiments were repeated twice (E–I) or three times (D).
We next determined whether Trim38 is essential for effective host defense against RNA virus in vivo. We intranasally infected Trim38+/+ and Trim38−/− mice with VSV or EMCV and monitored the survival of the mice for 2 wk. The results indicated that Trim38−/− mice were more susceptible to VSV- and EMCV-induced death (Fig. 2 H). Consistently, viral titers in the brains were much higher in Trim38−/− mice at 2 d after viral infection (Fig. 2 I). Collectively, these results suggest that Trim38 plays important roles in efficient host defense against RNA virus infection in vivo.
TRIM38 catalyzes sumoylation of MDA5 and RIG-I
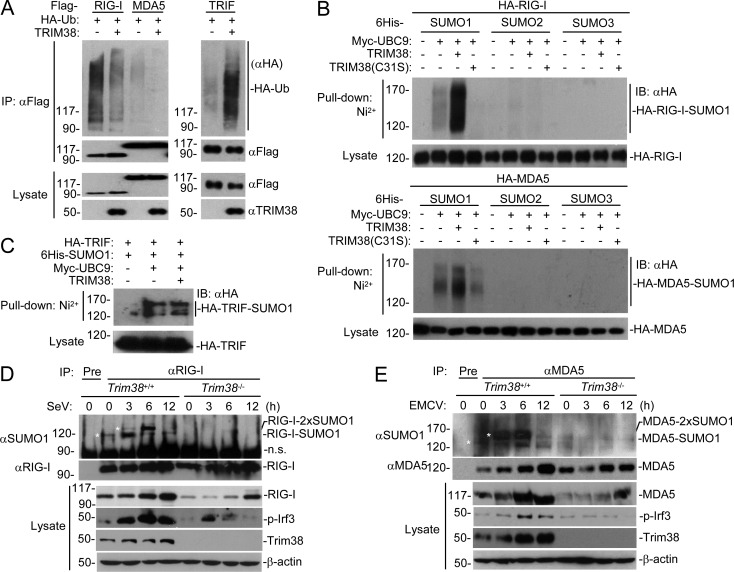
We next investigated the mechanisms of TRIM38 in the regulation of RIG-I and MDA5. Because TRIM38 is an E3 ubiquitin ligase (Hu et al., 2015), we next examined whether TRIM38 could catalyze polyubiquitination of MDA5 and RIG-I. However, overexpression of TRIM38 inhibited but not promoted the polyubiquitination of MDA5 and RIG-I (Fig. 3 A). In similar experiments, TRIM38 promotes polyubiquitination of TRIF (Fig. 3 A), which is consistent with our previous results (Hu et al., 2015). Previously, it has been demonstrated that certain TRIM protein family members, including PML, TRIM 27, TRIM28, and TRIM38, can act as E3 SUMO ligases (Sternsdorf et al., 1999; Muller et al., 2000; Chu and Yang, 2011; Liang et al., 2011; Hu et al., 2016). Interestingly, TRIM38 but not its enzyme-inactive mutant TRIM38(C31S) could promote SUMO1 but not SUMO2 or SUMO3 modification of RIG-I and MDA5 (Fig. 3 B). In similar experiments, TRIM38 failed to promote sumoylation of TRIF (Fig. 3 C). We further examined sumoylation of endogenous RIG-I and MDA5 in both Trim38+/+ and Trim38−/− MLFs. As shown in Fig. 3 F, a sumoylated RIG-I species of ∼115 kD was detected in uninfected wild-type but not Trim38−/− MLFs. Interestingly, another sumoylated RIG-I species of ∼130 kD was undetectable in uninfected wild-type MLFs, but markedly appeared at early phase (3–6 h) and decreased at late phase (12 h) of SeV infection (Fig. 3 D). These sumoylated RIG-I bands were not detected in Trim38−/− MLFs either before or after SeV infection (Fig. 3 D). Considering that the molecule weights of RIG-I and SUMO1 are ∼100 and ∼15–20 kD in SDS-PAGE, respectively, our results suggest that endogenous RIG-I is dynamically modified with one or two SUMO1 moieties before and after viral infection, respectively in a Trim38-dependent manner. Similar experiments also showed that a weak sumoylated MDA5 species of ∼135 kD was detected in uninfected wild-type but not Trim38−/− MLFs (Fig. 3 E). In addition, another sumoylated MDA5 species of ∼150 kD was undetectable in uninfected wild-type MLFs but markedly appeared at early phase (3–6 h) and decreased at late phase (12 h) of EMCV infection (Fig. 3 E). These sumoylated MDA5 bands were not detected in Trim38−/− MLFs either before or after EMCV infection (Fig. 3 E). Because the molecule weights of MDA5 and SUMO1 are ∼120 and ∼15–20 kD in SDS-PAGE, respectively, we deduce that endogenous MDA5 is dynamically modified with one or two SUMO1 moieties before and after viral infection, respectively, in a Trim38-dependent manner.
Figure 3.
TRIM38 catalyzes sumoylation of RIG-I and MDA5. (A) Effects of TRIM38 on polyubiquitination of RIG-I, MDA5, and TRIF. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by immunoprecipitation and immunoblotting analysis with the indicated antibodies. (B and C) Effects of TRIM38 on sumoylation of RIG-I, MDA5, and TRIF. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by Ni2+ pull-down assays and immunoblotting analysis. (D) Effects of Trim38 deficiency on sumoylation of endogenous RIG-I. Trim38+/+ and Trim38−/− MLFs were left uninfected or infected with SeV for the indicated times, followed by immunoprecipitation and immunoblotting analysis. *, sumoylation bands of RIG-I. (E) Effects of Trim38 deficiency on sumoylation of endogenous MDA5. Trim38+/+ and Trim38−/− MLFs were left uninfected or infected with EMCV for the indicated times, followed by immunoprecipitation and immunoblotting analysis. *, sumoylation bands of MDA5. Data are from one representative experiment. All experiments were repeated three times.
RIG-I and MDA5 are sumoylated at K96/K888 and K43/K865, respectively
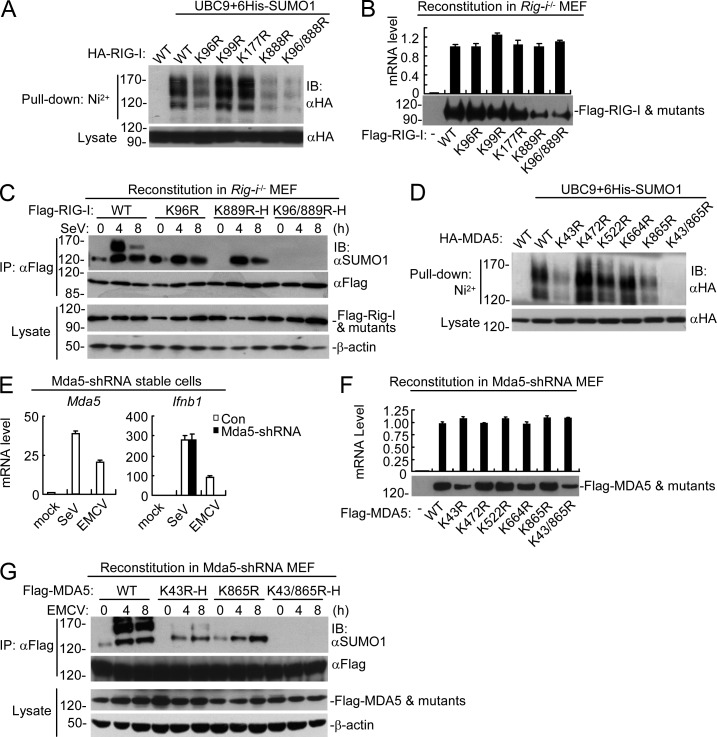
Using the sumoylation site prediction program (Xue et al., 2006), we identified four candidate motifs that are conserved between human RIG-I and its murine orthologue, including the motifs containing K96, K99, K177, and K888 in human RIG-I, which correspond to K96, K99, K177, and K889 in murine RIG-I, respectively. Mutagenesis indicated that mutation of either K96 or K888 but not K99 or K177 reduced sumoylation of human RIG-I, whereas simultaneous mutation of both K96 and K888 abolished its sumoylation (Fig. 4 A). To further confirm the sumoylation residues of RIG-I, we reconstituted wild-type RIG-I and its various mutants into Rig-i−/− MEFs via the pseudotyped retroviral-mediated gene transfer approach. Interestingly, when Rig-i−/− MEFs were infected with equal titers of pseudotyped retroviruses so that the mRNA levels of RIG-I and its mutants were comparable in the reconstituted cells, we observed that mutation of K889 (corresponding to K888 in human RIG-I) but not other residues dramatically decreased the stability of RIG-I (Fig. 4 B). To further analyze the sumoylation residues of RIG-I, we adjusted the titers of the pseudotyped retroviruses so that comparable protein levels of RIG-I and its mutants were expressed in the reconstituted cells. Immunoprecipitation experiments with these reconstituted cells indicated that mutation of K889 but not K96 abolished sumoylation of RIG-I in uninfected cells. Furthermore, mutation of either K96 or K889 attenuated SeV-induced sumoylation of RIG-I, and simultaneous mutation of K96 and K889 abolished sumoylation of RIG-I in both uninfected and infected cells (Fig. 4 C). These results suggest that RIG-I is sumoylated at K889 in uninfected cells and SeV infection induced further sumoylation of RIG-I at both K96 and K889, and that sumoylation at K889 of RIG-I increases its stability in resting cells.
