Abstract
RNA silencing is a conserved regulatory mechanism in fungi, plants and animals that regulates gene expression and defence against viruses and transgenes1. Small silencing RNAs of ~20–30 nucleotides and their associated effector proteins, the Argonaute family proteins, are the central components in RNA silencing2. A subset of small RNAs, such as microRNAs and small interfering RNAs (siRNAs) in plants, Piwi-interacting RNAs in animals and siRNAs in Drosophila, requires an additional crucial step for their maturation; that is, 2′-O-methylation on the 3′ terminal nucleotide3–6. A conserved S-adenosyl-l-methionine-dependent RNA methyltransferase, HUA ENHANCER 1 (HEN1), and its homologues are responsible for this specific modification3–5,7,8. Here we report the 3.1 Å crystal structure of full-length HEN1 from Arabidopsis in complex with a 22-nucleotide small RNA duplex and cofactor product S-adenosyl-l-homocysteine. Highly cooperative recognition of the small RNA substrate by multiple RNA binding domains and the methyltransferase domain in HEN1 measures the length of the RNA duplex and determines the substrate specificity. Metal ion coordination by both 2′ and 3′ hydroxyls on the 3′-terminal nucleotide and four invariant residues in the active site of the methyltransferase domain suggests a novel Mg2+-dependent 2′-O-methylation mechanism.
HEN1 was first identified in a genetic screen as a floral pattering gene and later found to be essential for Arabidopsis microRNA (miRNA) accumulation in vivo9,10. Subsequently, HEN1 was demonstrated to be a methyltransferase for miRNAs and all types of siRNAs in plants3,11. The 2′-O-methylation protects miRNAs and siRNAs from 3′-end uridylation and 3′-to-5′ exonuclease-mediated degradation in Arabidopsis12,13. The plant HEN1 and its animal homologues share a highly conserved methyltransferase (MTase) domain14 (Fig. 1e) that is not closely related to any known RNA 2′-O-MTases according to a phylogenetic analysis15. Two putative RNA binding modules, a double-stranded RNA binding domain (dsRBD) and a La motif have been identified in the amino-terminal region of HEN1 (ref. 15). To understand the specific recognition of small RNA substrates and the molecular mechanism of the 3′-end 2′-OH-specific methylation by HEN1 and its homologues, we determined the crystal structure of full-length Arabidopsis HEN1 in complex with a small RNA duplex in the presence of the cofactor product adenosyl-L-homocysteine (AdoHcy).
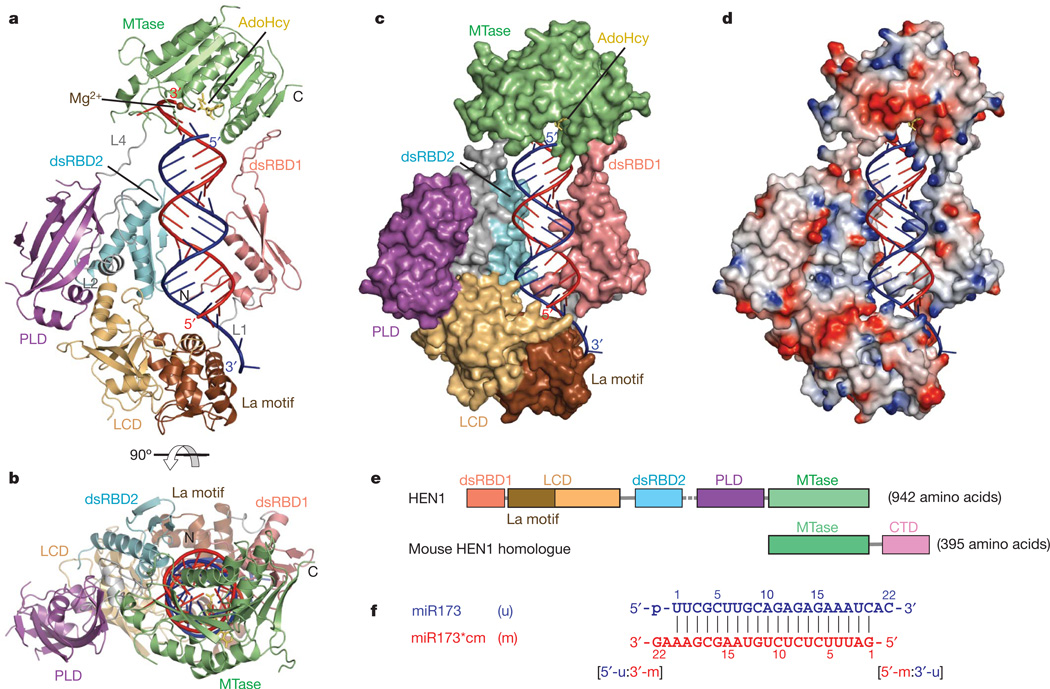
Figure 1. Structures of HEN1 in complex with a small RNA duplex and AdoHcy.
