Significance
Whereas most of the more than 130 described mycobacterial species are harmless saprophytes, Mycobacterium tuberculosis, the human tuberculosis-causing agent, represents one of the deadliest bacterial pathogens in the history of humankind. To explore the mechanisms behind this spectacular evolutionary trajectory toward pathogenicity, we have experimentally investigated the faculty of different tuberculosis-causing mycobacteria in conducting horizontal gene transfer (HGT). Our studies identified unique chromosomal DNA transfer between strains of the Mycobacterium canettii clade, which resemble most closely the putative common ancestor of the M. tuberculosis complex. This outstanding feature suggests that during the evolution of M. tuberculosis, HGT might have represented the major mechanism for acquisition of genes that helped these mycobacteria to increasingly resist host defenses and become major pathogens.
Keywords: recombination, DNA transfer, tuberculosis, Mycobacterium canettii, evolution
Abstract
Horizontal gene transfer (HGT) is a major driving force of bacterial diversification and evolution. For tuberculosis-causing mycobacteria, the impact of HGT in the emergence and distribution of dominant lineages remains a matter of debate. Here, by using fluorescence-assisted mating assays and whole genome sequencing, we present unique experimental evidence of chromosomal DNA transfer between tubercle bacilli of the early-branching Mycobacterium canettii clade. We found that the obtained recombinants had received multiple donor-derived DNA fragments in the size range of 100 bp to 118 kbp, fragments large enough to contain whole operons. Although the transfer frequency between M. canettii strains was low and no transfer could be observed among classical Mycobacterium tuberculosis complex (MTBC) strains, our study provides the proof of concept for genetic exchange in tubercle bacilli. This outstanding, now experimentally validated phenomenon presumably played a key role in the early evolution of the MTBC toward pathogenicity. Moreover, our findings also provide important information for the risk evaluation of potential transfer of drug resistance and fitness mutations among clinically relevant mycobacterial strains.
Horizontal gene transfer (HGT) is a major molecular mechanism responsible for generating genetic diversity, which plays a particularly important role for driving evolution in otherwise clonal bacterial populations. However, the contribution of HGT to the (recent) evolution of mycobacteria and the impact on the most dominant pathogenic species, Mycobacterium tuberculosis, the agent of human tuberculosis, remains controversial. Several studies provide overall congruence that the classical M. tuberculosis complex (MTBC) shows a perfectly clonal population structure (1–7), yet, other studies have proposed that frequent genetic exchange among MTBC strains might exist (8, 9). As potential interstrain recombination in tubercle bacilli influences the risk evaluation for transfer of drug resistance-associated mutations between clinical M. tuberculosis isolates, novel insights into the question of HGT among tubercle bacilli are of utmost importance.
Recent whole genome sequence (WGS) analyses of early branching, smooth tubercle bacilli (STB) of the Mycobacterium canettii clade revealed genome-wide traces of putative interstrain recombination events (10, 11), raising questions on the existence and functionality of particular HGT mechanisms in tubercle bacilli. An interesting observation in this respect was made for the nonvirulent, fast-growing model organism Mycobacterium smegmatis, which shows an unconventional conjugal DNA transfer (12, 13), distinct from classical plasmid-mediated DNA transfer described for other bacteria (14, 15). M. smegmatis transconjugants typically display highly mosaic genomes reminiscent of products of eukaryotic meiosis, composed of multiple donor-derived sequence segments that are distributed without apparent regional bias across the genome. Consequently, this type of HGT was termed distributive conjugal transfer (DCT) (16). Detection of mosaic sequence arrangements in M. canettii genomes (10, 11, 17, 18) raised the issue whether similar molecular mechanisms might be active in tuberculosis-causing mycobacteria and/or in mycobacteria, in general.
Here, we experimentally explored this question by establishing a mycobacterial mating assay with both fluorescent and antibiotic resistance markers, which allowed us to provide a unique experimental evidence of interstrain DNA exchange between M. canettii donor and recipient strains and, hence, show that HGT in slow-growing mycobacteria resembles DCT of phylogenetically distant fast-growing species. HGT thus appears to be an important instrument shaping mycobacterial evolution. However, this faculty might also have declined in some groups, such as extant members of the classical MTBC, for which we could not detect any signs of functional HGT despite extensive attempts and analyses. Our findings provide unique insights into the evolutionary processes that shaped the genome of one of the most successful human pathogens, M. tuberculosis.
Results
Setup of a Fluorescence-Mediated Mating Assay.