Figure 4.
Identification of sumoylation sites of RIG-I and MDA5. (A) Identification of sumoylation residues of RIG-I. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by Ni2+ pull-down assays and immunoblotting analysis. (B) Expression of RIG-I and its mutants in reconstituted Rig-i−/− MEFs. Reconstitution of RIG-I or its mutants into Rig-i−/− MEFs, followed by analysis of their mRNA and protein levels. (C) Sumoylation of RIG-I and its mutants in reconstituted cells. Rig-i−/− MEFs reconstituted with RIG-I or its mutants were left uninfected or infected with SeV for the indicated times before immunoprecipitation and immunoblotting analysis. To ensure comparable protein levels of RIG-I and its mutants in all reconstituted MEFs, the titers of retroviruses were adjusted for reconstitution. More retroviruses containing RIG-I K889R or K96/889R mutants were used to superinfect the cells, which were designated as K889R-H and K96/889R-H, respectively. (D) Identification of sumoylation residues of MDA5. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by Ni2+ pull-down assays and immunoblotting analysis. (E) Knockdown efficiency of MDA5-shRNA in stably transduced MEFs. The cells were left uninfected or infected with EMCV or SeV for 6 h before qPCR analysis. (F) Expression of MDA5 and its mutants in reconstituted MDA5-shRNA MEFs. Reconstitution of MDA5 and its mutants into MDA5-shRNA MEFs, followed by analysis of their mRNA and protein levels. (G) Sumoylation of MDA5 and its mutants in reconstituted cells. MDA5-shRNA MEFs reconstituted with MDA5 or its mutants were left uninfected or infected with EMCV for the indicated times before immunoprecipitation and immunoblotting analysis. To ensure comparable protein levels of MDA5 and its mutants in all reconstituted MEFs, the titers of retroviruses were adjusted for reconstitution. More retroviruses containing the K43R or K43/865R mutants were used to superinfect the cells, which were designated as K43R-H and K43/865R-H, respectively. All the experiments were repeated for three times.
We also identified five candidate motifs that are conserved between human MDA5 and its murine orthologue, including the motifs containing K43, K472, K522, K664, and K865 in human MDA5, which are corresponding to K43, K473, K523, K664, and K865 in murine MDA5, respectively. Mutagenesis indicated that mutation of either K43 or K865 but not K472, K522, or K664 reduced sumoylation of human MDA5, whereas simultaneous mutation of both K43 and K865 abolished its sumoylation (Fig. 4 D). To further confirm the sumoylation residues of murine MDA5, we established MDA5-shRNA MEFs in which basal and viral infection–induced expression of MDA5 was almost abolished (Fig. 4 E). We then reconstituted MDA5 and its various mutants into MDA5-shRNA MEFs via a pseudotyped retroviral-mediated gene transfer approach. Interestingly, when MDA5-shRNA MEFs were infected with equal titers of pseudotyped retroviruses so that the mRNA levels of MDA5 and its mutants were comparable in these reconstituted cells, we observed that mutation of K43 but not other residues dramatically decreased the stability of MDA5 (Fig. 4 F). We next infected MDA5-shRNA MEFs with proper titers of pseudotyped retroviruses so that comparable protein levels of MDA5 and its mutants were expressed in the reconstituted cells. Immunoprecipitation experiments with these reconstituted cells indicated that mutation of K43 but not K865 abolished the sumoylation of MDA5 in uninfected cells. Furthermore, mutation of either K43 or K865 attenuated EMCV-induced sumoylation of MDA5, and simultaneous mutation of K43 and K865 abolished sumoylation of MDA5 in both uninfected and infected cells (Fig. 4 G). These results suggest that murine MDA5 is sumoylated at K43 in uninfected cells and further sumoylated at K865 after EMCV infection, and sumoylation at K43 of MDA5 maintains its stability in resting cells.
Sumoylation regulates K48-linked polyubiquitination and degradation of RIG-I and MDA5
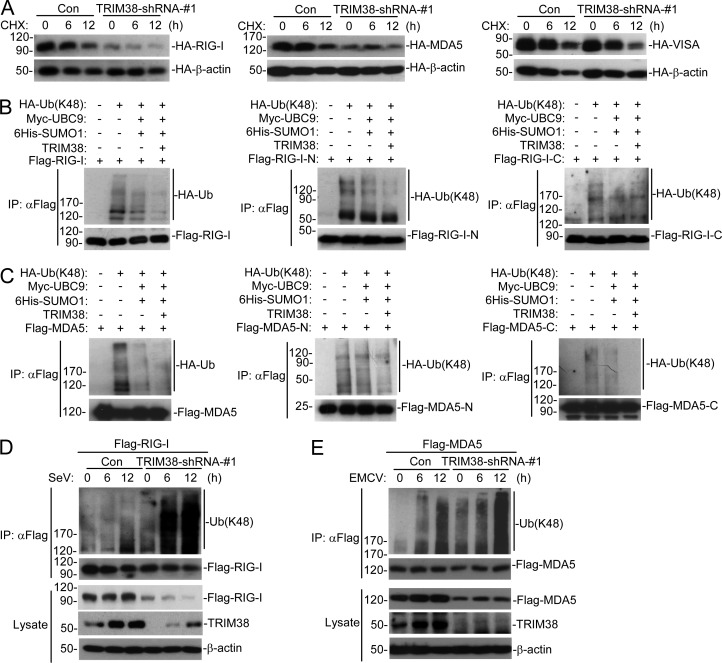
In our experiments, we routinely found that the protein levels of RIG-I and MDA5 were decreased in Trim38−/− MLFs (Fig. 3, D and E), suggesting that Trim38-mediated sumoylation of RIG-I and MDA5 promotes their stability. Consistently, knockdown of TRIM38 in human cells severely impaired the stability of overexpressed RIG-I and MDA5 but not VISA (Fig. 5 A). Previously, it has been reported that RNF125- and c-Cbl–mediated K48-linked polyubiquitination regulates degradation of RIG-I and MDA5 (Arimoto et al., 2007; Chen et al., 2013). We investigated whether sumoylation of RIG-I and MDA5 regulates their K48-linked polyubiquitination. The results indicated that sumoylation of RIG-I and MDA5 markedly inhibited their K48-linked polyubiquitination (Fig. 5, B and C). Interestingly, sumoylation of both N-terminal and C-terminal domains of RIG-I/MDA5 suppressed their respective K48-linked polyubiquitination of these mutants (Fig. 5, B and C), suggesting that sumoylation at both K96/K888 of RIG-I and K43/K865 of MDA5 regulates their K48-linked polyubiquitination. Consistently, knockdown of TRIM38 in human cells dramatically increased K48-linked polyubiquitination of RIG-I and MDA5 (Fig. 5, D and E). These data suggested that sumoylation of RIG-I and MDA5 regulates their K48-linked polyubiquitination and degradation.
Figure 5.
Sumoylation regulates K48-linked polyubiquitination and degradation of RIG-I and MDA5. (A) Effects of knockdown of TRIM38 on stabilities of RIG-I, MDA5, and VISA. HEK293 cells were transfected with the indicated plasmids for 36 h, followed by cycloheximide (CHX; 100 µg/ml) treatment for the indicated times before immunoblotting analysis. (B and C) Effect of sumoylation of RIG-I and MDA5 on their K48-linked polyubiquitination in mammalian overexpression system. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by immunoprecipitation and immunoblotting analysis. (D and E) Effects of knockdown of TRIM38 on K48-linked polyubiquitination of RIG-I and MDA5. HEK293 cells were transfected with the indicated plasmids for 36 h, and then left uninfected or infected with SeV (D) or EMCV (E) for the indicated times before immunoprecipitation and immunoblotting analysis. All the experiments were repeated three times.