a, Ribbon diagram of the complex. dsRBD, violet; La motif, chocolate; LCD, wheat; dsRDB2, cyan; PLD, purple; MTase, green; linkers including L1, L2 and L4, grey; RNA strand to be methylated (m strand), red; RNA strand not to be methylated (u strand), blue; AdoHcy, yellow; Mg2+, brown. b, Ribbon diagram of the complex rotated by 90° about the horizontal axis relative to a. c, d, Surface and surface charge views of HEN1 in the complex in the same orientation as a. e, Schematic representation of the domains in HEN1 with the same colour codes as in a. The disordered L3 is represented by the dashed line. f, Sequences of the small RNA duplex used in the co-crystallization. The m strand and u strand are coloured red and blue, respectively, and the two termini, [5′-m:3′-u] and [5′-u:3′-m], are indicated.
The recombinant full-length Arabidopsis HEN1 (residue 1–942) was co-crystallized with AdoHcy and a 22-nucleotide small RNA duplex containing a fully complementary 20-nucleotide segment (Fig. 1f) derived from a natural substrate of HEN1, miR173/miR173* (refs 3, 11) (Supplementary Fig. 2c). The crystal structure was determined at 3.1 Å as described in Methods. The structure revealed that Arabidopsis HEN1 binds to the small RNA substrate as a monomer (Fig. 1), which is supported by results from gel filtration experiments (Supplementary Fig. 2). The small RNA substrate exhibits an A-form conformation in the ternary complex structure and both duplex termini are specifically recognized by HEN1. The HEN1 protein consists of five structural domains (Fig. 1e), four of which directly interact with the small RNA substrate (Fig. 1a–c) with the exception of the PPIase-like domain (PLD) which shows a high degree of structural similarity to well characterized FK506-binding proteins16. The A-form duplex of the small RNA substrate is bound by two double-stranded RNA (dsRNA)-specific binding domains17, dsRBD1 and dsRBD2. The [5′-m:3′-u] terminus containing the 3′-end 2-nucleotide overhang of the strand that is not methylated (u strand) (Fig. 1f) is bound by the La-motif-containing domain (LCD). Meanwhile, the 3′-end 2-nucleotide overhang of the strand that is methylated (m strand) (Fig. 1f) is deeply buried into the active site of the MTase domain (Fig. 1c). The interface between HEN1 and the small RNA substrate buries a total solvent-accessible surface area of ~5,000 Å2 (Fig. 1c), of which dsRBD1, dsRBD2, LCD and the MTase domain each contributes 31%, 13%, 17% and 33%, respectively.
Structure-based sequence alignment and structural superimposition revealed that both dsRBDs contain distinct long insertions in the loop between β1 and β2 (Supplementary Fig. 4). The insertion in dsRBD1 is well defined (Supplementary Fig. 4b), in which a conserved hydrophobic patch stacks over the carboxy-terminal β-strand in the MTase domain (Supplementary Fig. 4c). The insertion in dsRBD2 is longer but less conserved than that in dsRBD1 in most plant HEN1 proteins (Supplementary Fig. 4a) and is completely disordered in the current structure (Supplementary Fig. 4b). Three conserved RNA binding motifs in canonical dsRBDs17 can be identified in dsRBD1 (Fig. 2a), whereas only two RNA binding motifs are identified in dsRBD2 owing to the disordered loop between β1 and β2 (Fig. 2b). As revealed by buried surface analysis, the interaction of dsRBD1 with the RNA substrate is more extensive than that of dsRBD2. Compared to dsRBD1, dsRBD2 shifts by approximately 3 Å away from the RNA duplex, which may favour binding small RNA duplexes with bulges that are common among miRNAs. The binding of the RNA duplex by dsRBD1 has a key role in substrate recognition, as deletion of dsRBD1 markedly reduced the substrate binding by HEN1, as determined by a cross-linking binding assay (Supplementary Fig. 5a), and its activity, as revealed by a small RNA methyltransferase assay (Supplementary Fig. 5b).
