Abstract
Heterochromatin mostly constitutes tightly packaged DNA, decorated with repressive histone marks, including histone H3 methylated at lysine 9, histone H4 methylated at lysine 20 and histone H3 methylated at lysine 27. Each of these marks is incorporated by specific histone lysine methyl transferases. While constitutive heterochromatin enriched with H3K9me3 and H4K20me3 occur within repetitive elements, including centromeres and telomeres, the facultative heterochromatin resides on the inactive X-chromosome and contains H3K27me3 mark. Origin recognition complex-associated (ORCA/LRWD1) protein is required for the initiation of DNA replication and also plays crucial roles in heterochromatin organization. ORCA associates with constitutive and facultative heterochromatin in human cells and binds to repressive histone marks. We demonstrate that ORCA binds to multiple repressive histone methyl transferases including G9a, GLP, Suv39h1 (H3K9me2/3), Suv420h1/h2 (H4K20me2/3) and EZH2 (H3K27me3). Removal of ORCA from human cells causes aberrations in the chromatin architecture. We propose that ORCA acts as a scaffold protein that enables the formation of multiple histone lysine methyltransferase complexes at heterochromatic sites thereby facilitating chromatin organization.
Keywords: histone methyl transferases, heterochromatin, ORCA/LRWD1, replication
Introduction
Every cycling cell needs to duplicate its genetic material accurately and then segregate the chromosomes faithfully to the daughter nuclei. Furthermore, the epigenetic information must also be restored from one cell generation to the next. Proteins involved in DNA replication play crucial roles in the inheritance of chromatin domains.1,2 However, the mechanism that ensures that chromatin architecture is reestablished once the replication has been accomplished remains to be elucidated and is an intense area of research.3,4 Several studies in budding yeast, fission yeast, Drosophila and humans have pointed out that DNA replication proteins coordinate heterochromatin organization and gene silencing either by facilitating nucleosome assembly of heterochromatin or by recruiting factors that are key to the establishment and maintenance of heterochromatin or by coordinating with the siRNA machinery to maintain heterochromatin.2 Whether the role of replication initiation factors in heterochromatin assembly is independent of their replication initiation function remained to be determined.
Initiation of DNA replication in eukaryotes requires the sequential assembly of a multiprotein complex at the origins of replication.5 Origin Recognition complex (ORC) consisting of 6 subunits serves as the landing pad for the establishment of pre-replication complex during G1 phase of the cell cycle.6 ORC-Associated (ORCA)/LRWD1 is an ORC interacting protein that is required for stabilizing ORC onto chromatin.7 In diploid fibroblasts, depletion of ORCA causes an accumulation of cells in G1 phase of the cell cycle.8 In addition, previous work from our laboratory has shown that ORCA regulates replication initiation by modulating the interaction between pre-replicative complex component Cdt1 and its inhibitor Geminin.8
While the role of ORC in replication initiation has been extensively studied and is relatively well understood, the role of ORC and ORCA in heterochromatin remained unclear. Others and we have previously shown that ORCA binds to heterochromatin, including at centromeres and telomeres in human and mouse cells.7,9-11 By using Stable Isotope Labeling in Cell Culture (SILAC) and modified N-terminal histone tails as baits, it was shown that ORCA binds specifically to repressive trimethylated H3K9, K27 and H4K20 marks.11 We recently conducted a study to investigate the function of ORCA at H3K9me3-containing heterochromatic domains.12 We found that ORCA interacts with multiple H3K9 KMTs G9a/GLP and Suv39H1. A multimeric complex containing all the H3K9 KMTs including G9a, GLP, Suv39h1 and SETDB1 is recruited to pericentric heterochromatin and aids in maintenance of H3K9me2 and me3.13 By using Single Molecule Pulldown assays we found that ORC-ORCA-H3K9 KMTs exist in a single complex. The existence of a complex containing ORC and H3K9 KMTs is very exciting as it reiterates the importance of a cross-talk between eukaryotic DNA replication proteins and the repressive epigenetic machinery. ORCA also directly binds to H3K9me2 and me3 with stronger binding to the trimethylated mark. The loss of ORCA resulted in the global reduction of H3K9me3, consistent with the observation that loss of ORCA also showed reduced association of Suv39H1 on chromatin. In addition to the reduction in the levels of H3K9me3, there was also a reduction of H3K9me2 upon depletion of ORCA. By using chromatin immunoprecipitation we further showed that ORCA binds to specific regions on the chromatin that are enriched for H3K9me2 and me3. Furthermore, the H3K9me2 and 3 are lost specifically from these regions when ORCA was depleted from human cells. In order to understand the role of ORCA in the complex containing G9a and Suv39H1, we investigated the stability of these complexes upon the loss of ORCA. We found that the loss of ORCA resulted in the reduction of the complexes containing G9a and Suv39H1.