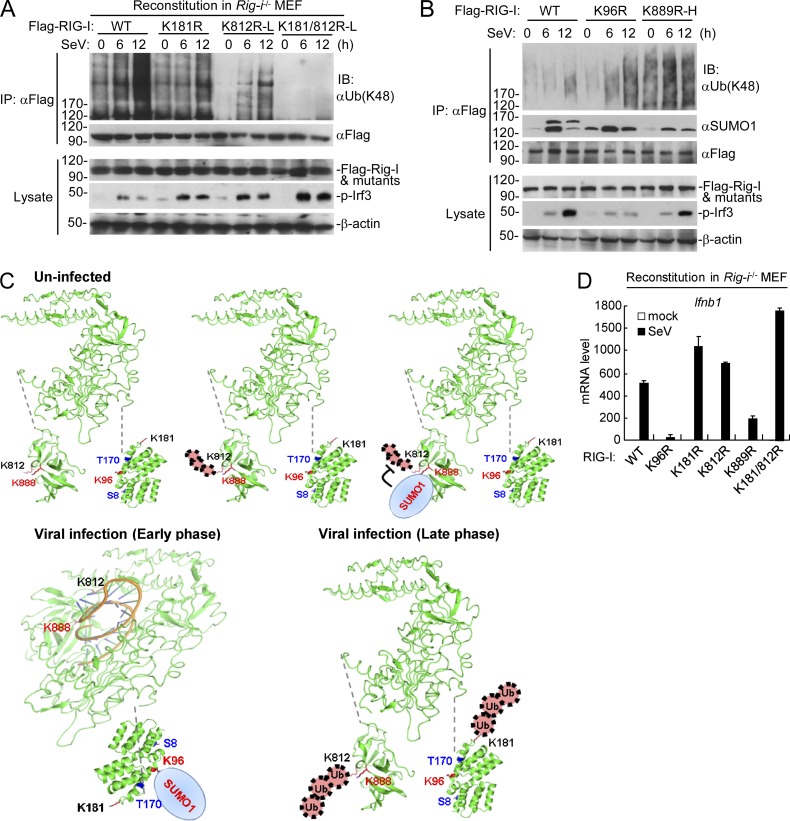
To further determine how the dynamic sumoylation of RIG-I and MDA5 mediated by TRIM38 regulates their K48-linked polyubiquitination and degradation, we examined endogenous sumoylation and K48-linked polyubiquitination of wild-type RIG-I/MDA5 and their mutants before and after viral infection. It has been reported that RNF125 and c-Cbl mediate K48-linked polyubiquitination of RIG-I at K181 and K812, respectively (Arimoto et al., 2007; Chen et al., 2013). Endogenous immunoprecipitation experiments with reconstitution cells indicated that mutation of K812 abolished the basal K48-linked polyubiquitination of RIG-I in uninfected cells, and mutation of K181 or K812 reduced SeV-induced K48-linked polyubiquitination of RIG-I, whereas simultaneous mutation of these two residues abolished its K48-linked polyubiquitination at the late phase of viral infection (12 h; Fig. 6 A). These results suggest that RIG-I undergoes K48-linked polyubiquitination at K812 and degradation in resting cells and SeV infection triggers dramatic K48-linked polyubiquitination at both K181 and K812 at the late phase of viral infection. In addition, mutation of K889 but not K96 to arginine abolished sumoylation of RIG-I in uninfected cells. In the same experiments, the basal K48-linked polyubiquitination of RIG-I, which mainly occurs at K812 was markedly increased by K889 mutation (Fig. 6 B), suggesting that sumoylation of RIG-I at K889 inhibits its K48-linked polyubiquitination at K812 and degradation in resting cells. In addition, mutation of K96 impaired SeV-induced sumoylation of RIG-I and increased its SeV-induced but not basal K48-linked polyubiquitination (Fig. 6 B), suggesting that SeV-induced sumoylation of RIG-I at K96 suppresses its K48-linked polyubiquitination at K181. Collectively, these results suggest that RIG-I is sumoylated at K889, which suppresses its K48-linked polyubiquitination at K812 and degradation in resting cells, and further sumoylated at K96, which suppresses its K48-linked polyubiquitination of K181 and degradation at the early phase of viral infection, followed by its desumoylation and K48-linked polyubiquitination at both K181 and K812 in late-infected cells (Fig. 6 C). Consistent with the biochemical results, mutation of K96 or K889 to arginine dramatically inhibited SeV-induced transcription of Ifnb1, whereas mutation of K181 or K812 markedly increased SeV-induced transcription of Ifnb1 in the reconstituted Rig-i−/− MEFs (Fig. 6 D).
Figure 6.
Physiological relationship of sumoylation and K48-linked polyubiquitination of RIG-I. (A) The K48-linked polyubiquitination of RIG-I and its mutants after SeV stimulation in reconstituted cells. Rig-i−/− MEFs reconstituted with RIG-I, or its mutants were infected with SeV for the indicated times, followed by immunoprecipitation and immunoblotting analysis. As described in Fig. 4 C, subinfection (K812R-L and K812/181R-L) with the RIG-I mutant-containing retroviruses was used to make the reconstituted RIG-I and mutants express at similar levels. (B) Dynamic sumoylation and K48-linked polyubiquitination of RIG-I and its mutants after SeV stimulation in reconstituted cells. Rig-i−/− MEFs reconstituted with RIG-I or its mutants were infected with SeV for the indicated times, followed by immunoprecipitation and immunoblotting analysis (top). As described in C, superinfection the RIG-I K889R mutant (K889R-H) containing retroviruses was used to make the reconstituted RIG-I and its mutants express at similar levels. (C) The schematic diagram for dynamic K48-linked polyubiquitination and sumoylation of RIG-I before and after viral infection. The crystal structures of RIG-I CARD (PDB:4P4H) and Helicase domains (PDB:5E3H) were obtained from the PDB database. Black, ubiquitination sites; red, sumoylation sites; blue, phosphorylation sites. (D) SeV-induced transcription of downstream antiviral genes in Rig-i−/− MEFs reconstituted with wild-type RIG-I or its mutants. The reconstituted cells were left uninfected or infected with SeV for 6 h before qPCR analysis. Data in D are from one representative experiment with three technical replicates (mean ± SD). All the experiments were repeated for three times.
