Abstract
While heme is an important cofactor for numerous proteins, it is highly toxic in its unbound form and can perpetuate the formation of reactive oxygen species. Heme oxygenase enzymes (HMOX1 and HMOX2) degrade heme into biliverdin and carbon monoxide, with biliverdin subsequently being converted to bilirubin by biliverdin reductase (BVRa or BVRb). As a result of the teleost-specific genome duplication event, zebrafish have paralogs of hmox1 (hmox1a and hmox1b) and hmox2 (hmox2a and hmox2b). Expression of all four hmox paralogs and two bvr isoforms were measured in adult tissues (gill, brain and liver) and sexually dimorphic differences were observed, most notably in the basal expression of hmox1a, hmox2a, hmox2b and bvrb in liver samples. hmox1a, hmox2a and hmox2b were significantly induced in male liver tissues in response to 96 hour cadmium exposure (20 µM). hmox2a and hmox2b were significantly induced in male brain samples, but only hmox2a was significantly reduced in male gill samples in response to the 96 hour cadmium exposure. hmox paralogs displayed significantly different levels of basal expression in most adult tissues, as well as during zebrafish development (24 to 120 hpf). Furthermore, hmox1a, hmox1b and bvrb were significantly induced in zebrafish eleutheroembryos in response to multiple pro-oxidants (cadmium, hemin and tert-butylhydroquinone). Knockdown of Nrf2a, a transcriptional regulator of hmox1a, was demonstrated to inhibit the Cd-mediated induction of hmox1b and bvrb. These results demonstrate distinct mechanisms of hmox and bvr transcriptional regulation in zebrafish, providing initial evidence of the partitioning of function of the hmox paralogs.
Keywords: biliverdin reductase, heme oxygenase, cadmium, pro-oxidant, Nrf2, zebrafish
1. Introduction
Heme molecules are essential for life, having roles in cellular differentiation, apoptosis and serving as a cofactor for numerous proteins (Larsen et al., 2012). However, unbound cellular heme is highly reactive and can generate damaging reactive oxygen species (ROS) via Fenton chemistry. Two isoforms of the heme-degrading enzyme, heme oxygenase 1 and 2 (HMOX1 and HMOX2), catalyze the breakdown of heme into carbon monoxide (CO), free iron, and the bile pigment biliverdin (Tenhunen et al., 1969; Abraham and Kappas, 2008). Biliverdin reductase a (BVRa) and biliverdin reductase b (BVRb) further reduce biliverdin to the proposed antioxidant bilirubin (Stocker et al., 1987; Yamaguchi et al., 1994). It has been suggested that BVRa functions to protect cellular lipophilic components from oxidative stress through the production of bilirubin via an antioxidant recycling pathway similar to the classic glutathione recycling pathway (Doré et al, 1999; Barañano et al, 2002; Sedlak and Snyder, 2004; Sedlak et al., 2009). The action of bilirubin as an effective antioxidant is notable for its potential benefits to cellular health. However, an excess of bilirubin can be problematic resulting in jaundice and toxic insult (Greenberg, 2002). Although heme and bilirubin have important cellular roles, the potential toxicity associated with these two molecules illustrates the necessity for tight regulation of this enzymatic pathway.
While the heme degradation pathway has been widely studied, much of the research has focused primarily on HMOX1 and BVRa in mammalian systems with little focus on developmental roles. To this end, HMOX1 has been shown to be highly inducible by a variety of stressors in mammalian systems, and is detected at high levels in tissues involved in heme metabolism including spleen and liver (Tenhunen et al, 1969), bone marrow (Brown et al., 1988; Abraham et al., 1989) and erythroid cells (Garcia-Santos et al., 2014). The other heme degrading enzyme, HMOX2, is expressed at high levels in the testes (Trakshel et al., 1986), brain (Maines et al., 1986; Trakshel et al., 1988; Ewing and Maines, 1992), and vascular tissues (Zakhary et al., 1996).
The BVR isoforms are distinct in their enzymatic actions and their evolutionary origins (Yamaguchi et al., 1993). BVRa has been shown to be highly expressed in rat kidney, liver, spleen and brain tissues (McCoubrey et al., 1995), as well as human liver (Maines and Trakshel, 1993) and the plasma membrane of macrophages (Wegiel et al., 2009). BVRb has been demonstrated to be abundant in adult bovine, rat, rainbow trout, and human erythroid cells, as well as rat and bovine liver (Xu et al., 1992; Shalloe et al., 1996; Saleh and McConkey, 2012). Although both BVRa and BVRb generate bilirubin, more recent published research is highly skewed toward BVRa while the BVRb isoform is typically not included in studies related to the antioxidant properties of this pathway. One explanation for this omission may be the result of the unique properties of BVRa. Although both BVR enzymes contain the reductase domain, BVRa’s domain has dual cofactor (NADH and NADPH) dual pH specificity. BVRa also has several regulatory domains not found in BVRb, such as a serine/threonine/tyrosine kinase (S/T/Y) domain (Kutty and Maines, 1981; Salim et al., 2001; Lerner-Marmarosh et al., 2005), a leucine zipper DNA-binding domain (Ahmad et al., 2002), as well as nuclear export and localization signals (Maines et al., 2001; Lerner-Marmarosh et al., 2008). Thus, BVRa may participate in cell signaling through a variety of mechanisms.
Due to their rapid transparent development and sequenced genome, zebrafish (Danio rerio) have become an excellent model for developmental and toxicological studies (Blechinger et al., 2002; Hill et al., 2005; Blechinger et al., 2007; Howe et al., 2013). Furthermore, zebrafish often have multiple homologues of human genes (Meyer and Van de Peer, 2005) resulting from a teleost-specific whole genome duplication event (Amores et al., 1998). Many of these homologues have undergone subfunction partitioning (Postlethwait et al., 2004), which makes zebrafish an advantageous model organism for ascertaining the myriad functions of a gene in which its human counterpart has multiple roles (Amores et al., 1998). In this regard, purported duplicates of hmox1 and hmox2 have been identified within the zebrafish genome (Nakajima et al., 2011). However, a full characterization of the expression patterns and response to cellular stress of these various isoforms is lacking.
As an initial step towards understanding novel developmental and protective roles of zebrafish heme degradation genes, we set out to quantitatively evaluate differences in their expression during early development and in adult tissues under normal and stressful conditions. We sought to determine how these genes responded to different inducers of oxidative stress during development (Cd, hemin, and tBHQ) or as adults (Cd). Interesting differences in developmental expression and adult tissue distribution, as well as responses to oxidative stress were noted. The results of these experiments and their implications are presented here.
Material and methods
Chemicals
Cadmium (Cd) chloride (CAS # 654054-66-7) and tert-butylhydoquinone (tBHQ) (CAS #1948-33-0) were purchased from Sigma-Aldrich (St. Louis, MO). Hemin chloride (CAS # 16009-13-5) was purchased from EMD Millipore.
Fish husbandry
The TL (Tupfel/Long fin mutations) wild-type strain of zebrafish was used for all experiments. Fertilized eggs were obtained from multiple group breedings from a Mass Embryo Production System (MEPS; Aquatic Habitats, Apopka, FL) with ~200 fish at a ratio of 2 female per 1 male fish. Procedures used in these experiments were approved by the Animal Care and Use Committee of the University of Alabama, Tuscaloosa, Alabama, USA.
Cadmium exposures in adult zebrafish
Adult male and female zebrafish that were 12 months of age were subjected to 20 µM Cd for 96 hours. Adult fish were grouped in a 4L beaker (3 male and 3 female) with 3L of fish water (360 mg Instant Ocean Sea Salt and 11 mg of NaHCO3 per liter of deionized water; 7.4 pH, 750 µ-siemens conductivity, 28.5 °C) or fish water with 20 µM Cd. Water was continuously aerated with a small air stone, and both control water and the water with Cd were renewed after 48 hours. The fish were fed once daily with live artemia. No overt signs of toxicity, such as changes in feeding or swimming behavior, were observed during the course of the Cd exposures. Adult zebrafish were anesthetized with 150 mM MS-222 in buffered fish water and euthanized by cervical transection. Brain, liver, and gill tissues were removed from male and female fish and flash frozen in liquid nitrogen. Tissues were stored at −80 °C until RNA isolation. Three biological replicates were collected for each sex and treatment group and consisted of equivalent amounts of total RNA pooled from three individual male or female fish.
Developmental time series
A large batch (>3000) of embryos was generated by active breeding of the fish in the MEPS system for 60 minutes. After 60 minutes embryos were collected and transferred to large Petri dishes (150 mm diameter) at a density of 100 embryos per 100 ml of 0.3× Danieau’s solution at 28.5 °C. Embryos were screened within 4 hours of fertilization for normal development and subsequently separated into 100 mm Petri dishes at a density of 20 embryos per dish in 25 ml of 0.3× Danieau’s solution at 28.5 °C under a 14 hour light/10 hour dark cycle with water changes every 24 hours. Three biological replicates of 20 pooled embryos were collected at several developmental timepoints (24, 48, 72, 96, and 120 hpf). Embryos were flash frozen in liquid nitrogen and stored at −80 °C until RNA isolation.
Toxicant exposures during zebrafish development
Acute 4 hour challenges with Cd (50 and 150 µM), hemin (100 µM or 150 µM), or tBHQ (1 and 5 µM) were performed starting at 72 hpf (3 replicates of 20 pooled embryos per treatment). An additional 8 hour Cd (50 µM) exposure was performed on 120 hpf larvae (3 replicates of 20 pooled embryos per treatment). Following all exposures, embryos were flash frozen in liquid nitrogen and stored at −80°C until RNA isolation.
NRF2a transient knockdown via morpholino
The previously described Nrf2a MO (5'CATTTCAATCTCCATCATGTCTCAG-3') was obtained from Gene Tools, LLC (Philomath, OR) (Kobayashi et al., 2002; Timme-Laragy et al., 2012). The standard control morpholino (5'-CCTCTTACCTCAGTTACAATTTATA-3') from Gene Tools was used to account for any nonspecific effects associated with microinjection. The control and NRF2a MOs are fluorescein tagged for screening purposes to guarantee that only successfully injected embryos were used for the subsequent experiments. The MOs were diluted to 0.18 mM in nuclease-free water. A Narishige IM-300 microinjector was used to inject 2.1 nl of morpholino into embryos at the two- to four-cell stage. Injection volumes were calibrated by injecting solutions into mineral oil and measuring the diameter of the sphere with a stage micrometer (volume = 4/3πr3; 160 µm diameter is equivalent to 2.1 nl). At 3 hpf, embryos were sorted to remove unfertilized eggs or abnormally developed embryos. Embryos were further screened at this time using a Nikon AZ100M fluorescent macroscope to confirm full distribution of the MOs.
Rapid amplification of cDNA ends for hmox1b transcript analysis
Total RNA was collected from embryos from several different developmental timepoints and pooled together for cDNA synthesis. A Marathon® cDNA Amplification Kit (BD Biosciences, Palo Alto, CA, U.S.A.) was used to synthesize double stranded zebrafish cDNA according to manufacturer’s protocols. Adaptor primers were used with a hmox1b gene specific primer (Table 1) with the following PCR parameters: 95 °C for 3 min, 95 °C for 1:00 min/58 °C for 45 s/68 °C for 1:00 min (30 cycles). The secondary PCR product was gel purified using a QIAGEN® QIAex II Gel Extraction Kit and cloned into the pGEM®-T Easy Vector (Promega, Madison, WI) for sequencing.