Furthermore, aberrant chromatin organization also resulted in defective DNA replication in cells depleted of ORCA. Specifically, we found that loss of ORCA showed a reduction of late replication patterns and aberrant replication timing. This was specifically at the regions that showed loss of H3K9me2 and me3. To tease out the role of ORCA in DNA replication initiation versus chromatin organization, we depleted ORCA at the G1/S transition and then probed for defects in DNA replication. We found that postG1 cells showed aberrant chromatin organization as evident by the mislocalization of HP1α and H3K9me3. Our results showed that ORCA regulates chromatin organization independent of its role in DNA replication initiation.
ORCA Interacts with Multiple Repressive Methyltransferases
Our work has demonstrated that ORCA interacts with repressive H3K9 KMTs G9a/GLP and Suv39H1. Since ORCA has been shown to bind to repressive histone lysine methylation marks, specifically H3K9me3, H3K27me3 and H4K20me3,9-12 we investigated whether ORCA also associates with the KMTs that catalyze H4K20 and H3K27 methylation. To investigate this we utilized U2OS 2–6–3 CLTon cells.14,15 This cell line has a tandem array of Lac operator (LacO) sequences inserted into a single locus in the cells. The locus is heterochromatic and can be visualized by the stable expression of mCherry-Lac repressor (LacI) in the cells. As observed earlier, tethering of H3K9 KMT G9a to the locus showed robust accumulation of ORCA at this site (Fig. 1A). In order to examine whether ORCA interacts with H3K27 KMT EZH2, we tethered YFP-LacI-EZH2 to the locus and found that CFP-ORCA is also recruited to the locus (Fig. 1A). This is interesting because a recent report shows that there is a functional cross-talk between H3K9 and H3K27 KMTs in mouse embryonic stem cells.16 H3K9 KMTs were found to cooperate with the H3K27 methylation complex, the Polycomb Repressive Complex 2 (PRC2).17 G9a and GLP double knockouts showed reduction in PRC2 recruitment and H3K27me3 levels. In addition, the authors showed that G9a monomethylates H3K27 and this aids PRC2-dependent trimethylation of H3K27. It would be interesting to investigate whether ORCA functions in a complex that contains both G9a and EZH2 and whether it regulates the function of this complex. Further, how would such a multimeric complex be recruited to chromatin sites? In addition, it would be important to investigate whether loss of ORCA leads to upregulation of PRC2-regulated genes. Previous work has shown that loss of ORCA in mouse cells leads to upregulation of centromeric transcription.10 It would be crucial to determine whether this transcriptional control by ORCA is restricted to repetitive heterochromatic regions or whether it extends to repressed euchromatic regions controlled by G9a/GLP and/or PRC2.
Figure 1.