DCT was originally described as a mechanism of DNA transfer between M. smegmatis donor and recipient trains that generated genome-wide mosaicism in the transconjugants (16). For the initial setup of our experimental mating assay, we thus rendered M. smegmatis strains mc2155 (NC_008596), known to be an efficient donor (16), GFP fluorescent, and hygromycin resistant (Hygr) by using the pYUB412-derived integrating cosmid F10 (19, 20). In parallel, red colonies of strain mc2874 (NZ_AOCJ01000000), known to be a good recipient (16), were obtained by transformation with a replicative, nonmobilizable plasmid, carrying a kanamycin resistance (Kmr) cassette and a gene encoding DsRed. After incubation on filter plates as previously described (12), potential recombinants were identified on Km/Hyg agar plates by using orange colony appearance as selection criterion. After PCR verifications (SI Appendix, Fig. S1A), 10 independent Kmr/Hygr strains were subjected to Illumina-based whole genome sequencing (WGS), generating reads that were de novo assembled and compared with the genomes of the donor and recipient strains (SI Appendix, Figs. S1B and S2). In accordance with previous reports (16), we observed marked genome mosaicism of the recombinant M. smegmatis strains, with 2–11 DNA blocks transferred from donor to recipient genomes. The sizes varied between 250 bp and 244 kb, with an average of 290 kb of total transferred DNA, and these sequence blocks were scattered around the genome without obvious regional bias. However, for one Kmr/Hygr variant (RC-Ms 10), unexpectedly, the observed genome sequence corresponded to the mc2155 donor strain, suggesting that it had received the nonmobilizable, kanamycin-resistance plasmid from the recipient (SI Appendix, Fig. S2). Overall, our results with M. smegmatis strains mc2155 and mc2874 confirm previous observations (16), but also suggest putative alternative mechanisms of plasmid transfer. Importantly, the results validated the used fluorescence/antibiotic markers in the experimental setup for use in HGT experiments on M. canettii and MTBC strains.
Interstrain HGT Identified in M. canettii Strains.
To assess various M. canettii and M. tuberculosis strains for their ability to exchange chromosomal DNA, green-fluorescent putative donor strains were constructed by transformation with pYUB412 or F10 integrating cosmids (20) as established before for M. smegmatis strains. Red-fluorescent potential M. canettii recipient strains were obtained by electroporation with plasmid pMRF1-DsRed, which was successful for all strains except M. canettii J (CIPT 140070017), which exclusively harbors an eptABCD gene cluster (10, 21), recently described as restricting plasmid maintenance in M. smegmatis (22).
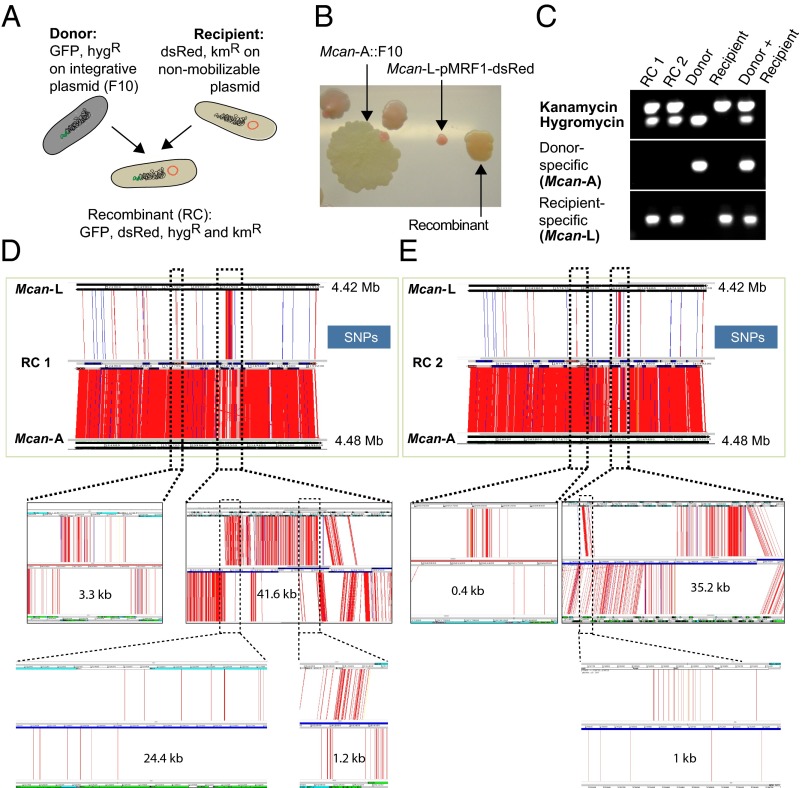
Mating of different Hygr strains (donors) with selected Kmr strains (recipients) for periods ranging from 3 to 7 d generated several hundred double-resistant colonies as a result of at least 44 mating combinations (Table 1) including donors carrying either cosmid F10 (SI Appendix, Table S1) or pYUB412 (SI Appendix, Table S2). Among all these clones, orange colonies originating from the mating pair M. canettii A::F10+M. canettii L-pMRF1 (Fig. 1 A and B and SI Appendix, Tables S1 and S4) revealed PCR results that were positive for both antibiotic cassettes and recipient-specific sequences (Fig. 1C). Subsequently, eight strains were selected for WGS analysis, as described below.
Table 1.