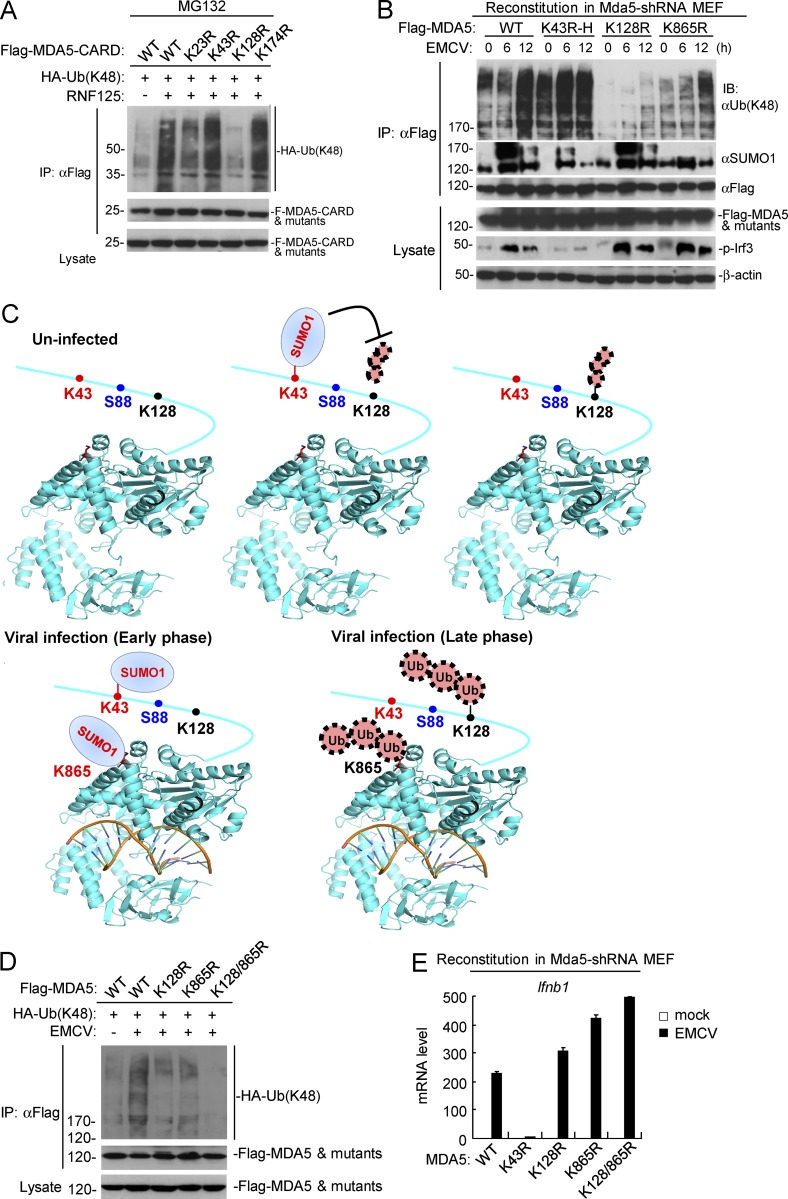
Mutagenesis and immunoprecipitation experiments indicated that RNF125-mediated K48-linked polyubiquitination of MDA5 at K128 (Fig. 7 A). As shown in Fig. 7 B, mutation of K43 but not K128 or K865 to arginine abolished the sumoylation of MDA5 in uninfected cells. In the same experiments, the basal K48-linked polyubiquitination of MDA5 was markedly increased by K43 mutation, abolished by K128 mutation and unchanged by K865 mutation, respectively, in uninfected cells (Fig. 7 B). These results suggest that K128 of MDA5 is modified by K48-linked polyubiquitination in resting cells, and sumoylation at K43 of MDA5 inhibits its K48-linked polyubiquitination at K128 and degradation in resting cells (Fig. 7 C). In addition, mutation of K128 of MDA5 attenuated EMCV-induced K48-linked polyubiquitination at the late phase of viral infection (12 h; Fig. 7 B), suggesting that MDA5 is modified with increased K48-linked polyubiquitination at K128 in late-infected cells. Interestingly, the K865 residue (as shown in K43R mutant) was markedly sumoylated at early phase (6 h), but decreased at late phase (12 h) after EMCV infection (Fig. 7 B). Mutation of K865 also markedly attenuated EMCV-induced K48-linked polyubiquitination of MDA5 at late phase of viral infection (12 h; Fig. 7 B), indicating that MDA5 is modified with vast K48-linked polyubiquitination at K865 in addition to K128 in late-infected cells. These results suggest that viral infection induces two-step sequential modifications of sumoylation and K48-linked polyubiquitination of MDA5 at K865 at early and late phase of viral infection, respectively (Fig. 7 C). Constantly, mutation of K128 and K865 individually or simultaneously markedly attenuated or completely abolished K48-linked polyubiquitination of MDA5, respectively (Fig. 7 D). These results suggest that viral infection induces K48-linked polyubiquitination of MDA5 at both K128 and K865. Consistent with the biochemical results, mutation of K43 to arginine dramatically inhibited EMCV-induced transcription of Ifnb1, whereas mutation of K128 or K865 markedly increased EMCV-induced transcription of Ifnb1 in the reconstituted MDA5-shRNA MEFs (Fig. 7 E).
Figure 7.
Physiological relationship of sumoylation and K48-linked polyubiquitination of MDA5. (A) Identification of the residue in MDA5-CARD targeted by RNF125 for K48-linked polyubiquitination. HEK293 cells were transfected with the indicated plasmids for 18 h, and then treated with MG132 for 6 h before immunoprecipitation and immunoblotting analysis. (B) Dynamic sumoylation and K48-linked polyubiquitination of MDA5 and its mutants after EMCV stimulation in reconstituted cells. MDA5-shRNA MEFs reconstituted with MDA5 or its mutants were infected with EMCV for the indicated times, followed by immunoprecipitation and immunoblotting analysis (top histogram). As described in Fig. 4 G, superinfection (K43R-H) or subinfection (K128R-L) of the MDA5 mutant-containing retroviruses was used to make the reconstituted MDA5 and its mutants express at similar levels. (C) The schematic diagram for dynamic K48-linked polyubiquitination and sumoylation of MDA5 before and after viral infection. The crystal structure of MDA5 Helicase domain (PDB: 4GL2) was obtained from the PDB database. Black, ubiquitination sites; red, sumoylation sites; blue, phosphorylation site. (D) Effects of simultaneous mutation of K128 and K865 on EMCV-induced K48-linked polyubiquitination of MDA5. HEK293 cells were transfected with the indicated plasmids for 24 h, and then cells were left uninfected or infected with EMCV for 12 h, followed by immunoprecipitation and immunoblotting analysis. (E) EMCV-induced transcription of downstream antiviral genes in MDA5-shRNA MEFs reconstituted with wild-type MDA5 or its mutants. The reconstituted cells were left uninfected or infected with EMCV for 6 h before qPCR analysis. Data in E are from one representative experiment with three technical replicates (mean ± SD). All the experiments were repeated three times.
Sumoylation of the CARDs of MDA5 and RIG-I facilitates their dephosphorylation by PP1 and activation
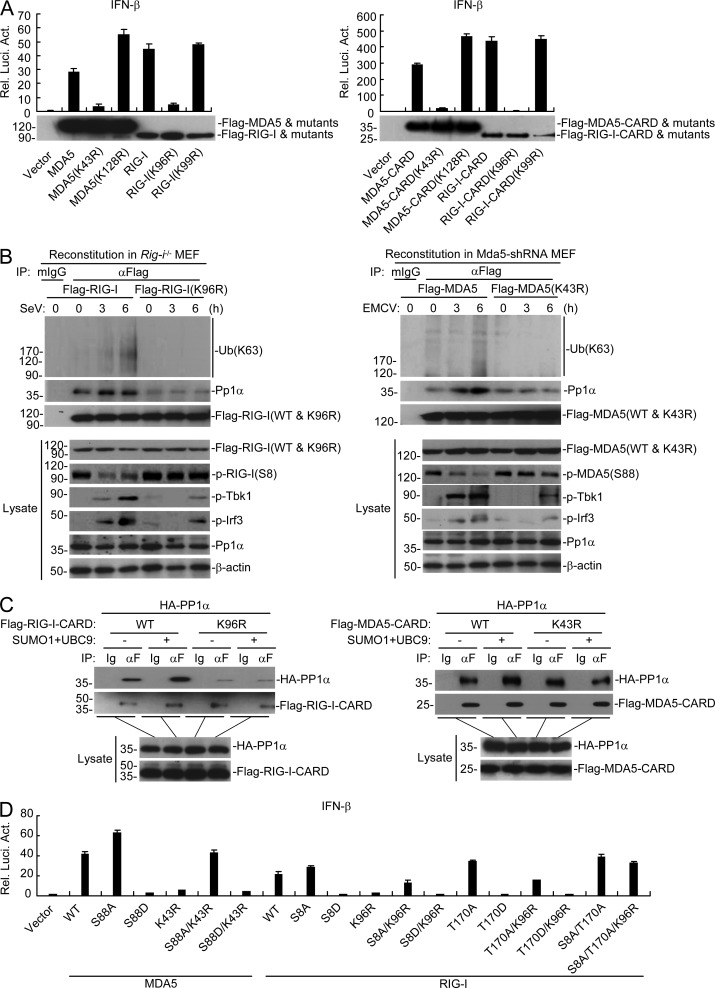
During our investigation on sumoylation of the CARDs of RIG-I and MDA5, we found that mutation of K96 of RIG-I–CARD or full-length RIG-I or mutation of K43 of MDA5-CARD or full-length MDA5 dramatically impaired their activation of the IFN-β promoter compared with their respective wild-type counterparts or other mutants in reporter assays (Fig. 8 A). These results suggest that sumoylation of RIG-I and MDA5 in their CARDs regulates their activation in addition to their stabilities. It has been reported that RIG-I and MDA5 are phosphorylated at S8/T170 and S88, respectively, to inhibit their activation in uninfected cells (Gack et al., 2010; Nistal-Villán et al., 2010; Maharaj et al., 2012; Wies et al., 2013), and viral infection induces their dephosphorylation mediated by PP1 and subsequent K63-linked polyubiquitination for their ultimate activation (Gack et al., 2007; Zeng et al., 2010; Wies et al., 2013). To determine which events are regulated by sumoylation of MDA5 and RIG-I, we examined their dephosphorylation and K63-linked polyubiquitination in reconstituted MEFs after SeV or EMCV infection, respectively. The results indicated that mutation of K96 of RIG-I or K43 of MDA5 had no marked effects on the basal phosphorylation of RIG-I or MDA5, but markedly impaired their dephosphorylation and K63-linked polyubiquitination induced by SeV or EMCV infection (Fig. 8 B). Consistently, mutation of K96 of RIG-I or K43 of MDA5 impaired their recruitment of PP1 after viral infection (Fig. 8 B), whereas sumoylation of RIG-I–CARD or MDA5-CARD promoted their interaction with PP1 in mammalian overexpression system (Fig. 8 C). Interestingly, mutation of K96 of RIG-I significantly impaired not only SeV-induced but also the constitutive association of RIG-I with PP1 (Fig. 8, B and C). Functionally, mutation of S8/T170 of RIG-I or S88 of MDA5 to aspartic acids, which mimic their constructive phosphorylated states abolished their activation of IFN-β, whereas mutation of S8/T170 of RIG-I and S88/MDA5 to alanines, which mimic their unphosphorylated states enhanced their activation of IFN-β, which is consistent with previous studies (Gack et al., 2010; Nistal-Villán et al., 2010; Maharaj et al., 2012; Wies et al., 2013). Interestingly, mutation of S8/T170 to alanines rescued the impaired activation of the IFN-β promoter mediated by RIG-I(K96R) (Fig. 8 D). Similarly, mutation of S88 of MDA5 to alanine rescued the impaired activation of the IFN-β promoter mediated by MDA5(K43R) (Fig. 8 D). These results suggest that sumoylation of the CARDs of RIG-I and MDA5 is required for their dephosphorylation and K63-linked polyubiquitination upon viral infection. Collectively, these results suggest that sumoylation of the CARDs of RIG-I and MDA5 potentiates their activation by facilitating their dephosphorylation mediated by PP1 and K63-linked polyubiquitination.