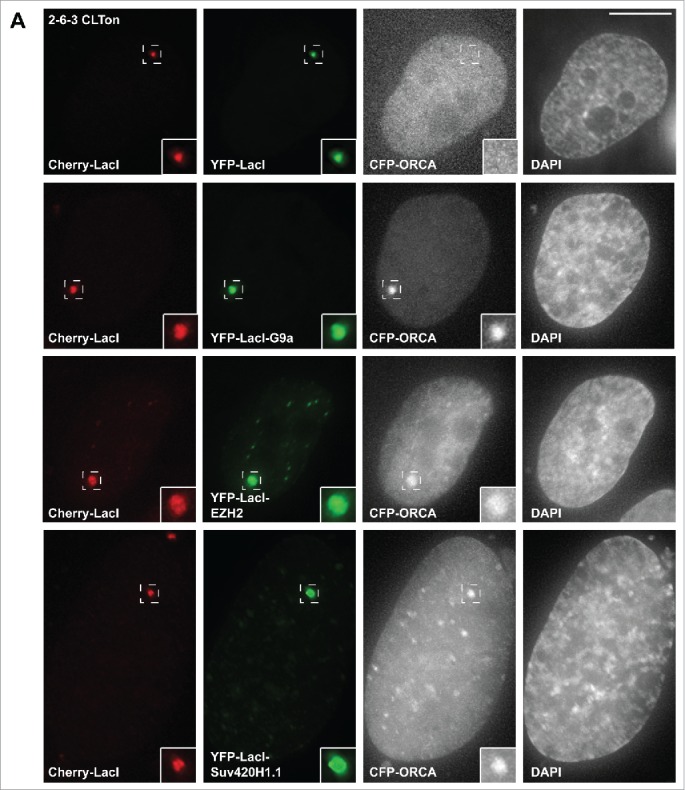
ORCA interacts with H3K9, K27 and H4K20 histone lysine methyltransferases. (A) U2OS 2–6–3 CLTon cells co-transfected with individual YFP-LacI-KMTs and CFP-ORCA. Inset represents 200% magnification of the boxed region.
H4K20me1 and H4K20me2 are shown to be involved in DNA replication, whereas H4K20me3 is required for pericentric heterochromatin organization.18 Suv4–20H1 and H2 catalyze the di- and tri methylation of H4K20.19 We tethered YFP-LacI-Suv420H1.1 to the CLTon locus and found that CFP-ORCA is recruited to the locus indicating that ORCA also interacts with Suv420H1.1 (Fig. 1A). The presence of H3K9me3 is essential for H4K20 trimethylation at pericentric heterochromatin.20-22
The mode of recruitment of different KMTs to specific target sites in mammalian cells is not clearly understood.23 None of the KMTs, except PRDM family members, can bind to DNA directly. The association of KMTs to specific DNA-binding proteins/chromatin binding proteins may provide a means by which they could be targeted to specific sites. The replication protein ORCA associates with specific chromatin marks and the histone modifying machinery via its WD domain. It remains to be determined if ORCA binds to DNA and the DNA modifying machinery directly.
Post-translational modifications on histones are preserved at specific genomic regions from one cell generation to another. It is largely believed that during DNA replication the modified histones present on the parental DNA are randomly segregated to the daughter strands and this is required for the further addition of modifications on the newly assembled histones. Post-translational modifications are transmitted with the parental histones to the newly formed DNA strand.24 Interestingly, H3K9me3 and H3K27me3 were found to propagate by modification of the parental and newly generated histones and this was found to extend over several cell generations.24 Recent evidence from Drosophila has pointed out that the histone modifications, specifically H3K4me3 and H3K27me3, are lost during DNA replication and that histone-modifying machinery associates with specific genomic loci that persists during DNA replication and enables the re-establishment of the histone modifications.25 During DNA replication, the DNA methyl transferase (DNMT1) and the H3K9 KMT G9a physically and functionally cooperate at the replication fork to coordinate DNA and histone methylation.26 The association of ORCA with the histone modifying machinery brings many interesting questions: Is this association occurring at specific stages of the cell cycle, is it occurring at specific origins, if this association is disrupted can this affect replication timing?