Overview of mating combinations between different M. canettii and M. tuberculosis strains
| M. canettii Hygr donor strains | Kmr receipient strains | MTBC Hygr donor strains | Kmr receipient strains |
| Mcan-A, | |||
| Mcan-K, | |||
| Mcan-K, | Mtb-H37Rv | Mcan-L, | |
| Mcan-A | Mcan-L, | Mcan-J, | |
| Mtb-H37Rv, | Mtb-79112, | ||
| H37RvΔRD1 | Mtb-Tb36 | ||
| Mcan-A, | |||
| Mcan-K, | |||
| Mcan-I | Mcan-L, | Mtb-79112 | Mtb-H37Rv |
| Mtb-H37Rv, | Mtb-Tb36 | ||
| H37RvΔRD1 | |||
| Mcan-A, | |||
| Mcan-K, | Mtb-Tb36 | Mtb-H37Rv | |
| Mcan-J | Mcan-L, | Mtb-79112 | |
| Mtb-H37Rv, | |||
| H37RvΔRD1 | |||
| Mcan-A, | Mcan-A, | ||
| Mcan-K | Mcan-L, | H37RvΔRD1 | Mcan-K, |
| Mtb-H37Rv, | Mcan-L, | ||
| H37RvΔRD1 | Mtb-H37Rv | ||
| Mcan-A, | |||
| Mcan-L | Mcan-K, | ||
| Mtb-H37Rv, | |||
| H37RvΔRD1, |
Donor strains carrying the pYUB412 or F10 cosmid were used, as well as recipient strains carrying the pMRF1-dsRed replicative plasmid, except for M. canettii J, which contains an integrative pMV306.Kan plasmid-mediated insertion, due to a putative restricted plasmid maintenance, linked to the presence of the eptABCD gene cluster. M. canettii donor strains A, K, L, and I additionally carried spontaneous rifampicin resistance mutations. Only for mating pair M. canettii A and M. canettii L recombinants (bold) were obtained.
Fig. 1.
DNA exchange between M. canettii A and M. canettii L. (A) Schematic representation of donor, recipient, and recombinant strains harboring plasmids with different antibiotic cassettes and genes encoding fluorochromes. (B) Differently colored KmR and HygR colonies corresponding to a spontaneously KmR M. canettii A::F10 donor strain, some spontaneously HygR M. canettii L pMRF-dsRed recipient strains, together with orange colored colonies that represent double-resistant putative recombinants, which have obtained the GFP-expressing gene cluster from the donor. (C) Results from PCR analysis of two recombinants, the donor M. canettii A and the recipient M. canettii L with oligonucleotides amplifying either the kanamycin or the hygromycin resistance cassettes as well as genes specific for the donor or the recipient strain. (D and E) ACT visualization of SNPs identified between two recombinant (RC) genomes (middle genome of each image) and either the donor M. canettii A (bottom) or the recipient M. canettii L genome (top). A selection of the transferred sequence blocks are enlarged (dotted lines) for better visualization of the donor-derived segments. SNPs are represented by red and indels by blue lines.
To test whether clinically relevant resistance mutations (23, 24) might also be subject to transfer, spontaneously rifampicin resistant (Rifr) donor strains (M. canettii A, I, K, and L) were included in the M. canettii mating experiments (Table 1 and SI Appendix, Tables S1 and S4). Although Hygr/Kmr resistant clones were obtained, PCR analysis did not confirm the presence of both cassettes, suggesting implication of spontaneous resistance mutations rather than DNA-transfer events. Moreover, none of these Hygr/Kmr double-resistant strains was resistant to rifampicin.
M. canettii Recombinants Show Genome Mosaicism.
For further genomic characterization, the above-described M. canettii A+L recombinants (RC) were repassaged two times on solid media to ensure isolated clones and subjected to DNA extraction and Illumina-based WGS. Comparison of the assembled sequences with those of reference genomes of M. canettii L (NC_019965) and M. canettii A (NC_015848) revealed several clearly defined interstrain recombination loci in all strains (Fig. 1 D and E and SI Appendix, Fig. S3). Additional PacBio sequencing (Macrogen) of RC1 and RC2 allowed us to obtain single contig chromosome assemblies of the recombinants, confirming the recombination sites on the respective RC1 and RC2 genomes (SI Appendix, Fig. S4). Moreover, this analysis pointed to a yet-undetected inversion of a 435-kb genomic region in strain M. canettii L that spans the origin of replication and is flanked by IS1557 transposase-encoding sequences (SI Appendix, Figs. S4 and S5). Together, these results provide unique experimental evidence that the two different M. canettii strains had exchanged DNA under our experimental conditions. The transferred DNA fragments were identified around the cosmid integration attB site, situated within the glyV-tRNA at position 2820.678 kb in the genome of M. canetti L and 2828.773 kb in M. canettii A. In addition, in all M. canettii recombinants with the exception of RC6, multiple donor-derived sequence blocks were identified around the genome without any regional bias (Table 2 and SI Appendix, Figs. S3 and S6). One to 13 individual sequence blocks were transferred, which varied remarkably in size, ranging between 100 bp to up to 118 kb (Table 2). In total, 32 to up to 335 kb of donor-derived DNA could be identified in the individual recombinants. Strikingly, as observed for M. smegmatis (SI Appendix, Fig. S2) (16), multiple homologous recombination events seem to have occurred in proximity to each other in the M. canettii recombinants, thus creating local genetic heterogeneity (Table 2). This effect was especially evident in M. canettii recombinants RC1 and RC4, where seven and five inherited donor fragments, respectively, were interspersed by stretches of recipient DNA in intervals of 1.2–15 kb (Fig. 1D and SI Appendix, Fig. S6 and Table S3). These observations thus offer a plausible explanation for the observed genome mosaicism (10) in M. canettii strains. Of note, however, the frequency of DNA transfer in M. canettii was found to be substantially lower than seen for in M. smegmatis, with <1 × 10−8 (recombinants per donor) compared with 5 × 10−4 to 1 × 10−6 in M. smegmatis, depending on the antibiotic marker used for selection (12).