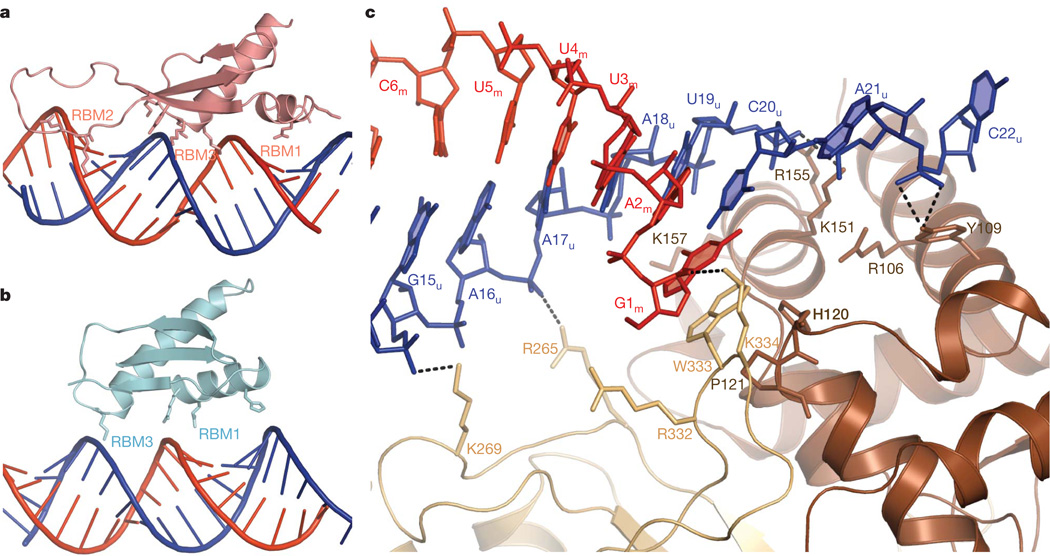
Figure 2. Small RNA substrate recognition by dsRBDs and LCD.
a, The duplex region of the small RNA substrate is bound by three RNA binding motifs in dsRBD1. b, The duplex region of the small RNA substrate is bound by two RNA binding motifs (RBM1 and RBM3) in dsRBD2. c, The LCD binds to the [5′-m:3′-u] terminus of the small RNA substrate. The 2-nucleotide 3′ overhang of the u strand is recognized by the La motif. The base of the 5′-terminal nucleotide G1m is end-capped by W333 in the C-terminal LCD.
As predicted by bioinformatics analysis15, the N-terminal half of the LCD contains a La motif fold (Supplementary Fig. 6) that has been shown to specifically bind RNA 3′ ends through synergistic cooperation with an RNA recognition motif in the La protein18,19. However, recognition of the [5′-m:3′-u] duplex terminus of the small RNA substrates by the La motif and the C-terminal portion of LCD in HEN1 (Fig. 1f) is different from that observed in the human La protein20. The 3′-terminal nucleotide binding pocket in the human La N-terminal domain (NTD)20 is occupied by two conserved residues—H120 and P121—within a HEN1-specific insertion (Supplementary Fig. 6c). The 2-nucleotide 3′ overhang of the u strand is looped out from the RNA duplex towards the first α-helix of the La motif and the phosphate of the overhang is bound by Y109 from the first α-helix of the La motif (Fig. 2c). Mutation of Y109 to alanine has no detectable effect on substrate binding or HEN1 activity (Supplementary Fig. 5a, b), indicating that this interaction is not essential for the interaction with small RNA substrates. This result is also consistent with a previous study showing that mutation of the 3′-end 2-nucleotide overhang of the u strand to either a 1-nucleotide or 3-nucleotide overhang has no effect on HEN1 activity on the m strand11.
Furthermore, W333, a conserved residue within a loop from the C-terminal portion of the LCD, stacks over the base of the 5′-terminal nucleotide G1m (Fig. 2c), and the side chain of W333 occupies the same position as the base of the antepenultimate nucleotide in the structure of the La NTD–RNA complex20 (Supplementary Fig. 6c). Therefore, W333 exactly stacks on H120 and P121 in the La motif of the LCD, which may stabilize the stacking interaction between W333 and the 5′-terminal nucleotide. This end-capping interaction has an essential role in the recognition of small RNA substrates, because the W333A mutant loses both RNA binding ability and small RNA methyltransferase activity (Supplementary Fig. 5a, b). A similar interaction between small RNA duplex and tryptophan residues has been observed in structures of the viral RNA silencing suppressor p19–small RNA complexes21,22. In fact, p19 interferes with small RNA 3′-end methylation by HEN1 (ref. 23). In addition, recognition of the [5′-m:3′-u] terminus by the LCD is also strengthened by a group of positively charged residues that project side chains into the major groove of the duplex terminus (Fig. 2c).