Figure 8.
Sumoylation of the CARDs of MDA5 and RIG-I is critical for their recruitment of PP1α. (A) Effects of MDA5, RIG-I, and their mutants on activation of the IFN-β promoter. HEK293 cells were transfected with the indicated plasmids for 24 h before luciferase assays. (B) Effects of sumoylation-defective mutation of the CARDs of RIG-I and MDA5 on their dephosphorylation, K63-linked polyubiquitination and recruitment of PP1α after viral infection. The reconstituted MEFs were left uninfected or infected with SeV or EMCV for the indicated times, followed by immunoprecipitation. The immunoprecipitates were divided into two equal portions, and one was used for immunoblot analysis with the PP1α antibody and the other was lysed in denaturing conditions and reimmunoprecipitated for endogenous ubiquitination detection. (C) Effects of sumoylation of RIG-I-CARD or MDA5-CARD on their interactions with PP1α. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by coimmunoprecipitation and immunoblotting analysis. (D) Effects of RIG-I-, MDA5- and their mutants on activation of the IFN-β promoter. HEK293 cells were transfected with the indicated plasmids for 24 h followed by luciferase assays. Data in A and d are from one representative experiment with three technical replicates (mean ± SD). All the experiments were repeated three times.
Desumoylation of MDA5 and RIG-I by SENP2 at the late phase of viral infection
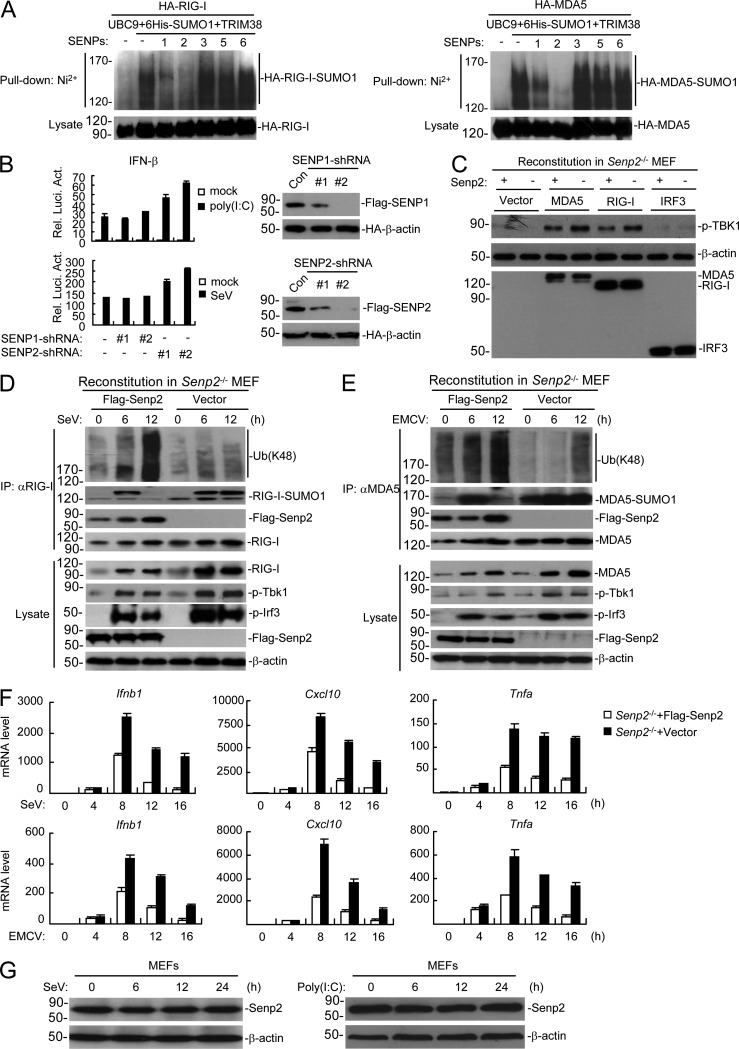
During our investigation of endogenous sumoylation of RIG-I and MDA5, we routinely observed that sumoylation of both RIG-I and MDA5 was decreased at the late phase of viral infection (Fig. 3, D and E). We hypothesized that RIG-I and MDA5 were desumoylated by a desumoylating enzyme. A screen of several SENP desumoylating enzymes indicated that SENP2 dramatically inhibited the sumoylation of RIG-I and MDA5 (Fig. 9 A). In these experiments, SENP1 had a much weaker ability, whereas other SENPs had no marked effects on the sumoylation of RIG-I and MDA5 (Fig. 9 A). Knockdown of SENP2 markedly potentiated SeV- or transfected poly(I:C)-induced activation of the IFN-β promoter, whereas knockdown of SENP1 had no marked effects (Fig. 9 B). Our previous work showed that SENP2 desumoylates IRF3 to inhibit virus-triggered IRF3 activation and type I IFNs induction (Ran et al., 2011). Interestingly, SENP2 deficiency potentiated both RIG-I– and MDA5-mediated phosphorylation and activation of TBK1, which functions upstream of IRF3 in RLR signaling pathways (Fig. 9 C). These results indicated that RIG-I and MDA5, in addition to IRF3, were indeed potential targets of SENP2. Furthermore, Senp2−/− MEFs showed sustained and increased sumoylation, dramatically reduced K48-linked polyubiquitination, and increased protein levels of RIG-I and MDA5, as well as sustained phosphorylation of Tbk1 and Irf3 in comparison to SENP2-reconstituted MEFs at the late phase of viral infection (Fig. 9, D and E). In these experiments, SENP2 was recruited to RIG-I and MDA5 at the late phase of viral infection in reconstituted cells (Fig. 9, D and E). Consistently, transcription of downstream antiviral genes including Ifnb1, Cxcl10, and Tnfa induced by SeV or EMCV infection was dramatically increased in Senp2−/− in comparison to SENP2-reconstituted MEFs at the late phase of viral infection (8–16 h; Fig. 9 F). Additional experiments indicated that the expression levels of SENP2 was not markedly changed after viral infection or poly(I:C) stimulation (Fig. 9 G), suggesting that the activity of SENP2 may be regulated by conformational changes, posttranslational modifications, or other factors rather than its expression level at the late phase of viral infection. Collectively, these results suggest that SENP2 desumoylates RIG-I and MDA5 and promotes their K48-linked polyubiquitination and degradation at the late phase of viral infection to avoid sustained activation of RIG-I and MDA5, as well as excessive innate immune response.
Figure 9.