Table 2.
Overview of exchanged fragments of the eight recombinants obtained after mapping against the genomes of M. canettii strains A and L
| F | RC1 | RC2 | RC3 | RC4 | RC6 | RC7 | RC9 | RC10 |
| NF | 8 | 4 | 3 | 13 | 1 | 7 | 7 | 5 |
| ST | 97 | 37 | 87 | 328 | 32 | 335 | 130 | 143 |
| SF | 0.3 | 0.1 | 26.6 | 1.0 | 32 | 15.1 | 0.19 | 3.5 |
| 0.7 | 0.4 | 28.5 | 5.7* | 16.3 | 3.0 | 6.3 | ||
| 1.2 | 1.0 | 31.4 | 6.9 | 26.4 | 3.5 | 16.2 | ||
| 3.3 | 35.2 | 7.5 | 35.2 | 8.6 | 20.2 | |||
| 11.5 | 11.2 | 42.4 | 18.0 | 96.5 | ||||
| 13.6 | 12.7 | 81.5 | 31.6 | |||||
| 24.4 | 21.2 | 117.6 | 65.3 | |||||
| 41.6 | 36.8 | |||||||
| 48.0 | ||||||||
| 54.9 | ||||||||
| 78.0 |
Patterns of RC1 and RC2 are shown in Fig. 1 and patterns of RC3, RC4, RC6, RC7, RC9, and RC10 in SI Appendix, Fig. S3. *Two fragments transferred with same size; F, features; NF, number of transferred fragments; SF, sizes of transferred fragments (in kilobases); ST, total size of transferred DNA (in kilobases).
Lack of HGT Among MTBC Strains.
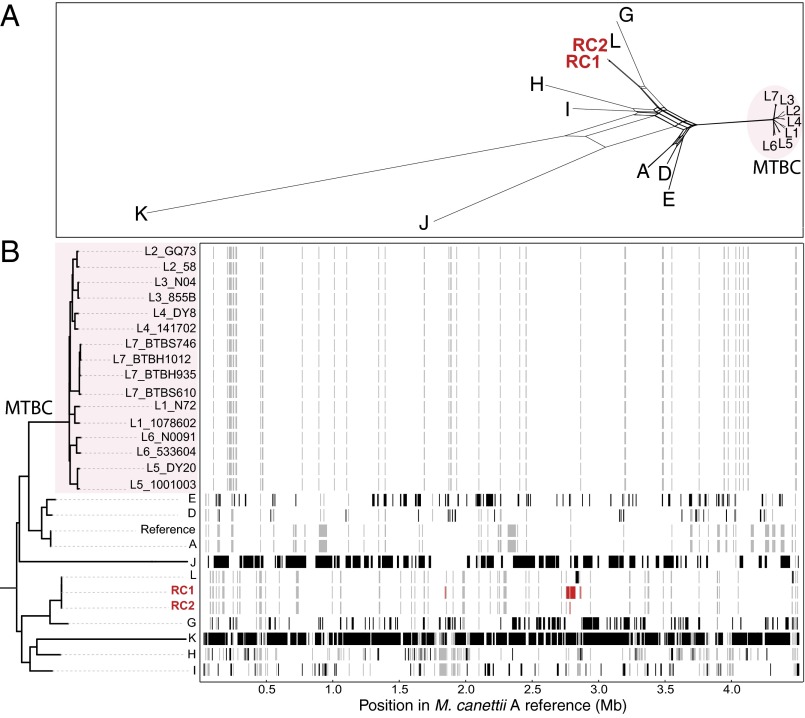
Having successfully established the mating conditions for M. smegmatis and M. canettii strains, we deemed our experimental setup as adequate to tackle the question whether DNA transfer mechanisms might be functional among M. tuberculosis strains. We undertook a series of mating experiments, using the same plasmids and protocols that had previously yielded positive transfer results both for M. smegmatis and/or M. canettii strains, in M. tuberculosis reference strain H37Rv and two M. tuberculosis strains of lineage 1. However, for none of the 12 mating pair combinations (summarized in Table 1 and SI Appendix, Tables S1 and S2), interstrain HGT could be detected under the conditions used. We also included M. tuberculosis H37Rv and several M. canettii strains (A, I, J, K, and L) as mating pairs in our experimental setup, as in silico genome comparison previously suggested potential transfer events from M. tuberculosis to M. canettii J (10), but could not isolate any M. tuberculosis/M. canettii recombinants. Finally, we also included M. tuberculosis H37RvΔRD1 showing a truncated esx-1 locus (25) into the series of mating experiments (Table 1), because previous data from M. smegmatis suggested that donor strains without a functional ESX-1 system yielded increased DCT (16). However, similar to the other tested M. tuberculosis strains, no recombinants were obtained (SI Appendix, Tables S1 and S2). We conclude from these experiments that M. tuberculosis strains seem to be less prone to HGT, in line with the predominantly clonal population structure of the MTBC that clearly contrasts with the recombinogenic M. canettii strain pool (Fig. 2).
Fig. 2.