The WD-40 Domain of ORCA Interacts with Suv420H2
WD repeat-containing proteins bind to histone and nucleosomes and function in a diverse array of cellular functions.27 ORCA consists of leucine rich repeats at its N-terminus and a WD-repeat domain at its C-terminus.7 We have found that the WD domain of ORCA associates with the modified histones and also mediates the association to H3K9 KMTs.12 In order to map the interaction of ORCA with H4K20 KMT-Suv420H2, we cotransfected HA-Suv420H2 and T7-ORCA in human U2OS cells. This was followed by immunoprecipitation with the T7 antibody (Fig. 2B). ORCA interacted robustly with Suv420H2 (Fig. 2Ab). This interaction could be indirect or meditated by multiple domains of Suv420H2. Different truncation mutants of ORCA including 1–127a (Spanning LRR), 1–270aa (LRR+linker), 128–647 (linker+WD) and 270–647aa (WD alone) were co-transfected with HA-Suv420H2 (Fig. 2Aa). Immunoprecipitation using T7 antibody revealed that the WD domain of ORCA mediates the association of ORCA to Suv420H2 (Fig. 2Ab). It is interesting to note that the Linker+WD mutant shows reduced association to Suv420H2 compared to the WD alone. Determination of the structure of ORCA would provide insights into the organization of these domains.
Figure 2.
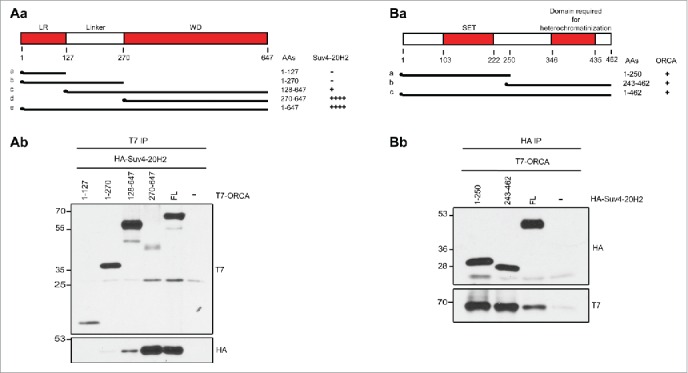
ORCA interacts with Suv420H2. (A) a. Schematic representation of various truncation mutants of ORCA containing a T7-epitope on the N-terminus. The specific domains that can associate with Suv420H2 are depicted as '+'. IP in U2OS cells expressing various T7-ORCA mutants and HA-Suv420H2 using T7 Ab and analysis by T7 and HA- IB. (B) a. Schematic representation of various truncation mutants of Suv420H2 containing a HA-epitope on the N-terminus. The interaction domains of Suv420H2 that interact with ORCA are denoted as '+'. b. IP in U2OS cells expressing various HA-Suv39H1 mutants and T7-G9a using HA Ab and analysis by T7 and HA IB.
Reciprocal immunoprecipitation from cells expressing HA-Suv420H2 and T7-ORCA using HA antibody showed the association of Suv420H2 with ORCA (Fig. 2Bb). To further determine the domain of Suv420H2 that interacts with ORCA, we made 2 truncation mutants of Suv420H2 (Fig. 2Ba), the N terminal fragment (1–250aa) containing the SET domain of Suv420H2 and the C-terminal fragment (250–462aa) containing the region required for heterochromatinization. Co-transfection of these HA tagged mutants with T7-ORCA followed by HA immunoprecipitation showed that both the N and C-terminal fragments independently interact with ORCA (Fig. 2B, lanes 1 and 2). In the future we will investigate whether the interaction between ORCA and Suv420H2 is direct by using purified proteins. Furthermore, we would also determine if this interaction is DNA-dependent.
ORCA was also found to interact with H4K20 monomethylase, PR-SET7 (data not shown). These observations that ORCA interacts with H4K20 KMTs raises many interesting possibilities. Work from Reinberg's lab28 pointed toward the possibility that during DNA replication initiation Orc1 and ORCA bind to H4K20me2 and me3 respectively, thereby establishing the origins poised for replication in S-phase. While this is an intriguing possibility, it would be crucial to conclusively determine whether this binding of ORC/ORCA to methylated H4K20 is related to the function of these proteins in DNA replication initiation as opposed to chromatin organization in repressive environments.