The MTase domain of HEN1 adopts a core α/β Rossmann structure, in which the cofactor product AdoHcy is bound as in classical S-adenosyl-l-methionine (AdoMet)-dependent MTases24 (Fig. 3b). The ribose ring of AdoHcy directly stacks over the 5′-terminal nucleotide U1u, and the 5′ phosphate of the u strand is hydrogen bonded to the side chain of S747 (Fig. 3b). Three conserved positively charged residues—K749, R753 and K756—interact with the major groove of the [5′-u:3′-m] terminus (Fig. 3b), enhancing the 5′ phosphate interaction. These three positively charged residues and S747 are only conserved in plant HEN1 proteins (Supplementary Fig. 7), indicating that recognition of the 5′-phosphate by the MTase domain is not applicable to animal HEN1 homologues. The backbone phosphate connecting the 2-nucleotide 3′ overhang and the RNA duplex segment is anchored by a loop formed by six residues (F692 to F697) within motif X of the MTase domain (Fig. 3a). In particular, two non-bridged phosphodiester oxygens form hydrogen bonds with main-chain amines of F692 and L697, respectively (Fig. 3a). The loop structure is further stabilized by hydrophobic stacking interactions between the side chains of F693 and L697 and by the hydrogen bond formed by the side-chain amide of the invariant residue Q700 and the carboxyl oxygen between tandem prolines P695 and P696 in the loop (Fig. 3a). Most residues in this loop are invariant in the MTase domains of both plant HEN1 and animal homologues (Supplementary Fig. 7), indicating that this specific interaction by the conserved loop in motif X may also be applicable to animal HEN1 homologues.
Figure 3. Small RNA substrate recognition by the MTase domain.
a, The phosphate connecting the 2-nucleotide 3′-overhang of the m strand with the duplex region is specifically recognized by a conserved loop (F692–L697). The penultimate nucleotide A21m is flipped out and its base is stacked on the side chains of L835 and R856. The phosphate of the 2-nucleotide overhang is hydrogen bonded by R701 and R856. b, The base of the 3′-terminal nucleotide of the m strand G22m is stacked on the terminal base pair formed by A20m and U1u and the 5′-phosphate of the u strand is recognized by S747. c, Both 2′ and 3′ hydroxyls of the 3′-terminal nucleotide G22m are coordinated to Mg2+ along with four invariant residues, E796, E799, H800 and H860. d, A stereo view of the Mg2+ coordination covered with Fo−Fc electron density omit map contoured at 3.0σ.
The penultimate nucleotide A21m of the 2-nucleotide 3′ overhang is flipped out from the duplex and the base of A21m is stacked on the side chains of the conserved residues R856 and L835 (Fig. 3a). The 3′-end nucleotide G22m is flipped back and its base is stacked over the terminal base pair of the duplex (Fig. 3b, c). There are no intermolecular hydrogen bonds between two bases of the 2-nucleotide 3′ overhang and the MTase domain, which is consistent with the non-sequence-specific methyltransferase activity of HEN1. The backbone phosphate of the 2-nucleotide 3′ overhang is secured by two invariant, positively charged residues R701 and R856 (Fig. 3). Mutation of either R701 or R856 to alanine attenuates the methyltransferase activity of HEN1 (Supplementary Fig. 5b, c), indicating that these two residues are important for the efficiency of HEN1 activity but are not essential.
The ribose ring of G22m is located in the centre of the active site of the MTase domain, where both the 2′ and 3′ hydroxyls of G22m and the side chains of four invariant residues (E796, E799, H800 and H860) are coordinated to a metal ion, Mg2+ (Fig. 3c, d and Supplementary Fig. 8). The highly organized Mg2+-mediated coordination precisely presents the 2′ hydroxyl of the 3′-terminal nucleotide towards the Sδ atom of AdoHcy (Fig. 3d), indicating that the 2′-O-methylation by HEN1 may be Mg2+-dependent (Supplementary Fig. 9). Treatment with increasing concentrations of EDTA that chelates Mg2+ in the reaction eventually eliminates HEN1 activity (Supplementary Fig. 5d, f), suggesting that HEN1 is indeed a Mg2+-dependent small RNA methyltransferase. Mutation of any one or two coordinated residues to alanines completely abolished HEN1 activity (Supplementary Fig. 5b, c).