Desumoylation of RIG-I and MDA5 by SENP2 at the late phase of viral infection. (A) Effects of SENPs on sumoylation of RIG-I and MDA5. HEK293 cells were transfected with the indicated plasmids for 24 h before Ni2+ pull-down assays and immunoblotting analysis. (B) Effects of knockdown of SENP1 and SENP2 on SeV- or poly(I:C)-induced activation of the IFN-β promoter. HEK293 cells were transfected with the indicated plasmids for 36 h, and then infected with SeV for 10 h or transfected with poly(I:C) for 18 h before luciferase assays. The knockdown efficiencies of SENP1 and SENP2 shRNAs are shown at the right panels. HEK293T cells were transfected with the indicated plasmids for 24 h followed by immunoblotting analysis. (C) Effects of SENP2 deficiency on RIG-I and MDA5-mediated activation of TBK1. The indicated proteins were transduced into SENP2- and vector-reconstituted SENP2−/− MEFs via retroviral approach, and then cells were harvested, followed by immunoblotting analysis with the indicated antibodies. (D and E) Effects of SENP2 deficiency on sumoylation and K48-linked polyubiquitination of RIG-I and MDA5. Senp2−/− or SENP2-reconstituted MEFs were left uninfected or infected with SeV (D) or EMCV (E) for the indicated times, followed by immunoprecipitation and immunoblotting analysis. (F) Effects of SENP2 deficiency on SeV- and EMCV-induced transcription of downstream antiviral genes. Senp2−/− or SENP2-reconstituted MEFs were left uninfected or infected with SeV or EMCV for the indicated times before qPCR analysis. (G) Effects of viral infection and poly(I:C)-transfected on expression of SENP2. Cells were infected with SeV (left) or transfected with poly(I:C) (right) for the indicated times before lysed for immunoblotting analysis with the indicated antibodies. Data in B and F are from one representative experiment with three technical replicates (mean ± SD). All the experiments were repeated three times.
Discussion
In this study, we identified TRIM38 as a RIG-I– and MDA5-associated protein. In contrast to its negative regulatory roles in TLR3/4-mediated or TNF/IL-1-triggered signaling (Hu et al., 2014, 2015), TRIM38 positively regulates RIG-I– and MDA5-mediated signaling and induction of downstream antiviral genes. Gene knockout in mice suggests that Trim38 is essential for efficient induction of type I IFNs, proinflammatory cytokines, and other downstream antiviral genes, as well as for host defense against RNA viruses in vivo.
Biochemical analysis suggests that Trim38 acts as a SUMO1 E3 ligase for RIG-I and MDA5. Mutagenesis indicates that Trim38 catalyzes the sumoylation of RIG-I at K96/K889 and MDA5 at K43/K865. These sumoylation are impaired in Trim38-deficient cells both before and after viral infection, further confirming that RIG-I and MDA5 are sumoylated by Trim38. Interestingly, reconstitution experiments indicate that the sumoylations of RIG-I and MDA5 are dynamically regulated. In uninfected cells, K889 of RIG-I is basally sumoylated. Upon viral infection, the sumoylation at K889 is enhanced and K96 is further sumoylated. Similarly, K43 of MDA5 is basally sumoylated in uninfected cells and K865 is further sumoylated upon viral infection.
The differential sumoylations of RIG-I and MDA5 are important for maintaining their stability in uninfected and early-infected cells, respectively. Trim38 deficiency markedly impairs stability of RIG-I and MDA5, suggesting an important role of Trim38-mediated sumoylation in the regulation of their stability. Several observations suggest that Trim38-mediated dynamic sumoylations of MDA5 and RIG-I maintain their stability by antagonizing their K48-linked polyubiquitination and degradation. First, sumoylations of MDA5, MDA5-CARD and MDA5-C markedly suppressed their respective K48-linked polyubiquitination. Second, mutation of K43 abolished the sumoylation of MDA5 in uninfected cells and attenuated EMCV-induced sumoylation in early-infected cells, but increased the basal and EMCV-induced K48-linked polyubiquitination of MDA5. Third, MDA5 was sumoylated at K865 in early-infected cells and desumoylated at the late phase of viral infection when K48-linked polyubiquitination of MDA5 at the same residue was readily detected. Fourth, reconstitution of SENP2 into Senp2−/− MEFs markedly abolished the sustained sumoylation of MDA5 but facilitated their K48-linked polyubiquitination at the late phase of viral infection. Similarly, our results suggest that dynamic sumoylation of RIG-I also maintains its stability by antagonizing its K48-linked polyubiquitination and degradation.
Previously, it has been reported that TRIM38 negatively regulates TLR- and RLR-mediated induction of inflammatory cytokines and type I IFNs by targeting TRAF6 and NAP1 for K48-linked polyubiquitination and proteasomal degradation in mouse cell line RAW264.7 (Zhao et al., 2012a,b). However, using genetic and biochemical approaches, we found that TRIM38 positively regulates RLR-mediated signaling, which is contradictory with the previous studies. In addition, two recent studies have demonstrated that TRIM38 targets TRIF but not TRAF6 or NAP1 for ubiquitination and degradation, leading to inhibition of TLR3/4-mediated innate immune responses (Xue et al., 2012; Hu et al., 2015).
Our experiments indicate that Trim38-mediated sumoylations of MDA5 and RIG-I are not only important for antagonizing their K48-linked polyubiquitination and degradation in uninfected and early-infected cells, they are also required for activation of RIG-I and MDA5. Previously, it has been demonstrated that dephosphorylation of RIG-I and MDA5 by the phosphatase PP1 after viral infection is critical for their activation (Wies et al., 2013). Our experiments indicated that mutation of K96 of RIG-I or K43 of MDA5 impaired their dephosphorylation triggered by viral infection. In addition, mutation of S8 and T170 to alanines rescued the impaired ability of RIG-I(K96R) to activate the IFN-β promoter, respectively. Similarly, mutation of S88 of MDA5 to alanines rescued the impaired ability of MDA5(K43R) to activate the IFN-β promoter. Consistently, mutation of K96 of RIG-I or K43 of MDA5 impaired their recruitments of PP1 upon viral infection, whereas sumoylation of the CARDs of RIG-I and MDA5 potentiated their interactions with PP1. Furthermore, mutation of K96 of RIG-I or K43 of MDA5 also abolished their K63-linked polyubiquitination and ultimate activation after viral infection.
Interestingly, although both RIG-I and MDA5 are sumoylated by Trim38 before and after viral infection, and both of their sumoylations contribute to their stability and activation, the patterns of their sumoylations are different. RIG-I is basally sumoylated in the CTT and further sumoylated in the CARD, whereas MDA5 is sumoylated in the opposite way. Previously, it has been demonstrated that RIG-I but not MDA5 forms “closed” conformation in the absence of viral infection (Saito et al., 2007), which may contribute to the distinct patterns of sumoylation of RIG-I and MDA5.
In our study, we also identified SENP2 as a desumoylating enzyme for RIG-I and MDA5. Senp2−/− MEFs showed sustained and increased sumoylation of RIG-I and MDA5, dramatically reduced K48-linked polyubiquitination of RIG-I and MDA5, increased protein levels of RIG-I and MDA5, sustained phosphorylations of Tbk1 and Irf3, and increased transcription of downstream antiviral genes in comparison to SENP2-reconstituted MEFs at the late phase of viral infection. These results suggest that SENP2 desumoylates RIG-I and MDA5 at the late phase of viral infection to avoid sustained activation of RIG-I and MDA5, as well as excessive innate immune response.
Based on our results, we propose a working model on the regulation of RIG-I– and MDA5-mediated innate immune responses to RNA viruses by divergent and dynamic posttranslational modifications. In the absence of viral infection, Trim38 catalyzes sumoylation of RIG-I at K889 or MDA5 at K43, which inhibits their K48-linked polyubiquitination at K812 or K128, respectively, as well as degradation by the ubiquitin–proteasomal pathways, and ensures proper levels of RIG-I and MDA5 for initiation of innate immune signaling upon viral infection. Meanwhile, RIG-I and MDA5 are phosphorylated in their CARDs to suppress their activation in resting cells (Gack et al., 2010; Nistal-Villán et al., 2010; Maharaj et al., 2012). Upon viral infection, Trim38 further catalyzes sumoylation of RIG-I at K96 or MDA5 at K43/K865. Sumoylation of RIG-I at K96 or MDA5 at K43 facilitates their recruitments of PP1 and dephosphorylation, followed by their K63-linked polyubiquitination and ultimate activation. Additionally, sumoylation of RIG-I at K96 suppresses its K48-linked polyubiquitination at K181 at the early phase of viral infection. Similarly, sumoylation of MDA5 at K43 and K865 suppresses its K48-linked polyubiquitination at K128 and K865, respectively, at the early phase of viral infection. These ensure that proper levels of RIG-I and MDA5 are activated for recruitment of the mitochondrial-associated adaptor protein VISA, followed by ultimate induction of downstream antiviral genes and efficient innate antiviral immune response. At the late phase of viral infection, SENP2 is recruited to RIG-I or MDA5 and desumoylates them at K43/K865 or K96/K889, respectively. The desumoylation of RIG-I and MDA5 leads to their K48-linked polyubiquitination at K181/K812 or K128/K865, respectively, and degradation by the ubiquitin-proteasomal pathways, and efficiently turning off viral RNA-triggered induction of downstream antiviral genes, as well as innate immune response.