Based on our results, we speculate that ORCA acts similar to HP1α and facilitates the establishment and maintenance of H3K9 methylation-containing heterochromatin. ORCA acts as a scaffold to hold together the H3K9 KMTs megacomplex and stabilizes them on chromatin. This leads to the establishment of methylated H3K9, thereby providing more binding sites for ORCA and this process continues with the end result of establishment of heterochromatin domains. It is quite possible that ORCA's binding to methylated H4K20 and interaction with PR-SET7 and Suv4–20H1/2 could be geared toward the exact same purpose at heterochromatin and could be independent of its role in DNA replication initiation. Another hypothesis is that ORCA regulates specific subsets of origins, namely late replicating ones that reside within heterochromatin. In that case, ORCA could have multiple, interdependent functions in heterochromatin, which can be broadly classified into those required for heterochromatin replication initiation and its organization. Determining the ORCA binding sites on the genome would provide important insights into its role in replication initiation and heterochromatin function. This would allow us to investigate whether ORCA associates with all the origins or predominantly with the later firing ones.
There is also increasing evidence for a functional cross-talk between multiple KMT complexes. For example, the G9a/GLP and the PRC2 interact physically as well as functionally.16 Further, the activity of G9a dictates the recruitment of PRC2 to specific target genes.16 Such a functional cross-talk may exist between multiple histone modifying complexes and further studies would be critical to determine mechanistic details of this interaction.
ORCA could Act as a Scaffolding Factor in Multiple Repressive Environments
We have previously shown that ORCA associates with constitutive heterochromatin present at centromeres and telomeres.7 In a genome wide RNAi screen to identify factors involved in Xi silencing, ORCA along with ORC was found to be involved in the maintenance of X-chromosome inactivation.29 ORCA was also uncovered in the Xist interactome as a Xist interacting protein.30 Since ORCA interacts with EZH2 and associates with H3K27me3,11 this could be the mechanism of association of ORCA with the inactive X chromosome and perhaps Xist and thereby facilitating silencing. In this context it is interesting to note that Orc2 localizes on Xi and impacts Xi silencing.29 It is likely that ORCA functions in the same pathway as Orc2 in mediating Xi silencing. Based on our previous data12 and data shown here we hypothesize that ORCA functions as a scaffolding factor for H3K9, H4K20 and H3K27 methyltransferases at multiple repressive environments such as centromeres, telomeres and inactive X, to name a few (Fig. 3). ORCA seems to be crucial for constitutive as well as facultative heterochromatin organization. Constitutive heterochromatin represents gene-poor pericentromeric regions enriched with H3K9me3 and H4K20me3.31 Our data supports the model that ORCA acts as a scaffold that enables the KMTs to carry out their function. Since facultative heterochromatin represents genomic regions that can adopt open or closed conformations depending on temporal and spatial contexts,32 it would be crucial to understand the role of ORCA in establishing or maintaining the facultative heterochromatin.
Figure 3.
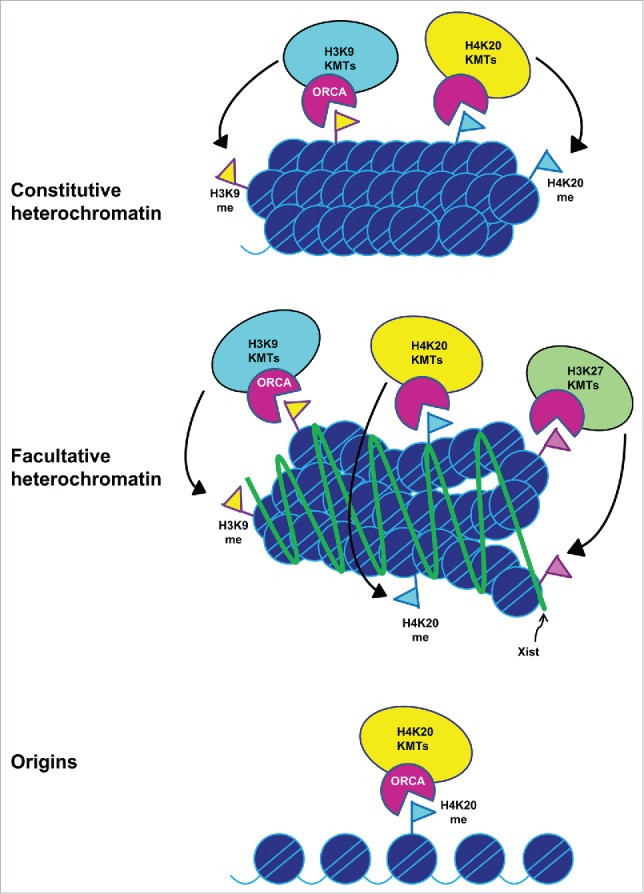
Model depicting the role of ORCA in organizing different chromatin domains. Model representing the mode of regulation of chromatin at constitutive and facultative heterochromatin and at origins by ORCA. Inactive X chromosome is depicted as an example of facultative heterochromatin.