Table 1.
qPCR and 5’ RACE PCR Primers
| Gene | qPCR | Sequence |
|---|---|---|
| hmox1a | Forward | 5′-GCTTCTGCTGTGCTCTCTATACG-3′ |
| Reverse | 5′-CAATCTCTCTCAGTCTCTGTGC -3′ | |
| hmox1b | Forward | 5’-GCAGTGATCTGTCTGAACAG-3′ |
| Reverse | 5′-GCTTGTACTGTGTTTGTGTG-3′ | |
| hmox2a | Forward | 5′-ATGGCGGTCAGTGGAAACACAACC-3′ |
| Reverse | 5′-GGCAACAGCAGCAACCAATGTGGC-3′ | |
| hmox2b | Forward | 5’TTTAGGAGGTTGAGTTGGAGTCAG-3′ |
| Reverse | 5′-TTCTGCCTTCTGGTGCACTTCT-3′ | |
| bvra | Forward | 5′-CAGGCAGTTTCTGGAGGCAGG-3′ |
| Reverse | 5′-CCAGACCCTTCTGTTGAGC-3′ | |
| bvrb | Forward | 5’GCATGTCAGCATTCCTCTTGTGG-3′ |
| Reverse | 5′-CACCAGCAATATGTGGAGG-3′ | |
| hpx | Forward | 5′-GATGGCCATTTCTACATGATCAAGGACA-3′ |
| Reverse | 5′-GCCCTCAATTCCCAGCACATCC-3′ | |
| hamp | Forward | 5’CACAGCCGTTCCCTTCATACAGCA-3′ |
| Reverse | 5′-GGTCTGCTAGTCTGTGTTCAGCTTC-3′ | |
| fpn1 | Forward | 5′-GTCCTACATTCATTCCTACAACTGAACC-3′ |
| Reverse | 5′-GTCAAGTCGAAGGACCAAAGACCAACT-3′ | |
| Gene | 5’ RACE PCR | Sequence |
| hmox1b | Reverse | 5′-CGCCTCGTAGATCTTGTAGAGC-3′ |
Phylogenetic analysis of zebrafish hmox isoforms
HMOX homologs were identified using mammalian HMOX protein sequences to search the zebrafish genome using BLAST. Proteins were aligned using ClustalW and a phylogenetic tree was constructed using the maximum likelihood method and the WAG model of amino acid substitution followed by likelihood calculation using the GAMMA model. To assess confidence in individual nodes, a bootstrap analysis with 1000 re-samplings was performed. The consensus bootstrap tree was rooted to the Drosophila melanogaster HMOX protein. The phylogenetic analysis was performed in MEGA7 (Kumar et al., 2013).
Promoter enhancer motif identification
The 4 kb promoter regions upstream of the transcription start site (exon 1) in the zebrafish hmox1a, hmox2a, hmox2b, bvra, and bvrb genes were searched for consensus metal-response element (MRE) MTF-1 binding motifs (TGCRCNC) (Chen et al., 1998), antioxidant-response element (ARE) NRF2 binding motifs (RTGAYNNNGC) (Venugopal and Jaiswal, 1998), and hypoxia-response element (HRE) (Semenza et al., 1996) HIF-1α binding motifs (RCGTG) using ApE software (http://biologylabs.utah.edu/jorgensen/wayned/ape/). The promoter region of zebrafish hmox1b is not available in the current D. rerio GRCz10 genome assembly (GCF_000002035.5, RefSeq; GCA_000002035.3, GenBank).
Gene expression assessment by real-time RT-PCR
Total RNA was prepared from embryos and adult tissues using the TRIzol® reagent (Life Technologies, Invitrogen) protocol. The qScript™ cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD) was used to generate cDNA from 1 µg total RNA per manufacturer’s instructions. PerfeCTa® SYBR® Green Supermix for iQ™ (Quanta Biosciences, Gaithersburg, MD) was used for real-time RT-PCR experiments in a MyiQ2 Two-Color Real-Time PCR Detection system (Bio-Rad, Hercules, CA) under the following conditions: 95°C for 3 min, 95°C for 15s/60°C for 30 sec (40 cycles). To ensure that only a single product was amplified, all real-time RT-PCR experiments were followed with a melt curve analysis. For all experiments, 18S ribosomal RNA was used as the ‘housekeeping’ gene for normalization. 18S ribosomal RNA levels were not affected by Cd treatment. Gene expression for the developmental time series, as well as the adult tissue analysis, was quantified by generating a standard curve using serially diluted plasmids containing full length copies of the transcripts. For the acute Cd, hemin and tBHQ exposures, as well as the MO experiment, changes in transcript expression were determined using the comparative Ct method (Schmittgen and Livak, 2008). All real-time RT-PCR primers are listed in Table 1.
Statistical analysis of gene expression profiling
Statistical analysis was performed using Prism 5 software (GraphPad Software Inc., San Diego, CA). For the developmental time series, statistical significance in transcript expression compared to 24 hpf embryos was determined using one-way analysis of variance (ANOVA) followed by a Dunnett’s post hoc test. For the adult tissue experiments, statistical significance in transcript expression between controls and treatment groups was determined using a two-way ANOVA followed by a Bonferroni post hoc test (p-value < 0.05). For Cd, tBHQ, and hemin exposures in 72 hpf larvae, statistical significance in comparison to control embryos was determined using one-way ANOVA followed by a Dunnett’s post hoc test. For Cd exposure in the 120 hpf larvae, statistical significance in comparison to control embryos was determined using standard Students’s t-test (p-value < 0.05). For the real-time RT-PCR analysis on Nrf2a morphants, statistical significance in transcript expression was determined using two-way ANOVA followed by a Bonferroni’s multiple comparisons test (*p-value < 0.05).
Results
5' RACE PCR and phylogenetic analysis of heme oxygenase
Initial protein alignments of zebrafish Hmox1a and Hmox1b showed a high degree of conservation at the C-terminal region of the protein. However, the Hmox1a isoform is 272 amino acid residues in length while the initial transcript (NCBI Ref Seq NM_205671.1) for Hmox1b only encoded for 247 amino acids, a result from missing part of the heme oxygenase domain found in the N-terminal region of the protein. Using 5' RACE PCR we were able to clone a novel full length transcript and deduce another 29 amino acids for the N-terminal region of the protein (NCBI Accn KX664458). Alignment with the Hmox1a sequence suggests that the Hmox1b NCBI reference sequence was deduced from a transcript that was missing the 5' untranslated region (UTR) and initial coding region that is found in exons 1 and 2, respectively (Figure 1).
Figure 1. Zebrafish Hmox1a and Hmox1b contain conserved heme oxygenase domains and heme signature motifs.
A) Schematic of the zebrafish hmox1a gene. Exons are denoted by boxes and introns are denoted by lines. The numbers represent the nucleotide length for each exon or intron. The heme oxygenase domain (HMOX Domain) spans exons 3–6 and is heighted by the dark grey line. B) Multiple sequence alignment of HO-1 proteins from fish (goldfish and zebrafish) and human. Sequence alignment was generated using Clustal W. Amino acids identical to zebrafish Hmox1a protein sequence are designated by dots. The HO Domain (grey line) and the HMOX Signature (light grey box) are denoted within their respective sequences. The new N-terminal region of the Hmox1b protein sequence is denoted by a box with a dashed grey line. Zebrafish Hmox1a: NP_001120988.1; Zebrafish Hmox1b: KX664458; Human HMOX1: NP_002124.1; Goldfish (Carassius auratus) Hmox1 GenBank: AHI15729.1
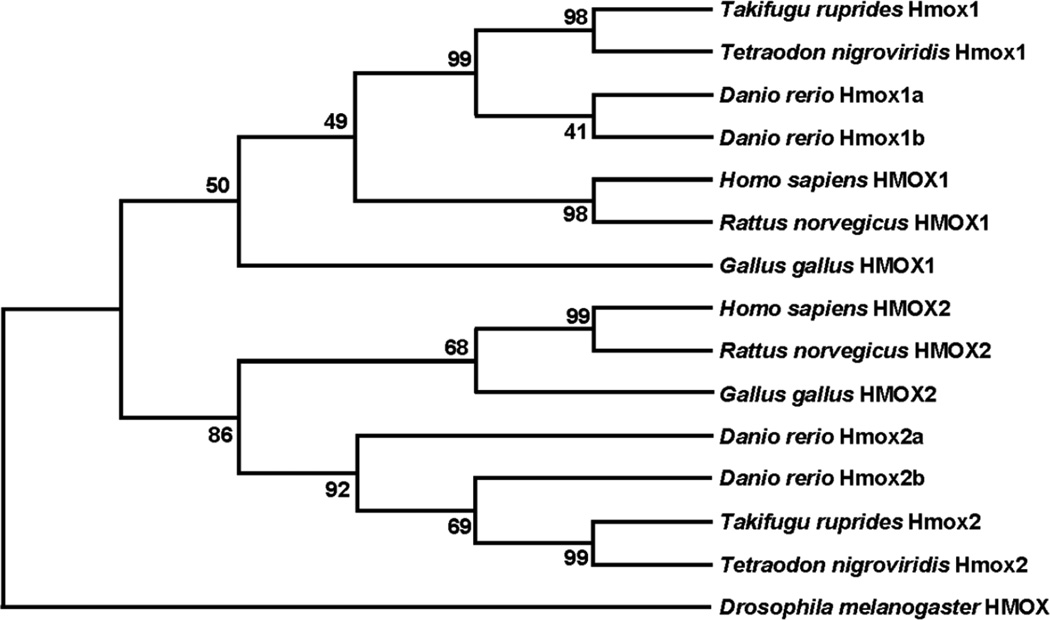
Using this new sequence for zebrafish Hmox1b, a phylogenetic analysis was performed to better understand the relationship between zebrafish and other vertebrate HMOX proteins. Phylogenetic analysis suggests conservation amongst HMOX1 proteins and HMOX2 proteins in humans, rats, and chickens (Figure 2). Hmox1a groups more closely to other fish Hmox1 proteins in comparison to vertebrate homologues. Likewise, zebrafish Hmox2a and Hmox2b show a closer relation in comparison to other vertebrate HMOX2 homologues.
Figure 2. Phylogenetic analysis of heme oxygenase proteins.
Protein alignments and tree construction were performed in MEGA6. Evolutionary history was inferred by using the Maximum Likelihood method. The Bootstrap method was used (1000 replicates) for the Test of Phylogeny. D. melanogaster) HMOX (NP_524321.1) was used as an out-group. HMOX1 proteins: D. rerio Hmox1a: NP_001120988.1; D. rerio Hmox1b: KX664458; T. ruprides Hmox1: UniProtKB - O73688; T. nigroviridis Hmox1: GenBank: CAF95107.1; H. sapiens HMOX1: NP_002124.1; R. norvegicus HMOX1: NP_036712.1; G. gallus HMOX1:NP_990675.1. Hmox2 proteins: D. rerio Hmox2a: NP_001096609.1; D. rerio Hmox2b: XP_002661145.1; T. nigroviridis Hmox2: GenBank: CAG00172.1; T. ruprides Hmox2: Ensemble: ENSTNIP00000012632.1m; H. sapiens HMOX2: NP_002125; R. norvegicus HMOX2: NP_077363.1; G. gallus HMOX2: XP_414960.1.