Recombinogenic vs. clonal strain cluster organization, distinguishing M. canettii and MTBC strains of lineages 1–7, respectively. Assessment of recombination among tubercle bacilli. (A) NeighborNet analysis of 9 M. canettii genomes and 16 MTBC genomes (representing each of the seven lineages), showing extensive recombination among the early branching tubercle bacilli relative to the MTBC. Relationships were inferred based on alignments of 53,603 variable nucleotides identified by whole genome comparisons against the M. canettii A reference sequence. (B) ClonalFrameML analysis of recombination of the same 27 genomes used in A above, showing the extent of recombination among M. canettii and the early-branching tubercle bacilli. The maximum likelihood phylogeny was inferred by using FastTree. Black horizontal bars indicate recombination events detected by the analysis in extant taxa. Red horizontal bars indicate recombination events detected in the laboratory-generated strains RC1 and RC2. Gray-shaded horizontal bars are inferred recombination events in a hypothetical common ancestor.
DNA Transfer Frequency Evaluated Under Different Mating Conditions.
To possibly increase the transfer frequency in tubercle bacilli, we performed a series of additional mating experiments with the above-mentioned successful mating pair of M. canettii A as donor and M. canettii L as recipient, using different conditions thought to favor HGT. However, neither the use of different mating temperatures (30 or 37 °C), nor different concentrations of the DNA cross-linker Mitomycin C (1.7 ng/mL, 30 ng/mL, and 100 ng/mL) yielded recombinants (SI Appendix, Table S4). Mitomycin C was used because it is known to induce competence for transformation in some bacteria, including Bacillus subtilis and Legionella pneumophila by inducing DNA damage (26). However, in another series of experiments, mating assays were performed in the presence of low concentrations of either oxygen or nitrogen radicals (hydrogen peroxide, H2O2 or nitrogen monoxide, NO), or slightly acidic pH (pH 6.1) to mimic putative stimuli encountered by pathogenic mycobacteria inside host cells. By this approach, several M. canettii A+L recombinants were obtained, whereas no particular impact of the corresponding conditions could be detected (SI Appendix, Table S5). Six selected clones were subjected to WGS (RC3, 4, 6, 7, 9, and 10) after PCR confirmation (Fig. 2 and SI Appendix, Fig. S3).
To evaluate whether DNA transfer could be triggered during intracellular stages of the bacterial lifecycle, we tested M. canettii, M. smegmatis, and M. canettii and M. tuberculosis mating combinations during infection of phagocytic amoeba of the species Acanthamoeba castellanii. Such amoeba were speculated to represent a host for environmental mycobacteria (10, 27). However, despite simultaneous uptake and survival of different bacteria inside single amoeba cells, even after prolonged incubation of up to 10 d (SI Appendix, Fig. S7) neither M. canettii, nor M. smegmatis, nor M. canettii/M. tuberculosis recombinants were obtained. These findings suggest that HGT does not occur under these experimental intracellular conditions or that it might occur at frequencies below detection thresholds.
Furthermore, extended cell-to-cell contact is required, because incubation of the different M. canettii mating combinations in liquid cultures using either the rich medium 7H9 supplemented with ADC or Sauton minimal medium did not yield any recombinants. Thus, phage-mediated DNA transfer can be excluded as a reason for the observed genome mosaicism. In addition, no prophage were detected in the genomes of M. canettii A and L, in contrast to M. canettii I, where a 55-kb prophage region was found (10). However, no recombinants were obtained when M. canettii I was used as a donor (SI Appendix, Table S1).
Finally, we also tested whether some of the results obtained might have been due to yet-unknown natural competence mechanisms of M. canettii. However, upon incubation of M. canettii strains A, J, K, and L with linear DNA fragments or vectors containing an apramycin resistance cassette flanked by DNA sequences homologous to a genomic region of M. tuberculosis H37Rv (genomic position at 1881.7–1886.5 kb), no PCR-confirmed clones were obtained, making natural competence an implausible mechanism for the observed DNA exchanges between M. canettii strains. These results also reflect the historic, long-lasting difficulties for the development of efficient tools to genetically manipulate mycobacteria, which only became practicable through the use of electroporation (28) or phage-assisted methods (29).
Discussion
Conjugation is besides transformation and transduction one of the three principal processes how bacteria may exchange DNA, thereby representing an important accelerator for evolution and adaptation to new environments (14, 15, 30). For mycobacteria, in contrast to other bacterial genera, knowledge about HGT processes remains scarce. Apart from the reported plasmid-mediated conjugal transfer in Mycobacterium marinum (31), a chromosomal transfer mechanism similar to eukaryotic meiotic-like recombination was described for M. smegmatis (16). Whereas the first mechanism involves a mega-plasmid, encoding elements of type IV and type VII secretion systems, present in similar form also in Mycobacterium kansasii and Mycobacterium yongonense (32), chromosomal HGT in M. smegmatis requires a set of genes with donor and recipient functions, as well as genes conferring mating identity linked to an ESX-1 type VII system (12, 16, 33, 34). In contrast, little is known on potential HGT in tuberculosis-causing mycobacteria.