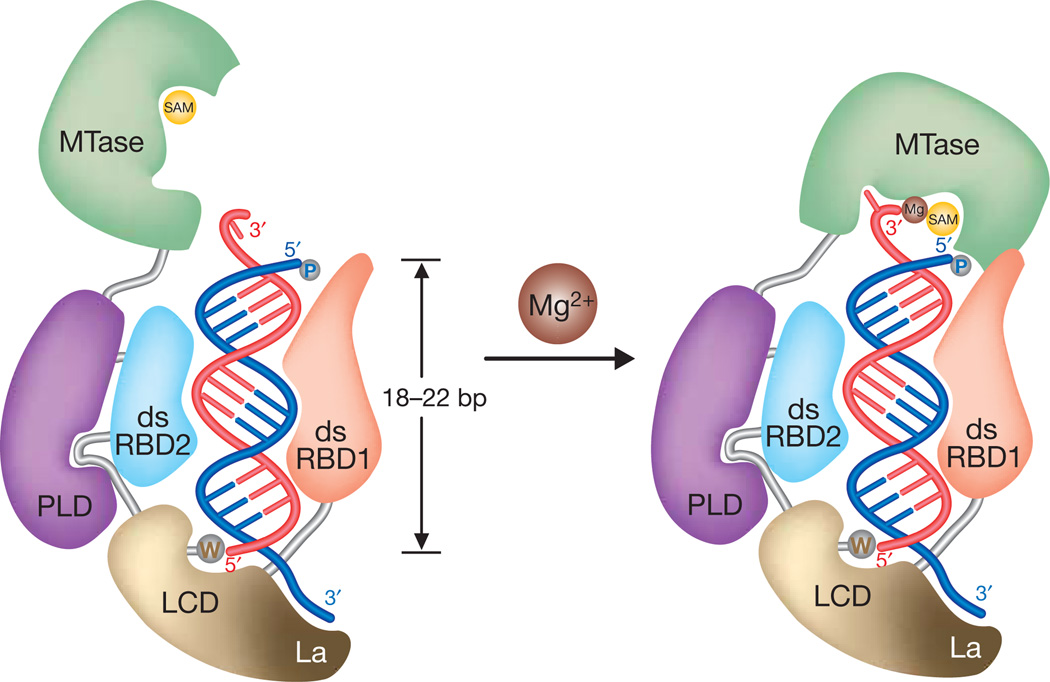
Previous biochemical studies3,11 defined the features of a small RNA substrate that are strictly required for HEN1 activity: a length of 19–25 nucleotides, a duplex with 2-nucleotide 3′ overhang, and free 2′ and 3′ hydroxyls on the 3′ terminal nucleotide. The structure of the HEN1–small RNA complex revealed that multiple domains in HEN1 cooperate to bind small RNA substrates, which precisely illuminates the RNA substrate specificity of HEN1 (Fig. 4). The RNA substrate may be initially targeted by the N-terminal domain dsRBD1 inHEN1, which allowsHEN1 to only act on double-stranded RNAs3. The recognition of an RNA duplex by a classical dsRBD spans about 16 bp25. Thus, the small RNA duplexes produced in plant, approximately 21–24 nucleotides long, are well targeted in the initial recognition. The end-capping interaction by LCD is synergized by dsRBD2, which, together with dsRBD1, forms a strong grip on the duplex region of the small RNA substrate, and these interactions help position the other duplex terminus towards the MTase domain. The recognition of the 2-nucleotide 3′ overhang by the MTase domain and the coordination of both the 2′ and 3′ hydroxyls of the 3′-terminal nucleotide to Mg2+ restrict the MTase domain within a limited range where it can efficiently methylate the 2′-hydroxyl on the 3′-end nucleotide. Overall, the preferred length of the small RNA substrates recognized by HEN1 is determined by the distance between the active site of the MTase domain and the 5′-end-capping site in the LCD (Fig. 4).
Figure 4. Proposed model for the specific recognition of small RNA substrates by HEN1 and the Mg2+-dependent 2′-O-methyltransferase mechanism.
A small RNA substrate is targeted by multiple RNA binding domains in HEN1. The duplex region is gripped by dsRBD1 and dsRBD2, and one terminus is projected towards the MTase domain that is located within a range of 18–22 bp from another terminus end-capped by a tryptophan residue in LCD. Consequently, the MTase domain preferably recognizes the 2-nucleotide 3′ overhang on the small RNA substrate of 20–24 nucleotides in length and methylates the 2′-hydroxyl of the 3′-terminal nucleotide in a Mg2+-dependent manner.
The mode by which HEN1 measures the length of the small RNA substrate is similar to that of the RNase Dicer, a molecular ruler cleaving the dsRNA substrate at a specified distance from the duplex terminus recognized by the PAZ domain26, although the 3′-end recognition by HEN1 is different compared with that by the PAZ domain27,28. Animal HEN1 homologues only act on single-stranded small RNAs4,5,8, and their small RNA methyltransferase activities are stimulated through interaction with Argonaute proteins4 (Y. Kirino, personal communication). Thus, it is possible that animal HEN1 homologues adopt an alternative mode to recognize small RNA substrates (Supplementary Fig. 10), but the mechanism of the Mg2+-dependent 2′-O-methylation by the MTase domain is expected to be conserved.