Comparative expression of hmox and bvr in adult tissues
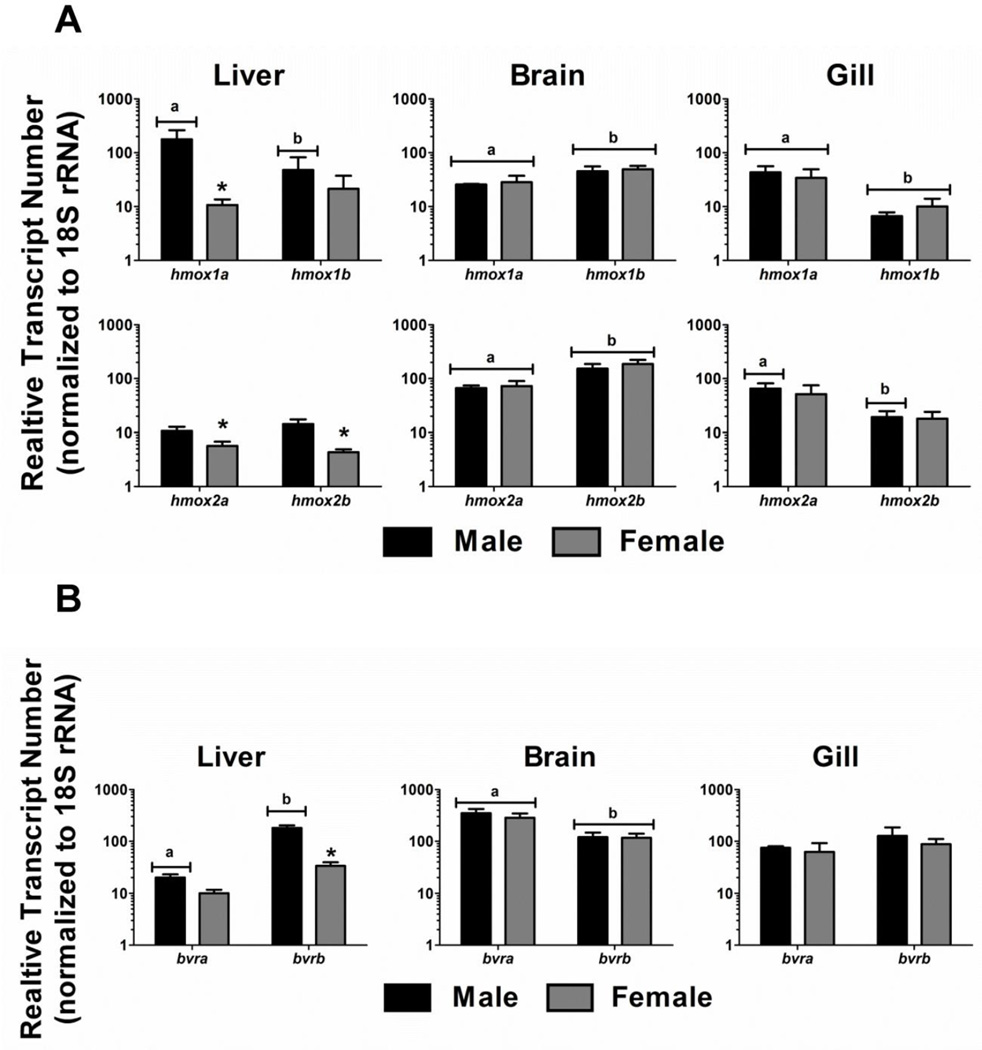
Real-time RT-PCR was used to characterize the differential expression of the hmox and bvr isoforms in liver, gill and brain tissues of adult male and female zebrafish. hmox1a transcript abundance was significantly different from hmox1b abundance in all three tissues of male zebrafish. In contrast, hmox1a and hmox1b transcript abundance was only significantly different in the brain and gills of females (Figure 3A). Furthermore, hmox1a transcript levels were an order of magnitude higher in male livers compared to females. hmox2a and hmox2b transcript levels were also significantly higher in male livers compared to females. There was a statistically significant difference in hmox2a and hmox2b expression in the brains of both male and female zebrafish. Finally, hmox2a and hmox2b expression levels were significantly different in the gill tissue of male zebrafish. Although a similar trend was observed in females, it was not a statistically significant difference (Figure 3A).
Figure 3. HO and BVR expression in adult zebrafish tissues.
Relative transcript abundance of hmox1 and hmox2 paralogs and bvr isoforms were determined in male and female liver, gill, and brain tissues. Statistical significance in transcript expression between males and females was determined using two-way ANOVA followed by a Bonferroni post hoc test (p-value < 0.05). The asterisk designates statistical significance between males and females. Different letters represent statistical significance between paralogous genes. All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates per treatment. A) Expression levels of hmox1a, hmox1b, hmox2a and hmox2b. B) Expression levels of bvra and bvrb.
There was no significant difference in either bvra or bvrb expression levels in gill tissues of either male or female zebrafish (Figure 3B). However, bvra expression in brain tissue was significantly higher in both males and females compared to bvrb expression. Although there was no statistically significant difference between bvra and bvrb expression in female liver tissue, bvrb was expressed at a significantly higher level compared to bvra in male liver tissues. Finally, there was also a significant difference in bvrb liver expression between males and females (Figure 3B).
Inducibility of hmox and bvr expression in adult tissues in response to Cd exposure
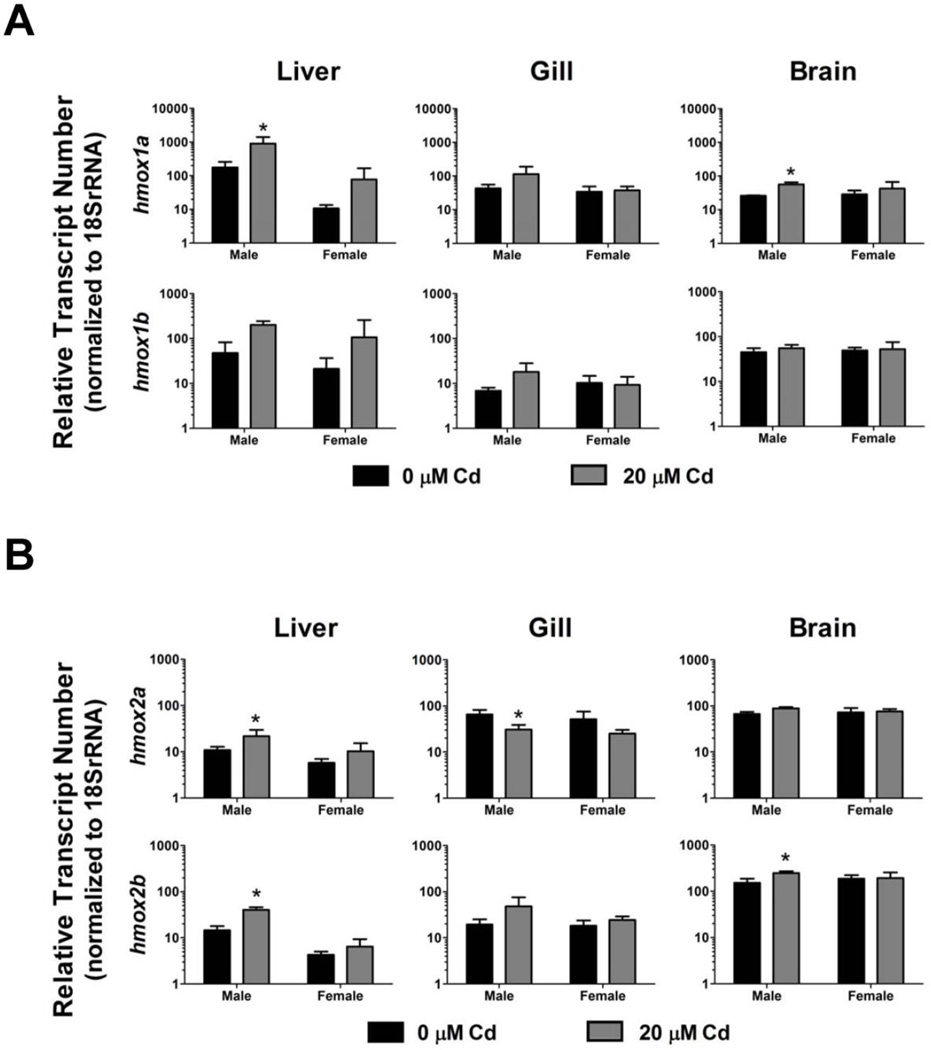
To compare induction levels of the hmox isoforms in adult tissues in response to pro-oxidant exposure, adult male and female zebrafish were continuously exposed to 20 µM Cd for a period of 96 hours. hmox1a was significantly induced in the liver and brain tissues of male zebrafish (Figure 4A). Although there was a trend toward induction of hmox1a in male gill tissue and female liver tissue, the differences were not statistically significant. Similar trends in induction were observed for hmox1b in male liver and gill tissue, and female liver tissue, although none of the changes were statistically significant (Figure 4A).
Figure 4. Induction of hmox1 and hmox2 paralogs in adult zebrafish in response to Cd exposure.
A) Relative fold induction of hmox1a and hmox1b in response to Cd exposure was determined in zebrafish liver, gill, and brain tissues (20 µM Cd for 96 hours). B) Relative fold induction of hmox2a and hmox2b in response to Cd exposure was determined in zebrafish liver, gill, and brain tissues (20 µM Cd for 96 hours). Statistical significance was determined using a two-way ANOVA followed by a Bonferroni post hoc test (*p-value < 0.05). All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates per treatment.
Similar trends in Cd-induced gene expression were also observable for the hmox2 isoforms in adult zebrafish tissues. hmox2a was significantly induced by Cd exposure in male liver samples, while a reduction of hmox2a expression was observed in the gill tissue of Cd-exposed male fish (Figure 4B). Similar trends for hmox2a were observed in the female liver and brain tissues although the reduction was not statistically significant. hmox2b was statistically induced in the brain and liver tissue of male zebrafish in response to Cd exposure (Figure 4B). Once again, there was a nonsignificant trend toward induction of hmox2b in the gill tissues of male zebrafish. Although the expression levels of hmox2b were generally higher in all three tissues of female zebrafish in response to Cd, the expression was neither statistically significant nor as prominent as the changes observed in male fish (Figure 4B).
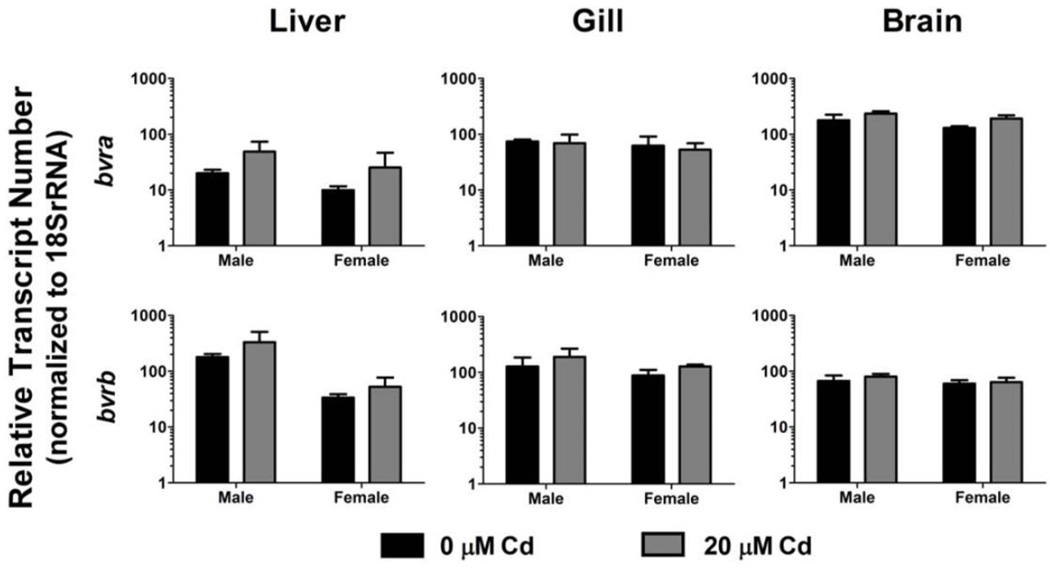
bvra and bvrb expression was also assessed in adult tissues under control and Cd-exposed conditions (Figure 5). Although there was a general trend toward bvra and bvrb induction in liver tissue from both male and female zebrafish in response to Cd exposure, the differences were not statistically significant. Expression of bvra and bvrb in control and Cd-treated fish was not significantly different in gill or brain tissue for either male or female zebrafish (Figure 5).