First hints came from putative recombination tracts identified by in silico analyses in genomes of smooth tubercle bacilli of the M. canettii clade (10, 11). M. canettii strains are remarkable, rare mycobacteria, which are almost exclusively isolated from patients in the region of the Horn of Africa (35, 36). Their mode of transmission seems to be different to M. tuberculosis because no human-to-human transmission of M. canettii strains was yet reported (18, 35, 37). M. canettii strains might have a yet-unknown, possibly aquatic, environmental reservoir where different strains can get in frequent direct contact, thereby enhancing the possibilities of HGT (10, 17, 38, 39). In a similar scenario, one could also imagine that M. tuberculosis and the MTBC originally emerged from an M. canettii-like strain pool and subsequently adapted to their respective host(s), followed by clonal expansion (21, 39) (Fig. 2). Our here-presented findings support this scenario, as we demonstrate that DNA transfer between two distinct M. canettii strains occurs and furthermore creates progeny containing several unlinked donor-derived DNA segments. These sequence fragments have sizes between 100 bp and 118 kb and are integrated at different loci on the chromosome. In M. canettii, the transferred sequences identified in the recipient strain contained in addition to the cosmid-vector sequences sections of donor-derived genomic regions flanking the attB vector integration site. These flanking regions were either integrated as a large segment as seen in recombinants RC2 (35.2-kb fragment), RC3 (31 kb), RC6 (32 kb), and RC10 (97 kb), or the integrated donor DNA was interrupted by stretches of recipient DNA, thus creating local genetic heterogeneity (e.g., recombinants RC1, 4, 7, and 9). Microheterogeneity was also observed in M. smegmatis transconjugants, where regions with short alternating donor and recipient DNA were created (16).
The molecular machinery involved in this process is yet unconfirmed. In M. smegmatis, ESX-1 and RecA are required in the recipient for DNA transfer to occur, the latter plausibly for homologous recombination events. An M. smegmatis recA donor mutant led to a higher rate of gene conversion in the recipient, likely caused by unrepaired DNA breaks in the donor and, hence, more potential DNA substrates to be transferred (13). For tubercle bacilli, previous genome-based in silico analyses suggested that the analyzed M. canettii strains contained M. canettii J and M. canettii K-derived recombination tracts, thus proposing that these two strains were efficient donors (11). Interestingly, sequence comparison of their recA genes revealed that both strains differ in their intein-encoding and predicted excised RecA sequences from other M. canettii and MTBC strains (SI Appendix, Fig. S8). However, under the conditions tested, we were not able to isolate any recombiants by using M. canettii strains J or K as donors. Instead, M. canettii A was found to be able to successfully transfer DNA into the recipient M. canettii L despite predictions that M. canettii A-derived sequences only made up a minor part of other M. canettii genomes in in silico analyses (11). The impact of inteins on recombination in selected mycobacterial strains might thus be limited, in accordance with previous findings (40).
Another pathway that may influence double-strand breaks and DNA transfer is represented by RecBCD (41), for which sequence analyses reveal a conservation of 97–99% amino acid (aa) identity among the different M. canettii and MTBC strains. Characteristic alterations were noted for the exoDNase V β-chain RecB, which in its N-terminal sequence shows 10 aa differences between M. canettii strains and M. tuberculosis/MTBC strains (SI Appendix, Fig. S9), as well as a C-terminal truncation in the M. bovis/M. bovis bacillus Calmette–Guérin lineage (42). These observations provide perspectives for future experiments aiming to determine the molecular elements of the observed HGT in tubercle bacilli.
Intriguingly, M. canettii A displayed an experimentally confirmed donor phenotype, yet showed a recipient genotype in previous in silico analyses (11), raising the question of a potential mating type switch or the possibility of having both donor and recipient properties. In M. smegmatis, transconjugants were reported to become donor-proficient through transfer of the so-called mid genes (mating identity), which are encoded within the esx-1 locus and comprise genes msmeg_0069-0078 (16). M. tuberculosis and M. canettii also contain a conserved ESX-1 locus, which likely is implicated in the observed DNA transfer among M. canettii A and L strains. However, sequence comparison showed that only three of six mid genes were conserved in tubercle bacilli (SI Appendix, Fig. S10 and Table S5), suggesting yet-unknown, additional mating type determinants. Comparison with M. smegmatis also revealed a 3- to 4-log lower frequency of DNA transfer in M. canettii strains, raising the question of whether the ability of DNA transfer declined in the course of evolution of slow growing mycobacteria or whether the conditions used in our study were not specifically favorable for DNA transfer between tubercle bacilli. The use of an M. tuberculosis ΔESX-1 deletion mutant (25) as donor did not lead to an increase in transfer frequency, unlike it was seen for M. smegmatis ΔESX-1 donor strains (33). It was rather the opposite, because we have obtained recombinants in M. canettii when using the F10 cosmid, which contains part of the M. tuberculosis ESX-1 locus in which the EsxA protein is fused to GFP (19). These results open interesting questions for future research on the potential involvement of the ESX-1 system in DNA transfer in slow growing mycobacteria. DNA transport coupled to protein transport via ESX secretion systems is reminiscent of type IV secretion, which in many bacterial species is involved in DNA transport. A distant connection between type IV and type VII secretion systems was recently also suggested by the description of mycobacterial plasmids that carry components with similarity to both systems, which might have played important roles in the evolution of specific chromosomal HGT mechanisms in mycobacteria (31, 32). It should be mentioned that our selection for recombinants was based on the transfer of the antibiotic resistance cassette, and as such, it is possible that some transfer events not involving the transfer of the selection marker escaped our selection screens. Thus, the actual frequency of HGT among M. canettii strains under natural environmental conditions might be higher, which could explain the extensive recombination patterns seen in the genomes of these strains (Fig. 2).