SILAC based proteomic studies has also revealed that ORCA/ORC can bind to methylated CpG DNA.9 The authors showed that ORCA/ORC bind to methylated DNA and histones in a cooperative fashion. It is interesting to note that the tethering of the repressive KMTs to the CLTon locus enhances ORCA's interaction with the locus in multiple ways. First, ORCA localizes to the locus by interacting with the KMTs. Second, ORCA can also bind to the methylated H3K9, K27 and H4K20 that are established by the KMTs. Third, the presence of methylated histones could aid ORCA's binding to methylated DNA. A natural segue to this would be investigating whether ORCA interacts with DNA methyltransferases. Another avenue of investigation that would be exciting to pursue in the light of this data would be to examine the possible role of ORCA in silencing of LINEs and SINEs. Finally, since bivalent (H3K27 me3 and H3K4me3 marked)33 and trivalent (H3K9me3, H3K27 me3 and H3K4me3 marked) domains33 exist in embryonic stem cells and regulate key events during differentiation, it would be important to investigate the role of ORCA during development.
Perspectives
ORCA is turning out to be a multifaceted protein playing key roles in heterochromatin organization and DNA replication. Determining genome wide association of ORCA will be crucial for gaining a better understanding of its function. In addition, it will be important to determine the full complement of histone modifications to which ORCA interacts with. Such studies will provide crucial insights into the possible role of ORCA in different chromatin environments. Finally, in the light of its association with EZH2, it will be exciting to investigate the function of ORCA in embryonic stem cells and in the context of development and differentiation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank Drs. M. Dundr, Ait-Si-Ali, D. Spector and B. Stillman for providing reagents and suggestions. We thank Dr. K. Prasanth, Ms. K. Dovalovsky and Mr. A. Matur for critical reading of the manuscript.
Funding
This work was supported by NSF-CMMB-IGERT and F31 (CA180616) NIH fellowship to SG; and NSF career (1243372) and NIH (1RO1GM099669) awards to SGP. The authors declare no competing financial interests.
References
- 1.Chakraborty A, Shen Z, Prasanth SG. "ORCanization" on heterochromatin: Linking DNA replication initiation to chromatin organization. Epigenetics 2011; 6:665-70; PMID:21586903; http://dx.doi.org/ 10.4161/epi.6.6.16179 [DOI] [PubMed] [Google Scholar]
- 2.Li Q, Zhang Z. Linking DNA replication to heterochromatin silencing and epigenetic inheritance. Acta Biochim Biophys Sin 2012; 44:3-13; PMID:22194009; http://dx.doi.org/ 10.1093/abbs/gmr107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abmayr SM, Workman JL. Holding on through DNA replication: histone modification or modifier? Cell 2012; 150:875-7; PMID:22939615; http://dx.doi.org/ 10.1016/j.cell.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 4.Giri S, Prasanth SG. Replicating and transcribing on twisted roads of chromatin. Brief Funct Genomics 2012; 11(3):188-204; PMID:22267489 [DOI] [PubMed] [Google Scholar]
- 5.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem 2002; 71:333-74; PMID:12045100; http://dx.doi.org/ 10.1146/annurev.biochem.71.110601.135425 [DOI] [PubMed] [Google Scholar]
- 6.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 1992; 357:128-34; PMID:1579162; http://dx.doi.org/ 10.1038/357128a0 [DOI] [PubMed] [Google Scholar]
- 7.Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell 2010; 40:99-111; PMID:20932478; http://dx.doi.org/ 10.1016/j.molcel.2010.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Z, Chakraborty A, Jain A, Giri S, Ha T, Prasanth KV, Prasanth SG. Dynamic association of ORCA with prereplicative complex components regulates DNA replication initiation. Mol Cell Biol 2012; 32:3107-20; PMID:22645314; http://dx.doi.org/ 10.1128/MCB.00362-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010; 143:470-84; PMID:21029866; http://dx.doi.org/ 10.1016/j.cell.2010.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan KM, Zhang Z. Leucine-rich repeat and WD repeat-containing protein 1 is recruited to pericentric heterochromatin by trimethylated lysine 9 of histone H3 and maintains heterochromatin silencing. J Biol Chem 2012; 287:15024-33; PMID:22427655; http://dx.doi.org/ 10.1074/jbc.M111.337980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al.. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010; 142:967-80; PMID:20850016; http://dx.doi.org/ 10.1016/j.cell.2010.08.020 [DOI] [PubMed] [Google Scholar]