Figure 5. bvr expression in in adult zebrafish in response to Cd exposure.
Relative fold induction of bvra and bvrb was determined in zebrafish liver, gill, and brain tissues in control and Cd-exposed fish (20 µM Cd for 96 hours). Statistical significance was determined using two-way ANOVA followed by a Bonferroni post hoc test (*p-value < 0.05). All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates per treatment.
Expression of hmox and bvr during zebrafish development
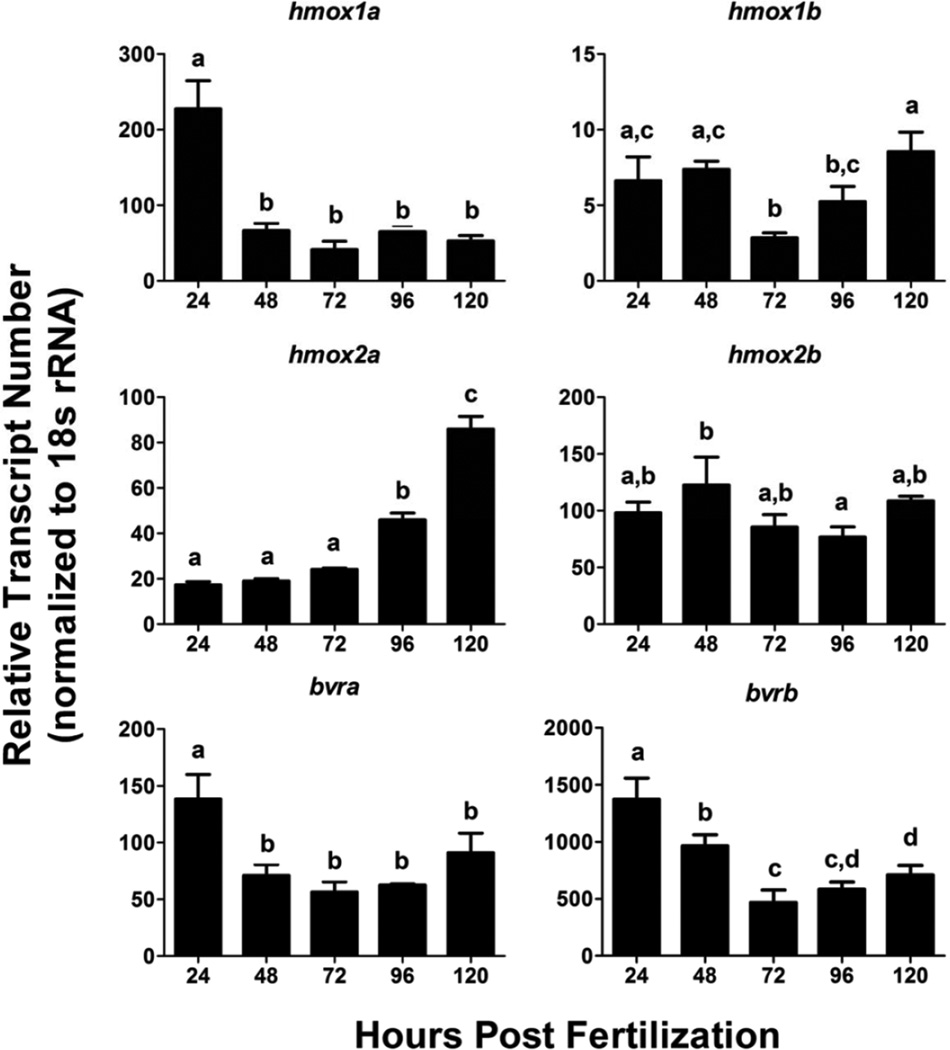
Quantitative expression profiling of hmox and bvr isoforms was performed in zebrafish embryos starting at 24 hpf and continuing in 24 hour intervals until 120 hpf (Figure 6). hmox1a was the most highly expressed isoform at 24 hpf, while hmox2b was the most highly expressed hmox isoform between 48 and 120 hpf. hmox1b transcript abundance was significantly lower at all time points, and did not have the same high level of exposure as seen for hmox1a at 24 hpf. hmox2a levels steadily increased during this developmental time period, with low levels being detected from 24 to 72 hpf followed by a statistically significant increase in expression at 96 and 120 hpf compared to 24 hpf (Figure 6). Conversely, hmox2b transcript abundance was generally higher than hmox2a and maintained relatively constant levels between 24–120 hpf. Additionally, bvra and bvrb displayed similar expression patterns between 24–120 hpf, with both isoforms having peaks in expression at 24 hpf which were statistically significant compared to the other timepoints. However, although the expression patterns shared the same trends, bvrb transcript levels were an order of magnitude higher in comparison to bvra at all of the assessed timepoints (Figure 6).
Figure 6. Changes in expression of hmox and bvr between 24–124 hpf.
Relative transcript abundance of each transcript was determined via real-time RT-PCR. Statistical significance in transcript expression in comparison to 24 hpf embryos was determined using one-way ANOVA followed by a Bonferroni post hoc test (*p-value < 0.05). Different letters represent statistical significance between developmental timepoints. All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates of 20 pooled embryos per treatment. hpf = hours past fertilization.
Effects of acute Cd exposure on heme degradation and iron homeostasis genes
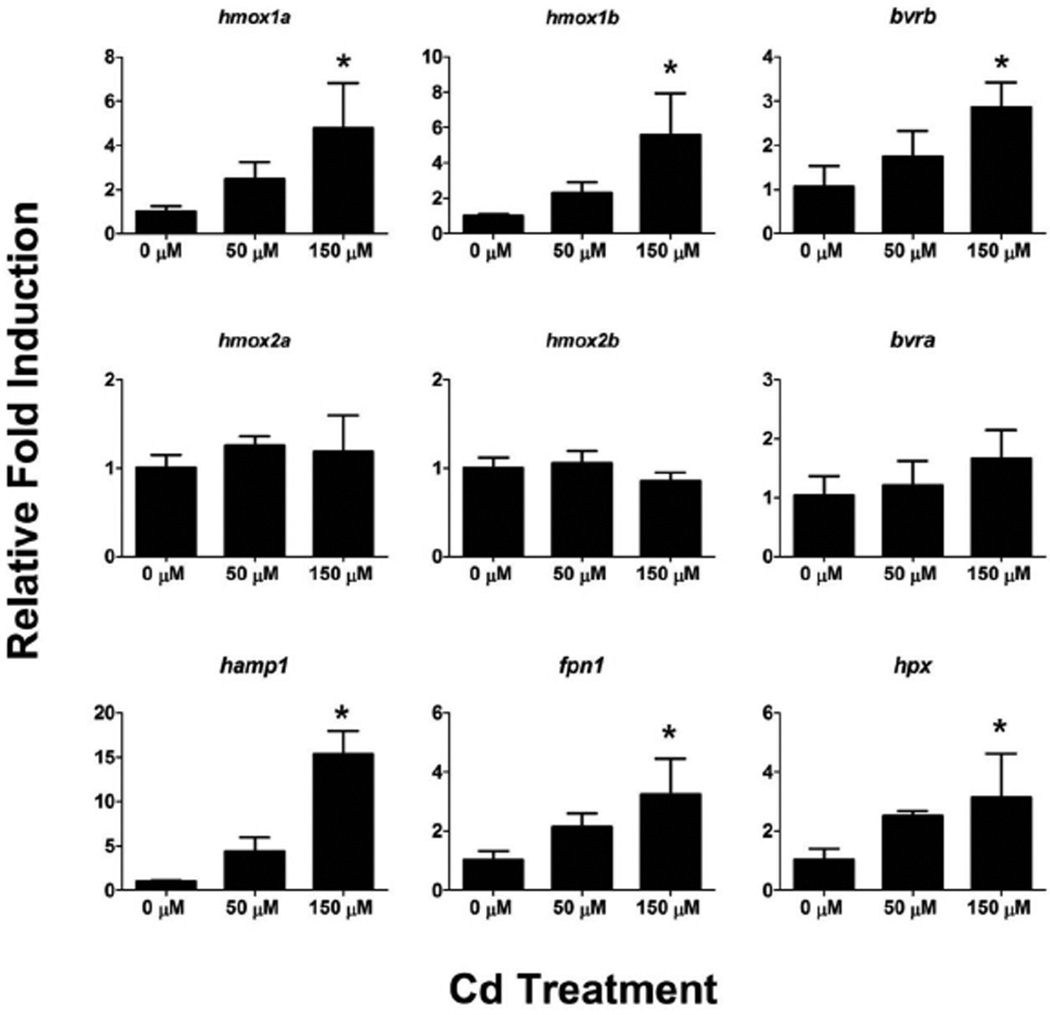
To investigate the inducibility of the hmox and bvr isoforms during zebrafish development, we performed 4 hour Cd challenges (50 and 150 µM Cd) on zebrafish eleutheroembryos starting at 72 hpf. hmox1a, hmox1b and bvrb were significantly upregulated by 150 µM Cd exposure at this timepoint (Figure 7). In contrast, there were no statistically significant changes in hmox2a, hmox2b or bvra expression in either Cd exposure at 72 hpf. To determine if related genes involved in maintaining heme and iron homeostasis responded to Cd exposure in the same manner as genes involved in heme degradation, we also assessed the expression levels of hemopexin (hpx), hepcidin (hamp) and ferroportin 1 (fpn1). fpn1, a major iron efflux transporter, and hamp, the peptide referred to as the master regulator of iron homeostasis for its role in regulating FPN1, were both significantly induced in the 150 µM Cd treatment. Furthermore, hpx, a heme binding protein, was also significantly induced in the 150 µM Cd treatment (Figure 7).
Figure 7. Effects of acute Cd exposure on gene expression in 76 hpf zebrafish.
Real-time RT-PCR was used to quantify changes in expression of heme degradation genes (hmox and bvr), as well as genes involved in heme and iron homeostasis, in whole zebrafish larvae at 76 hpf after a 4 hour exposure to Cd. Statistical significance in comparison to control embryos was determined using a one-way ANOVA followed by a Dunnett’s post hoc test (*p-value < 0.05). All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates of 20 pooled embryos per treatment.