It was also hypothesized that free-living amoeba might represent a niche where environmental mycobacteria might encounter conditions allowing DNA transfer, as is the case for other amoeba-resisting microorganisms (43). However, the lack of isolation of any recombinants after coinfection of A. castellanii suggests no particular role of amoeba as potential hosts of mating encounters of tubercle bacilli, at least under the conditions used. Our results thus open research questions on the potential triggers and the natural environment needed for optimal DNA transfer.
In conclusion, we want to emphasize that despite the low observed frequency, our study strongly suggests that the major mode of HGT among tubercle bacilli such as M. canettii, is similar to the single cell-to-cell conjugal transfer processes seen in M. smegmatis, involving the transfer of larger segments of donor-derived DNA by close cell-to-cell contact. However, we cannot entirely exclude the possibility of a distinct, yet unknown mechanism of DNA transfer operating in M. canettii strains. Nonetheless, it is plausible that such types of genome blending seen in our M. canettii recombinants, in addition to particular reductive evolution events, such as the loss of lipooligosaccharide production through recombination between two pks5 genes (21), or the loss of elements from the cobalamin-vitamin B12 synthesis pathway (10, 18, 44), have accelerated adaptation of an ancestral generalist Mycobacterium to the challenging conditions imposed by new hosts. Because the different exchanged genome segments can be large enough to contain (multiple) whole operons, HGT can also boost simultaneous creation of new functions. The sizes of the transferred DNA sequences might explain the existence of a series of strain-specific gene clusters, such as the mce5 operon or the eptABCD operon of strain M. canettii strain J (STB-J) (10, 21, 22), or the gene cluster encoding a second CRISPR-Cas system specific to M. canettii strain K (STB-K) (10), as well as genes that are specifically shared by a whole group of strains, such as the pe_pgrs33 (rv1818) gene of MTBC members (10, 39). Whereas the M. canettii strain-specific genes might be the result of recent HGT among these strains and other environmental bacteria, the supposed transfer of pe_pgrs33 to the common ancestor of the MTBC might represent a more ancient occurrence, at the beginning of the clonal emergence of the MTBC from M. canettii-like generalist bacteria toward pathogens of mammalian hosts. However, unlike previously postulated (8, 9), the ability of extensive HGT seems to have been lost during the development of this lineage into the extant MTBC, as is suggested by the here-reported failure of obtaining recombinants among different M. tuberculosis strains despite extensive attempts, and the observed clonal strain cluster organization within lineage 1–7 strains (Fig. 2). These results thus provide the scientific basis to unravel the molecular determinants underlying the HGT differences observed between M. canettii and the MTBC and provide insights into the risk evaluation of potential transfer of drug resistance and/or fitness mutations among clinical strains.
Materials and Methods
The materials and methods are described at length in SI Appendix, SI Materials and Methods. They include mating assays, cloning, bacterial strain and amoeba models, and sequence- and recombination-analysis details.
Supplementary Material
Acknowledgments
We thank William R. Jacobs for providing M. smegmatis mc2874 and M. tuberculosis ΔRD1 strains, Denis A. Mitchison for providing M. tuberculosis strain 79112, and Dick van Soolingen for M. tuberculosis strain Tb36. We thank Louis Jones for setup of the CanettiiList server. Whole-genome sequencing was performed at the Institut Pasteur Genomics Platform, member of the “France Génomique” consortium (ANR10-INBS-09-08). We thank the students of the Institut Pasteur genome analysis course for contributing one M. smegmatis clone for further analysis. This study was in part supported by the European Community’s Grants 260872 and 643381, European Union - European Federation of Pharmaceutical Industries and Associations Innovative Medicines Initiative Grant 115337, Agence National de Recherche Grant ANR-14-JAMR-001-02, and Fondation pour la Recherche Médicale FRM Grant DEQ20130326471. R.B. is a member of the LabEx consortium ANR-10-LABX-62-IBEID at the Institut Pasteur. E.C.B. was supported by a stipend from the Pasteur–Paris University International PhD program and the Institut Carnot Pasteur Maladies Infectieuses.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: DNA sequencing data for M. canettii recombinants RC1, RC2, RC3, RC4, RC6, RC7, RC9, and RC10 have been deposited in the European Nucleotide Archive under the accession nos. ERS896599, ERS896600, ERS1092931, ERS1092932, ERS1092934, ERS1092935, ERS1092936, and ERS1092937, respectively. Annotated M. canettii reference sequences are available via the CanettiiList server genolist.pasteur.fr/CanettiiList/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604921113/-/DCSupplemental.
