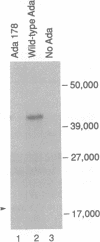
Abstract
The adaptive response of Escherichia coli protects cells against the mutagenic and toxic effects of alkylating agents. This response is controlled by the Ada protein, which not only functions as the transcriptional activator of the ada and alkA genes but also possesses two DNA methyltransferae activities. Ada is converted into an efficient transcriptional activator by transferring a methyl group from a DNA methylphosphotriester to its own Cys-69 residue and then binds to a DNA sequence (the Ada box) present in both the ada and alkA promoters. Although the Ada protein initially appeared to regulate the ada and alkA genes in a similar fashion, our studies show that the wild-type Ada protein and its truncated derivatives can differentially regulate ada and alkA transcription. In vivo, lower levels of wild-type methylated Ada are needed to activate ada transcription than alkA transcription. In cells exposed to alkylating agents, the N-terminal half of Ada, which contains the DNA-binding domain, is sufficient for efficient activation of alkA, but not ada, transcription. Moreover, truncated derivatives containing 80-90% of Ada are extremely strong constitutive activators of ada but are only inducible activators of alkA transcription. These results suggest that the mechanism by which Ada activates ada transcription differs from that by which it activates alkA transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Demple B., Jacobsson A., Olsson M., Robins P., Lindahl T. Repair of alkylated DNA in Escherichia coli. Physical properties of O6-methylguanine-DNA methyltransferase. J Biol Chem. 1982 Nov 25;257(22):13776–13780. [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Godowski P. J., Rusconi S., Miesfeld R., Yamamoto K. R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987 Jan 22;325(6102):365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- Hughes S. J., Sedgwick B. The adaptive response to alkylation damage. Constitutive expression through a mutation in the coding region of the ada gene. J Biol Chem. 1989 Dec 15;264(35):21369–21375. [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987 Jul 3;50(1):143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Lemotte P. K., Walker G. C. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985 Mar;161(3):888–895. doi: 10.1128/jb.161.3.888-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Long S., McCune S., Walker G. C. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J Bacteriol. 1988 Sep;170(9):4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- McCarthy T. V., Karran P., Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984 Mar;3(3):545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985 Apr 25;13(8):2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Lee N. L. Activation of ara operons by a truncated AraC protein does not require inducer. Proc Natl Acad Sci U S A. 1990 May;87(10):3708–3712. doi: 10.1073/pnas.87.10.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tokumoto Y., Sakumi K., Koike G., Nakabeppu Y., Sekiguchi M. Expression of the ada gene of Escherichia coli in response to alkylating agents. Identification of transcriptional regulatory elements. J Mol Biol. 1988 Aug 5;202(3):483–494. doi: 10.1016/0022-2836(88)90280-x. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol Gen Genet. 1983;191(3):466–472. doi: 10.1007/BF00425764. [DOI] [PubMed] [Google Scholar]
- Sedgwick B., Robins P., Totty N., Lindahl T. Functional domains and methyl acceptor sites of the Escherichia coli ada protein. J Biol Chem. 1988 Mar 25;263(9):4430–4433. [PubMed] [Google Scholar]
- Shevell D. E., Friedman B. M., Walker G. C. Resistance to alkylation damage in Escherichia coli: role of the Ada protein in induction of the adaptive response. Mutat Res. 1990 Nov-Dec;233(1-2):53–72. doi: 10.1016/0027-5107(90)90151-s. [DOI] [PubMed] [Google Scholar]
- Shevell D. E., LeMotte P. K., Walker G. C. Alteration of the carboxyl-terminal domain of Ada protein influences its inducibility, specificity, and strength as a transcriptional activator. J Bacteriol. 1988 Nov;170(11):5263–5271. doi: 10.1128/jb.170.11.5263-5271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K., Nakabeppu Y., Sekiguchi M. Functional sites of the Ada regulatory protein of Escherichia coli. Analysis by amino acid substitutions. J Mol Biol. 1988 May 20;201(2):261–271. doi: 10.1016/0022-2836(88)90137-4. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Volkert M. R., Nguyen D. C., Beard K. C. Escherichia coli gene induction by alkylation treatment. Genetics. 1986 Jan;112(1):11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikai T., Nakabeppu Y., Sekiguchi M. Proteolytic cleavage of Ada protein that carries methyltransferase and transcriptional regulator activities. J Biol Chem. 1988 Dec 15;263(35):19174–19180. [PubMed] [Google Scholar]