Induction of hmox1b and bvrb by Cd exposure is dependent on NRF2a
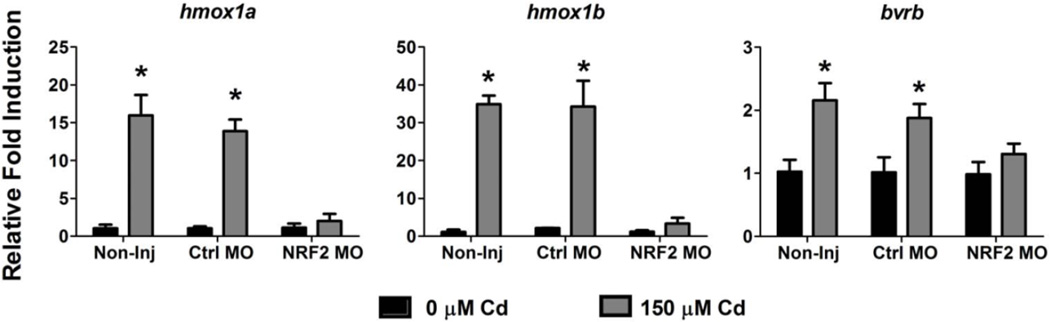
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a transcription factor most well known for its primary role in the regulation of a large battery of antioxidant genes, including hmox1. While the zebrafish Nrf2a paralog has been demonstrated to regulate the induction of hmox1a in response to oxidative stress (Alam et al., 1999; Nakajima et al., 2011; Timme-Laragy et al., 2012), its regulatory role in the induction of hmox1b and bvrb has not been confirmed. To determine if the induction of hmox1b and bvrb by Cd is Nrf2a dependent, embryos were injected with a standard Ctrl MO or the validated Nrf2a MO and exposed to 150 µM Cd for 4 hours starting at 72 hpf. To confirm the effectiveness of the Nrf2a knockdown, hmox1a expression was also assessed. Both the non-injected and Ctrl MO-injected embryos displayed significant induction of hmox1a, hmox1b and bvrb (Figure 8). In contrast, knockdown of Nrf2a resulted in the loss of Cd-mediated induction of all three genes: hmox1a (~85% reduction), hmox1b (~90% reduction), and bvrb (31% reduction) (Figure 8).
Figure 8. Effect of NRF2a knockdown on hmox1a, hmox1b and bvrb expression in response to Cd exposure.
Nrf2a translation was blocked by an Nrf2a MO injected at the two- to four-cell stage in zebrafish embryos. Real-time RT-PCR was used to quantify transcript abundance in whole zebrafish larvae at 76 hpf after a 4 hour exposure to 150 µM Cd. Relative fold change was determined by comparing all changes in gene expression to the non-injected controls (0 µM Cd). Statistically significant induction in response to Cd exposure was determined using a two-way ANOVA followed by a Bonferroni post hoc test (*p-value < 0.05). All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates of 20 pooled embryos per treatment.
Transcriptional response to additional pro-oxidant exposures
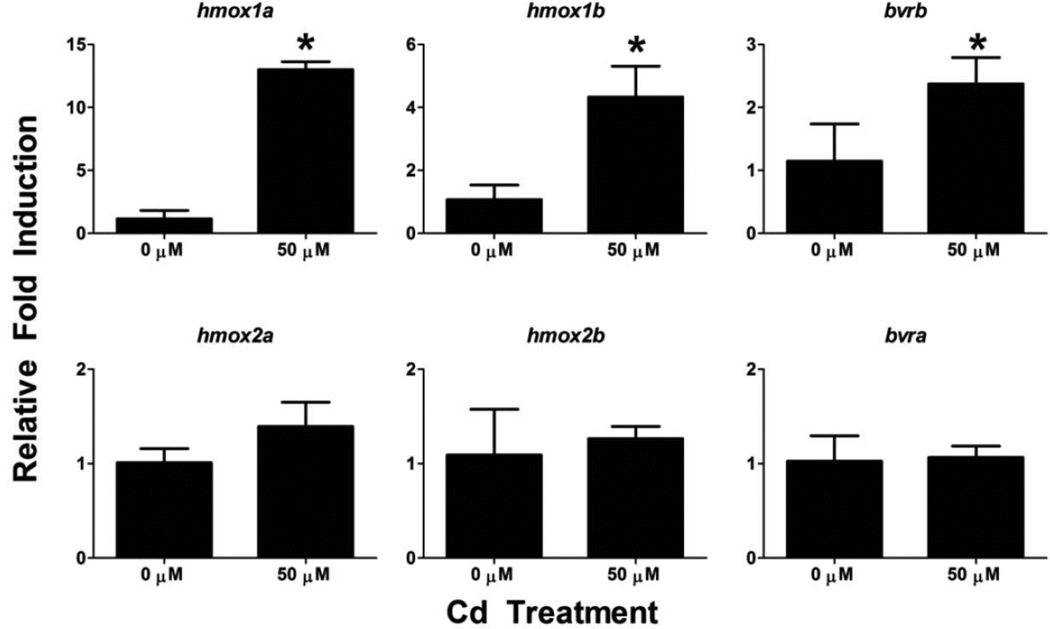
To determine if temporal changes in sensitivity to Cd exposure or if exposure to other compounds capable of generating oxidative stress through different mechanisms can affect the expression profiles of the hmox or bvr transcripts, additional chemical exposure treatments were performed. Since zebrafish eleutheroembryos become more sensitive to Cd exposure with age, 120 hpf larvae were acutely exposed to 50 µM Cd for 4 hours prior to characterization of HO and BVR expression. hmox1a, hmox1b and bvrb were significantly upregulated by 50 µM Cd exposure at this timepoint (Figure 9). However, there were still no statistically significant changes in hmox2a, hmox2b or bvra expression.
Figure 9. Effects of acute Cd exposure on gene expression in 124 hpf zebrafish.
Real-time RT-PCR was used to quantify changes in expression of heme degradation genes (hmox and bvr) following a 4 hour exposure to Cd starting at 120 hpf. Statistical significance in comparison to control embryos was determined using a Student’s t-test (*p-value < 0.05). All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates of 20 pooled embryos per treatment.
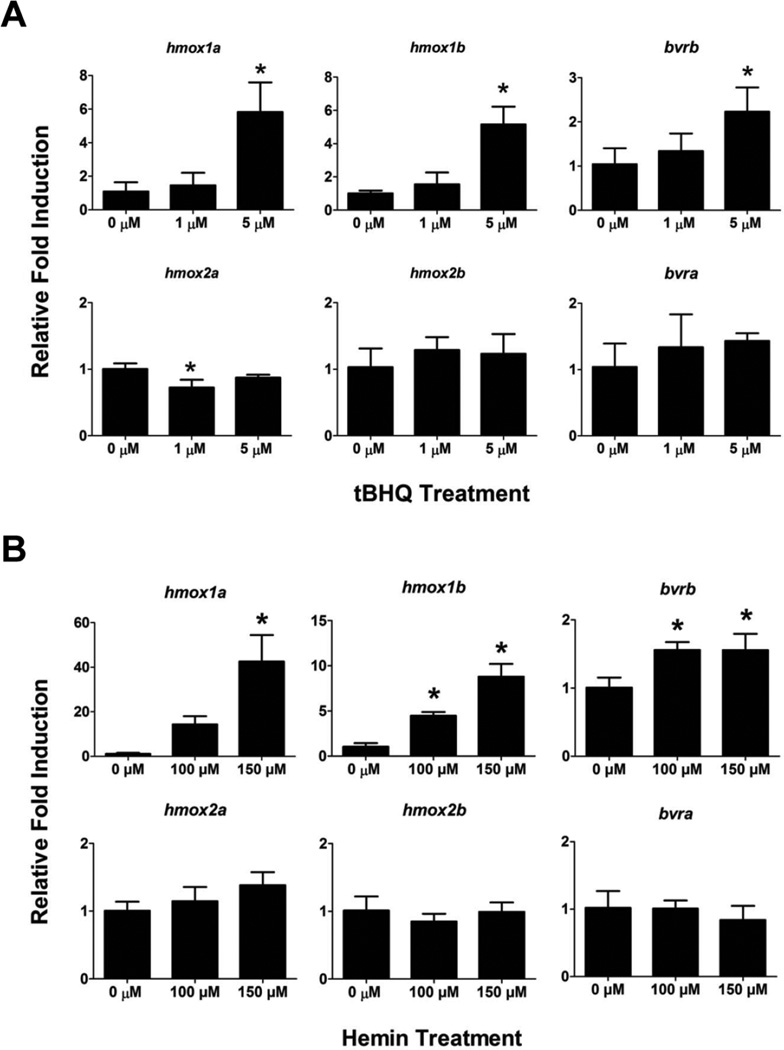
To determine if the hmox or bvr genes are induced in response to other pro-oxidants, 72 hpf eleutheroembryos were acutely exposed to tBHQ (1 and 5 µM) or hemin (100 and 150 µM) for four hours prior to characterization of hmox and bvr expression. Hemin is a porphyrin which can participate in Fenton reactions by releasing redox-active iron, whereas tBHQ is a quinone compound that is capable of producing oxidative stress via redox cycling. As seen with the Cd exposures, hmox1a, hmox1b and bvrb were only significantly upregulated by the higher tBHQ exposure (5 µM), while there were no statistically significant changes in hmox2a, hmox2b or bvra expression in either tBHQ treatment (Figure 10A). Similarly, hmox2a, hmox2b or bvra was not induced by either of the hemin exposures. However, while hmox1a was only significantly induced in the highest hemin treatment (150 µM), both hmox1b and bvrb were significantly induced by both hemin treatments (Figure 10B).
Figure 10. Effects of multiple pro-oxidant exposure on gene expression in 76 hpf zebrafish.
Real-time RT-PCR was used to quantify changes in expression of heme degradation genes (hmox and bvr) in 76 hpf larvae following a 4 hour exposure to either A) tBHQ or B) hemin. Statistical significance in comparison to control embryos was determined using a one-way ANOVA followed by a Dunnett’s post hoc test (*p-value < 0.05). All values are normalized to 18S ribosomal RNA. Error bars represent one standard deviation; n = 3 biological replicates of 20 pooled embryos per treatment.
Stress-responsive enhancer motifs in the hmox and bvr gene promoters
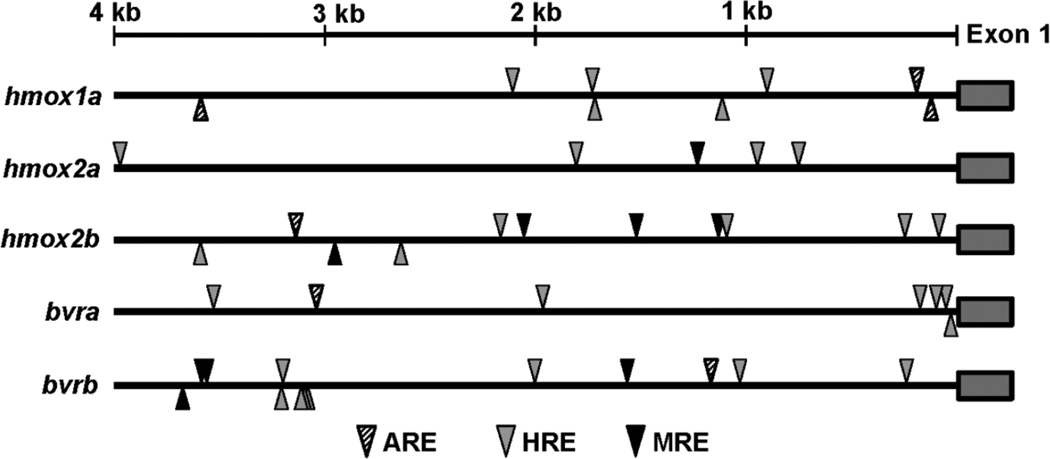
To gain further insight into the transcriptional regulation of the hmox and bvr genes in response to stress, the 4 kb region upstream of exon 1 for each gene was searched for DNA-binding motifs targeted by Nrf2 (ARE motif), Mtf-1 (MRE motif) and Hif-1α (HRE motif) (Figure 11). Although a previous study identified three AREs in the hmox1a promoter (Nakajima et al., 2011) using a slightly less conservative ARE consensus motif (TGASNNNGC), the two ARE motifs we identified within the proximal promoter region (within 250 base pairs of the transcription start site) are consistent with the previous study. A single ARE was identified in the promoter regions of hmox2b, bvra and bvrb, while hmox2a did not appear to contain the ARE motif. A single MRE was identified in the promoter region of hmox2a, while bvrb and hmox2b both had multiple MREs within the 4 kb promoter regions. All five genes had multiple HRE motifs spread throughout the promoter region, with bvra and hmox2b containing multiple HRE motifs within their proximal promoter regions (Figure 11).