References
- 1.Sreevatsan S, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94(18):9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosch R, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis. 2002;186(1):74–80. doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 4.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7(5):328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 5.Supply P, et al. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol Microbiol. 2003;47(2):529–538. doi: 10.1046/j.1365-2958.2003.03315.x. [DOI] [PubMed] [Google Scholar]
- 6.Baker L, Brown T, Maiden MC, Drobniewski F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis. 2004;10(9):1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42(6):498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Gutacker MM, Musser JM, Fu YX. Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol. 2006;188(23):8169–8177. doi: 10.1128/JB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namouchi A, Didelot X, Schöck U, Gicquel B, Rocha EP. After the bottleneck: Genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 2012;22(4):721–734. doi: 10.1101/gr.129544.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Supply P, et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45(2):172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortimer TD, Pepperell CS. Genomic signatures of distributive conjugal transfer among mycobacteria. Genome Biol Evol. 2014;6(9):2489–2500. doi: 10.1093/gbe/evu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons LM, Jankowski CS, Derbyshire KM. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol Microbiol. 1998;28(3):571–582. doi: 10.1046/j.1365-2958.1998.00818.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Derbyshire KM. Plasmid DNA transfer in Mycobacterium smegmatis involves novel DNA rearrangements in the recipient, which can be exploited for molecular genetic studies. Mol Microbiol. 2004;53(4):1233–1241. doi: 10.1111/j.1365-2958.2004.04201.x. [DOI] [PubMed] [Google Scholar]
- 14.Wollman EL, Jacob F, Hayes W. Conjugation and genetic recombination in Escherichia coli K-12. Cold Spring Harb Symp Quant Biol. 1956;21:141–162. doi: 10.1101/sqb.1956.021.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3(9):711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 16.Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol. 2013;11(7):e1001602. doi: 10.1371/journal.pbio.1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez MC, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1(1):e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blouin Y, et al. Progenitor “Mycobacterium canettii” clone responsible for lymph node tuberculosis epidemic, Djibouti. Emerg Infect Dis. 2014;20(1):21–28. doi: 10.3201/eid2001.130652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodin P, et al. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J Biol Chem. 2005;280(40):33953–33959. doi: 10.1074/jbc.M503515200. [DOI] [PubMed] [Google Scholar]
- 20.Bange FC, Collins FM, Jacobs WR., Jr Survival of mice infected with Mycobacterium smegmatis containing large DNA fragments from Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79(3):171–180. doi: 10.1054/tuld.1998.0201. [DOI] [PubMed] [Google Scholar]
- 21.Boritsch EC, et al. pks5-recombination-mediated surface remodelling in Mycobacterium tuberculosis emergence. Nat Microbiol. 2016;1:15019. doi: 10.1038/nmicrobiol.2015.19. [DOI] [PubMed] [Google Scholar]
- 22.Panas MW, et al. Noncanonical SMC protein in Mycobacterium smegmatis restricts maintenance of Mycobacterium fortuitum plasmids. Proc Natl Acad Sci USA. 2014;111(37):13264–13271. doi: 10.1073/pnas.1414207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heym B, et al. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: A molecular study. Lancet. 1994;344(8918):293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 24.Telenti A, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341(8846):647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 25.Hsu T, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USA. 2003;100(21):12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charpentier X, Kay E, Schneider D, Shuman HA. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol. 2011;193(5):1114–1121. doi: 10.1128/JB.01146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mba Medie F, Ben Salah I, Henrissat B, Raoult D, Drancourt M. Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS One. 2011;6(6):e20499. doi: 10.1371/journal.pone.0020499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelicic V, et al. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94(20):10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bardarov S, et al. Conditionally replicating mycobacteriophages: A system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94(20):10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brochet M, et al. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci USA. 2008;105(41):15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ummels R, et al. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. MBio 5(5):e01744-14.
- 32.Dumas E, et al. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol Evol. 2016;8(2):387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci USA. 2004;101(34):12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coros A, Callahan B, Battaglioli E, Derbyshire KM. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol. 2008;69(4):794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Soolingen D, et al. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: Characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47(4):1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 36.Koeck JL, et al. [Epidemiology of resistance to antituberculosis drugs in Mycobacterium tuberculosis complex strains isolated from adenopathies in Djibouti. Prospective study carried out in 1999] Med Trop (Mars) 2002;62(1):70–72. [PubMed] [Google Scholar]
- 37.Koeck JL, et al. Clinical characteristics of the smooth tubercle bacilli ‘Mycobacterium canettii’ infection suggest the existence of an environmental reservoir. Clin Microbiol Infect. 2011;17(7):1013–1019. doi: 10.1111/j.1469-0691.2010.03347.x. [DOI] [PubMed] [Google Scholar]
- 38.Fabre M, et al. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canettii” strains indicates that the M. tuberculosis complex is a recently emerged clone of “M. canettii”. J Clin Microbiol. 2004;42(7):3248–3255. doi: 10.1128/JCM.42.7.3248-3255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boritsch EC, et al. A glimpse into the past and predictions for the future: The molecular evolution of the tuberculosis agent. Mol Microbiol. 2014;93(5):835–852. doi: 10.1111/mmi.12720. [DOI] [PubMed] [Google Scholar]
- 40.Frischkorn K, et al. Investigation of mycobacterial recA function: Protein introns in the RecA of pathogenic mycobacteria do not affect competency for homologous recombination. Mol Microbiol. 1998;29(5):1203–1214. doi: 10.1046/j.1365-2958.1998.01003.x. [DOI] [PubMed] [Google Scholar]
- 41.Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol. 1998;29(6):1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 42.Garnier T, et al. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci USA. 2003;100(13):7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertelli C, Greub G. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front Cell Infect Microbiol. 2012;2:110. doi: 10.3389/fcimb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gopinath K, Moosa A, Mizrahi V, Warner DF. Vitamin B(12) metabolism in Mycobacterium tuberculosis. Future Microbiol. 2013;8(11):1405–1418. doi: 10.2217/fmb.13.113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.