- 12.Giri S, Aggarwal V, Pontis J, Shen Z, Chakraborty A, Khan A, Mizzen C, Prasanth KV, Ait-Si-Ali S, Ha T, et al.. The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. eLife 2015; 4; PMID:25922909; http://dx.doi.org/ 10.7554/eLife.06496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 2010; 37:46-56; PMID:20129054; http://dx.doi.org/ 10.1016/j.molcel.2009.12.017 [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty A, Prasanth KV, Prasanth SG. Dynamic phosphorylation of HP1alpha regulates mitotic progression in human cells. Nat Commun 2014; 5:3445; PMID:24619172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al.. From silencing to gene expression: real-time analysis in single cells. Cell 2004; 116:683-98; PMID:15006351; http://dx.doi.org/ 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell 2014; 53:277-89; PMID:24389103; http://dx.doi.org/ 10.1016/j.molcel.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 17.Mozzetta C, Pontis J, Ait-Si-Ali S. Functional Crosstalk Between Lysine Methyltransferases on Histone Substrates: The Case of G9A/GLP and Polycomb Repressive Complex 2. Antioxid Redox Signal 2015; 22:1365-81; PMID:25365549; http://dx.doi.org/ 10.1089/ars.2014.6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 2013; 41:2797-806; PMID:23345616; http://dx.doi.org/ 10.1093/nar/gkt012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18:1251-62; PMID:15145825; http://dx.doi.org/ 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005; 438:1116-22; PMID:16222246; http://dx.doi.org/ 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- 21.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410:116-20; PMID:11242053; http://dx.doi.org/ 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- 22.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 2005; 25:2525-38; PMID:15767660; http://dx.doi.org/ 10.1128/MCB.25.7.2525-2538.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol 2015; 16:499-513; PMID:26204160; http://dx.doi.org/ 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- 24.Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 2015; 29:585-90; PMID:25792596; http://dx.doi.org/ 10.1101/gad.256354.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 2012; 150:922-33; PMID:22921915; http://dx.doi.org/ 10.1016/j.cell.2012.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 2006; 20:3089-103; PMID:17085482; http://dx.doi.org/ 10.1101/gad.1463706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suganuma T, Pattenden SG, Workman JL. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev 2008; 22:1265-8; PMID:18483215; http://dx.doi.org/ 10.1101/gad.1676208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beck DB, Burton A, Oda H, Ziegler-Birling C, Torres-Padilla ME, Reinberg D. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev 2012; 26:2580-9; PMID:23152447; http://dx.doi.org/ 10.1101/gad.195636.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan KM, Zhang H, Malureanu L, van Deursen J, Zhang Z. Diverse factors are involved in maintaining X chromosome inactivation. Proc Natl Acad Sci U S A 2011; 108:16699-704; PMID:21940502; http://dx.doi.org/ 10.1073/pnas.1107616108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al.. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015; 349.31; PMID:26138965; http://dx.doi.org/ 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saksouk N, Simboeck E, Dejardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 2015; 8:3; PMID:25788984; http://dx.doi.org/ 10.1186/1756-8935-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 2007; 28:1-13; PMID:17936700; http://dx.doi.org/ 10.1016/j.molcel.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 33.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315-26; PMID:16630819; http://dx.doi.org/ 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]