Figure 11. Schematic of enhancer motifs in hmox and bvr gene promoters.
Diversity and position of the antioxidant-responsive elements (ARE), hypoxia-responsive elements (HRE) and metal-responsive elements (MRE) in the 4 kb upstream region of the hmox and bvr genes. Arrowheads above the line represent motifs on the sense DNA strand, while arrowheads below the line represent motifs on the antisense DNA strand.
Discussion
HMOX1 and HMOX2 catabolize unbound heme into biliverdin, CO and free iron, while biliverdin can be further reduced into bilirubin by either BVRa or BVRb. In addition to their basic role in heme metabolism, potential roles related to the amelioration of oxidative stress, erythropoiesis (Garcia-Santos et al., 2014) and cell signaling (HMOX2, BVRa) (Verma et al., 1993; Maines, 1997) have also been suggested for these enzymes. However, very little is known regarding the ontogenetic expression of these heme degradation genes. Furthermore, as a result of the teleost-specific genome duplication, zebrafish have multiple hmox1 (hmox1a and hmox1b) and hmox2 (hmox2a and hmox2b) genes. This study is the first to not only evaluate the ontogenetic expression of all four zebrafish hmox paralogs (hmox1a, hmox1b, hmox2a and hmox2b) and both bvr isoforms, but also the first to evaluate transcriptional responses of all the zebrafish hmox paralogs and bvr isoforms to pro-oxidant exposures within the same treatment context.
Differential expression of the hmox genes in adult tissues and during zebrafish development may be indicative of subfunction partitioning
Given the established observations of high levels of gene expression for HMOX1 in liver tissue (Tenhunen et al., 1969), and HMOX2 in brain (Trakshel et al., 1988; Maines et al., 1986; Ewing and Maine, 1992) and vascular tissues (Zakhary et al., 1996) of mammalian models, we chose to focus our attention on characterizing expression of the heme degradation enzymes in the liver, brain and gill tissues of adult zebrafish. It was also important to characterize any significant differences in hmox and bvr expression during zebrafish development. Significant differences in constitutive expression patterns of the hmox1 and hmox2 paralogs were observed in both brain and gill tissue (Figure 3A), whereas only hmox1a and hmox1b displayed significant differences in basal expression in male liver tissue samples. The most significant difference in basal hmox expression in adult tissues was the sexually dimorphic expression of hmox1a, hmox2a and hmox2b in liver (Figure 3A). Interestingly, all four hmox paralogs differed significantly in their basal expression patterns during early development (Figure 6). The duplication, degeneration, and complementation model (DDC) suggests that duplicated genes can gain a new function, delegate their original function (subfunction partitioning) or become pseudogenes which may be lost through selection if the redundancy is unnecessary (Amores et al., 1998). Many duplicated genes in zebrafish have undergone subfunctionalization, resulting in paralogs with complementary functions or expression patterns (Postlethwait et al., 2004) compared to the single mammalian homolog. Sexually dimorphic gene expression patterns, as seen for multiple hmox genes in adult liver tissue samples (Figure 3), are not uncommon and usually explained by differences in hormonally-mediated regulation. However, the contrasting basal expression of the hmox1 and hmox2 paralogs in adult brain and gill tissues (Figure 3A), as well as during zebrafish development (Figure 6), is indicative of differences in transcriptional regulation that may represent the first step in the determination of any partitioning of function between these duplicated genes.
The most significant difference in the expression of the hmox1 paralogs was the significantly high levels of hmox1a expression in 24 hpf embryos compared to hmox1b (Figure 6). These results may suggest that Hmox1a, and not Hmox1b, is required for an early developmental function that may or may not be related to a role in the oxidative stress response. For example, a previous study has demonstrated that the cellular redox environment is highly oxidized during early zebrafish development (~3–24 hpf) but transitions to a more reduced environment starting around 36 to 48 hpf (Timme-Laragy et al., 2013). During the earlier developmental period total glutathione levels are low and the ratio of reduced glutathione (GSH) to oxidized gluthathione (GSSG) is significantly lowered suggesting that GSH’s role as a major antioxidant may be limited during this early developmental period. Interestingly, a previous study demonstrated that depletion of BVR’s enzymatic product bilirubin was more detrimental than GSH depletion treatment with buthionine sulfoxime, and resulted in a greater incidence of oxidative stress and apoptosis (Barañano et al., 2002). Thus, the Hmox/Bvr enzymatic pathway may play a major antioxidant role during early zebrafish development. However, multiple studies have linked expression of HMOX enzymes and its production of the carbon monoxide by-product with angiogenesis and enhanced vascularization during mammalian development and under times of cellular stress (Deramaudt et al., 1998; Lin et al., 2008; Grochot-Przeczek et al., 2009; Zhao et al., 2011). Thus, the high expression of hmox1a during early zebrafish development may play a more fundamental role in cellular signaling.
The hmox2 paralogs also had contrasting expression patterns. hmox2a expression was quite low at 24 hpf but started to significantly increase around 96 hpf, while hmox2b expression was fairly consistent throughout development (Figure 6). HMOX2 is known to be highly expressed in mammalian brain tissues where it is thought to participate in cell signaling through the generation of CO (Verma et al., 1993; Maines, 1997). Neural tissue is one of the most prominent tissues during early zebrafish development and the embryonic brain is well established prior to the initiation of the rest of organogenesis. The constitutive expression of hmox2b during development and its prominent expression in the brains of adult zebrafish may be indicative of a more prominent signaling role of Hmox2b in neural tissues. In contrast, the significant increase in hmox2a expression during later larval development and its higher expression in gill tissues of adults may be indicative of other tissue-specific roles. While purely speculation at this time, such possibilities highlight the importance of considering the different physiological roles of various tissues when investigating the potential for novel or partitioned function of the hmox genes.
hmox and bvr expression in response to pro-oxidant exposure
Although the induction of Hmox1 by oxidative stress is well established (Alam et al., 1999; Ryter et al., 2006; Alam and Cook, 2007), not as many studies have focused on the transcriptional response of the BVR enzymes to pro-oxidant exposure. Furthermore, much of the research focusing on the antioxidant cycling of bilirubin via biliverdin reductase has primarily focused on BVRa (Stocker et al., 1987; Maines et al., 2001; Sedlak and Snyder, 2004; Maines, 2007; Lerner-Marmarosh et al., 2008; Sedlak et al., 2009). Although neither bvra nor bvrb displayed any significant induction in adult tissues in response to the Cd exposure, bvrb was significantly induced after exposure to all three pro-oxidants in zebrafish eleutheroembryos at various developmental timepoints (Figures 7, 9 and 10). bvrb was also shown to be expressed at nearly an order of magnitude higher than bvra throughout early zebrafish development 24–120 hpf (Figure 6). These observations suggest that Bvrb may also be playing a significant role in the oxidative stress response, as well as heme metabolism.
While exposure to Cd resulted in a general increase in hmox1a and hmox1b liver expression in both males and females (Figure 4), it is of interest that basal expression of hmox1a is nearly 20 times greater in male liver tissue than females, and that hmox1b expression is more than double that of females of the same tissue (Figure 3). A similar sexual dimorphic expression pattern was noted for bvrb in liver tissue, and bvrb is generally expressed at a higher level in liver tissues compared to bvra (Figure 3). Interestingly, females are known to be more susceptible to Cd toxicity than males, an observation believed to be primarily mediated through the estrogen receptor (Johnson et al., 2003). Since the liver serves as the primary detoxification organ, the strong dimorphic expression pattern of the hmox1 paralogs and bvrb may serve as an additional mechanism that contributes to the gender-specific sensitivity to Cd. Future studies would be required to confirm this hypothesis.
Although HMOX2 is generally regarded as being constitutively expressed and non-inducible (Maines, 1986; Ryter et al., 2006), this isoform has been shown to be activated by corticosterone, opiates and menadione (Liu et al., 2000; Maines and Gibbs, 2005; Vukomanovic et al., 2011). Consistent with known expression of HMOX2 in other vertebrates (Ewing and Maines, 1992; Maines and Trakshel, 1993; Ewing and Maines, 1997), both hmox2 paralogs were more highly expressed compared to the hmox1 paralogs in brain tissue, with hmox2b showing the greatest levels of expression (Figure 3). In addition, hmox2b was upregulated in response to Cd in brain and liver tissues of males (Figure 4). Interestingly, bvra was observed to be the predominant isoform in adult brain tissue consistent with a previous vertebrate study (McCoubrey et al., 1995). The differential expression of bvra and bvrb under basal conditions and in response to pro-oxidant exposure raises some interesting questions about the physiological requirements that necessitate the need for two biliverdin reducing enzymes. Furthermore, the functional and structural differences between the two BVR enzymes requires us to consider functions related to heme metabolism, as well as other mechanisms related to cell signaling and the oxidative stress response. For example, while both enzymes contain the reductase domain, BVRa’s domain has dual cofactor (NADH and NADPH) dual pH specificity (Kutty and Maines, 1981). In addition, BVRa also has several regulatory domains and motifs not found in BVRb, such as a serine, threonine, tyrosine (S/T/Y) kinase domain, a basic leucine zipper DNA binding domain (bZip), nuclear export and localization signals, and Src Homology 2 (SH2) domains, which collectively allow BVRa to interact with other proteins and participate in insulin signaling and mitogen-activated protein kinase (MAPK) pathways (Kapitulnik and Maines, 2008; Lerner-Marmarosh et al., 2005; Lerner-Marmarosh et al., 2008). Furthermore, while there appears to be a high degree of sequence similarity between BVRa proteins throughout nature (Maines, 2007), the extent to which these unique functional attributes of BVRa are conserved across vertebrates has not been rigorously examined. These discrepancies highlight the need for further studies in other model systems, such as zebrafish.
Potential regulators of HO and BVR expression in response to oxidative stress
NRF2, the master regulator of the oxidative stress response, is known to upregulate HMOX1 in response to oxidative stress (Alam, 1994; Alam et al., 1999; Alam and Cook, 2007), and while it has been implicated in the regulation of BVRb this has not been confirmed (Wu et al., 2011; Moon et al., 2012). Two paralogs of nrf2 in zebrafish resulting from the teleost-specific genome duplication event have undergone subfunction partitioning, with Nrf2a thought to act primarily as a transcriptional activator and Nrf2b acting as a transcriptional repressor (Timme-Laragy et al., 2012). While the induction of hmox1a was expected, we also documented novel observations regarding hmox1b and bvrb induction in response to Cd, tBHQ or hemin at different zebrafish developmental timepoints (Figures 7, 9 and 10). Furthermore, we also demonstrated the induction of several genes that produce proteins that play important roles in heme and iron homeostasis (Figure 7). Of these, both FPN1 and HAMP have been shown to be regulated by NRF2 in response to pro-oxidant exposure (Marro et al., 2010) or as a mechanism related to iron homeostasis (Harada et al., 2011; Tanaka et al., 2012; Bayele et al., 2015). However, the metal-responsive transcription factor 1 (MTF-1), the master regulator of metals homeostasis, is also activated in response to Cd (Stuart et al., 1985; Günther et al., 2012) and has been demonstrated to regulate HAMP, FPN1 and HPX (Troadec et al., 2010; Troadec et al., 2010; Balesaria et al., 2010; O'Shields et al., 2014). Future studies will address the co-regulation of these heme and iron homeostasis genes in by NRF2 and MTF-1 in response to pro-oxidant exposure.