METHODS SUMMARY
The cDNA of the full-length Arabidopsis HEN1 was cloned into the vector pET28 to result in an N-terminal 6× His tag and expressed in Escherichia coli BL21-Gold(DE3). The protein was purified by affinity, ion exchange and gel filtration chromatography and concentrated. HEN1 mutants were obtained with the QuickChange site-directed mutagenesis kit (Stratagene) or a PCR-based method, and verified by sequencing. RNA oligonucleotides used in the crystallization and assays were ordered from Dharmacon or Integrated DNA Technologies and purified by PAGE or HPLC. Small RNA duplexes were annealed before use. Crystals of HEN1 in complex with the small RNA duplex and AdoHyc were obtained by vapour diffusion with the reservoir solution of 15% PEG3350, 0.2 M sodium chloride, 0.01 M sodium bromide and 0.1 M phosphate-citrate, pH4.8. The 3.1 Å native data and the 3.4 Å MAD data were collected at beamlines 19BM and 23ID of Argonne National Laboratory, respectively (Supplementary Table 1). The final model was refined on 3.1 Å native data to Rfree 28.8% and Rfactor 26.0% with good stereochemistry. Figures were prepared with Pymol (http://www.pymol.org).
The in vitro small RNA methyltransferase assay was performed as previously described29 with minor modifications. Briefly, 100 µl methyltransferase reactions were set up for annealed small RNA substrates and HEN1 mutants and monitored by incorporation of the [14C]methyl group. To assay the Mg2+-dependent methyltransferase activity, Mg2+ was omitted in all annealing buffer and reactions except as indicated. Different amounts of Mg2+ and EDTA were added into individual reactions containing the HEN1 protein and were incubated at room temperature for 15 min before adding small RNA substrates and [14C]-labelled AdoMet. The in vitro RNA–protein crosslinking assay was carried out using iodouridine-labelled small RNA substrates as described30.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Protein expression and purification
DNA fragments corresponding to full-length HEN1 were amplified from the cDNA and inserted into the pET28a vector (Novagen) under NcoI and XhoI sites to result in an N-terminal His tag (MGHHHHHH). A double point mutation L604P/K640R was introduced into HEN1 during PCR amplification and L604P was reversed by site-directed mutagenesis to result in the single mutant K640R. Because no differences in methyltransferase activity were observed among the double mutant L604P/K640R, the single mutant K640R and wild-type HEN1 (data not shown), if not specifically indicated, the double mutant L604P/K640R and the single mutant K640R were treated as wild-type HEN1 for protein purification and crystallization in this study. Mutants W333A, E799A/H800A, H860A, R701A and R856A were generated by a PCR-based overlap extension method. E796A, H860Q, Y109A, H800Q as well as the correction of L604P were generated using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). The N-terminal deletion mutant ΔN89 (90–942) was generated by PCR amplification and subcloning. Primers for cloning and mutagenesis are listed in Supplementary Table 2. The presence of the mutations was confirmed by sequencing. The recombinant HEN1 proteins were expressed in E. coli BL21-Gold(DE3) (Stratagene). After induction with 0.2 mM IPTG, the cells were allowed to grow at 17 °C for 20 h. Collected cells were lysed by a C-3 cell disruptor (Avestin) at 4 °C. Proteins were purified by affinity His-Trap column, ion-exchange Q column, heparin column and size-exclusive column Superdex 200 (GE Healthcare). Further chromatography on a Mono-Q column (GE Healthcare) was required to obtain high-quality proteins for crystallization. Purified proteins were concentrated to 15 mg ml−1 in a buffer containing 10 mM HEPES (pH 7.5), 50 mM KCl and 2 mM dithiothreitol (DTT), and flash frozen in liquid nitrogen before storing at −80 °C. SeMet-labelled proteins were produced by inhibiting endogenous methionine biosynthesis31 in M9 minimial media supplemented with specific amino acids as well as SeMet, and purified as for the native protein. All mutants were expressed and purified as described above.
RNA preparations
Sequences of RNAs used in this study are listed in Supplementary Table 3. All RNA oligonucleotides were synthesized from Dharmacon or Integrated DNA Technologies and further purified with PAGE or HPLC. The concentrations of the RNAs were measured by ultraviolet spectrometry at 260 nm; RNA duplexes used for crystallization and the cross-linking assay were first annealed in a buffer of 30 mM HEPES-K, pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate. RNA duplexes used for small RNA methyltransferase assay were annealed in a buffer of 50 mM Tris-HCl, pH 7.6 and 100 mM KCl. Annealing was performed by heating the mixture for 5 min at 95 °C and slowly cooling it to 37 °C followed by incubation for 2 h at 37 °C and 1 h at 24 °C in a thermal cycler. Annealing efficiency was examined by running the anneal products on a 15% polyacrylamide native gel.