Materials and methods
Reagents, antibodies, viruses, and cells
The following reagents were used: GM-CSF (PeproTech); poly(I:C) and poly(I:C)-LMW (InvivoGen); cycloheximide (CHX), MG132, N-ethylmaleimide (NEM; Sigma-Aldrich); Lipofectamine 2000 (Invitrogen); polybrene (EMD Millipore); SYBR (Bio-Rad laboratories); RNase inhibitor (Thermo Fisher Scientific); ELISA kit for murine Ifn-β (PBL); ELISA kit for murine Tnfα (BioLegend); mouse monoclonal antibodies against HA (Covance); Flag and β-actin (Sigma-Aldrich); phospho-IκBα (S536; Cell Signaling Technology); rabbit polyclonal antibodies against phospho-IRF3(S396; Cell Signaling Technology), phospho-TBK1(S172; Abcam), SUMO1 (Abclone Biotechnology), K63-lined polyubiquitin and K48-linked polyubiquitin (EMD Millipore) were purchased from the indicated manufacturers. The phosphorylation antibodies including phospho-MDA5(S88), phospho-RIG-I(S8) and phospho-RIG-I(T170) were provided by M. Gack (The University of Chicago, Chicago, IL). Mouse antisera against RIG-I and MDA5 were raised using recombinant RIG-I(1–200) and MDA5(1–200), respectively. EMCV was provided by H.-C. Yang (China Agricultural University, Beijing, China). NDV, VSV, and SeV were previously described (Zhou et al., 2014). HEK293 cells and HFFs were obtained from ATCC. HEK293T cells were originally provided by Dr. Gary Johnson (National Jewish Health). Rig-i−/− and Senp2−/− MEFs were previously described (Ran et al., 2011; Li et al., 2012). Primary Trim38+/+ and Trim38−/− BMDMs, BMDCs and MLFs were prepared as previously described (Hu et al., 2015).
Constructs
Expression plasmids for RIG-I, MDA5, VISA, TBK1, TRIM38, SENP2, UBC9 and SUMO1 were previously described (Li et al., 2009; Chen et al., 2010; Zhong et al., 2010; Ran et al., 2011; Hu et al., 2014). Expression plasmids for other SENPs except SENP2 as well as SUMO2 and SUMO3 were provided by Dr. Chen Wang (Shanghai Institutes of Biological Sciences, Shanghai, China). Flag- or HA-tagged human and murine MDA5 and RIG-I mutants were constructed by standard molecular biology techniques.
Identification of MDA5-associated proteins
The empty vector- and MDA5-Flag–transduced stable HEK293 cells (5 × 107) were lysed, and then the cell lysates were subjected to immunoprecipitation with anti-Flag antibody. The immunoprecipitates were eluted with 3xFlag peptides, and then subjected to reimmunoprecipitation with anti-MDA5. The anti-MDA5–associated proteins were eluted and digested by trypsin in solution. The tryptic peptides were analyzed by HPLC-ESI/MS/MS with a Finnigan LTQ (Thermo Fisher Scientific) adapted for nanospray ionization. The tandem spectra were searched against Homo sapiens National Center for Biotechnology Information reference database using the SEQUEST. Results was filtered by Xcorr +1 > 1.9, +2 > 2.2, +3 > 3.5, sp > 500, Deltcn > 0.1, Rsp ≤ 5. The candidates were identified by subtraction of the identified proteins of the empty vector–transfected cells from that of the MDA5-Flag–transduced cells.
Trim38 knockout mice
Trim38 knockout mice and the genotyping methods were previously described (Hu et al., 2015). All animal experiments were performed in accordance with the Wuhan University animal care and use committee guidelines.
Transfection
HEK293T cells were transfected by standard calcium phosphate precipitation method. HFFs, BMDMs, L929 cells, and MLFs were transfected by Lipofectamine 2000 according to procedures recommended by the manufacturer.
Pseudotyped retroviral-mediated gene transfer
Establishment of MDA5-shRNA stable cell line in MEFs, reconstitution of MDA5 and its mutants into established MDA5-shRNA MEFs or reconstitution of RIG-I and its mutants into Rig-i−/− MLFs was performed by pseudotyped retroviral-mediated gene transfer. In brief, HEK293T cells plated on 100-mm dishes were transfected with the indicated retroviral MDA5-shRNA or expression plasmid (10 µg) together with the pGag-pol (10 µg) and the pVSV-G (3 µg) plasmids. 2 d after transfection, the viruses were harvested and used to infect the targeted cells in the presence of polybrene (8 µg/ml). The infected cells were selected with puromycin (1 µg/ml) for at least 2 d. During reconstitution, pseudotyped retroviruses containing the empty vectors were used for control infection to ensure that all of the cells receive identical amounts of retroviral infection.
RNAi or shRNA
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the pSuper.Retro-RNAi plasmid (Oligoengine). The following sequence was targeted for MDA5 mRNA: 5′-GCTTCAGGAATCTCATCTTAT-3′. TRIM38-, SENP1-, and SENP2-shRNA plasmids were previously described (Ran et al., 2011; Hu et al., 2014).
Ni2+ pull-down assays
Cells cultured in 6-cm plates were transfected with the indicated plasmids. 24 h after transfection, cells from each plate were collected and divided into two aliquots. One aliquot was lysed in lysis buffer and analyzed by immunoblotting analysis to examine the expression of transfected proteins. Another aliquot was lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-Cl, pH 8.0, 5 mM imidazole, and 10 mM β-mercaptoethanol), and incubated with Ni2+-NTA beads (QIAGEN) for 4 h at room temperature or overnight at 4°C. The beads were washed sequentially with buffers A, B (8 M urea, 0.1 M Na2PO4/NaH2PO4, 10 mM Tris-HCl, pH 8.0, and 10 mM β-mercaptoethanol), and C (same as B except pH 6.3). Beads with bound proteins were then boiled in SDS sample buffer, and the proteins were fractioned by SDS-PAGE and analyzed by immunoblotting.
Coimmunoprecipitation, endogenous sumoylation, and ubiquitination
Cells were lysed with RIPA buffer plus complete protease inhibitors and 20 mM NEM, and lysates were sonicated for 1 min. The lysates were centrifuged at 14,000 rpm for 20 min at 4°C. The supernatants were incubated with respective antibodies at 4°C overnight before protein G beads were added for 2 h. The beads were washed with cold PBS plus 0.5 M NaCl for three times followed by an additional wash with PBS. Proteins were separated by 8% SDS–PAGE, followed by immunoblotting analysis with the indicated antibodies.
To detect endogenous sumoylation and ubiquitination, the immunoprecipitates were reextracted in lysis buffer containing 1% SDS and denatured by heating for 5 min. The supernatants were diluted with regular lysis buffer until the concentration of SDS was decreased to 0.1%, followed by reimmunoprecipitation with the indicated antibodies. The immunoprecipitates were analyzed by immunoblotting with the ubiquitin antibody.
qPCR
Total RNA was isolated for qPCR analysis to measure mRNA levels of the indicated genes. Data shown are the relative abundance of the indicated mRNA normalized to that of Gapdh. The sequences of the primers used for qPCR analysis of murine Ifnb1, Il6, Tnfa, Cxcl10, Trim38, and Gapdh mRNAs have been previously described (Hu et al., 2015).
Plaque assays
The HEK293 cells (1 × 105) were transfected with the indicated plasmids for 24 h followed by VSV infection (MOI of 0.1), and then cells were washed with PBS for three times for removal of uninfected virus, followed by replacement of new complete medium 1 h after viral infection. After 24 h, the supernatants were collected and diluted to a series of concentrates, followed by subjection to infect confluent Vero cells cultured on 24-well plates. At 1 h after infection, supernatant was removed and 3% methylcellulose was overplayed. At 3 d after infection, overlay was removed, cells were fixed with 4% formaldehyde for 1 h and stained with 0.2% Crystal violet in 20% methanol. Plaques were counted, averaged and multiplied by the dilution factor to determine viral titer as pfu/ml.
Statistics
For mouse experiments, no specific blinding method was used, but mice in each sample group were selected randomly. The sample size (n) of each experimental group is described in each corresponding figure legend. GraphPad Prism software was used for all statistical analyses. Quantitative data displayed as histograms are expressed as means ± SD. Data were analyzed using an unpaired Student’s t test.
Acknowledgments
We thank Dr. Lei Yin for analysis of crystal structures of RIG-I and MDA5, and Yu Zhao for technical assistance on animal experiments.
This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0502102 and 2014CB910103), the National Natural Science Foundation of China (31630045, 31521091, and 91429304), and National Postdoctoral Program for Innovative Talents (BX201600116).