Since Nrf2a has been demonstrated to positively regulate the battery of antioxidant genes in zebrafish, including hmox1a (Nakajima et al., 2011; Fuse et al., 2015), we sought to confirm its role in regulating hmox1b and bvrb in response to Cd exposure in zebrafish eleutheroembryos. While no significant differences in the Cd-mediated induction of hmox1a, hmox1b or bvrb were observed between non-injected and Ctrl MO-injected embryos, expression of these transcripts were significantly reduced in the Nrf2a morphants (Figure 8). Thus, while Nrf2a does not appear to play a role in regulating the constitutive expression of bvrb in zebrafish at 72 hpf, it does appear to play a role in mediating the response to Cd treatment. These results are consistent with our observation of ARE motifs in the promoter regions of hmox1a and bvrb (Figure 11). Unfortunately, the current zebrafish genome assembly precludes the opportunity to identify enhancer motifs in the hmox1b genes, but our data suggests that there is likely some conserved ARE motifs within its proximal promoter.
Although bvra was not induced during development or in adult tissues in response to pro-oxidant exposures, previous studies suggest that it can be regulated by other well-known stress response pathways. For example, both rat and human BVRa gene promoters contain regulatory elements for NFκB and HIF-1α (Gibbs et al., 2010) which is in agreement with human BVRa being activated by hypoxia (Salim et al., 2001) and inhibited by NFκB (Gibbs et al., 2010). These results are consistent with the observation of four HRE motifs in the proximal promoter region of the zebrafish bvra gene (Figure 11). Additionally, computer analysis of the human BVRa promoter suggested numerous candidate regulatory elements including the aryl hydrocarbon receptor (AHR) and heat shock factor 1 (HSF-1) (Gibbs et al., 2010). Interestingly, both bilirubin and biliverdin have also been implicated in the activation of the AHR pathway (Sinal and Bend, 1997; Phelan et al., 1998). Thus, future studies are required to fully characterize the transcriptional response and regulation of bvra and bvrb under both basal and stressful cellular conditions.
Furthermore, these data confirm the role of Nrf2a in regulating hmox1b in response to Cd treatment. However, it should be noted that a previous study demonstrated a transient induction of hmox1a in the liver of 120 hpf zebrafish by diethyl maleate (DEM), while the DEM-mediated induction of hmox1b persisted for a much longer period of time (Fuse et al., 2015). This transient, liver-specific expression of hmox1a was attributed to regulation by the Bach1 transcriptional repressor. Interestingly, hemin treatment resulted in the prolonged induced of hmox1a in 120 hpf zebrafish livers, presumably mediated through hemin-dependent inhibition of Bach1. Thus, while Nrf2a does play a role in regulating hmox1a and hmox1b, other transcription factors may contribute to hmox1 paralog-specific differences in expression.
Conclusion
These differences in hmox1 and hmox2 developmental expression, adult tissue distribution, and responses to oxidative stress are consistent with potential subfunction partitioning of the paralogous isoforms. Furthermore, with respect to BVRa and BVRb, it is interesting that these two proteins of distinct evolutionary origin have evolved to have similar reductase activity. However, the presence of a diversity of other functional domains within the BVRa isoform offers future opportunities to evaluate the potential differences in function during development or within adult tissues. Future studies employing the powerful reverse genetics techniques, such as targeted mutagenesis using the CRISPR-Cas system, may be used to characterize the functional differences between the Hmox paralogs and BVR isoforms.
Highlights.
hmox1a, hmox2a, hmox2b and bvrb are sexually dimorphic in expression
hmox paralogs were induced in adult tissues by cadmium exposure
hmox1a, hmox1b and bvrb were induced by multiple pro-oxidants zebrafish embryos
Differential expression of zebrafish hmox paralogs suggest partitioning of function
Nrf2a mediates the induction of hmox1b and bvrb by cadmium in zebrafish embryos
Acknowledgments
This work was supported by the National Institutes of Health Pathway to Independence grant [R00ES017044] from the National Institute of Environmental Health Sciences awarded to M.J. Jenny. This work was also partially supported by a Grant-in-Aid of Research from Sigma Xi, The Scientific Research Society, to A. Holowiecki. The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
There are no competing interests.
References
- Abraham NG, Nelson JC, Ahmed T, Konwalinka G, Levere RD. Erythropoietin controls heme metabolic enzymes in normal human bone marrow culture. Exp. Hematol. 1989;17:908–913. [PubMed] [Google Scholar]
- Abraham NG, Kappas A. Pharmacological and clinical aspects of heme oxygenase. Pharmacol. Rev. 2008;60:79–127. doi: 10.1124/pr.107.07104. [DOI] [PubMed] [Google Scholar]
- Ahmad Z, Salim M, Maines MD. Human biliverdin reductase is a leucine zipper-like DNA-binding protein and functions in transcriptional activation of heme oxygenase-1 by oxidative stress. J. Biol. Chem. 2002;277:9226–9232. doi: 10.1074/jbc.M108239200. [DOI] [PubMed] [Google Scholar]
- Alam J. Multiple elements within the 5' distal enhancer of the mouse heme oxygenase-1 gene mediate induction by heavy metals. J. Biol. Chem. 1994;269:25049–25056. [PubMed] [Google Scholar]
- Alam J, Cook JL. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am. J. Respir. Cell. Mol. Biol. 2007;36:166–174. doi: 10.1165/rcmb.2006-0340TR. [DOI] [PubMed] [Google Scholar]
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Balesaria S, Ramesh B, McArdle H, Bayele HK, Srai SK. Divalent metal-dependent regulation of hepcidin expression by MTF-1. FEBS Lett. 2010;584:719–725. doi: 10.1016/j.febslet.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Barañano DE, Rao M, Ferris CD, Snyder SH. Biliverdin reductase: a major physiologic cytoprotectant. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16093–16098. doi: 10.1073/pnas.252626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayele HK, Balesaria B, Srai SK. Phytoestrogens modulate hepcidin expression by Nrf2: Implications for dietary control of iron absorption. Free Radic. Biol. Med. 2015;89:1192–1202. doi: 10.1016/j.freeradbiomed.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechinger S, Kusch R, Haugo K, Matz C, Chivers D, Krone P. Brief embryonic cadmium exposure induces a stress response and cell death in the developing olfactory system followed by long-term olfactory deficits in juvenile zebrafish. Toxicol. Appl. Pharmacol. 2007;224:72–80. doi: 10.1016/j.taap.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Blechinger S, Warren J, Jr, Kuwada J, Krone P. Developmental toxicology of cadmium in living embryos of a stable transgenic zebrafish line. Environ. Health Persp. 2002;110:1041. doi: 10.1289/ehp.021101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AC, Lutton JD, Pearson HA, Nelson JC, Levere RD, Abraham NG. Heme metabolism and in vitro erythropoiesis in anemia associated with hypochromic microcytosis. Am. J. Hematol. 1988;27:1–6. doi: 10.1002/ajh.2830270102. [DOI] [PubMed] [Google Scholar]
- Chen X, Agarwal A, Giedroc DP. Structural and functional heterogeneity among the zinc fingers of human MRE-binding transcription factor-1. Biochemistry. 1998;37:11152–11161. doi: 10.1021/bi980843r. [DOI] [PubMed] [Google Scholar]
- Deramaudt BM, Braunstein S, Remy P, Abraham NG. Gene transfer of human heme oxygenase into coronary endothelial cells potentially promotes angiogenesis. J. Cell. Biochem. 1998;68:121–127. doi: 10.1002/(sici)1097-4644(19980101)68:1<121::aid-jcb12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Doré S, Takahashi M, Ferris CD, Zakhary R, Hester LD, Guastella D, Snyder SH. Bilirubin, formed by activation of heme oxygenase-2, protects neurons against oxidative stress injury. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2445–2450. doi: 10.1073/pnas.96.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing JF, Maines MD. In situ hybridization and immunohistochemical localization of heme oxygenase-2 mRNA and protein in normal rat brain: Differential distribution of isozyme 1 and 2. Mol. Cell. Neurosci. 1992;3:559–570. doi: 10.1016/1044-7431(92)90068-d. [DOI] [PubMed] [Google Scholar]
- Ewing JF, Maines MD. Histochemical localization of heme oxygenase-2 protein and mRNA expression in rat brain. Brain Res. Protoc. 1997;1:165–174. doi: 10.1016/s1385-299x(96)00027-x. [DOI] [PubMed] [Google Scholar]
- Fuse Y, Nakajima H, Nakajima-Takagi Y, Nakajima O, Kobyashi M. Heme-mediated inhibition of Bach1 regulates the liver specificity and transience of the Nrf2-dependent induction of zebrafish heme oxygenase 1. Genes Cells. 2015;20:590–600. doi: 10.1111/gtc.12249. [DOI] [PubMed] [Google Scholar]
- Garcia-Santos D, Schranzhofer M, Horvathova M, Jaberi MM, Bogo Chies JA, Sheftel A, Ponka P. Heme oxygenase 1 is expressed in murine erythroid cells where it controls the level of regulatory heme. Blood. 2014;123:2269–2277. doi: 10.1182/blood-2013-04-496760. [DOI] [PubMed] [Google Scholar]
- Gibbs PE, Miralem T, Maines MD. Characterization of the human biliverdin reductase gene structure and regulatory elements: promoter activity is enhanced by hypoxia and suppressed by TNF-alpha-activated NF-kappaB. FASEB. J. 2010;24:3239–3254. doi: 10.1096/fj.09-144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA. The jaundice of the cell. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15837–15839. doi: 10.1073/pnas.012685199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochot-Przeczek A, Lach R, Mis J, Skrzypek K, Gozdecka M, Sroczynska P, Dubiel M, Rutkowski A, Kozakowska M, Zagorska A, Walczynski J, Was H, Kotlinowski J, Drukala J, Kurowski K, Kieda C, Herault Y, Dulak J, Jozkowicz A. Heme oxygenase-1 accelerates cutaneous wound healing in mice. PLoS One. 2009;4:e5803. doi: 10.1371/journal.pone.0005803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther V, Lindert U, Schaffner W. The taste of heavy metals: gene regulation by MTF-1. BBA-Mol. Cell Res. 2012;1823:1416–1425. doi: 10.1016/j.bbamcr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Harada N, Kanayama M, Maruyama A, Yoshida A, Tazumi K, Hosoya T, Mimura J, Toki T, Maher JM, Yamamoto M, Itoh K. Nrf2 regulates ferroportin 1-mediated iron efflux and counteracts lipopolysaccharide-induced ferroportin 1 mRNA suppression in macrophages. Arch. Biochem. Biophys. 2011;508:101–109. doi: 10.1016/j.abb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Hill A, Teraoka H, Heideman W, Peterson R. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol. Sci. 2005;86:6. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assuncao JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Threadgold G, Harden G, Ware D, Begum S, Mortimore B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Lloyd C, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Urun Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberlander M, Rudolph-Geiger S, Teucke M, Lanz C, Raddatz G, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Schuster SC, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nusslein-Volhard C, Hubbard TJ, Roest Crollius H, Rogers J, Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Kenney N, Stoica A, Hilakivi-Clarke L, Singh B, Chepko G, Clarke R, Sholler PF, Lirio AA, Foss C, Reiter R, Trock B, Paik S, Martin MB. Cadmium mimics the in vivo effects of estrogen in the uterus and mammary gland. Nat. Med. 2003;9:1081–1084. doi: 10.1038/nm902. [DOI] [PubMed] [Google Scholar]