Crystallization and data collection
Both miR173/miR173* and miR173/miR173*cm RNA duplexes were used to co-crystallize with HEN1 in the presence of AdoHcy. Only miR173/miR173*cm gave out crystals with enough quality for data collection. The ternary complex used for crystallization was prepared by adding 20-fold excess of AdoHcy to HEN1 protein and incubating on ice for 0.5 h followed by the addition of twofold excess of RNA. The final concentration of HEN1 in the complex is about 5 mg ml−1. The initial screening was carried out with commercial crystallization kits using Phoenix crystallization robot (Art Robbins Instruments) and detected using Rock Imager 2 and Rock Maker automated imaging system (Formulatrix). The preliminary hit was obtained at condition number 36 of Wizard II Screen (Emerald Biosystems), 0.1 M phosphate-citrate, pH4.2, 10% PEG 3000 and 0.2 M NaCl. The crystals were optimized using the hanging-drop vapour diffusion method at 20 °C and Additive Screen (Hampton Research) was used during the optimization of initial condition. Addition of NaBr (Additive Screen, No. 29) markedly improved the quality of the crystals. Finally, the crystals were grown in the solution containing 0.1 M phosphate-citrate, pH4.8, 15% PEG 3350 and 0.2 M NaCl and 0.01 M NaBr. SeMet-labelled crystals were obtained under the same condition as for the native crystals. Crystals were transferred into cryoprotectant solution with 20% glycerol and then flash-frozen in liquid nitrogen. Diffraction tests of collected crystals were performed at 100 K using a Rigaku X-ray generator equipped with R-AXIS IV++ detectors. A multi-wavelength anomalous dispersion (MAD) data set to 3.4 Å was collected on a SeMet-labelled crystal at the Argonne National Laboratory beamline 23ID-B. A native data set to 3.1 Å was collected at beamline 19BM, the Structural Biology Center at the Applied Photon Source. The diffraction data were processed and scaled with the HKL2000 package32. The data collection and processing statistics are summarized in Supplementary Table 1.
Structure determination and refinement
Phase was determined by the multiple-wavelength anomalous dispersion method33 using 3.4 Å MAD data by PHENIX package34. Out of a total of 26 sites, 18 selenium atoms were located using the program HYSS in PHENIX34. Heavy atom refinement and MAD phasing were carried out using programs SOLVE and RESOLVE in PHENIX34, and the figure of merit after phasing improvement by program RESOLVE was increased to 0.74 from initial 0.36. An initial model of HEN1 was manually built with the programs O35 and Coot36 using the locations of SeMet positions as guides. The model of the small RNA duplex was built based on the position of the 5′-phosphate that only exits in the miR173 strand. The initial model of the complex was refined through alternating cycles using the program phenix.refine in PHENIX. Non-crystallographic symmetry was used to restrain the core of two domains in the asymmetric unit while more variation was allowed in the loop regions. The final model was refined to the native data in the resolution range 20–3.1 Å using CNS version 1.2 (ref. 37) until the R/Rfree were 26.0/28.8 with good stereochemistry. Ramachandran analysis showed that 87.2% of residues are in most favoured regions, 12.8% of residues are in additional allowed regions, and no residues in the generally allowed or disallowed regions. The final model contains two double mutant L604P/K660R HEN1 molecules including residues 1–6, 213–215, 290–301, 411–454, 501–534, 542–551, 572–599, 839–850, 912–916 and 934–942 of chain A and residues 1–6, 292–303, 410–452, 501–519, 529–534, 540–546, 574–598, 840–851, 910–917 and 934–942 of chain D (Supplementary Fig. 1), two miR173/miR173*cm duplexes, two AdoHcy, two Mg2+, and 37 waters (Supplementary Table 1). Residue 535–541 of chain A and residue 535–539 of chain D are not certain owing to the poor electron density in the middle of a long disordered region. The refined structure was validated using PROCHECK38. Structural figures were prepared with PyMol (http://www.pymol.org).