The authors declare no competing financial interests.
Author contributions: H.-B. Shu and M.-M. Hu conceived and designed the study; M.-M. Hu, C.-Y. Liao, Q. Yang, and X.-Q. Xie performed the experiments; H.-B. Shu and M.-M. Hu analyzed the data. H.-B. Shu and M.-M. Hu wrote the manuscript.
Footnotes
Abbreviations:
- CARD
- caspase recruitment domain
- EMCV
- encephalomyocarditis virus
- IRF3
- IFN regulatory factor 3
- MITA
- mitochondrial mediator of IRF3 activation
- MDA5
- melanoma differentiation-associated gene 5
- MLF
- mouse lung fibroblast
- NDV
- Newcastle disease virus
- PAMP
- pathogen-associated molecular pattern
- PRR
- pathogen recognition receptor
- RLR
- RIG-I-like receptor
- RIG-I
- retinoic acid-inducible gene 1 protein
- SeV
- Sendai virus
- TRAF
- TNF receptor–associated factor
- VISA
- virus-induced signaling adaptor
- VSV
- vesicular stomatitis virus
References
- Akira S., Uematsu S., and Takeuchi O.. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., and Shimotohno K.. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA. 104:7500–7505. 10.1073/pnas.0611551104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S., Ricci E.P., Mercier B.C., Moore M.J., and Fitzgerald K.A.. 2014. Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14:361–376. 10.1038/nri3682 [DOI] [PubMed] [Google Scholar]
- Chen R., Zhang L., Zhong B., Tan B., Liu Y., and Shu H.B.. 2010. The ubiquitin-specific protease 17 is involved in virus-triggered type I IFN signaling. Cell Res. 20:802–811. 10.1038/cr.2010.41 [DOI] [PubMed] [Google Scholar]
- Chen W., Han C., Xie B., Hu X., Yu Q., Shi L., Wang Q., Li D., Wang J., Zheng P., et al. 2013. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 152:467–478. 10.1016/j.cell.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Chu Y., and Yang X.. 2011. SUMO E3 ligase activity of TRIM proteins. Oncogene. 30:1108–1116. 10.1038/onc.2010.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., and Jung J.U.. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 446:916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- Gack M.U., Nistal-Villán E., Inn K.S., García-Sastre A., and Jung J.U.. 2010. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J. Virol. 84:3220–3229. 10.1128/JVI.02241-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürtler C., and Bowie A.G.. 2013. Innate immune detection of microbial nucleic acids. Trends Microbiol. 21:413–420. 10.1016/j.tim.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Jiao S., Shi Z., Li C., Meng X., Zhang Z., Wang Y., Song X., Wang W., Zhang R., et al. 2015. A non-canonical role of the p97 complex in RIG-I antiviral signaling. EMBO J. 34:2903–2920. 10.15252/embj.201591888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18:483–490. 10.1016/j.cytogfr.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., and Chen Z.J.. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 146:448–461. 10.1016/j.cell.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M.M., Yang Q., Zhang J., Liu S.M., Zhang Y., Lin H., Huang Z.F., Wang Y.Y., Zhang X.D., Zhong B., and Shu H.B.. 2014. TRIM38 inhibits TNFα- and IL-1β-triggered NF-κB activation by mediating lysosome-dependent degradation of TAB2/3. Proc. Natl. Acad. Sci. USA. 111:1509–1514. 10.1073/pnas.1318227111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M.M., Xie X.Q., Yang Q., Liao C.Y., Ye W., Lin H., and Shu H.B.. 2015. TRIM38 negatively regulates TLR3/4-mediated innate immune and inflammatory responses by two sequential and distinct mechanisms. J. Immunol. 195:4415–4425. 10.4049/jimmunol.1500859 [DOI] [PubMed] [Google Scholar]
- Hu M.M., Yang Q., Xie X.Q., Liao C.Y., Lin H., Liu T.T., Yin L., and Shu H.B.. 2016. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 45:555–569. 10.1016/j.immuni.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., and Akira S.. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- Lei C.Q., Zhong B., Zhang Y., Zhang J., Wang S., and Shu H.B.. 2010. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 33:878–889. 10.1016/j.immuni.2010.11.021 [DOI] [PubMed] [Google Scholar]
- Li Y., Li C., Xue P., Zhong B., Mao A.P., Ran Y., Chen H., Wang Y.Y., Yang F., and Shu H.B.. 2009. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. USA. 106:7945–7950. 10.1073/pnas.0900818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen R., Zhou Q., Xu Z., Li C., Wang S., Mao A., Zhang X., He W., and Shu H.B.. 2012. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc. Natl. Acad. Sci. USA. 109:11770–11775. 10.1073/pnas.1203405109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Deng H., Li X., Wu X., Tang Q., Chang T.H., Peng H., Rauscher F.J. III, Ozato K., and Zhu F.. 2011. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 187:4754–4763. 10.4049/jimmunol.1101704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y.T., Sun L., and Chen Z.J.. 2013. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife. 2:e00785 10.7554/eLife.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., and Gale M. Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity. 34:680–692. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharaj N.P., Wies E., Stoll A., and Gack M.U.. 2012. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J. Virol. 86:1358–1371. 10.1128/JVI.06543-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., and Tschopp J.. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 437:1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Muller S., Berger M., Lehembre F., Seeler J.S., Haupt Y., and Dejean A.. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275:13321–13329. 10.1074/jbc.275.18.13321 [DOI] [PubMed] [Google Scholar]
- Nistal-Villán E., Gack M.U., Martínez-Delgado G., Maharaj N.P., Inn K.S., Yang H., Wang R., Aggarwal A.K., Jung J.U., and García-Sastre A.. 2010. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J. Biol. Chem. 285:20252–20261. 10.1074/jbc.M109.089912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y., Liu T.T., Zhou Q., Li S., Mao A.P., Li Y., Liu L.J., Cheng J.K., and Shu H.B.. 2011. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J. Mol. Cell Biol. 3:283–292. 10.1093/jmcb/mjr020 [DOI] [PubMed] [Google Scholar]
- Saito T., Hirai R., Loo Y.M., Owen D., Johnson C.L., Sinha S.C., Akira S., Fujita T., and Gale M. Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA. 104:582–587. 10.1073/pnas.0606699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., and Chen Z.J.. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 122:669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Reich B., and Will H.. 1999. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274:12555–12566. 10.1074/jbc.274.18.12555 [DOI] [PubMed] [Google Scholar]
- Versteeg G.A., Rajsbaum R., Sánchez-Aparicio M.T., Maestre A.M., Valdiviezo J., Shi M., Inn K.S., Fernandez-Sesma A., Jung J., and García-Sastre A.. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 38:384–398. 10.1016/j.immuni.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies E., Wang M.K., Maharaj N.P., Chen K., Zhou S., Finberg R.W., and Gack M.U.. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 38:437–449. 10.1016/j.immuni.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., and Shu H.B.. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 19:727–740. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Xue Q., Zhou Z., Lei X., Liu X., He B., Wang J., and Hung T.. 2012. TRIM38 negatively regulates TLR3-mediated IFN-β signaling by targeting TRIF for degradation. PLoS One. 7:e46825 10.1371/journal.pone.0046825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhou F., Fu C., Xu Y., and Yao X.. 2006. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 34(Web Server):W254-7 10.1093/nar/gkl207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Li Q., Mao A.P., Hu M.M., and Shu H.B.. 2014. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 6:154–163. 10.1093/jmcb/mju005 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., and Fujita T.. 2008. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 29:178–181. 10.1016/j.immuni.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., and Chen Z.J.. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 141:315–330. 10.1016/j.cell.2010.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Wang L., Zhang M., Wang P., Yuan C., Qi J., Meng H., and Gao C.. 2012a Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-β production and antiviral response by targeting NAP1. J. Immunol. 188:5311–5318. 10.4049/jimmunol.1103506 [DOI] [PubMed] [Google Scholar]
- Zhao W., Wang L., Zhang M., Yuan C., and Gao C.. 2012b E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J. Immunol. 188:2567–2574. 10.4049/jimmunol.1103255 [DOI] [PubMed] [Google Scholar]
- Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H.B.. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 29:538–550. 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Zhong B., Zhang Y., Tan B., Liu T.T., Wang Y.Y., and Shu H.B.. 2010. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J. Immunol. 184:6249–6255. 10.4049/jimmunol.0903748 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Lin H., Wang S., Wang S., Ran Y., Liu Y., Ye W., Xiong X., Zhong B., Shu H.B., and Wang Y.Y.. 2014. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe. 16:450–461. 10.1016/j.chom.2014.09.006 [DOI] [PubMed] [Google Scholar]