- Kapitulnik J, Maines MD. Pleiotropic functions of biliverdin reductase: cellular signaling and generation of cytoprotective and cytotoxic bilirubin. Trend Pharmacol. Sci. 2008;30:129–137. doi: 10.1016/j.tips.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty R, Maines MD. Purification and characterization of biliverdin reductase from rat liver. J. Biol. Chem. 1981;256:3956–3962. [PubMed] [Google Scholar]
- Larsen R, Gouveia Z, Soares MP, Gozzelino R. Heme cytotoxicity and the pathogenesis of immune-mediated inflammatory diseases. Front. Pharmacol. 2012;3:1–17. doi: 10.3389/fphar.2012.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner-Marmarosh N, Miralem T, Gibbs PE, Maines MD. Human biliverdin reductase is an ERK activator; hBVR is an ERK nuclear transporter and is required for MAPK signaling. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6870–6875. doi: 10.1073/pnas.0800750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner-Marmarosh N, Shen J, Torno M, Kravets A, Hu Z, Maines MD. Human biliverdin reductase: a member of the insulin receptor substrate family with serine/threonine/tyrosine kinase activity. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7109–7114. doi: 10.1073/pnas.0502173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Chen YH, Chang PF, Lee YT, Yet SF, Chau LY. Heme oxygenase-1 promotes neovascularization in ischemic heart by coinduction of VEGF and SDF-1. J. Mol. Cell. Cardiol. 2008;45:44–55. doi: 10.1016/j.yjmcc.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Liu N, Wang X, McCoubrey W, Maines MD. Developmentally regulated expression of two transcripts for heme oxygenase-2 with a first exon unique to rat testis: control by corticosterone of the oxygenase protein expression. Gene. 2000;241:175–183. doi: 10.1016/s0378-1119(99)00439-4. [DOI] [PubMed] [Google Scholar]
- Maines MD. The heme oxygenase system: a regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- Maines MD. Biliverdin reductase: PKC interaction at the cross-talk of MAPK and PI3K signaling pathways. Antioxid. Redox. Signal. 2007;9:2187–2195. doi: 10.1089/ars.2007.1805. [DOI] [PubMed] [Google Scholar]
- Maines MD, Ewing JF, Huang TJ, Panahian N. Nuclear localization of biliverdin reductase in the rat kidney: response to nephrotoxins that induce heme oxygenase-1. J. Pharmacol. Exp. Ther. 2001;296:1091–1097. [PubMed] [Google Scholar]
- Maines MD, Gibbs PE. 30 Some years of heme oxygenase: from a "molecular wrecking ball" to a "mesmerizing" trigger of cellular events. Biochem. Biophys. Res. Commun. 2005;338:568–577. doi: 10.1016/j.bbrc.2005.08.121. [DOI] [PubMed] [Google Scholar]
- Maines MD, Trakshel G, Kutty R. Characterization of two constitutive forms of rat liver microsomal heme oxygenase. Only one molecular species of the enzyme is inducible. J. Biol. Chem. 1986;261:411–419. [PubMed] [Google Scholar]
- Maines MD, Trakshel GM. Purification and characterization of human biliverdin reductase. Arch. Biochem. Biophys. 1993;300:320–326. doi: 10.1006/abbi.1993.1044. [DOI] [PubMed] [Google Scholar]
- Marro S, Chiabrando D, Messaba E, Stolte J, Turco E, Tolosano E, Muckenthaler MU. Heme controls ferroportin1 (FPN1) transcription involving Bach1, NRF2 and a MARE/ARE sequence motif at position −7007 of the FPN1 promoter. Haematologica. 2010;95:1261–1268. doi: 10.3324/haematol.2009.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoubrey WK, Jr, Cooklis MA, Maines MD. The structure, organization and differential expression of the rat gene encoding biliverdin reductase. Gene. 1995;160:235–240. doi: 10.1016/0378-1119(95)00112-j. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Moon M, McDevitt E, Zhu J, Stanley B, Krzeminski J, Amin S, Aliaga C, Miller T, Isom HC. Elevated hepatic iron activates NF-E2-related factor 2-regulated pathway in a dietary iron overload mouse model. Toxicol. Sci. 2012;129:74–85. doi: 10.1093/toxsci/kfs193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Nakajima-Takagi Y, Tsujita T, Akiyama S, Wakasa T, Mukaigasa K, Kaneko H, Tamaru Y, Yamamoto M, Kobayashi M. Tissue-restricted expression of Nrf2 and its target genes in zebrafish with gene-specific variations in the induction profiles. PLoS One. 2011;6:e26884. doi: 10.1371/journal.pone.0026884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shields B, McArthur AG, Holowiecki A, Kamper M, Tapley J, Jenny MJ. Inhibition of endogenous MTF-1 signaling in zebrafish embryos identifies novel roles for MTF-1 in development. BBA- Mol. Cell Res. 2014;1843:1818–1833. doi: 10.1016/j.bbamcr.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan D, Winter GM, Rogers WJ, Lam JC, Denison MS. Activation of the Ah receptor signal transduction pathway by bilirubin and biliverdin. Arch. Biochem. Biophys. 1998;357:155–163. doi: 10.1006/abbi.1998.0814. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol. Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- Saleh MC, McConkey S. NADH-dependent cytochrome b5 reductase and NADPH methemoglobin reductase activity in the erythrocytes of Oncorhynchus mykiss. Fish Physiol. Biochem. 2012;38:1807–1813. doi: 10.1007/s10695-012-9677-2. [DOI] [PubMed] [Google Scholar]
- Salim M, Brown-Kipphut B, Maines MD. Human biliverdin reductase is autophosphorylated, and phosphorylation is required for bilirubin formation. J. Biol. Chem. 2001;276:10929–10934. doi: 10.1074/jbc.M010753200. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak K. Analyzing real-time PCR data by the comparative CT method. Nature Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sedlak TW, Saleh M, Higginson DS, Paul BD, Juluri KR, Snyder SH. Bilirubin and glutathione have complementary antioxidant and cytoprotective roles. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5171–5176. doi: 10.1073/pnas.0813132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak TW, Snyder SH. Bilirubin benefits: cellular protection by a biliverdin reductase antioxidant cycle. Pediatrics. 2004;113:1776–1782. doi: 10.1542/peds.113.6.1776. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Keung SW, Passantino R, Concordet JP, Maire P, Giallonog A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Shalloe F, Gordon E, Ennis O, Mantle T. Evidence that biliverdin-IXb reductase and flavin reductase are identical. Biochem. J. 1996;316:385–387. doi: 10.1042/bj3160385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinal CJ, Bend JR. Aryl hydrocarbon receptor-dependent induction of cyp1a1 by bilirubin in mouse hepatoma heap 1c1c7 cells. Mol. Pharmacol. 1997;52:590–599. doi: 10.1124/mol.52.4.590. [DOI] [PubMed] [Google Scholar]
- Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Searle Peter F, Palmiter Richard D. Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. Nature. 1985;317:828–831. doi: 10.1038/317828a0. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Ikeda T, Yamamoto K, Ogawa H, Kamisako T. Dysregulation of fatty acid oxidation enzymes and iron-regulatory genes in livers of Nrf2-null mice. J. Gastroenterol. Hepatol. 2012;27:1711–1717. doi: 10.1111/j.1440-1746.2012.07180.x. [DOI] [PubMed] [Google Scholar]
- Tenhunen R, Marver HS, Schmid R. Microsomal Heme Oxygenase: Characterization of the Enzyme. J. Biol. Chem. 1969;244:6388–6394. [PubMed] [Google Scholar]
- Timme-Laragy AR, Goldstone JV, Imhoff BR, Stegeman JJ, Hahn ME, Hansen JM. Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic. Biol. Med. 2013;65:89–101. doi: 10.1016/j.freeradbiomed.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy AR, Karchner SI, Franks DG, Jenny MJ, Harbeitner RC, Goldstone JV, McArthur AG, Hahn ME. Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2) J. Biol. Chem. 2012;287:4609–4627. doi: 10.1074/jbc.M111.260125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakshel GM, Kutty RK, Maines MD. Purification and characterization of the major constitutive form of testicular heme oxygenase. The noninducible isoform. J. Biol. Chem. 1986;261:11131–11137. [PubMed] [Google Scholar]
- Trakshel GM, Kutty RK, Maines MD. Resolution of the rat brain heme oxygenase activity: absence of a detectable amount of the inducible form (HO-1) Arch. Biochem. Biophys. 1988;260:732–739. doi: 10.1016/0003-9861(88)90503-6. [DOI] [PubMed] [Google Scholar]
- Troadec MB, Ward DM, Lo E, Kaplan J, De Domenico I. Induction of FPN1 transcription by MTF-1 reveals a role for ferroportin in transition metal efflux. Blood. 2010;116:4657–4664. doi: 10.1182/blood-2010-04-278614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- Verma A, Hirsch DJ, Glatt CE, Ronnett GV, Snyder SH. Carbon monoxide: a putative neural messenger. Science. 1993;259:381–384. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- Vukomanovic D, McLaughlin BE, Rahman MN, Szarek WA, Brien JF, Jia Z, Nakatsu K. Selective activation of heme oxygenase-2 by menadione. Can. J. Physiol. Pharmacol. 2011;89:861–864. doi: 10.1139/y11-091. [DOI] [PubMed] [Google Scholar]
- Wegiel B, Baty CJ, Gallo D, Csizmadia E, Scott JR, Akhavan A, Chin BY, Kaczmarek E, Alam J, Bach FH, Zuckerbraun BS, Otterbein LE. Cell surface biliverdin reductase mediates biliverdin-induced anti-inflammatory effects via phosphatidylinositol 3-kinase and akt. J. Biol. Chem. 2009;284:21369–21378. doi: 10.1074/jbc.M109.027433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Cui J, Klaassen C. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol. Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Quandt K, Hultquist D. Characterization of NADPH-dependent methemoglobin reductase as a heme-binding protein present in erythrocytes and liver. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2130–2134. doi: 10.1073/pnas.89.6.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Komoda Y, Nakajima H. Biliverdin-IX alpha reductase and biliverdin-IX beta reductase from human liver. Purification and characterization. J. Biol. Chem. 1994;269:24343. [PubMed] [Google Scholar]
- Yamaguchi T, Komuro A, Nakano Y, Tomita M, Nakajima H. Complete amino acid sequence of biliverdin-IXβ reductase from human liver. Biochem. Biophys. Res. Commun. 1993;197:1518–1523. doi: 10.1006/bbrc.1993.2649. [DOI] [PubMed] [Google Scholar]
- Zakhary R, Gaine SP, Dinerman JL, Ruat M, Flavahan NA, Snyder SH. Heme oxygenase 2: endothelial and neuronal localization and role in endothelium-dependent relaxation. Proc. Natl. Acad. Sci. U.S.A. 1996;93:795–798. doi: 10.1073/pnas.93.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Azuma J, Kalish F, Wong RJ, Stevenson DK. Maternal heme oxygenase 1 regulates placental vasculature development via angiogenic factors in mice. Biol. Reprod. 2011;85:1005–1012. doi: 10.1095/biolreprod.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]