Small RNA methyltransferase assay
The in vitro small RNA methyltransferese assay monitored by the incorporation of the [14C]-methyl group was performed as previously described29 with minor modifications. A 100-µl methyltransferase reaction was set up for the annealed small RNA substrate miR173/miR173* and HEN1 mutants. The reaction mixture contained 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 5% glycerol, 2 µl RNasin (40Uµl−1; Promega), 0.5 µCi S-adenosyl-l-[methyl-14C] methionine (58.0 mCi mmol−1; Amersham Pharmacia Biosciences), 5 µg purified protein, and 1 nmol RNA substrate. After incubation at 37 °C for 2 h, the reaction was stopped by adding 100-µl 2× proteinase K solution (100 mM Tris-HCl, pH 8.0, 10 mM EDTA, 150 mM NaCl, 2% SDS, and 0.4 mg ml−1 proteinase K) followed by incubation at 65 °C for 15 min. The reaction was then extracted with phenol/chloroform. To precipitate the small RNAs, 1 ml glycogen (5 mg ml−1), 0.1 vol of 3 M NaOAc (pH 5.2), and 2.5 vol of ice-cold 100% ethanol were added to the reaction. The mixture was stored at −80 °C for 2 h and centrifuged at 4 °C for 30 min. The pellet was washed with 100 ml 70% cold ethanol. The RNAs in the pellet were dissolved with 1× RNA loading buffer, heated at 95 °C for 5 min, immediately put on ice, and loaded on to a 15% denaturing polyacrylamide gel with 7 Murea. After electrophoresis, the gel was treated with an autoradiography enhancer (En3hance from Perkin Elmer) following the manufacturer’s instructions and exposed to X-ray film at −80 °C. To assay the Mg2+-dependent methyltransferase activity, Mg2+ was omitted in all annealing buffer and reactions except as indicated. Different amounts of Mg2+ and EDTA were added into individual reactions containing the HEN1 protein and were incubated at room temperature for 15 min before adding small RNA substrates and [14C]-labelled AdoMet.
The in vitro RNA–protein photochemical crosslinking assay
Small RNA miR173 with a 5-iodouracil (5IU) substitute at U1 or U21 (mutated from A21) (Supplementary Table 3) was 5′-end labelled using γ-32P-ATP (NEN), and annealed with miR173*cm or used as a single-stranded substrate. The photochemical crosslinking assay was performed as described30. Typically, 20-µl reactions containing 0.2 µMRNA and 2 µMHEN1 or mutants were placed in a 1.5-ml microtube and incubated for 20 min on ice. Exposure to the ultraviolet light source (Spectroline, λmax = 312, 330 µWcm−2) was at a distance of ,2.5 cm, filtered through a polystyrene Petri dish for 10 min. Crosslinking products added with 2× loading buffer were separated on 12% SDS-polyacrylamide gels, which were exposed to Storage Phosphor Screen (GE Healthcare) and visualized using a Storm PhosphorImager (GE Healthcare).
Acknowledgments
We thank K. Sergiy for assistance with the in-house X-ray generator operation, and the staff at Advanced Photon Source beamlines 19ID and 23ID, Argonne National Laboratory, for help with data collection. We thank T. Townes and H. Wang for critical reading of the manuscript. This work was supported by a start-up fund (to J.-B.M.) and partly by a grant from the V Foundation for Cancer Research (to J.-B.M.) and a grant from the National Science Foundation (MCB-0718029 to X.C.). D.G.V. is supported by grants from National Institutes of Health (R01 GM074252 and R01 GM074840).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Y.H. expressed and purified proteins, grew crystals, solved structure and wrote the manuscript. J.-B.M. collected data, solved structure, performed crosslinking assays and wrote the manuscript. L.J. performed small RNA methyltransferase assays and wrote the manuscript. Q.H. and D.G.V. were involved in data processing and refinement. X.C. analysed data and wrote the manuscript. The overall project management and manuscript preparation were by Y.H., X.C. and J.-B.M.
Atomic coordinates and structural factors for the reported crystal structure have been deposited in the Protein Data Bank under access code 3HTX.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 3.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 5.Saito K, et al. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of Piwi-interacting RNAs at their 39 ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2’-O-methylated at their 39 termini. Nature Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 7.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurth HM, Mochizuki K. 2’-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2’ OH of the 39 terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 39-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X. A marked end. Nature Struct. Mol. Biol. 2007;14:259–260. doi: 10.1038/nsmb0407-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkaczuk K, Obarska A, Bujnicki J. Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol. Biol. 2006;6:6. doi: 10.1186/1471-2148-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang CB, Dhe-Paganon S, Yoon HS. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals. 2008;16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- 17.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded- RNA-binding motif: interference and much more. Nature Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 18.Curry S, Conte MR. A terminal affair: 39-end recognition by the human La protein. Trends Biochem. Sci. 2006;31:303–305. doi: 10.1016/j.tibs.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Maraia RJ, Bayfield MA. The La protein-RNA complex surfaces. Mol. Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Teplova M, et al. Structural basis for recognition and sequestration of UUUOH 39 temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vargason JM, Szittya G, Burgyán J, Hall TMT. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 22.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu B, Chapman EJ, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacRae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 27.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nature Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 28.Ma J-B, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, et al. Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Methods Enzymol. 2007;427:139–154. doi: 10.1016/S0076-6879(07)27008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stump WT, Hall KB. Crosslinking of an iodo-uridine-RNA hairpin to a single site on the human U1A N-terminal RNA binding domain. RNA. 1995;1:55–63. [PMC free article] [PubMed] [Google Scholar]
References
- 31.Doublié S, Carter CW., Jr Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W, Carter CW., Jr Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synthrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in eletron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature Protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]